XSB1183 : PREDICTED: similar to neurofibromin 2 isoform 6 isoform 7 [Canis familiaris]
[ CaMP Format ]
This entry is computationally expanded from SB0009
* Basic Information
| Organism | Canis lupus familiaris (dog) |
| Protein Names | similar to neurofibromin 2 isoform 6 isoform 7 |
| Gene Names | LOC477535; Derived by automated computational analysis using gene prediction method: GNOMON. Supporting evidence includes similarity to: 11 mRNAs, 31 ESTs, 107 Proteins |
| Gene Locus | Not available |
| GO Function | Not available |
* Information From OMIM
Not Available.
* Structure Information
1. Primary Information
Length: 550 aa
Average Mass: 64.310 kDa
Monoisotopic Mass: 64.269 kDa
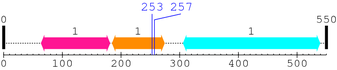
2. Domain Information
Annotated Domains: Not Available.
Computationally Assigned Domains (Pfam+HMMER):
| domain name | begin | end | score | e-value |
|---|---|---|---|---|
| FERM_M 1. | 64 | 181 | 100.7 | 5e-27 |
| FERM_C 1. | 185 | 274 | 177.6 | 3.5e-50 |
| --- cleavage 253 (inside FERM_C 185..274) --- | ||||
| --- cleavage 257 (inside FERM_C 185..274) --- | ||||
| ERM 1. | 306 | 539 | 294.2 | 2.8e-85 |
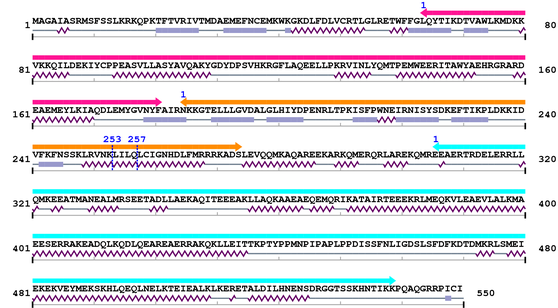
3. Sequence Information
Fasta Sequence: XSB1183.fasta
Amino Acid Sequence and Secondary Structures (PsiPred):
4. 3D Information
Not Available.
* Cleavage Information
2 [sites]
Cleavage sites (±10aa)
[Site 1] FNSSKLRVNK253-LILQLCIGNH
Lys253  Leu
Leu
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Phe244 | Asn245 | Ser246 | Ser247 | Lys248 | Leu249 | Arg250 | Val251 | Asn252 | Lys253 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Leu254 | Ile255 | Leu256 | Gln257 | Leu258 | Cys259 | Ile260 | Gly261 | Asn262 | His263 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| SYSDKEFTIKPLDKKIDVFKFNSSKLRVNKLILQLCIGNHDLFMRRRKADSLEVQQMKAQ |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | Homo sapiens | 121.00 | 35 | neurofibromin 2 isoform 6 |
| 2 | Canis familiaris | 121.00 | 32 | PREDICTED: similar to neurofibromin 2 isoform 6 is |
| 3 | Mus musculus | 121.00 | 25 | unnamed protein product |
| 4 | N/A | 121.00 | 24 | - |
| 5 | Rattus norvegicus | 121.00 | 8 | MERL_RAT Merlin (Moesin-ezrin-radixin-like protein |
| 6 | Xenopus tropicalis | 121.00 | 6 | neurofibromin 2 |
| 7 | Monodelphis domestica | 121.00 | 5 | PREDICTED: hypothetical protein |
| 8 | Gallus gallus | 121.00 | 5 | neurofibromatosis 2 |
| 9 | Macaca mulatta | 121.00 | 5 | PREDICTED: similar to neurofibromin 2 isoform 1 is |
| 10 | Bos taurus | 121.00 | 4 | PREDICTED: similar to merlin isoform 1 |
| 11 | Papio anubis anubis | 121.00 | 1 | MERL_PAPAN Merlin (Moesin-ezrin-radixin-like prote |
| 12 | Xenopus laevis | 119.00 | 6 | neurofibromin 2 (bilateral acoustic neuroma) |
| 13 | Danio rerio | 110.00 | 14 | PREDICTED: hypothetical protein |
| 14 | Tetraodon nigroviridis | 109.00 | 7 | unnamed protein product |
| 15 | Strongylocentrotus purpuratus | 87.00 | 4 | PREDICTED: similar to neurofibromin 2 (bilateral a |
| 16 | Oryctolagus cuniculus | 86.70 | 1 | villin 2 (ezrin) |
| 17 | Tribolium castaneum | 84.70 | 6 | PREDICTED: similar to neurofibromatosis 2 |
| 18 | Molgula tectiformis | 84.70 | 2 | Mt-ezrin/radixin/moesin |
| 19 | Pan troglodytes | 84.30 | 4 | PREDICTED: moesin |
| 20 | Sus scrofa | 84.30 | 2 | MOESIN protein |
| 21 | Biomphalaria glabrata | 84.30 | 1 | ezrin/radixin/moesin |
| 22 | mice, keratinocytes, Balb/MK, Peptide, 583 aa | 84.30 | 1 | RADI_MOUSE Radixin (ESP10) gb |
| 23 | Macaca fascicularis | 84.30 | 1 | unnamed protein product |
| 24 | Aplysia californica | 84.00 | 1 | ezrin/radixin/moesin |
| 25 | Ciona intestinalis | 83.60 | 1 | ezrin/radixin/moesin (ERM)-like protein |
| 26 | Caenorhabditis elegans | 82.40 | 6 | Ezrin/Radixin/Moesin family member (erm-1) |
| 27 | Caenorhabditis briggsae | 82.40 | 2 | Hypothetical protein CBG12867 |
| 28 | Drosophila melanogaster | 81.30 | 7 | Moesin CG10701-PD, isoform D |
| 29 | Drosophila pseudoobscura | 81.30 | 2 | GA10507-PA |
| 30 | Spodoptera frugiperda | 80.50 | 1 | A unnamed protein product |
| 31 | Aedes aegypti | 80.10 | 4 | moesin/ezrin/radixin |
| 32 | Anopheles gambiae str. PEST | 80.10 | 2 | ENSANGP00000017331 |
| 33 | Apis mellifera | 79.30 | 1 | PREDICTED: similar to Moesin CG10701-PD, isoform D |
| 34 | Taenia saginata | 76.30 | 1 | myosin-like protein |
| 35 | Taenia solium | 76.30 | 1 | H17g protein, tegumental antigen |
| 36 | Echinococcus multilocularis | 75.90 | 1 | tegument protein gb |
| 37 | Echinococcus granulosus | 75.90 | 1 | EG10 |
| 38 | Schistosoma japonicum | 53.10 | 1 | SJCHGC06288 protein |
Top-ranked sequences
| organism | matching |
|---|---|
| Homo sapiens | Query 224 SYSDKEFTIKPLDKKIDVFKFNSSKLRVNK#LILQLCIGNHDLFMRRRKADSLEVQQMKAQ 283 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 224 SYSDKEFTIKPLDKKIDVFKFNSSKLRVNK#LILQLCIGNHDLFMRRRKADSLEVQQMKAQ 283 |
| Canis familiaris | Query 224 SYSDKEFTIKPLDKKIDVFKFNSSKLRVNK#LILQLCIGNHDLFMRRRKADSLEVQQMKAQ 283 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 224 SYSDKEFTIKPLDKKIDVFKFNSSKLRVNK#LILQLCIGNHDLFMRRRKADSLEVQQMKAQ 283 |
| Mus musculus | Query 224 SYSDKEFTIKPLDKKIDVFKFNSSKLRVNK#LILQLCIGNHDLFMRRRKADSLEVQQMKAQ 283 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 224 SYSDKEFTIKPLDKKIDVFKFNSSKLRVNK#LILQLCIGNHDLFMRRRKADSLEVQQMKAQ 283 |
| N/A | Query 224 SYSDKEFTIKPLDKKIDVFKFNSSKLRVNK#LILQLCIGNHDLFMRRRKADSLEVQQMKAQ 283 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 224 SYSDKEFTIKPLDKKIDVFKFNSSKLRVNK#LILQLCIGNHDLFMRRRKADSLEVQQMKAQ 283 |
| Rattus norvegicus | Query 224 SYSDKEFTIKPLDKKIDVFKFNSSKLRVNK#LILQLCIGNHDLFMRRRKADSLEVQQMKAQ 283 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 224 SYSDKEFTIKPLDKKIDVFKFNSSKLRVNK#LILQLCIGNHDLFMRRRKADSLEVQQMKAQ 283 |
| Xenopus tropicalis | Query 224 SYSDKEFTIKPLDKKIDVFKFNSSKLRVNK#LILQLCIGNHDLFMRRRKADSLEVQQMKAQ 283 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 224 SYSDKEFTIKPLDKKIDVFKFNSSKLRVNK#LILQLCIGNHDLFMRRRKADSLEVQQMKAQ 283 |
| Monodelphis domestica | Query 224 SYSDKEFTIKPLDKKIDVFKFNSSKLRVNK#LILQLCIGNHDLFMRRRKADSLEVQQMKAQ 283 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 224 SYSDKEFTIKPLDKKIDVFKFNSSKLRVNK#LILQLCIGNHDLFMRRRKADSLEVQQMKAQ 283 |
| Gallus gallus | Query 224 SYSDKEFTIKPLDKKIDVFKFNSSKLRVNK#LILQLCIGNHDLFMRRRKADSLEVQQMKAQ 283 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 224 SYSDKEFTIKPLDKKIDVFKFNSSKLRVNK#LILQLCIGNHDLFMRRRKADSLEVQQMKAQ 283 |
| Macaca mulatta | Query 224 SYSDKEFTIKPLDKKIDVFKFNSSKLRVNK#LILQLCIGNHDLFMRRRKADSLEVQQMKAQ 283 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 224 SYSDKEFTIKPLDKKIDVFKFNSSKLRVNK#LILQLCIGNHDLFMRRRKADSLEVQQMKAQ 283 |
| Bos taurus | Query 224 SYSDKEFTIKPLDKKIDVFKFNSSKLRVNK#LILQLCIGNHDLFMRRRKADSLEVQQMKAQ 283 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 224 SYSDKEFTIKPLDKKIDVFKFNSSKLRVNK#LILQLCIGNHDLFMRRRKADSLEVQQMKAQ 283 |
| Papio anubis anubis | Query 224 SYSDKEFTIKPLDKKIDVFKFNSSKLRVNK#LILQLCIGNHDLFMRRRKADSLEVQQMKAQ 283 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 224 SYSDKEFTIKPLDKKIDVFKFNSSKLRVNK#LILQLCIGNHDLFMRRRKADSLEVQQMKAQ 283 |
| Xenopus laevis | Query 224 SYSDKEFTIKPLDKKIDVFKFNSSKLRVNK#LILQLCIGNHDLFMRRRKADSLEVQQMKAQ 283 ||||||||||||+|||||||||||||||||#|||||||||||||||||||||||||||||| Sbjct 265 SYSDKEFTIKPLEKKIDVFKFNSSKLRVNK#LILQLCIGNHDLFMRRRKADSLEVQQMKAQ 324 |
| Danio rerio | Query 224 SYSDKEFTIKPLDKKIDVFKFNSSKLRVNK#LILQLCIGNHDLFMRRRKADSLEVQQMKAQ 283 ||||||| |||+||+ ||||||||||||||#|||||||||||||||||+ ||||||||| | Sbjct 264 SYSDKEFAIKPVDKRADVFKFNSSKLRVNK#LILQLCIGNHDLFMRRRRVDSLEVQQMKTQ 323 |
| Tetraodon nigroviridis | Query 224 SYSDKEFTIKPLDKKIDVFKFNSSKLRVNK#LILQLCIGNHDLFMRRRKADSLEVQQMKAQ 283 ||||||||||||+|| |||||||+|| ||#|||||||||||||||||+ ||||||||||| Sbjct 159 SYSDKEFTIKPLEKKTKVFKFNSSRLRANK#LILQLCIGNHDLFMRRRRVDSLEVQQMKAQ 218 |
| Strongylocentrotus purpuratus | Query 224 SYSDKEFTIKPLDKKIDVFKFNSSKLRVNK#LILQLCIGNHDLFMRRRKADSLEVQQMKAQ 283 |+ ||+||||| || | | | |||+||#||| ||+|||+|||+||+|||+|||||||| Sbjct 275 SFRDKKFTIKPTAKKAPNFCFISPKLRMNK#LILDLCVGNHELFMQRRRADSMEVQQMKAQ 334 |
| Oryctolagus cuniculus | Query 224 SYSDKEFTIKPLDKKIDVFKFNSSKLRVNK#LILQLCIGNHDLFMRRRKADSLEVQQMKAQ 283 |++||+| |||+||| | | + +||+||# |||||+|||+|+||||| |++|||||||| Sbjct 249 SFNDKKFVIKPIDKKAPDFVFYAPRLRINK#RILQLCMGNHELYMRRRKPDTIEVQQMKAQ 308 |
| Tribolium castaneum | Query 224 SYSDKEFTIKPLDKKIDVFKFNSSKLRVNK#LILQLCIGNHDLFMRRRKADSLEVQQMKA 282 |+ ||+| |||+|| | | | |+|+||#||| ||+||||||||||| ||+|+||||| Sbjct 259 SFDDKKFIIKPVDKNSPNFVFFSQKVRMNK#LILDLCMGNHDLFMRRRKPDSMELQQMKA 317 |
| Molgula tectiformis | Query 224 SYSDKEFTIKPLDKKIDVFKFNSSKLRVNK#LILQLCIGNHDLFMRRRKADSLEVQQMKAQ 283 |++||+| |||+||| | | +||+||# || ||+|||+|+||||| ||+|||||||| Sbjct 249 SFNDKKFVIKPIDKKAPDFVFYVERLRINK#RILALCMGNHELYMRRRKPDSIEVQQMKAQ 308 |
| Pan troglodytes | Query 224 SYSDKEFTIKPLDKKIDVFKFNSSKLRVNK#LILQLCIGNHDLFMRRRKADSLEVQQMKAQ 283 |++||+| |||+||| | | + +||+||# || ||+|||+|+||||| |++|||||||| Sbjct 221 SFNDKKFVIKPIDKKAPDFVFYAPRLRINK#RILALCMGNHELYMRRRKPDTIEVQQMKAQ 280 |
| Sus scrofa | Query 224 SYSDKEFTIKPLDKKIDVFKFNSSKLRVNK#LILQLCIGNHDLFMRRRKADSLEVQQMKAQ 283 |++||+| |||+||| | | + +||+||# || ||+|||+|+||||| |++|||||||| Sbjct 249 SFNDKKFVIKPIDKKAPDFVFYAPRLRINK#RILALCMGNHELYMRRRKPDTIEVQQMKAQ 308 |
| Biomphalaria glabrata | Query 224 SYSDKEFTIKPLDKKIDVFKFNSSKLRVNK#LILQLCIGNHDLFMRRRKADSLEVQQMKAQ 283 |++||+| |||+||| | | + +||+||# || ||+|||+|+||||| |++|||||||| Sbjct 250 SFNDKKFVIKPIDKKAPDFVFYAPRLRINK#RILALCMGNHELYMRRRKPDTIEVQQMKAQ 309 |
| mice, keratinocytes, Balb/MK, Peptide, 583 aa | Query 224 SYSDKEFTIKPLDKKIDVFKFNSSKLRVNK#LILQLCIGNHDLFMRRRKADSLEVQQMKAQ 283 |++||+| |||+||| | | + +||+||# || ||+|||+|+||||| |++|||||||| Sbjct 249 SFNDKKFVIKPIDKKAPDFVFYAPRLRINK#RILALCMGNHELYMRRRKPDTIEVQQMKAQ 308 |
| Macaca fascicularis | Query 224 SYSDKEFTIKPLDKKIDVFKFNSSKLRVNK#LILQLCIGNHDLFMRRRKADSLEVQQMKAQ 283 |++||+| |||+||| | | + +||+||# || ||+|||+|+||||| |++|||||||| Sbjct 249 SFNDKKFVIKPIDKKAPDFVFYAPRLRINK#RILALCMGNHELYMRRRKPDTIEVQQMKAQ 308 |
| Aplysia californica | Query 224 SYSDKEFTIKPLDKKIDVFKFNSSKLRVNK#LILQLCIGNHDLFMRRRKADSLEVQQMKAQ 283 |++||+| |||+||| | | + +||+||# || ||+|||+|+||||| |++|||||||| Sbjct 250 SFNDKKFIIKPIDKKAPDFVFYAPRLRINK#RILALCMGNHELYMRRRKPDTIEVQQMKAQ 309 |
| Ciona intestinalis | Query 224 SYSDKEFTIKPLDKKIDVFKFNSSKLRVNK#LILQLCIGNHDLFMRRRKADSLEVQQMKAQ 283 |++||+| |||+||| | | +||+||# || ||+|||+|+||||| |++|||||||| Sbjct 273 SFNDKKFVIKPIDKKAPDFVFYVERLRINK#RILALCMGNHELYMRRRKPDTIEVQQMKAQ 332 |
| Caenorhabditis elegans | Query 224 SYSDKEFTIKPLDKKIDVFKFNSSKLRVNK#LILQLCIGNHDLFMRRRKADSLEVQQMKAQ 283 |++||+| |||+||| | | + +||+||# || ||+|||+|+||||| |++|||||| | Sbjct 250 SFNDKKFVIKPIDKKAHDFVFYAPRLRINK#RILALCMGNHELYMRRRKPDTIEVQQMKQQ 309 |
| Caenorhabditis briggsae | Query 224 SYSDKEFTIKPLDKKIDVFKFNSSKLRVNK#LILQLCIGNHDLFMRRRKADSLEVQQMKAQ 283 |++||+| |||+||| | | + +||+||# || ||+|||+|+||||| |++|||||| | Sbjct 245 SFNDKKFVIKPIDKKAHDFVFYAPRLRINK#RILALCMGNHELYMRRRKPDTIEVQQMKQQ 304 |
| Drosophila melanogaster | Query 224 SYSDKEFTIKPLDKKIDVFKFNSSKLRVNK#LILQLCIGNHDLFMRRRKADSLEVQQMKAQ 283 |+|+|+| |||+||| | | + ++|+||# || ||+|||+|+||||| |+++||||||| Sbjct 250 SFSEKKFIIKPIDKKAPDFMFFAPRVRINK#RILALCMGNHELYMRRRKPDTIDVQQMKAQ 309 |
| Drosophila pseudoobscura | Query 224 SYSDKEFTIKPLDKKIDVFKFNSSKLRVNK#LILQLCIGNHDLFMRRRKADSLEVQQMKAQ 283 |+|+|+| |||+||| | | + ++|+||# || ||+|||+|+||||| |+++||||||| Sbjct 245 SFSEKKFIIKPIDKKAPDFMFFAPRVRINK#RILALCMGNHELYMRRRKPDTIDVQQMKAQ 304 |
| Spodoptera frugiperda | Query 224 SYSDKEFTIKPLDKKIDVFKFNSSKLRVNK#LILQLCIGNHDLFMRRRKADSLEVQQMKAQ 283 |++|++| |||+||| | | + ++||||# || ||+|||+|+||||| |+++||||||| Sbjct 249 SFNDRKFIIKPIDKKAPDFVFFAPRVRVNK#RILALCMGNHELYMRRRKPDTIDVQQMKAQ 308 |
| Aedes aegypti | Query 224 SYSDKEFTIKPLDKKIDVFKFNSSKLRVNK#LILQLCIGNHDLFMRRRKADSLEVQQMKAQ 283 |++|++| |||+||| | | + ++|+||# || ||+|||+|+||||| |+++||||||| Sbjct 255 SFNDRKFIIKPIDKKAPDFVFFAPRVRINK#RILALCMGNHELYMRRRKPDTIDVQQMKAQ 314 |
| Anopheles gambiae str. PEST | Query 224 SYSDKEFTIKPLDKKIDVFKFNSSKLRVNK#LILQLCIGNHDLFMRRRKADSLEVQQMKAQ 283 |++|++| |||+||| | | + ++|+||# || ||+|||+|+||||| |+++||||||| Sbjct 296 SFNDRKFIIKPIDKKAPDFVFFAPRVRINK#RILALCMGNHELYMRRRKPDTIDVQQMKAQ 355 |
| Apis mellifera | Query 224 SYSDKEFTIKPLDKKIDVFKFNSSKLRVNK#LILQLCIGNHDLFMRRRKADSLEVQQMKAQ 283 |+++|+| |||+||| | | ++++++||# || ||+|||+|+||||| |+++||||||| Sbjct 184 SFNEKKFIIKPIDKKAPDFVFFATRVKINK#RILALCMGNHELYMRRRKPDTIDVQQMKAQ 243 |
| Taenia saginata | Query 224 SYSDKEFTIKPLDKKIDVFKFNSSKLRVNK#LILQLCIGNHDLFMRRRKADSLEVQQMKAQ 283 |+ ||+| ||| || | | | ++||# || || |||+|+|||||+||+|||||| | Sbjct 251 SFHDKKFIIKPADKSAKEFYFLVEKSKINK#RILALCTGNHELYMRRRKSDSIEVQQMKIQ 310 |
| Taenia solium | Query 224 SYSDKEFTIKPLDKKIDVFKFNSSKLRVNK#LILQLCIGNHDLFMRRRKADSLEVQQMKAQ 283 |+ ||+| ||| || | | | ++||# || || |||+|+|||||+||+|||||| | Sbjct 244 SFHDKKFIIKPADKSAKEFYFLVEKSKINK#RILALCTGNHELYMRRRKSDSIEVQQMKIQ 303 |
| Echinococcus multilocularis | Query 224 SYSDKEFTIKPLDKKIDVFKFNSSKLRVNK#LILQLCIGNHDLFMRRRKADSLEVQQMKAQ 283 |+ ||+| ||| || | | | ++||# || || |||+|+|||||+||+|||||| | Sbjct 251 SFHDKKFIIKPADKSAKEFFFLVEKSKINK#RILALCTGNHELYMRRRKSDSIEVQQMKIQ 310 |
| Echinococcus granulosus | Query 224 SYSDKEFTIKPLDKKIDVFKFNSSKLRVNK#LILQLCIGNHDLFMRRRKADSLEVQQMKAQ 283 |+ ||+| ||| || | | | ++||# || || |||+|+|||||+||+|||||| | Sbjct 251 SFHDKKFIIKPADKSAKEFFFLVEKSKINK#RILALCTGNHELYMRRRKSDSIEVQQMKIQ 310 |
| Schistosoma japonicum | Query 224 SYSDKEFTIKPLDKKIDVFKFNSSKLRVNK#LILQLCIGNHDLFMRRRKADSLEVQQMKAQ 283 ||| +| +||+ +| + ++#||| | +||| |+ ||+ ||+|||||| + Sbjct 249 SYSQNKFYVKPVGASGEVLTLYTDSTHTSR#LILNLSMGNHKLYAVRRQPDSIEVQQMKVK 308 |
[Site 2] KLRVNKLILQ257-LCIGNHDLFM
Gln257  Leu
Leu
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Lys248 | Leu249 | Arg250 | Val251 | Asn252 | Lys253 | Leu254 | Ile255 | Leu256 | Gln257 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Leu258 | Cys259 | Ile260 | Gly261 | Asn262 | His263 | Asp264 | Leu265 | Phe266 | Met267 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| KEFTIKPLDKKIDVFKFNSSKLRVNKLILQLCIGNHDLFMRRRKADSLEVQQMKAQAREE |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | Homo sapiens | 120.00 | 35 | neurofibromin 2 isoform 6 |
| 2 | Canis familiaris | 120.00 | 32 | PREDICTED: similar to neurofibromin 2 isoform 6 is |
| 3 | Mus musculus | 120.00 | 25 | neurofibromatosis 2 |
| 4 | N/A | 120.00 | 24 | - |
| 5 | Rattus norvegicus | 120.00 | 8 | MERL_RAT Merlin (Moesin-ezrin-radixin-like protein |
| 6 | Xenopus tropicalis | 120.00 | 6 | neurofibromin 2 |
| 7 | Gallus gallus | 120.00 | 5 | neurofibromatosis 2 |
| 8 | Monodelphis domestica | 120.00 | 5 | PREDICTED: hypothetical protein |
| 9 | Macaca mulatta | 120.00 | 5 | PREDICTED: similar to neurofibromin 2 isoform 1 is |
| 10 | Bos taurus | 120.00 | 4 | PREDICTED: similar to merlin isoform 1 |
| 11 | Papio anubis anubis | 120.00 | 1 | MERL_PAPAN Merlin (Moesin-ezrin-radixin-like prote |
| 12 | Xenopus laevis | 119.00 | 6 | neurofibromin 2 (bilateral acoustic neuroma) |
| 13 | Danio rerio | 110.00 | 15 | PREDICTED: hypothetical protein |
| 14 | Tetraodon nigroviridis | 108.00 | 7 | unnamed protein product |
| 15 | Strongylocentrotus purpuratus | 89.70 | 4 | PREDICTED: similar to neurofibromin 2 (bilateral a |
| 16 | Oryctolagus cuniculus | 88.60 | 1 | villin 2 (ezrin) |
| 17 | Pan troglodytes | 86.30 | 4 | PREDICTED: moesin |
| 18 | Sus scrofa | 86.30 | 2 | MOESIN protein |
| 19 | Macaca fascicularis | 86.30 | 1 | unnamed protein product |
| 20 | mice, keratinocytes, Balb/MK, Peptide, 583 aa | 86.30 | 1 | RADI_MOUSE Radixin (ESP10) gb |
| 21 | Tribolium castaneum | 85.50 | 6 | PREDICTED: similar to neurofibromatosis 2 |
| 22 | Ciona intestinalis | 85.50 | 1 | ezrin/radixin/moesin (ERM)-like protein |
| 23 | Aplysia californica | 84.70 | 1 | ezrin/radixin/moesin |
| 24 | Caenorhabditis briggsae | 84.30 | 2 | Hypothetical protein CBG12867 |
| 25 | Drosophila melanogaster | 83.60 | 7 | Moesin CG10701-PD, isoform D |
| 26 | Drosophila pseudoobscura | 83.60 | 2 | GA10507-PA |
| 27 | Molgula tectiformis | 83.60 | 2 | Mt-ezrin/radixin/moesin |
| 28 | Caenorhabditis elegans | 83.20 | 6 | Ezrin/Radixin/Moesin family member (erm-1) |
| 29 | Apis mellifera | 82.80 | 1 | PREDICTED: similar to Moesin CG10701-PD, isoform D |
| 30 | Spodoptera frugiperda | 82.40 | 1 | A unnamed protein product |
| 31 | Biomphalaria glabrata | 82.40 | 1 | ezrin/radixin/moesin |
| 32 | Aedes aegypti | 82.00 | 4 | moesin/ezrin/radixin |
| 33 | Anopheles gambiae str. PEST | 80.90 | 2 | ENSANGP00000017331 |
| 34 | Taenia saginata | 77.80 | 1 | myosin-like protein |
| 35 | Taenia solium | 77.80 | 1 | H17g protein, tegumental antigen |
| 36 | Echinococcus multilocularis | 77.40 | 1 | tegument protein gb |
| 37 | Echinococcus granulosus | 77.40 | 1 | EG10 |
| 38 | Schistosoma japonicum | 51.60 | 1 | SJCHGC06288 protein |
Top-ranked sequences
| organism | matching |
|---|---|
| Homo sapiens | Query 228 KEFTIKPLDKKIDVFKFNSSKLRVNKLILQ#LCIGNHDLFMRRRKADSLEVQQMKAQAREE 287 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 228 KEFTIKPLDKKIDVFKFNSSKLRVNKLILQ#LCIGNHDLFMRRRKADSLEVQQMKAQAREE 287 |
| Canis familiaris | Query 228 KEFTIKPLDKKIDVFKFNSSKLRVNKLILQ#LCIGNHDLFMRRRKADSLEVQQMKAQAREE 287 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 228 KEFTIKPLDKKIDVFKFNSSKLRVNKLILQ#LCIGNHDLFMRRRKADSLEVQQMKAQAREE 287 |
| Mus musculus | Query 228 KEFTIKPLDKKIDVFKFNSSKLRVNKLILQ#LCIGNHDLFMRRRKADSLEVQQMKAQAREE 287 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 228 KEFTIKPLDKKIDVFKFNSSKLRVNKLILQ#LCIGNHDLFMRRRKADSLEVQQMKAQAREE 287 |
| N/A | Query 228 KEFTIKPLDKKIDVFKFNSSKLRVNKLILQ#LCIGNHDLFMRRRKADSLEVQQMKAQAREE 287 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 228 KEFTIKPLDKKIDVFKFNSSKLRVNKLILQ#LCIGNHDLFMRRRKADSLEVQQMKAQAREE 287 |
| Rattus norvegicus | Query 228 KEFTIKPLDKKIDVFKFNSSKLRVNKLILQ#LCIGNHDLFMRRRKADSLEVQQMKAQAREE 287 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 228 KEFTIKPLDKKIDVFKFNSSKLRVNKLILQ#LCIGNHDLFMRRRKADSLEVQQMKAQAREE 287 |
| Xenopus tropicalis | Query 228 KEFTIKPLDKKIDVFKFNSSKLRVNKLILQ#LCIGNHDLFMRRRKADSLEVQQMKAQAREE 287 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 228 KEFTIKPLDKKIDVFKFNSSKLRVNKLILQ#LCIGNHDLFMRRRKADSLEVQQMKAQAREE 287 |
| Gallus gallus | Query 228 KEFTIKPLDKKIDVFKFNSSKLRVNKLILQ#LCIGNHDLFMRRRKADSLEVQQMKAQAREE 287 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 228 KEFTIKPLDKKIDVFKFNSSKLRVNKLILQ#LCIGNHDLFMRRRKADSLEVQQMKAQAREE 287 |
| Monodelphis domestica | Query 228 KEFTIKPLDKKIDVFKFNSSKLRVNKLILQ#LCIGNHDLFMRRRKADSLEVQQMKAQAREE 287 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 228 KEFTIKPLDKKIDVFKFNSSKLRVNKLILQ#LCIGNHDLFMRRRKADSLEVQQMKAQAREE 287 |
| Macaca mulatta | Query 228 KEFTIKPLDKKIDVFKFNSSKLRVNKLILQ#LCIGNHDLFMRRRKADSLEVQQMKAQAREE 287 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 228 KEFTIKPLDKKIDVFKFNSSKLRVNKLILQ#LCIGNHDLFMRRRKADSLEVQQMKAQAREE 287 |
| Bos taurus | Query 228 KEFTIKPLDKKIDVFKFNSSKLRVNKLILQ#LCIGNHDLFMRRRKADSLEVQQMKAQAREE 287 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 228 KEFTIKPLDKKIDVFKFNSSKLRVNKLILQ#LCIGNHDLFMRRRKADSLEVQQMKAQAREE 287 |
| Papio anubis anubis | Query 228 KEFTIKPLDKKIDVFKFNSSKLRVNKLILQ#LCIGNHDLFMRRRKADSLEVQQMKAQAREE 287 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 228 KEFTIKPLDKKIDVFKFNSSKLRVNKLILQ#LCIGNHDLFMRRRKADSLEVQQMKAQAREE 287 |
| Xenopus laevis | Query 228 KEFTIKPLDKKIDVFKFNSSKLRVNKLILQ#LCIGNHDLFMRRRKADSLEVQQMKAQAREE 287 ||||||||+|||||||||||||||||||||#|||||||||||||||||||||||||||||| Sbjct 269 KEFTIKPLEKKIDVFKFNSSKLRVNKLILQ#LCIGNHDLFMRRRKADSLEVQQMKAQAREE 328 |
| Danio rerio | Query 228 KEFTIKPLDKKIDVFKFNSSKLRVNKLILQ#LCIGNHDLFMRRRKADSLEVQQMKAQAREE 287 ||| |||+||+ ||||||||||||||||||#|||||||||||||+ ||||||||| ||||| Sbjct 268 KEFAIKPVDKRADVFKFNSSKLRVNKLILQ#LCIGNHDLFMRRRRVDSLEVQQMKTQAREE 327 |
| Tetraodon nigroviridis | Query 228 KEFTIKPLDKKIDVFKFNSSKLRVNKLILQ#LCIGNHDLFMRRRKADSLEVQQMKAQAREE 287 ||||||||+|| |||||||+|| ||||||#|||||||||||||+ ||||||||||||||| Sbjct 163 KEFTIKPLEKKTKVFKFNSSRLRANKLILQ#LCIGNHDLFMRRRRVDSLEVQQMKAQAREE 222 |
| Strongylocentrotus purpuratus | Query 228 KEFTIKPLDKKIDVFKFNSSKLRVNKLILQ#LCIGNHDLFMRRRKADSLEVQQMKAQAREE 287 |+||||| || | | | |||+||||| #||+|||+|||+||+|||+|||||||||||| Sbjct 279 KKFTIKPTAKKAPNFCFISPKLRMNKLILD#LCVGNHELFMQRRRADSMEVQQMKAQAREE 338 |
| Oryctolagus cuniculus | Query 228 KEFTIKPLDKKIDVFKFNSSKLRVNKLILQ#LCIGNHDLFMRRRKADSLEVQQMKAQAREE 287 |+| |||+||| | | + +||+|| |||#||+|||+|+||||| |++|||||||||||| Sbjct 253 KKFVIKPIDKKAPDFVFYAPRLRINKRILQ#LCMGNHELYMRRRKPDTIEVQQMKAQAREE 312 |
| Pan troglodytes | Query 228 KEFTIKPLDKKIDVFKFNSSKLRVNKLILQ#LCIGNHDLFMRRRKADSLEVQQMKAQAREE 287 |+| |||+||| | | + +||+|| || #||+|||+|+||||| |++|||||||||||| Sbjct 225 KKFVIKPIDKKAPDFVFYAPRLRINKRILA#LCMGNHELYMRRRKPDTIEVQQMKAQAREE 284 |
| Sus scrofa | Query 228 KEFTIKPLDKKIDVFKFNSSKLRVNKLILQ#LCIGNHDLFMRRRKADSLEVQQMKAQAREE 287 |+| |||+||| | | + +||+|| || #||+|||+|+||||| |++|||||||||||| Sbjct 253 KKFVIKPIDKKAPDFVFYAPRLRINKRILA#LCMGNHELYMRRRKPDTIEVQQMKAQAREE 312 |
| Macaca fascicularis | Query 228 KEFTIKPLDKKIDVFKFNSSKLRVNKLILQ#LCIGNHDLFMRRRKADSLEVQQMKAQAREE 287 |+| |||+||| | | + +||+|| || #||+|||+|+||||| |++|||||||||||| Sbjct 253 KKFVIKPIDKKAPDFVFYAPRLRINKRILA#LCMGNHELYMRRRKPDTIEVQQMKAQAREE 312 |
| mice, keratinocytes, Balb/MK, Peptide, 583 aa | Query 228 KEFTIKPLDKKIDVFKFNSSKLRVNKLILQ#LCIGNHDLFMRRRKADSLEVQQMKAQAREE 287 |+| |||+||| | | + +||+|| || #||+|||+|+||||| |++|||||||||||| Sbjct 253 KKFVIKPIDKKAPDFVFYAPRLRINKRILA#LCMGNHELYMRRRKPDTIEVQQMKAQAREE 312 |
| Tribolium castaneum | Query 228 KEFTIKPLDKKIDVFKFNSSKLRVNKLILQ#LCIGNHDLFMRRRKADSLEVQQMKAQAREE 287 |+| |||+|| | | | |+|+||||| #||+||||||||||| ||+|+||||| |+|| Sbjct 263 KKFIIKPVDKNSPNFVFFSQKVRMNKLILD#LCMGNHDLFMRRRKPDSMELQQMKAAAKEE 322 |
| Ciona intestinalis | Query 228 KEFTIKPLDKKIDVFKFNSSKLRVNKLILQ#LCIGNHDLFMRRRKADSLEVQQMKAQAREE 287 |+| |||+||| | | +||+|| || #||+|||+|+||||| |++|||||||||||| Sbjct 277 KKFVIKPIDKKAPDFVFYVERLRINKRILA#LCMGNHELYMRRRKPDTIEVQQMKAQAREE 336 |
| Aplysia californica | Query 228 KEFTIKPLDKKIDVFKFNSSKLRVNKLILQ#LCIGNHDLFMRRRKADSLEVQQMKAQAREE 287 |+| |||+||| | | + +||+|| || #||+|||+|+||||| |++|||||||||||+ Sbjct 254 KKFIIKPIDKKAPDFVFYAPRLRINKRILA#LCMGNHELYMRRRKPDTIEVQQMKAQARED 313 |
| Caenorhabditis briggsae | Query 228 KEFTIKPLDKKIDVFKFNSSKLRVNKLILQ#LCIGNHDLFMRRRKADSLEVQQMKAQAREE 287 |+| |||+||| | | + +||+|| || #||+|||+|+||||| |++|||||| ||||| Sbjct 249 KKFVIKPIDKKAHDFVFYAPRLRINKRILA#LCMGNHELYMRRRKPDTIEVQQMKQQAREE 308 |
| Drosophila melanogaster | Query 228 KEFTIKPLDKKIDVFKFNSSKLRVNKLILQ#LCIGNHDLFMRRRKADSLEVQQMKAQAREE 287 |+| |||+||| | | + ++|+|| || #||+|||+|+||||| |+++||||||||||| Sbjct 254 KKFIIKPIDKKAPDFMFFAPRVRINKRILA#LCMGNHELYMRRRKPDTIDVQQMKAQAREE 313 |
| Drosophila pseudoobscura | Query 228 KEFTIKPLDKKIDVFKFNSSKLRVNKLILQ#LCIGNHDLFMRRRKADSLEVQQMKAQAREE 287 |+| |||+||| | | + ++|+|| || #||+|||+|+||||| |+++||||||||||| Sbjct 249 KKFIIKPIDKKAPDFMFFAPRVRINKRILA#LCMGNHELYMRRRKPDTIDVQQMKAQAREE 308 |
| Molgula tectiformis | Query 228 KEFTIKPLDKKIDVFKFNSSKLRVNKLILQ#LCIGNHDLFMRRRKADSLEVQQMKAQARE 286 |+| |||+||| | | +||+|| || #||+|||+|+||||| ||+|||||||||+| Sbjct 253 KKFVIKPIDKKAPDFVFYVERLRINKRILA#LCMGNHELYMRRRKPDSIEVQQMKAQAKE 311 |
| Caenorhabditis elegans | Query 228 KEFTIKPLDKKIDVFKFNSSKLRVNKLILQ#LCIGNHDLFMRRRKADSLEVQQMKAQAREE 287 |+| |||+||| | | + +||+|| || #||+|||+|+||||| |++|||||| ||||+ Sbjct 254 KKFVIKPIDKKAHDFVFYAPRLRINKRILA#LCMGNHELYMRRRKPDTIEVQQMKQQARED 313 |
| Apis mellifera | Query 228 KEFTIKPLDKKIDVFKFNSSKLRVNKLILQ#LCIGNHDLFMRRRKADSLEVQQMKAQAREE 287 |+| |||+||| | | ++++++|| || #||+|||+|+||||| |+++||||||||||| Sbjct 188 KKFIIKPIDKKAPDFVFFATRVKINKRILA#LCMGNHELYMRRRKPDTIDVQQMKAQAREE 247 |
| Spodoptera frugiperda | Query 228 KEFTIKPLDKKIDVFKFNSSKLRVNKLILQ#LCIGNHDLFMRRRKADSLEVQQMKAQAREE 287 ++| |||+||| | | + ++|||| || #||+|||+|+||||| |+++||||||||||| Sbjct 253 RKFIIKPIDKKAPDFVFFAPRVRVNKRILA#LCMGNHELYMRRRKPDTIDVQQMKAQAREE 312 |
| Biomphalaria glabrata | Query 228 KEFTIKPLDKKIDVFKFNSSKLRVNKLILQ#LCIGNHDLFMRRRKADSLEVQQMKAQAR 285 |+| |||+||| | | + +||+|| || #||+|||+|+||||| |++|||||||||| Sbjct 254 KKFVIKPIDKKAPDFVFYAPRLRINKRILA#LCMGNHELYMRRRKPDTIEVQQMKAQAR 311 |
| Aedes aegypti | Query 228 KEFTIKPLDKKIDVFKFNSSKLRVNKLILQ#LCIGNHDLFMRRRKADSLEVQQMKAQAREE 287 ++| |||+||| | | + ++|+|| || #||+|||+|+||||| |+++||||||||||| Sbjct 259 RKFIIKPIDKKAPDFVFFAPRVRINKRILA#LCMGNHELYMRRRKPDTIDVQQMKAQAREE 318 |
| Anopheles gambiae str. PEST | Query 228 KEFTIKPLDKKIDVFKFNSSKLRVNKLILQ#LCIGNHDLFMRRRKADSLEVQQMKAQAREE 287 ++| |||+||| | | + ++|+|| || #||+|||+|+||||| |+++|||||||||+| Sbjct 300 RKFIIKPIDKKAPDFVFFAPRVRINKRILA#LCMGNHELYMRRRKPDTIDVQQMKAQARDE 359 |
| Taenia saginata | Query 228 KEFTIKPLDKKIDVFKFNSSKLRVNKLILQ#LCIGNHDLFMRRRKADSLEVQQMKAQAREE 287 |+| ||| || | | | ++|| || #|| |||+|+|||||+||+|||||| ||+|| Sbjct 255 KKFIIKPADKSAKEFYFLVEKSKINKRILA#LCTGNHELYMRRRKSDSIEVQQMKIQAKEE 314 |
| Taenia solium | Query 228 KEFTIKPLDKKIDVFKFNSSKLRVNKLILQ#LCIGNHDLFMRRRKADSLEVQQMKAQAREE 287 |+| ||| || | | | ++|| || #|| |||+|+|||||+||+|||||| ||+|| Sbjct 248 KKFIIKPADKSAKEFYFLVEKSKINKRILA#LCTGNHELYMRRRKSDSIEVQQMKIQAKEE 307 |
| Echinococcus multilocularis | Query 228 KEFTIKPLDKKIDVFKFNSSKLRVNKLILQ#LCIGNHDLFMRRRKADSLEVQQMKAQAREE 287 |+| ||| || | | | ++|| || #|| |||+|+|||||+||+|||||| ||+|| Sbjct 255 KKFIIKPADKSAKEFFFLVEKSKINKRILA#LCTGNHELYMRRRKSDSIEVQQMKIQAKEE 314 |
| Echinococcus granulosus | Query 228 KEFTIKPLDKKIDVFKFNSSKLRVNKLILQ#LCIGNHDLFMRRRKADSLEVQQMKAQAREE 287 |+| ||| || | | | ++|| || #|| |||+|+|||||+||+|||||| ||+|| Sbjct 255 KKFIIKPADKSAKEFFFLVEKSKINKRILA#LCTGNHELYMRRRKSDSIEVQQMKIQAKEE 314 |
| Schistosoma japonicum | Query 229 EFTIKPLDKKIDVFKFNSSKLRVNKLILQ#LCIGNHDLFMRRRKADSLEVQQMKAQARE 286 +| +||+ +| + ++||| #| +||| |+ ||+ ||+|||||| +|+| Sbjct 254 KFYVKPVGASGEVLTLYTDSTHTSRLILN#LSMGNHKLYAVRRQPDSIEVQQMKVKAKE 311 |
* References
None.