| # |
organism |
max score |
hits |
top seq |
| 1 |
Rattus norvegicus |
89.40 |
2 |
PREDICTED: similar to Beta crystallin A3 |
| 2 |
Cavia porcellus |
88.20 |
2 |
beta A3-crystallin |
| 3 |
Mus musculus |
88.20 |
2 |
crystallin, beta A1 |
| 4 |
Oryctolagus cuniculus |
86.70 |
2 |
beta A3-crystallin |
| 5 |
Canis lupus familiaris |
85.50 |
1 |
crystallin, beta A1 |
| 6 |
N/A |
84.70 |
5 |
- |
| 7 |
Bos taurus |
84.70 |
2 |
beta A3 crystallin |
| 8 |
Contains: Beta crystallin A3, isoform A1, Delta4 form; Beta crystallin A3, isoform A1, Delta7 form; Beta crystallin A3, isoform A1, Delta8 form |
84.70 |
1 |
CRBA1_BOVIN Beta crystallin A3 |
| 9 |
Macaca mulatta |
81.30 |
1 |
PREDICTED: similar to crystallin, beta A3 |
| 10 |
Pan troglodytes |
80.10 |
1 |
PREDICTED: similar to Beta crystallin A3 |
| 11 |
Homo sapiens |
80.10 |
1 |
crystallin, beta A3 |
| 12 |
Monodelphis domestica |
77.80 |
1 |
PREDICTED: similar to beta A3-crystallin |
| 13 |
Rana catesbeiana |
57.40 |
4 |
CRB32_RANCA Beta crystallin A3-2 emb |
| 14 |
Xenopus laevis |
50.80 |
2 |
crystallin, beta A1 |
| 15 |
Gallus gallus |
50.10 |
1 |
crystallin, beta A1 |
| 16 |
Tetraodon nigroviridis |
49.70 |
1 |
unnamed protein product |
| 17 |
rats, Sprague-Dawley, lens, Peptide Partial, 20 aa |
48.50 |
1 |
beta-crystallin isoform A3 {internal fragment} |
| 18 |
Rana temporaria |
46.60 |
2 |
unnamed protein product |
| 19 |
Danio rerio |
43.50 |
3 |
hypothetical protein LOC436859 |
| 20 |
Xenopus tropicalis |
42.70 |
1 |
Unknown (protein for MGC:122148) |
| organism | matching |
|---|
| Rattus norvegicus |
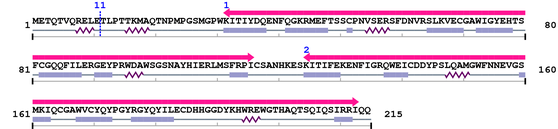
Query 1 METQTVQRELE#TLPTTKMAQTNPMPGSMGPWKITIYDQENF 41
|||||||||||#||||||||||||||||||||||||||||||
Query 1 METQTVQRELE#TLPTTKMAQTNPMPGSMGPWKITIYDQENF 41
|
| Cavia porcellus |
Query 1 METQTVQRELE#TLPTTKMAQTNPMPGSMGPWKITIYDQENF 41
|||||||||||#||||||||||||||||+|||||||||||||
Sbjct 1 METQTVQRELE#TLPTTKMAQTNPMPGSLGPWKITIYDQENF 41
|
| Mus musculus |
Query 1 METQTVQRELE#TLPTTKMAQTNPMPGSMGPWKITIYDQENF 41
|||||||||||#||||||||||||||||+|||||||||||||
Sbjct 1 METQTVQRELE#TLPTTKMAQTNPMPGSLGPWKITIYDQENF 41
|
| Oryctolagus cuniculus |
Query 1 METQTVQRELE#TLPTTKMAQTNPMPGSMGPWKITIYDQENF 41
|||||||+|||#||||||||||||||||+|||||||||||||
Sbjct 1 METQTVQQELE#TLPTTKMAQTNPMPGSLGPWKITIYDQENF 41
|
| Canis lupus familiaris |
Query 1 METQTVQRELE#TLPTTKMAQTNPMPGSMGPWKITIYDQENF 41
|||||||+|||#|||||||||||||||| |||||||||||||
Sbjct 1 METQTVQQELE#TLPTTKMAQTNPMPGSAGPWKITIYDQENF 41
|
| N/A |
Query 1 METQTVQRELE#TLPTTKMAQTNPMPGSMGPWKITIYDQENF 41
|||||||+|||#+|||||||||||||||+|||||||||||||
Sbjct 1 METQTVQQELE#SLPTTKMAQTNPMPGSVGPWKITIYDQENF 41
|
| Bos taurus |
Query 1 METQTVQRELE#TLPTTKMAQTNPMPGSMGPWKITIYDQENF 41
|||||||+|||#+|||||||||||||||+|||||||||||||
Sbjct 1 METQTVQQELE#SLPTTKMAQTNPMPGSVGPWKITIYDQENF 41
|
| Contains: Beta crystallin A3, isoform A1, Delta4 form; Beta crystallin A3, isoform A1, Delta7 form; Beta crystallin A3, isoform A1, Delta8 form |
Query 1 METQTVQRELE#TLPTTKMAQTNPMPGSMGPWKITIYDQENF 41
|||||||+|||#+|||||||||||||||+|||||||||||||
Sbjct 1 METQTVQQELE#SLPTTKMAQTNPMPGSVGPWKITIYDQENF 41
|
| Macaca mulatta |
Query 1 METQTVQRELE#TLPTTKMAQTNPMPGSMGPWKITIYDQENF 41
|||||||+|||#|||||||||||| ||+|||||||||||||
Sbjct 1 METQTVQQELE#TLPTTKMAQTNPTQGSLGPWKITIYDQENF 41
|
| Pan troglodytes |
Query 1 METQTVQRELE#TLPTTKMAQTNPMPGSMGPWKITIYDQENF 41
|||| |+|||#|||||||||||| |||+|||||||||||||
Sbjct 1 METQAEQQELE#TLPTTKMAQTNPTPGSLGPWKITIYDQENF 41
|
| Homo sapiens |
Query 1 METQTVQRELE#TLPTTKMAQTNPMPGSMGPWKITIYDQENF 41
|||| |+|||#|||||||||||| |||+|||||||||||||
Sbjct 1 METQAEQQELE#TLPTTKMAQTNPTPGSLGPWKITIYDQENF 41
|
| Monodelphis domestica |
Query 1 METQTVQRELE#TLPTTKMAQTNPMPGSMGPWKITIYDQENF 41
|||| ||||||#|+| +||||||||| +||||+||||||||
Sbjct 1 METQAVQRELE#TVPAARMAQTNPMPGPLGPWKVTIYDQENF 41
|
| Rana catesbeiana |
Query 1 METQTVQRELE#TLPTTKMAQTNPMPGSMGPWKITIYDQENF 41
|| +| | |# + + |||| ||+| +||||||+||||||
Sbjct 1 MEIPAIQTERE#DITSEKMAQINPLPVPLGPWKITVYDQENF 41
|
| Xenopus laevis |
Query 1 METQTVQRELE#TLPTTKMAQTNPMPGSMGPWKITIYDQENF 41
|| || | |# | |||+|||+ || ||||+||||||
Sbjct 1 MEISPVQTERE#NF-TKKMAETNPLSMRMGSWKITVYDQENF 40
|
| Gallus gallus |
Query 18 MAQTNPMPGSMGPWKITIYDQENF 41
||||||+| |||||||+||||||
Sbjct 1 MAQTNPLPVPMGPWKITVYDQENF 24
|
| Tetraodon nigroviridis |
Query 16 TKMAQTNPMPGSMGPWKITIYDQENF 41
|||||||||| |||||||+||||+|
Sbjct 1 TKMAQTNPMP--MGPWKITVYDQEHF 24
|
| rats, Sprague-Dawley, lens, Peptide Partial, 20 aa |
Query 12 TLPTTKMAQTNPMPGSMGPW 31
||||||||||||||||||||
Query 12 TLPTTKMAQTNPMPGSMGPW 31
|
| Rana temporaria |
Query 18 MAQTNPMPGSMGPWKITIYDQENF 41
||| ||+| +||||||+||||||
Sbjct 1 MAQINPLPVPLGPWKITVYDQENF 24
|
| Danio rerio |
Query 18 MAQTNPMPGSMGPWKITIYDQENF 41
|| |||| |||||||+||||+|
Sbjct 1 MALTNPMSMPMGPWKITVYDQEHF 24
|
| Xenopus tropicalis |
Query 18 MAQTNPMPGSMGPWKITIYDQENF 41
||+|||+ || ||||+||||||
Sbjct 1 MAETNPLSMRMGSWKITVYDQENF 24
|
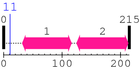
 Thr
Thr

 Sequence conservation (by blast)
Sequence conservation (by blast)