XSB2179 : PREDICTED: hypothetical protein [Ornithorhynchus anatinus]
[ CaMP Format ]
This entry is computationally expanded from SB0055
* Basic Information
| Organism | Ornithorhynchus anatinus (platypus) |
| Protein Names | hypothetical protein |
| Gene Names | LOC100079107; Derived by automated computational analysis using gene prediction method: GNOMON. Supporting evidence includes similarity to: 14 Proteins |
| Gene Locus | Not available |
| GO Function | Not available |
| Entrez Protein | Entrez Nucleotide | Entrez Gene | UniProt | OMIM | HGNC | HPRD | KEGG |
|---|---|---|---|---|---|---|---|
| XP_001510097 | XM_001510047 | 100079107 | N/A | N/A | N/A | N/A | N/A |
* Information From OMIM
Not Available.
* Structure Information
1. Primary Information
Length: 675 aa
Average Mass: 76.862 kDa
Monoisotopic Mass: 76.812 kDa
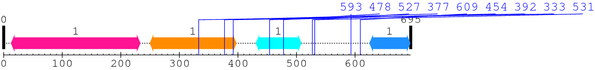
2. Domain Information
Annotated Domains: Not Available.
Computationally Assigned Domains (Pfam+HMMER):
| domain name | begin | end | score | e-value |
|---|---|---|---|---|
| C1_1 1. | 38 | 90 | 83.2 | 9.4e-22 |
| C1_1 2. | 103 | 155 | 92.5 | 1.5e-24 |
| C2 1. | 174 | 261 | 136.7 | 7.3e-38 |
| --- cleavage 310 --- | ||||
| --- cleavage 325 --- | ||||
| --- cleavage 317 --- | ||||
| Pkinase 1. | 342 | 600 | 290.8 | 3.1e-84 |
| Pkinase_C 1. | 620 | 665 | 57.0 | 7.2e-14 |
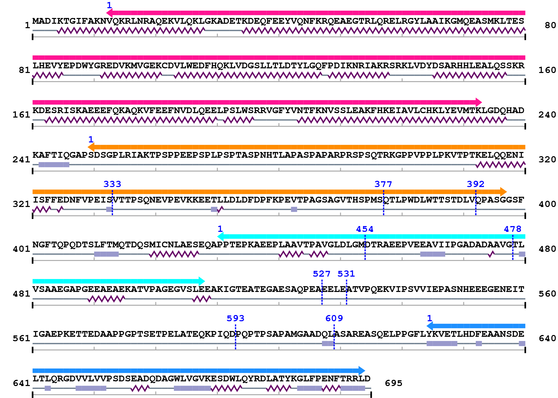
3. Sequence Information
Fasta Sequence: XSB2179.fasta
Amino Acid Sequence and Secondary Structures (PsiPred):
4. 3D Information
Not Available.
* Cleavage Information
3 [sites]
Cleavage sites (±10aa)
[Site 1] ELRQKFEKAK310-LGPAGNKVIG
Lys310  Leu
Leu
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Glu301 | Leu302 | Arg303 | Gln304 | Lys305 | Phe306 | Glu307 | Lys308 | Ala309 | Lys310 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Leu311 | Gly312 | Pro313 | Ala314 | Gly315 | Asn316 | Lys317 | Val318 | Ile319 | Gly320 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| QEEGEYYNVPIPEGDEEGNVELRQKFEKAKLGPAGNKVIGPSEDRKSSVPSNNLDRVKLT |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | Gallus gallus | 113.00 | 3 | protein kinase C, alpha |
| 2 | Rattus norvegicus | 112.00 | 4 | PREDICTED: similar to Protein kinase C alpha type |
| 3 | Homo sapiens | 111.00 | 11 | protein kinase C, alpha |
| 4 | N/A | 111.00 | 11 | - |
| 5 | Mus musculus | 111.00 | 6 | protein kinase C |
| 6 | Oryctolagus cuniculus | 111.00 | 5 | KPCA_RABIT Protein kinase C alpha type (PKC-alpha) |
| 7 | Bos taurus | 111.00 | 4 | protein kinase C, alpha |
| 8 | Canis familiaris | 111.00 | 3 | PREDICTED: similar to Protein kinase C, alpha type |
| 9 | synthetic construct | 111.00 | 1 | protein kinase C alpha |
| 10 | Monodelphis domestica | 104.00 | 4 | PREDICTED: similar to protein kinase C |
| 11 | Latimeria chalumnae | 102.00 | 2 | protein kinase C alpha |
| 12 | Xenopus tropicalis | 101.00 | 1 | hypothetical protein LOC780209 |
| 13 | Xenopus laevis | 95.50 | 1 | protein kinase C, alpha |
| 14 | Protopterus dolloi | 87.80 | 2 | protein kinase C alpha |
| 15 | Takifugu rubripes | 80.10 | 1 | protein kinase C, alpha type |
| 16 | Scyliorhinus canicula | 70.10 | 2 | protein kinase C alpha |
| 17 | Danio rerio | 67.00 | 7 | protein kinase C, beta 1 |
| 18 | Tetraodon nigroviridis | 67.00 | 3 | unnamed protein product |
| 19 | Macaca mulatta | 58.90 | 8 | PREDICTED: protein kinase C, beta 1 isoform 5 |
| 20 | Petromyzon marinus | 56.20 | 1 | protein kinase C |
| 21 | Myxine glutinosa | 48.90 | 1 | protein kinase C |
| 22 | Macaca fascicularis | 39.70 | 1 | KPCG_MACFA Protein kinase C gamma type (PKC-gamma) |
| 23 | Branchiostoma lanceolatum | 36.60 | 1 | protein kinase C |
| 24 | Aedes aegypti | 33.90 | 1 | protein kinase c |
| 25 | Anopheles gambiae str. PEST | 33.50 | 1 | ENSANGP00000009078 |
| 26 | Streptococcus pyogenes MGAS10750 | 32.70 | 1 | DNA mismatch repair protein mutL |
| 27 | Streptococcus pyogenes MGAS10270 | 32.70 | 1 | DNA mismatch repair protein mutL |
| 28 | Streptococcus pyogenes MGAS9429 | 32.70 | 1 | DNA mismatch repair protein |
| 29 | Apis mellifera | 32.30 | 1 | PREDICTED: similar to Protein C kinase 53E CG6622- |
| 30 | Streptococcus pyogenes MGAS315 | 32.30 | 1 | DNA mismatch repair protein |
| 31 | Bombyx mori | 32.30 | 1 | conventional protein kinase C |
Top-ranked sequences
| organism | matching |
|---|---|
| Gallus gallus | Query 281 QEEGEYYNVPIPEGDEEGNVELRQKFEKAK#LGPAGNKVIGPSEDRKSSVPSNNLDRVKLT 340 ||||||||||||+ ||+|| ||||||||||#||||||||| ||||| |||||||||||||| Sbjct 280 QEEGEYYNVPIPDADEDGNAELRQKFEKAK#LGPAGNKVITPSEDRNSSVPSNNLDRVKLT 339 |
| Rattus norvegicus | Query 281 QEEGEYYNVPIPEGDEEGNVELRQKFEKAK#LGPAGNKVIGPSEDRKSSVPSNNLDRVKLT 340 ||||||||||||||||||||||||||||||#||||||||| |||||| ||||||||||| Sbjct 280 QEEGEYYNVPIPEGDEEGNVELRQKFEKAK#LGPAGNKVISPSEDRKQ--PSNNLDRVKLT 337 |
| Homo sapiens | Query 281 QEEGEYYNVPIPEGDEEGNVELRQKFEKAK#LGPAGNKVIGPSEDRKSSVPSNNLDRVKLT 340 |||||||||||||||||||+||||||||||#||||||||| |||||| ||||||||||| Sbjct 280 QEEGEYYNVPIPEGDEEGNMELRQKFEKAK#LGPAGNKVISPSEDRKQ--PSNNLDRVKLT 337 |
| N/A | Query 281 QEEGEYYNVPIPEGDEEGNVELRQKFEKAK#LGPAGNKVIGPSEDRKSSVPSNNLDRVKLT 340 |||||||||||||||||||+||||||||||#||||||||| |||||| ||||||||||| Sbjct 266 QEEGEYYNVPIPEGDEEGNMELRQKFEKAK#LGPAGNKVISPSEDRKQ--PSNNLDRVKLT 323 |
| Mus musculus | Query 281 QEEGEYYNVPIPEGDEEGNVELRQKFEKAK#LGPAGNKVIGPSEDRKSSVPSNNLDRVKLT 340 |||||||||||||||||||+||||||||||#||||||||| |||||| ||||||||||| Sbjct 280 QEEGEYYNVPIPEGDEEGNMELRQKFEKAK#LGPAGNKVISPSEDRKQ--PSNNLDRVKLT 337 |
| Oryctolagus cuniculus | Query 281 QEEGEYYNVPIPEGDEEGNVELRQKFEKAK#LGPAGNKVIGPSEDRKSSVPSNNLDRVKLT 340 ||||||||||||||||+|||||||||||||#||||||||| |||||| ||||||||||| Sbjct 280 QEEGEYYNVPIPEGDEDGNVELRQKFEKAK#LGPAGNKVISPSEDRKQ--PSNNLDRVKLT 337 |
| Bos taurus | Query 281 QEEGEYYNVPIPEGDEEGNVELRQKFEKAK#LGPAGNKVIGPSEDRKSSVPSNNLDRVKLT 340 ||||||||||||||||||||||||||||||#||||||||| |||||+ ||||||||||| Sbjct 280 QEEGEYYNVPIPEGDEEGNVELRQKFEKAK#LGPAGNKVISPSEDRRQ--PSNNLDRVKLT 337 |
| Canis familiaris | Query 281 QEEGEYYNVPIPEGDEEGNVELRQKFEKAK#LGPAGNKVIGPSEDRKSSVPSNNLDRVKLT 340 ||||||||||||||||||||||||||||||#||||||||| |||||+ ||||||||||| Sbjct 198 QEEGEYYNVPIPEGDEEGNVELRQKFEKAK#LGPAGNKVISPSEDRRQ--PSNNLDRVKLT 255 |
| synthetic construct | Query 281 QEEGEYYNVPIPEGDEEGNVELRQKFEKAK#LGPAGNKVIGPSEDRKSSVPSNNLDRVKLT 340 |||||||||||||||||||+||||||||||#||||||||| |||||| ||||||||||| Sbjct 280 QEEGEYYNVPIPEGDEEGNMELRQKFEKAK#LGPAGNKVISPSEDRKQ--PSNNLDRVKLT 337 |
| Monodelphis domestica | Query 281 QEEGEYYNVPIPEGDEEGNVELRQKFEKAK#LGPAGNKVIGPSEDRKSSVPSNNLDRVKLT 340 |||||||||||||||||||+||||||||||#||| ||||| |+||| |+||||||||| Sbjct 280 QEEGEYYNVPIPEGDEEGNMELRQKFEKAK#LGPVGNKVISSSDDRK---PTNNLDRVKLT 336 |
| Latimeria chalumnae | Query 281 QEEGEYYNVPIPEGDEEGNVELRQKFEKAK#LGPAGNKVIGPSEDRKSSVPSNNLDRVKLT 340 ||||||||||| | |+ |||||||||||||#|||||+||| | |+|||| ||||||||||| Sbjct 202 QEEGEYYNVPISEADD-GNVELRQKFEKAK#LGPAGSKVISPVEERKSSTPSNNLDRVKLT 260 |
| Xenopus tropicalis | Query 281 QEEGEYYNVPIPEGDEEGNVELRQKFEKAK#LGPAGNKVIGPSEDRKSSVPSNNLDRVKLT 340 ||||||||||||| +++||+||||||||||#||||||||| |+++|+ |||||+| |+|| Sbjct 284 QEEGEYYNVPIPEEEDDGNIELRQKFEKAK#LGPAGNKVISPTDERRPYVPSNNIDSVRLT 343 |
| Xenopus laevis | Query 281 QEEGEYYNVPIPEGDEEGNVELRQKFEKAK#LGPAGNKVIGPSEDRKSSVPSNNLDRVKLT 340 ||||||||||||| |+ ||+||||||||||#||||||||| |+ +|+ +||||+| ++|| Sbjct 284 QEEGEYYNVPIPEADD-GNLELRQKFEKAK#LGPAGNKVINPTGERRPYIPSNNIDSIRLT 342 |
| Protopterus dolloi | Query 281 QEEGEYYNVPIPEGDEEGNVELRQKFEKAK#LGPAGNKVIGPSEDRKSSVPSNNLDRVKL 339 ||||||||||||+ |+ ||| ||||||||+#||||| ||| | +||||+| ||||| || Sbjct 200 QEEGEYYNVPIPDADD-GNVGLRQKFEKAR#LGPAGKKVISP--ERKSSLPLNNLDRFKL 255 |
| Takifugu rubripes | Query 281 QEEGEYYNVPIPEGDEEGNVELRQKFEKAK#LGPAGNKVIGPSEDRKSSVPSNNLDRVKL 339 ||||||||||||| |+ |+||||||||||#|| | ||| ||+ |+ |+|| |+|||+| Sbjct 278 QEEGEYYNVPIPEVDDV-NLELRQKFEKAK#LGQ-GKKVITPSDHRRFSLPSGNMDRVRL 334 |
| Scyliorhinus canicula | Query 281 QEEGEYYNVPIPEGDEEGNVELRQKFEKAK#LGPAGNKVIGPSEDRKSSVPSNNLDRVKL 339 ||||||||||| | ||+ |||+||||||#||||| | | ++| | |+|+|| | | Sbjct 213 QEEGEYYNVPIIEDDEDS--ELRKKFEKAK#LGPAGKKAI----EKKPSSPTNDLDHVHL 265 |
| Danio rerio | Query 281 QEEGEYYNVPIPEGDEEGNVELRQKFEKAK#LGPAGNKVIGPSEDRKSSVPSN-NLDRVKL 339 ||||||+|||+| |||| |||||||+||#+|| +| | | + | || | ||+|| Sbjct 279 QEEGEYFNVPVPPEGEEGNEELRQKFERAK#IGP--SKTDGSSSNAISKFDSNGNRDRMKL 336 Query 340 T 340 + Sbjct 337 S 337 |
| Tetraodon nigroviridis | Query 281 QEEGEYYNVPIPEGDEEGNVELRQKFEKAK#LGPAGNKVIGPSEDRKSSVPSN-NLDRVKL 339 ||||||+|||+| |||| |||||||+||#+|| | | | + | || | ||+|| Sbjct 216 QEEGEYFNVPVPPEGEEGNEELRQKFERAK#IGP-GKNTDGKSANAASRFDSNGNQDRMKL 274 |
| Macaca mulatta | Query 281 QEEGEYYNVPIPEGDEEGNVELRQKFEKAK#LGPAGNKVIGPSEDRKSSVP----SNNLDR 336 ||||||+|||+| ||| |||||||+||#+ | || | | ++| + | || Sbjct 280 QEEGEYFNVPVPPEGSEGNEELRQKFERAK#IS-QGTKV--PEEKTTNTVSKFDNNGNRDR 336 Query 337 VKLT 340 +||| Sbjct 337 MKLT 340 |
| Petromyzon marinus | Query 281 QEEGEYYNVPIPEGDEEGNVELRQKFEKAK#LGPAGNKVIGPSEDRKSSVP----SNNLDR 336 ||||||||||| |+|+ |+|+|||||+#|||+ | | | +|+ + |+|| Sbjct 210 QEEGEYYNVPIAPDIEDGDTEMRRKFEKAR#LGPS-VKPRASDERRANSLSAILNNANVDR 268 Query 337 VK 338 || Query 337 VK 338 |
| Myxine glutinosa | Query 281 QEEGEYYNVPI-PEGDEEGNVELRQKFEKAK#LGPAGNKVIGPSEDRKS-SVPSNNLDRVK 338 ||||||||||+ |||+| +||||+ ++++#+ + |++|+ | | + |||| Sbjct 201 QEEGEYYNVPVAPEGEE--GLELRQRLQRSQ#IARSDKNKQSPAKDQPSDSASLSGLDRVN 258 Query 339 L 339 | Query 339 L 339 |
| Macaca fascicularis | Query 281 QEEGEYYNVPIPEGDEEGNVELRQKF----------EKAK#LGPAGNKVIGPS 322 ||||||||||+ + | | | ||| |+ +#+||+ + + || Sbjct 280 QEEGEYYNVPVADAD---NCSLLQKFEACNYPLELYERVR#MGPSSSPIPSPS 328 |
| Branchiostoma lanceolatum | Query 281 QEEGEYYNVPIPEGDEEGNV-ELRQKFEKAK#L 311 ||||||||||| ||| || +|++ +| |#| Sbjct 211 QEEGEYYNVPI--ADEEENVAKLKEHLQKQK#L 240 |
| Aedes aegypti | Query 281 QEEGEYYNVPIPEGDEEGN--VELRQKFEKAK#L 311 ||||||||||+| ||| |+|+ + | #+ Sbjct 228 QEEGEYYNVPVP---EEGTDLVQLKSQMRKTS#I 257 |
| Anopheles gambiae str. PEST | Query 281 QEEGEYYNVPIPEGDEEGNVELRQKFEKAK#L 311 ||||||||||+|| + |+|+ + | #+ Sbjct 220 QEEGEYYNVPVPE-EGADLVQLKSQMRKTS#I 249 |
| Streptococcus pyogenes MGAS10750 | Query 291 IPEGD-EEGNVELRQKFEKAK#LGPAGNKVIGPSEDRKSSVPSN 332 ||| | | |+ | |||+#| | +||||| + +| | | Sbjct 395 IPETDFYSGAVDNSVKVEKAE#LLPHSEEVIGPSSVKHASRPQN 437 |
| Streptococcus pyogenes MGAS10270 | Query 291 IPEGD-EEGNVELRQKFEKAK#LGPAGNKVIGPSEDRKSSVPSN 332 ||| | | |+ | |||+#| | +||||| + +| | | Sbjct 395 IPETDFYSGAVDNSVKVEKAE#LLPHSEEVIGPSSVKHASRPQN 437 |
| Streptococcus pyogenes MGAS9429 | Query 291 IPEGD-EEGNVELRQKFEKAK#LGPAGNKVIGPSEDRKSSVPSN 332 ||| | | |+ | |||+#| | +||||| + +| | | Sbjct 382 IPETDFYSGAVDNSVKVEKAE#LLPHSEEVIGPSSVKHASRPQN 424 |
| Apis mellifera | Query 281 QEEGEYYNVPIPE 293 ||||||||||+|| Sbjct 297 QEEGEYYNVPVPE 309 |
| Streptococcus pyogenes MGAS315 | Query 291 IPEGDEE-GNVELRQKFEKAK#LGPAGNKVIGPSEDRKSSVPSN 332 ||| | | |+ | |||+#| | +||||| + +| | | Sbjct 382 IPETDFYFGTVDNSVKVEKAE#LLPHSEEVIGPSSVKHASRPQN 424 |
| Bombyx mori | Query 281 QEEGEYYNVPIPEGDEEGNVELRQKFEKAK#LGPAGNKVIGPSEDRKSSVPSN 332 |||||+||||+| ||| +| | + +# | + | ||+ || | Sbjct 283 QEEGEFYNVPVP---EEG-ADLVQLKNQMR#ATTVGARRPPPPPDRE--VPHN 328 |
[Site 2] NKVIGPSEDR325-KSSVPSNNLD
Arg325  Lys
Lys
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Asn316 | Lys317 | Val318 | Ile319 | Gly320 | Pro321 | Ser322 | Glu323 | Asp324 | Arg325 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Lys326 | Ser327 | Ser328 | Val329 | Pro330 | Ser331 | Asn332 | Asn333 | Leu334 | Asp335 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| EEGNVELRQKFEKAKLGPAGNKVIGPSEDRKSSVPSNNLDRVKLTDFNFLMVLGKGSFGK |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | Gallus gallus | 114.00 | 8 | protein kinase C, alpha |
| 2 | Rattus norvegicus | 110.00 | 8 | PREDICTED: similar to Protein kinase C alpha type |
| 3 | Latimeria chalumnae | 110.00 | 2 | protein kinase C alpha |
| 4 | Homo sapiens | 109.00 | 24 | aging-associated gene 6 protein |
| 5 | Mus musculus | 109.00 | 17 | protein kinase C |
| 6 | N/A | 109.00 | 16 | - |
| 7 | Canis familiaris | 109.00 | 13 | PREDICTED: similar to Protein kinase C, alpha type |
| 8 | Bos taurus | 109.00 | 8 | protein kinase C, alpha |
| 9 | Oryctolagus cuniculus | 109.00 | 5 | KPCA_RABIT Protein kinase C alpha type (PKC-alpha) |
| 10 | synthetic construct | 109.00 | 4 | protein kinase C alpha |
| 11 | Monodelphis domestica | 102.00 | 6 | PREDICTED: similar to protein kinase C |
| 12 | Xenopus tropicalis | 102.00 | 4 | hypothetical protein LOC780209 |
| 13 | Xenopus laevis | 97.40 | 4 | protein kinase C, alpha |
| 14 | Protopterus dolloi | 92.80 | 2 | protein kinase C alpha |
| 15 | Takifugu rubripes | 82.80 | 1 | protein kinase C, alpha type |
| 16 | Scyliorhinus canicula | 76.60 | 2 | protein kinase C alpha |
| 17 | Danio rerio | 74.70 | 14 | protein kinase C, beta 1 |
| 18 | Tetraodon nigroviridis | 74.70 | 7 | unnamed protein product |
| 19 | Macaca mulatta | 66.60 | 15 | PREDICTED: protein kinase C, beta 1 isoform 5 |
| 20 | Petromyzon marinus | 64.70 | 1 | protein kinase C |
| 21 | Myxine glutinosa | 51.60 | 1 | protein kinase C |
| 22 | Branchiostoma lanceolatum | 45.10 | 1 | protein kinase C |
| 23 | Macaca fascicularis | 44.30 | 4 | KPCG_MACFA Protein kinase C gamma type (PKC-gamma) |
| 24 | Caenorhabditis elegans | 42.40 | 8 | Protein Kinase C family member (pkc-2) |
| 25 | Caenorhabditis briggsae | 42.40 | 3 | Hypothetical protein CBG16150 |
| 26 | Aplysia californica | 41.60 | 2 | KPC2_APLCA Calcium-independent protein kinase C (A |
| 27 | Bombyx mori | 41.60 | 1 | conventional protein kinase C |
| 28 | Drosophila pseudoobscura | 41.20 | 3 | GA19732-PA |
| 29 | Anopheles gambiae str. PEST | 40.40 | 3 | ENSANGP00000009078 |
| 30 | Schizosaccharomyces pombe | 40.40 | 2 | SPAC17G8.14c |
| 31 | Yarrowia lipolytica | 40.40 | 1 | hypothetical protein |
| 32 | Schizosaccharomyces pombe 972h- | 40.40 | 1 | hypothetical protein SPAC17G8.14c |
| 33 | Aedes aegypti | 40.00 | 2 | protein kinase c |
| 34 | Drosophila melanogaster | 39.70 | 8 | Protein C kinase 53E CG6622-PB, isoform B |
| 35 | Apis mellifera | 39.70 | 2 | PREDICTED: similar to protein kinase C, delta |
| 36 | Suberites domuncula | 39.70 | 2 | Serine/Threonine protein kinase |
| 37 | Strongylocentrotus purpuratus | 38.50 | 5 | PREDICTED: similar to protein kinase C, partial |
| 38 | Tribolium castaneum | 38.10 | 4 | PREDICTED: similar to CG6622-PA, isoform A |
| 39 | Botryotinia fuckeliana | 36.60 | 1 | protein kinase C |
| 40 | Aspergillus clavatus NRRL 1 | 36.20 | 1 | protein kinase c |
| 41 | Asparagus officinalis | 35.80 | 1 | S6 ribosomal protein kinase |
| 42 | Magnaporthe grisea 70-15 | 35.80 | 1 | hypothetical protein MGG_08689 |
| 43 | Hydractinia echinata | 35.80 | 1 | putative calcium dependent protein kinase C |
| 44 | Pan troglodytes | 35.40 | 4 | PREDICTED: serum/glucocorticoid regulated kinase 3 |
| 45 | Squalus acanthias | 35.40 | 2 | s-sgk1 |
| 46 | Emericella nidulans | 35.40 | 1 | protein kinase C |
| 47 | Aspergillus nidulans FGSC A4 | 35.40 | 1 | protein kinase C-like |
| 48 | Magnaporthe grisea | 35.40 | 1 | AF136600_1 protein kinase C |
| 49 | Leptosphaeria maculans | 35.40 | 1 | AF487263_9 protein kinase C |
| 50 | Paramecium tetraurelia | 35.00 | 1 | hypothetical protein |
| 51 | Phaeosphaeria nodorum SN15 | 34.70 | 2 | hypothetical protein SNOG_07073 |
| 52 | Ancylostoma caninum | 34.70 | 1 | AF301263_1 protein kinase C |
| 53 | Sycon raphanus | 34.70 | 1 | Serine/Threonine protein kinase |
| 54 | Coprinopsis cinerea okayama7#130 | 34.30 | 1 | hypothetical protein CC1G_07190 |
| 55 | Cryptococcus neoformans var. neoformans B-3501A | 34.30 | 1 | hypothetical protein CNBC3910 |
| 56 | Cryptococcus neoformans var. neoformans | 34.30 | 1 | protein kinase C 1 |
| 57 | Cryptococcus neoformans var. grubii | 34.30 | 1 | protein kinase C 1 |
| 58 | Lodderomyces elongisporus NRRL YB-4239 | 34.30 | 1 | hypothetical protein LELG_02660 |
| 59 | Calliphora vicina | 34.30 | 1 | eye-specific protein kinase C |
| 60 | Cryptococcus neoformans var. neoformans JEC21 | 34.30 | 1 | protein kinase C |
| 61 | Plasmodium falciparum 3D7 | 33.90 | 2 | PKA |
| 62 | Tetrahymena thermophila SB210 | 33.90 | 1 | Protein kinase domain containing protein |
| 63 | Fundulus heteroclitus | 33.50 | 1 | serum and glucocorticoid-regulated kinase |
| 64 | Candida albicans | 33.50 | 1 | KPC1_CANAL Protein kinase C-like 1 (PKC 1) emb |
| 65 | Blumeria graminis | 33.50 | 1 | AF247001_1 protein kinase C |
| 66 | Sporothrix schenckii | 33.50 | 1 | AF124792_1 protein kinase C; serine/threonine prot |
| 67 | Aspergillus oryzae | 33.10 | 1 | unnamed protein product |
Top-ranked sequences
| organism | matching |
|---|---|
| Gallus gallus | Query 296 EEGNVELRQKFEKAKLGPAGNKVIGPSEDR#KSSVPSNNLDRVKLTDFNFLMVLGKGSFGK 355 |+|| ||||||||||||||||||| |||||# ||||||||||||||||||||||||||||| Sbjct 295 EDGNAELRQKFEKAKLGPAGNKVITPSEDR#NSSVPSNNLDRVKLTDFNFLMVLGKGSFGK 354 |
| Rattus norvegicus | Query 296 EEGNVELRQKFEKAKLGPAGNKVIGPSEDR#KSSVPSNNLDRVKLTDFNFLMVLGKGSFGK 355 |||||||||||||||||||||||| |||||#| |||||||||||||||||||||||||| Sbjct 295 EEGNVELRQKFEKAKLGPAGNKVISPSEDR#KQ--PSNNLDRVKLTDFNFLMVLGKGSFGK 352 |
| Latimeria chalumnae | Query 296 EEGNVELRQKFEKAKLGPAGNKVIGPSEDR#KSSVPSNNLDRVKLTDFNFLMVLGKGSFGK 355 ++||||||||||||||||||+||| | |+|#||| |||||||||||||||||||||||||| Sbjct 216 DDGNVELRQKFEKAKLGPAGSKVISPVEER#KSSTPSNNLDRVKLTDFNFLMVLGKGSFGK 275 |
| Homo sapiens | Query 296 EEGNVELRQKFEKAKLGPAGNKVIGPSEDR#KSSVPSNNLDRVKLTDFNFLMVLGKGSFGK 355 ||||+||||||||||||||||||| |||||#| |||||||||||||||||||||||||| Sbjct 295 EEGNMELRQKFEKAKLGPAGNKVISPSEDR#KQ--PSNNLDRVKLTDFNFLMVLGKGSFGK 352 |
| Mus musculus | Query 296 EEGNVELRQKFEKAKLGPAGNKVIGPSEDR#KSSVPSNNLDRVKLTDFNFLMVLGKGSFGK 355 ||||+||||||||||||||||||| |||||#| |||||||||||||||||||||||||| Sbjct 295 EEGNMELRQKFEKAKLGPAGNKVISPSEDR#KQ--PSNNLDRVKLTDFNFLMVLGKGSFGK 352 |
| N/A | Query 296 EEGNVELRQKFEKAKLGPAGNKVIGPSEDR#KSSVPSNNLDRVKLTDFNFLMVLGKGSFGK 355 ||||+||||||||||||||||||| |||||#| |||||||||||||||||||||||||| Sbjct 281 EEGNMELRQKFEKAKLGPAGNKVISPSEDR#KQ--PSNNLDRVKLTDFNFLMVLGKGSFGK 338 |
| Canis familiaris | Query 296 EEGNVELRQKFEKAKLGPAGNKVIGPSEDR#KSSVPSNNLDRVKLTDFNFLMVLGKGSFGK 355 |||||||||||||||||||||||| |||||#+ |||||||||||||||||||||||||| Sbjct 213 EEGNVELRQKFEKAKLGPAGNKVISPSEDR#RQ--PSNNLDRVKLTDFNFLMVLGKGSFGK 270 |
| Bos taurus | Query 296 EEGNVELRQKFEKAKLGPAGNKVIGPSEDR#KSSVPSNNLDRVKLTDFNFLMVLGKGSFGK 355 |||||||||||||||||||||||| |||||#+ |||||||||||||||||||||||||| Sbjct 295 EEGNVELRQKFEKAKLGPAGNKVISPSEDR#RQ--PSNNLDRVKLTDFNFLMVLGKGSFGK 352 |
| Oryctolagus cuniculus | Query 296 EEGNVELRQKFEKAKLGPAGNKVIGPSEDR#KSSVPSNNLDRVKLTDFNFLMVLGKGSFGK 355 |+|||||||||||||||||||||| |||||#| |||||||||||||||||||||||||| Sbjct 295 EDGNVELRQKFEKAKLGPAGNKVISPSEDR#KQ--PSNNLDRVKLTDFNFLMVLGKGSFGK 352 |
| synthetic construct | Query 296 EEGNVELRQKFEKAKLGPAGNKVIGPSEDR#KSSVPSNNLDRVKLTDFNFLMVLGKGSFGK 355 ||||+||||||||||||||||||| |||||#| |||||||||||||||||||||||||| Sbjct 295 EEGNMELRQKFEKAKLGPAGNKVISPSEDR#KQ--PSNNLDRVKLTDFNFLMVLGKGSFGK 352 |
| Monodelphis domestica | Query 296 EEGNVELRQKFEKAKLGPAGNKVIGPSEDR#KSSVPSNNLDRVKLTDFNFLMVLGKGSFGK 355 ||||+||||||||||||| ||||| |+||#| |+|||||||||||||||||||||||| Sbjct 295 EEGNMELRQKFEKAKLGPVGNKVISSSDDR#K---PTNNLDRVKLTDFNFLMVLGKGSFGK 351 |
| Xenopus tropicalis | Query 296 EEGNVELRQKFEKAKLGPAGNKVIGPSEDR#KSSVPSNNLDRVKLTDFNFLMVLGKGSFGK 355 ++||+||||||||||||||||||| |+++|#+ |||||+| |+||||+|||||||||||| Sbjct 299 DDGNIELRQKFEKAKLGPAGNKVISPTDER#RPYVPSNNIDSVRLTDFSFLMVLGKGSFGK 358 |
| Xenopus laevis | Query 296 EEGNVELRQKFEKAKLGPAGNKVIGPSEDR#KSSVPSNNLDRVKLTDFNFLMVLGKGSFGK 355 ++||+||||||||||||||||||| |+ +|#+ +||||+| ++|||| |||||||||||| Sbjct 298 DDGNLELRQKFEKAKLGPAGNKVINPTGER#RPYIPSNNIDSIRLTDFCFLMVLGKGSFGK 357 |
| Protopterus dolloi | Query 296 EEGNVELRQKFEKAKLGPAGNKVIGPSEDR#KSSVPSNNLDRVKLTDFNFLMVLGKGSFGK 355 ++||| ||||||||+||||| ||| | +|#|||+| ||||| || ||||+|||||||||| Sbjct 214 DDGNVGLRQKFEKARLGPAGKKVISP--ER#KSSLPLNNLDRFKLEDFNFVMVLGKGSFGK 271 |
| Takifugu rubripes | Query 296 EEGNVELRQKFEKAKLGPAGNKVIGPSEDR#KSSVPSNNLDRVKLTDFNFLMVLGKGSFGK 355 ++ |+|||||||||||| | ||| ||+ |#+ |+|| |+|||+| ||||| +|||||||| Sbjct 292 DDVNLELRQKFEKAKLGQ-GKKVITPSDHR#RFSLPSGNMDRVRLNDFNFLALLGKGSFGK 350 |
| Scyliorhinus canicula | Query 296 EEGNVELRQKFEKAKLGPAGNKVIGPSEDR#KSSVPSNNLDRVKLTDFNFLMVLGKGSFGK 355 ++ + |||+||||||||||| | | ++#| | |+|+|| | | ||||||||||||||| Sbjct 226 DDEDSELRKKFEKAKLGPAGKKAI----EK#KPSSPTNDLDHVHLEDFNFLMVLGKGSFGK 281 |
| Danio rerio | Query 296 EEGNVELRQKFEKAKLGPAGNKVIGPSEDR#KSSVPSN-NLDRVKLTDFNFLMVLGKGSFG 354 |||| |||||||+||+||+ | | | + # | || | ||+||+|||||||||||||| Sbjct 294 EEGNEELRQKFERAKIGPS--KTDGSSSNA#ISKFDSNGNRDRMKLSDFNFLMVLGKGSFG 351 Query 355 K 355 | Query 355 K 355 |
| Tetraodon nigroviridis | Query 296 EEGNVELRQKFEKAKLGPAGNKVIGPSEDR#KSSVPSN-NLDRVKLTDFNFLMVLGKGSFG 354 |||| |||||||+||+|| | | | + # | || | ||+|| |||||||||||||| Sbjct 231 EEGNEELRQKFERAKIGP-GKNTDGKSANA#ASRFDSNGNQDRMKLADFNFLMVLGKGSFG 289 Query 355 K 355 | Query 355 K 355 |
| Macaca mulatta | Query 297 EGNVELRQKFEKAKLGPAGNKVIGPSEDR#KSSVP----SNNLDRVKLTDFNFLMVLGKGS 352 ||| |||||||+||+ | || | | # ++| + | ||+||||||||||||||| Sbjct 296 EGNEELRQKFERAKISQ-GTKV--PEEKT#TNTVSKFDNNGNRDRMKLTDFNFLMVLGKGS 352 Query 353 FGK 355 ||| Query 353 FGK 355 |
| Petromyzon marinus | Query 296 EEGNVELRQKFEKAKLGPAGNKVIGPSEDR#K----SSVPSN-NLDRVKLTDFNFLMVLGK 350 |+|+ |+|+|||||+|||+ |++|#+ |++ +| |+|||| |||||||||| Sbjct 225 EDGDTEMRRKFEKARLGPSVKP--RASDER#RANSLSAILNNANVDRVKADDFNFLMVLGK 282 Query 351 GSFGK 355 ||||| Query 351 GSFGK 355 |
| Myxine glutinosa | Query 296 EEGNVELRQKFEKAKLGPAGNKVIGPSEDR#KSSVPS-NNLDRVKLTDFNFLMVLGKGSFG 354 ||| +||||+ +++++ + |++|+# | | + |||| | || || |||||||| Sbjct 216 EEG-LELRQRLQRSQIARSDKNKQSPAKDQ#PSDSASLSGLDRVNLEDFVFLTVLGKGSFG 274 Query 355 K 355 | Query 355 K 355 |
| Branchiostoma lanceolatum | Query 296 EEGNVELRQKFEKAKLGPAGNKVIGPSEDR#KSSVPSNNLDRVKLTDFNFLMVLGKGSFGK 355 || +|++ +| || + |# +|| +| ||||||||||||||| Sbjct 225 EENVAKLKEHLQKQKLDEQRKQ----KSKR#SEECNIGSLDHMKAADFNFLMVLGKGSFGK 280 |
| Macaca fascicularis | Query 306 FEKAKLGPAGNKVIGPSE---DR#KSSVPSNNLDRVKLTDFNFLMVLGKGSFGK 355 +|+ ++||+ + + || | #| + |+ ++||+|||||||||||| Sbjct 312 YERVRMGPSSSPIPSPSPSPTDP#KRCFFGASPGRLHISDFSFLMVLGKGSFGK 364 |
| Caenorhabditis elegans | Query 314 AGNKVIGPSEDR#KSSVP--SNNLDRVKLTDFNFLMVLGKGSFGK 355 + +| |+ |# |++| |+| + +| +||||| ||||||||| Sbjct 318 SSSKPKDPAAPR#ASTLPLGSSNHNVIKASDFNFLTVLGKGSFGK 361 |
| Caenorhabditis briggsae | Query 296 EEGNVELRQKFEKAKLGPAGNKVIGPSEDR#KSSVPSNNLDRVKLTDFNFLMVLGKGSFGK 355 |+ ++ |+ ++ || # +|+ + | | +| +||||| ||||||||| Sbjct 369 EKVRKKMNDNFKISRDSSAGKPRESTPRAT#STSLTNTNRDVIKASDFNFLTVLGKGSFGK 428 |
| Aplysia californica | Query 315 GNKVIGPSEDR#KSSVPSNNLDRVKLTDFNFLMVLGKGSFGK 355 |++ || ||# | | |+ | ||||+ ||||||||| Sbjct 380 GSRTRSPSSDR#SRSHHS----RISLHDFNFIKVLGKGSFGK 416 |
| Bombyx mori | Query 296 EEGNVELRQKFEKAKLGPAGNKVIGPSEDR#KSSVPSN--NLDRVKLTDFNFLMVLGKGSF 353 ||| +| | + + | + | ||#+ || | | ++ |||||+|||||||| Sbjct 295 EEG-ADLVQLKNQMRATTVGARRPPPPPDR#E--VPHNVAAADVIRATDFNFIMVLGKGSF 351 Query 354 GK 355 || Query 354 GK 355 |
| Drosophila pseudoobscura | Query 304 QKFEKAKLGPAGNKVIGPSEDR#KSSVPSNNLDRVKLTDFNFLMVLGKGSFGK 355 | | | |+ | + | # + + | ++ |||||+ ||||||||| Sbjct 190 QDLLKLKQKPSQKKPLVMRSDT#NTHSSISKKDMIRATDFNFIKVLGKGSFGK 241 |
| Anopheles gambiae str. PEST | Query 296 EEGN--VELRQKFEKAKLGPAGNKVIGPSEDR#KSSVPSNNLDRVKLTDFNFLMVLGKGSF 353 ||| |+|+ + | + | | |+# | ++ |||||||||||||| Sbjct 232 EEGADLVQLKSQMRKTSI----TKKIPMMGDK#DVPHNMTKKDVIRATDFNFLMVLGKGSF 287 Query 354 GK 355 || Query 354 GK 355 |
| Schizosaccharomyces pombe | Query 311 LGPAGNKVIGPSEDR#KSSVPSN----NLDRVKLTDFNFLMVLGKGSFGK 355 | | + + |+| # ||| + +|| | || || |||||+||| Sbjct 537 LAPPSSASLSSSKDA#NRSVPESPRREKKNRVTLDDFTFLAVLGKGNFGK 585 |
| Yarrowia lipolytica | Query 321 PSEDR#KSSVPSNNLDRVKLTDFNFLMVLGKGSFGK 355 |+ +|# | | |+ | ||||| |||||+||| Sbjct 904 PASNR#HSVVGKKKRPRIGLDDFNFLAVLGKGNFGK 938 |
| Schizosaccharomyces pombe 972h- | Query 311 LGPAGNKVIGPSEDR#KSSVPSN----NLDRVKLTDFNFLMVLGKGSFGK 355 | | + + |+| # ||| + +|| | || || |||||+||| Sbjct 629 LAPPSSASLSSSKDA#NRSVPESPRREKKNRVTLDDFTFLAVLGKGNFGK 677 |
| Aedes aegypti | Query 329 VPSN--NLDRVKLTDFNFLMVLGKGSFGK 355 || | | ++ |||||||||||||||| Sbjct 269 VPHNMGKKDVIRATDFNFLMVLGKGSFGK 297 |
| Drosophila melanogaster | Query 304 QKFEKAKLGPAGNKVIGPSEDR#KSSVPSNNLDRVKLTDFNFLMVLGKGSFGK 355 | | | |+ | + | # + | | ++ |||||+ ||||||||| Sbjct 313 QDLLKLKQKPSQKKPMVMRSDT#NTHTSSKK-DMIRATDFNFIKVLGKGSFGK 363 |
| Apis mellifera | Query 330 PSNNLDRVK---LTDFNFLMVLGKGSFGK 355 |+ || | | +|||||| ||||||||| Sbjct 287 PATNLPRFKKYNVTDFNFLKVLGKGSFGK 315 |
| Suberites domuncula | Query 296 EEGNVELRQKFEKAK-LGPAGNKVIGPSEDR#KSS--VP-SNNLDRVKLTDFNFLMVLGKG 351 +| +| ++| || | | | | |#+ | || + ++ ++|| ||| |+||||| Sbjct 293 DEEATKLIEEFRKATILDKAKNLVTDVPIMR#RGSELVPGAKDMPKMKLEDFNLLVVLGKG 352 Query 352 SFGK 355 |||| Query 352 SFGK 355 |
| Strongylocentrotus purpuratus | Query 315 GNKVIGPSEDR#KSSVPSNN---LDRVKLTDFNFLMVLGKGSFGK 355 | |+ |+ + # | ||+ + |+ +||||| ||||||||| Sbjct 10 GAKLPMPTREA#SKSQNSNSNSSMGLVRASDFNFLSVLGKGSFGK 53 |
| Tribolium castaneum | Query 335 DRVKLTDFNFLMVLGKGSFGK 355 | ++ +||||||||||||||| Sbjct 330 DIIRASDFNFLMVLGKGSFGK 350 |
| Botryotinia fuckeliana | Query 313 PAGNKVIGPSEDR#KSSVPSNNLD---RVKLTDFNFLMVLGKGSFGK 355 | +|+ + |#| +|+ |+ | |||| |||||+||| Sbjct 813 PGSQQVVPQGQQR#KQVPAANDAGTGRRIGLDHFNFLAVLGKGNFGK 858 |
| Aspergillus clavatus NRRL 1 | Query 326 KSSVPSNNLDRVKLTDFNFLMVLGKGSFGK 355 | || |+ | |||| |||||+||| Sbjct 771 KEEVPQQPKARIGLDHFNFLSVLGKGNFGK 800 |
| Asparagus officinalis | Query 331 SNNLDRVKLTDFNFLMVLGKGSFGK 355 | |++|| | ||+|| |+|+|+||| Sbjct 114 SENVERVGLEDFDFLKVVGQGAFGK 138 |
| Magnaporthe grisea 70-15 | Query 303 RQKFEKAKLGPAGNKVIGPSEDR#K--SSVPSNNLDRVKLTDFNFLMVLGKGSFGK 355 +| |+ + | || | |# |+ |+ | |||| |||||+||| Sbjct 816 QQHQEQQIISPTAGTVIPTSAKR#PLPSATDPGTGQRIGLDHFNFLAVLGKGNFGK 870 |
| Hydractinia echinata | Query 335 DRVKLTDFNFLMVLGKGSFGK 355 +|+|| || || |+||||||| Sbjct 191 ERLKLEDFTFLKVIGKGSFGK 211 |
| Pan troglodytes | Query 296 EEGNVELRQKFEKAKLGPAGNKVIGPSEDR#KSSVPSNNLDRVKLTDFNFLMVLGKGSFGK 355 | + +| + |||+|| # | |||+|| |+||||||| Sbjct 134 ERSSQKLHSTSQNINLGPSGNP--------#----------HAKPTDFDFLKVIGKGSFGK 175 |
| Squalus acanthias | Query 330 PSNNLDRVKLTDFNFLMVLGKGSFGK 355 ||+| + | +||||| |+||||||| Sbjct 89 PSSN-PQAKPSDFNFLKVIGKGSFGK 113 |
| Emericella nidulans | Query 326 KSSVPSNNLDRVKLTDFNFLMVLGKGSFGK 355 | +| |+ | |||| |||||+||| Sbjct 744 KEDIPQQPKVRIGLDHFNFLAVLGKGNFGK 773 |
| Aspergillus nidulans FGSC A4 | Query 326 KSSVPSNNLDRVKLTDFNFLMVLGKGSFGK 355 | +| |+ | |||| |||||+||| Sbjct 742 KEDIPQQPKVRIGLDHFNFLAVLGKGNFGK 771 |
| Magnaporthe grisea | Query 303 RQKFEKAKLGPAGNKVIGPSEDR#K--SSVPSNNLDRVKLTDFNFLMVLGKGSFGK 355 +| |+ + | || | |# |+ |+ | |||| |||||+||| Sbjct 816 QQHQEQQIISPTAGTVIPTSARR#PLPSATDPGTGQRIGLDHFNFLAVLGKGNFGK 870 |
| Leptosphaeria maculans | Query 316 NKVIGPSEDR#KSSVPSNNLD----RVKLTDFNFLMVLGKGSFGK 355 +| + |+ # || | |+ | |||| |||||+||| Sbjct 830 SKQLPPANPE#NKVTPSANTQGSGKRIGLDHFNFLAVLGKGNFGK 873 |
| Paramecium tetraurelia | Query 296 EEGNVELRQKFEKAKLGPAGNKVIGPSEDR#KSSVPSNNLDRVKLTDFNFLMVLGKGSFGK 355 +|| ++ +| || | #+ | | |||||| | |||+||||| Sbjct 32 KEGEQQMNDEFMKA---PTATFEQIEETGE#QVFVDDNREAIVKLTDFQFEKVLGRGSFGK 88 |
| Phaeosphaeria nodorum SN15 | Query 321 PSEDR#KSSV-PSNNLD----RVKLTDFNFLMVLGKGSFGK 355 |+| #|+ | || | |+ | |||| |||||+||| Sbjct 692 PTEAP#KNKVTPSANTQGTGRRIGLDHFNFLAVLGKGNFGK 731 |
| Ancylostoma caninum | Query 324 DR#KSSVPSNNLDRVKLTDFNFLMVLGKGSFGK 355 | #| |+ || +|| |+ ||||||||| Sbjct 13 DS#KHSLSGAQSQIVKESDFTFITVLGKGSFGK 44 |
| Sycon raphanus | Query 338 KLTDFNFLMVLGKGSFGK 355 ||+||+|| ||||||||| Sbjct 362 KLSDFSFLKVLGKGSFGK 379 |
| Coprinopsis cinerea okayama7#130 | Query 336 RVKLTDFNFLMVLGKGSFGK 355 +| | ||||| |||||+||| Sbjct 711 KVGLDDFNFLAVLGKGNFGK 730 |
| Cryptococcus neoformans var. neoformans B-3501A | Query 336 RVKLTDFNFLMVLGKGSFGK 355 +| | ||||| |||||+||| Sbjct 754 KVGLDDFNFLAVLGKGNFGK 773 |
| Cryptococcus neoformans var. neoformans | Query 336 RVKLTDFNFLMVLGKGSFGK 355 +| | ||||| |||||+||| Sbjct 751 KVGLDDFNFLAVLGKGNFGK 770 |
| Cryptococcus neoformans var. grubii | Query 336 RVKLTDFNFLMVLGKGSFGK 355 +| | ||||| |||||+||| Sbjct 751 KVGLDDFNFLAVLGKGNFGK 770 |
| Lodderomyces elongisporus NRRL YB-4239 | Query 326 KSSVPSNNLDRVKLTDFNFLMVLGKGSFGK 355 ||| +| | || || |||||+||| Sbjct 814 KSSKSKRRRRKVGLDDFQFLAVLGKGNFGK 843 |
| Calliphora vicina | Query 335 DRVKLTDFNFLMVLGKGSFGK 355 | ++ ||||+ ||||||+|| Sbjct 348 DMIRAADFNFIKVLGKGSYGK 368 |
| Cryptococcus neoformans var. neoformans JEC21 | Query 336 RVKLTDFNFLMVLGKGSFGK 355 +| | ||||| |||||+||| Sbjct 751 KVGLDDFNFLAVLGKGNFGK 770 |
| Plasmodium falciparum 3D7 | Query 324 DR#KSSVPSNNLDRVKLTDFNFLMVLGKGSFGK 355 | # | +| +++| ||||+ || ||||+ Sbjct 16 DS#SEQVLTNKKNKMKYEDFNFICTLGTGSFGR 47 |
| Tetrahymena thermophila SB210 | Query 317 KVIGPSEDR#KSSVPSNNLDRVKLTDFNFLMVLGKGSFGK 355 ++ | +|| #+ || +| |+|| | |+|+||||| Sbjct 52 QIKGNNEDE#Q-----NNQQKVVLSDFEILKVIGEGSFGK 85 |
| Fundulus heteroclitus | Query 321 PSEDR#KSSVPSNNLDRVKLTDFNFLMVLGKGSFGK 355 || +#+ ++ ++ | +||+|| |+||||||| Sbjct 77 PSPSQ#QINLGPSSNPSAKPSDFHFLKVIGKGSFGK 111 |
| Candida albicans | Query 296 EEGNVELRQKFEKAKLGPAGNKVIGPSEDR#KSSVPSNNLDRVKLTDFNFLMVLGKGSFGK 355 ++ | +++| ++ +|| | #|| +| | || || |||||+||| Sbjct 729 KQQNQQVQQVQQQEELGHQRTHSSG-----#KSGKSKRRKRKVGLDDFQFLAVLGKGNFGK 783 |
| Blumeria graminis | Query 321 PSEDR#KSSVPSNNLD---RVKLTDFNFLMVLGKGSFGK 355 |+ |#|++ +| |+ | |||| |||||+||| Sbjct 808 PAMTR#KAAPQANEPGTGRRIGLDHFNFLAVLGKGNFGK 845 |
| Sporothrix schenckii | Query 300 VELRQKFEKAKLGPAGNKVI-----GPSEDR#KSSVPSNNLDRVKLTDFNFLMVLGKGSFG 354 |+ +| | | + + | + # |+ |+ | +|||| |||||+|| Sbjct 822 VKTQQSEPPVSLPPVSSTAVDVTGPGVKKPL#PSATDPGTGQRIGLDNFNFLAVLGKGNFG 881 Query 355 K 355 | Query 355 K 355 |
| Aspergillus oryzae | Query 325 R#KSSVPSNNLDRVKLTDFNFLMVLGKGSFGK 355 +#+ + | |+ | |||| |||||+||| Sbjct 750 K#EDAPPQQPKVRIGLDHFNFLAVLGKGNFGK 780 |
[Site 3] KAKLGPAGNK317-VIGPSEDRKS
Lys317  Val
Val
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Lys308 | Ala309 | Lys310 | Leu311 | Gly312 | Pro313 | Ala314 | Gly315 | Asn316 | Lys317 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Val318 | Ile319 | Gly320 | Pro321 | Ser322 | Glu323 | Asp324 | Arg325 | Lys326 | Ser327 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| NVPIPEGDEEGNVELRQKFEKAKLGPAGNKVIGPSEDRKSSVPSNNLDRVKLTDFNFLMV |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | Gallus gallus | 112.00 | 3 | protein kinase C, alpha |
| 2 | Rattus norvegicus | 111.00 | 4 | PREDICTED: similar to Protein kinase C alpha type |
| 3 | Homo sapiens | 110.00 | 9 | protein kinase C, alpha |
| 4 | N/A | 110.00 | 8 | - |
| 5 | Mus musculus | 110.00 | 4 | protein kinase C |
| 6 | Oryctolagus cuniculus | 110.00 | 4 | KPCA_RABIT Protein kinase C alpha type (PKC-alpha) |
| 7 | Canis familiaris | 110.00 | 3 | PREDICTED: similar to Protein kinase C, alpha type |
| 8 | Bos taurus | 110.00 | 3 | protein kinase C, alpha |
| 9 | synthetic construct | 110.00 | 1 | protein kinase C alpha |
| 10 | Monodelphis domestica | 103.00 | 4 | PREDICTED: similar to protein kinase C |
| 11 | Latimeria chalumnae | 100.00 | 2 | protein kinase C alpha |
| 12 | Xenopus tropicalis | 98.60 | 1 | hypothetical protein LOC780209 |
| 13 | Xenopus laevis | 90.90 | 1 | protein kinase C, alpha |
| 14 | Protopterus dolloi | 85.10 | 2 | protein kinase C alpha |
| 15 | Takifugu rubripes | 75.50 | 1 | protein kinase C, alpha type |
| 16 | Scyliorhinus canicula | 68.60 | 2 | protein kinase C alpha |
| 17 | Danio rerio | 67.40 | 7 | protein kinase C, beta 1 |
| 18 | Tetraodon nigroviridis | 67.40 | 3 | unnamed protein product |
| 19 | Macaca mulatta | 59.30 | 6 | PREDICTED: protein kinase C, beta 1 isoform 5 |
| 20 | Petromyzon marinus | 54.30 | 1 | protein kinase C |
| 21 | Myxine glutinosa | 41.60 | 1 | protein kinase C |
| 22 | Branchiostoma lanceolatum | 33.90 | 1 | protein kinase C |
| 23 | Streptococcus pyogenes MGAS10750 | 32.70 | 1 | DNA mismatch repair protein mutL |
| 24 | Streptococcus pyogenes MGAS10270 | 32.70 | 1 | DNA mismatch repair protein mutL |
| 25 | Streptococcus pyogenes MGAS9429 | 32.70 | 1 | DNA mismatch repair protein |
| 26 | Streptococcus pyogenes MGAS315 | 32.30 | 1 | DNA mismatch repair protein |
Top-ranked sequences
| organism | matching |
|---|---|
| Gallus gallus | Query 288 NVPIPEGDEEGNVELRQKFEKAKLGPAGNK#VIGPSEDRKSSVPSNNLDRVKLTDFNFLMV 347 |||||+ ||+|| |||||||||||||||||#|| ||||| ||||||||||||||||||||| Sbjct 287 NVPIPDADEDGNAELRQKFEKAKLGPAGNK#VITPSEDRNSSVPSNNLDRVKLTDFNFLMV 346 |
| Rattus norvegicus | Query 288 NVPIPEGDEEGNVELRQKFEKAKLGPAGNK#VIGPSEDRKSSVPSNNLDRVKLTDFNFLMV 347 ||||||||||||||||||||||||||||||#|| |||||| |||||||||||||||||| Sbjct 287 NVPIPEGDEEGNVELRQKFEKAKLGPAGNK#VISPSEDRKQ--PSNNLDRVKLTDFNFLMV 344 |
| Homo sapiens | Query 288 NVPIPEGDEEGNVELRQKFEKAKLGPAGNK#VIGPSEDRKSSVPSNNLDRVKLTDFNFLMV 347 ||||||||||||+|||||||||||||||||#|| |||||| |||||||||||||||||| Sbjct 287 NVPIPEGDEEGNMELRQKFEKAKLGPAGNK#VISPSEDRKQ--PSNNLDRVKLTDFNFLMV 344 |
| N/A | Query 288 NVPIPEGDEEGNVELRQKFEKAKLGPAGNK#VIGPSEDRKSSVPSNNLDRVKLTDFNFLMV 347 ||||||||||||+|||||||||||||||||#|| |||||| |||||||||||||||||| Sbjct 273 NVPIPEGDEEGNMELRQKFEKAKLGPAGNK#VISPSEDRKQ--PSNNLDRVKLTDFNFLMV 330 |
| Mus musculus | Query 288 NVPIPEGDEEGNVELRQKFEKAKLGPAGNK#VIGPSEDRKSSVPSNNLDRVKLTDFNFLMV 347 ||||||||||||+|||||||||||||||||#|| |||||| |||||||||||||||||| Sbjct 287 NVPIPEGDEEGNMELRQKFEKAKLGPAGNK#VISPSEDRKQ--PSNNLDRVKLTDFNFLMV 344 |
| Oryctolagus cuniculus | Query 288 NVPIPEGDEEGNVELRQKFEKAKLGPAGNK#VIGPSEDRKSSVPSNNLDRVKLTDFNFLMV 347 |||||||||+||||||||||||||||||||#|| |||||| |||||||||||||||||| Sbjct 287 NVPIPEGDEDGNVELRQKFEKAKLGPAGNK#VISPSEDRKQ--PSNNLDRVKLTDFNFLMV 344 |
| Canis familiaris | Query 288 NVPIPEGDEEGNVELRQKFEKAKLGPAGNK#VIGPSEDRKSSVPSNNLDRVKLTDFNFLMV 347 ||||||||||||||||||||||||||||||#|| |||||+ |||||||||||||||||| Sbjct 205 NVPIPEGDEEGNVELRQKFEKAKLGPAGNK#VISPSEDRRQ--PSNNLDRVKLTDFNFLMV 262 |
| Bos taurus | Query 288 NVPIPEGDEEGNVELRQKFEKAKLGPAGNK#VIGPSEDRKSSVPSNNLDRVKLTDFNFLMV 347 ||||||||||||||||||||||||||||||#|| |||||+ |||||||||||||||||| Sbjct 287 NVPIPEGDEEGNVELRQKFEKAKLGPAGNK#VISPSEDRRQ--PSNNLDRVKLTDFNFLMV 344 |
| synthetic construct | Query 288 NVPIPEGDEEGNVELRQKFEKAKLGPAGNK#VIGPSEDRKSSVPSNNLDRVKLTDFNFLMV 347 ||||||||||||+|||||||||||||||||#|| |||||| |||||||||||||||||| Sbjct 287 NVPIPEGDEEGNMELRQKFEKAKLGPAGNK#VISPSEDRKQ--PSNNLDRVKLTDFNFLMV 344 |
| Monodelphis domestica | Query 288 NVPIPEGDEEGNVELRQKFEKAKLGPAGNK#VIGPSEDRKSSVPSNNLDRVKLTDFNFLMV 347 ||||||||||||+||||||||||||| |||#|| |+||| |+|||||||||||||||| Sbjct 287 NVPIPEGDEEGNMELRQKFEKAKLGPVGNK#VISSSDDRK---PTNNLDRVKLTDFNFLMV 343 |
| Latimeria chalumnae | Query 288 NVPIPEGDEEGNVELRQKFEKAKLGPAGNK#VIGPSEDRKSSVPSNNLDRVKLTDFNFLMV 347 |||| | |+ ||||||||||||||||||+|#|| | |+|||| |||||||||||||||||| Sbjct 209 NVPISEADD-GNVELRQKFEKAKLGPAGSK#VISPVEERKSSTPSNNLDRVKLTDFNFLMV 267 |
| Xenopus tropicalis | Query 288 NVPIPEGDEEGNVELRQKFEKAKLGPAGNK#VIGPSEDRKSSVPSNNLDRVKLTDFNFLMV 347 |||||| +++||+|||||||||||||||||#|| |+++|+ |||||+| |+||||+|||| Sbjct 291 NVPIPEEEDDGNIELRQKFEKAKLGPAGNK#VISPTDERRPYVPSNNIDSVRLTDFSFLMV 350 |
| Xenopus laevis | Query 288 NVPIPEGDEEGNVELRQKFEKAKLGPAGNK#VIGPSEDRKSSVPSNNLDRVKLTDFNFLMV 347 |||||| |+ ||+|||||||||||||||||#|| |+ +|+ +||||+| ++|||| |||| Sbjct 291 NVPIPEADD-GNLELRQKFEKAKLGPAGNK#VINPTGERRPYIPSNNIDSIRLTDFCFLMV 349 |
| Protopterus dolloi | Query 288 NVPIPEGDEEGNVELRQKFEKAKLGPAGNK#VIGPSEDRKSSVPSNNLDRVKLTDFNFLMV 347 |||||+ |+ ||| ||||||||+||||| |#|| | +||||+| ||||| || ||||+|| Sbjct 207 NVPIPDADD-GNVGLRQKFEKARLGPAGKK#VISP--ERKSSLPLNNLDRFKLEDFNFVMV 263 |
| Takifugu rubripes | Query 288 NVPIPEGDEEGNVELRQKFEKAKLGPAGNK#VIGPSEDRKSSVPSNNLDRVKLTDFNFL 345 |||||| |+ |+|||||||||||| | |#|| ||+ |+ |+|| |+|||+| ||||| Sbjct 285 NVPIPEVDDV-NLELRQKFEKAKLGQ-GKK#VITPSDHRRFSLPSGNMDRVRLNDFNFL 340 |
| Scyliorhinus canicula | Query 288 NVPIPEGDEEGNVELRQKFEKAKLGPAGNK#VIGPSEDRKSSVPSNNLDRVKLTDFNFLMV 347 |||| | ||+ |||+||||||||||| |# | ++| | |+|+|| | | ||||||| Sbjct 220 NVPIIEDDEDS--ELRKKFEKAKLGPAGKK#AI----EKKPSSPTNDLDHVHLEDFNFLMV 273 |
| Danio rerio | Query 288 NVPIPEGDEEGNVELRQKFEKAKLGPAGNK#VIGPSEDRKSSVPSN-NLDRVKLTDFNFLM 346 |||+| |||| |||||||+||+|| +|# | | + | || | ||+||+|||||| Sbjct 286 NVPVPPEGEEGNEELRQKFERAKIGP--SK#TDGSSSNAISKFDSNGNRDRMKLSDFNFLM 343 Query 347 V 347 | Query 347 V 347 |
| Tetraodon nigroviridis | Query 288 NVPIPEGDEEGNVELRQKFEKAKLGPAGNK#VIGPSEDRKSSVPSN-NLDRVKLTDFNFLM 346 |||+| |||| |||||||+||+|| | # | | + | || | ||+|| |||||| Sbjct 223 NVPVPPEGEEGNEELRQKFERAKIGP-GKN#TDGKSANAASRFDSNGNQDRMKLADFNFLM 281 Query 347 V 347 | Query 347 V 347 |
| Macaca mulatta | Query 288 NVPIPEGDEEGNVELRQKFEKAKLGPAGNK#VIGPSEDRKSSVP----SNNLDRVKLTDFN 343 |||+| ||| |||||||+||+ | |#| | | ++| + | ||+|||||| Sbjct 287 NVPVPPEGSEGNEELRQKFERAKIS-QGTK#V--PEEKTTNTVSKFDNNGNRDRMKLTDFN 343 Query 344 FLMV 347 |||| Query 344 FLMV 347 |
| Petromyzon marinus | Query 288 NVPIPEGDEEGNVELRQKFEKAKLGPAGNK#VIGPSEDRKSSVP----SNNLDRVKLTDFN 343 |||| |+|+ |+|+|||||+|||+ |# | | +|+ + |+|||| ||| Sbjct 217 NVPIAPDIEDGDTEMRRKFEKARLGPS-VK#PRASDERRANSLSAILNNANVDRVKADDFN 275 Query 344 FLMV 347 |||| Query 344 FLMV 347 |
| Myxine glutinosa | Query 288 NVPI-PEGDEEGNVELRQKFEKAKLGPAGNK#VIGPSEDRKSSVPS-NNLDRVKLTDFNFL 345 |||+ |||+| +||||+ +++++ + # |++|+ | | + |||| | || || Sbjct 208 NVPVAPEGEE--GLELRQRLQRSQIARSDKN#KQSPAKDQPSDSASLSGLDRVNLEDFVFL 265 Query 346 MV 347 | Sbjct 266 TV 267 |
| Branchiostoma lanceolatum | Query 288 NVPIPEGDEEGNV-ELRQKFEKAKLGPAGNK#VIGPSEDRKSSVPSNNLDRVKLTDFNFLM 346 |||| ||| || +|++ +| || +# | +|| +| |||||| Sbjct 218 NVPI--ADEEENVAKLKEHLQKQKLDEQRKQ#----KSKRSEECNIGSLDHMKAADFNFLM 271 Query 347 V 347 | Query 347 V 347 |
| Streptococcus pyogenes MGAS10750 | Query 291 IPEGD-EEGNVELRQKFEKAKLGPAGNK#VIGPSEDRKSSVPSN 332 ||| | | |+ | |||+| | +#||||| + +| | | Sbjct 395 IPETDFYSGAVDNSVKVEKAELLPHSEE#VIGPSSVKHASRPQN 437 |
| Streptococcus pyogenes MGAS10270 | Query 291 IPEGD-EEGNVELRQKFEKAKLGPAGNK#VIGPSEDRKSSVPSN 332 ||| | | |+ | |||+| | +#||||| + +| | | Sbjct 395 IPETDFYSGAVDNSVKVEKAELLPHSEE#VIGPSSVKHASRPQN 437 |
| Streptococcus pyogenes MGAS9429 | Query 291 IPEGD-EEGNVELRQKFEKAKLGPAGNK#VIGPSEDRKSSVPSN 332 ||| | | |+ | |||+| | +#||||| + +| | | Sbjct 382 IPETDFYSGAVDNSVKVEKAELLPHSEE#VIGPSSVKHASRPQN 424 |
| Streptococcus pyogenes MGAS315 | Query 291 IPEGDEE-GNVELRQKFEKAKLGPAGNK#VIGPSEDRKSSVPSN 332 ||| | | |+ | |||+| | +#||||| + +| | | Sbjct 382 IPETDFYFGTVDNSVKVEKAELLPHSEE#VIGPSSVKHASRPQN 424 |
* References
None.