XSB1203 : protein kinase C gamma [Homo sapiens]
[ CaMP Format ]
This entry is computationally expanded from SB0089
* Basic Information
| Organism | Homo sapiens (human) |
| Protein Names | Protein kinase C gamma type; PKC-gamma; 2.7.11.13; protein kinase C gamma |
| Gene Names | PKCG |
| Gene Locus | Not available |
| GO Function | Not available |
* Information From OMIM
Not Available.
* Structure Information
1. Primary Information
Length: 536 aa
Average Mass: 60.414 kDa
Monoisotopic Mass: 60.375 kDa
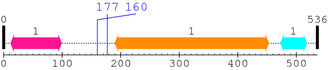
2. Domain Information
Annotated Domains: interpro / pfam / smart / prosite
Computationally Assigned Domains (Pfam+HMMER):
| domain name | begin | end | score | e-value |
|---|---|---|---|---|
| C2 1. | 12 | 99 | 140.6 | 4.7e-39 |
| --- cleavage 177 --- | ||||
| --- cleavage 160 --- | ||||
| Pkinase 1. | 190 | 453 | 274.6 | 2.2e-79 |
| Pkinase_C 1. | 473 | 518 | 52.4 | 1.7e-12 |
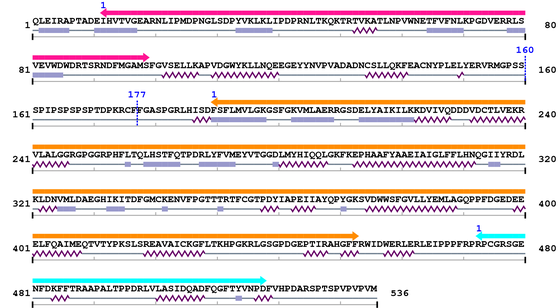
3. Sequence Information
Fasta Sequence: XSB1203.fasta
Amino Acid Sequence and Secondary Structures (PsiPred):
* Cleavage Information
2 [sites]
Cleavage sites (±10aa)
[Site 1] PSPTDPKRCF177-FGASPGRLHI
Phe177  Phe
Phe
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Pro168 | Ser169 | Pro170 | Thr171 | Asp172 | Pro173 | Lys174 | Arg175 | Cys176 | Phe177 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Phe178 | Gly179 | Ala180 | Ser181 | Pro182 | Gly183 | Arg184 | Leu185 | His186 | Ile187 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| LELYERVRMGPSSSPIPSPSPSPTDPKRCFFGASPGRLHISDFSFLMVLGKGSFGKVMLA |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | Homo sapiens | 94.00 | 17 | protein kinase C gamma |
| 2 | Macaca mulatta | 94.00 | 15 | PREDICTED: similar to protein kinase C, gamma isof |
| 3 | Macaca fascicularis | 94.00 | 3 | KPCG_MACFA Protein kinase C gamma type (PKC-gamma) |
| 4 | Mus musculus | 90.90 | 12 | protein kinase C, gamma |
| 5 | Oryctolagus cuniculus | 90.90 | 6 | protein kinase C, gamma |
| 6 | N/A | 88.60 | 19 | K |
| 7 | Bos taurus | 88.60 | 10 | PREDICTED: similar to Protein kinase C gamma type |
| 8 | Canis familiaris | 84.30 | 15 | PREDICTED: similar to Protein kinase C, gamma type |
| 9 | Gallus gallus | 44.70 | 6 | PREDICTED: similar to protein kinase C beta-1 |
| 10 | Danio rerio | 44.30 | 11 | protein kinase C |
| 11 | Tetraodon nigroviridis | 43.50 | 5 | unnamed protein product |
| 12 | Anopheles gambiae str. PEST | 43.50 | 3 | ENSANGP00000022036 |
| 13 | Scyliorhinus canicula | 43.10 | 2 | protein kinase C |
| 14 | Rattus norvegicus | 42.70 | 9 | PREDICTED: similar to Protein kinase C alpha type |
| 15 | Monodelphis domestica | 42.70 | 7 | PREDICTED: similar to protein kinase C |
| 16 | synthetic construct | 42.70 | 3 | protein kinase C alpha |
| 17 | Latimeria chalumnae | 42.70 | 2 | protein kinase C alpha |
| 18 | Petromyzon marinus | 42.70 | 1 | protein kinase C |
| 19 | Tribolium castaneum | 42.40 | 4 | PREDICTED: similar to CG1954-PA |
| 20 | Xenopus tropicalis | 42.40 | 3 | hypothetical protein LOC780209 |
| 21 | Aedes aegypti | 42.40 | 2 | protein kinase c |
| 22 | Hydractinia echinata | 42.40 | 1 | putative calcium dependent protein kinase C |
| 23 | Strongylocentrotus purpuratus | 41.60 | 6 | PREDICTED: similar to protein kinase C |
| 24 | Drosophila melanogaster | 41.60 | 6 | GH13631p |
| 25 | Apis mellifera | 41.60 | 2 | PREDICTED: similar to Protein C kinase 53E CG6622- |
| 26 | Caenorhabditis briggsae | 41.60 | 1 | Hypothetical protein CBG23354 |
| 27 | Drosophila pseudoobscura | 41.60 | 1 | GA15149-PA |
| 28 | Protopterus dolloi | 41.20 | 2 | protein kinase C beta 1 |
| 29 | Caenorhabditis elegans | 40.80 | 5 | Protein Kinase C family member (pkc-1) |
| 30 | Xenopus laevis | 40.80 | 5 | protein kinase C, alpha |
| 31 | Sycon raphanus | 40.80 | 2 | Serine/Threonine protein kinase |
| 32 | Branchiostoma lanceolatum | 40.40 | 1 | protein kinase C |
| 33 | Pan troglodytes | 39.70 | 5 | PREDICTED: protein kinase C, eta isoform 3 |
| 34 | Myxine glutinosa | 39.70 | 1 | protein kinase C |
| 35 | Takifugu rubripes | 39.70 | 1 | protein kinase C, alpha type |
| 36 | Bombyx mori | 39.70 | 1 | conventional protein kinase C |
| 37 | Aplysia californica | 39.30 | 2 | KPC2_APLCA Calcium-independent protein kinase C (A |
| 38 | Schizosaccharomyces pombe | 38.90 | 3 | protein kinase |
| 39 | Schizosaccharomyces pombe 972h- | 38.90 | 2 | hypothetical protein SPBC12D12.04c |
| 40 | Yarrowia lipolytica | 38.90 | 1 | hypothetical protein |
| 41 | Ancylostoma caninum | 38.10 | 1 | AF301263_1 protein kinase C |
| 42 | Tuber excavatum | 37.70 | 2 | protein kinase C-like protein |
| 43 | Coccidioides immitis RS | 37.70 | 1 | hypothetical protein CIMG_09206 |
| 44 | Tuber indicum | 37.40 | 6 | protein kinase C-like protein |
| 45 | Tuber brumale | 37.40 | 2 | protein kinase C-like protein |
| 46 | Hydra vulgaris | 37.40 | 2 | protein kinase C |
| 47 | Cryptococcus neoformans var. neoformans B-3501A | 37.40 | 1 | hypothetical protein CNBC3910 |
| 48 | Cryptococcus neoformans var. neoformans JEC21 | 37.40 | 1 | protein kinase C |
| 49 | Tuber pseudoexcavatum | 37.40 | 1 | protein kinase C-like protein |
| 50 | Debaryomyces hansenii CBS767 | 37.40 | 1 | hypothetical protein DEHA0G14773g |
| 51 | Cryptococcus neoformans var. neoformans | 37.40 | 1 | protein kinase C 1 |
| 52 | Coprinopsis cinerea okayama7#130 | 37.40 | 1 | hypothetical protein CC1G_07190 |
| 53 | Cryptococcus neoformans var. grubii | 37.40 | 1 | protein kinase C 1 |
| 54 | Pichia guilliermondii ATCC 6260 | 37.40 | 1 | hypothetical protein PGUG_04383 |
| 55 | Candida albicans SC5314 | 37.40 | 1 | protein kinase C |
| 56 | Tuber aestivum | 37.00 | 2 | protein kinase C-like protein |
| 57 | Pichia stipitis CBS 6054 | 37.00 | 1 | predicted protein |
| 58 | Lodderomyces elongisporus NRRL YB-4239 | 37.00 | 1 | hypothetical protein LELG_02660 |
| 59 | Squalus acanthias | 37.00 | 1 | s-sgk1 |
| 60 | Sporothrix schenckii | 37.00 | 1 | AF124792_1 protein kinase C; serine/threonine prot |
| 61 | Tuber rufum | 37.00 | 1 | protein kinase C-like protein |
| 62 | Tuber magnatum | 37.00 | 1 | protein kinase C homologue |
| 63 | Tuber borchii | 37.00 | 1 | protein kinase C homologue |
| 64 | Candida albicans | 37.00 | 1 | KPC1_CANAL Protein kinase C-like 1 (PKC 1) emb |
| 65 | Aspergillus clavatus NRRL 1 | 36.20 | 1 | protein kinase c |
| 66 | Dictyostelium discoideum | 36.20 | 1 | rac serine/threonine kinase homolog |
| 67 | Phaeosphaeria nodorum SN15 | 36.20 | 1 | hypothetical protein SNOG_07073 |
| 68 | Aspergillus oryzae | 36.20 | 1 | unnamed protein product |
| 69 | Neosartorya fischeri NRRL 181 | 36.20 | 1 | protein kinase c |
| 70 | Fundulus heteroclitus | 36.20 | 1 | serum and glucocorticoid-regulated kinase |
| 71 | Botryotinia fuckeliana | 36.20 | 1 | protein kinase C |
| 72 | Leptosphaeria maculans | 36.20 | 1 | AF487263_9 protein kinase C |
| 73 | Magnaporthe grisea | 36.20 | 1 | AF136600_1 protein kinase C |
| 74 | Magnaporthe grisea 70-15 | 36.20 | 1 | hypothetical protein MGG_08689 |
| 75 | Neurospora crassa | 36.20 | 1 | protein kinase C homologue |
| 76 | Aspergillus terreus NIH2624 | 36.20 | 1 | hypothetical protein ATEG_02078 |
| 77 | Blumeria graminis | 36.20 | 1 | AF247001_1 protein kinase C |
Top-ranked sequences
| organism | matching |
|---|---|
| Homo sapiens | Query 148 LELYERVRMGXXXXXXXXXXXXXTDPKRCF#FGASPGRLHISDFSFLMVLGKGSFGKVMLA 207 |||||||||| |||||||#|||||||||||||||||||||||||||||| Sbjct 148 LELYERVRMGPSSSPIPSPSPSPTDPKRCF#FGASPGRLHISDFSFLMVLGKGSFGKVMLA 207 |
| Macaca mulatta | Query 148 LELYERVRMGXXXXXXXXXXXXXTDPKRCF#FGASPGRLHISDFSFLMVLGKGSFGKVMLA 207 |||||||||| |||||||#|||||||||||||||||||||||||||||| Sbjct 307 LELYERVRMGPSSSPIPSPSPSPTDPKRCF#FGASPGRLHISDFSFLMVLGKGSFGKVMLA 366 |
| Macaca fascicularis | Query 148 LELYERVRMGXXXXXXXXXXXXXTDPKRCF#FGASPGRLHISDFSFLMVLGKGSFGKVMLA 207 |||||||||| |||||||#|||||||||||||||||||||||||||||| Sbjct 309 LELYERVRMGPSSSPIPSPSPSPTDPKRCF#FGASPGRLHISDFSFLMVLGKGSFGKVMLA 368 |
| Mus musculus | Query 148 LELYERVRMGXXXXXXXXXXXXXTDPKRCF#FGASPGRLHISDFSFLMVLGKGSFGKVMLA 207 |||||||||| || ||||#|||||||||||||||||||||||||||||| Sbjct 309 LELYERVRMGPSSSPIPSPSPSPTDSKRCF#FGASPGRLHISDFSFLMVLGKGSFGKVMLA 368 |
| Oryctolagus cuniculus | Query 148 LELYERVRMGXXXXXXXXXXXXXTDPKRCF#FGASPGRLHISDFSFLMVLGKGSFGKVMLA 207 |||||||||| || ||||#|||||||||||||||||||||||||||||| Sbjct 309 LELYERVRMGPSSSPIPSPSPSPTDSKRCF#FGASPGRLHISDFSFLMVLGKGSFGKVMLA 368 |
| N/A | Query 148 LELYERVRMGXXXXXXXXXXXXXTDPKRCF#FGASPGRLHISDFSFLMVLGKGSFGKVMLA 207 |||||||| | || ||||#|||||||||||||||||||||||||||||| Sbjct 294 LELYERVRTGPSSSPIPSPSPSPTDSKRCF#FGASPGRLHISDFSFLMVLGKGSFGKVMLA 353 |
| Bos taurus | Query 148 LELYERVRMGXXXXXXXXXXXXXTDPKRCF#FGASPGRLHISDFSFLMVLGKGSFGKVMLA 207 |||||||| | || ||||#|||||||||||||||||||||||||||||| Sbjct 309 LELYERVRTGPSSSPIPSPSPSPTDSKRCF#FGASPGRLHISDFSFLMVLGKGSFGKVMLA 368 |
| Canis familiaris | Query 148 LELYERVRMGXXXXXXXXXXXXXTDPKRCF#FGASPGRLHISDFSFLMVLGKGSFGKVMLA 207 |+ +|||||| || ||||#|| ||||||||||||||||||||||||||| Sbjct 285 LQKFERVRMGPSSSPIPSPSPSPTDSKRCF#FGTSPGRLHISDFSFLMVLGKGSFGKVMLA 344 |
| Gallus gallus | Query 181 SPGRLHISDFSFLMVLGKGSFGKVMLA 207 | |+ +|||+|||||||||||||||| Sbjct 332 SRDRMKLSDFNFLMVLGKGSFGKVMLA 358 |
| Danio rerio | Query 184 RLHISDFSFLMVLGKGSFGKVMLA 207 |+ +|||+|||||||||||||||| Sbjct 265 RMKLSDFNFLMVLGKGSFGKVMLA 288 |
| Tetraodon nigroviridis | Query 184 RLHISDFSFLMVLGKGSFGKVMLA 207 || ||||+|| ||||||||||||| Sbjct 433 RLGISDFTFLQVLGKGSFGKVMLA 456 |
| Anopheles gambiae str. PEST | Query 182 PGRLHISDFSFLMVLGKGSFGKVMLA 207 ||++ ++||+|+ ||||||||||||| Sbjct 392 PGKVFLNDFNFIKVLGKGSFGKVMLA 417 |
| Scyliorhinus canicula | Query 184 RLHISDFSFLMVLGKGSFGKVMLA 207 |+ ++||+|||||||||||||||| Sbjct 256 RMKLADFNFLMVLGKGSFGKVMLA 279 |
| Rattus norvegicus | Query 184 RLHISDFSFLMVLGKGSFGKVMLA 207 |+ ++||+|||||||||||||||| Sbjct 273 RVKLTDFNFLMVLGKGSFGKVMLA 296 |
| Monodelphis domestica | Query 184 RLHISDFSFLMVLGKGSFGKVMLA 207 |+ ++||+|||||||||||||||| Sbjct 332 RVKLTDFNFLMVLGKGSFGKVMLA 355 |
| synthetic construct | Query 184 RLHISDFSFLMVLGKGSFGKVMLA 207 |+ ++||+|||||||||||||||| Sbjct 333 RVKLTDFNFLMVLGKGSFGKVMLA 356 |
| Latimeria chalumnae | Query 184 RLHISDFSFLMVLGKGSFGKVMLA 207 |+ ++||+|||||||||||||||| Sbjct 256 RVKLTDFNFLMVLGKGSFGKVMLA 279 |
| Petromyzon marinus | Query 151 YERVRMGXXXXXXXXXXXXXTDPKRCF#FGASPGRLHISDFSFLMVLGKGSFGKVMLA 207 +|+ |+| # |+ |+ ||+|||||||||||||||| Sbjct 235 FEKARLGPSVKPRASDERRANSLSAIL#NNANVDRVKADDFNFLMVLGKGSFGKVMLA 291 |
| Tribolium castaneum | Query 181 SPGRLHISDFSFLMVLGKGSFGKVMLA 207 || ++ + ||+|+ ||||||||||||| Sbjct 352 SPNKIGLDDFNFIKVLGKGSFGKVMLA 378 |
| Xenopus tropicalis | Query 185 LHISDFSFLMVLGKGSFGKVMLA 207 + ++||||||||||||||||||| Sbjct 340 VRLTDFSFLMVLGKGSFGKVMLA 362 |
| Aedes aegypti | Query 172 DPKRCF#FGASPGRLHISDFSFLMVLGKGSFGKVMLA 207 | + # | + |++ ++||+|+ ||||||||||||| Sbjct 400 DDRAAS#GGQTVGKMCLNDFNFIKVLGKGSFGKVMLA 435 |
| Hydractinia echinata | Query 182 PGRLHISDFSFLMVLGKGSFGKVMLA 207 | || + ||+|| |+||||||||||| Sbjct 190 PERLKLEDFTFLKVIGKGSFGKVMLA 215 |
| Strongylocentrotus purpuratus | Query 180 ASPGRLHISDFSFLMVLGKGSFGKVMLA 207 +| | + |||+|| ||||||||||||| Sbjct 315 SSMGLVRASDFNFLSVLGKGSFGKVMLA 342 |
| Drosophila melanogaster | Query 182 PGRLHISDFSFLMVLGKGSFGKVMLA 207 ||+ + ||+|+ ||||||||||||| Sbjct 215 PGKCSLLDFNFIKVLGKGSFGKVMLA 240 |
| Apis mellifera | Query 185 LHISDFSFLMVLGKGSFGKVMLA 207 + |||+|||||||||||||||| Sbjct 350 IRASDFNFLMVLGKGSFGKVMLA 372 |
| Caenorhabditis briggsae | Query 183 GRLHISDFSFLMVLGKGSFGKVMLA 207 | | |+||+|+ ||||||||||||| Sbjct 372 GTLSINDFTFMKVLGKGSFGKVMLA 396 |
| Drosophila pseudoobscura | Query 182 PGRLHISDFSFLMVLGKGSFGKVMLA 207 ||+ + ||+|+ ||||||||||||| Sbjct 396 PGKCSLLDFNFIKVLGKGSFGKVMLA 421 |
| Protopterus dolloi | Query 184 RLHISDFSFLMVLGKGSFGKVML 206 |+ +|||+|||||||||||||+| Sbjct 266 RVKLSDFNFLMVLGKGSFGKVLL 288 |
| Caenorhabditis elegans | Query 183 GRLHISDFSFLMVLGKGSFGKVMLA 207 | | | ||+|+ ||||||||||||| Sbjct 371 GTLSIHDFTFMKVLGKGSFGKVMLA 395 |
| Xenopus laevis | Query 185 LHISDFSFLMVLGKGSFGKVMLA 207 + ++|| |||||||||||||||| Sbjct 339 IRLTDFCFLMVLGKGSFGKVMLA 361 |
| Sycon raphanus | Query 187 ISDFSFLMVLGKGSFGKVMLA 207 +|||||| ||||||||||||| Sbjct 363 LSDFSFLKVLGKGSFGKVMLA 383 |
| Branchiostoma lanceolatum | Query 174 KRCF#FGASPGRLHISDFSFLMVLGKGSFGKVMLA 207 + | # | | + +||+|||||||||||||||| Sbjct 252 EECN#IG-SLDHMKAADFNFLMVLGKGSFGKVMLA 284 |
| Pan troglodytes | Query 181 SPGRLHISDFSFLMVLGKGSFGKVMLA 207 | || | +| |+ ||||||||||||| Sbjct 185 SSNRLGIDNFEFIRVLGKGSFGKVMLA 211 |
| Myxine glutinosa | Query 184 RLHISDFSFLMVLGKGSFGKVMLA 207 |+++ || || ||||||||||||| Sbjct 256 RVNLEDFVFLTVLGKGSFGKVMLA 279 |
| Takifugu rubripes | Query 184 RLHISDFSFLMVLGKGSFGKVMLA 207 |+ ++||+|| +|||||||||||| Sbjct 331 RVRLNDFNFLALLGKGSFGKVMLA 354 |
| Bombyx mori | Query 185 LHISDFSFLMVLGKGSFGKVMLA 207 + +||+|+|||||||||||||| Sbjct 335 IRATDFNFIMVLGKGSFGKVMLA 357 |
| Aplysia californica | Query 184 RLHISDFSFLMVLGKGSFGKVMLA 207 |+ + ||+|+ ||||||||||||| Sbjct 397 RISLHDFNFIKVLGKGSFGKVMLA 420 |
| Schizosaccharomyces pombe | Query 184 RLHISDFSFLMVLGKGSFGKVMLA 207 |+ + ||+|| |||||+||||||| Sbjct 677 RIGLEDFTFLSVLGKGNFGKVMLA 700 |
| Schizosaccharomyces pombe 972h- | Query 184 RLHISDFSFLMVLGKGSFGKVMLA 207 |+ + ||+|| |||||+||||||| Sbjct 677 RIGLEDFTFLSVLGKGNFGKVMLA 700 |
| Yarrowia lipolytica | Query 184 RLHISDFSFLMVLGKGSFGKVMLA 207 |+ + ||+|| |||||+||||||| Sbjct 919 RIGLDDFNFLAVLGKGNFGKVMLA 942 |
| Ancylostoma caninum | Query 171 TDPKRCF#FGASPGRLHISDFSFLMVLGKGSFGKV 204 || | # || + |||+|+ |||||||||| Sbjct 12 TDSKHSL#SGAQSQIVKESDFTFITVLGKGSFGKV 45 |
| Tuber excavatum | Query 182 PGRLHISDFSFLMVLGKGSFGKVMLA 207 | ++ + |+|| |||||+||||||| Sbjct 323 PAKIGLDHFNFLAVLGKGNFGKVMLA 348 |
| Coccidioides immitis RS | Query 182 PGRLHISDFSFLMVLGKGSFGKVMLA 207 | |+ + |+|| |||||+||||||| Sbjct 738 PKRVGLDHFNFLAVLGKGNFGKVMLA 763 |
| Tuber indicum | Query 180 ASPGRLHISDFSFLMVLGKGSFGKVMLA 207 + | ++ + |+|| |||||+||||||| Sbjct 322 SKPTKIGLDHFNFLAVLGKGNFGKVMLA 349 |
| Tuber brumale | Query 180 ASPGRLHISDFSFLMVLGKGSFGKVMLA 207 + | ++ + |+|| |||||+||||||| Sbjct 321 SKPTKIGLDHFNFLAVLGKGNFGKVMLA 348 |
| Hydra vulgaris | Query 187 ISDFSFLMVLGKGSFGKVMLA 207 + ||+|| |+||||||||||| Sbjct 339 LEDFTFLKVIGKGSFGKVMLA 359 |
| Cryptococcus neoformans var. neoformans B-3501A | Query 184 RLHISDFSFLMVLGKGSFGKVMLA 207 ++ + ||+|| |||||+||||||| Sbjct 754 KVGLDDFNFLAVLGKGNFGKVMLA 777 |
| Cryptococcus neoformans var. neoformans JEC21 | Query 184 RLHISDFSFLMVLGKGSFGKVMLA 207 ++ + ||+|| |||||+||||||| Sbjct 751 KVGLDDFNFLAVLGKGNFGKVMLA 774 |
| Tuber pseudoexcavatum | Query 180 ASPGRLHISDFSFLMVLGKGSFGKVMLA 207 + | ++ + |+|| |||||+||||||| Sbjct 321 SKPTKIGLDHFNFLAVLGKGNFGKVMLA 348 |
| Debaryomyces hansenii CBS767 | Query 184 RLHISDFSFLMVLGKGSFGKVMLA 207 ++ + || || |||||+||||||| Sbjct 757 KIGLDDFQFLAVLGKGNFGKVMLA 780 |
| Cryptococcus neoformans var. neoformans | Query 184 RLHISDFSFLMVLGKGSFGKVMLA 207 ++ + ||+|| |||||+||||||| Sbjct 751 KVGLDDFNFLAVLGKGNFGKVMLA 774 |
| Coprinopsis cinerea okayama7#130 | Query 184 RLHISDFSFLMVLGKGSFGKVMLA 207 ++ + ||+|| |||||+||||||| Sbjct 711 KVGLDDFNFLAVLGKGNFGKVMLA 734 |
| Cryptococcus neoformans var. grubii | Query 184 RLHISDFSFLMVLGKGSFGKVMLA 207 ++ + ||+|| |||||+||||||| Sbjct 751 KVGLDDFNFLAVLGKGNFGKVMLA 774 |
| Pichia guilliermondii ATCC 6260 | Query 184 RLHISDFSFLMVLGKGSFGKVMLA 207 ++ + || || |||||+||||||| Sbjct 714 KIGLDDFQFLAVLGKGNFGKVMLA 737 |
| Candida albicans SC5314 | Query 178 FGASPGR---LHISDFSFLMVLGKGSFGKVMLA 207 || | | + + || || |||||+||||||| Sbjct 755 FGKSKRRKRKVGLDDFQFLAVLGKGNFGKVMLA 787 |
| Tuber aestivum | Query 182 PGRLHISDFSFLMVLGKGSFGKVMLA 207 | ++ + |+|| |||||+||||||| Sbjct 325 PTKIGLDHFNFLAVLGKGNFGKVMLA 350 |
| Pichia stipitis CBS 6054 | Query 184 RLHISDFSFLMVLGKGSFGKVMLA 207 ++ + || || |||||+||||||| Sbjct 747 KVGLDDFQFLAVLGKGNFGKVMLA 770 |
| Lodderomyces elongisporus NRRL YB-4239 | Query 184 RLHISDFSFLMVLGKGSFGKVMLA 207 ++ + || || |||||+||||||| Sbjct 824 KVGLDDFQFLAVLGKGNFGKVMLA 847 |
| Squalus acanthias | Query 188 SDFSFLMVLGKGSFGKVMLA 207 |||+|| |+||||||||+|| Sbjct 98 SDFNFLKVIGKGSFGKVLLA 117 |
| Sporothrix schenckii | Query 184 RLHISDFSFLMVLGKGSFGKVMLA 207 |+ + +|+|| |||||+||||||| Sbjct 863 RIGLDNFNFLAVLGKGNFGKVMLA 886 |
| Tuber rufum | Query 182 PGRLHISDFSFLMVLGKGSFGKVMLA 207 | ++ + |+|| |||||+||||||| Sbjct 323 PTKIGLDHFNFLAVLGKGNFGKVMLA 348 |
| Tuber magnatum | Query 182 PGRLHISDFSFLMVLGKGSFGKVMLA 207 | ++ + |+|| |||||+||||||| Sbjct 668 PTKIGLDHFNFLAVLGKGNFGKVMLA 693 |
| Tuber borchii | Query 182 PGRLHISDFSFLMVLGKGSFGKVMLA 207 | ++ + |+|| |||||+||||||| Sbjct 803 PTKIGLDHFNFLAVLGKGNFGKVMLA 828 |
| Candida albicans | Query 184 RLHISDFSFLMVLGKGSFGKVMLA 207 ++ + || || |||||+||||||| Sbjct 764 KVGLDDFQFLAVLGKGNFGKVMLA 787 |
| Aspergillus clavatus NRRL 1 | Query 184 RLHISDFSFLMVLGKGSFGKVMLA 207 |+ + |+|| |||||+||||||| Sbjct 781 RIGLDHFNFLSVLGKGNFGKVMLA 804 |
| Dictyostelium discoideum | Query 179 GASPGRLHISDFSFLMVLGKGSFGKVM 205 |++ |+ + || | |+||||||||| Sbjct 122 GSTQQRISVEDFDLLKVIGKGSFGKVM 148 |
| Phaeosphaeria nodorum SN15 | Query 184 RLHISDFSFLMVLGKGSFGKVMLA 207 |+ + |+|| |||||+||||||| Sbjct 712 RIGLDHFNFLAVLGKGNFGKVMLA 735 |
| Aspergillus oryzae | Query 184 RLHISDFSFLMVLGKGSFGKVMLA 207 |+ + |+|| |||||+||||||| Sbjct 761 RIGLDHFNFLAVLGKGNFGKVMLA 784 |
| Neosartorya fischeri NRRL 181 | Query 184 RLHISDFSFLMVLGKGSFGKVMLA 207 |+ + |+|| |||||+||||||| Sbjct 777 RIGLDHFNFLAVLGKGNFGKVMLA 800 |
| Fundulus heteroclitus | Query 188 SDFSFLMVLGKGSFGKVMLA 207 ||| || |+||||||||+|| Sbjct 96 SDFHFLKVIGKGSFGKVLLA 115 |
| Botryotinia fuckeliana | Query 184 RLHISDFSFLMVLGKGSFGKVMLA 207 |+ + |+|| |||||+||||||| Sbjct 839 RIGLDHFNFLAVLGKGNFGKVMLA 862 |
| Leptosphaeria maculans | Query 184 RLHISDFSFLMVLGKGSFGKVMLA 207 |+ + |+|| |||||+||||||| Sbjct 854 RIGLDHFNFLAVLGKGNFGKVMLA 877 |
| Magnaporthe grisea | Query 184 RLHISDFSFLMVLGKGSFGKVMLA 207 |+ + |+|| |||||+||||||| Sbjct 851 RIGLDHFNFLAVLGKGNFGKVMLA 874 |
| Magnaporthe grisea 70-15 | Query 184 RLHISDFSFLMVLGKGSFGKVMLA 207 |+ + |+|| |||||+||||||| Sbjct 851 RIGLDHFNFLAVLGKGNFGKVMLA 874 |
| Neurospora crassa | Query 184 RLHISDFSFLMVLGKGSFGKVMLA 207 |+ + |+|| |||||+||||||| Sbjct 811 RIGLDHFNFLAVLGKGNFGKVMLA 834 |
| Aspergillus terreus NIH2624 | Query 184 RLHISDFSFLMVLGKGSFGKVMLA 207 |+ + |+|| |||||+||||||| Sbjct 767 RIGLDHFNFLAVLGKGNFGKVMLA 790 |
| Blumeria graminis | Query 184 RLHISDFSFLMVLGKGSFGKVMLA 207 |+ + |+|| |||||+||||||| Sbjct 826 RIGLDHFNFLAVLGKGNFGKVMLA 849 |
[Site 2] YERVRMGPSS160-SPIPSPSPSP
Ser160  Ser
Ser
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Tyr151 | Glu152 | Arg153 | Val154 | Arg155 | Met156 | Gly157 | Pro158 | Ser159 | Ser160 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Ser161 | Pro162 | Ile163 | Pro164 | Ser165 | Pro166 | Ser167 | Pro168 | Ser169 | Pro170 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| DADNCSLLQKFEACNYPLELYERVRMGPSSSPIPSPSPSPTDPKRCFFGASPGRLHISDF |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | Macaca mulatta | 99.80 | 2 | PREDICTED: similar to protein kinase C, gamma isof |
| 2 | Homo sapiens | 99.80 | 2 | protein kinase C gamma |
| 3 | Macaca fascicularis | 99.80 | 1 | KPCG_MACFA Protein kinase C gamma type (PKC-gamma) |
| 4 | Mus musculus | 96.70 | 1 | protein kinase C, gamma |
| 5 | Oryctolagus cuniculus | 96.70 | 1 | protein kinase C, gamma |
| 6 | N/A | 93.20 | 2 | K |
| 7 | Bos taurus | 93.20 | 1 | PREDICTED: similar to Protein kinase C gamma type |
| 8 | Canis familiaris | 64.70 | 1 | PREDICTED: similar to Protein kinase C, gamma type |
Top-ranked sequences
| organism | matching |
|---|---|
| Macaca mulatta | Query 131 DADNCSLLQKFEACNYPLELYERVRMGXXX#XXXXXXXXXXTDPKRCFFGASPGRLHISDF 190 ||||||||||||||||||||||||||| # |||||||||||||||||||| Sbjct 290 DADNCSLLQKFEACNYPLELYERVRMGPSS#SPIPSPSPSPTDPKRCFFGASPGRLHISDF 349 |
| Homo sapiens | Query 131 DADNCSLLQKFEACNYPLELYERVRMGXXX#XXXXXXXXXXTDPKRCFFGASPGRLHISDF 190 ||||||||||||||||||||||||||| # |||||||||||||||||||| Sbjct 131 DADNCSLLQKFEACNYPLELYERVRMGPSS#SPIPSPSPSPTDPKRCFFGASPGRLHISDF 190 |
| Macaca fascicularis | Query 131 DADNCSLLQKFEACNYPLELYERVRMGXXX#XXXXXXXXXXTDPKRCFFGASPGRLHISDF 190 ||||||||||||||||||||||||||| # |||||||||||||||||||| Sbjct 292 DADNCSLLQKFEACNYPLELYERVRMGPSS#SPIPSPSPSPTDPKRCFFGASPGRLHISDF 351 |
| Mus musculus | Query 131 DADNCSLLQKFEACNYPLELYERVRMGXXX#XXXXXXXXXXTDPKRCFFGASPGRLHISDF 190 ||||||||||||||||||||||||||| # || ||||||||||||||||| Sbjct 292 DADNCSLLQKFEACNYPLELYERVRMGPSS#SPIPSPSPSPTDSKRCFFGASPGRLHISDF 351 |
| Oryctolagus cuniculus | Query 131 DADNCSLLQKFEACNYPLELYERVRMGXXX#XXXXXXXXXXTDPKRCFFGASPGRLHISDF 190 ||||||||||||||||||||||||||| # || ||||||||||||||||| Sbjct 292 DADNCSLLQKFEACNYPLELYERVRMGPSS#SPIPSPSPSPTDSKRCFFGASPGRLHISDF 351 |
| N/A | Query 131 DADNCSLLQKFEACNYPLELYERVRMGXXX#XXXXXXXXXXTDPKRCFFGASPGRLHISDF 190 |||||+||||||||||||||||||| | # || ||||||||||||||||| Sbjct 277 DADNCNLLQKFEACNYPLELYERVRTGPSS#SPIPSPSPSPTDSKRCFFGASPGRLHISDF 336 |
| Bos taurus | Query 131 DADNCSLLQKFEACNYPLELYERVRMGXXX#XXXXXXXXXXTDPKRCFFGASPGRLHISDF 190 |||||+||||||||||||||||||| | # || ||||||||||||||||| Sbjct 292 DADNCNLLQKFEACNYPLELYERVRTGPSS#SPIPSPSPSPTDSKRCFFGASPGRLHISDF 351 |
| Canis familiaris | Query 131 DADNCSLLQKFEACNYPLELYERVRMGXXX#XXXXXXXXXXTDPKRCFFGASPGRLHISDF 190 |||||||||||| ||||| # || |||||| |||||||||| Sbjct 278 DADNCSLLQKFE----------RVRMGPSS#SPIPSPSPSPTDSKRCFFGTSPGRLHISDF 327 |
* References
[PubMed ID: 8375396] Kochs G, Hummel R, Meyer D, Hug H, Marme D, Sarre TF, Activation and substrate specificity of the human protein kinase C alpha and zeta isoenzymes. Eur J Biochem. 1993 Sep 1;216(2):597-606.