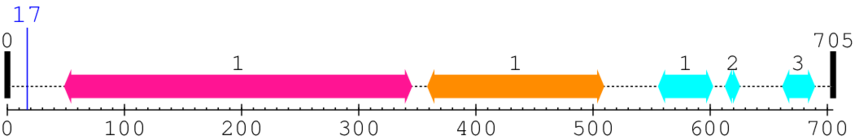
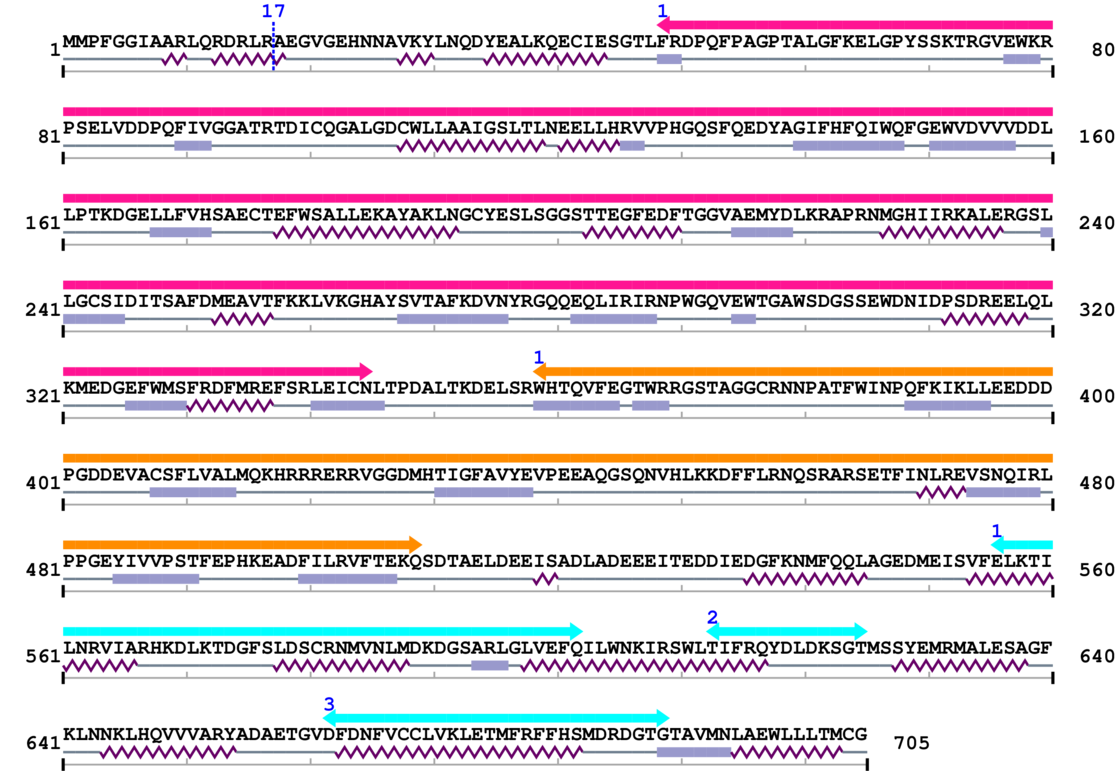
* Cleavage Information
1 [sites]
Source Reference: [PubMed ID: 2829834] Crawford C, Willis AC, Gagnon J, The effects of autolysis on the structure of chicken calpain II. Biochem J. 1987 Dec 1;248(2):579-88.
Cleavage sites (±10aa)
[Site 1] AARLQRDRLR17-AEGVGEHNNA
Arg17  Ala
Ala
 |
| P10 | P9 | P8 | P7 | P6 |
P5 | P4 | P3 | P2 | P1 |
| Ala8 | Ala9 | Arg10 | Leu11 | Gln12 | Arg13 | Asp14 | Arg15 | Leu16 | Arg17 |
 |
| P1' | P2' | P3' | P4' | P5' |
P6' | P7' | P8' | P9' | P10' |
| Ala18 | Glu19 | Gly20 | Val21 | Gly22 | Glu23 | His24 | Asn25 | Asn26 | Ala27 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
|
MMPFGGIAARLQRDRLRAEGVGEHNNAVKYLNQDYEALKQECIESGT
|
Summary
| # |
organism |
max score |
hits |
top seq |
| 1 |
Gallus gallus |
102.00 |
2 |
calpain 1, (mu/I) large subunit |
| 2 |
Coturnix coturnix |
98.20 |
1 |
quail calpain |
| 3 |
Xenopus laevis |
77.00 |
4 |
hypothetical protein LOC398772 |
| 4 |
Xenopus laevis |
76.60 |
1 |
mu/m-calpain large subunit |
| 5 |
Xenopus tropicalis |
72.80 |
4 |
CAPN2 protein |
| 6 |
N/A |
66.20 |
12 |
neutral protease large subunit |
| 7 |
Homo sapiens |
66.20 |
6 |
AF261089_1 calpain large polypeptide L2 |
| 8 |
Homo sapiens |
66.20 |
2 |
calpain 2, large subunit |
| 9 |
Macaca mulatta |
65.90 |
2 |
PREDICTED: calpain 2, large subunit |
| 10 |
Danio rerio |
62.00 |
6 |
PREDICTED: similar to Calpain 1, (mu/I) large sub |
| 11 |
Rattus norvegicus |
60.80 |
10 |
calpain 2 |
| 12 |
Mus musculus |
60.50 |
15 |
m-calpain large subunit |
| 13 |
Tetraodon nigroviridis |
59.30 |
7 |
unnamed protein product |
| 14 |
synthetic construct |
56.60 |
1 |
Homo sapiens calpain 1, (mu/I) large subunit |
| 15 |
Pongo pygmaeus |
56.60 |
1 |
CAN1_PONPY Calpain-1 catalytic subunit (Calpain-1 |
| 16 |
Stizostedion vitreum vitreum |
56.20 |
1 |
putative calpain-like protein |
| 17 |
Gallus gallus |
53.10 |
1 |
mu-calpain large subunit |
| 18 |
Sus scrofa |
52.40 |
2 |
calpain 1, large subunit |
| 19 |
Canis familiaris |
52.00 |
3 |
PREDICTED: similar to Calpain-1 catalytic subunit |
| 20 |
Bos taurus |
51.20 |
3 |
calpain 1, (mu/I) large subunit |
| 21 |
Rattus norvegicus |
51.20 |
1 |
calpain 1, large subunit |
| 22 |
Oncorhynchus mykiss |
49.30 |
1 |
m-calpain |
| 23 |
Canis familiaris |
45.80 |
1 |
PREDICTED: similar to Calpain-11 (Calcium-activat |
| 24 |
Danio rerio |
42.00 |
1 |
calpain 1, large subunit |
| 25 |
Oryctolagus cuniculus |
31.60 |
1 |
calpain 3, (p94) |
| 26 |
Shewanella denitrificans OS217 |
31.20 |
1 |
Phosphoenolpyruvate-protein phosphotransferase |
Top-ranked sequences
| organism | matching |
|---|
| Gallus gallus |
Query 1 MMPFGGIAARLQRDRLR#AEGVGEHNNAVKYLNQDYEALKQECIESGT 47
|||||||||||||||||#||||||||||||||||||||||||||||||
Query 1 MMPFGGIAARLQRDRLR#AEGVGEHNNAVKYLNQDYEALKQECIESGT 47
|
| Coturnix coturnix |
Query 1 MMPFGGIAARLQRDRLR#AEGVGEHNNAVKYLNQDYEALKQECIESG 46
|||||||||||||||||#||||||||||||||||||||||| |||||
Sbjct 1 MMPFGGIAARLQRDRLR#AEGVGEHNNAVKYLNQDYEALKQACIESG 46
|
| Xenopus laevis |
Query 1 MMPFGGIAARLQRDRLR#AEGVGEHNNAVKYLNQDYEALKQECIESG 46
||||||||++|+ |||+#| ||| |+ |+|| |||||+|||+|+|||
Sbjct 1 MMPFGGIASKLKIDRLK#ASGVGSHDCAIKYQNQDYESLKQQCLESG 46
|
| Xenopus laevis |
Query 1 MMPFGGIAARLQRDRLR#AEGVGEHNNAVKYLNQDYEALKQECIESG 46
|+||||||++|+ |||+#| ||| |+ |||| |||||+|||+|+|||
Sbjct 1 MIPFGGIASKLKIDRLK#ASGVGSHDCAVKYQNQDYESLKQQCVESG 46
|
| Xenopus tropicalis |
Query 1 MMPFGGIAARLQRDRLR#AEGVGEHNNAVKYLNQDYEALKQECIESGT 47
||||||||++|+ +||+#| ||| |+ ||+| |||||+|||+| |||+
Sbjct 1 MMPFGGIASKLKIERLK#ASGVGSHDCAVRYQNQDYESLKQQCRESGS 47
|
| N/A |
Query 5 GGIAARLQRDRLR#AEGVGEHNNAVKYLNQDYEALKQECIESGT 47
||||+| +|| #|||+| | |+||||||||||+ ||+|+||
Sbjct 2 AGIAAKLAKDREA#AEGLGSHERAIKYLNQDYEALRNECLEAGT 44
|
| Homo sapiens |
Query 5 GGIAARLQRDRLR#AEGVGEHNNAVKYLNQDYEALKQECIESGT 47
||||+| +|| #|||+| | |+||||||||||+ ||+|+||
Sbjct 2 AGIAAKLAKDREA#AEGLGSHERAIKYLNQDYEALRNECLEAGT 44
|
| Homo sapiens |
Query 5 GGIAARLQRDRLR#AEGVGEHNNAVKYLNQDYEALKQECIESGT 47
||||+| +|| #|||+| | |+||||||||||+ ||+|+||
Sbjct 2 AGIAAKLAKDREA#AEGLGSHERAIKYLNQDYEALRNECLEAGT 44
|
| Macaca mulatta |
Query 5 GGIAARLQRDRLR#AEGVGEHNNAVKYLNQDYEALKQECIESGT 47
||||+| +|| #|||+| | |+||||||||||+ ||+|+||
Sbjct 2 AGIAAKLVKDREA#AEGLGSHERAIKYLNQDYEALRNECLEAGT 44
|
| Danio rerio |
Query 1 MMPFGGIAARLQRDRLR#AEGVGEHNNAVKYLNQDYEALKQECIESG 46
| ||++ |+ ++||+#|||+| ++ |+|+|||||| |++||+|||
Sbjct 1 MFSIGGVSTRIYKERLQ#AEGMGANDLAIKFLNQDYEELRRECVESG 46
|
| Rattus norvegicus |
Query 5 GGIAARLQRDRLR#AEGVGEHNNAVKYLNQDYEALKQECIESG 46
||| +| +|| #|||+| | |+|||||||| |+ ||+|+|
Sbjct 2 AGIAMKLAKDREA#AEGLGSHERAIKYLNQDYETLRNECLEAG 43
|
| Mus musculus |
Query 5 GGIAARLQRDRLR#AEGVGEHNNAVKYLNQDYEALKQECIESG 46
||| +| +|| #|||+| | |+|||||||| |+ ||+|+|
Sbjct 2 AGIAIKLAKDREA#AEGLGSHERAIKYLNQDYETLRNECLEAG 43
|
| Tetraodon nigroviridis |
Query 6 GIAARLQRDRLR#AEGVGEHNNAVKYLNQDYEALKQECIESG 46
||+||+ +|||#|||+| + || + +||||||||+|+|||
Sbjct 1 GISARIYAERLR#AEGMGSNQQAVPFCSQDYEALKQQCVESG 41
|
| synthetic construct |
Query 6 GIAARLQRDRLR#AEGVGEHNNAVKYLNQDYEALKQECIESGT 47
|++|++|+ | |# |+| | ||+||| |||| |+ |++|||
Sbjct 13 GVSAQVQKQRAR#ELGLGRHENAIKYLGQDYEQLRVRCLQSGT 54
|
| Pongo pygmaeus |
Query 6 GIAARLQRDRLR#AEGVGEHNNAVKYLNQDYEALKQECIESGT 47
|++|++|+ | |# |+| | ||+||| |||| |+ |++|||
Sbjct 13 GVSAQVQKQRAR#ELGLGRHENAIKYLGQDYEQLRVRCLQSGT 54
|
| Stizostedion vitreum vitreum |
Query 6 GIAARLQRDRLR#AEGVGEHNNAVKYLNQDYEALKQECIES 45
|||| ||| | +# +|+| + ||||||||||||++ |+||
Sbjct 3 GIAATLQRRREK#EQGIGSCSQAVKYLNQDYEALRRRCLES 42
|
| Gallus gallus |
Query 6 GIAARLQRDRLR#AEGVGEHNNAVKYLNQDYEALKQECIESGT 47
|++|++|| | +#| |+|+| |||++ ||| ||+ +|+ ||+
Sbjct 13 GVSAQVQRQRAK#ALGLGQHQNAVRFRGQDYAALRDDCLRSGS 54
|
| Sus scrofa |
Query 6 GIAARLQRDRLR#AEGVGEHNNAVKYLNQDYEALKQECIESGT 47
|++|++|+ | +# |+| | ||+||| |||| |+ |++||+
Sbjct 13 GVSAQVQKLRAK#ELGLGRHENAIKYLGQDYEQLRAHCLQSGS 54
|
| Canis familiaris |
Query 6 GIAARLQRDRLR#AEGVGEHNNAVKYLNQDYEALKQECIESG 46
|++|++|+ | +# |+| | ||+||+ ||+| |+ |++||
Sbjct 13 GVSAQVQKQRAK#ELGLGRHENAIKYMGQDFEQLRAHCLQSG 53
|
| Bos taurus |
Query 6 GIAARLQRDRLR#AEGVGEHNNAVKYLNQDYEALKQECIESG 46
|++|++|+ | +# |+| | ||+||| |||| |+ |++ |
Sbjct 13 GVSAQVQKQRAK#ELGLGRHENAIKYLGQDYEQLRVHCLQRG 53
|
| Rattus norvegicus |
Query 6 GIAARLQRDRLR#AEGVGEHNNAVKYLNQDYEALKQECIESG 46
|++|++|+ | +# |+| | ||+||| |||| |+ |+++|
Sbjct 13 GVSAQVQKQRDK#ELGLGRHENAIKYLGQDYENLRARCLQNG 53
|
| Oncorhynchus mykiss |
Query 6 GIAARLQRDRLR#AEGVGEHNNAVKYLNQDYEALKQECIESG 46
|+|+ | + | #| | | + |||||||||+|||+ +|+ |
Sbjct 3 GMASNLAKKRAL#AAGFGTNANAVKYLNQDFEALRAQCLNRG 43
|
| Canis familiaris |
Query 6 GIAARLQRDRLR#AEGVGEHNNAVKYLNQDYEALKQECIESG 46
|+|| + ||+#|+|+|+|+|| | ||++| |+ |+ |
Sbjct 85 GMAAHISHSRLK#AKGLGQHHNAQNYNNQNFEDLQAACLRKG 125
|
| Danio rerio |
Query 6 GIAARLQRDRLR#AEGVGEHNNAVKYLNQDYEALKQECIES 45
|+||||+ |# |+|+++||||+| |||| |+ + +|
Sbjct 8 GMAARLRSQWDR#DAGLGQNHNAVKFLGQDYETLRAQSQQS 47
|
| Oryctolagus cuniculus |
Query 2 MPF---GGIAARLQRDRLR#AEGVGEHNNAVKYLNQDYEALKQECI 43
||+ | |+ |+| |# | | +|| ||+ |+| |+
Sbjct 1 MPYLLSGFFCDRVIRERDR#RNGEGTVTQPIKYEGQDFVVLRQRCL 45
|
| Shewanella denitrificans OS217 |
Query 1 MMPFGGIAARLQRDRLR#AEGVGEHNNAVKYLNQDYEALKQEC 42
|+ ||+ +|| # + | + || || | |+ || ||
Sbjct 571 MIQAGGVGTQLQILLPM#VSNLDEIDQAVGYLEQAYQELKSEC 612
|
* References
[PubMed ID: 18727211] Zhang ZR, Liu YP, Jiang X, Du HR, Zhu Q, Study on association of single nucleotide polymorphism of CAPN1 gene with muscle fibre and carcass traits in quality chicken populations. J Anim Breed Genet. 2008 Aug;125(4):258-64.
» More (5) « Hide Below
[PubMed ID: 17681928] ... Zhang ZR, Zhu Q, Jiang XS, Du HR, [Study on correlation between single nucleotide polymorphism of CAPN1 gene and muscle tenderness and carcass traits in chicken]. Yi Chuan. 2007 Aug;29(8):982-8.
[PubMed ID: 7742367] ... Sorimachi H, Tsukahara T, Okada-Ban M, Sugita H, Ishiura S, Suzuki K, Identification of a third ubiquitous calpain species--chicken muscle expresses four distinct calpains. Biochim Biophys Acta. 1995 Apr 26;1261(3):381-93.
[PubMed ID: 3038855] ... Minami Y, Emori Y, Kawasaki H, Suzuki K, E-F hand structure-domain of calcium-activated neutral protease (CANP) can bind Ca2+ ions. J Biochem. 1987 Apr;101(4):889-95.
[PubMed ID: 3000828] ... Emori Y, Ohno S, Tobita M, Suzuki K, Gene structure of calcium-dependent protease retains the ancestral organization of the calcium-binding protein gene. FEBS Lett. 1986 Jan 6;194(2):249-52.
[PubMed ID: 6095110] ... Ohno S, Emori Y, Imajoh S, Kawasaki H, Kisaragi M, Suzuki K, Evolutionary origin of a calcium-dependent protease by fusion of genes for a thiol protease and a calcium-binding protein? Nature. 1984 Dec 6-12;312(5994):566-70.
 Ala
Ala

 Sequence conservation (by blast)
Sequence conservation (by blast)