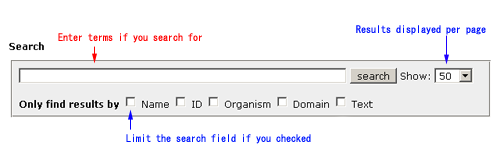
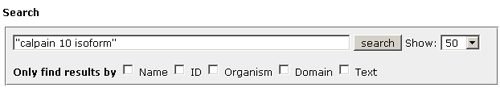
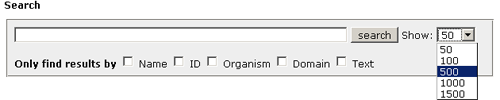
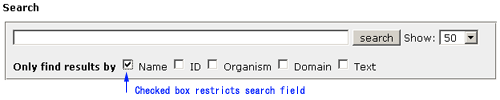
Introduction
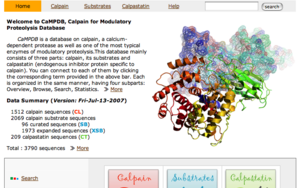
CaMPDB is a database on calpain, a calcium-dependent protease as well as one of the most typical enzymes of modulatory proteolysis. This
database consists of three parts: calpain, its substrates and calpastatin (endogenous inhibitor protein specific to calpain).
You can go to each of them by clicking the corresponding term provided in the above bar (or the pixtured boxes in the toppage). Each part is organized in the same manner, having four subparts: Overview, Browse, Search, Statistics. This page helps you to use these contents more effectively.
Calpain
Overview
This page gives you a detailed latest review with a lot of colored figures. It starts with Introduction, followed by four sections and two appendices:
- 1. History and nomenclature
- 2. Structure and functions of conventional calpains
- 3. Calpain superfamily and its members
- 4. References
- A1. Glossary
- A2. Crosseye view of m-calpain
In default setting, the above contents are hidden except
Introduction. All sections can be revealed by clicking "open" and can
be hided by clicking "close". Each section can be shown by clicking
its title and be hided by clicking it again.
- Reveal all sections
- Click "open" from the "All sections" menu
- Click "close" from the "All sections" menu if you want

- Reveal one section
- Click "section title"
- Click "section title" again if you want to close

Statistics
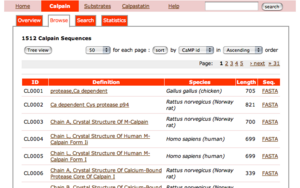
This page shows the classification of calpain sequences.
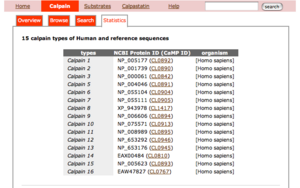
- The first table shows the reference sequences of 15 calpain classes.
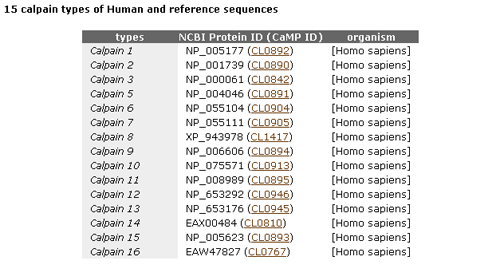
- The second table shows, for each species, the number of sequences which are classified into 15 classes.
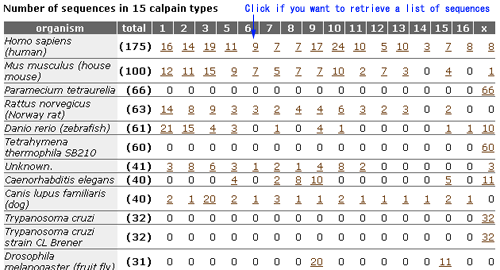
- The classification procedure is described at the bottom of this page.

- You can retreive a list of sequences by clicking the number of this table.
Calpain Viewer
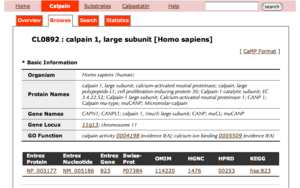
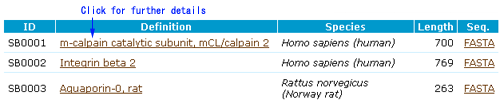
- The title of each entry shows its CaMPID and definition
- The first two letters of CaMPID shows the entry type: "CL" indicates a calpain sequence.

- Basic information has the following five parts with gene IDs of other major databases: Organism, Protein names, Gene names, Gene Locus, GO Function.
- By clicking a gene ID, you can see the information of the corresponding database.
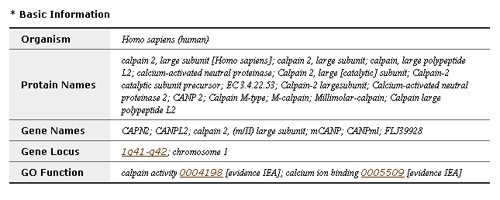
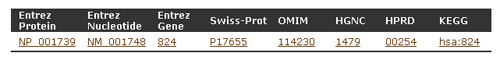
- Information from OMIM has three parts: Description, Function and Reference.

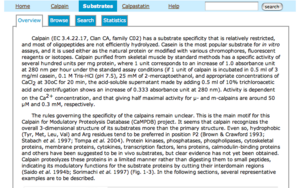
- Structure Information has the following four parts: Primary Information, Domain Information, Sequence Information and 3D Information.
- Primary Information of each entry has its sequence length, average mass and monoisotopic mass, which are computed by using a table given by ExPASy.

- Domain Information of each entry has the links to the corresponding protein of four major domain databases: interpro, pfam, smart and prosite. In addition to this, you can see the colored domain structure which is computed by using HMMER.
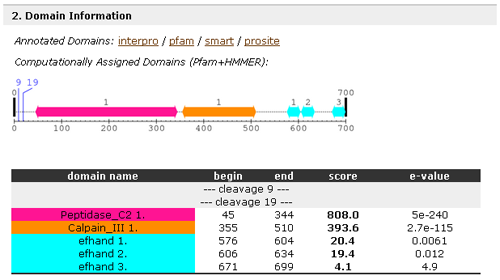
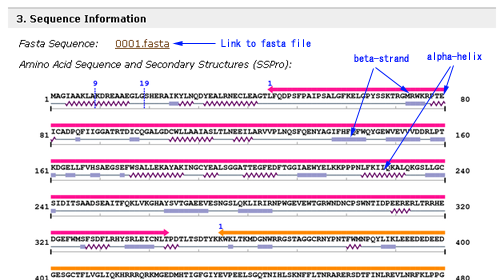
- Sequence Information of each entry has the link to its amino acid sequence in the fasta format and its actual amino acid sequence and secondary structures which are predicted by using a software called SSPro4 developed by P. Baldi's group. In addition to them, the domain structure assigned in the above is also shown.
- 3D information (if any) has the link to the PDB (Protein Data Bank) database.
- References are those related with each entry and retrieved from PubMed. You can click "More" to see more references.

substrates
Overview
This page gives you a detailed latest review with a lot of colored figures. It starts with Introduction, followed by five sections:
- 1. Activity of calpain
- 2. Substrates of calpains in brain function
- 3. p53 and calpain
- 4. Calpain and apoptosis
- 5. References
In default setting, the above contents are hidden except
Introduction. All sections can be revealed by clicking "open" and can
be hided by clicking "close". Each section can be shown by clicking
its title and be hided by clicking it again.
- Reveal all sections
- Click "open" from the "All sections" menu
- Click "close" from the "All sections" menu if you want

- Reveal one section
- Click "section title"
- Click "section title" again if you want to close

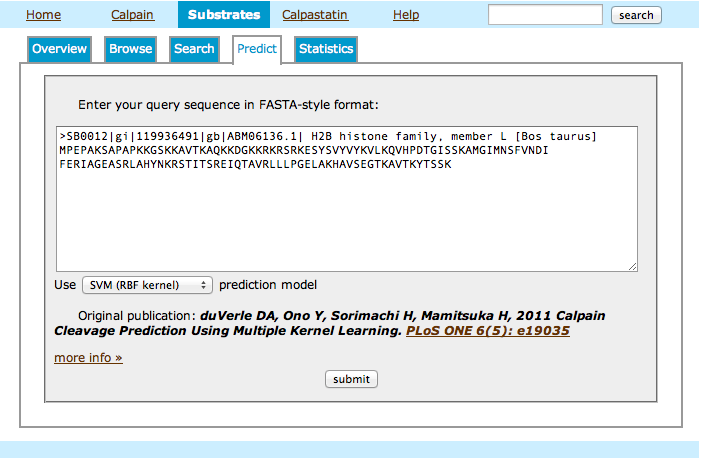
Prediction Tool
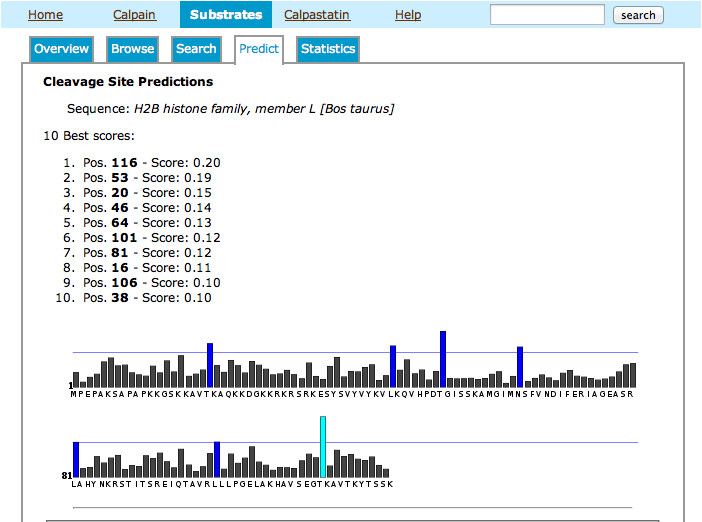
This page allows you to run a simple PSSM-based predictor on a putative substrate sequence in order to evaluate likelihood of cleavage sites by calpain.
- Explanation on the prediction tool usage:
- Enter your query sequence, either in FASTA format or as a raw string of single-letter-coded amino acids.
- Along with scores for the best 10 positions in the sequence, you will get a bar chart displaying all positive scores along the sequence. Top score is indicated in cyan, while scores above the top percentile are shown in dark blue.
- Example of prediction results:
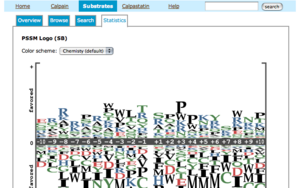
Statistics
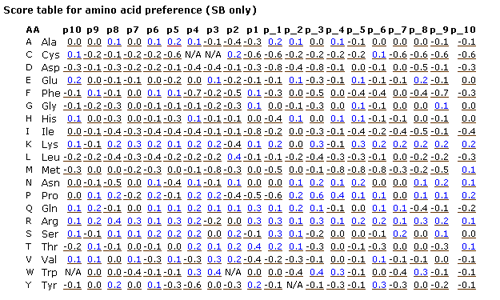
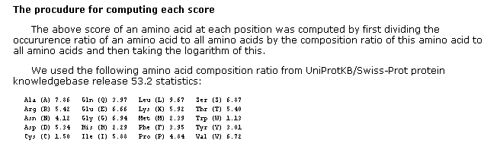
This page shows amino acid preferences of twenty positions near the proteolytic sites. They are computed in two different datasets, i.e. the
sequences from literature only (SB) and not only them but also the sequences computationally retrieved from the database (SB+XSB).
- the sequences from literature only (SB)
- the sequences from not only SB but also computationally retrieved(SB+XSP)

- The computation procedure is described at the bottom of this page.
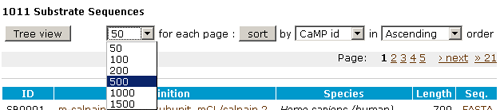
- You can retreive a list of sequences by clicking the number of each
table.
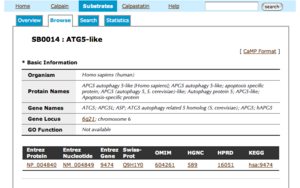
Substrates Viewer
- The title of each entry shows its CaMPID and definition
- The first two letters of CaMPID shows the entry type: "SB" indicates a substrate sequence derived from literature and "XSB" means a substrate sequence computationally retrieved from a database based on a SB sequence.
- Basic information has the following five parts with gene IDs of other major databases: Organism, Protein names, Gene names, Gene Locus, GO Function.
- By clicking a gene ID, you can see the information of the corresponding database.


- Information from OMIM has three parts: Description, Function and Reference.

- Structure Information has the following four parts: Primary Information, Domain Information, Sequence Information and 3D Information.
- Primary Information of each entry has its sequence length, average mass and monoisotopic mass, which are computed by using a table given by ExPASy.

- Domain Information of each entry has the links to the corresponding protein of four major domain databases: interpro, pfam, smart and prosite. In addition to this, you can see the colored domain structure which is computed by using HMMER.

- Sequence Information of each entry has the link to its amino acid sequence in the fasta format and its actual amino acid sequence and secondary structures which are predicted by using a software called SSPro4 developed by P. Baldi's group. In addition to them, the domain structure assigned in the above is also shown.

- 3D information (if any) has the link to the PDB (Protein Data Bank) database.

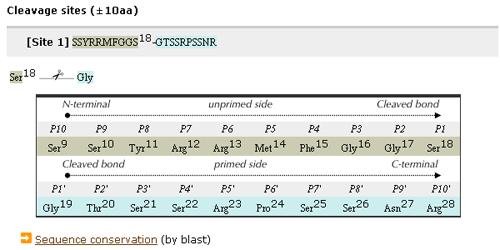
- Cleavage Information consists of three parts: the number of cleavages, their references and the detail of each cleavage site.
- The number of cleavage sites and their references are shown.

- The detail shows the twenty (upstream and downstream) amino acids around the cleavage site.
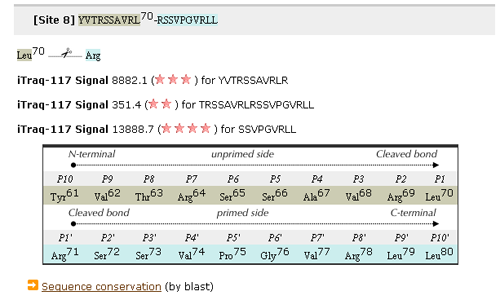
- In addition to this information, the result of our original experiment on the proteolysis by calpain on the peptide of the twenty amino acids is shown (if any). The result is given by a score indicating the strength of proteolysis. For example, "iTraq-117 Signal 8882.1 (***) for YVTRSSAVRLR" indicates that the peptide YVTRSSAVRLRSSVPOVRLL was split into YVTRSSAVRLR and SSVPOVRLL with a score of 8882.1. The larger the score is, the stronger the proteolysis is. Accompanying with the original score, stars are shown by taking the logarithm of the original score, because of the noisiness of experiments.
- References are those related with each entry and retrieved from PubMed. You can click "More" to see more references.

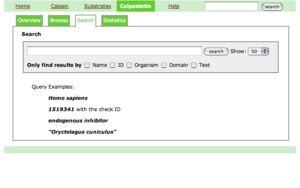
calpastatin
Overview
This page gives you a detailed latest review with a lot of colored figures. It starts with Introduction, followed by three sections:
- 1. Structure of calpastatin
- 2. Other synthetic inhibitors of calpain
- 3. References
In default setting, the above contents are hidden except
Introduction. All sections can be revealed by clicking "open" and can
be hided by clicking "close". Each section can be shown by clicking
its title and be hided by clicking it again.
- Reveal all sections
- Click "open" from the "All sections" menu
- Click "close" from the "All sections" menu if you want

- Reveal one section
- Click "section title"
- Click "section title" again if you want to close

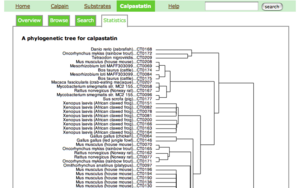
Statistics
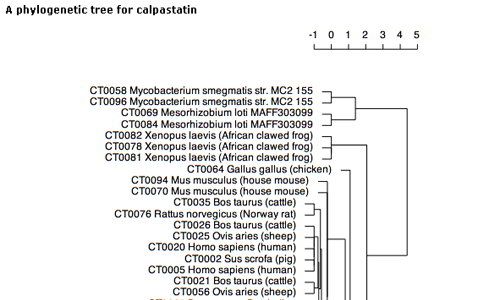
This page shows a phylogenetic tree of calpain sequences.
- The computation procedure is shown at the bottom of this page.

- You can click each sequence in the tree to see the detail of the corresponding entry.
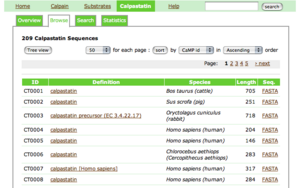
Calpastatin viewer
- The title of each entry shows its CaMPID and definition
- The first two letters of CaMPID shows the entry type: "CT" indicates a calpastatin sequence.

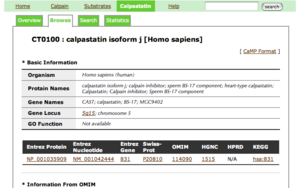
- Basic information has the following five parts with gene IDs of other major databases: Organism, Protein names, Gene names, Gene Locus, GO Function.
- By clicking a gene ID, you can see the information of the corresponding database.


- Information from OMIM has three parts: Description, Function and Reference.

- Structure Information has the following four parts: Primary Information, Domain Information, Sequence Information and 3D Information.
- Primary Information of each entry has its sequence length, average mass and monoisotopic mass, which are computed by using a table given by ExPASy.

- Domain Information of each entry has the links to the corresponding protein of four major domain databases: interpro, pfam, smart and prosite. In addition to this, you can see the colored domain structure which is computed by using HMMER.

- Sequence Information of each entry has the link to its amino acid sequence in the fasta format and its actual amino acid sequence and secondary structures which are predicted by using a software called SSPro4 developed by P. Baldi's group. In addition to them, the domain structure assigned in the above is also shown.

- 3D information (if any) has the link to the PDB (Protein Data Bank) database.

- References are those related with each entry and retrieved from PubMed. You can click "More" to see more references.

|