XSB2079 : unknown [Homo sapiens]
[ CaMP Format ]
This entry is computationally expanded from SB0093
* Basic Information
| Organism | Homo sapiens (human) |
| Protein Names | Amphiphysin; unknown |
| Gene Names | AMPH; AMPH1 |
| Gene Locus | Not available |
| GO Function | Not available |
* Information From OMIM
Not Available.
* Structure Information
1. Primary Information
Length: 563 aa
Average Mass: 60.857 kDa
Monoisotopic Mass: 60.820 kDa
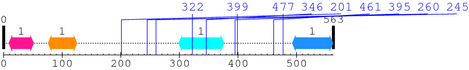
2. Domain Information
Annotated Domains: interpro / pfam / smart / prosite
Computationally Assigned Domains (Pfam+HMMER):
| domain name | begin | end | score | e-value |
|---|---|---|---|---|
| ATP-synt_DE 1. | 9 | 51 | -0.8 | 5.4 |
| GnsAB 1. | 76 | 125 | -3.6 | 4.1 |
| --- cleavage 201 --- | ||||
| --- cleavage 260 --- | ||||
| --- cleavage 245 --- | ||||
| Ribosomal_60s 1. | 299 | 376 | -34.9 | 6.9 |
| --- cleavage 322 (inside Ribosomal_60s 299..376) --- | ||||
| --- cleavage 346 (inside Ribosomal_60s 299..376) --- | ||||
| --- cleavage 399 --- | ||||
| --- cleavage 477 --- | ||||
| --- cleavage 461 --- | ||||
| --- cleavage 395 --- | ||||
| SH3_1 1. | 493 | 562 | 24.1 | 0.00017 |
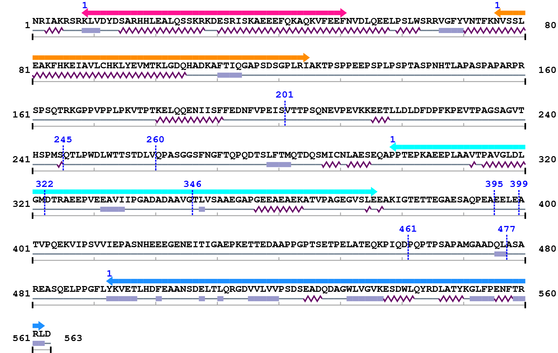
3. Sequence Information
Fasta Sequence: XSB2079.fasta
Amino Acid Sequence and Secondary Structures (PsiPred):
* Cleavage Information
9 [sites]
Cleavage sites (±10aa)
[Site 1] TPAVGLDLGM322-DTRAEEPVEE
Met322  Asp
Asp
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Thr313 | Pro314 | Ala315 | Val316 | Gly317 | Leu318 | Asp319 | Leu320 | Gly321 | Met322 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Asp323 | Thr324 | Arg325 | Ala326 | Glu327 | Glu328 | Pro329 | Val330 | Glu331 | Glu332 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| AESEQAPPTEPKAEEPLAAVTPAVGLDLGMDTRAEEPVEEAVIIPGADADAAVGTLVSAA |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | Homo sapiens | 117.00 | 9 | unknown |
| 2 | synthetic construct | 117.00 | 1 | amphiphysin |
| 3 | Macaca mulatta | 116.00 | 4 | PREDICTED: amphiphysin isoform 2 |
| 4 | Bos taurus | 95.10 | 2 | hypothetical protein LOC614722 |
| 5 | Rattus norvegicus | 61.20 | 1 | amphiphysin 1 |
| 6 | Mus musculus | 57.80 | 2 | amphiphysin |
| 7 | Pan troglodytes | 37.40 | 1 | PREDICTED: amphiphysin |
| 8 | Nocardioides sp. JS614 | 35.80 | 1 | protein of unknown function DUF224, cysteine-rich |
| 9 | Human immunodeficiency virus 1 | 35.40 | 1 | gag-pol polyprotein |
| 10 | Salinispora tropica CNB-440 | 35.40 | 1 | hypothetical protein Strop_0153 |
| 11 | Burkholderia cenocepacia PC184 | 33.50 | 2 | Chemotaxis protein histidine kinase |
| 12 | Oryza sativa (indica cultivar-group) | 33.50 | 2 | hypothetical protein OsI_004520 |
| 13 | Oryza sativa (japonica cultivar-group) | 33.50 | 1 | Os01g0874700 |
| 14 | Anaeromyxobacter sp. Fw109-5 | 33.10 | 1 | conserved hypothetical protein |
| 15 | Burkholderia xenovorans LB400 | 33.10 | 1 | ATP synthase F1, beta subunit |
| 16 | Canis familiaris | 32.70 | 1 | PREDICTED: hypothetical protein XP_536863 |
| 17 | Coccidioides immitis RS | 32.30 | 1 | hypothetical protein CIMG_09416 |
| 18 | Burkholderia cenocepacia MC0-3 | 32.30 | 1 | CheA signal transduction histidine kinases |
Top-ranked sequences
| organism | matching |
|---|---|
| Homo sapiens | Query 293 AESEQAPPTEPKAEEPLAAVTPAVGLDLGM#DTRAEEPVEEAVIIPGADADAAVGTLVSAA 352 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 293 AESEQAPPTEPKAEEPLAAVTPAVGLDLGM#DTRAEEPVEEAVIIPGADADAAVGTLVSAA 352 |
| synthetic construct | Query 293 AESEQAPPTEPKAEEPLAAVTPAVGLDLGM#DTRAEEPVEEAVIIPGADADAAVGTLVSAA 352 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 293 AESEQAPPTEPKAEEPLAAVTPAVGLDLGM#DTRAEEPVEEAVIIPGADADAAVGTLVSAA 352 |
| Macaca mulatta | Query 293 AESEQAPPTEPKAEEPLAAVTPAVGLDLGM#DTRAEEPVEEAVIIPGADADAAVGTLVSAA 352 |||||||||||||||||||||| |||||||#|||||||||||||||||||||||||||||| Sbjct 425 AESEQAPPTEPKAEEPLAAVTPGVGLDLGM#DTRAEEPVEEAVIIPGADADAAVGTLVSAA 484 |
| Bos taurus | Query 293 AESEQAPPTEPKAEEPLAAVTPAVGLDLGM#DTRAEEPVEEAVIIPGADADAAVGTLVSAA 352 |||||||||||||||| ||| || |||||+#||||||| | | |||| +|| |||| || Sbjct 422 AESEQAPPTEPKAEEPAAAVAPATGLDLGL#DTRAEEPEEGAAIIPGTEADVTAGTLVPAA 481 |
| Rattus norvegicus | Query 293 AESEQAPPTEPKAEEP-LAAVTPAVGLDLGM#DTRAEEPVEEAVIIPGADADAAVGT 347 ||+||| ||||+|||| | | |||||+#+ ||| ||| | || || ||| Sbjct 421 AETEQALPTEPQAEEPPTTAAAPTAGLDLGL#EM--EEPKEEAAIPPGTDAGETVGT 474 |
| Mus musculus | Query 293 AESEQAPPTEPKAEEPLA-AVTPAVGLDLGM#DTRAEEPVEEAVIIPGADADAAVGTLV 349 ||+||| ||||+|||| | | | |||||+#+ ||| ||||| | | | | | Sbjct 421 AETEQALPTEPQAEEPPATAAAPTAGLDLGL#E--MEEPKEEAVIPPATDTGETVETAV 476 |
| Pan troglodytes | Query 334 VIIPGADADAAVGTLVSAA 352 +|||||||||||||||||| Sbjct 701 LIIPGADADAAVGTLVSAA 719 |
| Nocardioides sp. JS614 | Query 293 AESEQAPPTEPKAE--EPLAAVTPAVGLDLG----M#DTRAEEPVEEA 333 || + | || | | || || ||| | ||| #| |+|| | Sbjct 861 AEPDAKPETESKEEAPEPPAATTPAAGADLGSGSLF#DIVADEPAASA 907 |
| Human immunodeficiency virus 1 | Query 294 ESEQAPPTEPKAEEPLAAVTPAVGLDLGM#DTRAEEPVEEAVIIPGAD 340 || || ||| |||||+ | | +# + | ++||++ ||| Sbjct 470 ESTPAPKQEPKDREPLAALKSLFGSD-PL#SQKIEGQIKEALLDTGAD 515 |
| Salinispora tropica CNB-440 | Query 306 EEPLAAVTPAVGLD----LGM#DTRAEEPVEEAVIIPGADA 341 ||| |+ +|+ +| +|+#| |||+| || +| |+| Sbjct 357 EEPTASPSPSTSIDAAPTVGV#DEETEEPIEAAVDVPVAEA 396 |
| Burkholderia cenocepacia PC184 | Query 293 AESEQAPPT---EPKAEEPLAAVTPAVGLDLGM#DTRAEEPVEEAVII--PGADADA 343 ||| || || |+||| ||| #| + ||+|| | ||||| Sbjct 128 AESGAGVPTAAAEPAPVAPVAAVAPAVEPAAAG#DRAPDHVVEQAVAAAHPAADADA 183 |
| Oryza sativa (indica cultivar-group) | Query 293 AESEQAPPTEPKAEEPLAAVTPAVGLDLGM#DTRAEEPVEEAVIIPGADADAAVGTLVSAA 352 | +|+| | ||+ || || # |+ | +|| | +| | | || | +|| Sbjct 200 APAEEAKPAEPEPEEKTVVVTEEEAA----#-TKTVEAIEETV-VPAAAAPAAAATEEAAA 253 |
| Oryza sativa (japonica cultivar-group) | Query 293 AESEQAPPTEPKAEEPLAAVTPAVGLDLGM#DTRAEEPVEEAVIIPGADADAAVGTLVSAA 352 | +|+| | ||+ || || # |+ | +|| | +| | | || | +|| Sbjct 203 APAEEAKPAEPEPEEKTVVVTEEEAA----#-TKTVEAIEETV-VPAAAAPAAAATEEAAA 256 |
| Anaeromyxobacter sp. Fw109-5 | Query 294 ESEQAPPTEPKAEEPLAAVTPAVGLDLGM#DTRAEEPVEEAVIIPGADADAAVGTLVSA 351 | | ||+||+ + |+ | + # |+ | ++| | | |+| | | Sbjct 36 EPAQQPPSEPEEQPPMDLSQPPASREPAA#AAEKEKGKEGRPLLPFGDEDVALGDRVKA 93 |
| Burkholderia xenovorans LB400 | Query 298 APPTEPKAEEPLAAVTPAVG--LDLGM#DTRAEEPVEEAVIIPGADADAAV 345 ||| + |+||+ || | +|+ #|| | +|+|++| | + Sbjct 10 APPAQAAADEPVGRVTAVRGAVIDVAF#DTGALPMIEDALVITTDDGQPII 59 |
| Canis familiaris | Query 293 AESEQA-----PPTEPKAEEPLAAV-TPAVGLDLGM#DTRAEEPVEEAVIIPG 338 || |+| || ||+| || | | | + | | +# |+| | | || Sbjct 69 AEGEEAVAVPGPPPEPRAPEPRAPVPEPGLDLSLSL#SPRSETP-ERQTCSPG 119 |
| Coccidioides immitis RS | Query 294 ESEQAPPTEPKAEEPLAAVTPAVGLDLGM#DTRAE--EPVEE--AVIIPGA 339 | + |+||||||| | | | + +#+ +|| || +| ||+ | | Sbjct 214 EQAEPEPSEPKAEEPKAPVEPEIQPTSEV#EKQAEATEPKDESPAVLNPEA 263 |
| Burkholderia cenocepacia MC0-3 | Query 298 APPTEPKAEEPLAAVTPAVGLDLGM#DTRAEEPVEEAVII--PGADADA 343 | || |+||| ||| #| + ||+|| | ||||| Sbjct 136 AAAAEPAPVAPVAAVAPAVDPAAAG#DRAPDHVVEQAVAAAHPAADADA 183 |
[Site 2] SAQPEAEELE399-ATVPQEKVIP
Glu399  Ala
Ala
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Ser390 | Ala391 | Gln392 | Pro393 | Glu394 | Ala395 | Glu396 | Glu397 | Leu398 | Glu399 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Ala400 | Thr401 | Val402 | Pro403 | Gln404 | Glu405 | Lys406 | Val407 | Ile408 | Pro409 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| GEGVSLEEAKIGTETTEGAESAQPEAEELEATVPQEKVIPSVVIEPASNHEEEGENEITI |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | Homo sapiens | 118.00 | 9 | unknown |
| 2 | synthetic construct | 118.00 | 1 | amphiphysin |
| 3 | Pan troglodytes | 114.00 | 1 | PREDICTED: amphiphysin |
| 4 | Macaca mulatta | 104.00 | 4 | PREDICTED: amphiphysin (Stiff-Man syndrome with br |
| 5 | Rattus norvegicus | 73.90 | 1 | amphiphysin 1 |
| 6 | Bos taurus | 68.60 | 1 | hypothetical protein LOC614722 |
| 7 | Canis familiaris | 66.60 | 1 | PREDICTED: similar to Amphiphysin |
| 8 | Monodelphis domestica | 62.40 | 1 | PREDICTED: similar to amphiphysin I variant CT2 |
| 9 | Mus musculus | 61.20 | 3 | amphiphysin |
| 10 | Gallus gallus | 51.20 | 1 | amphiphysin (Stiff-Man syndrome with breast cancer |
| 11 | Xenopus tropicalis | 47.80 | 1 | Unknown (protein for MGC:122074) |
| 12 | Xenopus laevis | 42.70 | 1 | hypothetical protein LOC432150 |
| 13 | Tetraodon nigroviridis | 39.70 | 1 | unnamed protein product |
| 14 | Plasmodium falciparum 3D7 | 35.00 | 1 | glutamate-rich protein |
| 15 | Plasmodium falciparum | 35.00 | 1 | AF247634_1 glutamate-rich protein |
| 16 | Magnetococcus sp. MC-1 | 34.70 | 1 | MJ0042 family finger-like protein |
| 17 | Candida glabrata | 34.30 | 1 | unnamed protein product |
| 18 | Chaetomium globosum CBS 148.51 | 34.30 | 1 | hypothetical protein CHGG_02690 |
| 19 | Aedes aegypti | 33.90 | 3 | troponin t, invertebrate |
| 20 | N/A | 33.90 | 2 | T |
| 21 | Caenorhabditis elegans | 33.90 | 1 | C53C9.2 |
| 22 | Debaryomyces hansenii CBS767 | 33.90 | 1 | hypothetical protein DEHA0C17446g |
| 23 | Oryza sativa (japonica cultivar-group) | 33.10 | 3 | hypothetical protein OsJ_020736 |
| 24 | Phaeosphaeria nodorum SN15 | 33.10 | 1 | hypothetical protein SNOG_02682 |
| 25 | Oryza sativa (indica cultivar-group) | 33.10 | 1 | hypothetical protein OsI_022473 |
| 26 | Myxococcus xanthus DK 1622 | 33.10 | 1 | hypothetical protein MXAN_0043 |
| 27 | Anopheles gambiae str. PEST | 32.70 | 6 | ENSANGP00000024163 |
| 28 | Strongylocentrotus purpuratus | 32.70 | 1 | PREDICTED: similar to Paf1, RNA polymerase II asso |
| 29 | Aspergillus niger | 32.70 | 1 | hypothetical protein An04g09080 |
| 30 | Caenorhabditis briggsae | 32.30 | 1 | Hypothetical protein CBG16761 |
Top-ranked sequences
| organism | matching |
|---|---|
| Homo sapiens | Query 370 GEGVSLEEAKIGTETTEGAESAQPEAEELE#ATVPQEKVIPSVVIEPASNHEEEGENEITI 429 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 370 GEGVSLEEAKIGTETTEGAESAQPEAEELE#ATVPQEKVIPSVVIEPASNHEEEGENEITI 429 |
| synthetic construct | Query 370 GEGVSLEEAKIGTETTEGAESAQPEAEELE#ATVPQEKVIPSVVIEPASNHEEEGENEITI 429 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 370 GEGVSLEEAKIGTETTEGAESAQPEAEELE#ATVPQEKVIPSVVIEPASNHEEEGENEITI 429 |
| Pan troglodytes | Query 370 GEGVSLEEAKIGTETTEGAESAQPEAEELE#ATVPQEKVIPSVVIEPASNHEEEGENEITI 429 ||||||||||||||||||||||||||||||# |||||||||||||||||||| |||||||| Sbjct 737 GEGVSLEEAKIGTETTEGAESAQPEAEELE#VTVPQEKVIPSVVIEPASNHEGEGENEITI 796 |
| Macaca mulatta | Query 370 GEGVSLEEAKIGTETTEGAESAQPEAEELE#ATVPQEKVIPSVVIEPASNHEEEGENEITI 429 ||||||||||||||| ||||||| ||||| #|||||||||||||||||||||||||+|||| Sbjct 460 GEGVSLEEAKIGTET-EGAESAQAEAEELA#ATVPQEKVIPSVVIEPASNHEEEGEHEITI 518 |
| Rattus norvegicus | Query 370 GEGVSLEEAKIGTETTEGAESAQPEAEELE#ATVPQEKVIPSVVIEPASNHEEEGENEIT 428 ||| | | ||| |+|| | | |+| |||#| |||||||||||||||||| | | | Sbjct 492 GEGESPEGAKIDVESTELASSESPQAAELE#AGAPQEKVIPSVVIEPASNHEGEEHQETT 550 |
| Bos taurus | Query 370 GEGVSLEEAKIGTETTEGAESAQPEAEELE#ATVPQEKVIPSVVIEPASNHEEEGENEITI 429 | | |||||| | || + + + || |#| ||||||||||+||||+| |||+| |+ Sbjct 499 GAGASLEEAKTDIEATEAIDGDRSQLEETE#AVAAQEKVIPSVVIQPASNNEGEGEHEGTV 558 |
| Canis familiaris | Query 370 GEGVSLEEAKIGTETTEGAESAQPEAEELE#ATVPQEKVIPSVVIEPASNHEEEGENE 426 ||| +|||+ || |+ + + + || |#| | ||||||||||||||+| ||++| Sbjct 653 GEGAGVEEARPDTEATQAVDGDRSQLEEAE#AVAPLEKVIPSVVIEPASNNEGEGDHE 709 |
| Monodelphis domestica | Query 371 EGVSLEEAKIGTETTEGAESAQPEAEELE#ATVPQEKVIPSVVIEPASNHEEEGENE 426 ||||+ | + || | || + ||+|#| ||||||||||||||||| | | | Sbjct 46 EGVSVVETQAVPETREATESDRAGTEEME#AVGIQEKVIPSVVIEPASNHEGEDEQE 101 |
| Mus musculus | Query 370 GEGVSLEEAKIGTETTEGAESAQPEAEELE#ATVPQEKVIPSVVIEPASNHEEEGENEIT 428 ||| | | ||| |+|| | | |+ | |#| || |||||||||||||| |||++ | Sbjct 496 GEGGSPEGAKIDGESTELAISESPQPVEPE#AGAPQ--VIPSVVIEPASNHEGEGEHQET 552 |
| Gallus gallus | Query 377 EAKIGTETTEGAESAQPEAEELE#ATVPQEKV--IPSVVIEPASNHEEEGENEITI 429 | + || || +|+ || +# | |||| |||||||||||+| ||| | Sbjct 500 EGAVRTEQEAAAEGDKPQGEEKD#VDVSQEKVSSIPSVVIEPASNNEGEGEEHHVI 554 |
| Xenopus tropicalis | Query 373 VSLEEAKIGTETTEGA---------ESAQPEAEELE#AT-VPQE-KVIPSVVIEPASNHEE 421 || | ++ |+ | | | +|| | +# | ||| |||||||||||||+ Sbjct 481 VSEIEVEVKEESAESAAEMADDIMEEQKEPEINEPK#ETEAPQEITCIPSVVIEPASNHED 540 Query 422 EGENEITI 429 | +|+||| Sbjct 541 EHDNDITI 548 |
| Xenopus laevis | Query 377 EAKIGTET-TEGAESAQPEAEELE#ATVPQEK-------VIPSVVIEPASNHEEEGENEIT 428 |+++ |+ | || | +| |# | | +|||||||||||||+| +++|| Sbjct 490 ESEVKQESPVEMAEDIMEEEKEPE#IIEPNETKAPPEITIIPSVVIEPASNHEDEHDDDIT 549 |
| Tetraodon nigroviridis | Query 372 GVSLEEAKIGTETTEGAESAQPEAEELE#ATVPQEKVIPSVVIE---PASNHEEEGENEIT 428 | | ++ | || ||||| +| || |#| | |+ || +| |+| | + |+| Sbjct 133 GFSFKKTK--KETGEGAESEEPAAEATE#AAAPAEEAKPSEAMEETSAAANDEAKPAEEVT 190 |
| Plasmodium falciparum 3D7 | Query 371 EGVSLEEAKIGTETTEGAESAQPEAEELE#ATVPQEKVIPSVVIEPASNHEE 421 | | +|| | | |+ +|| ++ |# +| +|| || + | | | Sbjct 1053 EIVEIEEVPSQTNNNENIETIKPEEKKNE#FSVVEEKAIPQEPVVPTLNENE 1103 |
| Plasmodium falciparum | Query 371 EGVSLEEAKIGTETTEGAESAQPEAEELE#ATVPQEKVIPSVVIEPASNHEE 421 | | +|| | | |+ +|| ++ |# +| +|| || + | | | Sbjct 1056 EIVEIEEVPSQTNNNENIETIKPEEKKNE#FSVVEEKAIPQEPVVPTLNENE 1106 |
| Magnetococcus sp. MC-1 | Query 376 EEAKIGTETTEGAESAQPEAEELE#ATVPQEKVIPSVVIEPASNHEEEGENEI 427 || | +| | | ||||| #| +|+ | | | ||| |+ Sbjct 323 EEESEAEEESEAEEEAAPEAEEEA#APEAEEEAAPEAEEEAAPEAEEEAAPEV 374 |
| Candida glabrata | Query 370 GEGVSLEEAKIGTETTEGAESAQPEAEELE#ATVPQEKVIPSVVIEPASNHE--EEGE--- 424 | |+ |||++ | || || +| | + # | + + ||| | ||| Sbjct 461 GREVTTEEAELAEEATEPAEETEPAKEVIT#EEAEPAKEVTAEEAEPAEETEPAEEGNASA 520 Query 425 NEITI 429 |+|+ Sbjct 521 EEVTV 525 |
| Chaetomium globosum CBS 148.51 | Query 370 GEGVSLEEAKIGTETTEGAESAQPEAEELE#ATVPQEKVI--PSVVIEPASNHEEEGENEI 427 || | |+ | ++ || ||# | | |+ + +| || || + | ++ Sbjct 1364 GENEHGTETKMLCSDAGGGQAQTPEGSGLE#GTAPSEQAVSAATVTAEPPLNHVAKTEGQV 1423 Query 428 T 428 | Query 428 T 428 |
| Aedes aegypti | Query 383 ETTEGAESAQPEAEELE#ATVPQEKVIPSVVIEPASNHEEEGENE 426 || || + +|| ||+|# |+|+ || | ||| | | Sbjct 326 ETPEGEDEVKPEDEEVE#EV---EEVVEEVVEEEEEEEEEEEEEE 366 |
| N/A | Query 371 EGVSLEEAKIGTETTEGAESAQPEAEELE#ATVPQEKVIPSVVIEPASNHEEEGENE 426 | | +++| +++ + +| +|| || |# +|| + |||+ ||| | | Sbjct 345 ERESSRQSEIDSQSVKASEPVEPEPEEEE#EEEEEEK-----IEEPAAKEEEEEEEE 395 |
| Caenorhabditis elegans | Query 371 EGVSLEEAKIGTETTEGAESAQPEAEELE#ATVPQEKVIPSVVIEPASNHEEEGENE 426 | | +++| +++ + +| +|| || |# +|| + |||+ ||| | | Sbjct 324 ERESSRQSEIDSQSVKASEPVEPEPEEEE#EEEEEEK-----IEEPAAKEEEEEEEE 374 |
| Debaryomyces hansenii CBS767 | Query 382 TETTEGAESAQPEAEELE#ATVPQEKVIPSVVIEPASNHEEE 422 | || |+| ||||+||# | + |+||++ | |+|| Sbjct 283 TRATESNETA-PEAEKLE#ENKPATEA-PTVVVDEADEHKEE 321 |
| Oryza sativa (japonica cultivar-group) | Query 376 EEAKIGTETTEGAESAQPEAEELE#ATVPQEKVI 408 ||||| | || | | ||+|# | | | + Sbjct 728 EEAKITTAATEEVEVTTPATEEVE#VTTPSTKEV 760 |
| Phaeosphaeria nodorum SN15 | Query 374 SLEEAKIGTETTEGAESAQPEAEELE#ATVPQEKVIPSVVIEPASNHEE 421 | ||||| |||| + ||+| # || |+ |+ + || || Sbjct 5484 SREEAKIFTETTTAEPAIQPDA----#PVVPVEQTQPTSLEEPVYPSEE 5527 |
| Oryza sativa (indica cultivar-group) | Query 376 EEAKIGTETTEGAESAQPEAEELE#ATVPQEKVI 408 ||||| | || | | ||+|# | | | + Sbjct 728 EEAKITTAATEEVEVTTPATEEVE#VTTPSTKEV 760 |
| Myxococcus xanthus DK 1622 | Query 386 EGAESAQPEAEELE#ATVPQEKVIPSVV 412 | || |+||||+|+#| +| +|+| | Sbjct 318 EDAEEAEPEAEDLQ#AWLPSPRVLPGEV 344 |
| Anopheles gambiae str. PEST | Query 383 ETTEGAESAQPEAEELE#ATVPQEKVIPSVVIEPASNHEEEGENE 426 || || + +|+ ||+|# |+|+ || | ||| | | Sbjct 331 ETPEGEDEVKPDDEEVE#EV---EEVVEEVVEEEEEEEEEEEEEE 371 |
| Strongylocentrotus purpuratus | Query 370 GEGVSLEEAKIGTETTEG-AESAQPEAEELE#ATVPQEKVIPSVVIEPASNHEEEGENE 426 ||| || + | | || ++ + + || |# |+ | | | ||| | | Sbjct 376 GEGEEQEEGEEGAEGVEGESQEEEEDGEEKE#TETEQQDSDAEQVEEAGSGEEEEQEAE 433 |
| Aspergillus niger | Query 373 VSLEEAKIGTETTEGAESAQPEAEELE#ATVPQEKVIPSVVIEPASNHEEEGENE 426 || + |++ ||| + +| ||| #| ||| ||| || | Sbjct 94 VSEQTAEVATETPVADKPTEPSAEEAH#AESTVEKVEEPKTEEPAEQEVAEGAVE 147 |
| Caenorhabditis briggsae | Query 371 EGVSLEEAKIGTETTEGAESAQPEAEELE#ATVPQEKVIPSVVIEPASNHEEEGENE 426 | | +++| ++ + | +|| || |# +|| + |||+ ||| | | Sbjct 324 ERESSRQSEIDAQSVKANEPVEPEPEEEE#EEEEEEK-----IEEPAAKEEEEEEEE 374 |
[Site 3] APAMGAADQL477-ASAREASQEL
Leu477  Ala
Ala
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Ala468 | Pro469 | Ala470 | Met471 | Gly472 | Ala473 | Ala474 | Asp475 | Gln476 | Leu477 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Ala478 | Ser479 | Ala480 | Arg481 | Glu482 | Ala483 | Ser484 | Gln485 | Glu486 | Leu487 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| ETPELATEQKPIQDPQPTPSAPAMGAADQLASAREASQELPPGFLYKVETLHDFEAANSD |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | Homo sapiens | 123.00 | 27 | unknown |
| 2 | Pan troglodytes | 123.00 | 13 | PREDICTED: amphiphysin |
| 3 | synthetic construct | 123.00 | 2 | amphiphysin |
| 4 | Macaca mulatta | 114.00 | 4 | PREDICTED: amphiphysin (Stiff-Man syndrome with br |
| 5 | Rattus norvegicus | 74.30 | 3 | amphiphysin 1 |
| 6 | Bos taurus | 70.90 | 2 | hypothetical protein LOC614722 |
| 7 | Mus musculus | 70.50 | 12 | amphiphysin |
| 8 | Canis familiaris | 68.20 | 3 | PREDICTED: similar to Amphiphysin |
| 9 | Gallus gallus | 64.30 | 2 | amphiphysin (Stiff-Man syndrome with breast cancer |
| 10 | Xenopus laevis | 42.40 | 3 | hypothetical protein LOC432150 |
| 11 | Xenopus tropicalis | 42.00 | 3 | Unknown (protein for MGC:122074) |
| 12 | Danio rerio | 40.80 | 3 | amphiphysin |
| 13 | Tetraodon nigroviridis | 40.80 | 2 | unnamed protein product |
| 14 | N/A | 35.80 | 4 | - |
| 15 | Monodelphis domestica | 34.30 | 2 | PREDICTED: similar to amphiphysin II |
| 16 | Lampetra fluviatilis | 33.90 | 1 | amphiphysin |
| 17 | Leishmania braziliensis | 33.90 | 1 | hypothetical protein, conserved |
| 18 | Sus scrofa | 33.10 | 1 | bridging integrator 1 |
| 19 | Tribolium castaneum | 33.10 | 1 | PREDICTED: similar to CG1943-PA, isoform A |
| 20 | Macaca fascicularis | 33.10 | 1 | unnamed protein product |
| 21 | Aspergillus fumigatus Af293 | 32.70 | 1 | topisomerase II associated protein (Pat1), putativ |
| 22 | Ostreococcus lucimarinus CCE9901 | 32.30 | 1 | predicted protein |
| 23 | Roseovarius sp. HTCC2601 | 32.30 | 1 | Possible TolA protein |
Top-ranked sequences
| organism | matching |
|---|---|
| Homo sapiens | Query 448 ETPELATEQKPIQDPQPTPSAPAMGAADQL#ASAREASQELPPGFLYKVETLHDFEAANSD 507 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 448 ETPELATEQKPIQDPQPTPSAPAMGAADQL#ASAREASQELPPGFLYKVETLHDFEAANSD 507 |
| Pan troglodytes | Query 448 ETPELATEQKPIQDPQPTPSAPAMGAADQL#ASAREASQELPPGFLYKVETLHDFEAANSD 507 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 448 ETPELATEQKPIQDPQPTPSAPAMGAADQL#ASAREASQELPPGFLYKVETLHDFEAANSD 507 |
| synthetic construct | Query 448 ETPELATEQKPIQDPQPTPSAPAMGAADQL#ASAREASQELPPGFLYKVETLHDFEAANSD 507 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 448 ETPELATEQKPIQDPQPTPSAPAMGAADQL#ASAREASQELPPGFLYKVETLHDFEAANSD 507 |
| Macaca mulatta | Query 448 ETPELATEQKPIQDPQPTPSAPAMGAADQL#ASAREASQELPPGFLYKVETLHDFEAANSD 507 |||||||| ||+||||| |||||+| ||||#|| ||||||||||||||||||||||||||| Sbjct 537 ETPELATEPKPLQDPQPMPSAPALGTADQL#ASVREASQELPPGFLYKVETLHDFEAANSD 596 |
| Rattus norvegicus | Query 448 ETPELATEQKPIQDPQPTPSAPAMGAADQL#ASAREASQELPPGFLYKVETLHDFEAANSD 507 | ||||| |+ || ||| || | # || +|+||||||||||||||||||||||| Sbjct 570 EKQELATEPTPLDSQAATP-APA-GAVDAS#LSAGDAAQELPPGFLYKVETLHDFEAANSD 627 |
| Bos taurus | Query 448 ETPELATE-QKPIQDPQPTPSAPAMGAADQL#ASAREASQELPPGFLYKVETLHDFEAANS 506 | ||||+| || + || |||| # || || ||+||||||||||||||||||| Sbjct 577 EMPELASEEQKAARASQPAPSAP--------#-SASEAFQEVPPGFLYKVETLHDFEAANS 627 Query 507 D 507 | Query 507 D 507 |
| Mus musculus | Query 448 ETPELATEQKPIQDPQPTPSAPAMGAADQL#ASAREASQELPPGFLYKVETLHDFEAANSD 507 | |+||| |+ | | | | | || | # || +|+||||||||||||||||||||||| Sbjct 573 EKQEVATEPTPL-DSQATLPASA-GAVDAS#LSAGDATQELPPGFLYKVETLHDFEAANSD 630 |
| Canis familiaris | Query 448 ETPELATEQKPIQDPQPTPSAPAMGAADQL#ASAREASQELPPGFLYKVETLHDFEAANSD 507 | |||| | | +| || ||| | # | | | |+|||||||||||||||||||| Sbjct 730 ELPELAPEPKAGEDSQPLPSALA-------#--ASEESLEVPPGFLYKVETLHDFEAANSD 780 |
| Gallus gallus | Query 448 ETPELATEQKPIQDPQPTPSAPAMGAADQL#ASAREASQELPPGFLYKVETLHDFEAANSD 507 || ++ +||| ++ | ||| || #||| + + ++|||||+||| |||||||||| Sbjct 573 ETSQIGSEQKATEEIQTTPSQ------DQP#ASAGDTASDMPPGFLFKVEVLHDFEAANSD 626 |
| Xenopus laevis | Query 478 ASAREASQELPPGFLYKVETLHDFEAANSD 507 | + +|| | ||||| | |||||||| | Sbjct 581 ADSISSSQSKPEGFLYKAEVLHDFEAANED 610 |
| Xenopus tropicalis | Query 478 ASAREASQELPPGFLYKVETLHDFEAANSD 507 | + || | ||||| | |||||||| | Sbjct 579 ADSLTTSQSKPEGFLYKAEALHDFEAANED 608 |
| Danio rerio | Query 484 SQELPPGFLYKVETLHDFEAANSD 507 | +|| ||+||||+||||||| | Sbjct 706 SSGMPPRFLFKVETMHDFEAANPD 729 |
| Tetraodon nigroviridis | Query 486 ELPPGFLYKVETLHDFEAANSD 507 +|| ||+||||+||||||||| Sbjct 761 QLPEDFLFKVETMHDFEAANSD 782 |
| N/A | Query 465 TPSAPAMGAADQL#ASAREASQELPPGFLYKVETLHDFEAANSD 507 | || || + # || +|||||++||+ ||+ | ++| Sbjct 338 TFSATVNGAVE--#GSAGTGRLDLPPGFMFKVQAQHDYTATDTD 378 |
| Monodelphis domestica | Query 486 ELPPGFLYKVETLHDFEAANSD 507 +|||||++||+ ||+ | +|| Sbjct 521 DLPPGFMFKVQAQHDYTATDSD 542 |
| Lampetra fluviatilis | Query 462 PQPTPSAPAM-------GAADQL#ASAR----EASQELPPGFLYKVETLHDFEAANSD 507 | | | | + | |+ #|| | ++||||||||+ ||+ ++| Sbjct 506 PTPVDSGPELDMAFVTNGDAEVE#ASKHVDTSSESTDMPPGFLYKVQAEHDYTPLDAD 562 |
| Leishmania braziliensis | Query 448 ETPELATEQKPIQDPQPTPSAPAMGAADQL#ASAREAS 484 ||||++| |+ ++ || +| | || # || |+ Sbjct 1548 ETPEVSTMQRSVRQPQQSPIIPGAAAARSF#TSATAAA 1584 |
| Sus scrofa | Query 486 ELPPGFLYKVETLHDFEAANSD 507 +|||||++||+ ||+ | ++| Sbjct 357 DLPPGFMFKVQAQHDYTATDTD 378 |
| Tribolium castaneum | Query 450 PELATEQKPIQDPQPTPSAPAMGAADQL#ASAREASQELPP 489 | ||+||+| + || | | + # + |+|| || Sbjct 66 PVQKTEEKPVQRAEETPKEPVKQATEAP#KPSAPANQEAPP 105 |
| Macaca fascicularis | Query 486 ELPPGFLYKVETLHDFEAANSD 507 +|||||++||+ ||+ | ++| Sbjct 362 DLPPGFMFKVQAQHDYTATDTD 383 |
| Aspergillus fumigatus Af293 | Query 452 LATEQKPIQDPQPTPSAPAMGAADQL#ASAREASQELPPGFL 492 +|+ | |+ | | | + ||# + || +||| || Sbjct 204 IASVQPPVSMPMPMPDLSHLQRTQQL#PNGPEAFSQLPPEFL 244 |
| Ostreococcus lucimarinus CCE9901 | Query 451 ELATEQKPIQDPQPTPSAPAMGAADQL#ASAREASQ-ELPPGFLYKVET 497 || |+ +| + ||+ +#++| ||| ++||||| +|+| Sbjct 186 ELDDPASKFMTPRERRKSPIVAAAEAI#SNAVEASHSDVPPGFLPRVQT 233 |
| Roseovarius sp. HTCC2601 | Query 450 PELATEQKPIQDPQPTPSAPAMGAADQL#ASAREASQELP 488 |+ | | +|+ +|+| | || | ++#++ |++ +| Sbjct 97 PQPAPEPEPVPEPEPDPLPPAAEAVPEV#SAPESATRPVP 135 |
[Site 4] PGADADAAVG346-TLVSAAEGAP
Gly346  Thr
Thr
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Pro337 | Gly338 | Ala339 | Asp340 | Ala341 | Asp342 | Ala343 | Ala344 | Val345 | Gly346 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Thr347 | Leu348 | Val349 | Ser350 | Ala351 | Ala352 | Glu353 | Gly354 | Ala355 | Pro356 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| GLDLGMDTRAEEPVEEAVIIPGADADAAVGTLVSAAEGAPGEEAEAEKATVPAGEGVSLE |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | Homo sapiens | 87.80 | 9 | unknown |
| 2 | Macaca mulatta | 86.70 | 3 | PREDICTED: amphiphysin isoform 2 |
| 3 | synthetic construct | 85.50 | 1 | amphiphysin |
| 4 | Bos taurus | 63.50 | 1 | hypothetical protein LOC614722 |
| 5 | Pan troglodytes | 53.50 | 1 | PREDICTED: amphiphysin |
| 6 | Rattus norvegicus | 42.40 | 1 | amphiphysin 1 |
| 7 | Mus musculus | 38.10 | 2 | amphiphysin |
Top-ranked sequences
| organism | matching |
|---|---|
| Homo sapiens | Query 317 GLDLGMDTRAEEPVEEAVIIPGADADAAVG#TLVSXXXXXXXXXXXXXKATVPAGEGVSLE 376 ||||||||||||||||||||||||||||||#|||| ||||||||||||| Sbjct 317 GLDLGMDTRAEEPVEEAVIIPGADADAAVG#TLVSAAEGAPGEEAEAEKATVPAGEGVSLE 376 |
| Macaca mulatta | Query 317 GLDLGMDTRAEEPVEEAVIIPGADADAAVG#TLVSXXXXXXXXXXXXXKATVPAGEGVSLE 376 ||||||||||||||||||||||||||||||#|||| |||+||||||||| Sbjct 449 GLDLGMDTRAEEPVEEAVIIPGADADAAVG#TLVSAAEGAPGEEAEAEKATLPAGEGVSLE 508 |
| synthetic construct | Query 317 GLDLGMDTRAEEPVEEAVIIPGADADAAVG#TLVSXXXXXXXXXXXXXKATVPAGEGVSLE 376 ||||||||||||||||||||||||||||||#|||| |||||||||||| Sbjct 449 GLDLGMDTRAEEPVEEAVIIPGADADAAVG#TLVSAAEGAPGEEAEAETATVPAGEGVSLE 508 |
| Bos taurus | Query 317 GLDLGMDTRAEEPVEEAVIIPGADADAAVG#TLVSXXXXXXXXXXXXXKATVPAGEGVSLE 376 |||||+||||||| | | |||| +|| |#||| || +||| | ||| Sbjct 446 GLDLGLDTRAEEPEEGAAIIPGTEADVTAG#TLVPAAEGVPVEEAETEKAALPAGAGASLE 505 |
| Pan troglodytes | Query 334 VIIPGADADAAVG#TLVSXXXXXXXXXXXXXKATVPAGEGVSLE 376 +||||||||||||#|||| ||||||||||||| Sbjct 701 LIIPGADADAAVG#TLVSAAEGAPGEEAEAEKATVPAGEGVSLE 743 |
| Rattus norvegicus | Query 317 GLDLGMDTRAEEPVEEAVIIPGADADAAVG#TLVSXXXXXXXXXXXXXKATVPAGEGVSLE 376 |||||++ ||| ||| | || || ||#| | || +||||| | | Sbjct 446 GLDLGLEM--EEPKEEAAIPPGTDAGETVG#TEGS-----TGEEAEAEKAALPAGEGESPE 498 |
| Mus musculus | Query 317 GLDLGMDTRAEEPVEEAVIIPGADADAAVG#TLVSXXXXXXXXXXXXXKATVPAGEGVSLE 376 |||||++ ||| ||||| | | | #| | || +||||| | | Sbjct 446 GLDLGLEM--EEPKEEAVIPPATDTGETVE#TAV-PTEGAPVEEAEAEKAALPAGEGGSPE 502 |
[Site 5] FEDNFVPEIS201-VTTPSQNEVP
Ser201  Val
Val
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Phe192 | Glu193 | Asp194 | Asn195 | Phe196 | Val197 | Pro198 | Glu199 | Ile200 | Ser201 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Val202 | Thr203 | Thr204 | Pro205 | Ser206 | Gln207 | Asn208 | Glu209 | Val210 | Pro211 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| PPLPKVTPTKELQQENIISFFEDNFVPEISVTTPSQNEVPEVKKEETLLDLDFDPFKPEV |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | Homo sapiens | 125.00 | 18 | unknown |
| 2 | Pan troglodytes | 125.00 | 6 | PREDICTED: amphiphysin |
| 3 | Canis familiaris | 125.00 | 2 | PREDICTED: similar to Amphiphysin |
| 4 | synthetic construct | 125.00 | 1 | amphiphysin |
| 5 | Macaca mulatta | 123.00 | 4 | PREDICTED: amphiphysin (Stiff-Man syndrome with br |
| 6 | Bos taurus | 123.00 | 2 | hypothetical protein LOC614722 |
| 7 | Rattus norvegicus | 117.00 | 3 | amphiphysin 1 |
| 8 | Monodelphis domestica | 117.00 | 2 | PREDICTED: similar to cytokine receptor common gam |
| 9 | Mus musculus | 116.00 | 4 | amphiphysin |
| 10 | Gallus gallus | 112.00 | 1 | amphiphysin (Stiff-Man syndrome with breast cancer |
| 11 | Xenopus tropicalis | 110.00 | 3 | Unknown (protein for MGC:122074) |
| 12 | Xenopus laevis | 108.00 | 1 | hypothetical protein LOC432150 |
| 13 | Danio rerio | 74.70 | 1 | amphiphysin |
| 14 | Lampetra fluviatilis | 64.70 | 1 | amphiphysin |
| 15 | N/A | 59.30 | 1 | A |
| 16 | Tetraodon nigroviridis | 55.80 | 1 | unnamed protein product |
Top-ranked sequences
| organism | matching |
|---|---|
| Homo sapiens | Query 172 PPLPKVTPTKELQQENIISFFEDNFVPEIS#VTTPSQNEVPEVKKEETLLDLDFDPFKPEV 231 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 172 PPLPKVTPTKELQQENIISFFEDNFVPEIS#VTTPSQNEVPEVKKEETLLDLDFDPFKPEV 231 |
| Pan troglodytes | Query 172 PPLPKVTPTKELQQENIISFFEDNFVPEIS#VTTPSQNEVPEVKKEETLLDLDFDPFKPEV 231 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 172 PPLPKVTPTKELQQENIISFFEDNFVPEIS#VTTPSQNEVPEVKKEETLLDLDFDPFKPEV 231 |
| Canis familiaris | Query 172 PPLPKVTPTKELQQENIISFFEDNFVPEIS#VTTPSQNEVPEVKKEETLLDLDFDPFKPEV 231 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 172 PPLPKVTPTKELQQENIISFFEDNFVPEIS#VTTPSQNEVPEVKKEETLLDLDFDPFKPEV 231 |
| synthetic construct | Query 172 PPLPKVTPTKELQQENIISFFEDNFVPEIS#VTTPSQNEVPEVKKEETLLDLDFDPFKPEV 231 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 172 PPLPKVTPTKELQQENIISFFEDNFVPEIS#VTTPSQNEVPEVKKEETLLDLDFDPFKPEV 231 |
| Macaca mulatta | Query 172 PPLPKVTPTKELQQENIISFFEDNFVPEIS#VTTPSQNEVPEVKKEETLLDLDFDPFKPEV 231 ||||||||||||+|||||||||||||||||#|||||||||||||||||||||||||||||| Sbjct 304 PPLPKVTPTKELRQENIISFFEDNFVPEIS#VTTPSQNEVPEVKKEETLLDLDFDPFKPEV 363 |
| Bos taurus | Query 172 PPLPKVTPTKELQQENIISFFEDNFVPEIS#VTTPSQNEVPEVKKEETLLDLDFDPFKPEV 231 ||||||||||||||||||||||||||||||#||||||||+|||||||||||||||||||+| Sbjct 304 PPLPKVTPTKELQQENIISFFEDNFVPEIS#VTTPSQNEIPEVKKEETLLDLDFDPFKPDV 363 |
| Rattus norvegicus | Query 172 PPLPKVTPTKELQQENIISFFEDNFVPEIS#VTTPSQNEVPEVKKEETLLDLDFDPFKPEV 231 ||||||||||||||||||+||||||||||+#||||||||| ||||||||||||||||||+| Sbjct 304 PPLPKVTPTKELQQENIINFFEDNFVPEIN#VTTPSQNEVLEVKKEETLLDLDFDPFKPDV 363 |
| Monodelphis domestica | Query 172 PPLPKVTPTKELQQENIISFFEDNFVPEIS#VTTPSQNEVPEVKKEETLLDLDFDPFKPEV 231 ||||||||||||||||||| ||||||||||#||||||||||| ||||+|||||||||||+| Sbjct 304 PPLPKVTPTKELQQENIISLFEDNFVPEIS#VTTPSQNEVPEQKKEESLLDLDFDPFKPDV 363 |
| Mus musculus | Query 172 PPLPKVTPTKELQQENIISFFEDNFVPEIS#VTTPSQNEVPEVKKEETLLDLDFDPFKPEV 231 ||||||||||||+|||||+||||||||||+#||||||||| ||||||||||||||||||+| Sbjct 304 PPLPKVTPTKELKQENIINFFEDNFVPEIN#VTTPSQNEVLEVKKEETLLDLDFDPFKPDV 363 |
| Gallus gallus | Query 172 PPLPKVTPTKELQQENIISFFEDNFVPEIS#VTTPSQNEVPEVKKEETLLDLDFDPFKPEV 231 |||||+||||||||||||+ |+|||||||+#||||||||+|| || |+||||||||||||| Sbjct 304 PPLPKLTPTKELQQENIINLFDDNFVPEIN#VTTPSQNEIPETKKVESLLDLDFDPFKPEV 363 |
| Xenopus tropicalis | Query 172 PPLPKVTPTKELQQENIISFFEDNFVPEIS#VTTPSQNEVPEVKKEETLLDLDFDPFKPE 230 |||||+||+||||||||||||||||||||+#|||||||| | ||||+|||||||||||| Sbjct 305 PPLPKLTPSKELQQENIISFFEDNFVPEIN#VTTPSQNEAVEEKKEESLLDLDFDPFKPE 363 |
| Xenopus laevis | Query 172 PPLPKVTPTKELQQENIISFFEDNFVPEIS#VTTPSQNEVPEVKKEETLLDLDFDPFKP 229 |||||+||+||||||||||||||||||||+#|||||||| | ||||+||||||||||| Sbjct 309 PPLPKLTPSKELQQENIISFFEDNFVPEIN#VTTPSQNEAVEEKKEESLLDLDFDPFKP 366 |
| Danio rerio | Query 172 PPLPKVTPTKELQQENIISFFEDNFVPEIS#VTTPSQNEVPEVKKEETLLDLDFDPFKPE 230 || ||||||+||||| || |+ | ||||#||+| || | |+|||||||||||+ Sbjct 304 PPPPKVTPTRELQQEQIIDLFDGGF-PEIS#VTSPQPNERP----GESLLDLDFDPFKPD 357 |
| Lampetra fluviatilis | Query 172 PPLPKVTPTKELQQENIISFFEDNFVPEIS#VTTPSQNEVPEVKKEETLLDLDFDPFKPE 230 || ||+||+||+||| | | |+|||||+ |# + |++ | | +|||+|||| ||+ Sbjct 303 PPPPKLTPSKEVQQEAIFSLFDDNFVPDFS#-SAPAE---AAKKDEGSLLDIDFDPLKPD 357 |
| N/A | Query 172 PPLPKVTPTKELQQENIISFFEDNFVPEIS#VTTPSQ 207 || || ||+||++|| |+| ||| ||||||#|||||| Sbjct 42 PPPPKHTPSKEVKQEQILSLFEDTFVPEIS#VTTPSQ 77 |
| Tetraodon nigroviridis | Query 172 PPLPKVTPTKELQQENIISFFEDNFVPEIS#VTTPSQNEVPEVKKEETLLDLDFDPFKPEV 231 || |||||+|+++ |||++ |+ |+||#||+|++ + | | +|||+| | | | Sbjct 212 PPPPKVTPSKDMKAENIVNLFDAAAAPDIS#VTSPTEFDRPAV---SSLLDVDLDSFTSTV 268 |
[Site 6] LATEQKPIQD461-PQPTPSAPAM
Asp461  Pro
Pro
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Leu452 | Ala453 | Thr454 | Glu455 | Gln456 | Lys457 | Pro458 | Ile459 | Gln460 | Asp461 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Pro462 | Gln463 | Pro464 | Thr465 | Pro466 | Ser467 | Ala468 | Pro469 | Ala470 | Met471 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| EPKETTEDAAPPGPTSETPELATEQKPIQDPQPTPSAPAMGAADQLASAREASQELPPGF |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | Homo sapiens | 125.00 | 12 | unknown |
| 2 | Pan troglodytes | 125.00 | 8 | PREDICTED: amphiphysin |
| 3 | synthetic construct | 122.00 | 1 | amphiphysin |
| 4 | Macaca mulatta | 108.00 | 13 | PREDICTED: amphiphysin (Stiff-Man syndrome with br |
| 5 | Rattus norvegicus | 61.60 | 8 | amphiphysin 1 |
| 6 | Bos taurus | 60.10 | 6 | hypothetical protein LOC614722 |
| 7 | Mus musculus | 52.80 | 10 | amphiphysin |
| 8 | Canis familiaris | 49.70 | 1 | PREDICTED: similar to Amphiphysin |
| 9 | Gallus gallus | 42.40 | 2 | amphiphysin (Stiff-Man syndrome with breast cancer |
| 10 | Thermobifida fusca YX | 41.60 | 1 | similar to DNA-directed RNA polymerase specialized |
| 11 | Ostreococcus tauri | 38.90 | 1 | unnamed protein product |
| 12 | Monodelphis domestica | 37.70 | 5 | PREDICTED: similar to calcium-binding tyrosine-pho |
| 13 | Caenorhabditis elegans | 37.00 | 2 | EGg Laying defective family member (egl-23) |
| 14 | Burkholderia cepacia AMMD | 37.00 | 1 | TonB family protein |
| 15 | Saccharopolyspora erythraea NRRL 2338 | 36.60 | 1 | hypothetical protein SACE_5459 |
| 16 | Mycobacterium vanbaalenii PYR-1 | 36.20 | 1 | hypothetical protein Mvan_1509 |
| 17 | Coprinopsis cinerea okayama7#130 | 35.80 | 6 | predicted protein |
| 18 | Aspergillus terreus NIH2624 | 35.80 | 3 | hypothetical protein ATEG_01252 |
| 19 | Gibberella zeae PH-1 | 35.80 | 2 | hypothetical protein FG08926.1 |
| 20 | Burkholderia dolosa AUO158 | 35.80 | 2 | Periplasmic protein TonB |
| 21 | Burkholderia ambifaria MC40-6 | 35.80 | 2 | TonB family protein |
| 22 | Apis mellifera | 35.80 | 2 | PREDICTED: similar to tenectin CG13648-PA, partial |
| 23 | Synechococcus sp. JA-2-3B'a(2-13) | 35.80 | 2 | TonB family protein |
| 24 | Roseobacter sp. SK209-2-6 | 35.80 | 1 | transcriptional regulator, MerR family protein |
| 25 | Roseovarius sp. HTCC2601 | 35.80 | 1 | Possible TolA protein |
| 26 | Theileria parva strain Muguga | 35.80 | 1 | hypothetical protein TP01_0380 |
| 27 | Rhodopseudomonas palustris BisB18 | 35.40 | 2 | TonB-like |
| 28 | Magnaporthe grisea 70-15 | 35.40 | 1 | hypothetical protein MGG_08761 |
| 29 | Sagittula stellata E-37 | 35.40 | 1 | hypothetical protein SSE37_03755 |
| 30 | Alcanivorax borkumensis SK2 | 35.40 | 1 | DNA polymerase III, gamma and tau subunits |
| 31 | N/A | 35.00 | 4 | - |
| 32 | Agrobacterium tumefaciens str. C58 | 35.00 | 2 | celB protein |
| 33 | Streptomyces ambofaciens ATCC 23877 | 35.00 | 1 | conserved hypothetical protein |
| 34 | Burkholderia cenocepacia PC184 | 35.00 | 1 | COG0810: Periplasmic protein TonB, links inner and |
| 35 | Burkholderia cenocepacia AU 1054 | 35.00 | 1 | TonB-like |
| 36 | Burkholderia cenocepacia MC0-3 | 35.00 | 1 | TonB family protein |
| 37 | Salinibacter ruber DSM 13855 | 35.00 | 1 | hypothetical protein SRU_0268 |
| 38 | Oryza sativa (japonica cultivar-group) | 34.70 | 4 | Os12g0165200 |
| 39 | Oryza sativa (indica cultivar-group) | 34.70 | 2 | hypothetical protein OsI_036340 |
| 40 | Crimean-Congo hemorrhagic fever virus | 34.70 | 2 | glycoprotein precursor |
| 41 | Yarrowia lipolytica | 34.70 | 2 | hypothetical protein |
| 42 | Cryptococcus neoformans var. neoformans JEC21 | 34.70 | 1 | general transcriptional repressor |
| 43 | Drosophila pseudoobscura | 34.70 | 1 | GA20650-PA |
| 44 | Anopheles gambiae str. PEST | 34.70 | 1 | ENSANGP00000010140 |
| 45 | Trichomonas vaginalis G3 | 34.70 | 1 | hypothetical protein TVAG_129770 |
| 46 | Cryptococcus neoformans var. neoformans B-3501A | 34.70 | 1 | hypothetical protein CNBK1120 |
| 47 | Mesorhizobium sp. BNC1 | 34.70 | 1 | Hemolysin-type calcium-binding region |
| 48 | Aedes aegypti | 34.70 | 1 | hypothetical protein AaeL_AAEL014367 |
| 49 | Chaetomium globosum CBS 148.51 | 34.30 | 4 | predicted protein |
| 50 | Tribolium castaneum | 34.30 | 3 | PREDICTED: similar to CG1943-PA, isoform A |
| 51 | Nocardia farcinica IFM 10152 | 34.30 | 3 | putative serine/threonine protein kinase |
| 52 | Leishmania braziliensis | 34.30 | 2 | hypothetical protein, conserved |
| 53 | Sulfitobacter sp. EE-36 | 34.30 | 2 | tonB domain protein, putative |
| 54 | Collinsella aerofaciens ATCC 25986 | 34.30 | 2 | Hypothetical protein COLAER_00357 |
| 55 | Sulfitobacter sp. NAS-14.1 | 34.30 | 2 | tonB domain protein, putative |
| 56 | Gluconobacter oxydans 621H | 34.30 | 1 | hypothetical protein GOX0962 |
| 57 | Rhodobacter sphaeroides 2.4.1 | 34.30 | 1 | conserved hypoothetical protein |
| 58 | Nostoc sp. PCC 7120 | 34.30 | 1 | hypothetical protein alr2745 |
| 59 | Lotus japonicus | 34.30 | 1 | putative arabinagalactan protein |
| 60 | Gloeobacter violaceus PCC 7421 | 34.30 | 1 | hypothetical protein glr3167 |
| 61 | Burkholderia vietnamiensis G4 | 34.30 | 1 | TonB family protein |
| 62 | Corynebacterium jeikeium K411 | 34.30 | 1 | signal recognition particle receptor |
| 63 | Anabaena variabilis ATCC 29413 | 34.30 | 1 | Peptidoglycan-binding domain 1 |
| 64 | Neurospora crassa OR74A | 34.30 | 1 | hypothetical protein |
| 65 | Yersinia bercovieri ATCC 43970 | 34.30 | 1 | COG3515: Uncharacterized protein conserved in bact |
| 66 | Caenorhabditis briggsae | 33.90 | 4 | Hypothetical protein CBG05727 |
| 67 | Arabidopsis thaliana | 33.90 | 3 | remorin family protein |
| 68 | Drosophila melanogaster | 33.90 | 3 | nahoda CG12781-PA, isoform A |
| 69 | Cenarchaeum symbiosum | 33.90 | 1 | hypothetical protein CENSYa_0842 |
| 70 | Rhodobacter sphaeroides ATCC 17025 | 33.90 | 1 | hypothetical protein Rsph17025_0561 |
| 71 | Leifsonia xyli subsp. xyli str. CTCB07 | 33.90 | 1 | hemagglutinin/hemolysin-related protein |
| 72 | Burkholderia sp. 383 | 33.90 | 1 | TonB-like protein |
| 73 | Fulvimarina pelagi HTCC2506 | 33.90 | 1 | hypothetical protein FP2506_03039 |
| 74 | Hosta virus X | 33.90 | 1 | replicase |
| 75 | Ralstonia solanacearum GMI1000 | 33.90 | 1 | PROBABLE OUTER MEMBRANE CHANNEL PROTEIN |
| 76 | Pichia pastoris | 33.90 | 1 | dihydrolipoamide acetyltransferase |
| 77 | Stenotrophomonas maltophilia R551-3 | 33.90 | 1 | TonB family protein |
| 78 | Erythrobacter sp. NAP1 | 33.90 | 1 | dihydrolipoamide acetyltransferase |
| 79 | Rhodobacter sphaeroides ATCC 17029 | 33.90 | 1 | hypothetical protein Rsph17029_0071 |
| 80 | Corynebacterium efficiens YS-314 | 33.90 | 1 | DNA polymerase III subunits gamma and tau |
| 81 | Leishmania infantum JPCM5 | 33.90 | 1 | hypothetical protein, conserved |
| 82 | Bacillus clausii KSM-K16 | 33.90 | 1 | peptidoglycan-binding domain-containing protein |
| 83 | Aspergillus fumigatus Af293 | 33.90 | 1 | transcriptional activator spt7 |
| 84 | Cyanothece sp. CCY0110 | 33.90 | 1 | hypothetical protein CY0110_24616 |
| 85 | Pichia guilliermondii ATCC 6260 | 33.90 | 1 | predicted protein |
| 86 | Aspergillus clavatus NRRL 1 | 33.50 | 3 | UV excision repair protein (RadW), putative |
| 87 | Danio rerio | 33.50 | 2 | PREDICTED: similar to LOC553397 protein |
| 88 | Myxococcus xanthus DK 1622 | 33.50 | 2 | serine/threonine protein kinase |
| 89 | Strongylocentrotus purpuratus | 33.50 | 2 | PREDICTED: similar to 5-amp-activated protein kina |
| 90 | Bacillus thuringiensis serovar konkukian str. 97-27 | 33.50 | 1 | hypothetical protein BT9727_3951 |
| 91 | Pyrobaculum arsenaticum DSM 13514 | 33.50 | 1 | extracellular solute-binding protein, family 1 |
| 92 | Schizosaccharomyces pombe 972h- | 33.50 | 1 | hypothetical protein SPAC23A1.17 |
| 93 | Murid herpesvirus 2 | 33.50 | 1 | pR48 |
| 94 | Corynebacterium glutamicum R | 33.50 | 1 | hypothetical protein cgR_1950 |
| 95 | Colwellia psychrerythraea 34H | 33.50 | 1 | glycosyl hydrolase, family 5 |
| 96 | Frankia alni ACN14a | 33.50 | 1 | Putative response regulator (Response regulator pr |
| 97 | Rhodobacterales bacterium HTCC2654 | 33.50 | 1 | tonB domain protein, putative |
| 98 | Tetraodon nigroviridis | 33.50 | 1 | unnamed protein product |
| 99 | Chromohalobacter salexigens DSM 3043 | 33.50 | 1 | cell divisionFtsK/SpoIIIE |
| 100 | Coccidioides immitis RS | 33.50 | 1 | predicted protein |
| 101 | Synechococcus sp. BL107 | 33.50 | 1 | Cell division transporter substrate-binding protei |
| 102 | Methylibium petroleiphilum PM1 | 33.50 | 1 | hypothetical protein Mpe_A2234 |
| 103 | Alnus glutinosa | 33.50 | 1 | ag13 |
| 104 | Lyngbya sp. PCC 8106 | 33.50 | 1 | putative hemagglutinin/hemolysin-related protein |
| 105 | Aspergillus niger | 33.50 | 1 | hypothetical protein An08g03930 |
| 106 | Ustilago maydis 521 | 33.50 | 1 | hypothetical protein UM05340.1 |
| 107 | Phaeosphaeria nodorum SN15 | 33.10 | 2 | predicted protein |
| 108 | Saccharomyces cerevisiae | 33.10 | 2 | Proline-rich actin-associated protein involved in |
| 109 | Streptococcus pneumoniae | 33.10 | 2 | AF071806_1 PspA |
| 110 | Neosartorya fischeri NRRL 181 | 33.10 | 2 | response regulator, putative |
| 111 | Vitis vinifera | 33.10 | 2 | hypothetical protein |
| 112 | Leishmania major | 33.10 | 1 | hypothetical protein, conserved |
| 113 | Mycobacterium sp. MCS | 33.10 | 1 | signal recognition particle-docking protein FtsY |
| 114 | Roseiflexus castenholzii DSM 13941 | 33.10 | 1 | serine/threonine protein kinase with FHA domain |
| 115 | Roseobacter denitrificans OCh 114 | 33.10 | 1 | signal recognition particle-docking protein FtsY |
| 116 | Desulfovibrio vulgaris subsp. vulgaris str. Hildenborough | 33.10 | 1 | chemotaxis protein CheA |
| 117 | Streptococcus pyogenes MGAS10750 | 33.10 | 1 | Fibronectin-binding protein |
| 118 | Mycobacterium sp. JLS | 33.10 | 1 | signal recognition particle-docking protein FtsY |
| 119 | Alcelaphine herpesvirus 1 | 33.10 | 1 | large tegument protein |
| 120 | Zea mays | 33.10 | 1 | AF159297_1 extensin-like protein |
| 121 | Streptococcus sobrinus | 33.10 | 1 | dextran-binding lectin |
| 122 | Bordetella bronchiseptica | 33.10 | 1 | pertactin |
| 123 | Sphingomonas sp. SKA58 | 33.10 | 1 | hypothetical protein SKA58_08876 |
| 124 | Dinoroseobacter shibae DFL 12 | 33.10 | 1 | hypothetical protein DshiDRAFT_2630 |
| 125 | Natronomonas pharaonis DSM 2160 | 33.10 | 1 | probable cell surface glycoprotein |
| 126 | Enterococcus faecalis | 33.10 | 1 | AF454824_10 EF0010 |
| 127 | Xanthobacter autotrophicus Py2 | 33.10 | 1 | hypothetical protein XautDRAFT_1507 |
| 128 | Enterococcus faecalis V583 | 33.10 | 1 | cell wall surface anchor family protein |
| 129 | Pyrobaculum islandicum DSM 4184 | 33.10 | 1 | LPXTG-motif cell wall anchor domain |
| 130 | Macaca fascicularis | 33.10 | 1 | unnamed protein product |
| 131 | Thiomicrospira crunogena XCL-2 | 32.70 | 1 | hypothetical protein Tcr_1483 |
| 132 | Chlamydomonas incerta | 32.70 | 1 | vegetative cell wall protein |
| 133 | Streptococcus sanguinis SK36 | 32.70 | 1 | CshA-like fibrillar surface protein C |
| 134 | Pichia stipitis CBS 6054 | 32.70 | 1 | predicted protein |
| 135 | Geobacter lovleyi SZ | 32.70 | 1 | von Willebrand factor, type A |
| 136 | Burkholderia cenocepacia HI2424 | 32.70 | 1 | TonB family protein |
| 137 | Polaromonas naphthalenivorans CJ2 | 32.70 | 1 | TonB family protein |
| 138 | Desulfovibrio vulgaris subsp. vulgaris DP4 | 32.70 | 1 | CheA signal transduction histidine kinases |
| 139 | Rhodopirellula baltica SH 1 | 32.70 | 1 | hypothetical protein-signal peptide and transmembr |
| 140 | Anaeromyxobacter sp. Fw109-5 | 32.70 | 1 | MJ0042 family finger-like protein |
| 141 | Pseudomonas aeruginosa UCBPP-PA14 | 32.70 | 1 | translocation protein in type III secretion |
Top-ranked sequences
| organism | matching |
|---|---|
| Homo sapiens | Query 432 EPKETTEDAAPPGPTSETPELATEQKPIQD#PQPTPSAPAMGAADQLASAREASQELPPGF 491 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 432 EPKETTEDAAPPGPTSETPELATEQKPIQD#PQPTPSAPAMGAADQLASAREASQELPPGF 491 |
| Pan troglodytes | Query 432 EPKETTEDAAPPGPTSETPELATEQKPIQD#PQPTPSAPAMGAADQLASAREASQELPPGF 491 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 432 EPKETTEDAAPPGPTSETPELATEQKPIQD#PQPTPSAPAMGAADQLASAREASQELPPGF 491 |
| synthetic construct | Query 432 EPKETTEDAAPPGPTSETPELATEQKPIQD#PQPTPSAPAMGAADQLASAREASQELPPGF 491 || |||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Sbjct 564 EPMETTEDAAPPGPTSETPELATEQKPIQD#PQPTPSAPAMGAADQLASAREASQELPPGF 623 |
| Macaca mulatta | Query 432 EPKETTEDAAPPGPTSETPELATEQKPIQD#PQPTPSAPAMGAADQLASAREASQELPPGF 491 ||+||||| || |||||||||||| ||+||#||| |||||+| |||||| ||||||||||| Sbjct 521 EPEETTEDTAPLGPTSETPELATEPKPLQD#PQPMPSAPALGTADQLASVREASQELPPGF 580 |
| Rattus norvegicus | Query 432 EPKETTEDAAPPGPTSETPELATEQKPIQD#PQPTPSAPAMGAADQLASAREASQELPPGF 491 | +| ||| || || | ||||| |+ # || ||| || | || +|+||||||| Sbjct 554 ETREATEDVAPQGPAGEKQELATEPTPLDS#QAATP-APA-GAVDASLSAGDAAQELPPGF 611 |
| Bos taurus | Query 432 EPKETTEDAAPPGPTSETPELAT-EQKPIQD#PQPTPSAPAMGAADQLASAREASQELPPG 490 | || |+| | |||||| ||||+ ||| + # || |||| || || ||+||| Sbjct 561 EYKEATDDTALPGPTSEMPELASEEQKAARA#SQPAPSAP---------SASEAFQEVPPG 611 Query 491 F 491 | Query 491 F 491 |
| Mus musculus | Query 432 EPKETTEDAAPPGPTSETPELATEQKPIQD#PQPTPSAPAMGAADQLASAREASQELPPGF 491 ||+| || | | | |+||| |+ |# | | | | || | || +|+||||||| Sbjct 557 EPREAAEDVAAQGSAGEKQEVATEPTPL-D#SQATLPASA-GAVDASLSAGDATQELPPGF 614 |
| Canis familiaris | Query 437 TEDAAPPGPTSETPELATEQKPIQD#PQPTPSAPAMGAADQLASAREASQELPPGF 491 | || ||||+| |||| | | +|# || ||| | | | | |+|||| Sbjct 719 TPAAAAPGPTTELPELAPEPKAGED#SQPLPSALA---------ASEESLEVPPGF 764 |
| Gallus gallus | Query 432 EPKETTEDAAPPGPTSETPELATEQKPIQD#PQPTPSAPAMGAADQLASAREASQELPPGF 491 | |+ + | ||| ++ +||| ++# | ||| || ||| + + ++|||| Sbjct 557 ESKDAAAEMGTQGTDSETSQIGSEQKATEE#IQTTPS------QDQPASAGDTASDMPPGF 610 |
| Thermobifida fusca YX | Query 437 TEDAA---PPGPTSETPELATEQKPIQD#PQPTPSAPAMGAADQLASAREASQELPP 489 +|||| || | + | | ++| #||| |+||| | | |+ | ++ || Sbjct 399 SEDAATVDPPAPAPQQPAPAPREEPASQ#PQPAPTAPA-APAPAPAPAQPAQEDPPP 453 |
| Ostreococcus tauri | Query 432 EPKETTEDAAPPGPTSE---TPELATEQKPIQD#PQPTPSAPAMGAADQLASAREAS 484 || | | | |||| ||| +| +#| | | + || +| +|| | Sbjct 341 EPTPTPEPTPTPEPTSEPTPTPEPTPTPEPTPE#PAPAPESTLGGAFSRLGLSRETS 396 |
| Monodelphis domestica | Query 432 EPKETTEDAAPPGPTSETPELATEQKPIQD#PQP----------TPSAPAMGAADQLASAR 481 ||+||+| + | | + ||+++| | #| | |+ || || || | Sbjct 132 EPEETSESVSEPSPKATTPKVST---PPSS#PSPLAAASPEFAYVPADPAQLAAQMLAIAA 188 Query 482 EASQELPP 489 + + || Sbjct 189 SEAGQPPP 196 |
| Caenorhabditis elegans | Query 432 EPKETTEDAAPPGPTSETPELAT-EQKPIQD#PQPTPSAPA----MGAADQLASAREASQE 486 || | || | || | | +|+++#| | | || + ||+ | |+| | Sbjct 539 EPSPVREPTPPPPPREPTPREPTPEPEPVRE#PTPPPPPPAKPRPLTAAEIAAQKRKAYSE 598 |
| Burkholderia cepacia AMMD | Query 432 EPKETTEDAAP-PGPTSETPELATEQKPIQD#PQPTPSAPAMGA-ADQLASAREASQELPP 489 ||| + | | | | +++| + | |#| | |+||| | | + ||| | | Sbjct 93 EPKPVQKAAKPAPQPVAQSPAPSPTPAPAAD#PTPAPAAPAAAAPAATPSPARETMQVSAP 152 |
| Saccharopolyspora erythraea NRRL 2338 | Query 432 EPKETTEDAAPPGPTS----ETPELATEQKPIQD#PQPTPSAPAMGAADQLASAREASQ 485 + + | || ||| + || | | +| +#|+| |+| + ||+ ++ + | Sbjct 812 QSEHAAEPAANPGPAARAQESAPEPAPEPEPAPE#PEPAPAASPLRLADRTSTDEDRKQ 869 |
| Mycobacterium vanbaalenii PYR-1 | Query 433 PKETTEDAAPPGPTSETPELATEQKPIQD#PQPTPS-AP---AMGAADQLASA 480 | ||| +| || | | | #| |||| || ++||| +|| Sbjct 249 PSETTSPTSPSETTSPAPSSETTSAPTSS#PSPTPSTAPTTTSVGAAPPSSSA 300 |
| Coprinopsis cinerea okayama7#130 | Query 433 PKETTEDAAPPGPTSETPELATEQKPIQD#PQPTPS---APAMGAADQLASAREASQE 486 | |+ | || || + |+ ++ |+ +#| |+ | | |+| | +| Sbjct 111 PASATDPANPPSPTQQGPDRSSSLSPVPE#PDQPPAEQQPQANGGAEQQPQREEEEEE 167 |
| Aspergillus terreus NIH2624 | Query 437 TEDAAPPGPTSETPELATEQKPIQD#PQPTPSAP--AMGAADQLASAREASQE 486 | || | | | +|+|+ #|||||+ | |+ + | ++ |+ Sbjct 220 TPQPNPPQPPSSQPPQQQQQQPVSQ#PQPTPNPPPQALPQSQQQVPQQQTPQQ 271 |
| Gibberella zeae PH-1 | Query 434 KETTEDAAPPGPTSETPELATE-------QKPIQD#PQPTPSAPAMGAADQLASAREASQE 486 ||| + | || + | +| + | |#||| ||| | | |++ Sbjct 326 KETQSYSPPSGPPPKAGEGSTSRPYDESAENPFAD#PQPGGSAPTTAAYQYSEEQRLATEP 385 Query 487 LPPGF 491 ||| Sbjct 386 FHPGF 390 |
| Burkholderia dolosa AUO158 | Query 432 EPKETTEDAAP-PGPTSETPELATEQKPIQD#PQPTPSAPAMGA-ADQLASAREASQELPP 489 ||| + | | | + +|| + | |#| | |+||| | | ||| | | Sbjct 93 EPKPVPKAVKPTPQPLAPSPEPSPTPAPAAD#PTPAPAAPAAAAPAAAPGPARETMQVSAP 152 |
| Burkholderia ambifaria MC40-6 | Query 432 EPKETTEDAAP-PGPTSETPELATEQKPIQD#PQPTPSAPAMGA-ADQLASAREASQELPP 489 ||| + | | | | +++| + | |#| | |+||+ | | + ||| | | Sbjct 93 EPKPVQKAAKPAPQPVAQSPAPSPTPAPAAD#PTPAPAAPSAAAPAATPSPARETMQVSAP 152 |
| Apis mellifera | Query 433 PKETTEDAAPPGPTSETPE--LATEQKPIQD#PQPTPSAPAMGAADQLASAREASQELPP 489 | | | | ||||| |+||| # | ||+ | +++|++ || +++ | Sbjct 2597 PTEVPEKMVPEEVPSETPEQALSTEQPVQLT#EAPEKMAPSTEAPEKMATSTEAPEKMAP 2655 |
| Synechococcus sp. JA-2-3B'a(2-13) | Query 432 EPKETTEDAAPPGPTSETPELATEQKPIQD---#PQPTP---SAPAMGAADQLASAREASQ 485 || + ++ || ||+| | | | | | #|+| | ||+ |+ | + + + Sbjct 154 EPDSESIESLPPEPTAEPPGPAPESPPADDLSP#PEPVPQEIEMPAIPEAESLLALQTPTP 213 Query 486 ELPP 489 || Sbjct 214 TPPP 217 |
| Roseobacter sp. SK209-2-6 | Query 432 EPKETTEDAAPPGPTSETPELATEQKPIQD#PQPTPSAPAMGAADQLASAREASQELPP 489 || | | | | | ||+ || || #||| | | + |+| ||+| | Sbjct 369 EPDEVTLDTATPAAPSESETLAPASKPA--#PQPEVSEP----IETAAAAAPQSQDLAP 420 |
| Roseovarius sp. HTCC2601 | Query 432 EPKETTEDAAP-PGPTSE---TPELATEQKPIQD#PQPTPSAPAMGAADQLASAREASQEL 487 ||+ | || | | | |+ | | +|+ +#|+| | || | ++++ |++ + Sbjct 75 EPQPVPEAPAPEPAPQPEPDPAPQPAPEPEPVPE#PEPDPLPPAAEAVPEVSAPESATRPV 134 Query 488 P 488 | Query 488 P 488 Query 433 PKETTEDAAPPGPTSETPELATEQKPIQD#PQPTPSAPAMGAADQ 476 | | | | | || | | | +| +#||| | ||| | | Sbjct 48 PVEAPEPAPQPEPTPE-PAPQPEPQPTPE#PQPVPEAPAPEPAPQ 90 |
| Theileria parva strain Muguga | Query 434 KETTEDAAPPGPTSETPELATEQKPIQD#PQPTP 466 | | || | ||+ || || | #||| | Sbjct 262 KPKPETKGPPVPKPRTPKSKTEPKPAQQ#PQPQP 294 |
| Rhodopseudomonas palustris BisB18 | Query 438 EDAAPPGPTSETPELATEQKPIQD#PQPTPSAPA 470 | | || | + ||+| |+| +#| | ||| Sbjct 103 EQAVPPTPLQDKPEVAAPQEPKAE#PAPQEPAPA 135 |
| Magnaporthe grisea 70-15 | Query 433 PKETTEDAAP-PGPTSETPELATEQKPIQD#PQPTPSAPA-MGAADQLASAR 481 || | || | | |+ | || + #| | | ||| | | +| |+ Sbjct 122 PKPAPEQPAPKPAPEQPAPKPAPEQPAPKQ#PVPKPPAPAETGKGDDMACAK 172 |
| Sagittula stellata E-37 | Query 432 EPKETTEDAAPPGPTSET---PELATEQKPIQD#PQPTPS---APAMGAADQLASAREASQ 485 +| | || | | +|| | || | ++#|+| || || || || |+ Sbjct 109 KPAEDTEPEPDPQPVAETAPVAEAATSDAP-EE#PEPAPSAQPAPESAEADAEDSADESGS 167 Query 486 ELPP 489 + | Sbjct 168 RVKP 171 |
| Alcanivorax borkumensis SK2 | Query 440 AAPPGPTSETPELATEQKPIQD#PQPTPSAPAMGAADQLASAREA 483 || | | | | |+ |+ #|+|+|+ | ||+| ||| +| Sbjct 393 AAAPAPVS-LPTPASASAPVTA#PEPSPTRAAPAAAEQAASADDA 435 |
| N/A | Query 433 PKETTEDAAPPGPTSETPELATEQKPIQD#PQPTPSAPAMGA--ADQLASAREASQELPP 489 |+ + ++ | | + | | |+| #| ||| |+ | | | | |+ ||| Sbjct 33 PERPAQTSSAPTPPAIPPVAPPPQPPVQV#PTPTPGQPSAPPPFAVQPAPQRPAAAPLPP 91 |
| Agrobacterium tumefaciens str. C58 | Query 433 PKETTEDAAPPGPTSETPELATEQKPIQD#PQPTPSAPAMGA--ADQLASAREASQELPP 489 |+ + + | | + | | |+| #| ||| |+ | | | | |+ ||| Sbjct 33 PERPAQTSPAPAPPAIPPVAPPPQPPVQV#PTPTPGQPSAPPPFAVQPAPQRPAAAPLPP 91 |
| Streptomyces ambofaciens ATCC 23877 | Query 433 PKETTEDAAPPGPTSETPELATEQKPIQD#PQPTPSAPAMGAADQLASAREASQEL 487 | | +| | || | || || | |# | + +| | ||| | Sbjct 65 PSEPSEPTEPTGPDSATPGETTENSPPPD#TDATDGRTSAPPPEQEAGIDEASTNL 119 |
| Burkholderia cenocepacia PC184 | Query 432 EPKETTEDAAP-PGPTSETPELATEQKPIQD#PQPTPSAPAMGA-ADQLASAREASQELPP 489 ||| + | | | | + +| + | |#| | |+||| | | ||| | | Sbjct 72 EPKPVQKAAKPTPQPVAPSPAPSPTPAPAAD#PTPAPAAPAPAAPAATPGPARETMQVSAP 131 |
| Burkholderia cenocepacia AU 1054 | Query 432 EPKETTEDAAP-PGPTSETPELATEQKPIQD#PQPTPSAPAMGA-ADQLASAREASQELPP 489 ||| + | | | | + +| + | |#| | |+||| | | ||| | | Sbjct 93 EPKPVQKAAKPTPQPVAPSPAPSPTPAPAAD#PTPAPAAPAPAAPAATPGPARETMQVSAP 152 |
| Burkholderia cenocepacia MC0-3 | Query 433 PKETTEDAAPPGPTSETPELATEQKPIQD#PQPTPSAPAMGAA 474 |+ | | | | | + || +| |+| #| | |+|| | Sbjct 137 PQATRE--AAPAPVTATPAVAAPAAPVQA#PAPAPAAPVRETA 176 |
| Salinibacter ruber DSM 13855 | Query 432 EPKETTEDAAPPGPTSETPELATEQKPIQD#PQPTPS-APAMG 472 || ++ +|||| ||| |++ | +# || || ||| | Sbjct 172 EPSPSSPSPSPPGPGDETPGSASD-SPTAN#GQPDPSTAPAAG 212 |
| Oryza sativa (japonica cultivar-group) | Query 432 EPKETTEDAAPPGPTSETPELATEQKPIQD#PQPTPSAPAMGAADQLASAREASQE 486 ||++ + || |||| | || |+|+#| || | ++ | + | Sbjct 352 EPEQEPQPETPP-VESETP--AEEQAPVQE#PPAADETPAAGEPEEQPKPRSSDSE 403 |
| Oryza sativa (indica cultivar-group) | Query 432 EPKETTEDAAPPGPTSETPELATEQKPIQD#PQPTPSAPAMGAADQLASAREASQE 486 ||++ + || |||| | || |+|+#| || | ++ | + | Sbjct 326 EPEQEPQPETPP-VESETP--AEEQAPVQE#PPAADETPAAGEPEEQPKPRSSDSE 377 |
| Crimean-Congo hemorrhagic fever virus | Query 434 KETTEDAAPPGPTSETPELAT-------EQKPIQD#PQPTPSA-PAMGAADQLASAREASQ 485 | +|| ++|| ||||| + | # |||||| ||| + || Sbjct 173 KTSTETSSPPPATSETPTPSPTTQVPTGNNNPNTS#RQPTPSAQPAMSNPATSPAQPNLSQ 232 Query 486 ELPP 489 || Sbjct 233 SAPP 236 |
| Yarrowia lipolytica | Query 435 ETTEDAAPPGPTSETPEL---ATEQKPIQD#PQPTPSAPAMGAADQLASAR-EASQELP 488 +|| | | || | ||+ | + | #|+|||+ || ++ + | + |+| Sbjct 143 QTTTPAVEPKPTPEVPEVKPEPTPEVPEVK#PEPTPAPPAPKPEPEVPEVKPEPTPEVP 200 |
| Cryptococcus neoformans var. neoformans JEC21 | Query 433 PKETTEDAAPPGPTSETPELATEQKPIQD#PQPTPSAPAMGAADQLASAREASQELPP 489 |+ |+ | | +| |+ |# |+|+||| || ++ |+|++ || Sbjct 1014 PQSPPRQASSPPPEAEKEASRPPTPPLPD#EPPSPAAPA--AAVRVESSRDSDPRTPP 1068 |
| Drosophila pseudoobscura | Query 432 EPKETTEDAA--PPGPTSETPELATEQKPIQD#PQPTPSAP 469 +|+ || | || |+| | | | | +#| | |+ | Sbjct 213 QPESTTAGAGDEPPATTTEEPALGTTLTPTPE#PAPVPTVP 252 |
| Anopheles gambiae str. PEST | Query 433 PKETTEDAAPPGPTSETPELATEQKPIQD#PQPTPSAPAMGAADQLASAREASQELPP 489 || || | ||+ ||+ | + #|+ ||+ | + | +|| | || Sbjct 489 PKPTT---TTPKPTTTTPKPTTTTTTTEK#PKTTPTPKATTTTPKATEAPKASTEEPP 542 |
| Trichomonas vaginalis G3 | Query 432 EPKE--TTEDAAPPGPTSETPELATEQKPIQD#PQPTPSAPAMGAADQLASAREASQELPP 489 |||+ +| | || | + |+ +|||+ #|||| | | ++ +|+ | Sbjct 313 EPKQQQSTLFAPPPKPVNNIPKPQPQQKPV--#PQPTVQQPQKAQESNLFGQKQPTQQKIP 370 |
| Cryptococcus neoformans var. neoformans B-3501A | Query 433 PKETTEDAAPPGPTSETPELATEQKPIQD#PQPTPSAPAMGAADQLASAREASQELPP 489 |+ |+ | | +| |+ |# |+|+||| || ++ |+|++ || Sbjct 1014 PQSPPRQASSPPPEAEKEASRPPTPPLPD#EPPSPAAPA--AAVRVESSRDSDPRTPP 1068 |
| Mesorhizobium sp. BNC1 | Query 434 KETTEDAAPP-GPTSETPELATEQKPIQD#PQPTPSAPAMGAADQLASAREASQELPP 489 || || | | ||| + |+| | +| | #| |+ | | + + +|| Sbjct 59 KEGTESAKEPVGPTEQQPDLVAESEPEQA#PVSEDEDEALPDKLPLGSGEQLADFIPP 115 |
| Aedes aegypti | Query 432 EPKETTEDAAPPGPTSET---PELATEQKPIQD#PQPTPSAPAMGAADQLASAREASQELP 488 || | |+ | | || || |+| +| +#|+|| |+ ++ + || Sbjct 594 EPTSEPEPASEPEPASEPASEPEPASEPEPASE#PEPTGEPEPSSASQSESTKEHTTTALP 653 Query 489 PG 490 | Sbjct 654 SG 655 Query 432 EPKETTEDAAPPGPTSETPELATEQKPIQD#PQPTP---SAPAMGAADQLASAREASQELP 488 || | |+ | |||| || |+| +| +#|+ | |+ |||+| +|+ Sbjct 674 EPTSEPEPASEPEPTSE-PEPASEPEPASE#PEATSTEHSSSDHSGKLQLAAAASTTQKTI 732 Query 489 P 489 | Query 489 P 489 |
| Chaetomium globosum CBS 148.51 | Query 432 EPKETTEDAAPPGPTSETP----ELATEQKPIQD#PQPTPSAPAMGAADQLASAREASQEL 487 || + |+ | | |||| | | |+| |+# + | || || | |+ Sbjct 100 EPTQEPEETAAPEPTSEPAPEPTEKVTTQEPAQE#TEQAPPAP---TEDQSPPGDEQQQQP 156 Query 488 PP 489 | Sbjct 157 AP 158 |
| Tribolium castaneum | Query 446 TSETPELATEQKPIQD#PQPTPSAPAMGAADQLASAREASQELPP 489 | | ||+||+| # + || | | + + |+|| || Sbjct 62 TEAKPVQKTEEKPVQR#AEETPKEPVKQATEAPKPSAPANQEAPP 105 |
| Nocardia farcinica IFM 10152 | Query 436 TTEDAAPPGPTSETPELATEQKPIQD#PQPTPSAP 469 || || | + || | | | #|+|| +|| Sbjct 418 TTRFVPPPPPQTSEPEPTTTQPPTTT#PEPTTAAP 451 |
| Leishmania braziliensis | Query 432 EPKETTEDAAPPGPTSETPELATEQKPIQD#PQPTPSAPAMGAADQLASAREAS 484 +| |+|+ ||||++| |+ ++ #|| +| | || || |+ Sbjct 1535 QPATTSEEDEED---EETPEVSTMQRSVRQ#PQQSPIIPGAAAARSFTSATAAA 1584 |
| Sulfitobacter sp. EE-36 | Query 432 EPKETTEDAAPPGPTSET---PELATEQKPIQD#PQPTPSAPAMGAADQ 476 +|+ | | | || | || | | +|+ +#|+| |+ +|| Sbjct 62 QPEPAPEPAPEPEPTPEPAPEPEPAPEPEPVPE#PEPIAPPPSEDVSDQ 109 |
| Collinsella aerofaciens ATCC 25986 | Query 432 EPKETTEDAAP--PGPTSETPELATEQKPIQD#PQPTPSAP 469 ||+ | | | | + ||| | | || #|+| |+ | Sbjct 747 EPEAAAETAVEKAPEPATATPEPAAEAKPTPA#PKPQPTTP 786 |
| Sulfitobacter sp. NAS-14.1 | Query 432 EPKETTEDAAPPGPTSET---PELATEQKPIQD#PQPTPSAPAMGAADQ 476 || | | | || | || | | +|+ +#|+| |+ +|| Sbjct 64 EPAPEPEPAPEPEPTPEPAPEPEPAPEPEPVPE#PEPIAPPPSEDVSDQ 111 |
| Gluconobacter oxydans 621H | Query 433 PKETTEDAAPPGPTSETPELATEQKPIQD#PQ-----PTPSAPAMGAADQLASAREASQEL 487 |+ | ||| |++++|| | + | #|| |||+ | +| | || Sbjct 34 PRARDEGGAPPPPSAQSPEETQENQ--QQ#PQKEQSTPTPAVPTPETPSAPDVPQEPSAEL 91 Query 488 P 488 | Query 488 P 488 |
| Rhodobacter sphaeroides 2.4.1 | Query 436 TTEDAAPPGPTSETPELATEQKPIQD#PQPTP-SAPAMGAADQLASAREA 483 ||| |||| |+ || || | +#| | | +||| | | | | | Sbjct 66 TTEPAAPPPATAPAPE-ATTPAP-AE#PAPAPEAAPATPAPDATAPAAPA 112 |
| Nostoc sp. PCC 7120 | Query 432 EPKETTEDAAPPGPTSETPELATEQKPIQD#PQ--PTPSA 468 ||+ || ||+ || | |+| #|+ ||||| Sbjct 181 EPRLANPRPTPPRPTTPTPRQQTPQRPSST#PRFGPTPSA 219 |
| Lotus japonicus | Query 433 PKETTEDAAPPGPTSETPELATEQKPIQD#PQPTPSAPAMGAADQLASAREA 483 | | || | ||+ || || #| |||+| || ||+ | Sbjct 47 PPATPPPAATPAPTT-TPPAATPAPSASP#PAPTPTASPTGAPTPSASSPPA 96 |
| Gloeobacter violaceus PCC 7421 | Query 433 PKETTEDAAPPGPTSETPELATEQKPIQD#PQ--PTPSAPAMGAADQLAS-------AREA 483 | | || | | | | || # | | ||| || + |+ | | Sbjct 168 PPPTVARAAAPDTPPPTASAAAESKPAPP#TPAPPPPPAPATAAAPETATNAAAPLEASSA 227 Query 484 SQELPPG 490 ||||| Sbjct 228 PTELPPG 234 |
| Burkholderia vietnamiensis G4 | Query 432 EPKETTEDAAP-PGPTSETPELATEQKPIQD#PQPTPSAPAMGA----ADQLASAREASQE 486 ||| + | | | | + +| + | |#| | |+||| | | | ||| | Sbjct 93 EPKPVQKAAKPTPQPLAPSPAPSPTPAPAAD#PTPAPAAPAAVAPAAPAATPAPARETMQV 152 Query 487 LPP 489 | Sbjct 153 SAP 155 |
| Corynebacterium jeikeium K411 | Query 432 EPKETTEDAAP-----PGPTSETPELATEQKPIQD#PQPTP 466 ||+ | | || | ||+| || | ||+ +#|+| | Sbjct 118 EPEPTPEPEAPKATPAPAPTAE-PEPVEEPKPVAE#PEPEP 156 |
| Anabaena variabilis ATCC 29413 | Query 432 EPKETTEDAAPPGPTSETPELATEQKPIQD#PQ--PTPSA 468 ||+ || ||+ || | |+| #|+ ||||| Sbjct 181 EPRLANPRPTPPRPTTPTPRQQTPQRPSST#PRFAPTPSA 219 |
| Neurospora crassa OR74A | Query 432 EPKETTEDAAPPGPTSETPE---LATEQKPIQD#PQPTPSAPAMGAADQLASAREASQEL 487 +|+| || ||| | |+| + |++ | +# |+ | | ++ | | ++| Sbjct 26 KPREAAEDVAPPKPNSDTTKNNVQASKDAPATN#GTTAPTEAAKAAPEKTLSPEEFERQL 84 |
| Yersinia bercovieri ATCC 43970 | Query 436 TTEDAAPPGPTSETPELATEQKPIQD#PQPTPSA 468 | | | |||+ | +||+| | #|| ||+| Sbjct 202 TIAQACHPPPTSQQPPMATQQLSILT#PQATPAA 234 |
| Caenorhabditis briggsae | Query 433 PKETTEDAAPPGPTSETPELATEQKPIQD#PQP-------TPSAPAMGAADQLASAREASQ 485 || | || | |+ |+ + #| | || | | ++| | ||+ Sbjct 121 PKATGSPPPPPPPPSDEPQEVVAGGASRR#PPPPPPRGTGTPPPPPTGVPEELNEANHASR 180 Query 486 ELPP 489 || Sbjct 181 RPPP 184 |
| Arabidopsis thaliana | Query 432 EPKETTEDAAPPGPTSETP--------ELATEQKPIQD#PQPTPS-APA 470 |||+ || + | || | | ++| ++||+ #| || ||| Sbjct 5 EPKKVTETVSEPTPTPEVPVEKPAAAADVAPQEKPVAP#PPVLPSPAPA 52 |
| Drosophila melanogaster | Query 432 EPKETTEDAAPPGPTSETPELATEQKPIQD#PQPTPSAPAMGAADQLASAREASQELPP 489 ||| | + | | || || +| +| +#|+|| | | +| + | | + | | Sbjct 461 EPKSEPEPKSEPEPKSE-PEPKSEPEPKSE#PEPT-SEPEPKSAPEPKSEPEPTSEPEP 516 |
| Cenarchaeum symbiosum | Query 432 EPKETTEDAAPPGPTSETPELATEQKPIQD#PQPTPSAP---AMGAADQLASAREA 483 || | | | | + || |+ | +|#| | | || | | | |+|++| Sbjct 3480 EPAGPAEPAVPEPPEARAPEPAS---PAED#PGPEPPAPQGVAPGEDTQDANAQDA 3531 |
| Rhodobacter sphaeroides ATCC 17025 | Query 432 EPKETTEDAAP---PGPTSET-PELATEQKPIQD#PQP--TPSAPAMGAADQLASAREASQ 485 +| |+||| | | +|| | || | +| #||| |+|| + + | || + Sbjct 70 QPAPPPEEAAPAPAPAPVAETPPPLAPEPRPEPA#PQPDAAPTAPEIAPVPEPEIA-EAPE 128 Query 486 ELPP 489 | | Sbjct 129 PLAP 132 |
| Leifsonia xyli subsp. xyli str. CTCB07 | Query 432 EPKETTEDAAPPGPTSETPELATEQKPIQD#PQPTPS-APAMGAADQLASAR 481 ||+ | ||| ||+||| || | #| | |+ || | +|| Sbjct 221 EPEPANEPAAPAEPTAETPAPAT--PPAAA#PLPAPAETPAADPATAPIAAR 269 |
| Burkholderia sp. 383 | Query 432 EPKETTEDAAPPGPTSETPELATEQKPIQD#PQPTPSAPAMGA-ADQLASAREASQELPP 489 ||| + | | |+ | | + |#| | |+||| | | ||| | | Sbjct 95 EPKPVQKAVLTPKPVSQAPS-PTPAPAVAD#PTPAPAAPAPAAPAAAPGPARETMQVSAP 152 |
| Fulvimarina pelagi HTCC2506 | Query 432 EPKETTEDAAPPGPTSE-TPELATEQKPIQD#PQPTPSAPAMGAADQLASAREASQELPP 489 || | | | | || | | +#|+| | + + |++ | | | + | | Sbjct 141 EPAPEPEPEPAPEPEPEPAPEPEPEPAPEPE#PEPAPESESQAPAEEAAPAEEPAPEPEP 199 |
| Hosta virus X | Query 436 TTEDAAPPGPTSETPELATEQKPIQD#PQPTPSAPAMGAADQLASA 480 ||++| || + | | | #|||| +| | | +|| Sbjct 588 TTQEAGTEGPPTTQPGKPTASSPRAA#PQPTANAETMEKGSQASSA 632 |
| Ralstonia solanacearum GMI1000 | Query 437 TEDAAPPGPTSETPELATEQKPIQD#PQPTPSAPAMGAA 474 | || || | +| || |#| | |+|| +| Sbjct 568 TVPAAAAGPASAASAIAAPAKPASD#PAPAPAAPKAPSA 605 |
| Pichia pastoris | Query 432 EPKETTEDAAPPGPTSETPELATEQKPIQD#PQPTPS 467 |||| ++|| | |+|| | +| || # || |+ Sbjct 144 EPKEEPKEAATPAPSSE--ESKSEAKPSSS#KQPRPA 177 |
| Stenotrophomonas maltophilia R551-3 | Query 432 EPKETTEDAAPPGPTSETPELATEQKPIQD#PQPTPSAPAMGAADQLASAREASQELPP 489 ||++ | | ||+ | | +#| | +||++ ++ || |+ + || Sbjct 81 EPRQQTVPTPLPTPTATVEEAQGIVLPAAE#PAPVQAAPSIDSSTPLAGAQLQYRSAPP 138 |
| Erythrobacter sp. NAP1 | Query 432 EPKETTEDAAPPGPTSETPELATEQKPIQD#PQPTP-SAPAMGAADQLAS 479 |||| + | | || ||| | | #| ||| | || || ||| Sbjct 102 EPKEAS--APMPDPTI-TPEPELEPAPAPT#PSPTPASEPAATAAKVLAS 147 |
| Rhodobacter sphaeroides ATCC 17029 | Query 436 TTEDAAPPGPTSETPELATEQKPIQD#PQPTPSAPAMGAADQLASAR 481 ||| |||| |+ || | #|+ |+ || | | |+ Sbjct 66 TTEPAAPPPATAPAPEATTPAPAEPA#PEAAPATPAPDATAPAAPAQ 111 |
| Corynebacterium efficiens YS-314 | Query 432 EPKETTEDAAPPGPTSETPELATEQKPIQD#P--QPTPSAPAMGAADQLASAREASQEL 487 || + | | | | | | | | |+ +#| +| |+ ||+ || | + | +| Sbjct 422 EPVKEPEPAPAPEPAPE-PVKAPEPAPVPE#PAPEPEPATPAVPTADLLPTIRSKWAQL 478 |
| Leishmania infantum JPCM5 | Query 433 PKETTEDAAPPGPTSETPELATEQKPIQD#PQPTPSAPAMGAADQLASAREASQELPPGF 491 | | | | || | +| # || |+|| +| |+ |+ +|| | Sbjct 1510 PSNATSGARLPAQTSGAPRTGASEK---S#SPPTSSSPAAASASSSTSSPVATSAVPPAF 1565 |
| Bacillus clausii KSM-K16 | Query 432 EPKETTEDAA--PPGPTSETPELATEQKPIQD#PQPTPSAPAMGAADQLASAREASQE 486 |||+ +|+ + | ++ | |++|+++#|| | | | | + | ++| Sbjct 185 EPKQQSEETSEEPKAKQNQEPLETVEEEPVEE#PQAEPQAKAEPETQQESKKEEPAEE 241 |
| Aspergillus fumigatus Af293 | Query 435 ETTEDAAPPGPTSETPELATEQKPIQD#PQPTPSAPAMGAADQLASAREASQE 486 | | || | |+| |+ | +| #|+ || | ||| + ++ + | Sbjct 144 EEEETEAPTPPKSKTSALSHEPSGVQS#PKHRPSIAASVAADNMKEVKKETLE 195 |
| Cyanothece sp. CCY0110 | Query 433 PKETTEDAAPPGPTSETPELATEQKPIQD#PQPTPSAP 469 |+ | || |+ ||+ | || #| ||| | Sbjct 400 PEPKTPTPAPENSTNSTPKTPIEPKPPIQ#PNPTPQTP 436 |
| Pichia guilliermondii ATCC 6260 | Query 433 PKETTEDAAPPGPTSETPELATEQKPI---QD#PQPTPSAP 469 | ||+| | || || + || || + #|+|+ | | Sbjct 718 PVETSETPETPEPTPETTPVGTETAPIETSES#PEPSESVP 757 |
| Aspergillus clavatus NRRL 1 | Query 432 EPKETTEDAAPPGPTSETPELATEQKPIQD#PQPTPSA----PAMGAADQLASAREASQEL 487 +|| |+ || | || | | #| | ||| ||+ | |+ + | + Sbjct 73 KPKATSA-AATPSQAPSTPSRAVASTPAAP#PAPAPSAATSTPAVPATPSPAAPAQPSADT 131 Query 488 PPGF 491 | | Sbjct 132 PVAF 135 |
| Danio rerio | Query 432 EPKETTEDAAPPGPTSETPELATEQK-------PIQD#PQPTPSAPAMGAADQLASAREAS 484 +| |+ |+ | +||| + | || | +#| | || ++ + |+++|+ Sbjct 169 DPLESA-DSVPSSSSSETSKKAAEQSPPPKPHTPASE#PDPLESADSLPKSSSRRSSKKAA 227 Query 485 QELPP 489 ++ || Sbjct 228 KQSPP 232 |
| Myxococcus xanthus DK 1622 | Query 433 PKETTEDAAPPGPTSETPELATEQKPIQD#PQPTPSAPAMGAADQLA 478 |+| +||| || | |++ #| | | | + | + | Sbjct 589 PREAKAEAAPVGPPPAMPIAPPVPVPVRP#PPPAPEAASRGGEEDAA 634 |
| Strongylocentrotus purpuratus | Query 432 EPKETTEDAAP-PGPTSE-TPELATEQKPIQD#PQPTPSAPAMGAADQLASAREASQELPP 489 ||+ | | | || | ||| | | #|+||| | + +| | +|| | Sbjct 242 EPEPEAAAAEPTPEPTPEPTPEPTPEPTPETT#PEPTPE-PEAPVVEPVAPVEEPAQEPTP 300 |
| Bacillus thuringiensis serovar konkukian str. 97-27 | Query 433 PKETTEDAAPPGPTSETPELATEQKPIQD#PQPTPSAPAMGAADQLASAREASQELP 488 || | | | ||| | || #| ||| ++ || +|++ +|+| Sbjct 130 PKAVTP-APKPVTRVETPATAPTPKPTPA#PTPTPKPVSVEAAVELSTPAPVKREVP 184 |
| Pyrobaculum arsenaticum DSM 13514 | Query 433 PKETTEDAAPPGPTSETPELATEQKPIQD#PQPTPS 467 | | ||| |||+ | | | | #| |||| Sbjct 49 PSPTQTPTAPPTPTSQ-PTTTTPTPPPQT#PSPTPS 82 |
| Schizosaccharomyces pombe 972h- | Query 432 EPKETTEDAAPPGPTSETPELATEQ-------KPIQD#PQPTPSAPAMGAADQLASAREAS 484 | | || |++|| +| | ||| | ++# +| || |+ + + Sbjct 469 EAKSTTNDSSPPKDSSSTSTQPTEQSNAQQAPSPKEE#ERPLPSEPSQNQPAEYRDTPDTP 528 Query 485 QELPP 489 + + | Sbjct 529 RNIMP 533 |
| Murid herpesvirus 2 | Query 432 EPKE--TTEDAAPPGPTSETPELATEQKPIQD#PQPTPSAPAMGAADQLASAREASQEL 487 ||| |||+ | + || | ++| |# | ||+| ++| || +| Sbjct 244 EPKTEPTTEEPTEPTTMATTPPKQTRKRPAPD#VQKTPAAKPTKHGRRMARIPEAMMDL 301 |
| Corynebacterium glutamicum R | Query 434 KETTEDAAPPGPTSETPELATEQKPIQD--#PQPTPS-----APAMGAADQLASAREASQE 486 +| || | |+| | | || |+ | #|+| |+ ||| | +| + || Sbjct 240 EEATEAAEAASVTAEFAEAAREQTPVPDTA#PEPAPAPIDDIAPASGRIGKLRTRLSRSQN 299 Query 487 L 487 + Sbjct 300 V 300 |
| Colwellia psychrerythraea 34H | Query 433 PKETTEDAAPPGPTSET-----PELATEQKPIQD#PQPTPSAPAMGAADQLASAREASQEL 487 | | | | +| ||| || +|| |#|+| | + ++| |+ | | Sbjct 451 PTSDLEPITAPEPITEPELITEPELITEPEPITD#PEPIPEIVSEPQPERLESSSAGSPGL 510 |
| Frankia alni ACN14a | Query 441 APPGPTSETPELATEQKPIQD#PQPTPSAPA--MGAADQLASAREA 483 | | | + ||+ + |+ #| |||+| | ||| + | || | Sbjct 258 ASPQPLASPQPLASARPPVST#PPPTPAATAATAGAAARTAPARPA 302 |
| Rhodobacterales bacterium HTCC2654 | Query 432 EPKET--TEDAAPPGPTSETPELATEQKPIQD#PQPTPSAPAMGAADQLAS---AREASQE 486 ||+| || || || | +|+ +#|+||| | | + | ++ Sbjct 59 EPEEAAPTETPVPPARPEPEPEPQPEPEPVPE#PEPTPLPEPPAPAPSLPTEDVQPETTEP 118 Query 487 LPP 489 || Sbjct 119 TPP 121 |
| Tetraodon nigroviridis | Query 434 KETTEDAAPPGPTSETPELATEQKPI-QD#PQPTPSA 468 || | + +|| ||||+ || | #|+|+||| Sbjct 207 KEQTPNVSPPRGAPATPELSPLPKPSNQS#PEPSPSA 242 |
| Chromohalobacter salexigens DSM 3043 | Query 433 PKETTEDAAPPGPTSETPELATEQKPIQD---#PQPTPSAPAMGAADQLAS---AREASQE 486 | +|||| | | ||| | + | #|| |||| + | + +||+ Sbjct 471 PPAASEDAASPAPVPETPSAAPRPRISWDDEA#PQAHSSAPASAPPEPEVSDAPSADASRS 530 Query 487 LPPG 490 | | Sbjct 531 APRG 534 |
| Coccidioides immitis RS | Query 438 EDAAPPGPTSETPELATEQKPIQD#PQPTPSAPAMGAADQLASAREASQEL 487 |+| || | + | || ||| #| | | + | || | ++ + Sbjct 42 EEAGPPPPPPKEPILAIEQKTEAK#PVAPESGPGLAAKACLAEAAASTNSI 91 |
| Synechococcus sp. BL107 | Query 432 EPKETTEDAAPPGPTSETPELATEQKPIQD#PQPTPSAPAMGAA--DQLASAREASQE 486 ||+ | || | |||++ | | #||||| || +| + +| |+ |+ |+ Sbjct 111 EPESAT---APNAPVPLTPEVS----PEQT#PQPTP-APTVGGSLLEQAAAQRQQRQQ 159 |
| Methylibium petroleiphilum PM1 | Query 445 PTSETPELATEQKPIQD#PQPTPSAPAMGAADQLASAREASQELPP 489 | | | | |+ | #|+| | ||| || +|+ || Sbjct 825 PRSLTTETASGAAPAAP#PEPAPPAPAAAAAASAPVVADAAPAAPP 869 |
| Alnus glutinosa | Query 433 PKETTEDAAP---PGPTSETPELATEQKP-IQD#PQPTPSAPAMGAADQLASAREASQELP 488 |||| |+ || | || | + ||+ | + +#|+| | | |+ | + |+| Sbjct 99 PKETPEEEAPKETPEPTVEETKEATDSAPAVPE#PKPEPEAEVPKKAE---VPEEVAAEVP 155 |
| Lyngbya sp. PCC 8106 | Query 432 EPKETTEDAAPPGPTSETPELATEQKPIQD#PQPTP 466 ||+ | | | || | || | +| +#|+||| Sbjct 1616 EPEPTPEPTPEPEPTPE-PEPTPEPEPTPE#PEPTP 1649 |
| Aspergillus niger | Query 433 PKETTEDAAPPGPTSETPELATEQKPIQD#PQPTPSAPA 470 | + |||| | | +| +|+ #| | ||| | Sbjct 263 PPAASAPAAPPPPPPSAPPVAPPSEPLSR#PSPLPSAIA 300 |
| Ustilago maydis 521 | Query 432 EPKETTEDAAPPGPTSETPELATEQKPIQD-----#PQPTPSAPAMGAADQLASAREASQE 486 ||+| ||| | + |+| + | #| | | || + | | | + Sbjct 502 EPEEEAAPPAPPAPPAPPAAAASETEEQDDQGIPP#PPPPPPAPPVAPAAPAAPALQEEAP 561 Query 487 LPP 489 || Sbjct 562 APP 564 |
| Phaeosphaeria nodorum SN15 | Query 432 EPKETTEDAAPPGPTSET---PELATEQKPIQD#PQPTPSAPAMGAA 474 +| || + | | | | | | | | #| | || |+ | | Sbjct 131 KPVETPQPAPAPAPVSSAAPAPAPAPEYTPAPA#PAPAPSKPSSGGA 176 |
| Saccharomyces cerevisiae | Query 432 EPKETTEDAAPPGPTSETPELATEQKPIQD#PQPTPSAPAMGAADQLASAREASQELPP 489 +| |||| || | | + #| | | || + + | + + || Sbjct 648 KPPSPPVAAAPPLPTFSAPSLPQQSVSTSI#PSPPPVAPTLSVRTETESISKNPTKSPP 705 |
| Streptococcus pneumoniae | Query 432 EPKETTEDAAPPGPTSETPELATEQKPIQD#PQPTPSAPAMGAADQLAS---AREASQE 486 ||++ |+ | | | | | + || #||| |+ || | || + +| Sbjct 310 EPEKPAEETPAPAPKPEQP--AEQPKPAPA#PQPAPAPKPEKTDDQQAEEDYARRSEEE 365 |
| Neosartorya fischeri NRRL 181 | Query 433 PKETTEDAAPPGPTSETPELATEQKP--IQD#PQPTPSAPAMGAADQLASA 480 | + |||| |+|+ | | + #|+| |+ | | | +| Sbjct 533 PVAAPQVPTPPGPPPESPQTKTHTPPARVAS#PRPRPTKPKKGGNGQSPNA 582 |
| Vitis vinifera | Query 440 AAPPGPTSETPELATEQK--PIQD#PQPTPSAPAMGAADQLASAREASQELPP 489 |||| | | | | +| |++ # | |+||+| | +++ |+ +|| Sbjct 842 AAPPRPASPVPPQAEQQDELPVES#VPPAPAAPSMPEA--ISTDPPATPLVPP 891 |
| Leishmania major | Query 433 PKETTEDAAPPGPTSETPELATEQKPIQD#PQPTPSAPAMGAADQLASAREASQELPPGF 491 | | | | || | +| # || |+|| +| |+ |+ || | Sbjct 1510 PSNATSGARLPAQTSSAPRAGASEK---S#SPPTLSSPAAASASSSTSSPVAASTAPPAF 1565 |
| Mycobacterium sp. MCS | Query 433 PKETTEDAAPPGPTSE---------TPELATEQKPIQD#PQPTPS---APAMGAADQLASA 480 | || || | +| ||| | +| +#|+| |+ || | |+| Sbjct 114 PTETAPPRAPERPVAEPEPAPEPEPTPEPIPEPEPAPE#PEPEPAEVIAPTHGRLDRLRGR 173 Query 481 REASQ 485 || Sbjct 174 LAKSQ 178 |
| Roseiflexus castenholzii DSM 13941 | Query 433 PKETTEDAAPPGPTSETPELATEQKPIQD#PQPTPSAPAMGAADQLASAREAS 484 | | | | ||| ||| + +| #|+|| + |+ | | | | + Sbjct 848 PSATVTPTATPLPTS-TPEPTSTPEPTNT#PEPTATPTALPTATPLPSPTETA 898 |
| Roseobacter denitrificans OCh 114 | Query 432 EPKETTEDAAPPGPTSETPELATEQKPIQD#PQPTPSAPAMGAADQLASAREASQELP 488 ||+ | | | | || |+ |+ # ||| | + | || + ||| Sbjct 142 EPEPEPEPEPEPEPEPE-PEPEPERTPVAH#APPTPEVPEVEVAPAQHSAEDDPFELP 197 |
| Desulfovibrio vulgaris subsp. vulgaris str. Hildenborough | Query 437 TEDAAPPGPTSETPELATEQKPIQD#PQPTPSAPAMGAADQLASAREASQELPPG 490 | |+ | | + +|| | #|+| |+||| +| +| +|+ | | Sbjct 564 TTSASAPESGSVAQGAPSSEKPAQK#PEPVPAAPAFQSA--ASSGAQAAASAPAG 615 |
| Streptococcus pyogenes MGAS10750 | Query 433 PKETTEDAAP-PGPTSETPELA--TEQKPIQD#PQPTPSAPAMGAADQLASAREASQELP 488 |+ | ||| | | || | +| +|+ | # |+||+ + | +|+| | Sbjct 193 PETPEEPAAPSPSPESEEPSVAASSEETPTPS#TPEEPAAPSPSPESEEPSVAASSEETP 251 |
| Mycobacterium sp. JLS | Query 433 PKETTEDAAPPGPTSE---------TPELATEQKPIQD#PQPTPS---APAMGAADQLASA 480 | || || | +| ||| | +| +#|+| |+ || | |+| Sbjct 106 PTETAPPRAPERPVAEPEPAPEPEPTPEPIPEPEPAPE#PEPEPAEVIAPTHGRLDRLRGR 165 Query 481 REASQ 485 || Sbjct 166 LAKSQ 170 |
| Alcelaphine herpesvirus 1 | Query 435 ETTEDAAPPGPTSETPELATEQKPIQD#PQPT-------PSAPAMGAADQLASAREASQEL 487 | |+ || |||++|| |+ | |||# | | | | | + ++ ++ | Sbjct 1024 EQTQAPAPKGPTADTP--VTDSKLIQD#SQQNDRHQKEKPLKPKKPQAISLPAVKDTTKHL 1081 Query 488 PP 489 | Sbjct 1082 KP 1083 |
| Zea mays | Query 433 PKETTEDAAPPGPTSETPELATEQKPIQD#PQPTPSAPAMGAADQLASAREASQELPP 489 | + + ++|| | | | | + |+ +# | |+ + ++| + + || Sbjct 738 PPQAPKSSSPPAPVSSPPPLKSSPPPVPE#SSPPPTPKSSPPLAPVSSPPQVEKTSPP 794 |
| Streptococcus sobrinus | Query 432 EPKETTEDAAPPGPTSETPELATEQKPIQD#PQPT----PSAPAMGAADQLASAREASQEL 487 |||+ | ||| | | |+ || + # ||| | | | + | | Sbjct 1040 EPKQPTPPAAPEPPKMTTVEI--PDKPTEP#KQPTPPVAPEPPKMTTVEIPDKPTEPKQPT 1097 Query 488 PP 489 || Query 488 PP 489 |
| Bordetella bronchiseptica | Query 433 PKETTEDAAPPGPTSETPELATEQKPIQD#PQPTPSAPAMGAADQLASAREAS 484 || + ||| + |+| | | #||| || | +|++| |+ Sbjct 546 PKPAPQPGPQPGP--QPPQLPQPQPPKPQ#PQPEAPAPQPPAGRELSAAANAA 595 |
| Sphingomonas sp. SKA58 | Query 441 APPGPTSETPELATEQKPIQD#PQPTP---SAPAMGAADQLASAREASQELP 488 || | +||| | |+ #| | | +||| || | | +|+ + | Sbjct 69 APAAPEAETPRAAA---PVSA#PPPPPRPAAAPAAPAAAPAAPAADAASDAP 116 |
| Dinoroseobacter shibae DFL 12 | Query 438 EDAAPPGPTSETPELATEQKPIQD#PQPTPSA 468 || || ||+ | || || |#| |+|+| Sbjct 3 EDTPPPQSTSKRPATPVEQSPI-D#PVPSPTA 32 |
| Natronomonas pharaonis DSM 2160 | Query 432 EPKETTEDAAPPGPT-SETPEL--ATEQKPI-QD#PQPTPSAPAMGAADQLASAR 481 +|+| |+ | |+ |||| | ++ | |#|+||| |+| | +| Sbjct 22 QPEEAPEETPEPEPSPEETPESEPAPDETPAPDD#PEPTPEDEPPSDAEQFAESR 75 |
| Enterococcus faecalis | Query 432 EPKETTEDAAPPGPTSET----PELATE-QKPIQD#PQP-TPSAPA 470 || | || | ||+ | || || ||++ # +| ||| || Sbjct 72 EPTEPTEPTTPTEPTTPTEPSEPEQPTEPSKPVEP#EKPVTPSKPA 116 |
| Xanthobacter autotrophicus Py2 | Query 433 PKETTEDAAPPGPTSETPELATEQKPI------QD#PQPTPSAPAMGAADQLASAREAS 484 | + ||| ||+ || | | | | # | |+|| || + |+ || Sbjct 158 PTPAAAEPAPPAPTAAAPEAAAPQAPAPQTSAPQA#AAPKPAAP-RAAAPAASEAKPAS 214 |
| Enterococcus faecalis V583 | Query 432 EPKETTEDAAPPGPTSET----PELATE-QKPIQD#PQP-TPSAPA 470 || | || | ||+ | || || ||++ # +| ||| || Sbjct 73 EPTEPTEPTTPTEPTTPTEPSEPEQPTEPSKPVEP#EKPVTPSKPA 117 |
| Pyrobaculum islandicum DSM 4184 | Query 440 AAPPGPTSETPELATEQKPIQD#PQPTPSAPAMGAADQLASA 480 | | || || || | | # | +||| | + ||+| Sbjct 798 ATTPTPTPTTPPPATTPTPPQP#TVVTTTAPATGTTEALAAA 838 |
| Macaca fascicularis | Query 432 EPKETTEDAAPPGPTSET-------PELATEQKPIQD#PQPTPSAPAMGAADQLASAREAS 484 +| | +|+|| || + || ++| #|+| | + | +| |+ | + Sbjct 8 DPPEGSEEAAEPGMDTPEDQDLPPCPEDIGKEKCTAA#PEPEPCEVSEPPAKRLRSSEEPT 67 Query 485 QELPPG 490 ++ ||| Sbjct 68 EKEPPG 73 |
| Thiomicrospira crunogena XCL-2 | Query 432 EPKETTEDAAPPGPTSETPELATEQKPIQD#PQPTPS---APAMGA 473 || | | | || | || | +| +#|+||| || || Sbjct 98 EPAPEPEPALEPEPTPE-PEPTPEPEPTPE#PEPTPEPEPTPAAGA 141 Query 432 EPKETTEDAAPPGPTSETPELATEQKPIQD#PQPTP 466 ||+ | | | | | || | | +| +#|+||| Sbjct 86 EPESEPEPAPEPEPAPE-PEPALEPEPTPE#PEPTP 119 |
| Chlamydomonas incerta | Query 433 PKETTEDAAPPGPTSETPELATEQKPIQD#PQPTPSAPA 470 | ||| || +|| + | #| | | +|| Sbjct 60 PSPAPPSPAPPSPTPPSPEPPSPAPPSPP#PSPAPPSPA 97 |
| Streptococcus sanguinis SK36 | Query 432 EPKETTEDAAPPGPTSETPELATEQ---KPIQD#PQPTPSAPAMGAADQLASAREASQEL 487 || ++ ++ +| || | +|| ||+++# || | + | +| ||+ +| Sbjct 87 EPSQSDDNPSPRSSESEKVEESTESTTTKPVEN#TQPITVRPEISNATVVAEKSEATADL 145 |
| Pichia stipitis CBS 6054 | Query 433 PKETTEDAAPPGPTSETPELATEQKPIQD#PQPTPSAPAMGAADQLASAREA----SQELP 488 | | || | | | |+ #| | | | ||| +| | |+|| Sbjct 355 PPPPNRGAVPPPPPSRVPGPTPPQRTGAP#PPPPPPRAARGAAPPPPPSRTARPAQPQQLP 414 Query 489 P 489 | Query 489 P 489 |
| Geobacter lovleyi SZ | Query 432 EPKETTEDAAPPGPTSETPELATEQKPIQD#PQPTPSAPAMGAADQLASAREASQEL---- 487 +||+ |+|++| | + | | # |+| | |++ +| || ||+ Sbjct 243 KPKQDTDDSSPDGDNTGTAAGDDSQSQAAG#-NPSPPVPKSGSSGELGGNPEALQEMLDCD 301 Query 488 PPGF 491 | || Sbjct 302 PGGF 305 |
| Burkholderia cenocepacia HI2424 | Query 433 PKETTEDAAPPGPTSETPELATEQKPIQD#PQPTPSAPAMGAA 474 |+ | | | | | || +| |+| #| | |+ | | Sbjct 137 PQATRE--AAPAPAPATPAVAAPAAPVQA#PAPAPAVPVRETA 176 |
| Polaromonas naphthalenivorans CJ2 | Query 432 EPKETTEDAAPPGPTSE--TPELATEQKP------------IQD#PQPTPSAPAMGAADQL 477 ||| ||| | || + + | | |#| |+|+|| | + Sbjct 86 EPKAEPAPPAPPAPPVRPVTPSVQKKVVPKPAARPQAQPLAIAD#PTPSPNAPTGSLAPAV 145 Query 478 ASAREASQELP 488 || |+ | | Sbjct 146 ASTPTAAPEAP 156 |
| Desulfovibrio vulgaris subsp. vulgaris DP4 | Query 437 TEDAAPPGPTSETPELATEQKPIQD#PQPTPSAPAMGAADQLASAREASQELPPG 490 | |+ | | + +|| | #|+| |+||| +| +| +|+ | | Sbjct 564 TTSASAPESGSVAQGAPSSEKPAQK#PEPVPAAPASQSA--ASSGAQAAASAPAG 615 |
| Rhodopirellula baltica SH 1 | Query 435 ETTEDAAPPGPTSETPELATEQKPIQD#PQPTPSAPAMGAADQLASAREASQELPP 489 |||++|||| | +| | + #| +||| ||+++ |+ + +| | Sbjct 202 ETTDEAAPPAP------VADEAPAAEA#PAAEEAAPAAPAAEEVKSSSDQVEEAAP 250 Query 432 EPKETTEDAAPPGPTS------ETPELATEQKPIQD#PQPTPSAPAMGAADQLASAREASQ 485 | ||+||| | + || + | |+ |# | || ||++ | | |++ Sbjct 180 EAAAATEEAAPAAPVADAAADAETTDEAAPPAPVAD#EAPAAEAP---AAEEAAPAAPAAE 236 Query 486 EL 487 |+ Sbjct 237 EV 238 |
| Anaeromyxobacter sp. Fw109-5 | Query 433 PKETTEDAAPPGPTSETPELATEQKPIQD#PQPTPSAPAMGAAD 475 |+| | || | + | |+ +#| | |+| ||+ Sbjct 353 PQEAVAAATPPAPPAAAPAEKAPPAPVAE#PAPKPAAETPRAAE 395 |
| Pseudomonas aeruginosa UCBPP-PA14 | Query 439 DAAPPGPTSETPELATEQKPIQD#PQPTPSAPAMGAADQLASAREASQELPP 489 | | | | |++| + #||| | || ||+ +|+ ++| || Sbjct 156 DEPPLVPVSSHPQIAGRTH--ER#PQPGPGFPAKAAAEVAPTAQASAQASPP 204 |
[Site 7] EGAESAQPEA395-EELEATVPQE
Ala395  Glu
Glu
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Glu386 | Gly387 | Ala388 | Glu389 | Ser390 | Ala391 | Gln392 | Pro393 | Glu394 | Ala395 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Glu396 | Glu397 | Leu398 | Glu399 | Ala400 | Thr401 | Val402 | Pro403 | Gln404 | Glu405 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| TVPAGEGVSLEEAKIGTETTEGAESAQPEAEELEATVPQEKVIPSVVIEPASNHEEEGEN |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | Homo sapiens | 119.00 | 14 | unknown |
| 2 | synthetic construct | 119.00 | 1 | amphiphysin |
| 3 | Pan troglodytes | 115.00 | 2 | PREDICTED: amphiphysin |
| 4 | Macaca mulatta | 104.00 | 4 | PREDICTED: amphiphysin (Stiff-Man syndrome with br |
| 5 | Rattus norvegicus | 75.90 | 1 | amphiphysin 1 |
| 6 | Bos taurus | 69.70 | 1 | hypothetical protein LOC614722 |
| 7 | Canis familiaris | 67.40 | 1 | PREDICTED: similar to Amphiphysin |
| 8 | Mus musculus | 64.30 | 3 | amphiphysin |
| 9 | Monodelphis domestica | 62.80 | 1 | PREDICTED: similar to amphiphysin I variant CT2 |
| 10 | Gallus gallus | 52.00 | 1 | amphiphysin (Stiff-Man syndrome with breast cancer |
| 11 | Xenopus laevis | 42.70 | 1 | hypothetical protein LOC432150 |
| 12 | Xenopus tropicalis | 42.40 | 1 | Unknown (protein for MGC:122074) |
| 13 | Tetraodon nigroviridis | 37.00 | 1 | unnamed protein product |
| 14 | Oryza sativa (indica cultivar-group) | 36.60 | 1 | hypothetical protein OsI_022473 |
| 15 | Oryza sativa (japonica cultivar-group) | 35.40 | 4 | hypothetical protein OsJ_020736 |
| 16 | Plasmodium falciparum 3D7 | 35.00 | 1 | glutamate-rich protein |
| 17 | Plasmodium falciparum | 35.00 | 1 | AF247634_1 glutamate-rich protein |
| 18 | Debaryomyces hansenii CBS767 | 33.90 | 1 | hypothetical protein DEHA0C17446g |
| 19 | Candida glabrata | 33.50 | 1 | unnamed protein product |
| 20 | Myxococcus xanthus DK 1622 | 33.10 | 1 | hypothetical protein MXAN_0043 |
| 21 | Phaeosphaeria nodorum SN15 | 33.10 | 1 | hypothetical protein SNOG_02682 |
| 22 | Magnetococcus sp. MC-1 | 32.70 | 1 | MJ0042 family finger-like protein |
| 23 | Aspergillus niger | 32.30 | 1 | hypothetical protein An04g09080 |
Top-ranked sequences
| organism | matching |
|---|---|
| Homo sapiens | Query 366 TVPAGEGVSLEEAKIGTETTEGAESAQPEA#EELEATVPQEKVIPSVVIEPASNHEEEGEN 425 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 366 TVPAGEGVSLEEAKIGTETTEGAESAQPEA#EELEATVPQEKVIPSVVIEPASNHEEEGEN 425 |
| synthetic construct | Query 366 TVPAGEGVSLEEAKIGTETTEGAESAQPEA#EELEATVPQEKVIPSVVIEPASNHEEEGEN 425 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 366 TVPAGEGVSLEEAKIGTETTEGAESAQPEA#EELEATVPQEKVIPSVVIEPASNHEEEGEN 425 |
| Pan troglodytes | Query 366 TVPAGEGVSLEEAKIGTETTEGAESAQPEA#EELEATVPQEKVIPSVVIEPASNHEEEGEN 425 ||||||||||||||||||||||||||||||#|||| |||||||||||||||||||| |||| Sbjct 733 TVPAGEGVSLEEAKIGTETTEGAESAQPEA#EELEVTVPQEKVIPSVVIEPASNHEGEGEN 792 |
| Macaca mulatta | Query 366 TVPAGEGVSLEEAKIGTETTEGAESAQPEA#EELEATVPQEKVIPSVVIEPASNHEEEGEN 425 |+||||||||||||||||| ||||||| ||#||| |||||||||||||||||||||||||+ Sbjct 456 TLPAGEGVSLEEAKIGTET-EGAESAQAEA#EELAATVPQEKVIPSVVIEPASNHEEEGEH 514 |
| Rattus norvegicus | Query 367 VPAGEGVSLEEAKIGTETTEGAESAQPEA#EELEATVPQEKVIPSVVIEPASNHEEE 422 +||||| | | ||| |+|| | | |+|# |||| |||||||||||||||||| | Sbjct 489 LPAGEGESPEGAKIDVESTELASSESPQA#AELEAGAPQEKVIPSVVIEPASNHEGE 544 |
| Bos taurus | Query 367 VPAGEGVSLEEAKIGTETTEGAESAQPEA#EELEATVPQEKVIPSVVIEPASNHEEEGEN 425 +||| | |||||| | || + + + #|| || ||||||||||+||||+| |||+ Sbjct 496 LPAGAGASLEEAKTDIEATEAIDGDRSQL#EETEAVAAQEKVIPSVVIQPASNNEGEGEH 554 |
| Canis familiaris | Query 367 VPAGEGVSLEEAKIGTETTEGAESAQPEA#EELEATVPQEKVIPSVVIEPASNHEEEGEN 425 +| ||| +|||+ || |+ + + + #|| || | ||||||||||||||+| ||++ Sbjct 650 LPPGEGAGVEEARPDTEATQAVDGDRSQL#EEAEAVAPLEKVIPSVVIEPASNNEGEGDH 708 |
| Mus musculus | Query 367 VPAGEGVSLEEAKIGTETTEGAESAQPEA#EELEATVPQEKVIPSVVIEPASNHEEEGEN 425 +||||| | | ||| |+|| | | |+ # | || || |||||||||||||| |||+ Sbjct 493 LPAGEGGSPEGAKIDGESTELAISESPQP#VEPEAGAPQ--VIPSVVIEPASNHEGEGEH 549 |
| Monodelphis domestica | Query 368 PAGEGVSLEEAKIGTETTEGAESAQPEA#EELEATVPQEKVIPSVVIEPASNHEEEGE 424 | ||||+ | + || | || + #||+|| ||||||||||||||||| | | Sbjct 43 PLAEGVSVVETQAVPETREATESDRAGT#EEMEAVGIQEKVIPSVVIEPASNHEGEDE 99 |
| Gallus gallus | Query 367 VPAGEGVSLEEAKIGTETTEGAESAQPEA#EELEATVPQEKV--IPSVVIEPASNHEEEGE 424 | || | | + || || +|+ #|| + | |||| |||||||||||+| ||| Sbjct 494 VAAGAG----EGAVRTEQEAAAEGDKPQG#EEKDVDVSQEKVSSIPSVVIEPASNNEGEGE 549 |
| Xenopus laevis | Query 366 TVPAGEGVSLEEAKIGTET-TEGAESAQPEA#EELEATVPQEK-------VIPSVVIEPAS 417 |+|+ + + |+++ |+ | || | #+| | | | +|||||||||| Sbjct 479 TLPSEQNEEVVESEVKQESPVEMAEDIMEEE#KEPEIIEPNETKAPPEITIIPSVVIEPAS 538 Query 418 NHEEEGEN 425 |||+| ++ Sbjct 539 NHEDEHDD 546 |
| Xenopus tropicalis | Query 369 AGEGVSLEEAKIGTETTEGA---------ESAQPEA#EELEAT-VPQE-KVIPSVVIEPAS 417 | + || | ++ |+ | | | +|| # | + | ||| |||||||||| Sbjct 477 AEKEVSEIEVEVKEESAESAAEMADDIMEEQKEPEI#NEPKETEAPQEITCIPSVVIEPAS 536 Query 418 NHEEEGEN 425 |||+| +| Sbjct 537 NHEDEHDN 544 |
| Tetraodon nigroviridis | Query 372 GVSLEEAKIGTETTEGAESAQPEA#EELEATVPQEKVIPSVVIEPAS 417 | | ++ | || ||||| +| |#| || | |+ || +| | Sbjct 133 GFSFKKTK--KETGEGAESEEPAA#EATEAAAPAEEAKPSEAMEETS 176 |
| Oryza sativa (indica cultivar-group) | Query 366 TVPAGEGVSL-----EEAKIGTETTEGAESAQPEA#EELEATVPQEKVI 408 |+|| | | + ||||| | || | | #||+| | | | + Sbjct 713 TIPATEKVEVTTPATEEAKITTAATEEVEVTTPAT#EEVEVTTPSTKEV 760 |
| Oryza sativa (japonica cultivar-group) | Query 366 TVPAGEGVSL-----EEAKIGTETTEGAESAQPEA#EELEATVPQEKVI 408 | || | | + ||||| | || | | #||+| | | | + Sbjct 713 TTPATEKVEVTTPATEEAKITTAATEEVEVTTPAT#EEVEVTTPSTKEV 760 |
| Plasmodium falciparum 3D7 | Query 371 EGVSLEEAKIGTETTEGAESAQPEA#EELEATVPQEKVIPSVVIEPASNHEE 421 | | +|| | | |+ +|| #++ | +| +|| || + | | | Sbjct 1053 EIVEIEEVPSQTNNNENIETIKPEE#KKNEFSVVEEKAIPQEPVVPTLNENE 1103 |
| Plasmodium falciparum | Query 371 EGVSLEEAKIGTETTEGAESAQPEA#EELEATVPQEKVIPSVVIEPASNHEE 421 | | +|| | | |+ +|| #++ | +| +|| || + | | | Sbjct 1056 EIVEIEEVPSQTNNNENIETIKPEE#KKNEFSVVEEKAIPQEPVVPTLNENE 1106 |
| Debaryomyces hansenii CBS767 | Query 382 TETTEGAESAQPEA#EELEATVPQEKVIPSVVIEPASNHEEE 422 | || |+| |||#|+|| | + |+||++ | |+|| Sbjct 283 TRATESNETA-PEA#EKLEENKPATEA-PTVVVDEADEHKEE 321 |
| Candida glabrata | Query 370 GEGVSLEEAKIGTETTEGAESAQPEA#EELEATVPQEKVIPSVVIEPASNHE--EEG 423 | |+ |||++ | || || +| #| + | + + ||| | ||| Sbjct 461 GREVTTEEAELAEEATEPAEETEPAK#EVITEEAEPAKEVTAEEAEPAEETEPAEEG 516 |
| Myxococcus xanthus DK 1622 | Query 386 EGAESAQPEA#EELEATVPQEKVIPSVV 412 | || |+|||#|+|+| +| +|+| | Sbjct 318 EDAEEAEPEA#EDLQAWLPSPRVLPGEV 344 |
| Phaeosphaeria nodorum SN15 | Query 374 SLEEAKIGTETTEGAESAQPEA#EELEATVPQEKVIPSVVIEPASNHEE 421 | ||||| |||| + ||+|# || |+ |+ + || || Sbjct 5484 SREEAKIFTETTTAEPAIQPDA#----PVVPVEQTQPTSLEEPVYPSEE 5527 |
| Magnetococcus sp. MC-1 | Query 376 EEAKIGTETTEGAESAQPEA#EELEATVPQEKVIPSVVIEPASNHEEE 422 || | +| | | |||#|| | +|+ | | | ||| Sbjct 323 EEESEAEEESEAEEEAAPEA#EEEAAPEAEEEAAPEAEEEAAPEAEEE 369 |
| Aspergillus niger | Query 373 VSLEEAKIGTETTEGAESAQPEA#EELEATVPQEKVIPSVVIEPASNHEEEG 423 || + |++ ||| + +| |#|| | ||| ||| || Sbjct 94 VSEQTAEVATETPVADKPTEPSA#EEAHAESTVEKVEEPKTEEPAEQEVAEG 144 |
[Site 8] DLWTTSTDLV260-QPASGGSFNG
Val260  Gln
Gln
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Asp251 | Leu252 | Trp253 | Thr254 | Thr255 | Ser256 | Thr257 | Asp258 | Leu259 | Val260 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Gln261 | Pro262 | Ala263 | Ser264 | Gly265 | Gly266 | Ser267 | Phe268 | Asn269 | Gly270 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| VTPAGSAGVTHSPMSQTLPWDLWTTSTDLVQPASGGSFNGFTQPQDTSLFTMQTDQSMIC |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | Homo sapiens | 129.00 | 11 | unknown |
| 2 | synthetic construct | 129.00 | 1 | amphiphysin |
| 3 | Macaca mulatta | 125.00 | 3 | PREDICTED: amphiphysin (Stiff-Man syndrome with br |
| 4 | Rattus norvegicus | 116.00 | 1 | amphiphysin 1 |
| 5 | Bos taurus | 112.00 | 2 | hypothetical protein LOC614722 |
| 6 | Mus musculus | 111.00 | 2 | amphiphysin |
| 7 | Monodelphis domestica | 97.80 | 1 | PREDICTED: similar to cytokine receptor common gam |
| 8 | Gallus gallus | 76.30 | 1 | amphiphysin (Stiff-Man syndrome with breast cancer |
| 9 | Xenopus tropicalis | 71.60 | 1 | Unknown (protein for MGC:122074) |
| 10 | Pan troglodytes | 63.50 | 1 | PREDICTED: amphiphysin |
| 11 | Xenopus laevis | 62.80 | 1 | hypothetical protein LOC432150 |
| 12 | Canis familiaris | 61.20 | 1 | PREDICTED: similar to Amphiphysin |
| 13 | Danio rerio | 43.10 | 1 | amphiphysin |
Top-ranked sequences
| organism | matching |
|---|---|
| Homo sapiens | Query 231 VTPAGSAGVTHSPMSQTLPWDLWTTSTDLV#QPASGGSFNGFTQPQDTSLFTMQTDQSMIC 290 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 231 VTPAGSAGVTHSPMSQTLPWDLWTTSTDLV#QPASGGSFNGFTQPQDTSLFTMQTDQSMIC 290 |
| synthetic construct | Query 231 VTPAGSAGVTHSPMSQTLPWDLWTTSTDLV#QPASGGSFNGFTQPQDTSLFTMQTDQSMIC 290 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 231 VTPAGSAGVTHSPMSQTLPWDLWTTSTDLV#QPASGGSFNGFTQPQDTSLFTMQTDQSMIC 290 |
| Macaca mulatta | Query 231 VTPAGSAGVTHSPMSQTLPWDLWTTSTDLV#QPASGGSFNGFTQPQDTSLFTMQTDQSMIC 290 |||||||||||||||||||||||||| |||#|||||||||||||||||||| ||||||||| Sbjct 363 VTPAGSAGVTHSPMSQTLPWDLWTTSADLV#QPASGGSFNGFTQPQDTSLFAMQTDQSMIC 422 |
| Rattus norvegicus | Query 231 VTPAGSAGVTHSPMSQTLPWDLWTTSTDLV#QPASGGSFNGFTQPQDTSLFTMQTDQSM 288 ||||||| |||||||||||||||||||||#||||||||| ||||||||||||||||+| Sbjct 363 VTPAGSAAATHSPMSQTLPWDLWTTSTDLV#QPASGGSFNDFTQPQDTSLFTMQTDQNM 420 |
| Bos taurus | Query 231 VTPAGSAGVTHSPMSQTLPWDLWTTSTDLV#QPASGGSFNGFTQPQDTSLFTMQTDQSM 288 ||||||||||||||||||||||||||||||#|||||||||||| ||||||||| |||| Sbjct 363 VTPAGSAGVTHSPMSQTLPWDLWTTSTDLV#QPASGGSFNGFT--QDTSLFTMQADQSM 418 |
| Mus musculus | Query 231 VTPAGSAGVTHSPMSQTLPWDLWTTSTDLV#QPASGGSFNGFTQPQDTSLFTMQTDQSM 288 | ||||| |||||||||||||||||||||#||||||||| ||| ||||||||||||+| Sbjct 363 VAPAGSAAATHSPMSQTLPWDLWTTSTDLV#QPASGGSFNDFTQAQDTSLFTMQTDQNM 420 |
| Monodelphis domestica | Query 231 VTPAGSAGVTHSPMSQTLPWDLWTTSTDLV#QPASGGSFNGFTQPQDTSLFTMQTDQS 287 ||| ||||+ |||||||||||||||++||#|||| |+||||||||||||| | |+ Sbjct 363 VTPVGSAGIAPSPMSQTLPWDLWTTSSELV#QPASSGTFNGFTQPQDTSLFDMPASQT 419 |
| Gallus gallus | Query 236 SAGVTHSPMSQTLPWDLWTTSTDLV#QPASGGSFNGFTQPQDTSLFTMQTDQSM 288 | ||||||||||||||||||+++||#|||| +|||| |||+ | +|+++++ Sbjct 365 STGVTHSPMSQTLPWDLWTTTSELV#QPASSTAFNGFA--QDTTAFAVQSNENV 415 |
| Xenopus tropicalis | Query 240 THSPMSQTLPWDLWTTSTDLV#QPASGGSFNGFTQPQDTS-LFTMQTDQS 287 +|||+|||||||||||+ ++|#|||| |+||||||| ++ |||+ +|+ Sbjct 372 SHSPLSQTLPWDLWTTNNEVV#QPASTGAFNGFTQPDASAPLFTLPLEQN 420 |
| Pan troglodytes | Query 231 VTPAGSAGVTHSPMSQTLPWDLWTTSTDLV#QP 262 ||||||||||||||||||||||||||||||#|| Query 231 VTPAGSAGVTHSPMSQTLPWDLWTTSTDLV#QP 262 Query 246 QTLPWDLWTTSTDLV#QPASGGSFNGFTQPQDTSLFTMQTDQSMIC 290 | + ||+ + #| |||||||||||||||||||||||||+|| Sbjct 656 QKIEDSLWS-GVEAC#QKASGGSFNGFTQPQDTSLFTMQTDQSVIC 699 |
| Xenopus laevis | Query 234 AGSAGV--THSPMSQTLPWDLWT-------TSTDLV#QPASGGSFNGFTQPQDTS-LFTMQ 283 | + |+ +|||+|||||||||| |+ ++|#|||| | ||||||| ++ |||+ Sbjct 368 AATVGIVQSHSPISQTLPWDLWTCFCPSTQTNNEVV#QPASTGVFNGFTQPDASAPLFTLP 427 Query 284 TDQS 287 ++++ Sbjct 428 SEKN 431 |
| Canis familiaris | Query 231 VTPAGSAGVTHSPMSQTLPWDLWTTSTDLV#QP 262 | | || |||||||||||||||||||||||#|| Sbjct 382 VAPVGSVGVTHSPMSQTLPWDLWTTSTDLV#QP 413 Query 246 QTLPWDLWTTSTDLV#QPASGGSFNGFTQPQDTSLFTMQTDQSMIC 290 | + +| + #+ ||||+||||||||||||||||||||||| Sbjct 572 QKIEDSVWA-GVETC#EKASGGTFNGFTQPQDTSLFTMQTDQSMIC 615 |
| Danio rerio | Query 238 GVTHSPMSQTLPWDLWTTSTDLV#QPASGGSFNGFT 272 | | ||+|||||||||| | #||| + ||| Sbjct 363 GQTQSPVSQTLPWDLWT--GDAA#QPAQPATDAGFT 395 |
[Site 9] SAGVTHSPMS245-QTLPWDLWTT
Ser245  Gln
Gln
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Ser236 | Ala237 | Gly238 | Val239 | Thr240 | His241 | Ser242 | Pro243 | Met244 | Ser245 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Gln246 | Thr247 | Leu248 | Pro249 | Trp250 | Asp251 | Leu252 | Trp253 | Thr254 | Thr255 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| EETLLDLDFDPFKPEVTPAGSAGVTHSPMSQTLPWDLWTTSTDLVQPASGGSFNGFTQPQ |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | Homo sapiens | 130.00 | 14 | unknown |
| 2 | synthetic construct | 130.00 | 1 | amphiphysin |
| 3 | Macaca mulatta | 128.00 | 3 | PREDICTED: amphiphysin (Stiff-Man syndrome with br |
| 4 | Bos taurus | 124.00 | 2 | hypothetical protein LOC614722 |
| 5 | Rattus norvegicus | 122.00 | 2 | amphiphysin 1 |
| 6 | Mus musculus | 117.00 | 3 | amphiphysin |
| 7 | Monodelphis domestica | 113.00 | 2 | PREDICTED: similar to cytokine receptor common gam |
| 8 | Canis familiaris | 98.20 | 2 | PREDICTED: similar to Amphiphysin |
| 9 | Gallus gallus | 94.00 | 1 | amphiphysin (Stiff-Man syndrome with breast cancer |
| 10 | Xenopus tropicalis | 92.00 | 1 | Unknown (protein for MGC:122074) |
| 11 | Xenopus laevis | 81.60 | 1 | hypothetical protein LOC432150 |
| 12 | Danio rerio | 64.70 | 1 | amphiphysin |
| 13 | Pan troglodytes | 40.80 | 4 | PREDICTED: bridging integrator 1 isoform 10 |
| 14 | Lampetra fluviatilis | 36.20 | 1 | amphiphysin |
Top-ranked sequences
| organism | matching |
|---|---|
| Homo sapiens | Query 216 EETLLDLDFDPFKPEVTPAGSAGVTHSPMS#QTLPWDLWTTSTDLVQPASGGSFNGFTQPQ 275 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 216 EETLLDLDFDPFKPEVTPAGSAGVTHSPMS#QTLPWDLWTTSTDLVQPASGGSFNGFTQPQ 275 |
| synthetic construct | Query 216 EETLLDLDFDPFKPEVTPAGSAGVTHSPMS#QTLPWDLWTTSTDLVQPASGGSFNGFTQPQ 275 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 216 EETLLDLDFDPFKPEVTPAGSAGVTHSPMS#QTLPWDLWTTSTDLVQPASGGSFNGFTQPQ 275 |
| Macaca mulatta | Query 216 EETLLDLDFDPFKPEVTPAGSAGVTHSPMS#QTLPWDLWTTSTDLVQPASGGSFNGFTQPQ 275 ||||||||||||||||||||||||||||||#||||||||||| |||||||||||||||||| Sbjct 348 EETLLDLDFDPFKPEVTPAGSAGVTHSPMS#QTLPWDLWTTSADLVQPASGGSFNGFTQPQ 407 |
| Bos taurus | Query 216 EETLLDLDFDPFKPEVTPAGSAGVTHSPMS#QTLPWDLWTTSTDLVQPASGGSFNGFTQ 273 ||||||||||||||+|||||||||||||||#|||||||||||||||||||||||||||| Sbjct 348 EETLLDLDFDPFKPDVTPAGSAGVTHSPMS#QTLPWDLWTTSTDLVQPASGGSFNGFTQ 405 |
| Rattus norvegicus | Query 216 EETLLDLDFDPFKPEVTPAGSAGVTHSPMS#QTLPWDLWTTSTDLVQPASGGSFNGFTQPQ 275 ||||||||||||||+||||||| ||||||#|||||||||||||||||||||||| ||||| Sbjct 348 EETLLDLDFDPFKPDVTPAGSAAATHSPMS#QTLPWDLWTTSTDLVQPASGGSFNDFTQPQ 407 |
| Mus musculus | Query 216 EETLLDLDFDPFKPEVTPAGSAGVTHSPMS#QTLPWDLWTTSTDLVQPASGGSFNGFTQPQ 275 ||||||||||||||+| ||||| ||||||#|||||||||||||||||||||||| ||| | Sbjct 348 EETLLDLDFDPFKPDVAPAGSAAATHSPMS#QTLPWDLWTTSTDLVQPASGGSFNDFTQAQ 407 |
| Monodelphis domestica | Query 216 EETLLDLDFDPFKPEVTPAGSAGVTHSPMS#QTLPWDLWTTSTDLVQPASGGSFNGFTQPQ 275 ||+|||||||||||+||| ||||+ ||||#|||||||||||++|||||| |+|||||||| Sbjct 348 EESLLDLDFDPFKPDVTPVGSAGIAPSPMS#QTLPWDLWTTSSELVQPASSGTFNGFTQPQ 407 |
| Canis familiaris | Query 216 EETLLDLDFDPFKPEVTPAGSAGVTHSPMS#QTLPWDLWTTSTDLVQP 262 |||||||||||||||| | || ||||||||#||||||||||||||||| Sbjct 367 EETLLDLDFDPFKPEVAPVGSVGVTHSPMS#QTLPWDLWTTSTDLVQP 413 |
| Gallus gallus | Query 217 ETLLDLDFDPFKPEVTPAGSAGVTHSPMS#QTLPWDLWTTSTDLVQPASGGSFNGFTQ 273 |+||||||||||||| | ||||||||#||||||||||+++|||||| +|||| | Sbjct 349 ESLLDLDFDPFKPEVV---STGVTHSPMS#QTLPWDLWTTTSELVQPASSTAFNGFAQ 402 |
| Xenopus tropicalis | Query 216 EETLLDLDFDPFKPEVTPAGSAGVTHSPMS#QTLPWDLWTTSTDLVQPASGGSFNGFTQP 274 ||+|||||||||||| | +|||+|#||||||||||+ ++||||| |+||||||| Sbjct 349 EESLLDLDFDPFKPESATVGIVQ-SHSPLS#QTLPWDLWTTNNEVVQPASTGAFNGFTQP 406 |
| Xenopus laevis | Query 216 EETLLDLDFDPFKPEVTPAGSAGVTHSPMS#QTLPWDLWT-------TSTDLVQPASGGSF 268 ||+||||||||||| | +|||+|#||||||||| |+ ++||||| | | Sbjct 353 EESLLDLDFDPFKPGAATVGIVQ-SHSPIS#QTLPWDLWTCFCPSTQTNNEVVQPASTGVF 411 Query 269 NGFTQP 274 |||||| Query 269 NGFTQP 274 |
| Danio rerio | Query 217 ETLLDLDFDPFKPEV-TPAGSAGVTHSPMS#QTLPWDLWTTSTDLVQPASGGSFNGFT 272 |+|||||||||||+ || | | ||+|#||||||||| | ||| + ||| Sbjct 344 ESLLDLDFDPFKPDTSTPIGQ---TQSPVS#QTLPWDLWT--GDAAQPAQPATDAGFT 395 |
| Pan troglodytes | Query 216 EETLLDLDFDPFKPEVTPAGSAGVTHSPMS#QTLPWDLW 253 + +|||||||| | +| + +| #|++||||| Sbjct 357 QASLLDLDFDPLPPVTSPVKAP----TPSG#QSIPWDLW 390 |
| Lampetra fluviatilis | Query 216 EETLLDLDFDPFKPE--VTPAGSAGVTHSPMS#QTLPWDL-WTTSTDLVQP 262 | +|||+|||| ||+ || | ++#||| +|| | | ||| Sbjct 343 EGSLLDIDFDPLKPDTATTPMSPA------IN#QTLLFDLAWEASPQQVQP 386 |
* References
[PubMed ID: 12853948] Hillier LW, Fulton RS, Fulton LA, Graves TA, Pepin KH, Wagner-McPherson C, Layman D, Maas J, Jaeger S, Walker R, Wylie K, Sekhon M, Becker MC, O'Laughlin MD, Schaller ME, Fewell GA, Delehaunty KD, Miner TL, Nash WE, Cordes M, Du H, Sun H, Edwards J, Bradshaw-Cordum H, Ali J, Andrews S, Isak A, Vanbrunt A, Nguyen C, Du F, Lamar B, Courtney L, Kalicki J, Ozersky P, Bielicki L, Scott K, Holmes A, Harkins R, Harris A, Strong CM, Hou S, Tomlinson C, Dauphin-Kohlberg S, Kozlowicz-Reilly A, Leonard S, Rohlfing T, Rock SM, Tin-Wollam AM, Abbott A, Minx P, Maupin R, Strowmatt C, Latreille P, Miller N, Johnson D, Murray J, Woessner JP, Wendl MC, Yang SP, Schultz BR, Wallis JW, Spieth J, Bieri TA, Nelson JO, Berkowicz N, Wohldmann PE, Cook LL, Hickenbotham MT, Eldred J, Williams D, Bedell JA, Mardis ER, Clifton SW, Chissoe SL, Marra MA, Raymond C, Haugen E, Gillett W, Zhou Y, James R, Phelps K, Iadanoto S, Bubb K, Simms E, Levy R, Clendenning J, Kaul R, Kent WJ, Furey TS, Baertsch RA, Brent MR, Keibler E, Flicek P, Bork P, Suyama M, Bailey JA, Portnoy ME, Torrents D, Chinwalla AT, Gish WR, Eddy SR, McPherson JD, Olson MV, Eichler EE, Green ED, Waterston RH, Wilson RK, The DNA sequence of human chromosome 7. Nature. 2003 Jul 10;424(6945):157-64.