| # |
organism |
max score |
hits |
top seq |
| 1 |
N/A |
120.00 |
21 |
A |
| 2 |
Homo sapiens |
120.00 |
15 |
unnamed protein product |
| 3 |
Mus musculus |
120.00 |
8 |
AF275367_1 epidermal growth factor receptor |
| 4 |
Rattus norvegicus |
120.00 |
7 |
epidermal growth factor receptor |
| 5 |
Pan troglodytes |
120.00 |
7 |
PREDICTED: epidermal growth factor receptor isofor |
| 6 |
synthetic construct |
120.00 |
4 |
unnamed protein product |
| 7 |
Monodelphis domestica |
120.00 |
3 |
PREDICTED: similar to Epidermal growth factor rece |
| 8 |
Canis familiaris |
120.00 |
3 |
epidermal growth factor receptor |
| 9 |
Macaca mulatta |
120.00 |
3 |
PREDICTED: epidermal growth factor receptor, parti |
| 10 |
Rous-associated virus type 1 |
119.00 |
2 |
S69372_2 v-erbB gene product |
| 11 |
Sus scrofa |
119.00 |
2 |
epidermal growth factor receptor |
| 12 |
Bos taurus |
119.00 |
1 |
PREDICTED: similar to epidermal growth factor rece |
| 13 |
Xenopus laevis |
115.00 |
3 |
epidermal growth factor receptor |
| 14 |
Avian erythroblastosis virus |
115.00 |
2 |
unnamed protein product |
| 15 |
Danio rerio |
112.00 |
4 |
epidermal growth factor receptor |
| 16 |
Tetraodon nigroviridis |
111.00 |
3 |
unnamed protein product |
| 17 |
Xiphophorus xiphidium |
110.00 |
2 |
epidermal growth factor receptor |
| 18 |
Gallus gallus |
107.00 |
4 |
erbB-2 |
| 19 |
Felis catus |
106.00 |
2 |
HER2 |
| 20 |
Mesocricetus auratus |
106.00 |
1 |
ERBB2_MESAU Receptor tyrosine-protein kinase erbB- |
| 21 |
Canis lupus familiaris |
106.00 |
1 |
erbB-2 |
| 22 |
Xiphophorus sp. |
102.00 |
1 |
XMRK_XIPMA Melanoma receptor tyrosine-protein kina |
| 23 |
Ovis aries |
94.00 |
1 |
erythroblastic leukemia viral oncogene-like 2 |
| 24 |
Strongylocentrotus purpuratus |
80.90 |
1 |
PREDICTED: similar to v-erb-a erythroblastic leuke |
| 25 |
Drosophila melanogaster |
80.10 |
147 |
mutant epidermal growth factor receptor |
| 26 |
Drosophila simulans |
78.20 |
1 |
epidermal growth factor receptor |
| organism | matching |
|---|
| N/A |
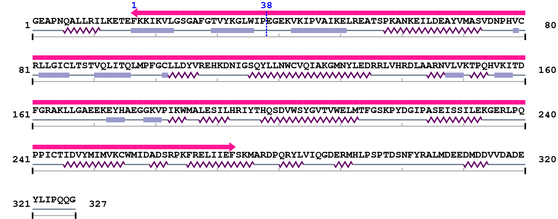
Query 9 LRILKETEFKKIKVLGSGAFGTVYKGLWIP#EGEKVKIPVAIKELREATSPKANKEILDEA 68
||||||||||||||||||||||||||||||#||||||||||||||||||||||||||||||
Query 9 LRILKETEFKKIKVLGSGAFGTVYKGLWIP#EGEKVKIPVAIKELREATSPKANKEILDEA 68
|
| Homo sapiens |
Query 9 LRILKETEFKKIKVLGSGAFGTVYKGLWIP#EGEKVKIPVAIKELREATSPKANKEILDEA 68
||||||||||||||||||||||||||||||#||||||||||||||||||||||||||||||
Query 9 LRILKETEFKKIKVLGSGAFGTVYKGLWIP#EGEKVKIPVAIKELREATSPKANKEILDEA 68
|
| Mus musculus |
Query 9 LRILKETEFKKIKVLGSGAFGTVYKGLWIP#EGEKVKIPVAIKELREATSPKANKEILDEA 68
||||||||||||||||||||||||||||||#||||||||||||||||||||||||||||||
Query 9 LRILKETEFKKIKVLGSGAFGTVYKGLWIP#EGEKVKIPVAIKELREATSPKANKEILDEA 68
|
| Rattus norvegicus |
Query 9 LRILKETEFKKIKVLGSGAFGTVYKGLWIP#EGEKVKIPVAIKELREATSPKANKEILDEA 68
||||||||||||||||||||||||||||||#||||||||||||||||||||||||||||||
Query 9 LRILKETEFKKIKVLGSGAFGTVYKGLWIP#EGEKVKIPVAIKELREATSPKANKEILDEA 68
|
| Pan troglodytes |
Query 9 LRILKETEFKKIKVLGSGAFGTVYKGLWIP#EGEKVKIPVAIKELREATSPKANKEILDEA 68
||||||||||||||||||||||||||||||#||||||||||||||||||||||||||||||
Query 9 LRILKETEFKKIKVLGSGAFGTVYKGLWIP#EGEKVKIPVAIKELREATSPKANKEILDEA 68
|
| synthetic construct |
Query 9 LRILKETEFKKIKVLGSGAFGTVYKGLWIP#EGEKVKIPVAIKELREATSPKANKEILDEA 68
||||||||||||||||||||||||||||||#||||||||||||||||||||||||||||||
Query 9 LRILKETEFKKIKVLGSGAFGTVYKGLWIP#EGEKVKIPVAIKELREATSPKANKEILDEA 68
|
| Monodelphis domestica |
Query 9 LRILKETEFKKIKVLGSGAFGTVYKGLWIP#EGEKVKIPVAIKELREATSPKANKEILDEA 68
||||||||||||||||||||||||||||||#||||||||||||||||||||||||||||||
Query 9 LRILKETEFKKIKVLGSGAFGTVYKGLWIP#EGEKVKIPVAIKELREATSPKANKEILDEA 68
|
| Canis familiaris |
Query 9 LRILKETEFKKIKVLGSGAFGTVYKGLWIP#EGEKVKIPVAIKELREATSPKANKEILDEA 68
||||||||||||||||||||||||||||||#||||||||||||||||||||||||||||||
Query 9 LRILKETEFKKIKVLGSGAFGTVYKGLWIP#EGEKVKIPVAIKELREATSPKANKEILDEA 68
|
| Macaca mulatta |
Query 9 LRILKETEFKKIKVLGSGAFGTVYKGLWIP#EGEKVKIPVAIKELREATSPKANKEILDEA 68
||||||||||||||||||||||||||||||#||||||||||||||||||||||||||||||
Query 9 LRILKETEFKKIKVLGSGAFGTVYKGLWIP#EGEKVKIPVAIKELREATSPKANKEILDEA 68
|
| Rous-associated virus type 1 |
Query 9 LRILKETEFKKIKVLGSGAFGTVYKGLWIP#EGEKVKIPVAIKELREATSPKANKEILDEA 68
|||||||||||+||||||||||||||||||#||||||||||||||||||||||||||||||
Sbjct 129 LRILKETEFKKVKVLGSGAFGTVYKGLWIP#EGEKVKIPVAIKELREATSPKANKEILDEA 188
|
| Sus scrofa |
Query 9 LRILKETEFKKIKVLGSGAFGTVYKGLWIP#EGEKVKIPVAIKELREATSPKANKEILDEA 68
|||||||||||+||||||||||||||||||#||||||||||||||||||||||||||||||
Sbjct 699 LRILKETEFKKVKVLGSGAFGTVYKGLWIP#EGEKVKIPVAIKELREATSPKANKEILDEA 758
|
| Bos taurus |
Query 9 LRILKETEFKKIKVLGSGAFGTVYKGLWIP#EGEKVKIPVAIKELREATSPKANKEILDEA 68
|||||||||||+||||||||||||||||||#||||||||||||||||||||||||||||||
Sbjct 681 LRILKETEFKKVKVLGSGAFGTVYKGLWIP#EGEKVKIPVAIKELREATSPKANKEILDEA 740
|
| Xenopus laevis |
Query 9 LRILKETEFKKIKVLGSGAFGTVYKGLWIP#EGEKVKIPVAIKELREATSPKANKEILDEA 68
||||||||||||||||||||||||+|||||#||| +|||||||||||||||||||||||||
Sbjct 289 LRILKETEFKKIKVLGSGAFGTVYQGLWIP#EGEGIKIPVAIKELREATSPKANKEILDEA 348
|
| Avian erythroblastosis virus |
Query 9 LRILKETEFKKIKVLGSGAFGTVYKGLWIP#EGEKVKIPVAIKELREATSPKANKEILDEA 68
|||||||||||+|||| |||||||||||||#||||| ||||||||||||||||||||||||
Sbjct 129 LRILKETEFKKVKVLGFGAFGTVYKGLWIP#EGEKVTIPVAIKELREATSPKANKEILDEA 188
|
| Danio rerio |
Query 9 LRILKETEFKKIKVLGSGAFGTVYKGLWIP#EGEKVKIPVAIKELREATSPKANKEILDEA 68
|||||||||||||||||||||||+||||+|#||| |||||||| |||||||||||||+|||
Sbjct 703 LRILKETEFKKIKVLGSGAFGTVHKGLWVP#EGENVKIPVAIKVLREATSPKANKEIMDEA 762
|
| Tetraodon nigroviridis |
Query 9 LRILKETEFKKIKVLGSGAFGTVYKGLWIP#EGEKVKIPVAIKELREATSPKANKEILDEA 68
|||||| |||||||||||||||||||||+|#||| |||||||| |||||||||||+|||||
Sbjct 674 LRILKEPEFKKIKVLGSGAFGTVYKGLWVP#EGEDVKIPVAIKVLREATSPKANKDILDEA 733
|
| Xiphophorus xiphidium |
Query 9 LRILKETEFKKIKVLGSGAFGTVYKGLWIP#EGEKVKIPVAIKELREATSPKANKEILDEA 68
|||||| |||||||||||||||||||||+|#||| |||||||| ||||||||||+||||||
Sbjct 714 LRILKEPEFKKIKVLGSGAFGTVYKGLWVP#EGEDVKIPVAIKVLREATSPKANQEILDEA 773
|
| Gallus gallus |
Query 9 LRILKETEFKKIKVLGSGAFGTVYKGLWIP#EGEKVKIPVAIKELREATSPKANKEILDEA 68
+||||||| ||+||||||||||||||+|||#+|| |||||||| ||| |||||||||||||
Sbjct 701 MRILKETELKKVKVLGSGAFGTVYKGIWIP#DGESVKIPVAIKVLRENTSPKANKEILDEA 760
|
| Felis catus |
Query 9 LRILKETEFKKIKVLGSGAFGTVYKGLWIP#EGEKVKIPVAIKELREATSPKANKEILDEA 68
+||||||| +|+||||||||||||||+|||#+|| |||||||| ||| |||||||||||||
Sbjct 65 MRILKETELRKVKVLGSGAFGTVYKGIWIP#DGENVKIPVAIKVLRENTSPKANKEILDEA 124
|
| Mesocricetus auratus |
Query 9 LRILKETEFKKIKVLGSGAFGTVYKGLWIP#EGEKVKIPVAIKELREATSPKANKEILDEA 68
+||||||| +|+||||||||||||||+|||#+|| |||||||| ||| |||||||||||||
Sbjct 712 MRILKETELRKVKVLGSGAFGTVYKGIWIP#DGENVKIPVAIKVLRENTSPKANKEILDEA 771
|
| Canis lupus familiaris |
Query 9 LRILKETEFKKIKVLGSGAFGTVYKGLWIP#EGEKVKIPVAIKELREATSPKANKEILDEA 68
+||||||| +|+||||||||||||||+|||#+|| |||||||| ||| |||||||||||||
Sbjct 711 MRILKETELRKVKVLGSGAFGTVYKGIWIP#DGENVKIPVAIKVLRENTSPKANKEILDEA 770
|
| Xiphophorus sp. |
Query 9 LRILKETEFKKIKVLGSGAFGTVYKGLWIP#EGEKVKIPVAIKELREATSPKANKEILDEA 68
||||||||||| +||||||||||||||| |#+|| ++|||||| |||||||| |+|+||||
Sbjct 702 LRILKETEFKKDRVLGSGAFGTVYKGLWNP#DGENIRIPVAIKVLREATSPKVNQEVLDEA 761
|
| Ovis aries |
Query 9 LRILKETEFKKIKVLGSGAFGTVYKGLWIP#EGEKVKIPVAIKELREATSPKAN 61
+||||||| +|+||||||||||||||+|||#+|| |||||||| ||| ||||||
Sbjct 63 MRILKETELRKVKVLGSGAFGTVYKGIWIP#DGENVKIPVAIKVLRENTSPKAN 115
|
| Strongylocentrotus purpuratus |
Query 9 LRILKETEFKKIKVLGSGAFGTVYKGLWIP#EGEKVKIPVAIKELREATSPKANKEILD 66
|+|+|+|| | |||||||||||||||||#+|||++|||||| ||| || | +|+|+
Sbjct 1012 LKIIKDTELKLGPVLGSGAFGTVYKGLWIP#DGEKIRIPVAIKALRE-VSPHAAEELLE 1068
|
| Drosophila melanogaster |
Query 9 LRILKETEFKKIKVLGSGAFGTVYKGLWIP#EGEKVKIPVAIKELREATSPKANKEILDEA 68
|||+|+|| +| ||| |||| ||||+|+|#||| |||||||||| ++| ++++| | ||
Sbjct 830 LRIVKDTELRKGGVLGMGAFGRVYKGVWVP#EGENVKIPVAIKELLKSTGAESSEEFLREA 889
|
| Drosophila simulans |
Query 9 LRILKETEFKKIKVLGSGAFGTVYKGLWIP#EGEKVKIPVAIKELREATSPKANKEILDEA 68
|||+|+ | +| ||| |||| ||||+|+|#||| |||||||||| ++| ++++| | ||
Sbjct 829 LRIVKDAELRKGGVLGMGAFGRVYKGVWVP#EGENVKIPVAIKELLKSTGAESSEEFLREA 888
|
 Glu
Glu

 Sequence conservation (by blast)
Sequence conservation (by blast)