XSB1914 : protein tyrosine phosphatase receptor type N [Mus musculus]
[ CaMP Format ]
This entry is computationally expanded from SB0024
* Basic Information
| Organism | Mus musculus (house mouse) |
| Protein Names | protein tyrosine phosphatase receptor type N |
| Gene Names | Ptprn |
| Gene Locus | Not available |
| GO Function | Not available |
* Information From OMIM
Not Available.
* Structure Information
1. Primary Information
Length: 873 aa
Average Mass: 94.681 kDa
Monoisotopic Mass: 94.622 kDa
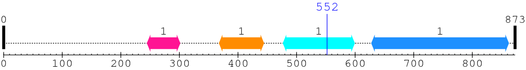
2. Domain Information
Annotated Domains: interpro / pfam / smart / prosite
Computationally Assigned Domains (Pfam+HMMER):
| domain name | begin | end | score | e-value |
|---|---|---|---|---|
| TrmB 1. | 245 | 301 | -30.4 | 4.9 |
| FDX-ACB 1. | 368 | 444 | -53.8 | 2.9 |
| FCD 1. | 477 | 599 | -29.2 | 7.7 |
| --- cleavage 552 (inside FCD 477..599) --- | ||||
| Y_phosphatase 1. | 628 | 862 | 386.2 | 5.8e-113 |
3. Sequence Information
Fasta Sequence: XSB1914.fasta
Amino Acid Sequence and Secondary Structures (PsiPred):
4. 3D Information
Not Available.
* Cleavage Information
1 [sites]
Cleavage sites (±10aa)
[Site 1] PEPSRVSSVS552-SQFSDAAQAS
Ser552  Ser
Ser
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Pro543 | Glu544 | Pro545 | Ser546 | Arg547 | Val548 | Ser549 | Ser550 | Val551 | Ser552 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Ser553 | Gln554 | Phe555 | Ser556 | Asp557 | Ala558 | Ala559 | Gln560 | Ala561 | Ser562 |
 Sequence conservation (by blast)
Sequence conservation (by blast)
* References
[PubMed ID: 11471062] Ehringer MA, Thompson J, Conroy O, Xu Y, Yang F, Canniff J, Beeson M, Gordon L, Bennett B, Johnson TE, Sikela JM, High-throughput sequence identification of gene coding variants within alcohol-related QTLs. Mamm Genome. 2001 Aug;12(8):657-63.