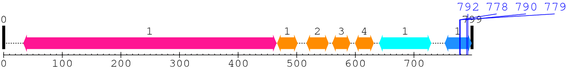
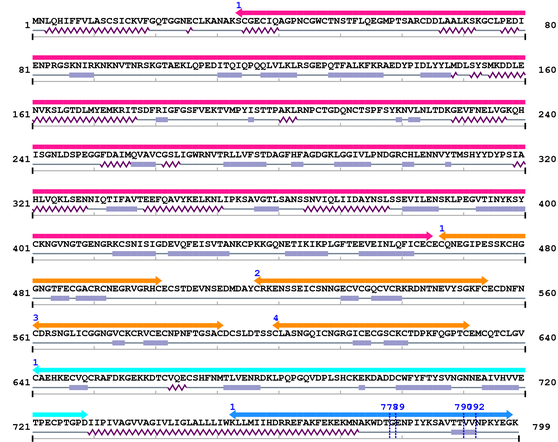
XSB2203 : PREDICTED: similar to integrin beta-1 subunit isoform 1 [Ornithorhynchus anatinus]
[ CaMP Format ]
This entry is computationally expanded from SB0063
* Basic Information
| Organism | Ornithorhynchus anatinus (platypus) |
| Protein Names | similar to integrin beta-1 subunit isoform 1 |
| Gene Names | LOC100073959; Derived by automated computational analysis using gene prediction method: GNOMON. Supporting evidence includes similarity to: 2 ESTs, 27 Proteins |
| Gene Locus | Not available |
| GO Function | Not available |
| Entrez Protein | Entrez Nucleotide | Entrez Gene | UniProt | OMIM | HGNC | HPRD | KEGG |
|---|---|---|---|---|---|---|---|
| XP_001508168 | XM_001508118 | 100073959 | N/A | N/A | N/A | N/A | N/A |
* Information From OMIM
Not Available.
* Structure Information
1. Primary Information
Length: 799 aa
Average Mass: 87.961 kDa
Monoisotopic Mass: 87.903 kDa
2. Domain Information
Annotated Domains: Not Available.
Computationally Assigned Domains (Pfam+HMMER):
| domain name | begin | end | score | e-value |
|---|---|---|---|---|
| Integrin_beta 1. | 34 | 465 | 1123.6 | 0 |
| EGF_2 1. | 467 | 501 | 12.0 | 0.13 |
| EGF_2 2. | 517 | 554 | 21.5 | 0.0036 |
| EGF_2 3. | 561 | 591 | 21.4 | 0.0038 |
| EGF_2 4. | 600 | 631 | 36.0 | 1.5e-07 |
| Integrin_B_tail 1. | 641 | 729 | 156.8 | 6.3e-44 |
| Integrin_b_cyt 1. | 753 | 798 | 112.3 | 1.6e-30 |
| --- cleavage 792 (inside Integrin_b_cyt 753..798) --- | ||||
| --- cleavage 778 (inside Integrin_b_cyt 753..798) --- | ||||
| --- cleavage 790 (inside Integrin_b_cyt 753..798) --- | ||||
| --- cleavage 779 (inside Integrin_b_cyt 753..798) --- | ||||
3. Sequence Information
Fasta Sequence: XSB2203.fasta
Amino Acid Sequence and Secondary Structures (PsiPred):
4. 3D Information
Not Available.
* Cleavage Information
4 [sites]
Cleavage sites (±10aa)
[Site 1] IYKSAVTTVV792-NPKYEGK
Val792  Asn
Asn
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Ile783 | Tyr784 | Lys785 | Ser786 | Ala787 | Val788 | Thr789 | Thr790 | Val791 | Val792 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Asn793 | Pro794 | Lys795 | Tyr796 | Glu797 | Gly798 | Lys799 | - | - | - |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| EFAKFEKEKMNAKWDTGENPIYKSAVTTVVNPKYEGK |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | N/A | 81.30 | 28 | I |
| 2 | Homo sapiens | 81.30 | 22 | integrin beta 1 isoform 1A precursor |
| 3 | Mus musculus | 81.30 | 14 | Fn receptor beta prechain |
| 4 | Bos taurus | 81.30 | 13 | integrin beta 1 |
| 5 | Pan troglodytes | 81.30 | 12 | PREDICTED: hypothetical protein |
| 6 | Rattus norvegicus | 81.30 | 10 | integrin beta 1 |
| 7 | Gallus gallus | 81.30 | 6 | integrin, beta 1 (fibronectin receptor, beta polyp |
| 8 | Monodelphis domestica | 81.30 | 6 | PREDICTED: similar to integrin beta-1 subunit isof |
| 9 | Sus scrofa | 81.30 | 6 | integrin beta-1 subunit |
| 10 | Tetraodon nigroviridis | 81.30 | 5 | unnamed protein product |
| 11 | Xenopus laevis | 81.30 | 5 | integrin beta 1 |
| 12 | synthetic construct | 81.30 | 5 | hypothetical protein |
| 13 | Oryctolagus cuniculus | 81.30 | 2 | integrin beta 1 |
| 14 | Pongo pygmaeus | 81.30 | 2 | ITB1_PONPY Integrin beta-1 precursor (Fibronectin |
| 15 | Xenopus tropicalis | 81.30 | 1 | integrin beta 1 (fibronectin receptor beta) |
| 16 | Felis catus | 81.30 | 1 | integrin beta 1 |
| 17 | Danio rerio | 80.90 | 12 | integrin beta1 subunit-like protein 1 |
| 18 | Ictalurus punctatus | 79.30 | 2 | beta-1 integrin |
| 19 | Drosophila melanogaster | 65.50 | 1 | myospheroid CG1560-PA |
| 20 | Anopheles gambiae str. PEST | 63.90 | 1 | ENSANGP00000021373 |
| 21 | Anopheles gambiae | 63.90 | 1 | integrin beta subunit |
| 22 | Aedes aegypti | 63.90 | 1 | integrin beta subunit |
| 23 | Pseudoplusia includens | 62.40 | 1 | integrin beta 1 |
| 24 | Ostrinia furnacalis | 62.40 | 1 | integrin beta 1 precursor |
| 25 | Tribolium castaneum | 61.20 | 2 | PREDICTED: similar to CG1560-PA |
| 26 | Biomphalaria glabrata | 60.80 | 1 | beta integrin subunit |
| 27 | Canis familiaris | 60.10 | 10 | PREDICTED: similar to integrin beta 1 isoform 1D p |
| 28 | Crassostrea gigas | 59.30 | 1 | integrin beta cgh |
| 29 | Podocoryne carnea | 57.40 | 1 | AF308652_1 integrin beta chain |
| 30 | Caenorhabditis briggsae | 56.20 | 1 | Hypothetical protein CBG03601 |
| 31 | Caenorhabditis elegans | 53.90 | 1 | Paralysed Arrest at Two-fold family member (pat-3) |
| 32 | Macaca mulatta | 50.80 | 12 | PREDICTED: similar to integrin beta chain, beta 3 |
| 33 | rats, Peptide Partial, 723 aa | 50.80 | 1 | beta 3 integrin, GPIIIA |
| 34 | Canis lupus familiaris | 50.80 | 1 | integrin beta chain, beta 3 |
| 35 | Equus caballus | 50.80 | 1 | integrin, beta 3 (platelet glycoprotein IIIa, anti |
| 36 | Mus sp. | 49.30 | 1 | beta 7 integrin |
| 37 | mice, Peptide Partial, 680 aa | 47.40 | 1 | beta 3 integrin, GPIIIA |
| 38 | Strongylocentrotus purpuratus | 47.00 | 5 | integrin beta-C subunit |
| 39 | Lytechinus variegatus | 47.00 | 1 | beta-C integrin subunit |
| 40 | Acropora millepora | 46.60 | 1 | integrin subunit betaCn1 |
| 41 | Oncorhynchus mykiss | 46.20 | 1 | CD18 protein |
| 42 | Ovis aries | 45.80 | 2 | antigen CD18 |
| 43 | Ovis canadensis | 45.80 | 1 | ITB2_OVICA Integrin beta-2 precursor (Cell surface |
| 44 | Bubalus bubalis | 45.80 | 1 | integrin beta-2 |
| 45 | Capra hircus | 45.80 | 1 | ITB2_CAPHI Integrin beta-2 precursor (Cell surface |
| 46 | Sigmodon hispidus | 45.10 | 1 | AF445415_1 integrin beta-2 precursor |
| 47 | Schistosoma japonicum | 40.40 | 1 | SJCHGC06221 protein |
| 48 | Pacifastacus leniusculus | 39.70 | 1 | integrin |
| 49 | Apis mellifera | 39.30 | 1 | PREDICTED: similar to myospheroid CG1560-PA |
| 50 | Halocynthia roretzi | 38.10 | 1 | integrin beta Hr2 precursor |
| 51 | Ophlitaspongia tenuis | 38.10 | 1 | integrin subunit betaPo1 |
| 52 | Cyprinus carpio | 35.80 | 2 | CD18 type 2 |
| 53 | Geodia cydonium | 34.30 | 1 | integrin beta subunit |
| 54 | Suberites domuncula | 33.50 | 1 | integrin beta subunit |
Top-ranked sequences
| organism | matching |
|---|---|
| N/A | Query 763 EFAKFEKEKMNAKWDTGENPIYKSAVTTVV#NPKYEGK 799 ||||||||||||||||||||||||||||||#||||||| Query 763 EFAKFEKEKMNAKWDTGENPIYKSAVTTVV#NPKYEGK 799 |
| Homo sapiens | Query 763 EFAKFEKEKMNAKWDTGENPIYKSAVTTVV#NPKYEGK 799 ||||||||||||||||||||||||||||||#||||||| Query 763 EFAKFEKEKMNAKWDTGENPIYKSAVTTVV#NPKYEGK 799 |
| Mus musculus | Query 763 EFAKFEKEKMNAKWDTGENPIYKSAVTTVV#NPKYEGK 799 ||||||||||||||||||||||||||||||#||||||| Query 763 EFAKFEKEKMNAKWDTGENPIYKSAVTTVV#NPKYEGK 799 |
| Bos taurus | Query 763 EFAKFEKEKMNAKWDTGENPIYKSAVTTVV#NPKYEGK 799 ||||||||||||||||||||||||||||||#||||||| Query 763 EFAKFEKEKMNAKWDTGENPIYKSAVTTVV#NPKYEGK 799 |
| Pan troglodytes | Query 763 EFAKFEKEKMNAKWDTGENPIYKSAVTTVV#NPKYEGK 799 ||||||||||||||||||||||||||||||#||||||| Query 763 EFAKFEKEKMNAKWDTGENPIYKSAVTTVV#NPKYEGK 799 |
| Rattus norvegicus | Query 763 EFAKFEKEKMNAKWDTGENPIYKSAVTTVV#NPKYEGK 799 ||||||||||||||||||||||||||||||#||||||| Query 763 EFAKFEKEKMNAKWDTGENPIYKSAVTTVV#NPKYEGK 799 |
| Gallus gallus | Query 763 EFAKFEKEKMNAKWDTGENPIYKSAVTTVV#NPKYEGK 799 ||||||||||||||||||||||||||||||#||||||| Query 763 EFAKFEKEKMNAKWDTGENPIYKSAVTTVV#NPKYEGK 799 |
| Monodelphis domestica | Query 763 EFAKFEKEKMNAKWDTGENPIYKSAVTTVV#NPKYEGK 799 ||||||||||||||||||||||||||||||#||||||| Query 763 EFAKFEKEKMNAKWDTGENPIYKSAVTTVV#NPKYEGK 799 |
| Sus scrofa | Query 763 EFAKFEKEKMNAKWDTGENPIYKSAVTTVV#NPKYEGK 799 ||||||||||||||||||||||||||||||#||||||| Query 763 EFAKFEKEKMNAKWDTGENPIYKSAVTTVV#NPKYEGK 799 |
| Tetraodon nigroviridis | Query 763 EFAKFEKEKMNAKWDTGENPIYKSAVTTVV#NPKYEGK 799 ||||||||||||||||||||||||||||||#||||||| Query 763 EFAKFEKEKMNAKWDTGENPIYKSAVTTVV#NPKYEGK 799 |
| Xenopus laevis | Query 763 EFAKFEKEKMNAKWDTGENPIYKSAVTTVV#NPKYEGK 799 ||||||||||||||||||||||||||||||#||||||| Query 763 EFAKFEKEKMNAKWDTGENPIYKSAVTTVV#NPKYEGK 799 |
| synthetic construct | Query 763 EFAKFEKEKMNAKWDTGENPIYKSAVTTVV#NPKYEGK 799 ||||||||||||||||||||||||||||||#||||||| Query 763 EFAKFEKEKMNAKWDTGENPIYKSAVTTVV#NPKYEGK 799 |
| Oryctolagus cuniculus | Query 763 EFAKFEKEKMNAKWDTGENPIYKSAVTTVV#NPKYEGK 799 ||||||||||||||||||||||||||||||#||||||| Query 763 EFAKFEKEKMNAKWDTGENPIYKSAVTTVV#NPKYEGK 799 |
| Pongo pygmaeus | Query 763 EFAKFEKEKMNAKWDTGENPIYKSAVTTVV#NPKYEGK 799 ||||||||||||||||||||||||||||||#||||||| Query 763 EFAKFEKEKMNAKWDTGENPIYKSAVTTVV#NPKYEGK 799 |
| Xenopus tropicalis | Query 763 EFAKFEKEKMNAKWDTGENPIYKSAVTTVV#NPKYEGK 799 ||||||||||||||||||||||||||||||#||||||| Query 763 EFAKFEKEKMNAKWDTGENPIYKSAVTTVV#NPKYEGK 799 |
| Felis catus | Query 763 EFAKFEKEKMNAKWDTGENPIYKSAVTTVV#NPKYEGK 799 ||||||||||||||||||||||||||||||#||||||| Query 763 EFAKFEKEKMNAKWDTGENPIYKSAVTTVV#NPKYEGK 799 |
| Danio rerio | Query 763 EFAKFEKEKMNAKWDTGENPIYKSAVTTVV#NPKYEGK 799 |||||||||||||||||||||||||||||+#||||||| Sbjct 762 EFAKFEKEKMNAKWDTGENPIYKSAVTTVI#NPKYEGK 798 |
| Ictalurus punctatus | Query 763 EFAKFEKEKMNAKWDTGENPIYKSAVTTVV#NPKYEGK 799 ||||||||||||||| ||||||||||||||#||||||| Sbjct 771 EFAKFEKEKMNAKWDAGENPIYKSAVTTVV#NPKYEGK 807 |
| Drosophila melanogaster | Query 763 EFAKFEKEKMNAKWDTGENPIYKSAVTTVV#NPKYEGK 799 |||+||||+|||||||||||||| | +| #|| | || Sbjct 810 EFARFEKERMNAKWDTGENPIYKQATSTFK#NPMYAGK 846 |
| Anopheles gambiae str. PEST | Query 763 EFAKFEKEKMNAKWDTGENPIYKSAVTTVV#NPKYEGK 799 |||+||||+| |||||||||||| | || #|| | || Sbjct 801 EFARFEKERMMAKWDTGENPIYKQATTTFK#NPTYAGK 837 |
| Anopheles gambiae | Query 763 EFAKFEKEKMNAKWDTGENPIYKSAVTTVV#NPKYEGK 799 |||+||||+| |||||||||||| | || #|| | || Sbjct 801 EFARFEKERMMAKWDTGENPIYKQATTTFK#NPTYAGK 837 |
| Aedes aegypti | Query 763 EFAKFEKEKMNAKWDTGENPIYKSAVTTVV#NPKYEGK 799 |||+||||+| |||||||||||| | || #|| | || Sbjct 799 EFARFEKERMMAKWDTGENPIYKQATTTFK#NPTYAGK 835 |
| Pseudoplusia includens | Query 763 EFAKFEKEKMNAKWDTGENPIYKSAVTTVV#NPKYEGK 799 |||+||||+| |||||||||||| | +| #|| | || Sbjct 801 EFARFEKERMMAKWDTGENPIYKQATSTFK#NPTYAGK 837 |
| Ostrinia furnacalis | Query 763 EFAKFEKEKMNAKWDTGENPIYKSAVTTVV#NPKYEGK 799 |||+||||+| |||||||||||| | +| #|| | || Sbjct 795 EFARFEKERMMAKWDTGENPIYKQATSTFK#NPTYAGK 831 |
| Tribolium castaneum | Query 763 EFAKFEKEKMNAKWDTGENPIYKSAVTTVV#NPKYEGK 799 |||||||| | |+|||||||||| | +| #|| | || Sbjct 793 EFAKFEKEAMMARWDTGENPIYKQATSTFK#NPTYAGK 829 |
| Biomphalaria glabrata | Query 763 EFAKFEKEKMNAKWDTGENPIYKSAVTTVV#NPKY 796 ||||||||+ ||||||||||||| | +| #|| | Sbjct 752 EFAKFEKERQNAKWDTGENPIYKQATSTFK#NPTY 785 |
| Canis familiaris | Query 763 EFAKFEKEKMNAKWDTGENPIYKSAVTTVV#NPKYEGK 799 |||||||||||||||| ||||||| + #|| | | Sbjct 785 EFAKFEKEKMNAKWDTQENPIYKSPINNFK#NPNYGRK 821 |
| Crassostrea gigas | Query 763 EFAKFEKEKMNAKWDTGENPIYKSAVTTVV#NPKY 796 | |+|||| |||+|||||||||| | +| |#|| | Sbjct 764 ELARFEKEAMNARWDTGENPIYKQATSTFV#NPTY 797 |
| Podocoryne carnea | Query 763 EFAKFEKEKMNAKWDTGENPIYKSAVTTVV#NPKYEGK 799 |||||||++ | |||+||||||| | +| #|| | | Sbjct 745 EFAKFEKDRQNPKWDSGENPIYKKATSTFQ#NPMYGNK 781 |
| Caenorhabditis briggsae | Query 763 EFAKFEKEKMNAKWDTGENPIYKSAVTTVV#NPKYEGK 799 |+||| |++ ||||| |||||| | || #|| | || Sbjct 772 EYAKFNNERLMAKWDTNENPIYKQATTTFK#NPVYAGK 808 |
| Caenorhabditis elegans | Query 763 EFAKFEKEKMNAKWDTGENPIYKSAVTTVV#NPKYEGK 799 |+| | |++ ||||| |||||| | || #|| | || Sbjct 771 EYATFNNERLMAKWDTNENPIYKQATTTFK#NPVYAGK 807 |
| Macaca mulatta | Query 763 EFAKFEKEKMNAKWDTGENPIYKSAVTTVV#NPKYEG 798 ||||||+|+ ||||| ||+|| | +| #| | | Sbjct 752 EFAKFEEERARAKWDTANNPLYKEATSTFT#NITYRG 787 |
| rats, Peptide Partial, 723 aa | Query 763 EFAKFEKEKMNAKWDTGENPIYKSAVTTVV#NPKYEG 798 ||||||+|+ ||||| ||+|| | +| #| | | Sbjct 687 EFAKFEEERARAKWDTANNPLYKEATSTFT#NITYRG 722 |
| Canis lupus familiaris | Query 763 EFAKFEKEKMNAKWDTGENPIYKSAVTTVV#NPKYEG 798 ||||||+|+ ||||| ||+|| | +| #| | | Sbjct 748 EFAKFEEERARAKWDTANNPLYKEATSTFT#NITYRG 783 |
| Equus caballus | Query 763 EFAKFEKEKMNAKWDTGENPIYKSAVTTVV#NPKYEG 798 ||||||+|+ ||||| ||+|| | +| #| | | Sbjct 748 EFAKFEEERARAKWDTANNPLYKEATSTFT#NITYRG 783 |
| Mus sp. | Query 763 EFAKFEKEKMNAKWDTGENPIYKSAVTTVV#NPKYEG 798 |+ +||||+ | ||+||||+|| |#||+++| Sbjct 756 EYRRFEKEQQQLNWKQDNNPLYKSAITTTV#NPRFQG 791 |
| mice, Peptide Partial, 680 aa | Query 763 EFAKFEKEKMNAKWDTGENPIYKSAVTTVV#N 793 ||||||+|+ ||||| ||+|| | +| #| Sbjct 648 EFAKFEEERARAKWDTANNPLYKEATSTFT#N 678 |
| Strongylocentrotus purpuratus | Query 763 EFAKFEKEKMNAKWDTGENPIYKSAVTTVV#NPKYEGK 799 || ||||+ || |+ ||||||| + + #|| | | Sbjct 770 EFQNFEKERANATWEGGENPIYKPSTSVFK#NPTYNIK 806 |
| Lytechinus variegatus | Query 763 EFAKFEKEKMNAKWDTGENPIYKSAVTTVV#NPKYEGK 799 || ||||+ || |+ ||||||| + + #|| | | Sbjct 770 EFQNFEKERANATWEGGENPIYKPSTSVFK#NPTYNIK 806 |
| Acropora millepora | Query 763 EFAKFEKEKMNAKWDTGENPIYKSAVTTVV#NPKYEG 798 |+ |||+|+|++|| +||+|++| || #|| | | Sbjct 754 EYQKFERERMHSKWTREKNPLYQAAKTTFE#NPTYAG 789 |
| Oncorhynchus mykiss | Query 763 EFAKFEKEKMNAKWDTGENPIYKSAVTTVV#NPKYEGK 799 || +|| |+ || |+||++ +| ||| #|| + |+ Sbjct 741 EFRRFENEQKKGKWSPGDNPLFMNATTTVA#NPTFTGE 777 |
| Ovis aries | Query 763 EFAKFEKEKMNAKWDTGENPIYKSAVTTVV#NPKY 796 |+ +|||||+ ++|+ +||++||| |||+#|||+ Sbjct 735 EYHRFEKEKLKSQWNN-DNPLFKSATTTVM#NPKF 767 |
| Ovis canadensis | Query 763 EFAKFEKEKMNAKWDTGENPIYKSAVTTVV#NPKY 796 |+ +|||||+ ++|+ +||++||| |||+#|||+ Sbjct 735 EYHRFEKEKLKSQWNN-DNPLFKSATTTVM#NPKF 767 |
| Bubalus bubalis | Query 763 EFAKFEKEKMNAKWDTGENPIYKSAVTTVV#NPKY 796 |+ +|||||+ ++|+ +||++||| |||+#|||+ Sbjct 734 EYHRFEKEKLKSQWNN-DNPLFKSATTTVM#NPKF 766 |
| Capra hircus | Query 763 EFAKFEKEKMNAKWDTGENPIYKSAVTTVV#NPKY 796 |+ +|||||+ ++|+ +||++||| |||+#|||+ Sbjct 735 EYHRFEKEKLKSQWNN-DNPLFKSATTTVM#NPKF 767 |
| Sigmodon hispidus | Query 763 EFAKFEKEKMNAKWDTGENPIYKSAVTTVV#NPKY 796 |+ |||||+ ++|+ +||++||| |||+#|||+ Sbjct 734 EYRHFEKEKLKSQWNN-DNPLFKSATTTVM#NPKF 766 |
| Schistosoma japonicum | Query 763 EFAKFEKEKMNAKWDTGENPIYKSAVTTVV#NPKYE 797 | | |+++ | +|+ ||||++| | |+#|| +| Sbjct 250 ELANFKQQGENMRWEMAENPIFESPTTNVL#NPTFE 284 |
| Pacifastacus leniusculus | Query 763 EFAKFEKEKMNAKWDTGENPIYKSAVTTVV#NPKY 796 |+||||||+ |||+|||| || #|| + Sbjct 764 EYAKFEKERKLPSGKRAENPLYKSAKTTFQ#NPAF 797 |
| Apis mellifera | Query 763 EFAKFEKEKMNA-KWDTGENPIYKSAVTTVV#NPKY 796 || |||+|++ | ||+ +||+|| | +| #|| + Sbjct 779 EFKKFERERILANKWNRRDNPLYKEATSTFK#NPTF 813 |
| Halocynthia roretzi | Query 763 EFAKFEKEKMNAKWDTGENPIYKSAVTTVV#NPKYEG 798 |+ ||+||+ || ||||+| +| + #|| + | Sbjct 798 EYEKFKKEEDLRKWTKGENPVYVNASSKFD#NPMFGG 833 |
| Ophlitaspongia tenuis | Query 763 EFAKFEKEKMNAKWDTGENPIYKSAVTTVV#NPKY 796 | |||+| |||+ |||+|+|| #|| | Sbjct 803 EVRKFEREIKNAKYTKNENPLYRSATKDYQ#NPLY 836 |
| Cyprinus carpio | Query 763 EFAKFEKEKMNAKWDTGENPIYKSAVTTVV#NPKYEG 798 |+ +|||++ + | +| ||++++| ||| #|| + | Sbjct 731 EWKRFEKDRKHEK-TSGTNPLFQNATTTVQ#NPTFSG 765 |
| Geodia cydonium | Query 763 EFAKFEKEKMNAKWDTGENPIYKSAVTTVV#NPKY 796 | ||| | |+ |||+| || | #|| | Sbjct 843 ELKKFENELAKVKYTKNENPLYHSATTEHK#NPIY 876 |
| Suberites domuncula | Query 763 EFAKFEKEKMNAKWDTGENPIYKSAVTTVV#NPKY 796 | ||||| |+ +||+|+|| #|| | Sbjct 729 EVKKFEKELAKTKYPKHQNPLYRSAAKDYQ#NPIY 762 |
[Site 2] KEKMNAKWDT778-GENPIYKSAV
Thr778  Gly
Gly
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Lys769 | Glu770 | Lys771 | Met772 | Asn773 | Ala774 | Lys775 | Trp776 | Asp777 | Thr778 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Gly779 | Glu780 | Asn781 | Pro782 | Ile783 | Tyr784 | Lys785 | Ser786 | Ala787 | Val788 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| LLIWKLLMIIHDRREFAKFEKEKMNAKWDTGENPIYKSAVTTVVNPKYEGK |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | N/A | 109.00 | 33 | I |
| 2 | Homo sapiens | 109.00 | 25 | ITB1_HUMAN Integrin beta-1 precursor (Fibronectin |
| 3 | Mus musculus | 109.00 | 18 | integrin beta 1 (fibronectin receptor beta) |
| 4 | Pan troglodytes | 109.00 | 14 | PREDICTED: hypothetical protein |
| 5 | Bos taurus | 109.00 | 13 | integrin beta 1 |
| 6 | Rattus norvegicus | 109.00 | 10 | integrin beta 1 |
| 7 | Tetraodon nigroviridis | 109.00 | 8 | unnamed protein product |
| 8 | Gallus gallus | 109.00 | 7 | integrin, beta 1 (fibronectin receptor, beta polyp |
| 9 | Monodelphis domestica | 109.00 | 6 | PREDICTED: similar to integrin beta-1 subunit isof |
| 10 | Xenopus laevis | 109.00 | 6 | integrin beta 1 |
| 11 | Sus scrofa | 109.00 | 6 | integrin beta-1 subunit |
| 12 | synthetic construct | 109.00 | 5 | hypothetical protein |
| 13 | Oryctolagus cuniculus | 109.00 | 2 | integrin beta 1 |
| 14 | Pongo pygmaeus | 109.00 | 2 | ITB1_PONPY Integrin beta-1 precursor (Fibronectin |
| 15 | Xenopus tropicalis | 109.00 | 1 | integrin beta 1 (fibronectin receptor beta) |
| 16 | Danio rerio | 108.00 | 14 | integrin beta1 subunit-like protein 1 |
| 17 | Felis catus | 107.00 | 1 | integrin beta 1 |
| 18 | Ictalurus punctatus | 105.00 | 2 | beta-1 integrin |
| 19 | Drosophila melanogaster | 88.60 | 1 | myospheroid CG1560-PA |
| 20 | Canis familiaris | 88.20 | 10 | PREDICTED: similar to integrin beta 1 isoform 1D p |
| 21 | Aedes aegypti | 87.00 | 1 | integrin beta subunit |
| 22 | Anopheles gambiae | 85.50 | 2 | integrin beta subunit |
| 23 | Anopheles gambiae str. PEST | 85.50 | 2 | ENSANGP00000021373 |
| 24 | Tribolium castaneum | 82.80 | 3 | PREDICTED: similar to CG1560-PA |
| 25 | Ostrinia furnacalis | 82.80 | 1 | integrin beta 1 precursor |
| 26 | Biomphalaria glabrata | 82.00 | 1 | beta integrin subunit |
| 27 | Pseudoplusia includens | 79.70 | 1 | integrin beta 1 |
| 28 | Podocoryne carnea | 78.20 | 1 | AF308652_1 integrin beta chain |
| 29 | Caenorhabditis briggsae | 77.80 | 1 | Hypothetical protein CBG03601 |
| 30 | Crassostrea gigas | 77.00 | 1 | integrin beta cgh |
| 31 | Caenorhabditis elegans | 75.50 | 1 | Paralysed Arrest at Two-fold family member (pat-3) |
| 32 | Macaca mulatta | 74.30 | 13 | PREDICTED: similar to integrin beta chain, beta 3 |
| 33 | Canis lupus familiaris | 74.30 | 1 | integrin beta chain, beta 3 |
| 34 | Equus caballus | 74.30 | 1 | integrin, beta 3 (platelet glycoprotein IIIa, anti |
| 35 | rats, Peptide Partial, 723 aa | 74.30 | 1 | beta 3 integrin, GPIIIA |
| 36 | mice, Peptide Partial, 680 aa | 70.90 | 1 | beta 3 integrin, GPIIIA |
| 37 | Strongylocentrotus purpuratus | 69.70 | 5 | integrin beta-C subunit |
| 38 | Lytechinus variegatus | 69.70 | 1 | beta-C integrin subunit |
| 39 | Ovis aries | 63.20 | 2 | ITB6_SHEEP Integrin beta-6 precursor emb |
| 40 | Pacifastacus leniusculus | 62.80 | 1 | integrin |
| 41 | Mus sp. | 60.80 | 1 | beta 7 integrin |
| 42 | Capra hircus | 58.90 | 1 | ITB2_CAPHI Integrin beta-2 precursor (Cell surface |
| 43 | Bubalus bubalis | 58.90 | 1 | integrin beta-2 |
| 44 | Ovis canadensis | 58.20 | 1 | ITB2_OVICA Integrin beta-2 precursor (Cell surface |
| 45 | Sigmodon hispidus | 57.80 | 1 | AF445415_1 integrin beta-2 precursor |
| 46 | Schistosoma japonicum | 57.00 | 1 | SJCHGC06221 protein |
| 47 | Apis mellifera | 56.20 | 1 | PREDICTED: similar to myospheroid CG1560-PA |
| 48 | Halocynthia roretzi | 54.70 | 2 | integrin beta Hr2 precursor |
| 49 | Oncorhynchus mykiss | 54.70 | 1 | CD18 protein |
| 50 | Acropora millepora | 52.80 | 1 | integrin subunit betaCn1 |
| 51 | Manduca sexta | 49.70 | 1 | plasmatocyte-specific integrin beta 1 |
| 52 | Spodoptera frugiperda | 47.40 | 1 | integrin beta 1 |
| 53 | Ophlitaspongia tenuis | 44.70 | 1 | integrin subunit betaPo1 |
| 54 | Cyprinus carpio | 44.30 | 2 | CD18 type 2 |
| 55 | Geodia cydonium | 43.10 | 1 | integrin beta subunit |
| 56 | Suberites domuncula | 41.20 | 1 | integrin beta subunit |
| 57 | Drosophila pseudoobscura | 33.10 | 1 | GA14581-PA |
Top-ranked sequences
| organism | matching |
|---|---|
| N/A | Query 749 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#GENPIYKSAVTTVVNPKYEGK 799 ||||||||||||||||||||||||||||||#||||||||||||||||||||| Query 749 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#GENPIYKSAVTTVVNPKYEGK 799 |
| Homo sapiens | Query 749 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#GENPIYKSAVTTVVNPKYEGK 799 ||||||||||||||||||||||||||||||#||||||||||||||||||||| Query 749 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#GENPIYKSAVTTVVNPKYEGK 799 |
| Mus musculus | Query 749 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#GENPIYKSAVTTVVNPKYEGK 799 ||||||||||||||||||||||||||||||#||||||||||||||||||||| Query 749 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#GENPIYKSAVTTVVNPKYEGK 799 |
| Pan troglodytes | Query 749 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#GENPIYKSAVTTVVNPKYEGK 799 ||||||||||||||||||||||||||||||#||||||||||||||||||||| Query 749 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#GENPIYKSAVTTVVNPKYEGK 799 |
| Bos taurus | Query 749 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#GENPIYKSAVTTVVNPKYEGK 799 ||||||||||||||||||||||||||||||#||||||||||||||||||||| Query 749 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#GENPIYKSAVTTVVNPKYEGK 799 |
| Rattus norvegicus | Query 749 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#GENPIYKSAVTTVVNPKYEGK 799 ||||||||||||||||||||||||||||||#||||||||||||||||||||| Query 749 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#GENPIYKSAVTTVVNPKYEGK 799 |
| Tetraodon nigroviridis | Query 749 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#GENPIYKSAVTTVVNPKYEGK 799 ||||||||||||||||||||||||||||||#||||||||||||||||||||| Query 749 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#GENPIYKSAVTTVVNPKYEGK 799 |
| Gallus gallus | Query 749 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#GENPIYKSAVTTVVNPKYEGK 799 ||||||||||||||||||||||||||||||#||||||||||||||||||||| Query 749 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#GENPIYKSAVTTVVNPKYEGK 799 |
| Monodelphis domestica | Query 749 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#GENPIYKSAVTTVVNPKYEGK 799 ||||||||||||||||||||||||||||||#||||||||||||||||||||| Query 749 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#GENPIYKSAVTTVVNPKYEGK 799 |
| Xenopus laevis | Query 749 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#GENPIYKSAVTTVVNPKYEGK 799 ||||||||||||||||||||||||||||||#||||||||||||||||||||| Query 749 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#GENPIYKSAVTTVVNPKYEGK 799 |
| Sus scrofa | Query 749 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#GENPIYKSAVTTVVNPKYEGK 799 ||||||||||||||||||||||||||||||#||||||||||||||||||||| Query 749 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#GENPIYKSAVTTVVNPKYEGK 799 |
| synthetic construct | Query 749 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#GENPIYKSAVTTVVNPKYEGK 799 ||||||||||||||||||||||||||||||#||||||||||||||||||||| Query 749 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#GENPIYKSAVTTVVNPKYEGK 799 |
| Oryctolagus cuniculus | Query 749 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#GENPIYKSAVTTVVNPKYEGK 799 ||||||||||||||||||||||||||||||#||||||||||||||||||||| Query 749 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#GENPIYKSAVTTVVNPKYEGK 799 |
| Pongo pygmaeus | Query 749 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#GENPIYKSAVTTVVNPKYEGK 799 ||||||||||||||||||||||||||||||#||||||||||||||||||||| Query 749 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#GENPIYKSAVTTVVNPKYEGK 799 |
| Xenopus tropicalis | Query 749 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#GENPIYKSAVTTVVNPKYEGK 799 ||||||||||||||||||||||||||||||#||||||||||||||||||||| Query 749 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#GENPIYKSAVTTVVNPKYEGK 799 |
| Danio rerio | Query 749 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#GENPIYKSAVTTVVNPKYEGK 799 ||||||||||||||||||||||||||||||#|||||||||||||+||||||| Sbjct 748 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#GENPIYKSAVTTVINPKYEGK 798 |
| Felis catus | Query 749 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#GENPIYKSAVTTVVNPKYEGK 799 |||||||||||| |||||||||||||||||#||||||||||||||||||||| Sbjct 748 LLIWKLLMIIHDTREFAKFEKEKMNAKWDT#GENPIYKSAVTTVVNPKYEGK 798 |
| Ictalurus punctatus | Query 749 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#GENPIYKSAVTTVVNPKYEGK 799 | ||||||||||||||||||||||||||| #||||||||||||||||||||| Sbjct 757 LFIWKLLMIIHDRREFAKFEKEKMNAKWDA#GENPIYKSAVTTVVNPKYEGK 807 |
| Drosophila melanogaster | Query 749 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#GENPIYKSAVTTVVNPKYEGK 799 ||+|||| ||||||||+||||+|||||||#||||||| | +| || | || Sbjct 796 LLLWKLLTTIHDRREFARFEKERMNAKWDT#GENPIYKQATSTFKNPMYAGK 846 |
| Canis familiaris | Query 749 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#GENPIYKSAVTTVVNPKYEGK 799 ||||||||||||||||||||||||||||||# ||||||| + || | | Sbjct 771 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#QENPIYKSPINNFKNPNYGRK 821 |
| Aedes aegypti | Query 749 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#GENPIYKSAVTTVVNPKYEGK 799 ||+|||| ||||||||+||||+| |||||#||||||| | || || | || Sbjct 785 LLLWKLLTTIHDRREFARFEKERMMAKWDT#GENPIYKQATTTFKNPTYAGK 835 |
| Anopheles gambiae | Query 749 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#GENPIYKSAVTTVVNPKYEGK 799 ||+||+| ||||||||+||||+| |||||#||||||| | || || | || Sbjct 787 LLLWKVLTSIHDRREFARFEKERMMAKWDT#GENPIYKQATTTFKNPTYAGK 837 |
| Anopheles gambiae str. PEST | Query 749 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#GENPIYKSAVTTVVNPKYEGK 799 ||+||+| ||||||||+||||+| |||||#||||||| | || || | || Sbjct 787 LLLWKVLTSIHDRREFARFEKERMMAKWDT#GENPIYKQATTTFKNPTYAGK 837 |
| Tribolium castaneum | Query 749 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#GENPIYKSAVTTVVNPKYEGK 799 ||+||| ||||||||||||| | |+|||#||||||| | +| || | || Sbjct 779 LLLWKLFTTIHDRREFAKFEKEAMMARWDT#GENPIYKQATSTFKNPTYAGK 829 |
| Ostrinia furnacalis | Query 749 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#GENPIYKSAVTTVVNPKYEGK 799 |++||++ ||||||||+||||+| |||||#||||||| | +| || | || Sbjct 781 LMLWKMVTTIHDRREFARFEKERMMAKWDT#GENPIYKQATSTFKNPTYAGK 831 |
| Biomphalaria glabrata | Query 749 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#GENPIYKSAVTTVVNPKY 796 ||+|||| ||| |||||||||+ ||||||#||||||| | +| || | Sbjct 738 LLLWKLLTFIHDTREFAKFEKERQNAKWDT#GENPIYKQATSTFKNPTY 785 |
| Pseudoplusia includens | Query 749 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#GENPIYKSAVTTVVNPKYEGK 799 |++||+ |||||||+||||+| |||||#||||||| | +| || | || Sbjct 787 LMLWKMATTSHDRREFARFEKERMMAKWDT#GENPIYKQATSTFKNPTYAGK 837 |
| Podocoryne carnea | Query 749 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#GENPIYKSAVTTVVNPKYEGK 799 ||||||| | ||||||||||++ | |||+#||||||| | +| || | | Sbjct 731 LLIWKLLATIQDRREFAKFEKDRQNPKWDS#GENPIYKKATSTFQNPMYGNK 781 |
| Caenorhabditis briggsae | Query 749 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#GENPIYKSAVTTVVNPKYEGK 799 ||+|||| ++||| |+||| |++ |||||# |||||| | || || | || Sbjct 758 LLLWKLLTVLHDRAEYAKFNNERLMAKWDT#NENPIYKQATTTFKNPVYAGK 808 |
| Crassostrea gigas | Query 749 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#GENPIYKSAVTTVVNPKY 796 |||||| | |+|| |+|||| |||+|||#||||||| | +| ||| | Sbjct 750 LLIWKLFTTISDKRELARFEKEAMNARWDT#GENPIYKQATSTFVNPTY 797 |
| Caenorhabditis elegans | Query 749 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#GENPIYKSAVTTVVNPKYEGK 799 ||+|||| ++||| |+| | |++ |||||# |||||| | || || | || Sbjct 757 LLLWKLLTVLHDRSEYATFNNERLMAKWDT#NENPIYKQATTTFKNPVYAGK 807 |
| Macaca mulatta | Query 749 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#GENPIYKSAVTTVVNPKYEG 798 |||||||+ ||||+||||||+|+ |||||# ||+|| | +| | | | Sbjct 738 LLIWKLLITIHDRKEFAKFEEERARAKWDT#ANNPLYKEATSTFTNITYRG 787 |
| Canis lupus familiaris | Query 749 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#GENPIYKSAVTTVVNPKYEG 798 |||||||+ ||||+||||||+|+ |||||# ||+|| | +| | | | Sbjct 734 LLIWKLLITIHDRKEFAKFEEERARAKWDT#ANNPLYKEATSTFTNITYRG 783 |
| Equus caballus | Query 749 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#GENPIYKSAVTTVVNPKYEG 798 |||||||+ ||||+||||||+|+ |||||# ||+|| | +| | | | Sbjct 734 LLIWKLLITIHDRKEFAKFEEERARAKWDT#ANNPLYKEATSTFTNITYRG 783 |
| rats, Peptide Partial, 723 aa | Query 749 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#GENPIYKSAVTTVVNPKYEG 798 |||||||+ ||||+||||||+|+ |||||# ||+|| | +| | | | Sbjct 673 LLIWKLLITIHDRKEFAKFEEERARAKWDT#ANNPLYKEATSTFTNITYRG 722 |
| mice, Peptide Partial, 680 aa | Query 749 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#GENPIYKSAVTTVVN 793 |||||||+ ||||+||||||+|+ |||||# ||+|| | +| | Sbjct 634 LLIWKLLITIHDRKEFAKFEEERARAKWDT#ANNPLYKEATSTFTN 678 |
| Strongylocentrotus purpuratus | Query 749 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#GENPIYKSAVTTVVNPKYEGK 799 ||||+|| ||||||| ||||+ || |+ #||||||| + + || | | Sbjct 756 LLIWRLLTYIHDRREFQNFEKERANATWEG#GENPIYKPSTSVFKNPTYNIK 806 |
| Lytechinus variegatus | Query 749 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#GENPIYKSAVTTVVNPKYEGK 799 ||||+|| ||||||| ||||+ || |+ #||||||| + + || | | Sbjct 756 LLIWRLLTYIHDRREFQNFEKERANATWEG#GENPIYKPSTSVFKNPTYNIK 806 |
| Ovis aries | Query 749 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#GENPIYKSAVTTVVNPKYEGK 799 | |||||+ |||+| |||| |+ ||| |#| ||+|+ + +| | |+ | Sbjct 726 LCIWKLLVSFHDRKEVAKFEAERSKAKWQT#GTNPLYRGSTSTFKNVTYKHK 776 |
| Pacifastacus leniusculus | Query 749 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#GENPIYKSAVTTVVNPKY 796 ||||||| +|||||+||||||+ # |||+|||| || || + Sbjct 750 LLIWKLLTTLHDRREYAKFEKERKLPSGKR#AENPLYKSAKTTFQNPAF 797 |
| Mus sp. | Query 749 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#GENPIYKSAVTTVVNPKYEG 798 +| ++| + |+||||+ +||||+ | # ||+||||+|| |||+++| Sbjct 742 VLAYRLSVEIYDRREYRRFEKEQQQLNWKQ#DNNPLYKSAITTTVNPRFQG 791 |
| Capra hircus | Query 749 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#GENPIYKSAVTTVVNPKY 796 |+||| | + | ||+ +|||||+ ++|+ # +||++||| |||+|||+ Sbjct 721 LVIWKALTHLSDLREYHRFEKEKLKSQWN-#NDNPLFKSATTTVMNPKF 767 |
| Bubalus bubalis | Query 749 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#GENPIYKSAVTTVVNPKY 796 |+||| | + | ||+ +|||||+ ++|+ # +||++||| |||+|||+ Sbjct 720 LVIWKALTHLSDLREYHRFEKEKLKSQWN-#NDNPLFKSATTTVMNPKF 766 |
| Ovis canadensis | Query 749 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#GENPIYKSAVTTVVNPKY 796 | ||| | + | ||+ +|||||+ ++|+ # +||++||| |||+|||+ Sbjct 721 LAIWKALTHLSDLREYHRFEKEKLKSQWN-#NDNPLFKSATTTVMNPKF 767 |
| Sigmodon hispidus | Query 749 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#GENPIYKSAVTTVVNPKY 796 |+||| | + | ||+ |||||+ ++|+ # +||++||| |||+|||+ Sbjct 720 LVIWKALTHLTDLREYRHFEKEKLKSQWN-#NDNPLFKSATTTVMNPKF 766 |
| Schistosoma japonicum | Query 749 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#GENPIYKSAVTTVVNPKYE 797 |||+||++ | |||| | |+++ | +|+ # ||||++| | |+|| +| Sbjct 236 LLIYKLVITIDDRRELANFKQQGENMRWEM#AENPIFESPTTNVLNPTFE 284 |
| Apis mellifera | Query 749 LLIWKLLMIIHDRREFAKFEKEKMNA-KWDT#GENPIYKSAVTTVVNPKY 796 || ||+| +| | ||| |||+|++ | ||+ # +||+|| | +| || + Sbjct 765 LLTWKILTMIRDNREFKKFERERILANKWNR#RDNPLYKEATSTFKNPTF 813 |
| Halocynthia roretzi | Query 749 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#GENPIYKSAVTTVVNPKYEG 798 |+|||++ + |+||+ ||+||+ || #||||+| +| + || + | Sbjct 784 LIIWKVIQTLRDKREYEKFKKEEDLRKWTK#GENPVYVNASSKFDNPMFGG 833 |
| Oncorhynchus mykiss | Query 749 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#GENPIYKSAVTTVVNPKYEGK 799 ||| | ++ + | +|| +|| |+ || #|+||++ +| ||| || + |+ Sbjct 727 LLIIKAILYMRDVKEFRRFENEQKKGKWSP#GDNPLFMNATTTVANPTFTGE 777 |
| Acropora millepora | Query 749 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#GENPIYKSAVTTVVNPKYEG 798 |++ | | + || |+ |||+|+|++|| # +||+|++| || || | | Sbjct 740 LIVIKGLFTMVDRIEYQKFERERMHSKWTR#EKNPLYQAAKTTFENPTYAG 789 |
| Manduca sexta | Query 749 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#GENPIYKSAVTTVVNPKY 796 |+ ||+|+ +||+||+||||+| + +| # ||+|+ || | Sbjct 714 LIAWKILVDLHDKREYAKFEEESRSRGFDV#SLNPLYQEPEINFSNPVY 761 |
| Spodoptera frugiperda | Query 749 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#GENPIYKSAVTTVVNPKYEGK 799 ++|||+|+ +||++|+ ||+ | + | +| # ||+|+ || | + Sbjct 729 VIIWKILVDMHDKKEYKKFQDEALAAGYDV#SLNPLYQDPSINFSNPVYNNQ 779 |
| Ophlitaspongia tenuis | Query 749 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#GENPIYKSAVTTVVNPKY 796 ||+ | |+++ | | |||+| |||+ # |||+|+|| || | Sbjct 789 LLLLKGLLLLWDVVEVRKFEREIKNAKYTK#NENPLYRSATKDYQNPLY 836 |
| Cyprinus carpio | Query 749 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#GENPIYKSAVTTVVNPKYEG 798 ||| | ++ | ||+ +|||++ + | +#| ||++++| ||| || + | Sbjct 717 LLIIKAVLYASDLREWKRFEKDRKHEK-TS#GTNPLFQNATTTVQNPTFSG 765 |
| Geodia cydonium | Query 749 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#GENPIYKSAVTTVVNPKY 796 |++ |+|+++ | | ||| | |+ # |||+| || | || | Sbjct 829 LILLKILLMVLDYTELKKFENELAKVKYTK#NENPLYHSATTEHKNPIY 876 |
| Suberites domuncula | Query 749 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#GENPIYKSAVTTVVNPKY 796 ||+ || + | | | ||||| |+ # +||+|+|| || | Sbjct 715 LLLLKLFLFIIDVIEVKKFEKELAKTKYPK#HQNPLYRSAAKDYQNPIY 762 |
| Drosophila pseudoobscura | Query 749 LLIWKLLMIIH--DRREFAKFEKEKMNAKWDT#GENPIYKSAVTTVVNPK 795 |||+ + | | ||+|+||+|+ || +# |||+|+ | | || Sbjct 713 LLIFLFIWCIRRKDAREYARFEEEQ--AKRVS#QENPLYRDPVGRYVVPK 759 |
[Site 3] NPIYKSAVTT790-VVNPKYEGK
Thr790  Val
Val
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Asn781 | Pro782 | Ile783 | Tyr784 | Lys785 | Ser786 | Ala787 | Val788 | Thr789 | Thr790 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Val791 | Val792 | Asn793 | Pro794 | Lys795 | Tyr796 | Glu797 | Gly798 | Lys799 | - |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| RREFAKFEKEKMNAKWDTGENPIYKSAVTTVVNPKYEGK |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | N/A | 85.10 | 31 | I |
| 2 | Homo sapiens | 85.10 | 25 | ITB1_HUMAN Integrin beta-1 precursor (Fibronectin |
| 3 | Mus musculus | 85.10 | 14 | integrin beta 1 (fibronectin receptor beta) |
| 4 | Pan troglodytes | 85.10 | 14 | PREDICTED: hypothetical protein |
| 5 | Bos taurus | 85.10 | 13 | integrin beta 1 |
| 6 | Rattus norvegicus | 85.10 | 10 | integrin beta 1 |
| 7 | Gallus gallus | 85.10 | 7 | integrin, beta 1 (fibronectin receptor, beta polyp |
| 8 | Monodelphis domestica | 85.10 | 6 | PREDICTED: similar to integrin beta-1 subunit isof |
| 9 | Sus scrofa | 85.10 | 6 | integrin beta-1 subunit |
| 10 | Xenopus laevis | 85.10 | 6 | integrin beta 1 |
| 11 | Tetraodon nigroviridis | 85.10 | 5 | unnamed protein product |
| 12 | synthetic construct | 85.10 | 5 | hypothetical protein |
| 13 | Oryctolagus cuniculus | 85.10 | 2 | integrin beta 1 |
| 14 | Pongo pygmaeus | 85.10 | 2 | ITB1_PONPY Integrin beta-1 precursor (Fibronectin |
| 15 | Xenopus tropicalis | 85.10 | 1 | integrin beta 1 (fibronectin receptor beta) |
| 16 | Danio rerio | 84.70 | 14 | integrin beta1 subunit-like protein 1 |
| 17 | Ictalurus punctatus | 83.20 | 2 | beta-1 integrin |
| 18 | Felis catus | 83.20 | 1 | integrin beta 1 |
| 19 | Drosophila melanogaster | 69.30 | 1 | myospheroid CG1560-PA |
| 20 | Anopheles gambiae str. PEST | 67.80 | 1 | ENSANGP00000021373 |
| 21 | Anopheles gambiae | 67.80 | 1 | integrin beta subunit |
| 22 | Aedes aegypti | 67.80 | 1 | integrin beta subunit |
| 23 | Pseudoplusia includens | 66.20 | 1 | integrin beta 1 |
| 24 | Ostrinia furnacalis | 66.20 | 1 | integrin beta 1 precursor |
| 25 | Tribolium castaneum | 65.10 | 2 | PREDICTED: similar to CG1560-PA |
| 26 | Canis familiaris | 63.90 | 10 | PREDICTED: similar to integrin beta 1 isoform 1D p |
| 27 | Biomphalaria glabrata | 62.80 | 1 | beta integrin subunit |
| 28 | Crassostrea gigas | 62.00 | 1 | integrin beta cgh |
| 29 | Podocoryne carnea | 61.20 | 1 | AF308652_1 integrin beta chain |
| 30 | Caenorhabditis briggsae | 57.80 | 1 | Hypothetical protein CBG03601 |
| 31 | Caenorhabditis elegans | 55.50 | 1 | Paralysed Arrest at Two-fold family member (pat-3) |
| 32 | Macaca mulatta | 53.50 | 13 | PREDICTED: similar to integrin beta chain, beta 3 |
| 33 | Equus caballus | 53.50 | 1 | integrin, beta 3 (platelet glycoprotein IIIa, anti |
| 34 | Canis lupus familiaris | 53.50 | 1 | integrin beta chain, beta 3 |
| 35 | rats, Peptide Partial, 723 aa | 53.50 | 1 | beta 3 integrin, GPIIIA |
| 36 | Mus sp. | 53.10 | 1 | beta 7 integrin |
| 37 | Strongylocentrotus purpuratus | 50.80 | 5 | integrin beta-C subunit |
| 38 | Lytechinus variegatus | 50.80 | 1 | beta-C integrin subunit |
| 39 | mice, Peptide Partial, 680 aa | 50.10 | 1 | beta 3 integrin, GPIIIA |
| 40 | Ovis aries | 47.80 | 2 | antigen CD18 |
| 41 | Bubalus bubalis | 47.80 | 1 | integrin beta-2 |
| 42 | Capra hircus | 47.80 | 1 | ITB2_CAPHI Integrin beta-2 precursor (Cell surface |
| 43 | Ovis canadensis | 47.80 | 1 | ITB2_OVICA Integrin beta-2 precursor (Cell surface |
| 44 | Acropora millepora | 47.40 | 1 | integrin subunit betaCn1 |
| 45 | Sigmodon hispidus | 47.00 | 1 | AF445415_1 integrin beta-2 precursor |
| 46 | Oncorhynchus mykiss | 47.00 | 1 | CD18 protein |
| 47 | Schistosoma japonicum | 44.30 | 1 | SJCHGC06221 protein |
| 48 | Pacifastacus leniusculus | 43.50 | 1 | integrin |
| 49 | Apis mellifera | 41.20 | 1 | PREDICTED: similar to myospheroid CG1560-PA |
| 50 | Halocynthia roretzi | 40.80 | 1 | integrin beta Hr2 precursor |
| 51 | Ophlitaspongia tenuis | 38.10 | 1 | integrin subunit betaPo1 |
| 52 | Cyprinus carpio | 37.70 | 2 | CD18 type 2 |
| 53 | Geodia cydonium | 34.30 | 1 | integrin beta subunit |
| 54 | Manduca sexta | 33.90 | 1 | plasmatocyte-specific integrin beta 1 |
Top-ranked sequences
| organism | matching |
|---|---|
| N/A | Query 761 RREFAKFEKEKMNAKWDTGENPIYKSAVTT#VVNPKYEGK 799 ||||||||||||||||||||||||||||||#||||||||| Query 761 RREFAKFEKEKMNAKWDTGENPIYKSAVTT#VVNPKYEGK 799 |
| Homo sapiens | Query 761 RREFAKFEKEKMNAKWDTGENPIYKSAVTT#VVNPKYEGK 799 ||||||||||||||||||||||||||||||#||||||||| Query 761 RREFAKFEKEKMNAKWDTGENPIYKSAVTT#VVNPKYEGK 799 |
| Mus musculus | Query 761 RREFAKFEKEKMNAKWDTGENPIYKSAVTT#VVNPKYEGK 799 ||||||||||||||||||||||||||||||#||||||||| Query 761 RREFAKFEKEKMNAKWDTGENPIYKSAVTT#VVNPKYEGK 799 |
| Pan troglodytes | Query 761 RREFAKFEKEKMNAKWDTGENPIYKSAVTT#VVNPKYEGK 799 ||||||||||||||||||||||||||||||#||||||||| Query 761 RREFAKFEKEKMNAKWDTGENPIYKSAVTT#VVNPKYEGK 799 |
| Bos taurus | Query 761 RREFAKFEKEKMNAKWDTGENPIYKSAVTT#VVNPKYEGK 799 ||||||||||||||||||||||||||||||#||||||||| Query 761 RREFAKFEKEKMNAKWDTGENPIYKSAVTT#VVNPKYEGK 799 |
| Rattus norvegicus | Query 761 RREFAKFEKEKMNAKWDTGENPIYKSAVTT#VVNPKYEGK 799 ||||||||||||||||||||||||||||||#||||||||| Query 761 RREFAKFEKEKMNAKWDTGENPIYKSAVTT#VVNPKYEGK 799 |
| Gallus gallus | Query 761 RREFAKFEKEKMNAKWDTGENPIYKSAVTT#VVNPKYEGK 799 ||||||||||||||||||||||||||||||#||||||||| Query 761 RREFAKFEKEKMNAKWDTGENPIYKSAVTT#VVNPKYEGK 799 |
| Monodelphis domestica | Query 761 RREFAKFEKEKMNAKWDTGENPIYKSAVTT#VVNPKYEGK 799 ||||||||||||||||||||||||||||||#||||||||| Query 761 RREFAKFEKEKMNAKWDTGENPIYKSAVTT#VVNPKYEGK 799 |
| Sus scrofa | Query 761 RREFAKFEKEKMNAKWDTGENPIYKSAVTT#VVNPKYEGK 799 ||||||||||||||||||||||||||||||#||||||||| Query 761 RREFAKFEKEKMNAKWDTGENPIYKSAVTT#VVNPKYEGK 799 |
| Xenopus laevis | Query 761 RREFAKFEKEKMNAKWDTGENPIYKSAVTT#VVNPKYEGK 799 ||||||||||||||||||||||||||||||#||||||||| Query 761 RREFAKFEKEKMNAKWDTGENPIYKSAVTT#VVNPKYEGK 799 |
| Tetraodon nigroviridis | Query 761 RREFAKFEKEKMNAKWDTGENPIYKSAVTT#VVNPKYEGK 799 ||||||||||||||||||||||||||||||#||||||||| Query 761 RREFAKFEKEKMNAKWDTGENPIYKSAVTT#VVNPKYEGK 799 |
| synthetic construct | Query 761 RREFAKFEKEKMNAKWDTGENPIYKSAVTT#VVNPKYEGK 799 ||||||||||||||||||||||||||||||#||||||||| Query 761 RREFAKFEKEKMNAKWDTGENPIYKSAVTT#VVNPKYEGK 799 |
| Oryctolagus cuniculus | Query 761 RREFAKFEKEKMNAKWDTGENPIYKSAVTT#VVNPKYEGK 799 ||||||||||||||||||||||||||||||#||||||||| Query 761 RREFAKFEKEKMNAKWDTGENPIYKSAVTT#VVNPKYEGK 799 |
| Pongo pygmaeus | Query 761 RREFAKFEKEKMNAKWDTGENPIYKSAVTT#VVNPKYEGK 799 ||||||||||||||||||||||||||||||#||||||||| Query 761 RREFAKFEKEKMNAKWDTGENPIYKSAVTT#VVNPKYEGK 799 |
| Xenopus tropicalis | Query 761 RREFAKFEKEKMNAKWDTGENPIYKSAVTT#VVNPKYEGK 799 ||||||||||||||||||||||||||||||#||||||||| Query 761 RREFAKFEKEKMNAKWDTGENPIYKSAVTT#VVNPKYEGK 799 |
| Danio rerio | Query 761 RREFAKFEKEKMNAKWDTGENPIYKSAVTT#VVNPKYEGK 799 ||||||||||||||||||||||||||||||#|+||||||| Sbjct 760 RREFAKFEKEKMNAKWDTGENPIYKSAVTT#VINPKYEGK 798 |
| Ictalurus punctatus | Query 761 RREFAKFEKEKMNAKWDTGENPIYKSAVTT#VVNPKYEGK 799 ||||||||||||||||| ||||||||||||#||||||||| Sbjct 769 RREFAKFEKEKMNAKWDAGENPIYKSAVTT#VVNPKYEGK 807 |
| Felis catus | Query 762 REFAKFEKEKMNAKWDTGENPIYKSAVTT#VVNPKYEGK 799 |||||||||||||||||||||||||||||#||||||||| Query 762 REFAKFEKEKMNAKWDTGENPIYKSAVTT#VVNPKYEGK 799 |
| Drosophila melanogaster | Query 761 RREFAKFEKEKMNAKWDTGENPIYKSAVTT#VVNPKYEGK 799 |||||+||||+|||||||||||||| | +|# || | || Sbjct 808 RREFARFEKERMNAKWDTGENPIYKQATST#FKNPMYAGK 846 |
| Anopheles gambiae str. PEST | Query 761 RREFAKFEKEKMNAKWDTGENPIYKSAVTT#VVNPKYEGK 799 |||||+||||+| |||||||||||| | ||# || | || Sbjct 799 RREFARFEKERMMAKWDTGENPIYKQATTT#FKNPTYAGK 837 |
| Anopheles gambiae | Query 761 RREFAKFEKEKMNAKWDTGENPIYKSAVTT#VVNPKYEGK 799 |||||+||||+| |||||||||||| | ||# || | || Sbjct 799 RREFARFEKERMMAKWDTGENPIYKQATTT#FKNPTYAGK 837 |
| Aedes aegypti | Query 761 RREFAKFEKEKMNAKWDTGENPIYKSAVTT#VVNPKYEGK 799 |||||+||||+| |||||||||||| | ||# || | || Sbjct 797 RREFARFEKERMMAKWDTGENPIYKQATTT#FKNPTYAGK 835 |
| Pseudoplusia includens | Query 761 RREFAKFEKEKMNAKWDTGENPIYKSAVTT#VVNPKYEGK 799 |||||+||||+| |||||||||||| | +|# || | || Sbjct 799 RREFARFEKERMMAKWDTGENPIYKQATST#FKNPTYAGK 837 |
| Ostrinia furnacalis | Query 761 RREFAKFEKEKMNAKWDTGENPIYKSAVTT#VVNPKYEGK 799 |||||+||||+| |||||||||||| | +|# || | || Sbjct 793 RREFARFEKERMMAKWDTGENPIYKQATST#FKNPTYAGK 831 |
| Tribolium castaneum | Query 761 RREFAKFEKEKMNAKWDTGENPIYKSAVTT#VVNPKYEGK 799 |||||||||| | |+|||||||||| | +|# || | || Sbjct 791 RREFAKFEKEAMMARWDTGENPIYKQATST#FKNPTYAGK 829 |
| Canis familiaris | Query 761 RREFAKFEKEKMNAKWDTGENPIYKSAVTT#VVNPKYEGK 799 |||||||||||||||||| ||||||| + # || | | Sbjct 783 RREFAKFEKEKMNAKWDTQENPIYKSPINN#FKNPNYGRK 821 |
| Biomphalaria glabrata | Query 762 REFAKFEKEKMNAKWDTGENPIYKSAVTT#VVNPKY 796 |||||||||+ ||||||||||||| | +|# || | Sbjct 751 REFAKFEKERQNAKWDTGENPIYKQATST#FKNPTY 785 |
| Crassostrea gigas | Query 761 RREFAKFEKEKMNAKWDTGENPIYKSAVTT#VVNPKY 796 +|| |+|||| |||+|||||||||| | +|# ||| | Sbjct 762 KRELARFEKEAMNARWDTGENPIYKQATST#FVNPTY 797 |
| Podocoryne carnea | Query 761 RREFAKFEKEKMNAKWDTGENPIYKSAVTT#VVNPKYEGK 799 |||||||||++ | |||+||||||| | +|# || | | Sbjct 743 RREFAKFEKDRQNPKWDSGENPIYKKATST#FQNPMYGNK 781 |
| Caenorhabditis briggsae | Query 761 RREFAKFEKEKMNAKWDTGENPIYKSAVTT#VVNPKYEGK 799 | |+||| |++ ||||| |||||| | ||# || | || Sbjct 770 RAEYAKFNNERLMAKWDTNENPIYKQATTT#FKNPVYAGK 808 |
| Caenorhabditis elegans | Query 761 RREFAKFEKEKMNAKWDTGENPIYKSAVTT#VVNPKYEGK 799 | |+| | |++ ||||| |||||| | ||# || | || Sbjct 769 RSEYATFNNERLMAKWDTNENPIYKQATTT#FKNPVYAGK 807 |
| Macaca mulatta | Query 761 RREFAKFEKEKMNAKWDTGENPIYKSAVTT#VVNPKYEG 798 |+||||||+|+ ||||| ||+|| | +|# | | | Sbjct 750 RKEFAKFEEERARAKWDTANNPLYKEATST#FTNITYRG 787 |
| Equus caballus | Query 761 RREFAKFEKEKMNAKWDTGENPIYKSAVTT#VVNPKYEG 798 |+||||||+|+ ||||| ||+|| | +|# | | | Sbjct 746 RKEFAKFEEERARAKWDTANNPLYKEATST#FTNITYRG 783 |
| Canis lupus familiaris | Query 761 RREFAKFEKEKMNAKWDTGENPIYKSAVTT#VVNPKYEG 798 |+||||||+|+ ||||| ||+|| | +|# | | | Sbjct 746 RKEFAKFEEERARAKWDTANNPLYKEATST#FTNITYRG 783 |
| rats, Peptide Partial, 723 aa | Query 761 RREFAKFEKEKMNAKWDTGENPIYKSAVTT#VVNPKYEG 798 |+||||||+|+ ||||| ||+|| | +|# | | | Sbjct 685 RKEFAKFEEERARAKWDTANNPLYKEATST#FTNITYRG 722 |
| Mus sp. | Query 761 RREFAKFEKEKMNAKWDTGENPIYKSAVTT#VVNPKYEG 798 |||+ +||||+ | ||+||||+||# |||+++| Sbjct 754 RREYRRFEKEQQQLNWKQDNNPLYKSAITT#TVNPRFQG 791 |
| Strongylocentrotus purpuratus | Query 761 RREFAKFEKEKMNAKWDTGENPIYKSAVTT#VVNPKYEGK 799 |||| ||||+ || |+ ||||||| + + # || | | Sbjct 768 RREFQNFEKERANATWEGGENPIYKPSTSV#FKNPTYNIK 806 |
| Lytechinus variegatus | Query 761 RREFAKFEKEKMNAKWDTGENPIYKSAVTT#VVNPKYEGK 799 |||| ||||+ || |+ ||||||| + + # || | | Sbjct 768 RREFQNFEKERANATWEGGENPIYKPSTSV#FKNPTYNIK 806 |
| mice, Peptide Partial, 680 aa | Query 761 RREFAKFEKEKMNAKWDTGENPIYKSAVTT#VVN 793 |+||||||+|+ ||||| ||+|| | +|# | Sbjct 646 RKEFAKFEEERARAKWDTANNPLYKEATST#FTN 678 |
| Ovis aries | Query 762 REFAKFEKEKMNAKWDTGENPIYKSAVTT#VVNPKY 796 ||+ +|||||+ ++|+ +||++||| ||#|+|||+ Sbjct 734 REYHRFEKEKLKSQWNN-DNPLFKSATTT#VMNPKF 767 |
| Bubalus bubalis | Query 762 REFAKFEKEKMNAKWDTGENPIYKSAVTT#VVNPKY 796 ||+ +|||||+ ++|+ +||++||| ||#|+|||+ Sbjct 733 REYHRFEKEKLKSQWNN-DNPLFKSATTT#VMNPKF 766 |
| Capra hircus | Query 762 REFAKFEKEKMNAKWDTGENPIYKSAVTT#VVNPKY 796 ||+ +|||||+ ++|+ +||++||| ||#|+|||+ Sbjct 734 REYHRFEKEKLKSQWNN-DNPLFKSATTT#VMNPKF 767 |
| Ovis canadensis | Query 762 REFAKFEKEKMNAKWDTGENPIYKSAVTT#VVNPKY 796 ||+ +|||||+ ++|+ +||++||| ||#|+|||+ Sbjct 734 REYHRFEKEKLKSQWNN-DNPLFKSATTT#VMNPKF 767 |
| Acropora millepora | Query 761 RREFAKFEKEKMNAKWDTGENPIYKSAVTT#VVNPKYEG 798 | |+ |||+|+|++|| +||+|++| ||# || | | Sbjct 752 RIEYQKFERERMHSKWTREKNPLYQAAKTT#FENPTYAG 789 |
| Sigmodon hispidus | Query 762 REFAKFEKEKMNAKWDTGENPIYKSAVTT#VVNPKY 796 ||+ |||||+ ++|+ +||++||| ||#|+|||+ Sbjct 733 REYRHFEKEKLKSQWNN-DNPLFKSATTT#VMNPKF 766 |
| Oncorhynchus mykiss | Query 762 REFAKFEKEKMNAKWDTGENPIYKSAVTT#VVNPKYEGK 799 +|| +|| |+ || |+||++ +| ||#| || + |+ Sbjct 740 KEFRRFENEQKKGKWSPGDNPLFMNATTT#VANPTFTGE 777 |
| Schistosoma japonicum | Query 761 RREFAKFEKEKMNAKWDTGENPIYKSAVTT#VVNPKYE 797 ||| | |+++ | +|+ ||||++| | #|+|| +| Sbjct 248 RRELANFKQQGENMRWEMAENPIFESPTTN#VLNPTFE 284 |
| Pacifastacus leniusculus | Query 761 RREFAKFEKEKMNAKWDTGENPIYKSAVTT#VVNPKY 796 |||+||||||+ |||+|||| ||# || + Sbjct 762 RREYAKFEKERKLPSGKRAENPLYKSAKTT#FQNPAF 797 |
| Apis mellifera | Query 762 REFAKFEKEKMNA-KWDTGENPIYKSAVTT#VVNPKY 796 ||| |||+|++ | ||+ +||+|| | +|# || + Sbjct 778 REFKKFERERILANKWNRRDNPLYKEATST#FKNPTF 813 |
| Halocynthia roretzi | Query 761 RREFAKFEKEKMNAKWDTGENPIYKSAVTT#VVNPKYEG 798 +||+ ||+||+ || ||||+| +| + # || + | Sbjct 796 KREYEKFKKEEDLRKWTKGENPVYVNASSK#FDNPMFGG 833 |
| Ophlitaspongia tenuis | Query 763 EFAKFEKEKMNAKWDTGENPIYKSAVTT#VVNPKY 796 | |||+| |||+ |||+|+|| # || | Sbjct 803 EVRKFEREIKNAKYTKNENPLYRSATKD#YQNPLY 836 |
| Cyprinus carpio | Query 762 REFAKFEKEKMNAKWDTGENPIYKSAVTT#VVNPKYEG 798 ||+ +|||++ + | +| ||++++| ||#| || + | Sbjct 730 REWKRFEKDRKHEK-TSGTNPLFQNATTT#VQNPTFSG 765 |
| Geodia cydonium | Query 763 EFAKFEKEKMNAKWDTGENPIYKSAVTT#VVNPKY 796 | ||| | |+ |||+| || | # || | Sbjct 843 ELKKFENELAKVKYTKNENPLYHSATTE#HKNPIY 876 |
| Manduca sexta | Query 761 RREFAKFEKEKMNAKWDTGENPIYKSAVTT#VVNPKY 796 +||+||||+| + +| ||+|+ # || | Sbjct 726 KREYAKFEEESRSRGFDVSLNPLYQEPEIN#FSNPVY 761 |
[Site 4] EKMNAKWDTG779-ENPIYKSAVT
Gly779  Glu
Glu
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Glu770 | Lys771 | Met772 | Asn773 | Ala774 | Lys775 | Trp776 | Asp777 | Thr778 | Gly779 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Glu780 | Asn781 | Pro782 | Ile783 | Tyr784 | Lys785 | Ser786 | Ala787 | Val788 | Thr789 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| LIWKLLMIIHDRREFAKFEKEKMNAKWDTGENPIYKSAVTTVVNPKYEGK |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | N/A | 107.00 | 33 | I |
| 2 | Homo sapiens | 107.00 | 25 | ITB1_HUMAN Integrin beta-1 precursor (Fibronectin |
| 3 | Mus musculus | 107.00 | 18 | integrin beta 1 (fibronectin receptor beta) |
| 4 | Pan troglodytes | 107.00 | 15 | PREDICTED: hypothetical protein |
| 5 | Danio rerio | 107.00 | 14 | integrin beta1 subunit-like protein 1 |
| 6 | Bos taurus | 107.00 | 13 | integrin beta 1 |
| 7 | Rattus norvegicus | 107.00 | 10 | integrin beta 1 |
| 8 | Tetraodon nigroviridis | 107.00 | 8 | unnamed protein product |
| 9 | Gallus gallus | 107.00 | 7 | integrin, beta 1 (fibronectin receptor, beta polyp |
| 10 | Sus scrofa | 107.00 | 6 | integrin beta-1 subunit |
| 11 | Monodelphis domestica | 107.00 | 6 | PREDICTED: similar to integrin beta-1 subunit isof |
| 12 | Xenopus laevis | 107.00 | 6 | integrin beta 1 |
| 13 | synthetic construct | 107.00 | 5 | hypothetical protein |
| 14 | Oryctolagus cuniculus | 107.00 | 2 | integrin beta 1 |
| 15 | Pongo pygmaeus | 107.00 | 2 | ITB1_PONPY Integrin beta-1 precursor (Fibronectin |
| 16 | Xenopus tropicalis | 107.00 | 1 | integrin beta 1 (fibronectin receptor beta) |
| 17 | Felis catus | 105.00 | 1 | integrin beta 1 |
| 18 | Ictalurus punctatus | 104.00 | 2 | beta-1 integrin |
| 19 | Drosophila melanogaster | 87.00 | 1 | myospheroid CG1560-PA |
| 20 | Canis familiaris | 86.70 | 10 | PREDICTED: similar to integrin beta 1 isoform 1D p |
| 21 | Aedes aegypti | 85.50 | 1 | integrin beta subunit |
| 22 | Anopheles gambiae | 84.00 | 1 | integrin beta subunit |
| 23 | Anopheles gambiae str. PEST | 84.00 | 1 | ENSANGP00000021373 |
| 24 | Tribolium castaneum | 81.30 | 3 | PREDICTED: similar to CG1560-PA |
| 25 | Ostrinia furnacalis | 81.30 | 1 | integrin beta 1 precursor |
| 26 | Biomphalaria glabrata | 80.50 | 1 | beta integrin subunit |
| 27 | Pseudoplusia includens | 78.20 | 1 | integrin beta 1 |
| 28 | Podocoryne carnea | 76.60 | 1 | AF308652_1 integrin beta chain |
| 29 | Caenorhabditis briggsae | 76.30 | 1 | Hypothetical protein CBG03601 |
| 30 | Crassostrea gigas | 75.50 | 1 | integrin beta cgh |
| 31 | Caenorhabditis elegans | 73.90 | 1 | Paralysed Arrest at Two-fold family member (pat-3) |
| 32 | Macaca mulatta | 72.80 | 13 | PREDICTED: similar to integrin beta chain, beta 3 |
| 33 | Equus caballus | 72.80 | 1 | integrin, beta 3 (platelet glycoprotein IIIa, anti |
| 34 | Canis lupus familiaris | 72.80 | 1 | integrin beta chain, beta 3 |
| 35 | rats, Peptide Partial, 723 aa | 72.80 | 1 | beta 3 integrin, GPIIIA |
| 36 | mice, Peptide Partial, 680 aa | 69.30 | 1 | beta 3 integrin, GPIIIA |
| 37 | Strongylocentrotus purpuratus | 68.20 | 5 | integrin beta-C subunit |
| 38 | Lytechinus variegatus | 68.20 | 1 | beta-C integrin subunit |
| 39 | Ovis aries | 62.00 | 2 | ITB6_SHEEP Integrin beta-6 precursor emb |
| 40 | Pacifastacus leniusculus | 61.20 | 1 | integrin |
| 41 | Mus sp. | 60.50 | 1 | beta 7 integrin |
| 42 | Bubalus bubalis | 57.40 | 1 | integrin beta-2 |
| 43 | Capra hircus | 57.40 | 1 | ITB2_CAPHI Integrin beta-2 precursor (Cell surface |
| 44 | Ovis canadensis | 57.00 | 1 | ITB2_OVICA Integrin beta-2 precursor (Cell surface |
| 45 | Sigmodon hispidus | 56.20 | 1 | AF445415_1 integrin beta-2 precursor |
| 46 | Schistosoma japonicum | 55.50 | 1 | SJCHGC06221 protein |
| 47 | Apis mellifera | 54.70 | 1 | PREDICTED: similar to myospheroid CG1560-PA |
| 48 | Halocynthia roretzi | 53.10 | 2 | integrin beta Hr2 precursor |
| 49 | Oncorhynchus mykiss | 53.10 | 1 | CD18 protein |
| 50 | Acropora millepora | 51.20 | 1 | integrin subunit betaCn1 |
| 51 | Manduca sexta | 48.10 | 1 | plasmatocyte-specific integrin beta 1 |
| 52 | Spodoptera frugiperda | 47.00 | 1 | integrin beta 1 |
| 53 | Ophlitaspongia tenuis | 43.10 | 1 | integrin subunit betaPo1 |
| 54 | Cyprinus carpio | 42.70 | 2 | CD18 type 2 |
| 55 | Geodia cydonium | 41.60 | 1 | integrin beta subunit |
| 56 | Suberites domuncula | 39.70 | 1 | integrin beta subunit |
Top-ranked sequences
| organism | matching |
|---|---|
| N/A | Query 750 LIWKLLMIIHDRREFAKFEKEKMNAKWDTG#ENPIYKSAVTTVVNPKYEGK 799 ||||||||||||||||||||||||||||||#|||||||||||||||||||| Query 750 LIWKLLMIIHDRREFAKFEKEKMNAKWDTG#ENPIYKSAVTTVVNPKYEGK 799 |
| Homo sapiens | Query 750 LIWKLLMIIHDRREFAKFEKEKMNAKWDTG#ENPIYKSAVTTVVNPKYEGK 799 ||||||||||||||||||||||||||||||#|||||||||||||||||||| Query 750 LIWKLLMIIHDRREFAKFEKEKMNAKWDTG#ENPIYKSAVTTVVNPKYEGK 799 |
| Mus musculus | Query 750 LIWKLLMIIHDRREFAKFEKEKMNAKWDTG#ENPIYKSAVTTVVNPKYEGK 799 ||||||||||||||||||||||||||||||#|||||||||||||||||||| Query 750 LIWKLLMIIHDRREFAKFEKEKMNAKWDTG#ENPIYKSAVTTVVNPKYEGK 799 |
| Pan troglodytes | Query 750 LIWKLLMIIHDRREFAKFEKEKMNAKWDTG#ENPIYKSAVTTVVNPKYEGK 799 ||||||||||||||||||||||||||||||#|||||||||||||||||||| Query 750 LIWKLLMIIHDRREFAKFEKEKMNAKWDTG#ENPIYKSAVTTVVNPKYEGK 799 |
| Danio rerio | Query 750 LIWKLLMIIHDRREFAKFEKEKMNAKWDTG#ENPIYKSAVTTVVNPKYEGK 799 ||||||||||||||||||||||||||||||#||||||||||||+||||||| Sbjct 749 LIWKLLMIIHDRREFAKFEKEKMNAKWDTG#ENPIYKSAVTTVINPKYEGK 798 |
| Bos taurus | Query 750 LIWKLLMIIHDRREFAKFEKEKMNAKWDTG#ENPIYKSAVTTVVNPKYEGK 799 ||||||||||||||||||||||||||||||#|||||||||||||||||||| Query 750 LIWKLLMIIHDRREFAKFEKEKMNAKWDTG#ENPIYKSAVTTVVNPKYEGK 799 |
| Rattus norvegicus | Query 750 LIWKLLMIIHDRREFAKFEKEKMNAKWDTG#ENPIYKSAVTTVVNPKYEGK 799 ||||||||||||||||||||||||||||||#|||||||||||||||||||| Query 750 LIWKLLMIIHDRREFAKFEKEKMNAKWDTG#ENPIYKSAVTTVVNPKYEGK 799 |
| Tetraodon nigroviridis | Query 750 LIWKLLMIIHDRREFAKFEKEKMNAKWDTG#ENPIYKSAVTTVVNPKYEGK 799 ||||||||||||||||||||||||||||||#|||||||||||||||||||| Query 750 LIWKLLMIIHDRREFAKFEKEKMNAKWDTG#ENPIYKSAVTTVVNPKYEGK 799 |
| Gallus gallus | Query 750 LIWKLLMIIHDRREFAKFEKEKMNAKWDTG#ENPIYKSAVTTVVNPKYEGK 799 ||||||||||||||||||||||||||||||#|||||||||||||||||||| Query 750 LIWKLLMIIHDRREFAKFEKEKMNAKWDTG#ENPIYKSAVTTVVNPKYEGK 799 |
| Sus scrofa | Query 750 LIWKLLMIIHDRREFAKFEKEKMNAKWDTG#ENPIYKSAVTTVVNPKYEGK 799 ||||||||||||||||||||||||||||||#|||||||||||||||||||| Query 750 LIWKLLMIIHDRREFAKFEKEKMNAKWDTG#ENPIYKSAVTTVVNPKYEGK 799 |
| Monodelphis domestica | Query 750 LIWKLLMIIHDRREFAKFEKEKMNAKWDTG#ENPIYKSAVTTVVNPKYEGK 799 ||||||||||||||||||||||||||||||#|||||||||||||||||||| Query 750 LIWKLLMIIHDRREFAKFEKEKMNAKWDTG#ENPIYKSAVTTVVNPKYEGK 799 |
| Xenopus laevis | Query 750 LIWKLLMIIHDRREFAKFEKEKMNAKWDTG#ENPIYKSAVTTVVNPKYEGK 799 ||||||||||||||||||||||||||||||#|||||||||||||||||||| Query 750 LIWKLLMIIHDRREFAKFEKEKMNAKWDTG#ENPIYKSAVTTVVNPKYEGK 799 |
| synthetic construct | Query 750 LIWKLLMIIHDRREFAKFEKEKMNAKWDTG#ENPIYKSAVTTVVNPKYEGK 799 ||||||||||||||||||||||||||||||#|||||||||||||||||||| Query 750 LIWKLLMIIHDRREFAKFEKEKMNAKWDTG#ENPIYKSAVTTVVNPKYEGK 799 |
| Oryctolagus cuniculus | Query 750 LIWKLLMIIHDRREFAKFEKEKMNAKWDTG#ENPIYKSAVTTVVNPKYEGK 799 ||||||||||||||||||||||||||||||#|||||||||||||||||||| Query 750 LIWKLLMIIHDRREFAKFEKEKMNAKWDTG#ENPIYKSAVTTVVNPKYEGK 799 |
| Pongo pygmaeus | Query 750 LIWKLLMIIHDRREFAKFEKEKMNAKWDTG#ENPIYKSAVTTVVNPKYEGK 799 ||||||||||||||||||||||||||||||#|||||||||||||||||||| Query 750 LIWKLLMIIHDRREFAKFEKEKMNAKWDTG#ENPIYKSAVTTVVNPKYEGK 799 |
| Xenopus tropicalis | Query 750 LIWKLLMIIHDRREFAKFEKEKMNAKWDTG#ENPIYKSAVTTVVNPKYEGK 799 ||||||||||||||||||||||||||||||#|||||||||||||||||||| Query 750 LIWKLLMIIHDRREFAKFEKEKMNAKWDTG#ENPIYKSAVTTVVNPKYEGK 799 |
| Felis catus | Query 750 LIWKLLMIIHDRREFAKFEKEKMNAKWDTG#ENPIYKSAVTTVVNPKYEGK 799 ||||||||||| ||||||||||||||||||#|||||||||||||||||||| Sbjct 749 LIWKLLMIIHDTREFAKFEKEKMNAKWDTG#ENPIYKSAVTTVVNPKYEGK 798 |
| Ictalurus punctatus | Query 751 IWKLLMIIHDRREFAKFEKEKMNAKWDTG#ENPIYKSAVTTVVNPKYEGK 799 ||||||||||||||||||||||||||| |#|||||||||||||||||||| Sbjct 759 IWKLLMIIHDRREFAKFEKEKMNAKWDAG#ENPIYKSAVTTVVNPKYEGK 807 |
| Drosophila melanogaster | Query 750 LIWKLLMIIHDRREFAKFEKEKMNAKWDTG#ENPIYKSAVTTVVNPKYEGK 799 |+|||| ||||||||+||||+||||||||#|||||| | +| || | || Sbjct 797 LLWKLLTTIHDRREFARFEKERMNAKWDTG#ENPIYKQATSTFKNPMYAGK 846 |
| Canis familiaris | Query 750 LIWKLLMIIHDRREFAKFEKEKMNAKWDTG#ENPIYKSAVTTVVNPKYEGK 799 ||||||||||||||||||||||||||||| #||||||| + || | | Sbjct 772 LIWKLLMIIHDRREFAKFEKEKMNAKWDTQ#ENPIYKSPINNFKNPNYGRK 821 |
| Aedes aegypti | Query 750 LIWKLLMIIHDRREFAKFEKEKMNAKWDTG#ENPIYKSAVTTVVNPKYEGK 799 |+|||| ||||||||+||||+| ||||||#|||||| | || || | || Sbjct 786 LLWKLLTTIHDRREFARFEKERMMAKWDTG#ENPIYKQATTTFKNPTYAGK 835 |
| Anopheles gambiae | Query 750 LIWKLLMIIHDRREFAKFEKEKMNAKWDTG#ENPIYKSAVTTVVNPKYEGK 799 |+||+| ||||||||+||||+| ||||||#|||||| | || || | || Sbjct 788 LLWKVLTSIHDRREFARFEKERMMAKWDTG#ENPIYKQATTTFKNPTYAGK 837 |
| Anopheles gambiae str. PEST | Query 750 LIWKLLMIIHDRREFAKFEKEKMNAKWDTG#ENPIYKSAVTTVVNPKYEGK 799 |+||+| ||||||||+||||+| ||||||#|||||| | || || | || Sbjct 788 LLWKVLTSIHDRREFARFEKERMMAKWDTG#ENPIYKQATTTFKNPTYAGK 837 |
| Tribolium castaneum | Query 750 LIWKLLMIIHDRREFAKFEKEKMNAKWDTG#ENPIYKSAVTTVVNPKYEGK 799 |+||| ||||||||||||| | |+||||#|||||| | +| || | || Sbjct 780 LLWKLFTTIHDRREFAKFEKEAMMARWDTG#ENPIYKQATSTFKNPTYAGK 829 |
| Ostrinia furnacalis | Query 750 LIWKLLMIIHDRREFAKFEKEKMNAKWDTG#ENPIYKSAVTTVVNPKYEGK 799 ++||++ ||||||||+||||+| ||||||#|||||| | +| || | || Sbjct 782 MLWKMVTTIHDRREFARFEKERMMAKWDTG#ENPIYKQATSTFKNPTYAGK 831 |
| Biomphalaria glabrata | Query 750 LIWKLLMIIHDRREFAKFEKEKMNAKWDTG#ENPIYKSAVTTVVNPKY 796 |+|||| ||| |||||||||+ |||||||#|||||| | +| || | Sbjct 739 LLWKLLTFIHDTREFAKFEKERQNAKWDTG#ENPIYKQATSTFKNPTY 785 |
| Pseudoplusia includens | Query 750 LIWKLLMIIHDRREFAKFEKEKMNAKWDTG#ENPIYKSAVTTVVNPKYEGK 799 ++||+ |||||||+||||+| ||||||#|||||| | +| || | || Sbjct 788 MLWKMATTSHDRREFARFEKERMMAKWDTG#ENPIYKQATSTFKNPTYAGK 837 |
| Podocoryne carnea | Query 750 LIWKLLMIIHDRREFAKFEKEKMNAKWDTG#ENPIYKSAVTTVVNPKYEGK 799 |||||| | ||||||||||++ | |||+|#|||||| | +| || | | Sbjct 732 LIWKLLATIQDRREFAKFEKDRQNPKWDSG#ENPIYKKATSTFQNPMYGNK 781 |
| Caenorhabditis briggsae | Query 750 LIWKLLMIIHDRREFAKFEKEKMNAKWDTG#ENPIYKSAVTTVVNPKYEGK 799 |+|||| ++||| |+||| |++ ||||| #|||||| | || || | || Sbjct 759 LLWKLLTVLHDRAEYAKFNNERLMAKWDTN#ENPIYKQATTTFKNPVYAGK 808 |
| Crassostrea gigas | Query 750 LIWKLLMIIHDRREFAKFEKEKMNAKWDTG#ENPIYKSAVTTVVNPKY 796 ||||| | |+|| |+|||| |||+||||#|||||| | +| ||| | Sbjct 751 LIWKLFTTISDKRELARFEKEAMNARWDTG#ENPIYKQATSTFVNPTY 797 |
| Caenorhabditis elegans | Query 750 LIWKLLMIIHDRREFAKFEKEKMNAKWDTG#ENPIYKSAVTTVVNPKYEGK 799 |+|||| ++||| |+| | |++ ||||| #|||||| | || || | || Sbjct 758 LLWKLLTVLHDRSEYATFNNERLMAKWDTN#ENPIYKQATTTFKNPVYAGK 807 |
| Macaca mulatta | Query 750 LIWKLLMIIHDRREFAKFEKEKMNAKWDTG#ENPIYKSAVTTVVNPKYEG 798 ||||||+ ||||+||||||+|+ ||||| # ||+|| | +| | | | Sbjct 739 LIWKLLITIHDRKEFAKFEEERARAKWDTA#NNPLYKEATSTFTNITYRG 787 |
| Equus caballus | Query 750 LIWKLLMIIHDRREFAKFEKEKMNAKWDTG#ENPIYKSAVTTVVNPKYEG 798 ||||||+ ||||+||||||+|+ ||||| # ||+|| | +| | | | Sbjct 735 LIWKLLITIHDRKEFAKFEEERARAKWDTA#NNPLYKEATSTFTNITYRG 783 |
| Canis lupus familiaris | Query 750 LIWKLLMIIHDRREFAKFEKEKMNAKWDTG#ENPIYKSAVTTVVNPKYEG 798 ||||||+ ||||+||||||+|+ ||||| # ||+|| | +| | | | Sbjct 735 LIWKLLITIHDRKEFAKFEEERARAKWDTA#NNPLYKEATSTFTNITYRG 783 |
| rats, Peptide Partial, 723 aa | Query 750 LIWKLLMIIHDRREFAKFEKEKMNAKWDTG#ENPIYKSAVTTVVNPKYEG 798 ||||||+ ||||+||||||+|+ ||||| # ||+|| | +| | | | Sbjct 674 LIWKLLITIHDRKEFAKFEEERARAKWDTA#NNPLYKEATSTFTNITYRG 722 |
| mice, Peptide Partial, 680 aa | Query 750 LIWKLLMIIHDRREFAKFEKEKMNAKWDTG#ENPIYKSAVTTVVN 793 ||||||+ ||||+||||||+|+ ||||| # ||+|| | +| | Sbjct 635 LIWKLLITIHDRKEFAKFEEERARAKWDTA#NNPLYKEATSTFTN 678 |
| Strongylocentrotus purpuratus | Query 750 LIWKLLMIIHDRREFAKFEKEKMNAKWDTG#ENPIYKSAVTTVVNPKYEGK 799 |||+|| ||||||| ||||+ || |+ |#|||||| + + || | | Sbjct 757 LIWRLLTYIHDRREFQNFEKERANATWEGG#ENPIYKPSTSVFKNPTYNIK 806 |
| Lytechinus variegatus | Query 750 LIWKLLMIIHDRREFAKFEKEKMNAKWDTG#ENPIYKSAVTTVVNPKYEGK 799 |||+|| ||||||| ||||+ || |+ |#|||||| + + || | | Sbjct 757 LIWRLLTYIHDRREFQNFEKERANATWEGG#ENPIYKPSTSVFKNPTYNIK 806 |
| Ovis aries | Query 751 IWKLLMIIHDRREFAKFEKEKMNAKWDTG#ENPIYKSAVTTVVNPKYEGK 799 |||||+ |||+| |||| |+ ||| ||# ||+|+ + +| | |+ | Sbjct 728 IWKLLVSFHDRKEVAKFEAERSKAKWQTG#TNPLYRGSTSTFKNVTYKHK 776 |
| Pacifastacus leniusculus | Query 750 LIWKLLMIIHDRREFAKFEKEKMNAKWDTG#ENPIYKSAVTTVVNPKY 796 |||||| +|||||+||||||+ #|||+|||| || || + Sbjct 751 LIWKLLTTLHDRREYAKFEKERKLPSGKRA#ENPLYKSAKTTFQNPAF 797 |
| Mus sp. | Query 750 LIWKLLMIIHDRREFAKFEKEKMNAKWDTG#ENPIYKSAVTTVVNPKYEG 798 | ++| + |+||||+ +||||+ | # ||+||||+|| |||+++| Sbjct 743 LAYRLSVEIYDRREYRRFEKEQQQLNWKQD#NNPLYKSAITTTVNPRFQG 791 |
| Bubalus bubalis | Query 750 LIWKLLMIIHDRREFAKFEKEKMNAKWDTG#ENPIYKSAVTTVVNPKY 796 +||| | + | ||+ +|||||+ ++|+ #+||++||| |||+|||+ Sbjct 721 VIWKALTHLSDLREYHRFEKEKLKSQWN-N#DNPLFKSATTTVMNPKF 766 |
| Capra hircus | Query 750 LIWKLLMIIHDRREFAKFEKEKMNAKWDTG#ENPIYKSAVTTVVNPKY 796 +||| | + | ||+ +|||||+ ++|+ #+||++||| |||+|||+ Sbjct 722 VIWKALTHLSDLREYHRFEKEKLKSQWN-N#DNPLFKSATTTVMNPKF 767 |
| Ovis canadensis | Query 751 IWKLLMIIHDRREFAKFEKEKMNAKWDTG#ENPIYKSAVTTVVNPKY 796 ||| | + | ||+ +|||||+ ++|+ #+||++||| |||+|||+ Sbjct 723 IWKALTHLSDLREYHRFEKEKLKSQWN-N#DNPLFKSATTTVMNPKF 767 |
| Sigmodon hispidus | Query 750 LIWKLLMIIHDRREFAKFEKEKMNAKWDTG#ENPIYKSAVTTVVNPKY 796 +||| | + | ||+ |||||+ ++|+ #+||++||| |||+|||+ Sbjct 721 VIWKALTHLTDLREYRHFEKEKLKSQWN-N#DNPLFKSATTTVMNPKF 766 |
| Schistosoma japonicum | Query 750 LIWKLLMIIHDRREFAKFEKEKMNAKWDTG#ENPIYKSAVTTVVNPKYE 797 ||+||++ | |||| | |+++ | +|+ #||||++| | |+|| +| Sbjct 237 LIYKLVITIDDRRELANFKQQGENMRWEMA#ENPIFESPTTNVLNPTFE 284 |
| Apis mellifera | Query 750 LIWKLLMIIHDRREFAKFEKEKMNA-KWDTG#ENPIYKSAVTTVVNPKY 796 | ||+| +| | ||| |||+|++ | ||+ #+||+|| | +| || + Sbjct 766 LTWKILTMIRDNREFKKFERERILANKWNRR#DNPLYKEATSTFKNPTF 813 |
| Halocynthia roretzi | Query 750 LIWKLLMIIHDRREFAKFEKEKMNAKWDTG#ENPIYKSAVTTVVNPKYEG 798 +|||++ + |+||+ ||+||+ || |#|||+| +| + || + | Sbjct 785 IIWKVIQTLRDKREYEKFKKEEDLRKWTKG#ENPVYVNASSKFDNPMFGG 833 |
| Oncorhynchus mykiss | Query 750 LIWKLLMIIHDRREFAKFEKEKMNAKWDTG#ENPIYKSAVTTVVNPKYEGK 799 || | ++ + | +|| +|| |+ || |#+||++ +| ||| || + |+ Sbjct 728 LIIKAILYMRDVKEFRRFENEQKKGKWSPG#DNPLFMNATTTVANPTFTGE 777 |
| Acropora millepora | Query 750 LIWKLLMIIHDRREFAKFEKEKMNAKWDTG#ENPIYKSAVTTVVNPKYEG 798 ++ | | + || |+ |||+|+|++|| #+||+|++| || || | | Sbjct 741 IVIKGLFTMVDRIEYQKFERERMHSKWTRE#KNPLYQAAKTTFENPTYAG 789 |
| Manduca sexta | Query 750 LIWKLLMIIHDRREFAKFEKEKMNAKWDTG#ENPIYKSAVTTVVNPKY 796 + ||+|+ +||+||+||||+| + +| # ||+|+ || | Sbjct 715 IAWKILVDLHDKREYAKFEEESRSRGFDVS#LNPLYQEPEINFSNPVY 761 |
| Spodoptera frugiperda | Query 750 LIWKLLMIIHDRREFAKFEKEKMNAKWDTG#ENPIYKSAVTTVVNPKYEGK 799 +|||+|+ +||++|+ ||+ | + | +| # ||+|+ || | + Sbjct 730 IIWKILVDMHDKKEYKKFQDEALAAGYDVS#LNPLYQDPSINFSNPVYNNQ 779 |
| Ophlitaspongia tenuis | Query 750 LIWKLLMIIHDRREFAKFEKEKMNAKWDTG#ENPIYKSAVTTVVNPKY 796 |+ | |+++ | | |||+| |||+ #|||+|+|| || | Sbjct 790 LLLKGLLLLWDVVEVRKFEREIKNAKYTKN#ENPLYRSATKDYQNPLY 836 |
| Cyprinus carpio | Query 750 LIWKLLMIIHDRREFAKFEKEKMNAKWDTG#ENPIYKSAVTTVVNPKYEG 798 || | ++ | ||+ +|||++ + | +|# ||++++| ||| || + | Sbjct 718 LIIKAVLYASDLREWKRFEKDRKHEK-TSG#TNPLFQNATTTVQNPTFSG 765 |
| Geodia cydonium | Query 750 LIWKLLMIIHDRREFAKFEKEKMNAKWDTG#ENPIYKSAVTTVVNPKY 796 ++ |+|+++ | | ||| | |+ #|||+| || | || | Sbjct 830 ILLKILLMVLDYTELKKFENELAKVKYTKN#ENPLYHSATTEHKNPIY 876 |
| Suberites domuncula | Query 750 LIWKLLMIIHDRREFAKFEKEKMNAKWDTG#ENPIYKSAVTTVVNPKY 796 |+ || + | | | ||||| |+ #+||+|+|| || | Sbjct 716 LLLKLFLFIIDVIEVKKFEKELAKTKYPKH#QNPLYRSAAKDYQNPIY 762 |
* References
None.