XSB2126 : PREDICTED: amphiphysin (Stiff-Man syndrome with breast cancer 128kDa autoantigen) isoform 1 [Macaca mulatta]
[ CaMP Format ]
This entry is computationally expanded from SB0093
* Basic Information
| Organism | Macaca mulatta (rhesus monkey) |
| Protein Names | amphiphysin (Stiff-Man syndrome with breast cancer 128kDa autoantigen) isoform 1 |
| Gene Names | AMPH; Derived by automated computational analysis using gene prediction method: GNOMON. Supporting evidence includes similarity to: 9 mRNAs, 54 ESTs, 8 Proteins |
| Gene Locus | Not available |
| GO Function | Not available |
| Entrez Protein | Entrez Nucleotide | Entrez Gene | UniProt | OMIM | HGNC | HPRD | KEGG |
|---|---|---|---|---|---|---|---|
| XP_001100038 | XM_001100038 | 704559 | N/A | N/A | N/A | N/A | N/A |
* Information From OMIM
Not Available.
* Structure Information
1. Primary Information
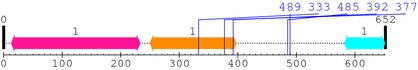
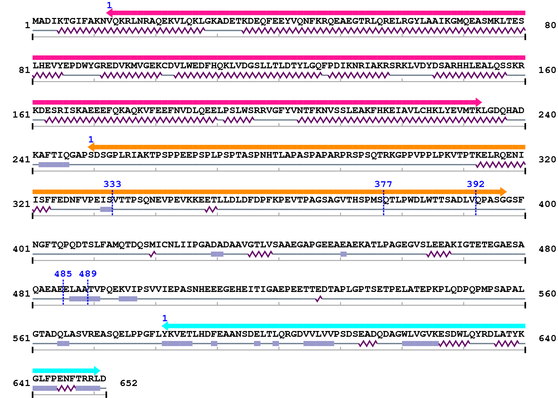
Length: 652 aa
Average Mass: 71.835 kDa
Monoisotopic Mass: 71.791 kDa
2. Domain Information
Annotated Domains: Not Available.
Computationally Assigned Domains (Pfam+HMMER):
| domain name | begin | end | score | e-value |
|---|---|---|---|---|
| BAR 1. | 13 | 233 | 288.1 | 1.9e-83 |
| Drf_FH1 1. | 250 | 397 | -39.0 | 3.6 |
| --- cleavage 333 (inside Drf_FH1 250..397) --- | ||||
| --- cleavage 392 (inside Drf_FH1 250..397) --- | ||||
| --- cleavage 377 (inside Drf_FH1 250..397) --- | ||||
| --- cleavage 489 --- | ||||
| --- cleavage 485 --- | ||||
| SH3_1 1. | 582 | 651 | 24.1 | 0.00017 |
3. Sequence Information
Fasta Sequence: XSB2126.fasta
Amino Acid Sequence and Secondary Structures (PsiPred):
4. 3D Information
Not Available.
* Cleavage Information
5 [sites]
Cleavage sites (±10aa)
[Site 1] AQAEAEELAA489-TVPQEKVIPS
Ala489  Thr
Thr
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Ala480 | Gln481 | Ala482 | Glu483 | Ala484 | Glu485 | Glu486 | Leu487 | Ala488 | Ala489 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Thr490 | Val491 | Pro492 | Gln493 | Glu494 | Lys495 | Val496 | Ile497 | Pro498 | Ser499 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| GEGVSLEEAKIGTETEGAESAQAEAEELAATVPQEKVIPSVVIEPASNHEEEGEHEITIG |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | Macaca mulatta | 51.60 | 3 | PREDICTED: amphiphysin (Stiff-Man syndrome with br |
| 2 | Homo sapiens | 47.00 | 8 | amphiphysin isoform 2 |
| 3 | Pan troglodytes | 47.00 | 1 | PREDICTED: amphiphysin |
| 4 | synthetic construct | 47.00 | 1 | amphiphysin |
| 5 | Rattus norvegicus | 35.00 | 1 | amphiphysin 1 |
| 6 | Bos taurus | 34.30 | 1 | hypothetical protein LOC614722 |
| 7 | Canis familiaris | 33.50 | 1 | PREDICTED: similar to Amphiphysin |
Top-ranked sequences
| organism | matching |
|---|---|
| Macaca mulatta | Query 460 GEGVSLEEAKIGXXXXXXXXXXXXXXXXXX#XVPQEKVIPSVVIEPASN 507 |||||||||||| # ||||||||||||||||| Sbjct 460 GEGVSLEEAKIGTETEGAESAQAEAEELAA#TVPQEKVIPSVVIEPASN 507 |
| Homo sapiens | Query 460 GEGVSLEEAKIGXXXXXXXXXXXXXXXXXX#X-VPQEKVIPSVVIEPASN 507 |||||||||||| # ||||||||||||||||| Sbjct 460 GEGVSLEEAKIGTETTEGAESAQPEAEELE#ATVPQEKVIPSVVIEPASN 508 |
| Pan troglodytes | Query 460 GEGVSLEEAKIGXXXXXXXXXXXXXXXXXX#X-VPQEKVIPSVVIEPASN 507 |||||||||||| # ||||||||||||||||| Sbjct 737 GEGVSLEEAKIGTETTEGAESAQPEAEELE#VTVPQEKVIPSVVIEPASN 785 |
| synthetic construct | Query 460 GEGVSLEEAKIGXXXXXXXXXXXXXXXXXX#X-VPQEKVIPSVVIEPASN 507 |||||||||||| # ||||||||||||||||| Sbjct 502 GEGVSLEEAKIGTETTEGAESAQPEAEELE#ATVPQEKVIPSVVIEPASN 550 |
| Rattus norvegicus | Query 492 PQEKVIPSVVIEPASN 507 |||||||||||||||| Query 492 PQEKVIPSVVIEPASN 507 |
| Bos taurus | Query 460 GEGVSLEEAKIGXXXXXXXXXXXXXXXXXX#XVP-QEKVIPSVVIEPASN 507 | | |||||| # | ||||||||||+|||| Sbjct 499 GAGASLEEAKTDIEATEAIDGDRSQLEETE#AVAAQEKVIPSVVIQPASN 547 |
| Canis familiaris | Query 460 GEGVSLEEAKIGXXXXXXXXXXXXXXXXXX#XV-PQEKVIPSVVIEPASN 507 ||| +|||+ # | | |||||||||||||| Sbjct 653 GEGAGVEEARPDTEATQAVDGDRSQLEEAE#AVAPLEKVIPSVVIEPASN 701 |
[Site 2] FEDNFVPEIS333-VTTPSQNEVP
Ser333  Val
Val
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Phe324 | Glu325 | Asp326 | Asn327 | Phe328 | Val329 | Pro330 | Glu331 | Ile332 | Ser333 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Val334 | Thr335 | Thr336 | Pro337 | Ser338 | Gln339 | Asn340 | Glu341 | Val342 | Pro343 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| PPLPKVTPTKELRQENIISFFEDNFVPEISVTTPSQNEVPEVKKEETLLDLDFDPFKPEV |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | Macaca mulatta | 125.00 | 4 | PREDICTED: amphiphysin (Stiff-Man syndrome with br |
| 2 | Homo sapiens | 123.00 | 18 | amphiphysin isoform 2 |
| 3 | Pan troglodytes | 123.00 | 6 | PREDICTED: amphiphysin |
| 4 | Canis familiaris | 123.00 | 2 | PREDICTED: similar to Amphiphysin |
| 5 | synthetic construct | 123.00 | 1 | amphiphysin |
| 6 | Bos taurus | 122.00 | 2 | hypothetical protein LOC614722 |
| 7 | Mus musculus | 116.00 | 4 | amphiphysin |
| 8 | Rattus norvegicus | 116.00 | 3 | amphiphysin 1 |
| 9 | Monodelphis domestica | 116.00 | 2 | PREDICTED: similar to cytokine receptor common gam |
| 10 | Gallus gallus | 110.00 | 1 | amphiphysin (Stiff-Man syndrome with breast cancer |
| 11 | Xenopus tropicalis | 109.00 | 3 | Unknown (protein for MGC:122074) |
| 12 | Xenopus laevis | 107.00 | 1 | hypothetical protein LOC432150 |
| 13 | Danio rerio | 73.20 | 1 | amphiphysin |
| 14 | Lampetra fluviatilis | 63.20 | 1 | amphiphysin |
| 15 | N/A | 59.70 | 1 | A |
| 16 | Tetraodon nigroviridis | 56.20 | 1 | unnamed protein product |
Top-ranked sequences
| organism | matching |
|---|---|
| Macaca mulatta | Query 304 PPLPKVTPTKELRQENIISFFEDNFVPEIS#VTTPSQNEVPEVKKEETLLDLDFDPFKPEV 363 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 304 PPLPKVTPTKELRQENIISFFEDNFVPEIS#VTTPSQNEVPEVKKEETLLDLDFDPFKPEV 363 |
| Homo sapiens | Query 304 PPLPKVTPTKELRQENIISFFEDNFVPEIS#VTTPSQNEVPEVKKEETLLDLDFDPFKPEV 363 ||||||||||||+|||||||||||||||||#|||||||||||||||||||||||||||||| Sbjct 304 PPLPKVTPTKELQQENIISFFEDNFVPEIS#VTTPSQNEVPEVKKEETLLDLDFDPFKPEV 363 |
| Pan troglodytes | Query 304 PPLPKVTPTKELRQENIISFFEDNFVPEIS#VTTPSQNEVPEVKKEETLLDLDFDPFKPEV 363 ||||||||||||+|||||||||||||||||#|||||||||||||||||||||||||||||| Sbjct 407 PPLPKVTPTKELQQENIISFFEDNFVPEIS#VTTPSQNEVPEVKKEETLLDLDFDPFKPEV 466 |
| Canis familiaris | Query 304 PPLPKVTPTKELRQENIISFFEDNFVPEIS#VTTPSQNEVPEVKKEETLLDLDFDPFKPEV 363 ||||||||||||+|||||||||||||||||#|||||||||||||||||||||||||||||| Sbjct 323 PPLPKVTPTKELQQENIISFFEDNFVPEIS#VTTPSQNEVPEVKKEETLLDLDFDPFKPEV 382 |
| synthetic construct | Query 304 PPLPKVTPTKELRQENIISFFEDNFVPEIS#VTTPSQNEVPEVKKEETLLDLDFDPFKPEV 363 ||||||||||||+|||||||||||||||||#|||||||||||||||||||||||||||||| Sbjct 304 PPLPKVTPTKELQQENIISFFEDNFVPEIS#VTTPSQNEVPEVKKEETLLDLDFDPFKPEV 363 |
| Bos taurus | Query 304 PPLPKVTPTKELRQENIISFFEDNFVPEIS#VTTPSQNEVPEVKKEETLLDLDFDPFKPEV 363 ||||||||||||+|||||||||||||||||#||||||||+|||||||||||||||||||+| Sbjct 304 PPLPKVTPTKELQQENIISFFEDNFVPEIS#VTTPSQNEIPEVKKEETLLDLDFDPFKPDV 363 |
| Mus musculus | Query 304 PPLPKVTPTKELRQENIISFFEDNFVPEIS#VTTPSQNEVPEVKKEETLLDLDFDPFKPEV 363 ||||||||||||+|||||+||||||||||+#||||||||| ||||||||||||||||||+| Sbjct 304 PPLPKVTPTKELKQENIINFFEDNFVPEIN#VTTPSQNEVLEVKKEETLLDLDFDPFKPDV 363 |
| Rattus norvegicus | Query 304 PPLPKVTPTKELRQENIISFFEDNFVPEIS#VTTPSQNEVPEVKKEETLLDLDFDPFKPEV 363 ||||||||||||+|||||+||||||||||+#||||||||| ||||||||||||||||||+| Sbjct 304 PPLPKVTPTKELQQENIINFFEDNFVPEIN#VTTPSQNEVLEVKKEETLLDLDFDPFKPDV 363 |
| Monodelphis domestica | Query 304 PPLPKVTPTKELRQENIISFFEDNFVPEIS#VTTPSQNEVPEVKKEETLLDLDFDPFKPEV 363 ||||||||||||+|||||| ||||||||||#||||||||||| ||||+|||||||||||+| Sbjct 304 PPLPKVTPTKELQQENIISLFEDNFVPEIS#VTTPSQNEVPEQKKEESLLDLDFDPFKPDV 363 |
| Gallus gallus | Query 304 PPLPKVTPTKELRQENIISFFEDNFVPEIS#VTTPSQNEVPEVKKEETLLDLDFDPFKPEV 363 |||||+||||||+|||||+ |+|||||||+#||||||||+|| || |+||||||||||||| Sbjct 304 PPLPKLTPTKELQQENIINLFDDNFVPEIN#VTTPSQNEIPETKKVESLLDLDFDPFKPEV 363 |
| Xenopus tropicalis | Query 304 PPLPKVTPTKELRQENIISFFEDNFVPEIS#VTTPSQNEVPEVKKEETLLDLDFDPFKPE 362 |||||+||+|||+||||||||||||||||+#|||||||| | ||||+|||||||||||| Sbjct 305 PPLPKLTPSKELQQENIISFFEDNFVPEIN#VTTPSQNEAVEEKKEESLLDLDFDPFKPE 363 |
| Xenopus laevis | Query 304 PPLPKVTPTKELRQENIISFFEDNFVPEIS#VTTPSQNEVPEVKKEETLLDLDFDPFKP 361 |||||+||+|||+||||||||||||||||+#|||||||| | ||||+||||||||||| Sbjct 309 PPLPKLTPSKELQQENIISFFEDNFVPEIN#VTTPSQNEAVEEKKEESLLDLDFDPFKP 366 |
| Danio rerio | Query 304 PPLPKVTPTKELRQENIISFFEDNFVPEIS#VTTPSQNEVPEVKKEETLLDLDFDPFKPE 362 || ||||||+||+|| || |+ | ||||#||+| || | |+|||||||||||+ Sbjct 304 PPPPKVTPTRELQQEQIIDLFDGGF-PEIS#VTSPQPNERP----GESLLDLDFDPFKPD 357 |
| Lampetra fluviatilis | Query 304 PPLPKVTPTKELRQENIISFFEDNFVPEIS#VTTPSQNEVPEVKKEETLLDLDFDPFKPE 362 || ||+||+||++|| | | |+|||||+ |# + |++ | | +|||+|||| ||+ Sbjct 303 PPPPKLTPSKEVQQEAIFSLFDDNFVPDFS#-SAPAE---AAKKDEGSLLDIDFDPLKPD 357 |
| N/A | Query 304 PPLPKVTPTKELRQENIISFFEDNFVPEIS#VTTPSQ 339 || || ||+||++|| |+| ||| ||||||#|||||| Sbjct 42 PPPPKHTPSKEVKQEQILSLFEDTFVPEIS#VTTPSQ 77 |
| Tetraodon nigroviridis | Query 304 PPLPKVTPTKELRQENIISFFEDNFVPEIS#VTTPSQNEVPEVKKEETLLDLDFDPFKPEV 363 || |||||+|+++ |||++ |+ |+||#||+|++ + | | +|||+| | | | Sbjct 212 PPPPKVTPSKDMKAENIVNLFDAAAAPDIS#VTSPTEFDRPAV---SSLLDVDLDSFTSTV 268 |
[Site 3] GAESAQAEAE485-ELAATVPQEK
Glu485  Glu
Glu
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Gly476 | Ala477 | Glu478 | Ser479 | Ala480 | Gln481 | Ala482 | Glu483 | Ala484 | Glu485 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Glu486 | Leu487 | Ala488 | Ala489 | Thr490 | Val491 | Pro492 | Gln493 | Glu494 | Lys495 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| TLPAGEGVSLEEAKIGTETEGAESAQAEAEELAATVPQEKVIPSVVIEPASNHEEEGEHE |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | Macaca mulatta | 77.40 | 3 | PREDICTED: amphiphysin (Stiff-Man syndrome with br |
| 2 | Homo sapiens | 68.90 | 8 | amphiphysin isoform 2 |
| 3 | synthetic construct | 68.90 | 1 | amphiphysin |
| 4 | Pan troglodytes | 66.20 | 1 | PREDICTED: amphiphysin |
| 5 | Bos taurus | 52.80 | 1 | hypothetical protein LOC614722 |
| 6 | Canis familiaris | 48.90 | 1 | PREDICTED: similar to Amphiphysin |
| 7 | Mus musculus | 48.50 | 3 | amphiphysin |
| 8 | Rattus norvegicus | 48.10 | 1 | amphiphysin 1 |
| 9 | Monodelphis domestica | 42.00 | 1 | PREDICTED: similar to amphiphysin I variant CT2 |
| 10 | Xenopus laevis | 36.20 | 1 | hypothetical protein LOC432150 |
| 11 | Gallus gallus | 36.20 | 1 | amphiphysin (Stiff-Man syndrome with breast cancer |
Top-ranked sequences
| organism | matching |
|---|---|
| Macaca mulatta | Query 456 TLPAGEGVSLEEAKIGXXXXXXXXXXXXXX#XXXXXVPQEKVIPSVVIEPASNHEEEGEHE 515 |||||||||||||||| # ||||||||||||||||||||||||| Sbjct 456 TLPAGEGVSLEEAKIGTETEGAESAQAEAE#ELAATVPQEKVIPSVVIEPASNHEEEGEHE 515 |
| Homo sapiens | Query 456 TLPAGEGVSLEEAKIGXXXXXXXXXXXXXX#XXXXX-VPQEKVIPSVVIEPASNHEEEGEH 514 |+|||||||||||||| # |||||||||||||||||||||||+ Sbjct 456 TVPAGEGVSLEEAKIGTETTEGAESAQPEA#EELEATVPQEKVIPSVVIEPASNHEEEGEN 515 Query 515 E 515 | Query 515 E 515 |
| synthetic construct | Query 456 TLPAGEGVSLEEAKIGXXXXXXXXXXXXXX#XXXXX-VPQEKVIPSVVIEPASNHEEEGEH 514 |+|||||||||||||| # |||||||||||||||||||||||+ Sbjct 498 TVPAGEGVSLEEAKIGTETTEGAESAQPEA#EELEATVPQEKVIPSVVIEPASNHEEEGEN 557 Query 515 E 515 | Query 515 E 515 |
| Pan troglodytes | Query 456 TLPAGEGVSLEEAKIGXXXXXXXXXXXXXX#XXXXX-VPQEKVIPSVVIEPASNHEEEGEH 514 |+|||||||||||||| # ||||||||||||||||||| |||+ Sbjct 733 TVPAGEGVSLEEAKIGTETTEGAESAQPEA#EELEVTVPQEKVIPSVVIEPASNHEGEGEN 792 Query 515 E 515 | Query 515 E 515 |
| Bos taurus | Query 457 LPAGEGVSLEEAKIGXXXXXXXXXXXXXX#XXXXXVP-QEKVIPSVVIEPASNHEEEGEHE 515 |||| | |||||| # | ||||||||||+||||+| ||||| Sbjct 496 LPAGAGASLEEAKTDIEATEAIDGDRSQL#EETEAVAAQEKVIPSVVIQPASNNEGEGEHE 555 |
| Canis familiaris | Query 457 LPAGEGVSLEEAKIGXXXXXXXXXXXXXX#XXXXXV-PQEKVIPSVVIEPASNHEEEGEHE 515 || ||| +|||+ # | | ||||||||||||||+| ||+|| Sbjct 650 LPPGEGAGVEEARPDTEATQAVDGDRSQL#EEAEAVAPLEKVIPSVVIEPASNNEGEGDHE 709 |
| Mus musculus | Query 457 LPAGEGVSLEEAKI-GXXXXXXXXXXXXXX#XXXXXVPQEKVIPSVVIEPASNHEEEGEHE 515 |||||| | | ||| | # || |||||||||||||| ||||+ Sbjct 493 LPAGEGGSPEGAKIDGESTELAISESPQPV#EPEAGAPQ--VIPSVVIEPASNHEGEGEHQ 550 |
| Rattus norvegicus | Query 457 LPAGEGVSLEEAKIGXXXXXXXXXXXXXX#XXXXX-VPQEKVIPSVVIEPASNHEEEGEHE 515 |||||| | | ||| # |||||||||||||||||| | ||+ Sbjct 489 LPAGEGESPEGAKIDVESTELASSESPQA#AELEAGAPQEKVIPSVVIEPASNHEGE-EHQ 547 |
| Monodelphis domestica | Query 493 QEKVIPSVVIEPASNHEEEGEHE 515 ||||||||||||||||| | | | Sbjct 79 QEKVIPSVVIEPASNHEGEDEQE 101 |
| Xenopus laevis | Query 492 PQEKVIPSVVIEPASNHEEEGEHE 515 |+ +|||||||||||||+| + + Sbjct 524 PEITIIPSVVIEPASNHEDEHDDD 547 |
| Gallus gallus | Query 491 VPQEKV--IPSVVIEPASNHEEEGE 513 | |||| |||||||||||+| ||| Sbjct 525 VSQEKVSSIPSVVIEPASNNEGEGE 549 |
[Site 4] DLWTTSADLV392-QPASGGSFNG
Val392  Gln
Gln
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Asp383 | Leu384 | Trp385 | Thr386 | Thr387 | Ser388 | Ala389 | Asp390 | Leu391 | Val392 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Gln393 | Pro394 | Ala395 | Ser396 | Gly397 | Gly398 | Ser399 | Phe400 | Asn401 | Gly402 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| VTPAGSAGVTHSPMSQTLPWDLWTTSADLVQPASGGSFNGFTQPQDTSLFAMQTDQSMIC |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | Macaca mulatta | 128.00 | 3 | PREDICTED: amphiphysin (Stiff-Man syndrome with br |
| 2 | Homo sapiens | 125.00 | 11 | amphiphysin isoform 2 |
| 3 | synthetic construct | 125.00 | 1 | amphiphysin |
| 4 | Rattus norvegicus | 112.00 | 1 | amphiphysin 1 |
| 5 | Bos taurus | 108.00 | 2 | hypothetical protein LOC614722 |
| 6 | Mus musculus | 107.00 | 2 | amphiphysin |
| 7 | Monodelphis domestica | 97.40 | 1 | PREDICTED: similar to cytokine receptor common gam |
| 8 | Gallus gallus | 77.80 | 1 | amphiphysin (Stiff-Man syndrome with breast cancer |
| 9 | Xenopus tropicalis | 68.90 | 1 | Unknown (protein for MGC:122074) |
| 10 | Pan troglodytes | 61.60 | 1 | PREDICTED: amphiphysin |
| 11 | Xenopus laevis | 60.10 | 1 | hypothetical protein LOC432150 |
| 12 | Canis familiaris | 59.30 | 1 | PREDICTED: similar to Amphiphysin |
| 13 | Danio rerio | 43.90 | 1 | amphiphysin |
Top-ranked sequences
| organism | matching |
|---|---|
| Macaca mulatta | Query 363 VTPAGSAGVTHSPMSQTLPWDLWTTSADLV#QPASGGSFNGFTQPQDTSLFAMQTDQSMIC 422 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 363 VTPAGSAGVTHSPMSQTLPWDLWTTSADLV#QPASGGSFNGFTQPQDTSLFAMQTDQSMIC 422 |
| Homo sapiens | Query 363 VTPAGSAGVTHSPMSQTLPWDLWTTSADLV#QPASGGSFNGFTQPQDTSLFAMQTDQSMIC 422 |||||||||||||||||||||||||| |||#|||||||||||||||||||| ||||||||| Sbjct 363 VTPAGSAGVTHSPMSQTLPWDLWTTSTDLV#QPASGGSFNGFTQPQDTSLFTMQTDQSMIC 422 |
| synthetic construct | Query 363 VTPAGSAGVTHSPMSQTLPWDLWTTSADLV#QPASGGSFNGFTQPQDTSLFAMQTDQSMIC 422 |||||||||||||||||||||||||| |||#|||||||||||||||||||| ||||||||| Sbjct 363 VTPAGSAGVTHSPMSQTLPWDLWTTSTDLV#QPASGGSFNGFTQPQDTSLFTMQTDQSMIC 422 |
| Rattus norvegicus | Query 363 VTPAGSAGVTHSPMSQTLPWDLWTTSADLV#QPASGGSFNGFTQPQDTSLFAMQTDQSM 420 ||||||| ||||||||||||||||| |||#||||||||| |||||||||| |||||+| Sbjct 363 VTPAGSAAATHSPMSQTLPWDLWTTSTDLV#QPASGGSFNDFTQPQDTSLFTMQTDQNM 420 |
| Bos taurus | Query 363 VTPAGSAGVTHSPMSQTLPWDLWTTSADLV#QPASGGSFNGFTQPQDTSLFAMQTDQSM 420 |||||||||||||||||||||||||| |||#|||||||||||| |||||| || |||| Sbjct 363 VTPAGSAGVTHSPMSQTLPWDLWTTSTDLV#QPASGGSFNGFT--QDTSLFTMQADQSM 418 |
| Mus musculus | Query 363 VTPAGSAGVTHSPMSQTLPWDLWTTSADLV#QPASGGSFNGFTQPQDTSLFAMQTDQSM 420 | ||||| ||||||||||||||||| |||#||||||||| ||| |||||| |||||+| Sbjct 363 VAPAGSAAATHSPMSQTLPWDLWTTSTDLV#QPASGGSFNDFTQAQDTSLFTMQTDQNM 420 |
| Monodelphis domestica | Query 363 VTPAGSAGVTHSPMSQTLPWDLWTTSADLV#QPASGGSFNGFTQPQDTSLFAMQTDQS 419 ||| ||||+ |||||||||||||||++||#|||| |+||||||||||||| | |+ Sbjct 363 VTPVGSAGIAPSPMSQTLPWDLWTTSSELV#QPASSGTFNGFTQPQDTSLFDMPASQT 419 |
| Gallus gallus | Query 368 SAGVTHSPMSQTLPWDLWTTSADLV#QPASGGSFNGFTQPQDTSLFAMQTDQSM 420 | ||||||||||||||||||+++||#|||| +|||| |||+ ||+|+++++ Sbjct 365 STGVTHSPMSQTLPWDLWTTTSELV#QPASSTAFNGFA--QDTTAFAVQSNENV 415 |
| Xenopus tropicalis | Query 372 THSPMSQTLPWDLWTTSADLV#QPASGGSFNGFTQPQDTS-LFAMQTDQS 419 +|||+|||||||||||+ ++|#|||| |+||||||| ++ || + +|+ Sbjct 372 SHSPLSQTLPWDLWTTNNEVV#QPASTGAFNGFTQPDASAPLFTLPLEQN 420 |
| Pan troglodytes | Query 363 VTPAGSAGVTHSPMSQTLPWDLWTTSADLV#QP 394 |||||||||||||||||||||||||| |||#|| Sbjct 466 VTPAGSAGVTHSPMSQTLPWDLWTTSTDLV#QP 497 Query 378 QTLPWDLWTTSADLV#QPASGGSFNGFTQPQDTSLFAMQTDQSMIC 422 | + ||+ + #| |||||||||||||||||| ||||||+|| Sbjct 656 QKIEDSLWS-GVEAC#QKASGGSFNGFTQPQDTSLFTMQTDQSVIC 699 |
| Xenopus laevis | Query 366 AGSAGV--THSPMSQTLPWDLWT-------TSADLV#QPASGGSFNGFTQPQDTS-LFAMQ 415 | + |+ +|||+|||||||||| |+ ++|#|||| | ||||||| ++ || + Sbjct 368 AATVGIVQSHSPISQTLPWDLWTCFCPSTQTNNEVV#QPASTGVFNGFTQPDASAPLFTLP 427 Query 416 TDQS 419 ++++ Sbjct 428 SEKN 431 |
| Canis familiaris | Query 363 VTPAGSAGVTHSPMSQTLPWDLWTTSADLV#QP 394 | | || ||||||||||||||||||| |||#|| Sbjct 382 VAPVGSVGVTHSPMSQTLPWDLWTTSTDLV#QP 413 Query 378 QTLPWDLWTTSADLV#QPASGGSFNGFTQPQDTSLFAMQTDQSMIC 422 | + +| + #+ ||||+||||||||||||| ||||||||| Sbjct 572 QKIEDSVWA-GVETC#EKASGGTFNGFTQPQDTSLFTMQTDQSMIC 615 |
| Danio rerio | Query 370 GVTHSPMSQTLPWDLWTTSADLV#QPASGGSFNGFT 404 | | ||+|||||||||| | #||| + ||| Sbjct 363 GQTQSPVSQTLPWDLWT--GDAA#QPAQPATDAGFT 395 |
[Site 5] SAGVTHSPMS377-QTLPWDLWTT
Ser377  Gln
Gln
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Ser368 | Ala369 | Gly370 | Val371 | Thr372 | His373 | Ser374 | Pro375 | Met376 | Ser377 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Gln378 | Thr379 | Leu380 | Pro381 | Trp382 | Asp383 | Leu384 | Trp385 | Thr386 | Thr387 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| EETLLDLDFDPFKPEVTPAGSAGVTHSPMSQTLPWDLWTTSADLVQPASGGSFNGFTQPQ |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | Macaca mulatta | 129.00 | 3 | PREDICTED: amphiphysin (Stiff-Man syndrome with br |
| 2 | Homo sapiens | 128.00 | 14 | amphiphysin isoform 2 |
| 3 | synthetic construct | 128.00 | 1 | amphiphysin |
| 4 | Bos taurus | 122.00 | 2 | hypothetical protein LOC614722 |
| 5 | Rattus norvegicus | 120.00 | 2 | amphiphysin 1 |
| 6 | Mus musculus | 115.00 | 3 | amphiphysin |
| 7 | Monodelphis domestica | 113.00 | 2 | PREDICTED: similar to cytokine receptor common gam |
| 8 | Canis familiaris | 96.30 | 2 | PREDICTED: similar to Amphiphysin |
| 9 | Gallus gallus | 94.00 | 1 | amphiphysin (Stiff-Man syndrome with breast cancer |
| 10 | Xenopus tropicalis | 91.30 | 1 | Unknown (protein for MGC:122074) |
| 11 | Xenopus laevis | 80.90 | 1 | hypothetical protein LOC432150 |
| 12 | Danio rerio | 65.50 | 1 | amphiphysin |
| 13 | Pan troglodytes | 40.80 | 4 | PREDICTED: bridging integrator 1 isoform 9 |
| 14 | Lampetra fluviatilis | 36.20 | 1 | amphiphysin |
| 15 | Santalum album | 32.30 | 1 | proline rich protein |
Top-ranked sequences
| organism | matching |
|---|---|
| Macaca mulatta | Query 348 EETLLDLDFDPFKPEVTPAGSAGVTHSPMS#QTLPWDLWTTSADLVQPASGGSFNGFTQPQ 407 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 348 EETLLDLDFDPFKPEVTPAGSAGVTHSPMS#QTLPWDLWTTSADLVQPASGGSFNGFTQPQ 407 |
| Homo sapiens | Query 348 EETLLDLDFDPFKPEVTPAGSAGVTHSPMS#QTLPWDLWTTSADLVQPASGGSFNGFTQPQ 407 ||||||||||||||||||||||||||||||#||||||||||| |||||||||||||||||| Sbjct 348 EETLLDLDFDPFKPEVTPAGSAGVTHSPMS#QTLPWDLWTTSTDLVQPASGGSFNGFTQPQ 407 |
| synthetic construct | Query 348 EETLLDLDFDPFKPEVTPAGSAGVTHSPMS#QTLPWDLWTTSADLVQPASGGSFNGFTQPQ 407 ||||||||||||||||||||||||||||||#||||||||||| |||||||||||||||||| Sbjct 348 EETLLDLDFDPFKPEVTPAGSAGVTHSPMS#QTLPWDLWTTSTDLVQPASGGSFNGFTQPQ 407 |
| Bos taurus | Query 348 EETLLDLDFDPFKPEVTPAGSAGVTHSPMS#QTLPWDLWTTSADLVQPASGGSFNGFTQ 405 ||||||||||||||+|||||||||||||||#||||||||||| |||||||||||||||| Sbjct 348 EETLLDLDFDPFKPDVTPAGSAGVTHSPMS#QTLPWDLWTTSTDLVQPASGGSFNGFTQ 405 |
| Rattus norvegicus | Query 348 EETLLDLDFDPFKPEVTPAGSAGVTHSPMS#QTLPWDLWTTSADLVQPASGGSFNGFTQPQ 407 ||||||||||||||+||||||| ||||||#||||||||||| |||||||||||| ||||| Sbjct 348 EETLLDLDFDPFKPDVTPAGSAAATHSPMS#QTLPWDLWTTSTDLVQPASGGSFNDFTQPQ 407 |
| Mus musculus | Query 348 EETLLDLDFDPFKPEVTPAGSAGVTHSPMS#QTLPWDLWTTSADLVQPASGGSFNGFTQPQ 407 ||||||||||||||+| ||||| ||||||#||||||||||| |||||||||||| ||| | Sbjct 348 EETLLDLDFDPFKPDVAPAGSAAATHSPMS#QTLPWDLWTTSTDLVQPASGGSFNDFTQAQ 407 |
| Monodelphis domestica | Query 348 EETLLDLDFDPFKPEVTPAGSAGVTHSPMS#QTLPWDLWTTSADLVQPASGGSFNGFTQPQ 407 ||+|||||||||||+||| ||||+ ||||#|||||||||||++|||||| |+|||||||| Sbjct 348 EESLLDLDFDPFKPDVTPVGSAGIAPSPMS#QTLPWDLWTTSSELVQPASSGTFNGFTQPQ 407 |
| Canis familiaris | Query 348 EETLLDLDFDPFKPEVTPAGSAGVTHSPMS#QTLPWDLWTTSADLVQP 394 |||||||||||||||| | || ||||||||#||||||||||| ||||| Sbjct 367 EETLLDLDFDPFKPEVAPVGSVGVTHSPMS#QTLPWDLWTTSTDLVQP 413 |
| Gallus gallus | Query 349 ETLLDLDFDPFKPEVTPAGSAGVTHSPMS#QTLPWDLWTTSADLVQPASGGSFNGFTQ 405 |+||||||||||||| | ||||||||#||||||||||+++|||||| +|||| | Sbjct 349 ESLLDLDFDPFKPEVV---STGVTHSPMS#QTLPWDLWTTTSELVQPASSTAFNGFAQ 402 |
| Xenopus tropicalis | Query 348 EETLLDLDFDPFKPEVTPAGSAGVTHSPMS#QTLPWDLWTTSADLVQPASGGSFNGFTQP 406 ||+|||||||||||| | +|||+|#||||||||||+ ++||||| |+||||||| Sbjct 349 EESLLDLDFDPFKPESATVGIVQ-SHSPLS#QTLPWDLWTTNNEVVQPASTGAFNGFTQP 406 |
| Xenopus laevis | Query 348 EETLLDLDFDPFKPEVTPAGSAGVTHSPMS#QTLPWDLWT-------TSADLVQPASGGSF 400 ||+||||||||||| | +|||+|#||||||||| |+ ++||||| | | Sbjct 353 EESLLDLDFDPFKPGAATVGIVQ-SHSPIS#QTLPWDLWTCFCPSTQTNNEVVQPASTGVF 411 Query 401 NGFTQP 406 |||||| Query 401 NGFTQP 406 |
| Danio rerio | Query 349 ETLLDLDFDPFKPEV-TPAGSAGVTHSPMS#QTLPWDLWTTSADLVQPASGGSFNGFT 404 |+|||||||||||+ || | | ||+|#||||||||| | ||| + ||| Sbjct 344 ESLLDLDFDPFKPDTSTPIGQ---TQSPVS#QTLPWDLWT--GDAAQPAQPATDAGFT 395 |
| Pan troglodytes | Query 348 EETLLDLDFDPFKPEVTPAGSAGVTHSPMS#QTLPWDLW 385 + +|||||||| | +| + +| #|++||||| Sbjct 386 QASLLDLDFDPLPPVTSPVKAP----TPSG#QSIPWDLW 419 |
| Lampetra fluviatilis | Query 348 EETLLDLDFDPFKPE--VTPAGSAGVTHSPMS#QTLPWDL-WTTSADLVQP 394 | +|||+|||| ||+ || | ++#||| +|| | | ||| Sbjct 343 EGSLLDIDFDPLKPDTATTPMSPA------IN#QTLLFDLAWEASPQQVQP 386 |
| Santalum album | Query 352 LDLDFDPFKPEVTPAGSAGVTHSPMS#QTLPWDLWTTSADLVQPAS 396 | | | | | |+| | +|| |# +|| | + | | | Sbjct 76 LGLPFPPLDPRKPPSGPFGNPNSPAS#-LIPWRRWDHTPPLFNPPS 119 |
* References
None.