XSB2125 : PREDICTED: amphiphysin [Pan troglodytes]
[ CaMP Format ]
This entry is computationally expanded from SB0093
* Basic Information
| Organism | Pan troglodytes (chimpanzee) |
| Protein Names | amphiphysin |
| Gene Names | AMPH; Derived by automated computational analysis using gene prediction method: GNOMON. Supporting evidence includes similarity to: 1 mRNA, 222 ESTs, 3 Proteins |
| Gene Locus | Not available |
| GO Function | Not available |
* Information From OMIM
Not Available.
* Structure Information
1. Primary Information
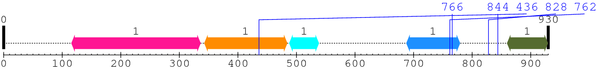
Length: 930 aa
Average Mass: 101.414 kDa
Monoisotopic Mass: 101.351 kDa
2. Domain Information
Annotated Domains: Not Available.
Computationally Assigned Domains (Pfam+HMMER):
| domain name | begin | end | score | e-value |
|---|---|---|---|---|
| BAR 1. | 116 | 336 | 288.1 | 1.9e-83 |
| Drf_FH1 1. | 343 | 484 | -45.7 | 9.2 |
| --- cleavage 436 (inside Drf_FH1 343..484) --- | ||||
| Attractin 1. | 488 | 538 | -13.6 | 5.1 |
| Cpn10 1. | 688 | 779 | -45.8 | 9.6 |
| --- cleavage 766 (inside Cpn10 688..779) --- | ||||
| --- cleavage 762 (inside Cpn10 688..779) --- | ||||
| --- cleavage 844 --- | ||||
| --- cleavage 828 --- | ||||
| SH3_1 1. | 860 | 929 | 24.1 | 0.00017 |
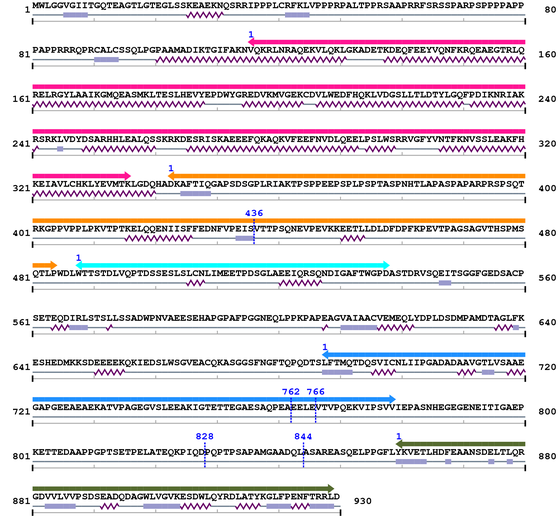
3. Sequence Information
Fasta Sequence: XSB2125.fasta
Amino Acid Sequence and Secondary Structures (PsiPred):
4. 3D Information
Not Available.
* Cleavage Information
5 [sites]
Cleavage sites (±10aa)
[Site 1] SAQPEAEELE766-VTVPQEKVIP
Glu766  Val
Val
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Ser757 | Ala758 | Gln759 | Pro760 | Glu761 | Ala762 | Glu763 | Glu764 | Leu765 | Glu766 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Val767 | Thr768 | Val769 | Pro770 | Gln771 | Glu772 | Lys773 | Val774 | Ile775 | Pro776 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| GEGVSLEEAKIGTETTEGAESAQPEAEELEVTVPQEKVIPSVVIEPASNHEGEGENEITI |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | Pan troglodytes | 119.00 | 1 | PREDICTED: amphiphysin |
| 2 | Homo sapiens | 114.00 | 9 | Amphiphysin (Stiff-Man syndrome with breast cancer |
| 3 | synthetic construct | 114.00 | 1 | amphiphysin |
| 4 | Macaca mulatta | 100.00 | 3 | PREDICTED: amphiphysin isoform 3 |
| 5 | Rattus norvegicus | 75.50 | 1 | amphiphysin 1 |
| 6 | Bos taurus | 70.10 | 1 | hypothetical protein LOC614722 |
| 7 | Canis familiaris | 68.20 | 1 | PREDICTED: similar to Amphiphysin |
| 8 | Monodelphis domestica | 63.90 | 1 | PREDICTED: similar to amphiphysin I variant CT2 |
| 9 | Mus musculus | 62.80 | 3 | amphiphysin |
| 10 | Gallus gallus | 55.80 | 1 | amphiphysin (Stiff-Man syndrome with breast cancer |
| 11 | Xenopus tropicalis | 46.60 | 1 | Unknown (protein for MGC:122074) |
| 12 | Xenopus laevis | 43.10 | 1 | hypothetical protein LOC432150 |
| 13 | Tetraodon nigroviridis | 38.50 | 2 | unnamed protein product |
| 14 | Oryza sativa (japonica cultivar-group) | 35.00 | 2 | Os06g0538200 |
| 15 | Oryza sativa (indica cultivar-group) | 35.00 | 1 | hypothetical protein OsI_022473 |
| 16 | Plasmodium falciparum | 34.30 | 1 | AF247634_1 glutamate-rich protein |
| 17 | Plasmodium falciparum 3D7 | 34.30 | 1 | glutamate-rich protein |
| 18 | Chaetomium globosum CBS 148.51 | 33.50 | 1 | hypothetical protein CHGG_02690 |
| 19 | Ustilago maydis 521 | 33.50 | 1 | hypothetical protein UM00553.1 |
| 20 | Danio rerio | 32.70 | 1 | amphiphysin |
Top-ranked sequences
| organism | matching |
|---|---|
| Pan troglodytes | Query 737 GEGVSLEEAKIGTETTEGAESAQPEAEELE#VTVPQEKVIPSVVIEPASNHEGEGENEITI 796 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 737 GEGVSLEEAKIGTETTEGAESAQPEAEELE#VTVPQEKVIPSVVIEPASNHEGEGENEITI 796 |
| Homo sapiens | Query 737 GEGVSLEEAKIGTETTEGAESAQPEAEELE#VTVPQEKVIPSVVIEPASNHEGEGENEITI 796 ||||||||||||||||||||||||||||||# |||||||||||||||||||| |||||||| Sbjct 502 GEGVSLEEAKIGTETTEGAESAQPEAEELE#ATVPQEKVIPSVVIEPASNHEEEGENEITI 561 |
| synthetic construct | Query 737 GEGVSLEEAKIGTETTEGAESAQPEAEELE#VTVPQEKVIPSVVIEPASNHEGEGENEITI 796 ||||||||||||||||||||||||||||||# |||||||||||||||||||| |||||||| Sbjct 502 GEGVSLEEAKIGTETTEGAESAQPEAEELE#ATVPQEKVIPSVVIEPASNHEEEGENEITI 561 |
| Macaca mulatta | Query 737 GEGVSLEEAKIGTETTEGAESAQPEAEELE#VTVPQEKVIPSVVIEPASNHEGEGENEITI 796 ||||||||||||||| ||||||| ||||| # |||||||||||||||||||| |||+|||| Sbjct 637 GEGVSLEEAKIGTET-EGAESAQAEAEELA#ATVPQEKVIPSVVIEPASNHEEEGEHEITI 695 |
| Rattus norvegicus | Query 737 GEGVSLEEAKIGTETTEGAESAQPEAEELE#VTVPQEKVIPSVVIEPASNHEGEGENEIT 795 ||| | | ||| |+|| | | |+| |||# |||||||||||||||||||| | | Sbjct 492 GEGESPEGAKIDVESTELASSESPQAAELE#AGAPQEKVIPSVVIEPASNHEGEEHQETT 550 |
| Bos taurus | Query 737 GEGVSLEEAKIGTETTEGAESAQPEAEELE#VTVPQEKVIPSVVIEPASNHEGEGENEITI 796 | | |||||| | || + + + || |# ||||||||||+||||+|||||+| |+ Sbjct 499 GAGASLEEAKTDIEATEAIDGDRSQLEETE#AVAAQEKVIPSVVIQPASNNEGEGEHEGTV 558 |
| Canis familiaris | Query 737 GEGVSLEEAKIGTETTEGAESAQPEAEELE#VTVPQEKVIPSVVIEPASNHEGEGENE 793 ||| +|||+ || |+ + + + || |# | ||||||||||||||+||||++| Sbjct 653 GEGAGVEEARPDTEATQAVDGDRSQLEEAE#AVAPLEKVIPSVVIEPASNNEGEGDHE 709 |
| Monodelphis domestica | Query 738 EGVSLEEAKIGTETTEGAESAQPEAEELE#VTVPQEKVIPSVVIEPASNHEGEGENE 793 ||||+ | + || | || + ||+|# ||||||||||||||||||| | | Sbjct 46 EGVSVVETQAVPETREATESDRAGTEEME#AVGIQEKVIPSVVIEPASNHEGEDEQE 101 |
| Mus musculus | Query 737 GEGVSLEEAKIGTETTEGAESAQPEAEELE#VTVPQEKVIPSVVIEPASNHEGEGENEIT 795 ||| | | ||| |+|| | | |+ | |# || ||||||||||||||||||++ | Sbjct 496 GEGGSPEGAKIDGESTELAISESPQPVEPE#AGAPQ--VIPSVVIEPASNHEGEGEHQET 552 |
| Gallus gallus | Query 744 EAKIGTETTEGAESAQPEAEELE#VTVPQEKV--IPSVVIEPASNHEGEGENEITI 796 | + || || +|+ || +#| | |||| |||||||||||+||||| | Sbjct 500 EGAVRTEQEAAAEGDKPQGEEKD#VDVSQEKVSSIPSVVIEPASNNEGEGEEHHVI 554 |
| Xenopus tropicalis | Query 740 VSLEEAKIGTETTEGA---------ESAQPEAEE-LE#VTVPQE-KVIPSVVIEPASNHEG 788 || | ++ |+ | | | +|| | |# ||| ||||||||||||| Sbjct 481 VSEIEVEVKEESAESAAEMADDIMEEQKEPEINEPKE#TEAPQEITCIPSVVIEPASNHED 540 Query 789 EGENEITI 796 | +|+||| Sbjct 541 EHDNDITI 548 |
| Xenopus laevis | Query 744 EAKIGTET-TEGAESAQPEAEELE#VTVPQEK-------VIPSVVIEPASNHEGEGENEIT 795 |+++ |+ | || | +| |#+ | | +||||||||||||| | +++|| Sbjct 490 ESEVKQESPVEMAEDIMEEEKEPE#IIEPNETKAPPEITIIPSVVIEPASNHEDEHDDDIT 549 |
| Tetraodon nigroviridis | Query 739 GVSLEEAKIGTETTEGAESAQPEAEELE#VTVPQEKVIPSVVIE---PASNHEGEGENEIT 795 | | ++ | || ||||| +| || |# | |+ || +| |+| | + |+| Sbjct 133 GFSFKKTK--KETGEGAESEEPAAEATE#AAAPAEEAKPSEAMEETSAAANDEAKPAEEVT 190 |
| Oryza sativa (japonica cultivar-group) | Query 743 EEAKIGTETTEGAESAQPEAEELE#VTVPQEKVIPSVVIEPASNHEGE 789 ||||| | || | | ||+|#|| | | | + |+ | | Sbjct 900 EEAKITTAATEEVEVTTPATEEVE#VTTPSTK---EVEVTIAATEEAE 943 |
| Oryza sativa (indica cultivar-group) | Query 743 EEAKIGTETTEGAESAQPEAEELE#VTVPQEKVIPSVVIEPASNHEGE 789 ||||| | || | | ||+|#|| | | | + |+ | | Sbjct 728 EEAKITTAATEEVEVTTPATEEVE#VTTPSTK---EVEVTIAATEEAE 771 |
| Plasmodium falciparum | Query 738 EGVSLEEAKIGTETTEGAESAQPEAEELE#VTVPQEKVIPSVVIEPASNHEGEGEN 792 | | +|| | | |+ +|| ++ |# +| +|| || + | | | || Sbjct 1056 EIVEIEEVPSQTNNNENIETIKPEEKKNE#FSVVEEKAIPQEPVVPTLN---ENEN 1107 |
| Plasmodium falciparum 3D7 | Query 738 EGVSLEEAKIGTETTEGAESAQPEAEELE#VTVPQEKVIPSVVIEPASNHEGEGEN 792 | | +|| | | |+ +|| ++ |# +| +|| || + | | | || Sbjct 1053 EIVEIEEVPSQTNNNENIETIKPEEKKNE#FSVVEEKAIPQEPVVPTLN---ENEN 1104 |
| Chaetomium globosum CBS 148.51 | Query 737 GEGVSLEEAKIGTETTEGAESAQPEAEELE#VTVPQEKVI--PSVVIEPASNHEGEGENEI 794 || | |+ | ++ || ||# | | |+ + +| || || + | ++ Sbjct 1364 GENEHGTETKMLCSDAGGGQAQTPEGSGLE#GTAPSEQAVSAATVTAEPPLNHVAKTEGQV 1423 Query 795 T 795 | Query 795 T 795 |
| Ustilago maydis 521 | Query 741 SLEEAKIGTETTEGAESAQPEAEELE#VTVPQEKVIPSVVIEPASNHEGEGENE 793 +|||| | | ||||+ | ++|# ||+| + || | | Sbjct 751 TLEEAVAAAENNEAAESARAEEAKME#EKEEQEEVAAAAAAAAEKTGNGETEEE 803 |
| Danio rerio | Query 737 GEGVSLEEAKIGTETT-EGAESAQPEAEELE#VTVPQEKVIPSVVIEPASNHEGEGENE 793 |+ | + | ||+ ||| + | |++ # ||||||||||++||+ + + Sbjct 633 GDAPDNVEPQPGDETSKEGAANNGNEEEKMP#--------IPSVVIEPASSNEGDDDRD 682 |
[Site 2] APAMGAADQL844-ASAREASQEL
Leu844  Ala
Ala
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Ala835 | Pro836 | Ala837 | Met838 | Gly839 | Ala840 | Ala841 | Asp842 | Gln843 | Leu844 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Ala845 | Ser846 | Ala847 | Arg848 | Glu849 | Ala850 | Ser851 | Gln852 | Glu853 | Leu854 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| ETPELATEQKPIQDPQPTPSAPAMGAADQLASAREASQELPPGFLYKVETLHDFEAANSD |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | Homo sapiens | 123.00 | 27 | amphiphysin isoform 1 |
| 2 | Pan troglodytes | 123.00 | 13 | PREDICTED: amphiphysin |
| 3 | synthetic construct | 123.00 | 2 | amphiphysin |
| 4 | Macaca mulatta | 114.00 | 4 | PREDICTED: amphiphysin isoform 3 |
| 5 | Rattus norvegicus | 74.30 | 3 | amphiphysin 1 |
| 6 | Bos taurus | 70.90 | 2 | hypothetical protein LOC614722 |
| 7 | Mus musculus | 70.50 | 12 | amphiphysin |
| 8 | Canis familiaris | 68.20 | 3 | PREDICTED: similar to Amphiphysin |
| 9 | Gallus gallus | 64.30 | 2 | amphiphysin (Stiff-Man syndrome with breast cancer |
| 10 | Xenopus laevis | 42.40 | 3 | hypothetical protein LOC432150 |
| 11 | Xenopus tropicalis | 42.00 | 3 | Unknown (protein for MGC:122074) |
| 12 | Danio rerio | 40.80 | 3 | amphiphysin |
| 13 | Tetraodon nigroviridis | 40.80 | 2 | unnamed protein product |
| 14 | N/A | 35.80 | 4 | - |
| 15 | Monodelphis domestica | 34.30 | 2 | PREDICTED: similar to amphiphysin II |
| 16 | Lampetra fluviatilis | 33.90 | 1 | amphiphysin |
| 17 | Leishmania braziliensis | 33.90 | 1 | hypothetical protein, conserved |
| 18 | Sus scrofa | 33.10 | 1 | bridging integrator 1 |
| 19 | Tribolium castaneum | 33.10 | 1 | PREDICTED: similar to CG1943-PA, isoform A |
| 20 | Macaca fascicularis | 33.10 | 1 | unnamed protein product |
| 21 | Aspergillus fumigatus Af293 | 32.70 | 1 | topisomerase II associated protein (Pat1), putativ |
| 22 | Ostreococcus lucimarinus CCE9901 | 32.30 | 1 | predicted protein |
| 23 | Roseovarius sp. HTCC2601 | 32.30 | 1 | Possible TolA protein |
Top-ranked sequences
| organism | matching |
|---|---|
| Homo sapiens | Query 815 ETPELATEQKPIQDPQPTPSAPAMGAADQL#ASAREASQELPPGFLYKVETLHDFEAANSD 874 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 815 ETPELATEQKPIQDPQPTPSAPAMGAADQL#ASAREASQELPPGFLYKVETLHDFEAANSD 874 |
| Pan troglodytes | Query 815 ETPELATEQKPIQDPQPTPSAPAMGAADQL#ASAREASQELPPGFLYKVETLHDFEAANSD 874 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 815 ETPELATEQKPIQDPQPTPSAPAMGAADQL#ASAREASQELPPGFLYKVETLHDFEAANSD 874 |
| synthetic construct | Query 815 ETPELATEQKPIQDPQPTPSAPAMGAADQL#ASAREASQELPPGFLYKVETLHDFEAANSD 874 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 815 ETPELATEQKPIQDPQPTPSAPAMGAADQL#ASAREASQELPPGFLYKVETLHDFEAANSD 874 |
| Macaca mulatta | Query 815 ETPELATEQKPIQDPQPTPSAPAMGAADQL#ASAREASQELPPGFLYKVETLHDFEAANSD 874 |||||||| ||+||||| |||||+| ||||#|| ||||||||||||||||||||||||||| Sbjct 714 ETPELATEPKPLQDPQPMPSAPALGTADQL#ASVREASQELPPGFLYKVETLHDFEAANSD 773 |
| Rattus norvegicus | Query 815 ETPELATEQKPIQDPQPTPSAPAMGAADQL#ASAREASQELPPGFLYKVETLHDFEAANSD 874 | ||||| |+ || ||| || | # || +|+||||||||||||||||||||||| Sbjct 570 EKQELATEPTPLDSQAATP-APA-GAVDAS#LSAGDAAQELPPGFLYKVETLHDFEAANSD 627 |
| Bos taurus | Query 815 ETPELATE-QKPIQDPQPTPSAPAMGAADQL#ASAREASQELPPGFLYKVETLHDFEAANS 873 | ||||+| || + || |||| # || || ||+||||||||||||||||||| Sbjct 577 EMPELASEEQKAARASQPAPSAP--------#-SASEAFQEVPPGFLYKVETLHDFEAANS 627 Query 874 D 874 | Query 874 D 874 |
| Mus musculus | Query 815 ETPELATEQKPIQDPQPTPSAPAMGAADQL#ASAREASQELPPGFLYKVETLHDFEAANSD 874 | |+||| |+ | | | | | || | # || +|+||||||||||||||||||||||| Sbjct 573 EKQEVATEPTPL-DSQATLPASA-GAVDAS#LSAGDATQELPPGFLYKVETLHDFEAANSD 630 |
| Canis familiaris | Query 815 ETPELATEQKPIQDPQPTPSAPAMGAADQL#ASAREASQELPPGFLYKVETLHDFEAANSD 874 | |||| | | +| || ||| | # | | | |+|||||||||||||||||||| Sbjct 730 ELPELAPEPKAGEDSQPLPSALA-------#--ASEESLEVPPGFLYKVETLHDFEAANSD 780 |
| Gallus gallus | Query 815 ETPELATEQKPIQDPQPTPSAPAMGAADQL#ASAREASQELPPGFLYKVETLHDFEAANSD 874 || ++ +||| ++ | ||| || #||| + + ++|||||+||| |||||||||| Sbjct 573 ETSQIGSEQKATEEIQTTPSQ------DQP#ASAGDTASDMPPGFLFKVEVLHDFEAANSD 626 |
| Xenopus laevis | Query 845 ASAREASQELPPGFLYKVETLHDFEAANSD 874 | + +|| | ||||| | |||||||| | Sbjct 581 ADSISSSQSKPEGFLYKAEVLHDFEAANED 610 |
| Xenopus tropicalis | Query 845 ASAREASQELPPGFLYKVETLHDFEAANSD 874 | + || | ||||| | |||||||| | Sbjct 579 ADSLTTSQSKPEGFLYKAEALHDFEAANED 608 |
| Danio rerio | Query 851 SQELPPGFLYKVETLHDFEAANSD 874 | +|| ||+||||+||||||| | Sbjct 706 SSGMPPRFLFKVETMHDFEAANPD 729 |
| Tetraodon nigroviridis | Query 853 ELPPGFLYKVETLHDFEAANSD 874 +|| ||+||||+||||||||| Sbjct 761 QLPEDFLFKVETMHDFEAANSD 782 |
| N/A | Query 832 TPSAPAMGAADQL#ASAREASQELPPGFLYKVETLHDFEAANSD 874 | || || + # || +|||||++||+ ||+ | ++| Sbjct 338 TFSATVNGAVE--#GSAGTGRLDLPPGFMFKVQAQHDYTATDTD 378 |
| Monodelphis domestica | Query 853 ELPPGFLYKVETLHDFEAANSD 874 +|||||++||+ ||+ | +|| Sbjct 521 DLPPGFMFKVQAQHDYTATDSD 542 |
| Lampetra fluviatilis | Query 829 PQPTPSAPAM-------GAADQL#ASAR----EASQELPPGFLYKVETLHDFEAANSD 874 | | | | + | |+ #|| | ++||||||||+ ||+ ++| Sbjct 506 PTPVDSGPELDMAFVTNGDAEVE#ASKHVDTSSESTDMPPGFLYKVQAEHDYTPLDAD 562 |
| Leishmania braziliensis | Query 815 ETPELATEQKPIQDPQPTPSAPAMGAADQL#ASAREAS 851 ||||++| |+ ++ || +| | || # || |+ Sbjct 1548 ETPEVSTMQRSVRQPQQSPIIPGAAAARSF#TSATAAA 1584 |
| Sus scrofa | Query 853 ELPPGFLYKVETLHDFEAANSD 874 +|||||++||+ ||+ | ++| Sbjct 357 DLPPGFMFKVQAQHDYTATDTD 378 |
| Tribolium castaneum | Query 817 PELATEQKPIQDPQPTPSAPAMGAADQL#ASAREASQELPP 856 | ||+||+| + || | | + # + |+|| || Sbjct 66 PVQKTEEKPVQRAEETPKEPVKQATEAP#KPSAPANQEAPP 105 |
| Macaca fascicularis | Query 853 ELPPGFLYKVETLHDFEAANSD 874 +|||||++||+ ||+ | ++| Sbjct 362 DLPPGFMFKVQAQHDYTATDTD 383 |
| Aspergillus fumigatus Af293 | Query 819 LATEQKPIQDPQPTPSAPAMGAADQL#ASAREASQELPPGFL 859 +|+ | |+ | | | + ||# + || +||| || Sbjct 204 IASVQPPVSMPMPMPDLSHLQRTQQL#PNGPEAFSQLPPEFL 244 |
| Ostreococcus lucimarinus CCE9901 | Query 818 ELATEQKPIQDPQPTPSAPAMGAADQL#ASAREASQ-ELPPGFLYKVET 864 || |+ +| + ||+ +#++| ||| ++||||| +|+| Sbjct 186 ELDDPASKFMTPRERRKSPIVAAAEAI#SNAVEASHSDVPPGFLPRVQT 233 |
| Roseovarius sp. HTCC2601 | Query 817 PELATEQKPIQDPQPTPSAPAMGAADQL#ASAREASQELP 855 |+ | | +|+ +|+| | || | ++#++ |++ +| Sbjct 97 PQPAPEPEPVPEPEPDPLPPAAEAVPEV#SAPESATRPVP 135 |
[Site 3] FEDNFVPEIS436-VTTPSQNEVP
Ser436  Val
Val
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Phe427 | Glu428 | Asp429 | Asn430 | Phe431 | Val432 | Pro433 | Glu434 | Ile435 | Ser436 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Val437 | Thr438 | Thr439 | Pro440 | Ser441 | Gln442 | Asn443 | Glu444 | Val445 | Pro446 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| PPLPKVTPTKELQQENIISFFEDNFVPEISVTTPSQNEVPEVKKEETLLDLDFDPFKPEV |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | Homo sapiens | 125.00 | 18 | amphiphysin isoform 1 |
| 2 | Pan troglodytes | 125.00 | 6 | PREDICTED: amphiphysin |
| 3 | Canis familiaris | 125.00 | 2 | PREDICTED: similar to Amphiphysin |
| 4 | synthetic construct | 125.00 | 1 | amphiphysin |
| 5 | Macaca mulatta | 123.00 | 4 | PREDICTED: amphiphysin isoform 3 |
| 6 | Bos taurus | 123.00 | 2 | hypothetical protein LOC614722 |
| 7 | Rattus norvegicus | 117.00 | 3 | amphiphysin 1 |
| 8 | Monodelphis domestica | 117.00 | 2 | PREDICTED: similar to cytokine receptor common gam |
| 9 | Mus musculus | 116.00 | 4 | amphiphysin |
| 10 | Gallus gallus | 112.00 | 1 | amphiphysin (Stiff-Man syndrome with breast cancer |
| 11 | Xenopus tropicalis | 110.00 | 3 | Unknown (protein for MGC:122074) |
| 12 | Xenopus laevis | 108.00 | 1 | hypothetical protein LOC432150 |
| 13 | Danio rerio | 74.70 | 1 | amphiphysin |
| 14 | Lampetra fluviatilis | 64.70 | 1 | amphiphysin |
| 15 | N/A | 59.30 | 1 | A |
| 16 | Tetraodon nigroviridis | 55.80 | 1 | unnamed protein product |
Top-ranked sequences
| organism | matching |
|---|---|
| Homo sapiens | Query 407 PPLPKVTPTKELQQENIISFFEDNFVPEIS#VTTPSQNEVPEVKKEETLLDLDFDPFKPEV 466 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 407 PPLPKVTPTKELQQENIISFFEDNFVPEIS#VTTPSQNEVPEVKKEETLLDLDFDPFKPEV 466 |
| Pan troglodytes | Query 407 PPLPKVTPTKELQQENIISFFEDNFVPEIS#VTTPSQNEVPEVKKEETLLDLDFDPFKPEV 466 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 407 PPLPKVTPTKELQQENIISFFEDNFVPEIS#VTTPSQNEVPEVKKEETLLDLDFDPFKPEV 466 |
| Canis familiaris | Query 407 PPLPKVTPTKELQQENIISFFEDNFVPEIS#VTTPSQNEVPEVKKEETLLDLDFDPFKPEV 466 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 407 PPLPKVTPTKELQQENIISFFEDNFVPEIS#VTTPSQNEVPEVKKEETLLDLDFDPFKPEV 466 |
| synthetic construct | Query 407 PPLPKVTPTKELQQENIISFFEDNFVPEIS#VTTPSQNEVPEVKKEETLLDLDFDPFKPEV 466 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 407 PPLPKVTPTKELQQENIISFFEDNFVPEIS#VTTPSQNEVPEVKKEETLLDLDFDPFKPEV 466 |
| Macaca mulatta | Query 407 PPLPKVTPTKELQQENIISFFEDNFVPEIS#VTTPSQNEVPEVKKEETLLDLDFDPFKPEV 466 ||||||||||||+|||||||||||||||||#|||||||||||||||||||||||||||||| Sbjct 307 PPLPKVTPTKELRQENIISFFEDNFVPEIS#VTTPSQNEVPEVKKEETLLDLDFDPFKPEV 366 |
| Bos taurus | Query 407 PPLPKVTPTKELQQENIISFFEDNFVPEIS#VTTPSQNEVPEVKKEETLLDLDFDPFKPEV 466 ||||||||||||||||||||||||||||||#||||||||+|||||||||||||||||||+| Sbjct 304 PPLPKVTPTKELQQENIISFFEDNFVPEIS#VTTPSQNEIPEVKKEETLLDLDFDPFKPDV 363 |
| Rattus norvegicus | Query 407 PPLPKVTPTKELQQENIISFFEDNFVPEIS#VTTPSQNEVPEVKKEETLLDLDFDPFKPEV 466 ||||||||||||||||||+||||||||||+#||||||||| ||||||||||||||||||+| Sbjct 304 PPLPKVTPTKELQQENIINFFEDNFVPEIN#VTTPSQNEVLEVKKEETLLDLDFDPFKPDV 363 |
| Monodelphis domestica | Query 407 PPLPKVTPTKELQQENIISFFEDNFVPEIS#VTTPSQNEVPEVKKEETLLDLDFDPFKPEV 466 ||||||||||||||||||| ||||||||||#||||||||||| ||||+|||||||||||+| Sbjct 304 PPLPKVTPTKELQQENIISLFEDNFVPEIS#VTTPSQNEVPEQKKEESLLDLDFDPFKPDV 363 |
| Mus musculus | Query 407 PPLPKVTPTKELQQENIISFFEDNFVPEIS#VTTPSQNEVPEVKKEETLLDLDFDPFKPEV 466 ||||||||||||+|||||+||||||||||+#||||||||| ||||||||||||||||||+| Sbjct 304 PPLPKVTPTKELKQENIINFFEDNFVPEIN#VTTPSQNEVLEVKKEETLLDLDFDPFKPDV 363 |
| Gallus gallus | Query 407 PPLPKVTPTKELQQENIISFFEDNFVPEIS#VTTPSQNEVPEVKKEETLLDLDFDPFKPEV 466 |||||+||||||||||||+ |+|||||||+#||||||||+|| || |+||||||||||||| Sbjct 304 PPLPKLTPTKELQQENIINLFDDNFVPEIN#VTTPSQNEIPETKKVESLLDLDFDPFKPEV 363 |
| Xenopus tropicalis | Query 407 PPLPKVTPTKELQQENIISFFEDNFVPEIS#VTTPSQNEVPEVKKEETLLDLDFDPFKPE 465 |||||+||+||||||||||||||||||||+#|||||||| | ||||+|||||||||||| Sbjct 305 PPLPKLTPSKELQQENIISFFEDNFVPEIN#VTTPSQNEAVEEKKEESLLDLDFDPFKPE 363 |
| Xenopus laevis | Query 407 PPLPKVTPTKELQQENIISFFEDNFVPEIS#VTTPSQNEVPEVKKEETLLDLDFDPFKP 464 |||||+||+||||||||||||||||||||+#|||||||| | ||||+||||||||||| Sbjct 309 PPLPKLTPSKELQQENIISFFEDNFVPEIN#VTTPSQNEAVEEKKEESLLDLDFDPFKP 366 |
| Danio rerio | Query 407 PPLPKVTPTKELQQENIISFFEDNFVPEIS#VTTPSQNEVPEVKKEETLLDLDFDPFKPE 465 || ||||||+||||| || |+ | ||||#||+| || | |+|||||||||||+ Sbjct 304 PPPPKVTPTRELQQEQIIDLFDGGF-PEIS#VTSPQPNERP----GESLLDLDFDPFKPD 357 |
| Lampetra fluviatilis | Query 407 PPLPKVTPTKELQQENIISFFEDNFVPEIS#VTTPSQNEVPEVKKEETLLDLDFDPFKPE 465 || ||+||+||+||| | | |+|||||+ |# + |++ | | +|||+|||| ||+ Sbjct 303 PPPPKLTPSKEVQQEAIFSLFDDNFVPDFS#-SAPAE---AAKKDEGSLLDIDFDPLKPD 357 |
| N/A | Query 407 PPLPKVTPTKELQQENIISFFEDNFVPEIS#VTTPSQ 442 || || ||+||++|| |+| ||| ||||||#|||||| Sbjct 42 PPPPKHTPSKEVKQEQILSLFEDTFVPEIS#VTTPSQ 77 |
| Tetraodon nigroviridis | Query 407 PPLPKVTPTKELQQENIISFFEDNFVPEIS#VTTPSQNEVPEVKKEETLLDLDFDPFKPEV 466 || |||||+|+++ |||++ |+ |+||#||+|++ + | | +|||+| | | | Sbjct 212 PPPPKVTPSKDMKAENIVNLFDAAAAPDIS#VTSPTEFDRPAV---SSLLDVDLDSFTSTV 268 |
[Site 4] LATEQKPIQD828-PQPTPSAPAM
Asp828  Pro
Pro
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Leu819 | Ala820 | Thr821 | Glu822 | Gln823 | Lys824 | Pro825 | Ile826 | Gln827 | Asp828 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Pro829 | Gln830 | Pro831 | Thr832 | Pro833 | Ser834 | Ala835 | Pro836 | Ala837 | Met838 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| EPKETTEDAAPPGPTSETPELATEQKPIQDPQPTPSAPAMGAADQLASAREASQELPPGF |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | Homo sapiens | 125.00 | 12 | Amphiphysin (Stiff-Man syndrome with breast cancer |
| 2 | Pan troglodytes | 125.00 | 8 | PREDICTED: amphiphysin |
| 3 | synthetic construct | 122.00 | 1 | amphiphysin |
| 4 | Macaca mulatta | 108.00 | 13 | PREDICTED: amphiphysin isoform 3 |
| 5 | Rattus norvegicus | 61.60 | 8 | amphiphysin 1 |
| 6 | Bos taurus | 60.10 | 6 | hypothetical protein LOC614722 |
| 7 | Mus musculus | 52.80 | 10 | amphiphysin |
| 8 | Canis familiaris | 49.70 | 1 | PREDICTED: similar to Amphiphysin |
| 9 | Gallus gallus | 42.40 | 2 | amphiphysin (Stiff-Man syndrome with breast cancer |
| 10 | Thermobifida fusca YX | 41.60 | 1 | similar to DNA-directed RNA polymerase specialized |
| 11 | Ostreococcus tauri | 38.90 | 1 | unnamed protein product |
| 12 | Monodelphis domestica | 37.70 | 5 | PREDICTED: similar to calcium-binding tyrosine-pho |
| 13 | Caenorhabditis elegans | 37.00 | 2 | EGg Laying defective family member (egl-23) |
| 14 | Burkholderia cepacia AMMD | 37.00 | 1 | TonB family protein |
| 15 | Saccharopolyspora erythraea NRRL 2338 | 36.60 | 1 | hypothetical protein SACE_5459 |
| 16 | Mycobacterium vanbaalenii PYR-1 | 36.20 | 1 | hypothetical protein Mvan_1509 |
| 17 | Coprinopsis cinerea okayama7#130 | 35.80 | 6 | predicted protein |
| 18 | Aspergillus terreus NIH2624 | 35.80 | 3 | hypothetical protein ATEG_01252 |
| 19 | Gibberella zeae PH-1 | 35.80 | 2 | hypothetical protein FG08926.1 |
| 20 | Burkholderia dolosa AUO158 | 35.80 | 2 | Periplasmic protein TonB |
| 21 | Burkholderia ambifaria MC40-6 | 35.80 | 2 | TonB family protein |
| 22 | Apis mellifera | 35.80 | 2 | PREDICTED: similar to tenectin CG13648-PA, partial |
| 23 | Synechococcus sp. JA-2-3B'a(2-13) | 35.80 | 2 | TonB family protein |
| 24 | Roseobacter sp. SK209-2-6 | 35.80 | 1 | transcriptional regulator, MerR family protein |
| 25 | Roseovarius sp. HTCC2601 | 35.80 | 1 | Possible TolA protein |
| 26 | Theileria parva strain Muguga | 35.80 | 1 | hypothetical protein TP01_0380 |
| 27 | Rhodopseudomonas palustris BisB18 | 35.40 | 2 | TonB-like |
| 28 | Magnaporthe grisea 70-15 | 35.40 | 1 | hypothetical protein MGG_08761 |
| 29 | Sagittula stellata E-37 | 35.40 | 1 | hypothetical protein SSE37_03755 |
| 30 | Alcanivorax borkumensis SK2 | 35.40 | 1 | DNA polymerase III, gamma and tau subunits |
| 31 | N/A | 35.00 | 4 | - |
| 32 | Agrobacterium tumefaciens str. C58 | 35.00 | 2 | hypothetical protein AGR_L_3024 |
| 33 | Streptomyces ambofaciens ATCC 23877 | 35.00 | 1 | conserved hypothetical protein |
| 34 | Burkholderia cenocepacia PC184 | 35.00 | 1 | COG0810: Periplasmic protein TonB, links inner and |
| 35 | Burkholderia cenocepacia AU 1054 | 35.00 | 1 | TonB-like |
| 36 | Burkholderia cenocepacia MC0-3 | 35.00 | 1 | TonB family protein |
| 37 | Salinibacter ruber DSM 13855 | 35.00 | 1 | hypothetical protein SRU_0268 |
| 38 | Oryza sativa (japonica cultivar-group) | 34.70 | 4 | hypothetical protein OsJ_033966 |
| 39 | Oryza sativa (indica cultivar-group) | 34.70 | 2 | hypothetical protein OsI_036340 |
| 40 | Crimean-Congo hemorrhagic fever virus | 34.70 | 2 | glycoprotein precursor |
| 41 | Yarrowia lipolytica | 34.70 | 2 | hypothetical protein |
| 42 | Cryptococcus neoformans var. neoformans JEC21 | 34.70 | 1 | general transcriptional repressor |
| 43 | Drosophila pseudoobscura | 34.70 | 1 | GA20650-PA |
| 44 | Anopheles gambiae str. PEST | 34.70 | 1 | ENSANGP00000010140 |
| 45 | Trichomonas vaginalis G3 | 34.70 | 1 | hypothetical protein TVAG_129770 |
| 46 | Cryptococcus neoformans var. neoformans B-3501A | 34.70 | 1 | hypothetical protein CNBK1120 |
| 47 | Mesorhizobium sp. BNC1 | 34.70 | 1 | Hemolysin-type calcium-binding region |
| 48 | Aedes aegypti | 34.70 | 1 | hypothetical protein AaeL_AAEL014367 |
| 49 | Chaetomium globosum CBS 148.51 | 34.30 | 4 | predicted protein |
| 50 | Tribolium castaneum | 34.30 | 3 | PREDICTED: similar to CG1943-PA, isoform A |
| 51 | Nocardia farcinica IFM 10152 | 34.30 | 3 | putative serine/threonine protein kinase |
| 52 | Leishmania braziliensis | 34.30 | 2 | hypothetical protein, conserved |
| 53 | Sulfitobacter sp. EE-36 | 34.30 | 2 | tonB domain protein, putative |
| 54 | Collinsella aerofaciens ATCC 25986 | 34.30 | 2 | Hypothetical protein COLAER_00357 |
| 55 | Sulfitobacter sp. NAS-14.1 | 34.30 | 2 | tonB domain protein, putative |
| 56 | Gluconobacter oxydans 621H | 34.30 | 1 | hypothetical protein GOX0962 |
| 57 | Rhodobacter sphaeroides 2.4.1 | 34.30 | 1 | conserved hypoothetical protein |
| 58 | Nostoc sp. PCC 7120 | 34.30 | 1 | hypothetical protein alr2745 |
| 59 | Lotus japonicus | 34.30 | 1 | putative arabinagalactan protein |
| 60 | Gloeobacter violaceus PCC 7421 | 34.30 | 1 | hypothetical protein glr3167 |
| 61 | Burkholderia vietnamiensis G4 | 34.30 | 1 | TonB family protein |
| 62 | Corynebacterium jeikeium K411 | 34.30 | 1 | signal recognition particle receptor |
| 63 | Anabaena variabilis ATCC 29413 | 34.30 | 1 | Peptidoglycan-binding domain 1 |
| 64 | Neurospora crassa OR74A | 34.30 | 1 | hypothetical protein |
| 65 | Yersinia bercovieri ATCC 43970 | 34.30 | 1 | COG3515: Uncharacterized protein conserved in bact |
| 66 | Caenorhabditis briggsae | 33.90 | 4 | Hypothetical protein CBG05727 |
| 67 | Arabidopsis thaliana | 33.90 | 3 | remorin family protein |
| 68 | Drosophila melanogaster | 33.90 | 3 | nahoda CG12781-PA, isoform A |
| 69 | Cenarchaeum symbiosum | 33.90 | 1 | hypothetical protein CENSYa_0842 |
| 70 | Rhodobacter sphaeroides ATCC 17025 | 33.90 | 1 | hypothetical protein Rsph17025_0561 |
| 71 | Leifsonia xyli subsp. xyli str. CTCB07 | 33.90 | 1 | hemagglutinin/hemolysin-related protein |
| 72 | Burkholderia sp. 383 | 33.90 | 1 | TonB-like protein |
| 73 | Fulvimarina pelagi HTCC2506 | 33.90 | 1 | hypothetical protein FP2506_03039 |
| 74 | Hosta virus X | 33.90 | 1 | replicase |
| 75 | Ralstonia solanacearum GMI1000 | 33.90 | 1 | PROBABLE OUTER MEMBRANE CHANNEL PROTEIN |
| 76 | Pichia pastoris | 33.90 | 1 | dihydrolipoamide acetyltransferase |
| 77 | Stenotrophomonas maltophilia R551-3 | 33.90 | 1 | TonB family protein |
| 78 | Erythrobacter sp. NAP1 | 33.90 | 1 | dihydrolipoamide acetyltransferase |
| 79 | Rhodobacter sphaeroides ATCC 17029 | 33.90 | 1 | hypothetical protein Rsph17029_0071 |
| 80 | Corynebacterium efficiens YS-314 | 33.90 | 1 | DNA polymerase III subunits gamma and tau |
| 81 | Leishmania infantum JPCM5 | 33.90 | 1 | hypothetical protein, conserved |
| 82 | Bacillus clausii KSM-K16 | 33.90 | 1 | peptidoglycan-binding domain-containing protein |
| 83 | Aspergillus fumigatus Af293 | 33.90 | 1 | transcriptional activator spt7 |
| 84 | Cyanothece sp. CCY0110 | 33.90 | 1 | hypothetical protein CY0110_24616 |
| 85 | Pichia guilliermondii ATCC 6260 | 33.90 | 1 | predicted protein |
| 86 | Aspergillus clavatus NRRL 1 | 33.50 | 3 | hypothetical protein ACLA_052560 |
| 87 | Danio rerio | 33.50 | 2 | PREDICTED: similar to LOC553397 protein |
| 88 | Myxococcus xanthus DK 1622 | 33.50 | 2 | serine/threonine protein kinase |
| 89 | Strongylocentrotus purpuratus | 33.50 | 2 | PREDICTED: similar to 5-amp-activated protein kina |
| 90 | Bacillus thuringiensis serovar konkukian str. 97-27 | 33.50 | 1 | hypothetical protein BT9727_3951 |
| 91 | Pyrobaculum arsenaticum DSM 13514 | 33.50 | 1 | extracellular solute-binding protein, family 1 |
| 92 | Schizosaccharomyces pombe 972h- | 33.50 | 1 | hypothetical protein SPAC23A1.17 |
| 93 | Murid herpesvirus 2 | 33.50 | 1 | pR48 |
| 94 | Corynebacterium glutamicum R | 33.50 | 1 | hypothetical protein cgR_1950 |
| 95 | Colwellia psychrerythraea 34H | 33.50 | 1 | glycosyl hydrolase, family 5 |
| 96 | Frankia alni ACN14a | 33.50 | 1 | Putative response regulator (Response regulator pr |
| 97 | Rhodobacterales bacterium HTCC2654 | 33.50 | 1 | tonB domain protein, putative |
| 98 | Tetraodon nigroviridis | 33.50 | 1 | unnamed protein product |
| 99 | Chromohalobacter salexigens DSM 3043 | 33.50 | 1 | cell divisionFtsK/SpoIIIE |
| 100 | Coccidioides immitis RS | 33.50 | 1 | predicted protein |
| 101 | Synechococcus sp. BL107 | 33.50 | 1 | Cell division transporter substrate-binding protei |
| 102 | Methylibium petroleiphilum PM1 | 33.50 | 1 | hypothetical protein Mpe_A2234 |
| 103 | Alnus glutinosa | 33.50 | 1 | ag13 |
| 104 | Lyngbya sp. PCC 8106 | 33.50 | 1 | putative hemagglutinin/hemolysin-related protein |
| 105 | Aspergillus niger | 33.50 | 1 | hypothetical protein An08g03930 |
| 106 | Ustilago maydis 521 | 33.50 | 1 | hypothetical protein UM05340.1 |
| 107 | Phaeosphaeria nodorum SN15 | 33.10 | 2 | predicted protein |
| 108 | Saccharomyces cerevisiae | 33.10 | 2 | Proline-rich actin-associated protein involved in |
| 109 | Streptococcus pneumoniae | 33.10 | 2 | AF071806_1 PspA |
| 110 | Neosartorya fischeri NRRL 181 | 33.10 | 2 | response regulator, putative |
| 111 | Vitis vinifera | 33.10 | 2 | hypothetical protein |
| 112 | Leishmania major | 33.10 | 1 | hypothetical protein, conserved |
| 113 | Mycobacterium sp. MCS | 33.10 | 1 | signal recognition particle-docking protein FtsY |
| 114 | Roseiflexus castenholzii DSM 13941 | 33.10 | 1 | serine/threonine protein kinase with FHA domain |
| 115 | Roseobacter denitrificans OCh 114 | 33.10 | 1 | signal recognition particle-docking protein FtsY |
| 116 | Desulfovibrio vulgaris subsp. vulgaris str. Hildenborough | 33.10 | 1 | chemotaxis protein CheA |
| 117 | Streptococcus pyogenes MGAS10750 | 33.10 | 1 | Fibronectin-binding protein |
| 118 | Mycobacterium sp. JLS | 33.10 | 1 | signal recognition particle-docking protein FtsY |
| 119 | Alcelaphine herpesvirus 1 | 33.10 | 1 | large tegument protein |
| 120 | Zea mays | 33.10 | 1 | AF159297_1 extensin-like protein |
| 121 | Streptococcus sobrinus | 33.10 | 1 | dextran-binding lectin |
| 122 | Bordetella bronchiseptica | 33.10 | 1 | pertactin |
| 123 | Sphingomonas sp. SKA58 | 33.10 | 1 | hypothetical protein SKA58_08876 |
| 124 | Dinoroseobacter shibae DFL 12 | 33.10 | 1 | hypothetical protein DshiDRAFT_2630 |
| 125 | Natronomonas pharaonis DSM 2160 | 33.10 | 1 | probable cell surface glycoprotein |
| 126 | Enterococcus faecalis | 33.10 | 1 | AF454824_10 EF0010 |
| 127 | Xanthobacter autotrophicus Py2 | 33.10 | 1 | hypothetical protein XautDRAFT_1507 |
| 128 | Enterococcus faecalis V583 | 33.10 | 1 | cell wall surface anchor family protein |
| 129 | Pyrobaculum islandicum DSM 4184 | 33.10 | 1 | LPXTG-motif cell wall anchor domain |
| 130 | Macaca fascicularis | 33.10 | 1 | unnamed protein product |
| 131 | Thiomicrospira crunogena XCL-2 | 32.70 | 1 | hypothetical protein Tcr_1483 |
| 132 | Chlamydomonas incerta | 32.70 | 1 | vegetative cell wall protein |
| 133 | Streptococcus sanguinis SK36 | 32.70 | 1 | CshA-like fibrillar surface protein C |
| 134 | Pichia stipitis CBS 6054 | 32.70 | 1 | predicted protein |
| 135 | Geobacter lovleyi SZ | 32.70 | 1 | von Willebrand factor, type A |
| 136 | Burkholderia cenocepacia HI2424 | 32.70 | 1 | TonB family protein |
| 137 | Polaromonas naphthalenivorans CJ2 | 32.70 | 1 | TonB family protein |
| 138 | Desulfovibrio vulgaris subsp. vulgaris DP4 | 32.70 | 1 | CheA signal transduction histidine kinases |
| 139 | Rhodopirellula baltica SH 1 | 32.70 | 1 | hypothetical protein-signal peptide and transmembr |
| 140 | Anaeromyxobacter sp. Fw109-5 | 32.70 | 1 | MJ0042 family finger-like protein |
| 141 | Pseudomonas aeruginosa UCBPP-PA14 | 32.70 | 1 | translocation protein in type III secretion |
Top-ranked sequences
| organism | matching |
|---|---|
| Homo sapiens | Query 799 EPKETTEDAAPPGPTSETPELATEQKPIQD#PQPTPSAPAMGAADQLASAREASQELPPGF 858 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 799 EPKETTEDAAPPGPTSETPELATEQKPIQD#PQPTPSAPAMGAADQLASAREASQELPPGF 858 |
| Pan troglodytes | Query 799 EPKETTEDAAPPGPTSETPELATEQKPIQD#PQPTPSAPAMGAADQLASAREASQELPPGF 858 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 799 EPKETTEDAAPPGPTSETPELATEQKPIQD#PQPTPSAPAMGAADQLASAREASQELPPGF 858 |
| synthetic construct | Query 799 EPKETTEDAAPPGPTSETPELATEQKPIQD#PQPTPSAPAMGAADQLASAREASQELPPGF 858 || |||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Sbjct 564 EPMETTEDAAPPGPTSETPELATEQKPIQD#PQPTPSAPAMGAADQLASAREASQELPPGF 623 |
| Macaca mulatta | Query 799 EPKETTEDAAPPGPTSETPELATEQKPIQD#PQPTPSAPAMGAADQLASAREASQELPPGF 858 ||+||||| || |||||||||||| ||+||#||| |||||+| |||||| ||||||||||| Sbjct 698 EPEETTEDTAPLGPTSETPELATEPKPLQD#PQPMPSAPALGTADQLASVREASQELPPGF 757 |
| Rattus norvegicus | Query 799 EPKETTEDAAPPGPTSETPELATEQKPIQD#PQPTPSAPAMGAADQLASAREASQELPPGF 858 | +| ||| || || | ||||| |+ # || ||| || | || +|+||||||| Sbjct 554 ETREATEDVAPQGPAGEKQELATEPTPLDS#QAATP-APA-GAVDASLSAGDAAQELPPGF 611 |
| Bos taurus | Query 799 EPKETTEDAAPPGPTSETPELAT-EQKPIQD#PQPTPSAPAMGAADQLASAREASQELPPG 857 | || |+| | |||||| ||||+ ||| + # || |||| || || ||+||| Sbjct 561 EYKEATDDTALPGPTSEMPELASEEQKAARA#SQPAPSAP---------SASEAFQEVPPG 611 Query 858 F 858 | Query 858 F 858 |
| Mus musculus | Query 799 EPKETTEDAAPPGPTSETPELATEQKPIQD#PQPTPSAPAMGAADQLASAREASQELPPGF 858 ||+| || | | | |+||| |+ |# | | | | || | || +|+||||||| Sbjct 557 EPREAAEDVAAQGSAGEKQEVATEPTPL-D#SQATLPASA-GAVDASLSAGDATQELPPGF 614 |
| Canis familiaris | Query 804 TEDAAPPGPTSETPELATEQKPIQD#PQPTPSAPAMGAADQLASAREASQELPPGF 858 | || ||||+| |||| | | +|# || ||| | | | | |+|||| Sbjct 719 TPAAAAPGPTTELPELAPEPKAGED#SQPLPSALA---------ASEESLEVPPGF 764 |
| Gallus gallus | Query 799 EPKETTEDAAPPGPTSETPELATEQKPIQD#PQPTPSAPAMGAADQLASAREASQELPPGF 858 | |+ + | ||| ++ +||| ++# | ||| || ||| + + ++|||| Sbjct 557 ESKDAAAEMGTQGTDSETSQIGSEQKATEE#IQTTPS------QDQPASAGDTASDMPPGF 610 |
| Thermobifida fusca YX | Query 804 TEDAA---PPGPTSETPELATEQKPIQD#PQPTPSAPAMGAADQLASAREASQELPP 856 +|||| || | + | | ++| #||| |+||| | | |+ | ++ || Sbjct 399 SEDAATVDPPAPAPQQPAPAPREEPASQ#PQPAPTAPA-APAPAPAPAQPAQEDPPP 453 |
| Ostreococcus tauri | Query 799 EPKETTEDAAPPGPTSE---TPELATEQKPIQD#PQPTPSAPAMGAADQLASAREAS 851 || | | | |||| ||| +| +#| | | + || +| +|| | Sbjct 341 EPTPTPEPTPTPEPTSEPTPTPEPTPTPEPTPE#PAPAPESTLGGAFSRLGLSRETS 396 |
| Monodelphis domestica | Query 799 EPKETTEDAAPPGPTSETPELATEQKPIQD#PQP----------TPSAPAMGAADQLASAR 848 ||+||+| + | | + ||+++| | #| | |+ || || || | Sbjct 132 EPEETSESVSEPSPKATTPKVST---PPSS#PSPLAAASPEFAYVPADPAQLAAQMLAIAA 188 Query 849 EASQELPP 856 + + || Sbjct 189 SEAGQPPP 196 |
| Caenorhabditis elegans | Query 799 EPKETTEDAAPPGPTSETPELAT-EQKPIQD#PQPTPSAPA----MGAADQLASAREASQE 853 || | || | || | | +|+++#| | | || + ||+ | |+| | Sbjct 539 EPSPVREPTPPPPPREPTPREPTPEPEPVRE#PTPPPPPPAKPRPLTAAEIAAQKRKAYSE 598 |
| Burkholderia cepacia AMMD | Query 799 EPKETTEDAAP-PGPTSETPELATEQKPIQD#PQPTPSAPAMGA-ADQLASAREASQELPP 856 ||| + | | | | +++| + | |#| | |+||| | | + ||| | | Sbjct 93 EPKPVQKAAKPAPQPVAQSPAPSPTPAPAAD#PTPAPAAPAAAAPAATPSPARETMQVSAP 152 |
| Saccharopolyspora erythraea NRRL 2338 | Query 799 EPKETTEDAAPPGPTS----ETPELATEQKPIQD#PQPTPSAPAMGAADQLASAREASQ 852 + + | || ||| + || | | +| +#|+| |+| + ||+ ++ + | Sbjct 812 QSEHAAEPAANPGPAARAQESAPEPAPEPEPAPE#PEPAPAASPLRLADRTSTDEDRKQ 869 |
| Mycobacterium vanbaalenii PYR-1 | Query 800 PKETTEDAAPPGPTSETPELATEQKPIQD#PQPTPS-AP---AMGAADQLASA 847 | ||| +| || | | | #| |||| || ++||| +|| Sbjct 249 PSETTSPTSPSETTSPAPSSETTSAPTSS#PSPTPSTAPTTTSVGAAPPSSSA 300 |
| Coprinopsis cinerea okayama7#130 | Query 800 PKETTEDAAPPGPTSETPELATEQKPIQD#PQPTPS---APAMGAADQLASAREASQE 853 | |+ | || || + |+ ++ |+ +#| |+ | | |+| | +| Sbjct 111 PASATDPANPPSPTQQGPDRSSSLSPVPE#PDQPPAEQQPQANGGAEQQPQREEEEEE 167 |
| Aspergillus terreus NIH2624 | Query 804 TEDAAPPGPTSETPELATEQKPIQD#PQPTPSAP--AMGAADQLASAREASQE 853 | || | | | +|+|+ #|||||+ | |+ + | ++ |+ Sbjct 220 TPQPNPPQPPSSQPPQQQQQQPVSQ#PQPTPNPPPQALPQSQQQVPQQQTPQQ 271 |
| Gibberella zeae PH-1 | Query 801 KETTEDAAPPGPTSETPELATE-------QKPIQD#PQPTPSAPAMGAADQLASAREASQE 853 ||| + | || + | +| + | |#||| ||| | | |++ Sbjct 326 KETQSYSPPSGPPPKAGEGSTSRPYDESAENPFAD#PQPGGSAPTTAAYQYSEEQRLATEP 385 Query 854 LPPGF 858 ||| Sbjct 386 FHPGF 390 |
| Burkholderia dolosa AUO158 | Query 799 EPKETTEDAAP-PGPTSETPELATEQKPIQD#PQPTPSAPAMGA-ADQLASAREASQELPP 856 ||| + | | | + +|| + | |#| | |+||| | | ||| | | Sbjct 93 EPKPVPKAVKPTPQPLAPSPEPSPTPAPAAD#PTPAPAAPAAAAPAAAPGPARETMQVSAP 152 |
| Burkholderia ambifaria MC40-6 | Query 799 EPKETTEDAAP-PGPTSETPELATEQKPIQD#PQPTPSAPAMGA-ADQLASAREASQELPP 856 ||| + | | | | +++| + | |#| | |+||+ | | + ||| | | Sbjct 93 EPKPVQKAAKPAPQPVAQSPAPSPTPAPAAD#PTPAPAAPSAAAPAATPSPARETMQVSAP 152 |
| Apis mellifera | Query 800 PKETTEDAAPPGPTSETPE--LATEQKPIQD#PQPTPSAPAMGAADQLASAREASQELPP 856 | | | | ||||| |+||| # | ||+ | +++|++ || +++ | Sbjct 2597 PTEVPEKMVPEEVPSETPEQALSTEQPVQLT#EAPEKMAPSTEAPEKMATSTEAPEKMAP 2655 |
| Synechococcus sp. JA-2-3B'a(2-13) | Query 799 EPKETTEDAAPPGPTSETPELATEQKPIQD---#PQPTP---SAPAMGAADQLASAREASQ 852 || + ++ || ||+| | | | | | #|+| | ||+ |+ | + + + Sbjct 154 EPDSESIESLPPEPTAEPPGPAPESPPADDLSP#PEPVPQEIEMPAIPEAESLLALQTPTP 213 Query 853 ELPP 856 || Sbjct 214 TPPP 217 |
| Roseobacter sp. SK209-2-6 | Query 799 EPKETTEDAAPPGPTSETPELATEQKPIQD#PQPTPSAPAMGAADQLASAREASQELPP 856 || | | | | | ||+ || || #||| | | + |+| ||+| | Sbjct 369 EPDEVTLDTATPAAPSESETLAPASKPA--#PQPEVSEP----IETAAAAAPQSQDLAP 420 |
| Roseovarius sp. HTCC2601 | Query 799 EPKETTEDAAP-PGPTSE---TPELATEQKPIQD#PQPTPSAPAMGAADQLASAREASQEL 854 ||+ | || | | | |+ | | +|+ +#|+| | || | ++++ |++ + Sbjct 75 EPQPVPEAPAPEPAPQPEPDPAPQPAPEPEPVPE#PEPDPLPPAAEAVPEVSAPESATRPV 134 Query 855 P 855 | Query 855 P 855 Query 800 PKETTEDAAPPGPTSETPELATEQKPIQD#PQPTPSAPAMGAADQ 843 | | | | | || | | | +| +#||| | ||| | | Sbjct 48 PVEAPEPAPQPEPTPE-PAPQPEPQPTPE#PQPVPEAPAPEPAPQ 90 |
| Theileria parva strain Muguga | Query 801 KETTEDAAPPGPTSETPELATEQKPIQD#PQPTP 833 | | || | ||+ || || | #||| | Sbjct 262 KPKPETKGPPVPKPRTPKSKTEPKPAQQ#PQPQP 294 |
| Rhodopseudomonas palustris BisB18 | Query 805 EDAAPPGPTSETPELATEQKPIQD#PQPTPSAPA 837 | | || | + ||+| |+| +#| | ||| Sbjct 103 EQAVPPTPLQDKPEVAAPQEPKAE#PAPQEPAPA 135 |
| Magnaporthe grisea 70-15 | Query 800 PKETTEDAAP-PGPTSETPELATEQKPIQD#PQPTPSAPA-MGAADQLASAR 848 || | || | | |+ | || + #| | | ||| | | +| |+ Sbjct 122 PKPAPEQPAPKPAPEQPAPKPAPEQPAPKQ#PVPKPPAPAETGKGDDMACAK 172 |
| Sagittula stellata E-37 | Query 799 EPKETTEDAAPPGPTSET---PELATEQKPIQD#PQPTPS---APAMGAADQLASAREASQ 852 +| | || | | +|| | || | ++#|+| || || || || |+ Sbjct 109 KPAEDTEPEPDPQPVAETAPVAEAATSDAP-EE#PEPAPSAQPAPESAEADAEDSADESGS 167 Query 853 ELPP 856 + | Sbjct 168 RVKP 171 |
| Alcanivorax borkumensis SK2 | Query 807 AAPPGPTSETPELATEQKPIQD#PQPTPSAPAMGAADQLASAREA 850 || | | | | |+ |+ #|+|+|+ | ||+| ||| +| Sbjct 393 AAAPAPVS-LPTPASASAPVTA#PEPSPTRAAPAAAEQAASADDA 435 |
| N/A | Query 800 PKETTEDAAPPGPTSETPELATEQKPIQD#PQPTPSAPAMGA--ADQLASAREASQELPP 856 |+ + ++ | | + | | |+| #| ||| |+ | | | | |+ ||| Sbjct 33 PERPAQTSSAPTPPAIPPVAPPPQPPVQV#PTPTPGQPSAPPPFAVQPAPQRPAAAPLPP 91 |
| Agrobacterium tumefaciens str. C58 | Query 800 PKETTEDAAPPGPTSETPELATEQKPIQD#PQPTPSAPAMGA--ADQLASAREASQELPP 856 |+ + + | | + | | |+| #| ||| |+ | | | | |+ ||| Sbjct 87 PERPAQTSPAPAPPAIPPVAPPPQPPVQV#PTPTPGQPSAPPPFAVQPAPQRPAAAPLPP 145 |
| Streptomyces ambofaciens ATCC 23877 | Query 800 PKETTEDAAPPGPTSETPELATEQKPIQD#PQPTPSAPAMGAADQLASAREASQEL 854 | | +| | || | || || | |# | + +| | ||| | Sbjct 65 PSEPSEPTEPTGPDSATPGETTENSPPPD#TDATDGRTSAPPPEQEAGIDEASTNL 119 |
| Burkholderia cenocepacia PC184 | Query 799 EPKETTEDAAP-PGPTSETPELATEQKPIQD#PQPTPSAPAMGA-ADQLASAREASQELPP 856 ||| + | | | | + +| + | |#| | |+||| | | ||| | | Sbjct 72 EPKPVQKAAKPTPQPVAPSPAPSPTPAPAAD#PTPAPAAPAPAAPAATPGPARETMQVSAP 131 |
| Burkholderia cenocepacia AU 1054 | Query 799 EPKETTEDAAP-PGPTSETPELATEQKPIQD#PQPTPSAPAMGA-ADQLASAREASQELPP 856 ||| + | | | | + +| + | |#| | |+||| | | ||| | | Sbjct 93 EPKPVQKAAKPTPQPVAPSPAPSPTPAPAAD#PTPAPAAPAPAAPAATPGPARETMQVSAP 152 |
| Burkholderia cenocepacia MC0-3 | Query 800 PKETTEDAAPPGPTSETPELATEQKPIQD#PQPTPSAPAMGAA 841 |+ | | | | | + || +| |+| #| | |+|| | Sbjct 137 PQATRE--AAPAPVTATPAVAAPAAPVQA#PAPAPAAPVRETA 176 |
| Salinibacter ruber DSM 13855 | Query 799 EPKETTEDAAPPGPTSETPELATEQKPIQD#PQPTPS-APAMG 839 || ++ +|||| ||| |++ | +# || || ||| | Sbjct 172 EPSPSSPSPSPPGPGDETPGSASD-SPTAN#GQPDPSTAPAAG 212 |
| Oryza sativa (japonica cultivar-group) | Query 799 EPKETTEDAAPPGPTSETPELATEQKPIQD#PQPTPSAPAMGAADQLASAREASQE 853 ||++ + || |||| | || |+|+#| || | ++ | + | Sbjct 352 EPEQEPQPETPP-VESETP--AEEQAPVQE#PPAADETPAAGEPEEQPKPRSSDSE 403 |
| Oryza sativa (indica cultivar-group) | Query 799 EPKETTEDAAPPGPTSETPELATEQKPIQD#PQPTPSAPAMGAADQLASAREASQE 853 ||++ + || |||| | || |+|+#| || | ++ | + | Sbjct 326 EPEQEPQPETPP-VESETP--AEEQAPVQE#PPAADETPAAGEPEEQPKPRSSDSE 377 |
| Crimean-Congo hemorrhagic fever virus | Query 801 KETTEDAAPPGPTSETPELAT-------EQKPIQD#PQPTPSA-PAMGAADQLASAREASQ 852 | +|| ++|| ||||| + | # |||||| ||| + || Sbjct 173 KTSTETSSPPPATSETPTPSPTTQVPTGNNNPNTS#RQPTPSAQPAMSNPATSPAQPNLSQ 232 Query 853 ELPP 856 || Sbjct 233 SAPP 236 |
| Yarrowia lipolytica | Query 802 ETTEDAAPPGPTSETPEL---ATEQKPIQD#PQPTPSAPAMGAADQLASAR-EASQELP 855 +|| | | || | ||+ | + | #|+|||+ || ++ + | + |+| Sbjct 143 QTTTPAVEPKPTPEVPEVKPEPTPEVPEVK#PEPTPAPPAPKPEPEVPEVKPEPTPEVP 200 |
| Cryptococcus neoformans var. neoformans JEC21 | Query 800 PKETTEDAAPPGPTSETPELATEQKPIQD#PQPTPSAPAMGAADQLASAREASQELPP 856 |+ |+ | | +| |+ |# |+|+||| || ++ |+|++ || Sbjct 1014 PQSPPRQASSPPPEAEKEASRPPTPPLPD#EPPSPAAPA--AAVRVESSRDSDPRTPP 1068 |
| Drosophila pseudoobscura | Query 799 EPKETTEDAA--PPGPTSETPELATEQKPIQD#PQPTPSAP 836 +|+ || | || |+| | | | | +#| | |+ | Sbjct 213 QPESTTAGAGDEPPATTTEEPALGTTLTPTPE#PAPVPTVP 252 |
| Anopheles gambiae str. PEST | Query 800 PKETTEDAAPPGPTSETPELATEQKPIQD#PQPTPSAPAMGAADQLASAREASQELPP 856 || || | ||+ ||+ | + #|+ ||+ | + | +|| | || Sbjct 489 PKPTT---TTPKPTTTTPKPTTTTTTTEK#PKTTPTPKATTTTPKATEAPKASTEEPP 542 |
| Trichomonas vaginalis G3 | Query 799 EPKE--TTEDAAPPGPTSETPELATEQKPIQD#PQPTPSAPAMGAADQLASAREASQELPP 856 |||+ +| | || | + |+ +|||+ #|||| | | ++ +|+ | Sbjct 313 EPKQQQSTLFAPPPKPVNNIPKPQPQQKPV--#PQPTVQQPQKAQESNLFGQKQPTQQKIP 370 |
| Cryptococcus neoformans var. neoformans B-3501A | Query 800 PKETTEDAAPPGPTSETPELATEQKPIQD#PQPTPSAPAMGAADQLASAREASQELPP 856 |+ |+ | | +| |+ |# |+|+||| || ++ |+|++ || Sbjct 1014 PQSPPRQASSPPPEAEKEASRPPTPPLPD#EPPSPAAPA--AAVRVESSRDSDPRTPP 1068 |
| Mesorhizobium sp. BNC1 | Query 801 KETTEDAAPP-GPTSETPELATEQKPIQD#PQPTPSAPAMGAADQLASAREASQELPP 856 || || | | ||| + |+| | +| | #| |+ | | + + +|| Sbjct 59 KEGTESAKEPVGPTEQQPDLVAESEPEQA#PVSEDEDEALPDKLPLGSGEQLADFIPP 115 |
| Aedes aegypti | Query 799 EPKETTEDAAPPGPTSET---PELATEQKPIQD#PQPTPSAPAMGAADQLASAREASQELP 855 || | |+ | | || || |+| +| +#|+|| |+ ++ + || Sbjct 594 EPTSEPEPASEPEPASEPASEPEPASEPEPASE#PEPTGEPEPSSASQSESTKEHTTTALP 653 Query 856 PG 857 | Sbjct 654 SG 655 Query 799 EPKETTEDAAPPGPTSETPELATEQKPIQD#PQPTP---SAPAMGAADQLASAREASQELP 855 || | |+ | |||| || |+| +| +#|+ | |+ |||+| +|+ Sbjct 674 EPTSEPEPASEPEPTSE-PEPASEPEPASE#PEATSTEHSSSDHSGKLQLAAAASTTQKTI 732 Query 856 P 856 | Query 856 P 856 |
| Chaetomium globosum CBS 148.51 | Query 799 EPKETTEDAAPPGPTSETP----ELATEQKPIQD#PQPTPSAPAMGAADQLASAREASQEL 854 || + |+ | | |||| | | |+| |+# + | || || | |+ Sbjct 100 EPTQEPEETAAPEPTSEPAPEPTEKVTTQEPAQE#TEQAPPAP---TEDQSPPGDEQQQQP 156 Query 855 PP 856 | Sbjct 157 AP 158 |
| Tribolium castaneum | Query 813 TSETPELATEQKPIQD#PQPTPSAPAMGAADQLASAREASQELPP 856 | | ||+||+| # + || | | + + |+|| || Sbjct 62 TEAKPVQKTEEKPVQR#AEETPKEPVKQATEAPKPSAPANQEAPP 105 |
| Nocardia farcinica IFM 10152 | Query 803 TTEDAAPPGPTSETPELATEQKPIQD#PQPTPSAP 836 || || | + || | | | #|+|| +|| Sbjct 418 TTRFVPPPPPQTSEPEPTTTQPPTTT#PEPTTAAP 451 |
| Leishmania braziliensis | Query 799 EPKETTEDAAPPGPTSETPELATEQKPIQD#PQPTPSAPAMGAADQLASAREAS 851 +| |+|+ ||||++| |+ ++ #|| +| | || || |+ Sbjct 1535 QPATTSEEDEED---EETPEVSTMQRSVRQ#PQQSPIIPGAAAARSFTSATAAA 1584 |
| Sulfitobacter sp. EE-36 | Query 799 EPKETTEDAAPPGPTSET---PELATEQKPIQD#PQPTPSAPAMGAADQ 843 +|+ | | | || | || | | +|+ +#|+| |+ +|| Sbjct 62 QPEPAPEPAPEPEPTPEPAPEPEPAPEPEPVPE#PEPIAPPPSEDVSDQ 109 |
| Collinsella aerofaciens ATCC 25986 | Query 799 EPKETTEDAAP--PGPTSETPELATEQKPIQD#PQPTPSAP 836 ||+ | | | | + ||| | | || #|+| |+ | Sbjct 747 EPEAAAETAVEKAPEPATATPEPAAEAKPTPA#PKPQPTTP 786 |
| Sulfitobacter sp. NAS-14.1 | Query 799 EPKETTEDAAPPGPTSET---PELATEQKPIQD#PQPTPSAPAMGAADQ 843 || | | | || | || | | +|+ +#|+| |+ +|| Sbjct 64 EPAPEPEPAPEPEPTPEPAPEPEPAPEPEPVPE#PEPIAPPPSEDVSDQ 111 |
| Gluconobacter oxydans 621H | Query 800 PKETTEDAAPPGPTSETPELATEQKPIQD#PQ-----PTPSAPAMGAADQLASAREASQEL 854 |+ | ||| |++++|| | + | #|| |||+ | +| | || Sbjct 34 PRARDEGGAPPPPSAQSPEETQENQ--QQ#PQKEQSTPTPAVPTPETPSAPDVPQEPSAEL 91 Query 855 P 855 | Query 855 P 855 |
| Rhodobacter sphaeroides 2.4.1 | Query 803 TTEDAAPPGPTSETPELATEQKPIQD#PQPTP-SAPAMGAADQLASAREA 850 ||| |||| |+ || || | +#| | | +||| | | | | | Sbjct 66 TTEPAAPPPATAPAPE-ATTPAP-AE#PAPAPEAAPATPAPDATAPAAPA 112 |
| Nostoc sp. PCC 7120 | Query 799 EPKETTEDAAPPGPTSETPELATEQKPIQD#PQ--PTPSA 835 ||+ || ||+ || | |+| #|+ ||||| Sbjct 181 EPRLANPRPTPPRPTTPTPRQQTPQRPSST#PRFGPTPSA 219 |
| Lotus japonicus | Query 800 PKETTEDAAPPGPTSETPELATEQKPIQD#PQPTPSAPAMGAADQLASAREA 850 | | || | ||+ || || #| |||+| || ||+ | Sbjct 47 PPATPPPAATPAPTT-TPPAATPAPSASP#PAPTPTASPTGAPTPSASSPPA 96 |
| Gloeobacter violaceus PCC 7421 | Query 800 PKETTEDAAPPGPTSETPELATEQKPIQD#PQ--PTPSAPAMGAADQLAS-------AREA 850 | | || | | | | || # | | ||| || + |+ | | Sbjct 168 PPPTVARAAAPDTPPPTASAAAESKPAPP#TPAPPPPPAPATAAAPETATNAAAPLEASSA 227 Query 851 SQELPPG 857 ||||| Sbjct 228 PTELPPG 234 |
| Burkholderia vietnamiensis G4 | Query 799 EPKETTEDAAP-PGPTSETPELATEQKPIQD#PQPTPSAPAMGA----ADQLASAREASQE 853 ||| + | | | | + +| + | |#| | |+||| | | | ||| | Sbjct 93 EPKPVQKAAKPTPQPLAPSPAPSPTPAPAAD#PTPAPAAPAAVAPAAPAATPAPARETMQV 152 Query 854 LPP 856 | Sbjct 153 SAP 155 |
| Corynebacterium jeikeium K411 | Query 799 EPKETTEDAAP-----PGPTSETPELATEQKPIQD#PQPTP 833 ||+ | | || | ||+| || | ||+ +#|+| | Sbjct 118 EPEPTPEPEAPKATPAPAPTAE-PEPVEEPKPVAE#PEPEP 156 |
| Anabaena variabilis ATCC 29413 | Query 799 EPKETTEDAAPPGPTSETPELATEQKPIQD#PQ--PTPSA 835 ||+ || ||+ || | |+| #|+ ||||| Sbjct 181 EPRLANPRPTPPRPTTPTPRQQTPQRPSST#PRFAPTPSA 219 |
| Neurospora crassa OR74A | Query 799 EPKETTEDAAPPGPTSETPE---LATEQKPIQD#PQPTPSAPAMGAADQLASAREASQEL 854 +|+| || ||| | |+| + |++ | +# |+ | | ++ | | ++| Sbjct 26 KPREAAEDVAPPKPNSDTTKNNVQASKDAPATN#GTTAPTEAAKAAPEKTLSPEEFERQL 84 |
| Yersinia bercovieri ATCC 43970 | Query 803 TTEDAAPPGPTSETPELATEQKPIQD#PQPTPSA 835 | | | |||+ | +||+| | #|| ||+| Sbjct 202 TIAQACHPPPTSQQPPMATQQLSILT#PQATPAA 234 |
| Caenorhabditis briggsae | Query 800 PKETTEDAAPPGPTSETPELATEQKPIQD#PQP-------TPSAPAMGAADQLASAREASQ 852 || | || | |+ |+ + #| | || | | ++| | ||+ Sbjct 121 PKATGSPPPPPPPPSDEPQEVVAGGASRR#PPPPPPRGTGTPPPPPTGVPEELNEANHASR 180 Query 853 ELPP 856 || Sbjct 181 RPPP 184 |
| Arabidopsis thaliana | Query 799 EPKETTEDAAPPGPTSETP--------ELATEQKPIQD#PQPTPS-APA 837 |||+ || + | || | | ++| ++||+ #| || ||| Sbjct 5 EPKKVTETVSEPTPTPEVPVEKPAAAADVAPQEKPVAP#PPVLPSPAPA 52 |
| Drosophila melanogaster | Query 799 EPKETTEDAAPPGPTSETPELATEQKPIQD#PQPTPSAPAMGAADQLASAREASQELPP 856 ||| | + | | || || +| +| +#|+|| | | +| + | | + | | Sbjct 461 EPKSEPEPKSEPEPKSE-PEPKSEPEPKSE#PEPT-SEPEPKSAPEPKSEPEPTSEPEP 516 |
| Cenarchaeum symbiosum | Query 799 EPKETTEDAAPPGPTSETPELATEQKPIQD#PQPTPSAP---AMGAADQLASAREA 850 || | | | | + || |+ | +|#| | | || | | | |+|++| Sbjct 3480 EPAGPAEPAVPEPPEARAPEPAS---PAED#PGPEPPAPQGVAPGEDTQDANAQDA 3531 |
| Rhodobacter sphaeroides ATCC 17025 | Query 799 EPKETTEDAAP---PGPTSET-PELATEQKPIQD#PQP--TPSAPAMGAADQLASAREASQ 852 +| |+||| | | +|| | || | +| #||| |+|| + + | || + Sbjct 70 QPAPPPEEAAPAPAPAPVAETPPPLAPEPRPEPA#PQPDAAPTAPEIAPVPEPEIA-EAPE 128 Query 853 ELPP 856 | | Sbjct 129 PLAP 132 |
| Leifsonia xyli subsp. xyli str. CTCB07 | Query 799 EPKETTEDAAPPGPTSETPELATEQKPIQD#PQPTPS-APAMGAADQLASAR 848 ||+ | ||| ||+||| || | #| | |+ || | +|| Sbjct 221 EPEPANEPAAPAEPTAETPAPAT--PPAAA#PLPAPAETPAADPATAPIAAR 269 |
| Burkholderia sp. 383 | Query 799 EPKETTEDAAPPGPTSETPELATEQKPIQD#PQPTPSAPAMGA-ADQLASAREASQELPP 856 ||| + | | |+ | | + |#| | |+||| | | ||| | | Sbjct 95 EPKPVQKAVLTPKPVSQAPS-PTPAPAVAD#PTPAPAAPAPAAPAAAPGPARETMQVSAP 152 |
| Fulvimarina pelagi HTCC2506 | Query 799 EPKETTEDAAPPGPTSE-TPELATEQKPIQD#PQPTPSAPAMGAADQLASAREASQELPP 856 || | | | | || | | +#|+| | + + |++ | | | + | | Sbjct 141 EPAPEPEPEPAPEPEPEPAPEPEPEPAPEPE#PEPAPESESQAPAEEAAPAEEPAPEPEP 199 |
| Hosta virus X | Query 803 TTEDAAPPGPTSETPELATEQKPIQD#PQPTPSAPAMGAADQLASA 847 ||++| || + | | | #|||| +| | | +|| Sbjct 588 TTQEAGTEGPPTTQPGKPTASSPRAA#PQPTANAETMEKGSQASSA 632 |
| Ralstonia solanacearum GMI1000 | Query 804 TEDAAPPGPTSETPELATEQKPIQD#PQPTPSAPAMGAA 841 | || || | +| || |#| | |+|| +| Sbjct 568 TVPAAAAGPASAASAIAAPAKPASD#PAPAPAAPKAPSA 605 |
| Pichia pastoris | Query 799 EPKETTEDAAPPGPTSETPELATEQKPIQD#PQPTPS 834 |||| ++|| | |+|| | +| || # || |+ Sbjct 144 EPKEEPKEAATPAPSSE--ESKSEAKPSSS#KQPRPA 177 |
| Stenotrophomonas maltophilia R551-3 | Query 799 EPKETTEDAAPPGPTSETPELATEQKPIQD#PQPTPSAPAMGAADQLASAREASQELPP 856 ||++ | | ||+ | | +#| | +||++ ++ || |+ + || Sbjct 81 EPRQQTVPTPLPTPTATVEEAQGIVLPAAE#PAPVQAAPSIDSSTPLAGAQLQYRSAPP 138 |
| Erythrobacter sp. NAP1 | Query 799 EPKETTEDAAPPGPTSETPELATEQKPIQD#PQPTP-SAPAMGAADQLAS 846 |||| + | | || ||| | | #| ||| | || || ||| Sbjct 102 EPKEAS--APMPDPTI-TPEPELEPAPAPT#PSPTPASEPAATAAKVLAS 147 |
| Rhodobacter sphaeroides ATCC 17029 | Query 803 TTEDAAPPGPTSETPELATEQKPIQD#PQPTPSAPAMGAADQLASAR 848 ||| |||| |+ || | #|+ |+ || | | |+ Sbjct 66 TTEPAAPPPATAPAPEATTPAPAEPA#PEAAPATPAPDATAPAAPAQ 111 |
| Corynebacterium efficiens YS-314 | Query 799 EPKETTEDAAPPGPTSETPELATEQKPIQD#P--QPTPSAPAMGAADQLASAREASQEL 854 || + | | | | | | | | |+ +#| +| |+ ||+ || | + | +| Sbjct 422 EPVKEPEPAPAPEPAPE-PVKAPEPAPVPE#PAPEPEPATPAVPTADLLPTIRSKWAQL 478 |
| Leishmania infantum JPCM5 | Query 800 PKETTEDAAPPGPTSETPELATEQKPIQD#PQPTPSAPAMGAADQLASAREASQELPPGF 858 | | | | || | +| # || |+|| +| |+ |+ +|| | Sbjct 1510 PSNATSGARLPAQTSGAPRTGASEK---S#SPPTSSSPAAASASSSTSSPVATSAVPPAF 1565 |
| Bacillus clausii KSM-K16 | Query 799 EPKETTEDAA--PPGPTSETPELATEQKPIQD#PQPTPSAPAMGAADQLASAREASQE 853 |||+ +|+ + | ++ | |++|+++#|| | | | | + | ++| Sbjct 185 EPKQQSEETSEEPKAKQNQEPLETVEEEPVEE#PQAEPQAKAEPETQQESKKEEPAEE 241 |
| Aspergillus fumigatus Af293 | Query 802 ETTEDAAPPGPTSETPELATEQKPIQD#PQPTPSAPAMGAADQLASAREASQE 853 | | || | |+| |+ | +| #|+ || | ||| + ++ + | Sbjct 144 EEEETEAPTPPKSKTSALSHEPSGVQS#PKHRPSIAASVAADNMKEVKKETLE 195 |
| Cyanothece sp. CCY0110 | Query 800 PKETTEDAAPPGPTSETPELATEQKPIQD#PQPTPSAP 836 |+ | || |+ ||+ | || #| ||| | Sbjct 400 PEPKTPTPAPENSTNSTPKTPIEPKPPIQ#PNPTPQTP 436 |
| Pichia guilliermondii ATCC 6260 | Query 800 PKETTEDAAPPGPTSETPELATEQKPI---QD#PQPTPSAP 836 | ||+| | || || + || || + #|+|+ | | Sbjct 718 PVETSETPETPEPTPETTPVGTETAPIETSES#PEPSESVP 757 |
| Aspergillus clavatus NRRL 1 | Query 803 TTEDAAPPGPTSETPELATEQKPIQD#PQPTPSAPAMGAADQLASAREASQELP 855 | || +| + | || |+ +|#| | || |++ |+ || +|+| Sbjct 900 TPEDPSPEPEPTATRELPAEEVISED#PAPAEDAP----AEEPATDRELVEEIP 948 |
| Danio rerio | Query 799 EPKETTEDAAPPGPTSETPELATEQK-------PIQD#PQPTPSAPAMGAADQLASAREAS 851 +| |+ |+ | +||| + | || | +#| | || ++ + |+++|+ Sbjct 169 DPLESA-DSVPSSSSSETSKKAAEQSPPPKPHTPASE#PDPLESADSLPKSSSRRSSKKAA 227 Query 852 QELPP 856 ++ || Sbjct 228 KQSPP 232 |
| Myxococcus xanthus DK 1622 | Query 800 PKETTEDAAPPGPTSETPELATEQKPIQD#PQPTPSAPAMGAADQLA 845 |+| +||| || | |++ #| | | | + | + | Sbjct 589 PREAKAEAAPVGPPPAMPIAPPVPVPVRP#PPPAPEAASRGGEEDAA 634 |
| Strongylocentrotus purpuratus | Query 799 EPKETTEDAAP-PGPTSE-TPELATEQKPIQD#PQPTPSAPAMGAADQLASAREASQELPP 856 ||+ | | | || | ||| | | #|+||| | + +| | +|| | Sbjct 242 EPEPEAAAAEPTPEPTPEPTPEPTPEPTPETT#PEPTPE-PEAPVVEPVAPVEEPAQEPTP 300 |
| Bacillus thuringiensis serovar konkukian str. 97-27 | Query 800 PKETTEDAAPPGPTSETPELATEQKPIQD#PQPTPSAPAMGAADQLASAREASQELP 855 || | | | ||| | || #| ||| ++ || +|++ +|+| Sbjct 130 PKAVTP-APKPVTRVETPATAPTPKPTPA#PTPTPKPVSVEAAVELSTPAPVKREVP 184 |
| Pyrobaculum arsenaticum DSM 13514 | Query 800 PKETTEDAAPPGPTSETPELATEQKPIQD#PQPTPS 834 | | ||| |||+ | | | | #| |||| Sbjct 49 PSPTQTPTAPPTPTSQ-PTTTTPTPPPQT#PSPTPS 82 |
| Schizosaccharomyces pombe 972h- | Query 799 EPKETTEDAAPPGPTSETPELATEQ-------KPIQD#PQPTPSAPAMGAADQLASAREAS 851 | | || |++|| +| | ||| | ++# +| || |+ + + Sbjct 469 EAKSTTNDSSPPKDSSSTSTQPTEQSNAQQAPSPKEE#ERPLPSEPSQNQPAEYRDTPDTP 528 Query 852 QELPP 856 + + | Sbjct 529 RNIMP 533 |
| Murid herpesvirus 2 | Query 799 EPKE--TTEDAAPPGPTSETPELATEQKPIQD#PQPTPSAPAMGAADQLASAREASQEL 854 ||| |||+ | + || | ++| |# | ||+| ++| || +| Sbjct 244 EPKTEPTTEEPTEPTTMATTPPKQTRKRPAPD#VQKTPAAKPTKHGRRMARIPEAMMDL 301 |
| Corynebacterium glutamicum R | Query 801 KETTEDAAPPGPTSETPELATEQKPIQD--#PQPTPS-----APAMGAADQLASAREASQE 853 +| || | |+| | | || |+ | #|+| |+ ||| | +| + || Sbjct 240 EEATEAAEAASVTAEFAEAAREQTPVPDTA#PEPAPAPIDDIAPASGRIGKLRTRLSRSQN 299 Query 854 L 854 + Sbjct 300 V 300 |
| Colwellia psychrerythraea 34H | Query 800 PKETTEDAAPPGPTSET-----PELATEQKPIQD#PQPTPSAPAMGAADQLASAREASQEL 854 | | | | +| ||| || +|| |#|+| | + ++| |+ | | Sbjct 451 PTSDLEPITAPEPITEPELITEPELITEPEPITD#PEPIPEIVSEPQPERLESSSAGSPGL 510 |
| Frankia alni ACN14a | Query 808 APPGPTSETPELATEQKPIQD#PQPTPSAPA--MGAADQLASAREA 850 | | | + ||+ + |+ #| |||+| | ||| + | || | Sbjct 258 ASPQPLASPQPLASARPPVST#PPPTPAATAATAGAAARTAPARPA 302 |
| Rhodobacterales bacterium HTCC2654 | Query 799 EPKET--TEDAAPPGPTSETPELATEQKPIQD#PQPTPSAPAMGAADQLAS---AREASQE 853 ||+| || || || | +|+ +#|+||| | | + | ++ Sbjct 59 EPEEAAPTETPVPPARPEPEPEPQPEPEPVPE#PEPTPLPEPPAPAPSLPTEDVQPETTEP 118 Query 854 LPP 856 || Sbjct 119 TPP 121 |
| Tetraodon nigroviridis | Query 801 KETTEDAAPPGPTSETPELATEQKPI-QD#PQPTPSA 835 || | + +|| ||||+ || | #|+|+||| Sbjct 207 KEQTPNVSPPRGAPATPELSPLPKPSNQS#PEPSPSA 242 |
| Chromohalobacter salexigens DSM 3043 | Query 800 PKETTEDAAPPGPTSETPELATEQKPIQD---#PQPTPSAPAMGAADQLAS---AREASQE 853 | +|||| | | ||| | + | #|| |||| + | + +||+ Sbjct 471 PPAASEDAASPAPVPETPSAAPRPRISWDDEA#PQAHSSAPASAPPEPEVSDAPSADASRS 530 Query 854 LPPG 857 | | Sbjct 531 APRG 534 |
| Coccidioides immitis RS | Query 805 EDAAPPGPTSETPELATEQKPIQD#PQPTPSAPAMGAADQLASAREASQEL 854 |+| || | + | || ||| #| | | + | || | ++ + Sbjct 42 EEAGPPPPPPKEPILAIEQKTEAK#PVAPESGPGLAAKACLAEAAASTNSI 91 |
| Synechococcus sp. BL107 | Query 799 EPKETTEDAAPPGPTSETPELATEQKPIQD#PQPTPSAPAMGAA--DQLASAREASQE 853 ||+ | || | |||++ | | #||||| || +| + +| |+ |+ |+ Sbjct 111 EPESAT---APNAPVPLTPEVS----PEQT#PQPTP-APTVGGSLLEQAAAQRQQRQQ 159 |
| Methylibium petroleiphilum PM1 | Query 812 PTSETPELATEQKPIQD#PQPTPSAPAMGAADQLASAREASQELPP 856 | | | | |+ | #|+| | ||| || +|+ || Sbjct 825 PRSLTTETASGAAPAAP#PEPAPPAPAAAAAASAPVVADAAPAAPP 869 |
| Alnus glutinosa | Query 800 PKETTEDAAP---PGPTSETPELATEQKP-IQD#PQPTPSAPAMGAADQLASAREASQELP 855 |||| |+ || | || | + ||+ | + +#|+| | | |+ | + |+| Sbjct 99 PKETPEEEAPKETPEPTVEETKEATDSAPAVPE#PKPEPEAEVPKKAE---VPEEVAAEVP 155 |
| Lyngbya sp. PCC 8106 | Query 799 EPKETTEDAAPPGPTSETPELATEQKPIQD#PQPTP 833 ||+ | | | || | || | +| +#|+||| Sbjct 1616 EPEPTPEPTPEPEPTPE-PEPTPEPEPTPE#PEPTP 1649 |
| Aspergillus niger | Query 800 PKETTEDAAPPGPTSETPELATEQKPIQD#PQPTPSAPA 837 | + |||| | | +| +|+ #| | ||| | Sbjct 263 PPAASAPAAPPPPPPSAPPVAPPSEPLSR#PSPLPSAIA 300 |
| Ustilago maydis 521 | Query 799 EPKETTEDAAPPGPTSETPELATEQKPIQD-----#PQPTPSAPAMGAADQLASAREASQE 853 ||+| ||| | + |+| + | #| | | || + | | | + Sbjct 502 EPEEEAAPPAPPAPPAPPAAAASETEEQDDQGIPP#PPPPPPAPPVAPAAPAAPALQEEAP 561 Query 854 LPP 856 || Sbjct 562 APP 564 |
| Phaeosphaeria nodorum SN15 | Query 799 EPKETTEDAAPPGPTSET---PELATEQKPIQD#PQPTPSAPAMGAA 841 +| || + | | | | | | | | #| | || |+ | | Sbjct 131 KPVETPQPAPAPAPVSSAAPAPAPAPEYTPAPA#PAPAPSKPSSGGA 176 |
| Saccharomyces cerevisiae | Query 799 EPKETTEDAAPPGPTSETPELATEQKPIQD#PQPTPSAPAMGAADQLASAREASQELPP 856 +| |||| || | | + #| | | || + + | + + || Sbjct 648 KPPSPPVAAAPPLPTFSAPSLPQQSVSTSI#PSPPPVAPTLSVRTETESISKNPTKSPP 705 |
| Streptococcus pneumoniae | Query 799 EPKETTEDAAPPGPTSETPELATEQKPIQD#PQPTPSAPAMGAADQLAS---AREASQE 853 ||++ |+ | | | | | + || #||| |+ || | || + +| Sbjct 310 EPEKPAEETPAPAPKPEQP--AEQPKPAPA#PQPAPAPKPEKTDDQQAEEDYARRSEEE 365 |
| Neosartorya fischeri NRRL 181 | Query 800 PKETTEDAAPPGPTSETPELATEQKP--IQD#PQPTPSAPAMGAADQLASA 847 | + |||| |+|+ | | + #|+| |+ | | | +| Sbjct 533 PVAAPQVPTPPGPPPESPQTKTHTPPARVAS#PRPRPTKPKKGGNGQSPNA 582 |
| Vitis vinifera | Query 807 AAPPGPTSETPELATEQK--PIQD#PQPTPSAPAMGAADQLASAREASQELPP 856 |||| | | | | +| |++ # | |+||+| | +++ |+ +|| Sbjct 842 AAPPRPASPVPPQAEQQDELPVES#VPPAPAAPSMPEA--ISTDPPATPLVPP 891 |
| Leishmania major | Query 800 PKETTEDAAPPGPTSETPELATEQKPIQD#PQPTPSAPAMGAADQLASAREASQELPPGF 858 | | | | || | +| # || |+|| +| |+ |+ || | Sbjct 1510 PSNATSGARLPAQTSSAPRAGASEK---S#SPPTLSSPAAASASSSTSSPVAASTAPPAF 1565 |
| Mycobacterium sp. MCS | Query 800 PKETTEDAAPPGPTSE---------TPELATEQKPIQD#PQPTPS---APAMGAADQLASA 847 | || || | +| ||| | +| +#|+| |+ || | |+| Sbjct 114 PTETAPPRAPERPVAEPEPAPEPEPTPEPIPEPEPAPE#PEPEPAEVIAPTHGRLDRLRGR 173 Query 848 REASQ 852 || Sbjct 174 LAKSQ 178 |
| Roseiflexus castenholzii DSM 13941 | Query 800 PKETTEDAAPPGPTSETPELATEQKPIQD#PQPTPSAPAMGAADQLASAREAS 851 | | | | ||| ||| + +| #|+|| + |+ | | | | + Sbjct 848 PSATVTPTATPLPTS-TPEPTSTPEPTNT#PEPTATPTALPTATPLPSPTETA 898 |
| Roseobacter denitrificans OCh 114 | Query 799 EPKETTEDAAPPGPTSETPELATEQKPIQD#PQPTPSAPAMGAADQLASAREASQELP 855 ||+ | | | | || |+ |+ # ||| | + | || + ||| Sbjct 142 EPEPEPEPEPEPEPEPE-PEPEPERTPVAH#APPTPEVPEVEVAPAQHSAEDDPFELP 197 |
| Desulfovibrio vulgaris subsp. vulgaris str. Hildenborough | Query 804 TEDAAPPGPTSETPELATEQKPIQD#PQPTPSAPAMGAADQLASAREASQELPPG 857 | |+ | | + +|| | #|+| |+||| +| +| +|+ | | Sbjct 564 TTSASAPESGSVAQGAPSSEKPAQK#PEPVPAAPAFQSA--ASSGAQAAASAPAG 615 |
| Streptococcus pyogenes MGAS10750 | Query 800 PKETTEDAAP-PGPTSETPELA--TEQKPIQD#PQPTPSAPAMGAADQLASAREASQELP 855 |+ | ||| | | || | +| +|+ | # |+||+ + | +|+| | Sbjct 193 PETPEEPAAPSPSPESEEPSVAASSEETPTPS#TPEEPAAPSPSPESEEPSVAASSEETP 251 |
| Mycobacterium sp. JLS | Query 800 PKETTEDAAPPGPTSE---------TPELATEQKPIQD#PQPTPS---APAMGAADQLASA 847 | || || | +| ||| | +| +#|+| |+ || | |+| Sbjct 106 PTETAPPRAPERPVAEPEPAPEPEPTPEPIPEPEPAPE#PEPEPAEVIAPTHGRLDRLRGR 165 Query 848 REASQ 852 || Sbjct 166 LAKSQ 170 |
| Alcelaphine herpesvirus 1 | Query 802 ETTEDAAPPGPTSETPELATEQKPIQD#PQPT-------PSAPAMGAADQLASAREASQEL 854 | |+ || |||++|| |+ | |||# | | | | | + ++ ++ | Sbjct 1024 EQTQAPAPKGPTADTP--VTDSKLIQD#SQQNDRHQKEKPLKPKKPQAISLPAVKDTTKHL 1081 Query 855 PP 856 | Sbjct 1082 KP 1083 |
| Zea mays | Query 800 PKETTEDAAPPGPTSETPELATEQKPIQD#PQPTPSAPAMGAADQLASAREASQELPP 856 | + + ++|| | | | | + |+ +# | |+ + ++| + + || Sbjct 738 PPQAPKSSSPPAPVSSPPPLKSSPPPVPE#SSPPPTPKSSPPLAPVSSPPQVEKTSPP 794 |
| Streptococcus sobrinus | Query 799 EPKETTEDAAPPGPTSETPELATEQKPIQD#PQPT----PSAPAMGAADQLASAREASQEL 854 |||+ | ||| | | |+ || + # ||| | | | + | | Sbjct 1040 EPKQPTPPAAPEPPKMTTVEI--PDKPTEP#KQPTPPVAPEPPKMTTVEIPDKPTEPKQPT 1097 Query 855 PP 856 || Query 855 PP 856 |
| Bordetella bronchiseptica | Query 800 PKETTEDAAPPGPTSETPELATEQKPIQD#PQPTPSAPAMGAADQLASAREAS 851 || + ||| + |+| | | #||| || | +|++| |+ Sbjct 546 PKPAPQPGPQPGP--QPPQLPQPQPPKPQ#PQPEAPAPQPPAGRELSAAANAA 595 |
| Sphingomonas sp. SKA58 | Query 808 APPGPTSETPELATEQKPIQD#PQPTP---SAPAMGAADQLASAREASQELP 855 || | +||| | |+ #| | | +||| || | | +|+ + | Sbjct 69 APAAPEAETPRAAA---PVSA#PPPPPRPAAAPAAPAAAPAAPAADAASDAP 116 |
| Dinoroseobacter shibae DFL 12 | Query 805 EDAAPPGPTSETPELATEQKPIQD#PQPTPSA 835 || || ||+ | || || |#| |+|+| Sbjct 3 EDTPPPQSTSKRPATPVEQSPI-D#PVPSPTA 32 |
| Natronomonas pharaonis DSM 2160 | Query 799 EPKETTEDAAPPGPT-SETPEL--ATEQKPI-QD#PQPTPSAPAMGAADQLASAR 848 +|+| |+ | |+ |||| | ++ | |#|+||| |+| | +| Sbjct 22 QPEEAPEETPEPEPSPEETPESEPAPDETPAPDD#PEPTPEDEPPSDAEQFAESR 75 |
| Enterococcus faecalis | Query 799 EPKETTEDAAPPGPTSET----PELATE-QKPIQD#PQP-TPSAPA 837 || | || | ||+ | || || ||++ # +| ||| || Sbjct 72 EPTEPTEPTTPTEPTTPTEPSEPEQPTEPSKPVEP#EKPVTPSKPA 116 |
| Xanthobacter autotrophicus Py2 | Query 800 PKETTEDAAPPGPTSETPELATEQKPI------QD#PQPTPSAPAMGAADQLASAREAS 851 | + ||| ||+ || | | | | # | |+|| || + |+ || Sbjct 158 PTPAAAEPAPPAPTAAAPEAAAPQAPAPQTSAPQA#AAPKPAAP-RAAAPAASEAKPAS 214 |
| Enterococcus faecalis V583 | Query 799 EPKETTEDAAPPGPTSET----PELATE-QKPIQD#PQP-TPSAPA 837 || | || | ||+ | || || ||++ # +| ||| || Sbjct 73 EPTEPTEPTTPTEPTTPTEPSEPEQPTEPSKPVEP#EKPVTPSKPA 117 |
| Pyrobaculum islandicum DSM 4184 | Query 807 AAPPGPTSETPELATEQKPIQD#PQPTPSAPAMGAADQLASA 847 | | || || || | | # | +||| | + ||+| Sbjct 798 ATTPTPTPTTPPPATTPTPPQP#TVVTTTAPATGTTEALAAA 838 |
| Macaca fascicularis | Query 799 EPKETTEDAAPPGPTSET-------PELATEQKPIQD#PQPTPSAPAMGAADQLASAREAS 851 +| | +|+|| || + || ++| #|+| | + | +| |+ | + Sbjct 8 DPPEGSEEAAEPGMDTPEDQDLPPCPEDIGKEKCTAA#PEPEPCEVSEPPAKRLRSSEEPT 67 Query 852 QELPPG 857 ++ ||| Sbjct 68 EKEPPG 73 |
| Thiomicrospira crunogena XCL-2 | Query 799 EPKETTEDAAPPGPTSETPELATEQKPIQD#PQPTPS---APAMGA 840 || | | | || | || | +| +#|+||| || || Sbjct 98 EPAPEPEPALEPEPTPE-PEPTPEPEPTPE#PEPTPEPEPTPAAGA 141 Query 799 EPKETTEDAAPPGPTSETPELATEQKPIQD#PQPTP 833 ||+ | | | | | || | | +| +#|+||| Sbjct 86 EPESEPEPAPEPEPAPE-PEPALEPEPTPE#PEPTP 119 |
| Chlamydomonas incerta | Query 800 PKETTEDAAPPGPTSETPELATEQKPIQD#PQPTPSAPA 837 | ||| || +|| + | #| | | +|| Sbjct 60 PSPAPPSPAPPSPTPPSPEPPSPAPPSPP#PSPAPPSPA 97 |
| Streptococcus sanguinis SK36 | Query 799 EPKETTEDAAPPGPTSETPELATEQ---KPIQD#PQPTPSAPAMGAADQLASAREASQEL 854 || ++ ++ +| || | +|| ||+++# || | + | +| ||+ +| Sbjct 87 EPSQSDDNPSPRSSESEKVEESTESTTTKPVEN#TQPITVRPEISNATVVAEKSEATADL 145 |
| Pichia stipitis CBS 6054 | Query 800 PKETTEDAAPPGPTSETPELATEQKPIQD#PQPTPSAPAMGAADQLASAREA----SQELP 855 | | || | | | |+ #| | | | ||| +| | |+|| Sbjct 355 PPPPNRGAVPPPPPSRVPGPTPPQRTGAP#PPPPPPRAARGAAPPPPPSRTARPAQPQQLP 414 Query 856 P 856 | Query 856 P 856 |
| Geobacter lovleyi SZ | Query 799 EPKETTEDAAPPGPTSETPELATEQKPIQD#PQPTPSAPAMGAADQLASAREASQEL---- 854 +||+ |+|++| | + | | # |+| | |++ +| || ||+ Sbjct 243 KPKQDTDDSSPDGDNTGTAAGDDSQSQAAG#-NPSPPVPKSGSSGELGGNPEALQEMLDCD 301 Query 855 PPGF 858 | || Sbjct 302 PGGF 305 |
| Burkholderia cenocepacia HI2424 | Query 800 PKETTEDAAPPGPTSETPELATEQKPIQD#PQPTPSAPAMGAA 841 |+ | | | | | || +| |+| #| | |+ | | Sbjct 137 PQATRE--AAPAPAPATPAVAAPAAPVQA#PAPAPAVPVRETA 176 |
| Polaromonas naphthalenivorans CJ2 | Query 799 EPKETTEDAAPPGPTSE--TPELATEQKP------------IQD#PQPTPSAPAMGAADQL 844 ||| ||| | || + + | | |#| |+|+|| | + Sbjct 86 EPKAEPAPPAPPAPPVRPVTPSVQKKVVPKPAARPQAQPLAIAD#PTPSPNAPTGSLAPAV 145 Query 845 ASAREASQELP 855 || |+ | | Sbjct 146 ASTPTAAPEAP 156 |
| Desulfovibrio vulgaris subsp. vulgaris DP4 | Query 804 TEDAAPPGPTSETPELATEQKPIQD#PQPTPSAPAMGAADQLASAREASQELPPG 857 | |+ | | + +|| | #|+| |+||| +| +| +|+ | | Sbjct 564 TTSASAPESGSVAQGAPSSEKPAQK#PEPVPAAPASQSA--ASSGAQAAASAPAG 615 |
| Rhodopirellula baltica SH 1 | Query 802 ETTEDAAPPGPTSETPELATEQKPIQD#PQPTPSAPAMGAADQLASAREASQELPP 856 |||++|||| | +| | + #| +||| ||+++ |+ + +| | Sbjct 202 ETTDEAAPPAP------VADEAPAAEA#PAAEEAAPAAPAAEEVKSSSDQVEEAAP 250 Query 799 EPKETTEDAAPPGPTS------ETPELATEQKPIQD#PQPTPSAPAMGAADQLASAREASQ 852 | ||+||| | + || + | |+ |# | || ||++ | | |++ Sbjct 180 EAAAATEEAAPAAPVADAAADAETTDEAAPPAPVAD#EAPAAEAP---AAEEAAPAAPAAE 236 Query 853 EL 854 |+ Sbjct 237 EV 238 |
| Anaeromyxobacter sp. Fw109-5 | Query 800 PKETTEDAAPPGPTSETPELATEQKPIQD#PQPTPSAPAMGAAD 842 |+| | || | + | |+ +#| | |+| ||+ Sbjct 353 PQEAVAAATPPAPPAAAPAEKAPPAPVAE#PAPKPAAETPRAAE 395 |
| Pseudomonas aeruginosa UCBPP-PA14 | Query 806 DAAPPGPTSETPELATEQKPIQD#PQPTPSAPAMGAADQLASAREASQELPP 856 | | | | |++| + #||| | || ||+ +|+ ++| || Sbjct 156 DEPPLVPVSSHPQIAGRTH--ER#PQPGPGFPAKAAAEVAPTAQASAQASPP 204 |
[Site 5] EGAESAQPEA762-EELEVTVPQE
Ala762  Glu
Glu
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Glu753 | Gly754 | Ala755 | Glu756 | Ser757 | Ala758 | Gln759 | Pro760 | Glu761 | Ala762 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Glu763 | Glu764 | Leu765 | Glu766 | Val767 | Thr768 | Val769 | Pro770 | Gln771 | Glu772 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| TVPAGEGVSLEEAKIGTETTEGAESAQPEAEELEVTVPQEKVIPSVVIEPASNHEGEGEN |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | Pan troglodytes | 119.00 | 2 | PREDICTED: amphiphysin |
| 2 | Homo sapiens | 115.00 | 14 | amphiphysin isoform 1 |
| 3 | synthetic construct | 115.00 | 1 | amphiphysin |
| 4 | Macaca mulatta | 100.00 | 4 | PREDICTED: amphiphysin isoform 3 |
| 5 | Rattus norvegicus | 77.40 | 1 | amphiphysin 1 |
| 6 | Bos taurus | 71.20 | 1 | hypothetical protein LOC614722 |
| 7 | Canis familiaris | 68.90 | 1 | PREDICTED: similar to Amphiphysin |
| 8 | Mus musculus | 65.90 | 3 | amphiphysin |
| 9 | Monodelphis domestica | 64.30 | 1 | PREDICTED: similar to amphiphysin I variant CT2 |
| 10 | Gallus gallus | 56.60 | 1 | amphiphysin (Stiff-Man syndrome with breast cancer |
| 11 | Xenopus laevis | 43.10 | 1 | hypothetical protein LOC432150 |
| 12 | Xenopus tropicalis | 41.20 | 1 | Unknown (protein for MGC:122074) |
| 13 | Oryza sativa (indica cultivar-group) | 38.50 | 1 | hypothetical protein OsI_022473 |
| 14 | Oryza sativa (japonica cultivar-group) | 37.40 | 2 | Os06g0538200 |
| 15 | Tetraodon nigroviridis | 35.40 | 2 | unnamed protein product |
| 16 | Plasmodium falciparum | 34.30 | 1 | AF247634_1 glutamate-rich protein |
| 17 | Plasmodium falciparum 3D7 | 34.30 | 1 | glutamate-rich protein |
Top-ranked sequences
| organism | matching |
|---|---|
| Pan troglodytes | Query 733 TVPAGEGVSLEEAKIGTETTEGAESAQPEA#EELEVTVPQEKVIPSVVIEPASNHEGEGEN 792 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 733 TVPAGEGVSLEEAKIGTETTEGAESAQPEA#EELEVTVPQEKVIPSVVIEPASNHEGEGEN 792 |
| Homo sapiens | Query 733 TVPAGEGVSLEEAKIGTETTEGAESAQPEA#EELEVTVPQEKVIPSVVIEPASNHEGEGEN 792 ||||||||||||||||||||||||||||||#|||| |||||||||||||||||||| |||| Sbjct 498 TVPAGEGVSLEEAKIGTETTEGAESAQPEA#EELEATVPQEKVIPSVVIEPASNHEEEGEN 557 |
| synthetic construct | Query 733 TVPAGEGVSLEEAKIGTETTEGAESAQPEA#EELEVTVPQEKVIPSVVIEPASNHEGEGEN 792 ||||||||||||||||||||||||||||||#|||| |||||||||||||||||||| |||| Sbjct 498 TVPAGEGVSLEEAKIGTETTEGAESAQPEA#EELEATVPQEKVIPSVVIEPASNHEEEGEN 557 |
| Macaca mulatta | Query 733 TVPAGEGVSLEEAKIGTETTEGAESAQPEA#EELEVTVPQEKVIPSVVIEPASNHEGEGEN 792 |+||||||||||||||||| ||||||| ||#||| |||||||||||||||||||| |||+ Sbjct 633 TLPAGEGVSLEEAKIGTET-EGAESAQAEA#EELAATVPQEKVIPSVVIEPASNHEEEGEH 691 |
| Rattus norvegicus | Query 734 VPAGEGVSLEEAKIGTETTEGAESAQPEA#EELEVTVPQEKVIPSVVIEPASNHEGE 789 +||||| | | ||| |+|| | | |+|# ||| |||||||||||||||||||| Sbjct 489 LPAGEGESPEGAKIDVESTELASSESPQA#AELEAGAPQEKVIPSVVIEPASNHEGE 544 |
| Bos taurus | Query 734 VPAGEGVSLEEAKIGTETTEGAESAQPEA#EELEVTVPQEKVIPSVVIEPASNHEGEGEN 792 +||| | |||||| | || + + + #|| | ||||||||||+||||+|||||+ Sbjct 496 LPAGAGASLEEAKTDIEATEAIDGDRSQL#EETEAVAAQEKVIPSVVIQPASNNEGEGEH 554 |
| Canis familiaris | Query 734 VPAGEGVSLEEAKIGTETTEGAESAQPEA#EELEVTVPQEKVIPSVVIEPASNHEGEGEN 792 +| ||| +|||+ || |+ + + + #|| | | ||||||||||||||+||||++ Sbjct 650 LPPGEGAGVEEARPDTEATQAVDGDRSQL#EEAEAVAPLEKVIPSVVIEPASNNEGEGDH 708 |
| Mus musculus | Query 734 VPAGEGVSLEEAKIGTETTEGAESAQPEA#EELEVTVPQEKVIPSVVIEPASNHEGEGEN 792 +||||| | | ||| |+|| | | |+ # | | || ||||||||||||||||||+ Sbjct 493 LPAGEGGSPEGAKIDGESTELAISESPQP#VEPEAGAPQ--VIPSVVIEPASNHEGEGEH 549 |
| Monodelphis domestica | Query 735 PAGEGVSLEEAKIGTETTEGAESAQPEA#EELEVTVPQEKVIPSVVIEPASNHEGEGE 791 | ||||+ | + || | || + #||+| ||||||||||||||||||| | Sbjct 43 PLAEGVSVVETQAVPETREATESDRAGT#EEMEAVGIQEKVIPSVVIEPASNHEGEDE 99 |
| Gallus gallus | Query 734 VPAGEGVSLEEAKIGTETTEGAESAQPEA#EELEVTVPQEKV--IPSVVIEPASNHEGEGE 791 | || | | + || || +|+ #|| +| | |||| |||||||||||+||||| Sbjct 494 VAAGAG----EGAVRTEQEAAAEGDKPQG#EEKDVDVSQEKVSSIPSVVIEPASNNEGEGE 549 |
| Xenopus laevis | Query 733 TVPAGEGVSLEEAKIGTET-TEGAESAQPEA#EELEVTVPQEK-------VIPSVVIEPAS 784 |+|+ + + |+++ |+ | || | #+| |+ | | +|||||||||| Sbjct 479 TLPSEQNEEVVESEVKQESPVEMAEDIMEEE#KEPEIIEPNETKAPPEITIIPSVVIEPAS 538 Query 785 NHEGEGEN 792 ||| | ++ Sbjct 539 NHEDEHDD 546 |
| Xenopus tropicalis | Query 736 AGEGVSLEEAKIGTETTEGA---------ESAQPEA#EE-LEVTVPQE-KVIPSVVIEPAS 784 | + || | ++ |+ | | | +|| # | | ||| |||||||||| Sbjct 477 AEKEVSEIEVEVKEESAESAAEMADDIMEEQKEPEI#NEPKETEAPQEITCIPSVVIEPAS 536 Query 785 NHEGEGEN 792 ||| | +| Sbjct 537 NHEDEHDN 544 |
| Oryza sativa (indica cultivar-group) | Query 733 TVPAGEGVSL-----EEAKIGTETTEGAESAQPEA#EELEVTVPQEKVIPSVVIEPASNHE 787 |+|| | | + ||||| | || | | #||+||| | | | + |+ | Sbjct 713 TIPATEKVEVTTPATEEAKITTAATEEVEVTTPAT#EEVEVTTPSTK---EVEVTIAATEE 769 Query 788 GE 789 | Sbjct 770 AE 771 |
| Oryza sativa (japonica cultivar-group) | Query 733 TVPAGEGVSL-----EEAKIGTETTEGAESAQPEA#EELEVTVPQEKVIPSVVIEPASNHE 787 | || | | + ||||| | || | | #||+||| | | | + |+ | Sbjct 885 TTPATEKVEVTTPATEEAKITTAATEEVEVTTPAT#EEVEVTTPSTK---EVEVTIAATEE 941 Query 788 GE 789 | Sbjct 942 AE 943 |
| Tetraodon nigroviridis | Query 739 GVSLEEAKIGTETTEGAESAQPEA#EELEVTVPQEKVIPSVVIEPAS 784 | | ++ | || ||||| +| |#| | | |+ || +| | Sbjct 133 GFSFKKTK--KETGEGAESEEPAA#EATEAAAPAEEAKPSEAMEETS 176 |
| Plasmodium falciparum | Query 738 EGVSLEEAKIGTETTEGAESAQPEA#EELEVTVPQEKVIPSVVIEPASNHEGEGEN 792 | | +|| | | |+ +|| #++ | +| +|| || + | | | || Sbjct 1056 EIVEIEEVPSQTNNNENIETIKPEE#KKNEFSVVEEKAIPQEPVVPTLN---ENEN 1107 |
| Plasmodium falciparum 3D7 | Query 738 EGVSLEEAKIGTETTEGAESAQPEA#EELEVTVPQEKVIPSVVIEPASNHEGEGEN 792 | | +|| | | |+ +|| #++ | +| +|| || + | | | || Sbjct 1053 EIVEIEEVPSQTNNNENIETIKPEE#KKNEFSVVEEKAIPQEPVVPTLN---ENEN 1104 |
* References
None.