XSB1083 : EGFR protein [Homo sapiens]
[ CaMP Format ]
This entry is computationally expanded from SB0050
* Basic Information
| Organism | Homo sapiens (human) |
| Protein Names | EGFR protein |
| Gene Names | EGFR |
| Gene Locus | Not available |
| GO Function | Not available |
* Information From OMIM
Description: EGFR and its ligands are cell signaling molecules involved in diverse cellular functions, including cell proliferation, differentiation, motility, and survival, and in tissue development (Wang et al., 2004).
Function: Carlin et al. (1982) showed that both the 145- and 165-kD EGFR proteins from A431 cells bound radiolabeled EGF, and both were phosphorylated upon EGF stimulation. Haley et al. (1987) showed that the activity of the EGFR promoter was modulated by adenovirus protein E1A. Stimulation with phorbol ester or fetal calf serum increased EGFR mRNA levels.
Function: EGFR signaling involves small GTPases of the Rho family, and EGFR trafficking involves small GTPases of the Rab family. Lanzetti et al. (2000) reported that the EPS8 (OMIM:600206) protein connects these signaling pathways. EPS8 is a substrate of EGFR that is held in a complex with SOS1 (OMIM:182530) by the adaptor protein E3B1 (OMIM:603050), thereby mediating activation of RAC (OMIM:602048). Through its SH3 domain, EPS8 interacts with RNTRE (OMIM:605405). Lanzetti et al. (2000) showed that RNTRE is a RAB5 (OMIM:179512) GTPase-activating protein whose activity is regulated by EGFR. By entering in a complex with EPS8, RNTRE acts on RAB5 and inhibits internalization of the EGFR. Furthermore, RNTRE diverts EPS8 from its RAC-activating function, resulting in the attenuation of RAC signaling. Thus, depending on its state of association with E3B1 or RNTRE, EPS8 participates in both EGFR signaling through RAC and EGFR trafficking through RAB5.
Function: Yang et al. (1996) demonstrated that treatment with genistein, an inhibitor of tyrosine kinase activity, inhibited EGF-induced tyrosine phosphorylation and degradation of EGFR in HepG2 cells, suggesting to the authors that tyrosine kinase activity is required for either the internalization or the degradation of EGF-EGFR receptor complexes.
Function: Downward et al. (1984) presented evidence that oncogene ERBB may be derived from the gene coding for EGFR. Spurr et al. (1984) assigned the oncogene ERBB to chromosome 7 by study of mouse-human somatic cell hybrids. The amino acid sequence of the protein encoded by v-erb B (deduced from the nucleotide sequence of the gene) displays strong homologies to tyrosine-specific protein kinases (Privalsky et al., 1984). Both ERBA and ERBB are on mouse chromosome 11, which carries alpha-globin genes and genes for colony-stimulating factor and interleukin-3 (Silver et al., 1985). Neither of the oncogenes is on chromosome 16, which carries alpha-globin genes in man. The most striking and consistent chromosomal finding in a series of human glioblastoma (OMIM:137800) cell lines was an increase in copy number of chromosome 7. Henn et al. (1986) found that in all of the cell lines ERBB-specific mRNA was increased to levels even higher than expected from the number of chromosomes 7 present. These changes were not found in benign astrocytomas.
Function: Using Southern blot analysis, Yamazaki et al. (1988) found in 2 cases of human glioblastoma multiforme that cells carried amplified ERBB genes which bore short deletion mutations within the ligand-binding domain of the EGF receptor. The products of these mutated ERBB genes were about 30 kD smaller than the normal 170-kD EGF receptor, and the tumor cell membrane fractions containing the 140-kD abnormal EGF receptor showed a significant elevation of tyrosine kinase activity without its ligand. In these 2 tumors, only the rearranged ERBB genes were amplified. This suggested that DNA rearrangement had occurred before gene amplification. Yamazaki et al. (1988) could not detect any abnormal bands of ERBB in other brain tumors tested.
Function: Maternal uniparental disomy (UPD) of chromosome 7 has been reported in approximately 10% of cases of Silver-Russell syndrome (SRS; OMIM:180860). This suggests that at least 1 gene on chromosome 7 is imprinted and involved in the pathogenesis of SRS. Wakeling et al. (1998) investigated the EGFR gene as a candidate for imprinting because the gene maps to 7p12, a region homologous to an imprinted region on mouse chromosome 11. Using a restriction fragment length polymorphism, they found, however, biallelic expression of EGFR in a range of normal human fetal tissues. Expression was also demonstrated in fibroblasts and lymphoblasts from SRS patients with maternal UPD7. Thus, no evidence that EGFR is imprinted was found, making its involvement in SRS unlikely. However, EGFR was shown to be widely expressed in the human fetus, providing evidence that it plays an important role in early development. The only gene known to be imprinted on chromosome 7 at that time was MEST, also called paternally expressed gene-1 (OMIM:601029), which maps to 7q32.
Function: Verveer et al. (2000) presented evidence for a novel signaling mechanism consisting of ligand-independent lateral propagation of receptor activation in the plasma membrane. They visualized the phosphorylation of green fluorescent protein-tagged ERBB1 receptors in cells focally stimulated with EGF covalently attached to beads. This was achieved by quantitative imaging of protein reaction states in cells by fluorescence resonance energy transfer (FRET) with global analysis of fluorescence lifetime imaging microscopy data. The rapid and extensive propagation of receptor phosphorylation over the entire cell after focal stimulation demonstrated a signaling wave at the plasma membrane resulting in full activation of all receptors.
Function: Activation of epidermal growth factor receptor triggers mitogenic signaling in gastrointestinal mucosa, and its expression is also upregulated in colon cancers and most neoplasms. Pai et al. (2002) investigated whether prostaglandins transactivate EGFR. Pai et al. (2002) demonstrated that prostaglandin E2 (PGE2; see OMIM:176804) rapidly phosphorylates EGFR and triggers the extracellular signal-regulated kinase 2 (ERK2; OMIM:176948)-mitogenic signaling pathway in normal gastric epithelial and colon cancer cell lines. Inactivation of EGFR kinase with selective inhibitors significantly reduced PGE2-induced ERK2 activation, c-fos mRNA expression, and cell proliferation. Inhibition of matrix metalloproteinases, TGFA, or c-Src (OMIM:190090) blocked PGE2-mediated EGFR transactivation and downstream signaling, indicating that PGE2-induced EGFR transactivation involves signaling transduced via TGF-alpha, an EGFR ligand, likely released by c-Src-activated MMPs.
Function: Using transgenic mice and inhibitor studies, Mak and Chan (2003) showed that EGFR signaling was indispensable for the initiation of hair growth in the anagen phase of the hair cycle, but continuous expression arrested follicular development at later stages.
Function: Schlessinger (2004) reviewed the signaling pathways that are activated by EGF and fibroblast growth factor (FGF) receptors (e.g., OMIM:136350). Both receptors stimulate a similar complement of intracellular signaling pathways. However, whereas activated EGF receptors function as the main platform for recruitment of signaling proteins, signaling through the FGF receptors is mediated primarily by assembly of a multidocking protein complex. Furthermore, FGF receptor signaling is subject to additional intracellular and extracellular control mechanisms that do not affect EGF receptor signaling.
Function: Tran et al. (2003) found that Caml (CAMLG; OMIM:601118)-deficient mouse epithelial cells expressed a functional EGFR. However, EGF (OMIM:131530) stimulation resulted in impaired Egfr recycling and cytoplasmic accumulation of Egfr. Immunoprecipitation analysis indicated a direct interaction between wildtype Caml and Egfr that was dependent on ligand binding. Mutation analysis indicated that Caml bound the kinase domain of Egfr, and the proteins colocalized in the ER. Tran et al. (2003) concluded that CAML may play a role in EGFR recycling during long-term proliferative responses.
Function: Wang et al. (2003) demonstrated that human cytomegalovirus (CMV) infects cells by interacting with EGFR and inducing signaling. Transfecting EGFR-negative cells with an EGFR cDNA renders nonsusceptible cells susceptible to human CMV. Ligand displacement and crosslinking analyses showed that human CMV interacts with EGFR through gB, its principal envelope glycoprotein. gB preferentially binds EGFR and EGFR-ERBB3 (OMIM:190151) with oligomeric molecules in CHO cells transfected with ERBB family cDNAs. Wang et al. (2003) concluded that, taken together, their data indicate that EGFR is a necessary component for human CMV-triggered signaling and viral entry.
Function: Koprivica et al. (2005) demonstrated that suppressing the kinase function of EGFR blocks the activities of both myelin inhibitors and chondroitin sulfate proteoglycans in inhibiting neurite outgrowth. In addition, regeneration inhibitors trigger the phosphorylation of EGFR in a calcium-dependent manner. Local administration of EGFR inhibitors promoted significant regeneration of injured mouse optic nerve fibers, pointing to a promising therapeutic avenue for enhancing axon regeneration after central nervous system (CNS) injury.
Function: Jamnongjit et al. (2005) found that EGFR signaling promoted steroidogenesis in mouse oocyte-granulosa cell complexes and luteinizing hormone (LH; see OMIM:152780)-induced steroidogenesis in a mouse Leydig cell line. Inhibition of metalloproteinase-mediated cleavage of membrane-bound Egf abrogated LH-induced steroidogenesis in ovarian follicles, but not in the Leydig cell line, suggesting that LH receptor (LHCGR; OMIM:152790) signaling activates EGFR by different mechanisms in these 2 systems.
Function: Using immunohistochemistry with a tissue microarray containing 406 NSCLC samples, Tai et al. (2006) documented overexpression of EGFR and FGF3 (OMIM:164950) in 69% and 61% of samples, respectively. They found a significant correlation (p less than 0.001) between overexpression of EGFR and of FGF3. Tai et al. (2006) suggested that co-overexpression of EGFR and FGF3 may play an important role in the pathogenesis of lung carcinoma.
Function: Jones et al. (2006) used microarrays comprising virtually every Src homology 2 (SH2) and phosphotyrosine-binding (PTB) domain encoded in the human genome to measure the equilibrium dissociation constant of each domain for 61 peptides representing physiologic sites of tyrosine phosphorylation on the 4 ErbB receptors. By slicing through the network at different affinity thresholds, Jones et al. (2006) found surprising differences between the receptors. Most notably, EGFR and ErbB2 (OMIM:164870) became markedly more promiscuous as the threshold was lowered, whereas ErbB3 (OMIM:190151) did not. Because EGFR and ErbB2 are overexpressed in many human cancers, Jones et al. (2006) concluded that the extent to which promiscuity changes with protein concentration may contribute to the oncogenic potential of receptor tyrosine kinases.
Function: Using healthy human skin fragments obtained as surgical residua, Sorensen et al. (2006) demonstrated that sterile wounding of human skin induces epidermal expression of the antimicrobial polypeptides beta-defensin-103 (DEFB103; OMIM:606611), lipocalin-2 (LCN2; OMIM:600181), and secretory leukocyte protease inhibitor (SLPI; OMIM:107285) through activation of EGFR by heparin-binding EGF (HBEGF; OMIM:126150). Studies in epidermal cultures showed that activation of EGFR generated antimicrobial concentrations of DEFB103 and increased activity of the cultures against Staphylococcus aureus. Sorensen et al. (2006) concluded that sterile wounding initiates an innate immune response that increases resistance to overt infection and microbial colonization.
* Structure Information
1. Primary Information
Length: 1091 aa
Average Mass: 120.692 kDa
Monoisotopic Mass: 120.614 kDa
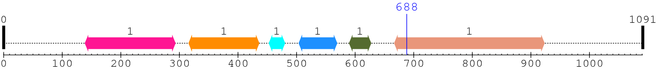
2. Domain Information
Annotated Domains: interpro / pfam / smart / prosite
Computationally Assigned Domains (Pfam+HMMER):
| domain name | begin | end | score | e-value |
|---|---|---|---|---|
| Furin-like 1. | 139 | 293 | 259.2 | 9.9e-75 |
| Recep_L_domain 1. | 316 | 436 | 155.1 | 2e-43 |
| CBM_10 1. | 453 | 480 | -4.6 | 9.2 |
| NinF 1. | 504 | 569 | 0.1 | 2.4 |
| Gram_pos_anchor 1. | 590 | 627 | 0.6 | 2.7 |
| Pkinase_Tyr 1. | 667 | 923 | 439.6 | 4.7e-129 |
| --- cleavage 688 (inside Pkinase_Tyr 667..923) --- | ||||
3. Sequence Information
Fasta Sequence: XSB1083.fasta
Amino Acid Sequence and Secondary Structures (PsiPred):
4. 3D Information
Not Available.
* Cleavage Information
1 [sites]
Cleavage sites (±10aa)
[Site 1] GTVYKGLWIP688-EGEKVKIPVA
Pro688  Glu
Glu
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Gly679 | Thr680 | Val681 | Tyr682 | Lys683 | Gly684 | Leu685 | Trp686 | Ile687 | Pro688 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Glu689 | Gly690 | Glu691 | Lys692 | Val693 | Lys694 | Ile695 | Pro696 | Val697 | Ala698 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| LRILKETEFKKIKVLGSGAFGTVYKGLWIPEGEKVKIPVAIKELREATSPKANKEILDEA |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | N/A | 120.00 | 21 | - |
| 2 | Homo sapiens | 120.00 | 15 | EGFR protein |
| 3 | Mus musculus | 120.00 | 8 | epidermal growth factor receptor isoform 1 |
| 4 | Rattus norvegicus | 120.00 | 7 | epidermal growth factor receptor |
| 5 | Pan troglodytes | 120.00 | 7 | PREDICTED: epidermal growth factor receptor isofor |
| 6 | synthetic construct | 120.00 | 4 | epidermal growth factor receptor |
| 7 | Monodelphis domestica | 120.00 | 3 | PREDICTED: similar to Epidermal growth factor rece |
| 8 | Canis familiaris | 120.00 | 3 | PREDICTED: similar to Epidermal growth factor rece |
| 9 | Macaca mulatta | 120.00 | 3 | PREDICTED: epidermal growth factor receptor, parti |
| 10 | Rous-associated virus type 1 | 119.00 | 2 | polyprotein |
| 11 | Sus scrofa | 119.00 | 2 | epidermal growth factor receptor |
| 12 | Bos taurus | 119.00 | 1 | PREDICTED: similar to epidermal growth factor rece |
| 13 | Xenopus laevis | 115.00 | 3 | epidermal growth factor receptor |
| 14 | Avian erythroblastosis virus | 115.00 | 2 | gag,v-erb-A,v-erb-B |
| 15 | Danio rerio | 112.00 | 4 | epidermal growth factor receptor |
| 16 | Tetraodon nigroviridis | 111.00 | 3 | unnamed protein product |
| 17 | Xiphophorus xiphidium | 110.00 | 2 | epidermal growth factor receptor |
| 18 | Gallus gallus | 107.00 | 4 | erbB-2 |
| 19 | Felis catus | 106.00 | 2 | v-erb-b2 erythroblastic leukemia viral oncogene ho |
| 20 | Mesocricetus auratus | 106.00 | 1 | ERBB2_MESAU Receptor tyrosine-protein kinase erbB- |
| 21 | Canis lupus familiaris | 106.00 | 1 | erbB-2 |
| 22 | Xiphophorus sp. | 102.00 | 1 | XMRK_XIPMA Melanoma receptor tyrosine-protein kina |
| 23 | Ovis aries | 94.00 | 1 | erythroblastic leukemia viral oncogene-like 2 |
| 24 | Strongylocentrotus purpuratus | 80.90 | 1 | PREDICTED: similar to v-erb-a erythroblastic leuke |
| 25 | Drosophila melanogaster | 80.10 | 147 | mutant epidermal growth factor receptor |
| 26 | Drosophila simulans | 78.20 | 1 | epidermal growth factor receptor |
Top-ranked sequences
| organism | matching |
|---|---|
| N/A | Query 659 LRILKETEFKKIKVLGSGAFGTVYKGLWIP#EGEKVKIPVAIKELREATSPKANKEILDEA 718 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 659 LRILKETEFKKIKVLGSGAFGTVYKGLWIP#EGEKVKIPVAIKELREATSPKANKEILDEA 718 |
| Homo sapiens | Query 659 LRILKETEFKKIKVLGSGAFGTVYKGLWIP#EGEKVKIPVAIKELREATSPKANKEILDEA 718 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 659 LRILKETEFKKIKVLGSGAFGTVYKGLWIP#EGEKVKIPVAIKELREATSPKANKEILDEA 718 |
| Mus musculus | Query 659 LRILKETEFKKIKVLGSGAFGTVYKGLWIP#EGEKVKIPVAIKELREATSPKANKEILDEA 718 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 659 LRILKETEFKKIKVLGSGAFGTVYKGLWIP#EGEKVKIPVAIKELREATSPKANKEILDEA 718 |
| Rattus norvegicus | Query 659 LRILKETEFKKIKVLGSGAFGTVYKGLWIP#EGEKVKIPVAIKELREATSPKANKEILDEA 718 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 659 LRILKETEFKKIKVLGSGAFGTVYKGLWIP#EGEKVKIPVAIKELREATSPKANKEILDEA 718 |
| Pan troglodytes | Query 659 LRILKETEFKKIKVLGSGAFGTVYKGLWIP#EGEKVKIPVAIKELREATSPKANKEILDEA 718 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 659 LRILKETEFKKIKVLGSGAFGTVYKGLWIP#EGEKVKIPVAIKELREATSPKANKEILDEA 718 |
| synthetic construct | Query 659 LRILKETEFKKIKVLGSGAFGTVYKGLWIP#EGEKVKIPVAIKELREATSPKANKEILDEA 718 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 659 LRILKETEFKKIKVLGSGAFGTVYKGLWIP#EGEKVKIPVAIKELREATSPKANKEILDEA 718 |
| Monodelphis domestica | Query 659 LRILKETEFKKIKVLGSGAFGTVYKGLWIP#EGEKVKIPVAIKELREATSPKANKEILDEA 718 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 659 LRILKETEFKKIKVLGSGAFGTVYKGLWIP#EGEKVKIPVAIKELREATSPKANKEILDEA 718 |
| Canis familiaris | Query 659 LRILKETEFKKIKVLGSGAFGTVYKGLWIP#EGEKVKIPVAIKELREATSPKANKEILDEA 718 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 659 LRILKETEFKKIKVLGSGAFGTVYKGLWIP#EGEKVKIPVAIKELREATSPKANKEILDEA 718 |
| Macaca mulatta | Query 659 LRILKETEFKKIKVLGSGAFGTVYKGLWIP#EGEKVKIPVAIKELREATSPKANKEILDEA 718 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 659 LRILKETEFKKIKVLGSGAFGTVYKGLWIP#EGEKVKIPVAIKELREATSPKANKEILDEA 718 |
| Rous-associated virus type 1 | Query 659 LRILKETEFKKIKVLGSGAFGTVYKGLWIP#EGEKVKIPVAIKELREATSPKANKEILDEA 718 |||||||||||+||||||||||||||||||#|||||||||||||||||||||||||||||| Sbjct 278 LRILKETEFKKVKVLGSGAFGTVYKGLWIP#EGEKVKIPVAIKELREATSPKANKEILDEA 337 |
| Sus scrofa | Query 659 LRILKETEFKKIKVLGSGAFGTVYKGLWIP#EGEKVKIPVAIKELREATSPKANKEILDEA 718 |||||||||||+||||||||||||||||||#|||||||||||||||||||||||||||||| Sbjct 699 LRILKETEFKKVKVLGSGAFGTVYKGLWIP#EGEKVKIPVAIKELREATSPKANKEILDEA 758 |
| Bos taurus | Query 659 LRILKETEFKKIKVLGSGAFGTVYKGLWIP#EGEKVKIPVAIKELREATSPKANKEILDEA 718 |||||||||||+||||||||||||||||||#|||||||||||||||||||||||||||||| Sbjct 681 LRILKETEFKKVKVLGSGAFGTVYKGLWIP#EGEKVKIPVAIKELREATSPKANKEILDEA 740 |
| Xenopus laevis | Query 659 LRILKETEFKKIKVLGSGAFGTVYKGLWIP#EGEKVKIPVAIKELREATSPKANKEILDEA 718 ||||||||||||||||||||||||+|||||#||| +||||||||||||||||||||||||| Sbjct 289 LRILKETEFKKIKVLGSGAFGTVYQGLWIP#EGEGIKIPVAIKELREATSPKANKEILDEA 348 |
| Avian erythroblastosis virus | Query 659 LRILKETEFKKIKVLGSGAFGTVYKGLWIP#EGEKVKIPVAIKELREATSPKANKEILDEA 718 |||||||||||+|||| |||||||||||||#||||| |||||||||||||||||||||||| Sbjct 546 LRILKETEFKKVKVLGFGAFGTVYKGLWIP#EGEKVTIPVAIKELREATSPKANKEILDEA 605 |
| Danio rerio | Query 659 LRILKETEFKKIKVLGSGAFGTVYKGLWIP#EGEKVKIPVAIKELREATSPKANKEILDEA 718 |||||||||||||||||||||||+||||+|#||| |||||||| |||||||||||||+||| Sbjct 703 LRILKETEFKKIKVLGSGAFGTVHKGLWVP#EGENVKIPVAIKVLREATSPKANKEIMDEA 762 |
| Tetraodon nigroviridis | Query 659 LRILKETEFKKIKVLGSGAFGTVYKGLWIP#EGEKVKIPVAIKELREATSPKANKEILDEA 718 |||||| |||||||||||||||||||||+|#||| |||||||| |||||||||||+||||| Sbjct 674 LRILKEPEFKKIKVLGSGAFGTVYKGLWVP#EGEDVKIPVAIKVLREATSPKANKDILDEA 733 |
| Xiphophorus xiphidium | Query 659 LRILKETEFKKIKVLGSGAFGTVYKGLWIP#EGEKVKIPVAIKELREATSPKANKEILDEA 718 |||||| |||||||||||||||||||||+|#||| |||||||| ||||||||||+|||||| Sbjct 714 LRILKEPEFKKIKVLGSGAFGTVYKGLWVP#EGEDVKIPVAIKVLREATSPKANQEILDEA 773 |
| Gallus gallus | Query 659 LRILKETEFKKIKVLGSGAFGTVYKGLWIP#EGEKVKIPVAIKELREATSPKANKEILDEA 718 +||||||| ||+||||||||||||||+|||#+|| |||||||| ||| ||||||||||||| Sbjct 701 MRILKETELKKVKVLGSGAFGTVYKGIWIP#DGESVKIPVAIKVLRENTSPKANKEILDEA 760 |
| Felis catus | Query 659 LRILKETEFKKIKVLGSGAFGTVYKGLWIP#EGEKVKIPVAIKELREATSPKANKEILDEA 718 +||||||| +|+||||||||||||||+|||#+|| |||||||| ||| ||||||||||||| Sbjct 712 MRILKETELRKVKVLGSGAFGTVYKGIWIP#DGENVKIPVAIKVLRENTSPKANKEILDEA 771 |
| Mesocricetus auratus | Query 659 LRILKETEFKKIKVLGSGAFGTVYKGLWIP#EGEKVKIPVAIKELREATSPKANKEILDEA 718 +||||||| +|+||||||||||||||+|||#+|| |||||||| ||| ||||||||||||| Sbjct 712 MRILKETELRKVKVLGSGAFGTVYKGIWIP#DGENVKIPVAIKVLRENTSPKANKEILDEA 771 |
| Canis lupus familiaris | Query 659 LRILKETEFKKIKVLGSGAFGTVYKGLWIP#EGEKVKIPVAIKELREATSPKANKEILDEA 718 +||||||| +|+||||||||||||||+|||#+|| |||||||| ||| ||||||||||||| Sbjct 711 MRILKETELRKVKVLGSGAFGTVYKGIWIP#DGENVKIPVAIKVLRENTSPKANKEILDEA 770 |
| Xiphophorus sp. | Query 659 LRILKETEFKKIKVLGSGAFGTVYKGLWIP#EGEKVKIPVAIKELREATSPKANKEILDEA 718 ||||||||||| +||||||||||||||| |#+|| ++|||||| |||||||| |+|+|||| Sbjct 702 LRILKETEFKKDRVLGSGAFGTVYKGLWNP#DGENIRIPVAIKVLREATSPKVNQEVLDEA 761 |
| Ovis aries | Query 659 LRILKETEFKKIKVLGSGAFGTVYKGLWIP#EGEKVKIPVAIKELREATSPKAN 711 +||||||| +|+||||||||||||||+|||#+|| |||||||| ||| |||||| Sbjct 63 MRILKETELRKVKVLGSGAFGTVYKGIWIP#DGENVKIPVAIKVLRENTSPKAN 115 |
| Strongylocentrotus purpuratus | Query 659 LRILKETEFKKIKVLGSGAFGTVYKGLWIP#EGEKVKIPVAIKELREATSPKANKEILD 716 |+|+|+|| | |||||||||||||||||#+|||++|||||| ||| || | +|+|+ Sbjct 1012 LKIIKDTELKLGPVLGSGAFGTVYKGLWIP#DGEKIRIPVAIKALRE-VSPHAAEELLE 1068 |
| Drosophila melanogaster | Query 659 LRILKETEFKKIKVLGSGAFGTVYKGLWIP#EGEKVKIPVAIKELREATSPKANKEILDEA 718 |||+|+|| +| ||| |||| ||||+|+|#||| |||||||||| ++| ++++| | || Sbjct 830 LRIVKDTELRKGGVLGMGAFGRVYKGVWVP#EGENVKIPVAIKELLKSTGAESSEEFLREA 889 |
| Drosophila simulans | Query 659 LRILKETEFKKIKVLGSGAFGTVYKGLWIP#EGEKVKIPVAIKELREATSPKANKEILDEA 718 |||+|+ | +| ||| |||| ||||+|+|#||| |||||||||| ++| ++++| | || Sbjct 829 LRIVKDAELRKGGVLGMGAFGRVYKGVWVP#EGENVKIPVAIKELLKSTGAESSEEFLREA 888 |
* References
[PubMed ID: 12477932] Strausberg RL, Feingold EA, Grouse LH, Derge JG, Klausner RD, Collins FS, Wagner L, Shenmen CM, Schuler GD, Altschul SF, Zeeberg B, Buetow KH, Schaefer CF, Bhat NK, Hopkins RF, Jordan H, Moore T, Max SI, Wang J, Hsieh F, Diatchenko L, Marusina K, Farmer AA, Rubin GM, Hong L, Stapleton M, Soares MB, Bonaldo MF, Casavant TL, Scheetz TE, Brownstein MJ, Usdin TB, Toshiyuki S, Carninci P, Prange C, Raha SS, Loquellano NA, Peters GJ, Abramson RD, Mullahy SJ, Bosak SA, McEwan PJ, McKernan KJ, Malek JA, Gunaratne PH, Richards S, Worley KC, Hale S, Garcia AM, Gay LJ, Hulyk SW, Villalon DK, Muzny DM, Sodergren EJ, Lu X, Gibbs RA, Fahey J, Helton E, Ketteman M, Madan A, Rodrigues S, Sanchez A, Whiting M, Madan A, Young AC, Shevchenko Y, Bouffard GG, Blakesley RW, Touchman JW, Green ED, Dickson MC, Rodriguez AC, Grimwood J, Schmutz J, Myers RM, Butterfield YS, Krzywinski MI, Skalska U, Smailus DE, Schnerch A, Schein JE, Jones SJ, Marra MA, Generation and initial analysis of more than 15,000 full-length human and mouse cDNA sequences. Proc Natl Acad Sci U S A. 2002 Dec 24;99(26):16899-903. Epub 2002 Dec 11.