SB0010 : Neurofilament medium polypeptide, Neurofilament 3
[ CaMP Format ]
* Basic Information
| Organism | Bos taurus (cattle) |
| Protein Names | PREDICTED: neurofilament, medium polypeptide [Bos taurus]; neurofilament, medium polypeptide |
| Gene Names | NEF3; Derived by automated computational analysis using gene prediction method: GNOMON. Supporting evidence includes similarity to: 1 mRNA, 94 ESTs, 40 Proteins |
| Gene Locus | Not available |
| GO Function | Not available |
* Information From OMIM
Not Available.
* Structure Information
1. Primary Information
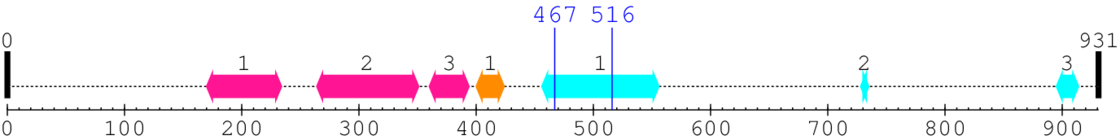
Length: 931 aa
Average Mass: 103.612 kDa
Monoisotopic Mass: 103.551 kDa
2. Domain Information
Annotated Domains: Not Available.
Computationally Assigned Domains (Pfam+HMMER):
| domain name | begin | end | score | e-value |
| Protein of unknown function (DUF1664) 1. | 170 | 234 | 56.0 | 3.4 |
| Protein of unknown function (DUF1664) 2. | 264 | 351 | 37.0 | 3.7 |
| Protein of unknown function (DUF1664) 3. | 360 | 394 | 47.0 | 0.1 |
| Intermediate filament head (DNA binding) region 1. | 400 | 424 | 5.0 | 0.0 |
| Intermediate filament protein 1. | 456 | 556 | 169.0 | 26.9 |
| --- cleavage 467 (inside Intermediate filament protein 456..556) --- |
| --- cleavage 516 (inside Intermediate filament protein 456..556) --- |
| Intermediate filament protein 2. | 728 | 735 | 86.0 | 4.2 |
| Intermediate filament protein 3. | 895 | 914 | 183.0 | 0.1 |
3. Sequence Information
Fasta Sequence: SB0010.fasta
Amino Acid Sequence and Secondary Structures (PsiPred):
4. 3D Information
Not Available.
* Cleavage Information
2 [sites] cleaved by Calpain 1
Source Reference: [PubMed ID: 15530438] Shaw G, Yang C, Zhang L, Cook P, Pike B, Hill WD, Characterization of the bovine neurofilament NF-M protein and cDNA sequence, and identification of in vitro and in vivo calpain cleavage sites. Biochem Biophys Res Commun. 2004 Dec 10;325(2):619-25.
Cleavage sites (±10aa)
[Site 1] IEETKVEDEK467-SEMEEALTAI
Lys467  Ser
Ser
 |
| P10 | P9 | P8 | P7 | P6 |
P5 | P4 | P3 | P2 | P1 |
| Ile458 | Glu459 | Glu460 | Thr461 | Lys462 | Val463 | Glu464 | Asp465 | Glu466 | Lys467 |
 |
| P1' | P2' | P3' | P4' | P5' |
P6' | P7' | P8' | P9' | P10' |
| Ser468 | Glu469 | Met470 | Glu471 | Glu472 | Ala473 | Leu474 | Thr475 | Ala476 | Ile477 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
|
QKTKVEAPKLKVQHKFVEEIIEETKVEDEKSEMEEALTAITEELAVSVKEEVKEEEAEEK
|
Summary
| # |
organism |
max score |
hits |
top seq |
| 1 |
Bos taurus |
106.00 |
1 |
neurofilament-M subunit |
| 2 |
Homo sapiens |
88.20 |
2 |
AF181990_1 neurofilament-3 (150 kD medium) |
| 3 |
Canis familiaris |
87.80 |
1 |
PREDICTED: similar to Neurofilament triplet M pro |
| 4 |
N/A |
87.40 |
4 |
NFM_RABIT Neurofilament medium polypeptide (NF-M) |
| 5 |
Mus musculus |
80.50 |
5 |
unnamed protein product |
| 6 |
Rattus norvegicus |
79.00 |
1 |
neurofilament 3, medium |
| 7 |
rats, Peptide Partial, 435 aa |
79.00 |
1 |
neurofilament protein M |
| 8 |
Gallus gallus |
76.60 |
2 |
neurofilament, medium polypeptide 150kDa |
| 9 |
Serinus canaria |
76.60 |
1 |
neurofilament medium subunit |
| 10 |
Xenopus laevis |
65.50 |
1 |
AF237379_1 neurofilament protein |
| 11 |
Xenopus laevis |
64.70 |
2 |
LOC397994 protein |
| 12 |
Carassius auratus gene, complete cds. |
63.90 |
1 |
- |
Top-ranked sequences
| organism | matching |
|---|
| Bos taurus |
Query 438 QKTKVEAPKLKVQHKFVEEIIEETKVEDEK#SEMEEALTAITEELAVSVKEEVKEEEAEEK 497
||||||||||||||||||||||||||||||#||||||||||||||||||||||||||||||
Query 438 QKTKVEAPKLKVQHKFVEEIIEETKVEDEK#SEMEEALTAITEELAVSVKEEVKEEEAEEK 497
|
| Homo sapiens |
Query 439 KTKVEAPKLKVQHKFVEEIIEETKVEDEK#SEMEEALTAITEELAVSV 485
|||||||||||||||||||||||||||||#|||||||||||||||||+
Sbjct 1 KTKVEAPKLKVQHKFVEEIIEETKVEDEK#SEMEEALTAITEELAVSM 47
|
| Canis familiaris |
Query 438 QKTKVEAPKLKVQHKFVEEIIEETKVEDEK#SEMEEALTAITEELAVSV 485
||||||||||||||||||||||||||||||#|||||||||| ||||||+
Sbjct 438 QKTKVEAPKLKVQHKFVEEIIEETKVEDEK#SEMEEALTAIAEELAVSM 485
|
| N/A |
Query 438 QKTKVEAPKLKVQHKFVEEIIEETKVEDEK#SEMEEALTAITEELAVSV 485
||||||||||||||||||||||||||||||#||||+||||| |||||||
Sbjct 223 QKTKVEAPKLKVQHKFVEEIIEETKVEDEK#SEMEDALTAIAEELAVSV 270
|
| Mus musculus |
Query 438 QKTKVEAPKLKVQHKFVEEIIEETKVEDEK#SEMEEALTAITEEL 481
||||||||||||||||||||||||||||||#||||| |||| |||
Sbjct 436 QKTKVEAPKLKVQHKFVEEIIEETKVEDEK#SEMEETLTAIAEEL 479
|
| Rattus norvegicus |
Query 438 QKTKVEAPKLKVQHKFVEEIIEETKVEDEK#SEMEEALTAITEEL 481
||||||||||||||||||||||||||||||#||||+||| | |||
Sbjct 436 QKTKVEAPKLKVQHKFVEEIIEETKVEDEK#SEMEDALTVIAEEL 479
|
| rats, Peptide Partial, 435 aa |
Query 438 QKTKVEAPKLKVQHKFVEEIIEETKVEDEK#SEMEEALTAITEEL 481
||||||||||||||||||||||||||||||#||||+||| | |||
Sbjct 27 QKTKVEAPKLKVQHKFVEEIIEETKVEDEK#SEMEDALTVIAEEL 70
|
| Gallus gallus |
Query 438 QKTKVEAPKLKVQHKFVEEIIEETKVEDEK#SEMEEALTAITEEL 481
||||+| |||||||||||||||||||||||#||||+||+|| ||+
Sbjct 434 QKTKIEPPKLKVQHKFVEEIIEETKVEDEK#SEMEDALSAIAEEM 477
|
| Serinus canaria |
Query 438 QKTKVEAPKLKVQHKFVEEIIEETKVEDEK#SEMEEALTAITEEL 481
||||+| |||||||||||||||||||||||#||||+||+|| ||+
Sbjct 63 QKTKIEPPKLKVQHKFVEEIIEETKVEDEK#SEMEDALSAIAEEM 106
|
| Xenopus laevis |
Query 438 QKTKVEAPKLKVQHKFVEEIIEETKVEDEK#SEMEEALTAITEELAVSV 485
|+ + | |++||||||||||||||||||++#|+|| || |+ +| |+
Sbjct 422 QQARAEPPRVKVQHKFVEEIIEETKVEDDR#SDMEAALAAMADEFALGF 469
|
| Xenopus laevis |
Query 438 QKTKVEAPKLKVQHKFVEEIIEETKVEDEK#SEMEEALTAITEELAVSV 485
| + | |++||||||||||||||||||++#|+|| || |+ +| | +
Sbjct 424 QHARSEPPRMKVQHKFVEEIIEETKVEDDR#SDMEAALAAMADEFASGL 471
|
| Carassius auratus gene, complete cds. |
Query 439 KTKVEAPKLKVQHKFVEEIIEETKVEDEK#SEMEEALTAITEELA 482
| | | |||||||||||||||||+|||||#|||+| | + |||+
Sbjct 409 KVKREPPKLKVQHKFVEEIIEETRVEDEK#SEMDEVLAEMAEELS 452
|
[Site 2] VVAAKKSPVK516-ATAPELKEEE
Lys516  Ala
Ala
 |
| P10 | P9 | P8 | P7 | P6 |
P5 | P4 | P3 | P2 | P1 |
| Val507 | Val508 | Ala509 | Ala510 | Lys511 | Lys512 | Ser513 | Pro514 | Val515 | Lys516 |
 |
| P1' | P2' | P3' | P4' | P5' |
P6' | P7' | P8' | P9' | P10' |
| Ala517 | Thr518 | Ala519 | Pro520 | Glu521 | Leu522 | Lys523 | Glu524 | Glu525 | Glu526 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
|
EEVKEEEAEEKEEKEEAEEEVVAAKKSPVKATAPELKEEEGEKEEEEGQEEEEEEEEAAK
|
No hits found.
 Ser
Ser

 Sequence conservation (by blast)
Sequence conservation (by blast) Ala
Ala

 Sequence conservation (by blast)
Sequence conservation (by blast)