XSB1187 : neurofibromin 2 isoform 2 [Homo sapiens]
[ CaMP Format ]
This entry is computationally expanded from SB0009
* Basic Information
| Organism | Homo sapiens (human) |
| Protein Names | Merlin; Moesin-ezrin-radixin-like protein; Neurofibromin-2; Schwannomin; Schwannomerlin; neurofibromin 2 isoform 2; moesin-ezrin-radizin-like protein; neurofibromin 2 (bilateral acoustic neuroma); moesin-ezrin-radixin like |
| Gene Names | NF2; SCH; neurofibromin 2 (merlin) |
| Gene Locus | 22q12.2; chromosome 22 |
| GO Function | Not available |
* Information From OMIM
Description: Cell-cell contact between normal cultured diploid cells results in inhibition of proliferation despite the unlimited availability of nutrients and growth-promoting factors. A similar phenomenon occurs in vivo in all adult solid tissues. NF2 is a critical regulator of contact-dependent inhibition of proliferation and functions at the interface between cell-cell adhesion, transmembrane signaling, and the actin cytoskeleton (Curto and McClatchey, 2008).
Function: Gutmann et al. (1999) studied rat schwannoma cell lines overexpressing wildtype merlin isoforms and mutant merlin proteins. Overexpression of wildtype merlin resulted in transient alterations in F-actin organization, cell spreading, and cell attachment, and in impaired cell motility as measured in an in vitro motility assay. These effects were observed only in cells overexpressing a merlin isoform capable of inhibiting cell growth and not with mutant merlin molecules (harboring NF2 patient mutations) or a merlin splice variant (isoform II) lacking growth-inhibitory activity. These data indicated that merlin may function to maintain normal cytoskeletal organization, and suggested that its influence on cell growth depends on specific cytoskeletal rearrangements.
Function: Gutmann et al. (2001) performed a detailed functional analysis of 8 naturally occurring nonconservative missense mutations in the NF2 gene. The authors analyzed proliferation, actin cytoskeleton-mediated events, and merlin folding in a regulatable expression system in rat schwannoma cells. They demonstrated that mutations clustered in the predicted alpha-helical region did not impair the function of merlin, whereas those in either the N- or C-terminus of the protein rendered merlin inactive as a negative growth regulator. The authors suggested that the key functional domains of merlin may lie within the highly conserved FERM domain and the unique C terminus of the protein.
Function: Gutmann et al. (2001) stated that the merlin protein functions as a negative growth regulator. They demonstrated that regulated overexpression of the hepatocyte growth factor-regulated tyrosine kinase substrate (HGS, or HRS; OMIM:604375) in rat schwannoma cells yielded effects similar to those seen with overexpression of merlin, including growth inhibition, decreased motility, and abnormalities in cell spreading. The HRS binding domain of merlin was mapped to residues 453-557. Overexpression of C-terminal merlin had no effect on HRS function, suggesting to the authors that merlin binding to HRS does not negatively regulate HRS growth suppressor activity, and that merlin and HRS may regulate cell growth in schwannoma cells through interacting pathways. Gutmann (2001) reviewed the functions of neurofibromin (OMIM:162200) and merlin in tumor suppression and cell-cell signaling, respectively.
Function: Using yeast 2-hybrid interaction cloning, Scoles et al. (2000) determined that schwannomin interacts with HRS. They demonstrated the interaction both in vivo, by immunoprecipitation of endogenous HRS with endogenous schwannomin, and in vitro, with a binding assay using bacterially purified HRS and schwannomin. The regions of interaction included schwannomin residues 256-579 and HRS residues from 480 to the end of either of 2 HRS isoforms. Schwannomin molecules with an L46R, L360P, L535P, or Q538P missense mutation demonstrated reduced affinity for HRS binding. Since HRS is associated with early endosomes and may mediate receptor translocation to the lysosome, the authors used indirect immunofluorescence to demonstrate that schwannomin and HRS colocalize at endosomes in STS26T Schwann cells. The authors hypothesized that schwannomin is involved in HRS-mediated cell signaling.
Function: Sun et al. (2002) generated a series of HRS truncation mutants to define the regions required for merlin binding and HRS growth suppression. The HRS domain required for merlin binding was narrowed to residues 470-497 (which contain the predicted coiled-coil domain), and the major domain responsible for HRS growth suppression was localized to residues 498-550. Merlin inhibited growth in Hrs +/+ but not Hrs -/- mouse embryonic fibroblast cells. In contrast, HRS could suppress cell growth in the absence of Nf2 expression. The authors concluded that merlin growth suppression requires HRS expression, and that the binding of merlin to HRS may facilitate its ability to function as a tumor suppressor.
Function: Using transient transfection methods, Scoles et al. (2002) showed that both schwannomin and HRS inhibited STAT3 (OMIM:102582) activation, and that schwannomin suppressed STAT3 activation mediated by IGF1 (OMIM:147440) treatment in a human schwannoma cell line. Schwannomin inhibited STAT3 and STAT5 (OMIM:601511) phosphorylation in a rat schwannoma cell line. Schwannomin with the pathogenic missense mutation Q538P (OMIM:607379.0006) failed to bind HRS and did not inhibit STAT5 phosphorylation. The authors hypothesized that schwannomin requires HRS interaction to be fully functionally active and to inhibit STAT activation.
Function: Scoles et al. (2002) noted that in addition to binding HRS, both of the major isoforms of schwannomin are involved in homodimerization and interact with beta II spectrin (OMIM:182790) and with EIF3S8 (OMIM:603916). Homodimerization and heterodimerization between isoforms occurs through the C-terminal half, as does interaction between schwannomin and HRS and beta II spectrin. Interaction with EIF3S8 occurs through the N-terminal half of schwannomin. Using yeast 2-hybrid assays to characterize further the effect of missense mutations on these interactions, Scoles et al. (2002) found that a mutation in the N-terminal half and a mutation in the C-terminal alpha helix significantly decreased dimerization and decreased the affinity between schwannomin and all interacting proteins. Several clinically significant mutations between amino acids 219 and 352 selectively enhanced interactions with their binding partners. Scoles et al. (2002) also determined that the sites for schwannomin self-interaction and their binding strengths differ between isoforms 1 and 2.
Function: To elucidate the properties of merlin that are critical for its tumor suppressor function, Stokowski and Cox (2000) expressed NF2-causing mutant merlin proteins in mammalian cells. They found that 80% of the merlin mutants significantly altered cell adhesion by causing cells to detach from the substratum. They stated that such changes in cell adhesion may be an initial step in the pathogenesis of NF2. In addition, they found that 4 missense mutations decreased the binding of merlin to the ERM-interacting phosphoprotein EBP-50. Some NF2 point mutations resembled dominant gain-of-function rather than loss-of-function alleles.
Function: Fernandez-Valle et al. (2002) noted that mice with conditional deletion of NF2 exon 2 in Schwann cells develop schwannomas, which confirms the crucial nature of exon 2 for growth control. They found that the molecular adaptor paxillin (OMIM:602505) binds directly to schwannomin in residues 50-70, which are encoded by exon 2. This interaction mediates the membrane localization of schwannomin to the plasma membrane, where it associates with beta-1-integrin (ITGB1; OMIM:135630) and ERBB2 (OMIM:164870). These studies defined a pathogenic mechanism for the development of neurofibromatosis II in humans with mutations in exon 2 of NF2.
Function: Schulze et al. (2002) used oncoretrovirus-mediated gene transfer of different merlin constructs to stably reexpress wildtype merlin in primary cells derived from human schwannomas. Using 2-parameter FACS analysis, they demonstrated that expression of wildtype merlin in NF2 cells led to significant reduction of proliferation and G0/G1 arrest in transduced schwannoma cells. In addition, there was increased apoptosis of schwannoma cells transduced with wildtype merlin. The authors concluded that merlin may act as a tumor suppressor.
Function: Bashour et al. (2002) observed that schwannoma-derived Schwann cells exhibited membrane ruffling and aberrant cell spreading when plated onto laminin (see OMIM:150240), indicative of fundamental F-actin cytoskeletal defects. They found that mutations in NF2 correlate with F-actin abnormalities. Using a protein transfer technique with primary human schwannoma cells containing NF2 mutations, they introduced the NF2 protein in an attempt to reverse the cytoskeletal abnormalities. They found that isoform-1 of merlin, the growth-suppressing isoform, reversed the abnormal ruffling and cell spreading and restored normal actin organization. Other isoforms of merlin and merlin containing a point mutation did not reverse the phenotype.
Function: Gautreau et al. (2002) noted that the N-terminal FERM domain of schwannomin is implicated in plasma membrane and filamentous actin binding and that mutations in this domain impair proper folding. They found that mutations in the FERM domain were unstable both in vivo and in vitro due to proteasome-mediated degradation. They hypothesized that loss of schwannomin through degradation could contribute to the pathophysiology of NF2.
Function: Shaw et al. (2001) concluded that NF2 functions in Rac (see OMIM:602048)-dependent signaling. Using electrophoretic mobility shift assays, they identified Rac-induced phosphorylation sites in NF2. They observed that expression of activated Rac-induced phosphorylation of the NF2 serine-518 residue inhibited NF2 self-association and decreased association of NF2 with the cytoskeleton. Using cell transfections, the authors showed that NF2 overexpression inhibited Rac-induced signaling in a phosphorylation-dependent manner. Also, NF2-deficient fibroblasts exhibited characteristics of cells overexpressing activated alleles of Rac. Shaw et al. (2001) hypothesized that NF2 functions as a tumor and metastasis suppressor through its ability to inhibit Rac-dependent signaling.
Function: Kressel and Schmucker (2002) showed that splicing out of exon 2 leads to unrestricted entry of merlin into the nucleus, yet skipping of adjacent exon 3 has no comparable effect. Exon 2 functioned as a cytoplasmic retention factor and was able to confer sole cytoplasmic localization to a GFP fusion protein. Merlin's ability to enter the nucleus is complemented by a nuclear-cytoplasmic shuttle protein sequence within exon 15 that facilitates export via the CRM1/exportin pathway. The authors proposed a cellular function different to the wildtype protein for naturally occurring splice variants lacking exon 2.
Function: Jin et al. (2006) identified MYPT1 (OMIM:602021) as the enzyme that activates the tumor suppressor function of merlin. The cellular MYPT1-PP1-delta (OMIM:600590)-specific inhibitor CPI17 (OMIM:608153) caused a loss of merlin function characterized by merlin phosphorylation, Ras activation, and transformation. Constitutively active merlin containing the mutation S518A reversed CPI17-induced transformation, showing that merlin is the decisive substrate of MYPT1-PP1-delta in tumor suppression. In addition, Jin et al. (2006) showed that CPI17 levels are raised in several human tumor cell lines and that the downregulation of CPI17 induces merlin dephosphorylation, inhibits Ras activation, and abolishes the transformed phenotype. Jin et al. (2006) concluded that MYPT1 and its substrate merlin are part of a previously undescribed tumor suppressor cascade that can be hindered in 2 ways, by mutation of the NF2 gene and by upregulation of the oncoprotein CPI17.
Function: Curto and McClatchey (2008) reviewed the mechanisms by which NF2 regulates contact-dependent inhibition of proliferation.
* Structure Information
1. Primary Information
Length: 590 aa
Average Mass: 69.090 kDa
Monoisotopic Mass: 69.046 kDa
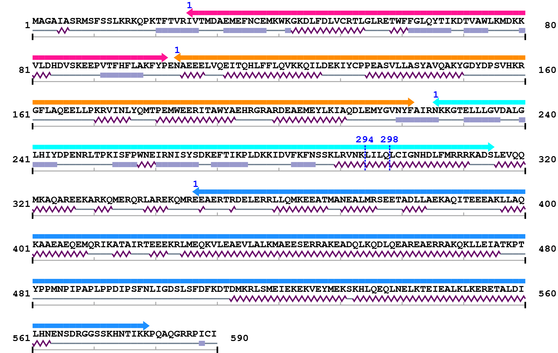
2. Domain Information
Annotated Domains: interpro / pfam / smart / prosite
Computationally Assigned Domains (Pfam+HMMER):
| domain name | begin | end | score | e-value |
|---|---|---|---|---|
| FERM_N 1. | 26 | 102 | 95.0 | 2.7e-25 |
| FERM_M 1. | 104 | 222 | 141.8 | 2.1e-39 |
| FERM_C 1. | 226 | 315 | 177.6 | 3.5e-50 |
| --- cleavage 294 (inside FERM_C 226..315) --- | ||||
| --- cleavage 298 (inside FERM_C 226..315) --- | ||||
| ERM 1. | 347 | 579 | 307.1 | 3.7e-89 |
3. Sequence Information
Fasta Sequence: XSB1187.fasta
Amino Acid Sequence and Secondary Structures (PsiPred):
4. 3D Information
Known Structures in PDB: 1H4R (X-ray; 180 A; A/B=1-313)
* Cleavage Information
2 [sites]
Cleavage sites (±10aa)
[Site 1] FNSSKLRVNK294-LILQLCIGNH
Lys294  Leu
Leu
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Phe285 | Asn286 | Ser287 | Ser288 | Lys289 | Leu290 | Arg291 | Val292 | Asn293 | Lys294 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Leu295 | Ile296 | Leu297 | Gln298 | Leu299 | Cys300 | Ile301 | Gly302 | Asn303 | His304 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| SYSDKEFTIKPLDKKIDVFKFNSSKLRVNKLILQLCIGNHDLFMRRRKADSLEVQQMKAQ |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | Homo sapiens | 121.00 | 35 | neurofibromin 2 isoform 2 |
| 2 | Canis familiaris | 121.00 | 32 | PREDICTED: similar to neurofibromin 2 isoform 2 is |
| 3 | Mus musculus | 121.00 | 25 | unnamed protein product |
| 4 | N/A | 121.00 | 24 | - |
| 5 | Rattus norvegicus | 121.00 | 8 | MERL_RAT Merlin (Moesin-ezrin-radixin-like protein |
| 6 | Xenopus tropicalis | 121.00 | 6 | neurofibromin 2 |
| 7 | Monodelphis domestica | 121.00 | 5 | PREDICTED: hypothetical protein |
| 8 | Gallus gallus | 121.00 | 5 | neurofibromatosis 2 |
| 9 | Macaca mulatta | 121.00 | 5 | PREDICTED: similar to neurofibromin 2 isoform 1 is |
| 10 | Bos taurus | 121.00 | 4 | PREDICTED: similar to merlin isoform 1 |
| 11 | Papio anubis anubis | 121.00 | 1 | MERL_PAPAN Merlin (Moesin-ezrin-radixin-like prote |
| 12 | Xenopus laevis | 119.00 | 6 | neurofibromin 2 (bilateral acoustic neuroma) |
| 13 | Danio rerio | 110.00 | 14 | PREDICTED: hypothetical protein |
| 14 | Tetraodon nigroviridis | 109.00 | 7 | unnamed protein product |
| 15 | Strongylocentrotus purpuratus | 87.00 | 4 | PREDICTED: similar to neurofibromin 2 (bilateral a |
| 16 | Oryctolagus cuniculus | 86.70 | 1 | villin 2 (ezrin) |
| 17 | Tribolium castaneum | 84.70 | 6 | PREDICTED: similar to neurofibromatosis 2 |
| 18 | Molgula tectiformis | 84.70 | 2 | Mt-ezrin/radixin/moesin |
| 19 | Pan troglodytes | 84.30 | 4 | PREDICTED: moesin |
| 20 | Sus scrofa | 84.30 | 2 | radixin |
| 21 | Biomphalaria glabrata | 84.30 | 1 | ezrin/radixin/moesin |
| 22 | mice, keratinocytes, Balb/MK, Peptide, 583 aa | 84.30 | 1 | RADI_MOUSE Radixin (ESP10) gb |
| 23 | Macaca fascicularis | 84.30 | 1 | unnamed protein product |
| 24 | Aplysia californica | 84.00 | 1 | ezrin/radixin/moesin |
| 25 | Ciona intestinalis | 83.60 | 1 | ezrin/radixin/moesin (ERM)-like protein |
| 26 | Caenorhabditis elegans | 82.40 | 6 | ERM-1Asv |
| 27 | Caenorhabditis briggsae | 82.40 | 2 | Hypothetical protein CBG12867 |
| 28 | Drosophila melanogaster | 81.30 | 7 | Moesin CG10701-PJ, isoform J |
| 29 | Drosophila pseudoobscura | 81.30 | 2 | GA10507-PA |
| 30 | Spodoptera frugiperda | 80.50 | 1 | A unnamed protein product |
| 31 | Aedes aegypti | 80.10 | 4 | moesin/ezrin/radixin |
| 32 | Anopheles gambiae str. PEST | 80.10 | 2 | ENSANGP00000017331 |
| 33 | Apis mellifera | 79.30 | 1 | PREDICTED: similar to Moesin CG10701-PD, isoform D |
| 34 | Taenia saginata | 76.30 | 1 | myosin-like protein |
| 35 | Taenia solium | 76.30 | 1 | H17g protein, tegumental antigen |
| 36 | Echinococcus multilocularis | 75.90 | 1 | tegument protein gb |
| 37 | Echinococcus granulosus | 75.90 | 1 | EG10 |
| 38 | Schistosoma japonicum | 53.10 | 1 | SJCHGC06288 protein |
Top-ranked sequences
| organism | matching |
|---|---|
| Homo sapiens | Query 265 SYSDKEFTIKPLDKKIDVFKFNSSKLRVNK#LILQLCIGNHDLFMRRRKADSLEVQQMKAQ 324 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 265 SYSDKEFTIKPLDKKIDVFKFNSSKLRVNK#LILQLCIGNHDLFMRRRKADSLEVQQMKAQ 324 |
| Canis familiaris | Query 265 SYSDKEFTIKPLDKKIDVFKFNSSKLRVNK#LILQLCIGNHDLFMRRRKADSLEVQQMKAQ 324 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 265 SYSDKEFTIKPLDKKIDVFKFNSSKLRVNK#LILQLCIGNHDLFMRRRKADSLEVQQMKAQ 324 |
| Mus musculus | Query 265 SYSDKEFTIKPLDKKIDVFKFNSSKLRVNK#LILQLCIGNHDLFMRRRKADSLEVQQMKAQ 324 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 265 SYSDKEFTIKPLDKKIDVFKFNSSKLRVNK#LILQLCIGNHDLFMRRRKADSLEVQQMKAQ 324 |
| N/A | Query 265 SYSDKEFTIKPLDKKIDVFKFNSSKLRVNK#LILQLCIGNHDLFMRRRKADSLEVQQMKAQ 324 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 265 SYSDKEFTIKPLDKKIDVFKFNSSKLRVNK#LILQLCIGNHDLFMRRRKADSLEVQQMKAQ 324 |
| Rattus norvegicus | Query 265 SYSDKEFTIKPLDKKIDVFKFNSSKLRVNK#LILQLCIGNHDLFMRRRKADSLEVQQMKAQ 324 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 265 SYSDKEFTIKPLDKKIDVFKFNSSKLRVNK#LILQLCIGNHDLFMRRRKADSLEVQQMKAQ 324 |
| Xenopus tropicalis | Query 265 SYSDKEFTIKPLDKKIDVFKFNSSKLRVNK#LILQLCIGNHDLFMRRRKADSLEVQQMKAQ 324 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 265 SYSDKEFTIKPLDKKIDVFKFNSSKLRVNK#LILQLCIGNHDLFMRRRKADSLEVQQMKAQ 324 |
| Monodelphis domestica | Query 265 SYSDKEFTIKPLDKKIDVFKFNSSKLRVNK#LILQLCIGNHDLFMRRRKADSLEVQQMKAQ 324 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 265 SYSDKEFTIKPLDKKIDVFKFNSSKLRVNK#LILQLCIGNHDLFMRRRKADSLEVQQMKAQ 324 |
| Gallus gallus | Query 265 SYSDKEFTIKPLDKKIDVFKFNSSKLRVNK#LILQLCIGNHDLFMRRRKADSLEVQQMKAQ 324 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 265 SYSDKEFTIKPLDKKIDVFKFNSSKLRVNK#LILQLCIGNHDLFMRRRKADSLEVQQMKAQ 324 |
| Macaca mulatta | Query 265 SYSDKEFTIKPLDKKIDVFKFNSSKLRVNK#LILQLCIGNHDLFMRRRKADSLEVQQMKAQ 324 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 265 SYSDKEFTIKPLDKKIDVFKFNSSKLRVNK#LILQLCIGNHDLFMRRRKADSLEVQQMKAQ 324 |
| Bos taurus | Query 265 SYSDKEFTIKPLDKKIDVFKFNSSKLRVNK#LILQLCIGNHDLFMRRRKADSLEVQQMKAQ 324 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 265 SYSDKEFTIKPLDKKIDVFKFNSSKLRVNK#LILQLCIGNHDLFMRRRKADSLEVQQMKAQ 324 |
| Papio anubis anubis | Query 265 SYSDKEFTIKPLDKKIDVFKFNSSKLRVNK#LILQLCIGNHDLFMRRRKADSLEVQQMKAQ 324 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 265 SYSDKEFTIKPLDKKIDVFKFNSSKLRVNK#LILQLCIGNHDLFMRRRKADSLEVQQMKAQ 324 |
| Xenopus laevis | Query 265 SYSDKEFTIKPLDKKIDVFKFNSSKLRVNK#LILQLCIGNHDLFMRRRKADSLEVQQMKAQ 324 ||||||||||||+|||||||||||||||||#|||||||||||||||||||||||||||||| Sbjct 265 SYSDKEFTIKPLEKKIDVFKFNSSKLRVNK#LILQLCIGNHDLFMRRRKADSLEVQQMKAQ 324 |
| Danio rerio | Query 265 SYSDKEFTIKPLDKKIDVFKFNSSKLRVNK#LILQLCIGNHDLFMRRRKADSLEVQQMKAQ 324 ||||||| |||+||+ ||||||||||||||#|||||||||||||||||+ ||||||||| | Sbjct 264 SYSDKEFAIKPVDKRADVFKFNSSKLRVNK#LILQLCIGNHDLFMRRRRVDSLEVQQMKTQ 323 |
| Tetraodon nigroviridis | Query 265 SYSDKEFTIKPLDKKIDVFKFNSSKLRVNK#LILQLCIGNHDLFMRRRKADSLEVQQMKAQ 324 ||||||||||||+|| |||||||+|| ||#|||||||||||||||||+ ||||||||||| Sbjct 159 SYSDKEFTIKPLEKKTKVFKFNSSRLRANK#LILQLCIGNHDLFMRRRRVDSLEVQQMKAQ 218 |
| Strongylocentrotus purpuratus | Query 265 SYSDKEFTIKPLDKKIDVFKFNSSKLRVNK#LILQLCIGNHDLFMRRRKADSLEVQQMKAQ 324 |+ ||+||||| || | | | |||+||#||| ||+|||+|||+||+|||+|||||||| Sbjct 275 SFRDKKFTIKPTAKKAPNFCFISPKLRMNK#LILDLCVGNHELFMQRRRADSMEVQQMKAQ 334 |
| Oryctolagus cuniculus | Query 265 SYSDKEFTIKPLDKKIDVFKFNSSKLRVNK#LILQLCIGNHDLFMRRRKADSLEVQQMKAQ 324 |++||+| |||+||| | | + +||+||# |||||+|||+|+||||| |++|||||||| Sbjct 249 SFNDKKFVIKPIDKKAPDFVFYAPRLRINK#RILQLCMGNHELYMRRRKPDTIEVQQMKAQ 308 |
| Tribolium castaneum | Query 265 SYSDKEFTIKPLDKKIDVFKFNSSKLRVNK#LILQLCIGNHDLFMRRRKADSLEVQQMKA 323 |+ ||+| |||+|| | | | |+|+||#||| ||+||||||||||| ||+|+||||| Sbjct 259 SFDDKKFIIKPVDKNSPNFVFFSQKVRMNK#LILDLCMGNHDLFMRRRKPDSMELQQMKA 317 |
| Molgula tectiformis | Query 265 SYSDKEFTIKPLDKKIDVFKFNSSKLRVNK#LILQLCIGNHDLFMRRRKADSLEVQQMKAQ 324 |++||+| |||+||| | | +||+||# || ||+|||+|+||||| ||+|||||||| Sbjct 249 SFNDKKFVIKPIDKKAPDFVFYVERLRINK#RILALCMGNHELYMRRRKPDSIEVQQMKAQ 308 |
| Pan troglodytes | Query 265 SYSDKEFTIKPLDKKIDVFKFNSSKLRVNK#LILQLCIGNHDLFMRRRKADSLEVQQMKAQ 324 |++||+| |||+||| | | + +||+||# || ||+|||+|+||||| |++|||||||| Sbjct 221 SFNDKKFVIKPIDKKAPDFVFYAPRLRINK#RILALCMGNHELYMRRRKPDTIEVQQMKAQ 280 |
| Sus scrofa | Query 265 SYSDKEFTIKPLDKKIDVFKFNSSKLRVNK#LILQLCIGNHDLFMRRRKADSLEVQQMKAQ 324 |++||+| |||+||| | | + +||+||# || ||+|||+|+||||| |++|||||||| Sbjct 249 SFNDKKFVIKPIDKKAPDFVFYAPRLRINK#RILALCMGNHELYMRRRKPDTIEVQQMKAQ 308 |
| Biomphalaria glabrata | Query 265 SYSDKEFTIKPLDKKIDVFKFNSSKLRVNK#LILQLCIGNHDLFMRRRKADSLEVQQMKAQ 324 |++||+| |||+||| | | + +||+||# || ||+|||+|+||||| |++|||||||| Sbjct 250 SFNDKKFVIKPIDKKAPDFVFYAPRLRINK#RILALCMGNHELYMRRRKPDTIEVQQMKAQ 309 |
| mice, keratinocytes, Balb/MK, Peptide, 583 aa | Query 265 SYSDKEFTIKPLDKKIDVFKFNSSKLRVNK#LILQLCIGNHDLFMRRRKADSLEVQQMKAQ 324 |++||+| |||+||| | | + +||+||# || ||+|||+|+||||| |++|||||||| Sbjct 249 SFNDKKFVIKPIDKKAPDFVFYAPRLRINK#RILALCMGNHELYMRRRKPDTIEVQQMKAQ 308 |
| Macaca fascicularis | Query 265 SYSDKEFTIKPLDKKIDVFKFNSSKLRVNK#LILQLCIGNHDLFMRRRKADSLEVQQMKAQ 324 |++||+| |||+||| | | + +||+||# || ||+|||+|+||||| |++|||||||| Sbjct 249 SFNDKKFVIKPIDKKAPDFVFYAPRLRINK#RILALCMGNHELYMRRRKPDTIEVQQMKAQ 308 |
| Aplysia californica | Query 265 SYSDKEFTIKPLDKKIDVFKFNSSKLRVNK#LILQLCIGNHDLFMRRRKADSLEVQQMKAQ 324 |++||+| |||+||| | | + +||+||# || ||+|||+|+||||| |++|||||||| Sbjct 250 SFNDKKFIIKPIDKKAPDFVFYAPRLRINK#RILALCMGNHELYMRRRKPDTIEVQQMKAQ 309 |
| Ciona intestinalis | Query 265 SYSDKEFTIKPLDKKIDVFKFNSSKLRVNK#LILQLCIGNHDLFMRRRKADSLEVQQMKAQ 324 |++||+| |||+||| | | +||+||# || ||+|||+|+||||| |++|||||||| Sbjct 273 SFNDKKFVIKPIDKKAPDFVFYVERLRINK#RILALCMGNHELYMRRRKPDTIEVQQMKAQ 332 |
| Caenorhabditis elegans | Query 265 SYSDKEFTIKPLDKKIDVFKFNSSKLRVNK#LILQLCIGNHDLFMRRRKADSLEVQQMKAQ 324 |++||+| |||+||| | | + +||+||# || ||+|||+|+||||| |++|||||| | Sbjct 250 SFNDKKFVIKPIDKKAHDFVFYAPRLRINK#RILALCMGNHELYMRRRKPDTIEVQQMKQQ 309 |
| Caenorhabditis briggsae | Query 265 SYSDKEFTIKPLDKKIDVFKFNSSKLRVNK#LILQLCIGNHDLFMRRRKADSLEVQQMKAQ 324 |++||+| |||+||| | | + +||+||# || ||+|||+|+||||| |++|||||| | Sbjct 245 SFNDKKFVIKPIDKKAHDFVFYAPRLRINK#RILALCMGNHELYMRRRKPDTIEVQQMKQQ 304 |
| Drosophila melanogaster | Query 265 SYSDKEFTIKPLDKKIDVFKFNSSKLRVNK#LILQLCIGNHDLFMRRRKADSLEVQQMKAQ 324 |+|+|+| |||+||| | | + ++|+||# || ||+|||+|+||||| |+++||||||| Sbjct 250 SFSEKKFIIKPIDKKAPDFMFFAPRVRINK#RILALCMGNHELYMRRRKPDTIDVQQMKAQ 309 |
| Drosophila pseudoobscura | Query 265 SYSDKEFTIKPLDKKIDVFKFNSSKLRVNK#LILQLCIGNHDLFMRRRKADSLEVQQMKAQ 324 |+|+|+| |||+||| | | + ++|+||# || ||+|||+|+||||| |+++||||||| Sbjct 245 SFSEKKFIIKPIDKKAPDFMFFAPRVRINK#RILALCMGNHELYMRRRKPDTIDVQQMKAQ 304 |
| Spodoptera frugiperda | Query 265 SYSDKEFTIKPLDKKIDVFKFNSSKLRVNK#LILQLCIGNHDLFMRRRKADSLEVQQMKAQ 324 |++|++| |||+||| | | + ++||||# || ||+|||+|+||||| |+++||||||| Sbjct 249 SFNDRKFIIKPIDKKAPDFVFFAPRVRVNK#RILALCMGNHELYMRRRKPDTIDVQQMKAQ 308 |
| Aedes aegypti | Query 265 SYSDKEFTIKPLDKKIDVFKFNSSKLRVNK#LILQLCIGNHDLFMRRRKADSLEVQQMKAQ 324 |++|++| |||+||| | | + ++|+||# || ||+|||+|+||||| |+++||||||| Sbjct 255 SFNDRKFIIKPIDKKAPDFVFFAPRVRINK#RILALCMGNHELYMRRRKPDTIDVQQMKAQ 314 |
| Anopheles gambiae str. PEST | Query 265 SYSDKEFTIKPLDKKIDVFKFNSSKLRVNK#LILQLCIGNHDLFMRRRKADSLEVQQMKAQ 324 |++|++| |||+||| | | + ++|+||# || ||+|||+|+||||| |+++||||||| Sbjct 296 SFNDRKFIIKPIDKKAPDFVFFAPRVRINK#RILALCMGNHELYMRRRKPDTIDVQQMKAQ 355 |
| Apis mellifera | Query 265 SYSDKEFTIKPLDKKIDVFKFNSSKLRVNK#LILQLCIGNHDLFMRRRKADSLEVQQMKAQ 324 |+++|+| |||+||| | | ++++++||# || ||+|||+|+||||| |+++||||||| Sbjct 184 SFNEKKFIIKPIDKKAPDFVFFATRVKINK#RILALCMGNHELYMRRRKPDTIDVQQMKAQ 243 |
| Taenia saginata | Query 265 SYSDKEFTIKPLDKKIDVFKFNSSKLRVNK#LILQLCIGNHDLFMRRRKADSLEVQQMKAQ 324 |+ ||+| ||| || | | | ++||# || || |||+|+|||||+||+|||||| | Sbjct 251 SFHDKKFIIKPADKSAKEFYFLVEKSKINK#RILALCTGNHELYMRRRKSDSIEVQQMKIQ 310 |
| Taenia solium | Query 265 SYSDKEFTIKPLDKKIDVFKFNSSKLRVNK#LILQLCIGNHDLFMRRRKADSLEVQQMKAQ 324 |+ ||+| ||| || | | | ++||# || || |||+|+|||||+||+|||||| | Sbjct 244 SFHDKKFIIKPADKSAKEFYFLVEKSKINK#RILALCTGNHELYMRRRKSDSIEVQQMKIQ 303 |
| Echinococcus multilocularis | Query 265 SYSDKEFTIKPLDKKIDVFKFNSSKLRVNK#LILQLCIGNHDLFMRRRKADSLEVQQMKAQ 324 |+ ||+| ||| || | | | ++||# || || |||+|+|||||+||+|||||| | Sbjct 251 SFHDKKFIIKPADKSAKEFFFLVEKSKINK#RILALCTGNHELYMRRRKSDSIEVQQMKIQ 310 |
| Echinococcus granulosus | Query 265 SYSDKEFTIKPLDKKIDVFKFNSSKLRVNK#LILQLCIGNHDLFMRRRKADSLEVQQMKAQ 324 |+ ||+| ||| || | | | ++||# || || |||+|+|||||+||+|||||| | Sbjct 251 SFHDKKFIIKPADKSAKEFFFLVEKSKINK#RILALCTGNHELYMRRRKSDSIEVQQMKIQ 310 |
| Schistosoma japonicum | Query 265 SYSDKEFTIKPLDKKIDVFKFNSSKLRVNK#LILQLCIGNHDLFMRRRKADSLEVQQMKAQ 324 ||| +| +||+ +| + ++#||| | +||| |+ ||+ ||+|||||| + Sbjct 249 SYSQNKFYVKPVGASGEVLTLYTDSTHTSR#LILNLSMGNHKLYAVRRQPDSIEVQQMKVK 308 |
[Site 2] KLRVNKLILQ298-LCIGNHDLFM
Gln298  Leu
Leu
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Lys289 | Leu290 | Arg291 | Val292 | Asn293 | Lys294 | Leu295 | Ile296 | Leu297 | Gln298 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Leu299 | Cys300 | Ile301 | Gly302 | Asn303 | His304 | Asp305 | Leu306 | Phe307 | Met308 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| KEFTIKPLDKKIDVFKFNSSKLRVNKLILQLCIGNHDLFMRRRKADSLEVQQMKAQAREE |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | Homo sapiens | 120.00 | 35 | neurofibromin 2 isoform 2 |
| 2 | Canis familiaris | 120.00 | 32 | PREDICTED: similar to neurofibromin 2 isoform 2 is |
| 3 | Mus musculus | 120.00 | 25 | unnamed protein product |
| 4 | N/A | 120.00 | 24 | - |
| 5 | Rattus norvegicus | 120.00 | 8 | MERL_RAT Merlin (Moesin-ezrin-radixin-like protein |
| 6 | Xenopus tropicalis | 120.00 | 6 | neurofibromin 2 |
| 7 | Gallus gallus | 120.00 | 5 | neurofibromatosis 2 |
| 8 | Monodelphis domestica | 120.00 | 5 | PREDICTED: hypothetical protein |
| 9 | Macaca mulatta | 120.00 | 5 | PREDICTED: similar to neurofibromin 2 isoform 1 is |
| 10 | Bos taurus | 120.00 | 4 | PREDICTED: similar to merlin isoform 1 |
| 11 | Papio anubis anubis | 120.00 | 1 | MERL_PAPAN Merlin (Moesin-ezrin-radixin-like prote |
| 12 | Xenopus laevis | 119.00 | 6 | neurofibromin 2 (bilateral acoustic neuroma) |
| 13 | Danio rerio | 110.00 | 15 | PREDICTED: hypothetical protein |
| 14 | Tetraodon nigroviridis | 108.00 | 7 | unnamed protein product |
| 15 | Strongylocentrotus purpuratus | 89.70 | 4 | PREDICTED: similar to neurofibromin 2 (bilateral a |
| 16 | Oryctolagus cuniculus | 88.60 | 1 | villin 2 (ezrin) |
| 17 | Pan troglodytes | 86.30 | 4 | PREDICTED: moesin |
| 18 | Sus scrofa | 86.30 | 2 | radixin |
| 19 | Macaca fascicularis | 86.30 | 1 | unnamed protein product |
| 20 | mice, keratinocytes, Balb/MK, Peptide, 583 aa | 86.30 | 1 | RADI_MOUSE Radixin (ESP10) gb |
| 21 | Tribolium castaneum | 85.50 | 6 | PREDICTED: similar to neurofibromatosis 2 |
| 22 | Ciona intestinalis | 85.50 | 1 | ezrin/radixin/moesin (ERM)-like protein |
| 23 | Aplysia californica | 84.70 | 1 | ezrin/radixin/moesin |
| 24 | Caenorhabditis briggsae | 84.30 | 2 | Hypothetical protein CBG12867 |
| 25 | Drosophila melanogaster | 83.60 | 7 | Moesin CG10701-PJ, isoform J |
| 26 | Drosophila pseudoobscura | 83.60 | 2 | GA10507-PA |
| 27 | Molgula tectiformis | 83.60 | 2 | Mt-ezrin/radixin/moesin |
| 28 | Caenorhabditis elegans | 83.20 | 6 | ERM-1Asv |
| 29 | Apis mellifera | 82.80 | 1 | PREDICTED: similar to Moesin CG10701-PD, isoform D |
| 30 | Spodoptera frugiperda | 82.40 | 1 | A unnamed protein product |
| 31 | Biomphalaria glabrata | 82.40 | 1 | ezrin/radixin/moesin |
| 32 | Aedes aegypti | 82.00 | 4 | moesin/ezrin/radixin |
| 33 | Anopheles gambiae str. PEST | 80.90 | 2 | ENSANGP00000017331 |
| 34 | Taenia saginata | 77.80 | 1 | myosin-like protein |
| 35 | Taenia solium | 77.80 | 1 | H17g protein, tegumental antigen |
| 36 | Echinococcus multilocularis | 77.40 | 1 | tegument protein gb |
| 37 | Echinococcus granulosus | 77.40 | 1 | EG10 |
| 38 | Schistosoma japonicum | 51.60 | 1 | SJCHGC06288 protein |
Top-ranked sequences
| organism | matching |
|---|---|
| Homo sapiens | Query 269 KEFTIKPLDKKIDVFKFNSSKLRVNKLILQ#LCIGNHDLFMRRRKADSLEVQQMKAQAREE 328 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 269 KEFTIKPLDKKIDVFKFNSSKLRVNKLILQ#LCIGNHDLFMRRRKADSLEVQQMKAQAREE 328 |
| Canis familiaris | Query 269 KEFTIKPLDKKIDVFKFNSSKLRVNKLILQ#LCIGNHDLFMRRRKADSLEVQQMKAQAREE 328 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 269 KEFTIKPLDKKIDVFKFNSSKLRVNKLILQ#LCIGNHDLFMRRRKADSLEVQQMKAQAREE 328 |
| Mus musculus | Query 269 KEFTIKPLDKKIDVFKFNSSKLRVNKLILQ#LCIGNHDLFMRRRKADSLEVQQMKAQAREE 328 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 269 KEFTIKPLDKKIDVFKFNSSKLRVNKLILQ#LCIGNHDLFMRRRKADSLEVQQMKAQAREE 328 |
| N/A | Query 269 KEFTIKPLDKKIDVFKFNSSKLRVNKLILQ#LCIGNHDLFMRRRKADSLEVQQMKAQAREE 328 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 269 KEFTIKPLDKKIDVFKFNSSKLRVNKLILQ#LCIGNHDLFMRRRKADSLEVQQMKAQAREE 328 |
| Rattus norvegicus | Query 269 KEFTIKPLDKKIDVFKFNSSKLRVNKLILQ#LCIGNHDLFMRRRKADSLEVQQMKAQAREE 328 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 269 KEFTIKPLDKKIDVFKFNSSKLRVNKLILQ#LCIGNHDLFMRRRKADSLEVQQMKAQAREE 328 |
| Xenopus tropicalis | Query 269 KEFTIKPLDKKIDVFKFNSSKLRVNKLILQ#LCIGNHDLFMRRRKADSLEVQQMKAQAREE 328 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 269 KEFTIKPLDKKIDVFKFNSSKLRVNKLILQ#LCIGNHDLFMRRRKADSLEVQQMKAQAREE 328 |
| Gallus gallus | Query 269 KEFTIKPLDKKIDVFKFNSSKLRVNKLILQ#LCIGNHDLFMRRRKADSLEVQQMKAQAREE 328 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 269 KEFTIKPLDKKIDVFKFNSSKLRVNKLILQ#LCIGNHDLFMRRRKADSLEVQQMKAQAREE 328 |
| Monodelphis domestica | Query 269 KEFTIKPLDKKIDVFKFNSSKLRVNKLILQ#LCIGNHDLFMRRRKADSLEVQQMKAQAREE 328 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 269 KEFTIKPLDKKIDVFKFNSSKLRVNKLILQ#LCIGNHDLFMRRRKADSLEVQQMKAQAREE 328 |
| Macaca mulatta | Query 269 KEFTIKPLDKKIDVFKFNSSKLRVNKLILQ#LCIGNHDLFMRRRKADSLEVQQMKAQAREE 328 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 269 KEFTIKPLDKKIDVFKFNSSKLRVNKLILQ#LCIGNHDLFMRRRKADSLEVQQMKAQAREE 328 |
| Bos taurus | Query 269 KEFTIKPLDKKIDVFKFNSSKLRVNKLILQ#LCIGNHDLFMRRRKADSLEVQQMKAQAREE 328 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 269 KEFTIKPLDKKIDVFKFNSSKLRVNKLILQ#LCIGNHDLFMRRRKADSLEVQQMKAQAREE 328 |
| Papio anubis anubis | Query 269 KEFTIKPLDKKIDVFKFNSSKLRVNKLILQ#LCIGNHDLFMRRRKADSLEVQQMKAQAREE 328 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 269 KEFTIKPLDKKIDVFKFNSSKLRVNKLILQ#LCIGNHDLFMRRRKADSLEVQQMKAQAREE 328 |
| Xenopus laevis | Query 269 KEFTIKPLDKKIDVFKFNSSKLRVNKLILQ#LCIGNHDLFMRRRKADSLEVQQMKAQAREE 328 ||||||||+|||||||||||||||||||||#|||||||||||||||||||||||||||||| Sbjct 269 KEFTIKPLEKKIDVFKFNSSKLRVNKLILQ#LCIGNHDLFMRRRKADSLEVQQMKAQAREE 328 |
| Danio rerio | Query 269 KEFTIKPLDKKIDVFKFNSSKLRVNKLILQ#LCIGNHDLFMRRRKADSLEVQQMKAQAREE 328 ||| |||+||+ ||||||||||||||||||#|||||||||||||+ ||||||||| ||||| Sbjct 268 KEFAIKPVDKRADVFKFNSSKLRVNKLILQ#LCIGNHDLFMRRRRVDSLEVQQMKTQAREE 327 |
| Tetraodon nigroviridis | Query 269 KEFTIKPLDKKIDVFKFNSSKLRVNKLILQ#LCIGNHDLFMRRRKADSLEVQQMKAQAREE 328 ||||||||+|| |||||||+|| ||||||#|||||||||||||+ ||||||||||||||| Sbjct 163 KEFTIKPLEKKTKVFKFNSSRLRANKLILQ#LCIGNHDLFMRRRRVDSLEVQQMKAQAREE 222 |
| Strongylocentrotus purpuratus | Query 269 KEFTIKPLDKKIDVFKFNSSKLRVNKLILQ#LCIGNHDLFMRRRKADSLEVQQMKAQAREE 328 |+||||| || | | | |||+||||| #||+|||+|||+||+|||+|||||||||||| Sbjct 279 KKFTIKPTAKKAPNFCFISPKLRMNKLILD#LCVGNHELFMQRRRADSMEVQQMKAQAREE 338 |
| Oryctolagus cuniculus | Query 269 KEFTIKPLDKKIDVFKFNSSKLRVNKLILQ#LCIGNHDLFMRRRKADSLEVQQMKAQAREE 328 |+| |||+||| | | + +||+|| |||#||+|||+|+||||| |++|||||||||||| Sbjct 253 KKFVIKPIDKKAPDFVFYAPRLRINKRILQ#LCMGNHELYMRRRKPDTIEVQQMKAQAREE 312 |
| Pan troglodytes | Query 269 KEFTIKPLDKKIDVFKFNSSKLRVNKLILQ#LCIGNHDLFMRRRKADSLEVQQMKAQAREE 328 |+| |||+||| | | + +||+|| || #||+|||+|+||||| |++|||||||||||| Sbjct 225 KKFVIKPIDKKAPDFVFYAPRLRINKRILA#LCMGNHELYMRRRKPDTIEVQQMKAQAREE 284 |
| Sus scrofa | Query 269 KEFTIKPLDKKIDVFKFNSSKLRVNKLILQ#LCIGNHDLFMRRRKADSLEVQQMKAQAREE 328 |+| |||+||| | | + +||+|| || #||+|||+|+||||| |++|||||||||||| Sbjct 253 KKFVIKPIDKKAPDFVFYAPRLRINKRILA#LCMGNHELYMRRRKPDTIEVQQMKAQAREE 312 |
| Macaca fascicularis | Query 269 KEFTIKPLDKKIDVFKFNSSKLRVNKLILQ#LCIGNHDLFMRRRKADSLEVQQMKAQAREE 328 |+| |||+||| | | + +||+|| || #||+|||+|+||||| |++|||||||||||| Sbjct 253 KKFVIKPIDKKAPDFVFYAPRLRINKRILA#LCMGNHELYMRRRKPDTIEVQQMKAQAREE 312 |
| mice, keratinocytes, Balb/MK, Peptide, 583 aa | Query 269 KEFTIKPLDKKIDVFKFNSSKLRVNKLILQ#LCIGNHDLFMRRRKADSLEVQQMKAQAREE 328 |+| |||+||| | | + +||+|| || #||+|||+|+||||| |++|||||||||||| Sbjct 253 KKFVIKPIDKKAPDFVFYAPRLRINKRILA#LCMGNHELYMRRRKPDTIEVQQMKAQAREE 312 |
| Tribolium castaneum | Query 269 KEFTIKPLDKKIDVFKFNSSKLRVNKLILQ#LCIGNHDLFMRRRKADSLEVQQMKAQAREE 328 |+| |||+|| | | | |+|+||||| #||+||||||||||| ||+|+||||| |+|| Sbjct 263 KKFIIKPVDKNSPNFVFFSQKVRMNKLILD#LCMGNHDLFMRRRKPDSMELQQMKAAAKEE 322 |
| Ciona intestinalis | Query 269 KEFTIKPLDKKIDVFKFNSSKLRVNKLILQ#LCIGNHDLFMRRRKADSLEVQQMKAQAREE 328 |+| |||+||| | | +||+|| || #||+|||+|+||||| |++|||||||||||| Sbjct 277 KKFVIKPIDKKAPDFVFYVERLRINKRILA#LCMGNHELYMRRRKPDTIEVQQMKAQAREE 336 |
| Aplysia californica | Query 269 KEFTIKPLDKKIDVFKFNSSKLRVNKLILQ#LCIGNHDLFMRRRKADSLEVQQMKAQAREE 328 |+| |||+||| | | + +||+|| || #||+|||+|+||||| |++|||||||||||+ Sbjct 254 KKFIIKPIDKKAPDFVFYAPRLRINKRILA#LCMGNHELYMRRRKPDTIEVQQMKAQARED 313 |
| Caenorhabditis briggsae | Query 269 KEFTIKPLDKKIDVFKFNSSKLRVNKLILQ#LCIGNHDLFMRRRKADSLEVQQMKAQAREE 328 |+| |||+||| | | + +||+|| || #||+|||+|+||||| |++|||||| ||||| Sbjct 249 KKFVIKPIDKKAHDFVFYAPRLRINKRILA#LCMGNHELYMRRRKPDTIEVQQMKQQAREE 308 |
| Drosophila melanogaster | Query 269 KEFTIKPLDKKIDVFKFNSSKLRVNKLILQ#LCIGNHDLFMRRRKADSLEVQQMKAQAREE 328 |+| |||+||| | | + ++|+|| || #||+|||+|+||||| |+++||||||||||| Sbjct 254 KKFIIKPIDKKAPDFMFFAPRVRINKRILA#LCMGNHELYMRRRKPDTIDVQQMKAQAREE 313 |
| Drosophila pseudoobscura | Query 269 KEFTIKPLDKKIDVFKFNSSKLRVNKLILQ#LCIGNHDLFMRRRKADSLEVQQMKAQAREE 328 |+| |||+||| | | + ++|+|| || #||+|||+|+||||| |+++||||||||||| Sbjct 249 KKFIIKPIDKKAPDFMFFAPRVRINKRILA#LCMGNHELYMRRRKPDTIDVQQMKAQAREE 308 |
| Molgula tectiformis | Query 269 KEFTIKPLDKKIDVFKFNSSKLRVNKLILQ#LCIGNHDLFMRRRKADSLEVQQMKAQARE 327 |+| |||+||| | | +||+|| || #||+|||+|+||||| ||+|||||||||+| Sbjct 253 KKFVIKPIDKKAPDFVFYVERLRINKRILA#LCMGNHELYMRRRKPDSIEVQQMKAQAKE 311 |
| Caenorhabditis elegans | Query 269 KEFTIKPLDKKIDVFKFNSSKLRVNKLILQ#LCIGNHDLFMRRRKADSLEVQQMKAQAREE 328 |+| |||+||| | | + +||+|| || #||+|||+|+||||| |++|||||| ||||+ Sbjct 254 KKFVIKPIDKKAHDFVFYAPRLRINKRILA#LCMGNHELYMRRRKPDTIEVQQMKQQARED 313 |
| Apis mellifera | Query 269 KEFTIKPLDKKIDVFKFNSSKLRVNKLILQ#LCIGNHDLFMRRRKADSLEVQQMKAQAREE 328 |+| |||+||| | | ++++++|| || #||+|||+|+||||| |+++||||||||||| Sbjct 188 KKFIIKPIDKKAPDFVFFATRVKINKRILA#LCMGNHELYMRRRKPDTIDVQQMKAQAREE 247 |
| Spodoptera frugiperda | Query 269 KEFTIKPLDKKIDVFKFNSSKLRVNKLILQ#LCIGNHDLFMRRRKADSLEVQQMKAQAREE 328 ++| |||+||| | | + ++|||| || #||+|||+|+||||| |+++||||||||||| Sbjct 253 RKFIIKPIDKKAPDFVFFAPRVRVNKRILA#LCMGNHELYMRRRKPDTIDVQQMKAQAREE 312 |
| Biomphalaria glabrata | Query 269 KEFTIKPLDKKIDVFKFNSSKLRVNKLILQ#LCIGNHDLFMRRRKADSLEVQQMKAQAR 326 |+| |||+||| | | + +||+|| || #||+|||+|+||||| |++|||||||||| Sbjct 254 KKFVIKPIDKKAPDFVFYAPRLRINKRILA#LCMGNHELYMRRRKPDTIEVQQMKAQAR 311 |
| Aedes aegypti | Query 269 KEFTIKPLDKKIDVFKFNSSKLRVNKLILQ#LCIGNHDLFMRRRKADSLEVQQMKAQAREE 328 ++| |||+||| | | + ++|+|| || #||+|||+|+||||| |+++||||||||||| Sbjct 259 RKFIIKPIDKKAPDFVFFAPRVRINKRILA#LCMGNHELYMRRRKPDTIDVQQMKAQAREE 318 |
| Anopheles gambiae str. PEST | Query 269 KEFTIKPLDKKIDVFKFNSSKLRVNKLILQ#LCIGNHDLFMRRRKADSLEVQQMKAQAREE 328 ++| |||+||| | | + ++|+|| || #||+|||+|+||||| |+++|||||||||+| Sbjct 300 RKFIIKPIDKKAPDFVFFAPRVRINKRILA#LCMGNHELYMRRRKPDTIDVQQMKAQARDE 359 |
| Taenia saginata | Query 269 KEFTIKPLDKKIDVFKFNSSKLRVNKLILQ#LCIGNHDLFMRRRKADSLEVQQMKAQAREE 328 |+| ||| || | | | ++|| || #|| |||+|+|||||+||+|||||| ||+|| Sbjct 255 KKFIIKPADKSAKEFYFLVEKSKINKRILA#LCTGNHELYMRRRKSDSIEVQQMKIQAKEE 314 |
| Taenia solium | Query 269 KEFTIKPLDKKIDVFKFNSSKLRVNKLILQ#LCIGNHDLFMRRRKADSLEVQQMKAQAREE 328 |+| ||| || | | | ++|| || #|| |||+|+|||||+||+|||||| ||+|| Sbjct 248 KKFIIKPADKSAKEFYFLVEKSKINKRILA#LCTGNHELYMRRRKSDSIEVQQMKIQAKEE 307 |
| Echinococcus multilocularis | Query 269 KEFTIKPLDKKIDVFKFNSSKLRVNKLILQ#LCIGNHDLFMRRRKADSLEVQQMKAQAREE 328 |+| ||| || | | | ++|| || #|| |||+|+|||||+||+|||||| ||+|| Sbjct 255 KKFIIKPADKSAKEFFFLVEKSKINKRILA#LCTGNHELYMRRRKSDSIEVQQMKIQAKEE 314 |
| Echinococcus granulosus | Query 269 KEFTIKPLDKKIDVFKFNSSKLRVNKLILQ#LCIGNHDLFMRRRKADSLEVQQMKAQAREE 328 |+| ||| || | | | ++|| || #|| |||+|+|||||+||+|||||| ||+|| Sbjct 255 KKFIIKPADKSAKEFFFLVEKSKINKRILA#LCTGNHELYMRRRKSDSIEVQQMKIQAKEE 314 |
| Schistosoma japonicum | Query 270 EFTIKPLDKKIDVFKFNSSKLRVNKLILQ#LCIGNHDLFMRRRKADSLEVQQMKAQARE 327 +| +||+ +| + ++||| #| +||| |+ ||+ ||+|||||| +|+| Sbjct 254 KFYVKPVGASGEVLTLYTDSTHTSRLILN#LSMGNHKLYAVRRQPDSIEVQQMKVKAKE 311 |
* References
[PubMed ID: 19351817] Blackford A, Parmigiani G, Kensler TW, Wolfgang C, Jones S, Zhang X, Parsons DW, Lin JC, Leary RJ, Eshleman JR, Goggins M, Jaffee EM, Iacobuzio-Donahue CA, Maitra A, Klein A, Cameron JL, Olino K, Schulick R, Winter J, Vogelstein B, Velculescu VE, Kinzler KW, Hruban RH, Genetic mutations associated with cigarette smoking in pancreatic cancer. Cancer Res. 2009 Apr 15;69(8):3681-8. Epub 2009 Apr 7.
[PubMed ID: 19144871] ... Seo PS, Quinn BJ, Khan AA, Zeng L, Takoudis CG, Hanada T, Bolis A, Bolino A, Chishti AH, Identification of erythrocyte p55/MPP1 as a binding partner of NF2 tumor suppressor protein/Merlin. Exp Biol Med (Maywood). 2009 Mar;234(3):255-62. Epub 2009 Jan 14.
[PubMed ID: 19029950] ... Lallemand D, Manent J, Couvelard A, Watilliaux A, Siena M, Chareyre F, Lampin A, Niwa-Kawakita M, Kalamarides M, Giovannini M, Merlin regulates transmembrane receptor accumulation and signaling at the plasma membrane in primary mouse Schwann cells and in human schwannomas. Oncogene. 2009 Feb 12;28(6):854-65. Epub 2008 Nov 24.
[PubMed ID: 18953429] ... Striedinger K, VandenBerg SR, Baia GS, McDermott MW, Gutmann DH, Lal A, The neurofibromatosis 2 tumor suppressor gene product, merlin, regulates human meningioma cell growth by signaling through YAP. Neoplasia. 2008 Nov;10(11):1204-12.
[PubMed ID: 18923024] ... Denisenko N, Cifuentes-Diaz C, Irinopoulou T, Carnaud M, Benoit E, Niwa-Kawakita M, Chareyre F, Giovannini M, Girault JA, Goutebroze L, Tumor suppressor schwannomin/merlin is critical for the organization of Schwann cell contacts in peripheral nerves. J Neurosci. 2008 Oct 15;28(42):10472-81.
[PubMed ID: 7485397] ... den Bakker MA, Tascilar M, Riegman PH, Hekman AC, Boersma W, Janssen PJ, de Jong TA, Hendriks W, van der Kwast TH, Zwarthoff EC, Neurofibromatosis type 2 protein co-localizes with elements of the cytoskeleton. Am J Pathol. 1995 Nov;147(5):1339-49.
[PubMed ID: 7666400] ... Evans DG, Bourn D, Wallace A, Ramsden RT, Mitchell JD, Strachan T, Diagnostic issues in a family with late onset type 2 neurofibromatosis. J Med Genet. 1995 Jun;32(6):470-4.
[PubMed ID: 7868131] ... Papi L, De Vitis LR, Vitelli F, Ammannati F, Mennonna P, Montali E, Bigozzi U, Somatic mutations in the neurofibromatosis type 2 gene in sporadic meningiomas. Hum Genet. 1995 Mar;95(3):347-51.
[PubMed ID: 7798645] ... Honda M, Arai E, Sawada S, Ohta A, Niimura M, Neurofibromatosis 2 and neurilemmomatosis gene are identical. J Invest Dermatol. 1995 Jan;104(1):74-7.
[PubMed ID: 1456285] ... Arai E, Ikeuchi T, Karasawa S, Tamura A, Yamamoto K, Kida M, Ichimura K, Yuasa Y, Tonomura A, Constitutional translocation t(4;22) (q12;q12.2) associated with neurofibromatosis type 2. Am J Med Genet. 1992 Sep 15;44(2):163-7.