XSB1983 : autophagy-related 5-like [Mus musculus]
[ CaMP Format ]
This entry is computationally expanded from SB0014
* Basic Information
| Organism | Mus musculus (house mouse) |
| Protein Names | Autophagy protein 5; APG5-like; autophagy-related 5-like; autophagy 5-like |
| Gene Names | Atg5; Apg5l; autophagy-related 5 (yeast) |
| Gene Locus | 10 26.0 cM; chromosome 10 |
| GO Function | Not available |
* Information From OMIM
Not Available.
* Structure Information
1. Primary Information
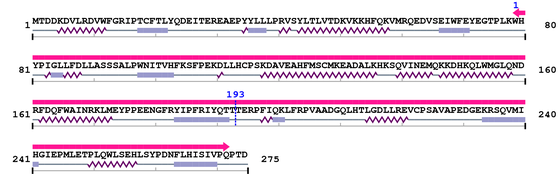
Length: 275 aa
Average Mass: 32.402 kDa
Monoisotopic Mass: 32.381 kDa
2. Domain Information
Annotated Domains: interpro / pfam / smart / prosite
Computationally Assigned Domains (Pfam+HMMER):
| domain name | begin | end | score | e-value |
|---|---|---|---|---|
| APG5 1. | 79 | 272 | 370.7 | 2.7e-108 |
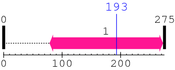
| --- cleavage 193 (inside APG5 79..272) --- | ||||
3. Sequence Information
Fasta Sequence: XSB1983.fasta
Amino Acid Sequence and Secondary Structures (PsiPred):
4. 3D Information
Not Available.
* Cleavage Information
1 [sites]
Cleavage sites (±10aa)
[Site 1] YIPFRIYQTT193-TERPFIQKLF
Thr193  Thr
Thr
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Tyr184 | Ile185 | Pro186 | Phe187 | Arg188 | Ile189 | Tyr190 | Gln191 | Thr192 | Thr193 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Thr194 | Glu195 | Arg196 | Pro197 | Phe198 | Ile199 | Gln200 | Lys201 | Leu202 | Phe203 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| QFWAINRKLMEYPPEENGFRYIPFRIYQTTTERPFIQKLFRPVAADGQLHTLGDLLREVC |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | Mus musculus | 130.00 | 2 | autophagy-related 5-like |
| 2 | Rattus norvegicus | 128.00 | 1 | ATG5_RAT Autophagy protein 5 emb |
| 3 | Sus scrofa | 127.00 | 1 | APG5 autophagy 5-like |
| 4 | Canis familiaris | 125.00 | 3 | PREDICTED: similar to Autophagy protein 5-like (AP |
| 5 | Homo sapiens | 125.00 | 2 | APG5 autophagy 5-like |
| 6 | Macaca mulatta | 125.00 | 2 | PREDICTED: ATG5 autophagy related 5 homolog (S. ce |
| 7 | Pongo pygmaeus | 125.00 | 1 | ATG5_PONPY Autophagy protein 5 (APG5-like) emb |
| 8 | Pan troglodytes | 125.00 | 1 | PREDICTED: hypothetical protein LOC462905 isoform |
| 9 | Bos taurus | 123.00 | 2 | autophagy protein 5 |
| 10 | Monodelphis domestica | 121.00 | 1 | PREDICTED: similar to Autophagy protein 5 |
| 11 | Gallus gallus | 118.00 | 1 | APG5 autophagy 5-like |
| 12 | Xenopus laevis | 108.00 | 2 | autophagy protein 5 |
| 13 | Ictalurus punctatus | 107.00 | 1 | autophagy protein 5 |
| 14 | Xenopus tropicalis | 107.00 | 1 | APG5 autophagy 5-like |
| 15 | Gasterosteus aculeatus | 105.00 | 1 | autophagy protein 5 |
| 16 | Oryzias latipes | 104.00 | 1 | autophagy protein 5 |
| 17 | Pimephales promelas | 103.00 | 1 | autophagy protein 5 |
| 18 | Tetraodon nigroviridis | 101.00 | 2 | autophagy protein 5 |
| 19 | Danio rerio | 100.00 | 2 | APG5 autophagy 5-like protein isoform 2 |
| 20 | Apis mellifera | 65.90 | 1 | PREDICTED: similar to APG5 autophagy 5-like protei |
| 21 | Callinectes sapidus | 61.20 | 1 | autophagy protein 5 |
| 22 | Tribolium castaneum | 59.70 | 1 | PREDICTED: similar to CG1643-PA |
| 23 | Aedes aegypti | 55.10 | 1 | Autophagy-specific protein, putative |
| 24 | Anopheles gambiae str. PEST | 54.30 | 1 | ENSANGP00000012467 |
| 25 | Drosophila erecta | 53.90 | 1 | autophagy protein 5 |
| 26 | Drosophila yakuba | 53.90 | 1 | autophagy protein 5 |
| 27 | Drosophila ananassae | 52.80 | 1 | autophagy protein 5 |
| 28 | Drosophila simulans | 52.40 | 1 | autophagy protein 5 |
| 29 | Drosophila melanogaster | 52.40 | 1 | Autophagy-specific gene 5 CG1643-PA |
| 30 | Drosophila persimilis | 51.20 | 1 | autophagy protein 5 |
| 31 | Drosophila pseudoobscura | 51.20 | 1 | GA14069-PA |
| 32 | Drosophila grimshawi | 50.40 | 1 | autophagy protein 5 |
| 33 | Strongylocentrotus purpuratus | 48.50 | 1 | PREDICTED: similar to autophagy protein 5, partial |
| 34 | Drosophila virilis | 48.10 | 1 | autophagy protein 5 |
| 35 | Drosophila mojavensis | 48.10 | 1 | autophagy protein 5 |
| 36 | Caenorhabditis briggsae | 40.80 | 1 | Hypothetical protein CBG22152 |
| 37 | Caenorhabditis elegans | 40.00 | 1 | ATG (AuTophaGy) Related family member (atgr-5) |
| 38 | Schistosoma japonicum | 39.30 | 1 | SJCHGC03143 protein |
| 39 | Ciona intestinalis | 36.60 | 1 | autophagy protein 5 |
| 40 | Ciona savignyi | 35.80 | 1 | autophagy protein 5 |
Top-ranked sequences
| organism | matching |
|---|---|
| Mus musculus | Query 164 QFWAINRKLMEYPPEENGFRYIPFRIYQTT#TERPFIQKLFRPVAADGQLHTLGDLLREVC 223 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 164 QFWAINRKLMEYPPEENGFRYIPFRIYQTT#TERPFIQKLFRPVAADGQLHTLGDLLREVC 223 |
| Rattus norvegicus | Query 164 QFWAINRKLMEYPPEENGFRYIPFRIYQTT#TERPFIQKLFRPVAADGQLHTLGDLLREVC 223 ||| ||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Sbjct 164 QFWTINRKLMEYPPEENGFRYIPFRIYQTT#TERPFIQKLFRPVAADGQLHTLGDLLREVC 223 |
| Sus scrofa | Query 164 QFWAINRKLMEYPPEENGFRYIPFRIYQTT#TERPFIQKLFRPVAADGQLHTLGDLLREVC 223 ||| |||||+||||||||||||||||||||#|||||||||||||||||||||||||||||| Sbjct 164 QFWTINRKLIEYPPEENGFRYIPFRIYQTT#TERPFIQKLFRPVAADGQLHTLGDLLREVC 223 |
| Canis familiaris | Query 164 QFWAINRKLMEYPPEENGFRYIPFRIYQTT#TERPFIQKLFRPVAADGQLHTLGDLLREVC 223 ||||||||||||| ||||||||||||||||#||||||||||||||||||||||||||+||| Sbjct 164 QFWAINRKLMEYPAEENGFRYIPFRIYQTT#TERPFIQKLFRPVAADGQLHTLGDLLKEVC 223 |
| Homo sapiens | Query 164 QFWAINRKLMEYPPEENGFRYIPFRIYQTT#TERPFIQKLFRPVAADGQLHTLGDLLREVC 223 ||||||||||||| ||||||||||||||||#||||||||||||||||||||||||||+||| Sbjct 164 QFWAINRKLMEYPAEENGFRYIPFRIYQTT#TERPFIQKLFRPVAADGQLHTLGDLLKEVC 223 |
| Macaca mulatta | Query 164 QFWAINRKLMEYPPEENGFRYIPFRIYQTT#TERPFIQKLFRPVAADGQLHTLGDLLREVC 223 ||||||||||||| ||||||||||||||||#||||||||||||||||||||||||||+||| Sbjct 164 QFWAINRKLMEYPAEENGFRYIPFRIYQTT#TERPFIQKLFRPVAADGQLHTLGDLLKEVC 223 |
| Pongo pygmaeus | Query 164 QFWAINRKLMEYPPEENGFRYIPFRIYQTT#TERPFIQKLFRPVAADGQLHTLGDLLREVC 223 ||||||||||||| ||||||||||||||||#||||||||||||||||||||||||||+||| Sbjct 164 QFWAINRKLMEYPAEENGFRYIPFRIYQTT#TERPFIQKLFRPVAADGQLHTLGDLLKEVC 223 |
| Pan troglodytes | Query 164 QFWAINRKLMEYPPEENGFRYIPFRIYQTT#TERPFIQKLFRPVAADGQLHTLGDLLREVC 223 ||||||||||||| ||||||||||||||||#||||||||||||||||||||||||||+||| Sbjct 126 QFWAINRKLMEYPAEENGFRYIPFRIYQTT#TERPFIQKLFRPVAADGQLHTLGDLLKEVC 185 |
| Bos taurus | Query 164 QFWAINRKLMEYPPEENGFRYIPFRIYQTT#TERPFIQKLFRPVAADGQLHTLGDLLREVC 223 ||||||||||||| ||||||||||||||||#|||||||||||||+ |||||||||||+||| Sbjct 164 QFWAINRKLMEYPAEENGFRYIPFRIYQTT#TERPFIQKLFRPVSTDGQLHTLGDLLKEVC 223 |
| Monodelphis domestica | Query 164 QFWAINRKLMEYPPEENGFRYIPFRIYQTT#TERPFIQKLFRPVAADGQLHTLGDLLREVC 223 ||||||||||||| |+|||||||||||| |#|||||||||||||| |||||||||||+||| Sbjct 164 QFWAINRKLMEYPAEDNGFRYIPFRIYQAT#TERPFIQKLFRPVATDGQLHTLGDLLKEVC 223 |
| Gallus gallus | Query 164 QFWAINRKLMEYPPEENGFRYIPFRIYQTT#TERPFIQKLFRPVAADGQLHTLGDLLREVC 223 |||||||||||||||++||||||||||| |#||||||||||||+|+ ||||||||||++|| Sbjct 164 QFWAINRKLMEYPPEDSGFRYIPFRIYQAT#TERPFIQKLFRPIASGGQLHTLGDLLKDVC 223 |
| Xenopus laevis | Query 164 QFWAINRKLMEYPPEENGFRYIPFRIYQTT#TERPFIQKLFRPVAADGQLHTLGDLLREVC 223 |||||||||||+ ||+ |||||||||||| # +|||||||||||| ||+ ||||||+||+| Sbjct 165 QFWAINRKLMEFSPEDGGFRYIPFRIYQTI#NDRPFIQKLFRPVANDGRPHTLGDLIREIC 224 |
| Ictalurus punctatus | Query 164 QFWAINRKLMEYPPEENGFRYIPFRIYQTT#TERPFIQKLFRPVAADGQLHTLGDLLREVC 223 ||||||||||||| || |||||||||||| #++|||||| ||||+++| |||||||+|+| Sbjct 164 QFWAINRKLMEYPTEEGGFRYIPFRIYQTL#SDRPFIQKPFRPVSSEGHAHTLGDLLKEMC 223 |
| Xenopus tropicalis | Query 164 QFWAINRKLMEYPPEENGFRYIPFRIYQTT#TERPFIQKLFRPVAADGQLHTLGDLLREVC 223 ||||||||||| |||+ ||||||||||| # ||||||||||||| ||+ +|||||+|||| Sbjct 164 QFWAINRKLMEVPPEDGGFRYIPFRIYQAI#NERPFIQKLFRPVANDGRPYTLGDLIREVC 223 |
| Gasterosteus aculeatus | Query 164 QFWAINRKLMEYPPEENGFRYIPFRIYQTT#TERPFIQKLFRPVAADGQLHTLGDLLREVC 223 |||||||||||| |+ |||||||||||||#++|||||||||| + +| |||||||+|+| Sbjct 164 QFWAINRKLMEYSSEDGGFRYIPFRIYQTT#SDRPFIQKLFRPTSPEGSAHTLGDLLKEMC 223 |
| Oryzias latipes | Query 164 QFWAINRKLMEYPPEENGFRYIPFRIYQTT#TERPFIQKLFRPVAADGQLHTLGDLLREVC 223 ||||+|||||||| || |||||||||||||# +|||||+|||||+ +| ||| |||+|+| Sbjct 164 QFWAMNRKLMEYPTEEGGFRYIPFRIYQTT#NDRPFIQRLFRPVSTEGNPHTLFDLLKEMC 223 |
| Pimephales promelas | Query 164 QFWAINRKLMEYPPEENGFRYIPFRIYQTT#TERPFIQKLFRPVAADGQLHTLGDLLREV 222 ||||+|||||||| |+ |||||||||||||#++|||||||||||+|+| ||||||+|+ Sbjct 164 QFWAMNRKLMEYPTEDGGFRYIPFRIYQTT#SDRPFIQKLFRPVSAEGHALTLGDLLKEL 222 |
| Tetraodon nigroviridis | Query 164 QFWAINRKLMEYPPEENGFRYIPFRIYQTT#TERPFIQKLFRPVAADGQLHTLGDLLREVC 223 ||||+||||||| || |||||||||||||# +|||||+|||||+ +| ||| |||+|+| Sbjct 164 QFWAMNRKLMEYSTEEGGFRYIPFRIYQTT#NDRPFIQRLFRPVSTEGNPHTLFDLLKEMC 223 |
| Danio rerio | Query 164 QFWAINRKLMEYPPEENGFRYIPFRIYQTT#TERPFIQKLFRPVAADGQLHTLGDLLREV 222 ||||+|||||||| || |||||||||||| #++||||| |||||+++|| ||||||+|+ Sbjct 166 QFWAMNRKLMEYPTEEGGFRYIPFRIYQTM#SDRPFIQTLFRPVSSEGQALTLGDLLKEL 224 |
| Apis mellifera | Query 164 QFWAINRKLMEYPPEENGFRYIPFRIYQTT#TERPFIQKLFRPVAADGQLHTLGDLLREV 222 |||++|||||| | ||+||||| | |#+| ++||| +|| +|| || || || Sbjct 167 QFWSVNRKLMEASNIEEGFKYIPFRCY--T#SEDKYVQKLVKPVNEEGQRKTLRHLLNEV 223 |
| Callinectes sapidus | Query 164 QFWAINRKLMEYPPEENGFRYIPFRIYQTT#TERPFIQKLFRPVAADGQLHTLGDLLRE 221 ||||||||||| ||| ||++||||++ # + | ||+| ||+ ||+ || ||| | Sbjct 166 QFWAINRKLMESTPEE-GFKFIPFRLH-CP#FQAP-IQRLIRPLTEDGRRVTLADLLME 220 |
| Tribolium castaneum | Query 164 QFWAINRKLMEYPPEENGFRYIPFRIYQTT#TERPFIQKLFRPVAADGQLHTLGDLLREV 222 ||||+|||||| |+ |+||||+ | # + + ||| +|| ||+ || ||+ |+ Sbjct 167 QFWAVNRKLMEVSQEQEHFKYIPFKCY---#LDDGYRQKLIKPVDEDGRRKTLQDLINEM 222 |
| Aedes aegypti | Query 164 QFWAINRKLMEYPPEENGFRYIPFRIYQTT#TERPFIQKLFRPVAADGQLHTLGDLLRE 221 ||||+||+||| |+++||++|| | | # + + ||| | || || ||| + Sbjct 165 QFWAVNRRLMEPIPDQDGFKHIPVRCY--A#EDGSYQQKLVAPSTETGQKRTLQDLLED 220 |
| Anopheles gambiae str. PEST | Query 164 QFWAINRKLMEYPPEENGFRYIPFRIYQTT#TERPFIQKLFRPVAADGQLHTLGDLL 219 ||||+||+||| |+++||++|| | | # + + ||| | | || | ||| Sbjct 165 QFWAVNRRLMEPIPDQDGFKHIPVRCY--A#EDGTYQQKLVAPSTASGQKRLLQDLL 218 |
| Drosophila erecta | Query 164 QFWAINRKLMEYPPEENGFRYIPFRIYQTT#TERPFIQKLFRPVAADGQLHTLGDLLREV 222 ||||+||+||| ++ |+ || ||| | # + + ||| |++ || +| ||+ |+ Sbjct 166 QFWAVNRRLMEPYGDQESFKNIPLRIY-TD#DDFTYTQKLISPISEGGQKKSLADLMAEL 223 |
| Drosophila yakuba | Query 164 QFWAINRKLMEYPPEENGFRYIPFRIYQTT#TERPFIQKLFRPVAADGQLHTLGDLLREV 222 ||||+||+||| ++ |+ || ||| | # + + ||| |++ || +| ||+ |+ Sbjct 166 QFWAVNRRLMEPYADQESFKNIPLRIY-TD#DDFTYSQKLISPISEGGQKKSLADLMAEL 223 |
| Drosophila ananassae | Query 164 QFWAINRKLMEYPPEENGFRYIPFRIYQTT#TERPFIQKLFRPVAADGQLHTLGDLLREV 222 ||||+||+||| ++ |++|| |+| # + + ||| |+ || +| ||+ |+ Sbjct 166 QFWAVNRRLMEPYGDQESFKHIPLRLY-AD#DDFTYTQKLISPITESGQKKSLADLMAEL 223 |
| Drosophila simulans | Query 164 QFWAINRKLMEYPPEENGFRYIPFRIYQTT#TERPFIQKLFRPVAADGQLHTLGDLLREV 222 ||||+||+||| + |+ || ||| | # + + ||| |++ || +| ||+ |+ Sbjct 166 QFWAVNRRLMEPYGDLESFKNIPLRIY-TD#DDFTYTQKLISPISVGGQKKSLADLMAEL 223 |
| Drosophila melanogaster | Query 164 QFWAINRKLMEYPPEENGFRYIPFRIYQTT#TERPFIQKLFRPVAADGQLHTLGDLLREV 222 ||||+||+||| + |+ || ||| | # + + ||| |++ || +| ||+ |+ Sbjct 166 QFWAVNRRLMEPYGDLESFKNIPLRIY-TD#DDFTYTQKLISPISVGGQKKSLADLMAEL 223 |
| Drosophila persimilis | Query 164 QFWAINRKLMEYPPEENGFRYIPFRIYQTT#TERPFIQKLFRPVAADGQLHTLGDLLREV 222 ||||+||+||| ++ |+ || |+| # + + ||| |++ || +| ||+ |+ Sbjct 165 QFWAVNRRLMEPYGDQESFKNIPVRLY-ND#DDFTYTQKLISPISESGQKKSLADLMAEL 222 |
| Drosophila pseudoobscura | Query 164 QFWAINRKLMEYPPEENGFRYIPFRIYQTT#TERPFIQKLFRPVAADGQLHTLGDLLREV 222 ||||+||+||| ++ |+ || |+| # + + ||| |++ || +| ||+ |+ Sbjct 166 QFWAVNRRLMEPYGDQESFKNIPVRLY-ND#DDFTYTQKLISPISESGQKKSLADLMAEL 223 |
| Drosophila grimshawi | Query 164 QFWAINRKLMEYPPEENGFRYIPFRIYQTT#TERPFIQKLFRPVAADGQLHTLGDLLREV 222 ||||+||+||| ++ |+ || | | # + ++||| |++ || | ||+ |+ Sbjct 164 QFWAVNRRLMEPYSDQESFKNIPIRFYY-D#DDFTYMQKLITPLSQSGQKKNLADLMAEL 221 |
| Strongylocentrotus purpuratus | Query 164 QFWAINRKLMEYPPEENGFRYIPFRIYQTT#TERPFIQKLFRPVAADGQLHTLGDLLRE 221 ||| ||+||| + |++|||+|+| # |+ |+||+|| +|+ || ||+| Sbjct 139 QFWTTNRRLMERL-NNDPFKFIPFKIFQVP#-EKLHTQRLFKPVNDEGEPRTLRALLKE 194 |
| Drosophila virilis | Query 164 QFWAINRKLMEYPPEENGFRYIPFRIYQTT#TERPFIQKLFRPVAADGQLHTLGDLLREV 222 ||||+||+||| ++ |+ || | | # + ++||| |+ || | ||+ |+ Sbjct 166 QFWAVNRRLMEPYSDQESFKNIPVRFYY-D#DDFTYMQKLITPLNESGQKKNLADLMIEL 223 |
| Drosophila mojavensis | Query 164 QFWAINRKLMEYPPEENGFRYIPFRIYQTT#TERPFIQKLFRPVAADGQLHTLGDLLREV 222 |||||||+||| ++ |+ || | | # + ++||| |+ || | ||+ |+ Sbjct 166 QFWAINRRLMEPYNDQESFKNIPVRFYY-D#DDFTYMQKLISPLNEAGQKKNLADLMAEL 223 |
| Caenorhabditis briggsae | Query 164 QFWAINRKLMEYPPEENGFRYIPFRIYQTT#TERPFIQKLFRPVAADGQLHTLGDLLREV 222 ++| + +|||+ | + | ++| |+| # +|| | | || | |+|| + |+ Sbjct 165 EYWGMIQKLMD-PKDGKDFLHVPLRVY--V#KNQPFKQALITVKHPDGSLRTIGDAVSEI 220 |
| Caenorhabditis elegans | Query 164 QFWAINRKLMEYPPEENGFRYIPFRIYQTT#TERPFIQKLFRPVAADGQLHTLGDLLREV 222 +|| | +|||| | | | +|| |+| # + | | | || | |+|+ + +| Sbjct 163 EFWTIVQKLME-TSEGNEFAHIPLRVY--V#KNQAFKQALITAKHPDGSLRTIGEAVSDV 218 |
| Schistosoma japonicum | Query 164 QFWAINRKLME--------YPPEE------------------------NGFRYIPFRIYQ 191 |||+|| |||| | || ||||| |+| Sbjct 66 QFWSINSKLMEPLPYNTKDLPSEEIPISEIPENKSKSANLDNISSSRCKTFRYIPCRLY- 124 Query 192 TT#TERP------FIQKLFRPVAADGQLHTLGDLLR 220 #+| | | ||| ||+ || | +| | +| Sbjct 125 CV#SENPNSAPSGFTQKLIRPLNDDGSLISLQDAIR 159 |
| Ciona intestinalis | Query 164 QFWAINRKLMEYPPEENGFRYIPFRIYQTT#TERPFIQKLFRP 205 +||+||++ | + ||+|| |+| # + |||||+| Sbjct 164 EFWSINKQFMSGFETTDYFRFIPLRVY---#FQNRIIQKLFKP 202 |
| Ciona savignyi | Query 164 QFWAINRKLMEYPPEENGFRYIPFRIYQTT#TERPFIQKLFRP 205 +||+||++ | || || ||| # + |||||+| Sbjct 162 EFWSINKQFMTDYEHSECFRCIPIRIY---#FQNQIIQKLFKP 200 |
* References
[PubMed ID: 19339967] Singh R, Kaushik S, Wang Y, Xiang Y, Novak I, Komatsu M, Tanaka K, Cuervo AM, Czaja MJ, Autophagy regulates lipid metabolism. Nature. 2009 Apr 30;458(7242):1131-5. Epub 2009 Apr 1.
[PubMed ID: 19299702] ... Pua HH, Guo J, Komatsu M, He YW, Autophagy is essential for mitochondrial clearance in mature T lymphocytes. J Immunol. 2009 Apr 1;182(7):4046-55.
[PubMed ID: 19167256] ... Takikita S, Myerowitz R, Zaal K, Raben N, Plotz PH, Murine muscle cell models for Pompe disease and their use in studying therapeutic approaches. Mol Genet Metab. 2009 Apr;96(4):208-17. Epub 2009 Jan 22.
[PubMed ID: 19139628] ... Cadwell K, Patel KK, Komatsu M, Virgin HW 4th, Stappenbeck TS, A common role for Atg16L1, Atg5 and Atg7 in small intestinal Paneth cells and Crohn disease. Autophagy. 2009 Feb;5(2):250-2. Epub 2009 Feb 8.
[PubMed ID: 19066443] ... Alirezaei M, Fox HS, Flynn CT, Moore CS, Hebb AL, Frausto RF, Bhan V, Kiosses WB, Whitton JL, Robertson GS, Crocker SJ, Elevated ATG5 expression in autoimmune demyelination and multiple sclerosis. Autophagy. 2009 Feb;5(2):152-8. Epub 2009 Feb 5.
[PubMed ID: 11890701] ... Tanida I, Tanida-Miyake E, Nishitani T, Komatsu M, Yamazaki H, Ueno T, Kominami E, Murine Apg12p has a substrate preference for murine Apg7p over three Apg8p homologs. Biochem Biophys Res Commun. 2002 Mar 22;292(1):256-62.
[PubMed ID: 11707774] ... Chalhoub N, Benachenhou N, Vacher J, Physical and transcriptional map of the mouse Chromosome 10 proximal region syntenic to human 6q16-q21. Mamm Genome. 2001 Dec;12(12):887-92.
[PubMed ID: 11266458] ... Mizushima N, Yamamoto A, Hatano M, Kobayashi Y, Kabeya Y, Suzuki K, Tokuhisa T, Ohsumi Y, Yoshimori T, Dissection of autophagosome formation using Apg5-deficient mouse embryonic stem cells. J Cell Biol. 2001 Feb 19;152(4):657-68.
[PubMed ID: 9626506] ... Pires-DaSilva A, Gruss P, Gene trap insertion into a novel gene expressed during mouse limb development. Dev Dyn. 1998 Jun;212(2):318-25.
[PubMed ID: 9563500] ... Hammond EM, Brunet CL, Johnson GD, Parkhill J, Milner AE, Brady G, Gregory CD, Grand RJ, Homology between a human apoptosis specific protein and the product of APG5, a gene involved in autophagy in yeast. FEBS Lett. 1998 Apr 3;425(3):391-5.