XSB1765 : Os12g0163700 [Oryza sativa (japonica cultivar-group)]
[ CaMP Format ]
This entry is computationally expanded from SB0071
* Basic Information
| Organism | Oryza sativa Japonica Group |
| Protein Names | Os12g0163700 protein; hypothetical protein |
| Gene Names | Os12g0163700 |
| Gene Locus | Not available |
| GO Function | Not available |
| Entrez Protein | Entrez Nucleotide | Entrez Gene | UniProt | OMIM | HGNC | HPRD | KEGG |
|---|---|---|---|---|---|---|---|
| NP_001066233 | NM_001072765 | 4351585 | Q0IPW3 | N/A | N/A | N/A | osa:4351585 |
* Information From OMIM
Not Available.
* Structure Information
1. Primary Information
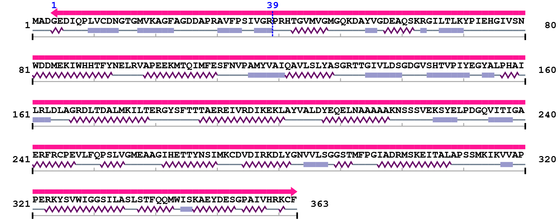
Length: 363 aa
Average Mass: 39.863 kDa
Monoisotopic Mass: 39.837 kDa
2. Domain Information
Annotated Domains: interpro / pfam / smart / prosite
Computationally Assigned Domains (Pfam+HMMER):
| domain name | begin | end | score | e-value |
|---|---|---|---|---|
| Actin 1. | 4 | 363 | 740.9 | 9.6e-220 |
| --- cleavage 39 (inside Actin 4..363) --- | ||||
3. Sequence Information
Fasta Sequence: XSB1765.fasta
Amino Acid Sequence and Secondary Structures (PsiPred):
4. 3D Information
Not Available.
* Cleavage Information
1 [sites]
Cleavage sites (±10aa)
[Site 1] RAVFPSIVGR39-PRHTGVMVGM
Arg39  Pro
Pro
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Arg30 | Ala31 | Val32 | Phe33 | Pro34 | Ser35 | Ile36 | Val37 | Gly38 | Arg39 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Pro40 | Arg41 | His42 | Thr43 | Gly44 | Val45 | Met46 | Val47 | Gly48 | Met49 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| LVCDNGTGMVKAGFAGDDAPRAVFPSIVGRPRHTGVMVGMGQKDAYVGDEAQSKRGILTL |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | N/A | 125.00 | 19 | - |
| 2 | Gossypium hirsutum | 125.00 | 16 | actin |
| 3 | Oryza sativa (japonica cultivar-group) | 125.00 | 13 | Os12g0163700 |
| 4 | Arabidopsis thaliana | 125.00 | 12 | actin 4 |
| 5 | Oryza sativa (indica cultivar-group) | 125.00 | 9 | hypothetical protein OsI_013714 |
| 6 | Solanum tuberosum | 125.00 | 6 | ACT13_SOLTU Actin-101 emb |
| 7 | Vitis vinifera | 125.00 | 5 | hypothetical protein |
| 8 | Pisum sativum | 125.00 | 5 | actin |
| 9 | Physcomitrella patens | 125.00 | 4 | actin 1 |
| 10 | Musa acuminata | 125.00 | 2 | AF246288_1 actin |
| 11 | Phalaenopsis sp. 'True Lady' | 125.00 | 2 | AF246714_1 actin-like protein |
| 12 | Striga asiatica | 125.00 | 2 | actin |
| 13 | Torenia fournieri | 125.00 | 2 | actin 1 |
| 14 | Nicotiana tabacum | 125.00 | 2 | actin |
| 15 | Brassica napus | 125.00 | 1 | actin |
| 16 | Stevia rebaudiana | 125.00 | 1 | actin |
| 17 | Picea rubens | 125.00 | 1 | AF172094_1 actin |
| 18 | Populus trichocarpa | 125.00 | 1 | actin |
| 19 | Malva pusilla | 125.00 | 1 | AF112538_1 actin |
| 20 | Elaeis oleifera | 125.00 | 1 | actin |
| 21 | Hordeum vulgare | 125.00 | 1 | actin |
| 22 | Medicago truncatula | 125.00 | 1 | Actin/actin-like |
| 23 | Trifolium pratense | 125.00 | 1 | actin |
| 24 | Ricinus communis | 125.00 | 1 | actin |
| 25 | Vigna radiata | 125.00 | 1 | AF143208_1 actin |
| 26 | Capsicum chinense | 125.00 | 1 | actin |
| 27 | Elaeis guineensis | 125.00 | 1 | actine |
| 28 | Musa x paradisiaca | 125.00 | 1 | AF285176_1 actin |
| 29 | Eucommia ulmoides | 125.00 | 1 | stem cambial region actin protein |
| 30 | Mimosa pudica | 125.00 | 1 | actin isoform B |
| 31 | Helianthus annuus | 125.00 | 1 | actin |
| 32 | Plantago major | 125.00 | 1 | actin 1 |
| 33 | Saccharum officinarum | 125.00 | 1 | actin |
| 34 | Triticum monococcum | 125.00 | 1 | AF326781_1 actin |
| 35 | Aegiceras corniculatum | 125.00 | 1 | actin |
| 36 | Sorghum bicolor | 125.00 | 1 | ACT1_SORBI Actin-1 emb |
| 37 | Phaseolus acutifolius | 125.00 | 1 | actin |
| 38 | Capsicum annuum | 125.00 | 1 | actin |
| 39 | Vicia faba | 125.00 | 1 | actin |
| 40 | Setaria italica | 125.00 | 1 | AF288226_1 actin |
| 41 | Phalaenopsis hybrid cultivar | 125.00 | 1 | actin |
| 42 | Brassica napus var. napus | 125.00 | 1 | actin |
| 43 | Anemia phyllitidis | 123.00 | 3 | actin 3 |
| 44 | Selaginella apoda | 123.00 | 2 | AF090969_1 actin |
| 45 | Glycine max | 123.00 | 2 | CAA23728.1 |
| 46 | Avena nuda | 123.00 | 1 | AF234528_2 actin 1 |
| 47 | Linum usitatissimum | 123.00 | 1 | actin |
| 48 | Chlamydomonas reinhardtii | 122.00 | 1 | ACT_CHLRE Actin dbj |
| 49 | Coleochaete scutata | 121.00 | 1 | ACT_COLSC Actin gb |
| 50 | Scherffelia dubia | 121.00 | 1 | ACT_SCHDU Actin gb |
| 51 | Branchiostoma belcheri | 120.00 | 1 | ACTM_BRABE Actin, muscle (BbMA1) dbj |
| 52 | Branchiostoma floridae | 120.00 | 1 | ACTM_BRAFL Actin, muscle dbj |
| 53 | Scyliorhinus retifer | 120.00 | 1 | AF388172_1 fast muscle actin |
| 54 | Mesostigma viride | 120.00 | 1 | ACT_MESVI Actin gb |
| 55 | Branchiostoma lanceolatum | 120.00 | 1 | ACTM_BRALA Actin, muscle emb |
| 56 | Danio rerio | 119.00 | 12 | hypothetical protein LOC415156 |
| 57 | Xenopus laevis | 119.00 | 8 | unnamed protein product |
| 58 | Homo sapiens | 119.00 | 8 | alpha 1 actin precursor |
| 59 | Mus musculus | 119.00 | 7 | unnamed protein product |
| 60 | Oreochromis niloticus | 119.00 | 7 | alpha actin |
| 61 | Bos taurus | 119.00 | 5 | actin, alpha 1, skeletal muscle |
| 62 | Xenopus tropicalis | 119.00 | 4 | actin, alpha 1, skeletal muscle |
| 63 | Cyprinus carpio | 119.00 | 4 | skeletal muscle actin mutant |
| 64 | synthetic construct | 119.00 | 4 | actin alpha 1 |
| 65 | Tetraodon nigroviridis | 119.00 | 3 | unnamed protein product |
| 66 | Rana catesbeiana | 119.00 | 2 | cardiac alpha actin 2 |
| 67 | Canis familiaris | 119.00 | 2 | PREDICTED: similar to Actin, alpha skeletal muscle |
| 68 | Molgula oculata | 119.00 | 2 | ACT2_MOLOC Actin, muscle-type (A2) dbj |
| 69 | Ictalurus punctatus | 119.00 | 2 | alpha actin |
| 70 | Macaca mulatta | 119.00 | 2 | PREDICTED: similar to Actin, alpha cardiac (Alpha- |
| 71 | Atractaspis microlepidota microlepidota | 119.00 | 1 | ACTS_ATRMM Actin, alpha skeletal muscle (Alpha-act |
| 72 | Nannochloris bacillaris | 119.00 | 1 | actin |
| 73 | Salmo trutta | 119.00 | 1 | AF267496_1 alpha actin |
| 74 | Gadus morhua | 119.00 | 1 | AF500273_1 fast skeletal muscle alpha-actin |
| 75 | Salmo salar | 119.00 | 1 | AF304406_1 fast myotomal muscle actin |
| 76 | Oxyuranus scutellatus scutellatus | 119.00 | 1 | alpha actin |
| 77 | Isatis tinctoria | 119.00 | 1 | actin |
| 78 | Halocynthia roretzi | 119.00 | 1 | muscle actin |
| 79 | Sphyraena idiastes | 119.00 | 1 | alpha actin |
| 80 | Oreochromis mossambicus | 119.00 | 1 | ACTSA_FUGRU Actin, alpha skeletal muscle A (Alpha- |
| 81 | Carassius auratus | 119.00 | 1 | ACTS_CARAU Actin, alpha skeletal muscle (Alpha-act |
| 82 | Sparus aurata | 119.00 | 1 | skeletal alpha-actin |
| 83 | Oryzias latipes | 119.00 | 1 | ACTS_ORYLA Actin, alpha skeletal muscle (Alpha-act |
| 84 | Ambystoma mexicanum | 119.00 | 1 | AF276076_1 cardiac actin |
| 85 | Litopenaeus vannamei | 118.00 | 1 | actin T2 |
| 86 | Haliotis iris | 118.00 | 1 | actin A1 |
| 87 | Gillichthys mirabilis | 118.00 | 1 | AF266233_1 skeletal alpha-actin |
| 88 | Notothenia coriiceps | 118.00 | 1 | alpha actin |
| 89 | Haliotis rufescens | 118.00 | 1 | actin |
| 90 | Morus alba | 118.00 | 1 | actin |
| 91 | Ostreococcus lucimarinus CCE9901 | 118.00 | 1 | predicted protein |
| 92 | Gallus gallus | 117.00 | 3 | smooth muscle gamma actin |
| 93 | Penaeus monodon | 117.00 | 1 | actin 1 |
| 94 | Bubalus bubalis | 117.00 | 1 | alpha 2 actin |
| 95 | Portunus pelagicus | 117.00 | 1 | actin |
| 96 | Coryphaenoides yaquinae | 117.00 | 1 | skeletal alpha-actin type-2b |
| 97 | Coryphaenoides armatus | 117.00 | 1 | skeletal alpha-actin type-2b |
| 98 | Monodelphis domestica | 117.00 | 1 | PREDICTED: hypothetical protein |
| 99 | Homarus americanus | 117.00 | 1 | AF399872_1 alpha actin |
| 100 | Ciona savignyi | 117.00 | 1 | ACTM_CIOSA Actin, muscle dbj |
| 101 | Pan troglodytes | 117.00 | 1 | PREDICTED: similar to actin alpha 1 skeletal muscl |
Top-ranked sequences
| organism | matching |
|---|---|
| N/A | Query 10 LVCDNGTGMVKAGFAGDDAPRAVFPSIVGR#PRHTGVMVGMGQKDAYVGDEAQSKRGILTL 69 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 10 LVCDNGTGMVKAGFAGDDAPRAVFPSIVGR#PRHTGVMVGMGQKDAYVGDEAQSKRGILTL 69 |
| Gossypium hirsutum | Query 10 LVCDNGTGMVKAGFAGDDAPRAVFPSIVGR#PRHTGVMVGMGQKDAYVGDEAQSKRGILTL 69 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 10 LVCDNGTGMVKAGFAGDDAPRAVFPSIVGR#PRHTGVMVGMGQKDAYVGDEAQSKRGILTL 69 |
| Oryza sativa (japonica cultivar-group) | Query 10 LVCDNGTGMVKAGFAGDDAPRAVFPSIVGR#PRHTGVMVGMGQKDAYVGDEAQSKRGILTL 69 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 10 LVCDNGTGMVKAGFAGDDAPRAVFPSIVGR#PRHTGVMVGMGQKDAYVGDEAQSKRGILTL 69 |
| Arabidopsis thaliana | Query 10 LVCDNGTGMVKAGFAGDDAPRAVFPSIVGR#PRHTGVMVGMGQKDAYVGDEAQSKRGILTL 69 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 10 LVCDNGTGMVKAGFAGDDAPRAVFPSIVGR#PRHTGVMVGMGQKDAYVGDEAQSKRGILTL 69 |
| Oryza sativa (indica cultivar-group) | Query 10 LVCDNGTGMVKAGFAGDDAPRAVFPSIVGR#PRHTGVMVGMGQKDAYVGDEAQSKRGILTL 69 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 10 LVCDNGTGMVKAGFAGDDAPRAVFPSIVGR#PRHTGVMVGMGQKDAYVGDEAQSKRGILTL 69 |
| Solanum tuberosum | Query 10 LVCDNGTGMVKAGFAGDDAPRAVFPSIVGR#PRHTGVMVGMGQKDAYVGDEAQSKRGILTL 69 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 10 LVCDNGTGMVKAGFAGDDAPRAVFPSIVGR#PRHTGVMVGMGQKDAYVGDEAQSKRGILTL 69 |
| Vitis vinifera | Query 10 LVCDNGTGMVKAGFAGDDAPRAVFPSIVGR#PRHTGVMVGMGQKDAYVGDEAQSKRGILTL 69 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 10 LVCDNGTGMVKAGFAGDDAPRAVFPSIVGR#PRHTGVMVGMGQKDAYVGDEAQSKRGILTL 69 |
| Pisum sativum | Query 10 LVCDNGTGMVKAGFAGDDAPRAVFPSIVGR#PRHTGVMVGMGQKDAYVGDEAQSKRGILTL 69 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 10 LVCDNGTGMVKAGFAGDDAPRAVFPSIVGR#PRHTGVMVGMGQKDAYVGDEAQSKRGILTL 69 |
| Physcomitrella patens | Query 10 LVCDNGTGMVKAGFAGDDAPRAVFPSIVGR#PRHTGVMVGMGQKDAYVGDEAQSKRGILTL 69 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 10 LVCDNGTGMVKAGFAGDDAPRAVFPSIVGR#PRHTGVMVGMGQKDAYVGDEAQSKRGILTL 69 |
| Musa acuminata | Query 10 LVCDNGTGMVKAGFAGDDAPRAVFPSIVGR#PRHTGVMVGMGQKDAYVGDEAQSKRGILTL 69 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 10 LVCDNGTGMVKAGFAGDDAPRAVFPSIVGR#PRHTGVMVGMGQKDAYVGDEAQSKRGILTL 69 |
| Phalaenopsis sp. 'True Lady' | Query 10 LVCDNGTGMVKAGFAGDDAPRAVFPSIVGR#PRHTGVMVGMGQKDAYVGDEAQSKRGILTL 69 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 10 LVCDNGTGMVKAGFAGDDAPRAVFPSIVGR#PRHTGVMVGMGQKDAYVGDEAQSKRGILTL 69 |
| Striga asiatica | Query 10 LVCDNGTGMVKAGFAGDDAPRAVFPSIVGR#PRHTGVMVGMGQKDAYVGDEAQSKRGILTL 69 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 10 LVCDNGTGMVKAGFAGDDAPRAVFPSIVGR#PRHTGVMVGMGQKDAYVGDEAQSKRGILTL 69 |
| Torenia fournieri | Query 10 LVCDNGTGMVKAGFAGDDAPRAVFPSIVGR#PRHTGVMVGMGQKDAYVGDEAQSKRGILTL 69 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 10 LVCDNGTGMVKAGFAGDDAPRAVFPSIVGR#PRHTGVMVGMGQKDAYVGDEAQSKRGILTL 69 |
| Nicotiana tabacum | Query 10 LVCDNGTGMVKAGFAGDDAPRAVFPSIVGR#PRHTGVMVGMGQKDAYVGDEAQSKRGILTL 69 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 10 LVCDNGTGMVKAGFAGDDAPRAVFPSIVGR#PRHTGVMVGMGQKDAYVGDEAQSKRGILTL 69 |
| Brassica napus | Query 10 LVCDNGTGMVKAGFAGDDAPRAVFPSIVGR#PRHTGVMVGMGQKDAYVGDEAQSKRGILTL 69 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 10 LVCDNGTGMVKAGFAGDDAPRAVFPSIVGR#PRHTGVMVGMGQKDAYVGDEAQSKRGILTL 69 |
| Stevia rebaudiana | Query 10 LVCDNGTGMVKAGFAGDDAPRAVFPSIVGR#PRHTGVMVGMGQKDAYVGDEAQSKRGILTL 69 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 10 LVCDNGTGMVKAGFAGDDAPRAVFPSIVGR#PRHTGVMVGMGQKDAYVGDEAQSKRGILTL 69 |
| Picea rubens | Query 10 LVCDNGTGMVKAGFAGDDAPRAVFPSIVGR#PRHTGVMVGMGQKDAYVGDEAQSKRGILTL 69 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 10 LVCDNGTGMVKAGFAGDDAPRAVFPSIVGR#PRHTGVMVGMGQKDAYVGDEAQSKRGILTL 69 |
| Populus trichocarpa | Query 10 LVCDNGTGMVKAGFAGDDAPRAVFPSIVGR#PRHTGVMVGMGQKDAYVGDEAQSKRGILTL 69 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 10 LVCDNGTGMVKAGFAGDDAPRAVFPSIVGR#PRHTGVMVGMGQKDAYVGDEAQSKRGILTL 69 |
| Malva pusilla | Query 10 LVCDNGTGMVKAGFAGDDAPRAVFPSIVGR#PRHTGVMVGMGQKDAYVGDEAQSKRGILTL 69 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 10 LVCDNGTGMVKAGFAGDDAPRAVFPSIVGR#PRHTGVMVGMGQKDAYVGDEAQSKRGILTL 69 |
| Elaeis oleifera | Query 10 LVCDNGTGMVKAGFAGDDAPRAVFPSIVGR#PRHTGVMVGMGQKDAYVGDEAQSKRGILTL 69 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 10 LVCDNGTGMVKAGFAGDDAPRAVFPSIVGR#PRHTGVMVGMGQKDAYVGDEAQSKRGILTL 69 |
| Hordeum vulgare | Query 10 LVCDNGTGMVKAGFAGDDAPRAVFPSIVGR#PRHTGVMVGMGQKDAYVGDEAQSKRGILTL 69 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 10 LVCDNGTGMVKAGFAGDDAPRAVFPSIVGR#PRHTGVMVGMGQKDAYVGDEAQSKRGILTL 69 |
| Medicago truncatula | Query 10 LVCDNGTGMVKAGFAGDDAPRAVFPSIVGR#PRHTGVMVGMGQKDAYVGDEAQSKRGILTL 69 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 10 LVCDNGTGMVKAGFAGDDAPRAVFPSIVGR#PRHTGVMVGMGQKDAYVGDEAQSKRGILTL 69 |
| Trifolium pratense | Query 10 LVCDNGTGMVKAGFAGDDAPRAVFPSIVGR#PRHTGVMVGMGQKDAYVGDEAQSKRGILTL 69 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 10 LVCDNGTGMVKAGFAGDDAPRAVFPSIVGR#PRHTGVMVGMGQKDAYVGDEAQSKRGILTL 69 |
| Ricinus communis | Query 10 LVCDNGTGMVKAGFAGDDAPRAVFPSIVGR#PRHTGVMVGMGQKDAYVGDEAQSKRGILTL 69 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 10 LVCDNGTGMVKAGFAGDDAPRAVFPSIVGR#PRHTGVMVGMGQKDAYVGDEAQSKRGILTL 69 |
| Vigna radiata | Query 10 LVCDNGTGMVKAGFAGDDAPRAVFPSIVGR#PRHTGVMVGMGQKDAYVGDEAQSKRGILTL 69 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 10 LVCDNGTGMVKAGFAGDDAPRAVFPSIVGR#PRHTGVMVGMGQKDAYVGDEAQSKRGILTL 69 |
| Capsicum chinense | Query 10 LVCDNGTGMVKAGFAGDDAPRAVFPSIVGR#PRHTGVMVGMGQKDAYVGDEAQSKRGILTL 69 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 10 LVCDNGTGMVKAGFAGDDAPRAVFPSIVGR#PRHTGVMVGMGQKDAYVGDEAQSKRGILTL 69 |
| Elaeis guineensis | Query 10 LVCDNGTGMVKAGFAGDDAPRAVFPSIVGR#PRHTGVMVGMGQKDAYVGDEAQSKRGILTL 69 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 10 LVCDNGTGMVKAGFAGDDAPRAVFPSIVGR#PRHTGVMVGMGQKDAYVGDEAQSKRGILTL 69 |
| Musa x paradisiaca | Query 10 LVCDNGTGMVKAGFAGDDAPRAVFPSIVGR#PRHTGVMVGMGQKDAYVGDEAQSKRGILTL 69 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 10 LVCDNGTGMVKAGFAGDDAPRAVFPSIVGR#PRHTGVMVGMGQKDAYVGDEAQSKRGILTL 69 |
| Eucommia ulmoides | Query 10 LVCDNGTGMVKAGFAGDDAPRAVFPSIVGR#PRHTGVMVGMGQKDAYVGDEAQSKRGILTL 69 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 10 LVCDNGTGMVKAGFAGDDAPRAVFPSIVGR#PRHTGVMVGMGQKDAYVGDEAQSKRGILTL 69 |
| Mimosa pudica | Query 10 LVCDNGTGMVKAGFAGDDAPRAVFPSIVGR#PRHTGVMVGMGQKDAYVGDEAQSKRGILTL 69 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 10 LVCDNGTGMVKAGFAGDDAPRAVFPSIVGR#PRHTGVMVGMGQKDAYVGDEAQSKRGILTL 69 |
| Helianthus annuus | Query 10 LVCDNGTGMVKAGFAGDDAPRAVFPSIVGR#PRHTGVMVGMGQKDAYVGDEAQSKRGILTL 69 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 10 LVCDNGTGMVKAGFAGDDAPRAVFPSIVGR#PRHTGVMVGMGQKDAYVGDEAQSKRGILTL 69 |
| Plantago major | Query 10 LVCDNGTGMVKAGFAGDDAPRAVFPSIVGR#PRHTGVMVGMGQKDAYVGDEAQSKRGILTL 69 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 10 LVCDNGTGMVKAGFAGDDAPRAVFPSIVGR#PRHTGVMVGMGQKDAYVGDEAQSKRGILTL 69 |
| Saccharum officinarum | Query 10 LVCDNGTGMVKAGFAGDDAPRAVFPSIVGR#PRHTGVMVGMGQKDAYVGDEAQSKRGILTL 69 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 10 LVCDNGTGMVKAGFAGDDAPRAVFPSIVGR#PRHTGVMVGMGQKDAYVGDEAQSKRGILTL 69 |
| Triticum monococcum | Query 10 LVCDNGTGMVKAGFAGDDAPRAVFPSIVGR#PRHTGVMVGMGQKDAYVGDEAQSKRGILTL 69 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 10 LVCDNGTGMVKAGFAGDDAPRAVFPSIVGR#PRHTGVMVGMGQKDAYVGDEAQSKRGILTL 69 |
| Aegiceras corniculatum | Query 10 LVCDNGTGMVKAGFAGDDAPRAVFPSIVGR#PRHTGVMVGMGQKDAYVGDEAQSKRGILTL 69 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 10 LVCDNGTGMVKAGFAGDDAPRAVFPSIVGR#PRHTGVMVGMGQKDAYVGDEAQSKRGILTL 69 |
| Sorghum bicolor | Query 10 LVCDNGTGMVKAGFAGDDAPRAVFPSIVGR#PRHTGVMVGMGQKDAYVGDEAQSKRGILTL 69 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 10 LVCDNGTGMVKAGFAGDDAPRAVFPSIVGR#PRHTGVMVGMGQKDAYVGDEAQSKRGILTL 69 |
| Phaseolus acutifolius | Query 10 LVCDNGTGMVKAGFAGDDAPRAVFPSIVGR#PRHTGVMVGMGQKDAYVGDEAQSKRGILTL 69 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 10 LVCDNGTGMVKAGFAGDDAPRAVFPSIVGR#PRHTGVMVGMGQKDAYVGDEAQSKRGILTL 69 |
| Capsicum annuum | Query 10 LVCDNGTGMVKAGFAGDDAPRAVFPSIVGR#PRHTGVMVGMGQKDAYVGDEAQSKRGILTL 69 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 10 LVCDNGTGMVKAGFAGDDAPRAVFPSIVGR#PRHTGVMVGMGQKDAYVGDEAQSKRGILTL 69 |
| Vicia faba | Query 10 LVCDNGTGMVKAGFAGDDAPRAVFPSIVGR#PRHTGVMVGMGQKDAYVGDEAQSKRGILTL 69 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 10 LVCDNGTGMVKAGFAGDDAPRAVFPSIVGR#PRHTGVMVGMGQKDAYVGDEAQSKRGILTL 69 |
| Setaria italica | Query 10 LVCDNGTGMVKAGFAGDDAPRAVFPSIVGR#PRHTGVMVGMGQKDAYVGDEAQSKRGILTL 69 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 10 LVCDNGTGMVKAGFAGDDAPRAVFPSIVGR#PRHTGVMVGMGQKDAYVGDEAQSKRGILTL 69 |
| Phalaenopsis hybrid cultivar | Query 10 LVCDNGTGMVKAGFAGDDAPRAVFPSIVGR#PRHTGVMVGMGQKDAYVGDEAQSKRGILTL 69 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 10 LVCDNGTGMVKAGFAGDDAPRAVFPSIVGR#PRHTGVMVGMGQKDAYVGDEAQSKRGILTL 69 |
| Brassica napus var. napus | Query 10 LVCDNGTGMVKAGFAGDDAPRAVFPSIVGR#PRHTGVMVGMGQKDAYVGDEAQSKRGILTL 69 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 10 LVCDNGTGMVKAGFAGDDAPRAVFPSIVGR#PRHTGVMVGMGQKDAYVGDEAQSKRGILTL 69 |
| Anemia phyllitidis | Query 10 LVCDNGTGMVKAGFAGDDAPRAVFPSIVGR#PRHTGVMVGMGQKDAYVGDEAQSKRGILTL 69 ||||||+|||||||||||||||||||||||#|||||||||||||||||||||||||||||| Sbjct 10 LVCDNGSGMVKAGFAGDDAPRAVFPSIVGR#PRHTGVMVGMGQKDAYVGDEAQSKRGILTL 69 |
| Selaginella apoda | Query 10 LVCDNGTGMVKAGFAGDDAPRAVFPSIVGR#PRHTGVMVGMGQKDAYVGDEAQSKRGILTL 69 ||||||+|||||||||||||||||||||||#|||||||||||||||||||||||||||||| Sbjct 1 LVCDNGSGMVKAGFAGDDAPRAVFPSIVGR#PRHTGVMVGMGQKDAYVGDEAQSKRGILTL 60 |
| Glycine max | Query 10 LVCDNGTGMVKAGFAGDDAPRAVFPSIVGR#PRHTGVMVGMGQKDAYVGDEAQSKRGILTL 69 ||||||||||||||||||||||||||||||#|||||||||||||| ||||||||||||||| Sbjct 10 LVCDNGTGMVKAGFAGDDAPRAVFPSIVGR#PRHTGVMVGMGQKDXYVGDEAQSKRGILTL 69 |
| Avena nuda | Query 11 VCDNGTGMVKAGFAGDDAPRAVFPSIVGR#PRHTGVMVGMGQKDAYVGDEAQSKRGILTL 69 |||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 11 VCDNGTGMVKAGFAGDDAPRAVFPSIVGR#PRHTGVMVGMGQKDAYVGDEAQSKRGILTL 69 |
| Linum usitatissimum | Query 11 VCDNGTGMVKAGFAGDDAPRAVFPSIVGR#PRHTGVMVGMGQKDAYVGDEAQSKRGILTL 69 |||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 11 VCDNGTGMVKAGFAGDDAPRAVFPSIVGR#PRHTGVMVGMGQKDAYVGDEAQSKRGILTL 69 |
| Chlamydomonas reinhardtii | Query 10 LVCDNGTGMVKAGFAGDDAPRAVFPSIVGR#PRHTGVMVGMGQKDAYVGDEAQSKRGILTL 69 ||||||+|||||||||||||||||||||||#||||||||||||||+||||||||||||||| Sbjct 10 LVCDNGSGMVKAGFAGDDAPRAVFPSIVGR#PRHTGVMVGMGQKDSYVGDEAQSKRGILTL 69 |
| Coleochaete scutata | Query 10 LVCDNGTGMVKAGFAGDDAPRAVFPSIVGR#PRHTGVMVGMGQKDAYVGDEAQSKRGILTL 69 ||||||+|||||||||||||||||||||||#||| |||||||||||||||||||||||||| Sbjct 10 LVCDNGSGMVKAGFAGDDAPRAVFPSIVGR#PRHQGVMVGMGQKDAYVGDEAQSKRGILTL 69 |
| Scherffelia dubia | Query 10 LVCDNGTGMVKAGFAGDDAPRAVFPSIVGR#PRHTGVMVGMGQKDAYVGDEAQSKRGILTL 69 ||||||+|||||||||||||||||||||||#||| |||||||||||||||||||||||||| Sbjct 11 LVCDNGSGMVKAGFAGDDAPRAVFPSIVGR#PRHQGVMVGMGQKDAYVGDEAQSKRGILTL 70 |
| Branchiostoma belcheri | Query 10 LVCDNGTGMVKAGFAGDDAPRAVFPSIVGR#PRHTGVMVGMGQKDAYVGDEAQSKRGILTL 69 ||||||+|+|||||||||||||||||||||#||| |||||||||||||||||||||||||| Sbjct 12 LVCDNGSGLVKAGFAGDDAPRAVFPSIVGR#PRHQGVMVGMGQKDAYVGDEAQSKRGILTL 71 |
| Branchiostoma floridae | Query 10 LVCDNGTGMVKAGFAGDDAPRAVFPSIVGR#PRHTGVMVGMGQKDAYVGDEAQSKRGILTL 69 ||||||+|+|||||||||||||||||||||#||| |||||||||||||||||||||||||| Sbjct 11 LVCDNGSGLVKAGFAGDDAPRAVFPSIVGR#PRHQGVMVGMGQKDAYVGDEAQSKRGILTL 70 |
| Scyliorhinus retifer | Query 10 LVCDNGTGMVKAGFAGDDAPRAVFPSIVGR#PRHTGVMVGMGQKDAYVGDEAQSKRGILTL 69 ||||||+|+|||||||||||||||||||||#||| |||||||||||||||||||||||||| Sbjct 10 LVCDNGSGLVKAGFAGDDAPRAVFPSIVGR#PRHQGVMVGMGQKDAYVGDEAQSKRGILTL 69 |
| Mesostigma viride | Query 10 LVCDNGTGMVKAGFAGDDAPRAVFPSIVGR#PRHTGVMVGMGQKDAYVGDEAQSKRGILTL 69 |+||||+|||||||||||||||||||||||#||| |||||||||||||||||||||||||| Sbjct 10 LMCDNGSGMVKAGFAGDDAPRAVFPSIVGR#PRHQGVMVGMGQKDAYVGDEAQSKRGILTL 69 |
| Branchiostoma lanceolatum | Query 10 LVCDNGTGMVKAGFAGDDAPRAVFPSIVGR#PRHTGVMVGMGQKDAYVGDEAQSKRGILTL 69 ||||||+|+|||||||||||||||||||||#||| |||||||||||||||||||||||||| Sbjct 13 LVCDNGSGLVKAGFAGDDAPRAVFPSIVGR#PRHQGVMVGMGQKDAYVGDEAQSKRGILTL 72 |
| Danio rerio | Query 10 LVCDNGTGMVKAGFAGDDAPRAVFPSIVGR#PRHTGVMVGMGQKDAYVGDEAQSKRGILTL 69 ||||||+|+|||||||||||||||||||||#||| ||||||||||+||||||||||||||| Sbjct 10 LVCDNGSGLVKAGFAGDDAPRAVFPSIVGR#PRHQGVMVGMGQKDSYVGDEAQSKRGILTL 69 |
| Xenopus laevis | Query 10 LVCDNGTGMVKAGFAGDDAPRAVFPSIVGR#PRHTGVMVGMGQKDAYVGDEAQSKRGILTL 69 ||||||+|+|||||||||||||||||||||#||| ||||||||||+||||||||||||||| Sbjct 10 LVCDNGSGLVKAGFAGDDAPRAVFPSIVGR#PRHQGVMVGMGQKDSYVGDEAQSKRGILTL 69 |
| Homo sapiens | Query 10 LVCDNGTGMVKAGFAGDDAPRAVFPSIVGR#PRHTGVMVGMGQKDAYVGDEAQSKRGILTL 69 ||||||+|+|||||||||||||||||||||#||| ||||||||||+||||||||||||||| Sbjct 10 LVCDNGSGLVKAGFAGDDAPRAVFPSIVGR#PRHQGVMVGMGQKDSYVGDEAQSKRGILTL 69 |
| Mus musculus | Query 10 LVCDNGTGMVKAGFAGDDAPRAVFPSIVGR#PRHTGVMVGMGQKDAYVGDEAQSKRGILTL 69 ||||||+|+|||||||||||||||||||||#||| ||||||||||+||||||||||||||| Sbjct 10 LVCDNGSGLVKAGFAGDDAPRAVFPSIVGR#PRHQGVMVGMGQKDSYVGDEAQSKRGILTL 69 |
| Oreochromis niloticus | Query 10 LVCDNGTGMVKAGFAGDDAPRAVFPSIVGR#PRHTGVMVGMGQKDAYVGDEAQSKRGILTL 69 ||||||+|+|||||||||||||||||||||#||| ||||||||||+||||||||||||||| Sbjct 5 LVCDNGSGLVKAGFAGDDAPRAVFPSIVGR#PRHQGVMVGMGQKDSYVGDEAQSKRGILTL 64 |
| Bos taurus | Query 10 LVCDNGTGMVKAGFAGDDAPRAVFPSIVGR#PRHTGVMVGMGQKDAYVGDEAQSKRGILTL 69 ||||||+|+|||||||||||||||||||||#||| ||||||||||+||||||||||||||| Sbjct 10 LVCDNGSGLVKAGFAGDDAPRAVFPSIVGR#PRHQGVMVGMGQKDSYVGDEAQSKRGILTL 69 |
| Xenopus tropicalis | Query 10 LVCDNGTGMVKAGFAGDDAPRAVFPSIVGR#PRHTGVMVGMGQKDAYVGDEAQSKRGILTL 69 ||||||+|+|||||||||||||||||||||#||| ||||||||||+||||||||||||||| Sbjct 10 LVCDNGSGLVKAGFAGDDAPRAVFPSIVGR#PRHQGVMVGMGQKDSYVGDEAQSKRGILTL 69 |
| Cyprinus carpio | Query 10 LVCDNGTGMVKAGFAGDDAPRAVFPSIVGR#PRHTGVMVGMGQKDAYVGDEAQSKRGILTL 69 ||||||+|+|||||||||||||||||||||#||| ||||||||||+||||||||||||||| Sbjct 10 LVCDNGSGLVKAGFAGDDAPRAVFPSIVGR#PRHQGVMVGMGQKDSYVGDEAQSKRGILTL 69 |
| synthetic construct | Query 10 LVCDNGTGMVKAGFAGDDAPRAVFPSIVGR#PRHTGVMVGMGQKDAYVGDEAQSKRGILTL 69 ||||||+|+|||||||||||||||||||||#||| ||||||||||+||||||||||||||| Sbjct 10 LVCDNGSGLVKAGFAGDDAPRAVFPSIVGR#PRHQGVMVGMGQKDSYVGDEAQSKRGILTL 69 |
| Tetraodon nigroviridis | Query 10 LVCDNGTGMVKAGFAGDDAPRAVFPSIVGR#PRHTGVMVGMGQKDAYVGDEAQSKRGILTL 69 ||||||+|+|||||||||||||||||||||#||| ||||||||||+||||||||||||||| Sbjct 10 LVCDNGSGLVKAGFAGDDAPRAVFPSIVGR#PRHQGVMVGMGQKDSYVGDEAQSKRGILTL 69 |
| Rana catesbeiana | Query 10 LVCDNGTGMVKAGFAGDDAPRAVFPSIVGR#PRHTGVMVGMGQKDAYVGDEAQSKRGILTL 69 ||||||+|+|||||||||||||||||||||#||| ||||||||||+||||||||||||||| Sbjct 10 LVCDNGSGLVKAGFAGDDAPRAVFPSIVGR#PRHQGVMVGMGQKDSYVGDEAQSKRGILTL 69 |
| Canis familiaris | Query 10 LVCDNGTGMVKAGFAGDDAPRAVFPSIVGR#PRHTGVMVGMGQKDAYVGDEAQSKRGILTL 69 ||||||+|+|||||||||||||||||||||#||| ||||||||||+||||||||||||||| Sbjct 10 LVCDNGSGLVKAGFAGDDAPRAVFPSIVGR#PRHQGVMVGMGQKDSYVGDEAQSKRGILTL 69 |
| Molgula oculata | Query 10 LVCDNGTGMVKAGFAGDDAPRAVFPSIVGR#PRHTGVMVGMGQKDAYVGDEAQSKRGILTL 69 ||||||+|+|||||||||||||||||||||#||| ||||||||||+||||||||||||||| Sbjct 11 LVCDNGSGLVKAGFAGDDAPRAVFPSIVGR#PRHQGVMVGMGQKDSYVGDEAQSKRGILTL 70 |
| Ictalurus punctatus | Query 10 LVCDNGTGMVKAGFAGDDAPRAVFPSIVGR#PRHTGVMVGMGQKDAYVGDEAQSKRGILTL 69 ||||||+|+|||||||||||||||||||||#||| ||||||||||+||||||||||||||| Sbjct 10 LVCDNGSGLVKAGFAGDDAPRAVFPSIVGR#PRHQGVMVGMGQKDSYVGDEAQSKRGILTL 69 |
| Macaca mulatta | Query 10 LVCDNGTGMVKAGFAGDDAPRAVFPSIVGR#PRHTGVMVGMGQKDAYVGDEAQSKRGILTL 69 ||||||+|+|||||||||||||||||||||#||| ||||||||||+||||||||||||||| Sbjct 10 LVCDNGSGLVKAGFAGDDAPRAVFPSIVGR#PRHQGVMVGMGQKDSYVGDEAQSKRGILTL 69 |
| Atractaspis microlepidota microlepidota | Query 10 LVCDNGTGMVKAGFAGDDAPRAVFPSIVGR#PRHTGVMVGMGQKDAYVGDEAQSKRGILTL 69 ||||||+|+|||||||||||||||||||||#||| ||||||||||+||||||||||||||| Sbjct 10 LVCDNGSGLVKAGFAGDDAPRAVFPSIVGR#PRHQGVMVGMGQKDSYVGDEAQSKRGILTL 69 |
| Nannochloris bacillaris | Query 10 LVCDNGTGMVKAGFAGDDAPRAVFPSIVGR#PRHTGVMVGMGQKDAYVGDEAQSKRGILTL 69 +|||||+|||||||||||||||||||||||#||| ||||||||||||||||||||||||+| Sbjct 11 IVCDNGSGMVKAGFAGDDAPRAVFPSIVGR#PRHQGVMVGMGQKDAYVGDEAQSKRGILSL 70 |
| Salmo trutta | Query 10 LVCDNGTGMVKAGFAGDDAPRAVFPSIVGR#PRHTGVMVGMGQKDAYVGDEAQSKRGILTL 69 ||||||+|+|||||||||||||||||||||#||| ||||||||||+||||||||||||||| Sbjct 10 LVCDNGSGLVKAGFAGDDAPRAVFPSIVGR#PRHQGVMVGMGQKDSYVGDEAQSKRGILTL 69 |
| Gadus morhua | Query 10 LVCDNGTGMVKAGFAGDDAPRAVFPSIVGR#PRHTGVMVGMGQKDAYVGDEAQSKRGILTL 69 ||||||+|+|||||||||||||||||||||#||| ||||||||||+||||||||||||||| Sbjct 10 LVCDNGSGLVKAGFAGDDAPRAVFPSIVGR#PRHQGVMVGMGQKDSYVGDEAQSKRGILTL 69 |
| Salmo salar | Query 10 LVCDNGTGMVKAGFAGDDAPRAVFPSIVGR#PRHTGVMVGMGQKDAYVGDEAQSKRGILTL 69 ||||||+|+|||||||||||||||||||||#||| ||||||||||+||||||||||||||| Sbjct 10 LVCDNGSGLVKAGFAGDDAPRAVFPSIVGR#PRHQGVMVGMGQKDSYVGDEAQSKRGILTL 69 |
| Oxyuranus scutellatus scutellatus | Query 10 LVCDNGTGMVKAGFAGDDAPRAVFPSIVGR#PRHTGVMVGMGQKDAYVGDEAQSKRGILTL 69 ||||||+|+|||||||||||||||||||||#||| ||||||||||+||||||||||||||| Sbjct 10 LVCDNGSGLVKAGFAGDDAPRAVFPSIVGR#PRHQGVMVGMGQKDSYVGDEAQSKRGILTL 69 |
| Isatis tinctoria | Query 10 LVCDNGTGMVKAGFAGDDAPRAVFPSIVGR#PRHTGVMVGMGQKDAYVGDEAQSKRGILTL 69 ||||||||||||||||||||||||||||||#|||||| | ||||||||||||||||||||| Sbjct 10 LVCDNGTGMVKAGFAGDDAPRAVFPSIVGR#PRHTGVTVWMGQKDAYVGDEAQSKRGILTL 69 |
| Halocynthia roretzi | Query 10 LVCDNGTGMVKAGFAGDDAPRAVFPSIVGR#PRHTGVMVGMGQKDAYVGDEAQSKRGILTL 69 ||||||+|+|||||||||||||||||||||#||| ||||||||||+||||||||||||||| Sbjct 11 LVCDNGSGLVKAGFAGDDAPRAVFPSIVGR#PRHQGVMVGMGQKDSYVGDEAQSKRGILTL 70 |
| Sphyraena idiastes | Query 10 LVCDNGTGMVKAGFAGDDAPRAVFPSIVGR#PRHTGVMVGMGQKDAYVGDEAQSKRGILTL 69 ||||||+|+|||||||||||||||||||||#||| ||||||||||+||||||||||||||| Sbjct 10 LVCDNGSGLVKAGFAGDDAPRAVFPSIVGR#PRHQGVMVGMGQKDSYVGDEAQSKRGILTL 69 |
| Oreochromis mossambicus | Query 10 LVCDNGTGMVKAGFAGDDAPRAVFPSIVGR#PRHTGVMVGMGQKDAYVGDEAQSKRGILTL 69 ||||||+|+|||||||||||||||||||||#||| ||||||||||+||||||||||||||| Sbjct 10 LVCDNGSGLVKAGFAGDDAPRAVFPSIVGR#PRHQGVMVGMGQKDSYVGDEAQSKRGILTL 69 |
| Carassius auratus | Query 10 LVCDNGTGMVKAGFAGDDAPRAVFPSIVGR#PRHTGVMVGMGQKDAYVGDEAQSKRGILTL 69 ||||||+|+|||||||||||||||||||||#||| ||||||||||+||||||||||||||| Sbjct 10 LVCDNGSGLVKAGFAGDDAPRAVFPSIVGR#PRHQGVMVGMGQKDSYVGDEAQSKRGILTL 69 |
| Sparus aurata | Query 10 LVCDNGTGMVKAGFAGDDAPRAVFPSIVGR#PRHTGVMVGMGQKDAYVGDEAQSKRGILTL 69 ||||||+|+|||||||||||||||||||||#||| ||||||||||+||||||||||||||| Sbjct 10 LVCDNGSGLVKAGFAGDDAPRAVFPSIVGR#PRHQGVMVGMGQKDSYVGDEAQSKRGILTL 69 |
| Oryzias latipes | Query 10 LVCDNGTGMVKAGFAGDDAPRAVFPSIVGR#PRHTGVMVGMGQKDAYVGDEAQSKRGILTL 69 ||||||+|+|||||||||||||||||||||#||| ||||||||||+||||||||||||||| Sbjct 10 LVCDNGSGLVKAGFAGDDAPRAVFPSIVGR#PRHQGVMVGMGQKDSYVGDEAQSKRGILTL 69 |
| Ambystoma mexicanum | Query 10 LVCDNGTGMVKAGFAGDDAPRAVFPSIVGR#PRHTGVMVGMGQKDAYVGDEAQSKRGILTL 69 ||||||+|+|||||||||||||||||||||#||| ||||||||||+||||||||||||||| Sbjct 10 LVCDNGSGLVKAGFAGDDAPRAVFPSIVGR#PRHQGVMVGMGQKDSYVGDEAQSKRGILTL 69 |
| Litopenaeus vannamei | Query 10 LVCDNGTGMVKAGFAGDDAPRAVFPSIVGR#PRHTGVMVGMGQKDAYVGDEAQSKRGILTL 69 ||||||+|||||||||||||||||||||||# || |||||||||||||||||||||||||| Sbjct 10 LVCDNGSGMVKAGFAGDDAPRAVFPSIVGR#ARHQGVMVGMGQKDAYVGDEAQSKRGILTL 69 |
| Haliotis iris | Query 10 LVCDNGTGMVKAGFAGDDAPRAVFPSIVGR#PRHTGVMVGMGQKDAYVGDEAQSKRGILTL 69 ||||||+|| ||||||||||||||||||||#||| ||||||||||+||||||||||||||| Sbjct 8 LVCDNGSGMCKAGFAGDDAPRAVFPSIVGR#PRHQGVMVGMGQKDSYVGDEAQSKRGILTL 67 |
| Gillichthys mirabilis | Query 10 LVCDNGTGMVKAGFAGDDAPRAVFPSIVGR#PRHTGVMVGMGQKDAYVGDEAQSKRGILTL 69 ||||||+|+|||||||||||||||||||||#||| ||||||||||+||||||||||||||| Sbjct 6 LVCDNGSGLVKAGFAGDBAPRAVFPSIVGR#PRHQGVMVGMGQKDSYVGDEAQSKRGILTL 65 |
| Notothenia coriiceps | Query 10 LVCDNGTGMVKAGFAGDDAPRAVFPSIVGR#PRHTGVMVGMGQKDAYVGDEAQSKRGILTL 69 ||||||+|+|||||||||||||||||||||#||| |||||||||| ||||||||||||||| Sbjct 10 LVCDNGSGLVKAGFAGDDAPRAVFPSIVGR#PRHQGVMVGMGQKDTYVGDEAQSKRGILTL 69 |
| Haliotis rufescens | Query 10 LVCDNGTGMVKAGFAGDDAPRAVFPSIVGR#PRHTGVMVGMGQKDAYVGDEAQSKRGILTL 69 ||||||+|| ||||||||||||||||||||#||| ||||||||||+||||||||||||||| Sbjct 8 LVCDNGSGMCKAGFAGDDAPRAVFPSIVGR#PRHQGVMVGMGQKDSYVGDEAQSKRGILTL 67 |
| Morus alba | Query 13 DNGTGMVKAGFAGDDAPRAVFPSIVGR#PRHTGVMVGMGQKDAYVGDEAQSKRGILTL 69 |||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 13 DNGTGMVKAGFAGDDAPRAVFPSIVGR#PRHTGVMVGMGQKDAYVGDEAQSKRGILTL 69 |
| Ostreococcus lucimarinus CCE9901 | Query 10 LVCDNGTGMVKAGFAGDDAPRAVFPSIVGR#PRHTGVMVGMGQKDAYVGDEAQSKRGILTL 69 ||||||+|||||||||||||+|||||||||#||| |||||||||| ||||||||||||||| Sbjct 10 LVCDNGSGMVKAGFAGDDAPKAVFPSIVGR#PRHHGVMVGMGQKDCYVGDEAQSKRGILTL 69 |
| Gallus gallus | Query 10 LVCDNGTGMVKAGFAGDDAPRAVFPSIVGR#PRHTGVMVGMGQKDAYVGDEAQSKRGILTL 69 ||||||+|+ ||||||||||||||||||||#||| ||||||||||+||||||||||||||| Sbjct 9 LVCDNGSGLCKAGFAGDDAPRAVFPSIVGR#PRHQGVMVGMGQKDSYVGDEAQSKRGILTL 68 |
| Penaeus monodon | Query 10 LVCDNGTGMVKAGFAGDDAPRAVFPSIVGR#PRHTGVMVGMGQKDAYVGDEAQSKRGILTL 69 || |||+|||||||||||||||||||||||#||| |||||||||||||||||||||||||| Sbjct 9 LVVDNGSGMVKAGFAGDDAPRAVFPSIVGR#PRHQGVMVGMGQKDAYVGDEAQSKRGILTL 68 |
| Bubalus bubalis | Query 10 LVCDNGTGMVKAGFAGDDAPRAVFPSIVGR#PRHTGVMVGMGQKDAYVGDEAQSKRGILTL 69 ||||||+|+ ||||||||||||||||||||#||| ||||||||||+||||||||||||||| Sbjct 10 LVCDNGSGLCKAGFAGDDAPRAVFPSIVGR#PRHQGVMVGMGQKDSYVGDEAQSKRGILTL 69 |
| Portunus pelagicus | Query 10 LVCDNGTGMVKAGFAGDDAPRAVFPSIVGR#PRHTGVMVGMGQKDAYVGDEAQSKRGILTL 69 || |||+|||||||||||||||||||||||#||| |||||||||||||||||||||||||| Sbjct 9 LVVDNGSGMVKAGFAGDDAPRAVFPSIVGR#PRHQGVMVGMGQKDAYVGDEAQSKRGILTL 68 |
| Coryphaenoides yaquinae | Query 10 LVCDNGTGMVKAGFAGDDAPRAVFPSIVGR#PRHTGVMVGMGQKDAYVGDEAQSKRGILT 68 ||||||+|+|||||||||||||||||||||#||| ||||||||||+|||||||||||||| Sbjct 10 LVCDNGSGLVKAGFAGDDAPRAVFPSIVGR#PRHQGVMVGMGQKDSYVGDEAQSKRGILT 68 |
| Coryphaenoides armatus | Query 10 LVCDNGTGMVKAGFAGDDAPRAVFPSIVGR#PRHTGVMVGMGQKDAYVGDEAQSKRGILTL 69 ||||||+|+|||||||||||||||||||||#||| ||||||||||+| ||||||||||||| Sbjct 10 LVCDNGSGLVKAGFAGDDAPRAVFPSIVGR#PRHQGVMVGMGQKDSYAGDEAQSKRGILTL 69 |
| Monodelphis domestica | Query 10 LVCDNGTGMVKAGFAGDDAPRAVFPSIVGR#PRHTGVMVGMGQKDAYVGDEAQSKRGILTL 69 ||||||+|+ ||||||||||||||||||||#||| ||||||||||+||||||||||||||| Sbjct 9 LVCDNGSGLCKAGFAGDDAPRAVFPSIVGR#PRHQGVMVGMGQKDSYVGDEAQSKRGILTL 68 |
| Homarus americanus | Query 10 LVCDNGTGMVKAGFAGDDAPRAVFPSIVGR#PRHTGVMVGMGQKDAYVGDEAQSKRGILTL 69 || |||+|||||||||||||||||||||||#||| |||||||||||||||||||||||||| Sbjct 10 LVVDNGSGMVKAGFAGDDAPRAVFPSIVGR#PRHQGVMVGMGQKDAYVGDEAQSKRGILTL 69 |
| Ciona savignyi | Query 10 LVCDNGTGMVKAGFAGDDAPRAVFPSIVGR#PRHTGVMVGMGQKDAYVGDEAQSKRGILTL 69 ||||||+|+||+||||||||||||||||||#||| ||||||||||+||||||||||||||| Sbjct 11 LVCDNGSGLVKSGFAGDDAPRAVFPSIVGR#PRHQGVMVGMGQKDSYVGDEAQSKRGILTL 70 |
| Pan troglodytes | Query 10 LVCDNGTGMVKAGFAGDDAPRAVFPSIVGR#PRHTGVMVGMGQKDAYVGDEAQSKRGILTL 69 ||||||+|+ ||||||||||||||||||||#||| ||||||||||+||||||||||||||| Sbjct 10 LVCDNGSGLCKAGFAGDDAPRAVFPSIVGR#PRHQGVMVGMGQKDSYVGDEAQSKRGILTL 69 |
* References
[PubMed ID: 16381971] Ohyanagi H, Tanaka T, Sakai H, Shigemoto Y, Yamaguchi K, Habara T, Fujii Y, Antonio BA, Nagamura Y, Imanishi T, Ikeo K, Itoh T, Gojobori T, Sasaki T, The Rice Annotation Project Database (RAP-DB): hub for Oryza sativa ssp. japonica genome information. Nucleic Acids Res. 2006 Jan 1;34(Database issue):D741-4.
[PubMed ID: 16100779] ... , The map-based sequence of the rice genome. Nature. 2005 Aug 11;436(7052):793-800.