SB0011 : IkB-[alpha], NFkB inhibitor [alpha]
[ CaMP Format ]
* Basic Information
| Organism | Homo sapiens (human) |
| Protein Names | NF-kappa-B inhibitor alpha [Homo sapiens]; NF-kappa-B inhibitor alpha; nuclear factor of kappa light chain gene enhancer in B-cells; IkappaBalpha; major histocompatibility complex enhancer-binding protein MAD3; ikB-alpha; I-kappa-B-alpha; IkB-alpha; Major histocompatibility complex enhancer-binding protein MAD3 |
| Gene Names | NFKBIA; IKBA, MAD3, NFKBI; IKBA; MAD3; NFKBI; nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, alpha |
| Gene Locus | 14q13; chromosome 14 |
| GO Function | Not available |
* Information From OMIM
Description: NFKB1 (OMIM:164011) or NFKB2 (OMIM:164012) is bound to REL (OMIM:164910), RELA (OMIM:164014), or RELB (OMIM:604758) to form the NFKB complex. The NFKB complex is inhibited by I-kappa-B proteins (NFKBIA or NFKBIB, OMIM:604495), which inactivate NF-kappa-B by trapping it in the cytoplasm. Phosphorylation of serine residues on the I-kappa-B proteins by kinases (IKBKA, OMIM:600664, or IKBKB, OMIM:603258) marks them for destruction via the ubiquitination pathway, thereby allowing activation of the NF-kappa-B complex. Activated NFKB complex translocates into the nucleus and binds DNA at kappa-B-binding motifs such as 5-prime GGGRNNYYCC 3-prime or 5-prime HGGARNYYCC 3-prime (where H is A, C, or T; R is an A or G purine; and Y is a C or T pyrimidine).
* Structure Information
1. Primary Information
Length: 317 aa
Average Mass: 35.609 kDa
Monoisotopic Mass: 35.587 kDa
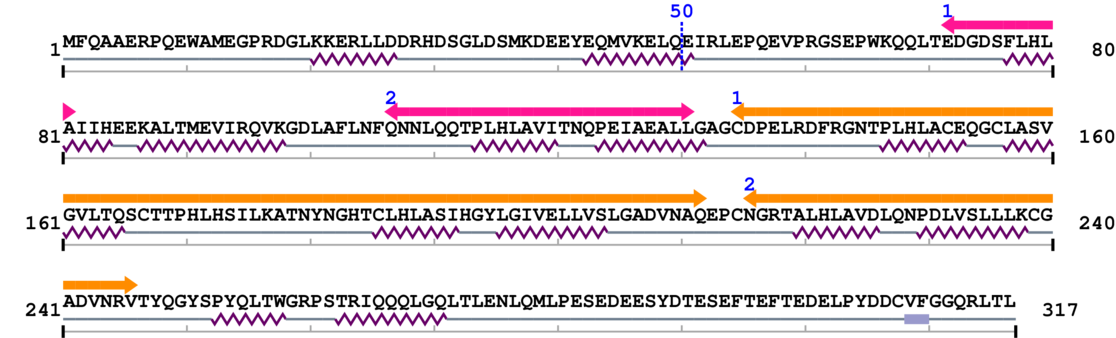
2. Domain Information
Annotated Domains: interpro / pfam / smart / prosite
Computationally Assigned Domains (Pfam+HMMER):
| domain name | begin | end | score | e-value |
|---|---|---|---|---|
| --- cleavage 50 --- | ||||
| Ankyrin repeats (many copies) 1. | 72 | 81 | 47.0 | 0.0 |
| Ankyrin repeats (many copies) 2. | 107 | 131 | 30.0 | 0.0 |
| Ankyrin repeats (3 copies) 1. | 135 | 212 | 20.0 | 0.3 |
| Ankyrin repeats (3 copies) 2. | 216 | 246 | 55.0 | 0.1 |
3. Sequence Information
Fasta Sequence: SB0011.fasta
Amino Acid Sequence and Secondary Structures (PsiPred):
* Cleavage Information
1 [sites] cleaved by Calpain 2
Source Reference: [PubMed ID: 15202778] Schaecher K, Goust JM, Banik NL, The effects of calpain inhibition on IkB alpha degradation after activation of PBMCs: identification of the calpain cleavage sites. Neurochem Res. 2004 Jul;29(7):1443-51.
Cleavage sites (±10aa)
[Site 1] EYEQMVKELQ50-EIRLEPQEVP
Gln50  Glu
Glu
iTraq-117 Signal 2255.1 (

 ) for EYEQMVKELQEIR
) for EYEQMVKELQEIR
iTraq-117 Signal 3623.7 (

 ) for EIRLEPQEVP
) for EIRLEPQEVP
iTraq-117 Signal 1737.0 (

 ) for LEPQEVP
) for LEPQEVP
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Glu41 | Tyr42 | Glu43 | Gln44 | Met45 | Val46 | Lys47 | Glu48 | Leu49 | Gln50 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Glu51 | Ile52 | Arg53 | Leu54 | Glu55 | Pro56 | Gln57 | Glu58 | Val59 | Pro60 |
 Sequence conservation (by blast)
Sequence conservation (by blast)
* References
[PubMed ID: 24457201] Stanborough T, Niederhauser J, Koch B, Bergler H, Pertschy B, Ribosomal protein S3 interacts with the NF-kappaB inhibitor IkappaBalpha. FEBS Lett. 2014 Mar 3;588(5):659-64. doi: 10.1016/j.febslet.2013.12.034. Epub