XSB1687 : PREDICTED: similar to Protein kinase C beta type (PKC-beta) (PKC-B) isoform 1 [Monodelphis domestica]
[ CaMP Format ]
This entry is computationally expanded from SB0039
* Basic Information
| Organism | Monodelphis domestica (gray short-tailed opossum) |
| Protein Names | similar to Protein kinase C beta type (PKC-beta) (PKC-B) isoform 1 |
| Gene Names | LOC100013346; Derived by automated computational analysis using gene prediction method: GNOMON. Supporting evidence includes similarity to: 8 Proteins |
| Gene Locus | Not available |
| GO Function | Not available |
| Entrez Protein | Entrez Nucleotide | Entrez Gene | UniProt | OMIM | HGNC | HPRD | KEGG |
|---|---|---|---|---|---|---|---|
| XP_001363066 | XM_001363029 | 100013346 | N/A | N/A | N/A | N/A | N/A |
* Information From OMIM
Not Available.
* Structure Information
1. Primary Information
Length: 668 aa
Average Mass: 76.425 kDa
Monoisotopic Mass: 76.375 kDa
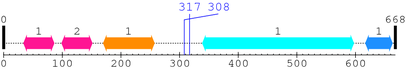
2. Domain Information
Annotated Domains: Not Available.
Computationally Assigned Domains (Pfam+HMMER):
| domain name | begin | end | score | e-value |
|---|---|---|---|---|
| C1_1 1. | 34 | 86 | 80.1 | 8.2e-21 |
| C1_1 2. | 99 | 151 | 97.3 | 5.4e-26 |
| C2 1. | 170 | 257 | 130.8 | 4.5e-36 |
| --- cleavage 317 --- | ||||
| --- cleavage 308 --- | ||||
| Pkinase 1. | 339 | 597 | 258.1 | 2e-74 |
| Pkinase_C 1. | 617 | 663 | 71.9 | 2.4e-18 |
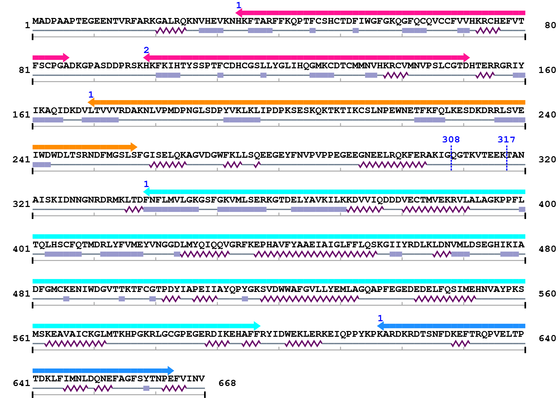
3. Sequence Information
Fasta Sequence: XSB1687.fasta
Amino Acid Sequence and Secondary Structures (PsiPred):
4. 3D Information
Not Available.
* Cleavage Information
2 [sites]
Cleavage sites (±10aa)
[Site 1] GQGTKVTEEK317-TANAISKIDN
Lys317  Thr
Thr
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Gly308 | Gln309 | Gly310 | Thr311 | Lys312 | Val313 | Thr314 | Glu315 | Glu316 | Lys317 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Thr318 | Ala319 | Asn320 | Ala321 | Ile322 | Ser323 | Lys324 | Ile325 | Asp326 | Asn327 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| PPEGEEGNEELRQKFERAKIGQGTKVTEEKTANAISKIDNNGNRDRMKLTDFNFLMVLGK |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | Monodelphis domestica | 123.00 | 3 | PREDICTED: similar to Protein kinase C beta type ( |
| 2 | N/A | 114.00 | 10 | K |
| 3 | Mus musculus | 114.00 | 4 | protein kinase C, beta 1 |
| 4 | Rattus norvegicus | 114.00 | 4 | unnamed protein product |
| 5 | Oryctolagus cuniculus | 113.00 | 3 | KPCB_RABIT Protein kinase C beta type (PKC-beta) ( |
| 6 | Macaca mulatta | 112.00 | 7 | PREDICTED: protein kinase C, beta isoform 1 |
| 7 | Canis familiaris | 111.00 | 3 | PREDICTED: similar to Protein kinase C, beta type |
| 8 | Homo sapiens | 109.00 | 8 | protein kinase C, beta isoform 1 |
| 9 | Gallus gallus | 108.00 | 3 | PREDICTED: similar to protein kinase C beta-1 |
| 10 | Bos taurus | 108.00 | 3 | Unknown (protein for MGC:148660) |
| 11 | Latimeria chalumnae | 100.00 | 2 | protein kinase C beta 1 |
| 12 | Protopterus dolloi | 99.00 | 2 | protein kinase C beta 1 |
| 13 | Tetraodon nigroviridis | 96.70 | 2 | unnamed protein product |
| 14 | Danio rerio | 94.70 | 6 | protein kinase C, beta 1 |
| 15 | Scyliorhinus canicula | 89.40 | 2 | protein kinase C |
| 16 | Petromyzon marinus | 66.60 | 1 | protein kinase C |
| 17 | synthetic construct | 60.10 | 1 | protein kinase C alpha |
| 18 | Takifugu rubripes | 58.20 | 1 | protein kinase C, alpha type |
| 19 | Xenopus laevis | 54.30 | 1 | protein kinase C, alpha |
| 20 | Xenopus tropicalis | 53.50 | 1 | hypothetical protein LOC780209 |
| 21 | Myxine glutinosa | 42.40 | 1 | protein kinase C |
| 22 | Caenorhabditis briggsae | 40.80 | 1 | Hypothetical protein CBG16150 |
| 23 | Branchiostoma lanceolatum | 37.70 | 1 | protein kinase C |
| 24 | Apis mellifera | 37.00 | 1 | PREDICTED: similar to Protein C kinase 53E CG6622- |
| 25 | Aedes aegypti | 35.80 | 1 | protein kinase c |
| 26 | Ostreococcus tauri | 35.00 | 1 | chloroplast SRP receptor cpFtsY precursor (ISS) |
| 27 | Aplysia californica | 34.70 | 1 | KPC1_APLCA Calcium-dependent protein kinase C (APL |
| 28 | Trichomonas vaginalis G3 | 33.10 | 1 | SMC family, C-terminal domain containing protein |
| 29 | Tribolium castaneum | 32.70 | 1 | PREDICTED: similar to CG1954-PA |
Top-ranked sequences
| organism | matching |
|---|---|
| Monodelphis domestica | Query 288 PPEGEEGNEELRQKFERAKIGQGTKVTEEK#TANAISKIDNNGNRDRMKLTDFNFLMVLGK 347 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 288 PPEGEEGNEELRQKFERAKIGQGTKVTEEK#TANAISKIDNNGNRDRMKLTDFNFLMVLGK 347 |
| N/A | Query 288 PPEGEEGNEELRQKFERAKIGQGTKVTEEK#TANAISKIDNNGNRDRMKLTDFNFLMVLGK 347 |||| |||||||||||||||||||| |||#||| ||| |||||||||||||||||||||| Sbjct 291 PPEGSEGNEELRQKFERAKIGQGTKAPEEK#TANTISKFDNNGNRDRMKLTDFNFLMVLGK 350 |
| Mus musculus | Query 288 PPEGEEGNEELRQKFERAKIGQGTKVTEEK#TANAISKIDNNGNRDRMKLTDFNFLMVLGK 347 |||| |||||||||||||||||||| |||#||| ||| |||||||||||||||||||||| Sbjct 291 PPEGSEGNEELRQKFERAKIGQGTKAPEEK#TANTISKFDNNGNRDRMKLTDFNFLMVLGK 350 |
| Rattus norvegicus | Query 288 PPEGEEGNEELRQKFERAKIGQGTKVTEEK#TANAISKIDNNGNRDRMKLTDFNFLMVLGK 347 |||| |||||||||||||||||||| |||#||| ||| |||||||||||||||||||||| Sbjct 291 PPEGSEGNEELRQKFERAKIGQGTKAPEEK#TANTISKFDNNGNRDRMKLTDFNFLMVLGK 350 |
| Oryctolagus cuniculus | Query 288 PPEGEEGNEELRQKFERAKIGQGTKVTEEK#TANAISKIDNNGNRDRMKLTDFNFLMVLGK 347 |||| |||||||||||||||||||| |||#| | ||| |||||||||||||||||||||| Sbjct 291 PPEGSEGNEELRQKFERAKIGQGTKTPEEK#TTNTISKFDNNGNRDRMKLTDFNFLMVLGK 350 |
| Macaca mulatta | Query 288 PPEGEEGNEELRQKFERAKIGQGTKVTEEK#TANAISKIDNNGNRDRMKLTDFNFLMVLGK 347 |||| ||||||||||||||| ||||| |||#| | +|| |||||||||||||||||||||| Sbjct 291 PPEGSEGNEELRQKFERAKISQGTKVPEEK#TTNTVSKFDNNGNRDRMKLTDFNFLMVLGK 350 |
| Canis familiaris | Query 288 PPEGEEGNEELRQKFERAKIGQGTKVTEEK#TANAISKIDNNGNRDRMKLTDFNFLMVLGK 347 |||| ||||||||||||||| |||| |||#| | |||+|||||||||||||||||||||| Sbjct 283 PPEGSEGNEELRQKFERAKISQGTKAPEEK#TTNTISKLDNNGNRDRMKLTDFNFLMVLGK 342 |
| Homo sapiens | Query 288 PPEGEEGNEELRQKFERAKIGQGTKVTEEK#TANAISKIDNNGNRDRMKLTDFNFLMVLGK 347 |||| | ||||||||||||| ||||| |||#| | +|| |||||||||||||||||||||| Sbjct 291 PPEGSEANEELRQKFERAKISQGTKVPEEK#TTNTVSKFDNNGNRDRMKLTDFNFLMVLGK 350 |
| Gallus gallus | Query 288 PPEGEEGNEELRQKFERAKIGQGTKVTEEK#TANAISKIDNNGNRDRMKLTDFNFLMVLGK 347 ||||||||||||||||||||| |+| +||#| ||||| ||||+||||||+|||||||||| Sbjct 290 PPEGEEGNEELRQKFERAKIGTGSKAADEK#TTNAISKFDNNGSRDRMKLSDFNFLMVLGK 349 |
| Bos taurus | Query 288 PPEGEEGNEELRQKFERAKIGQGTKVTEEK#TANAISKIDNNGNRDRMKLTDFNFLMVLGK 347 |||| |||||||||||||||| | | |||#| | ||| |||||||||||||||||||||| Sbjct 291 PPEGSEGNEELRQKFERAKIGPGPKTPEEK#TTNTISKFDNNGNRDRMKLTDFNFLMVLGK 350 |
| Latimeria chalumnae | Query 288 PPEGEEGNEELRQKFERAKIGQGTKVTEEK#TANAISKIDNNGNRDRMKLTDFNFLMVLGK 347 ||||||||||||+|||||||| |+| ||+|# |+ || |||||||+||+|||||||||| Sbjct 213 PPEGEEGNEELRKKFERAKIGPGSKATEDK#VKNSSSKFGNNGNRDRIKLSDFNFLMVLGK 272 |
| Protopterus dolloi | Query 288 PPEGEEGNEELRQKFERAKIGQGTKVTEEK#TANAISKIDNNGNRDRMKLTDFNFLMVLGK 347 ||||||||||||||||||||| | + |+|# ||| +| ||||||||+||+|||||||||| Sbjct 222 PPEGEEGNEELRQKFERAKIGPGAEAAEDK#IANA-AKFDNNGNRDRVKLSDFNFLMVLGK 280 |
| Tetraodon nigroviridis | Query 288 PPEGEEGNEELRQKFERAKIGQGTKVTEEK#TANAISKIDNNGNRDRMKLTDFNFLMVLGK 347 ||||||||||||||||||||| | | |+ |#+||| |+ |+|||+||||| |||||||||| Sbjct 227 PPEGEEGNEELRQKFERAKIGPG-KNTDGK#SANAASRFDSNGNQDRMKLADFNFLMVLGK 285 |
| Danio rerio | Query 288 PPEGEEGNEELRQKFERAKIGQGTKVTEEK#TANAISKIDNNGNRDRMKLTDFNFLMVLGK 347 ||||||||||||||||||||| |+ #++||||| |+|||||||||+|||||||||| Sbjct 290 PPEGEEGNEELRQKFERAKIGPSK--TDGS#SSNAISKFDSNGNRDRMKLSDFNFLMVLGK 347 |
| Scyliorhinus canicula | Query 288 PPEGEEGNEELRQKFERAKIGQGTKVTEEK#TANAISKIDNNGNRDRMKLTDFNFLMVLGK 347 | | |||||||||||||+|| |+| |||# +++ |||| ||||||||| |||||||||| Sbjct 212 PEEDSEGNEELRQKFERARIGPGSKAAEEK#RSSSWSKID-NGNRDRMKLADFNFLMVLGK 270 |
| Petromyzon marinus | Query 289 PEGEEGNEELRQKFERAKIGQGTK--VTEEK#TANAISKIDNNGNRDRMKLTDFNFLMVLG 346 |+ |+|+ |+|+|||+|++| | ++|+# ||++| | || | ||+| ||||||||| Sbjct 222 PDIEDGDTEMRRKFEKARLGPSVKPRASDER#RANSLSAILNNANVDRVKADDFNFLMVLG 281 Query 347 K 347 | Query 347 K 347 |
| synthetic construct | Query 289 PEG-EEGNEELRQKFERAKIGQ-GTKVTEEK#TANAISKIDNNGNRDRMKLTDFNFLMVLG 346 ||| |||| |||||||+||+| | || # + + | ||+|||||||||||| Sbjct 291 PEGDEEGNMELRQKFEKAKLGPAGNKVISPS#E----DRKQPSNNLDRVKLTDFNFLMVLG 346 Query 347 K 347 | Query 347 K 347 |
| Takifugu rubripes | Query 289 PEGEEGNEELRQKFERAKIGQGTKVTEEK#TANAISKIDNNGNRDRMKLTDFNFLMVLGK 347 || ++ | |||||||+||+||| || # | +|| ||++| ||||| +||| Sbjct 289 PEVDDVNLELRQKFEKAKLGQGKKVITPS#DHRRFSL--PSGNMDRVRLNDFNFLALLGK 345 |
| Xenopus laevis | Query 289 PEGEEGNEELRQKFERAKIGQGTKVTEEK#TANAISKIDNNGNRDRMKLTDFNFLMVLGK 347 || ++|| |||||||+||+| #| | +| | | ++|||| ||||||| Sbjct 295 PEADDGNLELRQKFEKAKLGPAGNKVINP#TGERRPYIPSN-NIDSIRLTDFCFLMVLGK 352 |
| Xenopus tropicalis | Query 288 PPEGEEGNEELRQKFERAKIGQGTKVTEEK#TANAISKIDNNGNRDRMKLTDFNFLMVLGK 347 | | ++|| |||||||+||+| #| + +| | | ++||||+||||||| Sbjct 295 PEEEDDGNIELRQKFEKAKLGPAGNKVISP#TDERRPYVPSN-NIDSVRLTDFSFLMVLGK 353 |
| Myxine glutinosa | Query 289 PEGEEGNEELRQKFERAKIGQGTKVTEEK#TANAISKIDNNGNRDRMKLTDFNFLMVLGK 347 |||||| | |||+ +|++| + | + # + | + ||+ | || || |||| Sbjct 213 PEGEEGLE-LRQRLQRSQIARSDKNKQSP#AKDQPSDSASLSGLDRVNLEDFVFLTVLGK 270 |
| Caenorhabditis briggsae | Query 289 PEGEEGNEELRQKF-ERAKIGQGTKVTE--EK#TANAISKIDNNGNRDRMKLTDFNFLMVL 345 || +| |++|+| + || + + + | #| | | | ||| +| +||||| || Sbjct 362 PEYDEDMEKVRKKMNDNFKISRDSSAGKPRES#TPRATSTSLTNTNRDVIKASDFNFLTVL 421 Query 346 GK 347 || Query 346 GK 347 |
| Branchiostoma lanceolatum | Query 292 EEGNEELRQKFERAKIGQGTKVTEEK#TANAISKIDNNGNRDRMKLTDFNFLMVLGK 347 || +|++ ++ |+ + | ++#+ | |+ | || |||||||||| Sbjct 225 EENVAKLKEHLQKQKLDEQRKQKSKR#SEEC-----NIGSLDHMKAADFNFLMVLGK 275 |
| Apis mellifera | Query 289 PEGEEGNEELRQKFERAKIGQGTKVTEEK#TANAISKIDNN-GNRDRMKLTDFNFLMVLGK 347 | ||| + | + + | | ||++ #| ++ +| | | ++ +|||||||||| Sbjct 306 PVPEEGVDLAELKMKPSP--QKTSVTKKT#TTTQDKEVPHNMGKSDLIRASDFNFLMVLGK 363 |
| Aedes aegypti | Query 289 PEGEEGNE--ELRQKFERAKIGQGTKVTEEK#TANAISKIDNNGNRDRMKLTDFNFLMVLG 346 | ||| + +|+ + + | + | +|# | | +| ++ |||||||||| Sbjct 237 PVPEEGTDLVQLKSQMRKTSISKKAPVLCDK#DVP-----HNMGKKDVIRATDFNFLMVLG 291 Query 347 K 347 | Query 347 K 347 |
| Ostreococcus tauri | Query 293 EGNEELRQKFERAK---IGQGTKVTEEK#TANAISKIDNNGNRDRMKLTDFNFLMVL 345 +| |++ ||+ +|+ || |#||| ++++ | + | +||| | ++ | Sbjct 15 DGTRARRRERTRARAFDVGKLAKVLRRK#TANDLARVFNGAEKTRERLTDVNDILAL 70 |
| Aplysia californica | Query 293 EGNEELRQKFERAKIGQGTKVTEEK#TANAISKIDNNGNRDRMKLTDFNFLMVLGK 347 | +|++ | |+ | + ||# |+ | +| ++ +||||| |||| Sbjct 280 ESIQEIKSKMHRSSI------SSEK#RYPEPDKVQNMSKQDIVRASDFNFLTVLGK 328 |
| Trichomonas vaginalis G3 | Query 290 EGEEGNEELRQKFERAKIGQGTKVTEEK#TANAISKIDNNG-NRDRMKLTD 338 | ++ ||+| |+ |+|| |+ |||#| | | |+ | + || | Sbjct 436 ENKKLNEKLNQELEKAKSDLSQKIEEEK#TLNGKSDIEEKTLNSLKEKLED 485 |
| Tribolium castaneum | Query 290 EGEEGNEELRQKFERAKIGQGTKVTEEK#TANAISKIDNNGNRDRMKLTDFNFLMVLGK 347 ||+ |+ + | + ||+ |||# || ++ ++ | ||||+ |||| Sbjct 311 EGQSENQAPEDEMENTD-ERLTKMMEEK#MANNGGDGAERKSKSKIGLEDFNFIKVLGK 367 |
[Site 2] RQKFERAKIG308-QGTKVTEEKT
Gly308  Gln
Gln
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Arg299 | Gln300 | Lys301 | Phe302 | Glu303 | Arg304 | Ala305 | Lys306 | Ile307 | Gly308 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Gln309 | Gly310 | Thr311 | Lys312 | Val313 | Thr314 | Glu315 | Glu316 | Lys317 | Thr318 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| EGEYFNVPVPPEGEEGNEELRQKFERAKIGQGTKVTEEKTANAISKIDNNGNRDRMKLTD |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | Monodelphis domestica | 89.70 | 3 | PREDICTED: similar to Protein kinase C beta type ( |
| 2 | N/A | 82.80 | 8 | K |
| 3 | Mus musculus | 82.80 | 4 | protein kinase C, beta 1 |
| 4 | Rattus norvegicus | 82.80 | 4 | unnamed protein product |
| 5 | Oryctolagus cuniculus | 81.30 | 3 | KPCB_RABIT Protein kinase C beta type (PKC-beta) ( |
| 6 | Homo sapiens | 80.10 | 8 | protein kinase C, beta isoform 1 |
| 7 | Macaca mulatta | 80.10 | 3 | PREDICTED: protein kinase C, beta isoform 1 |
| 8 | Canis familiaris | 79.70 | 2 | PREDICTED: similar to Protein kinase C, beta type |
| 9 | Bos taurus | 76.60 | 3 | Unknown (protein for MGC:148660) |
| 10 | Gallus gallus | 74.70 | 3 | PREDICTED: similar to protein kinase C beta-1 |
| 11 | Latimeria chalumnae | 67.00 | 1 | protein kinase C beta 1 |
| 12 | Protopterus dolloi | 65.10 | 1 | protein kinase C beta 1 |
| 13 | Tetraodon nigroviridis | 62.80 | 1 | unnamed protein product |
| 14 | Danio rerio | 60.80 | 5 | protein kinase C, beta 1 |
| 15 | Scyliorhinus canicula | 57.40 | 1 | protein kinase C |
| 16 | Petromyzon marinus | 44.70 | 1 | protein kinase C |
| 17 | Takifugu rubripes | 37.40 | 1 | protein kinase C, alpha type |
| 18 | synthetic construct | 35.00 | 1 | protein kinase C alpha |
Top-ranked sequences
| organism | matching |
|---|---|
| Monodelphis domestica | Query 279 EGEYFXXXXXXXXXXXXXXLRQKFERAKIG#QGTKVTEEKTANAISKIDNNGNRDRMKLTD 338 ||||| |||||||||||#|||||||||||||||||||||||||||||| Sbjct 279 EGEYFNVPVPPEGEEGNEELRQKFERAKIG#QGTKVTEEKTANAISKIDNNGNRDRMKLTD 338 |
| N/A | Query 279 EGEYFXXXXXXXXXXXXXXLRQKFERAKIG#QGTKVTEEKTANAISKIDNNGNRDRMKLTD 338 ||||| |||||||||||#|||| |||||| ||| ||||||||||||| Sbjct 282 EGEYFNVPVPPEGSEGNEELRQKFERAKIG#QGTKAPEEKTANTISKFDNNGNRDRMKLTD 341 |
| Mus musculus | Query 279 EGEYFXXXXXXXXXXXXXXLRQKFERAKIG#QGTKVTEEKTANAISKIDNNGNRDRMKLTD 338 ||||| |||||||||||#|||| |||||| ||| ||||||||||||| Sbjct 282 EGEYFNVPVPPEGSEGNEELRQKFERAKIG#QGTKAPEEKTANTISKFDNNGNRDRMKLTD 341 |
| Rattus norvegicus | Query 279 EGEYFXXXXXXXXXXXXXXLRQKFERAKIG#QGTKVTEEKTANAISKIDNNGNRDRMKLTD 338 ||||| |||||||||||#|||| |||||| ||| ||||||||||||| Sbjct 282 EGEYFNVPVPPEGSEGNEELRQKFERAKIG#QGTKAPEEKTANTISKFDNNGNRDRMKLTD 341 |
| Oryctolagus cuniculus | Query 279 EGEYFXXXXXXXXXXXXXXLRQKFERAKIG#QGTKVTEEKTANAISKIDNNGNRDRMKLTD 338 ||||| |||||||||||#|||| |||| | ||| ||||||||||||| Sbjct 282 EGEYFNVPVPPEGSEGNEELRQKFERAKIG#QGTKTPEEKTTNTISKFDNNGNRDRMKLTD 341 |
| Homo sapiens | Query 279 EGEYFXXXXXXXXXXXXXXLRQKFERAKIG#QGTKVTEEKTANAISKIDNNGNRDRMKLTD 338 ||||| |||||||||| #||||| |||| | +|| ||||||||||||| Sbjct 282 EGEYFNVPVPPEGSEANEELRQKFERAKIS#QGTKVPEEKTTNTVSKFDNNGNRDRMKLTD 341 |
| Macaca mulatta | Query 279 EGEYFXXXXXXXXXXXXXXLRQKFERAKIG#QGTKVTEEKTANAISKIDNNGNRDRMKLTD 338 ||||| |||||||||| #||||| |||| | +|| ||||||||||||| Sbjct 282 EGEYFNVPVPPEGSEGNEELRQKFERAKIS#QGTKVPEEKTTNTVSKFDNNGNRDRMKLTD 341 |
| Canis familiaris | Query 279 EGEYFXXXXXXXXXXXXXXLRQKFERAKIG#QGTKVTEEKTANAISKIDNNGNRDRMKLTD 338 ||||| |||||||||| #|||| |||| | |||+||||||||||||| Sbjct 274 EGEYFNVPVPPEGSEGNEELRQKFERAKIS#QGTKAPEEKTTNTISKLDNNGNRDRMKLTD 333 |
| Bos taurus | Query 279 EGEYFXXXXXXXXXXXXXXLRQKFERAKIG#QGTKVTEEKTANAISKIDNNGNRDRMKLTD 338 ||||| |||||||||||# | | |||| | ||| ||||||||||||| Sbjct 282 EGEYFNVPVPPEGSEGNEELRQKFERAKIG#PGPKTPEEKTTNTISKFDNNGNRDRMKLTD 341 |
| Gallus gallus | Query 279 EGEYFXXXXXXXXXXXXXXLRQKFERAKIG#QGTKVTEEKTANAISKIDNNGNRDRMKLTD 338 ||||| |||||||||||# |+| +||| ||||| ||||+||||||+| Sbjct 281 EGEYFNVPVPPEGEEGNEELRQKFERAKIG#TGSKAADEKTTNAISKFDNNGSRDRMKLSD 340 |
| Latimeria chalumnae | Query 279 EGEYFXXXXXXXXXXXXXXLRQKFERAKIG#QGTKVTEEKTANAISKIDNNGNRDRMKLTD 338 ||||| ||+||||||||# |+| ||+| |+ || |||||||+||+| Sbjct 204 EGEYFNVPVPPEGEEGNEELRKKFERAKIG#PGSKATEDKVKNSSSKFGNNGNRDRIKLSD 263 |
| Protopterus dolloi | Query 279 EGEYFXXXXXXXXXXXXXXLRQKFERAKIG#QGTKVTEEKTANAISKIDNNGNRDRMKLTD 338 ||||| |||||||||||# | + |+| ||| +| ||||||||+||+| Sbjct 213 EGEYFNVPVPPEGEEGNEELRQKFERAKIG#PGAEAAEDKIANA-AKFDNNGNRDRVKLSD 271 |
| Tetraodon nigroviridis | Query 279 EGEYFXXXXXXXXXXXXXXLRQKFERAKIG#QGTKVTEEKTANAISKIDNNGNRDRMKLTD 338 ||||| |||||||||||# | | |+ |+||| |+ |+|||+||||| | Sbjct 218 EGEYFNVPVPPEGEEGNEELRQKFERAKIG#PG-KNTDGKSANAASRFDSNGNQDRMKLAD 276 |
| Danio rerio | Query 279 EGEYFXXXXXXXXXXXXXXLRQKFERAKIG#QGTKVTEEKTANAISKIDNNGNRDRMKLTD 338 ||||| |||||||||||# |+ ++||||| |+|||||||||+| Sbjct 281 EGEYFNVPVPPEGEEGNEELRQKFERAKIG#PSK--TDGSSSNAISKFDSNGNRDRMKLSD 338 |
| Scyliorhinus canicula | Query 298 LRQKFERAKIG#QGTKVTEEKTANAISKIDNNGNRDRMKLTD 338 ||||||||+||# |+| ||| +++ |||| ||||||||| | Sbjct 222 LRQKFERARIG#PGSKAAEEKRSSSWSKID-NGNRDRMKLAD 261 |
| Petromyzon marinus | Query 279 EGEYFXXXXXXXXXXXXXXLRQKFERAKIG#QGTK--VTEEKTANAISKIDNNGNRDRMKL 336 ||||+ +|+|||+|++|# | ++|+ ||++| | || | ||+| Sbjct 212 EGEYYNVPIAPDIEDGDTEMRRKFEKARLG#PSVKPRASDERRANSLSAILNNANVDRVKA 271 Query 337 TD 338 | Sbjct 272 DD 273 |
| Takifugu rubripes | Query 279 EGEYFXXXXXXXXXXXXXXLRQKFERAKIG#QGTKVTEEKTANAISKIDNNGNRDRMKLTD 338 ||||+ ||||||+||+|#|| || | +|| ||++| | Sbjct 280 EGEYYNVPIPEVDDVNLE-LRQKFEKAKLG#QGKKVITPSDHRRFSL--PSGNMDRVRLND 336 |
| synthetic construct | Query 279 EGEYFXXXXXXXXXXXXXXLRQKFERAKIG-#QGTKVTEEKTANAISKIDNNGNRDRMKLT 337 ||||+ ||||||+||+| # | || + + | ||+||| Sbjct 282 EGEYYNVPIPEGDEEGNMELRQKFEKAKLGP#AGNKVISPSE----DRKQPSNNLDRVKLT 337 Query 338 D 338 | Query 338 D 338 |
* References
None.