SB0020 : CAPN1, [mu]-calpain catalytic subunit, [mu]CL
[ CaMP Format ]
* Basic Information
| Organism | Homo sapiens (human) |
| Protein Names | calpain-1 catalytic subunit [Homo sapiens]; calpain-1 catalytic subunit; calpain, large polypeptide L1; cell proliferation-inducing protein 30; CANP 1; calpain mu-type; micromolar-calpain; calpain-1 large subunit; calcium-activated neutral proteinase 1; cell proliferation-inducing gene 30 protein; Calpain-1 catalytic subunit; 3.4.22.52; Calcium-activated neutral proteinase 1; Calpain mu-type; Calpain-1 large subunit; Cell proliferation-inducing gene 30 protein; Micromolar-calpain; muCANP |
| Gene Names | CAPN1; PIG30; CANPL1; calpain 1, (mu/I) large subunit |
| Gene Locus | 11q13; chromosome 11 |
| GO Function | Not available |
* Information From OMIM
Description: Calpain (calcium-dependent protease; EC 3.4.22.17) is an intracellular protease that requires calcium for its catalytic activity. Two isozymes, calpain I (mu-calpain) and calpain II (m-calpain), with different calcium requirements, have been identified. Both are heterodimers composed of L (large, catalytic, 80 kD) and S (small, regulatory, 30 kD) subunits. The isozymes share an identical S subunit (CAPNS1; OMIM:114170), with the differences arising from the L subunits, L1 (CAPN1) and L2 (CAPN2; OMIM:114230) (summary by Ohno et al., 1990).
* Structure Information
1. Primary Information
Length: 714 aa
Average Mass: 81.889 kDa
Monoisotopic Mass: 81.838 kDa
2. Domain Information
Annotated Domains: interpro / pfam / smart / prosite
Computationally Assigned Domains (Pfam+HMMER):
| domain name | begin | end | score | e-value |
|---|---|---|---|---|
| --- cleavage 15 --- | ||||
| --- cleavage 27 --- | ||||
| Calpain large subunit, domain III 1. | 87 | 100 | 22.0 | 0.0 |
| Calpain family cysteine protease 1. | 496 | 532 | 74.0 | 0.0 |
| EF-hand domain pair 1. | 536 | 548 | 21.0 | 0.0 |
| EF-hand domain pair 2. | 561 | 611 | 16.0 | 0.1 |
| EF-hand domain pair 3. | 622 | 634 | 30.0 | 0.0 |
| EF-hand domain pair 4. | 686 | 703 | 29.0 | 0.6 |
3. Sequence Information
Fasta Sequence: SB0020.fasta
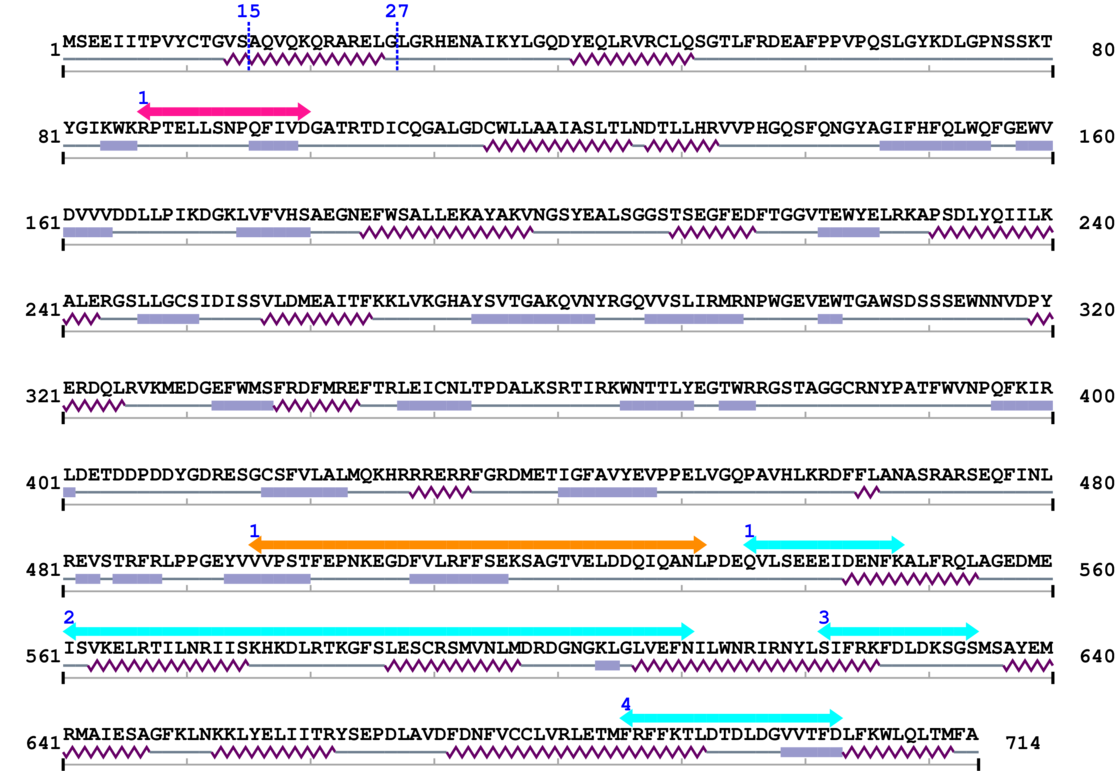
Amino Acid Sequence and Secondary Structures (PsiPred):
* Cleavage Information
2 [sites] cleaved by Calpain 1
Source Reference: [PubMed ID: 2065086] Zimmerman UJ, Schlaepfer WW, Two-stage autolysis of the catalytic subunit initiates activation of calpain I. Biochim Biophys Acta. 1991 Jun 24;1078(2):192-8.
Cleavage sites (±10aa)
[Site 1] ITPVYCTGVS15-AQVQKQRARE
Ser15  Ala
Ala
iTraq-117 Signal 83088.7 (


 ) for AQVQKQRARE
) for AQVQKQRARE
iTraq-117 Signal 2676.5 (

 ) for QVQKQRARE
) for QVQKQRARE
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Ile6 | Thr7 | Pro8 | Val9 | Tyr10 | Cys11 | Thr12 | Gly13 | Val14 | Ser15 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Ala16 | Gln17 | Val18 | Gln19 | Lys20 | Gln21 | Arg22 | Ala23 | Arg24 | Glu25 |
 Sequence conservation (by blast)
Sequence conservation (by blast)
[Site 2] VQKQRARELG27-LGRHENAIKY
Gly27  Leu
Leu
iTraq-117 Signal 11038.2 (


 ) for VQKQRARELG
) for VQKQRARELG
iTraq-117 Signal 151277.7 (



 ) for LGRHENAIKY
) for LGRHENAIKY
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Val18 | Gln19 | Lys20 | Gln21 | Arg22 | Ala23 | Arg24 | Glu25 | Leu26 | Gly27 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Leu28 | Gly29 | Arg30 | His31 | Glu32 | Asn33 | Ala34 | Ile35 | Lys36 | Tyr37 |
 Sequence conservation (by blast)
Sequence conservation (by blast)
* References
[PubMed ID: 24416790] Kim H, Kang AY, Ko AR, Park HC, So I, Park JH, Cheong HI, Hwang YH, Ahn C, Calpain-mediated proteolysis of polycystin-1 C-terminus induces JAK2 and ERK signal alterations. Exp Cell Res. 2014 Jan 1;320(1):62-8.