XSB1246 : protein kinase C, alpha [Bos taurus]
[ CaMP Format ]
This entry is computationally expanded from SB0055
* Basic Information
| Organism | Bos taurus (cattle) |
| Protein Names | Protein kinase C alpha type; PKC-alpha; PKC-A; 2.7.11.13; protein kinase C; alpha; protein kinase; C alpha |
| Gene Names | PRKCA; protein kinase C, alpha |
| Gene Locus | 19; chromosome 19 |
| GO Function | Not available |
| Entrez Protein | Entrez Nucleotide | Entrez Gene | UniProt | OMIM | HGNC | HPRD | KEGG |
|---|---|---|---|---|---|---|---|
| NP_776860 | NM_174435 | 282001 | P04409 | N/A | N/A | N/A | bta:282001 |
* Information From OMIM
Not Available.
* Structure Information
1. Primary Information
Length: 672 aa
Average Mass: 76.837 kDa
Monoisotopic Mass: 76.787 kDa
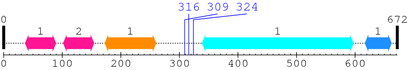
2. Domain Information
Annotated Domains: interpro / pfam / smart / prosite
Computationally Assigned Domains (Pfam+HMMER):
| domain name | begin | end | score | e-value |
|---|---|---|---|---|
| C1_1 1. | 37 | 89 | 83.8 | 6e-22 |
| C1_1 2. | 102 | 154 | 92.5 | 1.5e-24 |
| C2 1. | 173 | 260 | 129.0 | 1.5e-35 |
| --- cleavage 316 --- | ||||
| --- cleavage 309 --- | ||||
| --- cleavage 324 --- | ||||
| Pkinase 1. | 339 | 597 | 288.9 | 1.1e-83 |
| Pkinase_C 1. | 617 | 662 | 55.8 | 1.6e-13 |
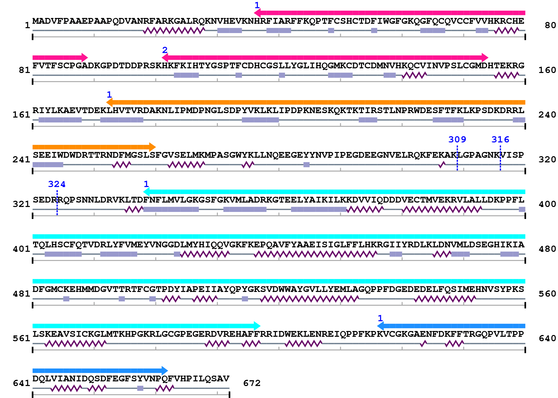
3. Sequence Information
Fasta Sequence: XSB1246.fasta
Amino Acid Sequence and Secondary Structures (PsiPred):
4. 3D Information
Not Available.
* Cleavage Information
3 [sites]
Cleavage sites (±10aa)
[Site 1] KAKLGPAGNK316-VISPSEDRRQ
Lys316  Val
Val
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Lys307 | Ala308 | Lys309 | Leu310 | Gly311 | Pro312 | Ala313 | Gly314 | Asn315 | Lys316 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Val317 | Ile318 | Ser319 | Pro320 | Ser321 | Glu322 | Asp323 | Arg324 | Arg325 | Gln326 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| NVPIPEGDEEGNVELRQKFEKAKLGPAGNKVISPSEDRRQPSNNLDRVKLTDFNFLMVLG |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | Canis familiaris | 124.00 | 5 | PREDICTED: similar to Protein kinase C, alpha type |
| 2 | Bos taurus | 124.00 | 4 | protein kinase C, alpha |
| 3 | Rattus norvegicus | 122.00 | 5 | PREDICTED: similar to Protein kinase C alpha type |
| 4 | N/A | 121.00 | 11 | - |
| 5 | Homo sapiens | 121.00 | 11 | protein kinase C, alpha |
| 6 | Mus musculus | 121.00 | 8 | protein kinase C |
| 7 | Oryctolagus cuniculus | 121.00 | 5 | KPCA_RABIT Protein kinase C alpha type (PKC-alpha) |
| 8 | synthetic construct | 121.00 | 1 | protein kinase C alpha |
| 9 | Monodelphis domestica | 108.00 | 4 | PREDICTED: similar to protein kinase C |
| 10 | Gallus gallus | 107.00 | 3 | protein kinase C, alpha |
| 11 | Xenopus tropicalis | 99.40 | 1 | hypothetical protein LOC780209 |
| 12 | Latimeria chalumnae | 97.10 | 2 | protein kinase C alpha |
| 13 | Xenopus laevis | 90.90 | 1 | protein kinase C, alpha |
| 14 | Protopterus dolloi | 85.50 | 2 | protein kinase C alpha |
| 15 | Takifugu rubripes | 74.70 | 1 | protein kinase C, alpha type |
| 16 | Scyliorhinus canicula | 68.60 | 2 | protein kinase C alpha |
| 17 | Danio rerio | 68.20 | 7 | protein kinase C, beta 1 |
| 18 | Tetraodon nigroviridis | 67.40 | 3 | unnamed protein product |
| 19 | Macaca mulatta | 60.50 | 9 | PREDICTED: protein kinase C, beta isoform 1 |
| 20 | Petromyzon marinus | 58.50 | 1 | protein kinase C |
| 21 | Myxine glutinosa | 45.10 | 1 | protein kinase C |
| 22 | Branchiostoma lanceolatum | 38.90 | 1 | protein kinase C |
| 23 | Macaca fascicularis | 38.90 | 1 | KPCG_MACFA Protein kinase C gamma type (PKC-gamma) |
| 24 | Bombyx mori | 37.00 | 1 | conventional protein kinase C |
| 25 | Anopheles gambiae str. PEST | 35.80 | 1 | ENSANGP00000009078 |
| 26 | Apis mellifera | 35.40 | 1 | PREDICTED: similar to Protein C kinase 53E CG6622- |
| 27 | Aedes aegypti | 34.30 | 1 | protein kinase c |
| 28 | Drosophila melanogaster | 33.50 | 4 | Protein C kinase 53E CG6622-PA, isoform A |
| 29 | Aplysia californica | 32.30 | 1 | KPC1_APLCA Calcium-dependent protein kinase C (APL |
Top-ranked sequences
| organism | matching |
|---|---|
| Canis familiaris | Query 287 NVPIPEGDEEGNVELRQKFEKAKLGPAGNK#VISPSEDRRQPSNNLDRVKLTDFNFLMVLG 346 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 287 NVPIPEGDEEGNVELRQKFEKAKLGPAGNK#VISPSEDRRQPSNNLDRVKLTDFNFLMVLG 346 |
| Bos taurus | Query 287 NVPIPEGDEEGNVELRQKFEKAKLGPAGNK#VISPSEDRRQPSNNLDRVKLTDFNFLMVLG 346 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 287 NVPIPEGDEEGNVELRQKFEKAKLGPAGNK#VISPSEDRRQPSNNLDRVKLTDFNFLMVLG 346 |
| Rattus norvegicus | Query 287 NVPIPEGDEEGNVELRQKFEKAKLGPAGNK#VISPSEDRRQPSNNLDRVKLTDFNFLMVLG 346 ||||||||||||||||||||||||||||||#||||||||+||||||||||||||||||||| Sbjct 287 NVPIPEGDEEGNVELRQKFEKAKLGPAGNK#VISPSEDRKQPSNNLDRVKLTDFNFLMVLG 346 |
| N/A | Query 287 NVPIPEGDEEGNVELRQKFEKAKLGPAGNK#VISPSEDRRQPSNNLDRVKLTDFNFLMVLG 346 ||||||||||||+|||||||||||||||||#||||||||+||||||||||||||||||||| Sbjct 273 NVPIPEGDEEGNMELRQKFEKAKLGPAGNK#VISPSEDRKQPSNNLDRVKLTDFNFLMVLG 332 |
| Homo sapiens | Query 287 NVPIPEGDEEGNVELRQKFEKAKLGPAGNK#VISPSEDRRQPSNNLDRVKLTDFNFLMVLG 346 ||||||||||||+|||||||||||||||||#||||||||+||||||||||||||||||||| Sbjct 287 NVPIPEGDEEGNMELRQKFEKAKLGPAGNK#VISPSEDRKQPSNNLDRVKLTDFNFLMVLG 346 |
| Mus musculus | Query 287 NVPIPEGDEEGNVELRQKFEKAKLGPAGNK#VISPSEDRRQPSNNLDRVKLTDFNFLMVLG 346 ||||||||||||+|||||||||||||||||#||||||||+||||||||||||||||||||| Sbjct 287 NVPIPEGDEEGNMELRQKFEKAKLGPAGNK#VISPSEDRKQPSNNLDRVKLTDFNFLMVLG 346 |
| Oryctolagus cuniculus | Query 287 NVPIPEGDEEGNVELRQKFEKAKLGPAGNK#VISPSEDRRQPSNNLDRVKLTDFNFLMVLG 346 |||||||||+||||||||||||||||||||#||||||||+||||||||||||||||||||| Sbjct 287 NVPIPEGDEDGNVELRQKFEKAKLGPAGNK#VISPSEDRKQPSNNLDRVKLTDFNFLMVLG 346 |
| synthetic construct | Query 287 NVPIPEGDEEGNVELRQKFEKAKLGPAGNK#VISPSEDRRQPSNNLDRVKLTDFNFLMVLG 346 ||||||||||||+|||||||||||||||||#||||||||+||||||||||||||||||||| Sbjct 287 NVPIPEGDEEGNMELRQKFEKAKLGPAGNK#VISPSEDRKQPSNNLDRVKLTDFNFLMVLG 346 |
| Monodelphis domestica | Query 287 NVPIPEGDEEGNVELRQKFEKAKLGPAGNK#VISPSEDRRQPSNNLDRVKLTDFNFLMVLG 346 ||||||||||||+||||||||||||| |||#||| |+||+ |+|||||||||||||||||| Sbjct 287 NVPIPEGDEEGNMELRQKFEKAKLGPVGNK#VISSSDDRK-PTNNLDRVKLTDFNFLMVLG 345 |
| Gallus gallus | Query 287 NVPIPEGDEEGNVELRQKFEKAKLGPAGNK#VISPSEDRRQ--PSNNLDRVKLTDFNFLMV 344 |||||+ ||+|| |||||||||||||||||#||+||||| |||||||||||||||||| Sbjct 287 NVPIPDADEDGNAELRQKFEKAKLGPAGNK#VITPSEDRNSSVPSNNLDRVKLTDFNFLMV 346 Query 345 LG 346 || Query 345 LG 346 |
| Xenopus tropicalis | Query 287 NVPIPEGDEEGNVELRQKFEKAKLGPAGNK#VISPSEDRRQ--PSNNLDRVKLTDFNFLMV 344 |||||| +++||+|||||||||||||||||#||||+++|| ||||+| |+||||+|||| Sbjct 291 NVPIPEEEDDGNIELRQKFEKAKLGPAGNK#VISPTDERRPYVPSNNIDSVRLTDFSFLMV 350 Query 345 LG 346 || Query 345 LG 346 |
| Latimeria chalumnae | Query 287 NVPIPEGDEEGNVELRQKFEKAKLGPAGNK#VISPSEDRRQ--PSNNLDRVKLTDFNFLMV 344 |||| | |+ ||||||||||||||||||+|#|||| |+|+ |||||||||||||||||| Sbjct 209 NVPISEADD-GNVELRQKFEKAKLGPAGSK#VISPVEERKSSTPSNNLDRVKLTDFNFLMV 267 Query 345 LG 346 || Query 345 LG 346 |
| Xenopus laevis | Query 287 NVPIPEGDEEGNVELRQKFEKAKLGPAGNK#VISPSEDRRQ--PSNNLDRVKLTDFNFLMV 344 |||||| |+ ||+|||||||||||||||||#||+|+ +|| ||||+| ++|||| |||| Sbjct 291 NVPIPEADD-GNLELRQKFEKAKLGPAGNK#VINPTGERRPYIPSNNIDSIRLTDFCFLMV 349 Query 345 LG 346 || Query 345 LG 346 |
| Protopterus dolloi | Query 287 NVPIPEGDEEGNVELRQKFEKAKLGPAGNK#VISPSEDRRQPSNNLDRVKLTDFNFLMVLG 346 |||||+ |+ ||| ||||||||+||||| |#|||| | ||||| || ||||+|||| Sbjct 207 NVPIPDADD-GNVGLRQKFEKARLGPAGKK#VISPERKSSLPLNNLDRFKLEDFNFVMVLG 265 |
| Takifugu rubripes | Query 287 NVPIPEGDEEGNVELRQKFEKAKLGPAGNK#VISPSEDRR--QPSNNLDRVKLTDFNFLMV 344 |||||| |+ |+|||||||||||| | |#||+||+ || || |+|||+| ||||| + Sbjct 285 NVPIPEVDDV-NLELRQKFEKAKLGQ-GKK#VITPSDHRRFSLPSGNMDRVRLNDFNFLAL 342 Query 345 LG 346 || Query 345 LG 346 |
| Scyliorhinus canicula | Query 287 NVPIPEGDEEGNVELRQKFEKAKLGPAGNK#VISPSEDRRQPSNNLDRVKLTDFNFLMVLG 346 |||| | ||+ |||+||||||||||| |# | |+|+|| | | ||||||||| Sbjct 220 NVPIIEDDEDS--ELRKKFEKAKLGPAGKK#AIEKKPS--SPTNDLDHVHLEDFNFLMVLG 275 |
| Danio rerio | Query 287 NVPIPEGDEEGNVELRQKFEKAKLGPAGNK#VISPSEDRRQPSN-NLDRVKLTDFNFLMVL 345 |||+| |||| |||||||+||+||+ # | + + || | ||+||+|||||||| Sbjct 286 NVPVPPEGEEGNEELRQKFERAKIGPSKTD#GSSSNAISKFDSNGNRDRMKLSDFNFLMVL 345 Query 346 G 346 | Query 346 G 346 |
| Tetraodon nigroviridis | Query 287 NVPIPEGDEEGNVELRQKFEKAKLGPAGNK--#VISPSEDRRQPSNNLDRVKLTDFNFLMV 344 |||+| |||| |||||||+||+|| | # + + | + | ||+|| ||||||| Sbjct 223 NVPVPPEGEEGNEELRQKFERAKIGPGKNTDG#KSANAASRFDSNGNQDRMKLADFNFLMV 282 Query 345 LG 346 || Query 345 LG 346 |
| Macaca mulatta | Query 287 NVPIPEGDEEGNVELRQKFEKAKLGPAGNK#V----ISPSEDRRQPSNNLDRVKLTDFNFL 342 |||+| ||| |||||||+||+ | |#| + + + + | ||+|||||||| Sbjct 287 NVPVPPEGSEGNEELRQKFERAKIS-QGTK#VPEEKTTNTVSKFDNNGNRDRMKLTDFNFL 345 Query 343 MVLG 346 |||| Query 343 MVLG 346 |
| Petromyzon marinus | Query 287 NVPIPEGDEEGNVELRQKFEKAKLGPAGNK#VISP--SEDRRQPS-------NNLDRVKLT 337 |||| |+|+ |+|+|||||+|||+ # + | |++|| | |+|||| Sbjct 217 NVPIAPDIEDGDTEMRRKFEKARLGPS---#-VKPRASDERRANSLSAILNNANVDRVKAD 272 Query 338 DFNFLMVLG 346 ||||||||| Query 338 DFNFLMVLG 346 |
| Myxine glutinosa | Query 287 NVPI-PEGDEEGNVELRQKFEKAKLGPAGNK#VISPSEDRRQPS---NNLDRVKLTDFNFL 342 |||+ |||+| +||||+ +++++ + # ||++|+ | + |||| | || || Sbjct 208 NVPVAPEGEE--GLELRQRLQRSQIARSDKN#KQSPAKDQPSDSASLSGLDRVNLEDFVFL 265 Query 343 MVLG 346 ||| Sbjct 266 TVLG 269 |
| Branchiostoma lanceolatum | Query 287 NVPIPEGDEEGNV-ELRQKFEKAKLGPAGNK#VISPSEDRRQPSNNLDRVKLTDFNFLMVL 345 |||| ||| || +|++ +| || +# ||+ | || +| |||||||| Sbjct 218 NVPI--ADEEENVAKLKEHLQKQKLDEQRKQ#KSKRSEECNIGS--LDHMKAADFNFLMVL 273 Query 346 G 346 | Query 346 G 346 |
| Macaca fascicularis | Query 287 NVPIPEGDEEGNVELRQKFE----------KAKLGPAGNK#VISPSEDRRQPSN-----NL 331 |||+ + | | | |||| + ++||+ + #+ ||| | + Sbjct 287 NVPVADAD---NCSLLQKFEACNYPLELYERVRMGPSSSP#IPSPSPSPTDPKRCFFGASP 343 Query 332 DRVKLTDFNFLMVLG 346 |+ ++||+|||||| Sbjct 344 GRLHISDFSFLMVLG 358 |
| Bombyx mori | Query 287 NVPIPEGDEEGNVELRQKFEKAKLGPAGNK#VISPSEDRRQPSN--NLDRVKLTDFNFLMV 344 |||+|| || +| | + + | +# | || | | | ++ |||||+|| Sbjct 290 NVPVPE---EG-ADLVQLKNQMRATTVGAR#RPPPPPDREVPHNVAAADVIRATDFNFIMV 345 Query 345 LG 346 || Query 345 LG 346 |
| Anopheles gambiae str. PEST | Query 287 NVPIPEGDEEGNVELRQKFEKAKLGPAGNK#VISPSEDRRQPSN--NLDRVKLTDFNFLMV 344 |||+|| + |+|+ + | + |# | |+ | | | ++ |||||||| Sbjct 227 NVPVPEEGAD-LVQLKSQMRKTSI----TK#KIPMMGDKDVPHNMTKKDVIRATDFNFLMV 281 Query 345 LG 346 || Query 345 LG 346 |
| Apis mellifera | Query 287 NVPIPE-GDEEGNVELRQKFEKAKLGPAGNK#VISPSEDRRQPSN--NLDRVKLTDFNFLM 343 |||+|| | + ++++ +| + |# + ++|+ | | | ++ +|||||| Sbjct 304 NVPVPEEGVDLAELKMKPSPQKTSV----TK#KTTTTQDKEVPHNMGKSDLIRASDFNFLM 359 Query 344 VLG 346 ||| Query 344 VLG 346 |
| Aedes aegypti | Query 287 NVPIPEGDEEGNVELRQKFEKAKLGPAGNK#VISPSEDRRQPSNNLDRVKLTDFNFLMVLG 346 |||+|| || ++ | + | + # + +| | ++ |||||||||| Sbjct 235 NVPVPE---EGTDLVQLKSQMRKTSISKKA#PVLCDKDVPHNMGKKDVIRATDFNFLMVLG 291 |
| Drosophila melanogaster | Query 287 NVPIPEGDEEGNVELRQKFEKAKLGPAGNK#VISPSEDRRQPSNNLDRVKLTDFNFLMVLG 346 ||| + ||+ ++|+|| + | #++ |+ |+ | ++ |||||+ ||| Sbjct 296 NVPCAD-DEQDLLKLKQKPSQKK------P#MVMRSDTNTHTSSKKDMIRATDFNFIKVLG 348 |
| Aplysia californica | Query 288 VPIPEGDEEGNVELRQKFEKAKLGPAGNK#VISPSEDRRQPSNNLDRVKLTDFNFLMVLG 346 ||+ + | |++ | ++ + ++# | |+ | + | |+ +||||| ||| Sbjct 272 VPVTDDITESIQEIKSKMHRSSIS---SE#KRYPEPDKVQNMSKQDIVRASDFNFLTVLG 327 |
[Site 2] ELRQKFEKAK309-LGPAGNKVIS
Lys309  Leu
Leu
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Glu300 | Leu301 | Arg302 | Gln303 | Lys304 | Phe305 | Glu306 | Lys307 | Ala308 | Lys309 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Leu310 | Gly311 | Pro312 | Ala313 | Gly314 | Asn315 | Lys316 | Val317 | Ile318 | Ser319 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| QEEGEYYNVPIPEGDEEGNVELRQKFEKAKLGPAGNKVISPSEDRRQPSNNLDRVKLTDF |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | Canis familiaris | 125.00 | 5 | PREDICTED: similar to Protein kinase C, alpha type |
| 2 | Bos taurus | 125.00 | 4 | protein kinase C, alpha |
| 3 | Rattus norvegicus | 124.00 | 5 | PREDICTED: similar to Protein kinase C alpha type |
| 4 | N/A | 123.00 | 13 | - |
| 5 | Homo sapiens | 123.00 | 11 | protein kinase C, alpha |
| 6 | Mus musculus | 123.00 | 9 | protein kinase C |
| 7 | Oryctolagus cuniculus | 123.00 | 5 | KPCA_RABIT Protein kinase C alpha type (PKC-alpha) |
| 8 | synthetic construct | 123.00 | 1 | protein kinase C alpha |
| 9 | Monodelphis domestica | 110.00 | 4 | PREDICTED: similar to protein kinase C |
| 10 | Gallus gallus | 109.00 | 3 | protein kinase C, alpha |
| 11 | Xenopus tropicalis | 103.00 | 1 | hypothetical protein LOC780209 |
| 12 | Latimeria chalumnae | 99.00 | 2 | protein kinase C alpha |
| 13 | Xenopus laevis | 96.30 | 1 | protein kinase C, alpha |
| 14 | Protopterus dolloi | 88.60 | 2 | protein kinase C alpha |
| 15 | Takifugu rubripes | 80.10 | 1 | protein kinase C, alpha type |
| 16 | Scyliorhinus canicula | 70.50 | 2 | protein kinase C alpha |
| 17 | Danio rerio | 68.60 | 7 | protein kinase C, beta 1 |
| 18 | Tetraodon nigroviridis | 67.80 | 3 | unnamed protein product |
| 19 | Macaca mulatta | 60.80 | 8 | PREDICTED: protein kinase C, beta 1 isoform 5 |
| 20 | Petromyzon marinus | 60.50 | 1 | protein kinase C |
| 21 | Myxine glutinosa | 52.80 | 1 | protein kinase C |
| 22 | Macaca fascicularis | 42.70 | 1 | KPCG_MACFA Protein kinase C gamma type (PKC-gamma) |
| 23 | Branchiostoma lanceolatum | 40.80 | 1 | protein kinase C |
| 24 | Bombyx mori | 38.10 | 1 | conventional protein kinase C |
| 25 | Anopheles gambiae str. PEST | 37.70 | 1 | ENSANGP00000009078 |
| 26 | Drosophila melanogaster | 37.40 | 4 | Protein C kinase 53E CG6622-PA, isoform A |
| 27 | Apis mellifera | 37.40 | 1 | PREDICTED: similar to Protein C kinase 53E CG6622- |
| 28 | Aedes aegypti | 36.20 | 1 | protein kinase c |
| 29 | Aplysia californica | 35.00 | 1 | KPC1_APLCA Calcium-dependent protein kinase C (APL |
| 30 | Drosophila pseudoobscura | 33.50 | 1 | GA19732-PA |
Top-ranked sequences
| organism | matching |
|---|---|
| Canis familiaris | Query 280 QEEGEYYNVPIPEGDEEGNVELRQKFEKAK#LGPAGNKVISPSEDRRQPSNNLDRVKLTDF 339 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 280 QEEGEYYNVPIPEGDEEGNVELRQKFEKAK#LGPAGNKVISPSEDRRQPSNNLDRVKLTDF 339 |
| Bos taurus | Query 280 QEEGEYYNVPIPEGDEEGNVELRQKFEKAK#LGPAGNKVISPSEDRRQPSNNLDRVKLTDF 339 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 280 QEEGEYYNVPIPEGDEEGNVELRQKFEKAK#LGPAGNKVISPSEDRRQPSNNLDRVKLTDF 339 |
| Rattus norvegicus | Query 280 QEEGEYYNVPIPEGDEEGNVELRQKFEKAK#LGPAGNKVISPSEDRRQPSNNLDRVKLTDF 339 ||||||||||||||||||||||||||||||#|||||||||||||||+|||||||||||||| Sbjct 280 QEEGEYYNVPIPEGDEEGNVELRQKFEKAK#LGPAGNKVISPSEDRKQPSNNLDRVKLTDF 339 |
| N/A | Query 280 QEEGEYYNVPIPEGDEEGNVELRQKFEKAK#LGPAGNKVISPSEDRRQPSNNLDRVKLTDF 339 |||||||||||||||||||+||||||||||#|||||||||||||||+|||||||||||||| Sbjct 266 QEEGEYYNVPIPEGDEEGNMELRQKFEKAK#LGPAGNKVISPSEDRKQPSNNLDRVKLTDF 325 |
| Homo sapiens | Query 280 QEEGEYYNVPIPEGDEEGNVELRQKFEKAK#LGPAGNKVISPSEDRRQPSNNLDRVKLTDF 339 |||||||||||||||||||+||||||||||#|||||||||||||||+|||||||||||||| Sbjct 280 QEEGEYYNVPIPEGDEEGNMELRQKFEKAK#LGPAGNKVISPSEDRKQPSNNLDRVKLTDF 339 |
| Mus musculus | Query 280 QEEGEYYNVPIPEGDEEGNVELRQKFEKAK#LGPAGNKVISPSEDRRQPSNNLDRVKLTDF 339 |||||||||||||||||||+||||||||||#|||||||||||||||+|||||||||||||| Sbjct 280 QEEGEYYNVPIPEGDEEGNMELRQKFEKAK#LGPAGNKVISPSEDRKQPSNNLDRVKLTDF 339 |
| Oryctolagus cuniculus | Query 280 QEEGEYYNVPIPEGDEEGNVELRQKFEKAK#LGPAGNKVISPSEDRRQPSNNLDRVKLTDF 339 ||||||||||||||||+|||||||||||||#|||||||||||||||+|||||||||||||| Sbjct 280 QEEGEYYNVPIPEGDEDGNVELRQKFEKAK#LGPAGNKVISPSEDRKQPSNNLDRVKLTDF 339 |
| synthetic construct | Query 280 QEEGEYYNVPIPEGDEEGNVELRQKFEKAK#LGPAGNKVISPSEDRRQPSNNLDRVKLTDF 339 |||||||||||||||||||+||||||||||#|||||||||||||||+|||||||||||||| Sbjct 280 QEEGEYYNVPIPEGDEEGNMELRQKFEKAK#LGPAGNKVISPSEDRKQPSNNLDRVKLTDF 339 |
| Monodelphis domestica | Query 280 QEEGEYYNVPIPEGDEEGNVELRQKFEKAK#LGPAGNKVISPSEDRRQPSNNLDRVKLTDF 339 |||||||||||||||||||+||||||||||#||| |||||| |+||+ |+||||||||||| Sbjct 280 QEEGEYYNVPIPEGDEEGNMELRQKFEKAK#LGPVGNKVISSSDDRK-PTNNLDRVKLTDF 338 |
| Gallus gallus | Query 280 QEEGEYYNVPIPEGDEEGNVELRQKFEKAK#LGPAGNKVISPSEDRRQ--PSNNLDRVKLT 337 ||||||||||||+ ||+|| ||||||||||#|||||||||+||||| ||||||||||| Sbjct 280 QEEGEYYNVPIPDADEDGNAELRQKFEKAK#LGPAGNKVITPSEDRNSSVPSNNLDRVKLT 339 Query 338 DF 339 || Query 338 DF 339 |
| Xenopus tropicalis | Query 280 QEEGEYYNVPIPEGDEEGNVELRQKFEKAK#LGPAGNKVISPSEDRRQ--PSNNLDRVKLT 337 ||||||||||||| +++||+||||||||||#|||||||||||+++|| ||||+| |+|| Sbjct 284 QEEGEYYNVPIPEEEDDGNIELRQKFEKAK#LGPAGNKVISPTDERRPYVPSNNIDSVRLT 343 Query 338 DF 339 || Query 338 DF 339 |
| Latimeria chalumnae | Query 280 QEEGEYYNVPIPEGDEEGNVELRQKFEKAK#LGPAGNKVISPSEDRRQ--PSNNLDRVKLT 337 ||||||||||| | |+ |||||||||||||#|||||+||||| |+|+ ||||||||||| Sbjct 202 QEEGEYYNVPISEADD-GNVELRQKFEKAK#LGPAGSKVISPVEERKSSTPSNNLDRVKLT 260 Query 338 DF 339 || Query 338 DF 339 |
| Xenopus laevis | Query 280 QEEGEYYNVPIPEGDEEGNVELRQKFEKAK#LGPAGNKVISPSEDRRQ--PSNNLDRVKLT 337 ||||||||||||| |+ ||+||||||||||#|||||||||+|+ +|| ||||+| ++|| Sbjct 284 QEEGEYYNVPIPEADD-GNLELRQKFEKAK#LGPAGNKVINPTGERRPYIPSNNIDSIRLT 342 Query 338 DF 339 || Query 338 DF 339 |
| Protopterus dolloi | Query 280 QEEGEYYNVPIPEGDEEGNVELRQKFEKAK#LGPAGNKVISPSEDRRQPSNNLDRVKLTDF 339 ||||||||||||+ |+ ||| ||||||||+#||||| ||||| | ||||| || || Sbjct 200 QEEGEYYNVPIPDADD-GNVGLRQKFEKAR#LGPAGKKVISPERKSSLPLNNLDRFKLEDF 258 |
| Takifugu rubripes | Query 280 QEEGEYYNVPIPEGDEEGNVELRQKFEKAK#LGPAGNKVISPSEDRR--QPSNNLDRVKLT 337 ||||||||||||| |+ |+||||||||||#|| | |||+||+ || || |+|||+| Sbjct 278 QEEGEYYNVPIPEVDDV-NLELRQKFEKAK#LG-QGKKVITPSDHRRFSLPSGNMDRVRLN 335 Query 338 DF 339 || Query 338 DF 339 |
| Scyliorhinus canicula | Query 280 QEEGEYYNVPIPEGDEEGNVELRQKFEKAK#LGPAGNKVISPSEDRRQPSNNLDRVKLTDF 339 ||||||||||| | ||+ |||+||||||#||||| | | |+|+|| | | || Sbjct 213 QEEGEYYNVPIIEDDEDS--ELRKKFEKAK#LGPAGKKAIEKKPS--SPTNDLDHVHLEDF 268 |
| Danio rerio | Query 280 QEEGEYYNVPIPEGDEEGNVELRQKFEKAK#LGPAGNKVISPSEDRRQPSN-NLDRVKLTD 338 ||||||+|||+| |||| |||||||+||#+||+ | + + || | ||+||+| Sbjct 279 QEEGEYFNVPVPPEGEEGNEELRQKFERAK#IGPSKTDGSSSNAISKFDSNGNRDRMKLSD 338 Query 339 F 339 | Query 339 F 339 |
| Tetraodon nigroviridis | Query 280 QEEGEYYNVPIPEGDEEGNVELRQKFEKAK#LGPAGNK--VISPSEDRRQPSNNLDRVKLT 337 ||||||+|||+| |||| |||||||+||#+|| | + + | + | ||+|| Sbjct 216 QEEGEYFNVPVPPEGEEGNEELRQKFERAK#IGPGKNTDGKSANAASRFDSNGNQDRMKLA 275 Query 338 DF 339 || Query 338 DF 339 |
| Macaca mulatta | Query 280 QEEGEYYNVPIPEGDEEGNVELRQKFEKAK#LGPAGNKV----ISPSEDRRQPSNNLDRVK 335 ||||||+|||+| ||| |||||||+||#+ | || + + + + | ||+| Sbjct 280 QEEGEYFNVPVPPEGSEGNEELRQKFERAK#IS-QGTKVPEEKTTNTVSKFDNNGNRDRMK 338 Query 336 LTDF 339 |||| Query 336 LTDF 339 |
| Petromyzon marinus | Query 280 QEEGEYYNVPIPEGDEEGNVELRQKFEKAK#LGPAGNKVISP--SEDRRQPS-------NN 330 ||||||||||| |+|+ |+|+|||||+#|||+ + | |++|| | | Sbjct 210 QEEGEYYNVPIAPDIEDGDTEMRRKFEKAR#LGPS----VKPRASDERRANSLSAILNNAN 265 Query 331 LDRVKLTDF 339 +|||| || Sbjct 266 VDRVKADDF 274 |
| Myxine glutinosa | Query 280 QEEGEYYNVPI-PEGDEEGNVELRQKFEKAK#LGPAGNKVISPSEDRRQPS---NNLDRVK 335 ||||||||||+ |||+| +||||+ ++++#+ + ||++|+ | + |||| Sbjct 201 QEEGEYYNVPVAPEGEE--GLELRQRLQRSQ#IARSDKNKQSPAKDQPSDSASLSGLDRVN 258 Query 336 LTDF 339 | || Sbjct 259 LEDF 262 |
| Macaca fascicularis | Query 280 QEEGEYYNVPIPEGDEEGNVELRQKF----------EKAK#LGPAGNKVISPSEDRRQPSN 329 ||||||||||+ + | | | ||| |+ +#+||+ + + ||| | Sbjct 280 QEEGEYYNVPVADAD---NCSLLQKFEACNYPLELYERVR#MGPSSSPIPSPSPSPTDPKR 336 Query 330 -----NLDRVKLTDF 339 + |+ ++|| Sbjct 337 CFFGASPGRLHISDF 351 |
| Branchiostoma lanceolatum | Query 280 QEEGEYYNVPIPEGDEEGNV-ELRQKFEKAK#LGPAGNKVISPSEDRRQPSNNLDRVKLTD 338 ||||||||||| ||| || +|++ +| |#| + ||+ +|| +| | Sbjct 211 QEEGEYYNVPI--ADEEENVAKLKEHLQKQK#LDEQRKQKSKRSEECN--IGSLDHMKAAD 266 Query 339 F 339 | Query 339 F 339 |
| Bombyx mori | Query 280 QEEGEYYNVPIPEGDEEGNVELRQKFEKAK#LGPAGNKVISPSEDRRQPSN--NLDRVKLT 337 |||||+||||+| ||| +| | + +# | + | || | | | ++ | Sbjct 283 QEEGEFYNVPVP---EEG-ADLVQLKNQMR#ATTVGARRPPPPPDREVPHNVAAADVIRAT 338 Query 338 DF 339 || Query 338 DF 339 |
| Anopheles gambiae str. PEST | Query 280 QEEGEYYNVPIPEGDEEGNVELRQKFEKAK#LGPAGNKVISPSEDRRQPSN--NLDRVKLT 337 ||||||||||+|| + |+|+ + | #+ | | |+ | | | ++ | Sbjct 220 QEEGEYYNVPVPE-EGADLVQLKSQMRKTS#I----TKKIPMMGDKDVPHNMTKKDVIRAT 274 Query 338 DF 339 || Query 338 DF 339 |
| Drosophila melanogaster | Query 280 QEEGEYYNVPIPEGDEEGNVELRQKFEKAK#LGPAGNKVISPSEDRRQPSNNLDRVKLTDF 339 |+|||||||| + ||+ ++|+|| + |# ++ |+ |+ | ++ ||| Sbjct 289 QDEGEYYNVPCAD-DEQDLLKLKQKPSQKK#------PMVMRSDTNTHTSSKKDMIRATDF 341 |
| Apis mellifera | Query 280 QEEGEYYNVPIP-EGDEEGNVELRQKFEKAK#LGPAGNKVISPSEDRRQPSN--NLDRVKL 336 ||||||||||+| || + ++++ +| #+ | + ++|+ | | | ++ Sbjct 297 QEEGEYYNVPVPEEGVDLAELKMKPSPQKTS#V----TKKTTTTQDKEVPHNMGKSDLIRA 352 Query 337 TDF 339 +|| Sbjct 353 SDF 355 |
| Aedes aegypti | Query 280 QEEGEYYNVPIPEGDEEGNVELRQKFEKAK#LGPAGNKVISPSEDRRQPSNNLDRVKLTDF 339 ||||||||||+| ||| ++ | + |# + + +| | ++ ||| Sbjct 228 QEEGEYYNVPVP---EEGTDLVQLKSQMRK#TSISKKAPVLCDKDVPHNMGKKDVIRATDF 284 |
| Aplysia californica | Query 280 QEEGEYYNVPIPEGDEEGNVELRQKFEKAK#LGPAGNKVISPSEDRRQPSNNLDRVKLTDF 339 |||||+| ||+ + | |++ | ++ #+ ++ | |+ | + | |+ +|| Sbjct 264 QEEGEFYGVPVTDDITESIQEIKSKMHRSS#I---SSEKRYPEPDKVQNMSKQDIVRASDF 320 |
| Drosophila pseudoobscura | Query 280 QEEGEYYNVPIPEGDEEGNVELRQKFEKAK#LGPAGNKVISPSEDRRQPS--NNLDRVKLT 337 |+|||||||| + ||+ ++|+|| # |+ | + | | + | ++ | Sbjct 175 QDEGEYYNVPCAD-DEQDLLKLKQK-----#--PSQKKPLVMRSDTNTHSSISKKDMIRAT 226 Query 338 DF 339 || Query 338 DF 339 |
[Site 3] NKVISPSEDR324-RQPSNNLDRV
Arg324  Arg
Arg
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Asn315 | Lys316 | Val317 | Ile318 | Ser319 | Pro320 | Ser321 | Glu322 | Asp323 | Arg324 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Arg325 | Gln326 | Pro327 | Ser328 | Asn329 | Asn330 | Leu331 | Asp332 | Arg333 | Val334 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| EEGNVELRQKFEKAKLGPAGNKVISPSEDRRQPSNNLDRVKLTDFNFLMVLGKGSFGKVM |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | Canis familiaris | 122.00 | 13 | PREDICTED: similar to Protein kinase C, alpha type |
| 2 | Bos taurus | 122.00 | 9 | protein kinase C, alpha |
| 3 | Rattus norvegicus | 121.00 | 8 | PREDICTED: similar to Protein kinase C alpha type |
| 4 | Homo sapiens | 120.00 | 26 | aging-associated gene 6 protein |
| 5 | N/A | 120.00 | 17 | - |
| 6 | Mus musculus | 120.00 | 10 | protein kinase C |
| 7 | Oryctolagus cuniculus | 120.00 | 6 | KPCA_RABIT Protein kinase C alpha type (PKC-alpha) |
| 8 | synthetic construct | 120.00 | 4 | protein kinase C alpha |
| 9 | Gallus gallus | 109.00 | 6 | protein kinase C, alpha |
| 10 | Monodelphis domestica | 106.00 | 7 | PREDICTED: similar to protein kinase C |
| 11 | Latimeria chalumnae | 106.00 | 2 | protein kinase C alpha |
| 12 | Xenopus tropicalis | 102.00 | 2 | hypothetical protein LOC780209 |
| 13 | Xenopus laevis | 97.10 | 2 | protein kinase C, alpha |
| 14 | Protopterus dolloi | 92.80 | 2 | protein kinase C alpha |
| 15 | Takifugu rubripes | 81.60 | 1 | protein kinase C, alpha type |
| 16 | Scyliorhinus canicula | 76.30 | 2 | protein kinase C alpha |
| 17 | Danio rerio | 75.10 | 14 | protein kinase C, beta 1 |
| 18 | Tetraodon nigroviridis | 74.30 | 5 | unnamed protein product |
| 19 | Petromyzon marinus | 68.60 | 1 | protein kinase C |
| 20 | Macaca mulatta | 67.40 | 15 | PREDICTED: protein kinase C, beta 1 isoform 5 |
| 21 | Myxine glutinosa | 54.70 | 1 | protein kinase C |
| 22 | Macaca fascicularis | 50.10 | 4 | KPCG_MACFA Protein kinase C gamma type (PKC-gamma) |
| 23 | Branchiostoma lanceolatum | 49.70 | 1 | protein kinase C |
| 24 | Bombyx mori | 47.80 | 1 | conventional protein kinase C |
| 25 | Aplysia californica | 46.60 | 2 | KPC2_APLCA Calcium-independent protein kinase C (A |
| 26 | Anopheles gambiae str. PEST | 46.20 | 3 | ENSANGP00000009078 |
| 27 | Apis mellifera | 45.80 | 2 | PREDICTED: similar to Protein C kinase 53E CG6622- |
| 28 | Aedes aegypti | 45.10 | 2 | protein kinase c |
| 29 | Drosophila melanogaster | 44.30 | 8 | Protein C kinase 53E CG6622-PA, isoform A |
| 30 | Tribolium castaneum | 43.10 | 3 | PREDICTED: similar to CG6622-PA, isoform A |
| 31 | Coprinopsis cinerea okayama7#130 | 42.40 | 1 | hypothetical protein CC1G_07190 |
| 32 | Drosophila pseudoobscura | 41.20 | 3 | GA19732-PA |
| 33 | Caenorhabditis briggsae | 41.20 | 1 | Hypothetical protein CBG16150 |
| 34 | Magnaporthe grisea 70-15 | 40.80 | 1 | hypothetical protein MGG_08689 |
| 35 | Caenorhabditis elegans | 40.40 | 5 | protein kinase C2 A isoform |
| 36 | Schizosaccharomyces pombe | 40.40 | 3 | SPAC17G8.14c |
| 37 | Schizosaccharomyces pombe 972h- | 40.40 | 2 | hypothetical protein SPAC17G8.14c |
| 38 | Magnaporthe grisea | 40.40 | 1 | AF136600_1 protein kinase C |
| 39 | Strongylocentrotus purpuratus | 40.00 | 4 | PREDICTED: similar to protein kinase C |
| 40 | Suberites domuncula | 40.00 | 2 | Serine/Threonine protein kinase |
| 41 | Hydractinia echinata | 39.30 | 1 | putative calcium dependent protein kinase C |
| 42 | Pan troglodytes | 38.90 | 8 | PREDICTED: serum/glucocorticoid regulated kinase 3 |
| 43 | Paramecium tetraurelia | 38.90 | 1 | hypothetical protein |
| 44 | Tuber excavatum | 38.50 | 2 | protein kinase C-like protein |
| 45 | Yarrowia lipolytica | 38.50 | 1 | hypothetical protein |
| 46 | Botryotinia fuckeliana | 38.50 | 1 | protein kinase C |
| 47 | Tuber aestivum | 38.10 | 2 | protein kinase C-like protein |
| 48 | Sycon raphanus | 38.10 | 1 | Serine/Threonine protein kinase |
| 49 | Asparagus officinalis | 38.10 | 1 | S6 ribosomal protein kinase |
| 50 | Leishmania braziliensis | 37.70 | 1 | zinc finger protein kinase-like |
| 51 | Leptosphaeria maculans | 37.70 | 1 | AF487263_9 protein kinase C |
| 52 | Squalus acanthias | 37.70 | 1 | s-sgk1 |
| 53 | Cryptococcus neoformans var. neoformans JEC21 | 37.70 | 1 | protein kinase C |
| 54 | Cryptococcus neoformans var. neoformans | 37.70 | 1 | protein kinase C 1 |
| 55 | Cryptococcus neoformans var. neoformans B-3501A | 37.70 | 1 | hypothetical protein CNBC3910 |
| 56 | Cryptococcus neoformans var. grubii | 37.70 | 1 | protein kinase C 1 |
| 57 | Calliphora vicina | 37.40 | 1 | eye-specific protein kinase C |
| 58 | Tuber indicum | 37.00 | 6 | protein kinase C-like protein |
| 59 | Tuber pseudoexcavatum | 37.00 | 2 | protein kinase C-like protein |
| 60 | Phaeosphaeria nodorum SN15 | 37.00 | 1 | hypothetical protein SNOG_13609 |
| 61 | Aspergillus oryzae | 36.60 | 1 | unnamed protein product |
| 62 | Sporothrix schenckii | 36.60 | 1 | AF124792_1 protein kinase C; serine/threonine prot |
| 63 | Cochliobolus heterostrophus | 36.60 | 1 | KPC1_COCHE Protein kinase C-like emb |
| 64 | Aspergillus clavatus NRRL 1 | 36.60 | 1 | protein kinase c |
| 65 | Medicago truncatula | 36.60 | 1 | Protein kinase |
| 66 | Debaryomyces hansenii CBS767 | 36.60 | 1 | hypothetical protein DEHA0G14773g |
| 67 | Hydra vulgaris | 36.20 | 2 | protein kinase C |
| 68 | Coccidioides immitis RS | 36.20 | 1 | hypothetical protein CIMG_09206 |
| 69 | Neosartorya fischeri NRRL 181 | 36.20 | 1 | protein kinase c |
| 70 | Pichia stipitis CBS 6054 | 35.80 | 1 | predicted protein |
| 71 | Trichomonas vaginalis G3 | 35.80 | 1 | AGC family protein kinase |
Top-ranked sequences
| organism | matching |
|---|---|
| Canis familiaris | Query 295 EEGNVELRQKFEKAKLGPAGNKVISPSEDR#RQPSNNLDRVKLTDFNFLMVLGKGSFGKVM 354 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 295 EEGNVELRQKFEKAKLGPAGNKVISPSEDR#RQPSNNLDRVKLTDFNFLMVLGKGSFGKVM 354 |
| Bos taurus | Query 295 EEGNVELRQKFEKAKLGPAGNKVISPSEDR#RQPSNNLDRVKLTDFNFLMVLGKGSFGKVM 354 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 295 EEGNVELRQKFEKAKLGPAGNKVISPSEDR#RQPSNNLDRVKLTDFNFLMVLGKGSFGKVM 354 |
| Rattus norvegicus | Query 295 EEGNVELRQKFEKAKLGPAGNKVISPSEDR#RQPSNNLDRVKLTDFNFLMVLGKGSFGKVM 354 ||||||||||||||||||||||||||||||#+||||||||||||||||||||||||||||| Sbjct 295 EEGNVELRQKFEKAKLGPAGNKVISPSEDR#KQPSNNLDRVKLTDFNFLMVLGKGSFGKVM 354 |
| Homo sapiens | Query 295 EEGNVELRQKFEKAKLGPAGNKVISPSEDR#RQPSNNLDRVKLTDFNFLMVLGKGSFGKVM 354 ||||+|||||||||||||||||||||||||#+||||||||||||||||||||||||||||| Sbjct 295 EEGNMELRQKFEKAKLGPAGNKVISPSEDR#KQPSNNLDRVKLTDFNFLMVLGKGSFGKVM 354 |
| N/A | Query 295 EEGNVELRQKFEKAKLGPAGNKVISPSEDR#RQPSNNLDRVKLTDFNFLMVLGKGSFGKVM 354 ||||+|||||||||||||||||||||||||#+||||||||||||||||||||||||||||| Sbjct 281 EEGNMELRQKFEKAKLGPAGNKVISPSEDR#KQPSNNLDRVKLTDFNFLMVLGKGSFGKVM 340 |
| Mus musculus | Query 295 EEGNVELRQKFEKAKLGPAGNKVISPSEDR#RQPSNNLDRVKLTDFNFLMVLGKGSFGKVM 354 ||||+|||||||||||||||||||||||||#+||||||||||||||||||||||||||||| Sbjct 295 EEGNMELRQKFEKAKLGPAGNKVISPSEDR#KQPSNNLDRVKLTDFNFLMVLGKGSFGKVM 354 |
| Oryctolagus cuniculus | Query 295 EEGNVELRQKFEKAKLGPAGNKVISPSEDR#RQPSNNLDRVKLTDFNFLMVLGKGSFGKVM 354 |+||||||||||||||||||||||||||||#+||||||||||||||||||||||||||||| Sbjct 295 EDGNVELRQKFEKAKLGPAGNKVISPSEDR#KQPSNNLDRVKLTDFNFLMVLGKGSFGKVM 354 |
| synthetic construct | Query 295 EEGNVELRQKFEKAKLGPAGNKVISPSEDR#RQPSNNLDRVKLTDFNFLMVLGKGSFGKVM 354 ||||+|||||||||||||||||||||||||#+||||||||||||||||||||||||||||| Sbjct 295 EEGNMELRQKFEKAKLGPAGNKVISPSEDR#KQPSNNLDRVKLTDFNFLMVLGKGSFGKVM 354 |
| Gallus gallus | Query 295 EEGNVELRQKFEKAKLGPAGNKVISPSEDR#RQ--PSNNLDRVKLTDFNFLMVLGKGSFGK 352 |+|| |||||||||||||||||||+|||||# |||||||||||||||||||||||||| Sbjct 295 EDGNAELRQKFEKAKLGPAGNKVITPSEDR#NSSVPSNNLDRVKLTDFNFLMVLGKGSFGK 354 Query 353 VM 354 || Query 353 VM 354 |
| Monodelphis domestica | Query 295 EEGNVELRQKFEKAKLGPAGNKVISPSEDR#RQPSNNLDRVKLTDFNFLMVLGKGSFGKVM 354 ||||+||||||||||||| |||||| |+||#+ |+|||||||||||||||||||||||||| Sbjct 295 EEGNMELRQKFEKAKLGPVGNKVISSSDDR#K-PTNNLDRVKLTDFNFLMVLGKGSFGKVM 353 |
| Latimeria chalumnae | Query 295 EEGNVELRQKFEKAKLGPAGNKVISPSEDR#RQ--PSNNLDRVKLTDFNFLMVLGKGSFGK 352 ++||||||||||||||||||+||||| |+|#+ |||||||||||||||||||||||||| Sbjct 216 DDGNVELRQKFEKAKLGPAGSKVISPVEER#KSSTPSNNLDRVKLTDFNFLMVLGKGSFGK 275 Query 353 VM 354 || Query 353 VM 354 |
| Xenopus tropicalis | Query 295 EEGNVELRQKFEKAKLGPAGNKVISPSEDR#RQ--PSNNLDRVKLTDFNFLMVLGKGSFGK 352 ++||+|||||||||||||||||||||+++|#| ||||+| |+||||+|||||||||||| Sbjct 299 DDGNIELRQKFEKAKLGPAGNKVISPTDER#RPYVPSNNIDSVRLTDFSFLMVLGKGSFGK 358 Query 353 VM 354 || Query 353 VM 354 |
| Xenopus laevis | Query 295 EEGNVELRQKFEKAKLGPAGNKVISPSEDR#RQ--PSNNLDRVKLTDFNFLMVLGKGSFGK 352 ++||+|||||||||||||||||||+|+ +|#| ||||+| ++|||| |||||||||||| Sbjct 298 DDGNLELRQKFEKAKLGPAGNKVINPTGER#RPYIPSNNIDSIRLTDFCFLMVLGKGSFGK 357 Query 353 VM 354 || Query 353 VM 354 |
| Protopterus dolloi | Query 295 EEGNVELRQKFEKAKLGPAGNKVISPSEDR#RQPSNNLDRVKLTDFNFLMVLGKGSFGKVM 354 ++||| ||||||||+||||| ||||| # | ||||| || ||||+|||||||||||| Sbjct 214 DDGNVGLRQKFEKARLGPAGKKVISPERKS#SLPLNNLDRFKLEDFNFVMVLGKGSFGKVM 273 |
| Takifugu rubripes | Query 295 EEGNVELRQKFEKAKLGPAGNKVISPSEDR#R--QPSNNLDRVKLTDFNFLMVLGKGSFGK 352 ++ |+|||||||||||| | |||+||+ |#| || |+|||+| ||||| +|||||||| Sbjct 292 DDVNLELRQKFEKAKLGQ-GKKVITPSDHR#RFSLPSGNMDRVRLNDFNFLALLGKGSFGK 350 Query 353 VM 354 || Query 353 VM 354 |
| Scyliorhinus canicula | Query 295 EEGNVELRQKFEKAKLGPAGNKVISPSEDR#RQPSNNLDRVKLTDFNFLMVLGKGSFGKVM 354 ++ + |||+||||||||||| | | # |+|+|| | | ||||||||||||||||| Sbjct 226 DDEDSELRKKFEKAKLGPAGKKAIEKKPS-#-SPTNDLDHVHLEDFNFLMVLGKGSFGKVM 283 |
| Danio rerio | Query 295 EEGNVELRQKFEKAKLGPAGNKVISPSEDR#RQPSN-NLDRVKLTDFNFLMVLGKGSFGKV 353 |||| |||||||+||+||+ | + #+ || | ||+||+|||||||||||||||| Sbjct 294 EEGNEELRQKFERAKIGPSKTDGSSSNAIS#KFDSNGNRDRMKLSDFNFLMVLGKGSFGKV 353 Query 354 M 354 | Query 354 M 354 |
| Tetraodon nigroviridis | Query 295 EEGNVELRQKFEKAKLGPAGNK--VISPSEDR#RQPSNNLDRVKLTDFNFLMVLGKGSFGK 352 |||| |||||||+||+|| | + + |# + | ||+|| ||||||||||||||| Sbjct 231 EEGNEELRQKFERAKIGPGKNTDGKSANAASR#FDSNGNQDRMKLADFNFLMVLGKGSFGK 290 Query 353 VM 354 || Query 353 VM 354 |
| Petromyzon marinus | Query 295 EEGNVELRQKFEKAKLGPAGNKVISP--SEDR#RQPS-------NNLDRVKLTDFNFLMVL 345 |+|+ |+|+|||||+|||+ + | |++|#| | |+|||| |||||||| Sbjct 225 EDGDTEMRRKFEKARLGPS----VKPRASDER#RANSLSAILNNANVDRVKADDFNFLMVL 280 Query 346 GKGSFGKVM 354 ||||||||| Query 346 GKGSFGKVM 354 |
| Macaca mulatta | Query 296 EGNVELRQKFEKAKLGPAGNKVISPSE------DR#RQPSNNLDRVKLTDFNFLMVLGKGS 349 ||| |||||||+||+ | || | | +# + | ||+||||||||||||||| Sbjct 296 EGNEELRQKFERAKISQ-GTKV--PEEKTTNTVSK#FDNNGNRDRMKLTDFNFLMVLGKGS 352 Query 350 FGKVM 354 ||||| Query 350 FGKVM 354 |
| Myxine glutinosa | Query 295 EEGNVELRQKFEKAKLGPAGNKVISPSEDR#RQPS---NNLDRVKLTDFNFLMVLGKGSFG 351 ||| +||||+ +++++ + ||++|+# | + |||| | || || |||||||| Sbjct 216 EEG-LELRQRLQRSQIARSDKNKQSPAKDQ#PSDSASLSGLDRVNLEDFVFLTVLGKGSFG 274 Query 352 KVM 354 ||| Query 352 KVM 354 |
| Macaca fascicularis | Query 305 FEKAKLGPAGNKVISPSEDR#RQPSN-----NLDRVKLTDFNFLMVLGKGSFGKVM 354 +|+ ++||+ + + ||| # | + |+ ++||+|||||||||||||| Sbjct 312 YERVRMGPSSSPIPSPSPSP#TDPKRCFFGASPGRLHISDFSFLMVLGKGSFGKVM 366 |
| Branchiostoma lanceolatum | Query 295 EEGNVELRQKFEKAKLGPAGNKVISPSEDR#RQPSNNLDRVKLTDFNFLMVLGKGSFGKVM 354 || +|++ +| || + ||+ # | || +| ||||||||||||||||| Sbjct 225 EENVAKLKEHLQKQKLDEQRKQKSKRSEEC#NIGS--LDHMKAADFNFLMVLGKGSFGKVM 282 |
| Bombyx mori | Query 295 EEGNVELRQKFEKAKLGPAGNKVISPSEDR#RQPSN--NLDRVKLTDFNFLMVLGKGSFGK 352 ||| +| | + + | + | ||# | | | ++ |||||+|||||||||| Sbjct 295 EEG-ADLVQLKNQMRATTVGARRPPPPPDR#EVPHNVAAADVIRATDFNFIMVLGKGSFGK 353 Query 353 VM 354 || Query 353 VM 354 |
| Aplysia californica | Query 314 GNKVISPSEDR#RQPSNNLDRVKLTDFNFLMVLGKGSFGKVM 354 |++ ||| ||# + ++ |+ | ||||+ ||||||||||| Sbjct 380 GSRTRSPSSDR#SRSHHS--RISLHDFNFIKVLGKGSFGKVM 418 |
| Anopheles gambiae str. PEST | Query 295 EEGN--VELRQKFEKAKLGPAGNKVISPSEDR#RQPSN--NLDRVKLTDFNFLMVLGKGSF 350 ||| |+|+ + | + | | |+# | | | ++ |||||||||||||| Sbjct 232 EEGADLVQLKSQMRKTSI----TKKIPMMGDK#DVPHNMTKKDVIRATDFNFLMVLGKGSF 287 Query 351 GKVM 354 |||| Query 351 GKVM 354 |
| Apis mellifera | Query 295 EEGNVELRQKFEKAKLGPAGNKVISPSEDR#RQPSN--NLDRVKLTDFNFLMVLGKGSFGK 352 ||| | + + + | + ++|+# | | | ++ +||||||||||||||| Sbjct 309 EEGVDLAELKMKPSPQKTSVTKKTTTTQDK#EVPHNMGKSDLIRASDFNFLMVLGKGSFGK 368 Query 353 VM 354 || Query 353 VM 354 |
| Aedes aegypti | Query 295 EEGNVELRQKFEKAKLGPAGNKVISPSEDR#RQPSNNLDRVKLTDFNFLMVLGKGSFGKVM 354 ||| ++ | + | + + +| # | ++ |||||||||||||||||| Sbjct 240 EEGTDLVQLKSQMRKTSISKKAPVLCDKDV#PHNMGKKDVIRATDFNFLMVLGKGSFGKVM 299 |
| Drosophila melanogaster | Query 303 QKFEKAKLGPAGNK-VISPSEDR#RQPSNNLDRVKLTDFNFLMVLGKGSFGKVM 354 | | | |+ | ++ |+ # |+ | ++ |||||+ ||||||||||+ Sbjct 304 QDLLKLKQKPSQKKPMVMRSDTN#THTSSKKDMIRATDFNFIKVLGKGSFGKVL 356 |
| Tribolium castaneum | Query 324 R#RQPSNN--LDRVKLTDFNFLMVLGKGSFGKVM 354 +#|+|| + | ++ +||||||||||||||||| Sbjct 320 Q#RRPSQHGQKDIIRASDFNFLMVLGKGSFGKVM 352 |
| Coprinopsis cinerea okayama7#130 | Query 306 EKAKLGPAGNKVISPSEDR#RQPSNNLDRVKLTDFNFLMVLGKGSFGKVM 354 ++ + | + + ++ +#|||| +| | ||||| |||||+||||| Sbjct 685 QQGMVPPQHQQQMIQAQPQ#RQPSKKR-KVGLDDFNFLAVLGKGNFGKVM 732 |
| Drosophila pseudoobscura | Query 303 QKFEKAKLGPAGNKVISPSEDR#RQPSN--NLDRVKLTDFNFLMVLGKGSFGKVM 354 | | | |+ | + | # |+ | ++ |||||+ ||||||||||+ Sbjct 190 QDLLKLKQKPSQKKPLVMRSDT#NTHSSISKKDMIRATDFNFIKVLGKGSFGKVL 243 |
| Caenorhabditis briggsae | Query 328 SNNLDRVKLTDFNFLMVLGKGSFGKVM 354 + | | +| +||||| ||||||||||+ Sbjct 404 NTNRDVIKASDFNFLTVLGKGSFGKVL 430 |
| Magnaporthe grisea 70-15 | Query 302 RQKFEKAKLGPAGNKVISPSEDR#RQPS----NNLDRVKLTDFNFLMVLGKGSFGKVM 354 +| |+ + | || | |# || |+ | |||| |||||+||||| Sbjct 816 QQHQEQQIISPTAGTVIPTSAKR#PLPSATDPGTGQRIGLDHFNFLAVLGKGNFGKVM 872 |
| Caenorhabditis elegans | Query 328 SNNLDRVKLTDFNFLMVLGKGSFGKVM 354 |+| + +| +||||| ||||||||||+ Sbjct 337 SSNHNVIKASDFNFLTVLGKGSFGKVL 363 |
| Schizosaccharomyces pombe | Query 320 PSEDR#RQPSNNLDRVKLTDFNFLMVLGKGSFGKVM 354 | |#|+ | || | || || |||||+||||| Sbjct 556 PESPR#REKKN---RVTLDDFTFLAVLGKGNFGKVM 587 |
| Schizosaccharomyces pombe 972h- | Query 320 PSEDR#RQPSNNLDRVKLTDFNFLMVLGKGSFGKVM 354 | |#|+ | || | || || |||||+||||| Sbjct 648 PESPR#REKKN---RVTLDDFTFLAVLGKGNFGKVM 679 |
| Magnaporthe grisea | Query 302 RQKFEKAKLGPAGNKVISPSEDR#RQPS----NNLDRVKLTDFNFLMVLGKGSFGKVM 354 +| |+ + | || | |# || |+ | |||| |||||+||||| Sbjct 816 QQHQEQQIISPTAGTVIPTSARR#PLPSATDPGTGQRIGLDHFNFLAVLGKGNFGKVM 872 |
| Strongylocentrotus purpuratus | Query 321 SEDR#RQPSNNLDRVKLTDFNFLMVLGKGSFGKVM 354 | # ++++ |+ +||||| ||||||||||| Sbjct 307 SSQN#SNSNSSMGLVRASDFNFLSVLGKGSFGKVM 340 |
| Suberites domuncula | Query 295 EEGNVELRQKFEKAKLGPAGNKVISPSEDR#RQPSN------NLDRVKLTDFNFLMVLGKG 348 +| +| ++| || + +++ #|+ | ++ ++|| ||| |+||||| Sbjct 293 DEEATKLIEEFRKATILDKAKNLVTDVPIM#RRGSELVPGAKDMPKMKLEDFNLLVVLGKG 352 Query 349 SFGKV 353 ||||| Query 349 SFGKV 353 |
| Hydractinia echinata | Query 332 DRVKLTDFNFLMVLGKGSFGKVM 354 +|+|| || || |+||||||||| Sbjct 191 ERLKLEDFTFLKVIGKGSFGKVM 213 |
| Pan troglodytes | Query 295 EEGNVELRQKFEKAKLGPAGNKVISPSEDR#RQPSNNLDRVKLTDFNFLMVLGKGSFGKVM 354 | + +| + |||+|| | # |||+|| |+||||||||+ Sbjct 134 ERSSQKLHSTSQNINLGPSGNPHAKP----#------------TDFDFLKVIGKGSFGKVL 177 |
| Paramecium tetraurelia | Query 295 EEGNVELRQKFEKAKLGPAGNKVISPSEDR#RQPSNNLDRVKLTDFNFLMVLGKGSFGKVM 354 +|| ++ +| || ++ | # | |||||| | |||+||||||| Sbjct 32 KEGEQQMNDEFMKAPTATF-EQIEETGEQV#FVDDNREAIVKLTDFQFEKVLGRGSFGKVM 90 |
| Tuber excavatum | Query 320 PSEDR#RQPSNNLDRVKLTDFNFLMVLGKGSFGKVM 354 |+ + #+|| ++ | |||| |||||+||||| Sbjct 312 PAPEP#QQPVIRPAKIGLDHFNFLAVLGKGNFGKVM 346 |
| Yarrowia lipolytica | Query 333 RVKLTDFNFLMVLGKGSFGKVM 354 |+ | ||||| |||||+||||| Sbjct 919 RIGLDDFNFLAVLGKGNFGKVM 940 |
| Botryotinia fuckeliana | Query 312 PAGNKVISPSEDR#RQPSNNLD-----RVKLTDFNFLMVLGKGSFGKVM 354 | +|+ + |#+| | |+ | |||| |||||+||||| Sbjct 813 PGSQQVVPQGQQR#KQVPAANDAGTGRRIGLDHFNFLAVLGKGNFGKVM 860 |
| Tuber aestivum | Query 320 PSEDR#RQPSNNLDRVKLTDFNFLMVLGKGSFGKVM 354 |+ + #+ | + ++ | |||| |||||+||||| Sbjct 314 PAPEP#QPPVSRPTKIGLDHFNFLAVLGKGNFGKVM 348 |
| Sycon raphanus | Query 335 KLTDFNFLMVLGKGSFGKVM 354 ||+||+|| ||||||||||| Sbjct 362 KLSDFSFLKVLGKGSFGKVM 381 |
| Asparagus officinalis | Query 321 SEDR#RQPSN-NLDRVKLTDFNFLMVLGKGSFGKV 353 +|+ # ||+ |++|| | ||+|| |+|+|+|||| Sbjct 106 AEEH#EVPSSENVERVGLEDFDFLKVVGQGAFGKV 139 |
| Leishmania braziliensis | Query 295 EEGNVELRQKFEKAKLGPAGNKVISPSEDR#RQPSNNLDRVKLTDFNFLMVLGKGSFGKVM 354 | | | +|+ + + ++|| # |+|| | | || | |+|+|+||||+ Sbjct 591 ESGQVMQQQRVINSTSPIPYSSLLSPRGSL#VSRSDNLSPVGLQDFQLLTVIGRGTFGKVL 650 |
| Leptosphaeria maculans | Query 308 AKLGPAGNKVISPSEDR#RQPSNNLD----RVKLTDFNFLMVLGKGSFGKVM 354 |++ |+ + |++# || | |+ | |||| |||||+||||| Sbjct 825 AQIPPSKQLPPANPENK#VTPSANTQGSGKRIGLDHFNFLAVLGKGNFGKVM 875 |
| Squalus acanthias | Query 327 PSNNLDRVKLTDFNFLMVLGKGSFGKVM 354 ||+| + | +||||| |+||||||||+ Sbjct 89 PSSN-PQAKPSDFNFLKVIGKGSFGKVL 115 |
| Cryptococcus neoformans var. neoformans JEC21 | Query 333 RVKLTDFNFLMVLGKGSFGKVM 354 +| | ||||| |||||+||||| Sbjct 751 KVGLDDFNFLAVLGKGNFGKVM 772 |
| Cryptococcus neoformans var. neoformans | Query 333 RVKLTDFNFLMVLGKGSFGKVM 354 +| | ||||| |||||+||||| Sbjct 751 KVGLDDFNFLAVLGKGNFGKVM 772 |
| Cryptococcus neoformans var. neoformans B-3501A | Query 333 RVKLTDFNFLMVLGKGSFGKVM 354 +| | ||||| |||||+||||| Sbjct 754 KVGLDDFNFLAVLGKGNFGKVM 775 |
| Cryptococcus neoformans var. grubii | Query 333 RVKLTDFNFLMVLGKGSFGKVM 354 +| | ||||| |||||+||||| Sbjct 751 KVGLDDFNFLAVLGKGNFGKVM 772 |
| Calliphora vicina | Query 320 PSEDR#RQPSNNL---DRVKLTDFNFLMVLGKGSFGKVM 354 | | #+ +|+ | ++ ||||+ ||||||+||++ Sbjct 333 PRIDN#KDMPHNMSKRDMIRAADFNFIKVLGKGSYGKIL 370 |
| Tuber indicum | Query 320 PSEDR#RQPSNNLDRVKLTDFNFLMVLGKGSFGKVM 354 |+ + #+| + ++ | |||| |||||+||||| Sbjct 313 PAPEP#QQVVSKPTKIGLDHFNFLAVLGKGNFGKVM 347 |
| Tuber pseudoexcavatum | Query 320 PSEDR#RQPSNNLDRVKLTDFNFLMVLGKGSFGKVM 354 |+ + #+| + ++ | |||| |||||+||||| Sbjct 312 PAPEP#QQVVSKPTKIGLDHFNFLAVLGKGNFGKVM 346 |
| Phaeosphaeria nodorum SN15 | Query 329 NNLDRVKLTDFNFLMVLGKGSFGKVM 354 | ||+ + || | |+||||||||| Sbjct 273 NRQDRLSIDDFELLKVVGKGSFGKVM 298 |
| Aspergillus oryzae | Query 312 PAGNKVISPSEDR#RQPSNNLDRVKLTDFNFLMVLGKGSFGKVM 354 | + ++ || # | |+ | |||| |||||+||||| Sbjct 741 PVMQQQVAMKEDA#-PPQQPKVRIGLDHFNFLAVLGKGNFGKVM 782 |
| Sporothrix schenckii | Query 333 RVKLTDFNFLMVLGKGSFGKVM 354 |+ | +|||| |||||+||||| Sbjct 863 RIGLDNFNFLAVLGKGNFGKVM 884 |
| Cochliobolus heterostrophus | Query 310 LGPAGNKVISPSEDR#RQPSNNLDRVKLTDFNFLMVLGKGSFGKVM 354 +| | ++|+ + #+ |+ | |||| |||||+||||| Sbjct 823 VGEVPGKKVTPAANT#QGTGK---RIGLDHFNFLAVLGKGNFGKVM 864 |
| Aspergillus clavatus NRRL 1 | Query 312 PAGNKVISPSEDR#RQPSNNLDRVKLTDFNFLMVLGKGSFGKVM 354 | +| | #+|| |+ | |||| |||||+||||| Sbjct 763 PMQQQVAVKEEVP#QQPKA---RIGLDHFNFLSVLGKGNFGKVM 802 |
| Medicago truncatula | Query 302 RQKFEKAKLGPAGNKVISPSEDR#RQPSNNLDRVKLTDFNFLMVLGKGSFGKV 353 +|| +|| + |+ +| | + +# | + + | +|| || ++|+| |||| Sbjct 540 KQKRQKAVVTPSNSKKRSSMKSK#HQKFSYTEIVNITD-NFKTIIGEGGFGKV 590 |
| Debaryomyces hansenii CBS767 | Query 326 QPSNNLDRVKLTDFNFLMVLGKGSFGKVM 354 +| ++ | || || |||||+||||| Sbjct 750 KPKRRRRKIGLDDFQFLAVLGKGNFGKVM 778 |
| Hydra vulgaris | Query 335 KLTDFNFLMVLGKGSFGKVM 354 || || || |+||||||||| Sbjct 338 KLEDFTFLKVIGKGSFGKVM 357 |
| Coccidioides immitis RS | Query 333 RVKLTDFNFLMVLGKGSFGKVM 354 || | |||| |||||+||||| Sbjct 740 RVGLDHFNFLAVLGKGNFGKVM 761 |
| Neosartorya fischeri NRRL 181 | Query 297 GNVELRQKFEKAKLGPAGNKVISPSEDR#RQPSNNLDRVKLTDFNFLMVLGKGSFGKVM 354 | + +|+ ++ + | +| | # | |+ | |||| |||||+||||| Sbjct 743 GMPQQQQQQQQQAVAPMQQQVAVKEEI-#-PPQQPKVRIGLDHFNFLAVLGKGNFGKVM 798 |
| Pichia stipitis CBS 6054 | Query 319 SPSEDR#RQPSNNLDRVKLTDFNFLMVLGKGSFGKVM 354 || + +#|+ +| | || || |||||+||||| Sbjct 737 SPHKSK#RRRR----KVGLDDFQFLAVLGKGNFGKVM 768 |
| Trichomonas vaginalis G3 | Query 325 RQPSNNLDRVKLTDFNFLMVLGKGSFGKVM 354 | + | +|+ + ||| + ||||| +|||| Sbjct 99 RSETYNGERLSMDDFNIISVLGKGKYGKVM 128 |
* References
[PubMed ID: 18977358] Venardos K, Enriquez C, Marshall T, Chin-Dusting JP, Ahlers B, Kaye DM, Protein kinase C mediated inhibition of endothelial L-arginine transport is mediated by MARCKS protein. J Mol Cell Cardiol. 2009 Jan;46(1):86-92. Epub 2008 Oct 14.
[PubMed ID: 18323518] ... Rask-Madsen C, King GL, Differential regulation of VEGF signaling by PKC-alpha and PKC-epsilon in endothelial cells. Arterioscler Thromb Vasc Biol. 2008 May;28(5):919-24. Epub 2008 Mar 6.
[PubMed ID: 16895917] ... Yeong SS, Zhu Y, Smith D, Verma C, Lim WG, Tan BJ, Li QT, Cheung NS, Cai M, Zhu YZ, Zhou SF, Tan SL, Duan W, The last 10 amino acid residues beyond the hydrophobic motif are critical for the catalytic competence and function of protein kinase Calpha. J Biol Chem. 2006 Oct 13;281(41):30768-81. Epub 2006 Aug 8.
[PubMed ID: 14563411] ... Aschrafi A, Franzen R, Shabahang S, Fabbro D, Pfeilschifter J, Huwiler A, Ceramide induces translocation of protein kinase C-alpha to the Golgi compartment of human embryonic kidney cells by interacting with the C2 domain. Biochim Biophys Acta. 2003 Oct 20;1634(1-2):30-9.
[PubMed ID: 9879008] ... Kuznetsov SB, Larkin DM, Kaftanovskaia EM, Ivanova EV, Astakhova NM, Cheriakene OV, Zhdanova NS, [Chromosome localization and analysis of synteny analysis of some genes in swine, cattle, and sheep (Artiodactyla)] Genetika. 1998 Sep;34(9):1200-4.
[PubMed ID: 9013603] ... Bornancin F, Parker PJ, Phosphorylation of protein kinase C-alpha on serine 657 controls the accumulation of active enzyme and contributes to its phosphatase-resistant state. J Biol Chem. 1997 Feb 7;272(6):3544-9.
[PubMed ID: 7844141] ... Staudinger J, Zhou J, Burgess R, Elledge SJ, Olson EN, PICK1: a perinuclear binding protein and substrate for protein kinase C isolated by the yeast two-hybrid system. J Cell Biol. 1995 Feb;128(3):263-71.
[PubMed ID: 8349635] ... Cazaubon SM, Parker PJ, Identification of the phosphorylated region responsible for the permissive activation of protein kinase C. J Biol Chem. 1993 Aug 15;268(23):17559-63.
[PubMed ID: 3045562] ... Nishizuka Y, The molecular heterogeneity of protein kinase C and its implications for cellular regulation. Nature. 1988 Aug 25;334(6184):661-5.
[PubMed ID: 3755547] ... Parker PJ, Coussens L, Totty N, Rhee L, Young S, Chen E, Stabel S, Waterfield MD, Ullrich A, The complete primary structure of protein kinase C--the major phorbol ester receptor. Science. 1986 Aug 22;233(4766):853-9.