XSB1905 : Islet Cell Antigen 512 [Homo sapiens]
[ CaMP Format ]
This entry is computationally expanded from SB0024
* Basic Information
| Organism | Homo sapiens (human) |
| Protein Names | Receptor-type tyrosine-protein phosphatase-like N; R-PTP-N; PTP IA-2; Islet cell antigen 512; ICA 512; Islet cell autoantigen 3 |
| Gene Names | ICA3, ICA512; ICA3; ICA512 |
| Gene Locus | Not available |
| GO Function | Not available |
* Information From OMIM
Not Available.
* Structure Information
1. Primary Information
Length: 591 aa
Average Mass: 64.763 kDa
Monoisotopic Mass: 64.723 kDa
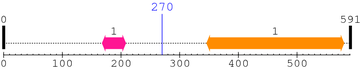
2. Domain Information
Annotated Domains: interpro / pfam / smart / prosite
Computationally Assigned Domains (Pfam+HMMER):
| domain name | begin | end | score | e-value |
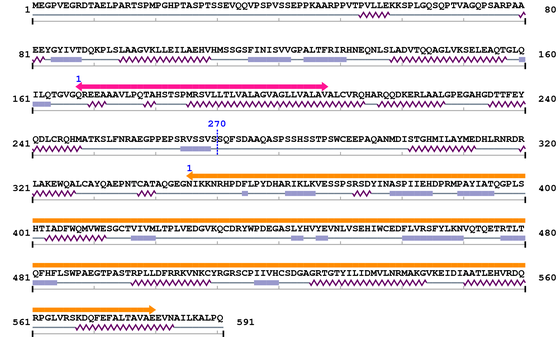
| Gram_pos_anchor 1. | 168 | 208 | 2.4 | 1.8 |
| --- cleavage 270 --- |
| Y_phosphatase 1. | 346 | 580 | 384.7 | 1.6e-112 |
3. Sequence Information
Fasta Sequence: XSB1905.fasta
Amino Acid Sequence and Secondary Structures (PsiPred):
4. 3D Information
Known Structures in PDB:
2I1Y (X-ray; 223 A; A/B=681-979), 2QT7 (X-ray; 130 A; A/B=468-558)
* Cleavage Information
1 [sites]
Cleavage sites (±10aa)
[Site 1] PEPSRVSSVS270-SQFSDAAQAS
Ser270  Ser
Ser
 |
| P10 | P9 | P8 | P7 | P6 |
P5 | P4 | P3 | P2 | P1 |
| Pro261 | Glu262 | Pro263 | Ser264 | Arg265 | Val266 | Ser267 | Ser268 | Val269 | Ser270 |
 |
| P1' | P2' | P3' | P4' | P5' |
P6' | P7' | P8' | P9' | P10' |
| Ser271 | Gln272 | Phe273 | Ser274 | Asp275 | Ala276 | Ala277 | Gln278 | Ala279 | Ser280 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
|
QDLCRQHMATKSLFNRAEGPPEPSRVSSVSSQFSDAAQASPSSHSSTPSWCEEPAQANMD
|
Summary
| # |
organism |
max score |
hits |
top seq |
| 1 |
Homo sapiens |
66.20 |
5 |
Islet Cell Antigen 512 |
| 2 |
N/A |
66.20 |
4 |
I |
| 3 |
Bos taurus |
66.20 |
1 |
protein tyrosine phosphatase, receptor type, N |
| 4 |
Pongo pygmaeus |
66.20 |
1 |
hypothetical protein |
| 5 |
Pan troglodytes |
66.20 |
1 |
PREDICTED: protein tyrosine phosphatase, receptor |
| 6 |
synthetic construct |
66.20 |
1 |
Homo sapiens protein tyrosine phosphatase, recepto |
| 7 |
Mus musculus |
63.20 |
9 |
unnamed protein product |
| 8 |
Rattus norvegicus |
63.20 |
2 |
brain-enriched membrane-associated protein tyrosin |
| 9 |
Rattus sp. |
63.20 |
1 |
BAA07397.1 |
| 10 |
Canis familiaris |
62.40 |
2 |
protein tyrosine phosphatase receptor type N |
Top-ranked sequences
| organism | matching |
|---|
| Homo sapiens |
Query 241 QDLCRQHMATKSLFNRAEGPPEXXXXXXXX#XXXXXXXXXXXXXXXXXXXWCEEPAQANMD 300
|||||||||||||||||||||| # |||||||||||
Sbjct 241 QDLCRQHMATKSLFNRAEGPPEPSRVSSVS#SQFSDAAQASPSSHSSTPSWCEEPAQANMD 300
|
| N/A |
Query 241 QDLCRQHMATKSLFNRAEGPPEXXXXXXXX#XXXXXXXXXXXXXXXXXXXWCEEPAQANMD 300
|||||||||||||||||||||| # |||||||||||
Sbjct 241 QDLCRQHMATKSLFNRAEGPPEPSRVSSVS#SQFSDAAQASPSSHSSTPSWCEEPAQANMD 300
|
| Bos taurus |
Query 241 QDLCRQHMATKSLFNRAEGPPEXXXXXXXX#XXXXXXXXXXXXXXXXXXXWCEEPAQANMD 300
|||||||||||||||||||||| # |||||||||||
Sbjct 629 QDLCRQHMATKSLFNRAEGPPEPSRVSSVS#SQFSDAAQASPSSHSSTPSWCEEPAQANMD 688
|
| Pongo pygmaeus |
Query 241 QDLCRQHMATKSLFNRAEGPPEXXXXXXXX#XXXXXXXXXXXXXXXXXXXWCEEPAQANMD 300
|||||||||||||||||||||| # |||||||||||
Sbjct 629 QDLCRQHMATKSLFNRAEGPPEPSRVSSVS#SQFSDAAQASPSSHSSTPSWCEEPAQANMD 688
|
| Pan troglodytes |
Query 241 QDLCRQHMATKSLFNRAEGPPEXXXXXXXX#XXXXXXXXXXXXXXXXXXXWCEEPAQANMD 300
|||||||||||||||||||||| # |||||||||||
Sbjct 629 QDLCRQHMATKSLFNRAEGPPEPSRVSSVS#SQFSDAAQASPSSHSSTPSWCEEPAQANMD 688
|
| synthetic construct |
Query 241 QDLCRQHMATKSLFNRAEGPPEXXXXXXXX#XXXXXXXXXXXXXXXXXXXWCEEPAQANMD 300
|||||||||||||||||||||| # |||||||||||
Sbjct 241 QDLCRQHMATKSLFNRAEGPPEPSRVSSVS#SQFSDAAQASPSSHSSTPSWCEEPAQANMD 300
|
| Mus musculus |
Query 241 QDLCRQHMATKSLFNRAEGPPEXXXXXXXX#XXXXXXXXXXXXXXXXXXXWCEEPAQANMD 300
||||||||||||||||||| || # |||||||||||
Sbjct 238 QDLCRQHMATKSLFNRAEGQPEPSRVSSVS#SQFSDAAQASPSSHSSTPSWCEEPAQANMD 297
|
| Rattus norvegicus |
Query 241 QDLCRQHMATKSLFNRAEGPPEXXXXXXXX#XXXXXXXXXXXXXXXXXXXWCEEPAQANMD 300
||||||||||||||||||| || # |||||||||||
Sbjct 238 QDLCRQHMATKSLFNRAEGQPEPSRVSSVS#SQFSDAAQASPSSHSSTPSWCEEPAQANMD 297
|
| Rattus sp. |
Query 241 QDLCRQHMATKSLFNRAEGPPEXXXXXXXX#XXXXXXXXXXXXXXXXXXXWCEEPAQANMD 300
||||||||||||||||||| || # |||||||||||
Sbjct 572 QDLCRQHMATKSLFNRAEGQPEPSRVSSVS#SQFSDAAQASPSSHSSTPSWCEEPAQANMD 631
|
| Canis familiaris |
Query 241 QDLCRQHMATKSLFNRAEGPPEXXXXXXXX#XXXXXXXXXXXXXXXXXXXWCEEPAQANMD 300
||||||||| ||||+||||||| # |||||||||||
Sbjct 26 QDLCRQHMAAKSLFSRAEGPPEPSRVSSVS#SQFSDAAQASPSSHSSTPSWCEEPAQANMD 85
|
 Ser
Ser

 Sequence conservation (by blast)
Sequence conservation (by blast)