SB0006 : Calcium/calmodulin-dependent protein kinase IV
[ CaMP Format ]
* Basic Information
| Organism | Mus musculus (house mouse) |
| Protein Names | calcium/calmodulin-dependent protein kinase IV [Mus musculus]; calcium/calmodulin-dependent protein kinase IV; Ca2+/calmodulin-dependent protein kinase type IV/Gr; Calcium/calmodulin-dependent protein kinase type IV; CaMK IV; 2.7.11.17; CaM kinase-GR |
| Gene Names | Camk4; calcium/calmodulin-dependent protein kinase IV |
| Gene Locus | 18 12.0 cM; chromosome 18 |
| GO Function | GO:0005524 - ATP binding [Evidence IEA]; GO:0016301 - kinase activity [Evidence IEA]; GO:0005515 - protein binding [Evidence IPI]; GO:0005516 - calmodulin binding [Evidence IEA]; GO:0000166 - nucleotide binding [Evidence IEA]; GO:0005509 - calcium ion binding [Evidence IEA]; GO:0016740 - transferase activity [Evidence IEA]; GO:0004672 - protein kinase activity [Evidence IEA]; GO:0016563 - transcriptional activator activity [Evidence IMP]; GO:0004674 - protein serine/threonine kinase activity [Evidence IEA]; GO:0004683 - calmodulin regulated protein kinase activity [Evidence TAS] |
* Information From OMIM
Not Available.
* Structure Information
1. Primary Information
Length: 469 aa
Average Mass: 52.627 kDa
Monoisotopic Mass: 52.594 kDa
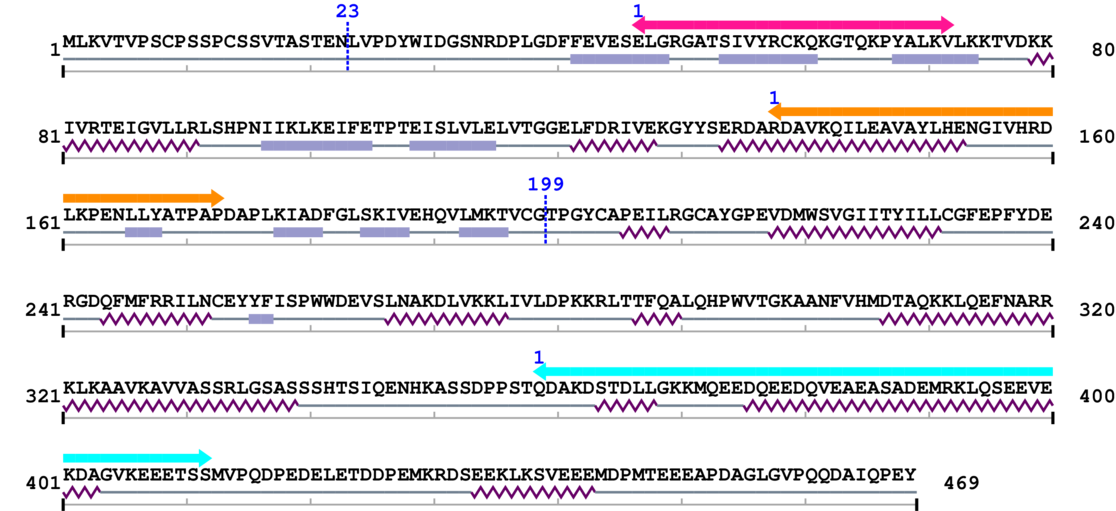
2. Domain Information
Annotated Domains: interpro / pfam / smart / prosite
Computationally Assigned Domains (Pfam+HMMER):
| domain name | begin | end | score | e-value |
|---|---|---|---|---|
| --- cleavage 23 --- | ||||
| Kinase-like 1. | 47 | 72 | 19.0 | 0.0 |
| Phosphotransferase enzyme family 1. | 138 | 173 | 148.0 | 0.0 |
| --- cleavage 199 --- | ||||
| Protein tyrosine kinase 1. | 359 | 412 | 23.0 | 0.1 |
3. Sequence Information
Fasta Sequence: SB0006.fasta
Amino Acid Sequence and Secondary Structures (PsiPred):
4. 3D Information
Not Available.
* Cleavage Information
2 [sites] cleaved by Calpain 2
Source Reference: [PubMed ID: 9685336] McGinnis KM, Whitton MM, Gnegy ME, Wang KK, Calcium/calmodulin-dependent protein kinase IV is cleaved by caspase-3 and calpain in SH-SY5Y human neuroblastoma cells undergoing apoptosis. J Biol Chem. 1998 Aug 7;273(32):19993-20000.
Cleavage sites (±10aa)
[Site 1] CSSVTASTEN23-LVPDYWIDGS
Asn23  Leu
Leu
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Cys14 | Ser15 | Ser16 | Val17 | Thr18 | Ala19 | Ser20 | Thr21 | Glu22 | Asn23 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Leu24 | Val25 | Pro26 | Asp27 | Tyr28 | Trp29 | Ile30 | Asp31 | Gly32 | Ser33 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| MLKVTVPSCPSSPCSSVTASTENLVPDYWIDGSNRDPLGDFFEVESELGRGAT |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | Mus musculus | 114.00 | 2 | KCC4_MOUSE Calcium/calmodulin-dependent protein k |
| 2 | N/A | 74.70 | 3 | calmodulin-dependent protein kinase |
| 3 | Rattus norvegicus | 74.70 | 2 | calcium/calmodulin-dependent protein kinase IV |
| 4 | Rattus sp. | 74.70 | 1 | Ca2+/calmodulin-dependent protein kinase IV beta |
| 5 | Homo sapiens | 69.70 | 2 | calcium/calmodulin-dependent protein kinase IV |
| 6 | synthetic construct | 69.70 | 1 | calcium/calmodulin-dependent protein kinase IV |
| 7 | Gallus gallus | 55.10 | 1 | calcium/calmodulin-dependent protein kinase IV |
| 8 | Danio rerio | 54.70 | 1 | calcium/calmodulin-dependent protein kinase IV |
| 9 | Xenopus laevis | 52.80 | 1 | calcium/calmodulin-dependent protein kinase IV |
| 10 | Theileria parva strain Muguga | 32.70 | 1 | hypothetical protein TP01_0299 |
| 11 | Tetraodon nigroviridis | 30.00 | 1 | unnamed protein product |
| 12 | Gadus morhua | 29.60 | 2 | MHC class Ia antigen |
Top-ranked sequences
| organism | matching |
|---|---|
| Mus musculus | Query 1 MLKVTVPSCPSSPCSSVTASTEN#LVPDYWIDGSNRDPLGDFFEVESELGRGAT 53 |||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 1 MLKVTVPSCPSSPCSSVTASTEN#LVPDYWIDGSNRDPLGDFFEVESELGRGAT 53 |
| N/A | Query 1 MLKVTVPSCPSSPCSSVTASTEN#LVPDYWIDGSNRDPLGDFFEVESELGRGAT 53 ||||||||||||||||||+||||#|||||||||| |||| |||||||||||||| Sbjct 1 MLKVTVPSCPSSPCSSVTSSTEN#LVPDYWIDGSKRDPLSDFFEVESELGRGAT 53 |
| Rattus norvegicus | Query 1 MLKVTVPSCPSSPCSSVTASTEN#LVPDYWIDGSNRDPLGDFFEVESELGRGAT 53 ||||||||||||||||||+||||#|||||||||| |||| |||||||||||||| Sbjct 1 MLKVTVPSCPSSPCSSVTSSTEN#LVPDYWIDGSKRDPLSDFFEVESELGRGAT 53 |
| Rattus sp. | Query 1 MLKVTVPSCPSSPCSSVTASTEN#LVPDYWIDGSNRDPLGDFFEVESELGRGAT 53 ||||||||||||||||||+||||#|||||||||| |||| |||||||||||||| Sbjct 29 MLKVTVPSCPSSPCSSVTSSTEN#LVPDYWIDGSKRDPLSDFFEVESELGRGAT 81 |
| Homo sapiens | Query 1 MLKVTVPSCPSSPCSSVTA----STEN#LVPDYWIDGSNRDPLGDFFEVESELGRGAT 53 ||||||||| +| |||||| | +#||||||||||||| | |||||||||||||| Sbjct 1 MLKVTVPSCSASSCSSVTASAAPGTAS#LVPDYWIDGSNRDALSDFFEVESELGRGAT 57 |
| synthetic construct | Query 1 MLKVTVPSCPSSPCSSVTA----STEN#LVPDYWIDGSNRDPLGDFFEVESELGRGAT 53 ||||||||| +| |||||| | +#||||||||||||| | |||||||||||||| Sbjct 1 MLKVTVPSCSASSCSSVTASAAPGTAS#LVPDYWIDGSNRDALSDFFEVESELGRGAT 57 |
| Gallus gallus | Query 13 PCSSVTASTEN#LVPDYWIDGSNRDPLGDFFEVESELGRGAT 53 | +| + | # |||||||||+| | +|++||||||||| Sbjct 2 PSTSASGSAGL#PAPDYWIDGSNKDTLTHYFDLESELGRGAT 42 |
| Danio rerio | Query 1 MLKVTVPSCPSSPCSSVTASTEN#LVPDYWIDGSNRDPLGDFFEVESELGRGAT 53 |||||+|+ ++| # +|||||| ++ | ||+|+||||||||| Sbjct 1 MLKVTMPASRAAP----------#---EYWIDGSKKETLADFYELESELGRGAT 40 |
| Xenopus laevis | Query 26 PDYWIDGSNRDPLGDFFEVESELGRGAT 53 |||||||||+| | ++|+||||||||| Sbjct 35 PDYWIDGSNKDTLAHYYELESELGRGAT 62 |
| Theileria parva strain Muguga | Query 28 YWIDGSNRDPLGDFFEVESEL 48 | || ||+|||||+ | || Sbjct 195 YVIDSSNQDPLGDYMSVRREL 215 |
| Tetraodon nigroviridis | Query 27 DYWIDGSNRD-PLGDFFEVESELGR 50 ++|+||| || + +|+ + ||||| Sbjct 12 EFWVDGSRRDRTVEEFYTLSSELGR 36 |
| Gadus morhua | Query 19 ASTEN#LVPDY-WIDGSNRDPLGDFFEVESELGRG 51 ++|+ #++| |+| +||| + |+ | |+| +| Sbjct 31 SNTQR#IIPKQDWVDQANRDKVPDYLERETENRKG 64 |
[Site 2] HQVLMKTVCG199-TPGYCAPEIL
Gly199  Thr
Thr
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| His190 | Gln191 | Val192 | Leu193 | Met194 | Lys195 | Thr196 | Val197 | Cys198 | Gly199 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Thr200 | Pro201 | Gly202 | Tyr203 | Cys204 | Ala205 | Pro206 | Glu207 | Ile208 | Leu209 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| TPAPDAPLKIADFGLSKIVEHQVLMKTVCGTPGYCAPEILRGCAYGPEVDMWSVGIITYI |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | N/A | 137.00 | 10 | calcium/calmodulin-dependent protein kinase |
| 2 | Mus musculus | 135.00 | 7 | calcium/calmodulin-dependent protein kinase IV |
| 3 | Rattus norvegicus | 135.00 | 6 | calcium/calmodulin-dependent protein kinase IV |
| 4 | Rattus sp. | 135.00 | 1 | Ca2+/calmodulin-dependent protein kinase IV beta |
| 5 | Homo sapiens | 130.00 | 7 | calcium/calmodulin-dependent protein kinase IV |
| 6 | synthetic construct | 130.00 | 2 | calcium/calmodulin-dependent protein kinase IV |
| 7 | Xenopus laevis | 128.00 | 6 | calcium/calmodulin-dependent protein kinase IV |
| 8 | Danio rerio | 125.00 | 3 | calcium/calmodulin-dependent protein kinase IV |
| 9 | Gallus gallus | 122.00 | 1 | calcium/calmodulin-dependent protein kinase IV |
| 10 | Tetraodon nigroviridis | 100.00 | 2 | unnamed protein product |
| 11 | Dictyostelium discoideum AX4 | 78.60 | 2 | myosin light chain kinase |
| 12 | Ciona intestinalis | 77.00 | 1 | calmodulin-dependent protein kinase homologue |
| 13 | Yarrowia lipolytica | 74.30 | 2 | hypothetical protein |
| 14 | Dictyostelium discoideum AX4 | 73.60 | 4 | putative protein serine/threonine kinase |
| 15 | Drosophila pseudoobscura | 73.20 | 1 | GA13377-PA |
| 16 | Drosophila melanogaster | 72.80 | 3 | Calcium/calmodulin-dependent protein kinase I CG1 |
| 17 | Entamoeba histolytica HM-1:IMSS | 72.80 | 2 | protein kinase |
| 18 | Glossina morsitans morsitans | 72.40 | 1 | calcium/ calmodulin-dependent protein kinase 1 |
| 19 | Aspergillus nidulans FGSC A4 | 72.00 | 2 | hypothetical protein AN3065.2 |
| 20 | Gibberella zeae PH-1 | 72.00 | 2 | hypothetical protein FG00337.1 |
| 21 | Aspergillus fumigatus Af293 | 72.00 | 2 | calcium/calmodulin dependent protein kinase |
| 22 | Aspergillus oryzae RIB40 | 72.00 | 1 | hypothetical protein |
| 23 | Bombyx mori | 71.60 | 1 | cell cycle checkpoint kinase 2 |
| 24 | Emericella nidulans | 71.60 | 1 | AF156027_1 calcium/calmodulin dependent protein k |
| 25 | Colletotrichum gloeosporioides | 71.20 | 1 | calmodulin-dependent protein kinase |
| 26 | Arthrobotrys dactyloides | 70.90 | 1 | calmodulin-binding protein kinase |
| 27 | Cryptococcus neoformans var. neoformans JEC21 | 70.50 | 1 | calmodulin-dependent protein kinase |
| 28 | Caenorhabditis elegans | 70.10 | 3 | CaM Kinase family member (cmk-1) |
| 29 | Caenorhabditis briggsae AF16 | 70.10 | 1 | Hypothetical protein CBG08406 |
| 30 | Ustilago maydis 521 | 70.10 | 1 | hypothetical protein UM05544.1 |
| 31 | Guillardia theta | 69.70 | 1 | SNF-related kinase |
| 32 | Schizophyllum commune | 69.70 | 1 | putative serine/threonine protein kinase |
| 33 | Arabidopsis thaliana | 69.30 | 5 | SnRK1.3 (SNF1-RELATED PROTEIN KINASE 1.3); kinase |
| 34 | Xenopus tropicalis | 69.30 | 2 | hypothetical protein LOC407849 |
| 35 | Bos taurus | 69.30 | 1 | pregnancy upregulated non-ubiquitously expressed |
| 36 | Candida albicans SC5314 | 68.90 | 2 | protein kinase |
| 37 | Schizosaccharomyces pombe 972h- | 68.60 | 2 | serine/threonine protein kinase Ssp2 |
| 38 | Schizosaccharomyces pombe | 68.60 | 2 | protein kinase |
| 39 | Arabidopsis thaliana | 68.60 | 2 | SNF1 family protein kinase |
| 40 | Candida glabrata CBS138 | 68.60 | 1 | hypothetical protein CAGL0M08910g |
| 41 | Kluyveromyces lactis | 68.60 | 1 | unnamed protein product |
| 42 | Cochliobolus carbonum | 68.60 | 1 | AF159253_1 serine threonine protein kinase SNF1p |
| 43 | Ashbya gossypii ATCC 10895 | 68.60 | 1 | AEL230Wp |
| 44 | Candida tropicalis | 68.60 | 1 | SNF1_CANTR Carbon catabolite-derepressing protein |
| 45 | Glycine max | 68.20 | 1 | AF128443_1 SNF-1-like serine/threonine protein ki |
| 46 | Cucumis sativus | 67.80 | 1 | SNF1-related protein kinase |
| 47 | Vicia faba | 67.80 | 1 | SNF1-related protein kinase |
| 48 | Canis familiaris | 67.80 | 1 | PREDICTED: similar to calcium/calmodulin-dependent |
| 49 | Solanum tuberosum | 67.80 | 1 | StubSNF1 protein |
| 50 | Nicotiana tabacum | 67.80 | 1 | protein kinase |
| 51 | Pisum sativum | 67.80 | 1 | SNF1-related protein kinase |
| 52 | Lycopersicon esculentum | 67.80 | 1 | AF143743_1 SNF1 |
| 53 | Oryza sativa | 67.40 | 2 | protein kinase SNF1 |
| 54 | Physcomitrella patens subsp. patens | 67.40 | 1 | snf1a Snf1-related kinase SNF1a |
| 55 | Sclerotinia sclerotiorum | 67.40 | 1 | serine threonine protein kinase |
| 56 | Entamoeba histolytica HM-1:IMSS | 67.00 | 2 | protein kinase, putative |
| 57 | Schistosoma japonicum | 67.00 | 1 | SJCHGC05776 protein |
| 58 | Chaetomium globosum CBS 148.51 | 65.10 | 1 | conserved hypothetical protein |
| 59 | Triticum aestivum | 64.70 | 1 | SNF1-related protein kinase |
Top-ranked sequences
| organism | matching |
|---|---|
| N/A | Query 170 TPAPDAPLKIADFGLSKIVEHQVLMKTVCG#TPGYCAPEILRGCAYGPEVDMWSVGIITYI 229 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 170 TPAPDAPLKIADFGLSKIVEHQVLMKTVCG#TPGYCAPEILRGCAYGPEVDMWSVGIITYI 229 |
| Mus musculus | Query 170 TPAPDAPLKIADFGLSKIVEHQVLMKTVCG#TPGYCAPEILRGCAYGPEVDMWSVGIITYI 229 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 170 TPAPDAPLKIADFGLSKIVEHQVLMKTVCG#TPGYCAPEILRGCAYGPEVDMWSVGIITYI 229 |
| Rattus norvegicus | Query 170 TPAPDAPLKIADFGLSKIVEHQVLMKTVCG#TPGYCAPEILRGCAYGPEVDMWSVGIITYI 229 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 170 TPAPDAPLKIADFGLSKIVEHQVLMKTVCG#TPGYCAPEILRGCAYGPEVDMWSVGIITYI 229 |
| Rattus sp. | Query 170 TPAPDAPLKIADFGLSKIVEHQVLMKTVCG#TPGYCAPEILRGCAYGPEVDMWSVGIITYI 229 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 170 TPAPDAPLKIADFGLSKIVEHQVLMKTVCG#TPGYCAPEILRGCAYGPEVDMWSVGIITYI 229 |
| Homo sapiens | Query 170 TPAPDAPLKIADFGLSKIVEHQVLMKTVCG#TPGYCAPEILRGCAYGPEVDMWSVGIITYI 229 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 170 TPAPDAPLKIADFGLSKIVEHQVLMKTVCG#TPGYCAPEILRGCAYGPEVDMWSVGIITYI 229 |
| synthetic construct | Query 170 TPAPDAPLKIADFGLSKIVEHQVLMKTVCG#TPGYCAPEILRGCAYGPEVDMWSVGIITYI 229 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 170 TPAPDAPLKIADFGLSKIVEHQVLMKTVCG#TPGYCAPEILRGCAYGPEVDMWSVGIITYI 229 |
| Xenopus laevis | Query 170 TPAPDAPLKIADFGLSKIVEHQVLMKTVCG#TPGYCAPEILRGCAYGPEVDMWSVGIITYI 229 |||||||||||||||||||+ || ||||||#|||||||||||||||||||||||||||||| Sbjct 178 TPAPDAPLKIADFGLSKIVDDQVTMKTVCG#TPGYCAPEILRGCAYGPEVDMWSVGIITYI 237 |
| Danio rerio | Query 170 TPAPDAPLKIADFGLSKIVEHQVLMKTVCG#TPGYCAPEILRGCAYGPEVDMWSVGIITYI 229 | |||||||||||||||||+ || ||||||#|||||||||||||||||||||||||+|||| Sbjct 157 TSAPDAPLKIADFGLSKIVDDQVTMKTVCG#TPGYCAPEILRGCAYGPEVDMWSVGVITYI 216 |
| Gallus gallus | Query 170 TPAPDAPLKIADFGLSKIVEHQVLMKTVCG#TPGYCAPEILRGCAYGPEVDMWSVGIITYI 229 |||||||||||||||||||| || ||||||#|||||||||||||||||||||||+|||||| Sbjct 159 TPAPDAPLKIADFGLSKIVEDQVTMKTVCG#TPGYCAPEILRGCAYGPEVDMWSLGIITYI 218 |
| Tetraodon nigroviridis | Query 180 ADFGLSKIVEHQVLMKTVCG#TPGYCAPEILRGCAYGPEVDMWSVGIITYI 229 ||||||||++ || ||||||#|||||||||||| ||||||||||||+| || Sbjct 1 ADFGLSKIIDDQVTMKTVCG#TPGYCAPEILRGNAYGPEVDMWSVGVILYI 50 |
| Dictyostelium discoideum AX4 | Query 177 LKIADFGLSKIVEHQVLMKTVCG#TPGYCAPEILRGCAYGPEVDMWSVGIITYI 229 + |||||||||+ ++|+| ||#|| | |||+| | ||||||+|+|||| Sbjct 147 VAIADFGLSKIIGQTLVMQTACG#TPSYVAPEVLNATGYDKEVDMWSIGVITYI 199 |
| Ciona intestinalis | Query 170 TPAPDAPLKIADFGLSKIVEHQVLMKTVCG#TPGYCAPEILRGCAYGPEVDMWSVGIITYI 229 + | |+ + |+||||||+ ++|| ||#|||| |||+|+ || |||+||+|+| || Sbjct 145 STAEDSKIMISDFGLSKVEVEGQMLKTACG#TPGYVAPEVLKQKPYGKEVDVWSIGVIAYI 204 |
| Yarrowia lipolytica | Query 170 TPAPDAPLKIADFGLSKIVEHQVL--MKTVCG#TPGYCAPEILRGCAYGPEVDMWSVGIIT 227 | | |+ | |||||||+|++ + +|| ||#|||| |||| +| ||||++|+|| Sbjct 143 TAAEDSDLLIADFGLSRIIDEERFRTLKTTCG#TPGYMAPEIFTKAGHGKPVDMWALGVIT 202 Query 228 Y 228 | Query 228 Y 228 |
| Dictyostelium discoideum AX4 | Query 174 DAPLKIADFGLSKIVEHQVLMKTVCG#TPGYCAPEILRGCAYGPEVDMWSVGIITYI 229 | +|||||||||| ++| ||#|| | |||+| | +| |||||+|+|||| Sbjct 192 DMTIKIADFGLSKIFGTGEALETSCG#TPDYVAPEVLTGGSYDNAVDMWSIGVITYI 247 |
| Drosophila pseudoobscura | Query 170 TPAPDAPLKIADFGLSKIVEHQVLMKTVCG#TPGYCAPEILRGCAYGPEVDMWSVGIITYI 229 +| |+ + |+||||||+ | +| | ||#|||| |||+| || ||+||+|+|+|| Sbjct 170 SPEDDSKIMISDFGLSKM-EDSGIMATACG#TPGYVAPEVLAQKPYGKAVDVWSIGVISYI 228 |
| Drosophila melanogaster | Query 170 TPAPDAPLKIADFGLSKIVEHQVLMKTVCG#TPGYCAPEILRGCAYGPEVDMWSVGIITYI 229 +| |+ + |+||||||+ | +| | ||#|||| |||+| || ||+||+|+|+|| Sbjct 178 SPDDDSKIMISDFGLSKM-EDSGIMATACG#TPGYVAPEVLAQKPYGKAVDVWSIGVISYI 236 |
| Entamoeba histolytica HM-1:IMSS | Query 170 TPAPDAPLKIADFGLSK-IVEHQVLMKTVCG#TPGYCAPEILRGCAYGPEVDMWSVGIITY 228 +| | ++||||| || | | ++ | ||#|| | |||+| || ||||||+|+||| Sbjct 138 SPDDDTDVRIADFGFSKMITEDAQILLTACG#TPVYVAPEVLNAKGYGMEVDMWSIGVITY 197 Query 229 I 229 + Sbjct 198 V 198 |
| Glossina morsitans morsitans | Query 170 TPAPDAPLKIADFGLSKIVEHQVLMKTVCG#TPGYCAPEILRGCAYGPEVDMWSVGIITYI 229 +| || + |+|||||| | | | ||#|||| |||+| || ||+||+|+|+|| Sbjct 179 SPDDDAKIMISDFGLSK-TEDSGTMATACG#TPGYVAPEVLAQKPYGKAVDVWSIGVISYI 237 |
| Aspergillus nidulans FGSC A4 | Query 170 TPAPDAPLKIADFGLSKIVEHQVL--MKTVCG#TPGYCAPEILRGCAYGPEVDMWSVGIIT 227 || +| | |||||||+|++ + | + | ||#|||| |||| +| ||+|++|+|| Sbjct 151 TPEDNADLLIADFGLSRIMDEEQLHVLTTTCG#TPGYMAPEIFDKSGHGKPVDIWAIGVIT 210 Query 228 Y 228 | Query 228 Y 228 |
| Gibberella zeae PH-1 | Query 170 TPAPDAPLKIADFGLSKIVEHQV--LMKTVCG#TPGYCAPEILRGCAYGPEVDMWSVGIIT 227 || +| | |||||||+|++ + ++ | ||#|||| |||| + +| ||+|++|+|| Sbjct 149 TPEDNADLLIADFGLSRIMDEEQFHVLTTTCG#TPGYMAPEIFKKTGHGKPVDLWALGVIT 208 Query 228 Y 228 | Query 228 Y 228 |
| Aspergillus fumigatus Af293 | Query 170 TPAPDAPLKIADFGLSKIVEHQV--LMKTVCG#TPGYCAPEILRGCAYGPEVDMWSVGIIT 227 || +| | |||||||+|++ + ++ | ||#|||| |||| + +| ||+|++|+|| Sbjct 151 TPEDNADLLIADFGLSRIMDEEQFHVLTTTCG#TPGYMAPEIFKKSGHGKPVDIWAIGVIT 210 Query 228 Y 228 | Query 228 Y 228 |
| Aspergillus oryzae RIB40 | Query 170 TPAPDAPLKIADFGLSKIVEHQV--LMKTVCG#TPGYCAPEILRGCAYGPEVDMWSVGIIT 227 || +| | |||||||+|++ + ++ | ||#|||| |||| + +| ||+|++|+|| Sbjct 177 TPEDNADLLIADFGLSRIMDEEQFHVLTTTCG#TPGYMAPEIFKKSGHGKPVDIWAIGVIT 236 Query 228 Y 228 | Query 228 Y 228 |
| Bombyx mori | Query 177 LKIADFGLSKIVEHQVLMKTVCG#TPGYCAPEILRGCA---YGPEVDMWSVGIITYI 229 +|| |||||| | |||+||#|| | |||+|| ||||||+||+|+| ++ Sbjct 318 VKITDFGLSKFVGEDSFMKTMCG#TPLYLAPEVLRANGQNTYGPEVDVWSLGVIFFV 373 |
| Emericella nidulans | Query 170 TPAPDAPLKIADFGLSKIVEHQVL--MKTVCG#TPGYCAPEILRGCAYGPEVDMWSVGIIT 227 || +| | |||||||+|++ + | + | ||#|||| |||| +| ||+|++|+|| Sbjct 151 TPEDNADLLIADFGLSRIMDEEQLHVLTTTCG#TPGYMAPEIFDKSGHGKPVDIWAIGLIT 210 Query 228 Y 228 | Query 228 Y 228 |
| Colletotrichum gloeosporioides | Query 170 TPAPDAPLKIADFGLSKIVEHQV--LMKTVCG#TPGYCAPEILRGCAYGPEVDMWSVGIIT 227 || +| | |||||||+|++ + ++ | ||#|||| |||| + +| ||+|++|+|| Sbjct 149 TPEDNADLLIADFGLSRIMDEEQFHVLTTTCG#TPGYMAPEIFKKIGHGKPVDVWALGVIT 208 Query 228 Y 228 | Query 228 Y 228 |
| Arthrobotrys dactyloides | Query 170 TPAPDAPLKIADFGLSKIVEHQV--LMKTVCG#TPGYCAPEILRGCAYGPEVDMWSVGIIT 227 || +| | |||||||+|++ + ++ | ||#|||| |||| + +| ||+|++|+|| Sbjct 160 TPEDNADLLIADFGLSRIMDEERFHVLTTTCG#TPGYMAPEIFKKTGHGKPVDIWAIGVIT 219 Query 228 Y 228 | Query 228 Y 228 |
| Cryptococcus neoformans var. neoformans JEC21 | Query 170 TPAPDAPLKIADFGLSKIVEHQ--VLMKTVCG#TPGYCAPEILRGCAYGPEVDMWSVGIIT 227 + | || | +|||||||+++ ++ | ||#|||| |||| + +| ||+|++|+|| Sbjct 143 SKAEDADLMLADFGLSKVLDDDKFAILTTTCG#TPGYMAPEIFKKAGHGKPVDIWAIGVIT 202 Query 228 Y 228 | Query 228 Y 228 |
| Caenorhabditis elegans | Query 174 DAPLKIADFGLSKIVEHQVLMKTVCG#TPGYCAPEILRGCAYGPEVDMWSVGIITYI 229 |+ + |+|||||| | +| | ||#|||| |||+|+ || ||+||+|+| || Sbjct 158 DSKIMISDFGLSK-TEDSGVMATACG#TPGYVAPEVLQQKPYGKAVDVWSIGVIAYI 212 |
| Caenorhabditis briggsae AF16 | Query 174 DAPLKIADFGLSKIVEHQVLMKTVCG#TPGYCAPEILRGCAYGPEVDMWSVGIITYI 229 |+ + |+|||||| | +| | ||#|||| |||+|+ || ||+||+|+| || Sbjct 158 DSKIMISDFGLSK-TEDSGVMATACG#TPGYVAPEVLQQKPYGKAVDVWSIGVIAYI 212 |
| Ustilago maydis 521 | Query 174 DAPLKIADFGLSKIVEHQ--VLMKTVCG#TPGYCAPEILRGCAYGPEVDMWSVGIITY 228 ++ | |||||||++|+ + ++ | ||#|||| |||| + +| ||||++|+||| Sbjct 150 ESDLLIADFGLSRVVDDEKITVLSTTCG#TPGYMAPEIFKKTGHGKPVDMWAIGVITY 206 |
| Guillardia theta | Query 177 LKIADFGLSKIVEHQVLMKTVCG#TPGYCAPEILRGCAY-GPEVDMWSVGIITY 228 +|||||||| |++ +|| ||#+| | |||++ | +| |||||+|| |+| | Sbjct 149 VKIADFGLSNIMKDGNFLKTSCG#SPNYAAPEVINGKSYLGPEVDVWSCGVIMY 201 |
| Schizophyllum commune | Query 170 TPAPDAPLKIADFGLSKIVEHQVL--MKTVCG#TPGYCAPEILRGCAYGPEVDMWSVGIIT 227 | | || + |||||||+++| + | + +||#|||| |||| + + ||+|++|+|| Sbjct 237 TQAEDADIMIADFGLSRVMEEEKLNMLTEICG#TPGYMAPEIFKKTGHSKPVDIWAMGVIT 296 Query 228 Y 228 | Query 228 Y 228 |
| Arabidopsis thaliana | Query 177 LKIADFGLSKIVEHQVLMKTVCG#TPGYCAPEILRGCAYGPEVDMWSVGIITY 228 +|| ||||| ++ +|| ||#+| | |||++ | |||+||+|| |+| | Sbjct 156 IKIVDFGLSNVMHDGHFLKTSCG#SPNYAAPEVISGKPYGPDVDIWSCGVILY 207 |
| Xenopus tropicalis | Query 173 PDAPLKIADFGLSKIVEHQVLMKTVCG#TPGYCAPEILRGCAYGPEVDMWSVGIITYI 229 | +|| ||||++ +| + |++||#|| | |||++ || ||||+|+|||| Sbjct 153 PHPKIKIIDFGLAQKIEDGTVFKSLCG#TPQYIAPEVINYEPLGPPTDMWSIGVITYI 209 |
| Bos taurus | Query 170 TPAPDAPLKIADFGLSKIVEHQVLMKTVCG#TPGYCAPEILRGCAYGPEVDMWSVGIITYI 229 || |+ + ++||||||| + ++ | ||#|||| |||+| || ||+|++|+|+|| Sbjct 146 TPFEDSKIMVSDFGLSKI-QAGNMLGTACG#TPGYVAPELLEQKPYGKAVDVWALGVISYI 204 |
| Candida albicans SC5314 | Query 177 LKIADFGLSKIVEHQVLMKTVCG#TPGYCAPEILRGCAY-GPEVDMWSVGIITYI 229 +|||||||| |+ +|| ||#+| | |||++ | | |||||+|| |+| |+ Sbjct 188 VKIADFGLSNIMTDGNFLKTSCG#SPNYAAPEVISGKLYAGPEVDVWSAGVILYV 241 |
| Schizosaccharomyces pombe 972h- | Query 177 LKIADFGLSKIVEHQVLMKTVCG#TPGYCAPEILRGCAY-GPEVDMWSVGIITYI 229 +|||||||| |+ +|| ||#+| | |||++ | | |||||+|| ||+ |+ Sbjct 170 VKIADFGLSNIMTDGNFLKTSCG#SPNYAAPEVINGKLYAGPEVDVWSCGIVLYV 223 |
| Schizosaccharomyces pombe | Query 177 LKIADFGLSKIVEHQVLMKTVCG#TPGYCAPEILRGCAY-GPEVDMWSVGIITYI 229 +|||||||| |+ +|| ||#+| | |||++ | | |||||+|| ||+ |+ Sbjct 170 VKIADFGLSNIMTDGNFLKTSCG#SPNYAAPEVINGKLYAGPEVDVWSCGIVLYV 223 |
| Arabidopsis thaliana | Query 177 LKIADFGLSKIVEHQVLMKTVCG#TPGYCAPEILRGCAYGPEVDMWSVGIITY 228 +|| ||||| ++ +|| ||#+| | |||++ | |||+||+|| |+| | Sbjct 156 IKIVDFGLSNVMHDGHFLKTSCG#SPNYAAPEVISGKPYGPDVDIWSCGVILY 207 |
| Candida glabrata CBS138 | Query 177 LKIADFGLSKIVEHQVLMKTVCG#TPGYCAPEILRGCAY-GPEVDMWSVGIITYI 229 +|||||||| |+ +|| ||#+| | |||++ | | |||||+|| |+| |+ Sbjct 175 VKIADFGLSNIMTDGNFLKTSCG#SPNYAAPEVISGKLYAGPEVDVWSCGVILYV 228 |
| Kluyveromyces lactis | Query 177 LKIADFGLSKIVEHQVLMKTVCG#TPGYCAPEILRGCAY-GPEVDMWSVGIITYI 229 +|||||||| |+ +|| ||#+| | |||++ | | |||||+|| |+| |+ Sbjct 171 VKIADFGLSNIMTDGNFLKTSCG#SPNYAAPEVISGKLYAGPEVDVWSSGVILYV 224 |
| Cochliobolus carbonum | Query 174 DAPLKIADFGLSKIVEHQVLMKTVCG#TPGYCAPEILRGCAY-GPEVDMWSVGIITYI 229 |+ +|||||||| |+ +|| ||#+| | |||++ | | |||||+|| |+| |+ Sbjct 197 DSNVKIADFGLSNIMTDGNFLKTSCG#SPNYAAPEVISGKLYAGPEVDVWSCGVILYV 253 |
| Ashbya gossypii ATCC 10895 | Query 177 LKIADFGLSKIVEHQVLMKTVCG#TPGYCAPEILRGCAY-GPEVDMWSVGIITYI 229 +|||||||| |+ +|| ||#+| | |||++ | | |||||+|| |+| |+ Sbjct 175 VKIADFGLSNIMTDGNFLKTSCG#SPNYAAPEVISGKLYAGPEVDVWSSGVILYV 228 |
| Candida tropicalis | Query 177 LKIADFGLSKIVEHQVLMKTVCG#TPGYCAPEILRGCAY-GPEVDMWSVGIITYI 229 +|||||||| |+ +|| ||#+| | |||++ | | |||||+|| |+| |+ Sbjct 188 VKIADFGLSNIMTDGNFLKTSCG#SPNYAAPEVISGKLYAGPEVDVWSSGVILYV 241 |
| Glycine max | Query 177 LKIADFGLSKIVEHQVLMKTVCG#TPGYCAPEILRGCAY-GPEVDMWSVGIITY 228 +|||||||| |+ +|| ||#+| | |||++ | | |||||+|| |+| | Sbjct 157 IKIADFGLSNIMRDGHFLKTSCG#SPNYAAPEVISGKLYAGPEVDVWSCGVILY 209 |
| Cucumis sativus | Query 177 LKIADFGLSKIVEHQVLMKTVCG#TPGYCAPEILRGCAY-GPEVDMWSVGIITY 228 +|||||||| |+ +|| ||#+| | |||++ | | |||||+|| |+| | Sbjct 145 VKIADFGLSNIMRDGHFLKTSCG#SPNYAAPEVISGKLYAGPEVDVWSCGVILY 197 |
| Vicia faba | Query 177 LKIADFGLSKIVEHQVLMKTVCG#TPGYCAPEILRGCAY-GPEVDMWSVGIITY 228 +|||||||| |+ +|| ||#+| | |||++ | | |||||+|| |+| | Sbjct 156 VKIADFGLSNIMRDGHFLKTSCG#SPNYAAPEVISGKLYAGPEVDVWSCGVILY 208 |
| Canis familiaris | Query 170 TPAPDAPLKIADFGLSKIVEHQVLMKTVCG#TPGYCAPEILRGCAYGPEVDMWSVGIITYI 229 || ++ + | ||||||+ | +| | ||#|||| |||+| | || ||+|+|||| Sbjct 186 TPEENSKIMITDFGLSKM-EQSGVMSTACG#TPGYVAPEVLAQKPYSKAVDCWSIGVITYI 244 |
| Solanum tuberosum | Query 177 LKIADFGLSKIVEHQVLMKTVCG#TPGYCAPEILRGCAY-GPEVDMWSVGIITY 228 +|||||||| |+ +|| ||#+| | |||++ | | |||||+|| |+| | Sbjct 156 VKIADFGLSNIMRDGHFLKTSCG#SPNYAAPEVISGKLYAGPEVDVWSCGVILY 208 |
| Nicotiana tabacum | Query 177 LKIADFGLSKIVEHQVLMKTVCG#TPGYCAPEILRGCAY-GPEVDMWSVGIITY 228 +|||||||| |+ +|| ||#+| | |||++ | | |||||+|| |+| | Sbjct 156 VKIADFGLSNIMRDGHFLKTSCG#SPNYAAPEVISGKLYAGPEVDVWSCGVILY 208 |
| Pisum sativum | Query 177 LKIADFGLSKIVEHQVLMKTVCG#TPGYCAPEILRGCAY-GPEVDMWSVGIITY 228 +|||||||| |+ +|| ||#+| | |||++ | | |||||+|| |+| | Sbjct 156 VKIADFGLSNIMRDGHFLKTSCG#SPNYAAPEVISGKLYAGPEVDVWSCGVILY 208 |
| Lycopersicon esculentum | Query 177 LKIADFGLSKIVEHQVLMKTVCG#TPGYCAPEILRGCAY-GPEVDMWSVGIITY 228 +|||||||| |+ +|| ||#+| | |||++ | | |||||+|| |+| | Sbjct 156 VKIADFGLSNIMRDGHFLKTSCG#SPNYAAPEVISGKLYAGPEVDVWSCGVILY 208 |
| Oryza sativa | Query 177 LKIADFGLSKIVEHQVLMKTVCG#TPGYCAPEILRGCAY-GPEVDMWSVGIITY 228 +|||||||| ++ +|| ||#+| | |||++ | | |||||+|| |+| | Sbjct 149 VKIADFGLSNVMRDGHFLKTSCG#SPNYAAPEVISGKLYAGPEVDVWSCGVILY 201 |
| Physcomitrella patens subsp. patens | Query 177 LKIADFGLSKIVEHQVLMKTVCG#TPGYCAPEILRGCAY-GPEVDMWSVGIITY 228 +|||||||| ++ +|| ||#+| | |||++ | | |||||+|| |+| | Sbjct 157 VKIADFGLSNVMRDGHFLKTSCG#SPNYAAPEVISGKLYAGPEVDVWSCGVILY 209 |
| Sclerotinia sclerotiorum | Query 177 LKIADFGLSKIVEHQVLMKTVCG#TPGYCAPEILRGCAY-GPEVDMWSVGIITYI 229 +|||||||| |+ +|| ||#+| | |||++ | | |||||+|| |+| |+ Sbjct 195 VKIADFGLSNIMTDGNFLKTSCG#SPNYAAPEVINGKLYAGPEVDVWSCGVILYV 248 |
| Entamoeba histolytica HM-1:IMSS | Query 177 LKIADFGLSKIVEHQVLMKTVCG#TPGYCAPEILRGCAY-GPEVDMWSVGIITYI 229 +|| |||||+|+ + | ||+||#|| | +|||| | | | +||+|| |++ |+ Sbjct 264 IKITDFGLSRIINPETLAKTMCG#TPLYVSPEILSGKPYNGSKVDVWSTGVVLYV 317 |
| Schistosoma japonicum | Query 177 LKIADFGLSKIVEHQVLMKTVCG#TPGYCAPEILRGCAY-GPEVDMWSVGIITY 228 +|||||||| |++ ++| ||#+| | |||++ | | |||||+|| |+| | Sbjct 152 VKIADFGLSNIMQDGEFLRTSCG#SPNYAAPEVISGKLYAGPEVDVWSCGVILY 204 |
| Chaetomium globosum CBS 148.51 | Query 177 LKIADFGLSKIVEHQVLMKTVCG#TPGYCAPEILRGCAY-GPEVDMWSVGIITYI 229 +|||||||| |+ +|| ||#+| | |||++ | | |||||+|| |+| |+ Sbjct 31 VKIADFGLSNIMTDGNFLKTSCG#SPNYAAPEVIGGKLYAGPEVDVWSCGVILYV 84 |
| Triticum aestivum | Query 177 LKIADFGLSKIVEHQVLMKTVCG#TPGYCAPEILRGCAY-GPEVDMWSVGIITY 228 +|||||||| |+ +|| ||#+| | |||++ | | |||||+|| |+| | Sbjct 110 VKIADFGLSNIMRDGHFLKTSCG#SPNYAAPEVISGKLYAGPEVDVWSCGVILY 162 |
* References
[PubMed ID: 17394138] Saino-Saito S, Cave JW, Akiba Y, Sasaki H, Goto K, Kobayashi K, Berlin R, Baker H, ER81 and CaMKIV identify anatomically and phenotypically defined subsets of mouse olfactory bulb interneurons. J Comp Neurol. 2007 Jun 1;502(4):485-96.
[PubMed ID: 17113785] ... Taylor DM, De Koninck P, Minotti S, Durham HD, Manipulation of protein kinases reveals different mechanisms for upregulation of heat shock proteins in motor neurons and non-neuronal cells. Mol Cell Neurosci. 2007 Jan;34(1):20-33. Epub 2006 Nov 20.
[PubMed ID: 17201924] ... Lemberger T, Parlato R, Dassesse D, Westphal M, Casanova E, Turiault M, Tronche F, Schiffmann SN, Schutz G, Expression of Cre recombinase in dopaminoceptive neurons. BMC Neurosci. 2007 Jan 3;8:4.
[PubMed ID: 16982426] ... Boyden ES, Katoh A, Pyle JL, Chatila TA, Tsien RW, Raymond JL, Selective engagement of plasticity mechanisms for motor memory storage. Neuron. 2006 Sep 21;51(6):823-34.
[PubMed ID: 16684769] ... Ohmae S, Takemoto-Kimura S, Okamura M, Adachi-Morishima A, Nonaka M, Fuse T, Kida S, Tanji M, Furuyashiki T, Arakawa Y, Narumiya S, Okuno H, Bito H, Molecular identification and characterization of a family of kinases with homology to Ca2+/calmodulin-dependent protein kinases I/IV. J Biol Chem. 2006 Jul 21;281(29):20427-39. Epub 2006 May 8.
[PubMed ID: 1358813] ... Mock BA, Contente S, Kenyon K, Friedman RM, Kozak CA, The gene for lysyl oxidase maps to mouse chromosome 18. Genomics. 1992 Nov;14(3):822-3.
[PubMed ID: 1319776] ... Danciger M, Kozak CA, Adamson MC, Farber DB, Chromosomal localization of the murine genes for the alpha- and beta-subunits of calcium/calmodulin-dependent protein kinase II. Mamm Genome. 1992;3(2):122-5.
[PubMed ID: 1893997] ... Jones DA, Glod J, Wilson-Shaw D, Hahn WE, Sikela JM, cDNA sequence and differential expression of the mouse Ca2+/calmodulin-dependent protein kinase IV gene. FEBS Lett. 1991 Sep 2;289(1):105-9.
[PubMed ID: 1981056] ... Sikela JM, Adamson MC, Wilson-Shaw D, Kozak CA, Genetic mapping of the gene for Ca2+/calmodulin-dependent protein kinase IV (Camk-4) to mouse chromosome 18. Genomics. 1990 Nov;8(3):579-82.
[PubMed ID: 2536634] ... Sikela JM, Law ML, Kao FT, Hartz JA, Wei Q, Hahn WE, Chromosomal localization of the human gene for brain Ca2+/calmodulin-dependent protein kinase type IV. Genomics. 1989 Jan;4(1):21-7.