XSB1868 : beta A3 crystallin [Bos taurus]
[ CaMP Format ]
This entry is computationally expanded from SB0069
* Basic Information
| Organism | Bos taurus (cattle) |
| Protein Names | Beta-crystallin A3; Beta-crystallin A3, isoform A1, Delta4 form; Beta-crystallin A3, isoform A1, Delta7 form; Beta-crystallin A3, isoform A1, Delta8 form; beta A3 crystallin |
| Gene Names | CRYBA1 |
| Gene Locus | Not available |
| GO Function | Not available |
| Entrez Protein | Entrez Nucleotide | Entrez Gene | UniProt | OMIM | HGNC | HPRD | KEGG |
|---|---|---|---|---|---|---|---|
| AAT72793 | N/A | N/A | P11843 | N/A | N/A | N/A | bta:282202 |
* Information From OMIM
Not Available.
* Structure Information
1. Primary Information
Length: 215 aa
Average Mass: 0.000 kDa
Monoisotopic Mass: 0.000 kDa
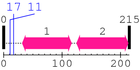
2. Domain Information
Annotated Domains: interpro / pfam / smart / prosite
Computationally Assigned Domains (Pfam+HMMER):
| domain name | begin | end | score | e-value |
|---|---|---|---|---|
| --- cleavage 17 --- | ||||
| --- cleavage 11 --- | ||||
| Crystall 1. | 32 | 116 | 177.1 | 5e-50 |
| Crystall 2. | 125 | 213 | 182.6 | 1.1e-51 |
3. Sequence Information
Fasta Sequence: XSB1868.fasta
Amino Acid Sequence and Secondary Structures (PsiPred):
4. 3D Information
Not Available.
* Cleavage Information
2 [sites]
Cleavage sites (±10aa)
[Site 1] QELESLPTTK17-MAQTNPMPGS
Lys17  Met
Met
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Gln8 | Glu9 | Leu10 | Glu11 | Ser12 | Leu13 | Pro14 | Thr15 | Thr16 | Lys17 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Met18 | Ala19 | Gln20 | Thr21 | Asn22 | Pro23 | Met24 | Pro25 | Gly26 | Ser27 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| METQTVQQELESLPTTKMAQTNPMPGSVGPWKITIYDQENFQGKRME |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | N/A | 100.00 | 8 | - |
| 2 | Bos taurus | 100.00 | 4 | beta A3 crystallin |
| 3 | Contains: Beta crystallin A3, isoform A1, Delta4 form; Beta crystallin A3, isoform A1, Delta7 form; Beta crystallin A3, isoform A1, Delta8 form | 100.00 | 1 | CRBA1_BOVIN Beta crystallin A3 |
| 4 | Oryctolagus cuniculus | 98.20 | 2 | beta A3-crystallin |
| 5 | Canis lupus familiaris | 97.80 | 2 | crystallin, beta A1 |
| 6 | Mus musculus | 96.70 | 5 | crystallin, beta A1 |
| 7 | Rattus norvegicus | 96.70 | 4 | PREDICTED: similar to Beta crystallin A3 |
| 8 | Cavia porcellus | 96.70 | 3 | beta A3-crystallin |
| 9 | Macaca mulatta | 92.80 | 1 | PREDICTED: similar to crystallin, beta A3 |
| 10 | Homo sapiens | 91.70 | 3 | crystallin, beta A3 |
| 11 | Pan troglodytes | 91.70 | 2 | PREDICTED: similar to Beta crystallin A3 |
| 12 | Monodelphis domestica | 86.30 | 2 | PREDICTED: similar to beta A3-crystallin |
| 13 | Rana catesbeiana | 69.30 | 4 | CRB32_RANCA Beta crystallin A3-2 emb |
| 14 | Xenopus laevis | 61.60 | 6 | crystallin, beta A1 |
| 15 | Gallus gallus | 60.50 | 1 | crystallin, beta A1 |
| 16 | Tetraodon nigroviridis | 58.90 | 1 | unnamed protein product |
| 17 | Rana temporaria | 58.20 | 2 | CRBA1_RANTE Beta crystallin A1 emb |
| 18 | Danio rerio | 54.70 | 6 | hypothetical protein LOC436683 |
| 19 | Xenopus tropicalis | 53.10 | 1 | Unknown (protein for MGC:122148) |
| 20 | rats, Sprague-Dawley, lens, Peptide Partial, 20 aa | 45.40 | 1 | beta-crystallin isoform A3 {internal fragment} |
| 21 | Canis familiaris | 35.00 | 1 | PREDICTED: similar to crystallin, beta B1 |
| 22 | Cynops pyrrhogaster | 33.10 | 1 | newt beta B1-crystallin |
| 23 | Sus scrofa | 32.70 | 1 | beta crystallin subunit beta B1 |
Top-ranked sequences
| organism | matching |
|---|---|
| N/A | Query 1 METQTVQQELESLPTTK#MAQTNPMPGSVGPWKITIYDQENFQGKRME 47 |||||||||||||||||#|||||||||||||||||||||||||||||| Query 1 METQTVQQELESLPTTK#MAQTNPMPGSVGPWKITIYDQENFQGKRME 47 |
| Bos taurus | Query 1 METQTVQQELESLPTTK#MAQTNPMPGSVGPWKITIYDQENFQGKRME 47 |||||||||||||||||#|||||||||||||||||||||||||||||| Query 1 METQTVQQELESLPTTK#MAQTNPMPGSVGPWKITIYDQENFQGKRME 47 |
| Contains: Beta crystallin A3, isoform A1, Delta4 form; Beta crystallin A3, isoform A1, Delta7 form; Beta crystallin A3, isoform A1, Delta8 form | Query 1 METQTVQQELESLPTTK#MAQTNPMPGSVGPWKITIYDQENFQGKRME 47 |||||||||||||||||#|||||||||||||||||||||||||||||| Query 1 METQTVQQELESLPTTK#MAQTNPMPGSVGPWKITIYDQENFQGKRME 47 |
| Oryctolagus cuniculus | Query 1 METQTVQQELESLPTTK#MAQTNPMPGSVGPWKITIYDQENFQGKRME 47 |||||||||||+|||||#||||||||||+||||||||||||||||||| Sbjct 1 METQTVQQELETLPTTK#MAQTNPMPGSLGPWKITIYDQENFQGKRME 47 |
| Canis lupus familiaris | Query 1 METQTVQQELESLPTTK#MAQTNPMPGSVGPWKITIYDQENFQGKRME 47 |||||||||||+|||||#|||||||||| ||||||||||||||||||| Sbjct 1 METQTVQQELETLPTTK#MAQTNPMPGSAGPWKITIYDQENFQGKRME 47 |
| Mus musculus | Query 1 METQTVQQELESLPTTK#MAQTNPMPGSVGPWKITIYDQENFQGKRME 47 |||||||+|||+|||||#||||||||||+||||||||||||||||||| Sbjct 1 METQTVQRELETLPTTK#MAQTNPMPGSLGPWKITIYDQENFQGKRME 47 |
| Rattus norvegicus | Query 1 METQTVQQELESLPTTK#MAQTNPMPGSVGPWKITIYDQENFQGKRME 47 |||||||+|||+|||||#||||||||||+||||||||||||||||||| Sbjct 1 METQTVQRELETLPTTK#MAQTNPMPGSMGPWKITIYDQENFQGKRME 47 |
| Cavia porcellus | Query 1 METQTVQQELESLPTTK#MAQTNPMPGSVGPWKITIYDQENFQGKRME 47 |||||||+|||+|||||#||||||||||+||||||||||||||||||| Sbjct 1 METQTVQRELETLPTTK#MAQTNPMPGSLGPWKITIYDQENFQGKRME 47 |
| Macaca mulatta | Query 1 METQTVQQELESLPTTK#MAQTNPMPGSVGPWKITIYDQENFQGKRME 47 |||||||||||+|||||#|||||| ||+||||||||||||||||||| Sbjct 1 METQTVQQELETLPTTK#MAQTNPTQGSLGPWKITIYDQENFQGKRME 47 |
| Homo sapiens | Query 1 METQTVQQELESLPTTK#MAQTNPMPGSVGPWKITIYDQENFQGKRME 47 |||| |||||+|||||#|||||| |||+||||||||||||||||||| Sbjct 1 METQAEQQELETLPTTK#MAQTNPTPGSLGPWKITIYDQENFQGKRME 47 |
| Pan troglodytes | Query 1 METQTVQQELESLPTTK#MAQTNPMPGSVGPWKITIYDQENFQGKRME 47 |||| |||||+|||||#|||||| |||+||||||||||||||||||| Sbjct 1 METQAEQQELETLPTTK#MAQTNPTPGSLGPWKITIYDQENFQGKRME 47 |
| Monodelphis domestica | Query 1 METQTVQQELESLPTTK#MAQTNPMPGSVGPWKITIYDQENFQGKRME 47 |||| ||+|||++| +#||||||||| +||||+|||||||||||||| Sbjct 1 METQAVQRELETVPAAR#MAQTNPMPGPLGPWKVTIYDQENFQGKRME 47 |
| Rana catesbeiana | Query 1 METQTVQQELESLPTTK#MAQTNPMPGSVGPWKITIYDQENFQGKRME 47 || +| | | + + |#||| ||+| +||||||+|||||||||||| Sbjct 1 MEIPAIQTEREDITSEK#MAQINPLPVPLGPWKITVYDQENFQGKRME 47 |
| Xenopus laevis | Query 1 METQTVQQELESLPTTK#MAQTNPMPGSVGPWKITIYDQENFQGKRME 47 || || | |+ | |#||+|||+ +| ||||+|||||||||||| Sbjct 1 MEISPVQTERENF-TKK#MAETNPLSMRMGSWKITVYDQENFQGKRME 46 |
| Gallus gallus | Query 18 MAQTNPMPGSVGPWKITIYDQENFQGKRME 47 ||||||+| +||||||+|||||||||||| Sbjct 1 MAQTNPLPVPMGPWKITVYDQENFQGKRME 30 |
| Tetraodon nigroviridis | Query 16 TK#MAQTNPMPGSVGPWKITIYDQENFQGKRME 47 ||#|||||||| +||||||+||||+|||+||| Sbjct 1 TK#MAQTNPMP--MGPWKITVYDQEHFQGRRME 30 |
| Rana temporaria | Query 18 MAQTNPMPGSVGPWKITIYDQENFQGKRME 47 ||| ||+| +||||||+|||||||||||| Sbjct 1 MAQINPLPVPLGPWKITVYDQENFQGKRME 30 |
| Danio rerio | Query 18 MAQTNPMPGSVGPWKITIYDQENFQGKRME 47 || ||||| +||||+|+|||||||||||| Sbjct 1 MALTNPMP--LGPWKLTVYDQENFQGKRME 28 |
| Xenopus tropicalis | Query 18 MAQTNPMPGSVGPWKITIYDQENFQGKRME 47 ||+|||+ +| ||||+|||||||||||| Sbjct 1 MAETNPLSMRMGSWKITVYDQENFQGKRME 30 |
| rats, Sprague-Dawley, lens, Peptide Partial, 20 aa | Query 12 SLPTTK#MAQTNPMPGSVGPW 31 +|||||#||||||||||+||| Sbjct 1 TLPTTK#MAQTNPMPGSMGPW 20 |
| Canis familiaris | Query 12 SLPTTK#MAQTNPMPGSVGPWKITIYDQENFQGKRME 47 ++||||# | | +|+ +++||||||+|+| Sbjct 41 TVPTTK#AGDLPP-----GSYKLVVFEQENFQGRRVE 71 |
| Cynops pyrrhogaster | Query 27 SVGPWKITIYDQENFQGKRME 47 ||| +|| |++||||||+ +| Sbjct 44 SVGTYKIIIFEQENFQGRHLE 64 |
| Sus scrofa | Query 19 AQTNPMPGSVGP-----WKITIYDQENFQGKRME 47 | ||| + | +|+ +++||||||+|+| Sbjct 39 APAQPMPAAKGDLPPGSYKLVVFEQENFQGRRVE 72 |
[Site 2] ETQTVQQELE11-SLPTTKMAQT
Glu11  Ser
Ser
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Glu2 | Thr3 | Gln4 | Thr5 | Val6 | Gln7 | Gln8 | Glu9 | Leu10 | Glu11 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Ser12 | Leu13 | Pro14 | Thr15 | Thr16 | Lys17 | Met18 | Ala19 | Gln20 | Thr21 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| METQTVQQELESLPTTKMAQTNPMPGSVGPWKITIYDQENF |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | N/A | 88.60 | 5 | - |
| 2 | Bos taurus | 88.60 | 2 | beta A3 crystallin |
| 3 | Contains: Beta crystallin A3, isoform A1, Delta4 form; Beta crystallin A3, isoform A1, Delta7 form; Beta crystallin A3, isoform A1, Delta8 form | 88.60 | 1 | CRBA1_BOVIN Beta crystallin A3 |
| 4 | Oryctolagus cuniculus | 86.30 | 2 | beta A3-crystallin |
| 5 | Canis lupus familiaris | 85.90 | 1 | crystallin, beta A1 |
| 6 | Cavia porcellus | 84.70 | 2 | beta A3-crystallin |
| 7 | Mus musculus | 84.70 | 2 | crystallin, beta A1 |
| 8 | Rattus norvegicus | 84.70 | 2 | PREDICTED: similar to Beta crystallin A3 |
| 9 | Macaca mulatta | 80.90 | 1 | PREDICTED: similar to crystallin, beta A3 |
| 10 | Homo sapiens | 79.70 | 1 | crystallin, beta A3 |
| 11 | Pan troglodytes | 79.70 | 1 | PREDICTED: similar to Beta crystallin A3 |
| 12 | Monodelphis domestica | 74.30 | 1 | PREDICTED: similar to beta A3-crystallin |
| 13 | Rana catesbeiana | 57.40 | 4 | CRB32_RANCA Beta crystallin A3-2 emb |
| 14 | Xenopus laevis | 49.70 | 2 | crystallin, beta A1 |
| 15 | Gallus gallus | 48.50 | 1 | crystallin, beta A1 |
| 16 | Tetraodon nigroviridis | 48.10 | 1 | unnamed protein product |
| 17 | Rana temporaria | 46.20 | 2 | unnamed protein product |
| 18 | rats, Sprague-Dawley, lens, Peptide Partial, 20 aa | 45.40 | 1 | beta-crystallin isoform A3 {internal fragment} |
| 19 | Danio rerio | 42.70 | 3 | hypothetical protein LOC436683 |
Top-ranked sequences
| organism | matching |
|---|---|
| N/A | Query 1 METQTVQQELE#SLPTTKMAQTNPMPGSVGPWKITIYDQENF 41 |||||||||||#|||||||||||||||||||||||||||||| Query 1 METQTVQQELE#SLPTTKMAQTNPMPGSVGPWKITIYDQENF 41 |
| Bos taurus | Query 1 METQTVQQELE#SLPTTKMAQTNPMPGSVGPWKITIYDQENF 41 |||||||||||#|||||||||||||||||||||||||||||| Query 1 METQTVQQELE#SLPTTKMAQTNPMPGSVGPWKITIYDQENF 41 |
| Contains: Beta crystallin A3, isoform A1, Delta4 form; Beta crystallin A3, isoform A1, Delta7 form; Beta crystallin A3, isoform A1, Delta8 form | Query 1 METQTVQQELE#SLPTTKMAQTNPMPGSVGPWKITIYDQENF 41 |||||||||||#|||||||||||||||||||||||||||||| Query 1 METQTVQQELE#SLPTTKMAQTNPMPGSVGPWKITIYDQENF 41 |
| Oryctolagus cuniculus | Query 1 METQTVQQELE#SLPTTKMAQTNPMPGSVGPWKITIYDQENF 41 |||||||||||#+|||||||||||||||+||||||||||||| Sbjct 1 METQTVQQELE#TLPTTKMAQTNPMPGSLGPWKITIYDQENF 41 |
| Canis lupus familiaris | Query 1 METQTVQQELE#SLPTTKMAQTNPMPGSVGPWKITIYDQENF 41 |||||||||||#+||||||||||||||| ||||||||||||| Sbjct 1 METQTVQQELE#TLPTTKMAQTNPMPGSAGPWKITIYDQENF 41 |
| Cavia porcellus | Query 1 METQTVQQELE#SLPTTKMAQTNPMPGSVGPWKITIYDQENF 41 |||||||+|||#+|||||||||||||||+||||||||||||| Sbjct 1 METQTVQRELE#TLPTTKMAQTNPMPGSLGPWKITIYDQENF 41 |
| Mus musculus | Query 1 METQTVQQELE#SLPTTKMAQTNPMPGSVGPWKITIYDQENF 41 |||||||+|||#+|||||||||||||||+||||||||||||| Sbjct 1 METQTVQRELE#TLPTTKMAQTNPMPGSLGPWKITIYDQENF 41 |
| Rattus norvegicus | Query 1 METQTVQQELE#SLPTTKMAQTNPMPGSVGPWKITIYDQENF 41 |||||||+|||#+|||||||||||||||+||||||||||||| Sbjct 1 METQTVQRELE#TLPTTKMAQTNPMPGSMGPWKITIYDQENF 41 |
| Macaca mulatta | Query 1 METQTVQQELE#SLPTTKMAQTNPMPGSVGPWKITIYDQENF 41 |||||||||||#+||||||||||| ||+||||||||||||| Sbjct 1 METQTVQQELE#TLPTTKMAQTNPTQGSLGPWKITIYDQENF 41 |
| Homo sapiens | Query 1 METQTVQQELE#SLPTTKMAQTNPMPGSVGPWKITIYDQENF 41 |||| |||||#+||||||||||| |||+||||||||||||| Sbjct 1 METQAEQQELE#TLPTTKMAQTNPTPGSLGPWKITIYDQENF 41 |
| Pan troglodytes | Query 1 METQTVQQELE#SLPTTKMAQTNPMPGSVGPWKITIYDQENF 41 |||| |||||#+||||||||||| |||+||||||||||||| Sbjct 1 METQAEQQELE#TLPTTKMAQTNPTPGSLGPWKITIYDQENF 41 |
| Monodelphis domestica | Query 1 METQTVQQELE#SLPTTKMAQTNPMPGSVGPWKITIYDQENF 41 |||| ||+|||#++| +||||||||| +||||+|||||||| Sbjct 1 METQAVQRELE#TVPAARMAQTNPMPGPLGPWKVTIYDQENF 41 |
| Rana catesbeiana | Query 1 METQTVQQELE#SLPTTKMAQTNPMPGSVGPWKITIYDQENF 41 || +| | |# + + |||| ||+| +||||||+|||||| Sbjct 1 MEIPAIQTERE#DITSEKMAQINPLPVPLGPWKITVYDQENF 41 |
| Xenopus laevis | Query 1 METQTVQQELE#SLPTTKMAQTNPMPGSVGPWKITIYDQENF 41 || || | |#+ | |||+|||+ +| ||||+|||||| Sbjct 1 MEISPVQTERE#NF-TKKMAETNPLSMRMGSWKITVYDQENF 40 |
| Gallus gallus | Query 18 MAQTNPMPGSVGPWKITIYDQENF 41 ||||||+| +||||||+|||||| Sbjct 1 MAQTNPLPVPMGPWKITVYDQENF 24 |
| Tetraodon nigroviridis | Query 16 TKMAQTNPMPGSVGPWKITIYDQENF 41 |||||||||| +||||||+||||+| Sbjct 1 TKMAQTNPMP--MGPWKITVYDQEHF 24 |
| Rana temporaria | Query 18 MAQTNPMPGSVGPWKITIYDQENF 41 ||| ||+| +||||||+|||||| Sbjct 1 MAQINPLPVPLGPWKITVYDQENF 24 |
| rats, Sprague-Dawley, lens, Peptide Partial, 20 aa | Query 12 SLPTTKMAQTNPMPGSVGPW 31 +|||||||||||||||+||| Sbjct 1 TLPTTKMAQTNPMPGSMGPW 20 |
| Danio rerio | Query 18 MAQTNPMPGSVGPWKITIYDQENF 41 || ||||| +||||+|+|||||| Sbjct 1 MALTNPMP--LGPWKLTVYDQENF 22 |
* References
None.