XSB1247 : protein kinase C, alpha [Homo sapiens]
[ CaMP Format ]
This entry is computationally expanded from SB0055
* Basic Information
| Organism | Homo sapiens (human) |
| Protein Names | Protein kinase C alpha type; PKC-alpha; PKC-A; 2.7.11.13; protein kinase C; alpha; aging-associated gene 6 |
| Gene Names | PRKCA; PKCA, PRKACA; PKCA; PRKACA; protein kinase C, alpha |
| Gene Locus | 17q22-q23.2; chromosome 17 |
| GO Function | Not available |
* Information From OMIM
Function: Ekinci and Shea (1999) stated that PKC-alpha is reversibly activated at the plasma membrane by transient generation of diacylglycerol (DAG), coupled with the release of Ca(2+) from intracellular stores, following receptor-mediated hydrolysis of inositol phospholipids. PKC-alpha is also irreversibly activated by calpain (see OMIM:114220)-mediated cleavage of its regulatory and catalytic subunits, resulting in a cofactor-independent, free PKC-alpha catalytic subunit termed PKM-alpha. Ekinci and Shea (1999) found that activation of PKC-alpha in human neuroblastoma cells by either the phorbol ester TPA or by ionophore-mediated calcium mobilization, which experimentally correspond to DAG-mediated and calpain-mediated activation, respectively, resulted in increased phosphorylation of the microtubule-associated protein tau (MAPT; OMIM:157140). Activation of PKC-alpha by calcium mobilization, but not TPA, generated PKM-alpha and resulted in calpain-dependent release of PKM-alpha from the plasma membrane. The TPA-mediated increase in tau phosphorylation was blocked by cotreatment with an MAP2K (see OMIM:176872) inhibitor, but ionophore-mediated tau phosphorylation was not.
Function: Lorenz et al. (2003) demonstrated that the RAF kinase inhibitor protein (RKIP; OMIM:604591) is a physiologic inhibitor of GRK2 (OMIM:109635). After stimulation of G protein-coupled receptors, RKIP dissociates from its known target, RAF1 (OMIM:164760), to associate with GRK2 and block its activity. This switch is triggered by a PKC-dependent phosphorylation of RKIP on serine-153. Lorenz et al. (2003) concluded that their data delineate a new principle in signal transduction: by activating PKC, the incoming receptor signal is enhanced both by removing an inhibitor from RAF1 and by blocking receptor internalization. A physiologic role for this mechanism is shown in cardiomyocytes in which the downregulation of RKIP restrains beta-adrenergic signaling and contractile activity.
Function: Birnbaum et al. (2004) tested the influence of PKC intracellular signaling on prefrontal cortical cognitive function in rats and monkeys and showed that high levels of PKC activity in prefrontal cortex, as seen for example during stress exposure, markedly impaired behavioral and electrophysiologic measures of working memory. Birnbaum et al. (2004) concluded that excessive PKC activation can disrupt prefrontal cortical regulation of behavior and thought, possibly contributing to signs of prefrontal cortical dysfunction such as distractibility, impaired judgment, impulsivity, and thought disorder.
Function: Bivona et al. (2006) found that the subcellular localization and function of Kras (see KRAS2; OMIM:190070) in mammalian cells was modulated by Pkc. Phosphorylation of Kras by Pkc agonists induced rapid translocation of Kras from the plasma membrane to several intracellular membranes, including the outer mitochondrial membrane, where Kras associated with Bclxl (BCL2L1; OMIM:600039). Phosphorylated Kras required Bclxl for induction of apoptosis.
Function: Among 304 Swiss individuals tested and genotyped, de Quervain and Papassotiropoulos (2006) found a significant association (p = 0.00008) between short-term episodic memory performance and genetic variations in a 7-gene cluster consisting of the ADCY8 (OMIM:103070), PRKACG (OMIM:176893), CAMK2G (OMIM:602123), GRIN2A (OMIM:138253), GRIN2B (OMIM:138252), GRM3 (OMIM:601115), and PRKCA genes, all of which have well-established molecular and biologic functions in animal memory. Functional MRI studies in an independent set of 32 individuals with similar memory performance showed a correlation between activation in memory-related brain regions, including the hippocampus and parahippocampal gyrus, and genetic variability in the 7-gene cluster. De Quervain and Papassotiropoulos (2006) concluded that these 7 genes encode proteins of the memory formation signaling cascade that are important for human memory function.
* Structure Information
1. Primary Information
Length: 672 aa
Average Mass: 76.764 kDa
Monoisotopic Mass: 76.714 kDa
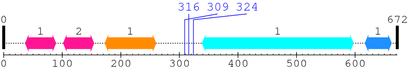
2. Domain Information
Annotated Domains: interpro / pfam / smart / prosite
Computationally Assigned Domains (Pfam+HMMER):
| domain name | begin | end | score | e-value |
|---|---|---|---|---|
| C1_1 1. | 37 | 89 | 83.2 | 9.4e-22 |
| C1_1 2. | 102 | 154 | 92.5 | 1.5e-24 |
| C2 1. | 173 | 260 | 137.9 | 3.3e-38 |
| --- cleavage 316 --- | ||||
| --- cleavage 309 --- | ||||
| --- cleavage 324 --- | ||||
| Pkinase 1. | 339 | 597 | 291.2 | 2.2e-84 |
| Pkinase_C 1. | 617 | 662 | 55.8 | 1.6e-13 |
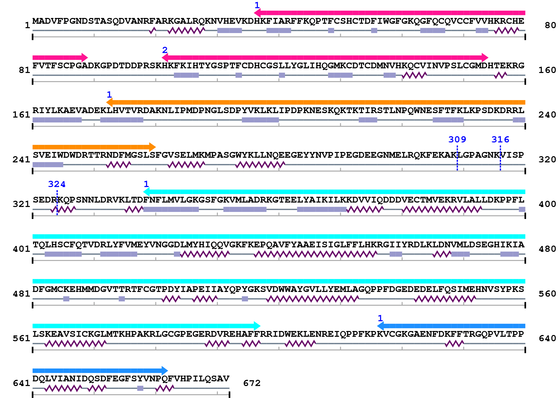
3. Sequence Information
Fasta Sequence: XSB1247.fasta
Amino Acid Sequence and Secondary Structures (PsiPred):
4. 3D Information
Known Structures in PDB: 2ELI (NMR; -; A=92-169)
* Cleavage Information
3 [sites]
Cleavage sites (±10aa)
[Site 1] KAKLGPAGNK316-VISPSEDRKQ
Lys316  Val
Val
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Lys307 | Ala308 | Lys309 | Leu310 | Gly311 | Pro312 | Ala313 | Gly314 | Asn315 | Lys316 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Val317 | Ile318 | Ser319 | Pro320 | Ser321 | Glu322 | Asp323 | Arg324 | Lys325 | Gln326 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| NVPIPEGDEEGNMELRQKFEKAKLGPAGNKVISPSEDRKQPSNNLDRVKLTDFNFLMVLG |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | N/A | 124.00 | 12 | - |
| 2 | Homo sapiens | 124.00 | 11 | protein kinase C, alpha |
| 3 | Mus musculus | 124.00 | 8 | protein kinase C |
| 4 | synthetic construct | 124.00 | 1 | protein kinase C alpha |
| 5 | Rattus norvegicus | 122.00 | 5 | PREDICTED: similar to Protein kinase C alpha type |
| 6 | Oryctolagus cuniculus | 121.00 | 5 | KPCA_RABIT Protein kinase C alpha type (PKC-alpha) |
| 7 | Canis familiaris | 121.00 | 4 | PREDICTED: similar to Protein kinase C, alpha type |
| 8 | Bos taurus | 121.00 | 4 | protein kinase C, alpha |
| 9 | Monodelphis domestica | 110.00 | 4 | PREDICTED: similar to protein kinase C |
| 10 | Gallus gallus | 107.00 | 3 | protein kinase C, alpha |
| 11 | Xenopus tropicalis | 97.40 | 1 | hypothetical protein LOC780209 |
| 12 | Latimeria chalumnae | 97.10 | 2 | protein kinase C alpha |
| 13 | Xenopus laevis | 90.10 | 1 | protein kinase C, alpha |
| 14 | Protopterus dolloi | 84.70 | 2 | protein kinase C alpha |
| 15 | Takifugu rubripes | 73.90 | 1 | protein kinase C, alpha type |
| 16 | Danio rerio | 69.30 | 7 | protein kinase C, beta 1 |
| 17 | Scyliorhinus canicula | 68.60 | 2 | protein kinase C alpha |
| 18 | Tetraodon nigroviridis | 67.40 | 3 | unnamed protein product |
| 19 | Macaca mulatta | 60.50 | 9 | PREDICTED: protein kinase C, beta isoform 1 |
| 20 | Petromyzon marinus | 57.00 | 1 | protein kinase C |
| 21 | Myxine glutinosa | 45.80 | 1 | protein kinase C |
| 22 | Macaca fascicularis | 38.90 | 1 | KPCG_MACFA Protein kinase C gamma type (PKC-gamma) |
| 23 | Branchiostoma lanceolatum | 38.10 | 1 | protein kinase C |
| 24 | Bombyx mori | 37.00 | 1 | conventional protein kinase C |
| 25 | Apis mellifera | 36.20 | 1 | PREDICTED: similar to Protein C kinase 53E CG6622- |
| 26 | Anopheles gambiae str. PEST | 35.00 | 1 | ENSANGP00000009078 |
| 27 | Aedes aegypti | 34.70 | 1 | protein kinase c |
| 28 | Drosophila melanogaster | 33.90 | 4 | Protein C kinase 53E CG6622-PA, isoform A |
| 29 | Aplysia californica | 33.50 | 1 | KPC1_APLCA Calcium-dependent protein kinase C (APL |
Top-ranked sequences
| organism | matching |
|---|---|
| N/A | Query 287 NVPIPEGDEEGNMELRQKFEKAKLGPAGNK#VISPSEDRKQPSNNLDRVKLTDFNFLMVLG 346 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 287 NVPIPEGDEEGNMELRQKFEKAKLGPAGNK#VISPSEDRKQPSNNLDRVKLTDFNFLMVLG 346 |
| Homo sapiens | Query 287 NVPIPEGDEEGNMELRQKFEKAKLGPAGNK#VISPSEDRKQPSNNLDRVKLTDFNFLMVLG 346 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 287 NVPIPEGDEEGNMELRQKFEKAKLGPAGNK#VISPSEDRKQPSNNLDRVKLTDFNFLMVLG 346 |
| Mus musculus | Query 287 NVPIPEGDEEGNMELRQKFEKAKLGPAGNK#VISPSEDRKQPSNNLDRVKLTDFNFLMVLG 346 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 287 NVPIPEGDEEGNMELRQKFEKAKLGPAGNK#VISPSEDRKQPSNNLDRVKLTDFNFLMVLG 346 |
| synthetic construct | Query 287 NVPIPEGDEEGNMELRQKFEKAKLGPAGNK#VISPSEDRKQPSNNLDRVKLTDFNFLMVLG 346 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 287 NVPIPEGDEEGNMELRQKFEKAKLGPAGNK#VISPSEDRKQPSNNLDRVKLTDFNFLMVLG 346 |
| Rattus norvegicus | Query 287 NVPIPEGDEEGNMELRQKFEKAKLGPAGNK#VISPSEDRKQPSNNLDRVKLTDFNFLMVLG 346 ||||||||||||+|||||||||||||||||#|||||||||||||||||||||||||||||| Sbjct 287 NVPIPEGDEEGNVELRQKFEKAKLGPAGNK#VISPSEDRKQPSNNLDRVKLTDFNFLMVLG 346 |
| Oryctolagus cuniculus | Query 287 NVPIPEGDEEGNMELRQKFEKAKLGPAGNK#VISPSEDRKQPSNNLDRVKLTDFNFLMVLG 346 |||||||||+||+|||||||||||||||||#|||||||||||||||||||||||||||||| Sbjct 287 NVPIPEGDEDGNVELRQKFEKAKLGPAGNK#VISPSEDRKQPSNNLDRVKLTDFNFLMVLG 346 |
| Canis familiaris | Query 287 NVPIPEGDEEGNMELRQKFEKAKLGPAGNK#VISPSEDRKQPSNNLDRVKLTDFNFLMVLG 346 ||||||||||||+|||||||||||||||||#||||||||+||||||||||||||||||||| Sbjct 205 NVPIPEGDEEGNVELRQKFEKAKLGPAGNK#VISPSEDRRQPSNNLDRVKLTDFNFLMVLG 264 |
| Bos taurus | Query 287 NVPIPEGDEEGNMELRQKFEKAKLGPAGNK#VISPSEDRKQPSNNLDRVKLTDFNFLMVLG 346 ||||||||||||+|||||||||||||||||#||||||||+||||||||||||||||||||| Sbjct 287 NVPIPEGDEEGNVELRQKFEKAKLGPAGNK#VISPSEDRRQPSNNLDRVKLTDFNFLMVLG 346 |
| Monodelphis domestica | Query 287 NVPIPEGDEEGNMELRQKFEKAKLGPAGNK#VISPSEDRKQPSNNLDRVKLTDFNFLMVLG 346 |||||||||||||||||||||||||| |||#||| |+||| |+|||||||||||||||||| Sbjct 287 NVPIPEGDEEGNMELRQKFEKAKLGPVGNK#VISSSDDRK-PTNNLDRVKLTDFNFLMVLG 345 |
| Gallus gallus | Query 287 NVPIPEGDEEGNMELRQKFEKAKLGPAGNK#VISPSEDRKQ--PSNNLDRVKLTDFNFLMV 344 |||||+ ||+|| |||||||||||||||||#||+||||| |||||||||||||||||| Sbjct 287 NVPIPDADEDGNAELRQKFEKAKLGPAGNK#VITPSEDRNSSVPSNNLDRVKLTDFNFLMV 346 Query 345 LG 346 || Query 345 LG 346 |
| Xenopus tropicalis | Query 287 NVPIPEGDEEGNMELRQKFEKAKLGPAGNK#VISPSEDRKQ--PSNNLDRVKLTDFNFLMV 344 |||||| +++||+|||||||||||||||||#||||+++|+ ||||+| |+||||+|||| Sbjct 291 NVPIPEEEDDGNIELRQKFEKAKLGPAGNK#VISPTDERRPYVPSNNIDSVRLTDFSFLMV 350 Query 345 LG 346 || Query 345 LG 346 |
| Latimeria chalumnae | Query 287 NVPIPEGDEEGNMELRQKFEKAKLGPAGNK#VISPSEDRKQ--PSNNLDRVKLTDFNFLMV 344 |||| | |+ ||+|||||||||||||||+|#|||| |+|| |||||||||||||||||| Sbjct 209 NVPISEADD-GNVELRQKFEKAKLGPAGSK#VISPVEERKSSTPSNNLDRVKLTDFNFLMV 267 Query 345 LG 346 || Query 345 LG 346 |
| Xenopus laevis | Query 287 NVPIPEGDEEGNMELRQKFEKAKLGPAGNK#VISPSEDRKQ--PSNNLDRVKLTDFNFLMV 344 |||||| |+ ||+|||||||||||||||||#||+|+ +|+ ||||+| ++|||| |||| Sbjct 291 NVPIPEADD-GNLELRQKFEKAKLGPAGNK#VINPTGERRPYIPSNNIDSIRLTDFCFLMV 349 Query 345 LG 346 || Query 345 LG 346 |
| Protopterus dolloi | Query 287 NVPIPEGDEEGNMELRQKFEKAKLGPAGNK#VISPSEDRKQPSNNLDRVKLTDFNFLMVLG 346 |||||+ |+ ||+ ||||||||+||||| |#|||| | ||||| || ||||+|||| Sbjct 207 NVPIPDADD-GNVGLRQKFEKARLGPAGKK#VISPERKSSLPLNNLDRFKLEDFNFVMVLG 265 |
| Takifugu rubripes | Query 287 NVPIPEGDEEGNMELRQKFEKAKLGPAGNK#VISPSEDRK--QPSNNLDRVKLTDFNFLMV 344 |||||| |+ |+|||||||||||| | |#||+||+ |+ || |+|||+| ||||| + Sbjct 285 NVPIPEVDDV-NLELRQKFEKAKLGQ-GKK#VITPSDHRRFSLPSGNMDRVRLNDFNFLAL 342 Query 345 LG 346 || Query 345 LG 346 |
| Danio rerio | Query 287 NVPIPEGDEEGNMELRQKFEKAKLGPAGNK#VISPSEDRKQPSN-NLDRVKLTDFNFLMVL 345 |||+| |||| |||||||+||+||+ # | + | || | ||+||+|||||||| Sbjct 286 NVPVPPEGEEGNEELRQKFERAKIGPSKTD#GSSSNAISKFDSNGNRDRMKLSDFNFLMVL 345 Query 346 G 346 | Query 346 G 346 |
| Scyliorhinus canicula | Query 287 NVPIPEGDEEGNMELRQKFEKAKLGPAGNK#VISPSEDRKQPSNNLDRVKLTDFNFLMVLG 346 |||| | ||+ |||+||||||||||| |# | + |+|+|| | | ||||||||| Sbjct 220 NVPIIEDDEDS--ELRKKFEKAKLGPAGKK#AI--EKKPSSPTNDLDHVHLEDFNFLMVLG 275 |
| Tetraodon nigroviridis | Query 287 NVPIPEGDEEGNMELRQKFEKAKLGPAGNK--#VISPSEDRKQPSNNLDRVKLTDFNFLMV 344 |||+| |||| |||||||+||+|| | # + + | + | ||+|| ||||||| Sbjct 223 NVPVPPEGEEGNEELRQKFERAKIGPGKNTDG#KSANAASRFDSNGNQDRMKLADFNFLMV 282 Query 345 LG 346 || Query 345 LG 346 |
| Macaca mulatta | Query 287 NVPIPEGDEEGNMELRQKFEKAKLGPAGNK#V----ISPSEDRKQPSNNLDRVKLTDFNFL 342 |||+| ||| |||||||+||+ | |#| + + + + | ||+|||||||| Sbjct 287 NVPVPPEGSEGNEELRQKFERAKIS-QGTK#VPEEKTTNTVSKFDNNGNRDRMKLTDFNFL 345 Query 343 MVLG 346 |||| Query 343 MVLG 346 |
| Petromyzon marinus | Query 287 NVPIPEGDEEGNMELRQKFEKAKLGPAGNK#VISP--SEDRKQPS-------NNLDRVKLT 337 |||| |+|+ |+|+|||||+|||+ # + | |++|+ | |+|||| Sbjct 217 NVPIAPDIEDGDTEMRRKFEKARLGPS---#-VKPRASDERRANSLSAILNNANVDRVKAD 272 Query 338 DFNFLMVLG 346 ||||||||| Query 338 DFNFLMVLG 346 |
| Myxine glutinosa | Query 287 NVPI-PEGDEEGNMELRQKFEKAKLGPAGNK#VISPSEDRKQPS---NNLDRVKLTDFNFL 342 |||+ |||+| +||||+ +++++ + # ||++|+ | + |||| | || || Sbjct 208 NVPVAPEGEE--GLELRQRLQRSQIARSDKN#KQSPAKDQPSDSASLSGLDRVNLEDFVFL 265 Query 343 MVLG 346 ||| Sbjct 266 TVLG 269 |
| Macaca fascicularis | Query 287 NVPIPEGDEEGNMELRQKFE----------KAKLGPAGNK#VISPSEDRKQPSN-----NL 331 |||+ + | | | |||| + ++||+ + #+ ||| | + Sbjct 287 NVPVADAD---NCSLLQKFEACNYPLELYERVRMGPSSSP#IPSPSPSPTDPKRCFFGASP 343 Query 332 DRVKLTDFNFLMVLG 346 |+ ++||+|||||| Sbjct 344 GRLHISDFSFLMVLG 358 |
| Branchiostoma lanceolatum | Query 287 NVPIPEGDEEGNMELRQKFEKAKLGPAGNK#VISPSEDRKQPSNNLDRVKLTDFNFLMVLG 346 |||| + +|| +|++ +| || +# ||+ | || +| ||||||||| Sbjct 218 NVPIAD-EEENVAKLKEHLQKQKLDEQRKQ#KSKRSEECNIGS--LDHMKAADFNFLMVLG 274 |
| Bombyx mori | Query 287 NVPIPEGDEEGNMELRQKFEKAKLGPAGNK#VISPSEDRKQPSN--NLDRVKLTDFNFLMV 344 |||+|| || +| | + + | +# | ||+ | | | ++ |||||+|| Sbjct 290 NVPVPE---EG-ADLVQLKNQMRATTVGAR#RPPPPPDREVPHNVAAADVIRATDFNFIMV 345 Query 345 LG 346 || Query 345 LG 346 |
| Apis mellifera | Query 287 NVPIPE-GDEEGNMELRQKFEKAKLGPAGNK#VISPSEDRKQPSN--NLDRVKLTDFNFLM 343 |||+|| | + ++++ +| + |# + ++|++ | | | ++ +|||||| Sbjct 304 NVPVPEEGVDLAELKMKPSPQKTSV----TK#KTTTTQDKEVPHNMGKSDLIRASDFNFLM 359 Query 344 VLG 346 ||| Query 344 VLG 346 |
| Anopheles gambiae str. PEST | Query 287 NVPIPEGDEEGNMELRQKFEKAKLGPAGNK#VISPSEDRKQPSN--NLDRVKLTDFNFLMV 344 |||+|| + ++|+ + | + |# | |+ | | | ++ |||||||| Sbjct 227 NVPVPEEGAD-LVQLKSQMRKTSI----TK#KIPMMGDKDVPHNMTKKDVIRATDFNFLMV 281 Query 345 LG 346 || Query 345 LG 346 |
| Aedes aegypti | Query 287 NVPIPEGDEEGNMELRQKFEKAKLGPAGNK#VISPSEDRKQPSNNLDRVKLTDFNFLMVLG 346 |||+|| || ++ | + | + # + +| | ++ |||||||||| Sbjct 235 NVPVPE---EGTDLVQLKSQMRKTSISKKA#PVLCDKDVPHNMGKKDVIRATDFNFLMVLG 291 |
| Drosophila melanogaster | Query 287 NVPIPEGDEEGNMELRQKFEKAKLGPAGNK#VISPSEDRKQPSNNLDRVKLTDFNFLMVLG 346 ||| + ||+ ++|+|| + | #++ |+ |+ | ++ |||||+ ||| Sbjct 296 NVPCAD-DEQDLLKLKQKPSQKK------P#MVMRSDTNTHTSSKKDMIRATDFNFIKVLG 348 |
| Aplysia californica | Query 288 VPIPEGDEEGNMELRQKFEKAKLGPAGNK#VISPSEDRKQPSNNLDRVKLTDFNFLMVLG 346 ||+ + | |++ | ++ + ++# | |+ | + | |+ +||||| ||| Sbjct 272 VPVTDDITESIQEIKSKMHRSSIS---SE#KRYPEPDKVQNMSKQDIVRASDFNFLTVLG 327 |
[Site 2] ELRQKFEKAK309-LGPAGNKVIS
Lys309  Leu
Leu
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Glu300 | Leu301 | Arg302 | Gln303 | Lys304 | Phe305 | Glu306 | Lys307 | Ala308 | Lys309 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Leu310 | Gly311 | Pro312 | Ala313 | Gly314 | Asn315 | Lys316 | Val317 | Ile318 | Ser319 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| QEEGEYYNVPIPEGDEEGNMELRQKFEKAKLGPAGNKVISPSEDRKQPSNNLDRVKLTDF |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | N/A | 126.00 | 13 | - |
| 2 | Homo sapiens | 126.00 | 11 | aging-associated gene 6 protein |
| 3 | Mus musculus | 126.00 | 9 | protein kinase C |
| 4 | synthetic construct | 126.00 | 1 | protein kinase C alpha |
| 5 | Rattus norvegicus | 124.00 | 5 | PREDICTED: similar to Protein kinase C alpha type |
| 6 | Oryctolagus cuniculus | 123.00 | 5 | KPCA_RABIT Protein kinase C alpha type (PKC-alpha) |
| 7 | Canis familiaris | 123.00 | 4 | PREDICTED: similar to Protein kinase C, alpha type |
| 8 | Bos taurus | 123.00 | 4 | protein kinase C, alpha |
| 9 | Monodelphis domestica | 112.00 | 4 | PREDICTED: similar to protein kinase C |
| 10 | Gallus gallus | 109.00 | 3 | protein kinase C, alpha |
| 11 | Xenopus tropicalis | 101.00 | 1 | hypothetical protein LOC780209 |
| 12 | Latimeria chalumnae | 99.00 | 2 | protein kinase C alpha |
| 13 | Xenopus laevis | 95.50 | 1 | protein kinase C, alpha |
| 14 | Protopterus dolloi | 87.80 | 2 | protein kinase C alpha |
| 15 | Takifugu rubripes | 79.30 | 1 | protein kinase C, alpha type |
| 16 | Scyliorhinus canicula | 70.50 | 2 | protein kinase C alpha |
| 17 | Danio rerio | 69.70 | 7 | protein kinase C, beta 1 |
| 18 | Tetraodon nigroviridis | 67.80 | 3 | unnamed protein product |
| 19 | Macaca mulatta | 60.80 | 9 | PREDICTED: protein kinase C, beta isoform 1 |
| 20 | Petromyzon marinus | 58.90 | 1 | protein kinase C |
| 21 | Myxine glutinosa | 53.50 | 1 | protein kinase C |
| 22 | Macaca fascicularis | 42.70 | 1 | KPCG_MACFA Protein kinase C gamma type (PKC-gamma) |
| 23 | Branchiostoma lanceolatum | 40.00 | 1 | protein kinase C |
| 24 | Apis mellifera | 38.10 | 1 | PREDICTED: similar to Protein C kinase 53E CG6622- |
| 25 | Bombyx mori | 38.10 | 1 | conventional protein kinase C |
| 26 | Drosophila melanogaster | 37.70 | 4 | Protein C kinase 53E CG6622-PA, isoform A |
| 27 | Anopheles gambiae str. PEST | 37.00 | 1 | ENSANGP00000009078 |
| 28 | Aedes aegypti | 36.60 | 1 | protein kinase c |
| 29 | Aplysia californica | 36.20 | 1 | KPC1_APLCA Calcium-dependent protein kinase C (APL |
| 30 | Drosophila pseudoobscura | 33.90 | 1 | GA19732-PA |
Top-ranked sequences
| organism | matching |
|---|---|
| N/A | Query 280 QEEGEYYNVPIPEGDEEGNMELRQKFEKAK#LGPAGNKVISPSEDRKQPSNNLDRVKLTDF 339 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 280 QEEGEYYNVPIPEGDEEGNMELRQKFEKAK#LGPAGNKVISPSEDRKQPSNNLDRVKLTDF 339 |
| Homo sapiens | Query 280 QEEGEYYNVPIPEGDEEGNMELRQKFEKAK#LGPAGNKVISPSEDRKQPSNNLDRVKLTDF 339 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 280 QEEGEYYNVPIPEGDEEGNMELRQKFEKAK#LGPAGNKVISPSEDRKQPSNNLDRVKLTDF 339 |
| Mus musculus | Query 280 QEEGEYYNVPIPEGDEEGNMELRQKFEKAK#LGPAGNKVISPSEDRKQPSNNLDRVKLTDF 339 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 280 QEEGEYYNVPIPEGDEEGNMELRQKFEKAK#LGPAGNKVISPSEDRKQPSNNLDRVKLTDF 339 |
| synthetic construct | Query 280 QEEGEYYNVPIPEGDEEGNMELRQKFEKAK#LGPAGNKVISPSEDRKQPSNNLDRVKLTDF 339 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 280 QEEGEYYNVPIPEGDEEGNMELRQKFEKAK#LGPAGNKVISPSEDRKQPSNNLDRVKLTDF 339 |
| Rattus norvegicus | Query 280 QEEGEYYNVPIPEGDEEGNMELRQKFEKAK#LGPAGNKVISPSEDRKQPSNNLDRVKLTDF 339 |||||||||||||||||||+||||||||||#|||||||||||||||||||||||||||||| Sbjct 280 QEEGEYYNVPIPEGDEEGNVELRQKFEKAK#LGPAGNKVISPSEDRKQPSNNLDRVKLTDF 339 |
| Oryctolagus cuniculus | Query 280 QEEGEYYNVPIPEGDEEGNMELRQKFEKAK#LGPAGNKVISPSEDRKQPSNNLDRVKLTDF 339 ||||||||||||||||+||+||||||||||#|||||||||||||||||||||||||||||| Sbjct 280 QEEGEYYNVPIPEGDEDGNVELRQKFEKAK#LGPAGNKVISPSEDRKQPSNNLDRVKLTDF 339 |
| Canis familiaris | Query 280 QEEGEYYNVPIPEGDEEGNMELRQKFEKAK#LGPAGNKVISPSEDRKQPSNNLDRVKLTDF 339 |||||||||||||||||||+||||||||||#|||||||||||||||+|||||||||||||| Sbjct 198 QEEGEYYNVPIPEGDEEGNVELRQKFEKAK#LGPAGNKVISPSEDRRQPSNNLDRVKLTDF 257 |
| Bos taurus | Query 280 QEEGEYYNVPIPEGDEEGNMELRQKFEKAK#LGPAGNKVISPSEDRKQPSNNLDRVKLTDF 339 |||||||||||||||||||+||||||||||#|||||||||||||||+|||||||||||||| Sbjct 280 QEEGEYYNVPIPEGDEEGNVELRQKFEKAK#LGPAGNKVISPSEDRRQPSNNLDRVKLTDF 339 |
| Monodelphis domestica | Query 280 QEEGEYYNVPIPEGDEEGNMELRQKFEKAK#LGPAGNKVISPSEDRKQPSNNLDRVKLTDF 339 ||||||||||||||||||||||||||||||#||| |||||| |+||| |+||||||||||| Sbjct 280 QEEGEYYNVPIPEGDEEGNMELRQKFEKAK#LGPVGNKVISSSDDRK-PTNNLDRVKLTDF 338 |
| Gallus gallus | Query 280 QEEGEYYNVPIPEGDEEGNMELRQKFEKAK#LGPAGNKVISPSEDRKQ--PSNNLDRVKLT 337 ||||||||||||+ ||+|| ||||||||||#|||||||||+||||| ||||||||||| Sbjct 280 QEEGEYYNVPIPDADEDGNAELRQKFEKAK#LGPAGNKVITPSEDRNSSVPSNNLDRVKLT 339 Query 338 DF 339 || Query 338 DF 339 |
| Xenopus tropicalis | Query 280 QEEGEYYNVPIPEGDEEGNMELRQKFEKAK#LGPAGNKVISPSEDRKQ--PSNNLDRVKLT 337 ||||||||||||| +++||+||||||||||#|||||||||||+++|+ ||||+| |+|| Sbjct 284 QEEGEYYNVPIPEEEDDGNIELRQKFEKAK#LGPAGNKVISPTDERRPYVPSNNIDSVRLT 343 Query 338 DF 339 || Query 338 DF 339 |
| Latimeria chalumnae | Query 280 QEEGEYYNVPIPEGDEEGNMELRQKFEKAK#LGPAGNKVISPSEDRKQ--PSNNLDRVKLT 337 ||||||||||| | |+ ||+||||||||||#|||||+||||| |+|| ||||||||||| Sbjct 202 QEEGEYYNVPISEADD-GNVELRQKFEKAK#LGPAGSKVISPVEERKSSTPSNNLDRVKLT 260 Query 338 DF 339 || Query 338 DF 339 |
| Xenopus laevis | Query 280 QEEGEYYNVPIPEGDEEGNMELRQKFEKAK#LGPAGNKVISPSEDRKQ--PSNNLDRVKLT 337 ||||||||||||| |+ ||+||||||||||#|||||||||+|+ +|+ ||||+| ++|| Sbjct 284 QEEGEYYNVPIPEADD-GNLELRQKFEKAK#LGPAGNKVINPTGERRPYIPSNNIDSIRLT 342 Query 338 DF 339 || Query 338 DF 339 |
| Protopterus dolloi | Query 280 QEEGEYYNVPIPEGDEEGNMELRQKFEKAK#LGPAGNKVISPSEDRKQPSNNLDRVKLTDF 339 ||||||||||||+ |+ ||+ ||||||||+#||||| ||||| | ||||| || || Sbjct 200 QEEGEYYNVPIPDADD-GNVGLRQKFEKAR#LGPAGKKVISPERKSSLPLNNLDRFKLEDF 258 |
| Takifugu rubripes | Query 280 QEEGEYYNVPIPEGDEEGNMELRQKFEKAK#LGPAGNKVISPSEDRK--QPSNNLDRVKLT 337 ||||||||||||| |+ |+||||||||||#|| | |||+||+ |+ || |+|||+| Sbjct 278 QEEGEYYNVPIPEVDDV-NLELRQKFEKAK#LG-QGKKVITPSDHRRFSLPSGNMDRVRLN 335 Query 338 DF 339 || Query 338 DF 339 |
| Scyliorhinus canicula | Query 280 QEEGEYYNVPIPEGDEEGNMELRQKFEKAK#LGPAGNKVISPSEDRKQPSNNLDRVKLTDF 339 ||||||||||| | ||+ |||+||||||#||||| | | + |+|+|| | | || Sbjct 213 QEEGEYYNVPIIEDDEDS--ELRKKFEKAK#LGPAGKKAI--EKKPSSPTNDLDHVHLEDF 268 |
| Danio rerio | Query 280 QEEGEYYNVPIPEGDEEGNMELRQKFEKAK#LGPAGNKVISPSEDRKQPSN-NLDRVKLTD 338 ||||||+|||+| |||| |||||||+||#+||+ | + | || | ||+||+| Sbjct 279 QEEGEYFNVPVPPEGEEGNEELRQKFERAK#IGPSKTDGSSSNAISKFDSNGNRDRMKLSD 338 Query 339 F 339 | Query 339 F 339 |
| Tetraodon nigroviridis | Query 280 QEEGEYYNVPIPEGDEEGNMELRQKFEKAK#LGPAGNK--VISPSEDRKQPSNNLDRVKLT 337 ||||||+|||+| |||| |||||||+||#+|| | + + | + | ||+|| Sbjct 216 QEEGEYFNVPVPPEGEEGNEELRQKFERAK#IGPGKNTDGKSANAASRFDSNGNQDRMKLA 275 Query 338 DF 339 || Query 338 DF 339 |
| Macaca mulatta | Query 280 QEEGEYYNVPIPEGDEEGNMELRQKFEKAK#LGPAGNKV----ISPSEDRKQPSNNLDRVK 335 ||||||+|||+| ||| |||||||+||#+ | || + + + + | ||+| Sbjct 280 QEEGEYFNVPVPPEGSEGNEELRQKFERAK#IS-QGTKVPEEKTTNTVSKFDNNGNRDRMK 338 Query 336 LTDF 339 |||| Query 336 LTDF 339 |
| Petromyzon marinus | Query 280 QEEGEYYNVPIPEGDEEGNMELRQKFEKAK#LGPAGNKVISP--SEDRKQPS-------NN 330 ||||||||||| |+|+ |+|+|||||+#|||+ + | |++|+ | | Sbjct 210 QEEGEYYNVPIAPDIEDGDTEMRRKFEKAR#LGPS----VKPRASDERRANSLSAILNNAN 265 Query 331 LDRVKLTDF 339 +|||| || Sbjct 266 VDRVKADDF 274 |
| Myxine glutinosa | Query 280 QEEGEYYNVPI-PEGDEEGNMELRQKFEKAK#LGPAGNKVISPSEDRKQPS---NNLDRVK 335 ||||||||||+ |||+| +||||+ ++++#+ + ||++|+ | + |||| Sbjct 201 QEEGEYYNVPVAPEGEE--GLELRQRLQRSQ#IARSDKNKQSPAKDQPSDSASLSGLDRVN 258 Query 336 LTDF 339 | || Sbjct 259 LEDF 262 |
| Macaca fascicularis | Query 280 QEEGEYYNVPIPEGDEEGNMELRQKF----------EKAK#LGPAGNKVISPSEDRKQPSN 329 ||||||||||+ + | | | ||| |+ +#+||+ + + ||| | Sbjct 280 QEEGEYYNVPVADAD---NCSLLQKFEACNYPLELYERVR#MGPSSSPIPSPSPSPTDPKR 336 Query 330 -----NLDRVKLTDF 339 + |+ ++|| Sbjct 337 CFFGASPGRLHISDF 351 |
| Branchiostoma lanceolatum | Query 280 QEEGEYYNVPIPEGDEEGNMELRQKFEKAK#LGPAGNKVISPSEDRKQPSNNLDRVKLTDF 339 ||||||||||| + +|| +|++ +| |#| + ||+ +|| +| || Sbjct 211 QEEGEYYNVPIAD-EEENVAKLKEHLQKQK#LDEQRKQKSKRSEECN--IGSLDHMKAADF 267 |
| Apis mellifera | Query 280 QEEGEYYNVPIP-EGDEEGNMELRQKFEKAK#LGPAGNKVISPSEDRKQPSN--NLDRVKL 336 ||||||||||+| || + ++++ +| #+ | + ++|++ | | | ++ Sbjct 297 QEEGEYYNVPVPEEGVDLAELKMKPSPQKTS#V----TKKTTTTQDKEVPHNMGKSDLIRA 352 Query 337 TDF 339 +|| Sbjct 353 SDF 355 |
| Bombyx mori | Query 280 QEEGEYYNVPIPEGDEEGNMELRQKFEKAK#LGPAGNKVISPSEDRKQPSN--NLDRVKLT 337 |||||+||||+| ||| +| | + +# | + | ||+ | | | ++ | Sbjct 283 QEEGEFYNVPVP---EEG-ADLVQLKNQMR#ATTVGARRPPPPPDREVPHNVAAADVIRAT 338 Query 338 DF 339 || Query 338 DF 339 |
| Drosophila melanogaster | Query 280 QEEGEYYNVPIPEGDEEGNMELRQKFEKAK#LGPAGNKVISPSEDRKQPSNNLDRVKLTDF 339 |+|||||||| + ||+ ++|+|| + |# ++ |+ |+ | ++ ||| Sbjct 289 QDEGEYYNVPCAD-DEQDLLKLKQKPSQKK#------PMVMRSDTNTHTSSKKDMIRATDF 341 |
| Anopheles gambiae str. PEST | Query 280 QEEGEYYNVPIPEGDEEGNMELRQKFEKAK#LGPAGNKVISPSEDRKQPSN--NLDRVKLT 337 ||||||||||+|| + ++|+ + | #+ | | |+ | | | ++ | Sbjct 220 QEEGEYYNVPVPE-EGADLVQLKSQMRKTS#I----TKKIPMMGDKDVPHNMTKKDVIRAT 274 Query 338 DF 339 || Query 338 DF 339 |
| Aedes aegypti | Query 280 QEEGEYYNVPIPEGDEEGNMELRQKFEKAK#LGPAGNKVISPSEDRKQPSNNLDRVKLTDF 339 ||||||||||+| ||| ++ | + |# + + +| | ++ ||| Sbjct 228 QEEGEYYNVPVP---EEGTDLVQLKSQMRK#TSISKKAPVLCDKDVPHNMGKKDVIRATDF 284 |
| Aplysia californica | Query 280 QEEGEYYNVPIPEGDEEGNMELRQKFEKAK#LGPAGNKVISPSEDRKQPSNNLDRVKLTDF 339 |||||+| ||+ + | |++ | ++ #+ ++ | |+ | + | |+ +|| Sbjct 264 QEEGEFYGVPVTDDITESIQEIKSKMHRSS#I---SSEKRYPEPDKVQNMSKQDIVRASDF 320 |
| Drosophila pseudoobscura | Query 280 QEEGEYYNVPIPEGDEEGNMELRQKFEKAK#LGPAGNKVISPSEDRKQPS--NNLDRVKLT 337 |+|||||||| + ||+ ++|+|| # |+ | + | | + | ++ | Sbjct 175 QDEGEYYNVPCAD-DEQDLLKLKQK-----#--PSQKKPLVMRSDTNTHSSISKKDMIRAT 226 Query 338 DF 339 || Query 338 DF 339 |
[Site 3] NKVISPSEDR324-KQPSNNLDRV
Arg324  Lys
Lys
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Asn315 | Lys316 | Val317 | Ile318 | Ser319 | Pro320 | Ser321 | Glu322 | Asp323 | Arg324 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Lys325 | Gln326 | Pro327 | Ser328 | Asn329 | Asn330 | Leu331 | Asp332 | Arg333 | Val334 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| EEGNMELRQKFEKAKLGPAGNKVISPSEDRKQPSNNLDRVKLTDFNFLMVLGKGSFGKVM |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | Homo sapiens | 122.00 | 24 | aging-associated gene 6 protein |
| 2 | N/A | 122.00 | 17 | - |
| 3 | Mus musculus | 122.00 | 11 | protein kinase C |
| 4 | synthetic construct | 122.00 | 4 | protein kinase C alpha |
| 5 | Rattus norvegicus | 121.00 | 8 | PREDICTED: similar to Protein kinase C alpha type |
| 6 | Canis familiaris | 120.00 | 13 | PREDICTED: similar to Protein kinase C, alpha type |
| 7 | Bos taurus | 120.00 | 7 | protein kinase C, alpha |
| 8 | Oryctolagus cuniculus | 120.00 | 6 | KPCA_RABIT Protein kinase C alpha type (PKC-alpha) |
| 9 | Gallus gallus | 109.00 | 7 | protein kinase C, alpha |
| 10 | Monodelphis domestica | 109.00 | 6 | PREDICTED: similar to protein kinase C |
| 11 | Latimeria chalumnae | 106.00 | 2 | protein kinase C alpha |
| 12 | Xenopus tropicalis | 100.00 | 3 | hypothetical protein LOC780209 |
| 13 | Xenopus laevis | 96.30 | 2 | protein kinase C, alpha |
| 14 | Protopterus dolloi | 92.00 | 2 | protein kinase C alpha |
| 15 | Takifugu rubripes | 80.90 | 1 | protein kinase C, alpha type |
| 16 | Scyliorhinus canicula | 76.60 | 2 | protein kinase C alpha |
| 17 | Danio rerio | 76.30 | 14 | protein kinase C, beta 1 |
| 18 | Tetraodon nigroviridis | 74.30 | 5 | unnamed protein product |
| 19 | Macaca mulatta | 67.40 | 15 | PREDICTED: protein kinase C, beta isoform 1 |
| 20 | Petromyzon marinus | 67.00 | 1 | protein kinase C |
| 21 | Myxine glutinosa | 55.50 | 1 | protein kinase C |
| 22 | Macaca fascicularis | 50.10 | 4 | KPCG_MACFA Protein kinase C gamma type (PKC-gamma) |
| 23 | Branchiostoma lanceolatum | 49.30 | 1 | protein kinase C |
| 24 | Bombyx mori | 47.80 | 1 | conventional protein kinase C |
| 25 | Aplysia californica | 47.00 | 2 | KPC2_APLCA Calcium-independent protein kinase C (A |
| 26 | Apis mellifera | 46.20 | 2 | PREDICTED: similar to Protein C kinase 53E CG6622- |
| 27 | Anopheles gambiae str. PEST | 45.40 | 3 | ENSANGP00000009078 |
| 28 | Aedes aegypti | 45.40 | 2 | protein kinase c |
| 29 | Drosophila melanogaster | 44.30 | 8 | Protein C kinase 53E CG6622-PA, isoform A |
| 30 | Tribolium castaneum | 42.00 | 3 | PREDICTED: similar to CG6622-PA, isoform A |
| 31 | Drosophila pseudoobscura | 41.20 | 3 | GA19732-PA |
| 32 | Caenorhabditis briggsae | 41.20 | 1 | Hypothetical protein CBG16150 |
| 33 | Coprinopsis cinerea okayama7#130 | 41.20 | 1 | hypothetical protein CC1G_07190 |
| 34 | Magnaporthe grisea 70-15 | 41.20 | 1 | hypothetical protein MGG_08689 |
| 35 | Magnaporthe grisea | 40.80 | 1 | AF136600_1 protein kinase C |
| 36 | Caenorhabditis elegans | 40.40 | 5 | protein kinase C2 A isoform |
| 37 | Strongylocentrotus purpuratus | 40.40 | 4 | PREDICTED: similar to protein kinase C |
| 38 | Pan troglodytes | 39.70 | 4 | PREDICTED: serum/glucocorticoid regulated kinase 3 |
| 39 | Botryotinia fuckeliana | 39.70 | 1 | protein kinase C |
| 40 | Paramecium tetraurelia | 39.70 | 1 | hypothetical protein |
| 41 | Schizosaccharomyces pombe | 39.30 | 3 | SPAC17G8.14c |
| 42 | Schizosaccharomyces pombe 972h- | 39.30 | 2 | hypothetical protein SPAC17G8.14c |
| 43 | Hydractinia echinata | 39.30 | 1 | putative calcium dependent protein kinase C |
| 44 | Leptosphaeria maculans | 38.90 | 1 | AF487263_9 protein kinase C |
| 45 | Yarrowia lipolytica | 38.90 | 1 | hypothetical protein |
| 46 | Tuber excavatum | 38.50 | 2 | protein kinase C-like protein |
| 47 | Suberites domuncula | 38.50 | 2 | Serine/Threonine protein kinase |
| 48 | Calliphora vicina | 38.50 | 1 | eye-specific protein kinase C |
| 49 | Asparagus officinalis | 38.50 | 1 | S6 ribosomal protein kinase |
| 50 | Tuber aestivum | 38.10 | 2 | protein kinase C-like protein |
| 51 | Sycon raphanus | 38.10 | 1 | Serine/Threonine protein kinase |
| 52 | Cryptococcus neoformans var. neoformans JEC21 | 37.70 | 1 | protein kinase C |
| 53 | Cryptococcus neoformans var. neoformans | 37.70 | 1 | protein kinase C 1 |
| 54 | Squalus acanthias | 37.70 | 1 | s-sgk1 |
| 55 | Cryptococcus neoformans var. neoformans B-3501A | 37.70 | 1 | hypothetical protein CNBC3910 |
| 56 | Cryptococcus neoformans var. grubii | 37.70 | 1 | protein kinase C 1 |
| 57 | Tuber indicum | 37.00 | 6 | protein kinase C-like protein |
| 58 | Phaeosphaeria nodorum SN15 | 37.00 | 2 | hypothetical protein SNOG_13609 |
| 59 | Tuber pseudoexcavatum | 37.00 | 2 | protein kinase C-like protein |
| 60 | Leishmania braziliensis | 37.00 | 1 | zinc finger protein kinase-like |
| 61 | Aspergillus clavatus NRRL 1 | 36.60 | 1 | protein kinase c |
| 62 | Debaryomyces hansenii CBS767 | 36.60 | 1 | hypothetical protein DEHA0G14773g |
| 63 | Cochliobolus heterostrophus | 36.60 | 1 | KPC1_COCHE Protein kinase C-like emb |
| 64 | Plasmodium berghei strain ANKA | 36.60 | 1 | rac-beta serine/threonine protein kinase |
| 65 | Ashbya gossypii ATCC 10895 | 36.60 | 1 | ACR191Cp |
| 66 | Aspergillus oryzae | 36.60 | 1 | unnamed protein product |
| 67 | Candida albicans | 36.60 | 1 | KPC1_CANAL Protein kinase C-like 1 (PKC 1) emb |
| 68 | Sporothrix schenckii | 36.60 | 1 | AF124792_1 protein kinase C; serine/threonine prot |
| 69 | Hydra vulgaris | 36.20 | 2 | protein kinase C |
| 70 | Coccidioides immitis RS | 36.20 | 1 | hypothetical protein CIMG_09206 |
| 71 | Plasmodium yoelii yoelii str. 17XNL | 36.20 | 1 | hypothetical protein PY00403 |
| 72 | Neosartorya fischeri NRRL 181 | 36.20 | 1 | protein kinase c |
| 73 | Aspergillus terreus NIH2624 | 36.20 | 1 | hypothetical protein ATEG_02078 |
| 74 | Medicago truncatula | 36.20 | 1 | Protein kinase |
| 75 | Blumeria graminis | 35.80 | 1 | AF247001_1 protein kinase C |
| 76 | Fundulus heteroclitus | 35.80 | 1 | serum and glucocorticoid-regulated kinase |
Top-ranked sequences
| organism | matching |
|---|---|
| Homo sapiens | Query 295 EEGNMELRQKFEKAKLGPAGNKVISPSEDR#KQPSNNLDRVKLTDFNFLMVLGKGSFGKVM 354 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 295 EEGNMELRQKFEKAKLGPAGNKVISPSEDR#KQPSNNLDRVKLTDFNFLMVLGKGSFGKVM 354 |
| N/A | Query 295 EEGNMELRQKFEKAKLGPAGNKVISPSEDR#KQPSNNLDRVKLTDFNFLMVLGKGSFGKVM 354 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 295 EEGNMELRQKFEKAKLGPAGNKVISPSEDR#KQPSNNLDRVKLTDFNFLMVLGKGSFGKVM 354 |
| Mus musculus | Query 295 EEGNMELRQKFEKAKLGPAGNKVISPSEDR#KQPSNNLDRVKLTDFNFLMVLGKGSFGKVM 354 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 295 EEGNMELRQKFEKAKLGPAGNKVISPSEDR#KQPSNNLDRVKLTDFNFLMVLGKGSFGKVM 354 |
| synthetic construct | Query 295 EEGNMELRQKFEKAKLGPAGNKVISPSEDR#KQPSNNLDRVKLTDFNFLMVLGKGSFGKVM 354 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 295 EEGNMELRQKFEKAKLGPAGNKVISPSEDR#KQPSNNLDRVKLTDFNFLMVLGKGSFGKVM 354 |
| Rattus norvegicus | Query 295 EEGNMELRQKFEKAKLGPAGNKVISPSEDR#KQPSNNLDRVKLTDFNFLMVLGKGSFGKVM 354 ||||+|||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Sbjct 295 EEGNVELRQKFEKAKLGPAGNKVISPSEDR#KQPSNNLDRVKLTDFNFLMVLGKGSFGKVM 354 |
| Canis familiaris | Query 295 EEGNMELRQKFEKAKLGPAGNKVISPSEDR#KQPSNNLDRVKLTDFNFLMVLGKGSFGKVM 354 ||||+|||||||||||||||||||||||||#+||||||||||||||||||||||||||||| Sbjct 213 EEGNVELRQKFEKAKLGPAGNKVISPSEDR#RQPSNNLDRVKLTDFNFLMVLGKGSFGKVM 272 |
| Bos taurus | Query 295 EEGNMELRQKFEKAKLGPAGNKVISPSEDR#KQPSNNLDRVKLTDFNFLMVLGKGSFGKVM 354 ||||+|||||||||||||||||||||||||#+||||||||||||||||||||||||||||| Sbjct 295 EEGNVELRQKFEKAKLGPAGNKVISPSEDR#RQPSNNLDRVKLTDFNFLMVLGKGSFGKVM 354 |
| Oryctolagus cuniculus | Query 295 EEGNMELRQKFEKAKLGPAGNKVISPSEDR#KQPSNNLDRVKLTDFNFLMVLGKGSFGKVM 354 |+||+|||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Sbjct 295 EDGNVELRQKFEKAKLGPAGNKVISPSEDR#KQPSNNLDRVKLTDFNFLMVLGKGSFGKVM 354 |
| Gallus gallus | Query 295 EEGNMELRQKFEKAKLGPAGNKVISPSEDR#KQ--PSNNLDRVKLTDFNFLMVLGKGSFGK 352 |+|| |||||||||||||||||||+|||||# |||||||||||||||||||||||||| Sbjct 295 EDGNAELRQKFEKAKLGPAGNKVITPSEDR#NSSVPSNNLDRVKLTDFNFLMVLGKGSFGK 354 Query 353 VM 354 || Query 353 VM 354 |
| Monodelphis domestica | Query 295 EEGNMELRQKFEKAKLGPAGNKVISPSEDR#KQPSNNLDRVKLTDFNFLMVLGKGSFGKVM 354 |||||||||||||||||| |||||| |+||#| |+|||||||||||||||||||||||||| Sbjct 295 EEGNMELRQKFEKAKLGPVGNKVISSSDDR#K-PTNNLDRVKLTDFNFLMVLGKGSFGKVM 353 |
| Latimeria chalumnae | Query 295 EEGNMELRQKFEKAKLGPAGNKVISPSEDR#KQ--PSNNLDRVKLTDFNFLMVLGKGSFGK 352 ++||+|||||||||||||||+||||| |+|#| |||||||||||||||||||||||||| Sbjct 216 DDGNVELRQKFEKAKLGPAGSKVISPVEER#KSSTPSNNLDRVKLTDFNFLMVLGKGSFGK 275 Query 353 VM 354 || Query 353 VM 354 |
| Xenopus tropicalis | Query 295 EEGNMELRQKFEKAKLGPAGNKVISPSEDR#KQ--PSNNLDRVKLTDFNFLMVLGKGSFGK 352 ++||+|||||||||||||||||||||+++|#+ ||||+| |+||||+|||||||||||| Sbjct 299 DDGNIELRQKFEKAKLGPAGNKVISPTDER#RPYVPSNNIDSVRLTDFSFLMVLGKGSFGK 358 Query 353 VM 354 || Query 353 VM 354 |
| Xenopus laevis | Query 295 EEGNMELRQKFEKAKLGPAGNKVISPSEDR#KQ--PSNNLDRVKLTDFNFLMVLGKGSFGK 352 ++||+|||||||||||||||||||+|+ +|#+ ||||+| ++|||| |||||||||||| Sbjct 298 DDGNLELRQKFEKAKLGPAGNKVINPTGER#RPYIPSNNIDSIRLTDFCFLMVLGKGSFGK 357 Query 353 VM 354 || Query 353 VM 354 |
| Protopterus dolloi | Query 295 EEGNMELRQKFEKAKLGPAGNKVISPSEDR#KQPSNNLDRVKLTDFNFLMVLGKGSFGKVM 354 ++||+ ||||||||+||||| ||||| # | ||||| || ||||+|||||||||||| Sbjct 214 DDGNVGLRQKFEKARLGPAGKKVISPERKS#SLPLNNLDRFKLEDFNFVMVLGKGSFGKVM 273 |
| Takifugu rubripes | Query 295 EEGNMELRQKFEKAKLGPAGNKVISPSEDR#K--QPSNNLDRVKLTDFNFLMVLGKGSFGK 352 ++ |+|||||||||||| | |||+||+ |#+ || |+|||+| ||||| +|||||||| Sbjct 292 DDVNLELRQKFEKAKLGQ-GKKVITPSDHR#RFSLPSGNMDRVRLNDFNFLALLGKGSFGK 350 Query 353 VM 354 || Query 353 VM 354 |
| Scyliorhinus canicula | Query 295 EEGNMELRQKFEKAKLGPAGNKVISPSEDR#KQPSNNLDRVKLTDFNFLMVLGKGSFGKVM 354 ++ + |||+||||||||||| | | # |+|+|| | | ||||||||||||||||| Sbjct 226 DDEDSELRKKFEKAKLGPAGKKAIEKKPS-#-SPTNDLDHVHLEDFNFLMVLGKGSFGKVM 283 |
| Danio rerio | Query 295 EEGNMELRQKFEKAKLGPAGNKVISPSEDR#KQPSN-NLDRVKLTDFNFLMVLGKGSFGKV 353 |||| |||||||+||+||+ | + #| || | ||+||+|||||||||||||||| Sbjct 294 EEGNEELRQKFERAKIGPSKTDGSSSNAIS#KFDSNGNRDRMKLSDFNFLMVLGKGSFGKV 353 Query 354 M 354 | Query 354 M 354 |
| Tetraodon nigroviridis | Query 295 EEGNMELRQKFEKAKLGPAGNK--VISPSEDR#KQPSNNLDRVKLTDFNFLMVLGKGSFGK 352 |||| |||||||+||+|| | + + |# + | ||+|| ||||||||||||||| Sbjct 231 EEGNEELRQKFERAKIGPGKNTDGKSANAASR#FDSNGNQDRMKLADFNFLMVLGKGSFGK 290 Query 353 VM 354 || Query 353 VM 354 |
| Macaca mulatta | Query 296 EGNMELRQKFEKAKLGPAGNKVISPSE------DR#KQPSNNLDRVKLTDFNFLMVLGKGS 349 ||| |||||||+||+ | || | | +# + | ||+||||||||||||||| Sbjct 296 EGNEELRQKFERAKISQ-GTKV--PEEKTTNTVSK#FDNNGNRDRMKLTDFNFLMVLGKGS 352 Query 350 FGKVM 354 ||||| Query 350 FGKVM 354 |
| Petromyzon marinus | Query 295 EEGNMELRQKFEKAKLGPAGNKVISPSEDR#KQPSN---------NLDRVKLTDFNFLMVL 345 |+|+ |+|+|||||+|||+ + | #++ +| |+|||| |||||||| Sbjct 225 EDGDTEMRRKFEKARLGPS----VKPRASD#ERRANSLSAILNNANVDRVKADDFNFLMVL 280 Query 346 GKGSFGKVM 354 ||||||||| Query 346 GKGSFGKVM 354 |
| Myxine glutinosa | Query 295 EEGNMELRQKFEKAKLGPAGNKVISPSEDR#KQPS---NNLDRVKLTDFNFLMVLGKGSFG 351 ||| +||||+ +++++ + ||++|+# | + |||| | || || |||||||| Sbjct 216 EEG-LELRQRLQRSQIARSDKNKQSPAKDQ#PSDSASLSGLDRVNLEDFVFLTVLGKGSFG 274 Query 352 KVM 354 ||| Query 352 KVM 354 |
| Macaca fascicularis | Query 305 FEKAKLGPAGNKVISPSEDR#KQPSN-----NLDRVKLTDFNFLMVLGKGSFGKVM 354 +|+ ++||+ + + ||| # | + |+ ++||+|||||||||||||| Sbjct 312 YERVRMGPSSSPIPSPSPSP#TDPKRCFFGASPGRLHISDFSFLMVLGKGSFGKVM 366 |
| Branchiostoma lanceolatum | Query 295 EEGNMELRQKFEKAKLGPAGNKVISPSEDR#KQPSNNLDRVKLTDFNFLMVLGKGSFGKVM 354 || +|++ +| || + ||+ # | || +| ||||||||||||||||| Sbjct 225 EENVAKLKEHLQKQKLDEQRKQKSKRSEEC#NIGS--LDHMKAADFNFLMVLGKGSFGKVM 282 |
| Bombyx mori | Query 295 EEGNMELRQKFEKAKLGPAGNKVISPSEDR#KQPSN--NLDRVKLTDFNFLMVLGKGSFGK 352 ||| +| | + + | + | ||#+ | | | ++ |||||+|||||||||| Sbjct 295 EEG-ADLVQLKNQMRATTVGARRPPPPPDR#EVPHNVAAADVIRATDFNFIMVLGKGSFGK 353 Query 353 VM 354 || Query 353 VM 354 |
| Aplysia californica | Query 314 GNKVISPSEDR#KQPSNNLDRVKLTDFNFLMVLGKGSFGKVM 354 |++ ||| ||# + ++ |+ | ||||+ ||||||||||| Sbjct 380 GSRTRSPSSDR#SRSHHS--RISLHDFNFIKVLGKGSFGKVM 418 |
| Apis mellifera | Query 295 EEGNMELRQKFEKAKLGPAGNKVISPSEDR#KQPSN--NLDRVKLTDFNFLMVLGKGSFGK 352 ||| | + + + | + ++|+#+ | | | ++ +||||||||||||||| Sbjct 309 EEGVDLAELKMKPSPQKTSVTKKTTTTQDK#EVPHNMGKSDLIRASDFNFLMVLGKGSFGK 368 Query 353 VM 354 || Query 353 VM 354 |
| Anopheles gambiae str. PEST | Query 295 EEGN--MELRQKFEKAKLGPAGNKVISPSEDR#KQPSN--NLDRVKLTDFNFLMVLGKGSF 350 ||| ++|+ + | + | | |+# | | | ++ |||||||||||||| Sbjct 232 EEGADLVQLKSQMRKTSI----TKKIPMMGDK#DVPHNMTKKDVIRATDFNFLMVLGKGSF 287 Query 351 GKVM 354 |||| Query 351 GKVM 354 |
| Aedes aegypti | Query 295 EEGNMELRQKFEKAKLGPAGNKVISPSEDR#KQPSNNLDRVKLTDFNFLMVLGKGSFGKVM 354 ||| ++ | + | + + +| # | ++ |||||||||||||||||| Sbjct 240 EEGTDLVQLKSQMRKTSISKKAPVLCDKDV#PHNMGKKDVIRATDFNFLMVLGKGSFGKVM 299 |
| Drosophila melanogaster | Query 303 QKFEKAKLGPAGNK-VISPSEDR#KQPSNNLDRVKLTDFNFLMVLGKGSFGKVM 354 | | | |+ | ++ |+ # |+ | ++ |||||+ ||||||||||+ Sbjct 304 QDLLKLKQKPSQKKPMVMRSDTN#THTSSKKDMIRATDFNFIKVLGKGSFGKVL 356 |
| Tribolium castaneum | Query 324 R#KQPSNN--LDRVKLTDFNFLMVLGKGSFGKVM 354 +#++|| + | ++ +||||||||||||||||| Sbjct 320 Q#RRPSQHGQKDIIRASDFNFLMVLGKGSFGKVM 352 |
| Drosophila pseudoobscura | Query 303 QKFEKAKLGPAGNKVISPSEDR#KQPSN--NLDRVKLTDFNFLMVLGKGSFGKVM 354 | | | |+ | + | # |+ | ++ |||||+ ||||||||||+ Sbjct 190 QDLLKLKQKPSQKKPLVMRSDT#NTHSSISKKDMIRATDFNFIKVLGKGSFGKVL 243 |
| Caenorhabditis briggsae | Query 328 SNNLDRVKLTDFNFLMVLGKGSFGKVM 354 + | | +| +||||| ||||||||||+ Sbjct 404 NTNRDVIKASDFNFLTVLGKGSFGKVL 430 |
| Coprinopsis cinerea okayama7#130 | Query 306 EKAKLGPAGNKVISPSEDR#KQPSNNLDRVKLTDFNFLMVLGKGSFGKVM 354 ++ + | + + ++ +#+||| +| | ||||| |||||+||||| Sbjct 685 QQGMVPPQHQQQMIQAQPQ#RQPSKKR-KVGLDDFNFLAVLGKGNFGKVM 732 |
| Magnaporthe grisea 70-15 | Query 302 RQKFEKAKLGPAGNKVISPSEDR#KQPS----NNLDRVKLTDFNFLMVLGKGSFGKVM 354 +| |+ + | || | |# || |+ | |||| |||||+||||| Sbjct 816 QQHQEQQIISPTAGTVIPTSAKR#PLPSATDPGTGQRIGLDHFNFLAVLGKGNFGKVM 872 |
| Magnaporthe grisea | Query 302 RQKFEKAKLGPAGNKVISPSEDR#KQPS----NNLDRVKLTDFNFLMVLGKGSFGKVM 354 +| |+ + | || | |# || |+ | |||| |||||+||||| Sbjct 816 QQHQEQQIISPTAGTVIPTSARR#PLPSATDPGTGQRIGLDHFNFLAVLGKGNFGKVM 872 |
| Caenorhabditis elegans | Query 328 SNNLDRVKLTDFNFLMVLGKGSFGKVM 354 |+| + +| +||||| ||||||||||+ Sbjct 337 SSNHNVIKASDFNFLTVLGKGSFGKVL 363 |
| Strongylocentrotus purpuratus | Query 321 SEDR#KQPSNNLDRVKLTDFNFLMVLGKGSFGKVM 354 | # ++++ |+ +||||| ||||||||||| Sbjct 307 SSQN#SNSNSSMGLVRASDFNFLSVLGKGSFGKVM 340 |
| Pan troglodytes | Query 295 EEGNMELRQKFEKAKLGPAGNKVISPSEDR#KQPSNNLDRVKLTDFNFLMVLGKGSFGKVM 354 | + +| + |||+|| | # |||+|| |+||||||||+ Sbjct 134 ERSSQKLHSTSQNINLGPSGNPHAKP----#------------TDFDFLKVIGKGSFGKVL 177 |
| Botryotinia fuckeliana | Query 312 PAGNKVISPSEDR#KQPSNNLD-----RVKLTDFNFLMVLGKGSFGKVM 354 | +|+ + |#|| | |+ | |||| |||||+||||| Sbjct 813 PGSQQVVPQGQQR#KQVPAANDAGTGRRIGLDHFNFLAVLGKGNFGKVM 860 |
| Paramecium tetraurelia | Query 295 EEGNMELRQKFEKAKLGPAGNKVISPSEDR#KQPSNNLDRVKLTDFNFLMVLGKGSFGKVM 354 +|| ++ +| || ++ | # | |||||| | |||+||||||| Sbjct 32 KEGEQQMNDEFMKAPTATF-EQIEETGEQV#FVDDNREAIVKLTDFQFEKVLGRGSFGKVM 90 |
| Schizosaccharomyces pombe | Query 320 PSEDR#KQPSNNLDRVKLTDFNFLMVLGKGSFGKVM 354 | |#++ | || | || || |||||+||||| Sbjct 556 PESPR#REKKN---RVTLDDFTFLAVLGKGNFGKVM 587 |
| Schizosaccharomyces pombe 972h- | Query 320 PSEDR#KQPSNNLDRVKLTDFNFLMVLGKGSFGKVM 354 | |#++ | || | || || |||||+||||| Sbjct 648 PESPR#REKKN---RVTLDDFTFLAVLGKGNFGKVM 679 |
| Hydractinia echinata | Query 332 DRVKLTDFNFLMVLGKGSFGKVM 354 +|+|| || || |+||||||||| Sbjct 191 ERLKLEDFTFLKVIGKGSFGKVM 213 |
| Leptosphaeria maculans | Query 299 MELRQKFEKAKLGPAGNKVISPSEDR#KQPSNNLD----RVKLTDFNFLMVLGKGSFGKVM 354 | +| |++ |+ + |++# || | |+ | |||| |||||+||||| Sbjct 816 MSQKQPEAIAQIPPSKQLPPANPENK#VTPSANTQGSGKRIGLDHFNFLAVLGKGNFGKVM 875 |
| Yarrowia lipolytica | Query 308 AKLGPAGNKVISPSEDR#KQPSNNLDRVKLTDFNFLMVLGKGSFGKVM 354 | || |+ | +#|+| |+ | ||||| |||||+||||| Sbjct 900 ASPAPASNRH-SVVGKK#KRP-----RIGLDDFNFLAVLGKGNFGKVM 940 |
| Tuber excavatum | Query 320 PSEDR#KQPSNNLDRVKLTDFNFLMVLGKGSFGKVM 354 |+ + #+|| ++ | |||| |||||+||||| Sbjct 312 PAPEP#QQPVIRPAKIGLDHFNFLAVLGKGNFGKVM 346 |
| Suberites domuncula | Query 295 EEGNMELRQKFEKAKLGPAGNKVISPSEDR#KQPSN------NLDRVKLTDFNFLMVLGKG 348 +| +| ++| || + +++ #++ | ++ ++|| ||| |+||||| Sbjct 293 DEEATKLIEEFRKATILDKAKNLVTDVPIM#RRGSELVPGAKDMPKMKLEDFNLLVVLGKG 352 Query 349 SFGKV 353 ||||| Query 349 SFGKV 353 |
| Calliphora vicina | Query 320 PSEDR#KQPSNNL---DRVKLTDFNFLMVLGKGSFGKVM 354 | | #| +|+ | ++ ||||+ ||||||+||++ Sbjct 333 PRIDN#KDMPHNMSKRDMIRAADFNFIKVLGKGSYGKIL 370 |
| Asparagus officinalis | Query 321 SEDR#KQPSN-NLDRVKLTDFNFLMVLGKGSFGKV 353 +|+ #+ ||+ |++|| | ||+|| |+|+|+|||| Sbjct 106 AEEH#EVPSSENVERVGLEDFDFLKVVGQGAFGKV 139 |
| Tuber aestivum | Query 320 PSEDR#KQPSNNLDRVKLTDFNFLMVLGKGSFGKVM 354 |+ + #+ | + ++ | |||| |||||+||||| Sbjct 314 PAPEP#QPPVSRPTKIGLDHFNFLAVLGKGNFGKVM 348 |
| Sycon raphanus | Query 335 KLTDFNFLMVLGKGSFGKVM 354 ||+||+|| ||||||||||| Sbjct 362 KLSDFSFLKVLGKGSFGKVM 381 |
| Cryptococcus neoformans var. neoformans JEC21 | Query 333 RVKLTDFNFLMVLGKGSFGKVM 354 +| | ||||| |||||+||||| Sbjct 751 KVGLDDFNFLAVLGKGNFGKVM 772 |
| Cryptococcus neoformans var. neoformans | Query 333 RVKLTDFNFLMVLGKGSFGKVM 354 +| | ||||| |||||+||||| Sbjct 751 KVGLDDFNFLAVLGKGNFGKVM 772 |
| Squalus acanthias | Query 327 PSNNLDRVKLTDFNFLMVLGKGSFGKVM 354 ||+| + | +||||| |+||||||||+ Sbjct 89 PSSN-PQAKPSDFNFLKVIGKGSFGKVL 115 |
| Cryptococcus neoformans var. neoformans B-3501A | Query 333 RVKLTDFNFLMVLGKGSFGKVM 354 +| | ||||| |||||+||||| Sbjct 754 KVGLDDFNFLAVLGKGNFGKVM 775 |
| Cryptococcus neoformans var. grubii | Query 333 RVKLTDFNFLMVLGKGSFGKVM 354 +| | ||||| |||||+||||| Sbjct 751 KVGLDDFNFLAVLGKGNFGKVM 772 |
| Tuber indicum | Query 320 PSEDR#KQPSNNLDRVKLTDFNFLMVLGKGSFGKVM 354 |+ + #+| + ++ | |||| |||||+||||| Sbjct 313 PAPEP#QQVVSKPTKIGLDHFNFLAVLGKGNFGKVM 347 |
| Phaeosphaeria nodorum SN15 | Query 329 NNLDRVKLTDFNFLMVLGKGSFGKVM 354 | ||+ + || | |+||||||||| Sbjct 273 NRQDRLSIDDFELLKVVGKGSFGKVM 298 |
| Tuber pseudoexcavatum | Query 320 PSEDR#KQPSNNLDRVKLTDFNFLMVLGKGSFGKVM 354 |+ + #+| + ++ | |||| |||||+||||| Sbjct 312 PAPEP#QQVVSKPTKIGLDHFNFLAVLGKGNFGKVM 346 |
| Leishmania braziliensis | Query 295 EEGNMELRQKFEKAKLGPAGNKVISPSEDR#KQPSNNLDRVKLTDFNFLMVLGKGSFGKVM 354 | | + +|+ + + ++|| # |+|| | | || | |+|+|+||||+ Sbjct 591 ESGQVMQQQRVINSTSPIPYSSLLSPRGSL#VSRSDNLSPVGLQDFQLLTVIGRGTFGKVL 650 |
| Aspergillus clavatus NRRL 1 | Query 312 PAGNKVISPSEDR#KQPSNNLDRVKLTDFNFLMVLGKGSFGKVM 354 | +| | #+|| |+ | |||| |||||+||||| Sbjct 763 PMQQQVAVKEEVP#QQPKA---RIGLDHFNFLSVLGKGNFGKVM 802 |
| Debaryomyces hansenii CBS767 | Query 326 QPSNNLDRVKLTDFNFLMVLGKGSFGKVM 354 +| ++ | || || |||||+||||| Sbjct 750 KPKRRRRKIGLDDFQFLAVLGKGNFGKVM 778 |
| Cochliobolus heterostrophus | Query 310 LGPAGNKVISPSEDR#KQPSNNLDRVKLTDFNFLMVLGKGSFGKVM 354 +| | ++|+ + #+ |+ | |||| |||||+||||| Sbjct 823 VGEVPGKKVTPAANT#QGTGK---RIGLDHFNFLAVLGKGNFGKVM 864 |
| Plasmodium berghei strain ANKA | Query 298 NMELRQKFEKAKLGPAGNKVISPSEDR#KQPSNNLDRVKLTDFNFLMVLGKGSFGKVM 354 |+ + +||+| | | | +#|+ |+| +|||| |+||||+|||+ Sbjct 259 NLYIHDMYEKSKKKRL-RKFIPLSNKK#KR------RIKPDNFNFLKVIGKGSYGKVL 308 |
| Ashbya gossypii ATCC 10895 | Query 299 MELRQKFEKAKLGPAGNKVISPSEDR-#KQPSNNLDRVKLTDFNFLMVLGKGSFGKVM 354 || + | | | + + |+ | #|+ + +| | || | |||||+||||+ Sbjct 781 MESIPQHPPAPLSPPKAQPVQPASGRH#KRKTPKRRKVSLDDFILLKVLGKGNFGKVL 837 |
| Aspergillus oryzae | Query 312 PAGNKVISPSEDR#KQPSNNLDRVKLTDFNFLMVLGKGSFGKVM 354 | + ++ || # | |+ | |||| |||||+||||| Sbjct 741 PVMQQQVAMKEDA#P-PQQPKVRIGLDHFNFLAVLGKGNFGKVM 782 |
| Candida albicans | Query 295 EEGNMELRQKFEKAKLGPAGNKVISPSEDR#KQPSNNLDRVKLTDFNFLMVLGKGSFGKVM 354 ++ | +++| ++ +|| | #|+ +| | || || |||||+||||| Sbjct 729 KQQNQQVQQVQQQEELGHQRTHSSGKSGKS#KRRKR---KVGLDDFQFLAVLGKGNFGKVM 785 |
| Sporothrix schenckii | Query 333 RVKLTDFNFLMVLGKGSFGKVM 354 |+ | +|||| |||||+||||| Sbjct 863 RIGLDNFNFLAVLGKGNFGKVM 884 |
| Hydra vulgaris | Query 335 KLTDFNFLMVLGKGSFGKVM 354 || || || |+||||||||| Sbjct 344 KLEDFTFLKVIGKGSFGKVM 363 |
| Coccidioides immitis RS | Query 333 RVKLTDFNFLMVLGKGSFGKVM 354 || | |||| |||||+||||| Sbjct 740 RVGLDHFNFLAVLGKGNFGKVM 761 |
| Plasmodium yoelii yoelii str. 17XNL | Query 300 ELRQKFEKAKLGPAGNKVISPSEDR#KQPSNNLDRVKLTDFNFLMVLGKGSFGKVM 354 ++ +| +| +| | | | +#|+ ||| +|||| |+||||+|||+ Sbjct 370 DMHEKSKKKRL----RKFIPLSNKK#KR------RVKPDNFNFLKVIGKGSYGKVL 414 |
| Neosartorya fischeri NRRL 181 | Query 297 GNMELRQKFEKAKLGPAGNKVISPSEDR#KQPSNNLDRVKLTDFNFLMVLGKGSFGKVM 354 | + +|+ ++ + | +| | # | |+ | |||| |||||+||||| Sbjct 743 GMPQQQQQQQQQAVAPMQQQVAVKEEI-#-PPQQPKVRIGLDHFNFLAVLGKGNFGKVM 798 |
| Aspergillus terreus NIH2624 | Query 312 PAGNKVISPSEDR#KQPSNNLDRVKLTDFNFLMVLGKGSFGKVM 354 | + ++ |+ # | + |+ | |||| |||||+||||| Sbjct 747 PVMQQQVAVKEEV#PPPQPKV-RIGLDHFNFLAVLGKGNFGKVM 788 |
| Medicago truncatula | Query 302 RQKFEKAKLGPAGNKVISPSEDR#KQPSNNLDRVKLTDFNFLMVLGKGSFGKV 353 +|| +|| + |+ +| | + +# | + + | +|| || ++|+| |||| Sbjct 540 KQKRQKAVVTPSNSKKRSSMKSK#HQKFSYTEIVNITD-NFKTIIGEGGFGKV 590 |
| Blumeria graminis | Query 333 RVKLTDFNFLMVLGKGSFGKVM 354 |+ | |||| |||||+||||| Sbjct 826 RIGLDHFNFLAVLGKGNFGKVM 847 |
| Fundulus heteroclitus | Query 319 SPSEDR#KQPSNNLDRVKLTDFNFLMVLGKGSFGKVM 354 |||+ # ++ | +||+|| |+||||||||+ Sbjct 78 SPSQQI#NLGPSSNPSAKPSDFHFLKVIGKGSFGKVL 113 |
* References
[PubMed ID: 19235902] Haughian JM, Bradford AP, Protein kinase C alpha (PKCalpha) regulates growth and invasion of endometrial cancer cells. J Cell Physiol. 2009 Jul;220(1):112-8.
[PubMed ID: 19357285] ... Kohutek ZA, diPierro CG, Redpath GT, Hussaini IM, ADAM-10-mediated N-cadherin cleavage is protein kinase C-alpha dependent and promotes glioblastoma cell migration. J Neurosci. 2009 Apr 8;29(14):4605-15.
[PubMed ID: 19158275] ... Nyman U, Vlachos P, Cascante A, Hermanson O, Zhivotovsky B, Joseph B, Protein kinase C-dependent phosphorylation regulates the cell cycle-inhibitory function of the p73 carboxy terminus transactivation domain. Mol Cell Biol. 2009 Apr;29(7):1814-25. Epub 2009 Jan 21.
[PubMed ID: 19291828] ... Wu DL, Sui FY, Du C, Zhang CW, Hui B, Xu SL, Lu HZ, Song GJ, Antisense expression of PKCalpha improved sensitivity of SGC7901/VCR cells to doxorubicin. World J Gastroenterol. 2009 Mar 14;15(10):1259-63.
[PubMed ID: 19176518] ... Fan QW, Cheng C, Knight ZA, Haas-Kogan D, Stokoe D, James CD, McCormick F, Shokat KM, Weiss WA, EGFR signals to mTOR through PKC and independently of Akt in glioma. Sci Signal. 2009 Jan 27;2(55):ra4.
[PubMed ID: 12881490] ... Kawakami Y, Kitaura J, Yao L, McHenry RW, Kawakami Y, Newton AC, Kang S, Kato RM, Leitges M, Rawlings DJ, Kawakami T, A Ras activation pathway dependent on Syk phosphorylation of protein kinase C. Proc Natl Acad Sci U S A. 2003 Aug 5;100(16):9470-5. Epub 2003 Jul 24.
[PubMed ID: 12665801] ... Gevaert K, Goethals M, Martens L, Van Damme J, Staes A, Thomas GR, Vandekerckhove J, Exploring proteomes and analyzing protein processing by mass spectrometric identification of sorted N-terminal peptides. Nat Biotechnol. 2003 May;21(5):566-9. Epub 2003 Mar 31.
[PubMed ID: 12162751] ... Hodgkinson CP, Sale EM, Sale GJ, Characterization of PDK2 activity against protein kinase B gamma. Biochemistry. 2002 Aug 13;41(32):10351-9.
[PubMed ID: 10339425] ... Ziegler WH, Parekh DB, Le Good JA, Whelan RD, Kelly JJ, Frech M, Hemmings BA, Parker PJ, Rapamycin-sensitive phosphorylation of PKC on a carboxy-terminal site by an atypical PKC complex. Curr Biol. 1999 May 20;9(10):522-9.
[PubMed ID: 8856079] ... Gysin S, Imber R, Replacement of Ser657 of protein kinase C-alpha by alanine leads to premature down regulation after phorbol-ester-induced translocation to the membrane. Eur J Biochem. 1996 Sep 15;240(3):747-50.
[PubMed ID: 1335366] ... Li L, Zhou J, James G, Heller-Harrison R, Czech MP, Olson EN, FGF inactivates myogenic helix-loop-helix proteins through phosphorylation of a conserved protein kinase C site in their DNA-binding domains. Cell. 1992 Dec 24;71(7):1181-94.
[PubMed ID: 1322130] ... Rider MH, Vandamme J, Lebeau E, Vertommen D, Vidal H, Rousseau GG, Vandekerckhove J, Hue L, The two forms of bovine heart 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase result from alternative splicing. Biochem J. 1992 Jul 15;285 ( Pt 2):405-11.
[PubMed ID: 1321150] ... Moss SJ, Doherty CA, Huganir RL, Identification of the cAMP-dependent protein kinase and protein kinase C phosphorylation sites within the major intracellular domains of the beta 1, gamma 2S, and gamma 2L subunits of the gamma-aminobutyric acid type A receptor. J Biol Chem. 1992 Jul 15;267(20):14470-6.
[PubMed ID: 1375933] ... Bredt DS, Ferris CD, Snyder SH, Nitric oxide synthase regulatory sites. Phosphorylation by cyclic AMP-dependent protein kinase, protein kinase C, and calcium/calmodulin protein kinase; identification of flavin and calmodulin binding sites. J Biol Chem. 1992 Jun 5;267(16):10976-81.
[PubMed ID: 1374067] ... Omary MB, Baxter GT, Chou CF, Riopel CL, Lin WY, Strulovici B, PKC epsilon-related kinase associates with and phosphorylates cytokeratin 8 and 18. J Cell Biol. 1992 May;117(3):583-93.