XSB2213 : PREDICTED: cyclin-dependent kinase inhibitor 2D (p19, inhibits CDK4) isoform 1 [Macaca mulatta]
[ CaMP Format ]
This entry is computationally expanded from SB0083
* Basic Information
| Organism | Macaca mulatta (rhesus monkey) |
| Protein Names | cyclin-dependent kinase inhibitor 2D (p19; inhibits CDK4) isoform 1 |
| Gene Names | CDKN2D; Derived by automated computational analysis using gene prediction method: GNOMON. Supporting evidence includes similarity to: 8 mRNAs, 113 ESTs, 2 Proteins |
| Gene Locus | Not available |
| GO Function | Not available |
| Entrez Protein | Entrez Nucleotide | Entrez Gene | UniProt | OMIM | HGNC | HPRD | KEGG |
|---|---|---|---|---|---|---|---|
| XP_001102138 | XM_001102138 | 713172 | N/A | N/A | N/A | N/A | N/A |
* Information From OMIM
Not Available.
* Structure Information
1. Primary Information
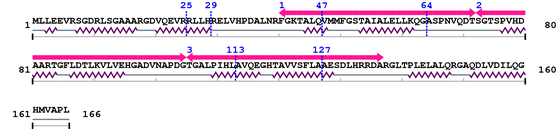
Length: 166 aa
Average Mass: 17.688 kDa
Monoisotopic Mass: 17.677 kDa
2. Domain Information
Annotated Domains: Not Available.
Computationally Assigned Domains (Pfam+HMMER):
| domain name | begin | end | score | e-value |
|---|---|---|---|---|
| --- cleavage 29 --- | ||||
| --- cleavage 25 --- | ||||
| Ank 1. | 41 | 72 | 17.7 | 0.049 |
| --- cleavage 47 (inside Ank 41..72) --- | ||||
| --- cleavage 64 (inside Ank 41..72) --- | ||||
| Ank 2. | 73 | 105 | 29.6 | 1.2e-05 |
| Ank 3. | 106 | 137 | 15.6 | 0.21 |
| --- cleavage 113 (inside Ank 106..137) --- | ||||
| --- cleavage 127 (inside Ank 106..137) --- | ||||
3. Sequence Information
Fasta Sequence: XSB2213.fasta
Amino Acid Sequence and Secondary Structures (PsiPred):
4. 3D Information
Not Available.
* Cleavage Information
6 [sites]
Cleavage sites (±10aa)
[Site 1] LNRFGKTALQ47-VMMFGSTAIA
Gln47  Val
Val
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Leu38 | Asn39 | Arg40 | Phe41 | Gly42 | Lys43 | Thr44 | Ala45 | Leu46 | Gln47 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Val48 | Met49 | Met50 | Phe51 | Gly52 | Ser53 | Thr54 | Ala55 | Ile56 | Ala57 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| RGDVQEVRRLLHRELVHPDALNRFGKTALQVMMFGSTAIALELLKQGASPNVQDTSGTSP |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | Homo sapiens | 120.00 | 26 | cyclin-dependent kinase inhibitor 2D |
| 2 | N/A | 120.00 | 25 | - |
| 3 | synthetic construct | 120.00 | 7 | cyclin-dependent kinase inhibitor 2D (p19, inhibit |
| 4 | Macaca mulatta | 120.00 | 5 | PREDICTED: cyclin-dependent kinase inhibitor 2D (p |
| 5 | Mus musculus | 115.00 | 35 | cyclin-dependent kinase inhibitor 2D (p19, inhibit |
| 6 | Rattus norvegicus | 115.00 | 14 | similar to cyclin-dependent kinase inhibitor 2D |
| 7 | Canis familiaris | 112.00 | 26 | PREDICTED: similar to Cyclin-dependent kinase 4 in |
| 8 | Bos taurus | 109.00 | 7 | cyclin-dependent kinase inhibitor 2D (p19, inhibit |
| 9 | Monodelphis domestica | 108.00 | 9 | PREDICTED: similar to Cyclin-dependent kinase inhi |
| 10 | Xenopus tropicalis | 99.00 | 2 | cyclin-dependent kinase inhibitor 2D (p19, inhibit |
| 11 | Xenopus laevis | 95.10 | 5 | ankyrin 2, neuronal |
| 12 | Xiphophorus maculatus | 68.60 | 2 | p19 |
| 13 | Tetraodon nigroviridis | 65.50 | 4 | unnamed protein product |
| 14 | Takifugu rubripes | 64.70 | 2 | p19 |
| 15 | Mesocricetus auratus | 53.50 | 3 | p15INK4b cyclin-dependent kinase inhibitor |
| 16 | Gallus gallus | 51.20 | 5 | PREDICTED: similar to Chain A, Crystal Structure O |
| 17 | Macaca fascicularis | 49.70 | 1 | unnamed protein product |
| 18 | Danio rerio | 48.10 | 9 | PREDICTED: similar to p13 |
| 19 | Sus scrofa | 47.80 | 2 | cyclin-dependent kinase inhibitor 2B |
| 20 | Pan troglodytes | 46.20 | 5 | PREDICTED: similar to p16-INK4 |
| 21 | human, brain tumors, Peptide, 156 aa | 46.20 | 1 | cyclin D-dependent kinases 4 and 6-binding protein |
| 22 | Xiphophorus helleri | 44.70 | 1 | p13CDKN2X |
| 23 | Myxococcus xanthus DK 1622 | 40.80 | 1 | ankyrin repeat protein |
| 24 | Apis mellifera | 39.30 | 2 | PREDICTED: similar to CG3104-PA, isoform A |
| 25 | Solibacter usitatus Ellin6076 | 38.90 | 1 | ankyrin-related protein |
| 26 | Drosophila melanogaster | 38.50 | 1 | CG3104-PA, isoform A |
| 27 | Tribolium castaneum | 38.10 | 2 | PREDICTED: similar to CG6896-PA |
| 28 | Trichomonas vaginalis G3 | 38.10 | 2 | hypothetical protein TVAG_185920 |
| 29 | Pseudomonas stutzeri A1501 | 38.10 | 1 | ankyrin domain protein |
| 30 | Medicago truncatula | 37.70 | 3 | Ankyrin |
| 31 | Equus caballus | 37.70 | 1 | cyclin-dependent kinase 2A inhibitor |
| 32 | Wolbachia endosymbiont of Culex pipiens | 37.00 | 1 | ankyrin domain protein ank12 |
| 33 | Aspergillus niger CBS 513.88 | 37.00 | 1 | hypothetical protein An10g00420 |
| 34 | Thermofilum pendens Hrk 5 | 36.60 | 1 | Ankyrin |
| 35 | Schistosoma japonicum | 36.60 | 1 | SJCHGC04635 protein |
| 36 | Methanosarcina barkeri str. fusaro | 36.20 | 1 | hypothetical protein Mbar_A2380 |
| 37 | Rickettsiella grylli | 35.80 | 2 | hypothetical protein Rgryl_01000812 |
| 38 | Ostreococcus lucimarinus CCE9901 | 35.80 | 1 | predicted protein |
| 39 | Cryptosporidium parvum Iowa II | 35.80 | 1 | conserved hypothetical protein |
| 40 | Aspergillus oryzae | 35.80 | 1 | unnamed protein product |
| 41 | Cryptosporidium parvum | 35.80 | 1 | acyl-CoA-binding protein |
| 42 | Cryptosporidium hominis TU502 | 35.80 | 1 | proteasome 26S subunit |
| 43 | Aedes aegypti | 35.40 | 1 | protein phosphatase 1 regulatory inhibitor subunit |
| 44 | Egeria densa | 35.40 | 1 | inward potassium channel alpha subunit |
| 45 | Anopheles gambiae str. PEST | 35.40 | 1 | ENSANGP00000028069 |
| 46 | Rickettsia felis URRWXCal2 | 35.40 | 1 | Ankyrin repeat |
| 47 | Caenorhabditis elegans | 35.00 | 7 | UNCoordinated family member (unc-44) |
| 48 | Orientia tsutsugamushi Boryong | 35.00 | 2 | ankyrin repeat protein with 5 ankyrin repeats |
| 49 | Caenorhabditis briggsae | 35.00 | 1 | Hypothetical protein CBG09456 |
| 50 | Dictyostelium discoideum AX4 | 34.70 | 1 | SecG |
| 51 | Leptospira borgpetersenii serovar Hardjo-bovis L550 | 34.70 | 1 | Ankyrin repeat protein |
| 52 | Bordetella pertussis Tohama I | 34.30 | 1 | hypothetical protein BP2881 |
| 53 | Bordetella parapertussis 12822 | 34.30 | 1 | hypothetical protein BPP0952 |
| 54 | Strongylocentrotus purpuratus | 33.90 | 5 | PREDICTED: similar to MGC80441 protein, partial |
| 55 | Orf virus | 33.90 | 1 | ORF126 ankyrin repeat protein |
| 56 | Neurospora crassa OR74A | 33.90 | 1 | hypothetical protein |
| 57 | Pseudomonas entomophila L48 | 33.90 | 1 | hypothetical protein PSEEN1535 |
| 58 | Aspergillus niger | 32.70 | 1 | hypothetical protein An08g11190 |
Top-ranked sequences
| organism | matching |
|---|---|
| Homo sapiens | Query 18 RGDVQEVRRLLHRELVHPDALNRFGKTALQ#VMMFGSTAIALELLKQGASPNVQDTSGTSP 77 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 18 RGDVQEVRRLLHRELVHPDALNRFGKTALQ#VMMFGSTAIALELLKQGASPNVQDTSGTSP 77 |
| N/A | Query 18 RGDVQEVRRLLHRELVHPDALNRFGKTALQ#VMMFGSTAIALELLKQGASPNVQDTSGTSP 77 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 18 RGDVQEVRRLLHRELVHPDALNRFGKTALQ#VMMFGSTAIALELLKQGASPNVQDTSGTSP 77 |
| synthetic construct | Query 18 RGDVQEVRRLLHRELVHPDALNRFGKTALQ#VMMFGSTAIALELLKQGASPNVQDTSGTSP 77 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 18 RGDVQEVRRLLHRELVHPDALNRFGKTALQ#VMMFGSTAIALELLKQGASPNVQDTSGTSP 77 |
| Macaca mulatta | Query 18 RGDVQEVRRLLHRELVHPDALNRFGKTALQ#VMMFGSTAIALELLKQGASPNVQDTSGTSP 77 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 18 RGDVQEVRRLLHRELVHPDALNRFGKTALQ#VMMFGSTAIALELLKQGASPNVQDTSGTSP 77 |
| Mus musculus | Query 18 RGDVQEVRRLLHRELVHPDALNRFGKTALQ#VMMFGSTAIALELLKQGASPNVQDTSGTSP 77 ||||||||||||||||||||||||||||||#|||||| |+||||||||||||||| ||||| Sbjct 18 RGDVQEVRRLLHRELVHPDALNRFGKTALQ#VMMFGSPAVALELLKQGASPNVQDASGTSP 77 |
| Rattus norvegicus | Query 18 RGDVQEVRRLLHRELVHPDALNRFGKTALQ#VMMFGSTAIALELLKQGASPNVQDTSGTSP 77 ||||||||||||||||||||||||||||||#|||||| |+||||||||||||||| ||||| Sbjct 18 RGDVQEVRRLLHRELVHPDALNRFGKTALQ#VMMFGSPAVALELLKQGASPNVQDASGTSP 77 |
| Canis familiaris | Query 18 RGDVQEVRRLLHRELVHPDALNRFGKTALQ#VMMFGSTAIALELLKQGASPNVQDTSGTSP 77 ||||||||||||||||||||||||||||||#|||||| |||||||||||||||| +||+| Sbjct 18 RGDVQEVRRLLHRELVHPDALNRFGKTALQ#VMMFGSPTIALELLKQGASPNVQDATGTTP 77 |
| Bos taurus | Query 18 RGDVQEVRRLLHRELVHPDALNRFGKTALQ#VMMFGSTAIALELLKQGASPNVQDTSGTSP 77 |||||||||||| |||||| ||||||||||#|||||| |||||||||||||||| |||+| Sbjct 18 RGDVQEVRRLLHSELVHPDVLNRFGKTALQ#VMMFGSPTIALELLKQGASPNVQDASGTTP 77 |
| Monodelphis domestica | Query 18 RGDVQEVRRLLHRELVHPDALNRFGKTALQ#VMMFGSTAIALELLKQGASPNVQDTSGTSP 77 |||| |||||||+| ||||+||||||||||#||||||+ |||||||||||||||| ||||| Sbjct 18 RGDVNEVRRLLHQEFVHPDSLNRFGKTALQ#VMMFGSSPIALELLKQGASPNVQDGSGTSP 77 |
| Xenopus tropicalis | Query 18 RGDVQEVRRLLHRELVHPDALNRFGKTALQ#VMMFGSTAIALELLKQGASPNVQDTSGTSP 77 |||+ ||+|||| | +||| ||||||||||#||||||| +| ||||||||||+|| ||+| Sbjct 18 RGDLLEVKRLLHEERIHPDCLNRFGKTALQ#VMMFGSTPVASELLKQGASPNIQDAYGTTP 77 |
| Xenopus laevis | Query 18 RGDVQEVRRLLHRELVHPDALNRFGKTALQ#VMMFGSTAIALELLKQGASPNVQDTSGTSP 77 |||+ ||+|||| | +||| || |||||||#||||||| +| |||||||+||+|| ||+| Sbjct 18 RGDLTEVKRLLHEERIHPDCLNCFGKTALQ#VMMFGSTPVASELLKQGATPNIQDAYGTTP 77 |
| Xiphophorus maculatus | Query 18 RGDVQEVRRLLHRELVHPDALNRFGKTALQ#VMMFGSTAIALELLKQGASPNVQDTSGTSP 77 +|+ ||+|+| |||| | ||+||||#||| |++ +| ||++|| ||||| | +| Sbjct 18 KGNADEVQRILEECRVHPDTPNEFGRTALQ#VMMMGNSKVARLLLEKGAEPNVQDKHGIAP 77 |
| Tetraodon nigroviridis | Query 18 RGDVQEVRRLLHRELVHPDALNRFGKTALQ#VMMFGSTAIALELLKQGASPNVQDTSGTSP 77 +| ||||+| | || | |||||||#||| |+ || ||++|| ||||| | +| Sbjct 18 KGRTAEVRRILEECRVPPDTPNEFGKTALQ#VMMLGNCKIASLLLEKGADPNVQDKHGIAP 77 |
| Takifugu rubripes | Query 18 RGDVQEVRRLLHRELVHPDALNRFGKTALQ#VMMFGSTAIALELLKQGASPNVQDTSGTSP 77 +| ||+|+| | || | |||||||#||| |+ || ||++|| ||||| | +| Sbjct 18 KGRTSEVQRILEECRVPPDTRNEFGKTALQ#VMMLGNCKIASLLLEKGADPNVQDKHGIAP 77 |
| Mesocricetus auratus | Query 18 RGDVQEVRRLLHRELVHPDALNRFGKTALQ#VMMFGSTAIALELLKQGASPNVQD 71 || |+ ||+|| | |+|+||||+ +|#||| ||| +| || || || | Sbjct 16 RGQVETVRQLLEAG-VDPNAVNRFGRRPIQ#VMMMGSTQVAELLLLHGAEPNCAD 68 |
| Gallus gallus | Query 18 RGDVQEVRRLLHRELVHPDALNRFGKTALQ#VMMFGSTAIALELLKQGASPNVQDTSG 74 +||+ ++ || + |+ +| | ||+||||#|| |+ || || ||+||++|++| Sbjct 15 KGDLVQLTNLLQKN-VNVNAQNGFGRTALQ#VMKLGNPEIARRLLMSGANPNLKDSTG 70 |
| Macaca fascicularis | Query 18 RGDVQEVRRLLHRELVHPDALNRFGKTALQ#VMMFGSTAIALELLKQGASPNVQD 71 || |++||+|| |+ +||||+ |+|#||| || +| || || || | Sbjct 24 RGLVEKVRQLLEAG-ADPNGVNRFGRRAIQ#VMMMGSARVAELLLLHGAEPNCAD 76 |
| Danio rerio | Query 19 GDVQEVRRLLHRELVHPDALNRFGKTALQ#VMMFGSTAIALELLKQGASPNVQD-TSGTSP 77 |++ |+ || |+ + +|+| +| +|#||| |+ +|| ||+||| ||| | +|++| Sbjct 15 GNISHVQFLLSNG-VNANVVNKFRRTPIQ#VMMMGNAPLALVLLEQGADPNVPDPDTGSTP 73 |
| Sus scrofa | Query 18 RGDVQEVRRLLHRELVHPDALNRFGKTALQ#VMMFGSTAIALELLKQGASPNVQD 71 || |+ ||+|| |+ || ||+ +|#||| || +| || || || | Sbjct 17 RGQVETVRQLLEAG-ADPNGLNHFGRRPIQ#VMMMGSARVAELLLLHGADPNCAD 69 |
| Pan troglodytes | Query 18 RGDVQEVRRLLHRELVHPDALNRFGKTALQ#VMMFGSTAIALELLKQGASPNVQD 71 || |+||| || + |+| | +|+ +|#||| || +| || || || | Sbjct 223 RGRVEEVRALLEAGAL-PNAPNSYGRRPIQ#VMMMGSARVAELLLLHGAEPNCAD 275 |
| human, brain tumors, Peptide, 156 aa | Query 18 RGDVQEVRRLLHRELVHPDALNRFGKTALQ#VMMFGSTAIALELLKQGASPNVQD 71 || |+||| || + |+| | +|+ +|#||| || +| || || || | Sbjct 22 RGRVEEVRALLEAVAL-PNAPNSYGRRPIQ#VMMMGSARVAELLLLHGAEPNCAD 74 |
| Xiphophorus helleri | Query 18 RGDVQEVRRLLHRELVHPDALNRFGKTALQ#VMMFGSTAIALELLKQGASPNVQDTS-GTS 76 +| || || + + +| ||+ |+|#||| ||+ +| || +|| ||| | | | + Sbjct 13 KGHTAEVEALLLQG-APVNGVNSFGRRAIQ#VMMMGSSEVARLLLTRGADPNVTDKSTGAT 71 Query 77 P 77 | Query 77 P 77 |
| Myxococcus xanthus DK 1622 | Query 27 LLHRELVHPDALNRFGKTALQ-#VMMFGSTAIALELLKQGASPNVQDTSGTS 76 |++++ | ||||+||| # +|| + +| +|||||+|+| || | Sbjct 127 LINQQPCAVDQSNRFGQTALMF#ASLFGRKEVVEQLRQQGASPDVRDGSGRS 177 |
| Apis mellifera | Query 18 RGDVQEVRRLLHRELVHPDALNRFGKTALQ#VMMFGSTAIAL-ELLKQGASPNVQDTSGTS 76 ||| + +| || || | ++ | | | #+ | |+ |||+||| || + +||+ Sbjct 19 RGDAKRLRVLLDSGRVHVDCKDKDGTTPLI#LAAAGGHIEAVTELLQQGADPNARRLTGTT 78 |
| Solibacter usitatus Ellin6076 | Query 18 RGDVQEVRRLLHRELVHPDALNRFGKTALQ#VMMFGSTAIALELLKQ-GASPNVQDTSGTS 76 | | |+ ||| + +|+| ||+| | | #+ | +||| | || || | + Sbjct 62 RQDDLEMSRLLLKAGANPNAANRYGMTVLA#LACTNGNAAIVELLLQAGADPNAALPGGET 121 Query 77 P 77 | Query 77 P 77 |
| Drosophila melanogaster | Query 18 RGDVQEVRRLLHRELVHPDALNRFGKTALQ#VMMFGS-TAIALELLKQGASPNVQDTSGTS 76 ||| + |+| || | + | | | #+ | | +||| ||| || + +||+ Sbjct 19 RGDEVALLRVLDSGKVHVDCKDEDGTTPLI#LAAAGGHTYCVMELLDQGADPNSRRLTGTT 78 Query 77 P 77 | Query 77 P 77 |
| Tribolium castaneum | Query 18 RGDVQEVRRLLHRELVHPDALNRFGKTAL-Q#VMMFGSTAIALELLKQGASPNVQDTSGTS 76 | |+ |||||| + | ||+ | | ||| |# + + |+ | ||+ |+ | +|+ + Sbjct 72 RNDIDEVRRLLMKG-VSPDSCNEDGLTALHQ#CCIDNNEAMLLLLLEYNANVNAEDSEKWT 130 Query 77 P 77 | Query 77 P 77 |
| Trichomonas vaginalis G3 | Query 26 RLLHRELVHP--------DALNRFGKTALQ#VMMFG-STAIALELLKQGASPNVQDTSG 74 || | ||| ||+| |+||| #| + ++ +| || |||||||| +| Sbjct 518 RLNHPSLVHALLEHDVNLDAINDEGQTALM#VAVVSWASEVAKLLLTFGASPNVQDKNG 575 |
| Pseudomonas stutzeri A1501 | Query 18 RGDVQEVRRLLHR--ELVHPDALNRFGKTALQ#VM-MFGSTAIALELLKQGASPNVQDTSG 74 +||+ || |+ | ++ | | ||||| #+ || ||| |+ ||| |+ | | Sbjct 93 KGDLPMVRLLVERGTDVESPCA---DGKTALM#MAAMFNHTAIVEWLVAQGADPHATDAGG 149 Query 75 TSP 77 |+| Sbjct 150 TTP 152 |
| Medicago truncatula | Query 36 DALNRFGKTALQ#VMMFGST-AIALELLKQGASPNVQDTSGTSP 77 |++|+ | ||| # + | |+ ||++||||++|| | +| Sbjct 183 DSINKEGLTALH#KAVIGKKEAVISHLLRKGASPHIQDKDGATP 225 |
| Equus caballus | Query 35 PDALNRFGKTALQ#VMMFGSTAIALELLKQGASPNVQD 71 |+ +| ||+ +|#||| || +| || || || | Sbjct 5 PNGVNGFGRRPIQ#VMMMGSVHVAELLLLHGADPNRAD 41 |
| Wolbachia endosymbiont of Culex pipiens | Query 19 GDVQEVRRLLHRELVHPDALNRFGKTALQ-#VMMFGSTAIALELLKQGASPNVQDTSGTSP 77 | |+ | | | |+ + +|++| ||| # +| | ||++||+||+ + | | Sbjct 64 GGCLEIVRFLIDEGVNVNIINKYGSTALHN#AAYYGDLRIIKFLLEKGANPNIINDDGKKP 123 |
| Aspergillus niger CBS 513.88 | Query 19 GDVQEVRRLLHRELVHPDALNRFGKTAL-Q#VMMFGSTAIALELLKQGASPNVQDTSGTSP 77 || | +|| | |+ || |+ +| #|+ +|| | ||+ | |||+ | +| Sbjct 47 GDTNAVVKLLLRYGADPNIRNRHGRVSLFW#VLNYGSGASVEALLRAGCDPNVRYRYGRTP 106 |
| Thermofilum pendens Hrk 5 | Query 19 GDVQEVRRLLHRELVHPDALNRFGKTALQ#VMMFGSTAIALELLKQGASPNVQDTSGTSP 77 | + |+ || | | |++ || | | # + | + +| ||++|| || +++|| +| Sbjct 230 GSAEVVKFLLERG-ADPCAVDAFGNTPLH#-LAFKNMEVAKLLLEKGADPNAKNSSGMTP 286 |
| Schistosoma japonicum | Query 18 RGDVQEVRRLLHRELVHPDALNRFGKTAL-Q#VMMFGSTAIALELLKQGASPNVQDTSGTS 76 |||+ ||| || | || | | |+| |# + + + ||+ ||+|| +|| + Sbjct 94 RGDLDEVRELLSSG-VCPDVANEDGLTSLHQ#CCIDNNLEMCRLLLRYGANPNSRDTELWT 152 Query 77 P 77 | Query 77 P 77 |
| Methanosarcina barkeri str. fusaro | Query 19 GDVQEVRRLLHRELVHPDALNRFGKTALQ#VMMFGSTAIALELL-KQGASPNVQDTSG 74 | ++ | + | + | ++ + +||||| # + +||| | || ||+|| +| Sbjct 23 GQIENVEKFLKSDKVDINSQDAYGKTALN#YAVNKGYRDVVELLIKGGADPNIQDENG 79 |
| Rickettsiella grylli | Query 22 QEVRRLLHRELVHPDALNRFGKTALQ#VMMFGSTAIALE-LLKQGASPNVQDTSGTSP 77 ||+ |++ + | +| +|+| | # ++| +|++ |++||| | | +||+| Sbjct 211 QEMLEKLYQAGIPIDTVNNYGETPLF#SAIYGINPLAVKFFLERGASVNHQSVNGTTP 267 |
| Ostreococcus lucimarinus CCE9901 | Query 19 GDVQEVRRLLHRELVHPDALNRFGKTALQ#VMM-FGSTAIALELLKQGASPNVQDTSGTSP 77 ||+ || +| | | ++ |+||| # + || +| ||| +||+ +| | +| Sbjct 54 GDLLGVRAMLDARTVDVDDVDAGGRTALH#FAVGFGREHVARELLDRGAALETRDDWGKAP 113 |
| Cryptosporidium parvum Iowa II | Query 18 RGDVQEVRRLLHRELVHPDALNRFGKTALQ#VMMF-GSTAIALELLKQGASPNVQDTSGTS 76 + +|+ ++++| | ++ + | || #+ | + ||| ||| ||+|||+| | | Sbjct 159 KNEVEFLKKILERYPFLSNSRSSEGSYALH#LACDKGYSEIALLLLKNGANPNVKDNFGDS 218 Query 77 P 77 | Query 77 P 77 |
| Aspergillus oryzae | Query 24 VRRLLHRELVHPDALNRFGKTALQ#V-MMFGSTAIALELLKQGASPNVQDTSGTSP 77 | ||| |+|+| | ||| #| +| |+ + |||++|| | + + +| | Sbjct 168 VMRLLLSYGADPNAVNSEGATALH#VGVMNGNYTMVAELLQRGADPTLTNAAGWLP 222 |
| Cryptosporidium parvum | Query 18 RGDVQEVRRLLHRELVHPDALNRFGKTALQ#VMMF-GSTAIALELLKQGASPNVQDTSGTS 76 + +|+ ++++| | ++ + | || #+ | + ||| ||| ||+|||+| | | Sbjct 159 KNEVEFLKKILERYPFLSNSRSSEGSYALH#LACDKGYSEIALLLLKNGANPNVKDNFGDS 218 Query 77 P 77 | Query 77 P 77 |
| Cryptosporidium hominis TU502 | Query 18 RGDVQEVRRLLHRELVHPDALNRFGKTALQ#VMMF-GSTAIALELLKQGASPNVQDTSGTS 76 + +|+ ++++| | ++ + | || #+ | + ||| ||| ||+|||+| | | Sbjct 159 KNEVEFLKKILERYPFLSNSRSSEGSYALH#LACDKGYSEIALLLLKNGANPNVKDNFGDS 218 Query 77 P 77 | Query 77 P 77 |
| Aedes aegypti | Query 18 RGDVQEVRRLLHRELVHPDALNRFGKTALQ#VMMFGSTAIALE-LLKQGASPNVQDTSGTS 76 | |+ || +|| + | ||| | | ||| # | | || ||+ | ||+ + Sbjct 86 RNDIAEVAKLLQKG-VTPDATNMDGLTALH#QCCIDDNAEMLNLLLDYGANVNAQDSERWT 144 Query 77 P 77 | Query 77 P 77 |
| Egeria densa | Query 18 RGDVQEVRRLLHRELVHPDALNRFGKTALQ#VMMF-GSTAIALELLKQGASPNVQDTSGTS 76 ||| | +|| |+ + |+||| #+ || || || +||+|| +|+ |++ Sbjct 433 RGDYMLVDKLLECGS-DPNEADSKGRTALH#IAARNGSERCALLLLDRGANPNSKDSDGST 491 Query 77 P 77 | Query 77 P 77 |
| Anopheles gambiae str. PEST | Query 18 RGDVQEVRRLLHRELVHPDALNRFGKTALQ#VMMFGSTAIALE-LLKQGASPNVQDTSGTS 76 | |+ || +|| + | ||| | | ||| # | | || ||+ | ||+ + Sbjct 26 RNDIAEVAKLLQKG-VTPDATNMDGLTALH#QCCIDDNAEMLNLLLDYGANVNAQDSERWT 84 Query 77 P 77 | Query 77 P 77 |
| Rickettsia felis URRWXCal2 | Query 19 GDVQEVRRLLHRELVHPDALNRFGKTALQ#VMMF-GSTAIALELLKQGASPNVQDTSG 74 ||+ ||+ || + + | |+||| # +| | | ||| ||||+ | || Sbjct 17 GDLSEVQALLKSGVNIDEHKNERGETALY#NTLFTGYMDRAAFLLKHKASPNIPDNSG 73 |
| Caenorhabditis elegans | Query 19 GDVQEVRRLLHRELVHPDALNRFGKTALQ#VM-MFGSTAIALELLKQGASPNVQDTSGTSP 77 | + || |+ |+ || | | ||| #+ + | + | |++ ||+ ||| +| +| Sbjct 77 GHSEVVRELIKRQ-AQVDAATRKGNTALH#IASLAGQSLIVTILVENGANVNVQSVNGFTP 135 |
| Orientia tsutsugamushi Boryong | Query 19 GDVQEVRRLLHRELVHPDALNRFGKTALQ#V-MMFGSTAIALELLKQGASPNVQDTSG 74 ||+++|++|++++ ++ + | ||| #| + + + || || ||+ | || | Sbjct 18 GDIEKVKQLINKDSTLVNSKYKDGTTALS#VALKYKNLPIAEILLNNGANVNAQDNDG 74 |
| Caenorhabditis briggsae | Query 19 GDVQEVRRLLHRELVHPDALNRFGKTALQ#VM-MFGSTAIALELLKQGASPNVQDTSGTSP 77 | + || |+ |+ || | | ||| #+ + | + | |++ ||+ ||| +| +| Sbjct 78 GHSEVVRELIKRQ-AQVDAATRKGNTALH#IASLAGQSLIVTILVENGANVNVQSVNGFTP 136 |
| Dictyostelium discoideum AX4 | Query 19 GDVQEVRRLLHRELVHPDALNRFGKTALQ#VMMF-GSTAIALELLKQGASPNVQDTSGTSP 77 ||++++ ||+ || + +| | # | || | || + |+ |++|++| +| Sbjct 46 GDIEKLSNLLNNSATSPDTPDSEKRTPLH#HAAFCGSAACVNFLLDKKANANIKDSAGNTP 105 |
| Leptospira borgpetersenii serovar Hardjo-bovis L550 | Query 18 RGDVQEVRRLLHRELVHPDALNRFGKTALQ#VMMF-GSTAIALELLKQGASPNVQDTSGTS 76 |||++ ++ + | | | || | | | #+ + | +| |+ ||| || | | | Sbjct 48 RGDIEALKAQIGT-LRHVDEKNRKGYTLLM#LAAYNGQEEASLFLISQGADPNSTDFEGNS 106 |
| Bordetella pertussis Tohama I | Query 18 RGDVQEVRRLLHRELVHPDALNRFGKTA-LQ#VMMFGS-----TAIALELLKQGASPNVQD 71 || |+ | || | | || +|| | | |+# ++ | || |++ || |+ | Sbjct 139 RGHVEVVDMLL-RAGVDPDHVNRLGWTGLLE#AIILGDGGTRHQAIVARLIEGGADVNLAD 197 Query 72 TSGTSP 77 | || Sbjct 198 GDGVSP 203 |
| Bordetella parapertussis 12822 | Query 18 RGDVQEVRRLLHRELVHPDALNRFGKTA-LQ#VMMFGS-----TAIALELLKQGASPNVQD 71 || |+ | || | | || +|| | | |+# ++ | || |++ || |+ | Sbjct 145 RGHVEVVDMLL-RAGVDPDHVNRLGWTGLLE#AIILGDGGTRHQAIVARLIEGGADVNLAD 203 Query 72 TSGTSP 77 | || Sbjct 204 GDGVSP 209 |
| Strongylocentrotus purpuratus | Query 39 NRFGKTALQ#VMMF-GSTAIALELLKQGASPNVQDTSGTSP 77 | |+||| #+ | + +|| |+ ||| |||| +| +| Sbjct 87 NERGETALH#MAAIKGDSQMALNLINQGAEVNVQDFAGWTP 126 |
| Orf virus | Query 27 LLHRELVHPDALNRFGKTALQ#VMMFG---STAIALELLKQGASPNVQDTSGTSP 77 || | +| + |+| | #+ + | | +|| |+ ||+|| | | +| Sbjct 136 LLAEHGAHVNAKDDLGRTPLH#IYLSGFFVSAPVALALIALGANPNATDAYGRTP 189 |
| Neurospora crassa OR74A | Query 36 DALNRFGKTAL-Q#VMMFGSTAIALELLKQGASPNVQDTSGTS 76 | |++|+||| |# + |+| |+++|| ||+|| | | Sbjct 939 DMANKYGETALMQ#AVSTRRWALAKLLIEKGADPNLQDGQGES 980 |
| Pseudomonas entomophila L48 | Query 18 RGDVQEVRRLLHRELVHPDALNRFGKTALQ#VM-MFGSTAIALELLKQGASPNVQDTSGTS 76 +||+ +| || | || |+||| #+ || | || ||| | +| +||+ Sbjct 93 KGDLAMIRLLLEHG-VPVDAAAADGRTALM#LAAMFNRLEILEHLLAQGADPAHRDAAGTT 151 |
| Aspergillus niger | Query 19 GDVQEVRRLLHRELVHPDALNRFGKTALQ#VMMF-GSTAIALELLKQGASPNVQDTSGTSP 77 | + | +|| + |+ + |+| | # + | |+| ||+||| || ||+|| +| Sbjct 734 GGHEAVAKLLLEQGADPNTPDSSGRTPLS#RASWRGHEALAKLLLEQGADPNTQDSSGRTP 793 Query 22 QEVRRLLHRELVHPDALNRFGKTALQ#VMMF-GSTAIALELLKQGASPNVQDTSGTSP 77 + + +|| + |+ + |+| | # + | |+| ||+||| || ||+|| +| Sbjct 770 EALAKLLLEQGADPNTQDSSGRTPLS#RASWRGHEALAKLLLEQGADPNTQDSSGWTP 826 Query 19 GDVQEVRRLLHRELVHPDALNRFGKTAL-Q#VMMFGSTAIALELLKQGASPNVQDTSGTSP 77 | + | +|| |+| + |+| | # | |+| ||+||| || +|+|| +| Sbjct 668 GGHEAVAKLLLEWGADPNARDSSGQTLLIW#ASEKGHEAVAKLLLEQGADPNARDSSGWTP 727 Query 52 GSTAIALELLKQGASPNVQDTSGTSP 77 | |+| ||+||| || ||+|| | Sbjct 636 GHEAVAKLLLEQGADPNTQDSSGQIP 661 |
[Site 2] DGTGALPIHL113-AVQEGHTAVV
Leu113  Ala
Ala
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Asp104 | Gly105 | Thr106 | Gly107 | Ala108 | Leu109 | Pro110 | Ile111 | His112 | Leu113 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Ala114 | Val115 | Gln116 | Glu117 | Gly118 | His119 | Thr120 | Ala121 | Val122 | Val123 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| TGFLDTLKVLVEHGADVNAPDGTGALPIHLAVQEGHTAVVSFLAAESDLHRRDARGLTPL |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | Macaca mulatta | 122.00 | 5 | PREDICTED: cyclin-dependent kinase inhibitor 2D (p |
| 2 | Homo sapiens | 120.00 | 21 | cyclin-dependent kinase inhibitor 2D |
| 3 | N/A | 120.00 | 17 | - |
| 4 | synthetic construct | 120.00 | 12 | cyclin-dependent kinase inhibitor 2D (p19, inhibit |
| 5 | Canis familiaris | 117.00 | 6 | PREDICTED: similar to Cyclin-dependent kinase 4 in |
| 6 | Bos taurus | 114.00 | 5 | cyclin-dependent kinase inhibitor 2D (p19, inhibit |
| 7 | Monodelphis domestica | 105.00 | 5 | PREDICTED: similar to Cyclin-dependent kinase inhi |
| 8 | Mus musculus | 104.00 | 22 | cyclin-dependent kinase inhibitor 2D (p19, inhibit |
| 9 | Rattus norvegicus | 101.00 | 5 | similar to cyclin-dependent kinase inhibitor 2D |
| 10 | Xenopus tropicalis | 79.70 | 3 | cyclin-dependent kinase inhibitor 2D (p19, inhibit |
| 11 | Xenopus laevis | 78.60 | 1 | ankyrin 2, neuronal |
| 12 | Takifugu rubripes | 76.60 | 1 | p19 |
| 13 | Xiphophorus maculatus | 74.30 | 2 | p19 |
| 14 | Tetraodon nigroviridis | 58.50 | 4 | unnamed protein product |
| 15 | Gallus gallus | 57.80 | 4 | PREDICTED: similar to Chain A, Crystal Structure O |
| 16 | Danio rerio | 57.80 | 1 | PREDICTED: similar to Chain A, Structure Of P18ink |
| 17 | Arabidopsis thaliana | 52.80 | 3 | putative potassium transporter/channel |
| 18 | Tribolium castaneum | 50.80 | 3 | PREDICTED: similar to UNCoordinated family member |
| 19 | Neosartorya fischeri NRRL 181 | 50.40 | 2 | Ankyrin repeat protein |
| 20 | Strongylocentrotus purpuratus | 50.10 | 99 | PREDICTED: similar to KIAA1255 protein |
| 21 | Pan troglodytes | 50.10 | 5 | PREDICTED: huntingtin interacting protein 14 |
| 22 | Macaca fascicularis | 48.10 | 1 | unnamed protein product |
| 23 | Trichomonas vaginalis G3 | 47.80 | 8 | hypothetical protein TVAG_180110 |
| 24 | Gibberella zeae PH-1 | 47.80 | 1 | hypothetical protein FG08155.1 |
| 25 | Canarypox virus | 47.40 | 1 | CNPV295 ankyrin repeat protein |
| 26 | Plasmodium berghei strain ANKA | 47.40 | 1 | hypothetical protein |
| 27 | Pseudomonas stutzeri A1501 | 47.00 | 1 | ankyrin domain protein |
| 28 | Aedes aegypti | 43.10 | 1 | conserved hypothetical protein |
| 29 | Anopheles gambiae str. PEST | 41.60 | 1 | ENSANGP00000006257 |
| 30 | Synechococcus sp. JA-2-3B'a(2-13) | 38.90 | 1 | ankyrin repeat protein |
| 31 | Synechococcus sp. JA-3-3Ab | 36.20 | 1 | ankyrin repeat protein |
| 32 | Thermofilum pendens Hrk 5 | 34.30 | 1 | Ankyrin |
| 33 | Wolbachia endosymbiont of Drosophila melanogaster | 33.10 | 1 | ankyrin repeat domain protein |
| 34 | Aspergillus terreus NIH2624 | 33.10 | 1 | predicted protein |
| 35 | Apis mellifera | 32.70 | 2 | PREDICTED: similar to ankyrin repeat domain 50 |
| 36 | Xiphophorus helleri | 32.30 | 1 | p13CDKN2X |
Top-ranked sequences
| organism | matching |
|---|---|
| Macaca mulatta | Query 84 TGFLDTLKVLVEHGADVNAPDGTGALPIHL#AVQEGHTAVVSFLAAESDLHRRDARGLTPL 143 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 84 TGFLDTLKVLVEHGADVNAPDGTGALPIHL#AVQEGHTAVVSFLAAESDLHRRDARGLTPL 143 |
| Homo sapiens | Query 84 TGFLDTLKVLVEHGADVNAPDGTGALPIHL#AVQEGHTAVVSFLAAESDLHRRDARGLTPL 143 |||||||||||||||||| |||||||||||#|||||||||||||||||||||||||||||| Sbjct 84 TGFLDTLKVLVEHGADVNVPDGTGALPIHL#AVQEGHTAVVSFLAAESDLHRRDARGLTPL 143 |
| N/A | Query 84 TGFLDTLKVLVEHGADVNAPDGTGALPIHL#AVQEGHTAVVSFLAAESDLHRRDARGLTPL 143 |||||||||||||||||| |||||||||||#|||||||||||||||||||||||||||||| Sbjct 84 TGFLDTLKVLVEHGADVNVPDGTGALPIHL#AVQEGHTAVVSFLAAESDLHRRDARGLTPL 143 |
| synthetic construct | Query 84 TGFLDTLKVLVEHGADVNAPDGTGALPIHL#AVQEGHTAVVSFLAAESDLHRRDARGLTPL 143 |||||||||||||||||| |||||||||||#|||||||||||||||||||||||||||||| Sbjct 84 TGFLDTLKVLVEHGADVNVPDGTGALPIHL#AVQEGHTAVVSFLAAESDLHRRDARGLTPL 143 |
| Canis familiaris | Query 84 TGFLDTLKVLVEHGADVNAPDGTGALPIHL#AVQEGHTAVVSFLAAESDLHRRDARGLTPL 143 ||||||||||||||||||||||||||||||#||+||||||||||| ||||| ||||||||| Sbjct 84 TGFLDTLKVLVEHGADVNAPDGTGALPIHL#AVREGHTAVVSFLATESDLHHRDARGLTPL 143 |
| Bos taurus | Query 84 TGFLDTLKVLVEHGADVNAPDGTGALPIHL#AVQEGHTAVVSFLAAESDLHRRDARGLTPL 143 ||||||||||||||||||||||||||||||#||+||||+|||||| ||||| ||| ||||| Sbjct 84 TGFLDTLKVLVEHGADVNAPDGTGALPIHL#AVREGHTSVVSFLATESDLHHRDATGLTPL 143 |
| Monodelphis domestica | Query 84 TGFLDTLKVLVEHGADVNAPDGTGALPIHL#AVQEGHTAVVSFLAAESDLHRRDARGLTPL 143 |||||||+|||||||||| |||+|||||||#||+||| ||||||| |||| ||| ||||| Sbjct 84 TGFLDTLRVLVEHGADVNVPDGSGALPIHL#AVREGHAAVVSFLAGESDLQHRDAGGLTPL 143 |
| Mus musculus | Query 84 TGFLDTLKVLVEHGADVNAPDGTGALPIHL#AVQEGHTAVVSFLAAESDLHRRDARGLTPL 143 ||||||||||||||||||| | ||+|||||#|++|||++|||||| ||||| ||| ||||| Sbjct 84 TGFLDTLKVLVEHGADVNALDSTGSLPIHL#AIREGHSSVVSFLAPESDLHHRDASGLTPL 143 |
| Rattus norvegicus | Query 84 TGFLDTLKVLVEHGADVNAPDGTGALPIHL#AVQEGHTAVVSFLAAESDLHRRDARGLTPL 143 |||||||||||||||||| | ||+|||||#|++|||++|||||| ||||| +|| ||||| Sbjct 84 TGFLDTLKVLVEHGADVNTLDSTGSLPIHL#AIREGHSSVVSFLAPESDLHHKDASGLTPL 143 |
| Xenopus tropicalis | Query 85 GFLDTLKVLVEHGADVNAPDGTGALPIHL#AVQEGHTAVVSFLAAESDLHRRDARGLTP 142 ||||||+|||+|||++| || +|+|||||#|++ || |+++|| ||| || | || Sbjct 85 GFLDTLQVLVQHGAEINTPDASGSLPIHL#ALKAGHVPVITYLALISDLQHRDREGHTP 142 |
| Xenopus laevis | Query 85 GFLDTLKVLVEHGADVNAPDGTGALPIHL#AVQEGHTAVVSFLAAESDLHRRDARGLTP 142 ||||||+|||+|||+|| || +|+|||||#|++ || |+++|| ||| + | | || Sbjct 85 GFLDTLQVLVQHGAEVNTPDASGSLPIHL#ALKAGHVPVITYLALISDLQQHDREGHTP 142 |
| Takifugu rubripes | Query 84 TGFLDTLKVLVEHGADVNAPDGTGALPIHL#AVQEGHTAVVSFLAAESDLHRRDARGLT 141 |||||||+||||+|| || || +||||||+#|++||| || ||| ||| + | | Sbjct 84 TGFLDTLQVLVEYGASVNLPDQSGALPIHI#AIREGHRDVVEFLAPRSDLKHANKSGQT 141 |
| Xiphophorus maculatus | Query 84 TGFLDTLKVLVEHGADVNAPDGTGALPIHL#AVQEGHTAVVSFLAAESDLHRRDARGLTPL 143 ||||+||+||||||| || | ||||||+#|++||| +| ||| ||| + | | + Sbjct 84 TGFLETLQVLVEHGASVNIQDQNGALPIHI#AIREGHRDIVEFLAPRSDLKHANVSGQTAI 143 |
| Tetraodon nigroviridis | Query 84 TGFLDTLKVLVEHGADVNAPDGTGALPI-HL#AVQEGHTAVVSFLA 127 |||||||+||||+|| || || +||||| | # ++|| || ||| Sbjct 84 TGFLDTLQVLVEYGASVNLPDQSGALPIPHC#QSEKGHRDVVEFLA 128 |
| Gallus gallus | Query 85 GFLDTLKVLVEHGADVNAPDGTGALPIHL#AVQEGHTAVVSFL--AAESDLHRRDARGLTP 142 ||||||+ |+| |||| | | ||+||#| |||| || || | + ++ || | Sbjct 81 GFLDTLQTLLEFHADVNIEDAEGNLPLHL#AAQEGHVRVVEFLLRRTPSRVAHQNRRGDTA 140 Query 143 L 143 | Query 143 L 143 |
| Danio rerio | Query 85 GFLDTLKVLVEHGADVNAPDGTGALPIHL#AVQEGHTAVVSFLAAE--SDLHRRDARGLTP 142 |+|||| || ++||||| | | ||+||#| +||| || || + +|+| || Sbjct 83 GYLDTLHVLAQNGADVNLLDNDGNLPLHL#AAREGHLDVVQFLVTHCVTQPFLANAKGYTP 142 |
| Arabidopsis thaliana | Query 87 LDTLKVLVEHGADVNAPDGTGALPIHL#AVQEGHTAVVSFLAAE-SDLHRRDARGLTP 142 || || ++++| || ||| | +| #|| ||| +| || + +|| |+ | || Sbjct 241 LDALKDIIKYGGDVTLPDGNGTTALHR#AVSEGHLEIVKFLLDQGADLDWPDSYGWTP 297 |
| Tribolium castaneum | Query 87 LDTLKVLVEHGADVNAPDGTGALPIHL#AVQEGHTAVVSFLAAE-SDLHRRDARGLTPL 143 |+ | |++ |||+|||| +|| |+| #||+ ++ | + +|++|+| | ||| Sbjct 126 LELCKFLIDSGADLNAPDSSGATPLHY#AVELNFPKIILLLVHKGADVNRQDLDGYTPL 183 |
| Neosartorya fischeri NRRL 181 | Query 84 TGFLDTLKVLVEHGADVNAPDGTGALPIHL#AVQEGHTAVVSFLAAES--DLHRRDARGLT 141 || ++ ++ |+ + +|| | | |+| #||++|| |||+ |+ |++| |+|| | Sbjct 200 TGDIEIIQRLLNYEYEVNTRDNRGRTPLHR#AVEKGHLAVVNLFLAQPNIDINRADSRGRT 259 Query 142 PL 143 |+ Sbjct 260 PM 261 |
| Strongylocentrotus purpuratus | Query 85 GFLDTLKVLVEHGADVNAPDGTGALPIHL#AVQEGHTAVVSFLAAES--DLHRRDARGLTP 142 | ++ |+|| ||+|| | | |+|+#|+ | ++| + + || || |||| Sbjct 642 GLEQVVQCLIEHQADINAKDAEGRTPLHI#AISNQHPTIISLIMSHPVLDLTLRDKGGLTP 701 |
| Pan troglodytes | Query 85 GFLDTLKVLVEHGADVNAPDGTGALPIHL#AVQEGHTAVVSFLAAE-SDLHRRDARGLTPL 143 | | + |+++||| + || | |||#| | |||++|++| |+ |+ | |+||| Sbjct 266 GHLSMVVQLMKYGADPSLIDGEGCSCIHL#AAQFGHTSIVAYLIAKGQDVDMMDQNGMTPL 325 |
| Macaca fascicularis | Query 93 LVEHGADVNAPDGTGALPIHL#AVQEGHTAVVSFLAAE-SDLHRRDARGLTPL 143 |+++||| + || | |||#| | |||++|++| |+ |+ | |+||| Sbjct 5 LMKYGADPSLIDGEGCSCIHL#AAQFGHTSIVAYLIAKGQDVDMMDQNGMTPL 56 |
| Trichomonas vaginalis G3 | Query 84 TGFLDTLKVLVEHGADVNAPDGTGALPIHL#AVQEGHTAVVSFLAAE-SDLHRRDARGLTP 142 +|+|+ +++||| ||||+ | | |+| #|| + +| || |+ +| + | | || Sbjct 2 SGYLEIIQILVEQGADVHCHDNHGWTPLHR#AVLWDYVPIVQFLLAKGADPNAIDLDGFTP 61 Query 143 L 143 | Query 143 L 143 |
| Gibberella zeae PH-1 | Query 85 GFLDTLKVLVEHGADVNAPDGTGALPIHL#AVQEGHTAVVSFLAAES--DLHRRDARGLTP 142 |+ |++||+||||| |+ + | + +#| +||| ++|| | | | ||+ +| Sbjct 843 GYTATVEVLLEHGADANSREKYGRTALSM#AAEEGHESIVSLLLATEGVDADSRDSEEQSP 902 Query 143 L 143 | Query 143 L 143 |
| Canarypox virus | Query 86 FLDTLKVLVEHGADVNAPDGTGALPIHL#AVQEGHTAVVSFLAAESDLHRRDARGLTPL 143 | + |+||++| |+| | |++|+| #| ++ |+ | + ++++ |+ | ||| Sbjct 145 FYEMTKLLVDNGIDINCKDNYGSIPLHY#AAKKSELPVIDLLISNTNINSRNNRNNTPL 202 |
| Plasmodium berghei strain ANKA | Query 85 GFLDTLKVLVEHGADVNAPDGTGALPIHL#AVQEGHTAVVSFLA-AESDLHRRDARGLT 141 |||| +| |++ ||++| | | +|+#| | ++ || | ++|+++| ||| Sbjct 251 GFLDIVKFLIKAGANINEEDSLGDSVLHI#AAYSGKMEIIKFLINAGVNIHKKNAEGLT 308 |
| Pseudomonas stutzeri A1501 | Query 85 GFLDTLKVLVEHGADVNAPDGTGALPIHL#AVQEGHTAVVSFLAAE-SDLHRRDARGLTPL 143 | | +++||| | || +| | + +#| |||+| +| |+ +| | || | ||| Sbjct 94 GDLPMVRLLVERGTDVESPCADGKTALMM#AAMFNHTAIVEWLVAQGADPHATDAGGTTPL 153 |
| Aedes aegypti | Query 85 GFLDTLKVLVEHGADVNAPDGTGALPIHL#AVQEGHTAVVSFLAAESDLHRR--DARGLTP 142 | + + |+++||| +| || | +|+#||+||| ||| | ||+|+ + +| | Sbjct 916 GTIGAVSALLQNGADFDAVDGDGNNALHI#AVREGHIAVVRELLTESELNAEVVNMKGRNP 975 Query 143 L 143 | Query 143 L 143 Query 85 GFLDTLKVLVEHGADVNAPDGTGALPIHL#AVQEGHTAVVSFLAAES--DLHRRDARGLTP 142 | + | |++|||+||| | |+|+#|++ | ++ | || || | || Sbjct 747 GLRNVLHTLIDHGANVNAVDCDLKTPLHV#AIENQHEEIIGILLCHPGIDLKIRDKTGNTP 806 |
| Anopheles gambiae str. PEST | Query 85 GFLDTLKVLVEHGADVNAPDGTGALPIHL#AVQEGHTAVVSFLAAESDLHRR--DARGLTP 142 | + + |+++||| +| || | +|+#||+||| ||| | ||+|+ + +| | Sbjct 915 GTVGAVSALLQNGADFDAVDGDGNNALHI#AVREGHVAVVRELLTESELNAETVNLKGRNP 974 Query 143 L 143 | Query 143 L 143 Query 85 GFLDTLKVLVEHGADVNAPDGTGALPIHL#AVQEGHTAVVSFLAAES--DLHRRDARGLTP 142 | + | |+ |||+||| | |+|+#|++ | ++ | || || | || Sbjct 746 GLRNVLHTLIGHGANVNAVDCDLKTPLHV#AIENQHEEIIGILLCHPGIDLKIRDKTGNTP 805 |
| Synechococcus sp. JA-2-3B'a(2-13) | Query 85 GFLDTLKVLVEHGADVNAPDGTGALPIHL#AVQEGHTAVVS-FLAAESDLHRRDARGLTPL 143 | |+ | +|+ ||| || | ||| |+||#| | +| | | ||+ | | ||| Sbjct 45 GDLELLDLLLAHGASPNALDPTGATPLHL#AAAVGQVKLVERLLRAGSDIDLPDNHGYTPL 104 Query 85 GFLDTLKVLVEHGADVNAPDGTGALPIHL#AVQEGHTAVVSFLAAE-SDLHRRDARGLTPL 143 | | + |+ +||+ || + || |+| #| ||| ++ | ++|+ |+ | ||| Sbjct 170 GDLGIARFLLLYGANANARNRFGATPMHW#AAWEGHIEILELLLENGAELNPRNEDGHTPL 229 |
| Synechococcus sp. JA-3-3Ab | Query 85 GFLDTLKVLVEHGADVNAPDGTGALPIHL#AVQEGHTAVVS-FLAAESDLHRRDARGLTPL 143 | |+ | +|+ +|| |++ | ||| |+||#| | || | | ||+ | | ||| Sbjct 45 GNLELLDLLLAYGASVDSMDSTGATPLHL#AAASGQVEVVERLLKAGSDIDLLDRYGYTPL 104 Query 85 GFLDTLKVLVEHGADVNAPDGTGALPIHL#AVQEGHTAVVSFLAAE-SDLHRRDARGLTPL 143 | | + |+ +|||||| + || |+| #| ||| ++ | ++|+ |+ | ||| Sbjct 170 GHLGIARFLILYGADVNARNRFGATPMHW#AAWEGHIQILELLLENGAELNPRNHDGHTPL 229 Query 84 TGFLDTLKVLVEHGADVNAPDGTGALPIHL#AVQEGHTAVVSFLAA-ESDLHRRDARGLTP 142 || | |+ || |+| | || |+| #| +|| + || +|++ |+ | || Sbjct 136 TGRKSVLARLLSKGAPVDAVDETGRTPLHE#AATQGHLGIARFLILYGADVNARNRFGATP 195 Query 143 L 143 + Sbjct 196 M 196 Query 90 LKVLVEHGADVNAPDGTGALPIHL#AVQEGHTAVVSFLAA-ESDLHRRDARGLTPL 143 | ++ | ||||| | |+| #|| |+ ++ | | + + |+ | ||| Sbjct 17 LLAYLQQGGHVNAPDSQGFTPLHK#AVLIGNLELLDLLLAYGASVDSMDSTGATPL 71 |
| Thermofilum pendens Hrk 5 | Query 84 TGFLDTLKVLVEHGADVNAPDGTGALPIHL#AVQEGHTAVVSFLAAE-SDLHRRDARGLTP 142 +| ++ +++|+||||||+| + | |+| #| || +| | +| + |++ | || Sbjct 668 SGNVEAVRLLLEHGADVDARNDFGGTPLHH#AAARGHLEIVRLLLKHGADSNARNSHGETP 727 Query 143 L 143 | Query 143 L 143 Query 85 GFLDTLKVLVEHGADVNAPDGTGALPIHL#AVQEGHTAVVSFLAAE-SDLHRRDARGLTPL 143 || | ++|++ |||||| + +| |+| #| ++| | | +| | | ||| Sbjct 133 GFADIARLLLDRGADVNAKNSSGKTPLHY#AAEQGSAEVAKLLLERGADPGATDTYGNTPL 192 Query 87 LDTLKVLVEHGADVNAPDGTGALPIHL#AVQEGHTAVVSFLAAE-SDLHRRDARGLTPL 143 ++ |+|+| |||||| + | |+| #| || || || +| || | ||| Sbjct 199 IEVSKLLLERGADVNARNNEGRTPLHR#AAMEGSAEVVKFLLERGADPCAVDAFGNTPL 256 Query 87 LDTLKVLVEHGADVNAPDGTGALPIHL#AVQEGHTAVVSFLAAE-SDLHRRDARGLTPL 143 ++ |+|+| ||| || + +| |+| #| | || | +|+ +| ||||| Sbjct 263 MEVAKLLLEKGADPNAKNSSGMTPLHF#AAGLGKVEVVELLLEHGADVDAKDNDGLTPL 320 Query 86 FLDTLKVLVEHGADVNAPDGTGALPIHL#AVQEGHTAVVSFLAAE-SDLHRRDARGLTPL 143 + +++|+| | | || | | |+| #| + | || | +| + |+ |+||| Sbjct 371 YAKVVRLLLEKGLDANAKDEYGRTPLHW#AAERGCPEVVELLLEHGADPNARNDSGMTPL 429 Query 85 GFLDTLKVLVEHGADVNAPDGTGALPIHL#AVQEGHTAVVSFLAAESDLHRRDARGLTPL 143 | + |+|+| ||| | | | |+||#||+ + + | +|++ |+ | ||| Sbjct 166 GSAEVAKLLLERGADPGATDTYGNTPLHL#AVRSIEVSKL-LLERGADVNARNNEGRTPL 223 Query 87 LDTLKVLVEHGADVNAPDGTGALPIHL#AVQ-----EGHTAVVSFLAAESDLHRRDARGLT 141 |+ + |+||||| || | | |+| # ++ | |+ | +| |++ ||+ Sbjct 783 LEVARWLLEHGADPNARDWEGNTPLHY#VIEHSFWRERREAIELLLEHGADPSIRNSEGLS 842 Query 142 PL 143 || Query 142 PL 143 Query 85 GFLDTLKVLVEHGADVNAPDGTGALPIHL#AVQEGHTAVVSFLAAE-SDLHRRDARGLTPL 143 | +||++ ||| || | | |+||#| | + | +|++ +++ | ||| Sbjct 100 GHFVVAEVLLDRGADPNATDEEGNTPLHL#AALLGFADIARLLLDRGADVNAKNSSGKTPL 159 Query 85 GFLDTLKVLVEHGADVNAPDGTGALPIHL#AVQEGHTAVVSFLAAE-SDLHRRDARGLTPL 143 | + +++|+||||| || + +| |+||#| | | +| + + | ||| Sbjct 403 GCPEVVELLLEHGADPNARNDSGMTPLHL#AATVKDTEAAKLLLEHGADPNAEEYGGSTPL 462 Query 85 GFLDTLKVLVEHGADVNAPDGTGALPIHL#AVQEGHTAVVSFL 126 | ++ +++|+| |||+|| + | |+|+#| + |+ | | Sbjct 543 GDVEVIEILLERGADINARNKFGETPLHV#AAERGNFEAVKLL 584 Query 91 KVLVEHGADVNAPDGTGALPIHL#AVQEG---HTAVVSFLAAE-SDLHRRDARGLTPL 143 ++|+ ||||||| | |+|+#|| | | | +| +| + || | ||| Sbjct 751 ELLLIHGADVNARDSRDQTPLHI#AVFFGSREHLEVARWLLEHGADPNARDWEGNTPL 807 Query 85 GFLDTLKVLVEHGADVNAPDGTGALPIHL#AVQEGHTAVVSFLAAESDLHRRDARGLTPL 143 | + +| |+| ||| | | | |+||#| + | + | +| + +++ |+||| Sbjct 230 GSAEVVKFLLERGADPCAVDAFGNTPLHL#AFKNMEVAKL-LLEKGADPNAKNSSGMTPL 287 Query 91 KVLVEHGADVNAPDGTGALPIHL#AVQEGHTAVVSFLAAE-SDLHRRDARGLTPL 143 + |+| |||+|| | |+| #| |+ | | +|+ |+ | ||| Sbjct 642 RFLIERGADINARTKDGETPLHK#ATSSGNVEAVRLLLEHGADVDARNDFGGTPL 695 Query 85 GFLDTLKVLVEHGADVNAPDGTGALPIHL#AVQEGHTAVVS-------------FLAAESD 131 | |+ +++|++|||| || + | |+| # + | + | | +| Sbjct 702 GHLEIVRLLLKHGADSNARNSHGETPLHY#VAE--HADMCSKNAWDNCLRIAELLLIHGAD 759 Query 132 LHRRDARGLTPL 143 ++ ||+| ||| Sbjct 760 VNARDSRDQTPL 771 Query 88 DTLKVLVEHGADVNAPDGTGALPIHL#AVQEGHTAVVSFLAAE-SDLHRRDARGLTPL 143 | +|+| |||+|| | |+| #| + || +|++ | | ||| Sbjct 606 DVFTLLLERGADINARDWFDRTPLHG#AAGCRDAGIARFLIERGADINARTKDGETPL 662 |
| Wolbachia endosymbiont of Drosophila melanogaster | Query 85 GFLDTLKVLVEHGADVNAPDGTGALPIHL#AVQEGHTAVVSFLAAE-SDLHRRDARGLTPL 143 | ++ +| |++ ||||| | |+| #| + ||| || | + +|++ +| | ||| Sbjct 190 GHIEVVKHLIKKGADVNVQSKVGRTPLHN#AAKHGHTQVVEVLLKKGADVNIQDRGGRTPL 249 Query 85 GFLDTLKVLVEHGADVNAPDGTGALPIHL#AVQEGHTAVVSFL----AAESDLHRRDA 137 | ++||++ ||||| | | |+| #||| + + | | | +|| | Sbjct 223 GHTQVVEVLLKKGADVNIQDRGGRTPLHY#AVQRRYPKLAKLLLNDGADPSFIHRSKA 279 Query 85 GFLDTLKVLVEHGADVNAPDG-TGALPIHL#AVQEGHTAVVSFLA-AESDLHRRDARGLTP 142 | + ++ |+++|| +| |+|+#| + || +| |+ |+|+ ++ | || Sbjct 90 GHIQIVENLLDNGAKTGIKNGYCKEAPLHV#AAKHGHIRIVEILSKKEADIDLKNRYGETP 149 Query 143 L 143 | Query 143 L 143 Query 85 GFLDTLKVLVEHGADVNAPDGTGALPIHL#AVQEGHTAVV-SFLAAESDLHRRDARGLTPL 143 | + +++| + ||++ + | |+| #| + ||| |+ + | ++++ + | ||| Sbjct 124 GHIRIVEILSKKEADIDLKNRYGETPLHY#AAKYGHTQVLENLLGRSTNVNVQSEVGRTPL 183 |
| Aspergillus terreus NIH2624 | Query 85 GFLDTLKVLVEHGADVNAPDGTGALPIHL#AVQEGHTAVVSFLAAESDL 132 | +||++ |+|||||| | | + +|+|+#| +||| |+ | ++ | Sbjct 903 GSVDTVQTLIEHGADVMAQDRSLDIPLHI#AAREGHRDVIDCLLKQNPL 950 Query 85 GFLDTLKVLVEHGADVNAPDGTGALPIHL#AVQEGHTAVVSFLAAE-SDLHRRDARGLTPL 143 | |||+ +|| + | + + | +| #| || |+ || +++ |+ | | ||| Sbjct 804 GDLDTVLLLVAYDACIRSVSNRGLTALHR#AAGRGHLEVMQFLIEHGAEIDRKTADGWTPL 863 Query 85 GFLDTLKVLVEHGADVNAPDGTGALPIHL#AVQEGHTAVVSFLAAE-SDLHRRDARGLTPL 143 | |+ ++ |+||||+++ | |+| #| | | | +|+ + | | | Sbjct 837 GHLEVMQFLIEHGAEIDRKTADGWTPLHG#ASSSGRAEAVKLLLKNGADIKHQSFDGSTAL 896 Query 88 DTLKVLVEHGADVNAPDGTGALPIHL#AVQEGHTAVVSFLAAESDLHRRDARGLTP 142 | |+ ++ |||+|+ | +| +||#| | || || ++ |+|| Sbjct 1385 DVLEYVLATGADINSADPSGRTALHL#ATYYPH-AVQIILAYGGNVKAHTDGGMTP 1438 |
| Apis mellifera | Query 85 GFLDTLKVLVEHGADVNAPDGTGALPIHL#AVQEGHTAVVSFLA--AESDLHRRDARGLTP 142 | ||++||+ | ||||| | | +++# | |+ || | +|+ ||+ | || Sbjct 922 GHYDTVRVLLAHNADVNAKDADGRSTLYI#LALENRLAMARFLLEHARADVESRDSEGRTP 981 Query 143 L 143 | Query 143 L 143 Query 85 GFLDTLKVLVEHGADVNAPDGTGALPIHL#AVQEGHTAVVSFLAA-ESDLHRRDARGLTPL 143 |+ + +|+| || |+ | | |+ +#| ||| | | |+|+ || | ||| Sbjct 723 GYAKVVTILLERGAAVDHQDKDGMTPLLV#AAFEGHRDVCELLLEYEADVDHCDATGRTPL 782 Query 85 GFLDTLKVLVEHGADVNAPDGTGALPIHL#AVQEGHTAVVSFL---AAESDLHRRDAR 138 | | +| |++|||||| | | + #| ||+ +| | || | | | Sbjct 652 GHEDIVKALLQHGADVNRTDDEGRTALIA#AAYMGHSEIVEHLLDFGAEIDHADNDGR 708 Query 85 GFLDTLKVLVEHGADVNAPDGTGALPIHL#AVQEGHTAVVSFL----AAESDLHRRDAR 138 | +| + |+| || ++ | | + |#| |||| |+| | | | | | + Sbjct 855 GHIDVCEALLEAGAKIDETDNDGKGALML#AAQEGHAALVERLLEQHGAPIDQHAHDGK 912 Query 85 GFLDTLKVLVEHGADVNAPDGTGALPIHL#AVQEGHTAVVSFL 126 | | ++|+|+ |||+ | || |+ #| || +||+ | Sbjct 756 GHRDVCELLLEYEADVDHCDATGRTPLWA#AASMGHGSVVALL 797 Query 91 KVLVEHG-ADVNAPDGTGALPIHL#AVQEGHTAVVSFLAAE--SDLHRRDARGLTPL 143 + |+|| ||| + | | |+|+#+ +|| +|+ | | + ++ | ||| Sbjct 961 RFLLEHARADVESRDSEGRTPLHV#SAWQGHVEMVALLLTEGGASVNACDNENRTPL 1016 Query 92 VLVEHGADVNAPDGTGALPIHL#AVQEGHTAVVSFL 126 +| | || ||| | |+| #| +|| |+| | Sbjct 997 LLTEGGASVNACDNENRTPLHS#AAWQGHAAIVRLL 1031 Query 85 GFLDTLKVLVEHGADVNAPDGTGALPIHL#AVQEGHTAVVSFL---AAESDLHRRDARG 139 | | +| |++ | | | +| |+| #| ||| | | |+ | | +| Sbjct 822 GGTDVVKQLLDRGLDEQHRDNSGWTPLHY#AAFEGHIDVCEALLEAGAKIDETDNDGKG 879 |
| Xiphophorus helleri | Query 84 TGFLDTLKVLVEHGADVNAPDGTGALPIHL#AVQEGHTAVVSFL 126 ||||||+++||| ||| | | ||| |#| | ||| ||+ | Sbjct 79 TGFLDTVQLLVEAGADPQARDKDNCLPIDL#ARQNGHTDVVAVL 121 Query 85 GFLDTLKVLVEHGADVNAPD-GTGALPIHL#AVQEGHTAVVSFLA-AESDLHRRDARGLTP 142 | + ++|+ ||| | | ||| |+| #| + | | | | +| || | Sbjct 46 GSSEVARLLLTRGADPNVTDKSTGATPLHD#AARTGFLDTVQLLVEAGADPQARDKDNCLP 105 Query 143 L 143 + Sbjct 106 I 106 |
[Site 3] DVQEVRRLLH29-RELVHPDALN
His29  Arg
Arg
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Asp20 | Val21 | Gln22 | Glu23 | Val24 | Arg25 | Arg26 | Leu27 | Leu28 | His29 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Arg30 | Glu31 | Leu32 | Val33 | His34 | Pro35 | Asp36 | Ala37 | Leu38 | Asn39 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| MLLEEVRSGDRLSGAAARGDVQEVRRLLHRELVHPDALNRFGKTALQVMMFGSTAIALE |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | Macaca mulatta | 115.00 | 6 | PREDICTED: cyclin-dependent kinase inhibitor 2D (p |
| 2 | N/A | 114.00 | 17 | - |
| 3 | Homo sapiens | 114.00 | 15 | cyclin-dependent kinase inhibitor 2D |
| 4 | synthetic construct | 114.00 | 7 | cyclin-dependent kinase inhibitor 2D (p19, inhibit |
| 5 | Canis familiaris | 110.00 | 2 | PREDICTED: similar to Cyclin-dependent kinase 4 in |
| 6 | Rattus norvegicus | 108.00 | 7 | similar to cyclin-dependent kinase inhibitor 2D |
| 7 | Mus musculus | 107.00 | 15 | cyclin-dependent kinase inhibitor 2D (p19, inhibit |
| 8 | Bos taurus | 105.00 | 5 | cyclin-dependent kinase inhibitor 2D (p19, inhibit |
| 9 | Monodelphis domestica | 102.00 | 5 | PREDICTED: similar to Cyclin-dependent kinase inhi |
| 10 | Xenopus tropicalis | 89.70 | 2 | cyclin-dependent kinase inhibitor 2D (p19, inhibit |
| 11 | Xenopus laevis | 87.00 | 2 | ankyrin 2, neuronal |
| 12 | Xiphophorus maculatus | 60.50 | 2 | p19 |
| 13 | Tetraodon nigroviridis | 55.80 | 4 | unnamed protein product |
| 14 | Takifugu rubripes | 55.10 | 2 | p19 |
| 15 | Mesocricetus auratus | 48.90 | 3 | p15INK4b cyclin-dependent kinase inhibitor |
| 16 | Macaca fascicularis | 47.00 | 1 | unnamed protein product |
| 17 | Gallus gallus | 46.60 | 3 | cyclin-dependent kinase inhibitor 2A (melanoma, p1 |
| 18 | Sus scrofa | 44.70 | 2 | cyclin dependant kinase inhibitor |
| 19 | human, brain tumors, Peptide, 156 aa | 44.30 | 1 | cyclin D-dependent kinases 4 and 6-binding protein |
| 20 | Pan troglodytes | 44.30 | 1 | PREDICTED: similar to p16-INK4 |
| 21 | Danio rerio | 42.40 | 3 | PREDICTED: similar to Chain A, Structure Of P18ink |
| 22 | Xiphophorus helleri | 37.40 | 1 | p13CDKN2X |
| 23 | Mus spretus | 35.00 | 1 | cyclin-dependent kinase inhibitor protein |
| 24 | Apis mellifera | 34.70 | 1 | PREDICTED: similar to MYPT-75D CG6896-PA |
| 25 | Tribolium castaneum | 33.50 | 1 | PREDICTED: similar to CG6896-PA |
| 26 | Canis lupus familiaris | 33.10 | 1 | uveal autoantigen with coiled-coil domains and ank |
| 27 | Populus tremula x Populus tremuloides | 32.30 | 1 | potassium channel 2 |
| 28 | Hyperthermus butylicus DSM 5456 | 32.30 | 1 | V-type ATP synthase alpha chain |
Top-ranked sequences
| organism | matching |
|---|---|
| Macaca mulatta | Query 1 MLLEEVRSGDRLSGAAARGDVQEVRRLLH#RELVHPDALNRFGKTALQVMMFGSTAIALE 59 |||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 1 MLLEEVRSGDRLSGAAARGDVQEVRRLLH#RELVHPDALNRFGKTALQVMMFGSTAIALE 59 |
| N/A | Query 1 MLLEEVRSGDRLSGAAARGDVQEVRRLLH#RELVHPDALNRFGKTALQVMMFGSTAIALE 59 |||||||+|||||||||||||||||||||#|||||||||||||||||||||||||||||| Sbjct 1 MLLEEVRAGDRLSGAAARGDVQEVRRLLH#RELVHPDALNRFGKTALQVMMFGSTAIALE 59 |
| Homo sapiens | Query 1 MLLEEVRSGDRLSGAAARGDVQEVRRLLH#RELVHPDALNRFGKTALQVMMFGSTAIALE 59 |||||||+|||||||||||||||||||||#|||||||||||||||||||||||||||||| Sbjct 1 MLLEEVRAGDRLSGAAARGDVQEVRRLLH#RELVHPDALNRFGKTALQVMMFGSTAIALE 59 |
| synthetic construct | Query 1 MLLEEVRSGDRLSGAAARGDVQEVRRLLH#RELVHPDALNRFGKTALQVMMFGSTAIALE 59 |||||||+|||||||||||||||||||||#|||||||||||||||||||||||||||||| Sbjct 1 MLLEEVRAGDRLSGAAARGDVQEVRRLLH#RELVHPDALNRFGKTALQVMMFGSTAIALE 59 |
| Canis familiaris | Query 1 MLLEEVRSGDRLSGAAARGDVQEVRRLLH#RELVHPDALNRFGKTALQVMMFGSTAIALE 59 |||||||+|||||||||||||||||||||#|||||||||||||||||||||||| |||| Sbjct 1 MLLEEVRAGDRLSGAAARGDVQEVRRLLH#RELVHPDALNRFGKTALQVMMFGSPTIALE 59 |
| Rattus norvegicus | Query 1 MLLEEVRSGDRLSGAAARGDVQEVRRLLH#RELVHPDALNRFGKTALQVMMFGSTAIALE 59 |||||| |||||||||||||||||||||#|||||||||||||||||||||||| |+||| Sbjct 1 MLLEEVSVGDRLSGAAARGDVQEVRRLLH#RELVHPDALNRFGKTALQVMMFGSPAVALE 59 |
| Mus musculus | Query 1 MLLEEVRSGDRLSGAAARGDVQEVRRLLH#RELVHPDALNRFGKTALQVMMFGSTAIALE 59 |||||| |||||||||||||||||||||#|||||||||||||||||||||||| |+||| Sbjct 1 MLLEEVCVGDRLSGAAARGDVQEVRRLLH#RELVHPDALNRFGKTALQVMMFGSPAVALE 59 |
| Bos taurus | Query 1 MLLEEVRSGDRLSGAAARGDVQEVRRLLH#RELVHPDALNRFGKTALQVMMFGSTAIALE 59 |||||| +|||||||||||||||||||||# |||||| |||||||||||||||| |||| Sbjct 1 MLLEEVHAGDRLSGAAARGDVQEVRRLLH#SELVHPDVLNRFGKTALQVMMFGSPTIALE 59 |
| Monodelphis domestica | Query 1 MLLEEVRSGDRLSGAAARGDVQEVRRLLH#RELVHPDALNRFGKTALQVMMFGSTAIALE 59 |||||+ +||||||||||||| |||||||#+| ||||+||||||||||||||||+ |||| Sbjct 1 MLLEEISAGDRLSGAAARGDVNEVRRLLH#QEFVHPDSLNRFGKTALQVMMFGSSPIALE 59 |
| Xenopus tropicalis | Query 1 MLLEEVRSGDRLSGAAARGDVQEVRRLLH#RELVHPDALNRFGKTALQVMMFGSTAIALE 59 |||+| +||||+ ||||||+ ||+||||# | +||| ||||||||||||||||| +| | Sbjct 1 MLLQETSAGDRLTRAAARGDLLEVKRLLH#EERIHPDCLNRFGKTALQVMMFGSTPVASE 59 |
| Xenopus laevis | Query 1 MLLEEVRSGDRLSGAAARGDVQEVRRLLH#RELVHPDALNRFGKTALQVMMFGSTAIALE 59 |||+| +||||+ ||||||+ ||+||||# | +||| || |||||||||||||| +| | Sbjct 1 MLLQETSAGDRLTRAAARGDLTEVKRLLH#EERIHPDCLNCFGKTALQVMMFGSTPVASE 59 |
| Xiphophorus maculatus | Query 1 MLLEEVRSGDRLSGAAARGDVQEVRRLLH#RELVHPDALNRFGKTALQVMMFGSTAIA 57 |+| ++ +| |+ |||+|+ ||+|+| # |||| | ||+||||||| |++ +| Sbjct 1 MVLSQMDAGKALTAAAAKGNADEVQRILE#ECRVHPDTPNEFGRTALQVMMMGNSKVA 57 |
| Tetraodon nigroviridis | Query 1 MLLEEVRSGDRLSGAAARGDVQEVRRLLH#RELVHPDALNRFGKTALQVMMFGSTAIA 57 |++ ++ +| |+ |||+| ||||+| # | || | |||||||||| |+ || Sbjct 1 MVIGQMDAGKALAAAAAKGRTAEVRRILE#ECRVPPDTPNEFGKTALQVMMLGNCKIA 57 |
| Takifugu rubripes | Query 1 MLLEEVRSGDRLSGAAARGDVQEVRRLLH#RELVHPDALNRFGKTALQVMMFGSTAIA 57 |++ ++ +| |+ |||+| ||+|+| # | || | |||||||||| |+ || Sbjct 1 MVIGQMDAGKALAAAAAKGRTSEVQRILE#ECRVPPDTRNEFGKTALQVMMLGNCKIA 57 |
| Mesocricetus auratus | Query 12 LSGAAARGDVQEVRRLLH#RELVHPDALNRFGKTALQVMMFGSTAIA 57 |+ ||||| |+ ||+|| # | |+|+||||+ +|||| ||| +| Sbjct 10 LATAAARGQVETVRQLLE#AG-VDPNAVNRFGRRPIQVMMMGSTQVA 54 |
| Macaca fascicularis | Query 8 SGDRLSGAAARGDVQEVRRLLH#RELVHPDALNRFGKTALQVMMFGSTAIA 57 | + |+ ||||| |++||+|| # |+ +||||+ |+|||| || +| Sbjct 14 SDEGLTSAAARGLVEKVRQLLE#AG-ADPNGVNRFGRRAIQVMMMGSARVA 62 |
| Gallus gallus | Query 8 SGDRLSGAAARGDVQEVRRLLH#RELVHPDALNRFGKTALQVMMFGSTAIA 57 + | |+ ||||||+ |+ || # |+|+| ||+| +|||| || +| Sbjct 9 AADELANAAARGDLLRVKELLD#GA-ADPNAVNSFGRTPIQVMMLGSPRVA 57 |
| Sus scrofa | Query 8 SGDRLSGAAARGDVQEVRRLLH#RELVHPDALNRFGKTALQVMMFGSTAIA 57 | | |+ ||||| ||| || # + +| ||+|+| +|||| ||| +| Sbjct 4 SADWLASAAARGREGEVRALLE#AGAL-ANAPNRYGRTPIQVMMMGSTRVA 52 |
| human, brain tumors, Peptide, 156 aa | Query 8 SGDRLSGAAARGDVQEVRRLLH#RELVHPDALNRFGKTALQVMMFGSTAIA 57 | | |+ ||||| |+||| || # + |+| | +|+ +|||| || +| Sbjct 12 SADWLATAAARGRVEEVRALLE#AVAL-PNAPNSYGRRPIQVMMMGSARVA 60 |
| Pan troglodytes | Query 8 SGDRLSGAAARGDVQEVRRLLH#RELVHPDALNRFGKTALQVMMFGSTAIA 57 | | |+ ||||| |+||| || # + |+| | +|+ +|||| || +| Sbjct 213 SADWLATAAARGRVEEVRALLE#AGAL-PNAPNSYGRRPIQVMMMGSARVA 261 |
| Danio rerio | Query 10 DRLSGAAARGDVQEVRRLLH#RELVHPDALNRFGKTALQVMMFGSTAIA 57 |||| ||| ||+ || + | # |+ + |++|+|||||| | +|| Sbjct 8 DRLSTAAAIGDLMEVEQTLQ#SN-VNVNEKNKYGRTALQVMKLGCPSIA 54 |
| Xiphophorus helleri | Query 10 DRLSGAAARGDVQEVRRLLH#RELVHPDALNRFGKTALQVMMFGSTAIA 57 | |+ |||+| || || #+ + +| ||+ |+|||| ||+ +| Sbjct 5 DELTTAAAKGHTAEVEALLL#QG-APVNGVNSFGRRAIQVMMMGSSEVA 51 |
| Mus spretus | Query 8 SGDRLSGAAARGDVQEVRRLLH#RELVHPDALNRFGKTALQ 47 + |||+ |||+| | +|| || # | | | | ||+| +| Sbjct 4 AADRLARAAAQGRVHDVRALLE#AG-VSPKAPNSFGRTPIQ 42 |
| Apis mellifera | Query 15 AAARGDVQEVRRLLH#RELVHPDALNRFGKTAL 46 |||| |+ |||||| #++ |+||+ | | ||| Sbjct 68 AAARNDIDEVRRLL-#KKGVNPDSTNEDGLTAL 98 |
| Tribolium castaneum | Query 15 AAARGDVQEVRRLLH#RELVHPDALNRFGKTAL 46 |||| |+ |||||| #+ | ||+ | | ||| Sbjct 69 AAARNDIDEVRRLLM#KG-VSPDSCNEDGLTAL 99 |
| Canis lupus familiaris | Query 10 DRLSGAAARGDVQEVRRLLH#RELVHPDALNRFGKTALQVM 49 ||| || ||||++| +| #++ ++| |+ |++| |+ Sbjct 38 DRLMKAAERGDVEKVSSILA#KKGINPGKLDVEGRSAFHVV 77 |
| Populus tremula x Populus tremuloides | Query 8 SGDRLSGAAARGDVQEVRRLLH#RELVHPDALNRFGKTALQVMM 50 +|| | || + |+ ++ || #+ | + |+ +| ||||||| | Sbjct 623 AGDLLCTAAKQNDLMVMKELLK#QGL-NVDSKDRHGKTALQVAM 664 |
| Hyperthermus butylicus DSM 5456 | Query 6 VRSGDRLSGAAARGDVQEVRRLLH#RELVHPDALNR 40 ++ ||++|| |+||| + |#+ || || | Sbjct 127 LKPGDKVSGGDVLGEVQETSLITH#KVLVPPDVHGR 161 |
[Site 4] AIALELLKQG64-ASPNVQDTSG
Gly64  Ala
Ala
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Ala55 | Ile56 | Ala57 | Leu58 | Glu59 | Leu60 | Leu61 | Lys62 | Gln63 | Gly64 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Ala65 | Ser66 | Pro67 | Asn68 | Val69 | Gln70 | Asp71 | Thr72 | Ser73 | Gly74 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| PDALNRFGKTALQVMMFGSTAIALELLKQGASPNVQDTSGTSPVHDAARTGFLDTLKVLV |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | Homo sapiens | 119.00 | 29 | cyclin-dependent kinase inhibitor 2D |
| 2 | N/A | 119.00 | 21 | - |
| 3 | synthetic construct | 119.00 | 10 | cyclin-dependent kinase inhibitor 2D (p19, inhibit |
| 4 | Macaca mulatta | 119.00 | 7 | PREDICTED: cyclin-dependent kinase inhibitor 2D (p |
| 5 | Mus musculus | 114.00 | 38 | cyclin-dependent kinase inhibitor 2D (p19, inhibit |
| 6 | Rattus norvegicus | 114.00 | 8 | similar to cyclin-dependent kinase inhibitor 2D |
| 7 | Monodelphis domestica | 109.00 | 12 | PREDICTED: similar to Cyclin-dependent kinase inhi |
| 8 | Bos taurus | 109.00 | 8 | cyclin-dependent kinase inhibitor 2D (p19, inhibit |
| 9 | Canis familiaris | 109.00 | 7 | PREDICTED: similar to Cyclin-dependent kinase 4 in |
| 10 | Xenopus tropicalis | 102.00 | 4 | cyclin-dependent kinase inhibitor 2D (p19, inhibit |
| 11 | Xenopus laevis | 97.80 | 6 | ankyrin 2, neuronal |
| 12 | Takifugu rubripes | 86.70 | 2 | p19 |
| 13 | Tetraodon nigroviridis | 86.30 | 6 | unnamed protein product |
| 14 | Xiphophorus maculatus | 82.40 | 2 | p19 |
| 15 | Danio rerio | 71.20 | 2 | PREDICTED: similar to p13 |
| 16 | Xiphophorus helleri | 71.20 | 1 | p13CDKN2X |
| 17 | Mesocricetus auratus | 69.70 | 3 | p15INK4b cyclin-dependent kinase inhibitor |
| 18 | Gallus gallus | 68.20 | 7 | PREDICTED: similar to Chain A, Crystal Structure O |
| 19 | Macaca fascicularis | 68.20 | 1 | unnamed protein product |
| 20 | Sus scrofa | 67.80 | 4 | cyclin dependant kinase inhibitor |
| 21 | Pan troglodytes | 62.80 | 9 | PREDICTED: similar to p16-INK4 |
| 22 | human, brain tumors, Peptide, 156 aa | 62.80 | 1 | cyclin D-dependent kinases 4 and 6-binding protein |
| 23 | Equus caballus | 53.90 | 1 | cyclin-dependent kinase 2A inhibitor |
| 24 | Felis catus | 51.20 | 2 | p16/CDKN2A/MTS1 |
| 25 | Medicago truncatula | 49.70 | 1 | Ankyrin |
| 26 | Mus spretus | 49.30 | 1 | cyclin-dependent kinase inhibitor protein |
| 27 | Strongylocentrotus purpuratus | 48.50 | 7 | PREDICTED: similar to MGC80441 protein, partial |
| 28 | Arabidopsis thaliana | 45.40 | 1 | EMB506 (EMBRYO DEFECTIVE 506); protein binding |
| 29 | Vitis vinifera | 45.40 | 1 | hypothetical protein |
| 30 | Oryza sativa (japonica cultivar-group) | 43.90 | 3 | hypothetical protein OsJ_019894 |
| 31 | Ustilago maydis 521 | 43.90 | 1 | hypothetical protein UM02100.1 |
| 32 | Rattus sp. | 43.90 | 1 | AAP31998.1 |
| 33 | Oryza sativa (indica cultivar-group) | 43.90 | 1 | hypothetical protein OsI_021528 |
| 34 | Theileria annulata strain Ankara | 43.50 | 1 | hypothetical protein TA20480 |
| 35 | Aedes aegypti | 43.10 | 4 | ankyrin repeat domain |
| 36 | Ashbya gossypii ATCC 10895 | 43.10 | 1 | ADL244Wp |
| 37 | Drosophila melanogaster | 43.10 | 1 | CG3104-PA, isoform A |
| 38 | Tribolium castaneum | 42.70 | 1 | PREDICTED: similar to CG18247-PA |
| 39 | Dictyostelium discoideum | 42.70 | 1 | homeobox-containing protein Wariai |
| 40 | Dictyostelium discoideum AX4 | 42.70 | 1 | putative homeobox transcription factor |
| 41 | Treponema denticola ATCC 35405 | 42.70 | 1 | ankyrin repeat protein |
| 42 | Neosartorya fischeri NRRL 181 | 42.70 | 1 | Ankyrin repeat protein |
| 43 | Aspergillus fumigatus Af293 | 42.40 | 1 | ankyrin repeat protein |
| 44 | Anopheles gambiae str. PEST | 42.00 | 4 | ENSANGP00000004013 |
| 45 | Trichomonas vaginalis G3 | 42.00 | 3 | hypothetical protein TVAG_580310 |
| 46 | Samanea saman | 42.00 | 1 | pulvinus inward-rectifying channel for potassium S |
| 47 | Myxococcus xanthus DK 1622 | 42.00 | 1 | ankyrin repeat protein |
| 48 | Legionella pneumophila subsp. pneumophila str. Philadelphia 1 | 41.60 | 1 | putative ankyrin-repeat containing protein |
| 49 | Aspergillus oryzae | 41.20 | 1 | unnamed protein product |
| 50 | Apis mellifera | 37.40 | 1 | PREDICTED: similar to no mechanoreceptor potential |
| 51 | Hemicentrotus pulcherrimus | 35.80 | 1 | inv-like protein |
| 52 | Rickettsiella grylli | 35.80 | 1 | hypothetical protein Rgryl_01000336 |
| 53 | Wolbachia endosymbiont of Drosophila melanogaster | 35.40 | 1 | ankyrin repeat domain protein |
| 54 | Caenorhabditis briggsae | 35.00 | 2 | Hypothetical protein CBG13617 |
| 55 | Wolbachia pipientis | 34.70 | 1 | ankyrin domain protein |
| 56 | Wolbachia endosymbiont of Drosophila willistoni TSC#14030-0811.24 | 34.70 | 1 | hypothetical protein Wendoof_01000724 |
| 57 | Caenorhabditis elegans | 33.10 | 8 | TRP (transient receptor potential) channel family |
| 58 | Thermofilum pendens Hrk 5 | 32.70 | 1 | Ankyrin |
Top-ranked sequences
| organism | matching |
|---|---|
| Homo sapiens | Query 35 PDALNRFGKTALQVMMFGSTAIALELLKQG#ASPNVQDTSGTSPVHDAARTGFLDTLKVLV 94 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 35 PDALNRFGKTALQVMMFGSTAIALELLKQG#ASPNVQDTSGTSPVHDAARTGFLDTLKVLV 94 |
| N/A | Query 35 PDALNRFGKTALQVMMFGSTAIALELLKQG#ASPNVQDTSGTSPVHDAARTGFLDTLKVLV 94 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 35 PDALNRFGKTALQVMMFGSTAIALELLKQG#ASPNVQDTSGTSPVHDAARTGFLDTLKVLV 94 |
| synthetic construct | Query 35 PDALNRFGKTALQVMMFGSTAIALELLKQG#ASPNVQDTSGTSPVHDAARTGFLDTLKVLV 94 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 35 PDALNRFGKTALQVMMFGSTAIALELLKQG#ASPNVQDTSGTSPVHDAARTGFLDTLKVLV 94 |
| Macaca mulatta | Query 35 PDALNRFGKTALQVMMFGSTAIALELLKQG#ASPNVQDTSGTSPVHDAARTGFLDTLKVLV 94 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 35 PDALNRFGKTALQVMMFGSTAIALELLKQG#ASPNVQDTSGTSPVHDAARTGFLDTLKVLV 94 |
| Mus musculus | Query 35 PDALNRFGKTALQVMMFGSTAIALELLKQG#ASPNVQDTSGTSPVHDAARTGFLDTLKVLV 94 ||||||||||||||||||| |+||||||||#||||||| |||||||||||||||||||||| Sbjct 35 PDALNRFGKTALQVMMFGSPAVALELLKQG#ASPNVQDASGTSPVHDAARTGFLDTLKVLV 94 |
| Rattus norvegicus | Query 35 PDALNRFGKTALQVMMFGSTAIALELLKQG#ASPNVQDTSGTSPVHDAARTGFLDTLKVLV 94 ||||||||||||||||||| |+||||||||#||||||| |||||||||||||||||||||| Sbjct 35 PDALNRFGKTALQVMMFGSPAVALELLKQG#ASPNVQDASGTSPVHDAARTGFLDTLKVLV 94 |
| Monodelphis domestica | Query 35 PDALNRFGKTALQVMMFGSTAIALELLKQG#ASPNVQDTSGTSPVHDAARTGFLDTLKVLV 94 ||+||||||||||||||||+ |||||||||#||||||| ||||| ||||||||||||+||| Sbjct 35 PDSLNRFGKTALQVMMFGSSPIALELLKQG#ASPNVQDGSGTSPAHDAARTGFLDTLRVLV 94 |
| Bos taurus | Query 35 PDALNRFGKTALQVMMFGSTAIALELLKQG#ASPNVQDTSGTSPVHDAARTGFLDTLKVLV 94 || |||||||||||||||| |||||||||#||||||| |||+| |||||||||||||||| Sbjct 35 PDVLNRFGKTALQVMMFGSPTIALELLKQG#ASPNVQDASGTTPAHDAARTGFLDTLKVLV 94 |
| Canis familiaris | Query 35 PDALNRFGKTALQVMMFGSTAIALELLKQG#ASPNVQDTSGTSPVHDAARTGFLDTLKVLV 94 ||||||||||||||||||| |||||||||#||||||| +||+| |||||||||||||||| Sbjct 35 PDALNRFGKTALQVMMFGSPTIALELLKQG#ASPNVQDATGTTPAHDAARTGFLDTLKVLV 94 |
| Xenopus tropicalis | Query 35 PDALNRFGKTALQVMMFGSTAIALELLKQG#ASPNVQDTSGTSPVHDAARTGFLDTLKVLV 94 || ||||||||||||||||| +| ||||||#||||+|| ||+| ||||| ||||||+||| Sbjct 35 PDCLNRFGKTALQVMMFGSTPVASELLKQG#ASPNIQDAYGTTPAHDAARCGFLDTLQVLV 94 |
| Xenopus laevis | Query 35 PDALNRFGKTALQVMMFGSTAIALELLKQG#ASPNVQDTSGTSPVHDAARTGFLDTLKVLV 94 || || |||||||||||||| +| ||||||#|+||+|| ||+| ||||| ||||||+||| Sbjct 35 PDCLNCFGKTALQVMMFGSTPVASELLKQG#ATPNIQDAYGTTPAHDAARCGFLDTLQVLV 94 |
| Takifugu rubripes | Query 35 PDALNRFGKTALQVMMFGSTAIALELLKQG#ASPNVQDTSGTSPVHDAARTGFLDTLKVLV 94 || | |||||||||| |+ || ||++|#| ||||| | +||||||||||||||+||| Sbjct 35 PDTRNEFGKTALQVMMLGNCKIASLLLEKG#ADPNVQDKHGIAPVHDAARTGFLDTLQVLV 94 |
| Tetraodon nigroviridis | Query 35 PDALNRFGKTALQVMMFGSTAIALELLKQG#ASPNVQDTSGTSPVHDAARTGFLDTLKVLV 94 || | |||||||||| |+ || ||++|#| ||||| | +||||||||||||||+||| Sbjct 35 PDTPNEFGKTALQVMMLGNCKIASLLLEKG#ADPNVQDKHGIAPVHDAARTGFLDTLQVLV 94 |
| Xiphophorus maculatus | Query 35 PDALNRFGKTALQVMMFGSTAIALELLKQG#ASPNVQDTSGTSPVHDAARTGFLDTLKVLV 94 || | ||+||||||| |++ +| ||++|#| ||||| | +||||||+||||+||+||| Sbjct 35 PDTPNEFGRTALQVMMMGNSKVARLLLEKG#AEPNVQDKHGIAPVHDAAQTGFLETLQVLV 94 |
| Danio rerio | Query 36 DALNRFGKTALQVMMFGSTAIALELLKQG#ASPNVQD-TSGTSPVHDAARTGFLDTLKVLV 94 + +|+| +| +|||| |+ +|| ||+||#| ||| | +|++|+||||||||+||+++|+ Sbjct 31 NVVNKFRRTPIQVMMMGNAPLALVLLEQG#ADPNVPDPDTGSTPLHDAARTGFIDTVRLLI 90 |
| Xiphophorus helleri | Query 36 DALNRFGKTALQVMMFGSTAIALELLKQG#ASPNVQDTS-GTSPVHDAARTGFLDTLKVLV 94 + +| ||+ |+|||| ||+ +| || +|#| ||| | | | +|+|||||||||||+++|| Sbjct 30 NGVNSFGRRAIQVMMMGSSEVARLLLTRG#ADPNVTDKSTGATPLHDAARTGFLDTVQLLV 89 |
| Mesocricetus auratus | Query 35 PDALNRFGKTALQVMMFGSTAIALELLKQG#ASPNVQDTSG-TSPVHDAARTGFLDTLKVL 93 |+|+||||+ +|||| ||| +| || |#| || | + | ||||||| |||||| || Sbjct 32 PNAVNRFGRRPIQVMMMGSTQVAELLLLHG#AEPNCADPNTLTRPVHDAAREGFLDTLVVL 91 |
| Gallus gallus | Query 36 DALNRFGKTALQVMMFGSTAIALELLKQG#ASPNVQDTSGTSPVHDAARTGFLDTLKVLV 94 +| | ||+|||||| |+ || || |#|+||++|++| + +|| || ||||||+ |+ Sbjct 32 NAQNGFGRTALQVMKLGNPEIARRLLMSG#ANPNLKDSTGFAVIHDVARAGFLDTLQTLL 90 |
| Macaca fascicularis | Query 35 PDALNRFGKTALQVMMFGSTAIALELLKQG#ASPNVQDTSG-TSPVHDAARTGFLDTLKVL 93 |+ +||||+ |+|||| || +| || |#| || | + | ||||||| |||||| || Sbjct 40 PNGVNRFGRRAIQVMMMGSARVAELLLLHG#AEPNCADPATLTRPVHDAAREGFLDTLVVL 99 |
| Sus scrofa | Query 36 DALNRFGKTALQVMMFGSTAIALELLKQG#ASPNVQDTSG-TSPVHDAARTGFLDTLKVL 93 +| ||+|+| +|||| ||| +| || |#| || +| + | ||||||| |||||| || Sbjct 31 NAPNRYGRTPIQVMMMGSTRVAELLLLHG#ADPNCEDPATLTRPVHDAAREGFLDTLVVL 89 |
| Pan troglodytes | Query 35 PDALNRFGKTALQVMMFGSTAIALELLKQG#ASPNVQDTSG-TSPVHDAARTGFLDTLKVL 93 |+| | +|+ +|||| || +| || |#| || | + | ||||||| |||||| || Sbjct 239 PNAPNSYGRRPIQVMMMGSARVAELLLLHG#AEPNCADPATLTRPVHDAAREGFLDTLVVL 298 |
| human, brain tumors, Peptide, 156 aa | Query 35 PDALNRFGKTALQVMMFGSTAIALELLKQG#ASPNVQDTSG-TSPVHDAARTGFLDTLKVL 93 |+| | +|+ +|||| || +| || |#| || | + | ||||||| |||||| || Sbjct 38 PNAPNSYGRRPIQVMMMGSARVAELLLLHG#AEPNCADPATLTRPVHDAAREGFLDTLVVL 97 |
| Equus caballus | Query 35 PDALNRFGKTALQVMMFGSTAIALELLKQG#ASPNVQDTSG-TSPVHDAARTGFL 87 |+ +| ||+ +|||| || +| || |#| || | | ||||||| ||| Sbjct 5 PNGVNGFGRRPIQVMMMGSVHVAELLLLHG#ADPNRADPDTLTRPVHDAAREGFL 58 |
| Felis catus | Query 48 VMMFGSTAIALELLKQG#ASPNVQDTSG-TSPVHDAARTGFLDTLKVL 93 ||| || +| || |#| || | + | ||||||| |||||| || Sbjct 1 VMMMGSARVAELLLLHG#ADPNCADPATLTRPVHDAAREGFLDTLVVL 47 |
| Medicago truncatula | Query 36 DALNRFGKTALQVMMFGST-AIALELLKQG#ASPNVQDTSGTSPVHDAARTGFLDTLKVLV 94 |++|+ | ||| + | |+ ||++|#|||++|| | +|+| | | |+|+|+ Sbjct 183 DSINKEGLTALHKAVIGKKEAVISHLLRKG#ASPHIQDKDGATPLHYAVEVGAKQTVKLLI 242 |
| Mus spretus | Query 48 VMMFGSTAIALELLKQG#ASPNVQD-TSGTSPVHDAARTGFLDTLKVL 93 ||| |+ +| || |#| | +| |+ + ||||||| |||||| || Sbjct 1 VMMMGNVHVAALLLNYG#ADSNCEDPTTFSRPVHDAAREGFLDTLVVL 47 |
| Strongylocentrotus purpuratus | Query 39 NRFGKTALQVMMF-GSTAIALELLKQG#ASPNVQDTSGTSPVHDAARTGFLDTLKVLV 94 | |+||| + | + +|| |+ ||#| |||| +| +|+|+| |+ + |||+ Sbjct 87 NERGETALHMAAIKGDSQMALNLINQG#AEVNVQDFAGWTPLHEACNHGYYEVAKVLI 143 |
| Arabidopsis thaliana | Query 36 DALNRFGKTALQVMMFGST-AIALELLKQG#ASPNVQDTSGTSPVHDAARTGFLDTLKVL 93 | +++ +||| + | |+ ||++|#|+|++|| | +|+| | + | | |+|+| Sbjct 179 DDVDKDNQTALHKAIIGKKEAVISHLLRKG#ANPHLQDRDGAAPIHYAVQVGALQTVKLL 237 |
| Vitis vinifera | Query 36 DALNRFGKTALQVMMFGST-AIALELLKQG#ASPNVQDTSGTSPVHDAARTGFLDTLKVLV 94 | +++ | ||| + | |+ ||++|#|+|+|+| | +|+| | + | + |+|+|+ Sbjct 185 DNVDKDGLTALHRAIIGKKEAVISHLLRKG#ANPHVRDRDGATPLHYAVQVGAMQTVKLLI 244 |
| Oryza sativa (japonica cultivar-group) | Query 38 LNRFGKTALQVMMFGST-AIALELLKQG#ASPNVQDTSGTSPVHDAARTGFLDTLKVLV 94 |++ | | | + | |+ ||++|#|+|+|+| | +|+| | + | | |+|+|+ Sbjct 165 LDKDGFTPLHKAVIGKKEAVISHLLRRG#ANPHVRDRDGATPLHYAVQVGALQTVKLLI 222 |
| Ustilago maydis 521 | Query 39 NRFGKTAL-QVMMFGSTAIALELLKQG#ASPNVQDTSGTSPVHDAARTGFLDTLKVLV 94 +| |+||| + | | ++| || |#| | || | + +|+| |+|| +++|| Sbjct 398 DREGETALHKAAMAGKLSVASLLLSHG#ADANAQDADGWTALHNACSRGYLDLVRLLV 454 |
| Rattus sp. | Query 49 MMFGSTAIALELLKQG#ASPNVQDTSG-TSPVHDAARTGFLD 88 || || +| || |#| || | + | ||||||| |||| Sbjct 1 MMMGSAQVAELLLLHG#AEPNCADPATLTRPVHDAAREGFLD 41 |
| Oryza sativa (indica cultivar-group) | Query 38 LNRFGKTALQVMMFGST-AIALELLKQG#ASPNVQDTSGTSPVHDAARTGFLDTLKVLV 94 |++ | | | + | |+ ||++|#|+|+|+| | +|+| | + | | |+|+|+ Sbjct 165 LDKDGFTPLHKAVIGKKEAVISHLLRRG#ANPHVRDRDGATPLHYAVQVGALQTVKLLI 222 |
| Theileria annulata strain Ankara | Query 52 GSTAIALELLKQG#ASPNVQDTSGTSPVHDAARTGFLDTLKVLV 94 |+| + +||+ |#| ||+ |+ | +|+| |+ +|+| |+|+ Sbjct 253 GNTGLVKKLLENG#ADPNICDSQGWTPLHCASESGYLSVCKLLI 295 |
| Aedes aegypti | Query 52 GSTAIALELLKQG#ASPNVQDTSGTSPVHDAARTGFLDTLKVLV 94 | || +|||+||#| || + +||+ + ||+ ||+| ++|+ Sbjct 22 GHTACVIELLEQG#ADPNARRVTGTTALFFAAQGGFVDVARILL 64 |
| Ashbya gossypii ATCC 10895 | Query 42 GKTALQVMMF-GSTAIALELLKQG#ASPNVQDTSGTSPVHDAARTGFLDTLKVLV 94 |+| ||| | +| +|+++|# | || +| ||+|+|| | |+ +|+|+ Sbjct 341 GRTRLQVACDKGKYDLARKLIEEG#YDVNDQDNAGNSPLHEAALNGHLEVVKLLI 394 |
| Drosophila melanogaster | Query 52 GSTAIALELLKQG#ASPNVQDTSGTSPVHDAARTGFLDTLKVLV 94 | | +||| ||#| || + +||+|+ ||+ | || +|+|+ Sbjct 54 GHTYCVMELLDQG#ADPNSRRLTGTTPLFFAAQGGHLDVVKILI 96 |
| Tribolium castaneum | Query 36 DALNRFGKTALQVM-MFGSTAIALELLKQG#ASPNVQDTSGTSPVHDAARTGFLDTLKVLV 94 +| |+ |+||+ + | | | +|+ |#|| |++||+| +|+| | + | |+++|| Sbjct 145 EAKNQNGQTAVHLASMNGKDDILRKLIDYG#ASVNLRDTAGYTPLHYACQNNFPSTVRLLV 204 |
| Dictyostelium discoideum | Query 36 DALNRFGKTALQVM-MFGSTAIALELLKQG#ASPNVQDTSGTSPVHDAARTGFLDTLKVLV 94 +|++ | | | + | | ||+ |#| ||+||+ | +|+| | | ++|+| |+ Sbjct 532 NAIDIDGHTPLHTSSLMGQYLITRLLLENG#ADPNIQDSEGYTPIHYAVRESRIETVKFLI 591 |
| Dictyostelium discoideum AX4 | Query 36 DALNRFGKTALQVM-MFGSTAIALELLKQG#ASPNVQDTSGTSPVHDAARTGFLDTLKVLV 94 +|++ | | | + | | ||+ |#| ||+||+ | +|+| | | ++|+| |+ Sbjct 535 NAIDIDGHTPLHTSSLMGHDLITRLLLENG#ADPNIQDSEGYTPIHYAVRESRIETVKFLI 594 |
| Treponema denticola ATCC 35405 | Query 35 PDALNRFGKTALQVMMFGSTAIAL--ELLKQG#ASPNVQDTSGTSPVHDAARTGFLDTL 90 |++ | | ||| ++| ++ | ||+ |#|+||++| | +|+| ||| | | + Sbjct 307 PNSRNSSGNTALHLVMPEASRSKLFNELITAG#ANPNLKDNYGETPLHVAARIGMNDDI 364 |
| Neosartorya fischeri NRRL 181 | Query 51 FGSTAIALELLKQG#ASPNVQDTSGTSPVHDAARTGFLDTLKVLV 94 || | || |#| ||| | ||++ ||+ | | ++|||| Sbjct 303 FGHPDAVLALLHHG#ADPNVPSVDGQSPIYSAAKLGQLSSVKVLV 346 |
| Aspergillus fumigatus Af293 | Query 51 FGSTAIALELLKQG#ASPNVQDTSGTSPVHDAARTGFLDTLKVLV 94 || | || |#| ||| | ||++ ||+ | | ++|||| Sbjct 302 FGHPDAVLALLHHG#ADPNVPSVDGQSPIYSAAKLGQLGSVKVLV 345 |
| Anopheles gambiae str. PEST | Query 52 GSTAIALELLKQG#ASPNVQDTSGTSPVHDAARTGFLDTLKVLV 94 | || +|||+||#| || + +||+ + ||+ |++| ++|+ Sbjct 13 GHTACVIELLEQG#ADPNARRVTGTTALFFAAQGGYVDVARILL 55 |
| Trichomonas vaginalis G3 | Query 36 DALNRFGKTALQVMMF-GSTAIALELLKQG#ASPNVQDTSGTSPVHDAARTGFLDTLKVLV 94 ++ + ||| | + | ||| |||||#| || +| | +|+ ||+ | | + +|| Sbjct 474 NSADNMGKTCLLLAAASGHTAIVDFLLKQG#AEPNAKDREGFTPLLAAAQNGRTDCVSLLV 533 |
| Samanea saman | Query 46 LQVMMFGSTAIALELLKQG#ASPNVQDTSGTSPVHDAARTGFLDTLKVLV 94 | | |+ | |||| |# |+| |+ | +|+| || | | +|||+ Sbjct 548 LTVASTGNAAFLEELLKAG#LDPDVGDSKGKTPLHIAASNGHEDCVKVLL 596 |
| Myxococcus xanthus DK 1622 | Query 36 DALNRFGKTALQ-VMMFGSTAIALELLKQG#ASPNVQDTSGTSPVHDAART 84 | ||||+||| +|| + +| +||#|||+|+| || | | ||| Sbjct 136 DQSNRFGQTALMFASLFGRKEVVEQLRQQG#ASPDVRDGSGRS-AQDWART 184 |
| Legionella pneumophila subsp. pneumophila str. Philadelphia 1 | Query 38 LNRFGKTALQ------VMMFGSTAIALELLKQG#ASPNVQDTSGTSPVHDAARTGFLDTLK 91 ||+ || | +| | | |+| |#|| ++|| || ||+| | + | +|+| Sbjct 462 LNKKDKTGNQYTALHWAVMQGHLDCARLLVKSG#ASVDIQDKSGKSPLHYAIKRGDKETIK 521 Query 92 VLV 94 +|| Sbjct 522 LLV 524 |
| Aspergillus oryzae | Query 35 PDALNRFGKTALQV-MMFGSTAIALELLKQG#ASPNVQDTSGTSPVHDAARTG 85 |+|+| | ||| | +| |+ + |||++|#| | + + +| |+| | | Sbjct 179 PNAVNSEGATALHVGVMNGNYTMVAELLQRG#ADPTLTNAAGWLPLHQAVHAG 230 |
| Apis mellifera | Query 39 NRFGKTALQVMM-FGSTAIALELLKQG#ASPNVQDTSGTSPVHDAARTGFLDTLKVLV 94 +|+||| | + | + || ||#| | | +| +|+| ||| |+|| +|+|| Sbjct 1025 DRYGKTGLHIAATHGHYQMVEVLLGQG#AEINATDKNGWTPLHCAARAGYLDVVKLLV 1081 Query 57 ALELLKQG#ASPNVQDTSGTSPVHDAARTGFLDTLKVLV 94 || ||| |#| ||+ | +||| || | | || +|+ Sbjct 469 ALMLLKSG#AGPNLTTDDGQTPVHVAASHGNLATLLLLL 506 |
| Hemicentrotus pulcherrimus | Query 35 PDALNRFGKTALQVMMFGSTAIALELL-KQG#ASPNVQDTSGTSPVHDAARTGFLDTLKVL 93 |+ + |+| || +| + || + |#| ||+|| | + +| | ||+|| ++| Sbjct 499 PNVQDNAGRTPLQCAAYGGFIRCMTLLLEHG#ADPNLQDNEGMTALHWACSTGYLDATRLL 558 Query 94 V 94 + Sbjct 559 L 559 Query 52 GSTAIALELLKQG#ASPNVQDTSGTSPVHDAARTGFLDTLKVLV 94 | | + |+ # |||||| +| +|+ || ||+ + +|+ Sbjct 484 GHTCVCYHLMTHD#ISPNVQDNAGRTPLQCAAYGGFIRCMTLLL 526 |
| Rickettsiella grylli | Query 39 NRFGKTALQVMM-FGSTAIALELLKQG#ASPNVQDTSGTSPVHDAARTGFLDTLKVLV 94 +|+|+| | | + || | + + |+++|#| | | | +|+| | | +++++++|+ Sbjct 652 DRYGRTPLHVAIWFGYTELVIYLVERG#ADVNSTDQLGNTPLHTAGITNYVNSIQILL 708 Query 36 DALNRFGKTAL-QVMMFGSTAIALELLKQG#ASPNVQDTSGTSPVHDAARTGFLDTLKVLV 94 +| | | | | + | | |+ + #|+ + ||++|+|+||| | || +| |+ Sbjct 458 EANNDSGSTPLHEAARNGHLDIVKYLIGKN#ATIEANNDSGSTPLHEAARNGHLDIVKYLI 517 Query 39 NRFGKTALQVMMF-GSTAIALELLKQG#ASPNVQDTSGTSPVHDAARTGFLDTLKVLV 94 |+ | | | | | + |+ + #|+ + ||++|+|+||| | || +| |+ Sbjct 428 NKMGVTPLYVASRNGHLDMVKYLIGKN#ATIEANNDSGSTPLHEAARNGHLDIVKYLI 484 |
| Wolbachia endosymbiont of Drosophila melanogaster | Query 36 DALNRFGKTALQ-VMMFGSTAIALELLKQG#ASPNVQDTSGTSPVHDAARTGFLDTLKVLV 94 | ||+|+| | +| | + || + # + ||| | +|+|||| | ++ +| |+ Sbjct 140 DLKNRYGETPLHYAAKYGHTQVLENLLGRS#TNVNVQSEVGRTPLHDAANNGHIEVVKHLI 199 Query 52 GSTAIALELLKQG#ASPNVQDTSGTSPVHDAARTGFLDTLKVLV 94 | + |+|+|#| ||| | +|+|+||+ | ++||+ Sbjct 190 GHIEVVKHLIKKG#ADVNVQSKVGRTPLHNAAKHGHTQVVEVLL 232 Query 39 NRFGKTALQ-VMMFGSTAIALELLKQG#ASPNVQDTSGTSPVHDAARTGFLDTLKVLV 94 ++ |+| | | | + |||+|#| |+|| | +|+| | + + |+|+ Sbjct 209 SKVGRTPLHNAAKHGHTQVVEVLLKKG#ADVNIQDRGGRTPLHYAVQRRYPKLAKLLL 265 |
| Caenorhabditis briggsae | Query 36 DALNRFGKTALQ-VMMFGSTAIALELLKQG#ASPNVQDTSGTSPVHDAARTGFLDTLKVLV 94 +|+ +||| || |++ || |#|+|| +| | +|+| || | | +|+ + Sbjct 951 EAITLDNQTALHFAAKFGQLAVSQTLLALG#ANPNARDDKGQTPLHLAAENDFPDVVKLFL 1010 Query 37 ALNRFGKTALQVMMF-GSTAIALELLKQG#ASPNVQDTSGTSPVHDAARTGFLDTLKVLV 94 | | | | | + | |+ || ||+|# + + | +| || || | +|+|+ Sbjct 557 ARTRDGSTLLHIAACSGHTSTALAFLKRG#VPLMMPNKKGALGLHSAAAAGFNDVVKMLI 615 Query 36 DALNRFGKTALQVMMF-GSTAIALELLKQG#ASPNVQDTSGTSPVHDAARTGFLDTLKVLV 94 | + |+||| + | | +| ||+ #| | + +| +|+| ||+ | + + ||| Sbjct 884 DVFDEMGRTALHLAAFNGHLSIVHLLLQHK#AFVNSKSKTGEAPLHLAAQHGHVKVVNVLV 943 |
| Wolbachia pipientis | Query 36 DALNRFGKTALQ-VMMFGSTAIALELLKQG#ASPNVQDTSGTSPVHDAARTGFLDTLKVLV 94 | ||+|+| | +| | + || + # + ||| | +|+|||| | ++ +| |+ Sbjct 139 DLKNRYGETPLHYAAKYGHTQVLENLLGRS#TNVNVQSEVGRTPLHDAANNGHIEVVKHLI 198 Query 52 GSTAIALELLKQG#ASPNVQDTSGTSPVHDAARTGFLDTLKVLV 94 | + |+|+|#| ||| | +|+|+|| |+++ +| |+ Sbjct 189 GHIEVVKHLIKKG#ADVNVQSKVGRTPLHNAANNGYIEVVKHLI 231 Query 39 NRFGKTALQ-VMMFGSTAIALELLKQG#ASPNVQDTSGTSPVHDAARTGFLDTLKVLV 94 ++ |+| | | | + |||+|#| |+|| | +|+| | + |+ |+|+ Sbjct 274 SKVGRTPLHNAAKHGHTQVVEVLLKKG#ADVNIQDRGGRTPLHYAVQRGYPKLAKLLL 330 Query 52 GSTAIALELLKQG#ASPNVQDTSGTSPVHDAARTGFLDTLKVLV 94 | + |+|+ #| || | | +|+||||+ | ++ +| |+ Sbjct 222 GYIEVVKHLIKKE#ADVNVVDQYGRTPLHDAAKHGRIEVVKHLI 264 Query 36 DALNRFGKTALQ-VMMFGSTAIALELLKQG#ASPNVQDTSGTSPVHDAARTGFLDTLKVLV 94 + ++++|+| | | + |+++ #| ||| | +|+|+||+ | ++||+ Sbjct 238 NVVDQYGRTPLHDAAKHGRIEVVKHLIEKE#ADVNVQSKVGRTPLHNAAKHGHTQVVEVLL 297 |
| Wolbachia endosymbiont of Drosophila willistoni TSC#14030-0811.24 | Query 36 DALNRFGKTALQ-VMMFGSTAIALELLKQG#ASPNVQDTSGTSPVHDAARTGFLDTLKVLV 94 | ||+|+| | +| | + || + # + ||| | +|+|||| | ++ +| |+ Sbjct 140 DLKNRYGETPLHYAAKYGHTQVLENLLGRS#TNVNVQSEVGRTPLHDAANNGHIEVVKHLI 199 Query 52 GSTAIALELLKQG#ASPNVQDTSGTSPVHDAARTGFLDTLKVLV 94 | + |+|+|#| ||| | +|+|+|| |+++ +| |+ Sbjct 190 GHIEVVKHLIKKG#ADVNVQSKVGRTPLHNAANNGYIEVVKHLI 232 Query 39 NRFGKTALQ-VMMFGSTAIALELLKQG#ASPNVQDTSGTSPVHDAARTGFLDTLKVLV 94 ++ |+| | | | + |||+|#| |+|| | +|+| | + |+ |+|+ Sbjct 275 SKVGRTPLHNAAKHGHTQVVEVLLKKG#ADVNIQDRGGRTPLHYAVQRGYPKLAKLLL 331 Query 52 GSTAIALELLKQG#ASPNVQDTSGTSPVHDAARTGFLDTLKVLV 94 | + |+|+ #| || | | +|+||||+ | ++ +| |+ Sbjct 223 GYIEVVKHLIKKE#ADVNVVDQYGRTPLHDAAKHGRIEVVKHLI 265 Query 36 DALNRFGKTALQ-VMMFGSTAIALELLKQG#ASPNVQDTSGTSPVHDAARTGFLDTLKVLV 94 + ++++|+| | | + |+++ #| ||| | +|+|+||+ | ++||+ Sbjct 239 NVVDQYGRTPLHDAAKHGRIEVVKHLIEKE#ADVNVQSKVGRTPLHNAAKHGHTQVVEVLL 298 |
| Caenorhabditis elegans | Query 36 DALNRFGKTALQ-VMMFGSTAIALELLKQG#ASPNVQDTSGTSPVHDAARTGFLDTLKVLV 94 +|+ +||| || |++ || |#|+|| +| | +|+| || | | +|+ + Sbjct 934 EAITLDNQTALHFAAKFGQLAVSQTLLALG#ANPNARDDKGQTPLHLAAENDFPDVVKLFL 993 Query 37 ALNRFGKTALQVMMF-GSTAIALELLKQG#ASPNVQDTSGTSPVHDAARTGFLDTLKVLV 94 | | | | | + | |+ || ||+|# + + | +| || || | +|+|+ Sbjct 541 ARTRDGSTLLHIAACSGHTSTALAFLKRG#VPLFMPNKKGALGLHSAAAAGFNDVVKMLI 599 Query 36 DALNRFGKTALQVMMF-GSTAIALELLKQG#ASPNVQDTSGTSPVHDAARTGFLDTLKVLV 94 | + |+||| + | | ++ ||+ #| | + +| +|+| ||+ | + + ||| Sbjct 867 DVFDEMGRTALHLAAFNGHLSLVHLLLQHK#AFVNSKSKTGEAPLHLAAQHGHVKVVNVLV 926 Query 42 GKTALQVMMFGSTA----IALELLKQG#ASPNVQDTSGTSPVHDAARTGFLDTLKVLV 94 |+||| + + | |+ ||| |# |+| | + +| |||+| | +++|+ Sbjct 646 GQTALHIAASLNGAESRDCAMMLLKSG#GQPDVAQMDGETCLHIAARSGNKDIMRLLL 702 |
| Thermofilum pendens Hrk 5 | Query 35 PDALNRFGKTALQVMMFGSTAIALELLKQG#ASPNVQDTSGTSPVHDAARTGFLDTLKVLV 94 | |++ || | | + | + +| ||++|#| || +++|| +|+| || | ++ +++|+ Sbjct 245 PCAVDAFGNTPLH-LAFKNMEVAKLLLEKG#ADPNAKNSSGMTPLHFAAGLGKVEVVELLL 303 Query 38 LNRFGKTALQ-VMMFGSTAIALELLKQG#ASPNVQDTSGTSPVHDAARTGFLDTLKVLV 94 | | |+| | ++| +| || +|#| || | | +|+| || || | ++|+ Sbjct 85 LGRKGRTPLHWAAVYGHFVVAEVLLDRG#ADPNATDEEGNTPLHLAALLGFADIARLLL 142 Query 35 PDALNRFGKTALQVM-MFGSTAIALELLKQG#ASPNVQDTSGTSPVHDAARTGFLDTLKVL 93 |+| + | | | + + | || || +|#| | +++|| +|+| || | + |+| Sbjct 115 PNATDEEGNTPLHLAALLGFADIARLLLDRG#ADVNAKNSSGKTPLHYAAEQGSAEVAKLL 174 Query 94 V 94 + Sbjct 175 L 175 Query 36 DALNRFGKTALQVMMF-GSTAIALELLKQG#ASPNVQDTSGTSPVHDAART 84 +| | ||| | || +| ||++|#| | || | +|+| | |+ Sbjct 149 NAKNSSGKTPLHYAAEQGSAEVAKLLLERG#ADPGATDTYGNTPLHLAVRS 198 Query 35 PDALNRFGKTALQVMMFGSTAIALELLKQG#ASPNVQDTSGTSPVHDAARTGFLDTLKVLV 94 | | + +| | | + + | ++ ||++|#| | ++ | +|+| || | + +| |+ Sbjct 181 PGATDTYGNTPLHLAV-RSIEVSKLLLERG#ADVNARNNEGRTPLHRAAMEGSAEVVKFLL 239 Query 36 DALNRFGKTALQ-VMMFGSTAIALELLKQG#ASPNVQDTSGTSPVHDAARTGFLDTLKVLV 94 +| + +|+| | | + ||+ |#| || ++ || +|+| || + |+|+ Sbjct 386 NAKDEYGRTPLHWAAERGCPEVVELLLEHG#ADPNARNDSGMTPLHLAATVKDTEAAKLLL 445 Query 35 PDALNRFGKTALQVMMFGSTAIALE-LLKQG#ASPNVQDTSGTSPVHDAARTGFLDTLKVL 93 |+ + | | | + +| ||++|#| | ++ | +|+| || | + +|+| Sbjct 525 PNTRDNDGNTLLHAAAWNGDVEVIEILLERG#ADINARNKFGETPLHVAAERGNFEAVKLL 584 Query 94 V 94 + Sbjct 585 L 585 |
[Site 5] GHTAVVSFLA127-AESDLHRRDA
Ala127  Ala
Ala
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Gly118 | His119 | Thr120 | Ala121 | Val122 | Val123 | Ser124 | Phe125 | Leu126 | Ala127 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Ala128 | Glu129 | Ser130 | Asp131 | Leu132 | His133 | Arg134 | Arg135 | Asp136 | Ala137 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| ADVNAPDGTGALPIHLAVQEGHTAVVSFLAAESDLHRRDARGLTPLELALQRGAQDLVDI |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | Macaca mulatta | 120.00 | 5 | PREDICTED: cyclin-dependent kinase inhibitor 2D (p |
| 2 | Homo sapiens | 118.00 | 22 | cyclin-dependent kinase inhibitor 2D |
| 3 | N/A | 118.00 | 16 | - |
| 4 | synthetic construct | 118.00 | 12 | cyclin-dependent kinase inhibitor 2D (p19, inhibit |
| 5 | Canis familiaris | 105.00 | 3 | PREDICTED: similar to Cyclin-dependent kinase 4 in |
| 6 | Bos taurus | 103.00 | 4 | cyclin-dependent kinase inhibitor 2D (p19, inhibit |
| 7 | Mus musculus | 96.70 | 19 | cyclin-dependent kinase inhibitor 2D (p19, inhibit |
| 8 | Rattus norvegicus | 94.00 | 8 | similar to cyclin-dependent kinase inhibitor 2D |
| 9 | Monodelphis domestica | 88.60 | 4 | PREDICTED: similar to Cyclin-dependent kinase inhi |
| 10 | Xenopus tropicalis | 59.70 | 1 | cyclin-dependent kinase inhibitor 2D (p19, inhibit |
| 11 | Xenopus laevis | 58.50 | 2 | ankyrin 2, neuronal |
| 12 | Takifugu rubripes | 58.50 | 1 | p19 |
| 13 | Xiphophorus maculatus | 55.80 | 1 | p19 |
| 14 | Tribolium castaneum | 51.60 | 3 | PREDICTED: similar to CG6742-PA, isoform A |
| 15 | Anopheles gambiae str. PEST | 49.70 | 1 | ENSANGP00000001193 |
| 16 | Drosophila melanogaster | 49.30 | 2 | TRPA1_DROME Transient receptor potential cation ch |
| 17 | Drosophila pseudoobscura | 49.30 | 2 | GA19105-PA |
| 18 | Tetraodon nigroviridis | 48.50 | 3 | unnamed protein product |
| 19 | Aedes aegypti | 48.50 | 1 | conserved hypothetical protein |
| 20 | Strongylocentrotus purpuratus | 47.80 | 86 | PREDICTED: similar to putative transient receptor |
| 21 | Aspergillus clavatus NRRL 1 | 45.80 | 2 | Ankyrin repeat protein |
| 22 | Danio rerio | 45.40 | 6 | PREDICTED: hypothetical protein |
| 23 | Pan troglodytes | 45.40 | 5 | PREDICTED: ankyrin repeat and FYVE domain containi |
| 24 | Gallus gallus | 45.40 | 4 | PREDICTED: similar to Chain A, Crystal Structure O |
| 25 | Arabidopsis thaliana | 45.40 | 3 | putative potassium transporter/channel |
| 26 | Neosartorya fischeri NRRL 181 | 45.10 | 1 | Ankyrin repeat protein |
| 27 | Egeria densa | 44.30 | 1 | inward potassium channel alpha subunit |
| 28 | Macaca fascicularis | 43.90 | 1 | unnamed protein product |
| 29 | Wolbachia endosymbiont strain TRS of Brugia malayi | 43.90 | 1 | Ankyrin repeat-containing protein |
| 30 | Trichomonas vaginalis G3 | 43.50 | 8 | ankyrin repeat protein, putative |
| 31 | Apis mellifera | 43.50 | 2 | PREDICTED: similar to centaurin beta 1A CG6742-PA, |
| 32 | Hordeum vulgare | 43.50 | 1 | inwardly rectifying potassium channel AKT1 |
| 33 | Phaeosphaeria nodorum SN15 | 37.70 | 1 | hypothetical protein SNOG_07152 |
| 34 | Aspergillus nidulans FGSC A4 | 36.60 | 2 | hypothetical protein AN0782.2 |
| 35 | Wolbachia endosymbiont of Drosophila melanogaster | 35.40 | 1 | ankyrin repeat domain protein |
| 36 | Wolbachia endosymbiont of Drosophila willistoni TSC#14030-0811.24 | 34.70 | 3 | hypothetical protein Wendoof_01000743 |
| 37 | Wolbachia pipientis | 34.70 | 3 | ankyrin domain protein |
| 38 | Magnaporthe grisea 70-15 | 33.90 | 1 | hypothetical protein MGG_05629 |
| 39 | Aspergillus niger | 33.90 | 1 | hypothetical protein An12g05680 |
| 40 | Caenorhabditis briggsae | 33.90 | 1 | Hypothetical protein CBG13617 |
| 41 | Caenorhabditis elegans | 33.90 | 1 | TRP (transient receptor potential) channel family |
| 42 | Wolbachia endosymbiont of Culex pipiens | 33.10 | 3 | ankyrin domain protein ank2 |
| 43 | Legionella pneumophila str. Lens | 33.10 | 1 | hypothetical protein lpl2058 |
Top-ranked sequences
| organism | matching |
|---|---|
| Macaca mulatta | Query 98 ADVNAPDGTGALPIHLAVQEGHTAVVSFLA#AESDLHRRDARGLTPLELALQRGAQDLVDI 157 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 98 ADVNAPDGTGALPIHLAVQEGHTAVVSFLA#AESDLHRRDARGLTPLELALQRGAQDLVDI 157 |
| Homo sapiens | Query 98 ADVNAPDGTGALPIHLAVQEGHTAVVSFLA#AESDLHRRDARGLTPLELALQRGAQDLVDI 157 |||| |||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Sbjct 98 ADVNVPDGTGALPIHLAVQEGHTAVVSFLA#AESDLHRRDARGLTPLELALQRGAQDLVDI 157 |
| N/A | Query 98 ADVNAPDGTGALPIHLAVQEGHTAVVSFLA#AESDLHRRDARGLTPLELALQRGAQDLVDI 157 |||| |||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Sbjct 98 ADVNVPDGTGALPIHLAVQEGHTAVVSFLA#AESDLHRRDARGLTPLELALQRGAQDLVDI 157 |
| synthetic construct | Query 98 ADVNAPDGTGALPIHLAVQEGHTAVVSFLA#AESDLHRRDARGLTPLELALQRGAQDLVDI 157 |||| |||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Sbjct 98 ADVNVPDGTGALPIHLAVQEGHTAVVSFLA#AESDLHRRDARGLTPLELALQRGAQDLVDI 157 |
| Canis familiaris | Query 98 ADVNAPDGTGALPIHLAVQEGHTAVVSFLA#AESDLHRRDARGLTPLELALQRGAQDLVDI 157 ||||||||||||||||||+|||||||||||# ||||| |||||||||||| |||||+|| Sbjct 98 ADVNAPDGTGALPIHLAVREGHTAVVSFLA#TESDLHHRDARGLTPLELAQGIGAQDLMDI 157 |
| Bos taurus | Query 98 ADVNAPDGTGALPIHLAVQEGHTAVVSFLA#AESDLHRRDARGLTPLELALQRGAQDLVDI 157 ||||||||||||||||||+||||+||||||# ||||| ||| |||||||| ||||+|+|| Sbjct 98 ADVNAPDGTGALPIHLAVREGHTSVVSFLA#TESDLHHRDATGLTPLELARGRGAQELMDI 157 |
| Mus musculus | Query 98 ADVNAPDGTGALPIHLAVQEGHTAVVSFLA#AESDLHRRDARGLTPLELALQRGAQDLVDI 157 ||||| | ||+||||||++|||++||||||# ||||| ||| |||||||| |||||+|+|| Sbjct 98 ADVNALDSTGSLPIHLAIREGHSSVVSFLA#PESDLHHRDASGLTPLELARQRGAQNLMDI 157 |
| Rattus norvegicus | Query 98 ADVNAPDGTGALPIHLAVQEGHTAVVSFLA#AESDLHRRDARGLTPLELALQRGAQDLVDI 157 |||| | ||+||||||++|||++||||||# ||||| +|| |||||||| |||||+|+|| Sbjct 98 ADVNTLDSTGSLPIHLAIREGHSSVVSFLA#PESDLHHKDASGLTPLELARQRGAQNLMDI 157 |
| Monodelphis domestica | Query 98 ADVNAPDGTGALPIHLAVQEGHTAVVSFLA#AESDLHRRDARGLTPLELALQRGAQDL 154 |||| |||+|||||||||+||| |||||||# |||| ||| |||||||| | || | Sbjct 98 ADVNVPDGSGALPIHLAVREGHAAVVSFLA#GESDLQHRDAGGLTPLELARQCGAGQL 154 |
| Xenopus tropicalis | Query 98 ADVNAPDGTGALPIHLAVQEGHTAVVSFLA#AESDLHRRDARGLTPLELA 146 |++| || +|+||||||++ || |+++||# ||| || | || +|| Sbjct 98 AEINTPDASGSLPIHLALKAGHVPVITYLA#LISDLQHRDREGHTPSQLA 146 |
| Xenopus laevis | Query 98 ADVNAPDGTGALPIHLAVQEGHTAVVSFLA#AESDLHRRDARGLTPLELA 146 |+|| || +|+||||||++ || |+++||# ||| + | | || +|| Sbjct 98 AEVNTPDASGSLPIHLALKAGHVPVITYLA#LISDLQQHDREGHTPSQLA 146 |
| Takifugu rubripes | Query 98 ADVNAPDGTGALPIHLAVQEGHTAVVSFLA#AESDLHRRDARGLTPLELALQRGAQDLVDI 157 | || || +||||||+|++||| || |||# ||| + | | ++| |++|+ Sbjct 98 ASVNLPDQSGALPIHIAIREGHRDVVEFLA#PRSDLKHANKSGQTAADVARASRVPDMMDL 157 |
| Xiphophorus maculatus | Query 98 ADVNAPDGTGALPIHLAVQEGHTAVVSFLA#AESDLHRRDARGLTPLELALQRGAQDLVD 156 | || | ||||||+|++||| +| |||# ||| + | | +++| | |+++ Sbjct 98 ASVNIQDQNGALPIHIAIREGHRDIVEFLA#PRSDLKHANVSGQTAIDVARSLGELDMMN 156 |
| Tribolium castaneum | Query 98 ADVNAPDGTGALPIHLAVQEGHTAVVSFLA#A-ESDLHRRDARGLTPLELALQRGAQDLVD 156 | +|| | | |+||| |||||| | | # +| | | | ||++| |+ |+| Sbjct 679 AKINAQDQNGKTPLHLATQEGHTAQVCLLL#KHRADQHLEDDEGKVPLQMAEQKEHADIVT 738 Query 157 I 157 + Sbjct 739 L 739 |
| Anopheles gambiae str. PEST | Query 100 VNAPDGTGALPIHLAVQEGHTAVVSFLA#AESDLHRRDARGLTPLELALQRGAQDLVDI 157 +| || | |+|+| |+||| || | # | || | || || | + +++ Sbjct 490 INESDGEGLTPLHIASQQGHTRVVQLLL#NRGALLHRDHNGRNPLHLAAMSGYRQTIEL 547 |
| Drosophila melanogaster | Query 100 VNAPDGTGALPIHLAVQEGHTAVVSFLA#AESDLHRRDARGLTPLELALQRGAQDLVDI 157 +| || | |+|++ |+||| || | # | || | ||+|| | + +++ Sbjct 506 INESDGAGMTPLHISSQQGHTRVVQLLL#NRGALLHRDHTGRNPLQLAAMSGYTETIEL 563 |
| Drosophila pseudoobscura | Query 100 VNAPDGTGALPIHLAVQEGHTAVVSFLA#AESDLHRRDARGLTPLELALQRGAQDLVDI 157 +| || | |+|++ |+||| || | # | || | ||+|| | + +++ Sbjct 519 INESDGAGMTPLHISSQQGHTRVVQLLL#NRGALLHRDHTGRNPLQLAAMSGYTETIEL 576 |
| Tetraodon nigroviridis | Query 98 ADVNAPDGTGALPIHLAVQEGHTAVVSFLA#AESDLHRRDARGLTPLELALQRGAQDLVDI 157 || | | || ||+|+| | |+ +|| | #| + | |++|| +| + |++++ Sbjct 231 ADANKPGKTGLLPLHVAAQTGNDTIVSMLI#AVTSKARVRRTGISPLHVAAEHNCDDVLEL 290 |
| Aedes aegypti | Query 100 VNAPDGTGALPIHLAVQEGHTAVVSFLA#AESDLHRRDARGLTPLELALQRGAQDLVDI 157 +| || | |+|+| +|||| || | # | || | || || | +++ Sbjct 197 INESDGEGLTPLHIASKEGHTRVVQLLL#NRGALLHRDHNGRNPLHLAAMSGYTQTIEL 254 |
| Strongylocentrotus purpuratus | Query 107 GALPIHLAVQEGHTAVVSFLA#AES--DLHRRDARGLTPLELALQRGAQDLV 155 | +|+||| | |||||| | #++| || +| || | | | | |+| Sbjct 1 GTIPLHLAAQNGHTAVVGLLL#SKSTTQLHVKDKRGRTGLHQAAANGHYDMV 51 |
| Aspergillus clavatus NRRL 1 | Query 98 ADVNAPDGTGALPIHLAVQEG----HTAVVSFLA-#AESDLHRRDARGLTPLELALQRGAQ 152 |||| || | |+|+ | | ||| | #+ + | +| ||||||++| + | Sbjct 208 ADVNVVDGNGRTPLHVCAATGNTADHVAVVRLLVN#SGASLSTKDHRGLTPLQVAAEAGNH 267 Query 153 DLVD 156 +++ Sbjct 268 RIIE 271 |
| Danio rerio | Query 107 GALPIHLAVQEGHTAVVSFLA--#AESDLHRRDARGLTPLELALQRGAQDLV 155 || |++|| |||| || || # ++|+| | |+||| | | || Sbjct 2 GATPLYLACQEGHLHVVEFLVKD#CQADVHLRAQDGMTPLHAAAHMGHHSLV 52 |
| Pan troglodytes | Query 98 ADVNAPDGTGALPIHLAVQEGHTAVVSFLA#AESDLHR--RDARGLTPLELAL 147 |+||| | | |||+|+ | ++ | #+ |+| || +|||| |+ Sbjct 736 ANVNAQDAEGRTPIHVAISSQHGVIIQLLV#SHPDIHLNVRDRQGLTPFACAM 787 |
| Gallus gallus | Query 98 ADVNAPDGTGALPIHLAVQEGHTAVVSFL--A#AESDLHRRDARGLTPLELA 146 |||| | | ||+||| |||| || || # | + ++ || | |++| Sbjct 94 ADVNIEDAEGNLPLHLAAQEGHVRVVEFLLRR#TPSRVAHQNRRGDTALDVA 144 |
| Arabidopsis thaliana | Query 99 DVNAPDGTGALPIHLAVQEGHTAVVSFLA#AE-SDLHRRDARGLTPLELALQRGAQDL 154 || ||| | +| || ||| +| || # + +|| |+ | || || +| +++ Sbjct 253 DVTLPDGNGTTALHRAVSEGHLEIVKFLL#DQGADLDWPDSYGWTPRGLADHQGNEEI 309 |
| Neosartorya fischeri NRRL 181 | Query 99 DVNAPDGTGALPIHLAVQEGHTAVVSFLA#AES--DLHRRDARGLTPL 143 +|| | | |+| ||++|| |||+ #|+ |++| |+|| ||+ Sbjct 215 EVNTRDNRGRTPLHRAVEKGHLAVVNLFL#AQPNIDINRADSRGRTPM 261 |
| Egeria densa | Query 98 ADVNAPDGTGALPIHLAVQEGHTAVVSFLA#AE-SDLHRRDARGLTPLELALQRGAQDL 154 ||+| | +|+ +| || +|+ +|||| # +| | | | || | | |+| |+ Sbjct 544 ADINLPKRSGSTALHSAVCDGNVEIVSFLL#QHGADPDRPDINGWTPREYADQQGHHDI 601 |
| Macaca fascicularis | Query 98 ADVNAPDGTGALPIHLAVQEGHTAVVSFLA#AE-SDLHRRDARGLTPLELALQR 149 || + || | |||| | |||++|++| #|+ |+ | |+||| | | Sbjct 10 ADPSLIDGEGCSCIHLAAQFGHTSIVAYLI#AKGQDVDMMDQNGMTPLMWAAYR 62 |
| Wolbachia endosymbiont strain TRS of Brugia malayi | Query 99 DVNAPDGTGALPIHLAVQEGHTAVV-SFLA#AESDLHRRDARGLTPLELALQRGAQDLVDI 157 |||| | +||||++|| +| | | # ++|++ | | ||| |+| | + +++ Sbjct 83 DVNAVDKDKHTSLHLAVEKGHIGIVNSLLE#VKADVNVEDENGKTPLYTAVQFGCKGEIEV 142 |
| Trichomonas vaginalis G3 | Query 99 DVNAPDGTGALPIHLAVQEGHTAVVSFLA#AES-DLHRRDARGLTPLELALQRGAQDLVDI 157 |+|| | | |+| || | +| || #+ |++ +| | ||| | ++ + |+|++ Sbjct 64 DINAKDKKGCTPLHYAVIRGLVKLVKFLV#SHGVDVNSKDFLGSTPLHYAAEQFSFDIVEV 123 |
| Apis mellifera | Query 98 ADVNAPDGTGALPIHLAVQEGHTAVVSFLA#A-ESDLHRRDARGLTPLELALQRGAQDLVD 156 | +| | | |++|| + |||| | | # +| | | |+ || +|++ |+| Sbjct 790 ARINCQDADGKTPLYLATELGHTAQVCLLL#KHRADQHIEDESGIKPLSIAVKEANADIVT 849 Query 157 I 157 + Sbjct 850 L 850 |
| Hordeum vulgare | Query 99 DVNAPDGTGALPIHLAVQEGHTAVVSFLA#AE-SDLHRRDARGLTPLELALQRGAQDLVDI 157 ||| | | +|+| || +|+ +| | # +|+ ++|+ | || || |+| +++ ++ Sbjct 664 DVNRPSKDGNIPLHRAVCDGNVEMVELLL#RHGADIDKQDSNGWTPRALAEQQGHEEIQNL 723 |
| Phaeosphaeria nodorum SN15 | Query 99 DVNAPDGTGALPIHLAVQEGHTAVVSFLA#AES--DLHRRDARGLTPLELALQRGAQDLVD 156 |||| | | + ||+|+| ++| | #+ |++ ++ | ||| |++ | +|+|| Sbjct 649 DVNAKDNAGRTAVVLAIQQGCKSIVELLL#STGRIDVNTKNITGRTPLMWAIRHGRKDMVD 708 Query 157 I 157 + Sbjct 709 L 709 Query 99 DVNAPDGTGALPIHLAVQEGHTAVVSFL--A#AESDLHRRDARGLTPLELALQRGAQDLVD 156 ||| + || |+ |++ | +| | # + +++ || | ||| |+|+ +|+++ Sbjct 683 DVNTKNITGRTPLMWAIRHGRKDMVDLLLNT#GKVEINAEDAYGWTPLMWAVQQARKDIIE 742 Query 157 I 157 + Sbjct 743 L 743 |
| Aspergillus nidulans FGSC A4 | Query 98 ADVNAPDGTGALPIHLAVQEGHTAVVSFLA#AE-SDLHRRDARGLTPLELALQRGAQDL 154 ||||| +| +|||| || +|+ | # +|++ | | ||| | ||++ + Sbjct 268 ADVNAVEGRNKTALHLAVAHGHERIVALLL#RHGADINARSDGGWTPLHNACDRGSESI 325 Query 98 ADVNAPDGTGALPIHLAVQEGHTAVVS-FLA#AESDLHRRDARGLTPLELALQRGAQDLVD 156 ||+|| | |+| | | ++ ||#| + ++ + |++|| || | | +++|+ Sbjct 301 ADINARSDGGWTPLHNACDRGSESIARRLLA#AGAKINAQLLNGISPLHLAAQGGHREVVE 360 Query 98 ADVNAPDGTGALPIHLAVQEGHTAVVSFLA#AESDLHR--RDARGLTPLELALQRGAQDLV 155 | +|| | |+||| | || || | # ||| + || | | | | +|+| Sbjct 334 AKINAQLLNGISPLHLAAQGGHREVVECLL#ERSDLKQRLRDNFGSTAFLRAAQFKRKDIV 393 |
| Wolbachia endosymbiont of Drosophila melanogaster | Query 99 DVNAPDGTGALPIHLAVQEGHTAVVSFLA#AESDLHRRDARGLTPLELALQRGAQDLVD 156 +||| | |+||| + | || | # ++|++ +|| ||| +| | +|+|+ Sbjct 162 NVNAEDDDRCTPLHLAAEANHIEVVKILV#EKADVNIKDADRWTPLHVAAANGHKDVVE 219 Query 99 DVNAPDGTGALPIHLAVQEGHTAVVSFLA#AESDLHRRDARGLTPLELALQRGAQDLV 155 +||| | |+||| + | || | # ++|++ +|| ||| +| | +|+| Sbjct 227 NVNAEDDDRCTPLHLAAEANHIEVVKILV#EKADVNIKDADRWTPLHVAAANGHEDVV 283 Query 98 ADVNAPDGTGALPIHLAVQEGHTAVVSFLA#AESDLH--RRDARGLTPLELALQRGAQDLV 155 ||+|| |+||| || +| |+# ++ +|+ ||||| || +|+| Sbjct 94 ADINAEHDNKITPLHLAAHYGHKEIVQVLS#KAEGINVDAKDSDGLTPLHLATANSHKDVV 153 Query 156 D 156 + Sbjct 154 E 154 Query 98 ADVNAPDGTGALPIHLAVQEGHTAVVS-FLA#AESDLHRRDARGLTPLELALQRGAQDLVD 156 | | | +| |+| | | || +| | #| +| +| | || +| +| |++ Sbjct 291 AKVKAKNGDRHTPLHFAAQNGHEGIVKVLLE#AGADPSLKDVDGKTPRDLTKDQGIIQLLE 350 Query 98 ADVNAPDGTGALPIHLAVQEGHTAVVSFLA#AE-SDLHRRDARGLTPLELALQRGAQDLVD 156 |||| | |+|+| || || | #|+ + + ++ ||| | | | + +| Sbjct 258 ADVNIKDADRWTPLHVAAANGHEDVVKTLI#AKGAKVKAKNGDRHTPLHFAAQNGHEGIVK 317 Query 157 I 157 + Sbjct 318 V 318 Query 98 ADVNAPDGTGALPIHLAVQEGHTAVVSFLA#AES-DLHRRDARGLTPLELALQRGAQDLVD 156 |||| | |+|+| || || | #| +++ | ||| || + ++| Sbjct 193 ADVNIKDADRWTPLHVAAANGHKDVVETLI#ANKVNVNAEDDDRCTPLHLAAEANHIEVVK 252 Query 157 I 157 | Query 157 I 157 |
| Wolbachia endosymbiont of Drosophila willistoni TSC#14030-0811.24 | Query 98 ADVNAPDGTGALPIHLAVQEGHTAVVS-FLA#AESDLHRRDARGLTPLELALQRGAQDLVD 156 |++|| | | |+| ||+ || ||+ + # +++++ +| +| ||| | +| ++|+ Sbjct 32 AEINAKDNQGMAPLHWAVKVGHINVVNGLIK#GKAEINAKDNQGRTPLHWAASKGGIEVVN 91 Query 98 ADVNAPDGTGALPIHLAVQEGHTAVVSFLA#AE-SDLHRRDARGLTPLELALQRGAQDLV 155 |++|| | | |+| | +| ||+ | # + +|++ + | || | + | |+| Sbjct 65 AEINAKDNQGRTPLHWAASKGGIEVVNALI#EKGADVNAVNKYGDAPLRFAARDGHIDIV 123 |
| Wolbachia pipientis | Query 98 ADVNAPDGTGALPIHLAVQEGHTAVVS-FLA#AESDLHRRDARGLTPLELALQRGAQDLVD 156 |++|| | | |+| ||+ || ||+ + # +++++ +| +| ||| | +| ++|+ Sbjct 105 AEINAKDNQGMAPLHWAVKVGHINVVNGLIK#GKAEINAKDNQGRTPLHWAASKGGIEVVN 164 Query 98 ADVNAPDGTGALPIHLAVQEGHTAVVSFLA#AE-SDLHRRDARGLTPLELALQRGAQDLV 155 |++|| | | |+| | +| ||+ | # + +|++ + | || | + | |+| Sbjct 138 AEINAKDNQGRTPLHWAASKGGIEVVNALI#EKGADVNAVNKYGDAPLRFAARDGHIDIV 196 |
| Magnaporthe grisea 70-15 | Query 99 DVNAPDGTGALPIHLAVQEGHTAVVSFLA#AES--DLHRRDARGLTPLELALQRGAQDLVD 156 |+|| | | |+||| | | || | #++| |++ || +| ||| | +| | |+ Sbjct 1458 DLNARDEDGRTPLHLAAANGETRVVKMLL#SQSGVDINARDDQGQTPLHRAAAQGYHDFVE 1517 Query 99 DVNAPDGTGALPIHLAVQEGH---------------TAVVSFLA#AES--DLHRRDARGLT 141 |+|| | | |+| | +|+ |+ + | #|+ +++ || ||| Sbjct 1492 DINARDDQGQTPLHRAAAQGYHDFVEAYSIKDYSDRTSALRLLL#ADDRIEVNARDKDGLT 1551 Query 142 PLELALQRGAQDLVDI 157 ||+ |+ | | + Sbjct 1552 PLQQAVAFGQPQSVQV 1567 |
| Aspergillus niger | Query 98 ADVNAPDGTGALPIHLAVQEGHTAVVSFLA#A--ESDLHRRDARGLTPLELALQRGAQDLV 155 ||+++ | +| |+ || | || ||| + #| | |++ +|+|| ||+ | + | + +| Sbjct 943 ADIDSRDSSGNSPLSLAAQFGHEAVVKIVL#ATGEVDVNNKDSRGKTPISWAAREGYEAVV 1002 Query 99 DVNAPDGTGALPIHLAVQEGHTAVVSFLA#A--ESDLHRRDARGLTPLELALQRGAQDLVD 156 |+++ | |+ |+ || + || +| | #| ++|+ ||+ | +|| || | | + +| Sbjct 910 DIDSKDVPGSTPLALAAENGHERIVRLLL#ATGKADIDSRDSSGNSPLSLAAQFGHEAVVK 969 Query 157 I 157 | Query 157 I 157 Query 98 ADVNAPDGT-GALPIHLAVQEGHTAVVSFLA#A--ESDLHRRDARGLTPLELALQRGAQDL 154 |++|+ | |+ | + || ||| | #| | |+ |++ | +|| || | | ++ Sbjct 841 ANLNSKDSDYDQTPLSWAAERGHVAVVKLLL#AAEEVDIDSRNSIGRSPLSLAAQNGHDEV 900 Query 155 VDI 157 | + Sbjct 901 VKL 903 Query 100 VNAPDGTGALPIHLAVQEGHTAVVSFLA#A--ESDLHRRDARGLTPLELALQRGAQDLVDI 157 ||+ | | |+ | + |+ ||+ | #| +|+ || | || + | + | +| | Sbjct 1151 VNSRDSEGRTPLSWAAENGNEAVIKLLI#ATGNADVSARDVNGRTPQDWAAEYDHQSVVKI 1210 Query 99 DVNAPDGT-GALPIHLAVQEGHTAVVSFLA#A--ESDLHRRDAR-GLTPLELALQRGAQDL 154 ||| | | |+ | ||| ||| | #+ | |+ || | | +|| | | +++ Sbjct 1046 DVNIRDRRYGREPLSWAAGEGHEAVVKLLL#STREVDVDVRDPRYGRSPLSWAAGEGHEEV 1105 Query 155 VDI 157 |++ Sbjct 1106 VEL 1108 |
| Caenorhabditis briggsae | Query 109 LPIHLAVQEGHTAVVSFLA#AES--DLHRRDARGLTPLELALQRGAQDLVDI 157 +|+||| |+|| ||| | #+ | | +| || ||| || | | ++| + Sbjct 1211 IPLHLAAQQGHIAVVGMLL#SRSTQQQHAKDWRGRTPLHLAAQNGHYEMVSL 1261 Query 101 NAPDGTGALPIHLAVQEGHTAVVSFLA#AE-SDLHRRDARGLTPLELALQRGAQDLVDI 157 +| | | |+||| | || +|| | #|+ |+++ | | | | | + | +| + Sbjct 1237 HAKDWRGRTPLHLAAQNGHYEMVSLLI#AQGSNINVMDQNGWTGLHFATRAGHLSVVKL 1294 Query 102 APDGTGALPIHLAVQEGHTAVVSFL---A#AESDLHRRDARGLTPLELALQRGAQDLV 155 | || |+|||||| + |+ +| | #++ + | | | | || + |+ + | Sbjct 421 AEDGEGSLPIHLAFKFGNVNIVELLLSGP#SDEQTRKADGNGDTLLHLAARSGSIEAV 477 Query 98 ADVNAPDGTGALPIHLAVQEGHTAVV---------------------------------- 123 | ||+ || |+||| | || || Sbjct 914 AFVNSKSKTGEAPLHLAAQHGHVKVVNVLVQDHGASLEAITLDNQTALHFAAKFGQLAVS 973 Query 124 -SFLA#AESDLHRRDARGLTPLELALQRGAQDLVDI 157 + ||# ++ + || +| ||| || + |+| + Sbjct 974 QTLLA#LGANPNARDDKGQTPLHLAAENDFPDVVKL 1008 |
| Caenorhabditis elegans | Query 109 LPIHLAVQEGHTAVVSFLA#AES--DLHRRDARGLTPLELALQRGAQDLVDI 157 +|+||| |+|| ||| | #+ | | +| || ||| || | | ++| + Sbjct 1194 IPLHLAAQQGHIAVVGMLL#SRSTQQQHAKDWRGRTPLHLAAQNGHYEMVSL 1244 Query 101 NAPDGTGALPIHLAVQEGHTAVVSFLA#AE-SDLHRRDARGLTPLELALQRGAQDLVDI 157 +| | | |+||| | || +|| | #|+ |+++ | | | | | + | +| + Sbjct 1220 HAKDWRGRTPLHLAAQNGHYEMVSLLI#AQGSNINVMDQNGWTGLHFATRAGHLSVVKL 1277 Query 102 APDGTGALPIHLAVQEGHTAVVSFL---A#AESDLHRRDARGLTPLELALQRG 150 | || |+|||||| + |+ +| | #++ + | | | | || + | Sbjct 405 AEDGDGSLPIHLAFKFGNVNIVELLLSGP#SDEQTRKADGNGDTLLHLAARSG 456 Query 98 ADVNAPDGTGALPIHLAVQEGHTAVV---------------------------------- 123 | ||+ || |+||| | || || Sbjct 897 AFVNSKSKTGEAPLHLAAQHGHVKVVNVLVQDHGAALEAITLDNQTALHFAAKFGQLAVS 956 Query 124 -SFLA#AESDLHRRDARGLTPLELALQRGAQDLVDI 157 + ||# ++ + || +| ||| || + |+| + Sbjct 957 QTLLA#LGANPNARDDKGQTPLHLAAENDFPDVVKL 991 |
| Wolbachia endosymbiont of Culex pipiens | Query 98 ADVNAPDGTGALPIHLAVQEGHTAVVS-FLA#AESDLHRRDARGLTPLELALQRGAQDLVD 156 |+||| | |+|+| + || +|| | #||++++ | ||| | | |+|+ Sbjct 62 ANVNAVGSEGWTPLHVAAENGHASVVEVLLK#AEANVNAVGIEGCTPLHFAAGNGHVDIVN 121 Query 157 I 157 + Sbjct 122 L 122 Query 98 ADVNAPDGTGAL-PIHLAVQEGHTAVVS-FLA#AESDLHRRDARGLTPLELALQRGAQDLV 155 |+||| | |+|+| + || +|| | #|+++++ + | ||| +| + | +| Sbjct 28 ANVNAVDSNKWFTPLHVAAENGHASVVEVLLK#AKANVNAVGSEGWTPLHVAAENGHASVV 87 Query 156 DI 157 ++ Sbjct 88 EV 89 Query 98 ADVNAPDGTGALPIHLAVQEGHTAVVSFLA#AE-SDLHRRDARGLTPLELA 146 |+||| | |+| | || +|+ | # + ++++ | | |||+ | Sbjct 95 ANVNAVGIEGCTPLHFAAGNGHVDIVNLLL#EKGANVNAVDRYGKTPLDYA 144 |
| Legionella pneumophila str. Lens | Query 99 DVNAPDGTGALPIHLAVQEGHTAVVSFLA#A-ESDLHRRDARGLTPLELALQRGAQDLVD 156 | | | |+ | ||| + || ||++ ||# ++||++ | | || +| | ++| Sbjct 460 DFNKKDINGSTPAHLAARNGHVAVIAELA#KHQADLNKADNDGFTPAHIAALTGQSMVLD 518 Query 99 DVNAPDGTGALPIHLAVQEGHTAVVSFLA#AES-DLHRRDARGLTPLELALQRG 150 | | + | | ||| + || ||++ ||# + ++ + | || || | | Sbjct 361 DFNKTNERGDTPAHLAARNGHVAVIAELA#KHGVNFNKANRSGDTPAHLAAQNG 413 Query 101 NAPDGTGALPIHLAVQEGHTAVVSFLA#AES-DLHRRDARGLTPLELALQRG 150 | +| | ||| | || | + ||# | ++ + || || || + | Sbjct 330 NKTSRSGNTPAHLAAQNGHVAAIIELA#KYGVDFNKTNERGDTPAHLAARNG 380 Query 101 NAPDGTGALPIHLAVQEGHTAVVSFLA#AES-DLHRRDARGLTPLELALQRG 150 | + +| | ||| | || ||++ ||# + ++|+++ | | +| | Sbjct 396 NKANRSGDTPAHLAAQNGHAAVITELA#KNNVNVHQQNEHGSTLAHIAALNG 446 Query 99 DVNAPDGTGALPIHLAVQEGHTAVVSFLA#A-ESDLHRRDARGLTPLELALQRG 150 |+| | | | |||| | +| ||# + | ++ |+| || |+| Sbjct 262 DLNKADKDGFTPAHLAVLHGDVTFISELA#RHQVDFNKVTKHGVTTAHLAAQQG 314 Query 99 DVNAPDGTGALPIHLAVQEGHTAVVSFLA#A-ESDLHRRDARGLTPLELALQRG 150 +|+ + |+ |+| || |++ ||# + | +++| | || || + | Sbjct 427 NVHQQNEHGSTLAHIAALNGHAVVIAELA#KHQVDFNKKDINGSTPAHLAARNG 479 |
[Site 6] AARGDVQEVR25-RLLHRELVHP
Arg25  Arg
Arg
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Ala16 | Ala17 | Arg18 | Gly19 | Asp20 | Val21 | Gln22 | Glu23 | Val24 | Arg25 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Arg26 | Leu27 | Leu28 | His29 | Arg30 | Glu31 | Leu32 | Val33 | His34 | Pro35 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| MLLEEVRSGDRLSGAAARGDVQEVRRLLHRELVHPDALNRFGKTALQVMMFGSTA |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | Macaca mulatta | 109.00 | 6 | PREDICTED: cyclin-dependent kinase inhibitor 2D (p |
| 2 | N/A | 108.00 | 17 | - |
| 3 | Homo sapiens | 108.00 | 15 | cyclin-dependent kinase inhibitor 2D |
| 4 | synthetic construct | 108.00 | 7 | cyclin-dependent kinase inhibitor 2D (p19, inhibit |
| 5 | Canis familiaris | 104.00 | 2 | PREDICTED: similar to Cyclin-dependent kinase 4 in |
| 6 | Rattus norvegicus | 102.00 | 7 | similar to cyclin-dependent kinase inhibitor 2D |
| 7 | Mus musculus | 101.00 | 15 | cyclin-dependent kinase inhibitor 2D (p19, inhibit |
| 8 | Bos taurus | 99.00 | 5 | cyclin-dependent kinase inhibitor 2D (p19, inhibit |
| 9 | Monodelphis domestica | 96.30 | 5 | PREDICTED: similar to Cyclin-dependent kinase inhi |
| 10 | Xenopus tropicalis | 86.30 | 2 | cyclin-dependent kinase inhibitor 2D (p19, inhibit |
| 11 | Xenopus laevis | 83.60 | 2 | ankyrin 2, neuronal |
| 12 | Xiphophorus maculatus | 58.20 | 2 | p19 |
| 13 | Tetraodon nigroviridis | 53.50 | 4 | unnamed protein product |
| 14 | Takifugu rubripes | 52.80 | 2 | p19 |
| 15 | Mesocricetus auratus | 46.60 | 3 | p15INK4b cyclin-dependent kinase inhibitor |
| 16 | Gallus gallus | 44.70 | 3 | cyclin-dependent kinase inhibitor 2A (melanoma, p1 |
| 17 | Macaca fascicularis | 44.70 | 1 | unnamed protein product |
| 18 | Sus scrofa | 42.40 | 2 | cyclin dependant kinase inhibitor |
| 19 | Pan troglodytes | 42.00 | 1 | PREDICTED: similar to p16-INK4 |
| 20 | human, brain tumors, Peptide, 156 aa | 42.00 | 1 | cyclin D-dependent kinases 4 and 6-binding protein |
| 21 | Danio rerio | 39.70 | 3 | PREDICTED: similar to Chain A, Structure Of P18ink |
| 22 | Xiphophorus helleri | 35.00 | 1 | p13CDKN2X |
| 23 | Mus spretus | 35.00 | 1 | cyclin-dependent kinase inhibitor protein |
| 24 | Apis mellifera | 34.70 | 1 | PREDICTED: similar to MYPT-75D CG6896-PA |
| 25 | Tribolium castaneum | 33.50 | 1 | PREDICTED: similar to CG6896-PA |
| 26 | Canis lupus familiaris | 33.10 | 1 | uveal autoantigen with coiled-coil domains and ank |
| 27 | Populus tremula x Populus tremuloides | 32.30 | 1 | potassium channel 2 |
| 28 | Hyperthermus butylicus DSM 5456 | 32.30 | 1 | V-type ATP synthase alpha chain |
Top-ranked sequences
| organism | matching |
|---|---|
| Macaca mulatta | Query 1 MLLEEVRSGDRLSGAAARGDVQEVR#RLLHRELVHPDALNRFGKTALQVMMFGSTA 55 |||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 1 MLLEEVRSGDRLSGAAARGDVQEVR#RLLHRELVHPDALNRFGKTALQVMMFGSTA 55 |
| N/A | Query 1 MLLEEVRSGDRLSGAAARGDVQEVR#RLLHRELVHPDALNRFGKTALQVMMFGSTA 55 |||||||+|||||||||||||||||#|||||||||||||||||||||||||||||| Sbjct 1 MLLEEVRAGDRLSGAAARGDVQEVR#RLLHRELVHPDALNRFGKTALQVMMFGSTA 55 |
| Homo sapiens | Query 1 MLLEEVRSGDRLSGAAARGDVQEVR#RLLHRELVHPDALNRFGKTALQVMMFGSTA 55 |||||||+|||||||||||||||||#|||||||||||||||||||||||||||||| Sbjct 1 MLLEEVRAGDRLSGAAARGDVQEVR#RLLHRELVHPDALNRFGKTALQVMMFGSTA 55 |
| synthetic construct | Query 1 MLLEEVRSGDRLSGAAARGDVQEVR#RLLHRELVHPDALNRFGKTALQVMMFGSTA 55 |||||||+|||||||||||||||||#|||||||||||||||||||||||||||||| Sbjct 1 MLLEEVRAGDRLSGAAARGDVQEVR#RLLHRELVHPDALNRFGKTALQVMMFGSTA 55 |
| Canis familiaris | Query 1 MLLEEVRSGDRLSGAAARGDVQEVR#RLLHRELVHPDALNRFGKTALQVMMFGS 53 |||||||+|||||||||||||||||#|||||||||||||||||||||||||||| Sbjct 1 MLLEEVRAGDRLSGAAARGDVQEVR#RLLHRELVHPDALNRFGKTALQVMMFGS 53 |
| Rattus norvegicus | Query 1 MLLEEVRSGDRLSGAAARGDVQEVR#RLLHRELVHPDALNRFGKTALQVMMFGSTA 55 |||||| |||||||||||||||||#|||||||||||||||||||||||||||| | Sbjct 1 MLLEEVSVGDRLSGAAARGDVQEVR#RLLHRELVHPDALNRFGKTALQVMMFGSPA 55 |
| Mus musculus | Query 1 MLLEEVRSGDRLSGAAARGDVQEVR#RLLHRELVHPDALNRFGKTALQVMMFGSTA 55 |||||| |||||||||||||||||#|||||||||||||||||||||||||||| | Sbjct 1 MLLEEVCVGDRLSGAAARGDVQEVR#RLLHRELVHPDALNRFGKTALQVMMFGSPA 55 |
| Bos taurus | Query 1 MLLEEVRSGDRLSGAAARGDVQEVR#RLLHRELVHPDALNRFGKTALQVMMFGS 53 |||||| +|||||||||||||||||#|||| |||||| |||||||||||||||| Sbjct 1 MLLEEVHAGDRLSGAAARGDVQEVR#RLLHSELVHPDVLNRFGKTALQVMMFGS 53 |
| Monodelphis domestica | Query 1 MLLEEVRSGDRLSGAAARGDVQEVR#RLLHRELVHPDALNRFGKTALQVMMFGST 54 |||||+ +||||||||||||| |||#||||+| ||||+||||||||||||||||+ Sbjct 1 MLLEEISAGDRLSGAAARGDVNEVR#RLLHQEFVHPDSLNRFGKTALQVMMFGSS 54 |
| Xenopus tropicalis | Query 1 MLLEEVRSGDRLSGAAARGDVQEVR#RLLHRELVHPDALNRFGKTALQVMMFGST 54 |||+| +||||+ ||||||+ ||+#|||| | +||| ||||||||||||||||| Sbjct 1 MLLQETSAGDRLTRAAARGDLLEVK#RLLHEERIHPDCLNRFGKTALQVMMFGST 54 |
| Xenopus laevis | Query 1 MLLEEVRSGDRLSGAAARGDVQEVR#RLLHRELVHPDALNRFGKTALQVMMFGST 54 |||+| +||||+ ||||||+ ||+#|||| | +||| || |||||||||||||| Sbjct 1 MLLQETSAGDRLTRAAARGDLTEVK#RLLHEERIHPDCLNCFGKTALQVMMFGST 54 |
| Xiphophorus maculatus | Query 1 MLLEEVRSGDRLSGAAARGDVQEVR#RLLHRELVHPDALNRFGKTALQVMMFGST 54 |+| ++ +| |+ |||+|+ ||+#|+| |||| | ||+||||||| |++ Sbjct 1 MVLSQMDAGKALTAAAAKGNADEVQ#RILEECRVHPDTPNEFGRTALQVMMMGNS 54 |
| Tetraodon nigroviridis | Query 1 MLLEEVRSGDRLSGAAARGDVQEVR#RLLHRELVHPDALNRFGKTALQVMMFGS 53 |++ ++ +| |+ |||+| |||#|+| | || | |||||||||| |+ Sbjct 1 MVIGQMDAGKALAAAAAKGRTAEVR#RILEECRVPPDTPNEFGKTALQVMMLGN 53 |
| Takifugu rubripes | Query 1 MLLEEVRSGDRLSGAAARGDVQEVR#RLLHRELVHPDALNRFGKTALQVMMFGS 53 |++ ++ +| |+ |||+| ||+#|+| | || | |||||||||| |+ Sbjct 1 MVIGQMDAGKALAAAAAKGRTSEVQ#RILEECRVPPDTRNEFGKTALQVMMLGN 53 |
| Mesocricetus auratus | Query 12 LSGAAARGDVQEVR#RLLHRELVHPDALNRFGKTALQVMMFGST 54 |+ ||||| |+ ||#+|| | |+|+||||+ +|||| ||| Sbjct 10 LATAAARGQVETVR#QLLEAG-VDPNAVNRFGRRPIQVMMMGST 51 |
| Gallus gallus | Query 8 SGDRLSGAAARGDVQEVR#RLLHRELVHPDALNRFGKTALQVMMFGS 53 + | |+ ||||||+ |+# || |+|+| ||+| +|||| || Sbjct 9 AADELANAAARGDLLRVK#ELLDGA-ADPNAVNSFGRTPIQVMMLGS 53 |
| Macaca fascicularis | Query 8 SGDRLSGAAARGDVQEVR#RLLHRELVHPDALNRFGKTALQVMMFGS 53 | + |+ ||||| |++||#+|| |+ +||||+ |+|||| || Sbjct 14 SDEGLTSAAARGLVEKVR#QLLEAG-ADPNGVNRFGRRAIQVMMMGS 58 |
| Sus scrofa | Query 8 SGDRLSGAAARGDVQEVR#RLLHRELVHPDALNRFGKTALQVMMFGST 54 | | |+ ||||| |||# || + +| ||+|+| +|||| ||| Sbjct 4 SADWLASAAARGREGEVR#ALLEAGAL-ANAPNRYGRTPIQVMMMGST 49 |
| Pan troglodytes | Query 8 SGDRLSGAAARGDVQEVR#RLLHRELVHPDALNRFGKTALQVMMFGS 53 | | |+ ||||| |+|||# || + |+| | +|+ +|||| || Sbjct 213 SADWLATAAARGRVEEVR#ALLEAGAL-PNAPNSYGRRPIQVMMMGS 257 |
| human, brain tumors, Peptide, 156 aa | Query 8 SGDRLSGAAARGDVQEVR#RLLHRELVHPDALNRFGKTALQVMMFGS 53 | | |+ ||||| |+|||# || + |+| | +|+ +|||| || Sbjct 12 SADWLATAAARGRVEEVR#ALL-EAVALPNAPNSYGRRPIQVMMMGS 56 |
| Danio rerio | Query 10 DRLSGAAARGDVQEVR#RLLHRELVHPDALNRFGKTALQVMMFG 52 |||| ||| ||+ || #+ | |+ + |++|+|||||| | Sbjct 8 DRLSTAAAIGDLMEVE#QTLQSN-VNVNEKNKYGRTALQVMKLG 49 |
| Xiphophorus helleri | Query 10 DRLSGAAARGDVQEVR#RLLHRELVHPDALNRFGKTALQVMMFGST 54 | |+ |||+| || # || + + +| ||+ |+|||| ||+ Sbjct 5 DELTTAAAKGHTAEVE#ALLLQG-APVNGVNSFGRRAIQVMMMGSS 48 |
| Mus spretus | Query 8 SGDRLSGAAARGDVQEVR#RLLHRELVHPDALNRFGKTALQ 47 + |||+ |||+| | +||# || | | | | ||+| +| Sbjct 4 AADRLARAAAQGRVHDVR#ALLEAG-VSPKAPNSFGRTPIQ 42 |
| Apis mellifera | Query 15 AAARGDVQEVR#RLLHRELVHPDALNRFGKTAL 46 |||| |+ |||#||| ++ |+||+ | | ||| Sbjct 68 AAARNDIDEVR#RLL-KKGVNPDSTNEDGLTAL 98 |
| Tribolium castaneum | Query 15 AAARGDVQEVR#RLLHRELVHPDALNRFGKTAL 46 |||| |+ |||#||| + | ||+ | | ||| Sbjct 69 AAARNDIDEVR#RLLMKG-VSPDSCNEDGLTAL 99 |
| Canis lupus familiaris | Query 10 DRLSGAAARGDVQEVR#RLLHRELVHPDALNRFGKTALQVM 49 ||| || ||||++| # +| ++ ++| |+ |++| |+ Sbjct 38 DRLMKAAERGDVEKVS#SILAKKGINPGKLDVEGRSAFHVV 77 |
| Populus tremula x Populus tremuloides | Query 8 SGDRLSGAAARGDVQEVR#RLLHRELVHPDALNRFGKTALQVMM 50 +|| | || + |+ ++# || + | + |+ +| ||||||| | Sbjct 623 AGDLLCTAAKQNDLMVMK#ELLKQGL-NVDSKDRHGKTALQVAM 664 |
| Hyperthermus butylicus DSM 5456 | Query 6 VRSGDRLSGAAARGDVQEVR#RLLHRELVHPDALNR 40 ++ ||++|| |+||| # + |+ || || | Sbjct 127 LKPGDKVSGGDVLGEVQETS#LITHKVLVPPDVHGR 161 |
* References
None.