SB0002 : Integrin [beta]2
[ CaMP Format ]
* Basic Information
| Organism | Homo sapiens (human) |
| Protein Names | integrin beta-2 precursor [Homo sapiens]; integrin beta-2 precursor; integrin beta-2; leukocyte-associated antigens CD18/11A, CD18/11B, CD18/11C; complement receptor C3 beta-subunit; cell surface adhesion glycoprotein (LFA-1/CR3/P150,959 beta subunit precursor); leukocyte cell adhesion molecule CD18; integrin beta chain, beta 2; Integrin beta-2; Cell surface adhesion glycoproteins LFA-1/CR3/p150,95 subunit beta; Complement receptor C3 subunit beta |
| Gene Names | ITGB2; CD18, MFI7; CD18; MFI7; integrin, beta 2 (complement component 3 receptor 3 and 4 subunit) |
| Gene Locus | 21q22.3; chromosome 21 |
| GO Function | Not available |
* Information From OMIM
Description: The leukocyte cell adhesion molecule belongs to the class of cell membrane glycoproteins known as integrins, which are alpha-beta heterodimers. The alpha subunits vary in size from 120 to 180 kD and each is noncovalently associated with a beta subunit (90 to 110 kD). There are 8 known beta subunits and 14 known alpha subunits. Although the alpha and beta subunits could in theory associate to give more than 100 integrin heterodimers, the diversity is restricted and different combinations are associated with different cell types (Hynes, 1992).
Function: By quantitative fluorescence flow cytometric analysis, Taylor et al. (1988) showed that the expression of CD18 was increased in lymphoblastoid cells from persons with Down syndrome, consistent with the location of the gene on chromosome 21.
* Structure Information
1. Primary Information
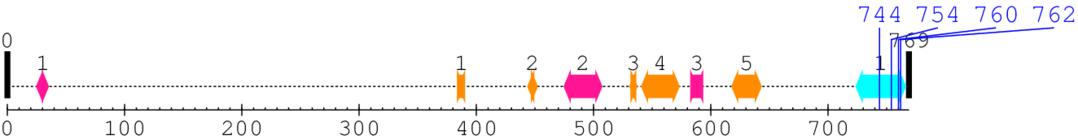
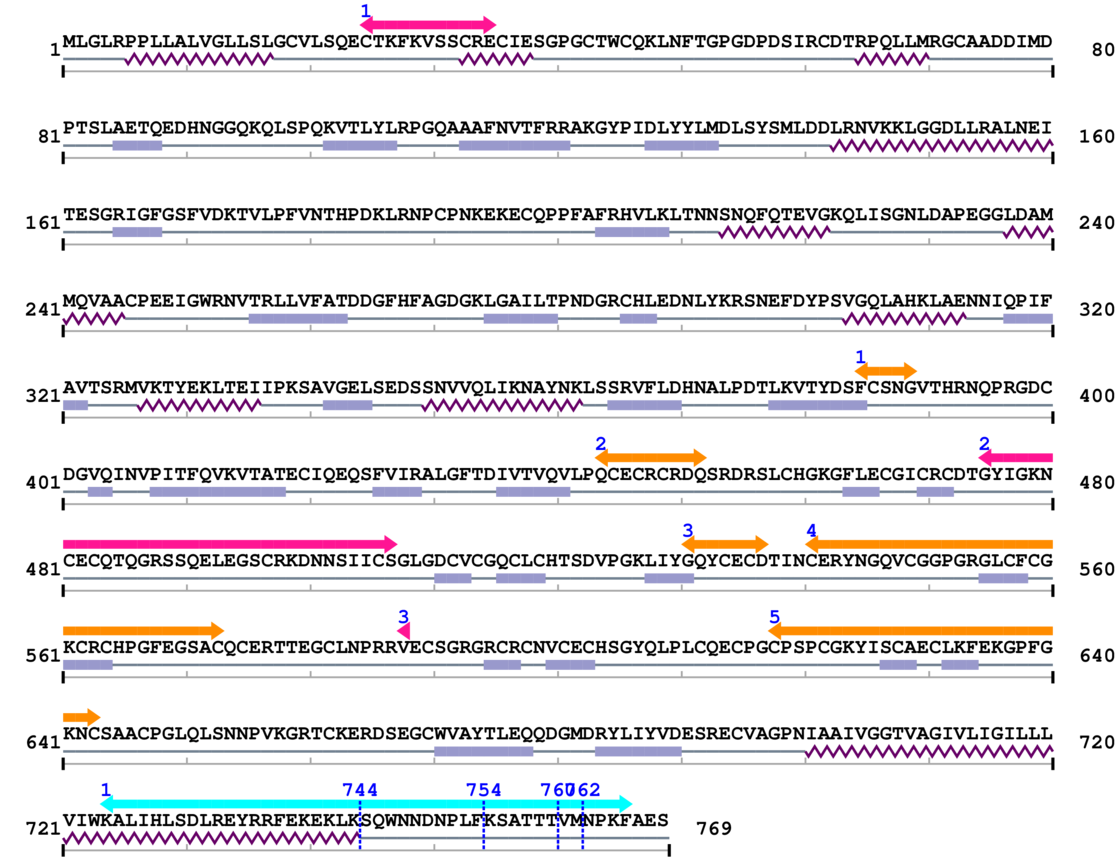
Length: 769 aa
Average Mass: 84.781 kDa
Monoisotopic Mass: 84.726 kDa
2. Domain Information
Annotated Domains: interpro / pfam / smart / prosite
Computationally Assigned Domains (Pfam+HMMER):
| domain name | begin | end | score | e-value |
|---|---|---|---|---|
| Integrin beta tail domain 1. | 25 | 35 | 10.0 | 2.8 |
| EGF-like domain 1. | 385 | 389 | 7.0 | 0.2 |
| EGF-like domain 2. | 444 | 452 | 17.0 | 2.7 |
| Integrin beta tail domain 2. | 475 | 507 | 16.0 | 13.0 |
| EGF-like domain 3. | 531 | 537 | 19.0 | 0.3 |
| EGF-like domain 4. | 541 | 573 | 1.0 | 17.1 |
| Integrin beta tail domain 3. | 588 | 588 | 46.0 | 12.4 |
| EGF-like domain 5. | 618 | 643 | 14.0 | 8.4 |
| Integrin beta cytoplasmic domain 1. | 724 | 766 | 1.0 | 0.5 |
| --- cleavage 744 (inside Integrin beta cytoplasmic domain 724..766) --- | ||||
| --- cleavage 754 (inside Integrin beta cytoplasmic domain 724..766) --- | ||||
| --- cleavage 760 (inside Integrin beta cytoplasmic domain 724..766) --- | ||||
| --- cleavage 762 (inside Integrin beta cytoplasmic domain 724..766) --- | ||||
3. Sequence Information
Fasta Sequence: SB0002.fasta
Amino Acid Sequence and Secondary Structures (PsiPred):
4. 3D Information
Known Structures in PDB: 1JX3 (Model; -; A=126-364), 1L3Y (NMR; -; A=535-574), 1YUK (X-ray; 180 A; A=23-125, B=365-482), 2JF1 (X-ray; 220 A; T=735-769), 2P26 (X-ray; 175 A; A=23-535), 2P28 (X-ray; 220 A; A=23-122, B=362-574), 2V7D (X-ray; 250 A; P/Q/R/S=755-764), 3K6S (X-ray; 350 A; B/D/F/H=23-699), 3K71 (X-ray; 395 A; B/D/F/H=23-699), 3K72 (X-ray; 370 A; B/D=23-699), 4NEH (X-ray; 275 A; B=23-695), 4NEN (X-ray; 290 A; B=23-696)
* Cleavage Information
4 [sites] cleaved by Calpain 2
Source Reference: [PubMed ID: 10571053] Pfaff M, Du X, Ginsberg MH, Calpain cleavage of integrin beta cytoplasmic domains. FEBS Lett. 1999 Oct 22;460(1):17-22.
Cleavage sites (±10aa)
[Site 1] YRRFEKEKLK744-SQWNNDNPLF
Lys744  Ser
Ser
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Tyr735 | Arg736 | Arg737 | Phe738 | Glu739 | Lys740 | Glu741 | Lys742 | Leu743 | Lys744 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Ser745 | Gln746 | Trp747 | Asn748 | Asn749 | Asp750 | Asn751 | Pro752 | Leu753 | Phe754 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| IGILLLVIWKALIHLSDLREYRRFEKEKLKSQWNNDNPLFKSATTTVMNPKFAES |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | Homo sapiens | 120.00 | 10 | leukocyte adhesion protein beta-subunit precursor |
| 2 | synthetic construct | 120.00 | 3 | integrin, beta 2 (antigen CD18 (p95), lymphocyte |
| 3 | Pan troglodytes | 120.00 | 2 | integrin, beta 2 |
| 4 | Homo sapiens | 120.00 | 1 | integrin, beta 2 precursor |
| 5 | Canis familiaris | 112.00 | 10 | AF181965_1 B-2 integrin |
| 6 | Sus scrofa | 108.00 | 3 | CD18 leukocyte adhesion molecule |
| 7 | Ovis aries | 104.00 | 2 | antigen CD18 |
| 8 | Ovis canadensis | 104.00 | 1 | ITB2_OVICA Integrin beta-2 precursor (Cell surfac |
| 9 | N/A | 103.00 | 23 | ITB2_MOUSE Integrin beta-2 precursor (Cell surfac |
| 10 | Mus musculus | 103.00 | 12 | integrin beta 2 |
| 11 | Sigmodon hispidus | 103.00 | 1 | AF445415_1 integrin beta-2 precursor |
| 12 | Bos taurus | 102.00 | 7 | integrin, beta 2 |
| 13 | Rattus norvegicus | 101.00 | 5 | Itgb2 protein |
| 14 | Gallus gallus | 67.80 | 3 | integrin, beta 2 |
| 15 | Xenopus laevis | 67.40 | 1 | hypothetical protein LOC379708 |
| 16 | Ictalurus punctatus | 67.00 | 2 | beta-1 integrin |
| 17 | Anopheles gambiae | 61.20 | 1 | integrin beta subunit |
| 18 | Crassostrea gigas | 61.20 | 1 | integrin beta cgh |
| 19 | Anopheles gambiae str. PEST | 61.20 | 1 | integrin beta subunit (AGAP000815-PA) |
| 20 | Mus sp. | 59.30 | 1 | beta 7 integrin |
| 21 | Pseudoplusia includens | 58.20 | 1 | integrin beta 1 |
| 22 | Cyprinus carpio | 56.60 | 2 | CD18 type 2 |
| 23 | Xenopus laevis | 56.60 | 2 | integrin beta-3 subunit |
| 24 | Oryctolagus cuniculus | 55.80 | 1 | glycoprotein IIIa |
| 25 | Tetraodon nigroviridis | 54.70 | 7 | unnamed protein product |
| 26 | Mus musculus | 54.30 | 1 | integrin beta 3 precursor |
| 27 | Canis lupus familiaris | 54.30 | 1 | integrin beta chain, beta 3 |
| 28 | Sus scrofa | 54.30 | 1 | integrin beta chain, beta 3 |
| 29 | mice, Peptide Partial, 680 aa | 54.30 | 1 | beta 3 integrin, GPIIIA |
| 30 | rats, Peptide Partial, 723 aa | 54.30 | 1 | beta 3 integrin, GPIIIA |
| 31 | Danio rerio | 52.80 | 6 | integrin beta 3b |
| 32 | Felis catus | 52.80 | 1 | integrin beta 1 |
| 33 | Pongo pygmaeus | 52.40 | 1 | ITB1_PONPY Integrin beta-1 precursor (Fibronectin |
| 34 | Gallus gallus | 50.80 | 2 | integrin, beta 3 precursor |
| 35 | Oryctolagus cuniculus | 50.80 | 1 | integrin beta 1 |
| 36 | Halocynthia roretzi | 50.10 | 2 | integrin beta Hr2 precursor |
| 37 | Strongylocentrotus purpuratus | 49.70 | 3 | integrin beta L subunit |
| 38 | Drosophila melanogaster | 48.10 | 1 | myospheroid CG1560-PA |
| 39 | Pacifastacus leniusculus | 48.10 | 1 | integrin |
| 40 | Acropora millepora | 46.60 | 1 | integrin subunit betaCn1 |
| 41 | Biomphalaria glabrata | 43.90 | 1 | beta integrin subunit |
| 42 | Schistosoma japonicum | 41.20 | 1 | SJCHGC06221 protein |
| 43 | Caenorhabditis briggsae | 39.70 | 1 | Hypothetical protein CBG03601 |
| 44 | Lytechinus variegatus | 39.30 | 1 | beta-C integrin subunit |
| 45 | Podocoryne carnea | 39.30 | 1 | AF308652_1 integrin beta chain |
| 46 | Caenorhabditis elegans | 38.50 | 1 | Paralysed Arrest at Two-fold family member (pat-3 |
Top-ranked sequences
| organism | matching |
|---|---|
| Homo sapiens | Query 715 IGILLLVIWKALIHLSDLREYRRFEKEKLK#SQWNNDNPLFKSATTTVMNPKFAES 769 ||||||||||||||||||||||||||||||#||||||||||||||||||||||||| Query 715 IGILLLVIWKALIHLSDLREYRRFEKEKLK#SQWNNDNPLFKSATTTVMNPKFAES 769 |
| synthetic construct | Query 715 IGILLLVIWKALIHLSDLREYRRFEKEKLK#SQWNNDNPLFKSATTTVMNPKFAES 769 ||||||||||||||||||||||||||||||#||||||||||||||||||||||||| Query 715 IGILLLVIWKALIHLSDLREYRRFEKEKLK#SQWNNDNPLFKSATTTVMNPKFAES 769 |
| Pan troglodytes | Query 715 IGILLLVIWKALIHLSDLREYRRFEKEKLK#SQWNNDNPLFKSATTTVMNPKFAES 769 ||||||||||||||||||||||||||||||#||||||||||||||||||||||||| Query 715 IGILLLVIWKALIHLSDLREYRRFEKEKLK#SQWNNDNPLFKSATTTVMNPKFAES 769 |
| Homo sapiens | Query 715 IGILLLVIWKALIHLSDLREYRRFEKEKLK#SQWNNDNPLFKSATTTVMNPKFAES 769 ||||||||||||||||||||||||||||||#||||||||||||||||||||||||| Query 715 IGILLLVIWKALIHLSDLREYRRFEKEKLK#SQWNNDNPLFKSATTTVMNPKFAES 769 |
| Canis familiaris | Query 715 IGILLLVIWKALIHLSDLREYRRFEKEKLK#SQWNNDNPLFKSATTTVMNPKFAES 769 |||||| ||||| |||||||++|||||||+#||||||||||||||||||||+|||| Sbjct 710 IGILLLAIWKALTHLSDLREFKRFEKEKLR#SQWNNDNPLFKSATTTVMNPRFAES 764 |
| Sus scrofa | Query 717 ILLLVIWKALIHLSDLREYRRFEKEKLK#SQWNNDNPLFKSATTTVMNPKFAE 768 | |||||| | ||||||||+||||||||#|||||||||||||||||||||||| Sbjct 717 IFLLVIWKVLTHLSDLREYKRFEKEKLK#SQWNNDNPLFKSATTTVMNPKFAE 768 |
| Ovis aries | Query 722 IWKALIHLSDLREYRRFEKEKLK#SQWNNDNPLFKSATTTVMNPKFAES 769 ||||| |||||||| ||||||||#||||||||||||||||||||||||| Sbjct 723 IWKALTHLSDLREYHRFEKEKLK#SQWNNDNPLFKSATTTVMNPKFAES 770 |
| Ovis canadensis | Query 722 IWKALIHLSDLREYRRFEKEKLK#SQWNNDNPLFKSATTTVMNPKFAES 769 ||||| |||||||| ||||||||#||||||||||||||||||||||||| Sbjct 723 IWKALTHLSDLREYHRFEKEKLK#SQWNNDNPLFKSATTTVMNPKFAES 770 |
| N/A | Query 723 WKALIHLSDLREYRRFEKEKLK#SQWNNDNPLFKSATTTVMNPKFAES 769 |||| ||+||||||||||||||#||||||||||||||||||||||||| Sbjct 725 WKALTHLTDLREYRRFEKEKLK#SQWNNDNPLFKSATTTVMNPKFAES 771 |
| Mus musculus | Query 723 WKALIHLSDLREYRRFEKEKLK#SQWNNDNPLFKSATTTVMNPKFAES 769 |||| ||+||||||||||||||#||||||||||||||||||||||||| Sbjct 724 WKALTHLTDLREYRRFEKEKLK#SQWNNDNPLFKSATTTVMNPKFAES 770 |
| Sigmodon hispidus | Query 722 IWKALIHLSDLREYRRFEKEKLK#SQWNNDNPLFKSATTTVMNPKFAES 769 ||||| ||+|||||| |||||||#||||||||||||||||||||||||| Sbjct 722 IWKALTHLTDLREYRHFEKEKLK#SQWNNDNPLFKSATTTVMNPKFAES 769 |
| Bos taurus | Query 723 WKALIHLSDLREYRRFEKEKLK#SQWNNDNPLFKSATTTVMNPKFAES 769 |||| |||||||| ||||||||#||||||||||||||||||||||||| Sbjct 723 WKALTHLSDLREYHRFEKEKLK#SQWNNDNPLFKSATTTVMNPKFAES 769 |
| Rattus norvegicus | Query 723 WKALIHLSDLREYRRFEKEKLK#SQWNNDNPLFKSATTTVMNPKFAES 769 |||| ||+|| |||||||||||#||||||||||||||||||||||||| Sbjct 270 WKALTHLTDLNEYRRFEKEKLK#SQWNNDNPLFKSATTTVMNPKFAES 316 |
| Gallus gallus | Query 731 DLREYRRFEKEKLK#SQWNN-DNPLFKSATTTVMNPKF 766 | |||||||||| |#++|| |||||||||||||||+| Sbjct 733 DRREYRRFEKEKSK#AKWNEADNPLFKSATTTVMNPRF 769 |
| Xenopus laevis | Query 715 IGILLLVIWKALIHLSDLREYRRFEKEKLK#SQWN-NDNPLFKSATTTVMNPKFA 767 ||+ |+||| + + | ||+|||||++ #|+|| |||+ +||||| || |+ Sbjct 717 IGLACLIIWKIITEIKDRNEYQRFEKERMN#SKWNPGHNPLYHNATTTVQNPNFS 770 |
| Ictalurus punctatus | Query 715 IGILLLVIWKALIHLSDLREYRRFEKEKLK#SQWN-NDNPLFKSATTTVMNPKF 766 ||+ || ||| |+ + | ||+ +|||||+ #++|+ +||++||| |||+|||+ Sbjct 752 IGLALLFIWKLLMIIHDRREFAKFEKEKMN#AKWDAGENPIYKSAVTTVVNPKY 804 |
| Anopheles gambiae | Query 716 GILLLVIWKALIHLSDLREYRRFEKEKLK#SQWNN-DNPLFKSATTTVMNPKFA 767 |+ +|++|| | + | ||+ |||||++ #++|+ +||++| |||| || +| Sbjct 783 GMAVLLLWKVLTSIHDRREFARFEKERMM#AKWDTGENPIYKQATTTFKNPTYA 835 |
| Crassostrea gigas | Query 716 GILLLVIWKALIHLSDLREYRRFEKEKLK#SQWNN-DNPLFKSATTTVMNPKFAE 768 ||+||+||| +|| || ||||| + #++|+ +||++| ||+| +|| + + Sbjct 746 GIILLLIWKLFTTISDKRELARFEKEAMN#ARWDTGENPIYKQATSTFVNPTYRQ 799 |
| Anopheles gambiae str. PEST | Query 716 GILLLVIWKALIHLSDLREYRRFEKEKLK#SQWNN-DNPLFKSATTTVMNPKFA 767 |+ +|++|| | + | ||+ |||||++ #++|+ +||++| |||| || +| Sbjct 783 GMAVLLLWKVLTSIHDRREFARFEKERMM#AKWDTGENPIYKQATTTFKNPTYA 835 |
| Mus sp. | Query 715 IGILLLVIWKALIHLSDLREYRRFEKEKLK#SQWNND-NPLFKSATTTVMNPKF 766 +|+ |++ ++ + + | |||||||||+ +# | | |||+||| || +||+| Sbjct 737 VGLGLVLAYRLSVEIYDRREYRRFEKEQQQ#LNWKQDNNPLYKSAITTTVNPRF 789 |
| Pseudoplusia includens | Query 715 IGILLLVIWKALIHLSDLREYRRFEKEKLK#SQWNN-DNPLFKSATTTVMNPKFA 767 +|+ ||++|| | ||+ |||||++ #++|+ +||++| ||+| || +| Sbjct 782 VGLALLMLWKMATTSHDRREFARFEKERMM#AKWDTGENPIYKQATSTFKNPTYA 835 |
| Cyprinus carpio | Query 724 KALIHLSDLREYRRFEKEKLK#SQWNNDNPLFKSATTTVMNPKFA 767 ||+++ |||||++||||++ # + + ||||++||||| || |+ Sbjct 721 KAVLYASDLREWKRFEKDRKH#EKTSGTNPLFQNATTTVQNPTFS 764 |
| Xenopus laevis | Query 715 IGILLLVIWKALIHLSDLREYRRFEKEKLK#SQWNN-DNPLFKSATTTVMN 763 ||++ |+||| || + | ||+ +||+|+ |#++|+ |||+| ||+| | Sbjct 731 IGLVALLIWKLLITIHDRREFAKFEEERAK#AKWDTAHNPLYKGATSTFTN 780 |
| Oryctolagus cuniculus | Query 715 IGILLLVIWKALIHLSDLREYRRFEKEKLK#SQWNN-DNPLFKSATTTVMN 763 || ||+||| || + | +|+ +||+|+ +#++|+ +|||+| ||+| | Sbjct 733 IGFALLLIWKLLITIHDRKEFAKFEEERAR#AKWDTANNPLYKEATSTFTN 782 |
| Tetraodon nigroviridis | Query 715 IGILLLVIWKALIHLSDLREYRRFEKEKLK#SQWNN-DNPLFKSATTTVMNPKF 766 +|+ |+||| || + | ||+ +||+|+ +#++|+ |||+| ||+| | | Sbjct 748 LGLAALLIWKLLITIHDRREFAKFEEERAR#AKWDTGHNPLYKGATSTFTNITF 800 |
| Mus musculus | Query 715 IGILLLVIWKALIHLSDLREYRRFEKEKLK#SQWNN-DNPLFKSATTTVMN 763 ||+ |+||| || + | +|+ +||+|+ +#++|+ +|||+| ||+| | Sbjct 732 IGLATLLIWKLLITIHDRKEFAKFEEERAR#AKWDTANNPLYKEATSTFTN 781 |
| Canis lupus familiaris | Query 715 IGILLLVIWKALIHLSDLREYRRFEKEKLK#SQWNN-DNPLFKSATTTVMN 763 ||+ |+||| || + | +|+ +||+|+ +#++|+ +|||+| ||+| | Sbjct 729 IGLATLLIWKLLITIHDRKEFAKFEEERAR#AKWDTANNPLYKEATSTFTN 778 |
| Sus scrofa | Query 715 IGILLLVIWKALIHLSDLREYRRFEKEKLK#SQWNN-DNPLFKSATTTVMN 763 ||+ |+||| || + | +|+ +||+|+ +#++|+ +|||+| ||+| | Sbjct 729 IGLATLLIWKLLITIHDRKEFAKFEEERAR#AKWDTANNPLYKEATSTFTN 778 |
| mice, Peptide Partial, 680 aa | Query 715 IGILLLVIWKALIHLSDLREYRRFEKEKLK#SQWNN-DNPLFKSATTTVMN 763 ||+ |+||| || + | +|+ +||+|+ +#++|+ +|||+| ||+| | Sbjct 629 IGLATLLIWKLLITIHDRKEFAKFEEERAR#AKWDTANNPLYKEATSTFTN 678 |
| rats, Peptide Partial, 723 aa | Query 715 IGILLLVIWKALIHLSDLREYRRFEKEKLK#SQWNN-DNPLFKSATTTVMN 763 ||+ |+||| || + | +|+ +||+|+ +#++|+ +|||+| ||+| | Sbjct 668 IGLATLLIWKLLITIHDRKEFAKFEEERAR#AKWDTANNPLYKEATSTFTN 717 |
| Danio rerio | Query 715 IGILLLVIWKALIHLSDLREYRRFEKEKLK#SQWNN-DNPLFKSATTTVMN 763 +|+ |+||| || + | ||+ +||+|+ +#++| |||+| ||+| | Sbjct 734 LGLAALLIWKLLITIHDRREFAKFEEERAR#AKWETGHNPLYKGATSTFTN 783 |
| Felis catus | Query 726 LIHLSDLREYRRFEKEKLK#SQWNN-DNPLFKSATTTVMNPKF 766 +|| | ||+ +|||||+ #++|+ +||++||| |||+|||+ Sbjct 756 IIH--DTREFAKFEKEKMN#AKWDTGENPIYKSAVTTVVNPKY 795 |
| Pongo pygmaeus | Query 726 LIHLSDLREYRRFEKEKLK#SQWNN-DNPLFKSATTTVMNPKF 766 +|| | ||+ +|||||+ #++|+ +||++||| |||+|||+ Sbjct 756 IIH--DRREFAKFEKEKMN#AKWDTGENPIYKSAVTTVVNPKY 795 |
| Gallus gallus | Query 723 WKALIHLSDLREYRRFEKEKLK#SQWNN-DNPLFKSATTTVMN 763 || || + | ||+ |||+|| +#++|+ +|||+| ||+| | Sbjct 733 WKLLITIHDRREFARFEEEKAR#AKWDTGNNPLYKEATSTFTN 774 |
| Oryctolagus cuniculus | Query 726 LIHLSDLREYRRFEKEKLK#SQWN-NDNPLFKSATTTVMNPKF 766 +|| | ||+ +|||||+ #++|+ +||++||| |||+|||+ Sbjct 21 IIH--DRREFAKFEKEKMN#AKWDTGENPIYKSAVTTVVNPKY 60 |
| Halocynthia roretzi | Query 715 IGILLLVIWKALIHLSDLREYRRFEKEKLK#SQWNN-DNPLFKSATTTVMNPKFA 767 |||+ |+||| + | | ||| +|+||+ # +| +||++ +|++ || | Sbjct 779 IGIIALIIWKVIQTLRDKREYEKFKKEEDL#RKWTKGENPVYVNASSKFDNPMFG 832 |
| Strongylocentrotus purpuratus | Query 717 ILLLVIWKALIHLSDLREYRRFEKEKLK#SQWN-NDNPLFKSATTTVMNPKFAE 768 ++| |+ ++ | ||| ++| + |#+||+ +|||++||+||| || + + Sbjct 750 MILARAWRLYTYVQDKREYAQWENDCKK#AQWDQSDNPIYKSSTTTFKNPTYGK 802 |
| Drosophila melanogaster | Query 729 LSDLREYRRFEKEKLK#SQWNN-DNPLFKSATTTVMNPKFA 767 + | ||+ |||||++ #++|+ +||++| ||+| || +| Sbjct 805 IHDRREFARFEKERMN#AKWDTGENPIYKQATSTFKNPMYA 844 |
| Pacifastacus leniusculus | Query 731 DLREYRRFEKE-KLK#SQWNNDNPLFKSATTTVMNPKFAES 769 | ||| +|||| || #| +|||+||| || || ||+| Sbjct 761 DRREYAKFEKERKLP#SGKRAENPLYKSAKTTFQNPAFAQS 800 |
| Acropora millepora | Query 724 KALIHLSDLREYRRFEKEKLK#SQWNND-NPLFKSATTTVMNPKFA 767 | | + | ||++||+|++ #|+| + |||+++| || || +| Sbjct 744 KGLFTMVDRIEYQKFERERMH#SKWTREKNPLYQAAKTTFENPTYA 788 |
| Biomphalaria glabrata | Query 729 LSDLREYRRFEKEKLK#SQWNN-DNPLFKSATTTVMNPKFA 767 + | ||+ +||||+ #++|+ +||++| ||+| || + Sbjct 747 IHDTREFAKFEKERQN#AKWDTGENPIYKQATSTFKNPTYG 786 |
| Schistosoma japonicum | Query 723 WKALIHLSDLREYRRFEKEKLK#SQWN-NDNPLFKSATTTVMNPKFAES 769 +| +| + | || |+++ # +| +||+|+| || |+|| | |+ Sbjct 239 YKLVITIDDRRELANFKQQGEN#MRWEMAENPIFESPTTNVLNPTFEEN 286 |
| Caenorhabditis briggsae | Query 722 IWKALIHLSDLREYRRFEKEKLK#SQWN-NDNPLFKSATTTVMNPKFA 767 +|| | | | || +| |+| #++|+ |+||++| |||| || +| Sbjct 760 LWKLLTVLHDRAEYAKFNNERLM#AKWDTNENPIYKQATTTFKNPVYA 806 |
| Lytechinus variegatus | Query 728 HLSDLREYRRFEKEKLK#SQWN-NDNPLFKSATTTVMNPKF 766 ++ | ||++ ||||+ #+ | +||++| +|+ || + Sbjct 764 YIHDRREFQNFEKERAN#ATWEGGENPIYKPSTSVFKNPTY 803 |
| Podocoryne carnea | Query 715 IGILLLVIWKALIHLSDLREYRRFEKEKLK#SQWNN-DNPLFKSATTTVMNPKFA 767 ||+ ||+||| | + | ||+ +|||++ # +|++ +||++| ||+| || + Sbjct 726 IGLALLLIWKLLATIQDRREFAKFEKDRQN#PKWDSGENPIYKKATSTFQNPMYG 779 |
| Caenorhabditis elegans | Query 722 IWKALIHLSDLREYRRFEKEKLK#SQWN-NDNPLFKSATTTVMNPKFA 767 +|| | | | || | |+| #++|+ |+||++| |||| || +| Sbjct 759 LWKLLTVLHDRSEYATFNNERLM#AKWDTNENPIYKQATTTFKNPVYA 805 |
[Site 2] SQWNNDNPLF754-KSATTTVMNP
Phe754  Lys
Lys
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Ser745 | Gln746 | Trp747 | Asn748 | Asn749 | Asp750 | Asn751 | Pro752 | Leu753 | Phe754 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Lys755 | Ser756 | Ala757 | Thr758 | Thr759 | Thr760 | Val761 | Met762 | Asn763 | Pro764 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| ALIHLSDLREYRRFEKEKLKSQWNNDNPLFKSATTTVMNPKFAES |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | Homo sapiens | 100.00 | 9 | leukocyte adhesion protein beta-subunit precursor |
| 2 | synthetic construct | 100.00 | 2 | integrin, beta 2 (antigen CD18 (p95), lymphocyte |
| 3 | Pan troglodytes | 100.00 | 2 | integrin, beta 2 |
| 4 | Homo sapiens | 100.00 | 1 | integrin, beta 2 precursor |
| 5 | N/A | 97.80 | 19 | ITB2_MOUSE Integrin beta-2 precursor (Cell surfac |
| 6 | Mus musculus | 97.80 | 9 | integrin beta 2 |
| 7 | Bos taurus | 96.30 | 5 | integrin, beta 2 |
| 8 | Ovis aries | 96.30 | 2 | antigen CD18 |
| 9 | Ovis canadensis | 96.30 | 1 | ITB2_OVICA Integrin beta-2 precursor (Cell surfac |
| 10 | Rattus norvegicus | 95.10 | 5 | Itgb2 protein |
| 11 | Sigmodon hispidus | 95.10 | 1 | AF445415_1 integrin beta-2 precursor |
| 12 | Canis familiaris | 94.00 | 9 | AF181965_1 B-2 integrin |
| 13 | Sus scrofa | 94.00 | 3 | CD18 leukocyte adhesion molecule |
| 14 | Gallus gallus | 67.80 | 3 | integrin, beta 2 |
| 15 | Ictalurus punctatus | 55.10 | 2 | CD18 |
| 16 | Cyprinus carpio | 54.70 | 2 | CD18 type 2 |
| 17 | Mus sp. | 54.70 | 1 | beta 7 integrin |
| 18 | Xenopus laevis | 53.50 | 1 | hypothetical protein LOC379708 |
| 19 | Danio rerio | 52.80 | 6 | integrin, beta 1a |
| 20 | Felis catus | 52.80 | 1 | integrin beta 1 |
| 21 | Tetraodon nigroviridis | 52.40 | 6 | unnamed protein product |
| 22 | Xenopus laevis | 52.40 | 2 | integrin beta-1 subunit |
| 23 | Pongo pygmaeus | 52.40 | 1 | ITB1_PONPY Integrin beta-1 precursor (Fibronectin |
| 24 | Oryctolagus cuniculus | 50.80 | 1 | integrin beta 1 |
| 25 | Anopheles gambiae | 50.10 | 1 | integrin beta subunit |
| 26 | Anopheles gambiae str. PEST | 50.10 | 1 | integrin beta subunit (AGAP000815-PA) |
| 27 | Drosophila melanogaster | 48.10 | 1 | myospheroid CG1560-PA |
| 28 | Pacifastacus leniusculus | 48.10 | 1 | integrin |
| 29 | Pseudoplusia includens | 47.40 | 1 | integrin beta 1 |
| 30 | Crassostrea gigas | 46.60 | 1 | integrin beta cgh |
| 31 | Strongylocentrotus purpuratus | 45.40 | 3 | integrin beta L subunit |
| 32 | Gallus gallus | 45.10 | 2 | integrin, beta 3 precursor |
| 33 | Acropora millepora | 44.70 | 1 | integrin subunit betaCn1 |
| 34 | Caenorhabditis briggsae | 44.30 | 1 | Hypothetical protein CBG03601 |
| 35 | Caenorhabditis elegans | 43.50 | 1 | Paralysed Arrest at Two-fold family member (pat-3 |
| 36 | Biomphalaria glabrata | 43.50 | 1 | beta integrin subunit |
| 37 | Podocoryne carnea | 42.00 | 1 | AF308652_1 integrin beta chain |
| 38 | Sus scrofa | 41.60 | 1 | integrin beta chain, beta 3 |
| 39 | Oryctolagus cuniculus | 41.60 | 1 | glycoprotein IIIa |
| 40 | mice, Peptide Partial, 680 aa | 41.60 | 1 | beta 3 integrin, GPIIIA |
| 41 | rats, Peptide Partial, 723 aa | 41.60 | 1 | beta 3 integrin, GPIIIA |
| 42 | Mus musculus | 41.60 | 1 | integrin beta 3 precursor |
| 43 | Canis lupus familiaris | 41.60 | 1 | integrin beta chain, beta 3 |
| 44 | Lytechinus variegatus | 39.30 | 1 | beta-C integrin subunit |
| 45 | Schistosoma japonicum | 38.90 | 1 | SJCHGC06221 protein |
| 46 | Ophlitaspongia tenuis | 35.80 | 1 | integrin subunit betaPo1 |
| 47 | Halocynthia roretzi | 35.00 | 2 | integrin beta Hr2 precursor |
| 48 | Geodia cydonium | 34.70 | 1 | integrin beta subunit |
| 49 | Suberites domuncula | 33.10 | 1 | integrin beta subunit |
| 50 | Bradyrhizobium japonicum USDA 110 | 31.60 | 1 | hypothetical protein bll6129 |
| 51 | Bradyrhizobium sp. BTAi1 | 31.20 | 1 | hypothetical protein BBta_5817 |
| 52 | Drosophila pseudoobscura | 31.20 | 1 | GA14581-PA |
Top-ranked sequences
| organism | matching |
|---|---|
| Homo sapiens | Query 725 ALIHLSDLREYRRFEKEKLKSQWNNDNPLF#KSATTTVMNPKFAES 769 ||||||||||||||||||||||||||||||#||||||||||||||| Query 725 ALIHLSDLREYRRFEKEKLKSQWNNDNPLF#KSATTTVMNPKFAES 769 |
| synthetic construct | Query 725 ALIHLSDLREYRRFEKEKLKSQWNNDNPLF#KSATTTVMNPKFAES 769 ||||||||||||||||||||||||||||||#||||||||||||||| Query 725 ALIHLSDLREYRRFEKEKLKSQWNNDNPLF#KSATTTVMNPKFAES 769 |
| Pan troglodytes | Query 725 ALIHLSDLREYRRFEKEKLKSQWNNDNPLF#KSATTTVMNPKFAES 769 ||||||||||||||||||||||||||||||#||||||||||||||| Query 725 ALIHLSDLREYRRFEKEKLKSQWNNDNPLF#KSATTTVMNPKFAES 769 |
| Homo sapiens | Query 725 ALIHLSDLREYRRFEKEKLKSQWNNDNPLF#KSATTTVMNPKFAES 769 ||||||||||||||||||||||||||||||#||||||||||||||| Query 725 ALIHLSDLREYRRFEKEKLKSQWNNDNPLF#KSATTTVMNPKFAES 769 |
| N/A | Query 725 ALIHLSDLREYRRFEKEKLKSQWNNDNPLF#KSATTTVMNPKFAES 769 || ||+||||||||||||||||||||||||#||||||||||||||| Sbjct 727 ALTHLTDLREYRRFEKEKLKSQWNNDNPLF#KSATTTVMNPKFAES 771 |
| Mus musculus | Query 725 ALIHLSDLREYRRFEKEKLKSQWNNDNPLF#KSATTTVMNPKFAES 769 || ||+||||||||||||||||||||||||#||||||||||||||| Sbjct 726 ALTHLTDLREYRRFEKEKLKSQWNNDNPLF#KSATTTVMNPKFAES 770 |
| Bos taurus | Query 725 ALIHLSDLREYRRFEKEKLKSQWNNDNPLF#KSATTTVMNPKFAES 769 || |||||||| ||||||||||||||||||#||||||||||||||| Sbjct 725 ALTHLSDLREYHRFEKEKLKSQWNNDNPLF#KSATTTVMNPKFAES 769 |
| Ovis aries | Query 725 ALIHLSDLREYRRFEKEKLKSQWNNDNPLF#KSATTTVMNPKFAES 769 || |||||||| ||||||||||||||||||#||||||||||||||| Sbjct 726 ALTHLSDLREYHRFEKEKLKSQWNNDNPLF#KSATTTVMNPKFAES 770 |
| Ovis canadensis | Query 725 ALIHLSDLREYRRFEKEKLKSQWNNDNPLF#KSATTTVMNPKFAES 769 || |||||||| ||||||||||||||||||#||||||||||||||| Sbjct 726 ALTHLSDLREYHRFEKEKLKSQWNNDNPLF#KSATTTVMNPKFAES 770 |
| Rattus norvegicus | Query 725 ALIHLSDLREYRRFEKEKLKSQWNNDNPLF#KSATTTVMNPKFAES 769 || ||+|| |||||||||||||||||||||#||||||||||||||| Sbjct 272 ALTHLTDLNEYRRFEKEKLKSQWNNDNPLF#KSATTTVMNPKFAES 316 |
| Sigmodon hispidus | Query 725 ALIHLSDLREYRRFEKEKLKSQWNNDNPLF#KSATTTVMNPKFAES 769 || ||+|||||| |||||||||||||||||#||||||||||||||| Sbjct 725 ALTHLTDLREYRHFEKEKLKSQWNNDNPLF#KSATTTVMNPKFAES 769 |
| Canis familiaris | Query 725 ALIHLSDLREYRRFEKEKLKSQWNNDNPLF#KSATTTVMNPKFAES 769 || |||||||++|||||||+||||||||||#||||||||||+|||| Sbjct 720 ALTHLSDLREFKRFEKEKLRSQWNNDNPLF#KSATTTVMNPRFAES 764 |
| Sus scrofa | Query 726 LIHLSDLREYRRFEKEKLKSQWNNDNPLF#KSATTTVMNPKFAE 768 | ||||||||+||||||||||||||||||#|||||||||||||| Sbjct 726 LTHLSDLREYKRFEKEKLKSQWNNDNPLF#KSATTTVMNPKFAE 768 |
| Gallus gallus | Query 731 DLREYRRFEKEKLKSQWNN-DNPLF#KSATTTVMNPKF 766 | |||||||||| |++|| |||||#||||||||||+| Sbjct 733 DRREYRRFEKEKSKAKWNEADNPLF#KSATTTVMNPRF 769 |
| Ictalurus punctatus | Query 725 ALIHLSDLREYRRFEKEKLKSQW-NNDNPLF#KSATTTVMNPKF 766 || + ||+|+++|||| + || +||||#++||||| || | Sbjct 726 ALFYFKDLKEWKKFEKEAQRRQWAKGENPLF#QNATTTVANPAF 768 |
| Cyprinus carpio | Query 725 ALIHLSDLREYRRFEKEKLKSQWNNDNPLF#KSATTTVMNPKFA 767 |+++ |||||++||||++ + + ||||#++||||| || |+ Sbjct 722 AVLYASDLREWKRFEKDRKHEKTSGTNPLF#QNATTTVQNPTFS 764 |
| Mus sp. | Query 727 IHLSDLREYRRFEKEKLKSQWNND-NPLF#KSATTTVMNPKF 766 + + | |||||||||+ + | | |||+#||| || +||+| Sbjct 749 VEIYDRREYRRFEKEQQQLNWKQDNNPLY#KSAITTTVNPRF 789 |
| Xenopus laevis | Query 726 LIHLSDLREYRRFEKEKLKSQWN-NDNPLF#KSATTTVMNPKFA 767 + + | ||+|||||++ |+|| |||+# +||||| || |+ Sbjct 728 ITEIKDRNEYQRFEKERMNSKWNPGHNPLY#HNATTTVQNPNFS 770 |
| Danio rerio | Query 726 LIHLSDLREYRRFEKEKLKSQWNN-DNPLF#KSATTTVMNPKF 766 +|| | ||+ +|||||+ ++|+ +||++#||| |||+|||+ Sbjct 756 IIH--DRREFAKFEKEKMNAKWDTGENPIY#KSAVTTVINPKY 795 |
| Felis catus | Query 726 LIHLSDLREYRRFEKEKLKSQWNN-DNPLF#KSATTTVMNPKF 766 +|| | ||+ +|||||+ ++|+ +||++#||| |||+|||+ Sbjct 756 IIH--DTREFAKFEKEKMNAKWDTGENPIY#KSAVTTVVNPKY 795 |
| Tetraodon nigroviridis | Query 726 LIHLSDLREYRRFEKEKLKSQWNN-DNPLF#KSATTTVMNPKF 766 +|| | ||+ +|||||+ ++|+ +||++#||| |||+|||+ Sbjct 749 IIH--DRREFAKFEKEKMNAKWDTGENPIY#KSAVTTVVNPKY 788 |
| Xenopus laevis | Query 726 LIHLSDLREYRRFEKEKLKSQWNN-DNPLF#KSATTTVMNPKF 766 +|| | ||+ +|||||+ ++|+ +||++#||| |||+|||+ Sbjct 756 IIH--DRREFAKFEKEKMNAKWDTGENPIY#KSAVTTVVNPKY 795 |
| Pongo pygmaeus | Query 726 LIHLSDLREYRRFEKEKLKSQWNN-DNPLF#KSATTTVMNPKF 766 +|| | ||+ +|||||+ ++|+ +||++#||| |||+|||+ Sbjct 756 IIH--DRREFAKFEKEKMNAKWDTGENPIY#KSAVTTVVNPKY 795 |
| Oryctolagus cuniculus | Query 726 LIHLSDLREYRRFEKEKLKSQWN-NDNPLF#KSATTTVMNPKF 766 +|| | ||+ +|||||+ ++|+ +||++#||| |||+|||+ Sbjct 21 IIH--DRREFAKFEKEKMNAKWDTGENPIY#KSAVTTVVNPKY 60 |
| Anopheles gambiae | Query 726 LIHLSDLREYRRFEKEKLKSQWNN-DNPLF#KSATTTVMNPKFA 767 | + | ||+ |||||++ ++|+ +||++#| |||| || +| Sbjct 793 LTSIHDRREFARFEKERMMAKWDTGENPIY#KQATTTFKNPTYA 835 |
| Anopheles gambiae str. PEST | Query 726 LIHLSDLREYRRFEKEKLKSQWNN-DNPLF#KSATTTVMNPKFA 767 | + | ||+ |||||++ ++|+ +||++#| |||| || +| Sbjct 793 LTSIHDRREFARFEKERMMAKWDTGENPIY#KQATTTFKNPTYA 835 |
| Drosophila melanogaster | Query 729 LSDLREYRRFEKEKLKSQWNN-DNPLF#KSATTTVMNPKFA 767 + | ||+ |||||++ ++|+ +||++#| ||+| || +| Sbjct 805 IHDRREFARFEKERMNAKWDTGENPIY#KQATSTFKNPMYA 844 |
| Pacifastacus leniusculus | Query 731 DLREYRRFEKE-KLKSQWNNDNPLF#KSATTTVMNPKFAES 769 | ||| +|||| || | +|||+#||| || || ||+| Sbjct 761 DRREYAKFEKERKLPSGKRAENPLY#KSAKTTFQNPAFAQS 800 |
| Pseudoplusia includens | Query 731 DLREYRRFEKEKLKSQWNN-DNPLF#KSATTTVMNPKFA 767 | ||+ |||||++ ++|+ +||++#| ||+| || +| Sbjct 798 DRREFARFEKERMMAKWDTGENPIY#KQATSTFKNPTYA 835 |
| Crassostrea gigas | Query 729 LSDLREYRRFEKEKLKSQWNN-DNPLF#KSATTTVMNPKFAE 768 +|| || ||||| + ++|+ +||++#| ||+| +|| + + Sbjct 759 ISDKRELARFEKEAMNARWDTGENPIY#KQATSTFVNPTYRQ 799 |
| Strongylocentrotus purpuratus | Query 728 HLSDLREYRRFEKEKLKSQWN-NDNPLF#KSATTTVMNPKFAE 768 ++ | ||| ++| + |+||+ +|||++#||+||| || + + Sbjct 761 YVQDKREYAQWENDCKKAQWDQSDNPIY#KSSTTTFKNPTYGK 802 |
| Gallus gallus | Query 726 LIHLSDLREYRRFEKEKLKSQWNN-DNPLF#KSATTTVMN 763 || + | ||+ |||+|| +++|+ +|||+#| ||+| | Sbjct 736 LITIHDRREFARFEEEKARAKWDTGNNPLY#KEATSTFTN 774 |
| Acropora millepora | Query 725 ALIHLSDLREYRRFEKEKLKSQWNND-NPLF#KSATTTVMNPKFA 767 | + | ||++||+|++ |+| + |||+#++| || || +| Sbjct 745 GLFTMVDRIEYQKFERERMHSKWTREKNPLY#QAAKTTFENPTYA 788 |
| Caenorhabditis briggsae | Query 731 DLREYRRFEKEKLKSQWN-NDNPLF#KSATTTVMNPKFA 767 | || +| |+| ++|+ |+||++#| |||| || +| Sbjct 769 DRAEYAKFNNERLMAKWDTNENPIY#KQATTTFKNPVYA 806 |
| Caenorhabditis elegans | Query 731 DLREYRRFEKEKLKSQWN-NDNPLF#KSATTTVMNPKFA 767 | || | |+| ++|+ |+||++#| |||| || +| Sbjct 768 DRSEYATFNNERLMAKWDTNENPIY#KQATTTFKNPVYA 805 |
| Biomphalaria glabrata | Query 729 LSDLREYRRFEKEKLKSQWNN-DNPLF#KSATTTVMNPKFA 767 + | ||+ +||||+ ++|+ +||++#| ||+| || + Sbjct 747 IHDTREFAKFEKERQNAKWDTGENPIY#KQATSTFKNPTYG 786 |
| Podocoryne carnea | Query 729 LSDLREYRRFEKEKLKSQWNN-DNPLF#KSATTTVMNPKFA 767 + | ||+ +|||++ +|++ +||++#| ||+| || + Sbjct 740 IQDRREFAKFEKDRQNPKWDSGENPIY#KKATSTFQNPMYG 779 |
| Sus scrofa | Query 726 LIHLSDLREYRRFEKEKLKSQWNN-DNPLF#KSATTTVMN 763 || + | +|+ +||+|+ +++|+ +|||+#| ||+| | Sbjct 740 LITIHDRKEFAKFEEERARAKWDTANNPLY#KEATSTFTN 778 |
| Oryctolagus cuniculus | Query 726 LIHLSDLREYRRFEKEKLKSQWNN-DNPLF#KSATTTVMN 763 || + | +|+ +||+|+ +++|+ +|||+#| ||+| | Sbjct 744 LITIHDRKEFAKFEEERARAKWDTANNPLY#KEATSTFTN 782 |
| mice, Peptide Partial, 680 aa | Query 726 LIHLSDLREYRRFEKEKLKSQWNN-DNPLF#KSATTTVMN 763 || + | +|+ +||+|+ +++|+ +|||+#| ||+| | Sbjct 640 LITIHDRKEFAKFEEERARAKWDTANNPLY#KEATSTFTN 678 |
| rats, Peptide Partial, 723 aa | Query 726 LIHLSDLREYRRFEKEKLKSQWNN-DNPLF#KSATTTVMN 763 || + | +|+ +||+|+ +++|+ +|||+#| ||+| | Sbjct 679 LITIHDRKEFAKFEEERARAKWDTANNPLY#KEATSTFTN 717 |
| Mus musculus | Query 726 LIHLSDLREYRRFEKEKLKSQWNN-DNPLF#KSATTTVMN 763 || + | +|+ +||+|+ +++|+ +|||+#| ||+| | Sbjct 743 LITIHDRKEFAKFEEERARAKWDTANNPLY#KEATSTFTN 781 |
| Canis lupus familiaris | Query 726 LIHLSDLREYRRFEKEKLKSQWNN-DNPLF#KSATTTVMN 763 || + | +|+ +||+|+ +++|+ +|||+#| ||+| | Sbjct 740 LITIHDRKEFAKFEEERARAKWDTANNPLY#KEATSTFTN 778 |
| Lytechinus variegatus | Query 728 HLSDLREYRRFEKEKLKSQWN-NDNPLF#KSATTTVMNPKF 766 ++ | ||++ ||||+ + | +||++#| +|+ || + Sbjct 764 YIHDRREFQNFEKERANATWEGGENPIY#KPSTSVFKNPTY 803 |
| Schistosoma japonicum | Query 726 LIHLSDLREYRRFEKEKLKSQWN-NDNPLF#KSATTTVMNPKFAES 769 +| + | || |+++ +| +||+|#+| || |+|| | |+ Sbjct 242 VITIDDRRELANFKQQGENMRWEMAENPIF#ESPTTNVLNPTFEEN 286 |
| Ophlitaspongia tenuis | Query 731 DLREYRRFEKEKLKSQWN-NDNPLF#KSATTTVMNPKFAE 768 |+ | |+||+| +++ |+|||+#+||| || + + Sbjct 800 DVVEVRKFEREIKNAKYTKNENPLY#RSATKDYQNPLYGK 838 |
| Halocynthia roretzi | Query 729 LSDLREYRRFEKEKLKSQWNN-DNPLF#KSATTTVMNPKFA 767 | | ||| +|+||+ +| +||++# +|++ || | Sbjct 793 LRDKREYEKFKKEEDLRKWTKGENPVY#VNASSKFDNPMFG 832 |
| Geodia cydonium | Query 731 DLREYRRFEKEKLKSQWN-NDNPLF#KSATTTVMNPKFAE 768 | | ++|| | | ++ |+|||+# |||| || + + Sbjct 840 DYTELKKFENELAKVKYTKNENPLY#HSATTEHKNPIYGQ 878 |
| Suberites domuncula | Query 731 DLREYRRFEKEKLKSQW-NNDNPLF#KSATTTVMNPKFAE 768 |+ | ++|||| |+++ + |||+#+|| || + + Sbjct 726 DVIEVKKFEKELAKTKYPKHQNPLY#RSAAKDYQNPIYGQ 764 |
| Bradyrhizobium japonicum USDA 110 | Query 726 LIHLSDLREYRRFEKEKLKSQWNNDNPLF#KSATTTVMNP 764 |+| +||+| | + + |+||+ + #+ | ++| | Sbjct 70 LVHATDLQEAMRIQSDFLRSQFTSAGDHM#RQMTGSLMQP 108 |
| Bradyrhizobium sp. BTAi1 | Query 726 LIHLSDLREYRRFEKEKLKSQWNNDNPLF#KSATTTVM 762 |+| ++| | | + | |+||+ | #|+ | +| Sbjct 70 LVHATELEEAMRIQSEFLRSQFTNAGEHM#KTITAGIM 106 |
| Drosophila pseudoobscura | Query 727 IHLSDLREYRRFEKEKLKSQWNNDNPLF#KSATTTVMNPK 765 | | ||| |||+|+ | + + +|||+#+ + || Sbjct 722 IRRKDAREYARFEEEQAK-RVSQENPLY#RDPVGRYVVPK 759 |
[Site 3] NPLFKSATTT760-VMNPKFAES
Thr760  Val
Val
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Asn751 | Pro752 | Leu753 | Phe754 | Lys755 | Ser756 | Ala757 | Thr758 | Thr759 | Thr760 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Val761 | Met762 | Asn763 | Pro764 | Lys765 | Phe766 | Ala767 | Glu768 | Ser769 | - |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| DLREYRRFEKEKLKSQWNNDNPLFKSATTTVMNPKFAES |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | N/A | 89.40 | 19 | complement receptor C3 beta-subunit |
| 2 | Mus musculus | 89.40 | 9 | integrin beta 2 |
| 3 | Homo sapiens | 89.40 | 9 | leukocyte adhesion protein beta-subunit precursor |
| 4 | synthetic construct | 89.40 | 2 | integrin, beta 2 (antigen CD18 (p95), lymphocyte |
| 5 | Pan troglodytes | 89.40 | 2 | integrin, beta 2 |
| 6 | Homo sapiens | 89.40 | 1 | integrin, beta 2 precursor |
| 7 | Rattus norvegicus | 87.00 | 5 | Itgb2 protein |
| 8 | Bos taurus | 87.00 | 5 | integrin, beta 2 |
| 9 | Ovis aries | 87.00 | 2 | antigen CD18 |
| 10 | Sigmodon hispidus | 87.00 | 1 | AF445415_1 integrin beta-2 precursor |
| 11 | Ovis canadensis | 87.00 | 1 | ITB2_OVICA Integrin beta-2 precursor (Cell surfac |
| 12 | Sus scrofa | 86.30 | 3 | CD18 leukocyte adhesion molecule |
| 13 | Canis familiaris | 84.30 | 9 | AF181965_1 B-2 integrin |
| 14 | Gallus gallus | 65.90 | 3 | integrin, beta 2 |
| 15 | Mus sp. | 53.90 | 1 | beta 7 integrin |
| 16 | Xenopus laevis | 52.00 | 1 | hypothetical protein LOC379708 |
| 17 | Xenopus laevis | 51.60 | 2 | integrin beta-1 subunit |
| 18 | Felis catus | 51.60 | 1 | integrin beta 1 |
| 19 | Tetraodon nigroviridis | 51.20 | 6 | unnamed protein product |
| 20 | Danio rerio | 50.80 | 6 | integrin, beta 1a |
| 21 | Ictalurus punctatus | 50.80 | 2 | CD18 |
| 22 | Pongo pygmaeus | 50.40 | 1 | ITB1_PONPY Integrin beta-1 precursor (Fibronectin |
| 23 | Cyprinus carpio | 49.70 | 2 | CD18 type 2 |
| 24 | Oryctolagus cuniculus | 49.30 | 1 | integrin beta 1 |
| 25 | Anopheles gambiae | 47.80 | 1 | integrin beta subunit |
| 26 | Pacifastacus leniusculus | 47.80 | 1 | integrin |
| 27 | Anopheles gambiae str. PEST | 47.80 | 1 | integrin beta subunit (AGAP000815-PA) |
| 28 | Pseudoplusia includens | 47.40 | 1 | integrin beta 1 |
| 29 | Drosophila melanogaster | 47.00 | 1 | myospheroid CG1560-PA |
| 30 | Strongylocentrotus purpuratus | 44.30 | 3 | integrin beta L subunit |
| 31 | Crassostrea gigas | 43.10 | 1 | integrin beta cgh |
| 32 | Caenorhabditis briggsae | 42.70 | 1 | Hypothetical protein CBG03601 |
| 33 | Gallus gallus | 42.40 | 2 | integrin, beta 3 precursor |
| 34 | Caenorhabditis elegans | 42.00 | 1 | Paralysed Arrest at Two-fold family member (pat-3 |
| 35 | Acropora millepora | 42.00 | 1 | integrin subunit betaCn1 |
| 36 | Biomphalaria glabrata | 41.60 | 1 | beta integrin subunit |
| 37 | Podocoryne carnea | 40.00 | 1 | AF308652_1 integrin beta chain |
| 38 | mice, Peptide Partial, 680 aa | 38.50 | 1 | beta 3 integrin, GPIIIA |
| 39 | Canis lupus familiaris | 38.50 | 1 | integrin beta chain, beta 3 |
| 40 | Oryctolagus cuniculus | 38.10 | 1 | glycoprotein IIIa |
| 41 | Mus musculus | 38.10 | 1 | integrin beta 3 precursor |
| 42 | Lytechinus variegatus | 38.10 | 1 | beta-C integrin subunit |
| 43 | rats, Peptide Partial, 723 aa | 38.10 | 1 | beta 3 integrin, GPIIIA |
| 44 | Sus scrofa | 38.10 | 1 | integrin beta chain, beta 3 |
| 45 | Schistosoma japonicum | 36.60 | 1 | SJCHGC06221 protein |
| 46 | Ophlitaspongia tenuis | 35.80 | 1 | integrin subunit betaPo1 |
| 47 | Geodia cydonium | 34.70 | 1 | integrin beta subunit |
| 48 | Halocynthia roretzi | 33.10 | 2 | integrin beta Hr1 precursor |
| 49 | Suberites domuncula | 33.10 | 1 | integrin beta subunit |
Top-ranked sequences
| organism | matching |
|---|---|
| N/A | Query 731 DLREYRRFEKEKLKSQWNNDNPLFKSATTT#VMNPKFAES 769 ||||||||||||||||||||||||||||||#||||||||| Query 731 DLREYRRFEKEKLKSQWNNDNPLFKSATTT#VMNPKFAES 769 |
| Mus musculus | Query 731 DLREYRRFEKEKLKSQWNNDNPLFKSATTT#VMNPKFAES 769 ||||||||||||||||||||||||||||||#||||||||| Query 731 DLREYRRFEKEKLKSQWNNDNPLFKSATTT#VMNPKFAES 769 |
| Homo sapiens | Query 731 DLREYRRFEKEKLKSQWNNDNPLFKSATTT#VMNPKFAES 769 ||||||||||||||||||||||||||||||#||||||||| Query 731 DLREYRRFEKEKLKSQWNNDNPLFKSATTT#VMNPKFAES 769 |
| synthetic construct | Query 731 DLREYRRFEKEKLKSQWNNDNPLFKSATTT#VMNPKFAES 769 ||||||||||||||||||||||||||||||#||||||||| Query 731 DLREYRRFEKEKLKSQWNNDNPLFKSATTT#VMNPKFAES 769 |
| Pan troglodytes | Query 731 DLREYRRFEKEKLKSQWNNDNPLFKSATTT#VMNPKFAES 769 ||||||||||||||||||||||||||||||#||||||||| Query 731 DLREYRRFEKEKLKSQWNNDNPLFKSATTT#VMNPKFAES 769 |
| Homo sapiens | Query 731 DLREYRRFEKEKLKSQWNNDNPLFKSATTT#VMNPKFAES 769 ||||||||||||||||||||||||||||||#||||||||| Query 731 DLREYRRFEKEKLKSQWNNDNPLFKSATTT#VMNPKFAES 769 |
| Rattus norvegicus | Query 731 DLREYRRFEKEKLKSQWNNDNPLFKSATTT#VMNPKFAES 769 || |||||||||||||||||||||||||||#||||||||| Sbjct 278 DLNEYRRFEKEKLKSQWNNDNPLFKSATTT#VMNPKFAES 316 |
| Bos taurus | Query 731 DLREYRRFEKEKLKSQWNNDNPLFKSATTT#VMNPKFAES 769 ||||| ||||||||||||||||||||||||#||||||||| Sbjct 731 DLREYHRFEKEKLKSQWNNDNPLFKSATTT#VMNPKFAES 769 |
| Ovis aries | Query 731 DLREYRRFEKEKLKSQWNNDNPLFKSATTT#VMNPKFAES 769 ||||| ||||||||||||||||||||||||#||||||||| Sbjct 732 DLREYHRFEKEKLKSQWNNDNPLFKSATTT#VMNPKFAES 770 |
| Sigmodon hispidus | Query 731 DLREYRRFEKEKLKSQWNNDNPLFKSATTT#VMNPKFAES 769 |||||| |||||||||||||||||||||||#||||||||| Sbjct 731 DLREYRHFEKEKLKSQWNNDNPLFKSATTT#VMNPKFAES 769 |
| Ovis canadensis | Query 731 DLREYRRFEKEKLKSQWNNDNPLFKSATTT#VMNPKFAES 769 ||||| ||||||||||||||||||||||||#||||||||| Sbjct 732 DLREYHRFEKEKLKSQWNNDNPLFKSATTT#VMNPKFAES 770 |
| Sus scrofa | Query 731 DLREYRRFEKEKLKSQWNNDNPLFKSATTT#VMNPKFAE 768 |||||+||||||||||||||||||||||||#|||||||| Sbjct 731 DLREYKRFEKEKLKSQWNNDNPLFKSATTT#VMNPKFAE 768 |
| Canis familiaris | Query 731 DLREYRRFEKEKLKSQWNNDNPLFKSATTT#VMNPKFAES 769 ||||++|||||||+||||||||||||||||#||||+|||| Sbjct 726 DLREFKRFEKEKLRSQWNNDNPLFKSATTT#VMNPRFAES 764 |
| Gallus gallus | Query 731 DLREYRRFEKEKLKSQWNN-DNPLFKSATTT#VMNPKF 766 | |||||||||| |++|| |||||||||||#||||+| Sbjct 733 DRREYRRFEKEKSKAKWNEADNPLFKSATTT#VMNPRF 769 |
| Mus sp. | Query 731 DLREYRRFEKEKLKSQWNND-NPLFKSATTT#VMNPKF 766 | |||||||||+ + | | |||+||| ||# +||+| Sbjct 753 DRREYRRFEKEQQQLNWKQDNNPLYKSAITT#TVNPRF 789 |
| Xenopus laevis | Query 731 DLREYRRFEKEKLKSQWN-NDNPLFKSATTT#VMNPKFA 767 | ||+|||||++ |+|| |||+ +||||#| || |+ Sbjct 733 DRNEYQRFEKERMNSKWNPGHNPLYHNATTT#VQNPNFS 770 |
| Xenopus laevis | Query 731 DLREYRRFEKEKLKSQWNN-DNPLFKSATTT#VMNPKF 766 | ||+ +|||||+ ++|+ +||++||| ||#|+|||+ Sbjct 759 DRREFAKFEKEKMNAKWDTGENPIYKSAVTT#VVNPKY 795 |
| Felis catus | Query 731 DLREYRRFEKEKLKSQWNN-DNPLFKSATTT#VMNPKF 766 | ||+ +|||||+ ++|+ +||++||| ||#|+|||+ Sbjct 759 DTREFAKFEKEKMNAKWDTGENPIYKSAVTT#VVNPKY 795 |
| Tetraodon nigroviridis | Query 731 DLREYRRFEKEKLKSQWNN-DNPLFKSATTT#VMNPKF 766 | ||+ +|||||+ ++|+ +||++||| ||#|+|||+ Sbjct 832 DRREFAKFEKEKMNAKWDTGENPIYKSAVTT#VVNPKY 868 |
| Danio rerio | Query 731 DLREYRRFEKEKLKSQWNN-DNPLFKSATTT#VMNPKF 766 | ||+ +|||||+ ++|+ +||++||| ||#|+|||+ Sbjct 759 DRREFAKFEKEKMNAKWDTGENPIYKSAVTT#VINPKY 795 |
| Ictalurus punctatus | Query 731 DLREYRRFEKEKLKSQW-NNDNPLFKSATTT#VMNPKF 766 ||+|+++|||| + || +||||++||||#| || | Sbjct 732 DLKEWKKFEKEAQRRQWAKGENPLFQNATTT#VANPAF 768 |
| Pongo pygmaeus | Query 731 DLREYRRFEKEKLKSQWNN-DNPLFKSATTT#VMNPKF 766 | ||+ +|||||+ ++|+ +||++||| ||#|+|||+ Sbjct 759 DRREFAKFEKEKMNAKWDTGENPIYKSAVTT#VVNPKY 795 |
| Cyprinus carpio | Query 731 DLREYRRFEKEKLKSQWNNDNPLFKSATTT#VMNPKFA 767 ||||++||||++ + + ||||++||||#| || |+ Sbjct 728 DLREWKRFEKDRKHEKTSGTNPLFQNATTT#VQNPTFS 764 |
| Oryctolagus cuniculus | Query 731 DLREYRRFEKEKLKSQWN-NDNPLFKSATTT#VMNPKF 766 | ||+ +|||||+ ++|+ +||++||| ||#|+|||+ Sbjct 24 DRREFAKFEKEKMNAKWDTGENPIYKSAVTT#VVNPKY 60 |
| Anopheles gambiae | Query 731 DLREYRRFEKEKLKSQWNN-DNPLFKSATTT#VMNPKFA 767 | ||+ |||||++ ++|+ +||++| ||||# || +| Sbjct 798 DRREFARFEKERMMAKWDTGENPIYKQATTT#FKNPTYA 835 |
| Pacifastacus leniusculus | Query 731 DLREYRRFEKE-KLKSQWNNDNPLFKSATTT#VMNPKFAES 769 | ||| +|||| || | +|||+||| ||# || ||+| Sbjct 761 DRREYAKFEKERKLPSGKRAENPLYKSAKTT#FQNPAFAQS 800 |
| Anopheles gambiae str. PEST | Query 731 DLREYRRFEKEKLKSQWNN-DNPLFKSATTT#VMNPKFA 767 | ||+ |||||++ ++|+ +||++| ||||# || +| Sbjct 798 DRREFARFEKERMMAKWDTGENPIYKQATTT#FKNPTYA 835 |
| Pseudoplusia includens | Query 731 DLREYRRFEKEKLKSQWNN-DNPLFKSATTT#VMNPKFA 767 | ||+ |||||++ ++|+ +||++| ||+|# || +| Sbjct 798 DRREFARFEKERMMAKWDTGENPIYKQATST#FKNPTYA 835 |
| Drosophila melanogaster | Query 731 DLREYRRFEKEKLKSQWNN-DNPLFKSATTT#VMNPKFA 767 | ||+ |||||++ ++|+ +||++| ||+|# || +| Sbjct 807 DRREFARFEKERMNAKWDTGENPIYKQATST#FKNPMYA 844 |
| Strongylocentrotus purpuratus | Query 731 DLREYRRFEKEKLKSQWN-NDNPLFKSATTT#VMNPKFAE 768 | ||| ++| + |+||+ +|||++||+|||# || + + Sbjct 764 DKREYAQWENDCKKAQWDQSDNPIYKSSTTT#FKNPTYGK 802 |
| Crassostrea gigas | Query 731 DLREYRRFEKEKLKSQWNN-DNPLFKSATTT#VMNPKFAE 768 | || ||||| + ++|+ +||++| ||+|# +|| + + Sbjct 761 DKRELARFEKEAMNARWDTGENPIYKQATST#FVNPTYRQ 799 |
| Caenorhabditis briggsae | Query 731 DLREYRRFEKEKLKSQWN-NDNPLFKSATTT#VMNPKFA 767 | || +| |+| ++|+ |+||++| ||||# || +| Sbjct 769 DRAEYAKFNNERLMAKWDTNENPIYKQATTT#FKNPVYA 806 |
| Gallus gallus | Query 731 DLREYRRFEKEKLKSQWNN-DNPLFKSATTT#VMN 763 | ||+ |||+|| +++|+ +|||+| ||+|# | Sbjct 741 DRREFARFEEEKARAKWDTGNNPLYKEATST#FTN 774 |
| Caenorhabditis elegans | Query 731 DLREYRRFEKEKLKSQWN-NDNPLFKSATTT#VMNPKFA 767 | || | |+| ++|+ |+||++| ||||# || +| Sbjct 768 DRSEYATFNNERLMAKWDTNENPIYKQATTT#FKNPVYA 805 |
| Acropora millepora | Query 731 DLREYRRFEKEKLKSQWNND-NPLFKSATTT#VMNPKFA 767 | ||++||+|++ |+| + |||+++| ||# || +| Sbjct 751 DRIEYQKFERERMHSKWTREKNPLYQAAKTT#FENPTYA 788 |
| Biomphalaria glabrata | Query 731 DLREYRRFEKEKLKSQWNN-DNPLFKSATTT#VMNPKFA 767 | ||+ +||||+ ++|+ +||++| ||+|# || + Sbjct 749 DTREFAKFEKERQNAKWDTGENPIYKQATST#FKNPTYG 786 |
| Podocoryne carnea | Query 731 DLREYRRFEKEKLKSQWNN-DNPLFKSATTT#VMNPKFA 767 | ||+ +|||++ +|++ +||++| ||+|# || + Sbjct 742 DRREFAKFEKDRQNPKWDSGENPIYKKATST#FQNPMYG 779 |
| mice, Peptide Partial, 680 aa | Query 731 DLREYRRFEKEKLKSQWNN-DNPLFKSATTT#VMN 763 | +|+ +||+|+ +++|+ +|||+| ||+|# | Sbjct 645 DRKEFAKFEEERARAKWDTANNPLYKEATST#FTN 678 |
| Canis lupus familiaris | Query 731 DLREYRRFEKEKLKSQWNN-DNPLFKSATTT#VMN 763 | +|+ +||+|+ +++|+ +|||+| ||+|# | Sbjct 745 DRKEFAKFEEERARAKWDTANNPLYKEATST#FTN 778 |
| Oryctolagus cuniculus | Query 731 DLREYRRFEKEKLKSQWNN-DNPLFKSATTT#VMN 763 | +|+ +||+|+ +++|+ +|||+| ||+|# | Sbjct 749 DRKEFAKFEEERARAKWDTANNPLYKEATST#FTN 782 |
| Mus musculus | Query 731 DLREYRRFEKEKLKSQWNN-DNPLFKSATTT#VMN 763 | +|+ +||+|+ +++|+ +|||+| ||+|# | Sbjct 748 DRKEFAKFEEERARAKWDTANNPLYKEATST#FTN 781 |
| Lytechinus variegatus | Query 731 DLREYRRFEKEKLKSQWN-NDNPLFKSATTT#VMNPKF 766 | ||++ ||||+ + | +||++| +|+ # || + Sbjct 767 DRREFQNFEKERANATWEGGENPIYKPSTSV#FKNPTY 803 |
| rats, Peptide Partial, 723 aa | Query 731 DLREYRRFEKEKLKSQWNN-DNPLFKSATTT#VMN 763 | +|+ +||+|+ +++|+ +|||+| ||+|# | Sbjct 684 DRKEFAKFEEERARAKWDTANNPLYKEATST#FTN 717 |
| Sus scrofa | Query 731 DLREYRRFEKEKLKSQWNN-DNPLFKSATTT#VMN 763 | +|+ +||+|+ +++|+ +|||+| ||+|# | Sbjct 745 DRKEFAKFEEERARAKWDTANNPLYKEATST#FTN 778 |
| Schistosoma japonicum | Query 731 DLREYRRFEKEKLKSQWN-NDNPLFKSATTT#VMNPKFAES 769 | || |+++ +| +||+|+| || #|+|| | |+ Sbjct 247 DRRELANFKQQGENMRWEMAENPIFESPTTN#VLNPTFEEN 286 |
| Ophlitaspongia tenuis | Query 731 DLREYRRFEKEKLKSQWN-NDNPLFKSATTT#VMNPKFAE 768 |+ | |+||+| +++ |+|||++||| # || + + Sbjct 800 DVVEVRKFEREIKNAKYTKNENPLYRSATKD#YQNPLYGK 838 |
| Geodia cydonium | Query 731 DLREYRRFEKEKLKSQWN-NDNPLFKSATTT#VMNPKFAE 768 | | ++|| | | ++ |+|||+ |||| # || + + Sbjct 840 DYTELKKFENELAKVKYTKNENPLYHSATTE#HKNPIYGQ 878 |
| Halocynthia roretzi | Query 731 DLREYRRFEKEKLKSQWNND-NPLFKSATTT#VMNPKFA 767 | +|+ ||+| +|+| | ||+| + + # || + Sbjct 812 DKKEWENFEREMKQSRWTKDINPVFVNPSAK#FENPSYG 849 |
| Suberites domuncula | Query 731 DLREYRRFEKEKLKSQW-NNDNPLFKSATTT#VMNPKFAE 768 |+ | ++|||| |+++ + |||++|| # || + + Sbjct 726 DVIEVKKFEKELAKTKYPKHQNPLYRSAAKD#YQNPIYGQ 764 |
[Site 4] LFKSATTTVM762-NPKFAES
Met762  Asn
Asn
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Leu753 | Phe754 | Lys755 | Ser756 | Ala757 | Thr758 | Thr759 | Thr760 | Val761 | Met762 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Asn763 | Pro764 | Lys765 | Phe766 | Ala767 | Glu768 | Ser769 | - | - | - |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| REYRRFEKEKLKSQWNNDNPLFKSATTTVMNPKFAES |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | Homo sapiens | 85.50 | 7 | leukocyte adhesion protein beta-subunit precursor |
| 2 | synthetic construct | 85.50 | 2 | integrin, beta 2 (antigen CD18 (p95), lymphocyte |
| 3 | Pan troglodytes | 85.50 | 2 | integrin, beta 2 |
| 4 | Homo sapiens | 85.50 | 1 | integrin, beta 2 precursor |
| 5 | N/A | 85.10 | 19 | complement receptor C3 beta-subunit |
| 6 | Mus musculus | 85.10 | 8 | integrin beta 2 |
| 7 | Rattus norvegicus | 83.20 | 4 | Itgb2 protein |
| 8 | Bos taurus | 82.80 | 4 | integrin, beta 2 |
| 9 | Ovis aries | 82.80 | 1 | antigen CD18 |
| 10 | Ovis canadensis | 82.80 | 1 | ITB2_OVICA Integrin beta-2 precursor (Cell surfac |
| 11 | Sigmodon hispidus | 82.80 | 1 | AF445415_1 integrin beta-2 precursor |
| 12 | Sus scrofa | 82.40 | 3 | CD18 leukocyte adhesion molecule |
| 13 | Canis familiaris | 80.50 | 4 | AF181965_1 B-2 integrin |
| 14 | Gallus gallus | 63.90 | 3 | integrin, beta 2 |
| 15 | Mus sp. | 52.40 | 1 | beta 7 integrin |
| 16 | Xenopus laevis | 50.10 | 1 | hypothetical protein LOC379708 |
| 17 | Xenopus laevis | 49.70 | 2 | integrin beta-1 subunit |
| 18 | Tetraodon nigroviridis | 49.30 | 6 | unnamed protein product |
| 19 | Ictalurus punctatus | 49.30 | 2 | beta-1 integrin |
| 20 | Felis catus | 49.30 | 1 | integrin beta 1 |
| 21 | Danio rerio | 48.90 | 6 | integrin, beta 1b |
| 22 | Pongo pygmaeus | 48.50 | 1 | ITB1_PONPY Integrin beta-1 precursor (Fibronectin |
| 23 | Oryctolagus cuniculus | 47.80 | 1 | integrin beta 1 |
| 24 | Pacifastacus leniusculus | 46.20 | 1 | integrin |
| 25 | Anopheles gambiae | 46.20 | 1 | integrin beta subunit |
| 26 | Anopheles gambiae str. PEST | 45.80 | 1 | integrin beta subunit (AGAP000815-PA) |
| 27 | Pseudoplusia includens | 45.80 | 1 | integrin beta 1 |
| 28 | Cyprinus carpio | 45.40 | 2 | CD18 type 2 |
| 29 | Drosophila melanogaster | 45.10 | 1 | myospheroid CG1560-PA |
| 30 | Strongylocentrotus purpuratus | 42.70 | 3 | integrin beta L subunit |
| 31 | Caenorhabditis briggsae | 42.00 | 1 | Hypothetical protein CBG03601 |
| 32 | Crassostrea gigas | 41.60 | 1 | integrin beta cgh |
| 33 | Acropora millepora | 41.60 | 1 | integrin subunit betaCn1 |
| 34 | Gallus gallus | 40.80 | 2 | integrin, beta 3 precursor |
| 35 | Caenorhabditis elegans | 40.40 | 1 | Paralysed Arrest at Two-fold family member (pat-3 |
| 36 | Biomphalaria glabrata | 39.70 | 1 | beta integrin subunit |
| 37 | Podocoryne carnea | 38.50 | 1 | AF308652_1 integrin beta chain |
| 38 | mice, Peptide Partial, 680 aa | 37.00 | 1 | beta 3 integrin, GPIIIA |
| 39 | Canis lupus familiaris | 37.00 | 1 | integrin beta chain, beta 3 |
| 40 | Oryctolagus cuniculus | 36.60 | 1 | glycoprotein IIIa |
| 41 | Mus musculus | 36.60 | 1 | integrin beta 3 precursor |
| 42 | Lytechinus variegatus | 36.60 | 1 | beta-C integrin subunit |
| 43 | rats, Peptide Partial, 723 aa | 36.60 | 1 | beta 3 integrin, GPIIIA |
| 44 | Sus scrofa | 36.60 | 1 | integrin beta chain, beta 3 |
| 45 | Schistosoma japonicum | 35.00 | 1 | SJCHGC06221 protein |
| 46 | Ophlitaspongia tenuis | 33.90 | 1 | integrin subunit betaPo1 |
| 47 | Geodia cydonium | 33.10 | 1 | integrin beta subunit |
| 48 | Suberites domuncula | 31.60 | 1 | integrin beta subunit |
| 49 | Halocynthia roretzi | 31.60 | 1 | integrin beta Hr1 precursor |
Top-ranked sequences
| organism | matching |
|---|---|
| Homo sapiens | Query 733 REYRRFEKEKLKSQWNNDNPLFKSATTTVM#NPKFAES 769 ||||||||||||||||||||||||||||||#||||||| Query 733 REYRRFEKEKLKSQWNNDNPLFKSATTTVM#NPKFAES 769 |
| synthetic construct | Query 733 REYRRFEKEKLKSQWNNDNPLFKSATTTVM#NPKFAES 769 ||||||||||||||||||||||||||||||#||||||| Query 733 REYRRFEKEKLKSQWNNDNPLFKSATTTVM#NPKFAES 769 |
| Pan troglodytes | Query 733 REYRRFEKEKLKSQWNNDNPLFKSATTTVM#NPKFAES 769 ||||||||||||||||||||||||||||||#||||||| Query 733 REYRRFEKEKLKSQWNNDNPLFKSATTTVM#NPKFAES 769 |
| Homo sapiens | Query 733 REYRRFEKEKLKSQWNNDNPLFKSATTTVM#NPKFAES 769 ||||||||||||||||||||||||||||||#||||||| Query 733 REYRRFEKEKLKSQWNNDNPLFKSATTTVM#NPKFAES 769 |
| N/A | Query 733 REYRRFEKEKLKSQWNNDNPLFKSATTTVM#NPKFAES 769 ||||||||||||||||||||||||||||||#||||||| Query 733 REYRRFEKEKLKSQWNNDNPLFKSATTTVM#NPKFAES 769 |
| Mus musculus | Query 733 REYRRFEKEKLKSQWNNDNPLFKSATTTVM#NPKFAES 769 ||||||||||||||||||||||||||||||#||||||| Query 733 REYRRFEKEKLKSQWNNDNPLFKSATTTVM#NPKFAES 769 |
| Rattus norvegicus | Query 734 EYRRFEKEKLKSQWNNDNPLFKSATTTVM#NPKFAES 769 |||||||||||||||||||||||||||||#||||||| Query 734 EYRRFEKEKLKSQWNNDNPLFKSATTTVM#NPKFAES 769 |
| Bos taurus | Query 733 REYRRFEKEKLKSQWNNDNPLFKSATTTVM#NPKFAES 769 ||| ||||||||||||||||||||||||||#||||||| Sbjct 733 REYHRFEKEKLKSQWNNDNPLFKSATTTVM#NPKFAES 769 |
| Ovis aries | Query 733 REYRRFEKEKLKSQWNNDNPLFKSATTTVM#NPKFAES 769 ||| ||||||||||||||||||||||||||#||||||| Sbjct 734 REYHRFEKEKLKSQWNNDNPLFKSATTTVM#NPKFAES 770 |
| Ovis canadensis | Query 733 REYRRFEKEKLKSQWNNDNPLFKSATTTVM#NPKFAES 769 ||| ||||||||||||||||||||||||||#||||||| Sbjct 734 REYHRFEKEKLKSQWNNDNPLFKSATTTVM#NPKFAES 770 |
| Sigmodon hispidus | Query 733 REYRRFEKEKLKSQWNNDNPLFKSATTTVM#NPKFAES 769 |||| |||||||||||||||||||||||||#||||||| Sbjct 733 REYRHFEKEKLKSQWNNDNPLFKSATTTVM#NPKFAES 769 |
| Sus scrofa | Query 733 REYRRFEKEKLKSQWNNDNPLFKSATTTVM#NPKFAE 768 |||+||||||||||||||||||||||||||#|||||| Sbjct 733 REYKRFEKEKLKSQWNNDNPLFKSATTTVM#NPKFAE 768 |
| Canis familiaris | Query 733 REYRRFEKEKLKSQWNNDNPLFKSATTTVM#NPKFAES 769 ||++|||||||+||||||||||||||||||#||+|||| Sbjct 728 REFKRFEKEKLRSQWNNDNPLFKSATTTVM#NPRFAES 764 |
| Gallus gallus | Query 733 REYRRFEKEKLKSQWNN-DNPLFKSATTTVM#NPKF 766 |||||||||| |++|| |||||||||||||#||+| Sbjct 735 REYRRFEKEKSKAKWNEADNPLFKSATTTVM#NPRF 769 |
| Mus sp. | Query 733 REYRRFEKEKLKSQWNND-NPLFKSATTTVM#NPKF 766 |||||||||+ + | | |||+||| || +#||+| Sbjct 755 REYRRFEKEQQQLNWKQDNNPLYKSAITTTV#NPRF 789 |
| Xenopus laevis | Query 734 EYRRFEKEKLKSQWN-NDNPLFKSATTTVM#NPKFA 767 ||+|||||++ |+|| |||+ +||||| #|| |+ Sbjct 736 EYQRFEKERMNSKWNPGHNPLYHNATTTVQ#NPNFS 770 |
| Xenopus laevis | Query 733 REYRRFEKEKLKSQWNN-DNPLFKSATTTVM#NPKF 766 ||+ +|||||+ ++|+ +||++||| |||+#|||+ Sbjct 761 REFAKFEKEKMNAKWDTGENPIYKSAVTTVV#NPKY 795 |
| Tetraodon nigroviridis | Query 733 REYRRFEKEKLKSQWNN-DNPLFKSATTTVM#NPKF 766 ||+ +|||||+ ++|+ +||++||| |||+#|||+ Sbjct 834 REFAKFEKEKMNAKWDTGENPIYKSAVTTVV#NPKY 868 |
| Ictalurus punctatus | Query 733 REYRRFEKEKLKSQWN-NDNPLFKSATTTVM#NPKF 766 ||+ +|||||+ ++|+ +||++||| |||+#|||+ Sbjct 770 REFAKFEKEKMNAKWDAGENPIYKSAVTTVV#NPKY 804 |
| Felis catus | Query 733 REYRRFEKEKLKSQWNN-DNPLFKSATTTVM#NPKF 766 ||+ +|||||+ ++|+ +||++||| |||+#|||+ Sbjct 761 REFAKFEKEKMNAKWDTGENPIYKSAVTTVV#NPKY 795 |
| Danio rerio | Query 733 REYRRFEKEKLKSQWN-NDNPLFKSATTTVM#NPKF 766 ||+ +|||||+ ++|+ +||++||| |||+#|||+ Sbjct 758 REFAKFEKEKMNAKWDAGENPIYKSAVTTVV#NPKY 792 |
| Pongo pygmaeus | Query 733 REYRRFEKEKLKSQWNN-DNPLFKSATTTVM#NPKF 766 ||+ +|||||+ ++|+ +||++||| |||+#|||+ Sbjct 761 REFAKFEKEKMNAKWDTGENPIYKSAVTTVV#NPKY 795 |
| Oryctolagus cuniculus | Query 733 REYRRFEKEKLKSQWN-NDNPLFKSATTTVM#NPKF 766 ||+ +|||||+ ++|+ +||++||| |||+#|||+ Sbjct 26 REFAKFEKEKMNAKWDTGENPIYKSAVTTVV#NPKY 60 |
| Pacifastacus leniusculus | Query 733 REYRRFEKE-KLKSQWNNDNPLFKSATTTVM#NPKFAES 769 ||| +|||| || | +|||+||| || #|| ||+| Sbjct 763 REYAKFEKERKLPSGKRAENPLYKSAKTTFQ#NPAFAQS 800 |
| Anopheles gambiae | Query 733 REYRRFEKEKLKSQWNN-DNPLFKSATTTVM#NPKFA 767 ||+ |||||++ ++|+ +||++| |||| #|| +| Sbjct 800 REFARFEKERMMAKWDTGENPIYKQATTTFK#NPTYA 835 |
| Anopheles gambiae str. PEST | Query 733 REYRRFEKEKLKSQWNN-DNPLFKSATTTVM#NPKFA 767 ||+ |||||++ ++|+ +||++| |||| #|| +| Sbjct 800 REFARFEKERMMAKWDTGENPIYKQATTTFK#NPTYA 835 |
| Pseudoplusia includens | Query 733 REYRRFEKEKLKSQWNN-DNPLFKSATTTVM#NPKFA 767 ||+ |||||++ ++|+ +||++| ||+| #|| +| Sbjct 800 REFARFEKERMMAKWDTGENPIYKQATSTFK#NPTYA 835 |
| Cyprinus carpio | Query 733 REYRRFEKEKLKSQWNNDNPLFKSATTTVM#NPKFA 767 ||++||||++ + + ||||++||||| #|| |+ Sbjct 730 REWKRFEKDRKHEKTSGTNPLFQNATTTVQ#NPTFS 764 |
| Drosophila melanogaster | Query 733 REYRRFEKEKLKSQWNN-DNPLFKSATTTVM#NPKFA 767 ||+ |||||++ ++|+ +||++| ||+| #|| +| Sbjct 809 REFARFEKERMNAKWDTGENPIYKQATSTFK#NPMYA 844 |
| Strongylocentrotus purpuratus | Query 733 REYRRFEKEKLKSQWN-NDNPLFKSATTTVM#NPKFAE 768 ||| ++| + |+||+ +|||++||+||| #|| + + Sbjct 766 REYAQWENDCKKAQWDQSDNPIYKSSTTTFK#NPTYGK 802 |
| Caenorhabditis briggsae | Query 734 EYRRFEKEKLKSQWN-NDNPLFKSATTTVM#NPKFA 767 || +| |+| ++|+ |+||++| |||| #|| +| Sbjct 772 EYAKFNNERLMAKWDTNENPIYKQATTTFK#NPVYA 806 |
| Crassostrea gigas | Query 733 REYRRFEKEKLKSQWNN-DNPLFKSATTTVM#NPKFAE 768 || ||||| + ++|+ +||++| ||+| +#|| + + Sbjct 763 RELARFEKEAMNARWDTGENPIYKQATSTFV#NPTYRQ 799 |
| Acropora millepora | Query 734 EYRRFEKEKLKSQWNND-NPLFKSATTTVM#NPKFA 767 ||++||+|++ |+| + |||+++| || #|| +| Sbjct 754 EYQKFERERMHSKWTREKNPLYQAAKTTFE#NPTYA 788 |
| Gallus gallus | Query 733 REYRRFEKEKLKSQWNN-DNPLFKSATTTVM#N 763 ||+ |||+|| +++|+ +|||+| ||+| #| Sbjct 743 REFARFEEEKARAKWDTGNNPLYKEATSTFT#N 774 |
| Caenorhabditis elegans | Query 734 EYRRFEKEKLKSQWN-NDNPLFKSATTTVM#NPKFA 767 || | |+| ++|+ |+||++| |||| #|| +| Sbjct 771 EYATFNNERLMAKWDTNENPIYKQATTTFK#NPVYA 805 |
| Biomphalaria glabrata | Query 733 REYRRFEKEKLKSQWNN-DNPLFKSATTTVM#NPKFA 767 ||+ +||||+ ++|+ +||++| ||+| #|| + Sbjct 751 REFAKFEKERQNAKWDTGENPIYKQATSTFK#NPTYG 786 |
| Podocoryne carnea | Query 733 REYRRFEKEKLKSQWNN-DNPLFKSATTTVM#NPKFA 767 ||+ +|||++ +|++ +||++| ||+| #|| + Sbjct 744 REFAKFEKDRQNPKWDSGENPIYKKATSTFQ#NPMYG 779 |
| mice, Peptide Partial, 680 aa | Query 733 REYRRFEKEKLKSQWNN-DNPLFKSATTTVM#N 763 +|+ +||+|+ +++|+ +|||+| ||+| #| Sbjct 647 KEFAKFEEERARAKWDTANNPLYKEATSTFT#N 678 |
| Canis lupus familiaris | Query 733 REYRRFEKEKLKSQWNN-DNPLFKSATTTVM#N 763 +|+ +||+|+ +++|+ +|||+| ||+| #| Sbjct 747 KEFAKFEEERARAKWDTANNPLYKEATSTFT#N 778 |
| Oryctolagus cuniculus | Query 733 REYRRFEKEKLKSQWNN-DNPLFKSATTTVM#N 763 +|+ +||+|+ +++|+ +|||+| ||+| #| Sbjct 751 KEFAKFEEERARAKWDTANNPLYKEATSTFT#N 782 |
| Mus musculus | Query 733 REYRRFEKEKLKSQWNN-DNPLFKSATTTVM#N 763 +|+ +||+|+ +++|+ +|||+| ||+| #| Sbjct 750 KEFAKFEEERARAKWDTANNPLYKEATSTFT#N 781 |
| Lytechinus variegatus | Query 733 REYRRFEKEKLKSQWN-NDNPLFKSATTTVM#NPKF 766 ||++ ||||+ + | +||++| +|+ #|| + Sbjct 769 REFQNFEKERANATWEGGENPIYKPSTSVFK#NPTY 803 |
| rats, Peptide Partial, 723 aa | Query 733 REYRRFEKEKLKSQWNN-DNPLFKSATTTVM#N 763 +|+ +||+|+ +++|+ +|||+| ||+| #| Sbjct 686 KEFAKFEEERARAKWDTANNPLYKEATSTFT#N 717 |
| Sus scrofa | Query 733 REYRRFEKEKLKSQWNN-DNPLFKSATTTVM#N 763 +|+ +||+|+ +++|+ +|||+| ||+| #| Sbjct 747 KEFAKFEEERARAKWDTANNPLYKEATSTFT#N 778 |
| Schistosoma japonicum | Query 733 REYRRFEKEKLKSQWN-NDNPLFKSATTTVM#NPKFAES 769 || |+++ +| +||+|+| || |+#|| | |+ Sbjct 249 RELANFKQQGENMRWEMAENPIFESPTTNVL#NPTFEEN 286 |
| Ophlitaspongia tenuis | Query 734 EYRRFEKEKLKSQWN-NDNPLFKSATTTVM#NPKFAE 768 | |+||+| +++ |+|||++||| #|| + + Sbjct 803 EVRKFEREIKNAKYTKNENPLYRSATKDYQ#NPLYGK 838 |
| Geodia cydonium | Query 734 EYRRFEKEKLKSQWN-NDNPLFKSATTTVM#NPKFAE 768 | ++|| | | ++ |+|||+ |||| #|| + + Sbjct 843 ELKKFENELAKVKYTKNENPLYHSATTEHK#NPIYGQ 878 |
| Suberites domuncula | Query 734 EYRRFEKEKLKSQW-NNDNPLFKSATTTVM#NPKFAE 768 | ++|||| |+++ + |||++|| #|| + + Sbjct 729 EVKKFEKELAKTKYPKHQNPLYRSAAKDYQ#NPIYGQ 764 |
| Halocynthia roretzi | Query 733 REYRRFEKEKLKSQWNND-NPLFKSATTTVM#NPKFA 767 +|+ ||+| +|+| | ||+| + + #|| + Sbjct 814 KEWENFEREMKQSRWTKDINPVFVNPSAKFE#NPSYG 849 |
* References
[PubMed ID: 24889201] Raftery MJ, Lalwani P, Krautkrmer E, Peters T, Scharffetter-Kochanek K, Kruger R, Hofmann J, Seeger K, Kruger DH, Schonrich G, beta2 integrin mediates hantavirus-induced release of neutrophil extracellular traps. J Exp Med. 2014 Jun 30;211(7):1485-97. doi: 10.1084/jem.20131092. Epub 2014 Jun
[PubMed ID: 24829201] ... Riches JC, O'Donovan CJ, Kingdon SJ, McClanahan F, Clear AJ, Neuberg DS, Werner L, Croce CM, Ramsay AG, Rassenti LZ, Kipps TJ, Gribben JG, Trisomy 12 chronic lymphocytic leukemia cells exhibit upregulation of integrin signaling that is modulated by NOTCH1 mutations. Blood. 2014 Jun 26;123(26):4101-10. doi: 10.1182/blood-2014-01-552307. Epub 2014
[PubMed ID: 24769233] ... Chiasserini D, van Weering JR, Piersma SR, Pham TV, Malekzadeh A, Teunissen CE, de Wit H, Jimenez CR, Proteomic analysis of cerebrospinal fluid extracellular vesicles: a comprehensive dataset. J Proteomics. 2014 Jun 25;106:191-204. doi: 10.1016/j.jprot.2014.04.028. Epub
[PubMed ID: 24500432] ... Gonzalez AM, Fragala MS, Jajtner AR, Townsend JR, Wells AJ, Beyer KS, Boone CH, Pruna GJ, Mangine GT, Bohner JD, Fukuda DH, Stout JR, Hoffman JR, Effects of beta-hydroxy-beta-methylbutyrate free acid and cold water immersion on expression of CR3 and MIP-1beta following resistance exercise. Am J Physiol Regul Integr Comp Physiol. 2014 Apr 1;306(7):R483-9. doi:
[PubMed ID: 24006483] ... Azcutia V, Routledge M, Williams MR, Newton G, Frazier WA, Manica A, Croce KJ, Parkos CA, Schmider AB, Turman MV, Soberman RJ, Luscinskas FW, CD47 plays a critical role in T-cell recruitment by regulation of LFA-1 and VLA-4 integrin adhesive functions. Mol Biol Cell. 2013 Nov;24(21):3358-68. doi: 10.1091/mbc.E13-01-0063. Epub 2013
[PubMed ID: 11700305] ... Fagerholm S, Morrice N, Gahmberg CG, Cohen P, Phosphorylation of the cytoplasmic domain of the integrin CD18 chain by protein kinase C isoforms in leukocytes. J Biol Chem. 2002 Jan 18;277(3):1728-38. Epub 2001 Nov 7.
[PubMed ID: 1352501] ... Corbi AL, Vara A, Ursa A, Garcia Rodriguez MC, Fontan G, Sanchez-Madrid F, Molecular basis for a severe case of leukocyte adhesion deficiency. Eur J Immunol. 1992 Jul;22(7):1877-81.
[PubMed ID: 1590804] ... Matsuura S, Kishi F, Tsukahara M, Nunoi H, Matsuda I, Kobayashi K, Kajii T, Leukocyte adhesion deficiency: identification of novel mutations in two Japanese patients with a severe form. Biochem Biophys Res Commun. 1992 May 15;184(3):1460-7.
[PubMed ID: 1347532] ... Back AL, Kwok WW, Hickstein DD, Identification of two molecular defects in a child with leukocyte adherence deficiency. J Biol Chem. 1992 Mar 15;267(8):5482-7.
[PubMed ID: 1346613] ... Nelson C, Rabb H, Arnaout MA, Genetic cause of leukocyte adhesion molecule deficiency. Abnormal splicing and a missense mutation in a conserved region of CD18 impair cell surface expression of beta 2 integrins. J Biol Chem. 1992 Feb 15;267(5):3351-7.
[PubMed ID: 1674496] ... Petersen MB, Slaugenhaupt SA, Lewis JG, Warren AC, Chakravarti A, Antonarakis SE, A genetic linkage map of 27 markers on human chromosome 21. Genomics. 1991 Mar;9(3):407-19.