SB0009 : Neurofibromin 2, Merlin
[ CaMP Format ]
* Basic Information
| Organism | Homo sapiens (human) |
| Protein Names | merlin isoform 1 [Homo sapiens]; merlin isoform 1; moesin-ezrin-radixin like; neurofibromin 2 (bilateral acoustic neuroma); schwannomin; moesin-ezrin-radizin-like protein; schwannomerlin; merlin; neurofibromin-2; moesin-ezrin-radixin-like protein; Merlin; Moesin-ezrin-radixin-like protein; Neurofibromin-2; Schwannomerlin; Schwannomin |
| Gene Names | NF2; SCH; neurofibromin 2 (merlin) |
| Gene Locus | 22q12.2; chromosome 22 |
| GO Function | Not available |
* Information From OMIM
Description: Cell-cell contact between normal cultured diploid cells results in inhibition of proliferation despite the unlimited availability of nutrients and growth-promoting factors. A similar phenomenon occurs in vivo in all adult solid tissues. NF2 is a critical regulator of contact-dependent inhibition of proliferation and functions at the interface between cell-cell adhesion, transmembrane signaling, and the actin cytoskeleton (Curto and McClatchey, 2008).
Function: Gutmann et al. (1999) studied rat schwannoma cell lines overexpressing wildtype merlin isoforms and mutant merlin proteins. Overexpression of wildtype merlin resulted in transient alterations in F-actin organization, cell spreading, and cell attachment, and in impaired cell motility as measured in an in vitro motility assay. These effects were observed only in cells overexpressing a merlin isoform capable of inhibiting cell growth and not with mutant merlin molecules (harboring NF2 patient mutations) or a merlin splice variant (isoform II) lacking growth-inhibitory activity. These data indicated that merlin may function to maintain normal cytoskeletal organization, and suggested that its influence on cell growth depends on specific cytoskeletal rearrangements.
* Structure Information
1. Primary Information
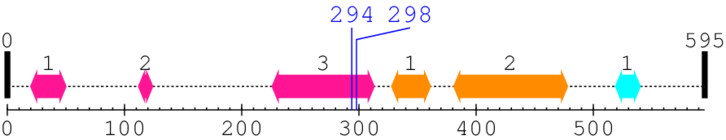
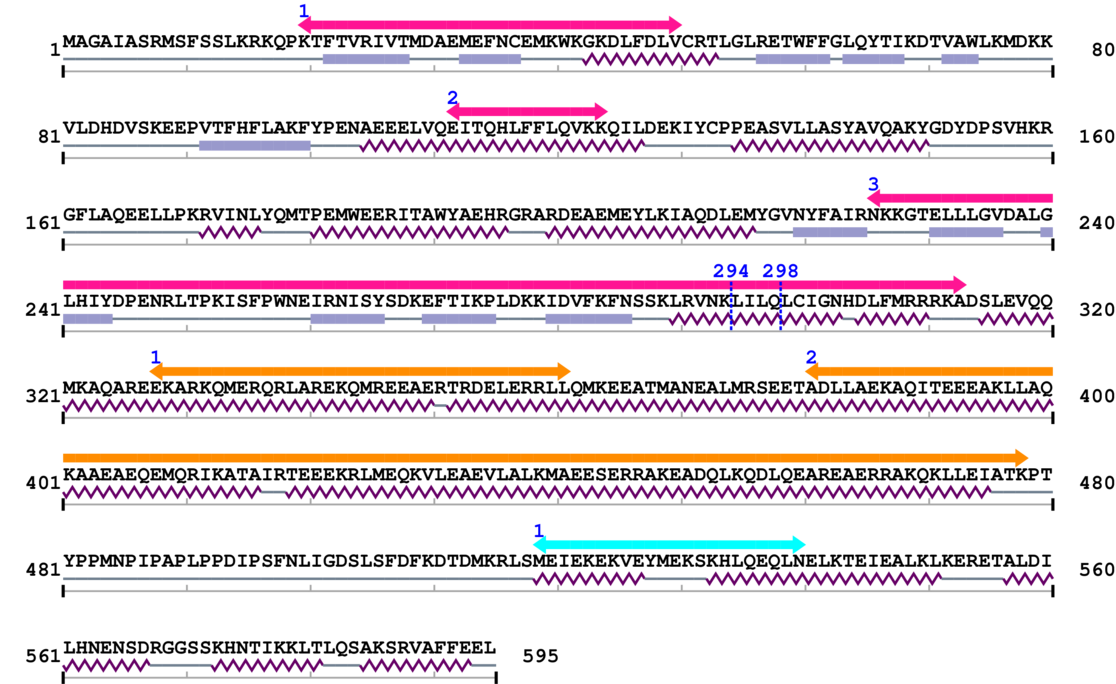
Length: 595 aa
Average Mass: 69.689 kDa
Monoisotopic Mass: 69.646 kDa
2. Domain Information
Annotated Domains: interpro / pfam / smart / prosite
Computationally Assigned Domains (Pfam+HMMER):
| domain name | begin | end | score | e-value |
|---|---|---|---|---|
| FERM C-terminal PH-like domain 1. | 20 | 50 | 43.0 | 0.0 |
| FERM C-terminal PH-like domain 2. | 112 | 124 | 78.0 | 0.1 |
| FERM C-terminal PH-like domain 3. | 226 | 313 | 1.0 | 0.0 |
| --- cleavage 294 (inside FERM C-terminal PH-like domain 226..313) --- | ||||
| --- cleavage 298 (inside FERM C-terminal PH-like domain 226..313) --- | ||||
| FERM central domain 1. | 328 | 361 | 67.0 | 1.2 |
| FERM central domain 2. | 381 | 478 | 20.0 | 17.5 |
| FERM N-terminal domain 1. | 519 | 540 | 43.0 | 0.2 |
3. Sequence Information
Fasta Sequence: SB0009.fasta
Amino Acid Sequence and Secondary Structures (PsiPred):
* Cleavage Information
2 [sites] cleaved by Calpain 1 and 2
Source Reference: [PubMed ID: 9701243] Kimura Y, Koga H, Araki N, Mugita N, Fujita N, Takeshima H, Nishi T, Yamashima T, Saido TC, Yamasaki T, Moritake K, Saya H, Nakao M, The involvement of calpain-dependent proteolysis of the tumor suppressor NF2 (merlin) in schwannomas and meningiomas. Nat Med. 1998 Aug;4(8):915-22.
Cleavage sites (±10aa)
[Site 1] FNSSKLRVNK294-LILQLCIGNH
Lys294  Leu
Leu
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Phe285 | Asn286 | Ser287 | Ser288 | Lys289 | Leu290 | Arg291 | Val292 | Asn293 | Lys294 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Leu295 | Ile296 | Leu297 | Gln298 | Leu299 | Cys300 | Ile301 | Gly302 | Asn303 | His304 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| SYSDKEFTIKPLDKKIDVFKFNSSKLRVNKLILQLCIGNHDLFMRRRKADSLEVQQMKAQ |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | Mus musculus | 127.00 | 21 | unnamed protein product |
| 2 | Homo sapiens | 127.00 | 4 | neurofibromin 2 isoform 7 |
| 3 | Homo sapiens | 126.00 | 21 | Neurofibromin 2 (bilateral acoustic neuroma) |
| 4 | N/A | 126.00 | 20 | 1915322A membrane-organizing protein |
| 5 | Rattus norvegicus | 126.00 | 6 | MERL_RAT Merlin (Moesin-ezrin-radixin-like protei |
| 6 | Papio anubis anubis | 126.00 | 1 | MERL_PAPAN Merlin (Moesin-ezrin-radixin-like prot |
| 7 | Canis familiaris | 125.00 | 14 | PREDICTED: similar to neurofibromin 2 isoform 9 i |
| 8 | Danio rerio | 115.00 | 4 | PREDICTED: hypothetical protein |
| 9 | Tetraodon nigroviridis | 114.00 | 7 | unnamed protein product |
| 10 | Bos taurus | 89.40 | 3 | ezrin |
| 11 | Gallus gallus | 89.00 | 2 | ezrin |
| 12 | Oryctolagus cuniculus | 89.00 | 1 | villin 2 (ezrin) |
| 13 | Canis familiaris | 87.00 | 5 | PREDICTED: similar to Moesin (Membrane-organizing |
| 14 | Sus scrofa | 87.00 | 2 | radixin |
| 15 | mice, keratinocytes, Balb/MK, Peptide, 583 aa | 87.00 | 1 | RADI_MOUSE Radixin (ESP10) gi |
| 16 | Xenopus laevis | 86.70 | 2 | U29763_1 moesin |
| 17 | Biomphalaria glabrata | 86.30 | 1 | ezrin/radixin/moesin |
| 18 | Aplysia californica | 85.90 | 1 | ezrin/radixin/moesin |
| 19 | Ciona intestinalis | 85.10 | 1 | ezrin/radixin/moesin (ERM)-like protein |
| 20 | Caenorhabditis elegans | 84.30 | 3 | ERM-1Asv |
| 21 | Caenorhabditis briggsae AF16 | 84.30 | 1 | Hypothetical protein CBG12867 |
| 22 | Drosophila melanogaster | 82.40 | 5 | Moesin CG10701-PJ, isoform J |
| 23 | Drosophila pseudoobscura | 82.40 | 2 | GA10507-PA |
| 24 | Drosophila melanogaster | 80.90 | 2 | GH01330p |
| 25 | Apis mellifera | 80.10 | 1 | PREDICTED: similar to Moesin CG10701-PD, isoform |
| 26 | Taenia solium | 76.30 | 1 | H17g protein, tegumental antigen |
| 27 | Taenia saginata | 76.30 | 1 | myosin-like protein |
| 28 | Echinococcus granulosus | 75.50 | 1 | EG10 |
| 29 | Echinococcus multilocularis | 75.50 | 1 | tegument protein gi |
Top-ranked sequences
| organism | matching |
|---|---|
| Mus musculus | Query 265 SYSDKEFTIKPLDKKIDVFKFNSSKLRVNK#LILQLCIGNHDLFMRRRKADSLEVQQMKAQ 324 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 265 SYSDKEFTIKPLDKKIDVFKFNSSKLRVNK#LILQLCIGNHDLFMRRRKADSLEVQQMKAQ 324 |
| Homo sapiens | Query 265 SYSDKEFTIKPLDKKIDVFKFNSSKLRVNK#LILQLCIGNHDLFMRRRKADSLEVQQMKAQ 324 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 265 SYSDKEFTIKPLDKKIDVFKFNSSKLRVNK#LILQLCIGNHDLFMRRRKADSLEVQQMKAQ 324 |
| Homo sapiens | Query 265 SYSDKEFTIKPLDKKIDVFKFNSSKLRVNK#LILQLCIGNHDLFMRRRKADSLEVQQMKAQ 324 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 265 SYSDKEFTIKPLDKKIDVFKFNSSKLRVNK#LILQLCIGNHDLFMRRRKADSLEVQQMKAQ 324 |
| N/A | Query 265 SYSDKEFTIKPLDKKIDVFKFNSSKLRVNK#LILQLCIGNHDLFMRRRKADSLEVQQMKAQ 324 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 265 SYSDKEFTIKPLDKKIDVFKFNSSKLRVNK#LILQLCIGNHDLFMRRRKADSLEVQQMKAQ 324 |
| Rattus norvegicus | Query 265 SYSDKEFTIKPLDKKIDVFKFNSSKLRVNK#LILQLCIGNHDLFMRRRKADSLEVQQMKAQ 324 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 265 SYSDKEFTIKPLDKKIDVFKFNSSKLRVNK#LILQLCIGNHDLFMRRRKADSLEVQQMKAQ 324 |
| Papio anubis anubis | Query 265 SYSDKEFTIKPLDKKIDVFKFNSSKLRVNK#LILQLCIGNHDLFMRRRKADSLEVQQMKAQ 324 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 265 SYSDKEFTIKPLDKKIDVFKFNSSKLRVNK#LILQLCIGNHDLFMRRRKADSLEVQQMKAQ 324 |
| Canis familiaris | Query 265 SYSDKEFTIKPLDKKIDVFKFNSSKLRVNK#LILQLCIGNHDLFMRRRKADSLEVQQMKAQ 324 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 265 SYSDKEFTIKPLDKKIDVFKFNSSKLRVNK#LILQLCIGNHDLFMRRRKADSLEVQQMKAQ 324 |
| Danio rerio | Query 265 SYSDKEFTIKPLDKKIDVFKFNSSKLRVNK#LILQLCIGNHDLFMRRRKADSLEVQQMKAQ 324 ||||||| |||+||+ ||||||||||||||#|||||||||||||||||+ ||||||||| | Sbjct 264 SYSDKEFAIKPVDKRADVFKFNSSKLRVNK#LILQLCIGNHDLFMRRRRVDSLEVQQMKTQ 323 |
| Tetraodon nigroviridis | Query 265 SYSDKEFTIKPLDKKIDVFKFNSSKLRVNK#LILQLCIGNHDLFMRRRKADSLEVQQMKAQ 324 ||||||||||||+|| |||||||+|| ||#|||||||||||||||||+ ||||||||||| Sbjct 159 SYSDKEFTIKPLEKKTKVFKFNSSRLRANK#LILQLCIGNHDLFMRRRRVDSLEVQQMKAQ 218 |
| Bos taurus | Query 265 SYSDKEFTIKPLDKKIDVFKFNSSKLRVNK#LILQLCIGNHDLFMRRRKADSLEVQQMKAQ 324 |++||+| |||+||| | | + +||+||# |||||+|||+|+||||| |++|||||||| Sbjct 249 SFNDKKFVIKPIDKKAPDFVFYAPRLRINK#RILQLCMGNHELYMRRRKPDTIEVQQMKAQ 308 |
| Gallus gallus | Query 265 SYSDKEFTIKPLDKKIDVFKFNSSKLRVNK#LILQLCIGNHDLFMRRRKADSLEVQQMKAQ 324 |++||+| |||+||| | | + +||+||# |||||+|||+|+||||| |++|||||||| Sbjct 249 SFNDKKFVIKPIDKKAPDFVFYAPRLRINK#RILQLCMGNHELYMRRRKPDTIEVQQMKAQ 308 |
| Oryctolagus cuniculus | Query 265 SYSDKEFTIKPLDKKIDVFKFNSSKLRVNK#LILQLCIGNHDLFMRRRKADSLEVQQMKAQ 324 |++||+| |||+||| | | + +||+||# |||||+|||+|+||||| |++|||||||| Sbjct 249 SFNDKKFVIKPIDKKAPDFVFYAPRLRINK#RILQLCMGNHELYMRRRKPDTIEVQQMKAQ 308 |
| Canis familiaris | Query 265 SYSDKEFTIKPLDKKIDVFKFNSSKLRVNK#LILQLCIGNHDLFMRRRKADSLEVQQMKAQ 324 |++||+| |||+||| | | + +||+||# || ||+|||+|+||||| |++|||||||| Sbjct 249 SFNDKKFVIKPIDKKAPDFVFYAPRLRINK#RILALCMGNHELYMRRRKPDTIEVQQMKAQ 308 |
| Sus scrofa | Query 265 SYSDKEFTIKPLDKKIDVFKFNSSKLRVNK#LILQLCIGNHDLFMRRRKADSLEVQQMKAQ 324 |++||+| |||+||| | | + +||+||# || ||+|||+|+||||| |++|||||||| Sbjct 249 SFNDKKFVIKPIDKKAPDFVFYAPRLRINK#RILALCMGNHELYMRRRKPDTIEVQQMKAQ 308 |
| mice, keratinocytes, Balb/MK, Peptide, 583 aa | Query 265 SYSDKEFTIKPLDKKIDVFKFNSSKLRVNK#LILQLCIGNHDLFMRRRKADSLEVQQMKAQ 324 |++||+| |||+||| | | + +||+||# || ||+|||+|+||||| |++|||||||| Sbjct 249 SFNDKKFVIKPIDKKAPDFVFYAPRLRINK#RILALCMGNHELYMRRRKPDTIEVQQMKAQ 308 |
| Xenopus laevis | Query 265 SYSDKEFTIKPLDKKIDVFKFNSSKLRVNK#LILQLCIGNHDLFMRRRKADSLEVQQMKAQ 324 |++||+| |||+||| | | + +||+||# || ||+|||+|+||||| |++|||||||| Sbjct 249 SFNDKKFVIKPIDKKAPDFVFYAPRLRINK#RILALCMGNHELYMRRRKPDTIEVQQMKAQ 308 |
| Biomphalaria glabrata | Query 265 SYSDKEFTIKPLDKKIDVFKFNSSKLRVNK#LILQLCIGNHDLFMRRRKADSLEVQQMKAQ 324 |++||+| |||+||| | | + +||+||# || ||+|||+|+||||| |++|||||||| Sbjct 250 SFNDKKFVIKPIDKKAPDFVFYAPRLRINK#RILALCMGNHELYMRRRKPDTIEVQQMKAQ 309 |
| Aplysia californica | Query 265 SYSDKEFTIKPLDKKIDVFKFNSSKLRVNK#LILQLCIGNHDLFMRRRKADSLEVQQMKAQ 324 |++||+| |||+||| | | + +||+||# || ||+|||+|+||||| |++|||||||| Sbjct 250 SFNDKKFIIKPIDKKAPDFVFYAPRLRINK#RILALCMGNHELYMRRRKPDTIEVQQMKAQ 309 |
| Ciona intestinalis | Query 265 SYSDKEFTIKPLDKKIDVFKFNSSKLRVNK#LILQLCIGNHDLFMRRRKADSLEVQQMKAQ 324 |++||+| |||+||| | | +||+||# || ||+|||+|+||||| |++|||||||| Sbjct 273 SFNDKKFVIKPIDKKAPDFVFYVERLRINK#RILALCMGNHELYMRRRKPDTIEVQQMKAQ 332 |
| Caenorhabditis elegans | Query 265 SYSDKEFTIKPLDKKIDVFKFNSSKLRVNK#LILQLCIGNHDLFMRRRKADSLEVQQMKAQ 324 |++||+| |||+||| | | + +||+||# || ||+|||+|+||||| |++|||||| | Sbjct 250 SFNDKKFVIKPIDKKAHDFVFYAPRLRINK#RILALCMGNHELYMRRRKPDTIEVQQMKQQ 309 |
| Caenorhabditis briggsae AF16 | Query 265 SYSDKEFTIKPLDKKIDVFKFNSSKLRVNK#LILQLCIGNHDLFMRRRKADSLEVQQMKAQ 324 |++||+| |||+||| | | + +||+||# || ||+|||+|+||||| |++|||||| | Sbjct 245 SFNDKKFVIKPIDKKAHDFVFYAPRLRINK#RILALCMGNHELYMRRRKPDTIEVQQMKQQ 304 |
| Drosophila melanogaster | Query 265 SYSDKEFTIKPLDKKIDVFKFNSSKLRVNK#LILQLCIGNHDLFMRRRKADSLEVQQMKAQ 324 |+|+|+| |||+||| | | + ++|+||# || ||+|||+|+||||| |+++||||||| Sbjct 250 SFSEKKFIIKPIDKKAPDFMFFAPRVRINK#RILALCMGNHELYMRRRKPDTIDVQQMKAQ 309 |
| Drosophila pseudoobscura | Query 265 SYSDKEFTIKPLDKKIDVFKFNSSKLRVNK#LILQLCIGNHDLFMRRRKADSLEVQQMKAQ 324 |+|+|+| |||+||| | | + ++|+||# || ||+|||+|+||||| |+++||||||| Sbjct 245 SFSEKKFIIKPIDKKAPDFMFFAPRVRINK#RILALCMGNHELYMRRRKPDTIDVQQMKAQ 304 |
| Drosophila melanogaster | Query 265 SYSDKEFTIKPLDKKIDVFKFNSSKLRVNK#LILQLCIGNHDLFMRRRKADSLEVQQMKAQ 324 |+ ||+|||+ +| |+ | | | | +||#+|| || |||||+||||| |++|+|||||| Sbjct 259 SFDDKKFTIRLVDAKVSNFIFYSQDLHINK#MILDLCKGNHDLYMRRRKPDTMEIQQMKAQ 318 |
| Apis mellifera | Query 265 SYSDKEFTIKPLDKKIDVFKFNSSKLRVNK#LILQLCIGNHDLFMRRRKADSLEVQQMKAQ 324 |+++|+| |||+||| | | ++++++||# || ||+|||+|+||||| |+++||||||| Sbjct 184 SFNEKKFIIKPIDKKAPDFVFFATRVKINK#RILALCMGNHELYMRRRKPDTIDVQQMKAQ 243 |
| Taenia solium | Query 265 SYSDKEFTIKPLDKKIDVFKFNSSKLRVNK#LILQLCIGNHDLFMRRRKADSLEVQQMKAQ 324 |+ ||+| ||| || | | | ++||# || || |||+|+|||||+||+|||||| | Sbjct 244 SFHDKKFIIKPADKSAKEFYFLVEKSKINK#RILALCTGNHELYMRRRKSDSIEVQQMKIQ 303 |
| Taenia saginata | Query 265 SYSDKEFTIKPLDKKIDVFKFNSSKLRVNK#LILQLCIGNHDLFMRRRKADSLEVQQMKAQ 324 |+ ||+| ||| || | | | ++||# || || |||+|+|||||+||+|||||| | Sbjct 251 SFHDKKFIIKPADKSAKEFYFLVEKSKINK#RILALCTGNHELYMRRRKSDSIEVQQMKIQ 310 |
| Echinococcus granulosus | Query 265 SYSDKEFTIKPLDKKIDVFKFNSSKLRVNK#LILQLCIGNHDLFMRRRKADSLEVQQMKAQ 324 |+ ||+| ||| || | | | ++||# || || |||+|+|||||+||+|||||| | Sbjct 251 SFHDKKFIIKPADKSAKEFFFLVEKSKINK#RILALCTGNHELYMRRRKSDSIEVQQMKIQ 310 |
| Echinococcus multilocularis | Query 265 SYSDKEFTIKPLDKKIDVFKFNSSKLRVNK#LILQLCIGNHDLFMRRRKADSLEVQQMKAQ 324 |+ ||+| ||| || | | | ++||# || || |||+|+|||||+||+|||||| | Sbjct 251 SFHDKKFIIKPADKSAKEFFFLVEKSKINK#RILALCTGNHELYMRRRKSDSIEVQQMKIQ 310 |
[Site 2] KLRVNKLILQ298-LCIGNHDLFM
Gln298  Leu
Leu
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Lys289 | Leu290 | Arg291 | Val292 | Asn293 | Lys294 | Leu295 | Ile296 | Leu297 | Gln298 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Leu299 | Cys300 | Ile301 | Gly302 | Asn303 | His304 | Asp305 | Leu306 | Phe307 | Met308 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| KEFTIKPLDKKIDVFKFNSSKLRVNKLILQLCIGNHDLFMRRRKADSLEVQQMKAQAREE |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | Mus musculus | 126.00 | 21 | unnamed protein product |
| 2 | Homo sapiens | 125.00 | 21 | NF2 protein |
| 3 | N/A | 125.00 | 19 | neurofibromatosis type 2 |
| 4 | Homo sapiens | 125.00 | 4 | neurofibromin 2 isoform 6 |
| 5 | Papio anubis anubis | 125.00 | 1 | MERL_PAPAN Merlin (Moesin-ezrin-radixin-like prot |
| 6 | Canis familiaris | 124.00 | 14 | PREDICTED: similar to neurofibromin 2 isoform 9 i |
| 7 | Rattus norvegicus | 124.00 | 6 | MERL_RAT Merlin (Moesin-ezrin-radixin-like protei |
| 8 | Danio rerio | 113.00 | 4 | PREDICTED: hypothetical protein |
| 9 | Tetraodon nigroviridis | 112.00 | 7 | unnamed protein product |
| 10 | Bos taurus | 90.90 | 3 | ezrin |
| 11 | Gallus gallus | 90.90 | 2 | ezrin |
| 12 | Oryctolagus cuniculus | 90.90 | 1 | villin 2 (ezrin) |
| 13 | Canis familiaris | 89.00 | 5 | PREDICTED: similar to Moesin (Membrane-organizing |
| 14 | Sus scrofa | 88.60 | 2 | radixin |
| 15 | Xenopus laevis | 88.60 | 2 | U29763_1 moesin |
| 16 | mice, keratinocytes, Balb/MK, Peptide, 583 aa | 88.60 | 1 | RADI_MOUSE Radixin (ESP10) gi |
| 17 | Ciona intestinalis | 87.00 | 1 | ezrin/radixin/moesin (ERM)-like protein |
| 18 | Caenorhabditis briggsae AF16 | 86.30 | 1 | Hypothetical protein CBG12867 |
| 19 | Aplysia californica | 86.30 | 1 | ezrin/radixin/moesin |
| 20 | Caenorhabditis elegans | 85.10 | 3 | ERM-1Asv |
| 21 | Drosophila melanogaster | 84.70 | 5 | Moesin CG10701-PJ, isoform J |
| 22 | Drosophila pseudoobscura | 84.70 | 2 | GA10507-PA |
| 23 | Biomphalaria glabrata | 84.70 | 1 | ezrin/radixin/moesin |
| 24 | Apis mellifera | 83.60 | 1 | PREDICTED: similar to Moesin CG10701-PD, isoform |
| 25 | Drosophila melanogaster | 82.80 | 2 | Merlin CG14228-PA |
| 26 | Taenia solium | 77.40 | 1 | H17g protein, tegumental antigen |
| 27 | Taenia saginata | 77.40 | 1 | myosin-like protein |
| 28 | Echinococcus granulosus | 76.60 | 1 | EG10 |
| 29 | Echinococcus multilocularis | 76.60 | 1 | tegument protein gi |
Top-ranked sequences
| organism | matching |
|---|---|
| Mus musculus | Query 269 KEFTIKPLDKKIDVFKFNSSKLRVNKLILQ#LCIGNHDLFMRRRKADSLEVQQMKAQAREE 328 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 269 KEFTIKPLDKKIDVFKFNSSKLRVNKLILQ#LCIGNHDLFMRRRKADSLEVQQMKAQAREE 328 |
| Homo sapiens | Query 269 KEFTIKPLDKKIDVFKFNSSKLRVNKLILQ#LCIGNHDLFMRRRKADSLEVQQMKAQAREE 328 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 269 KEFTIKPLDKKIDVFKFNSSKLRVNKLILQ#LCIGNHDLFMRRRKADSLEVQQMKAQAREE 328 |
| N/A | Query 269 KEFTIKPLDKKIDVFKFNSSKLRVNKLILQ#LCIGNHDLFMRRRKADSLEVQQMKAQAREE 328 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 269 KEFTIKPLDKKIDVFKFNSSKLRVNKLILQ#LCIGNHDLFMRRRKADSLEVQQMKAQAREE 328 |
| Homo sapiens | Query 269 KEFTIKPLDKKIDVFKFNSSKLRVNKLILQ#LCIGNHDLFMRRRKADSLEVQQMKAQAREE 328 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 269 KEFTIKPLDKKIDVFKFNSSKLRVNKLILQ#LCIGNHDLFMRRRKADSLEVQQMKAQAREE 328 |
| Papio anubis anubis | Query 269 KEFTIKPLDKKIDVFKFNSSKLRVNKLILQ#LCIGNHDLFMRRRKADSLEVQQMKAQAREE 328 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 269 KEFTIKPLDKKIDVFKFNSSKLRVNKLILQ#LCIGNHDLFMRRRKADSLEVQQMKAQAREE 328 |
| Canis familiaris | Query 269 KEFTIKPLDKKIDVFKFNSSKLRVNKLILQ#LCIGNHDLFMRRRKADSLEVQQMKAQAREE 328 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 269 KEFTIKPLDKKIDVFKFNSSKLRVNKLILQ#LCIGNHDLFMRRRKADSLEVQQMKAQAREE 328 |
| Rattus norvegicus | Query 269 KEFTIKPLDKKIDVFKFNSSKLRVNKLILQ#LCIGNHDLFMRRRKADSLEVQQMKAQAREE 328 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 269 KEFTIKPLDKKIDVFKFNSSKLRVNKLILQ#LCIGNHDLFMRRRKADSLEVQQMKAQAREE 328 |
| Danio rerio | Query 269 KEFTIKPLDKKIDVFKFNSSKLRVNKLILQ#LCIGNHDLFMRRRKADSLEVQQMKAQAREE 328 ||| |||+||+ ||||||||||||||||||#|||||||||||||+ ||||||||| ||||| Sbjct 268 KEFAIKPVDKRADVFKFNSSKLRVNKLILQ#LCIGNHDLFMRRRRVDSLEVQQMKTQAREE 327 |
| Tetraodon nigroviridis | Query 269 KEFTIKPLDKKIDVFKFNSSKLRVNKLILQ#LCIGNHDLFMRRRKADSLEVQQMKAQAREE 328 ||||||||+|| |||||||+|| ||||||#|||||||||||||+ ||||||||||||||| Sbjct 163 KEFTIKPLEKKTKVFKFNSSRLRANKLILQ#LCIGNHDLFMRRRRVDSLEVQQMKAQAREE 222 |
| Bos taurus | Query 269 KEFTIKPLDKKIDVFKFNSSKLRVNKLILQ#LCIGNHDLFMRRRKADSLEVQQMKAQAREE 328 |+| |||+||| | | + +||+|| |||#||+|||+|+||||| |++|||||||||||| Sbjct 253 KKFVIKPIDKKAPDFVFYAPRLRINKRILQ#LCMGNHELYMRRRKPDTIEVQQMKAQAREE 312 |
| Gallus gallus | Query 269 KEFTIKPLDKKIDVFKFNSSKLRVNKLILQ#LCIGNHDLFMRRRKADSLEVQQMKAQAREE 328 |+| |||+||| | | + +||+|| |||#||+|||+|+||||| |++|||||||||||| Sbjct 253 KKFVIKPIDKKAPDFVFYAPRLRINKRILQ#LCMGNHELYMRRRKPDTIEVQQMKAQAREE 312 |
| Oryctolagus cuniculus | Query 269 KEFTIKPLDKKIDVFKFNSSKLRVNKLILQ#LCIGNHDLFMRRRKADSLEVQQMKAQAREE 328 |+| |||+||| | | + +||+|| |||#||+|||+|+||||| |++|||||||||||| Sbjct 253 KKFVIKPIDKKAPDFVFYAPRLRINKRILQ#LCMGNHELYMRRRKPDTIEVQQMKAQAREE 312 |
| Canis familiaris | Query 269 KEFTIKPLDKKIDVFKFNSSKLRVNKLILQ#LCIGNHDLFMRRRKADSLEVQQMKAQAREE 328 |+| |||+||| | | + +||+|| || #||+|||+|+||||| |++|||||||||||| Sbjct 253 KKFVIKPIDKKAPDFVFYAPRLRINKRILA#LCMGNHELYMRRRKPDTIEVQQMKAQAREE 312 |
| Sus scrofa | Query 269 KEFTIKPLDKKIDVFKFNSSKLRVNKLILQ#LCIGNHDLFMRRRKADSLEVQQMKAQAREE 328 |+| |||+||| | | + +||+|| || #||+|||+|+||||| |++|||||||||||| Sbjct 253 KKFVIKPIDKKAPDFVFYAPRLRINKRILA#LCMGNHELYMRRRKPDTIEVQQMKAQAREE 312 |
| Xenopus laevis | Query 269 KEFTIKPLDKKIDVFKFNSSKLRVNKLILQ#LCIGNHDLFMRRRKADSLEVQQMKAQAREE 328 |+| |||+||| | | + +||+|| || #||+|||+|+||||| |++|||||||||||| Sbjct 253 KKFVIKPIDKKAPDFVFYAPRLRINKRILA#LCMGNHELYMRRRKPDTIEVQQMKAQAREE 312 |
| mice, keratinocytes, Balb/MK, Peptide, 583 aa | Query 269 KEFTIKPLDKKIDVFKFNSSKLRVNKLILQ#LCIGNHDLFMRRRKADSLEVQQMKAQAREE 328 |+| |||+||| | | + +||+|| || #||+|||+|+||||| |++|||||||||||| Sbjct 253 KKFVIKPIDKKAPDFVFYAPRLRINKRILA#LCMGNHELYMRRRKPDTIEVQQMKAQAREE 312 |
| Ciona intestinalis | Query 269 KEFTIKPLDKKIDVFKFNSSKLRVNKLILQ#LCIGNHDLFMRRRKADSLEVQQMKAQAREE 328 |+| |||+||| | | +||+|| || #||+|||+|+||||| |++|||||||||||| Sbjct 277 KKFVIKPIDKKAPDFVFYVERLRINKRILA#LCMGNHELYMRRRKPDTIEVQQMKAQAREE 336 |
| Caenorhabditis briggsae AF16 | Query 269 KEFTIKPLDKKIDVFKFNSSKLRVNKLILQ#LCIGNHDLFMRRRKADSLEVQQMKAQAREE 328 |+| |||+||| | | + +||+|| || #||+|||+|+||||| |++|||||| ||||| Sbjct 249 KKFVIKPIDKKAHDFVFYAPRLRINKRILA#LCMGNHELYMRRRKPDTIEVQQMKQQAREE 308 |
| Aplysia californica | Query 269 KEFTIKPLDKKIDVFKFNSSKLRVNKLILQ#LCIGNHDLFMRRRKADSLEVQQMKAQAREE 328 |+| |||+||| | | + +||+|| || #||+|||+|+||||| |++|||||||||||+ Sbjct 254 KKFIIKPIDKKAPDFVFYAPRLRINKRILA#LCMGNHELYMRRRKPDTIEVQQMKAQARED 313 |
| Caenorhabditis elegans | Query 269 KEFTIKPLDKKIDVFKFNSSKLRVNKLILQ#LCIGNHDLFMRRRKADSLEVQQMKAQAREE 328 |+| |||+||| | | + +||+|| || #||+|||+|+||||| |++|||||| ||||+ Sbjct 254 KKFVIKPIDKKAHDFVFYAPRLRINKRILA#LCMGNHELYMRRRKPDTIEVQQMKQQARED 313 |
| Drosophila melanogaster | Query 269 KEFTIKPLDKKIDVFKFNSSKLRVNKLILQ#LCIGNHDLFMRRRKADSLEVQQMKAQAREE 328 |+| |||+||| | | + ++|+|| || #||+|||+|+||||| |+++||||||||||| Sbjct 254 KKFIIKPIDKKAPDFMFFAPRVRINKRILA#LCMGNHELYMRRRKPDTIDVQQMKAQAREE 313 |
| Drosophila pseudoobscura | Query 269 KEFTIKPLDKKIDVFKFNSSKLRVNKLILQ#LCIGNHDLFMRRRKADSLEVQQMKAQAREE 328 |+| |||+||| | | + ++|+|| || #||+|||+|+||||| |+++||||||||||| Sbjct 249 KKFIIKPIDKKAPDFMFFAPRVRINKRILA#LCMGNHELYMRRRKPDTIDVQQMKAQAREE 308 |
| Biomphalaria glabrata | Query 269 KEFTIKPLDKKIDVFKFNSSKLRVNKLILQ#LCIGNHDLFMRRRKADSLEVQQMKAQAR 326 |+| |||+||| | | + +||+|| || #||+|||+|+||||| |++|||||||||| Sbjct 254 KKFVIKPIDKKAPDFVFYAPRLRINKRILA#LCMGNHELYMRRRKPDTIEVQQMKAQAR 311 |
| Apis mellifera | Query 269 KEFTIKPLDKKIDVFKFNSSKLRVNKLILQ#LCIGNHDLFMRRRKADSLEVQQMKAQAREE 328 |+| |||+||| | | ++++++|| || #||+|||+|+||||| |+++||||||||||| Sbjct 188 KKFIIKPIDKKAPDFVFFATRVKINKRILA#LCMGNHELYMRRRKPDTIDVQQMKAQAREE 247 |
| Drosophila melanogaster | Query 269 KEFTIKPLDKKIDVFKFNSSKLRVNKLILQ#LCIGNHDLFMRRRKADSLEVQQMKAQAREE 328 |+|||+ +| |+ | | | | +||+|| #|| |||||+||||| |++|+|||||||+|| Sbjct 263 KKFTIRLVDAKVSNFIFYSQDLHINKMILD#LCKGNHDLYMRRRKPDTMEIQQMKAQAKEE 322 |
| Taenia solium | Query 269 KEFTIKPLDKKIDVFKFNSSKLRVNKLILQ#LCIGNHDLFMRRRKADSLEVQQMKAQAREE 328 |+| ||| || | | | ++|| || #|| |||+|+|||||+||+|||||| ||+|| Sbjct 248 KKFIIKPADKSAKEFYFLVEKSKINKRILA#LCTGNHELYMRRRKSDSIEVQQMKIQAKEE 307 |
| Taenia saginata | Query 269 KEFTIKPLDKKIDVFKFNSSKLRVNKLILQ#LCIGNHDLFMRRRKADSLEVQQMKAQAREE 328 |+| ||| || | | | ++|| || #|| |||+|+|||||+||+|||||| ||+|| Sbjct 255 KKFIIKPADKSAKEFYFLVEKSKINKRILA#LCTGNHELYMRRRKSDSIEVQQMKIQAKEE 314 |
| Echinococcus granulosus | Query 269 KEFTIKPLDKKIDVFKFNSSKLRVNKLILQ#LCIGNHDLFMRRRKADSLEVQQMKAQAREE 328 |+| ||| || | | | ++|| || #|| |||+|+|||||+||+|||||| ||+|| Sbjct 255 KKFIIKPADKSAKEFFFLVEKSKINKRILA#LCTGNHELYMRRRKSDSIEVQQMKIQAKEE 314 |
| Echinococcus multilocularis | Query 269 KEFTIKPLDKKIDVFKFNSSKLRVNKLILQ#LCIGNHDLFMRRRKADSLEVQQMKAQAREE 328 |+| ||| || | | | ++|| || #|| |||+|+|||||+||+|||||| ||+|| Sbjct 255 KKFIIKPADKSAKEFFFLVEKSKINKRILA#LCTGNHELYMRRRKSDSIEVQQMKIQAKEE 314 |
* References
[PubMed ID: 24309211] Schulz A, Kyselyova A, Baader SL, Jung MJ, Zoch A, Mautner VF, Hagel C, Morrison H, Neuronal merlin influences ERBB2 receptor expression on Schwann cells through neuregulin 1 type III signalling. Brain. 2014 Feb;137(Pt 2):420-32. doi: 10.1093/brain/awt327. Epub 2013 Dec 5.
[PubMed ID: 24249803] ... Plotkin SR, Ardern-Holmes SL, Barker FG 2nd, Blakeley JO, Evans DG, Ferner RE, Hadlock TA, Halpin C, Hearing and facial function outcomes for neurofibromatosis 2 clinical trials. Neurology. 2013 Nov 19;81(21 Suppl 1):S25-32. doi:
[PubMed ID: 23921927] ... Lassaletta L, Torres-Martin M, Pena-Granero C, Roda JM, Santa-Cruz-Ruiz S, Castresana JS, Gavilan J, Rey JA, NF2 genetic alterations in sporadic vestibular schwannomas: clinical implications. Otol Neurotol. 2013 Sep;34(7):1355-61. doi: 10.1097/MAO.0b013e318298ac79.
[PubMed ID: 23666797] ... Pecina-Slaus N, Merlin, the NF2 gene product. Pathol Oncol Res. 2013 Jul;19(3):365-73. doi: 10.1007/s12253-013-9644-y. Epub
[PubMed ID: 24171707] ... Tabernero M, Jara-Acevedo M, Nieto AB, Caballero AR, Otero A, Sousa P, Goncalves J, Domingues PH, Orfao A, Association between mutation of the NF2 gene and monosomy 22 in menopausal women with sporadic meningiomas. BMC Med Genet. 2013 Oct 30;14:114. doi: 10.1186/1471-2350-14-114.
[PubMed ID: 17322306] ... Sugiyama N, Masuda T, Shinoda K, Nakamura A, Tomita M, Ishihama Y, Phosphopeptide enrichment by aliphatic hydroxy acid-modified metal oxide chromatography for nano-LC-MS/MS in proteomics applications. Mol Cell Proteomics. 2007 Jun;6(6):1103-9. Epub 2007 Feb 23.
[PubMed ID: 11719502] ... Xiao GH, Beeser A, Chernoff J, Testa JR, p21-activated kinase links Rac/Cdc42 signaling to merlin. J Biol Chem. 2002 Jan 11;277(2):883-6. Epub 2001 Nov 21.
[PubMed ID: 7485397] ... den Bakker MA, Tascilar M, Riegman PH, Hekman AC, Boersma W, Janssen PJ, de Jong TA, Hendriks W, van der Kwast TH, Zwarthoff EC, Neurofibromatosis type 2 protein co-localizes with elements of the cytoskeleton. Am J Pathol. 1995 Nov;147(5):1339-49.
[PubMed ID: 7666400] ... Evans DG, Bourn D, Wallace A, Ramsden RT, Mitchell JD, Strachan T, Diagnostic issues in a family with late onset type 2 neurofibromatosis. J Med Genet. 1995 Jun;32(6):470-4.
[PubMed ID: 7798645] ... Honda M, Arai E, Sawada S, Ohta A, Niimura M, Neurofibromatosis 2 and neurilemmomatosis gene are identical. J Invest Dermatol. 1995 Jan;104(1):74-7.
[PubMed ID: 20301380] ... Evans DG, Neurofibromatosis 2
[PubMed ID: 1456285] ... Arai E, Ikeuchi T, Karasawa S, Tamura A, Yamamoto K, Kida M, Ichimura K, Yuasa Y, Tonomura A, Constitutional translocation t(4;22) (q12;q12.2) associated with neurofibromatosis type 2. Am J Med Genet. 1992 Sep 15;44(2):163-7.