SB0012 : Histone H2B type 1
[ CaMP Format ]
* Basic Information
| Organism | Bos taurus (cattle) |
| Protein Names | H2B histone family, member L [Bos taurus]; H2B histone family, member L; Histone H2B type 1 |
| Gene Names | HIST1H2BC |
| Gene Locus | Not available |
| GO Function | Not available |
| Entrez Protein | Entrez Nucleotide | Entrez Gene | UniProt | OMIM | HGNC | HPRD | KEGG |
|---|---|---|---|---|---|---|---|
| ABM06136 | N/A | N/A | P62808 | N/A | N/A | N/A | bta:787465 |
* Information From OMIM
Not Available.
* Structure Information
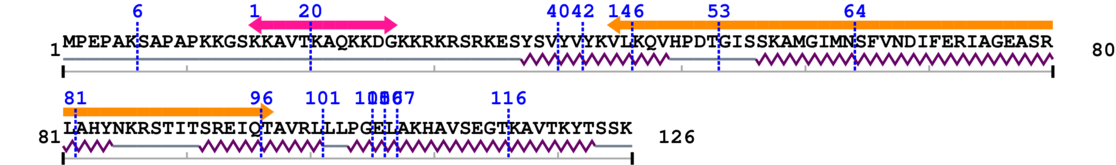
1. Primary Information
Length: 126 aa
Average Mass: 13.906 kDa
Monoisotopic Mass: 13.898 kDa
2. Domain Information
Annotated Domains: interpro / pfam / smart / prosite
Computationally Assigned Domains (Pfam+HMMER):
| domain name | begin | end | score | e-value |
|---|---|---|---|---|
| --- cleavage 6 --- | ||||
| Histone-like transcription factor (CBF/NF-Y) and archaeal histone 1. | 16 | 27 | 42.0 | 0.4 |
| --- cleavage 20 (inside Histone-like transcription factor (CBF/NF-Y) and archaeal histone 16..27) --- | ||||
| --- cleavage 40 --- | ||||
| --- cleavage 42 --- | ||||
| Transcription initiation factor TFIID subunit A 1. | 45 | 97 | 8.0 | 0.0 |
| --- cleavage 46 (inside Transcription initiation factor TFIID subunit A 45..97) --- | ||||
| --- cleavage 53 (inside Transcription initiation factor TFIID subunit A 45..97) --- | ||||
| --- cleavage 64 (inside Transcription initiation factor TFIID subunit A 45..97) --- | ||||
| --- cleavage 81 (inside Transcription initiation factor TFIID subunit A 45..97) --- | ||||
| --- cleavage 96 (inside Transcription initiation factor TFIID subunit A 45..97) --- | ||||
3. Sequence Information
Fasta Sequence: SB0012.fasta
Amino Acid Sequence and Secondary Structures (PsiPred):
4. 3D Information
Not Available.
* Cleavage Information
14 [sites]
Source Reference: [PubMed ID: 3038857] Sakai K, Akanuma H, Imahori K, Kawashima S, A unique specificity of a calcium activated neutral protease indicated in histone hydrolysis. J Biochem. 1987 Apr;101(4):911-8.
Cleavage sites (±10aa)
[Site 1] MPEPAK6-SAPAPKKGSK
Lys6  Ser
Ser
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| - | - | - | - | Met1 | Pro2 | Glu3 | Pro4 | Ala5 | Lys6 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Ser7 | Ala8 | Pro9 | Ala10 | Pro11 | Lys12 | Lys13 | Gly14 | Ser15 | Lys16 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| MPEPAKSAPAPKKGSKKAVTKAQKKDGKKRKRSRKE |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | Homo sapiens | 68.90 | 23 | histone cluster 2, H2be |
| 2 | Mus musculus | 68.90 | 15 | histone cluster 1, H2bj |
| 3 | Canis familiaris | 68.90 | 7 | PREDICTED: similar to H2B histone family, member |
| 4 | Rattus norvegicus | 68.90 | 3 | histone 1, H2bn (predicted) |
| 5 | Macaca fascicularis | 68.60 | 1 | H2B1K_MACFA Histone H2B type 1-K (H2B K) gi |
| 6 | Bos taurus | 67.40 | 14 | PREDICTED: similar to Hist1h2bj protein |
| 7 | Bos taurus | 67.40 | 1 | histone cluster 1, H2bn |
| 8 | Rattus norvegicus | 66.20 | 5 | PREDICTED: similar to Histone H2B 291B |
| 9 | Homo sapiens | 66.20 | 1 | histone cluster 2, H2bf |
| 10 | N/A | 65.90 | 10 | 0506206A histone H2B |
| 11 | Pongo pygmaeus | 65.50 | 1 | H2B2E_PONPY Histone H2B type 2-E gi |
| 12 | Xenopus laevis | 64.70 | 4 | histone H2B |
| 13 | Xenopus tropicalis | 62.80 | 3 | hypothetical protein LOC550005 |
| 14 | Rhacophorus schlegelii | 60.50 | 1 | H2B_RHASC Histone H2B gi |
| 15 | Gallus gallus | 59.70 | 10 | H2B7_CHICK Histone H2B 7 (H2B VII) gi |
| 16 | Danio rerio | 57.40 | 6 | histone 2, H2, like |
| 17 | Expression vector pET3-H2B | 53.90 | 1 | H Chain H, 2.9 Angstrom X-Ray Structure Of Hybrid |
| 18 | Tetraodon nigroviridis | 52.40 | 9 | unnamed protein product |
| 19 | Oncorhynchus mykiss | 47.80 | 1 | unnamed protein product |
| 20 | Cairina moschata | 45.10 | 1 | H2B_CAIMO Histone H2B gi |
| 21 | Pan troglodytes | 39.70 | 1 | PREDICTED: hypothetical protein |
| 22 | cattle, Peptide Partial, 18 aa | 31.20 | 1 | thymic peptide, TP=0.58 kda peptide {N-terminal} |
| 23 | Desulfotalea psychrophila LSv54 | 30.80 | 1 | hypothetical protein DP2470 |
Top-ranked sequences
| organism | matching |
|---|---|
| Homo sapiens | Query 1 MPEPAK#SAPAPKKGSKKAVTKAQKKDGKKRKRSRKE 36 ||||||#|||||||||||||||||||||||||||||| Query 1 MPEPAK#SAPAPKKGSKKAVTKAQKKDGKKRKRSRKE 36 |
| Mus musculus | Query 1 MPEPAK#SAPAPKKGSKKAVTKAQKKDGKKRKRSRKE 36 ||||||#|||||||||||||||||||||||||||||| Query 1 MPEPAK#SAPAPKKGSKKAVTKAQKKDGKKRKRSRKE 36 |
| Canis familiaris | Query 1 MPEPAK#SAPAPKKGSKKAVTKAQKKDGKKRKRSRKE 36 ||||||#|||||||||||||||||||||||||||||| Query 1 MPEPAK#SAPAPKKGSKKAVTKAQKKDGKKRKRSRKE 36 |
| Rattus norvegicus | Query 1 MPEPAK#SAPAPKKGSKKAVTKAQKKDGKKRKRSRKE 36 ||||||#|||||||||||||||||||||||||||||| Query 1 MPEPAK#SAPAPKKGSKKAVTKAQKKDGKKRKRSRKE 36 |
| Macaca fascicularis | Query 1 MPEPAK#SAPAPKKGSKKAVTKAQKKDGKKRKRSRKE 36 ||||||#|||||||||||||||||||||||||||||| Query 1 MPEPAK#SAPAPKKGSKKAVTKAQKKDGKKRKRSRKE 36 |
| Bos taurus | Query 1 MPEPAK#SAPAPKKGSKKAVTKAQKKDGKKRKRSRKE 36 ||||+|#||||||||||||+||||||||||||||||| Sbjct 1 MPEPSK#SAPAPKKGSKKAITKAQKKDGKKRKRSRKE 36 |
| Bos taurus | Query 1 MPEPAK#SAPAPKKGSKKAVTKAQKKDGKKRKRSRKE 36 ||||+|#|||||||||||||||||||||||||||||| Sbjct 1 MPEPSK#SAPAPKKGSKKAVTKAQKKDGKKRKRSRKE 36 |
| Rattus norvegicus | Query 1 MPEPAK#SAPAPKKGSKKAVTKAQKKDGKKRKRSRKE 36 ||| ||#|||||||||||||||||||||||||||||| Sbjct 1 MPETAK#SAPAPKKGSKKAVTKAQKKDGKKRKRSRKE 36 |
| Homo sapiens | Query 1 MPEPAK#SAPAPKKGSKKAVTKAQKKDGKKRKRSRKE 36 ||+|||#||||||||||||||| |||||||||||||| Sbjct 1 MPDPAK#SAPAPKKGSKKAVTKVQKKDGKKRKRSRKE 36 |
| N/A | Query 2 PEPAK#SAPAPKKGSKKAVTKAQKKDGKKRKRSRKE 36 |||||#|||||||||||||||||||||||||||||| Query 2 PEPAK#SAPAPKKGSKKAVTKAQKKDGKKRKRSRKE 36 |
| Pongo pygmaeus | Query 1 MPEPAK#SAPAPKKGSKKAVTKAQKKDGKKRKRSRKE 36 ||||||#||||||||||||||||||||||||||| || Sbjct 1 MPEPAK#SAPAPKKGSKKAVTKAQKKDGKKRKRSCKE 36 |
| Xenopus laevis | Query 1 MPEPAK#SAPAPKKGSKKAVTKAQKKDGKKRKRSRKE 36 ||||||#||||||||||||||| ||||||||++|||| Sbjct 1 MPEPAK#SAPAPKKGSKKAVTKTQKKDGKKRRKSRKE 36 |
| Xenopus tropicalis | Query 1 MPEPAK#SAPAPKKGSKKAVTKAQKKDGKKRKRSRKE 36 ||||||#|||||||||||||+| ||||||||++|||| Sbjct 1 MPEPAK#SAPAPKKGSKKAVSKTQKKDGKKRRKSRKE 36 |
| Rhacophorus schlegelii | Query 1 MPEPAK#SAPAPKKGSKKAVTKAQKKDGKKRKRSRKE 36 ||||||#|||| ||||||||+| ||||||||++|||| Sbjct 1 MPEPAK#SAPAAKKGSKKAVSKVQKKDGKKRRKSRKE 36 |
| Gallus gallus | Query 1 MPEPAK#SAPAPKKGSKKAVTKAQKKDGKKRKRSRKE 36 ||||||#||||||||||||||| ||| |||||+||| Sbjct 1 MPEPAK#SAPAPKKGSKKAVTKTQKKGDKKRKRARKE 36 |
| Danio rerio | Query 1 MPEPAK#SAPAPKKGSKKAVTKAQKKDGKKRKRSRKE 36 ||||||#||||||||||||||| ||| |||+++||| Sbjct 1 MPEPAK#SAPAPKKGSKKAVTKTQKKGDKKRRKTRKE 36 |
| Expression vector pET3-H2B | Query 5 AK#SAPAPKKGSKKAVTKAQKKDGKKRKRSRKE 36 ||#||||||||||||||| ||||||||+++||| Sbjct 2 AK#SAPAPKKGSKKAVTKTQKKDGKKRRKTRKE 33 |
| Tetraodon nigroviridis | Query 1 MPEPAK#SAPAPKKGSKKAVTKAQKKDGKKRKRSRKE 36 || |||#|||| |||||||+|||||| |||+++||| Sbjct 1 MPGPAK#SAPAAKKGSKKAITKAQKKGDKKRRKNRKE 36 |
| Oncorhynchus mykiss | Query 1 MPEPAK#SAPAPKKGSKKAVTKAQKKDGKKRKRSRKE 36 ||||||#||| |||||||||| | ||||++|||| Sbjct 1 MPEPAK#SAP--KKGSKKAVTKTAGKGGKKRRKSRKE 34 |
| Cairina moschata | Query 1 MPEPAK#SAPAPKKGSKKAVTKAQ 23 ||||||#||||||||||||||| | Sbjct 1 MPEPAK#SAPAPKKGSKKAVTKTQ 23 |
| Pan troglodytes | Query 1 MPEPAK#SAPAPKKGSKKAVTKAQKK 25 ||||++#+||| |||||||+|||||| Sbjct 1 MPEPSR#AAPASKKGSKKAITKAQKK 25 |
| cattle, Peptide Partial, 18 aa | Query 2 PEPAK#SAPAPKKGSKKAV 19 |||||#||||||||||+ | Sbjct 1 PEPAK#SAPAPKKGSKEEV 18 |
| Desulfotalea psychrophila LSv54 | Query 4 PAK#SAPAPKKGSKKAVTKAQKKDGKKRKRSR 34 || # +|||| +++ + |+|+||+| +| Sbjct 19 PAA#LSPAPKPPTRRTMKDAEKRDGRKVYATR 49 |
[Site 2] PKKGSKKAVT20-KAQKKDGKKR
Thr20  Lys
Lys
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Pro11 | Lys12 | Lys13 | Gly14 | Ser15 | Lys16 | Lys17 | Ala18 | Val19 | Thr20 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Lys21 | Ala22 | Gln23 | Lys24 | Lys25 | Asp26 | Gly27 | Lys28 | Lys29 | Arg30 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| MPEPAKSAPAPKKGSKKAVTKAQKKDGKKRKRSRKESYSVYVYKVLKQVH |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | Mus musculus | 98.60 | 15 | H2b histone family, member A |
| 2 | Homo sapiens | 97.80 | 23 | histone cluster 1, H2bg |
| 3 | Canis familiaris | 97.80 | 7 | PREDICTED: similar to H2B histone family, member |
| 4 | Rattus norvegicus | 97.40 | 3 | histone 1, H2bn (predicted) |
| 5 | Bos taurus | 96.70 | 1 | histone cluster 1, H2bn |
| 6 | Bos taurus | 96.30 | 14 | PREDICTED: similar to Hist1h2bj protein |
| 7 | Macaca fascicularis | 95.90 | 1 | H2B1K_MACFA Histone H2B type 1-K (H2B K) gi |
| 8 | N/A | 95.10 | 15 | 0506206A histone H2B |
| 9 | Rattus norvegicus | 95.10 | 5 | PREDICTED: similar to Histone H2B 291B |
| 10 | Homo sapiens | 94.70 | 1 | histone cluster 2, H2bf |
| 11 | Pongo pygmaeus | 94.40 | 1 | H2B2E_PONPY Histone H2B type 2-E gi |
| 12 | Xenopus laevis | 92.40 | 4 | histone H2B |
| 13 | Xenopus tropicalis | 90.90 | 3 | hypothetical protein LOC550005 |
| 14 | Gallus gallus | 89.00 | 9 | H2B7_CHICK Histone H2B 7 (H2B VII) gi |
| 15 | Rhacophorus schlegelii | 87.80 | 1 | H2B_RHASC Histone H2B gi |
| 16 | Danio rerio | 85.50 | 6 | histone 2, H2, like |
| 17 | Expression vector pET3-H2B | 81.60 | 1 | H Chain H, 2.9 Angstrom X-Ray Structure Of Hybrid |
| 18 | Tetraodon nigroviridis | 80.90 | 10 | unnamed protein product |
| 19 | Oncorhynchus mykiss | 75.90 | 1 | unnamed protein product |
| 20 | Cairina moschata | 71.60 | 1 | H2B_CAIMO Histone H2B gi |
| 21 | Pan troglodytes | 67.00 | 1 | PREDICTED: hypothetical protein |
| 22 | Psammechinus miliaris | 54.30 | 1 | H2BE2_PSAMI Histone H2B.2, embryonic gi |
| 23 | Strongylocentrotus purpuratus | 54.30 | 1 | PREDICTED: similar to histone H2B |
| 24 | Strongylocentrotus purpuratus | 53.50 | 2 | unnamed protein product |
| 25 | Psammechinus miliaris | 52.40 | 1 | H2BE1_PSAMI Histone H2B.1, embryonic gi |
| 26 | Caenorhabditis briggsae AF16 | 50.40 | 3 | Hypothetical protein CBG03777 |
| 27 | Caenorhabditis elegans | 50.10 | 6 | HIStone family member (his-41) |
| 28 | Candida glabrata | 48.10 | 2 | unnamed protein product |
| 29 | Aspergillus nidulans FGSC A4 | 47.80 | 1 | H2B_EMENI Histone H2B |
Top-ranked sequences
| organism | matching |
|---|---|
| Mus musculus | Query 1 MPEPAKSAPAPKKGSKKAVT#KAQKKDGKKRKRSRKESYSVYVYKVLKQVH 50 ||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 1 MPEPAKSAPAPKKGSKKAVT#KAQKKDGKKRKRSRKESYSVYVYKVLKQVH 50 |
| Homo sapiens | Query 1 MPEPAKSAPAPKKGSKKAVT#KAQKKDGKKRKRSRKESYSVYVYKVLKQVH 50 ||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 1 MPEPAKSAPAPKKGSKKAVT#KAQKKDGKKRKRSRKESYSVYVYKVLKQVH 50 |
| Canis familiaris | Query 1 MPEPAKSAPAPKKGSKKAVT#KAQKKDGKKRKRSRKESYSVYVYKVLKQVH 50 ||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 1 MPEPAKSAPAPKKGSKKAVT#KAQKKDGKKRKRSRKESYSVYVYKVLKQVH 50 |
| Rattus norvegicus | Query 1 MPEPAKSAPAPKKGSKKAVT#KAQKKDGKKRKRSRKESYSVYVYKVLKQVH 50 ||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 1 MPEPAKSAPAPKKGSKKAVT#KAQKKDGKKRKRSRKESYSVYVYKVLKQVH 50 |
| Bos taurus | Query 1 MPEPAKSAPAPKKGSKKAVT#KAQKKDGKKRKRSRKESYSVYVYKVLKQVH 50 ||||+|||||||||||||||#|||||||||||||||||||||||||||||| Sbjct 1 MPEPSKSAPAPKKGSKKAVT#KAQKKDGKKRKRSRKESYSVYVYKVLKQVH 50 |
| Bos taurus | Query 1 MPEPAKSAPAPKKGSKKAVT#KAQKKDGKKRKRSRKESYSVYVYKVLKQVH 50 ||||+|||||||||||||+|#|||||||||||||||||||+|||||||||| Sbjct 1 MPEPSKSAPAPKKGSKKAIT#KAQKKDGKKRKRSRKESYSIYVYKVLKQVH 50 |
| Macaca fascicularis | Query 1 MPEPAKSAPAPKKGSKKAVT#KAQKKDGKKRKRSRKESYSVYVYKVLKQVH 50 ||||||||||||||||||||#|||||||||||||||||||||||||||+|| Sbjct 1 MPEPAKSAPAPKKGSKKAVT#KAQKKDGKKRKRSRKESYSVYVYKVLKRVH 50 |
| N/A | Query 2 PEPAKSAPAPKKGSKKAVT#KAQKKDGKKRKRSRKESYSVYVYKVLKQVH 50 |||||||||||||||||||#|||||||||||||||||||||||||||||| Query 2 PEPAKSAPAPKKGSKKAVT#KAQKKDGKKRKRSRKESYSVYVYKVLKQVH 50 |
| Rattus norvegicus | Query 1 MPEPAKSAPAPKKGSKKAVT#KAQKKDGKKRKRSRKESYSVYVYKVLKQVH 50 ||| ||||||||||||||||#|||||||||||||||||||||||||||||| Sbjct 1 MPETAKSAPAPKKGSKKAVT#KAQKKDGKKRKRSRKESYSVYVYKVLKQVH 50 |
| Homo sapiens | Query 1 MPEPAKSAPAPKKGSKKAVT#KAQKKDGKKRKRSRKESYSVYVYKVLKQVH 50 ||+|||||||||||||||||#| |||||||||||||||||||||||||||| Sbjct 1 MPDPAKSAPAPKKGSKKAVT#KVQKKDGKKRKRSRKESYSVYVYKVLKQVH 50 |
| Pongo pygmaeus | Query 1 MPEPAKSAPAPKKGSKKAVT#KAQKKDGKKRKRSRKESYSVYVYKVLKQVH 50 ||||||||||||||||||||#||||||||||||| |||||+|||||||||| Sbjct 1 MPEPAKSAPAPKKGSKKAVT#KAQKKDGKKRKRSCKESYSIYVYKVLKQVH 50 |
| Xenopus laevis | Query 1 MPEPAKSAPAPKKGSKKAVT#KAQKKDGKKRKRSRKESYSVYVYKVLKQVH 50 ||||||||||||||||||||#| ||||||||++||||||++|||||||||| Sbjct 1 MPEPAKSAPAPKKGSKKAVT#KTQKKDGKKRRKSRKESYAIYVYKVLKQVH 50 |
| Xenopus tropicalis | Query 1 MPEPAKSAPAPKKGSKKAVT#KAQKKDGKKRKRSRKESYSVYVYKVLKQVH 50 |||||||||||||||||||+#| ||||||||++||||||++|||||||||| Sbjct 1 MPEPAKSAPAPKKGSKKAVS#KTQKKDGKKRRKSRKESYAIYVYKVLKQVH 50 |
| Gallus gallus | Query 1 MPEPAKSAPAPKKGSKKAVT#KAQKKDGKKRKRSRKESYSVYVYKVLKQVH 50 ||||||||||||||||||||#| ||| |||||+||||||+|||||||||| Sbjct 1 MPEPAKSAPAPKKGSKKAVT#KTQKKGDKKRKRARKESYSIYVYKVLKQVH 50 |
| Rhacophorus schlegelii | Query 1 MPEPAKSAPAPKKGSKKAVT#KAQKKDGKKRKRSRKESYSVYVYKVLKQVH 50 |||||||||| ||||||||+#| ||||||||++||||||++|||||||||| Sbjct 1 MPEPAKSAPAAKKGSKKAVS#KVQKKDGKKRRKSRKESYAIYVYKVLKQVH 50 |
| Danio rerio | Query 1 MPEPAKSAPAPKKGSKKAVT#KAQKKDGKKRKRSRKESYSVYVYKVLKQVH 50 ||||||||||||||||||||#| ||| |||+++|||||++|||||||||| Sbjct 1 MPEPAKSAPAPKKGSKKAVT#KTQKKGDKKRRKTRKESYAIYVYKVLKQVH 50 |
| Expression vector pET3-H2B | Query 5 AKSAPAPKKGSKKAVT#KAQKKDGKKRKRSRKESYSVYVYKVLKQVH 50 ||||||||||||||||#| ||||||||+++|||||++|||||||||| Sbjct 2 AKSAPAPKKGSKKAVT#KTQKKDGKKRRKTRKESYAIYVYKVLKQVH 47 |
| Tetraodon nigroviridis | Query 1 MPEPAKSAPAPKKGSKKAVT#KAQKKDGKKRKRSRKESYSVYVYKVLKQVH 50 || ||||||| |||||||+|#||||| |||+++|||||++|||||||||| Sbjct 1 MPGPAKSAPAAKKGSKKAIT#KAQKKGDKKRRKNRKESYAIYVYKVLKQVH 50 |
| Oncorhynchus mykiss | Query 1 MPEPAKSAPAPKKGSKKAVT#KAQKKDGKKRKRSRKESYSVYVYKVLKQVH 50 ||||||||| |||||||||#| | ||||++||||||++|||||||||| Sbjct 1 MPEPAKSAP--KKGSKKAVT#KTAGKGGKKRRKSRKESYAIYVYKVLKQVH 48 |
| Cairina moschata | Query 1 MPEPAKSAPAPKKGSKKAVT#KAQKKDGKKRKRSRKESYSVYVYKVLKQVH 50 ||||||||||||||||||||#| ||| ||||+|||||||+|||||||||| Sbjct 1 MPEPAKSAPAPKKGSKKAVT#KTQKKGDKKRKKSRKESYSIYVYKVLKQVH 50 |
| Pan troglodytes | Query 1 MPEPAKSAPAPKKGSKKAVT#KAQKKDGKKRKRSRKESYSVYVYKVLKQVH 50 ||||+++||| |||||||+|#|||||||||||| ||||||+|||||||||| Sbjct 1 MPEPSRAAPASKKGSKKAIT#KAQKKDGKKRKRGRKESYSIYVYKVLKQVH 50 |
| Psammechinus miliaris | Query 5 AKSAPAPKKGSKKAVT#KAQKKDGKKRKRSRKESYSVYVYKVLKQVH 50 | +| |||||||| # + |||| | ||||| +|+|||||||| Sbjct 2 APTAQVAKKGSKKAVK#APRPSGGKKRNRKRKESYGIYIYKVLKQVH 47 |
| Strongylocentrotus purpuratus | Query 5 AKSAPAPKKGSKKAVT#KAQKKDGKKRKRSRKESYSVYVYKVLKQVH 50 | +| |||||||| # + |||| | ||||| +|+|||||||| Sbjct 2 APTAQVAKKGSKKAVK#APRPSGGKKRNRKRKESYGIYIYKVLKQVH 47 |
| Strongylocentrotus purpuratus | Query 5 AKSAPAPKKGSKKAVT#KAQKKDGKKRKRSRKESYSVYVYKVLKQVH 50 | +| |||||||| # + |||| | ||||| +|+|||||||| Sbjct 2 APTAQVAKKGSKKAVK#APRPSGGKKRNRKRKESYGIYIYKVLKQVH 47 |
| Psammechinus miliaris | Query 5 AKSAPAPKKGSKKAVT#KAQKKDGKKRKRSRKESYSVYVYKVLKQVH 50 | + |||||||| # + |||| | ||||| +|+|||||||| Sbjct 2 APTGQVAKKGSKKAVK#PPRASGGKKRHRKRKESYGIYIYKVLKQVH 47 |
| Caenorhabditis briggsae AF16 | Query 5 AKSAPAPKKGSKKAVT#KAQKKDGKKRKRSRKESYSVYVYKVLKQVH 50 | |+ | | | |# + ||||||+ ||||||||+|+|||||| Sbjct 2 APPKPSAKGAKKAAKT#VTKPKDGKKRRAHRKESYSVYIYRVLKQVH 47 |
| Caenorhabditis elegans | Query 20 T#KAQKKDGKKRKRSRKESYSVYVYKVLKQVH 50 |# ++ ||||||| +||||||||+|+|||||| Sbjct 17 T#VSKPKDGKKRKHARKESYSVYIYRVLKQVH 47 |
| Candida glabrata | Query 4 PAKSAPAPKKGSKKAVT#KAQKKDGKKRKRSRKESYSVYVYKVLKQVH 50 || ||| || + | # | ||||| ++|||+|| |+|||||| | Sbjct 9 PASKAPAEKKPAAKKT-#-APSSDGKKRTKARKETYSSYIYKVLKQTH 53 |
| Aspergillus nidulans FGSC A4 | Query 4 PAKSAPAPKKGSKKAVT#KAQKKDGKKRKRSRKESYSVYVYKVLKQVH 50 || ||| || + | # | + ||| ++|||+|| |+|||||||| Sbjct 17 PAGKAPAEKKEAGKKTA#AAASGEKKKRGKTRKETYSSYIYKVLKQVH 63 |
[Site 3] KRSRKESYSV40-YVYKVLKQVH
Val40  Tyr
Tyr
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Lys31 | Arg32 | Ser33 | Arg34 | Lys35 | Glu36 | Ser37 | Tyr38 | Ser39 | Val40 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Tyr41 | Val42 | Tyr43 | Lys44 | Val45 | Leu46 | Lys47 | Gln48 | Val49 | His50 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| PKKGSKKAVTKAQKKDGKKRKRSRKESYSVYVYKVLKQVHPDTGISSKAMGIMNSFVNDI |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | Homo sapiens | 122.00 | 23 | histone cluster 1, H2bl |
| 2 | Canis familiaris | 122.00 | 8 | PREDICTED: similar to Histone H2B 291B, partial |
| 3 | Mus musculus | 121.00 | 15 | H2B1P_MOUSE Histone H2B type 1-P gi |
| 4 | Bos taurus | 121.00 | 14 | histone H2B-like |
| 5 | N/A | 121.00 | 12 | 0506206A histone H2B |
| 6 | Rattus norvegicus | 121.00 | 3 | PREDICTED: similar to Histone H2B 291B |
| 7 | Rattus norvegicus | 120.00 | 3 | histone cluster 1, H2bl |
| 8 | Macaca fascicularis | 119.00 | 1 | H2B1K_MACFA Histone H2B type 1-K (H2B K) gi |
| 9 | Homo sapiens | 119.00 | 1 | histone cluster 2, H2bf |
| 10 | Bos taurus | 118.00 | 1 | histone cluster 1, H2bn |
| 11 | Pongo pygmaeus | 117.00 | 1 | H2B2E_PONPY Histone H2B type 2-E gi |
| 12 | Xenopus laevis | 112.00 | 4 | histone H2B |
| 13 | Expression vector pET3-H2B | 112.00 | 1 | H Chain H, 2.9 Angstrom X-Ray Structure Of Hybrid |
| 14 | Xenopus tropicalis | 111.00 | 3 | histone 1, H2bk |
| 15 | Gallus gallus | 110.00 | 9 | PREDICTED: similar to histone H2B |
| 16 | Danio rerio | 110.00 | 6 | PREDICTED: hypothetical protein |
| 17 | Rhacophorus schlegelii | 108.00 | 1 | H2B_RHASC Histone H2B gi |
| 18 | Oncorhynchus mykiss | 108.00 | 1 | unnamed protein product |
| 19 | Tetraodon nigroviridis | 105.00 | 10 | unnamed protein product |
| 20 | Cairina moschata | 94.40 | 1 | H2B_CAIMO Histone H2B gi |
| 21 | Pan troglodytes | 93.20 | 1 | PREDICTED: hypothetical protein |
| 22 | Strongylocentrotus purpuratus | 92.00 | 2 | PREDICTED: similar to histone H2B |
| 23 | Psammechinus miliaris | 92.00 | 2 | H2BE2_PSAMI Histone H2B.2, embryonic gi |
| 24 | Psammechinus miliaris | 92.00 | 1 | H2BE1_PSAMI Histone H2B.1, embryonic gi |
| 25 | Caenorhabditis elegans | 89.70 | 6 | HIStone family member (his-41) |
| 26 | Caenorhabditis briggsae AF16 | 88.20 | 3 | Hypothetical protein CBG10818 |
| 27 | Holothuria tubulosa | 83.60 | 1 | H2B_HOLTU Histone H2B gi |
| 28 | Strongylocentrotus purpuratus | 83.60 | 1 | histone H2B |
| 29 | Acropora formosa | 83.20 | 1 | H2B_ACRFO Histone H2B gi |
| 30 | Drosophila yakuba | 82.80 | 1 | histone 2B |
Top-ranked sequences
| organism | matching |
|---|---|
| Homo sapiens | Query 11 PKKGSKKAVTKAQKKDGKKRKRSRKESYSV#YVYKVLKQVHPDTGISSKAMGIMNSFVNDI 70 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 11 PKKGSKKAVTKAQKKDGKKRKRSRKESYSV#YVYKVLKQVHPDTGISSKAMGIMNSFVNDI 70 |
| Canis familiaris | Query 11 PKKGSKKAVTKAQKKDGKKRKRSRKESYSV#YVYKVLKQVHPDTGISSKAMGIMNSFVNDI 70 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 11 PKKGSKKAVTKAQKKDGKKRKRSRKESYSV#YVYKVLKQVHPDTGISSKAMGIMNSFVNDI 70 |
| Mus musculus | Query 11 PKKGSKKAVTKAQKKDGKKRKRSRKESYSV#YVYKVLKQVHPDTGISSKAMGIMNSFVNDI 70 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 11 PKKGSKKAVTKAQKKDGKKRKRSRKESYSV#YVYKVLKQVHPDTGISSKAMGIMNSFVNDI 70 |
| Bos taurus | Query 11 PKKGSKKAVTKAQKKDGKKRKRSRKESYSV#YVYKVLKQVHPDTGISSKAMGIMNSFVNDI 70 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 11 PKKGSKKAVTKAQKKDGKKRKRSRKESYSV#YVYKVLKQVHPDTGISSKAMGIMNSFVNDI 70 |
| N/A | Query 11 PKKGSKKAVTKAQKKDGKKRKRSRKESYSV#YVYKVLKQVHPDTGISSKAMGIMNSFVNDI 70 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 11 PKKGSKKAVTKAQKKDGKKRKRSRKESYSV#YVYKVLKQVHPDTGISSKAMGIMNSFVNDI 70 |
| Rattus norvegicus | Query 11 PKKGSKKAVTKAQKKDGKKRKRSRKESYSV#YVYKVLKQVHPDTGISSKAMGIMNSFVNDI 70 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 11 PKKGSKKAVTKAQKKDGKKRKRSRKESYSV#YVYKVLKQVHPDTGISSKAMGIMNSFVNDI 70 |
| Rattus norvegicus | Query 11 PKKGSKKAVTKAQKKDGKKRKRSRKESYSV#YVYKVLKQVHPDTGISSKAMGIMNSFVNDI 70 ||||||||||||||||||+|||||||||||#|||||||||||||||||||||||||||||| Sbjct 11 PKKGSKKAVTKAQKKDGKERKRSRKESYSV#YVYKVLKQVHPDTGISSKAMGIMNSFVNDI 70 |
| Macaca fascicularis | Query 11 PKKGSKKAVTKAQKKDGKKRKRSRKESYSV#YVYKVLKQVHPDTGISSKAMGIMNSFVNDI 70 ||||||||||||||||||||||||||||||#|||||||+|||||||||||||||||||||| Sbjct 11 PKKGSKKAVTKAQKKDGKKRKRSRKESYSV#YVYKVLKRVHPDTGISSKAMGIMNSFVNDI 70 |
| Homo sapiens | Query 11 PKKGSKKAVTKAQKKDGKKRKRSRKESYSV#YVYKVLKQVHPDTGISSKAMGIMNSFVNDI 70 ||||||||||| ||||||||||||||||||#|||||||||||||||||||||||||||||| Sbjct 11 PKKGSKKAVTKVQKKDGKKRKRSRKESYSV#YVYKVLKQVHPDTGISSKAMGIMNSFVNDI 70 |
| Bos taurus | Query 11 PKKGSKKAVTKAQKKDGKKRKRSRKESYSV#YVYKVLKQVHPDTGISSKAMGIMNSFVNDI 70 ||||||||||||||||||||||||||||||#||||||||||||||||||||| |||||||| Sbjct 11 PKKGSKKAVTKAQKKDGKKRKRSRKESYSV#YVYKVLKQVHPDTGISSKAMGNMNSFVNDI 70 |
| Pongo pygmaeus | Query 11 PKKGSKKAVTKAQKKDGKKRKRSRKESYSV#YVYKVLKQVHPDTGISSKAMGIMNSFVNDI 70 ||||||||||||||||||||||| |||||+#|||||||||||||||||||||||||||||| Sbjct 11 PKKGSKKAVTKAQKKDGKKRKRSCKESYSI#YVYKVLKQVHPDTGISSKAMGIMNSFVNDI 70 |
| Xenopus laevis | Query 11 PKKGSKKAVTKAQKKDGKKRKRSRKESYSV#YVYKVLKQVHPDTGISSKAMGIMNSFVNDI 70 ||||||||||| ||||||||++||||||++#|||||||||||||||||||| ||||||||+ Sbjct 11 PKKGSKKAVTKTQKKDGKKRRKSRKESYAI#YVYKVLKQVHPDTGISSKAMSIMNSFVNDV 70 |
| Expression vector pET3-H2B | Query 11 PKKGSKKAVTKAQKKDGKKRKRSRKESYSV#YVYKVLKQVHPDTGISSKAMGIMNSFVNDI 70 ||||||||||| ||||||||+++|||||++#|||||||||||||||||||| ||||||||+ Sbjct 8 PKKGSKKAVTKTQKKDGKKRRKTRKESYAI#YVYKVLKQVHPDTGISSKAMSIMNSFVNDV 67 |
| Xenopus tropicalis | Query 11 PKKGSKKAVTKAQKKDGKKRKRSRKESYSV#YVYKVLKQVHPDTGISSKAMGIMNSFVNDI 70 ||||||||||| ||||||||+++|||||++#|||||||||||||||||||| ||||||||+ Sbjct 11 PKKGSKKAVTKTQKKDGKKRRKTRKESYAI#YVYKVLKQVHPDTGISSKAMSIMNSFVNDV 70 |
| Gallus gallus | Query 11 PKKGSKKAVTKAQKKDGKKRKRSRKESYSV#YVYKVLKQVHPDTGISSKAMGIMNSFVNDI 70 ||||||||||| ||| ||| +|||||||+#|||||||||||||||||||||||||||||| Sbjct 11 PKKGSKKAVTKTQKKGDKKRHKSRKESYSI#YVYKVLKQVHPDTGISSKAMGIMNSFVNDI 70 |
| Danio rerio | Query 11 PKKGSKKAVTKAQKKDGKKRKRSRKESYSV#YVYKVLKQVHPDTGISSKAMGIMNSFVNDI 70 |||||||||||+ | ||||||+|||||++#|||||||||||||||||||||||||||||| Sbjct 9 PKKGSKKAVTKSAGKGGKKRKRTRKESYAI#YVYKVLKQVHPDTGISSKAMGIMNSFVNDI 68 |
| Rhacophorus schlegelii | Query 12 KKGSKKAVTKAQKKDGKKRKRSRKESYSV#YVYKVLKQVHPDTGISSKAMGIMNSFVNDI 70 ||||||||+| ||||||||++||||||++#|||||||||||||||||||| ||||||||| Sbjct 12 KKGSKKAVSKVQKKDGKKRRKSRKESYAI#YVYKVLKQVHPDTGISSKAMSIMNSFVNDI 70 |
| Oncorhynchus mykiss | Query 11 PKKGSKKAVTKAQKKDGKKRKRSRKESYSV#YVYKVLKQVHPDTGISSKAMGIMNSFVNDI 70 ||||||||||| | ||||++||||||++#|||||||||||||||||||||||||||||| Sbjct 9 PKKGSKKAVTKTAGKGGKKRRKSRKESYAI#YVYKVLKQVHPDTGISSKAMGIMNSFVNDI 68 |
| Tetraodon nigroviridis | Query 11 PKKGSKKAVTKAQKKDGKKRKRSRKESYSV#YVYKVLKQVHPDTGISSKAMGIMNSFVNDI 70 ||||||||||| | |||++|+|||||++#|||||||||||||||||||| ||||||||| Sbjct 9 PKKGSKKAVTKTASKGGKKKRRTRKESYAI#YVYKVLKQVHPDTGISSKAMSIMNSFVNDI 68 |
| Cairina moschata | Query 11 PKKGSKKAVTKAQKKDGKKRKRSRKESYSV#YVYKVLKQVHPDTGISSKAMGIMNSFVNDI 70 ||||||||||| ||| ||||+|||||||+#|||||||||||||||||||||||||||||| Sbjct 11 PKKGSKKAVTKTQKKGDKKRKKSRKESYSI#YVYKVLKQVHPDTGISSKAMGIMNSFVNDI 70 |
| Pan troglodytes | Query 12 KKGSKKAVTKAQKKDGKKRKRSRKESYSV#YVYKVLKQVHPDTGISSKAMGIMNSFVNDI 70 |||||||+||||||||||||| ||||||+#|||||||||||||||||||||||||||||| Sbjct 12 KKGSKKAITKAQKKDGKKRKRGRKESYSI#YVYKVLKQVHPDTGISSKAMGIMNSFVNDI 70 |
| Strongylocentrotus purpuratus | Query 12 KKGSKKAVTKAQKKDGKKRKRSRKESYSV#YVYKVLKQVHPDTGISSKAMGIMNSFVNDI 70 |||||||| + |||| | ||||| +#|+|||||||||||||||+|| ||||||||| Sbjct 9 KKGSKKAVKAPRPSGGKKRNRKRKESYGI#YIYKVLKQVHPDTGISSRAMVIMNSFVNDI 67 |
| Psammechinus miliaris | Query 12 KKGSKKAVTKAQKKDGKKRKRSRKESYSV#YVYKVLKQVHPDTGISSKAMGIMNSFVNDI 70 |||||||| + |||| | ||||| +#|+|||||||||||||||+|| ||||||||| Sbjct 9 KKGSKKAVKAPRPSGGKKRNRKRKESYGI#YIYKVLKQVHPDTGISSRAMIIMNSFVNDI 67 |
| Psammechinus miliaris | Query 12 KKGSKKAVTKAQKKDGKKRKRSRKESYSV#YVYKVLKQVHPDTGISSKAMGIMNSFVNDI 70 |||||||| + |||| | ||||| +#|+||||||||||||+||+|| ||||||||| Sbjct 9 KKGSKKAVKPPRASGGKKRHRKRKESYGI#YIYKVLKQVHPDTGVSSRAMTIMNSFVNDI 67 |
| Caenorhabditis elegans | Query 20 TKAQKKDGKKRKRSRKESYSV#YVYKVLKQVHPDTGISSKAMGIMNSFVNDI 70 | ++ ||||||| +|||||||#|+|+||||||||||+||||| ||||||||+ Sbjct 17 TVSKPKDGKKRKHARKESYSV#YIYRVLKQVHPDTGVSSKAMSIMNSFVNDV 67 |
| Caenorhabditis briggsae AF16 | Query 20 TKAQKKDGKKRKRSRKESYSV#YVYKVLKQVHPDTGISSKAMGIMNSFVNDI 70 | + ||||||| |||||||#|+|+||||||||||+||||| ||||||||+ Sbjct 17 TVTKPKDGKKRKAHRKESYSV#YIYRVLKQVHPDTGVSSKAMSIMNSFVNDV 67 |
| Holothuria tubulosa | Query 28 KKRKRSRKESYSV#YVYKVLKQVHPDTGISSKAMGIMNSFVNDI 70 +||||+|+||||+#|+|||+|||||||||||+|| ||||||||| Sbjct 25 RKRKRTRRESYSI#YIYKVMKQVHPDTGISSRAMSIMNSFVNDI 67 |
| Strongylocentrotus purpuratus | Query 12 KKGSKKAV--TKAQKKDGKKRKRSRKESYSV#YVYKVLKQVHPDTGISSKAMGIMNSFVND 69 |||||||| || |||| | ||||| +#|+|||||||||||||||+|| |||| |+| Sbjct 9 KKGSKKAVKGTKTAX-GGKKRNRKRKESYGI#YIYKVLKQVHPDTGISSRAMVIMNSXVBD 67 Query 70 I 70 | Query 70 I 70 |
| Acropora formosa | Query 12 KKGSKKAVTKAQK---KDGKKRKRSRKESYSV#YVYKVLKQVHPDTGISSKAMGIMNSFVN 68 ||| |+ | ||+ + |+|+ |||||++#|+|||||||||||||||||||||||||| Sbjct 9 KKGEKR-VGKAKSGTAETAKRRRGKRKESYAI#YIYKVLKQVHPDTGISSKAMGIMNSFVN 67 Query 69 DI 70 || Query 69 DI 70 |
| Drosophila yakuba | Query 25 KDGKKRKRSRKESYSV#YVYKVLKQVHPDTGISSKAMGIMNSFVNDI 70 | || || |||||++#|+|||||||||||||||||| ||||||||| Sbjct 22 KTDKKMKRKRKESYAI#YIYKVLKQVHPDTGISSKAMSIMNSFVNDI 67 |
[Site 4] SRKESYSVYV42-YKVLKQVHPD
Val42  Tyr
Tyr
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Ser33 | Arg34 | Lys35 | Glu36 | Ser37 | Tyr38 | Ser39 | Val40 | Tyr41 | Val42 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Tyr43 | Lys44 | Val45 | Leu46 | Lys47 | Gln48 | Val49 | His50 | Pro51 | Asp52 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| KGSKKAVTKAQKKDGKKRKRSRKESYSVYVYKVLKQVHPDTGISSKAMGIMNSFVNDIFE |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | Canis familiaris | 124.00 | 8 | PREDICTED: similar to Histone H2B 291B, partial |
| 2 | Homo sapiens | 123.00 | 23 | histone cluster 1, H2bl |
| 3 | Mus musculus | 123.00 | 15 | histone cluster 1, H2bj |
| 4 | Bos taurus | 122.00 | 13 | PREDICTED: similar to Hist1h2bj protein |
| 5 | N/A | 122.00 | 12 | 0506206A histone H2B |
| 6 | Rattus norvegicus | 122.00 | 3 | PREDICTED: similar to Histone H2B 291B |
| 7 | Rattus norvegicus | 122.00 | 3 | histone 1, H2bn (predicted) |
| 8 | Macaca fascicularis | 120.00 | 1 | H2B1K_MACFA Histone H2B type 1-K (H2B K) gi |
| 9 | Homo sapiens | 120.00 | 1 | histone cluster 2, H2bf |
| 10 | Bos taurus | 119.00 | 1 | histone cluster 1, H2bn |
| 11 | Pongo pygmaeus | 119.00 | 1 | H2B2E_PONPY Histone H2B type 2-E gi |
| 12 | Xenopus laevis | 114.00 | 4 | histone H2B |
| 13 | Expression vector pET3-H2B | 113.00 | 1 | H Chain H, 2.9 Angstrom X-Ray Structure Of Hybrid |
| 14 | Gallus gallus | 112.00 | 9 | hypothetical protein LOC417957 |
| 15 | Xenopus tropicalis | 112.00 | 3 | histone 1, H2bk |
| 16 | Rhacophorus schlegelii | 112.00 | 1 | H2B_RHASC Histone H2B gi |
| 17 | Danio rerio | 111.00 | 6 | PREDICTED: hypothetical protein |
| 18 | Tetraodon nigroviridis | 108.00 | 10 | unnamed protein product |
| 19 | Oncorhynchus mykiss | 108.00 | 1 | unnamed protein product |
| 20 | Pan troglodytes | 97.10 | 1 | PREDICTED: hypothetical protein |
| 21 | Psammechinus miliaris | 96.70 | 3 | H2BE2_PSAMI Histone H2B.2, embryonic gi |
| 22 | Strongylocentrotus purpuratus | 96.70 | 2 | PREDICTED: similar to histone H2B |
| 23 | Psammechinus miliaris | 96.70 | 1 | H2BE1_PSAMI Histone H2B.1, embryonic gi |
| 24 | Caenorhabditis elegans | 95.50 | 6 | HIStone family member (his-41) |
| 25 | Cairina moschata | 95.50 | 1 | H2B_CAIMO Histone H2B gi |
| 26 | Caenorhabditis briggsae AF16 | 93.60 | 3 | Hypothetical protein CBG10818 |
| 27 | Holothuria tubulosa | 88.60 | 1 | H2B_HOLTU Histone H2B gi |
| 28 | Drosophila yakuba | 88.20 | 1 | histone 2B |
| 29 | Acropora formosa | 88.20 | 1 | H2B_ACRFO Histone H2B gi |
| 30 | Strongylocentrotus purpuratus | 87.80 | 1 | histone H2B |
| 31 | Candida glabrata | 87.00 | 1 | unnamed protein product |
Top-ranked sequences
| organism | matching |
|---|---|
| Canis familiaris | Query 13 KGSKKAVTKAQKKDGKKRKRSRKESYSVYV#YKVLKQVHPDTGISSKAMGIMNSFVNDIFE 72 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 13 KGSKKAVTKAQKKDGKKRKRSRKESYSVYV#YKVLKQVHPDTGISSKAMGIMNSFVNDIFE 72 |
| Homo sapiens | Query 13 KGSKKAVTKAQKKDGKKRKRSRKESYSVYV#YKVLKQVHPDTGISSKAMGIMNSFVNDIFE 72 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 13 KGSKKAVTKAQKKDGKKRKRSRKESYSVYV#YKVLKQVHPDTGISSKAMGIMNSFVNDIFE 72 |
| Mus musculus | Query 13 KGSKKAVTKAQKKDGKKRKRSRKESYSVYV#YKVLKQVHPDTGISSKAMGIMNSFVNDIFE 72 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 13 KGSKKAVTKAQKKDGKKRKRSRKESYSVYV#YKVLKQVHPDTGISSKAMGIMNSFVNDIFE 72 |
| Bos taurus | Query 13 KGSKKAVTKAQKKDGKKRKRSRKESYSVYV#YKVLKQVHPDTGISSKAMGIMNSFVNDIFE 72 ||||||+||||||||||||||||||||+||#|||||||||||||||||||||||||||||| Sbjct 13 KGSKKAITKAQKKDGKKRKRSRKESYSIYV#YKVLKQVHPDTGISSKAMGIMNSFVNDIFE 72 |
| N/A | Query 13 KGSKKAVTKAQKKDGKKRKRSRKESYSVYV#YKVLKQVHPDTGISSKAMGIMNSFVNDIFE 72 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||+ Sbjct 12 KGSKKAVTKAQKKDGKKRKRSRKESYSVYV#YKVLKQVHPDTGISSKAMGIMNSFVNDIFZ 71 |
| Rattus norvegicus | Query 13 KGSKKAVTKAQKKDGKKRKRSRKESYSVYV#YKVLKQVHPDTGISSKAMGIMNSFVNDIFE 72 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 13 KGSKKAVTKAQKKDGKKRKRSRKESYSVYV#YKVLKQVHPDTGISSKAMGIMNSFVNDIFE 72 |
| Rattus norvegicus | Query 13 KGSKKAVTKAQKKDGKKRKRSRKESYSVYV#YKVLKQVHPDTGISSKAMGIMNSFVNDIFE 72 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 13 KGSKKAVTKAQKKDGKKRKRSRKESYSVYV#YKVLKQVHPDTGISSKAMGIMNSFVNDIFE 72 |
| Macaca fascicularis | Query 13 KGSKKAVTKAQKKDGKKRKRSRKESYSVYV#YKVLKQVHPDTGISSKAMGIMNSFVNDIFE 72 ||||||||||||||||||||||||||||||#|||||+|||||||||||||||||||||||| Sbjct 13 KGSKKAVTKAQKKDGKKRKRSRKESYSVYV#YKVLKRVHPDTGISSKAMGIMNSFVNDIFE 72 |
| Homo sapiens | Query 13 KGSKKAVTKAQKKDGKKRKRSRKESYSVYV#YKVLKQVHPDTGISSKAMGIMNSFVNDIFE 72 ||||||||| ||||||||||||||||||||#|||||||||||||||||||||||||||||| Sbjct 13 KGSKKAVTKVQKKDGKKRKRSRKESYSVYV#YKVLKQVHPDTGISSKAMGIMNSFVNDIFE 72 |
| Bos taurus | Query 13 KGSKKAVTKAQKKDGKKRKRSRKESYSVYV#YKVLKQVHPDTGISSKAMGIMNSFVNDIFE 72 ||||||||||||||||||||||||||||||#||||||||||||||||||| |||||||||| Sbjct 13 KGSKKAVTKAQKKDGKKRKRSRKESYSVYV#YKVLKQVHPDTGISSKAMGNMNSFVNDIFE 72 |
| Pongo pygmaeus | Query 13 KGSKKAVTKAQKKDGKKRKRSRKESYSVYV#YKVLKQVHPDTGISSKAMGIMNSFVNDIFE 72 ||||||||||||||||||||| |||||+||#|||||||||||||||||||||||||||||| Sbjct 13 KGSKKAVTKAQKKDGKKRKRSCKESYSIYV#YKVLKQVHPDTGISSKAMGIMNSFVNDIFE 72 |
| Xenopus laevis | Query 13 KGSKKAVTKAQKKDGKKRKRSRKESYSVYV#YKVLKQVHPDTGISSKAMGIMNSFVNDIFE 72 ||||||||| ||||||||++||||||++||#|||||||||||||||||| ||||||||+|| Sbjct 13 KGSKKAVTKTQKKDGKKRRKSRKESYAIYV#YKVLKQVHPDTGISSKAMSIMNSFVNDVFE 72 |
| Expression vector pET3-H2B | Query 13 KGSKKAVTKAQKKDGKKRKRSRKESYSVYV#YKVLKQVHPDTGISSKAMGIMNSFVNDIFE 72 ||||||||| ||||||||+++|||||++||#|||||||||||||||||| ||||||||+|| Sbjct 10 KGSKKAVTKTQKKDGKKRRKTRKESYAIYV#YKVLKQVHPDTGISSKAMSIMNSFVNDVFE 69 |
| Gallus gallus | Query 13 KGSKKAVTKAQKKDGKKRKRSRKESYSVYV#YKVLKQVHPDTGISSKAMGIMNSFVNDIFE 72 ||||||||| ||| |||++|||||||+||#|||||||||||||||||||||||||||||| Sbjct 13 KGSKKAVTKTQKKGDKKRRKSRKESYSIYV#YKVLKQVHPDTGISSKAMGIMNSFVNDIFE 72 |
| Xenopus tropicalis | Query 13 KGSKKAVTKAQKKDGKKRKRSRKESYSVYV#YKVLKQVHPDTGISSKAMGIMNSFVNDIFE 72 ||||||||| ||||||||+++|||||++||#|||||||||||||||||| ||||||||+|| Sbjct 13 KGSKKAVTKTQKKDGKKRRKTRKESYAIYV#YKVLKQVHPDTGISSKAMSIMNSFVNDVFE 72 |
| Rhacophorus schlegelii | Query 13 KGSKKAVTKAQKKDGKKRKRSRKESYSVYV#YKVLKQVHPDTGISSKAMGIMNSFVNDIFE 72 |||||||+| ||||||||++||||||++||#|||||||||||||||||| ||||||||||| Sbjct 13 KGSKKAVSKVQKKDGKKRRKSRKESYAIYV#YKVLKQVHPDTGISSKAMSIMNSFVNDIFE 72 |
| Danio rerio | Query 13 KGSKKAVTKAQKKDGKKRKRSRKESYSVYV#YKVLKQVHPDTGISSKAMGIMNSFVNDIFE 72 |||||||||+ | ||||||+|||||++||#|||||||||||||||||||||||||||||| Sbjct 11 KGSKKAVTKSAGKGGKKRKRTRKESYAIYV#YKVLKQVHPDTGISSKAMGIMNSFVNDIFE 70 |
| Tetraodon nigroviridis | Query 13 KGSKKAVTKAQKKDGKKRKRSRKESYSVYV#YKVLKQVHPDTGISSKAMGIMNSFVNDIFE 72 ||||||+|||||| |||+++|||||++||#|||||||||||||||||| ||||||||||| Sbjct 13 KGSKKAITKAQKKGDKKRRKNRKESYAIYV#YKVLKQVHPDTGISSKAMSIMNSFVNDIFE 72 |
| Oncorhynchus mykiss | Query 13 KGSKKAVTKAQKKDGKKRKRSRKESYSVYV#YKVLKQVHPDTGISSKAMGIMNSFVNDIFE 72 ||||||||| | ||||++||||||++||#|||||||||||||||||||||||||||||| Sbjct 11 KGSKKAVTKTAGKGGKKRRKSRKESYAIYV#YKVLKQVHPDTGISSKAMGIMNSFVNDIFE 70 |
| Pan troglodytes | Query 13 KGSKKAVTKAQKKDGKKRKRSRKESYSVYV#YKVLKQVHPDTGISSKAMGIMNSFVNDIFE 72 ||||||+||||||||||||| ||||||+||#|||||||||||||||||||||||||||||| Sbjct 13 KGSKKAITKAQKKDGKKRKRGRKESYSIYV#YKVLKQVHPDTGISSKAMGIMNSFVNDIFE 72 |
| Psammechinus miliaris | Query 13 KGSKKAVTKAQKKDGKKRKRSRKESYSVYV#YKVLKQVHPDTGISSKAMGIMNSFVNDIFE 72 ||||||| + |||| | ||||| +|+#|||||||||||||||+|| ||||||||||| Sbjct 10 KGSKKAVKAPRPSGGKKRNRKRKESYGIYI#YKVLKQVHPDTGISSRAMIIMNSFVNDIFE 69 |
| Strongylocentrotus purpuratus | Query 13 KGSKKAVTKAQKKDGKKRKRSRKESYSVYV#YKVLKQVHPDTGISSKAMGIMNSFVNDIFE 72 ||||||| + |||| | ||||| +|+#|||||||||||||||+|| ||||||||||| Sbjct 10 KGSKKAVKAPRPSGGKKRNRKRKESYGIYI#YKVLKQVHPDTGISSRAMVIMNSFVNDIFE 69 |
| Psammechinus miliaris | Query 13 KGSKKAVTKAQKKDGKKRKRSRKESYSVYV#YKVLKQVHPDTGISSKAMGIMNSFVNDIFE 72 ||||||| + |||| | ||||| +|+#||||||||||||+||+|| ||||||||||| Sbjct 10 KGSKKAVKPPRASGGKKRHRKRKESYGIYI#YKVLKQVHPDTGVSSRAMTIMNSFVNDIFE 69 |
| Caenorhabditis elegans | Query 20 TKAQKKDGKKRKRSRKESYSVYV#YKVLKQVHPDTGISSKAMGIMNSFVNDIFE 72 | ++ ||||||| +||||||||+#|+||||||||||+||||| ||||||||+|| Sbjct 17 TVSKPKDGKKRKHARKESYSVYI#YRVLKQVHPDTGVSSKAMSIMNSFVNDVFE 69 |
| Cairina moschata | Query 13 KGSKKAVTKAQKKDGKKRKRSRKESYSVYV#YKVLKQVHPDTGISSKAMGIMNSFVNDIFE 72 ||||||||| ||| ||||+|||||||+||#|||||||||||||||||||||||||||||| Sbjct 13 KGSKKAVTKTQKKGDKKRKKSRKESYSIYV#YKVLKQVHPDTGISSKAMGIMNSFVNDIFE 72 |
| Caenorhabditis briggsae AF16 | Query 20 TKAQKKDGKKRKRSRKESYSVYV#YKVLKQVHPDTGISSKAMGIMNSFVNDIFE 72 | + ||||||| ||||||||+#|+||||||||||+||||| ||||||||+|| Sbjct 17 TVTKPKDGKKRKAHRKESYSVYI#YRVLKQVHPDTGVSSKAMSIMNSFVNDVFE 69 |
| Holothuria tubulosa | Query 28 KKRKRSRKESYSVYV#YKVLKQVHPDTGISSKAMGIMNSFVNDIFE 72 +||||+|+||||+|+#|||+|||||||||||+|| ||||||||||| Sbjct 25 RKRKRTRRESYSIYI#YKVMKQVHPDTGISSRAMSIMNSFVNDIFE 69 |
| Drosophila yakuba | Query 25 KDGKKRKRSRKESYSVYV#YKVLKQVHPDTGISSKAMGIMNSFVNDIFE 72 | || || |||||++|+#|||||||||||||||||| ||||||||||| Sbjct 22 KTDKKMKRKRKESYAIYI#YKVLKQVHPDTGISSKAMSIMNSFVNDIFE 69 |
| Acropora formosa | Query 27 GKKRKRSRKESYSVYV#YKVLKQVHPDTGISSKAMGIMNSFVNDIFE 72 |+|+ |||||++|+#|||||||||||||||||||||||||||||| Sbjct 26 AKRRRGKRKESYAIYI#YKVLKQVHPDTGISSKAMGIMNSFVNDIFE 71 |
| Strongylocentrotus purpuratus | Query 13 KGSKKAV--TKAQKKDGKKRKRSRKESYSVYV#YKVLKQVHPDTGISSKAMGIMNSFVNDI 70 ||||||| || |||| | ||||| +|+#|||||||||||||||+|| |||| |+|| Sbjct 10 KGSKKAVKGTKTAX-GGKKRNRKRKESYGIYI#YKVLKQVHPDTGISSRAMVIMNSXVBDI 68 Query 71 FE 72 || Query 71 FE 72 |
| Candida glabrata | Query 22 AQKKDGKKRKRSRKESYSVYV#YKVLKQVHPDTGISSKAMGIMNSFVNDIFE 72 | ||||| ++|||+|| |+#|||||| ||||||| |+| |+||||||||| Sbjct 25 APSSDGKKRTKARKETYSSYI#YKVLKQTHPDTGISQKSMSILNSFVNDIFE 75 |
[Site 5] SYSVYVYKVL46-KQVHPDTGIS
Leu46  Lys
Lys
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Ser37 | Tyr38 | Ser39 | Val40 | Tyr41 | Val42 | Tyr43 | Lys44 | Val45 | Leu46 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Lys47 | Gln48 | Val49 | His50 | Pro51 | Asp52 | Thr53 | Gly54 | Ile55 | Ser56 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| KAVTKAQKKDGKKRKRSRKESYSVYVYKVLKQVHPDTGISSKAMGIMNSFVNDIFERIAG |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | Canis familiaris | 125.00 | 8 | PREDICTED: similar to Histone H2B 291B, partial |
| 2 | Homo sapiens | 124.00 | 23 | histone cluster 1, H2bk |
| 3 | N/A | 124.00 | 17 | 0506206A histone H2B |
| 4 | Bos taurus | 124.00 | 13 | histone H2B-like |
| 5 | Rattus norvegicus | 124.00 | 3 | PREDICTED: similar to Histone H2B 291B |
| 6 | Mus musculus | 123.00 | 15 | histone cluster 1, H2bh |
| 7 | Rattus norvegicus | 123.00 | 3 | histone 1, H2bn (predicted) |
| 8 | Macaca fascicularis | 122.00 | 1 | H2B1K_MACFA Histone H2B type 1-K (H2B K) gi |
| 9 | Homo sapiens | 122.00 | 1 | histone cluster 2, H2bf |
| 10 | Bos taurus | 121.00 | 1 | histone cluster 1, H2bn |
| 11 | Pongo pygmaeus | 120.00 | 1 | H2B2E_PONPY Histone H2B type 2-E gi |
| 12 | Xenopus laevis | 115.00 | 4 | histone H2B |
| 13 | Xenopus tropicalis | 114.00 | 3 | histone 2, H2bf |
| 14 | Rhacophorus schlegelii | 114.00 | 1 | H2B_RHASC Histone H2B gi |
| 15 | Expression vector pET3-H2B | 114.00 | 1 | H Chain H, 2.9 Angstrom X-Ray Structure Of Hybrid |
| 16 | Gallus gallus | 113.00 | 9 | hypothetical protein LOC417957 |
| 17 | Danio rerio | 112.00 | 6 | PREDICTED: hypothetical protein |
| 18 | Oncorhynchus mykiss | 110.00 | 1 | unnamed protein product |
| 19 | Tetraodon nigroviridis | 107.00 | 3 | unnamed protein product |
| 20 | Caenorhabditis elegans | 100.00 | 6 | HIStone family member (his-41) |
| 21 | Caenorhabditis briggsae AF16 | 99.40 | 3 | Hypothetical protein CBG10818 |
| 22 | Psammechinus miliaris | 97.80 | 1 | H2BE1_PSAMI Histone H2B.1, embryonic gi |
| 23 | Strongylocentrotus purpuratus | 97.80 | 1 | PREDICTED: similar to histone H2B |
| 24 | Psammechinus miliaris | 97.40 | 3 | H2BE2_PSAMI Histone H2B.2, embryonic gi |
| 25 | Cairina moschata | 96.30 | 1 | H2B_CAIMO Histone H2B gi |
| 26 | Pan troglodytes | 95.50 | 1 | PREDICTED: hypothetical protein |
| 27 | Holothuria tubulosa | 94.00 | 1 | H2B_HOLTU Histone H2B gi |
| 28 | Drosophila yakuba | 94.00 | 1 | histone 2B |
| 29 | Acropora formosa | 93.20 | 1 | H2B_ACRFO Histone H2B gi |
| 30 | Candida glabrata | 92.40 | 2 | unnamed protein product |
| 31 | Tigriopus californicus | 92.40 | 1 | H2B1_TIGCA Histone H2B.1/H2B.2 gi |
| 32 | Strongylocentrotus purpuratus | 91.70 | 1 | histone H2B |
Top-ranked sequences
| organism | matching |
|---|---|
| Canis familiaris | Query 17 KAVTKAQKKDGKKRKRSRKESYSVYVYKVL#KQVHPDTGISSKAMGIMNSFVNDIFERIAG 76 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 17 KAVTKAQKKDGKKRKRSRKESYSVYVYKVL#KQVHPDTGISSKAMGIMNSFVNDIFERIAG 76 |
| Homo sapiens | Query 17 KAVTKAQKKDGKKRKRSRKESYSVYVYKVL#KQVHPDTGISSKAMGIMNSFVNDIFERIAG 76 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 17 KAVTKAQKKDGKKRKRSRKESYSVYVYKVL#KQVHPDTGISSKAMGIMNSFVNDIFERIAG 76 |
| N/A | Query 17 KAVTKAQKKDGKKRKRSRKESYSVYVYKVL#KQVHPDTGISSKAMGIMNSFVNDIFERIAG 76 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||+|||| Sbjct 16 KAVTKAQKKDGKKRKRSRKESYSVYVYKVL#KQVHPDTGISSKAMGIMNSFVNDIFZRIAG 75 |
| Bos taurus | Query 17 KAVTKAQKKDGKKRKRSRKESYSVYVYKVL#KQVHPDTGISSKAMGIMNSFVNDIFERIAG 76 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 17 KAVTKAQKKDGKKRKRSRKESYSVYVYKVL#KQVHPDTGISSKAMGIMNSFVNDIFERIAG 76 |
| Rattus norvegicus | Query 17 KAVTKAQKKDGKKRKRSRKESYSVYVYKVL#KQVHPDTGISSKAMGIMNSFVNDIFERIAG 76 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 17 KAVTKAQKKDGKKRKRSRKESYSVYVYKVL#KQVHPDTGISSKAMGIMNSFVNDIFERIAG 76 |
| Mus musculus | Query 17 KAVTKAQKKDGKKRKRSRKESYSVYVYKVL#KQVHPDTGISSKAMGIMNSFVNDIFERIAG 76 ||+|||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Sbjct 17 KALTKAQKKDGKKRKRSRKESYSVYVYKVL#KQVHPDTGISSKAMGIMNSFVNDIFERIAG 76 |
| Rattus norvegicus | Query 17 KAVTKAQKKDGKKRKRSRKESYSVYVYKVL#KQVHPDTGISSKAMGIMNSFVNDIFERIAG 76 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 17 KAVTKAQKKDGKKRKRSRKESYSVYVYKVL#KQVHPDTGISSKAMGIMNSFVNDIFERIAG 76 |
| Macaca fascicularis | Query 17 KAVTKAQKKDGKKRKRSRKESYSVYVYKVL#KQVHPDTGISSKAMGIMNSFVNDIFERIAG 76 ||||||||||||||||||||||||||||||#|+|||||||||||||||||||||||||||| Sbjct 17 KAVTKAQKKDGKKRKRSRKESYSVYVYKVL#KRVHPDTGISSKAMGIMNSFVNDIFERIAG 76 |
| Homo sapiens | Query 17 KAVTKAQKKDGKKRKRSRKESYSVYVYKVL#KQVHPDTGISSKAMGIMNSFVNDIFERIAG 76 ||||| ||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Sbjct 17 KAVTKVQKKDGKKRKRSRKESYSVYVYKVL#KQVHPDTGISSKAMGIMNSFVNDIFERIAG 76 |
| Bos taurus | Query 17 KAVTKAQKKDGKKRKRSRKESYSVYVYKVL#KQVHPDTGISSKAMGIMNSFVNDIFERIAG 76 ||||||||||||||||||||||||||||||#||||||||||||||| |||||||||||||| Sbjct 17 KAVTKAQKKDGKKRKRSRKESYSVYVYKVL#KQVHPDTGISSKAMGNMNSFVNDIFERIAG 76 |
| Pongo pygmaeus | Query 17 KAVTKAQKKDGKKRKRSRKESYSVYVYKVL#KQVHPDTGISSKAMGIMNSFVNDIFERIAG 76 ||||||||||||||||| |||||+||||||#|||||||||||||||||||||||||||||| Sbjct 17 KAVTKAQKKDGKKRKRSCKESYSIYVYKVL#KQVHPDTGISSKAMGIMNSFVNDIFERIAG 76 |
| Xenopus laevis | Query 17 KAVTKAQKKDGKKRKRSRKESYSVYVYKVL#KQVHPDTGISSKAMGIMNSFVNDIFERIAG 76 ||||| ||||||||++||||||++||||||#|||||||||||||| ||||||||+|||||| Sbjct 17 KAVTKTQKKDGKKRRKSRKESYAIYVYKVL#KQVHPDTGISSKAMSIMNSFVNDVFERIAG 76 |
| Xenopus tropicalis | Query 17 KAVTKAQKKDGKKRKRSRKESYSVYVYKVL#KQVHPDTGISSKAMGIMNSFVNDIFERIAG 76 ||||| ||||||||+++|||||++||||||#|||||||||||||| ||||||||+|||||| Sbjct 17 KAVTKTQKKDGKKRRKTRKESYAIYVYKVL#KQVHPDTGISSKAMSIMNSFVNDVFERIAG 76 |
| Rhacophorus schlegelii | Query 17 KAVTKAQKKDGKKRKRSRKESYSVYVYKVL#KQVHPDTGISSKAMGIMNSFVNDIFERIAG 76 |||+| ||||||||++||||||++||||||#|||||||||||||| ||||||||||||||| Sbjct 17 KAVSKVQKKDGKKRRKSRKESYAIYVYKVL#KQVHPDTGISSKAMSIMNSFVNDIFERIAG 76 |
| Expression vector pET3-H2B | Query 17 KAVTKAQKKDGKKRKRSRKESYSVYVYKVL#KQVHPDTGISSKAMGIMNSFVNDIFERIAG 76 ||||| ||||||||+++|||||++||||||#|||||||||||||| ||||||||+|||||| Sbjct 14 KAVTKTQKKDGKKRRKTRKESYAIYVYKVL#KQVHPDTGISSKAMSIMNSFVNDVFERIAG 73 |
| Gallus gallus | Query 17 KAVTKAQKKDGKKRKRSRKESYSVYVYKVL#KQVHPDTGISSKAMGIMNSFVNDIFERIAG 76 ||||| ||| |||++|||||||+||||||#|||||||||||||||||||||||||||||| Sbjct 17 KAVTKTQKKGDKKRRKSRKESYSIYVYKVL#KQVHPDTGISSKAMGIMNSFVNDIFERIAG 76 |
| Danio rerio | Query 17 KAVTKAQKKDGKKRKRSRKESYSVYVYKVL#KQVHPDTGISSKAMGIMNSFVNDIFERIAG 76 ||||| | ||||||+|||||++||||||#|||||||||||||||||||||||||||||| Sbjct 15 KAVTKTAGKGGKKRKRTRKESYAIYVYKVL#KQVHPDTGISSKAMGIMNSFVNDIFERIAG 74 |
| Oncorhynchus mykiss | Query 17 KAVTKAQKKDGKKRKRSRKESYSVYVYKVL#KQVHPDTGISSKAMGIMNSFVNDIFERIAG 76 ||||| | ||||++||||||++||||||#|||||||||||||||||||||||||||||| Sbjct 15 KAVTKTAGKGGKKRRKSRKESYAIYVYKVL#KQVHPDTGISSKAMGIMNSFVNDIFERIAG 74 |
| Tetraodon nigroviridis | Query 17 KAVTKAQKKDGKKRKRSRKESYSVYVYKVL#KQVHPDTGISSKAMGIMNSFVNDIFERIA 75 ||+|||||| |||+++|||||++||||||#|||||||||||||| |||||||||||||| Sbjct 17 KAITKAQKKGDKKRRKNRKESYAIYVYKVL#KQVHPDTGISSKAMSIMNSFVNDIFERIA 75 |
| Caenorhabditis elegans | Query 20 TKAQKKDGKKRKRSRKESYSVYVYKVL#KQVHPDTGISSKAMGIMNSFVNDIFERIAG 76 | ++ ||||||| +||||||||+|+||#||||||||+||||| ||||||||+||||| Sbjct 17 TVSKPKDGKKRKHARKESYSVYIYRVL#KQVHPDTGVSSKAMSIMNSFVNDVFERIAS 73 |
| Caenorhabditis briggsae AF16 | Query 20 TKAQKKDGKKRKRSRKESYSVYVYKVL#KQVHPDTGISSKAMGIMNSFVNDIFERIAG 76 | + ||||||| ||||||||+|+||#||||||||+||||| ||||||||+||||| Sbjct 17 TVTKPKDGKKRKAHRKESYSVYIYRVL#KQVHPDTGVSSKAMSIMNSFVNDVFERIAA 73 |
| Psammechinus miliaris | Query 17 KAVTKAQKKDGKKRKRSRKESYSVYVYKVL#KQVHPDTGISSKAMGIMNSFVNDIFERIAG 76 ||| + |||| | ||||| +|+||||#||||||||+||+|| ||||||||||||||| Sbjct 14 KAVKPPRASGGKKRHRKRKESYGIYIYKVL#KQVHPDTGVSSRAMTIMNSFVNDIFERIAG 73 |
| Strongylocentrotus purpuratus | Query 17 KAVTKAQKKDGKKRKRSRKESYSVYVYKVL#KQVHPDTGISSKAMGIMNSFVNDIFERIAG 76 ||| + |||| | ||||| +|+||||#|||||||||||+|| ||||||||||||||| Sbjct 14 KAVKAPRPSGGKKRNRKRKESYGIYIYKVL#KQVHPDTGISSRAMVIMNSFVNDIFERIAG 73 |
| Psammechinus miliaris | Query 17 KAVTKAQKKDGKKRKRSRKESYSVYVYKVL#KQVHPDTGISSKAMGIMNSFVNDIFERIAG 76 ||| + |||| | ||||| +|+||||#|||||||||||+|| ||||||||||||||| Sbjct 14 KAVKAPRPSGGKKRNRKRKESYGIYIYKVL#KQVHPDTGISSRAMIIMNSFVNDIFERIAG 73 |
| Cairina moschata | Query 17 KAVTKAQKKDGKKRKRSRKESYSVYVYKVL#KQVHPDTGISSKAMGIMNSFVNDIFERIAG 76 ||||| ||| ||||+|||||||+||||||#|||||||||||||||||||||||||||||| Sbjct 17 KAVTKTQKKGDKKRKKSRKESYSIYVYKVL#KQVHPDTGISSKAMGIMNSFVNDIFERIAG 76 |
| Pan troglodytes | Query 17 KAVTKAQKKDGKKRKRSRKESYSVYVYKVL#KQVHPDTGISSKAMGIMNSFVNDIFERIAG 76 ||+||||||||||||| ||||||+||||||#||||||||||||||||||||||||||||| Sbjct 17 KAITKAQKKDGKKRKRGRKESYSIYVYKVL#KQVHPDTGISSKAMGIMNSFVNDIFERIAS 76 |
| Holothuria tubulosa | Query 28 KKRKRSRKESYSVYVYKVL#KQVHPDTGISSKAMGIMNSFVNDIFERIAG 76 +||||+|+||||+|+|||+#|||||||||||+|| |||||||||||||| Sbjct 25 RKRKRTRRESYSIYIYKVM#KQVHPDTGISSRAMSIMNSFVNDIFERIAA 73 |
| Drosophila yakuba | Query 25 KDGKKRKRSRKESYSVYVYKVL#KQVHPDTGISSKAMGIMNSFVNDIFERIAG 76 | || || |||||++|+||||#|||||||||||||| |||||||||||||| Sbjct 22 KTDKKMKRKRKESYAIYIYKVL#KQVHPDTGISSKAMSIMNSFVNDIFERIAA 73 |
| Acropora formosa | Query 27 GKKRKRSRKESYSVYVYKVL#KQVHPDTGISSKAMGIMNSFVNDIFERIA 75 |+|+ |||||++|+||||#||||||||||||||||||||||||||||| Sbjct 26 AKRRRGKRKESYAIYIYKVL#KQVHPDTGISSKAMGIMNSFVNDIFERIA 74 |
| Candida glabrata | Query 22 AQKKDGKKRKRSRKESYSVYVYKVL#KQVHPDTGISSKAMGIMNSFVNDIFERIAG 76 | ||||| ++|||+|| |+||||#|| ||||||| |+| |+|||||||||||| Sbjct 23 APSADGKKRTKARKETYSSYIYKVL#KQTHPDTGISQKSMSILNSFVNDIFERIAS 77 |
| Tigriopus californicus | Query 25 KDGKKRKRSRKESYSVYVYKVL#KQVHPDTGISSKAMGIMNSFVNDIFERIAG 76 | ||+ | |||||++|+||||#|||||||||||||| |||||||||||||| Sbjct 22 KGDKKKNRKRKESYAIYIYKVL#KQVHPDTGISSKAMSIMNSFVNDIFERIAS 73 |
| Strongylocentrotus purpuratus | Query 27 GKKRKRSRKESYSVYVYKVL#KQVHPDTGISSKAMGIMNSFVNDIFERIAG 76 |||| | ||||| +|+||||#|||||||||||+|| |||| |+|||||||| Sbjct 25 GKKRNRKRKESYGIYIYKVL#KQVHPDTGISSRAMVIMNSXVBDIFERIAG 74 |
[Site 6] KVLKQVHPDT53-GISSKAMGIM
Thr53  Gly
Gly
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Lys44 | Val45 | Leu46 | Lys47 | Gln48 | Val49 | His50 | Pro51 | Asp52 | Thr53 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Gly54 | Ile55 | Ser56 | Ser57 | Lys58 | Ala59 | Met60 | Gly61 | Ile62 | Met63 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| KKDGKKRKRSRKESYSVYVYKVLKQVHPDTGISSKAMGIMNSFVNDIFERIAGEASRLAH |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | Canis familiaris | 127.00 | 8 | PREDICTED: similar to Histone H2B 291B, partial |
| 2 | Homo sapiens | 126.00 | 23 | histone cluster 1, H2bk |
| 3 | Rattus norvegicus | 126.00 | 3 | PREDICTED: similar to Histone H2B 291B |
| 4 | N/A | 125.00 | 17 | 0506206A histone H2B |
| 5 | Mus musculus | 125.00 | 14 | unnamed protein product |
| 6 | Bos taurus | 125.00 | 12 | histone H2B-like |
| 7 | Rattus norvegicus | 125.00 | 3 | histone 1, H2bn (predicted) |
| 8 | Homo sapiens | 125.00 | 1 | histone cluster 2, H2bf |
| 9 | Macaca fascicularis | 124.00 | 1 | H2B1K_MACFA Histone H2B type 1-K (H2B K) gi |
| 10 | Pongo pygmaeus | 122.00 | 1 | H2B2E_PONPY Histone H2B type 2-E gi |
| 11 | Bos taurus | 122.00 | 1 | histone cluster 1, H2bn |
| 12 | Xenopus laevis | 120.00 | 4 | H2B12_XENLA Histone H2B 1.2 (H2B1.2) gi |
| 13 | Danio rerio | 119.00 | 6 | PREDICTED: hypothetical protein |
| 14 | Xenopus tropicalis | 119.00 | 3 | hypothetical protein LOC550005 |
| 15 | Rhacophorus schlegelii | 119.00 | 1 | H2B_RHASC Histone H2B gi |
| 16 | Expression vector pET3-H2B | 118.00 | 1 | H Chain H, 2.9 Angstrom X-Ray Structure Of Hybrid |
| 17 | Gallus gallus | 116.00 | 9 | hypothetical protein LOC417957 |
| 18 | Oncorhynchus mykiss | 115.00 | 1 | unnamed protein product |
| 19 | Caenorhabditis elegans | 113.00 | 6 | HIStone family member (his-41) |
| 20 | Caenorhabditis briggsae AF16 | 112.00 | 4 | Hypothetical protein CBG10818 |
| 21 | Tetraodon nigroviridis | 111.00 | 3 | unnamed protein product |
| 22 | Holothuria tubulosa | 107.00 | 1 | H2B_HOLTU Histone H2B gi |
| 23 | Drosophila yakuba | 107.00 | 1 | histone 2B |
| 24 | Tigriopus californicus | 105.00 | 1 | H2B1_TIGCA Histone H2B.1/H2B.2 gi |
| 25 | Psammechinus miliaris | 104.00 | 3 | H2BE2_PSAMI Histone H2B.2, embryonic gi |
| 26 | Strongylocentrotus purpuratus | 104.00 | 2 | PREDICTED: similar to histone H2B |
| 27 | Cairina moschata | 103.00 | 1 | H2B_CAIMO Histone H2B gi |
| 28 | Psammechinus miliaris | 103.00 | 1 | H2BE1_PSAMI Histone H2B.1, embryonic gi |
| 29 | Candida glabrata | 101.00 | 2 | unnamed protein product |
| 30 | Pan troglodytes | 101.00 | 1 | PREDICTED: hypothetical protein |
| 31 | Strongylocentrotus purpuratus | 101.00 | 1 | histone H2B |
Top-ranked sequences
| organism | matching |
|---|---|
| Canis familiaris | Query 24 KKDGKKRKRSRKESYSVYVYKVLKQVHPDT#GISSKAMGIMNSFVNDIFERIAGEASRLAH 83 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 24 KKDGKKRKRSRKESYSVYVYKVLKQVHPDT#GISSKAMGIMNSFVNDIFERIAGEASRLAH 83 |
| Homo sapiens | Query 24 KKDGKKRKRSRKESYSVYVYKVLKQVHPDT#GISSKAMGIMNSFVNDIFERIAGEASRLAH 83 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 24 KKDGKKRKRSRKESYSVYVYKVLKQVHPDT#GISSKAMGIMNSFVNDIFERIAGEASRLAH 83 |
| Rattus norvegicus | Query 24 KKDGKKRKRSRKESYSVYVYKVLKQVHPDT#GISSKAMGIMNSFVNDIFERIAGEASRLAH 83 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 24 KKDGKKRKRSRKESYSVYVYKVLKQVHPDT#GISSKAMGIMNSFVNDIFERIAGEASRLAH 83 |
| N/A | Query 24 KKDGKKRKRSRKESYSVYVYKVLKQVHPDT#GISSKAMGIMNSFVNDIFERIAGEASRLAH 83 ||||||||||||||||||||||||||||||#||||||||||||||||||+||||||||||| Sbjct 23 KKDGKKRKRSRKESYSVYVYKVLKQVHPDT#GISSKAMGIMNSFVNDIFZRIAGEASRLAH 82 |
| Mus musculus | Query 24 KKDGKKRKRSRKESYSVYVYKVLKQVHPDT#GISSKAMGIMNSFVNDIFERIAGEASRLAH 83 ||||||||||||||||||||||||||||||#||||+||||||||||||||||||||||||| Sbjct 24 KKDGKKRKRSRKESYSVYVYKVLKQVHPDT#GISSRAMGIMNSFVNDIFERIAGEASRLAH 83 |
| Bos taurus | Query 24 KKDGKKRKRSRKESYSVYVYKVLKQVHPDT#GISSKAMGIMNSFVNDIFERIAGEASRLAH 83 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 24 KKDGKKRKRSRKESYSVYVYKVLKQVHPDT#GISSKAMGIMNSFVNDIFERIAGEASRLAH 83 |
| Rattus norvegicus | Query 24 KKDGKKRKRSRKESYSVYVYKVLKQVHPDT#GISSKAMGIMNSFVNDIFERIAGEASRLAH 83 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 24 KKDGKKRKRSRKESYSVYVYKVLKQVHPDT#GISSKAMGIMNSFVNDIFERIAGEASRLAH 83 |
| Homo sapiens | Query 24 KKDGKKRKRSRKESYSVYVYKVLKQVHPDT#GISSKAMGIMNSFVNDIFERIAGEASRLAH 83 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 24 KKDGKKRKRSRKESYSVYVYKVLKQVHPDT#GISSKAMGIMNSFVNDIFERIAGEASRLAH 83 |
| Macaca fascicularis | Query 24 KKDGKKRKRSRKESYSVYVYKVLKQVHPDT#GISSKAMGIMNSFVNDIFERIAGEASRLAH 83 ||||||||||||||||||||||||+|||||#|||||||||||||||||||||||||||||| Sbjct 24 KKDGKKRKRSRKESYSVYVYKVLKRVHPDT#GISSKAMGIMNSFVNDIFERIAGEASRLAH 83 |
| Pongo pygmaeus | Query 24 KKDGKKRKRSRKESYSVYVYKVLKQVHPDT#GISSKAMGIMNSFVNDIFERIAGEASRLAH 83 |||||||||| |||||+|||||||||||||#|||||||||||||||||||||||||||||| Sbjct 24 KKDGKKRKRSCKESYSIYVYKVLKQVHPDT#GISSKAMGIMNSFVNDIFERIAGEASRLAH 83 |
| Bos taurus | Query 24 KKDGKKRKRSRKESYSVYVYKVLKQVHPDT#GISSKAMGIMNSFVNDIFERIAGEASRLAH 83 ||||||||||||||||||||||||||||||#|||||||| ||||||||||||||||||||| Sbjct 24 KKDGKKRKRSRKESYSVYVYKVLKQVHPDT#GISSKAMGNMNSFVNDIFERIAGEASRLAH 83 |
| Xenopus laevis | Query 24 KKDGKKRKRSRKESYSVYVYKVLKQVHPDT#GISSKAMGIMNSFVNDIFERIAGEASRLAH 83 |||||||++||||||++|||||+|||||||#|||||||||||||||||||||||||||||| Sbjct 24 KKDGKKRRKSRKESYAIYVYKVMKQVHPDT#GISSKAMGIMNSFVNDIFERIAGEASRLAH 83 |
| Danio rerio | Query 25 KDGKKRKRSRKESYSVYVYKVLKQVHPDT#GISSKAMGIMNSFVNDIFERIAGEASRLAH 83 | ||||||+|||||++|||||||||||||#|||||||||||||||||||||||||||||| Sbjct 23 KGGKKRKRTRKESYAIYVYKVLKQVHPDT#GISSKAMGIMNSFVNDIFERIAGEASRLAH 81 |
| Xenopus tropicalis | Query 24 KKDGKKRKRSRKESYSVYVYKVLKQVHPDT#GISSKAMGIMNSFVNDIFERIAGEASRLAH 83 |||||||++||||||++|||||||||||||#||||||| ||||||||+||||||||||||| Sbjct 24 KKDGKKRRKSRKESYAIYVYKVLKQVHPDT#GISSKAMSIMNSFVNDVFERIAGEASRLAH 83 |
| Rhacophorus schlegelii | Query 24 KKDGKKRKRSRKESYSVYVYKVLKQVHPDT#GISSKAMGIMNSFVNDIFERIAGEASRLAH 83 |||||||++||||||++|||||||||||||#||||||| |||||||||||||||||||||| Sbjct 24 KKDGKKRRKSRKESYAIYVYKVLKQVHPDT#GISSKAMSIMNSFVNDIFERIAGEASRLAH 83 |
| Expression vector pET3-H2B | Query 24 KKDGKKRKRSRKESYSVYVYKVLKQVHPDT#GISSKAMGIMNSFVNDIFERIAGEASRLAH 83 |||||||+++|||||++|||||||||||||#||||||| ||||||||+||||||||||||| Sbjct 21 KKDGKKRRKTRKESYAIYVYKVLKQVHPDT#GISSKAMSIMNSFVNDVFERIAGEASRLAH 80 |
| Gallus gallus | Query 24 KKDGKKRKRSRKESYSVYVYKVLKQVHPDT#GISSKAMGIMNSFVNDIFERIAGEASRLAH 83 || |||++|||||||+|||||||||||||#|||||||||||||||||||||||||||||| Sbjct 24 KKGDKKRRKSRKESYSIYVYKVLKQVHPDT#GISSKAMGIMNSFVNDIFERIAGEASRLAH 83 |
| Oncorhynchus mykiss | Query 25 KDGKKRKRSRKESYSVYVYKVLKQVHPDT#GISSKAMGIMNSFVNDIFERIAGEASRLAH 83 | ||||++||||||++|||||||||||||#||||||||||||||||||||||||+||||| Sbjct 23 KGGKKRRKSRKESYAIYVYKVLKQVHPDT#GISSKAMGIMNSFVNDIFERIAGESSRLAH 81 |
| Caenorhabditis elegans | Query 25 KDGKKRKRSRKESYSVYVYKVLKQVHPDT#GISSKAMGIMNSFVNDIFERIAGEASRLAH 83 ||||||| +||||||||+|+|||||||||#|+||||| ||||||||+||||| ||||||| Sbjct 22 KDGKKRKHARKESYSVYIYRVLKQVHPDT#GVSSKAMSIMNSFVNDVFERIASEASRLAH 80 |
| Caenorhabditis briggsae AF16 | Query 25 KDGKKRKRSRKESYSVYVYKVLKQVHPDT#GISSKAMGIMNSFVNDIFERIAGEASRLAH 83 ||||||| ||||||||+|+|||||||||#|+||||| ||||||||+||||| ||||||| Sbjct 22 KDGKKRKAHRKESYSVYIYRVLKQVHPDT#GVSSKAMSIMNSFVNDVFERIAAEASRLAH 80 |
| Tetraodon nigroviridis | Query 25 KDGKKRKRSRKESYSVYVYKVLKQVHPDT#GISSKAMGIMNSFVNDIFERIAGEASRLAH 83 | |||++|+|||||++|||||||||||||#||||||| |||||||||||||| ||||||| Sbjct 23 KGGKKKRRTRKESYAIYVYKVLKQVHPDT#GISSKAMSIMNSFVNDIFERIASEASRLAH 81 |
| Holothuria tubulosa | Query 28 KKRKRSRKESYSVYVYKVLKQVHPDT#GISSKAMGIMNSFVNDIFERIAGEASRLAH 83 +||||+|+||||+|+|||+|||||||#||||+|| |||||||||||||| ||||||| Sbjct 25 RKRKRTRRESYSIYIYKVMKQVHPDT#GISSRAMSIMNSFVNDIFERIAAEASRLAH 80 |
| Drosophila yakuba | Query 25 KDGKKRKRSRKESYSVYVYKVLKQVHPDT#GISSKAMGIMNSFVNDIFERIAGEASRLAH 83 | || || |||||++|+|||||||||||#||||||| |||||||||||||| ||||||| Sbjct 22 KTDKKMKRKRKESYAIYIYKVLKQVHPDT#GISSKAMSIMNSFVNDIFERIAAEASRLAH 80 |
| Tigriopus californicus | Query 25 KDGKKRKRSRKESYSVYVYKVLKQVHPDT#GISSKAMGIMNSFVNDIFERIAGEASRLAH 83 | ||+ | |||||++|+|||||||||||#||||||| |||||||||||||| ||||||| Sbjct 22 KGDKKKNRKRKESYAIYIYKVLKQVHPDT#GISSKAMSIMNSFVNDIFERIASEASRLAH 80 |
| Psammechinus miliaris | Query 27 GKKRKRSRKESYSVYVYKVLKQVHPDT#GISSKAMGIMNSFVNDIFERIAGEASRLAH 83 |||| | ||||| +|+|||||||||||#||||+|| ||||||||||||||||+|||| Sbjct 24 GKKRNRKRKESYGIYIYKVLKQVHPDT#GISSRAMIIMNSFVNDIFERIAGESSRLAQ 80 |
| Strongylocentrotus purpuratus | Query 27 GKKRKRSRKESYSVYVYKVLKQVHPDT#GISSKAMGIMNSFVNDIFERIAGEASRLAH 83 |||| | ||||| +|+|||||||||||#||||+|| ||||||||||||||||+|||| Sbjct 24 GKKRNRKRKESYGIYIYKVLKQVHPDT#GISSRAMVIMNSFVNDIFERIAGESSRLAQ 80 |
| Cairina moschata | Query 36 ESYSVYVYKVLKQVHPDT#GISSKAMGIMNSFVNDIFERIAGEASRLAH 83 ||||+|||||||||||||#|||||||||||||||||||||||||||||| Sbjct 36 ESYSIYVYKVLKQVHPDT#GISSKAMGIMNSFVNDIFERIAGEASRLAH 83 |
| Psammechinus miliaris | Query 27 GKKRKRSRKESYSVYVYKVLKQVHPDT#GISSKAMGIMNSFVNDIFERIAGEASRLAH 83 |||| | ||||| +|+|||||||||||#|+||+|| |||||||||||||||||||| Sbjct 24 GKKRHRKRKESYGIYIYKVLKQVHPDT#GVSSRAMTIMNSFVNDIFERIAGEASRLTQ 80 |
| Candida glabrata | Query 26 DGKKRKRSRKESYSVYVYKVLKQVHPDT#GISSKAMGIMNSFVNDIFERIAGEASRLA 82 ||||| ++|||+|| |+|||||| ||||#||| |+| |+|||||||||||| |||+|| Sbjct 29 DGKKRTKARKETYSSYIYKVLKQTHPDT#GISQKSMSILNSFVNDIFERIASEASKLA 85 |
| Pan troglodytes | Query 36 ESYSVYVYKVLKQVHPDT#GISSKAMGIMNSFVNDIFERIAGEASRLAH 83 ||||+|||||||||||||#|||||||||||||||||||||| ||||||| Sbjct 36 ESYSIYVYKVLKQVHPDT#GISSKAMGIMNSFVNDIFERIASEASRLAH 83 |
| Strongylocentrotus purpuratus | Query 27 GKKRKRSRKESYSVYVYKVLKQVHPDT#GISSKAMGIMNSFVNDIFERIAGEASRLAH 83 |||| | ||||| +|+|||||||||||#||||+|| |||| |+|||||||||+|||| Sbjct 25 GKKRNRKRKESYGIYIYKVLKQVHPDT#GISSRAMVIMNSXVBDIFERIAGESSRLAQ 81 |
[Site 7] ISSKAMGIMN64-SFVNDIFERI
Asn64  Ser
Ser
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Ile55 | Ser56 | Ser57 | Lys58 | Ala59 | Met60 | Gly61 | Ile62 | Met63 | Asn64 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Ser65 | Phe66 | Val67 | Asn68 | Asp69 | Ile70 | Phe71 | Glu72 | Arg73 | Ile74 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| KESYSVYVYKVLKQVHPDTGISSKAMGIMNSFVNDIFERIAGEASRLAHYNKRSTITSRE |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | Canis familiaris | 128.00 | 9 | PREDICTED: similar to Histone H2B 291B, partial |
| 2 | Homo sapiens | 127.00 | 20 | histone cluster 1, H2bk |
| 3 | N/A | 127.00 | 15 | B Chain B, Histone Octamer (Chicken), Chromosomal |
| 4 | Mus musculus | 127.00 | 13 | histone cluster 1, H2bh |
| 5 | Bos taurus | 127.00 | 10 | PREDICTED: similar to histone H2b-616 |
| 6 | Gallus gallus | 127.00 | 9 | hypothetical protein LOC417956 |
| 7 | Rattus norvegicus | 127.00 | 3 | PREDICTED: similar to Histone H2B 291B |
| 8 | Homo sapiens | 127.00 | 1 | histone cluster 2, H2bf |
| 9 | Cairina moschata | 127.00 | 1 | H2B_CAIMO Histone H2B gi |
| 10 | Pongo pygmaeus | 127.00 | 1 | H2B2E_PONPY Histone H2B type 2-E gi |
| 11 | Rattus norvegicus | 126.00 | 3 | histone 1, H2bn (predicted) |
| 12 | Danio rerio | 125.00 | 6 | histone 2, H2, like |
| 13 | Xenopus laevis | 125.00 | 4 | H2B12_XENLA Histone H2B 1.2 (H2B1.2) gi |
| 14 | Macaca fascicularis | 125.00 | 1 | H2B1K_MACFA Histone H2B type 1-K (H2B K) gi |
| 15 | Oncorhynchus mykiss | 124.00 | 1 | unnamed protein product |
| 16 | Bos taurus | 124.00 | 1 | histone cluster 1, H2bn |
| 17 | Pan troglodytes | 124.00 | 1 | PREDICTED: hypothetical protein |
| 18 | Rhacophorus schlegelii | 123.00 | 1 | H2B_RHASC Histone H2B gi |
| 19 | Xenopus tropicalis | 122.00 | 3 | histone 2, H2bf |
| 20 | Expression vector pET3-H2B | 122.00 | 1 | H Chain H, 2.9 Angstrom X-Ray Structure Of Hybrid |
| 21 | Tetraodon nigroviridis | 120.00 | 6 | unnamed protein product |
| 22 | Drosophila yakuba | 120.00 | 2 | histone 2B |
| 23 | Tigriopus californicus | 119.00 | 1 | H2B1_TIGCA Histone H2B.1/H2B.2 gi |
| 24 | Drosophila hydei | 118.00 | 2 | H2B_DROHY Histone H2B gi |
| 25 | Anopheles gambiae str. PEST | 118.00 | 1 | AGAP012894-PA |
| 26 | Asellus aquaticus | 118.00 | 1 | putative H2B histone |
| 27 | Platynereis dumerilii | 118.00 | 1 | H2B_PLADU Histone H2B gi |
| 28 | Drosophila melanogaster | 118.00 | 1 | His2B:CG17949 CG17949-PA |
| 29 | Caenorhabditis elegans | 117.00 | 6 | HIStone family member (his-8) |
| 30 | Caenorhabditis briggsae AF16 | 117.00 | 3 | Hypothetical protein CBG10818 |
| 31 | Rhynchosciara americana | 117.00 | 1 | AF378198_4 histone H2B |
| 32 | Urechis caupo | 117.00 | 1 | H2B_URECA Histone H2B gi |
| 33 | Chaetopterus variopedatus | 117.00 | 1 | histone H2B |
| 34 | Chironomus thummi | 116.00 | 1 | H2B_CHITH Histone H2B gi |
| 35 | Mytilus edulis | 116.00 | 1 | histone H2B |
| 36 | Drosophila sechellia | 115.00 | 1 | histone 2B |
| 37 | Mytilus galloprovincialis | 115.00 | 1 | histone H2B |
Top-ranked sequences
| organism | matching |
|---|---|
| Canis familiaris | Query 35 KESYSVYVYKVLKQVHPDTGISSKAMGIMN#SFVNDIFERIAGEASRLAHYNKRSTITSRE 94 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 35 KESYSVYVYKVLKQVHPDTGISSKAMGIMN#SFVNDIFERIAGEASRLAHYNKRSTITSRE 94 |
| Homo sapiens | Query 35 KESYSVYVYKVLKQVHPDTGISSKAMGIMN#SFVNDIFERIAGEASRLAHYNKRSTITSRE 94 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 35 KESYSVYVYKVLKQVHPDTGISSKAMGIMN#SFVNDIFERIAGEASRLAHYNKRSTITSRE 94 |
| N/A | Query 35 KESYSVYVYKVLKQVHPDTGISSKAMGIMN#SFVNDIFERIAGEASRLAHYNKRSTITSRE 94 |||||+||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Sbjct 34 KESYSIYVYKVLKQVHPDTGISSKAMGIMN#SFVNDIFERIAGEASRLAHYNKRSTITSRE 93 |
| Mus musculus | Query 35 KESYSVYVYKVLKQVHPDTGISSKAMGIMN#SFVNDIFERIAGEASRLAHYNKRSTITSRE 94 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 35 KESYSVYVYKVLKQVHPDTGISSKAMGIMN#SFVNDIFERIAGEASRLAHYNKRSTITSRE 94 |
| Bos taurus | Query 35 KESYSVYVYKVLKQVHPDTGISSKAMGIMN#SFVNDIFERIAGEASRLAHYNKRSTITSRE 94 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 35 KESYSVYVYKVLKQVHPDTGISSKAMGIMN#SFVNDIFERIAGEASRLAHYNKRSTITSRE 94 |
| Gallus gallus | Query 35 KESYSVYVYKVLKQVHPDTGISSKAMGIMN#SFVNDIFERIAGEASRLAHYNKRSTITSRE 94 |||||+||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Sbjct 35 KESYSIYVYKVLKQVHPDTGISSKAMGIMN#SFVNDIFERIAGEASRLAHYNKRSTITSRE 94 |
| Rattus norvegicus | Query 35 KESYSVYVYKVLKQVHPDTGISSKAMGIMN#SFVNDIFERIAGEASRLAHYNKRSTITSRE 94 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 35 KESYSVYVYKVLKQVHPDTGISSKAMGIMN#SFVNDIFERIAGEASRLAHYNKRSTITSRE 94 |
| Homo sapiens | Query 35 KESYSVYVYKVLKQVHPDTGISSKAMGIMN#SFVNDIFERIAGEASRLAHYNKRSTITSRE 94 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 35 KESYSVYVYKVLKQVHPDTGISSKAMGIMN#SFVNDIFERIAGEASRLAHYNKRSTITSRE 94 |
| Cairina moschata | Query 35 KESYSVYVYKVLKQVHPDTGISSKAMGIMN#SFVNDIFERIAGEASRLAHYNKRSTITSRE 94 |||||+||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Sbjct 35 KESYSIYVYKVLKQVHPDTGISSKAMGIMN#SFVNDIFERIAGEASRLAHYNKRSTITSRE 94 |
| Pongo pygmaeus | Query 35 KESYSVYVYKVLKQVHPDTGISSKAMGIMN#SFVNDIFERIAGEASRLAHYNKRSTITSRE 94 |||||+||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Sbjct 35 KESYSIYVYKVLKQVHPDTGISSKAMGIMN#SFVNDIFERIAGEASRLAHYNKRSTITSRE 94 |
| Rattus norvegicus | Query 35 KESYSVYVYKVLKQVHPDTGISSKAMGIMN#SFVNDIFERIAGEASRLAHYNKRSTITSRE 94 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 35 KESYSVYVYKVLKQVHPDTGISSKAMGIMN#SFVNDIFERIAGEASRLAHYNKRSTITSRE 94 |
| Danio rerio | Query 35 KESYSVYVYKVLKQVHPDTGISSKAMGIMN#SFVNDIFERIAGEASRLAHYNKRSTITSRE 94 ||||++||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Sbjct 35 KESYAIYVYKVLKQVHPDTGISSKAMGIMN#SFVNDIFERIAGEASRLAHYNKRSTITSRE 94 |
| Xenopus laevis | Query 35 KESYSVYVYKVLKQVHPDTGISSKAMGIMN#SFVNDIFERIAGEASRLAHYNKRSTITSRE 94 ||||++|||||+||||||||||||||||||#|||||||||||||||||||||||||||||| Sbjct 35 KESYAIYVYKVMKQVHPDTGISSKAMGIMN#SFVNDIFERIAGEASRLAHYNKRSTITSRE 94 |
| Macaca fascicularis | Query 35 KESYSVYVYKVLKQVHPDTGISSKAMGIMN#SFVNDIFERIAGEASRLAHYNKRSTITSRE 94 |||||||||||||+||||||||||||||||#|||||||||||||||||||||||||||||| Sbjct 35 KESYSVYVYKVLKRVHPDTGISSKAMGIMN#SFVNDIFERIAGEASRLAHYNKRSTITSRE 94 |
| Oncorhynchus mykiss | Query 35 KESYSVYVYKVLKQVHPDTGISSKAMGIMN#SFVNDIFERIAGEASRLAHYNKRSTITSRE 94 ||||++||||||||||||||||||||||||#|||||||||||||+|||||||||||||||| Sbjct 33 KESYAIYVYKVLKQVHPDTGISSKAMGIMN#SFVNDIFERIAGESSRLAHYNKRSTITSRE 92 |
| Bos taurus | Query 35 KESYSVYVYKVLKQVHPDTGISSKAMGIMN#SFVNDIFERIAGEASRLAHYNKRSTITSRE 94 ||||||||||||||||||||||||||| ||#|||||||||||||||||||||||||||||| Sbjct 35 KESYSVYVYKVLKQVHPDTGISSKAMGNMN#SFVNDIFERIAGEASRLAHYNKRSTITSRE 94 |
| Pan troglodytes | Query 35 KESYSVYVYKVLKQVHPDTGISSKAMGIMN#SFVNDIFERIAGEASRLAHYNKRSTITSRE 94 |||||+||||||||||||||||||||||||#||||||||||| |||||||||||||||||| Sbjct 35 KESYSIYVYKVLKQVHPDTGISSKAMGIMN#SFVNDIFERIASEASRLAHYNKRSTITSRE 94 |
| Rhacophorus schlegelii | Query 35 KESYSVYVYKVLKQVHPDTGISSKAMGIMN#SFVNDIFERIAGEASRLAHYNKRSTITSRE 94 ||||++|||||||||||||||||||| |||#|||||||||||||||||||||||||||||| Sbjct 35 KESYAIYVYKVLKQVHPDTGISSKAMSIMN#SFVNDIFERIAGEASRLAHYNKRSTITSRE 94 |
| Xenopus tropicalis | Query 35 KESYSVYVYKVLKQVHPDTGISSKAMGIMN#SFVNDIFERIAGEASRLAHYNKRSTITSRE 94 ||||++|||||||||||||||||||| |||#|||||+|||||||||||||||||||||||| Sbjct 35 KESYAIYVYKVLKQVHPDTGISSKAMSIMN#SFVNDVFERIAGEASRLAHYNKRSTITSRE 94 |
| Expression vector pET3-H2B | Query 35 KESYSVYVYKVLKQVHPDTGISSKAMGIMN#SFVNDIFERIAGEASRLAHYNKRSTITSRE 94 ||||++|||||||||||||||||||| |||#|||||+|||||||||||||||||||||||| Sbjct 32 KESYAIYVYKVLKQVHPDTGISSKAMSIMN#SFVNDVFERIAGEASRLAHYNKRSTITSRE 91 |
| Tetraodon nigroviridis | Query 35 KESYSVYVYKVLKQVHPDTGISSKAMGIMN#SFVNDIFERIAGEASRLAHYNKRSTITSRE 94 ||||++|||||||||||||||||||| |||#||||||||||| |||||||||||||||||| Sbjct 33 KESYAIYVYKVLKQVHPDTGISSKAMSIMN#SFVNDIFERIASEASRLAHYNKRSTITSRE 92 |
| Drosophila yakuba | Query 35 KESYSVYVYKVLKQVHPDTGISSKAMGIMN#SFVNDIFERIAGEASRLAHYNKRSTITSRE 94 ||||++|+|||||||||||||||||| |||#||||||||||| |||||||||||||||||| Sbjct 32 KESYAIYIYKVLKQVHPDTGISSKAMSIMN#SFVNDIFERIAAEASRLAHYNKRSTITSRE 91 |
| Tigriopus californicus | Query 35 KESYSVYVYKVLKQVHPDTGISSKAMGIMN#SFVNDIFERIAGEASRLAHYNKRSTITSRE 94 ||||++|+|||||||||||||||||| |||#||||||||||| |||||||||||||||||| Sbjct 32 KESYAIYIYKVLKQVHPDTGISSKAMSIMN#SFVNDIFERIASEASRLAHYNKRSTITSRE 91 |
| Drosophila hydei | Query 36 ESYSVYVYKVLKQVHPDTGISSKAMGIMN#SFVNDIFERIAGEASRLAHYNKRSTITSRE 94 |||++|+|||||||||||||||||| |||#||||||||||| |||||||||||||||||| Sbjct 33 ESYAIYIYKVLKQVHPDTGISSKAMSIMN#SFVNDIFERIAAEASRLAHYNKRSTITSRE 91 |
| Anopheles gambiae str. PEST | Query 36 ESYSVYVYKVLKQVHPDTGISSKAMGIMN#SFVNDIFERIAGEASRLAHYNKRSTITSRE 94 |||++|+|||||||||||||||||| |||#||||||||||| |||||||||||||||||| Sbjct 34 ESYAIYIYKVLKQVHPDTGISSKAMSIMN#SFVNDIFERIAAEASRLAHYNKRSTITSRE 92 |
| Asellus aquaticus | Query 36 ESYSVYVYKVLKQVHPDTGISSKAMGIMN#SFVNDIFERIAGEASRLAHYNKRSTITSRE 94 ||||+|+|||||||||||||||||| |||#||||||||||| |||||||||||||||||| Sbjct 33 ESYSIYIYKVLKQVHPDTGISSKAMSIMN#SFVNDIFERIAAEASRLAHYNKRSTITSRE 91 |
| Platynereis dumerilii | Query 36 ESYSVYVYKVLKQVHPDTGISSKAMGIMN#SFVNDIFERIAGEASRLAHYNKRSTITSRE 94 ||||+|+|||||||||||||||||| |||#||||||||||| |||||||||||||||||| Sbjct 33 ESYSIYIYKVLKQVHPDTGISSKAMSIMN#SFVNDIFERIAAEASRLAHYNKRSTITSRE 91 |
| Drosophila melanogaster | Query 36 ESYSVYVYKVLKQVHPDTGISSKAMGIMN#SFVNDIFERIAGEASRLAHYNKRSTITSRE 94 |||++|+|||||||||||||||||| |||#||||||||||| |||||||||||||||||| Sbjct 33 ESYAIYIYKVLKQVHPDTGISSKAMSIMN#SFVNDIFERIAAEASRLAHYNKRSTITSRE 91 |
| Caenorhabditis elegans | Query 35 KESYSVYVYKVLKQVHPDTGISSKAMGIMN#SFVNDIFERIAGEASRLAHYNKRSTITSRE 94 |||||||+|+||||||||||+||||| |||#|||||+||||| ||||||||||||||+||| Sbjct 32 KESYSVYIYRVLKQVHPDTGVSSKAMSIMN#SFVNDVFERIAAEASRLAHYNKRSTISSRE 91 |
| Caenorhabditis briggsae AF16 | Query 35 KESYSVYVYKVLKQVHPDTGISSKAMGIMN#SFVNDIFERIAGEASRLAHYNKRSTITSRE 94 |||||||+|+||||||||||+||||| |||#|||||+||||| ||||||||||||||+||| Sbjct 32 KESYSVYIYRVLKQVHPDTGVSSKAMSIMN#SFVNDVFERIAAEASRLAHYNKRSTISSRE 91 |
| Rhynchosciara americana | Query 36 ESYSVYVYKVLKQVHPDTGISSKAMGIMN#SFVNDIFERIAGEASRLAHYNKRSTITSRE 94 |||++|+|||||||||||||||||| |||#||||||||||| |||||||||||||||||| Sbjct 33 ESYAIYIYKVLKQVHPDTGISSKAMSIMN#SFVNDIFERIAAEASRLAHYNKRSTITSRE 91 |
| Urechis caupo | Query 36 ESYSVYVYKVLKQVHPDTGISSKAMGIMN#SFVNDIFERIAGEASRLAHYNKRSTITSRE 94 ||||+|+|||||||||||||||||| |||#||||||||||| |||||||||+|||||||| Sbjct 33 ESYSIYIYKVLKQVHPDTGISSKAMSIMN#SFVNDIFERIAAEASRLAHYNRRSTITSRE 91 |
| Chaetopterus variopedatus | Query 36 ESYSVYVYKVLKQVHPDTGISSKAMGIMN#SFVNDIFERIAGEASRLAHYNKRSTITSRE 94 ||||+|||||+|||||||||||||| |||#|||||+||||| |||||||||||||||||| Sbjct 33 ESYSIYVYKVMKQVHPDTGISSKAMSIMN#SFVNDLFERIASEASRLAHYNKRSTITSRE 91 |
| Chironomus thummi | Query 36 ESYSVYVYKVLKQVHPDTGISSKAMGIMN#SFVNDIFERIAGEASRLAHYNKRSTITSRE 94 |||++|++|||||||||||+||||| |||#||||||||||| |||||||||||||||||| Sbjct 35 ESYAIYIFKVLKQVHPDTGVSSKAMSIMN#SFVNDIFERIAAEASRLAHYNKRSTITSRE 93 |
| Mytilus edulis | Query 36 ESYSVYVYKVLKQVHPDTGISSKAMGIMN#SFVNDIFERIAGEASRLAHYNKRSTITSRE 94 |||++|+||||+|||||||+||||| |||#||||||||||| |||||||||||||||||| Sbjct 34 ESYAIYIYKVLRQVHPDTGVSSKAMSIMN#SFVNDIFERIAAEASRLAHYNKRSTITSRE 92 |
| Drosophila sechellia | Query 36 ESYSVYVYKVLKQVHPDTGISSKAMGIMN#SFVNDIFERIAGEASRLAHYNKRSTITSRE 94 |||++|+|||||||||||||| ||| |||#||||||||||| |||||||||||||||||| Sbjct 33 ESYAIYIYKVLKQVHPDTGISLKAMSIMN#SFVNDIFERIAAEASRLAHYNKRSTITSRE 91 |
| Mytilus galloprovincialis | Query 36 ESYSVYVYKVLKQVHPDTGISSKAMGIMN#SFVNDIFERIAGEASRLAHYNKRSTITSRE 94 |||++|+||||+|||||||+||||| |||#||||||||||| |||||||||||||||||| Sbjct 34 ESYAIYIYKVLRQVHPDTGVSSKAMSIMN#SFVNDIFERIAAEASRLAHYNKRSTITSRE 92 |
[Site 8] ERIAGEASRL81-AHYNKRSTIT
Leu81  Ala
Ala
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Glu72 | Arg73 | Ile74 | Ala75 | Gly76 | Glu77 | Ala78 | Ser79 | Arg80 | Leu81 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Ala82 | His83 | Tyr84 | Asn85 | Lys86 | Arg87 | Ser88 | Thr89 | Ile90 | Thr91 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| DTGISSKAMGIMNSFVNDIFERIAGEASRLAHYNKRSTITSREIQTAVRLLLPGELAKHA |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | Homo sapiens | 127.00 | 19 | histone cluster 2, H2be |
| 2 | N/A | 127.00 | 16 | 0506206A histone H2B |
| 3 | Mus musculus | 127.00 | 13 | histone cluster 1, H2bh |
| 4 | Canis familiaris | 127.00 | 9 | PREDICTED: similar to H2B histone family, member |
| 5 | Gallus gallus | 127.00 | 9 | histone H2B |
| 6 | Bos taurus | 127.00 | 8 | PREDICTED: similar to histone H2b-616 |
| 7 | Danio rerio | 127.00 | 6 | histone 2, H2, like |
| 8 | Xenopus laevis | 127.00 | 4 | H2B12_XENLA Histone H2B 1.2 (H2B1.2) gi |
| 9 | Rattus norvegicus | 127.00 | 3 | PREDICTED: similar to Histone H2B 291B |
| 10 | Pongo pygmaeus | 127.00 | 1 | H2B2E_PONPY Histone H2B type 2-E gi |
| 11 | Macaca fascicularis | 127.00 | 1 | H2B1K_MACFA Histone H2B type 1-K (H2B K) gi |
| 12 | Homo sapiens | 127.00 | 1 | histone cluster 2, H2bf |
| 13 | Rattus norvegicus | 126.00 | 3 | histone 1, H2bn (predicted) |
| 14 | Oncorhynchus mykiss | 125.00 | 1 | unnamed protein product |
| 15 | Xenopus tropicalis | 124.00 | 3 | histone 2, H2bf |
| 16 | Cairina moschata | 124.00 | 1 | H2B_CAIMO Histone H2B gi |
| 17 | Bos taurus | 124.00 | 1 | histone cluster 1, H2bn |
| 18 | Expression vector pET3-H2B | 124.00 | 1 | H Chain H, 2.9 Angstrom X-Ray Structure Of Hybrid |
| 19 | Rhacophorus schlegelii | 124.00 | 1 | H2B_RHASC Histone H2B gi |
| 20 | Pan troglodytes | 123.00 | 1 | PREDICTED: hypothetical protein |
| 21 | Tetraodon nigroviridis | 122.00 | 7 | unnamed protein product |
| 22 | Drosophila sechellia | 122.00 | 2 | histone 2B |
| 23 | Drosophila hydei | 122.00 | 2 | histone H2b |
| 24 | Drosophila yakuba | 122.00 | 2 | H2B_DROYA Histone H2B gi |
| 25 | Rhynchosciara americana | 122.00 | 1 | AF378198_4 histone H2B |
| 26 | Platynereis dumerilii | 122.00 | 1 | H2B_PLADU Histone H2B gi |
| 27 | Anopheles gambiae str. PEST | 122.00 | 1 | AGAP012894-PA |
| 28 | Drosophila melanogaster | 122.00 | 1 | His2B:CG17949 CG17949-PA |
| 29 | Tigriopus californicus | 121.00 | 1 | H2B1_TIGCA Histone H2B.1/H2B.2 gi |
| 30 | Asellus aquaticus | 121.00 | 1 | putative H2B histone |
| 31 | Mytilus galloprovincialis | 120.00 | 1 | histone H2B |
| 32 | Chironomus thummi | 120.00 | 1 | H2B_CHITH Histone H2B gi |
| 33 | Chaetopterus variopedatus | 120.00 | 1 | histone H2B |
| 34 | Mytilus chilensis | 120.00 | 1 | histone H2B |
| 35 | Urechis caupo | 120.00 | 1 | H2B_URECA Histone H2B gi |
| 36 | Mytilus edulis | 120.00 | 1 | histone H2B |
| 37 | Caenorhabditis elegans | 118.00 | 6 | HIStone family member (his-8) |
| 38 | Caenorhabditis briggsae AF16 | 118.00 | 3 | Hypothetical protein CBG10818 |
Top-ranked sequences
| organism | matching |
|---|---|
| Homo sapiens | Query 52 DTGISSKAMGIMNSFVNDIFERIAGEASRL#AHYNKRSTITSREIQTAVRLLLPGELAKHA 111 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 52 DTGISSKAMGIMNSFVNDIFERIAGEASRL#AHYNKRSTITSREIQTAVRLLLPGELAKHA 111 |
| N/A | Query 52 DTGISSKAMGIMNSFVNDIFERIAGEASRL#AHYNKRSTITSREIQTAVRLLLPGELAKHA 111 ||||||||||||||||||||+|||||||||#|||||||||||||||||||||||||||||| Sbjct 51 DTGISSKAMGIMNSFVNDIFZRIAGEASRL#AHYNKRSTITSREIQTAVRLLLPGELAKHA 110 |
| Mus musculus | Query 52 DTGISSKAMGIMNSFVNDIFERIAGEASRL#AHYNKRSTITSREIQTAVRLLLPGELAKHA 111 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 52 DTGISSKAMGIMNSFVNDIFERIAGEASRL#AHYNKRSTITSREIQTAVRLLLPGELAKHA 111 |
| Canis familiaris | Query 52 DTGISSKAMGIMNSFVNDIFERIAGEASRL#AHYNKRSTITSREIQTAVRLLLPGELAKHA 111 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 52 DTGISSKAMGIMNSFVNDIFERIAGEASRL#AHYNKRSTITSREIQTAVRLLLPGELAKHA 111 |
| Gallus gallus | Query 52 DTGISSKAMGIMNSFVNDIFERIAGEASRL#AHYNKRSTITSREIQTAVRLLLPGELAKHA 111 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 52 DTGISSKAMGIMNSFVNDIFERIAGEASRL#AHYNKRSTITSREIQTAVRLLLPGELAKHA 111 |
| Bos taurus | Query 52 DTGISSKAMGIMNSFVNDIFERIAGEASRL#AHYNKRSTITSREIQTAVRLLLPGELAKHA 111 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 52 DTGISSKAMGIMNSFVNDIFERIAGEASRL#AHYNKRSTITSREIQTAVRLLLPGELAKHA 111 |
| Danio rerio | Query 52 DTGISSKAMGIMNSFVNDIFERIAGEASRL#AHYNKRSTITSREIQTAVRLLLPGELAKHA 111 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 52 DTGISSKAMGIMNSFVNDIFERIAGEASRL#AHYNKRSTITSREIQTAVRLLLPGELAKHA 111 |
| Xenopus laevis | Query 52 DTGISSKAMGIMNSFVNDIFERIAGEASRL#AHYNKRSTITSREIQTAVRLLLPGELAKHA 111 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 52 DTGISSKAMGIMNSFVNDIFERIAGEASRL#AHYNKRSTITSREIQTAVRLLLPGELAKHA 111 |
| Rattus norvegicus | Query 52 DTGISSKAMGIMNSFVNDIFERIAGEASRL#AHYNKRSTITSREIQTAVRLLLPGELAKHA 111 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 52 DTGISSKAMGIMNSFVNDIFERIAGEASRL#AHYNKRSTITSREIQTAVRLLLPGELAKHA 111 |
| Pongo pygmaeus | Query 52 DTGISSKAMGIMNSFVNDIFERIAGEASRL#AHYNKRSTITSREIQTAVRLLLPGELAKHA 111 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 52 DTGISSKAMGIMNSFVNDIFERIAGEASRL#AHYNKRSTITSREIQTAVRLLLPGELAKHA 111 |
| Macaca fascicularis | Query 52 DTGISSKAMGIMNSFVNDIFERIAGEASRL#AHYNKRSTITSREIQTAVRLLLPGELAKHA 111 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 52 DTGISSKAMGIMNSFVNDIFERIAGEASRL#AHYNKRSTITSREIQTAVRLLLPGELAKHA 111 |
| Homo sapiens | Query 52 DTGISSKAMGIMNSFVNDIFERIAGEASRL#AHYNKRSTITSREIQTAVRLLLPGELAKHA 111 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 52 DTGISSKAMGIMNSFVNDIFERIAGEASRL#AHYNKRSTITSREIQTAVRLLLPGELAKHA 111 |
| Rattus norvegicus | Query 52 DTGISSKAMGIMNSFVNDIFERIAGEASRL#AHYNKRSTITSREIQTAVRLLLPGELAKHA 111 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 52 DTGISSKAMGIMNSFVNDIFERIAGEASRL#AHYNKRSTITSREIQTAVRLLLPGELAKHA 111 |
| Oncorhynchus mykiss | Query 52 DTGISSKAMGIMNSFVNDIFERIAGEASRL#AHYNKRSTITSREIQTAVRLLLPGELAKHA 111 ||||||||||||||||||||||||||+|||#|||||||||||||||||||||||||||||| Sbjct 50 DTGISSKAMGIMNSFVNDIFERIAGESSRL#AHYNKRSTITSREIQTAVRLLLPGELAKHA 109 |
| Xenopus tropicalis | Query 52 DTGISSKAMGIMNSFVNDIFERIAGEASRL#AHYNKRSTITSREIQTAVRLLLPGELAKHA 111 ||||||||| ||||||||+|||||||||||#|||||||||||||||||||||||||||||| Sbjct 52 DTGISSKAMSIMNSFVNDVFERIAGEASRL#AHYNKRSTITSREIQTAVRLLLPGELAKHA 111 |
| Cairina moschata | Query 52 DTGISSKAMGIMNSFVNDIFERIAGEASRL#AHYNKRSTITSREIQTAVRLLLPGELAKHA 111 ||||||||||||||||||||||||||||||#||||||||||||||||||| |||||||||| Sbjct 52 DTGISSKAMGIMNSFVNDIFERIAGEASRL#AHYNKRSTITSREIQTAVRSLLPGELAKHA 111 |
| Bos taurus | Query 52 DTGISSKAMGIMNSFVNDIFERIAGEASRL#AHYNKRSTITSREIQTAVRLLLPGELAKHA 111 |||||||||| |||||||||||||||||||#|||||||||||||||||||||||||||||| Sbjct 52 DTGISSKAMGNMNSFVNDIFERIAGEASRL#AHYNKRSTITSREIQTAVRLLLPGELAKHA 111 |
| Expression vector pET3-H2B | Query 52 DTGISSKAMGIMNSFVNDIFERIAGEASRL#AHYNKRSTITSREIQTAVRLLLPGELAKHA 111 ||||||||| ||||||||+|||||||||||#|||||||||||||||||||||||||||||| Sbjct 49 DTGISSKAMSIMNSFVNDVFERIAGEASRL#AHYNKRSTITSREIQTAVRLLLPGELAKHA 108 |
| Rhacophorus schlegelii | Query 52 DTGISSKAMGIMNSFVNDIFERIAGEASRL#AHYNKRSTITSREIQTAVRLLLPGELAKHA 111 ||||||||| ||||||||||||||||||||#|||||||||||||||||||||||||||||| Sbjct 52 DTGISSKAMSIMNSFVNDIFERIAGEASRL#AHYNKRSTITSREIQTAVRLLLPGELAKHA 111 |
| Pan troglodytes | Query 52 DTGISSKAMGIMNSFVNDIFERIAGEASRL#AHYNKRSTITSREIQTAVRLLLPGELAKHA 111 |||||||||||||||||||||||| |||||#|||||||||||||+|||||||||||||||| Sbjct 52 DTGISSKAMGIMNSFVNDIFERIASEASRL#AHYNKRSTITSREMQTAVRLLLPGELAKHA 111 |
| Tetraodon nigroviridis | Query 52 DTGISSKAMGIMNSFVNDIFERIAGEASRL#AHYNKRSTITSREIQTAVRLLLPGELAKHA 111 ||||||||| |||||||||||||| |||||#|||||||||||||||||||||||||||||| Sbjct 50 DTGISSKAMSIMNSFVNDIFERIASEASRL#AHYNKRSTITSREIQTAVRLLLPGELAKHA 109 |
| Drosophila sechellia | Query 52 DTGISSKAMGIMNSFVNDIFERIAGEASRL#AHYNKRSTITSREIQTAVRLLLPGELAKHA 111 ||||||||| |||||||||||||| |||||#|||||||||||||||||||||||||||||| Sbjct 45 DTGISSKAMSIMNSFVNDIFERIAAEASRL#AHYNKRSTITSREIQTAVRLLLPGELAKHA 104 |
| Drosophila hydei | Query 52 DTGISSKAMGIMNSFVNDIFERIAGEASRL#AHYNKRSTITSREIQTAVRLLLPGELAKHA 111 ||||||||| |||||||||||||| |||||#|||||||||||||||||||||||||||||| Sbjct 49 DTGISSKAMSIMNSFVNDIFERIAAEASRL#AHYNKRSTITSREIQTAVRLLLPGELAKHA 108 |
| Drosophila yakuba | Query 52 DTGISSKAMGIMNSFVNDIFERIAGEASRL#AHYNKRSTITSREIQTAVRLLLPGELAKHA 111 ||||||||| |||||||||||||| |||||#|||||||||||||||||||||||||||||| Sbjct 49 DTGISSKAMSIMNSFVNDIFERIAAEASRL#AHYNKRSTITSREIQTAVRLLLPGELAKHA 108 |
| Rhynchosciara americana | Query 52 DTGISSKAMGIMNSFVNDIFERIAGEASRL#AHYNKRSTITSREIQTAVRLLLPGELAKHA 111 ||||||||| |||||||||||||| |||||#|||||||||||||||||||||||||||||| Sbjct 49 DTGISSKAMSIMNSFVNDIFERIAAEASRL#AHYNKRSTITSREIQTAVRLLLPGELAKHA 108 |
| Platynereis dumerilii | Query 52 DTGISSKAMGIMNSFVNDIFERIAGEASRL#AHYNKRSTITSREIQTAVRLLLPGELAKHA 111 ||||||||| |||||||||||||| |||||#|||||||||||||||||||||||||||||| Sbjct 49 DTGISSKAMSIMNSFVNDIFERIAAEASRL#AHYNKRSTITSREIQTAVRLLLPGELAKHA 108 |
| Anopheles gambiae str. PEST | Query 52 DTGISSKAMGIMNSFVNDIFERIAGEASRL#AHYNKRSTITSREIQTAVRLLLPGELAKHA 111 ||||||||| |||||||||||||| |||||#|||||||||||||||||||||||||||||| Sbjct 50 DTGISSKAMSIMNSFVNDIFERIAAEASRL#AHYNKRSTITSREIQTAVRLLLPGELAKHA 109 |
| Drosophila melanogaster | Query 52 DTGISSKAMGIMNSFVNDIFERIAGEASRL#AHYNKRSTITSREIQTAVRLLLPGELAKHA 111 ||||||||| |||||||||||||| |||||#|||||||||||||||||||||||||||||| Sbjct 49 DTGISSKAMSIMNSFVNDIFERIAAEASRL#AHYNKRSTITSREIQTAVRLLLPGELAKHA 108 |
| Tigriopus californicus | Query 52 DTGISSKAMGIMNSFVNDIFERIAGEASRL#AHYNKRSTITSREIQTAVRLLLPGELAKHA 111 ||||||||| |||||||||||||| |||||#|||||||||||||||||||||||||||||| Sbjct 49 DTGISSKAMSIMNSFVNDIFERIASEASRL#AHYNKRSTITSREIQTAVRLLLPGELAKHA 108 |
| Asellus aquaticus | Query 52 DTGISSKAMGIMNSFVNDIFERIAGEASRL#AHYNKRSTITSREIQTAVRLLLPGELAKHA 111 ||||||||| |||||||||||||| |||||#|||||||||||||||||||||||||||||| Sbjct 49 DTGISSKAMSIMNSFVNDIFERIAAEASRL#AHYNKRSTITSREIQTAVRLLLPGELAKHA 108 |
| Mytilus galloprovincialis | Query 52 DTGISSKAMGIMNSFVNDIFERIAGEASRL#AHYNKRSTITSREIQTAVRLLLPGELAKHA 111 |||+||||| |||||||||||||| |||||#|||||||||||||||||||||||||||||| Sbjct 50 DTGVSSKAMSIMNSFVNDIFERIAAEASRL#AHYNKRSTITSREIQTAVRLLLPGELAKHA 109 |
| Chironomus thummi | Query 52 DTGISSKAMGIMNSFVNDIFERIAGEASRL#AHYNKRSTITSREIQTAVRLLLPGELAKHA 111 |||+||||| |||||||||||||| |||||#|||||||||||||||||||||||||||||| Sbjct 51 DTGVSSKAMSIMNSFVNDIFERIAAEASRL#AHYNKRSTITSREIQTAVRLLLPGELAKHA 110 |
| Chaetopterus variopedatus | Query 52 DTGISSKAMGIMNSFVNDIFERIAGEASRL#AHYNKRSTITSREIQTAVRLLLPGELAKHA 111 ||||||||| ||||||||+||||| |||||#|||||||||||||||||||||||||||||| Sbjct 49 DTGISSKAMSIMNSFVNDLFERIASEASRL#AHYNKRSTITSREIQTAVRLLLPGELAKHA 108 |
| Mytilus chilensis | Query 52 DTGISSKAMGIMNSFVNDIFERIAGEASRL#AHYNKRSTITSREIQTAVRLLLPGELAKHA 111 |||+||||| |||||||||||||| |||||#|||||||||||||||||||||||||||||| Sbjct 50 DTGVSSKAMSIMNSFVNDIFERIAAEASRL#AHYNKRSTITSREIQTAVRLLLPGELAKHA 109 |
| Urechis caupo | Query 52 DTGISSKAMGIMNSFVNDIFERIAGEASRL#AHYNKRSTITSREIQTAVRLLLPGELAKHA 111 ||||||||| |||||||||||||| |||||#||||+||||||||||||||||||||||||| Sbjct 49 DTGISSKAMSIMNSFVNDIFERIAAEASRL#AHYNRRSTITSREIQTAVRLLLPGELAKHA 108 |
| Mytilus edulis | Query 52 DTGISSKAMGIMNSFVNDIFERIAGEASRL#AHYNKRSTITSREIQTAVRLLLPGELAKHA 111 |||+||||| |||||||||||||| |||||#|||||||||||||||||||||||||||||| Sbjct 50 DTGVSSKAMSIMNSFVNDIFERIAAEASRL#AHYNKRSTITSREIQTAVRLLLPGELAKHA 109 |
| Caenorhabditis elegans | Query 52 DTGISSKAMGIMNSFVNDIFERIAGEASRL#AHYNKRSTITSREIQTAVRLLLPGELAKHA 111 |||+||||| ||||||||+||||| |||||#|||||||||+||||||||||+||||||||| Sbjct 49 DTGVSSKAMSIMNSFVNDVFERIAAEASRL#AHYNKRSTISSREIQTAVRLILPGELAKHA 108 |
| Caenorhabditis briggsae AF16 | Query 52 DTGISSKAMGIMNSFVNDIFERIAGEASRL#AHYNKRSTITSREIQTAVRLLLPGELAKHA 111 |||+||||| ||||||||+||||| |||||#|||||||||+||||||||||+||||||||| Sbjct 49 DTGVSSKAMSIMNSFVNDVFERIAAEASRL#AHYNKRSTISSREIQTAVRLILPGELAKHA 108 |
[Site 9] RSTITSREIQ96-TAVRLLLPGE
Gln96  Thr
Thr
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Arg87 | Ser88 | Thr89 | Ile90 | Thr91 | Ser92 | Arg93 | Glu94 | Ile95 | Gln96 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Thr97 | Ala98 | Val99 | Arg100 | Leu101 | Leu102 | Leu103 | Pro104 | Gly105 | Glu106 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| VNDIFERIAGEASRLAHYNKRSTITSREIQTAVRLLLPGELAKHAVSEGTKAVTKYTSSK |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | Homo sapiens | 123.00 | 20 | histone cluster 1, H2bb |
| 2 | N/A | 123.00 | 17 | 701196A histone H2B |
| 3 | Mus musculus | 123.00 | 13 | histone cluster 1, H2bh |
| 4 | Bos taurus | 123.00 | 8 | PREDICTED: similar to histone H2b-616 |
| 5 | Canis familiaris | 123.00 | 8 | PREDICTED: similar to histone 1, H2ai (predicted) |
| 6 | Gallus gallus | 123.00 | 8 | histone H2B |
| 7 | Danio rerio | 123.00 | 6 | histone 2, H2, like |
| 8 | Rattus norvegicus | 123.00 | 3 | PREDICTED: similar to Histone H2B 291B |
| 9 | Rattus norvegicus | 123.00 | 2 | histone 1, H2bn (predicted) |
| 10 | Homo sapiens | 123.00 | 1 | histone cluster 2, H2bf |
| 11 | Pongo pygmaeus | 123.00 | 1 | H2B2E_PONPY Histone H2B type 2-E gi |
| 12 | Bos taurus | 123.00 | 1 | histone cluster 1, H2bn |
| 13 | Xenopus laevis | 122.00 | 4 | H2B12_XENLA Histone H2B 1.2 (H2B1.2) gi |
| 14 | Macaca fascicularis | 122.00 | 1 | H2B1K_MACFA Histone H2B type 1-K (H2B K) gi |
| 15 | Rhacophorus schlegelii | 122.00 | 1 | H2B_RHASC Histone H2B gi |
| 16 | Xenopus tropicalis | 121.00 | 3 | histone 1, H2bk |
| 17 | Expression vector pET3-H2B | 121.00 | 1 | H Chain H, 2.9 Angstrom X-Ray Structure Of Hybrid |
| 18 | Oncorhynchus mykiss | 121.00 | 1 | unnamed protein product |
| 19 | Tetraodon nigroviridis | 120.00 | 9 | unnamed protein product |
| 20 | Mytilus edulis | 120.00 | 2 | histone H2B |
| 21 | Drosophila hydei | 120.00 | 2 | H2B_DROHY Histone H2B gi |
| 22 | Drosophila sechellia | 120.00 | 2 | histone 2B |
| 23 | Drosophila yakuba | 120.00 | 2 | histone 2B |
| 24 | Mytilus chilensis | 120.00 | 1 | histone H2B |
| 25 | Drosophila virilis | 120.00 | 1 | histone H2B |
| 26 | Platynereis dumerilii | 120.00 | 1 | H2B_PLADU Histone H2B gi |
| 27 | Tigriopus californicus | 120.00 | 1 | H2B1_TIGCA Histone H2B.1/H2B.2 gi |
| 28 | Anopheles gambiae str. PEST | 120.00 | 1 | AGAP012894-PA |
| 29 | Rhynchosciara americana | 120.00 | 1 | AF378198_4 histone H2B |
| 30 | Chironomus thummi | 120.00 | 1 | H2B_CHITH Histone H2B gi |
| 31 | Cairina moschata | 120.00 | 1 | H2B_CAIMO Histone H2B gi |
| 32 | Drosophila melanogaster | 120.00 | 1 | His2B:CG17949 CG17949-PA |
| 33 | Chaetopterus variopedatus | 119.00 | 1 | histone H2B |
| 34 | Mytilus galloprovincialis | 118.00 | 1 | histone H2B |
| 35 | Caenorhabditis elegans | 117.00 | 6 | HIStone family member (his-41) |
| 36 | Caenorhabditis briggsae AF16 | 117.00 | 3 | Hypothetical protein CBG10818 |
| 37 | Urechis caupo | 117.00 | 1 | H2B_URECA Histone H2B gi |
| 38 | Pan troglodytes | 115.00 | 1 | PREDICTED: hypothetical protein |
Top-ranked sequences
| organism | matching |
|---|---|
| Homo sapiens | Query 67 VNDIFERIAGEASRLAHYNKRSTITSREIQ#TAVRLLLPGELAKHAVSEGTKAVTKYTSSK 126 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 67 VNDIFERIAGEASRLAHYNKRSTITSREIQ#TAVRLLLPGELAKHAVSEGTKAVTKYTSSK 126 |
| N/A | Query 67 VNDIFERIAGEASRLAHYNKRSTITSREIQ#TAVRLLLPGELAKHAVSEGTKAVTKYTSSK 126 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 67 VNDIFERIAGEASRLAHYNKRSTITSREIQ#TAVRLLLPGELAKHAVSEGTKAVTKYTSSK 126 |
| Mus musculus | Query 67 VNDIFERIAGEASRLAHYNKRSTITSREIQ#TAVRLLLPGELAKHAVSEGTKAVTKYTSSK 126 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 67 VNDIFERIAGEASRLAHYNKRSTITSREIQ#TAVRLLLPGELAKHAVSEGTKAVTKYTSSK 126 |
| Bos taurus | Query 67 VNDIFERIAGEASRLAHYNKRSTITSREIQ#TAVRLLLPGELAKHAVSEGTKAVTKYTSSK 126 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 67 VNDIFERIAGEASRLAHYNKRSTITSREIQ#TAVRLLLPGELAKHAVSEGTKAVTKYTSSK 126 |
| Canis familiaris | Query 67 VNDIFERIAGEASRLAHYNKRSTITSREIQ#TAVRLLLPGELAKHAVSEGTKAVTKYTSSK 126 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 67 VNDIFERIAGEASRLAHYNKRSTITSREIQ#TAVRLLLPGELAKHAVSEGTKAVTKYTSSK 126 |
| Gallus gallus | Query 67 VNDIFERIAGEASRLAHYNKRSTITSREIQ#TAVRLLLPGELAKHAVSEGTKAVTKYTSSK 126 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 67 VNDIFERIAGEASRLAHYNKRSTITSREIQ#TAVRLLLPGELAKHAVSEGTKAVTKYTSSK 126 |
| Danio rerio | Query 67 VNDIFERIAGEASRLAHYNKRSTITSREIQ#TAVRLLLPGELAKHAVSEGTKAVTKYTSSK 126 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 67 VNDIFERIAGEASRLAHYNKRSTITSREIQ#TAVRLLLPGELAKHAVSEGTKAVTKYTSSK 126 |
| Rattus norvegicus | Query 67 VNDIFERIAGEASRLAHYNKRSTITSREIQ#TAVRLLLPGELAKHAVSEGTKAVTKYTSSK 126 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 67 VNDIFERIAGEASRLAHYNKRSTITSREIQ#TAVRLLLPGELAKHAVSEGTKAVTKYTSSK 126 |
| Rattus norvegicus | Query 67 VNDIFERIAGEASRLAHYNKRSTITSREIQ#TAVRLLLPGELAKHAVSEGTKAVTKYTSSK 126 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 67 VNDIFERIAGEASRLAHYNKRSTITSREIQ#TAVRLLLPGELAKHAVSEGTKAVTKYTSSK 126 |
| Homo sapiens | Query 67 VNDIFERIAGEASRLAHYNKRSTITSREIQ#TAVRLLLPGELAKHAVSEGTKAVTKYTSSK 126 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 67 VNDIFERIAGEASRLAHYNKRSTITSREIQ#TAVRLLLPGELAKHAVSEGTKAVTKYTSSK 126 |
| Pongo pygmaeus | Query 67 VNDIFERIAGEASRLAHYNKRSTITSREIQ#TAVRLLLPGELAKHAVSEGTKAVTKYTSSK 126 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 67 VNDIFERIAGEASRLAHYNKRSTITSREIQ#TAVRLLLPGELAKHAVSEGTKAVTKYTSSK 126 |
| Bos taurus | Query 67 VNDIFERIAGEASRLAHYNKRSTITSREIQ#TAVRLLLPGELAKHAVSEGTKAVTKYTSSK 126 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 67 VNDIFERIAGEASRLAHYNKRSTITSREIQ#TAVRLLLPGELAKHAVSEGTKAVTKYTSSK 126 |
| Xenopus laevis | Query 67 VNDIFERIAGEASRLAHYNKRSTITSREIQ#TAVRLLLPGELAKHAVSEGTKAVTKYTSSK 126 ||||||||||||||||||||||||||||||#||||||||||||||||||||||||||||+| Sbjct 67 VNDIFERIAGEASRLAHYNKRSTITSREIQ#TAVRLLLPGELAKHAVSEGTKAVTKYTSAK 126 |
| Macaca fascicularis | Query 67 VNDIFERIAGEASRLAHYNKRSTITSREIQ#TAVRLLLPGELAKHAVSEGTKAVTKYTSSK 126 ||||||||||||||||||||||||||||||#||||||||||||||||||||||||||||+| Sbjct 67 VNDIFERIAGEASRLAHYNKRSTITSREIQ#TAVRLLLPGELAKHAVSEGTKAVTKYTSAK 126 |
| Rhacophorus schlegelii | Query 67 VNDIFERIAGEASRLAHYNKRSTITSREIQ#TAVRLLLPGELAKHAVSEGTKAVTKYTSSK 126 ||||||||||||||||||||||||||||||#||||||||||||||||||||||||||||+| Sbjct 67 VNDIFERIAGEASRLAHYNKRSTITSREIQ#TAVRLLLPGELAKHAVSEGTKAVTKYTSAK 126 |
| Xenopus tropicalis | Query 67 VNDIFERIAGEASRLAHYNKRSTITSREIQ#TAVRLLLPGELAKHAVSEGTKAVTKYTSSK 126 |||+||||||||||||||||||||||||||#||||||||||||||||||||||||||||+| Sbjct 67 VNDVFERIAGEASRLAHYNKRSTITSREIQ#TAVRLLLPGELAKHAVSEGTKAVTKYTSAK 126 |
| Expression vector pET3-H2B | Query 67 VNDIFERIAGEASRLAHYNKRSTITSREIQ#TAVRLLLPGELAKHAVSEGTKAVTKYTSSK 126 |||+||||||||||||||||||||||||||#||||||||||||||||||||||||||||+| Sbjct 64 VNDVFERIAGEASRLAHYNKRSTITSREIQ#TAVRLLLPGELAKHAVSEGTKAVTKYTSAK 123 |
| Oncorhynchus mykiss | Query 67 VNDIFERIAGEASRLAHYNKRSTITSREIQ#TAVRLLLPGELAKHAVSEGTKAVTKYTSSK 126 |||||||||||+||||||||||||||||||#|||||||||||||||||||||||||||||| Sbjct 65 VNDIFERIAGESSRLAHYNKRSTITSREIQ#TAVRLLLPGELAKHAVSEGTKAVTKYTSSK 124 |
| Tetraodon nigroviridis | Query 67 VNDIFERIAGEASRLAHYNKRSTITSREIQ#TAVRLLLPGELAKHAVSEGTKAVTKYTSSK 126 ||||||||| ||||||||||||||||||||#|||||||||||||||||||||||||||||| Sbjct 65 VNDIFERIASEASRLAHYNKRSTITSREIQ#TAVRLLLPGELAKHAVSEGTKAVTKYTSSK 124 |
| Mytilus edulis | Query 67 VNDIFERIAGEASRLAHYNKRSTITSREIQ#TAVRLLLPGELAKHAVSEGTKAVTKYTSSK 126 ||||||||| ||||||||||||||||||||#|||||||||||||||||||||||||||||| Sbjct 65 VNDIFERIAAEASRLAHYNKRSTITSREIQ#TAVRLLLPGELAKHAVSEGTKAVTKYTSSK 124 |
| Drosophila hydei | Query 67 VNDIFERIAGEASRLAHYNKRSTITSREIQ#TAVRLLLPGELAKHAVSEGTKAVTKYTSSK 126 ||||||||| ||||||||||||||||||||#|||||||||||||||||||||||||||||| Sbjct 64 VNDIFERIAAEASRLAHYNKRSTITSREIQ#TAVRLLLPGELAKHAVSEGTKAVTKYTSSK 123 |
| Drosophila sechellia | Query 67 VNDIFERIAGEASRLAHYNKRSTITSREIQ#TAVRLLLPGELAKHAVSEGTKAVTKYTSSK 126 ||||||||| ||||||||||||||||||||#|||||||||||||||||||||||||||||| Sbjct 64 VNDIFERIAAEASRLAHYNKRSTITSREIQ#TAVRLLLPGELAKHAVSEGTKAVTKYTSSK 123 |
| Drosophila yakuba | Query 67 VNDIFERIAGEASRLAHYNKRSTITSREIQ#TAVRLLLPGELAKHAVSEGTKAVTKYTSSK 126 ||||||||| ||||||||||||||||||||#|||||||||||||||||||||||||||||| Sbjct 64 VNDIFERIAAEASRLAHYNKRSTITSREIQ#TAVRLLLPGELAKHAVSEGTKAVTKYTSSK 123 |
| Mytilus chilensis | Query 67 VNDIFERIAGEASRLAHYNKRSTITSREIQ#TAVRLLLPGELAKHAVSEGTKAVTKYTSSK 126 ||||||||| ||||||||||||||||||||#|||||||||||||||||||||||||||||| Sbjct 65 VNDIFERIAAEASRLAHYNKRSTITSREIQ#TAVRLLLPGELAKHAVSEGTKAVTKYTSSK 124 |
| Drosophila virilis | Query 67 VNDIFERIAGEASRLAHYNKRSTITSREIQ#TAVRLLLPGELAKHAVSEGTKAVTKYTSSK 126 ||||||||| ||||||||||||||||||||#|||||||||||||||||||||||||||||| Sbjct 10 VNDIFERIAAEASRLAHYNKRSTITSREIQ#TAVRLLLPGELAKHAVSEGTKAVTKYTSSK 69 |
| Platynereis dumerilii | Query 67 VNDIFERIAGEASRLAHYNKRSTITSREIQ#TAVRLLLPGELAKHAVSEGTKAVTKYTSSK 126 ||||||||| ||||||||||||||||||||#|||||||||||||||||||||||||||||| Sbjct 64 VNDIFERIAAEASRLAHYNKRSTITSREIQ#TAVRLLLPGELAKHAVSEGTKAVTKYTSSK 123 |
| Tigriopus californicus | Query 67 VNDIFERIAGEASRLAHYNKRSTITSREIQ#TAVRLLLPGELAKHAVSEGTKAVTKYTSSK 126 ||||||||| ||||||||||||||||||||#|||||||||||||||||||||||||||||| Sbjct 64 VNDIFERIASEASRLAHYNKRSTITSREIQ#TAVRLLLPGELAKHAVSEGTKAVTKYTSSK 123 |
| Anopheles gambiae str. PEST | Query 67 VNDIFERIAGEASRLAHYNKRSTITSREIQ#TAVRLLLPGELAKHAVSEGTKAVTKYTSSK 126 ||||||||| ||||||||||||||||||||#|||||||||||||||||||||||||||||| Sbjct 65 VNDIFERIAAEASRLAHYNKRSTITSREIQ#TAVRLLLPGELAKHAVSEGTKAVTKYTSSK 124 |
| Rhynchosciara americana | Query 67 VNDIFERIAGEASRLAHYNKRSTITSREIQ#TAVRLLLPGELAKHAVSEGTKAVTKYTSSK 126 ||||||||| ||||||||||||||||||||#|||||||||||||||||||||||||||||| Sbjct 64 VNDIFERIAAEASRLAHYNKRSTITSREIQ#TAVRLLLPGELAKHAVSEGTKAVTKYTSSK 123 |
| Chironomus thummi | Query 67 VNDIFERIAGEASRLAHYNKRSTITSREIQ#TAVRLLLPGELAKHAVSEGTKAVTKYTSSK 126 ||||||||| ||||||||||||||||||||#|||||||||||||||||||||||||||||| Sbjct 66 VNDIFERIAAEASRLAHYNKRSTITSREIQ#TAVRLLLPGELAKHAVSEGTKAVTKYTSSK 125 |
| Cairina moschata | Query 67 VNDIFERIAGEASRLAHYNKRSTITSREIQ#TAVRLLLPGELAKHAVSEGTKAVTKYTSSK 126 ||||||||||||||||||||||||||||||#|||| ||||||||||||||||||||||||| Sbjct 67 VNDIFERIAGEASRLAHYNKRSTITSREIQ#TAVRSLLPGELAKHAVSEGTKAVTKYTSSK 126 |
| Drosophila melanogaster | Query 67 VNDIFERIAGEASRLAHYNKRSTITSREIQ#TAVRLLLPGELAKHAVSEGTKAVTKYTSSK 126 ||||||||| ||||||||||||||||||||#|||||||||||||||||||||||||||||| Sbjct 64 VNDIFERIAAEASRLAHYNKRSTITSREIQ#TAVRLLLPGELAKHAVSEGTKAVTKYTSSK 123 |
| Chaetopterus variopedatus | Query 67 VNDIFERIAGEASRLAHYNKRSTITSREIQ#TAVRLLLPGELAKHAVSEGTKAVTKYTSSK 126 |||+||||| ||||||||||||||||||||#|||||||||||||||||||||||||||||| Sbjct 64 VNDLFERIASEASRLAHYNKRSTITSREIQ#TAVRLLLPGELAKHAVSEGTKAVTKYTSSK 123 |
| Mytilus galloprovincialis | Query 67 VNDIFERIAGEASRLAHYNKRSTITSREIQ#TAVRLLLPGELAKHAVSEGTKAVTKYTSSK 126 ||||||||| ||||||||||||||||||||#||||||||||||||||||||+||||||||| Sbjct 65 VNDIFERIAAEASRLAHYNKRSTITSREIQ#TAVRLLLPGELAKHAVSEGTEAVTKYTSSK 124 |
| Caenorhabditis elegans | Query 67 VNDIFERIAGEASRLAHYNKRSTITSREIQ#TAVRLLLPGELAKHAVSEGTKAVTKYTSSK 126 |||+||||| ||||||||||||||+|||||#|||||+|||||||||||||||||||||||| Sbjct 64 VNDVFERIASEASRLAHYNKRSTISSREIQ#TAVRLILPGELAKHAVSEGTKAVTKYTSSK 123 |
| Caenorhabditis briggsae AF16 | Query 67 VNDIFERIAGEASRLAHYNKRSTITSREIQ#TAVRLLLPGELAKHAVSEGTKAVTKYTSSK 126 |||+||||| ||||||||||||||+|||||#|||||+|||||||||||||||||||||||| Sbjct 64 VNDVFERIAAEASRLAHYNKRSTISSREIQ#TAVRLILPGELAKHAVSEGTKAVTKYTSSK 123 |
| Urechis caupo | Query 67 VNDIFERIAGEASRLAHYNKRSTITSREIQ#TAVRLLLPGELAKHAVSEGTKAVTKYTSSK 126 ||||||||| |||||||||+||||||||||#||||||||||||||||||||||||||| || Sbjct 64 VNDIFERIAAEASRLAHYNRRSTITSREIQ#TAVRLLLPGELAKHAVSEGTKAVTKYTQSK 123 |
| Pan troglodytes | Query 67 VNDIFERIAGEASRLAHYNKRSTITSREIQ#TAVRLLLPGELAKHAVSEGTKAVTKYTSSK 126 ||||||||| ||||||||||||||||||+|#|||||||||||||||||| ||||||||||| Sbjct 67 VNDIFERIASEASRLAHYNKRSTITSREMQ#TAVRLLLPGELAKHAVSERTKAVTKYTSSK 126 |
[Site 10] SREIQTAVRL101-LLPGELAKHA
Leu101  Leu
Leu
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Ser92 | Arg93 | Glu94 | Ile95 | Gln96 | Thr97 | Ala98 | Val99 | Arg100 | Leu101 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Leu102 | Leu103 | Pro104 | Gly105 | Glu106 | Leu107 | Ala108 | Lys109 | His110 | Ala111 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| ERIAGEASRLAHYNKRSTITSREIQTAVRLLLPGELAKHAVSEGTKAVTKYTSSK |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | Homo sapiens | 110.00 | 20 | histone cluster 2, H2be |
| 2 | N/A | 110.00 | 17 | B Chain B, Histone Octamer (Chicken), Chromosomal |
| 3 | Mus musculus | 110.00 | 13 | unnamed protein product |
| 4 | Bos taurus | 110.00 | 10 | PREDICTED: similar to histone H2b-616 |
| 5 | Canis familiaris | 110.00 | 8 | PREDICTED: similar to testis-specific histone 2b |
| 6 | Gallus gallus | 110.00 | 8 | hypothetical protein LOC417956 |
| 7 | Danio rerio | 110.00 | 6 | histone 2, H2, like |
| 8 | Rattus norvegicus | 110.00 | 3 | PREDICTED: similar to Histone H2B 291B |
| 9 | Rattus norvegicus | 110.00 | 2 | histone cluster 1, H2ba |
| 10 | Pongo pygmaeus | 110.00 | 1 | H2B2E_PONPY Histone H2B type 2-E gi |
| 11 | Bos taurus | 110.00 | 1 | histone cluster 1, H2bn |
| 12 | Homo sapiens | 110.00 | 1 | histone cluster 2, H2bf |
| 13 | Xenopus laevis | 109.00 | 4 | histone H2B |
| 14 | Xenopus tropicalis | 109.00 | 3 | histone 1, H2bk |
| 15 | Rhacophorus schlegelii | 109.00 | 1 | H2B_RHASC Histone H2B gi |
| 16 | Expression vector pET3-H2B | 109.00 | 1 | H Chain H, 2.9 Angstrom X-Ray Structure Of Hybrid |
| 17 | Macaca fascicularis | 109.00 | 1 | H2B1K_MACFA Histone H2B type 1-K (H2B K) gi |
| 18 | Tetraodon nigroviridis | 108.00 | 9 | unnamed protein product |
| 19 | Drosophila hydei | 108.00 | 2 | H2B_DROHY Histone H2B gi |
| 20 | Drosophila sechellia | 108.00 | 2 | histone 2B |
| 21 | Drosophila yakuba | 108.00 | 2 | histone 2B |
| 22 | Anopheles gambiae str. PEST | 108.00 | 1 | AGAP012894-PA |
| 23 | Cairina moschata | 108.00 | 1 | H2B_CAIMO Histone H2B gi |
| 24 | Drosophila virilis | 108.00 | 1 | histone H2B |
| 25 | Platynereis dumerilii | 108.00 | 1 | H2B_PLADU Histone H2B gi |
| 26 | Oncorhynchus mykiss | 108.00 | 1 | unnamed protein product |
| 27 | Tigriopus californicus | 108.00 | 1 | H2B1_TIGCA Histone H2B.1/H2B.2 gi |
| 28 | Rhynchosciara americana | 108.00 | 1 | AF378198_4 histone H2B |
| 29 | Chironomus thummi | 108.00 | 1 | H2B_CHITH Histone H2B gi |
| 30 | Chaetopterus variopedatus | 108.00 | 1 | histone H2B |
| 31 | Drosophila melanogaster | 108.00 | 1 | His2B:CG17949 CG17949-PA |
| 32 | Mytilus edulis | 107.00 | 1 | histone H2B |
| 33 | Mytilus chilensis | 107.00 | 1 | histone H2B |
| 34 | Caenorhabditis elegans | 106.00 | 6 | HIStone family member (his-52) |
| 35 | Mytilus galloprovincialis | 106.00 | 1 | histone H2B |
| 36 | Caenorhabditis briggsae AF16 | 105.00 | 3 | Hypothetical protein CBG10818 |
| 37 | Urechis caupo | 105.00 | 1 | H2B_URECA Histone H2B gi |
| 38 | Bombyx mori | 104.00 | 1 | Histone H2b |
Top-ranked sequences
| organism | matching |
|---|---|
| Homo sapiens | Query 72 ERIAGEASRLAHYNKRSTITSREIQTAVRL#LLPGELAKHAVSEGTKAVTKYTSSK 126 ||||||||||||||||||||||||||||||#||||||||||||||||||||||||| Query 72 ERIAGEASRLAHYNKRSTITSREIQTAVRL#LLPGELAKHAVSEGTKAVTKYTSSK 126 |
| N/A | Query 72 ERIAGEASRLAHYNKRSTITSREIQTAVRL#LLPGELAKHAVSEGTKAVTKYTSSK 126 ||||||||||||||||||||||||||||||#||||||||||||||||||||||||| Query 72 ERIAGEASRLAHYNKRSTITSREIQTAVRL#LLPGELAKHAVSEGTKAVTKYTSSK 126 |
| Mus musculus | Query 72 ERIAGEASRLAHYNKRSTITSREIQTAVRL#LLPGELAKHAVSEGTKAVTKYTSSK 126 ||||||||||||||||||||||||||||||#||||||||||||||||||||||||| Query 72 ERIAGEASRLAHYNKRSTITSREIQTAVRL#LLPGELAKHAVSEGTKAVTKYTSSK 126 |
| Bos taurus | Query 72 ERIAGEASRLAHYNKRSTITSREIQTAVRL#LLPGELAKHAVSEGTKAVTKYTSSK 126 ||||||||||||||||||||||||||||||#||||||||||||||||||||||||| Query 72 ERIAGEASRLAHYNKRSTITSREIQTAVRL#LLPGELAKHAVSEGTKAVTKYTSSK 126 |
| Canis familiaris | Query 72 ERIAGEASRLAHYNKRSTITSREIQTAVRL#LLPGELAKHAVSEGTKAVTKYTSSK 126 ||||||||||||||||||||||||||||||#||||||||||||||||||||||||| Query 72 ERIAGEASRLAHYNKRSTITSREIQTAVRL#LLPGELAKHAVSEGTKAVTKYTSSK 126 |
| Gallus gallus | Query 72 ERIAGEASRLAHYNKRSTITSREIQTAVRL#LLPGELAKHAVSEGTKAVTKYTSSK 126 ||||||||||||||||||||||||||||||#||||||||||||||||||||||||| Query 72 ERIAGEASRLAHYNKRSTITSREIQTAVRL#LLPGELAKHAVSEGTKAVTKYTSSK 126 |
| Danio rerio | Query 72 ERIAGEASRLAHYNKRSTITSREIQTAVRL#LLPGELAKHAVSEGTKAVTKYTSSK 126 ||||||||||||||||||||||||||||||#||||||||||||||||||||||||| Query 72 ERIAGEASRLAHYNKRSTITSREIQTAVRL#LLPGELAKHAVSEGTKAVTKYTSSK 126 |
| Rattus norvegicus | Query 72 ERIAGEASRLAHYNKRSTITSREIQTAVRL#LLPGELAKHAVSEGTKAVTKYTSSK 126 ||||||||||||||||||||||||||||||#||||||||||||||||||||||||| Query 72 ERIAGEASRLAHYNKRSTITSREIQTAVRL#LLPGELAKHAVSEGTKAVTKYTSSK 126 |
| Rattus norvegicus | Query 72 ERIAGEASRLAHYNKRSTITSREIQTAVRL#LLPGELAKHAVSEGTKAVTKYTSSK 126 ||||||||||||||||||||||||||||||#||||||||||||||||||||||||| Query 72 ERIAGEASRLAHYNKRSTITSREIQTAVRL#LLPGELAKHAVSEGTKAVTKYTSSK 126 |
| Pongo pygmaeus | Query 72 ERIAGEASRLAHYNKRSTITSREIQTAVRL#LLPGELAKHAVSEGTKAVTKYTSSK 126 ||||||||||||||||||||||||||||||#||||||||||||||||||||||||| Query 72 ERIAGEASRLAHYNKRSTITSREIQTAVRL#LLPGELAKHAVSEGTKAVTKYTSSK 126 |
| Bos taurus | Query 72 ERIAGEASRLAHYNKRSTITSREIQTAVRL#LLPGELAKHAVSEGTKAVTKYTSSK 126 ||||||||||||||||||||||||||||||#||||||||||||||||||||||||| Query 72 ERIAGEASRLAHYNKRSTITSREIQTAVRL#LLPGELAKHAVSEGTKAVTKYTSSK 126 |
| Homo sapiens | Query 72 ERIAGEASRLAHYNKRSTITSREIQTAVRL#LLPGELAKHAVSEGTKAVTKYTSSK 126 ||||||||||||||||||||||||||||||#||||||||||||||||||||||||| Query 72 ERIAGEASRLAHYNKRSTITSREIQTAVRL#LLPGELAKHAVSEGTKAVTKYTSSK 126 |
| Xenopus laevis | Query 72 ERIAGEASRLAHYNKRSTITSREIQTAVRL#LLPGELAKHAVSEGTKAVTKYTSSK 126 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||+| Sbjct 72 ERIAGEASRLAHYNKRSTITSREIQTAVRL#LLPGELAKHAVSEGTKAVTKYTSAK 126 |
| Xenopus tropicalis | Query 72 ERIAGEASRLAHYNKRSTITSREIQTAVRL#LLPGELAKHAVSEGTKAVTKYTSSK 126 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||+| Sbjct 72 ERIAGEASRLAHYNKRSTITSREIQTAVRL#LLPGELAKHAVSEGTKAVTKYTSAK 126 |
| Rhacophorus schlegelii | Query 72 ERIAGEASRLAHYNKRSTITSREIQTAVRL#LLPGELAKHAVSEGTKAVTKYTSSK 126 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||+| Sbjct 72 ERIAGEASRLAHYNKRSTITSREIQTAVRL#LLPGELAKHAVSEGTKAVTKYTSAK 126 |
| Expression vector pET3-H2B | Query 72 ERIAGEASRLAHYNKRSTITSREIQTAVRL#LLPGELAKHAVSEGTKAVTKYTSSK 126 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||+| Sbjct 69 ERIAGEASRLAHYNKRSTITSREIQTAVRL#LLPGELAKHAVSEGTKAVTKYTSAK 123 |
| Macaca fascicularis | Query 72 ERIAGEASRLAHYNKRSTITSREIQTAVRL#LLPGELAKHAVSEGTKAVTKYTSSK 126 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||+| Sbjct 72 ERIAGEASRLAHYNKRSTITSREIQTAVRL#LLPGELAKHAVSEGTKAVTKYTSAK 126 |
| Tetraodon nigroviridis | Query 72 ERIAGEASRLAHYNKRSTITSREIQTAVRL#LLPGELAKHAVSEGTKAVTKYTSSK 126 |||| |||||||||||||||||||||||||#||||||||||||||||||||||||| Sbjct 70 ERIASEASRLAHYNKRSTITSREIQTAVRL#LLPGELAKHAVSEGTKAVTKYTSSK 124 |
| Drosophila hydei | Query 72 ERIAGEASRLAHYNKRSTITSREIQTAVRL#LLPGELAKHAVSEGTKAVTKYTSSK 126 |||| |||||||||||||||||||||||||#||||||||||||||||||||||||| Sbjct 69 ERIAAEASRLAHYNKRSTITSREIQTAVRL#LLPGELAKHAVSEGTKAVTKYTSSK 123 |
| Drosophila sechellia | Query 72 ERIAGEASRLAHYNKRSTITSREIQTAVRL#LLPGELAKHAVSEGTKAVTKYTSSK 126 |||| |||||||||||||||||||||||||#||||||||||||||||||||||||| Sbjct 69 ERIAAEASRLAHYNKRSTITSREIQTAVRL#LLPGELAKHAVSEGTKAVTKYTSSK 123 |
| Drosophila yakuba | Query 72 ERIAGEASRLAHYNKRSTITSREIQTAVRL#LLPGELAKHAVSEGTKAVTKYTSSK 126 |||| |||||||||||||||||||||||||#||||||||||||||||||||||||| Sbjct 69 ERIAAEASRLAHYNKRSTITSREIQTAVRL#LLPGELAKHAVSEGTKAVTKYTSSK 123 |
| Anopheles gambiae str. PEST | Query 72 ERIAGEASRLAHYNKRSTITSREIQTAVRL#LLPGELAKHAVSEGTKAVTKYTSSK 126 |||| |||||||||||||||||||||||||#||||||||||||||||||||||||| Sbjct 70 ERIAAEASRLAHYNKRSTITSREIQTAVRL#LLPGELAKHAVSEGTKAVTKYTSSK 124 |
| Cairina moschata | Query 72 ERIAGEASRLAHYNKRSTITSREIQTAVRL#LLPGELAKHAVSEGTKAVTKYTSSK 126 ||||||||||||||||||||||||||||| #||||||||||||||||||||||||| Sbjct 72 ERIAGEASRLAHYNKRSTITSREIQTAVRS#LLPGELAKHAVSEGTKAVTKYTSSK 126 |
| Drosophila virilis | Query 72 ERIAGEASRLAHYNKRSTITSREIQTAVRL#LLPGELAKHAVSEGTKAVTKYTSSK 126 |||| |||||||||||||||||||||||||#||||||||||||||||||||||||| Sbjct 15 ERIAAEASRLAHYNKRSTITSREIQTAVRL#LLPGELAKHAVSEGTKAVTKYTSSK 69 |
| Platynereis dumerilii | Query 72 ERIAGEASRLAHYNKRSTITSREIQTAVRL#LLPGELAKHAVSEGTKAVTKYTSSK 126 |||| |||||||||||||||||||||||||#||||||||||||||||||||||||| Sbjct 69 ERIAAEASRLAHYNKRSTITSREIQTAVRL#LLPGELAKHAVSEGTKAVTKYTSSK 123 |
| Oncorhynchus mykiss | Query 72 ERIAGEASRLAHYNKRSTITSREIQTAVRL#LLPGELAKHAVSEGTKAVTKYTSSK 126 ||||||+|||||||||||||||||||||||#||||||||||||||||||||||||| Sbjct 70 ERIAGESSRLAHYNKRSTITSREIQTAVRL#LLPGELAKHAVSEGTKAVTKYTSSK 124 |
| Tigriopus californicus | Query 72 ERIAGEASRLAHYNKRSTITSREIQTAVRL#LLPGELAKHAVSEGTKAVTKYTSSK 126 |||| |||||||||||||||||||||||||#||||||||||||||||||||||||| Sbjct 69 ERIASEASRLAHYNKRSTITSREIQTAVRL#LLPGELAKHAVSEGTKAVTKYTSSK 123 |
| Rhynchosciara americana | Query 72 ERIAGEASRLAHYNKRSTITSREIQTAVRL#LLPGELAKHAVSEGTKAVTKYTSSK 126 |||| |||||||||||||||||||||||||#||||||||||||||||||||||||| Sbjct 69 ERIAAEASRLAHYNKRSTITSREIQTAVRL#LLPGELAKHAVSEGTKAVTKYTSSK 123 |
| Chironomus thummi | Query 72 ERIAGEASRLAHYNKRSTITSREIQTAVRL#LLPGELAKHAVSEGTKAVTKYTSSK 126 |||| |||||||||||||||||||||||||#||||||||||||||||||||||||| Sbjct 71 ERIAAEASRLAHYNKRSTITSREIQTAVRL#LLPGELAKHAVSEGTKAVTKYTSSK 125 |
| Chaetopterus variopedatus | Query 72 ERIAGEASRLAHYNKRSTITSREIQTAVRL#LLPGELAKHAVSEGTKAVTKYTSSK 126 |||| |||||||||||||||||||||||||#||||||||||||||||||||||||| Sbjct 69 ERIASEASRLAHYNKRSTITSREIQTAVRL#LLPGELAKHAVSEGTKAVTKYTSSK 123 |
| Drosophila melanogaster | Query 72 ERIAGEASRLAHYNKRSTITSREIQTAVRL#LLPGELAKHAVSEGTKAVTKYTSSK 126 |||| |||||||||||||||||||||||||#||||||||||||||||||||||||| Sbjct 69 ERIAAEASRLAHYNKRSTITSREIQTAVRL#LLPGELAKHAVSEGTKAVTKYTSSK 123 |
| Mytilus edulis | Query 72 ERIAGEASRLAHYNKRSTITSREIQTAVRL#LLPGELAKHAVSEGTKAVTKYTSSK 126 |||| |||||||||||||||||||||||||#||||||||||||||||||||||||| Sbjct 70 ERIAAEASRLAHYNKRSTITSREIQTAVRL#LLPGELAKHAVSEGTKAVTKYTSSK 124 |
| Mytilus chilensis | Query 72 ERIAGEASRLAHYNKRSTITSREIQTAVRL#LLPGELAKHAVSEGTKAVTKYTSSK 126 |||| |||||||||||||||||||||||||#||||||||||||||||||||||||| Sbjct 70 ERIAAEASRLAHYNKRSTITSREIQTAVRL#LLPGELAKHAVSEGTKAVTKYTSSK 124 |
| Caenorhabditis elegans | Query 72 ERIAGEASRLAHYNKRSTITSREIQTAVRL#LLPGELAKHAVSEGTKAVTKYTSSK 126 |||| ||||||||||||||+||||||||||#+|||||||||||||||||||||||| Sbjct 87 ERIAAEASRLAHYNKRSTISSREIQTAVRL#ILPGELAKHAVSEGTKAVTKYTSSK 141 |
| Mytilus galloprovincialis | Query 72 ERIAGEASRLAHYNKRSTITSREIQTAVRL#LLPGELAKHAVSEGTKAVTKYTSSK 126 |||| |||||||||||||||||||||||||#|||||||||||||||+||||||||| Sbjct 70 ERIAAEASRLAHYNKRSTITSREIQTAVRL#LLPGELAKHAVSEGTEAVTKYTSSK 124 |
| Caenorhabditis briggsae AF16 | Query 72 ERIAGEASRLAHYNKRSTITSREIQTAVRL#LLPGELAKHAVSEGTKAVTKYTSSK 126 |||| ||||||||||||||+||||||||||#+|||||||||||||||||||||||| Sbjct 69 ERIAAEASRLAHYNKRSTISSREIQTAVRL#ILPGELAKHAVSEGTKAVTKYTSSK 123 |
| Urechis caupo | Query 72 ERIAGEASRLAHYNKRSTITSREIQTAVRL#LLPGELAKHAVSEGTKAVTKYTSSK 126 |||| |||||||||+|||||||||||||||#|||||||||||||||||||||| || Sbjct 69 ERIAAEASRLAHYNRRSTITSREIQTAVRL#LLPGELAKHAVSEGTKAVTKYTQSK 123 |
| Bombyx mori | Query 72 ERIAGEASRLAHYNKRSTITSREIQTAVRL#LLPGELAKHAVSEGTKAVTKYTSSK 126 |||| ||||||||||||||||||+||+|||#||||||||||||||||+|||||||| Sbjct 9 ERIAAEASRLAHYNKRSTITSREVQTSVRL#LLPGELAKHAVSEGTKSVTKYTSSK 63 |
[Site 11] QTAVRLLLPG105-ELAKHAVSEG
Gly105  Glu
Glu
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Gln96 | Thr97 | Ala98 | Val99 | Arg100 | Leu101 | Leu102 | Leu103 | Pro104 | Gly105 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Glu106 | Leu107 | Ala108 | Lys109 | His110 | Ala111 | Val112 | Ser113 | Glu114 | Gly115 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| GEASRLAHYNKRSTITSREIQTAVRLLLPGELAKHAVSEGTKAVTKYTSSK |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | Homo sapiens | 103.00 | 20 | histone cluster 1, H2bo |
| 2 | N/A | 103.00 | 16 | 0506206A histone H2B |
| 3 | Mus musculus | 103.00 | 13 | histone cluster 1, H2bh |
| 4 | Bos taurus | 103.00 | 10 | PREDICTED: similar to histone H2b-616 |
| 5 | Canis familiaris | 103.00 | 8 | PREDICTED: similar to histone 1, H2ai (predicted) |
| 6 | Gallus gallus | 103.00 | 8 | hypothetical protein LOC417957 |
| 7 | Danio rerio | 103.00 | 6 | histone 2, H2, like |
| 8 | Rattus norvegicus | 103.00 | 3 | PREDICTED: similar to Histone H2B 291B |
| 9 | Rattus norvegicus | 103.00 | 2 | histone 1, H2bn (predicted) |
| 10 | Homo sapiens | 103.00 | 1 | histone cluster 2, H2bf |
| 11 | Pongo pygmaeus | 103.00 | 1 | H2B2E_PONPY Histone H2B type 2-E gi |
| 12 | Bos taurus | 103.00 | 1 | histone cluster 1, H2bn |
| 13 | Xenopus laevis | 102.00 | 4 | histone H2B |
| 14 | Xenopus tropicalis | 102.00 | 3 | hypothetical protein LOC550005 |
| 15 | Rhacophorus schlegelii | 102.00 | 1 | H2B_RHASC Histone H2B gi |
| 16 | Macaca fascicularis | 102.00 | 1 | H2B1K_MACFA Histone H2B type 1-K (H2B K) gi |
| 17 | Expression vector pET3-H2B | 102.00 | 1 | H Chain H, 2.9 Angstrom X-Ray Structure Of Hybrid |
| 18 | Drosophila hydei | 101.00 | 2 | H2B_DROHY Histone H2B gi |
| 19 | Drosophila yakuba | 101.00 | 2 | histone 2B |
| 20 | Drosophila sechellia | 101.00 | 2 | histone 2B |
| 21 | Drosophila virilis | 101.00 | 2 | histone H2B |
| 22 | Oncorhynchus mykiss | 101.00 | 1 | unnamed protein product |
| 23 | Drosophila melanogaster | 101.00 | 1 | His2B:CG17949 CG17949-PA |
| 24 | Chironomus thummi | 101.00 | 1 | H2B_CHITH Histone H2B gi |
| 25 | Anopheles gambiae str. PEST | 101.00 | 1 | AGAP012894-PA |
| 26 | Tetraodon nigroviridis | 100.00 | 9 | unnamed protein product |
| 27 | Rhynchosciara americana | 100.00 | 1 | AF378198_4 histone H2B |
| 28 | Tigriopus californicus | 100.00 | 1 | H2B1_TIGCA Histone H2B.1/H2B.2 gi |
| 29 | Mytilus edulis | 100.00 | 1 | histone H2B |
| 30 | Mytilus chilensis | 100.00 | 1 | histone H2B |
| 31 | Chaetopterus variopedatus | 100.00 | 1 | histone H2B |
| 32 | Platynereis dumerilii | 100.00 | 1 | H2B_PLADU Histone H2B gi |
| 33 | Cairina moschata | 100.00 | 1 | H2B_CAIMO Histone H2B gi |
| 34 | Caenorhabditis elegans | 99.00 | 6 | HIStone family member (his-4) |
| 35 | Caenorhabditis briggsae AF16 | 98.60 | 3 | Hypothetical protein CBG10818 |
| 36 | Mytilus galloprovincialis | 98.60 | 1 | histone H2B |
| 37 | Urechis caupo | 98.20 | 1 | H2B_URECA Histone H2B gi |
| 38 | Mytilus californianus | 97.40 | 1 | histone H2B |
| 39 | Bombyx mori | 97.10 | 1 | Histone H2b |
Top-ranked sequences
| organism | matching |
|---|---|
| Homo sapiens | Query 76 GEASRLAHYNKRSTITSREIQTAVRLLLPG#ELAKHAVSEGTKAVTKYTSSK 126 ||||||||||||||||||||||||||||||#||||||||||||||||||||| Query 76 GEASRLAHYNKRSTITSREIQTAVRLLLPG#ELAKHAVSEGTKAVTKYTSSK 126 |
| N/A | Query 76 GEASRLAHYNKRSTITSREIQTAVRLLLPG#ELAKHAVSEGTKAVTKYTSSK 126 ||||||||||||||||||||||||||||||#||||||||||||||||||||| Query 76 GEASRLAHYNKRSTITSREIQTAVRLLLPG#ELAKHAVSEGTKAVTKYTSSK 126 |
| Mus musculus | Query 76 GEASRLAHYNKRSTITSREIQTAVRLLLPG#ELAKHAVSEGTKAVTKYTSSK 126 ||||||||||||||||||||||||||||||#||||||||||||||||||||| Query 76 GEASRLAHYNKRSTITSREIQTAVRLLLPG#ELAKHAVSEGTKAVTKYTSSK 126 |
| Bos taurus | Query 76 GEASRLAHYNKRSTITSREIQTAVRLLLPG#ELAKHAVSEGTKAVTKYTSSK 126 ||||||||||||||||||||||||||||||#||||||||||||||||||||| Query 76 GEASRLAHYNKRSTITSREIQTAVRLLLPG#ELAKHAVSEGTKAVTKYTSSK 126 |
| Canis familiaris | Query 76 GEASRLAHYNKRSTITSREIQTAVRLLLPG#ELAKHAVSEGTKAVTKYTSSK 126 ||||||||||||||||||||||||||||||#||||||||||||||||||||| Query 76 GEASRLAHYNKRSTITSREIQTAVRLLLPG#ELAKHAVSEGTKAVTKYTSSK 126 |
| Gallus gallus | Query 76 GEASRLAHYNKRSTITSREIQTAVRLLLPG#ELAKHAVSEGTKAVTKYTSSK 126 ||||||||||||||||||||||||||||||#||||||||||||||||||||| Query 76 GEASRLAHYNKRSTITSREIQTAVRLLLPG#ELAKHAVSEGTKAVTKYTSSK 126 |
| Danio rerio | Query 76 GEASRLAHYNKRSTITSREIQTAVRLLLPG#ELAKHAVSEGTKAVTKYTSSK 126 ||||||||||||||||||||||||||||||#||||||||||||||||||||| Query 76 GEASRLAHYNKRSTITSREIQTAVRLLLPG#ELAKHAVSEGTKAVTKYTSSK 126 |
| Rattus norvegicus | Query 76 GEASRLAHYNKRSTITSREIQTAVRLLLPG#ELAKHAVSEGTKAVTKYTSSK 126 ||||||||||||||||||||||||||||||#||||||||||||||||||||| Query 76 GEASRLAHYNKRSTITSREIQTAVRLLLPG#ELAKHAVSEGTKAVTKYTSSK 126 |
| Rattus norvegicus | Query 76 GEASRLAHYNKRSTITSREIQTAVRLLLPG#ELAKHAVSEGTKAVTKYTSSK 126 ||||||||||||||||||||||||||||||#||||||||||||||||||||| Query 76 GEASRLAHYNKRSTITSREIQTAVRLLLPG#ELAKHAVSEGTKAVTKYTSSK 126 |
| Homo sapiens | Query 76 GEASRLAHYNKRSTITSREIQTAVRLLLPG#ELAKHAVSEGTKAVTKYTSSK 126 ||||||||||||||||||||||||||||||#||||||||||||||||||||| Query 76 GEASRLAHYNKRSTITSREIQTAVRLLLPG#ELAKHAVSEGTKAVTKYTSSK 126 |
| Pongo pygmaeus | Query 76 GEASRLAHYNKRSTITSREIQTAVRLLLPG#ELAKHAVSEGTKAVTKYTSSK 126 ||||||||||||||||||||||||||||||#||||||||||||||||||||| Query 76 GEASRLAHYNKRSTITSREIQTAVRLLLPG#ELAKHAVSEGTKAVTKYTSSK 126 |
| Bos taurus | Query 76 GEASRLAHYNKRSTITSREIQTAVRLLLPG#ELAKHAVSEGTKAVTKYTSSK 126 ||||||||||||||||||||||||||||||#||||||||||||||||||||| Query 76 GEASRLAHYNKRSTITSREIQTAVRLLLPG#ELAKHAVSEGTKAVTKYTSSK 126 |
| Xenopus laevis | Query 76 GEASRLAHYNKRSTITSREIQTAVRLLLPG#ELAKHAVSEGTKAVTKYTSSK 126 ||||||||||||||||||||||||||||||#|||||||||||||||||||+| Sbjct 76 GEASRLAHYNKRSTITSREIQTAVRLLLPG#ELAKHAVSEGTKAVTKYTSAK 126 |
| Xenopus tropicalis | Query 76 GEASRLAHYNKRSTITSREIQTAVRLLLPG#ELAKHAVSEGTKAVTKYTSSK 126 ||||||||||||||||||||||||||||||#|||||||||||||||||||+| Sbjct 76 GEASRLAHYNKRSTITSREIQTAVRLLLPG#ELAKHAVSEGTKAVTKYTSAK 126 |
| Rhacophorus schlegelii | Query 76 GEASRLAHYNKRSTITSREIQTAVRLLLPG#ELAKHAVSEGTKAVTKYTSSK 126 ||||||||||||||||||||||||||||||#|||||||||||||||||||+| Sbjct 76 GEASRLAHYNKRSTITSREIQTAVRLLLPG#ELAKHAVSEGTKAVTKYTSAK 126 |
| Macaca fascicularis | Query 76 GEASRLAHYNKRSTITSREIQTAVRLLLPG#ELAKHAVSEGTKAVTKYTSSK 126 ||||||||||||||||||||||||||||||#|||||||||||||||||||+| Sbjct 76 GEASRLAHYNKRSTITSREIQTAVRLLLPG#ELAKHAVSEGTKAVTKYTSAK 126 |
| Expression vector pET3-H2B | Query 76 GEASRLAHYNKRSTITSREIQTAVRLLLPG#ELAKHAVSEGTKAVTKYTSSK 126 ||||||||||||||||||||||||||||||#|||||||||||||||||||+| Sbjct 73 GEASRLAHYNKRSTITSREIQTAVRLLLPG#ELAKHAVSEGTKAVTKYTSAK 123 |
| Drosophila hydei | Query 76 GEASRLAHYNKRSTITSREIQTAVRLLLPG#ELAKHAVSEGTKAVTKYTSSK 126 |||||||||||||||||||||||||||||#||||||||||||||||||||| Sbjct 73 AEASRLAHYNKRSTITSREIQTAVRLLLPG#ELAKHAVSEGTKAVTKYTSSK 123 |
| Drosophila yakuba | Query 76 GEASRLAHYNKRSTITSREIQTAVRLLLPG#ELAKHAVSEGTKAVTKYTSSK 126 |||||||||||||||||||||||||||||#||||||||||||||||||||| Sbjct 73 AEASRLAHYNKRSTITSREIQTAVRLLLPG#ELAKHAVSEGTKAVTKYTSSK 123 |
| Drosophila sechellia | Query 76 GEASRLAHYNKRSTITSREIQTAVRLLLPG#ELAKHAVSEGTKAVTKYTSSK 126 |||||||||||||||||||||||||||||#||||||||||||||||||||| Sbjct 73 AEASRLAHYNKRSTITSREIQTAVRLLLPG#ELAKHAVSEGTKAVTKYTSSK 123 |
| Drosophila virilis | Query 76 GEASRLAHYNKRSTITSREIQTAVRLLLPG#ELAKHAVSEGTKAVTKYTSSK 126 |||||||||||||||||||||||||||||#||||||||||||||||||||| Sbjct 19 AEASRLAHYNKRSTITSREIQTAVRLLLPG#ELAKHAVSEGTKAVTKYTSSK 69 |
| Oncorhynchus mykiss | Query 76 GEASRLAHYNKRSTITSREIQTAVRLLLPG#ELAKHAVSEGTKAVTKYTSSK 126 ||+|||||||||||||||||||||||||||#||||||||||||||||||||| Sbjct 74 GESSRLAHYNKRSTITSREIQTAVRLLLPG#ELAKHAVSEGTKAVTKYTSSK 124 |
| Drosophila melanogaster | Query 76 GEASRLAHYNKRSTITSREIQTAVRLLLPG#ELAKHAVSEGTKAVTKYTSSK 126 |||||||||||||||||||||||||||||#||||||||||||||||||||| Sbjct 73 AEASRLAHYNKRSTITSREIQTAVRLLLPG#ELAKHAVSEGTKAVTKYTSSK 123 |
| Chironomus thummi | Query 76 GEASRLAHYNKRSTITSREIQTAVRLLLPG#ELAKHAVSEGTKAVTKYTSSK 126 |||||||||||||||||||||||||||||#||||||||||||||||||||| Sbjct 75 AEASRLAHYNKRSTITSREIQTAVRLLLPG#ELAKHAVSEGTKAVTKYTSSK 125 |
| Anopheles gambiae str. PEST | Query 76 GEASRLAHYNKRSTITSREIQTAVRLLLPG#ELAKHAVSEGTKAVTKYTSSK 126 |||||||||||||||||||||||||||||#||||||||||||||||||||| Sbjct 74 AEASRLAHYNKRSTITSREIQTAVRLLLPG#ELAKHAVSEGTKAVTKYTSSK 124 |
| Tetraodon nigroviridis | Query 77 EASRLAHYNKRSTITSREIQTAVRLLLPG#ELAKHAVSEGTKAVTKYTSSK 126 |||||||||||||||||||||||||||||#||||||||||||||||||||| Query 77 EASRLAHYNKRSTITSREIQTAVRLLLPG#ELAKHAVSEGTKAVTKYTSSK 126 |
| Rhynchosciara americana | Query 76 GEASRLAHYNKRSTITSREIQTAVRLLLPG#ELAKHAVSEGTKAVTKYTSSK 126 |||||||||||||||||||||||||||||#||||||||||||||||||||| Sbjct 73 AEASRLAHYNKRSTITSREIQTAVRLLLPG#ELAKHAVSEGTKAVTKYTSSK 123 |
| Tigriopus californicus | Query 77 EASRLAHYNKRSTITSREIQTAVRLLLPG#ELAKHAVSEGTKAVTKYTSSK 126 |||||||||||||||||||||||||||||#||||||||||||||||||||| Query 77 EASRLAHYNKRSTITSREIQTAVRLLLPG#ELAKHAVSEGTKAVTKYTSSK 126 |
| Mytilus edulis | Query 77 EASRLAHYNKRSTITSREIQTAVRLLLPG#ELAKHAVSEGTKAVTKYTSSK 126 |||||||||||||||||||||||||||||#||||||||||||||||||||| Query 77 EASRLAHYNKRSTITSREIQTAVRLLLPG#ELAKHAVSEGTKAVTKYTSSK 126 |
| Mytilus chilensis | Query 77 EASRLAHYNKRSTITSREIQTAVRLLLPG#ELAKHAVSEGTKAVTKYTSSK 126 |||||||||||||||||||||||||||||#||||||||||||||||||||| Query 77 EASRLAHYNKRSTITSREIQTAVRLLLPG#ELAKHAVSEGTKAVTKYTSSK 126 |
| Chaetopterus variopedatus | Query 77 EASRLAHYNKRSTITSREIQTAVRLLLPG#ELAKHAVSEGTKAVTKYTSSK 126 |||||||||||||||||||||||||||||#||||||||||||||||||||| Query 77 EASRLAHYNKRSTITSREIQTAVRLLLPG#ELAKHAVSEGTKAVTKYTSSK 126 |
| Platynereis dumerilii | Query 76 GEASRLAHYNKRSTITSREIQTAVRLLLPG#ELAKHAVSEGTKAVTKYTSSK 126 |||||||||||||||||||||||||||||#||||||||||||||||||||| Sbjct 73 AEASRLAHYNKRSTITSREIQTAVRLLLPG#ELAKHAVSEGTKAVTKYTSSK 123 |
| Cairina moschata | Query 76 GEASRLAHYNKRSTITSREIQTAVRLLLPG#ELAKHAVSEGTKAVTKYTSSK 126 ||||||||||||||||||||||||| ||||#||||||||||||||||||||| Sbjct 76 GEASRLAHYNKRSTITSREIQTAVRSLLPG#ELAKHAVSEGTKAVTKYTSSK 126 |
| Caenorhabditis elegans | Query 76 GEASRLAHYNKRSTITSREIQTAVRLLLPG#ELAKHAVSEGTKAVTKYTSSK 126 ||||||||||||||+||||||||||+|||#||||||||||||||||||||| Sbjct 91 AEASRLAHYNKRSTISSREIQTAVRLILPG#ELAKHAVSEGTKAVTKYTSSK 141 |
| Caenorhabditis briggsae AF16 | Query 76 GEASRLAHYNKRSTITSREIQTAVRLLLPG#ELAKHAVSEGTKAVTKYTSSK 126 ||||||||||||||+||||||||||+|||#||||||||||||||||||||| Sbjct 73 AEASRLAHYNKRSTISSREIQTAVRLILPG#ELAKHAVSEGTKAVTKYTSSK 123 |
| Mytilus galloprovincialis | Query 77 EASRLAHYNKRSTITSREIQTAVRLLLPG#ELAKHAVSEGTKAVTKYTSSK 126 |||||||||||||||||||||||||||||#|||||||||||+||||||||| Sbjct 75 EASRLAHYNKRSTITSREIQTAVRLLLPG#ELAKHAVSEGTEAVTKYTSSK 124 |
| Urechis caupo | Query 76 GEASRLAHYNKRSTITSREIQTAVRLLLPG#ELAKHAVSEGTKAVTKYTSSK 126 |||||||||+|||||||||||||||||||#|||||||||||||||||| || Sbjct 73 AEASRLAHYNRRSTITSREIQTAVRLLLPG#ELAKHAVSEGTKAVTKYTQSK 123 |
| Mytilus californianus | Query 78 ASRLAHYNKRSTITSREIQTAVRLLLPG#ELAKHAVSEGTKAVTKYTSSK 126 |||||||||||||||||+||||||||||#||||||||||||||||||||| Sbjct 76 ASRLAHYNKRSTITSREVQTAVRLLLPG#ELAKHAVSEGTKAVTKYTSSK 124 |
| Bombyx mori | Query 76 GEASRLAHYNKRSTITSREIQTAVRLLLPG#ELAKHAVSEGTKAVTKYTSSK 126 ||||||||||||||||||+||+|||||||#||||||||||||+|||||||| Sbjct 13 AEASRLAHYNKRSTITSREVQTSVRLLLPG#ELAKHAVSEGTKSVTKYTSSK 63 |
[Site 12] TAVRLLLPGE106-LAKHAVSEGT
Glu106  Leu
Leu
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Thr97 | Ala98 | Val99 | Arg100 | Leu101 | Leu102 | Leu103 | Pro104 | Gly105 | Glu106 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Leu107 | Ala108 | Lys109 | His110 | Ala111 | Val112 | Ser113 | Glu114 | Gly115 | Thr116 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| EASRLAHYNKRSTITSREIQTAVRLLLPGELAKHAVSEGTKAVTKYTSSK |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | N/A | 101.00 | 16 | H2B3_TIGCA Histone H2B.3 gi |
| 2 | Tetraodon nigroviridis | 101.00 | 9 | unnamed protein product |
| 3 | Drosophila virilis | 101.00 | 2 | histone H2B |
| 4 | Drosophila sechellia | 101.00 | 2 | histone 2B |
| 5 | Drosophila melanogaster | 101.00 | 1 | His2B:CG17949 CG17949-PA |
| 6 | Anopheles gambiae str. PEST | 101.00 | 1 | AGAP012894-PA |
| 7 | Chironomus thummi | 101.00 | 1 | H2B_CHITH Histone H2B gi |
| 8 | Homo sapiens | 100.00 | 21 | histone cluster 1, H2bn |
| 9 | Mus musculus | 100.00 | 13 | histone cluster 1, H2bb |
| 10 | Bos taurus | 100.00 | 10 | PREDICTED: similar to histone H2b-616 |
| 11 | Gallus gallus | 100.00 | 8 | PREDICTED: similar to histone H2B |
| 12 | Canis familiaris | 100.00 | 8 | PREDICTED: similar to histone 3, H2ba |
| 13 | Danio rerio | 100.00 | 6 | histone 2, H2, like |
| 14 | Rattus norvegicus | 100.00 | 3 | PREDICTED: similar to Histone H2B 291B |
| 15 | Mytilus edulis | 100.00 | 2 | histone H2B |
| 16 | Drosophila hydei | 100.00 | 2 | histone H2b |
| 17 | Drosophila yakuba | 100.00 | 2 | histone 2B |
| 18 | Rattus norvegicus | 100.00 | 2 | histone cluster 1, H2ba |
| 19 | Bos taurus | 100.00 | 1 | histone cluster 1, H2bn |
| 20 | Chaetopterus variopedatus | 100.00 | 1 | histone H2B |
| 21 | Mytilus chilensis | 100.00 | 1 | histone H2B |
| 22 | Rhynchosciara americana | 100.00 | 1 | AF378198_4 histone H2B |
| 23 | Tigriopus californicus | 100.00 | 1 | H2B1_TIGCA Histone H2B.1/H2B.2 gi |
| 24 | Homo sapiens | 100.00 | 1 | histone cluster 2, H2bf |
| 25 | Pongo pygmaeus | 100.00 | 1 | H2B2E_PONPY Histone H2B type 2-E gi |
| 26 | Platynereis dumerilii | 100.00 | 1 | H2B_PLADU Histone H2B gi |
| 27 | Xenopus laevis | 99.80 | 4 | histone H2B |
| 28 | Xenopus tropicalis | 99.80 | 3 | hypothetical protein LOC550005 |
| 29 | Rhacophorus schlegelii | 99.80 | 1 | H2B_RHASC Histone H2B gi |
| 30 | Expression vector pET3-H2B | 99.80 | 1 | H Chain H, 2.9 Angstrom X-Ray Structure Of Hybrid |
| 31 | Macaca fascicularis | 99.80 | 1 | H2B1K_MACFA Histone H2B type 1-K (H2B K) gi |
| 32 | Oncorhynchus mykiss | 99.40 | 1 | unnamed protein product |
| 33 | Caenorhabditis elegans | 99.00 | 6 | HIStone family member (his-4) |
| 34 | Caenorhabditis briggsae AF16 | 98.60 | 3 | Hypothetical protein CBG10818 |
| 35 | Mytilus galloprovincialis | 98.60 | 1 | histone H2B |
| 36 | Cairina moschata | 98.20 | 1 | H2B_CAIMO Histone H2B gi |
| 37 | Mytilus californianus | 97.80 | 1 | histone H2B |
| 38 | Urechis caupo | 97.80 | 1 | H2B_URECA Histone H2B gi |
| 39 | Anopheles gambiae | 97.40 | 1 | histone H2B |
| 40 | Bombyx mori | 97.10 | 1 | Histone H2b |
| 41 | Holothuria tubulosa | 96.30 | 1 | H2B_HOLTU Histone H2B gi |
Top-ranked sequences
| organism | matching |
|---|---|
| N/A | Query 77 EASRLAHYNKRSTITSREIQTAVRLLLPGE#LAKHAVSEGTKAVTKYTSSK 126 ||||||||||||||||||||||||||||||#|||||||||||||||||||| Query 77 EASRLAHYNKRSTITSREIQTAVRLLLPGE#LAKHAVSEGTKAVTKYTSSK 126 |
| Tetraodon nigroviridis | Query 77 EASRLAHYNKRSTITSREIQTAVRLLLPGE#LAKHAVSEGTKAVTKYTSSK 126 ||||||||||||||||||||||||||||||#|||||||||||||||||||| Query 77 EASRLAHYNKRSTITSREIQTAVRLLLPGE#LAKHAVSEGTKAVTKYTSSK 126 |
| Drosophila virilis | Query 77 EASRLAHYNKRSTITSREIQTAVRLLLPGE#LAKHAVSEGTKAVTKYTSSK 126 ||||||||||||||||||||||||||||||#|||||||||||||||||||| Query 77 EASRLAHYNKRSTITSREIQTAVRLLLPGE#LAKHAVSEGTKAVTKYTSSK 126 |
| Drosophila sechellia | Query 77 EASRLAHYNKRSTITSREIQTAVRLLLPGE#LAKHAVSEGTKAVTKYTSSK 126 ||||||||||||||||||||||||||||||#|||||||||||||||||||| Query 77 EASRLAHYNKRSTITSREIQTAVRLLLPGE#LAKHAVSEGTKAVTKYTSSK 126 |
| Drosophila melanogaster | Query 77 EASRLAHYNKRSTITSREIQTAVRLLLPGE#LAKHAVSEGTKAVTKYTSSK 126 ||||||||||||||||||||||||||||||#|||||||||||||||||||| Query 77 EASRLAHYNKRSTITSREIQTAVRLLLPGE#LAKHAVSEGTKAVTKYTSSK 126 |
| Anopheles gambiae str. PEST | Query 77 EASRLAHYNKRSTITSREIQTAVRLLLPGE#LAKHAVSEGTKAVTKYTSSK 126 ||||||||||||||||||||||||||||||#|||||||||||||||||||| Query 77 EASRLAHYNKRSTITSREIQTAVRLLLPGE#LAKHAVSEGTKAVTKYTSSK 126 |
| Chironomus thummi | Query 77 EASRLAHYNKRSTITSREIQTAVRLLLPGE#LAKHAVSEGTKAVTKYTSSK 126 ||||||||||||||||||||||||||||||#|||||||||||||||||||| Query 77 EASRLAHYNKRSTITSREIQTAVRLLLPGE#LAKHAVSEGTKAVTKYTSSK 126 |
| Homo sapiens | Query 77 EASRLAHYNKRSTITSREIQTAVRLLLPGE#LAKHAVSEGTKAVTKYTSSK 126 ||||||||||||||||||||||||||||||#|||||||||||||||||||| Query 77 EASRLAHYNKRSTITSREIQTAVRLLLPGE#LAKHAVSEGTKAVTKYTSSK 126 |
| Mus musculus | Query 77 EASRLAHYNKRSTITSREIQTAVRLLLPGE#LAKHAVSEGTKAVTKYTSSK 126 ||||||||||||||||||||||||||||||#|||||||||||||||||||| Query 77 EASRLAHYNKRSTITSREIQTAVRLLLPGE#LAKHAVSEGTKAVTKYTSSK 126 |
| Bos taurus | Query 77 EASRLAHYNKRSTITSREIQTAVRLLLPGE#LAKHAVSEGTKAVTKYTSSK 126 ||||||||||||||||||||||||||||||#|||||||||||||||||||| Query 77 EASRLAHYNKRSTITSREIQTAVRLLLPGE#LAKHAVSEGTKAVTKYTSSK 126 |
| Gallus gallus | Query 77 EASRLAHYNKRSTITSREIQTAVRLLLPGE#LAKHAVSEGTKAVTKYTSSK 126 ||||||||||||||||||||||||||||||#|||||||||||||||||||| Query 77 EASRLAHYNKRSTITSREIQTAVRLLLPGE#LAKHAVSEGTKAVTKYTSSK 126 |
| Canis familiaris | Query 77 EASRLAHYNKRSTITSREIQTAVRLLLPGE#LAKHAVSEGTKAVTKYTSSK 126 ||||||||||||||||||+|||||||||||#|||||||||||||||||||| Sbjct 77 EASRLAHYNKRSTITSREVQTAVRLLLPGE#LAKHAVSEGTKAVTKYTSSK 126 |
| Danio rerio | Query 77 EASRLAHYNKRSTITSREIQTAVRLLLPGE#LAKHAVSEGTKAVTKYTSSK 126 ||||||||||||||||||||||||||||||#|||||||||||||||||||| Query 77 EASRLAHYNKRSTITSREIQTAVRLLLPGE#LAKHAVSEGTKAVTKYTSSK 126 |
| Rattus norvegicus | Query 77 EASRLAHYNKRSTITSREIQTAVRLLLPGE#LAKHAVSEGTKAVTKYTSSK 126 ||||||||||||||||||||||||||||||#|||||||||||||||||||| Query 77 EASRLAHYNKRSTITSREIQTAVRLLLPGE#LAKHAVSEGTKAVTKYTSSK 126 |
| Mytilus edulis | Query 77 EASRLAHYNKRSTITSREIQTAVRLLLPGE#LAKHAVSEGTKAVTKYTSSK 126 ||||||||||||||||||||||||||||||#|||||||||||||||||||| Query 77 EASRLAHYNKRSTITSREIQTAVRLLLPGE#LAKHAVSEGTKAVTKYTSSK 126 |
| Drosophila hydei | Query 77 EASRLAHYNKRSTITSREIQTAVRLLLPGE#LAKHAVSEGTKAVTKYTSSK 126 ||||||||||||||||||||||||||||||#|||||||||||||||||||| Query 77 EASRLAHYNKRSTITSREIQTAVRLLLPGE#LAKHAVSEGTKAVTKYTSSK 126 |
| Drosophila yakuba | Query 77 EASRLAHYNKRSTITSREIQTAVRLLLPGE#LAKHAVSEGTKAVTKYTSSK 126 ||||||||||||||||||||||||||||||#|||||||||||||||||||| Query 77 EASRLAHYNKRSTITSREIQTAVRLLLPGE#LAKHAVSEGTKAVTKYTSSK 126 |
| Rattus norvegicus | Query 77 EASRLAHYNKRSTITSREIQTAVRLLLPGE#LAKHAVSEGTKAVTKYTSSK 126 ||||||||||||||||||||||||||||||#|||||||||||||||||||| Query 77 EASRLAHYNKRSTITSREIQTAVRLLLPGE#LAKHAVSEGTKAVTKYTSSK 126 |
| Bos taurus | Query 77 EASRLAHYNKRSTITSREIQTAVRLLLPGE#LAKHAVSEGTKAVTKYTSSK 126 ||||||||||||||||||||||||||||||#|||||||||||||||||||| Query 77 EASRLAHYNKRSTITSREIQTAVRLLLPGE#LAKHAVSEGTKAVTKYTSSK 126 |
| Chaetopterus variopedatus | Query 77 EASRLAHYNKRSTITSREIQTAVRLLLPGE#LAKHAVSEGTKAVTKYTSSK 126 ||||||||||||||||||||||||||||||#|||||||||||||||||||| Query 77 EASRLAHYNKRSTITSREIQTAVRLLLPGE#LAKHAVSEGTKAVTKYTSSK 126 |
| Mytilus chilensis | Query 77 EASRLAHYNKRSTITSREIQTAVRLLLPGE#LAKHAVSEGTKAVTKYTSSK 126 ||||||||||||||||||||||||||||||#|||||||||||||||||||| Query 77 EASRLAHYNKRSTITSREIQTAVRLLLPGE#LAKHAVSEGTKAVTKYTSSK 126 |
| Rhynchosciara americana | Query 77 EASRLAHYNKRSTITSREIQTAVRLLLPGE#LAKHAVSEGTKAVTKYTSSK 126 ||||||||||||||||||||||||||||||#|||||||||||||||||||| Query 77 EASRLAHYNKRSTITSREIQTAVRLLLPGE#LAKHAVSEGTKAVTKYTSSK 126 |
| Tigriopus californicus | Query 77 EASRLAHYNKRSTITSREIQTAVRLLLPGE#LAKHAVSEGTKAVTKYTSSK 126 ||||||||||||||||||||||||||||||#|||||||||||||||||||| Query 77 EASRLAHYNKRSTITSREIQTAVRLLLPGE#LAKHAVSEGTKAVTKYTSSK 126 |
| Homo sapiens | Query 77 EASRLAHYNKRSTITSREIQTAVRLLLPGE#LAKHAVSEGTKAVTKYTSSK 126 ||||||||||||||||||||||||||||||#|||||||||||||||||||| Query 77 EASRLAHYNKRSTITSREIQTAVRLLLPGE#LAKHAVSEGTKAVTKYTSSK 126 |
| Pongo pygmaeus | Query 77 EASRLAHYNKRSTITSREIQTAVRLLLPGE#LAKHAVSEGTKAVTKYTSSK 126 ||||||||||||||||||||||||||||||#|||||||||||||||||||| Query 77 EASRLAHYNKRSTITSREIQTAVRLLLPGE#LAKHAVSEGTKAVTKYTSSK 126 |
| Platynereis dumerilii | Query 77 EASRLAHYNKRSTITSREIQTAVRLLLPGE#LAKHAVSEGTKAVTKYTSSK 126 ||||||||||||||||||||||||||||||#|||||||||||||||||||| Query 77 EASRLAHYNKRSTITSREIQTAVRLLLPGE#LAKHAVSEGTKAVTKYTSSK 126 |
| Xenopus laevis | Query 77 EASRLAHYNKRSTITSREIQTAVRLLLPGE#LAKHAVSEGTKAVTKYTSSK 126 ||||||||||||||||||||||||||||||#||||||||||||||||||+| Sbjct 77 EASRLAHYNKRSTITSREIQTAVRLLLPGE#LAKHAVSEGTKAVTKYTSAK 126 |
| Xenopus tropicalis | Query 77 EASRLAHYNKRSTITSREIQTAVRLLLPGE#LAKHAVSEGTKAVTKYTSSK 126 ||||||||||||||||||||||||||||||#||||||||||||||||||+| Sbjct 77 EASRLAHYNKRSTITSREIQTAVRLLLPGE#LAKHAVSEGTKAVTKYTSAK 126 |
| Rhacophorus schlegelii | Query 77 EASRLAHYNKRSTITSREIQTAVRLLLPGE#LAKHAVSEGTKAVTKYTSSK 126 ||||||||||||||||||||||||||||||#||||||||||||||||||+| Sbjct 77 EASRLAHYNKRSTITSREIQTAVRLLLPGE#LAKHAVSEGTKAVTKYTSAK 126 |
| Expression vector pET3-H2B | Query 77 EASRLAHYNKRSTITSREIQTAVRLLLPGE#LAKHAVSEGTKAVTKYTSSK 126 ||||||||||||||||||||||||||||||#||||||||||||||||||+| Sbjct 74 EASRLAHYNKRSTITSREIQTAVRLLLPGE#LAKHAVSEGTKAVTKYTSAK 123 |
| Macaca fascicularis | Query 77 EASRLAHYNKRSTITSREIQTAVRLLLPGE#LAKHAVSEGTKAVTKYTSSK 126 ||||||||||||||||||||||||||||||#||||||||||||||||||+| Sbjct 77 EASRLAHYNKRSTITSREIQTAVRLLLPGE#LAKHAVSEGTKAVTKYTSAK 126 |
| Oncorhynchus mykiss | Query 77 EASRLAHYNKRSTITSREIQTAVRLLLPGE#LAKHAVSEGTKAVTKYTSSK 126 |+||||||||||||||||||||||||||||#|||||||||||||||||||| Sbjct 75 ESSRLAHYNKRSTITSREIQTAVRLLLPGE#LAKHAVSEGTKAVTKYTSSK 124 |
| Caenorhabditis elegans | Query 77 EASRLAHYNKRSTITSREIQTAVRLLLPGE#LAKHAVSEGTKAVTKYTSSK 126 ||||||||||||||+||||||||||+||||#|||||||||||||||||||| Sbjct 92 EASRLAHYNKRSTISSREIQTAVRLILPGE#LAKHAVSEGTKAVTKYTSSK 141 |
| Caenorhabditis briggsae AF16 | Query 77 EASRLAHYNKRSTITSREIQTAVRLLLPGE#LAKHAVSEGTKAVTKYTSSK 126 ||||||||||||||+||||||||||+||||#|||||||||||||||||||| Sbjct 74 EASRLAHYNKRSTISSREIQTAVRLILPGE#LAKHAVSEGTKAVTKYTSSK 123 |
| Mytilus galloprovincialis | Query 77 EASRLAHYNKRSTITSREIQTAVRLLLPGE#LAKHAVSEGTKAVTKYTSSK 126 ||||||||||||||||||||||||||||||#||||||||||+||||||||| Sbjct 75 EASRLAHYNKRSTITSREIQTAVRLLLPGE#LAKHAVSEGTEAVTKYTSSK 124 |
| Cairina moschata | Query 77 EASRLAHYNKRSTITSREIQTAVRLLLPGE#LAKHAVSEGTKAVTKYTSSK 126 |||||||||||||||||||||||| |||||#|||||||||||||||||||| Sbjct 77 EASRLAHYNKRSTITSREIQTAVRSLLPGE#LAKHAVSEGTKAVTKYTSSK 126 |
| Mytilus californianus | Query 78 ASRLAHYNKRSTITSREIQTAVRLLLPGE#LAKHAVSEGTKAVTKYTSSK 126 |||||||||||||||||+|||||||||||#|||||||||||||||||||| Sbjct 76 ASRLAHYNKRSTITSREVQTAVRLLLPGE#LAKHAVSEGTKAVTKYTSSK 124 |
| Urechis caupo | Query 77 EASRLAHYNKRSTITSREIQTAVRLLLPGE#LAKHAVSEGTKAVTKYTSSK 126 |||||||||+||||||||||||||||||||#||||||||||||||||| || Sbjct 74 EASRLAHYNRRSTITSREIQTAVRLLLPGE#LAKHAVSEGTKAVTKYTQSK 123 |
| Anopheles gambiae | Query 79 SRLAHYNKRSTITSREIQTAVRLLLPGE#LAKHAVSEGTKAVTKYTSSK 126 ||||||||||||||||||||||||||||#|||||||||||||||||||| Query 79 SRLAHYNKRSTITSREIQTAVRLLLPGE#LAKHAVSEGTKAVTKYTSSK 126 |
| Bombyx mori | Query 77 EASRLAHYNKRSTITSREIQTAVRLLLPGE#LAKHAVSEGTKAVTKYTSSK 126 ||||||||||||||||||+||+||||||||#|||||||||||+|||||||| Sbjct 14 EASRLAHYNKRSTITSREVQTSVRLLLPGE#LAKHAVSEGTKSVTKYTSSK 63 |
| Holothuria tubulosa | Query 77 EASRLAHYNKRSTITSREIQTAVRLLLPGE#LAKHAVSEGTKAVTKYTSSK 126 |||||||||++|||||||+|||||||||||#|||||||||||||||||+|| Sbjct 74 EASRLAHYNRKSTITSREVQTAVRLLLPGE#LAKHAVSEGTKAVTKYTTSK 123 |
[Site 13] AVRLLLPGEL107-AKHAVSEGTK
Leu107  Ala
Ala
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Ala98 | Val99 | Arg100 | Leu101 | Leu102 | Leu103 | Pro104 | Gly105 | Glu106 | Leu107 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Ala108 | Lys109 | His110 | Ala111 | Val112 | Ser113 | Glu114 | Gly115 | Thr116 | Lys117 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| ASRLAHYNKRSTITSREIQTAVRLLLPGELAKHAVSEGTKAVTKYTSSK |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | Tetraodon nigroviridis | 99.40 | 9 | unnamed protein product |
| 2 | Drosophila sechellia | 99.40 | 2 | histone 2B |
| 3 | Anopheles gambiae str. PEST | 99.40 | 1 | AGAP012894-PA |
| 4 | Homo sapiens | 99.00 | 21 | histone cluster 1, H2bm |
| 5 | N/A | 99.00 | 16 | 701196A histone H2B |
| 6 | Mus musculus | 99.00 | 13 | histone cluster 1, H2bb |
| 7 | Bos taurus | 99.00 | 10 | PREDICTED: similar to Hist1h2bj protein |
| 8 | Rattus norvegicus | 99.00 | 3 | histone 1, H2bn (predicted) |
| 9 | Drosophila yakuba | 99.00 | 2 | H2B_DROYA Histone H2B gi |
| 10 | Drosophila virilis | 99.00 | 2 | histone H2B |
| 11 | Drosophila hydei | 99.00 | 2 | H2B_DROHY Histone H2B gi |
| 12 | Bos taurus | 99.00 | 1 | histone cluster 1, H2bn |
| 13 | Rhynchosciara americana | 99.00 | 1 | AF378198_4 histone H2B |
| 14 | Platynereis dumerilii | 99.00 | 1 | H2B_PLADU Histone H2B gi |
| 15 | Drosophila melanogaster | 99.00 | 1 | His2B:CG17949 CG17949-PA |
| 16 | Tigriopus californicus | 99.00 | 1 | H2B1_TIGCA Histone H2B.1/H2B.2 gi |
| 17 | Chaetopterus variopedatus | 99.00 | 1 | histone H2B |
| 18 | Chironomus thummi | 99.00 | 1 | H2B_CHITH Histone H2B gi |
| 19 | Homo sapiens | 99.00 | 1 | histone cluster 2, H2bf |
| 20 | Canis familiaris | 98.60 | 8 | PREDICTED: similar to testis-specific histone 2b |
| 21 | Gallus gallus | 98.60 | 8 | hypothetical protein LOC417957 |
| 22 | Danio rerio | 98.60 | 6 | histone 2, H2, like |
| 23 | Rattus norvegicus | 98.60 | 3 | PREDICTED: similar to Histone H2B 291B |
| 24 | Pongo pygmaeus | 98.60 | 1 | H2B2E_PONPY Histone H2B type 2-E gi |
| 25 | Mytilus edulis | 98.20 | 2 | histone H2B |
| 26 | Mytilus chilensis | 98.20 | 1 | histone H2B |
| 27 | Xenopus laevis | 97.80 | 4 | histone H2B |
| 28 | Xenopus tropicalis | 97.80 | 3 | histone 2, H2bf |
| 29 | Expression vector pET3-H2B | 97.80 | 1 | H Chain H, 2.9 Angstrom X-Ray Structure Of Hybrid |
| 30 | Rhacophorus schlegelii | 97.80 | 1 | H2B_RHASC Histone H2B gi |
| 31 | Mytilus californianus | 97.80 | 1 | histone H2B |
| 32 | Macaca fascicularis | 97.40 | 1 | H2B1K_MACFA Histone H2B type 1-K (H2B K) gi |
| 33 | Anopheles gambiae | 97.40 | 1 | histone H2B |
| 34 | Oncorhynchus mykiss | 97.10 | 1 | unnamed protein product |
| 35 | Caenorhabditis elegans | 96.70 | 6 | HIStone family member (his-52) |
| 36 | Mytilus galloprovincialis | 96.70 | 1 | histone H2B |
| 37 | Caenorhabditis briggsae AF16 | 96.30 | 3 | Hypothetical protein CBG10818 |
| 38 | Urechis caupo | 95.90 | 1 | H2B_URECA Histone H2B gi |
| 39 | Cairina moschata | 95.90 | 1 | H2B_CAIMO Histone H2B gi |
| 40 | Bombyx mori | 95.10 | 1 | Histone H2b |
Top-ranked sequences
| organism | matching |
|---|---|
| Tetraodon nigroviridis | Query 78 ASRLAHYNKRSTITSREIQTAVRLLLPGEL#AKHAVSEGTKAVTKYTSSK 126 ||||||||||||||||||||||||||||||#||||||||||||||||||| Query 78 ASRLAHYNKRSTITSREIQTAVRLLLPGEL#AKHAVSEGTKAVTKYTSSK 126 |
| Drosophila sechellia | Query 78 ASRLAHYNKRSTITSREIQTAVRLLLPGEL#AKHAVSEGTKAVTKYTSSK 126 ||||||||||||||||||||||||||||||#||||||||||||||||||| Query 78 ASRLAHYNKRSTITSREIQTAVRLLLPGEL#AKHAVSEGTKAVTKYTSSK 126 |
| Anopheles gambiae str. PEST | Query 78 ASRLAHYNKRSTITSREIQTAVRLLLPGEL#AKHAVSEGTKAVTKYTSSK 126 ||||||||||||||||||||||||||||||#||||||||||||||||||| Query 78 ASRLAHYNKRSTITSREIQTAVRLLLPGEL#AKHAVSEGTKAVTKYTSSK 126 |
| Homo sapiens | Query 78 ASRLAHYNKRSTITSREIQTAVRLLLPGEL#AKHAVSEGTKAVTKYTSSK 126 ||||||||||||||||||||||||||||||#||||||||||||||||||| Query 78 ASRLAHYNKRSTITSREIQTAVRLLLPGEL#AKHAVSEGTKAVTKYTSSK 126 |
| N/A | Query 78 ASRLAHYNKRSTITSREIQTAVRLLLPGEL#AKHAVSEGTKAVTKYTSSK 126 ||||||||||||||||||||||||||||||#||||||||||||||||||| Query 78 ASRLAHYNKRSTITSREIQTAVRLLLPGEL#AKHAVSEGTKAVTKYTSSK 126 |
| Mus musculus | Query 78 ASRLAHYNKRSTITSREIQTAVRLLLPGEL#AKHAVSEGTKAVTKYTSSK 126 ||||||||||||||||||||||||||||||#||||||||||||||||||| Query 78 ASRLAHYNKRSTITSREIQTAVRLLLPGEL#AKHAVSEGTKAVTKYTSSK 126 |
| Bos taurus | Query 78 ASRLAHYNKRSTITSREIQTAVRLLLPGEL#AKHAVSEGTKAVTKYTSSK 126 ||||||||||||||||||||||||||||||#||||||||||||||||||| Query 78 ASRLAHYNKRSTITSREIQTAVRLLLPGEL#AKHAVSEGTKAVTKYTSSK 126 |
| Rattus norvegicus | Query 78 ASRLAHYNKRSTITSREIQTAVRLLLPGEL#AKHAVSEGTKAVTKYTSSK 126 ||||||||||||||||||||||||||||||#||||||||||||||||||| Query 78 ASRLAHYNKRSTITSREIQTAVRLLLPGEL#AKHAVSEGTKAVTKYTSSK 126 |
| Drosophila yakuba | Query 78 ASRLAHYNKRSTITSREIQTAVRLLLPGEL#AKHAVSEGTKAVTKYTSSK 126 ||||||||||||||||||||||||||||||#||||||||||||||||||| Query 78 ASRLAHYNKRSTITSREIQTAVRLLLPGEL#AKHAVSEGTKAVTKYTSSK 126 |
| Drosophila virilis | Query 78 ASRLAHYNKRSTITSREIQTAVRLLLPGEL#AKHAVSEGTKAVTKYTSSK 126 ||||||||||||||||||||||||||||||#||||||||||||||||||| Query 78 ASRLAHYNKRSTITSREIQTAVRLLLPGEL#AKHAVSEGTKAVTKYTSSK 126 |
| Drosophila hydei | Query 78 ASRLAHYNKRSTITSREIQTAVRLLLPGEL#AKHAVSEGTKAVTKYTSSK 126 ||||||||||||||||||||||||||||||#||||||||||||||||||| Query 78 ASRLAHYNKRSTITSREIQTAVRLLLPGEL#AKHAVSEGTKAVTKYTSSK 126 |
| Bos taurus | Query 78 ASRLAHYNKRSTITSREIQTAVRLLLPGEL#AKHAVSEGTKAVTKYTSSK 126 ||||||||||||||||||||||||||||||#||||||||||||||||||| Query 78 ASRLAHYNKRSTITSREIQTAVRLLLPGEL#AKHAVSEGTKAVTKYTSSK 126 |
| Rhynchosciara americana | Query 78 ASRLAHYNKRSTITSREIQTAVRLLLPGEL#AKHAVSEGTKAVTKYTSSK 126 ||||||||||||||||||||||||||||||#||||||||||||||||||| Query 78 ASRLAHYNKRSTITSREIQTAVRLLLPGEL#AKHAVSEGTKAVTKYTSSK 126 |
| Platynereis dumerilii | Query 78 ASRLAHYNKRSTITSREIQTAVRLLLPGEL#AKHAVSEGTKAVTKYTSSK 126 ||||||||||||||||||||||||||||||#||||||||||||||||||| Query 78 ASRLAHYNKRSTITSREIQTAVRLLLPGEL#AKHAVSEGTKAVTKYTSSK 126 |
| Drosophila melanogaster | Query 78 ASRLAHYNKRSTITSREIQTAVRLLLPGEL#AKHAVSEGTKAVTKYTSSK 126 ||||||||||||||||||||||||||||||#||||||||||||||||||| Query 78 ASRLAHYNKRSTITSREIQTAVRLLLPGEL#AKHAVSEGTKAVTKYTSSK 126 |
| Tigriopus californicus | Query 78 ASRLAHYNKRSTITSREIQTAVRLLLPGEL#AKHAVSEGTKAVTKYTSSK 126 ||||||||||||||||||||||||||||||#||||||||||||||||||| Query 78 ASRLAHYNKRSTITSREIQTAVRLLLPGEL#AKHAVSEGTKAVTKYTSSK 126 |
| Chaetopterus variopedatus | Query 78 ASRLAHYNKRSTITSREIQTAVRLLLPGEL#AKHAVSEGTKAVTKYTSSK 126 ||||||||||||||||||||||||||||||#||||||||||||||||||| Query 78 ASRLAHYNKRSTITSREIQTAVRLLLPGEL#AKHAVSEGTKAVTKYTSSK 126 |
| Chironomus thummi | Query 78 ASRLAHYNKRSTITSREIQTAVRLLLPGEL#AKHAVSEGTKAVTKYTSSK 126 ||||||||||||||||||||||||||||||#||||||||||||||||||| Query 78 ASRLAHYNKRSTITSREIQTAVRLLLPGEL#AKHAVSEGTKAVTKYTSSK 126 |
| Homo sapiens | Query 78 ASRLAHYNKRSTITSREIQTAVRLLLPGEL#AKHAVSEGTKAVTKYTSSK 126 ||||||||||||||||||||||||||||||#||||||||||||||||||| Query 78 ASRLAHYNKRSTITSREIQTAVRLLLPGEL#AKHAVSEGTKAVTKYTSSK 126 |
| Canis familiaris | Query 78 ASRLAHYNKRSTITSREIQTAVRLLLPGEL#AKHAVSEGTKAVTKYTSSK 126 ||||||||||||||||||||||||||||||#||||||||||||||||||| Query 78 ASRLAHYNKRSTITSREIQTAVRLLLPGEL#AKHAVSEGTKAVTKYTSSK 126 |
| Gallus gallus | Query 78 ASRLAHYNKRSTITSREIQTAVRLLLPGEL#AKHAVSEGTKAVTKYTSSK 126 ||||||||||||||||||||||||||||||#||||||||||||||||||| Query 78 ASRLAHYNKRSTITSREIQTAVRLLLPGEL#AKHAVSEGTKAVTKYTSSK 126 |
| Danio rerio | Query 78 ASRLAHYNKRSTITSREIQTAVRLLLPGEL#AKHAVSEGTKAVTKYTSSK 126 ||||||||||||||||||||||||||||||#||||||||||||||||||| Query 78 ASRLAHYNKRSTITSREIQTAVRLLLPGEL#AKHAVSEGTKAVTKYTSSK 126 |
| Rattus norvegicus | Query 78 ASRLAHYNKRSTITSREIQTAVRLLLPGEL#AKHAVSEGTKAVTKYTSSK 126 ||||||||||||||||||||||||||||||#||||||||||||||||||| Query 78 ASRLAHYNKRSTITSREIQTAVRLLLPGEL#AKHAVSEGTKAVTKYTSSK 126 |
| Pongo pygmaeus | Query 78 ASRLAHYNKRSTITSREIQTAVRLLLPGEL#AKHAVSEGTKAVTKYTSSK 126 ||||||||||||||||||||||||||||||#||||||||||||||||||| Query 78 ASRLAHYNKRSTITSREIQTAVRLLLPGEL#AKHAVSEGTKAVTKYTSSK 126 |
| Mytilus edulis | Query 78 ASRLAHYNKRSTITSREIQTAVRLLLPGEL#AKHAVSEGTKAVTKYTSSK 126 ||||||||||||||||||||||||||||||#||||||||||||||||||| Query 78 ASRLAHYNKRSTITSREIQTAVRLLLPGEL#AKHAVSEGTKAVTKYTSSK 126 |
| Mytilus chilensis | Query 78 ASRLAHYNKRSTITSREIQTAVRLLLPGEL#AKHAVSEGTKAVTKYTSSK 126 ||||||||||||||||||||||||||||||#||||||||||||||||||| Query 78 ASRLAHYNKRSTITSREIQTAVRLLLPGEL#AKHAVSEGTKAVTKYTSSK 126 |
| Xenopus laevis | Query 78 ASRLAHYNKRSTITSREIQTAVRLLLPGEL#AKHAVSEGTKAVTKYTSSK 126 ||||||||||||||||||||||||||||||#|||||||||||||||||+| Sbjct 78 ASRLAHYNKRSTITSREIQTAVRLLLPGEL#AKHAVSEGTKAVTKYTSAK 126 |
| Xenopus tropicalis | Query 78 ASRLAHYNKRSTITSREIQTAVRLLLPGEL#AKHAVSEGTKAVTKYTSSK 126 ||||||||||||||||||||||||||||||#|||||||||||||||||+| Sbjct 78 ASRLAHYNKRSTITSREIQTAVRLLLPGEL#AKHAVSEGTKAVTKYTSAK 126 |
| Expression vector pET3-H2B | Query 78 ASRLAHYNKRSTITSREIQTAVRLLLPGEL#AKHAVSEGTKAVTKYTSSK 126 ||||||||||||||||||||||||||||||#|||||||||||||||||+| Sbjct 75 ASRLAHYNKRSTITSREIQTAVRLLLPGEL#AKHAVSEGTKAVTKYTSAK 123 |
| Rhacophorus schlegelii | Query 78 ASRLAHYNKRSTITSREIQTAVRLLLPGEL#AKHAVSEGTKAVTKYTSSK 126 ||||||||||||||||||||||||||||||#|||||||||||||||||+| Sbjct 78 ASRLAHYNKRSTITSREIQTAVRLLLPGEL#AKHAVSEGTKAVTKYTSAK 126 |
| Mytilus californianus | Query 78 ASRLAHYNKRSTITSREIQTAVRLLLPGEL#AKHAVSEGTKAVTKYTSSK 126 |||||||||||||||||+||||||||||||#||||||||||||||||||| Sbjct 76 ASRLAHYNKRSTITSREVQTAVRLLLPGEL#AKHAVSEGTKAVTKYTSSK 124 |
| Macaca fascicularis | Query 78 ASRLAHYNKRSTITSREIQTAVRLLLPGEL#AKHAVSEGTKAVTKYTSSK 126 ||||||||||||||||||||||||||||||#|||||||||||||||||+| Sbjct 78 ASRLAHYNKRSTITSREIQTAVRLLLPGEL#AKHAVSEGTKAVTKYTSAK 126 |
| Anopheles gambiae | Query 79 SRLAHYNKRSTITSREIQTAVRLLLPGEL#AKHAVSEGTKAVTKYTSSK 126 |||||||||||||||||||||||||||||#||||||||||||||||||| Query 79 SRLAHYNKRSTITSREIQTAVRLLLPGEL#AKHAVSEGTKAVTKYTSSK 126 |
| Oncorhynchus mykiss | Query 78 ASRLAHYNKRSTITSREIQTAVRLLLPGEL#AKHAVSEGTKAVTKYTSSK 126 +|||||||||||||||||||||||||||||#||||||||||||||||||| Sbjct 76 SSRLAHYNKRSTITSREIQTAVRLLLPGEL#AKHAVSEGTKAVTKYTSSK 124 |
| Caenorhabditis elegans | Query 78 ASRLAHYNKRSTITSREIQTAVRLLLPGEL#AKHAVSEGTKAVTKYTSSK 126 |||||||||||||+||||||||||+|||||#||||||||||||||||||| Sbjct 93 ASRLAHYNKRSTISSREIQTAVRLILPGEL#AKHAVSEGTKAVTKYTSSK 141 |
| Mytilus galloprovincialis | Query 78 ASRLAHYNKRSTITSREIQTAVRLLLPGEL#AKHAVSEGTKAVTKYTSSK 126 ||||||||||||||||||||||||||||||#|||||||||+||||||||| Sbjct 76 ASRLAHYNKRSTITSREIQTAVRLLLPGEL#AKHAVSEGTEAVTKYTSSK 124 |
| Caenorhabditis briggsae AF16 | Query 78 ASRLAHYNKRSTITSREIQTAVRLLLPGEL#AKHAVSEGTKAVTKYTSSK 126 |||||||||||||+||||||||||+|||||#||||||||||||||||||| Sbjct 75 ASRLAHYNKRSTISSREIQTAVRLILPGEL#AKHAVSEGTKAVTKYTSSK 123 |
| Urechis caupo | Query 78 ASRLAHYNKRSTITSREIQTAVRLLLPGEL#AKHAVSEGTKAVTKYTSSK 126 ||||||||+|||||||||||||||||||||#|||||||||||||||| || Sbjct 75 ASRLAHYNRRSTITSREIQTAVRLLLPGEL#AKHAVSEGTKAVTKYTQSK 123 |
| Cairina moschata | Query 78 ASRLAHYNKRSTITSREIQTAVRLLLPGEL#AKHAVSEGTKAVTKYTSSK 126 ||||||||||||||||||||||| ||||||#||||||||||||||||||| Sbjct 78 ASRLAHYNKRSTITSREIQTAVRSLLPGEL#AKHAVSEGTKAVTKYTSSK 126 |
| Bombyx mori | Query 78 ASRLAHYNKRSTITSREIQTAVRLLLPGEL#AKHAVSEGTKAVTKYTSSK 126 |||||||||||||||||+||+|||||||||#||||||||||+|||||||| Sbjct 15 ASRLAHYNKRSTITSREVQTSVRLLLPGEL#AKHAVSEGTKSVTKYTSSK 63 |
[Site 14] LAKHAVSEGT116-KAVTKYTSSK
Thr116  Lys
Lys
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Leu107 | Ala108 | Lys109 | His110 | Ala111 | Val112 | Ser113 | Glu114 | Gly115 | Thr116 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Lys117 | Ala118 | Val119 | Thr120 | Lys121 | Tyr122 | Thr123 | Ser124 | Ser125 | Lys126 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| RSTITSREIQTAVRLLLPGELAKHAVSEGTKAVTKYTSSK |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | N/A | 80.50 | 14 | H2B3_TIGCA Histone H2B.3 gi |
| 2 | Tetraodon nigroviridis | 80.50 | 11 | unnamed protein product |
| 3 | Drosophila virilis | 80.50 | 2 | histone H2B |
| 4 | Drosophila hydei | 80.50 | 2 | H2B_DROHY Histone H2B gi |
| 5 | Drosophila sechellia | 80.50 | 2 | histone 2B |
| 6 | Anopheles gambiae str. PEST | 80.50 | 1 | AGAP012894-PA |
| 7 | Homo sapiens | 80.10 | 22 | histone cluster 1, H2bh |
| 8 | Mus musculus | 80.10 | 13 | histone cluster 1, H2bj |
| 9 | Gallus gallus | 80.10 | 9 | hypothetical protein LOC417956 |
| 10 | Bos taurus | 80.10 | 8 | histone H2B |
| 11 | Drosophila yakuba | 80.10 | 2 | H2B_DROYA Histone H2B gi |
| 12 | Drosophila melanogaster | 80.10 | 1 | His2B:CG17949 CG17949-PA |
| 13 | Platynereis dumerilii | 80.10 | 1 | H2B_PLADU Histone H2B gi |
| 14 | Chironomus thummi | 80.10 | 1 | H2B_CHITH Histone H2B gi |
| 15 | Chaetopterus variopedatus | 80.10 | 1 | histone H2B |
| 16 | Rhynchosciara americana | 80.10 | 1 | AF378198_4 histone H2B |
| 17 | Tigriopus californicus | 80.10 | 1 | H2B1_TIGCA Histone H2B.1/H2B.2 gi |
| 18 | Bos taurus | 80.10 | 1 | histone cluster 1, H2bn |
| 19 | Anopheles gambiae | 80.10 | 1 | histone H2B |
| 20 | Homo sapiens | 80.10 | 1 | histone cluster 2, H2bf |
| 21 | Canis familiaris | 79.70 | 7 | PREDICTED: similar to testis-specific histone 2b |
| 22 | Danio rerio | 79.70 | 6 | histone 2, H2, like |
| 23 | Rattus norvegicus | 79.70 | 3 | histone cluster 1, H2ba |
| 24 | Rattus norvegicus | 79.70 | 3 | PREDICTED: similar to Histone H2B 291B |
| 25 | Mytilus edulis | 79.70 | 2 | histone H2B |
| 26 | Pongo pygmaeus | 79.70 | 1 | H2B2E_PONPY Histone H2B type 2-E gi |
| 27 | Oncorhynchus mykiss | 79.70 | 1 | unnamed protein product |
| 28 | Mytilus chilensis | 79.30 | 1 | histone H2B |
| 29 | Xenopus laevis | 79.00 | 3 | histone H2B |
| 30 | Xenopus tropicalis | 79.00 | 3 | histone 2, H2bf |
| 31 | Rhacophorus schlegelii | 79.00 | 1 | H2B_RHASC Histone H2B gi |
| 32 | Expression vector pET3-H2B | 79.00 | 1 | H Chain H, 2.9 Angstrom X-Ray Structure Of Hybrid |
| 33 | Mytilus californianus | 79.00 | 1 | histone H2B |
| 34 | Urechis caupo | 78.60 | 1 | H2B_URECA Histone H2B gi |
| 35 | Macaca fascicularis | 78.60 | 1 | H2B1K_MACFA Histone H2B type 1-K (H2B K) gi |
| 36 | Caenorhabditis elegans | 77.80 | 6 | HIStone family member (his-48) |
| 37 | Caenorhabditis briggsae AF16 | 77.80 | 3 | Hypothetical protein CBG10818 |
| 38 | Mytilus galloprovincialis | 77.80 | 1 | histone H2B |
| 39 | Cairina moschata | 77.00 | 1 | H2B_CAIMO Histone H2B gi |
| 40 | Bombyx mori | 76.60 | 1 | Histone H2b |
Top-ranked sequences
| organism | matching |
|---|---|
| N/A | Query 87 RSTITSREIQTAVRLLLPGELAKHAVSEGT#KAVTKYTSSK 126 ||||||||||||||||||||||||||||||#|||||||||| Query 87 RSTITSREIQTAVRLLLPGELAKHAVSEGT#KAVTKYTSSK 126 |
| Tetraodon nigroviridis | Query 87 RSTITSREIQTAVRLLLPGELAKHAVSEGT#KAVTKYTSSK 126 ||||||||||||||||||||||||||||||#|||||||||| Query 87 RSTITSREIQTAVRLLLPGELAKHAVSEGT#KAVTKYTSSK 126 |
| Drosophila virilis | Query 87 RSTITSREIQTAVRLLLPGELAKHAVSEGT#KAVTKYTSSK 126 ||||||||||||||||||||||||||||||#|||||||||| Query 87 RSTITSREIQTAVRLLLPGELAKHAVSEGT#KAVTKYTSSK 126 |
| Drosophila hydei | Query 87 RSTITSREIQTAVRLLLPGELAKHAVSEGT#KAVTKYTSSK 126 ||||||||||||||||||||||||||||||#|||||||||| Query 87 RSTITSREIQTAVRLLLPGELAKHAVSEGT#KAVTKYTSSK 126 |
| Drosophila sechellia | Query 87 RSTITSREIQTAVRLLLPGELAKHAVSEGT#KAVTKYTSSK 126 ||||||||||||||||||||||||||||||#|||||||||| Query 87 RSTITSREIQTAVRLLLPGELAKHAVSEGT#KAVTKYTSSK 126 |
| Anopheles gambiae str. PEST | Query 87 RSTITSREIQTAVRLLLPGELAKHAVSEGT#KAVTKYTSSK 126 ||||||||||||||||||||||||||||||#|||||||||| Query 87 RSTITSREIQTAVRLLLPGELAKHAVSEGT#KAVTKYTSSK 126 |
| Homo sapiens | Query 87 RSTITSREIQTAVRLLLPGELAKHAVSEGT#KAVTKYTSSK 126 ||||||||||||||||||||||||||||||#|||||||||| Query 87 RSTITSREIQTAVRLLLPGELAKHAVSEGT#KAVTKYTSSK 126 |
| Mus musculus | Query 87 RSTITSREIQTAVRLLLPGELAKHAVSEGT#KAVTKYTSSK 126 ||||||||||||||||||||||||||||||#|||||||||| Query 87 RSTITSREIQTAVRLLLPGELAKHAVSEGT#KAVTKYTSSK 126 |
| Gallus gallus | Query 87 RSTITSREIQTAVRLLLPGELAKHAVSEGT#KAVTKYTSSK 126 ||||||||||||||||||||||||||||||#|||||||||| Query 87 RSTITSREIQTAVRLLLPGELAKHAVSEGT#KAVTKYTSSK 126 |
| Bos taurus | Query 87 RSTITSREIQTAVRLLLPGELAKHAVSEGT#KAVTKYTSSK 126 ||||||||||||||||||||||||||||||#|||||||||| Query 87 RSTITSREIQTAVRLLLPGELAKHAVSEGT#KAVTKYTSSK 126 |
| Drosophila yakuba | Query 87 RSTITSREIQTAVRLLLPGELAKHAVSEGT#KAVTKYTSSK 126 ||||||||||||||||||||||||||||||#|||||||||| Query 87 RSTITSREIQTAVRLLLPGELAKHAVSEGT#KAVTKYTSSK 126 |
| Drosophila melanogaster | Query 87 RSTITSREIQTAVRLLLPGELAKHAVSEGT#KAVTKYTSSK 126 ||||||||||||||||||||||||||||||#|||||||||| Query 87 RSTITSREIQTAVRLLLPGELAKHAVSEGT#KAVTKYTSSK 126 |
| Platynereis dumerilii | Query 87 RSTITSREIQTAVRLLLPGELAKHAVSEGT#KAVTKYTSSK 126 ||||||||||||||||||||||||||||||#|||||||||| Query 87 RSTITSREIQTAVRLLLPGELAKHAVSEGT#KAVTKYTSSK 126 |
| Chironomus thummi | Query 87 RSTITSREIQTAVRLLLPGELAKHAVSEGT#KAVTKYTSSK 126 ||||||||||||||||||||||||||||||#|||||||||| Query 87 RSTITSREIQTAVRLLLPGELAKHAVSEGT#KAVTKYTSSK 126 |
| Chaetopterus variopedatus | Query 87 RSTITSREIQTAVRLLLPGELAKHAVSEGT#KAVTKYTSSK 126 ||||||||||||||||||||||||||||||#|||||||||| Query 87 RSTITSREIQTAVRLLLPGELAKHAVSEGT#KAVTKYTSSK 126 |
| Rhynchosciara americana | Query 87 RSTITSREIQTAVRLLLPGELAKHAVSEGT#KAVTKYTSSK 126 ||||||||||||||||||||||||||||||#|||||||||| Query 87 RSTITSREIQTAVRLLLPGELAKHAVSEGT#KAVTKYTSSK 126 |
| Tigriopus californicus | Query 87 RSTITSREIQTAVRLLLPGELAKHAVSEGT#KAVTKYTSSK 126 ||||||||||||||||||||||||||||||#|||||||||| Query 87 RSTITSREIQTAVRLLLPGELAKHAVSEGT#KAVTKYTSSK 126 |
| Bos taurus | Query 87 RSTITSREIQTAVRLLLPGELAKHAVSEGT#KAVTKYTSSK 126 ||||||||||||||||||||||||||||||#|||||||||| Query 87 RSTITSREIQTAVRLLLPGELAKHAVSEGT#KAVTKYTSSK 126 |
| Anopheles gambiae | Query 87 RSTITSREIQTAVRLLLPGELAKHAVSEGT#KAVTKYTSSK 126 ||||||||||||||||||||||||||||||#|||||||||| Query 87 RSTITSREIQTAVRLLLPGELAKHAVSEGT#KAVTKYTSSK 126 |
| Homo sapiens | Query 87 RSTITSREIQTAVRLLLPGELAKHAVSEGT#KAVTKYTSSK 126 ||||||||||||||||||||||||||||||#|||||||||| Query 87 RSTITSREIQTAVRLLLPGELAKHAVSEGT#KAVTKYTSSK 126 |
| Canis familiaris | Query 87 RSTITSREIQTAVRLLLPGELAKHAVSEGT#KAVTKYTSSK 126 ||||||||||||||||||||||||||||||#|||||||||| Query 87 RSTITSREIQTAVRLLLPGELAKHAVSEGT#KAVTKYTSSK 126 |
| Danio rerio | Query 87 RSTITSREIQTAVRLLLPGELAKHAVSEGT#KAVTKYTSSK 126 ||||||||||||||||||||||||||||||#|||||||||| Query 87 RSTITSREIQTAVRLLLPGELAKHAVSEGT#KAVTKYTSSK 126 |
| Rattus norvegicus | Query 87 RSTITSREIQTAVRLLLPGELAKHAVSEGT#KAVTKYTSSK 126 ||||||||||||||||||||||||||||||#|||||||||| Query 87 RSTITSREIQTAVRLLLPGELAKHAVSEGT#KAVTKYTSSK 126 |
| Rattus norvegicus | Query 87 RSTITSREIQTAVRLLLPGELAKHAVSEGT#KAVTKYTSSK 126 ||||||||||||||||||||||||||||||#|||||||||| Query 87 RSTITSREIQTAVRLLLPGELAKHAVSEGT#KAVTKYTSSK 126 |
| Mytilus edulis | Query 87 RSTITSREIQTAVRLLLPGELAKHAVSEGT#KAVTKYTSSK 126 ||||||||||||||||||||||||||||||#|||||||||| Query 87 RSTITSREIQTAVRLLLPGELAKHAVSEGT#KAVTKYTSSK 126 |
| Pongo pygmaeus | Query 87 RSTITSREIQTAVRLLLPGELAKHAVSEGT#KAVTKYTSSK 126 ||||||||||||||||||||||||||||||#|||||||||| Query 87 RSTITSREIQTAVRLLLPGELAKHAVSEGT#KAVTKYTSSK 126 |
| Oncorhynchus mykiss | Query 87 RSTITSREIQTAVRLLLPGELAKHAVSEGT#KAVTKYTSSK 126 ||||||||||||||||||||||||||||||#|||||||||| Query 87 RSTITSREIQTAVRLLLPGELAKHAVSEGT#KAVTKYTSSK 126 |
| Mytilus chilensis | Query 87 RSTITSREIQTAVRLLLPGELAKHAVSEGT#KAVTKYTSSK 126 ||||||||||||||||||||||||||||||#|||||||||| Query 87 RSTITSREIQTAVRLLLPGELAKHAVSEGT#KAVTKYTSSK 126 |
| Xenopus laevis | Query 87 RSTITSREIQTAVRLLLPGELAKHAVSEGT#KAVTKYTSSK 126 ||||||||||||||||||||||||||||||#||||||||+| Sbjct 87 RSTITSREIQTAVRLLLPGELAKHAVSEGT#KAVTKYTSAK 126 |
| Xenopus tropicalis | Query 87 RSTITSREIQTAVRLLLPGELAKHAVSEGT#KAVTKYTSSK 126 ||||||||||||||||||||||||||||||#||||||||+| Sbjct 87 RSTITSREIQTAVRLLLPGELAKHAVSEGT#KAVTKYTSAK 126 |
| Rhacophorus schlegelii | Query 87 RSTITSREIQTAVRLLLPGELAKHAVSEGT#KAVTKYTSSK 126 ||||||||||||||||||||||||||||||#||||||||+| Sbjct 87 RSTITSREIQTAVRLLLPGELAKHAVSEGT#KAVTKYTSAK 126 |
| Expression vector pET3-H2B | Query 87 RSTITSREIQTAVRLLLPGELAKHAVSEGT#KAVTKYTSSK 126 ||||||||||||||||||||||||||||||#||||||||+| Sbjct 84 RSTITSREIQTAVRLLLPGELAKHAVSEGT#KAVTKYTSAK 123 |
| Mytilus californianus | Query 87 RSTITSREIQTAVRLLLPGELAKHAVSEGT#KAVTKYTSSK 126 ||||||||+|||||||||||||||||||||#|||||||||| Sbjct 85 RSTITSREVQTAVRLLLPGELAKHAVSEGT#KAVTKYTSSK 124 |
| Urechis caupo | Query 87 RSTITSREIQTAVRLLLPGELAKHAVSEGT#KAVTKYTSSK 126 ||||||||||||||||||||||||||||||#||||||| || Sbjct 84 RSTITSREIQTAVRLLLPGELAKHAVSEGT#KAVTKYTQSK 123 |
| Macaca fascicularis | Query 87 RSTITSREIQTAVRLLLPGELAKHAVSEGT#KAVTKYTSSK 126 ||||||||||||||||||||||||||||||#||||||||+| Sbjct 87 RSTITSREIQTAVRLLLPGELAKHAVSEGT#KAVTKYTSAK 126 |
| Caenorhabditis elegans | Query 87 RSTITSREIQTAVRLLLPGELAKHAVSEGT#KAVTKYTSSK 126 ||||+||||||||||+||||||||||||||#|||||||||| Sbjct 84 RSTISSREIQTAVRLILPGELAKHAVSEGT#KAVTKYTSSK 123 |
| Caenorhabditis briggsae AF16 | Query 87 RSTITSREIQTAVRLLLPGELAKHAVSEGT#KAVTKYTSSK 126 ||||+||||||||||+||||||||||||||#|||||||||| Sbjct 84 RSTISSREIQTAVRLILPGELAKHAVSEGT#KAVTKYTSSK 123 |
| Mytilus galloprovincialis | Query 87 RSTITSREIQTAVRLLLPGELAKHAVSEGT#KAVTKYTSSK 126 ||||||||||||||||||||||||||||||#+||||||||| Sbjct 85 RSTITSREIQTAVRLLLPGELAKHAVSEGT#EAVTKYTSSK 124 |
| Cairina moschata | Query 87 RSTITSREIQTAVRLLLPGELAKHAVSEGT#KAVTKYTSSK 126 |||||||||||||| |||||||||||||||#|||||||||| Sbjct 87 RSTITSREIQTAVRSLLPGELAKHAVSEGT#KAVTKYTSSK 126 |
| Bombyx mori | Query 87 RSTITSREIQTAVRLLLPGELAKHAVSEGT#KAVTKYTSSK 126 ||||||||+||+||||||||||||||||||#|+|||||||| Sbjct 24 RSTITSREVQTSVRLLLPGELAKHAVSEGT#KSVTKYTSSK 63 |
* References
[PubMed ID: 12140684] Sonstegard TS, Capuco AV, White J, Van Tassell CP, Connor EE, Cho J, Sultana R, Shade L, Wray JE, Wells KD, Quackenbush J, Analysis of bovine mammary gland EST and functional annotation of the Bos taurus gene index. Mamm Genome. 2002 Jul;13(7):373-9.
[PubMed ID: 16305752] ... Harhay GP, Sonstegard TS, Keele JW, Heaton MP, Clawson ML, Snelling WM, Wiedmann RT, Van Tassell CP, Smith TP, Characterization of 954 bovine full-CDS cDNA sequences. BMC Genomics. 2005 Nov 23;6:166.