XSB1653 : calpain 2 [Rattus norvegicus]
[ CaMP Format ]
This entry is computationally expanded from SB0001
* Basic Information
| Organism | Rattus norvegicus (Norway rat) |
| Protein Names | Calpain-2 catalytic subunit; 3.4.22.53; Calpain-2 large subunit; Calcium-activated neutral proteinase 2; CANP 2; Calpain M-type; M-calpain; Millimolar-calpain; calpain 2 |
| Gene Names | Capn2; calpain 2 |
| Gene Locus | 13q26; chromosome 13 |
| GO Function | Not available |
* Information From OMIM
Not Available.
* Structure Information
1. Primary Information
Length: 700 aa
Average Mass: 79.919 kDa
Monoisotopic Mass: 79.869 kDa
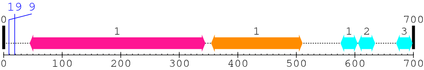
2. Domain Information
Annotated Domains: interpro / pfam / smart / prosite
Computationally Assigned Domains (Pfam+HMMER):
| domain name | begin | end | score | e-value |
|---|---|---|---|---|
| --- cleavage 19 --- | ||||
| --- cleavage 9 --- | ||||
| Peptidase_C2 1. | 45 | 344 | 812.0 | 3.8e-241 |
| Calpain_III 1. | 355 | 510 | 385.6 | 8.7e-113 |
| efhand 1. | 576 | 604 | 20.4 | 0.0074 |
| efhand 2. | 606 | 634 | 19.4 | 0.015 |
| efhand 3. | 671 | 698 | 3.2 | 9.5 |
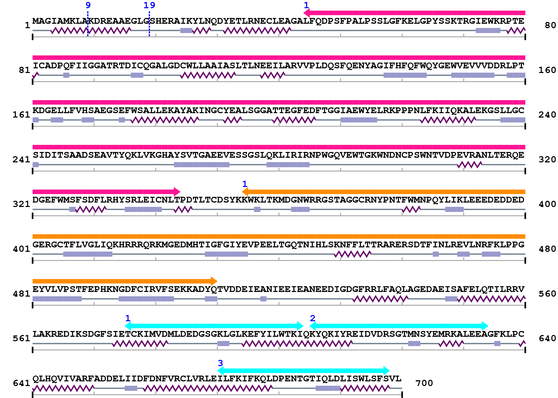
3. Sequence Information
Fasta Sequence: XSB1653.fasta
Amino Acid Sequence and Secondary Structures (PsiPred):
* Cleavage Information
2 [sites]
Cleavage sites (±10aa)
[Site 1] KDREAAEGLG19-SHERAIKYLN
Gly19  Ser
Ser
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Lys10 | Asp11 | Arg12 | Glu13 | Ala14 | Ala15 | Glu16 | Gly17 | Leu18 | Gly19 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Ser20 | His21 | Glu22 | Arg23 | Ala24 | Ile25 | Lys26 | Tyr27 | Leu28 | Asn29 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| MAGIAMKLAKDREAAEGLGSHERAIKYLNQDYETLRNECLEAGALFQDP |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | N/A | 101.00 | 14 | A |
| 2 | Rattus norvegicus | 101.00 | 9 | calpain 2 |
| 3 | Mus musculus | 99.80 | 20 | m-calpain |
| 4 | Homo sapiens | 95.50 | 25 | calpain 2, large subunit |
| 5 | catalytic | 95.50 | 1 | Calpain 2, large |
| 6 | Macaca mulatta | 94.00 | 3 | PREDICTED: calpain 2, large subunit |
| 7 | Bos taurus | 92.40 | 7 | Unknown (protein for MGC:138930) |
| 8 | Monodelphis domestica | 80.10 | 2 | PREDICTED: similar to calpain 2 |
| 9 | Xenopus laevis | 69.70 | 10 | hypothetical protein LOC399075 |
| 10 | Gallus gallus | 68.90 | 6 | calpain 1, (mu/I) large subunit |
| 11 | Coturnix coturnix | 68.20 | 1 | quail calpain |
| 12 | Xenopus tropicalis | 67.80 | 6 | calpain 2, (m/II) large subunit |
| 13 | Canis familiaris | 67.40 | 6 | PREDICTED: similar to calpain 8 |
| 14 | Danio rerio | 64.30 | 31 | hypothetical protein LOC550505 |
| 15 | Oncorhynchus mykiss | 60.50 | 3 | m-calpain |
| 16 | Tetraodon nigroviridis | 60.10 | 7 | unnamed protein product |
| 17 | Pan troglodytes | 59.70 | 6 | PREDICTED: similar to calpain, partial |
| 18 | Stizostedion vitreum vitreum | 59.70 | 1 | putative calpain-like protein |
| 19 | Pongo pygmaeus | 55.80 | 1 | CAN1_PONPY Calpain-1 catalytic subunit (Calpain-1 |
| 20 | synthetic construct | 55.80 | 1 | Homo sapiens calpain 1, (mu/I) large subunit |
| 21 | Sus scrofa | 55.10 | 3 | micromolar calcium-activated neutral protease 1 is |
| 22 | Macaca fascicularis | 40.80 | 1 | unnamed protein product |
| 23 | Bos grunniens | 35.40 | 1 | micromolar calcium-activated neutral protease 1 la |
| 24 | Drosophila melanogaster | 35.00 | 6 | CAA55297.1 |
| 25 | Aedes aegypti | 35.00 | 1 | calpain, putative |
| 26 | Oryctolagus cuniculus | 34.70 | 1 | calpain 3, (p94) |
Top-ranked sequences
| organism | matching |
|---|---|
| N/A | Query 1 MAGIAMKLAKDREAAEGLG#SHERAIKYLNQDYETLRNECLEAGALFQDP 49 |||||||||||||||||||#|||||||||||||||||||||||||||||| Query 1 MAGIAMKLAKDREAAEGLG#SHERAIKYLNQDYETLRNECLEAGALFQDP 49 |
| Rattus norvegicus | Query 1 MAGIAMKLAKDREAAEGLG#SHERAIKYLNQDYETLRNECLEAGALFQDP 49 |||||||||||||||||||#|||||||||||||||||||||||||||||| Query 1 MAGIAMKLAKDREAAEGLG#SHERAIKYLNQDYETLRNECLEAGALFQDP 49 |
| Mus musculus | Query 1 MAGIAMKLAKDREAAEGLG#SHERAIKYLNQDYETLRNECLEAGALFQDP 49 |||||+|||||||||||||#|||||||||||||||||||||||||||||| Sbjct 1 MAGIAIKLAKDREAAEGLG#SHERAIKYLNQDYETLRNECLEAGALFQDP 49 |
| Homo sapiens | Query 1 MAGIAMKLAKDREAAEGLG#SHERAIKYLNQDYETLRNECLEAGALFQDP 49 ||||| |||||||||||||#|||||||||||||| ||||||||| ||||| Sbjct 1 MAGIAAKLAKDREAAEGLG#SHERAIKYLNQDYEALRNECLEAGTLFQDP 49 |
| catalytic | Query 1 MAGIAMKLAKDREAAEGLG#SHERAIKYLNQDYETLRNECLEAGALFQDP 49 ||||| |||||||||||||#|||||||||||||| ||||||||| ||||| Sbjct 30 MAGIAAKLAKDREAAEGLG#SHERAIKYLNQDYEALRNECLEAGTLFQDP 78 |
| Macaca mulatta | Query 1 MAGIAMKLAKDREAAEGLG#SHERAIKYLNQDYETLRNECLEAGALFQDP 49 ||||| || ||||||||||#|||||||||||||| ||||||||| ||||| Sbjct 1 MAGIAAKLVKDREAAEGLG#SHERAIKYLNQDYEALRNECLEAGTLFQDP 49 |
| Bos taurus | Query 1 MAGIAMKLAKDREAAEGLG#SHERAIKYLNQDYETLRNECLEAGALFQDP 49 ||||| |||||||||||||#|||||+||||||| ||+|||||||||||| Sbjct 1 MAGIAAKLAKDREAAEGLG#SHERAVKYLNQDYAALRDECLEAGALFQDP 49 |
| Monodelphis domestica | Query 1 MAGIAMKLAKDREAAEGLG#SHERAIKYLNQDYETLRNECLEAGALFQDP 49 |+||| |+ +||||| |||#+||+|+|||||||||| +|||| | ||||| Sbjct 1 MSGIAAKVVRDREAAAGLG#THEQAVKYLNQDYETLLSECLEEGKLFQDP 49 |
| Xenopus laevis | Query 1 MAGIAMKLAKDREAAEGLG#SHERAIKYLNQDYETLRNECLEAGALFQDP 49 |+|+| +|||+| | | |#++|+|+ |||||+| ||||||++|+||+|| Sbjct 1 MSGVADRLAKERARASGFG#TNEKALPYLNQDFEALRNECLQSGSLFKDP 49 |
| Gallus gallus | Query 3 GIAMKLAKDREAAEGLG#SHERAIKYLNQDYETLRNECLEAGALFQDP 49 ||| +| +|| |||+|# | |+|||||||| |+ ||+|+| ||+|| Sbjct 6 GIAARLQRDRLRAEGVG#EHNNAVKYLNQDYEALKQECIESGTLFRDP 52 |
| Coturnix coturnix | Query 3 GIAMKLAKDREAAEGLG#SHERAIKYLNQDYETLRNECLEAGALFQDP 49 ||| +| +|| |||+|# | |+|||||||| |+ |+|+||||+|| Sbjct 6 GIAARLQRDRLRAEGVG#EHNNAVKYLNQDYEALKQACIESGALFRDP 52 |
| Xenopus tropicalis | Query 1 MAGIAMKLAKDREAAEGLG#SHERAIKYLNQDYETLRNECLEAGALFQD 48 |+|+| +|||+| | |||#++|+|+ ||+|||| ||||||++| ||+| Sbjct 1 MSGVADRLAKERALASGLG#TNEKALPYLSQDYEALRNECLQSGTLFKD 48 |
| Canis familiaris | Query 1 MAGIAMKLAKDREAAEGLG#SHERAIKYLNQDYETLRNECLEAGALFQDP 49 |+ +| ++|| + || |||#||++|+||| ||+| || +||+|| ||+|| Sbjct 94 MSALAARIAKQQAAAGGLG#SHQKAVKYLGQDFEKLRQQCLDAGVLFKDP 142 |
| Danio rerio | Query 1 MAGIAMKLAKDREAAEGLG#SHERAIKYLNQDYETLRNECLEAGALFQD 48 |+||| || |+| | |+|#++ +| |+|||||| |+ ||||+| |||| Sbjct 1 MSGIATKLQKNRARAAGIG#TNAQAAKFLNQDYEALKRECLESGRLFQD 48 |
| Oncorhynchus mykiss | Query 1 MAGIAMKLAKDREAAEGLG#SHERAIKYLNQDYETLRNECLEAGALFQDP 49 |+|+| ||| | | | |#++ |+||||||+| || +|| | || || Sbjct 1 MSGMASNLAKKRALAAGFG#TNANAVKYLNQDFEALRAQCLNRGGLFSDP 49 |
| Tetraodon nigroviridis | Query 1 MAGIAMKLAKDREAAEGLG#SHERAIKYLNQDYETLRNECLEAGALFQD 48 |+||| || |+ |+| |#+ +| ||||||+|+|| ||| | |||| Sbjct 1 MSGIAAKLQHHRDRAQGFG#TASQAQKYLNQDFESLRESCLERGELFQD 48 |
| Pan troglodytes | Query 1 MAGIAMKLAKDREAAEGLG#SHERAIKYLNQDYETLRNECLEAGALFQDP 49 || | +++ | | +|||#|++ |+||| ||++||| +||++| ||+|| Sbjct 1 MAAQAAGVSRQRAATQGLG#SNQNALKYLGQDFKTLRQQCLDSGVLFKDP 49 |
| Stizostedion vitreum vitreum | Query 1 MAGIAMKLAKDREAAEGLG#SHERAIKYLNQDYETLRNECLEAGALFQD 48 |+||| | + || +|+|#| +|+|||||||| || |||+ +||+| Sbjct 1 MSGIAATLQRRREKEQGIG#SCSQAVKYLNQDYEALRRRCLESESLFKD 48 |
| Pongo pygmaeus | Query 3 GIAMKLAKDREAAEGLG#SHERAIKYLNQDYETLRNECLEAGALFQD 48 |++ ++ | | |||# || ||||| |||| || ||++| ||+| Sbjct 13 GVSAQVQKQRARELGLG#RHENAIKYLGQDYEQLRVRCLQSGTLFRD 58 |
| synthetic construct | Query 3 GIAMKLAKDREAAEGLG#SHERAIKYLNQDYETLRNECLEAGALFQD 48 |++ ++ | | |||# || ||||| |||| || ||++| ||+| Sbjct 13 GVSAQVQKQRARELGLG#RHENAIKYLGQDYEQLRVRCLQSGTLFRD 58 |
| Sus scrofa | Query 3 GIAMKLAKDREAAEGLG#SHERAIKYLNQDYETLRNECLEAGALFQD 48 |++ ++ | | |||# || ||||| |||| || ||++|+||+| Sbjct 13 GVSAQVQKLRAKELGLG#RHENAIKYLGQDYEQLRAHCLQSGSLFRD 58 |
| Macaca fascicularis | Query 3 GIAMKLAKDREAAEGLG#SHERAIKYLNQDYETLRNECLEAGALFQDP 49 |+ + | | |+|# | | + || +| || || | ||+|| Sbjct 36 GMVAHINYSRLKASGVG#QHHNAHNFGNQSFEELRAVCLRKGELFEDP 82 |
| Bos grunniens | Query 27 YLNQDYETLRNECLEAGALFQD 48 || |||| || ||+ ||||+| Sbjct 1 YLGQDYEQLRVHCLQRGALFRD 22 |
| Drosophila melanogaster | Query 30 QDYETLRNECLEAGALFQDP 49 |||||+ | || +|+||+|| Sbjct 50 QDYETILNSCLASGSLFEDP 69 |
| Aedes aegypti | Query 30 QDYETLRNECLEAGALFQDP 49 ||| ||++|||+|+||+|| Sbjct 115 QDYYVLRSQCLESGSLFEDP 134 |
| Oryctolagus cuniculus | Query 7 KLAKDREAAEGLG#SHERAIKYLNQDYETLRNECLEAGALFQD 48 ++ ++|+ | |#+ + ||| ||+ || || ||+| Sbjct 12 RVIRERDRRNGEG#TVTQPIKYEGQDFVVLRQRCLAQKCLFED 53 |
[Site 2] MAGIAMKLA9-KDREAAEGLG
Ala9  Lys
Lys
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| - | Met1 | Ala2 | Gly3 | Ile4 | Ala5 | Met6 | Lys7 | Leu8 | Ala9 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Lys10 | Asp11 | Arg12 | Glu13 | Ala14 | Ala15 | Glu16 | Gly17 | Leu18 | Gly19 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| MAGIAMKLAKDREAAEGLGSHERAIKYLNQDYETLRNEC |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | N/A | 81.60 | 12 | A |
| 2 | Rattus norvegicus | 81.60 | 5 | calpain 2 |
| 3 | Mus musculus | 80.10 | 15 | unnamed protein product |
| 4 | Homo sapiens | 77.40 | 17 | AF261089_1 calpain large polypeptide L2 |
| 5 | catalytic | 77.40 | 1 | Calpain 2, large |
| 6 | Macaca mulatta | 75.90 | 3 | PREDICTED: calpain 2, large subunit |
| 7 | Bos taurus | 72.80 | 6 | Unknown (protein for MGC:138930) |
| 8 | Monodelphis domestica | 64.30 | 2 | PREDICTED: similar to calpain 2 |
| 9 | Xenopus tropicalis | 56.20 | 5 | calpain 2, (m/II) large subunit |
| 10 | Xenopus laevis | 55.10 | 9 | hypothetical protein LOC399075 |
| 11 | Gallus gallus | 54.70 | 5 | PREDICTED: similar to calpain 8 (nCL-2) |
| 12 | Canis familiaris | 52.00 | 3 | PREDICTED: similar to calpain 8 |
| 13 | Coturnix coturnix | 52.00 | 1 | quail calpain |
| 14 | Danio rerio | 51.20 | 23 | hypothetical protein LOC541374 |
| 15 | Oncorhynchus mykiss | 50.40 | 3 | calpain 2 catalytic subunit |
| 16 | Stizostedion vitreum vitreum | 49.70 | 1 | putative calpain-like protein |
| 17 | Tetraodon nigroviridis | 47.00 | 6 | unnamed protein product |
| 18 | Pan troglodytes | 45.40 | 5 | PREDICTED: similar to calpain, partial |
| 19 | Pongo pygmaeus | 44.30 | 1 | CAN1_PONPY Calpain-1 catalytic subunit (Calpain-1 |
| 20 | synthetic construct | 44.30 | 1 | Homo sapiens calpain 1, (mu/I) large subunit |
| 21 | Sus scrofa | 43.10 | 2 | micromolar calcium-activated neutral protease 1 is |
Top-ranked sequences
| organism | matching |
|---|---|
| N/A | Query 1 MAGIAMKLA#KDREAAEGLGSHERAIKYLNQDYETLRNEC 39 |||||||||#|||||||||||||||||||||||||||||| Query 1 MAGIAMKLA#KDREAAEGLGSHERAIKYLNQDYETLRNEC 39 |
| Rattus norvegicus | Query 1 MAGIAMKLA#KDREAAEGLGSHERAIKYLNQDYETLRNEC 39 |||||||||#|||||||||||||||||||||||||||||| Query 1 MAGIAMKLA#KDREAAEGLGSHERAIKYLNQDYETLRNEC 39 |
| Mus musculus | Query 1 MAGIAMKLA#KDREAAEGLGSHERAIKYLNQDYETLRNEC 39 |||||+|||#|||||||||||||||||||||||||||||| Sbjct 1 MAGIAIKLA#KDREAAEGLGSHERAIKYLNQDYETLRNEC 39 |
| Homo sapiens | Query 1 MAGIAMKLA#KDREAAEGLGSHERAIKYLNQDYETLRNEC 39 ||||| |||#|||||||||||||||||||||||| ||||| Sbjct 1 MAGIAAKLA#KDREAAEGLGSHERAIKYLNQDYEALRNEC 39 |
| catalytic | Query 1 MAGIAMKLA#KDREAAEGLGSHERAIKYLNQDYETLRNEC 39 ||||| |||#|||||||||||||||||||||||| ||||| Sbjct 30 MAGIAAKLA#KDREAAEGLGSHERAIKYLNQDYEALRNEC 68 |
| Macaca mulatta | Query 1 MAGIAMKLA#KDREAAEGLGSHERAIKYLNQDYETLRNEC 39 ||||| || #|||||||||||||||||||||||| ||||| Sbjct 1 MAGIAAKLV#KDREAAEGLGSHERAIKYLNQDYEALRNEC 39 |
| Bos taurus | Query 1 MAGIAMKLA#KDREAAEGLGSHERAIKYLNQDYETLRNEC 39 ||||| |||#|||||||||||||||+||||||| ||+|| Sbjct 1 MAGIAAKLA#KDREAAEGLGSHERAVKYLNQDYAALRDEC 39 |
| Monodelphis domestica | Query 1 MAGIAMKLA#KDREAAEGLGSHERAIKYLNQDYETLRNEC 39 |+||| |+ #+||||| |||+||+|+|||||||||| +|| Sbjct 1 MSGIAAKVV#RDREAAAGLGTHEQAVKYLNQDYETLLSEC 39 |
| Xenopus tropicalis | Query 1 MAGIAMKLA#KDREAAEGLGSHERAIKYLNQDYETLRNEC 39 |+|+| +||#|+| | |||++|+|+ ||+|||| ||||| Sbjct 1 MSGVADRLA#KERALASGLGTNEKALPYLSQDYEALRNEC 39 |
| Xenopus laevis | Query 1 MAGIAMKLA#KDREAAEGLGSHERAIKYLNQDYETLRNEC 39 |+|+| +||#|+| | | |++|+|+ |||||+| ||||| Sbjct 1 MSGVADRLA#KERARASGFGTNEKALPYLNQDFEALRNEC 39 |
| Gallus gallus | Query 1 MAGIAMKLA#KDREAAEGLGSHERAIKYLNQDYETLRNEC 39 |||+| +| #++| |||||||+ +|| || |||| || | Sbjct 1 MAGLAARLC#QERAAAEGLGSYRKAISYLGQDYEALRQRC 39 |
| Canis familiaris | Query 1 MAGIAMKLA#KDREAAEGLGSHERAIKYLNQDYETLRNEC 39 |+ +| ++|#| + || |||||++|+||| ||+| || +| Sbjct 94 MSALAARIA#KQQAAAGGLGSHQKAVKYLGQDFEKLRQQC 132 |
| Coturnix coturnix | Query 3 GIAMKLA#KDREAAEGLGSHERAIKYLNQDYETLRNEC 39 ||| +| #+|| |||+| | |+|||||||| |+ | Sbjct 6 GIAARLQ#RDRLRAEGVGEHNNAVKYLNQDYEALKQAC 42 |
| Danio rerio | Query 1 MAGIAMKLA#KDREAAEGLGSHERAIKYLNQDYETLRNEC 39 |+|+| ||#| | | | |++ |+|||||++||||+|| Sbjct 1 MSGVASTLA#KKRALAAGFGTNSNAVKYLNQNFETLRSEC 39 |
| Oncorhynchus mykiss | Query 1 MAGIAMKLA#KDREAAEGLGSHERAIKYLNQDYETLRNEC 39 |||+| ||#| | | | |++ | ||||||+|+||+|| Sbjct 1 MAGVASTLA#KKRALAAGFGTNANASKYLNQDFESLRSEC 39 |
| Stizostedion vitreum vitreum | Query 1 MAGIAMKLA#KDREAAEGLGSHERAIKYLNQDYETLRNEC 39 |+||| | #+ || +|+|| +|+|||||||| || | Sbjct 1 MSGIAATLQ#RRREKEQGIGSCSQAVKYLNQDYEALRRRC 39 |
| Tetraodon nigroviridis | Query 1 MAGIAMKLA#KDREAAEGLGSHERAIKYLNQDYETLRNEC 39 |+||| || # |+ |+| |+ +| ||||||+|+|| | Sbjct 1 MSGIAAKLQ#HHRDRAQGFGTASQAQKYLNQDFESLRESC 39 |
| Pan troglodytes | Query 1 MAGIAMKLA#KDREAAEGLGSHERAIKYLNQDYETLRNEC 39 || | ++#+ | | +||||++ |+||| ||++||| +| Sbjct 1 MAAQAAGVS#RQRAATQGLGSNQNALKYLGQDFKTLRQQC 39 |
| Pongo pygmaeus | Query 3 GIAMKLA#KDREAAEGLGSHERAIKYLNQDYETLRNEC 39 |++ ++ #| | ||| || ||||| |||| || | Sbjct 13 GVSAQVQ#KQRARELGLGRHENAIKYLGQDYEQLRVRC 49 |
| synthetic construct | Query 3 GIAMKLA#KDREAAEGLGSHERAIKYLNQDYETLRNEC 39 |++ ++ #| | ||| || ||||| |||| || | Sbjct 13 GVSAQVQ#KQRARELGLGRHENAIKYLGQDYEQLRVRC 49 |
| Sus scrofa | Query 3 GIAMKLA#KDREAAEGLGSHERAIKYLNQDYETLRNEC 39 |++ ++ #| | ||| || ||||| |||| || | Sbjct 13 GVSAQVQ#KLRAKELGLGRHENAIKYLGQDYEQLRAHC 49 |
* References
[PubMed ID: 19020623] Hanna RA, Campbell RL, Davies PL, Calcium-bound structure of calpain and its mechanism of inhibition by calpastatin. Nature. 2008 Nov 20;456(7220):409-12.
[PubMed ID: 19020622] ... Moldoveanu T, Gehring K, Green DR, Concerted multi-pronged attack by calpastatin to occlude the catalytic cleft of heterodimeric calpains. Nature. 2008 Nov 20;456(7220):404-8.
[PubMed ID: 17712625] ... Sun M, Xu C, Neuroprotective mechanism of taurine due to up-regulating calpastatin and down-regulating calpain and caspase-3 during focal cerebral ischemia. Cell Mol Neurobiol. 2008 Jun;28(4):593-611. Epub 2007 Aug 22.
[PubMed ID: 18258589] ... Sprague CR, Fraley TS, Jang HS, Lal S, Greenwood JA, Phosphoinositide binding to the substrate regulates susceptibility to proteolysis by calpain. J Biol Chem. 2008 Apr 4;283(14):9217-23. Epub 2008 Feb 6.
[PubMed ID: 17543955] ... Hanna RA, Garcia-Diaz BE, Davies PL, Calpastatin simultaneously binds four calpains with different kinetic constants. FEBS Lett. 2007 Jun 26;581(16):2894-8. Epub 2007 May 25.
[PubMed ID: 11102442] ... Hosfield CM, Moldoveanu T, Davies PL, Elce JS, Jia Z, Calpain mutants with increased Ca2+ sensitivity and implications for the role of the C(2)-like domain. J Biol Chem. 2001 Mar 9;276(10):7404-7. Epub 2000 Dec 1.
[PubMed ID: 11342050] ... Moldoveanu T, Hosfield CM, Jia Z, Elce JS, Davies PL, Ca(2+)-induced structural changes in rat m-calpain revealed by partial proteolysis. Biochim Biophys Acta. 2001 Feb 9;1545(1-2):245-54.
[PubMed ID: 10601010] ... Hosfield CM, Elce JS, Davies PL, Jia Z, Crystal structure of calpain reveals the structural basis for Ca(2+)-dependent protease activity and a novel mode of enzyme activation. EMBO J. 1999 Dec 15;18(24):6880-9.
[PubMed ID: 7635186] ... Arthur JS, Gauthier S, Elce JS, Active site residues in m-calpain: identification by site-directed mutagenesis. FEBS Lett. 1995 Jul 24;368(3):397-400.
[PubMed ID: 8218419] ... DeLuca CI, Davies PL, Samis JA, Elce JS, Molecular cloning and bacterial expression of cDNA for rat calpain II 80 kDa subunit. Biochim Biophys Acta. 1993 Oct 19;1216(1):81-93.