SB0022 : Cortactin
[ CaMP Format ]
* Basic Information
| Organism | Homo sapiens (human) |
| Protein Names | src substrate cortactin isoform a [Homo sapiens]; src substrate cortactin isoform a; oncogene EMS1; ems1 sequence (mammary tumor and squamous cell carcinoma-associated (p80/85 src substrate); 1110020L01Rik; amplaxin; Src substrate cortactin; Amplaxin; Oncogene EMS1 |
| Gene Names | CTTN; EMS1; cortactin |
| Gene Locus | 11q13; chromosome 11 |
| GO Function | Not available |
* Information From OMIM
Description: Cortactin is a multidomain protein that functions as a key regulator of the actin cytoskeleton. It has roles in many actin-based cellular processes, including cell migration and invasion, tumor cell metastasis, and endocytosis (summary by Tegtmeyer et al., 2011).
Function: Human cortactin is overexpressed in carcinoma cells with an amplification of 11q13 and is found in 2 forms, designated p80 and p85. Van Damme et al. (1997) found that in carcinoma cells with the 11q13 amplification, p85 was produced from p80 by posttranslational modification. Also, treatment of these cells with epidermal growth factor (OMIM:131530) or vanadate caused conversion of p80 to p85 and enhanced phosphorylation of the p85 form. Both overexpression and posttranslational modification of cortactin coincided with its redistribution from the cytoplasm to cell-matrix contact sites, implying a role for cortactin in the modulation of cellular adhesive properties.
* Structure Information
1. Primary Information
Length: 550 aa
Average Mass: 61.585 kDa
Monoisotopic Mass: 61.549 kDa
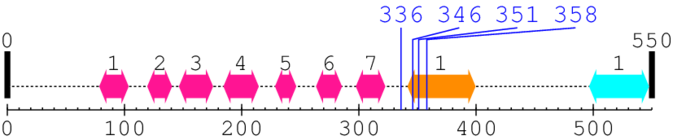
2. Domain Information
Annotated Domains: interpro / pfam / smart / prosite
Computationally Assigned Domains (Pfam+HMMER):
| domain name | begin | end | score | e-value |
|---|---|---|---|---|
| TMEM210 family 1. | 79 | 103 | 50.0 | 0.0 |
| TMEM210 family 2. | 120 | 140 | 54.0 | 0.1 |
| TMEM210 family 3. | 147 | 175 | 44.0 | 0.0 |
| TMEM210 family 4. | 185 | 214 | 45.0 | 0.0 |
| TMEM210 family 5. | 229 | 246 | 52.0 | 0.0 |
| TMEM210 family 6. | 264 | 285 | 50.0 | 0.0 |
| TMEM210 family 7. | 298 | 322 | 47.0 | 0.0 |
| --- cleavage 336 --- | ||||
| Protein of unknown function (DUF1682) 1. | 342 | 399 | 251.0 | 15.6 |
| --- cleavage 346 (inside Protein of unknown function (DUF1682) 342..399) --- | ||||
| --- cleavage 351 (inside Protein of unknown function (DUF1682) 342..399) --- | ||||
| --- cleavage 358 (inside Protein of unknown function (DUF1682) 342..399) --- | ||||
| Variant SH3 domain 1. | 497 | 547 | 2.0 | 0.0 |
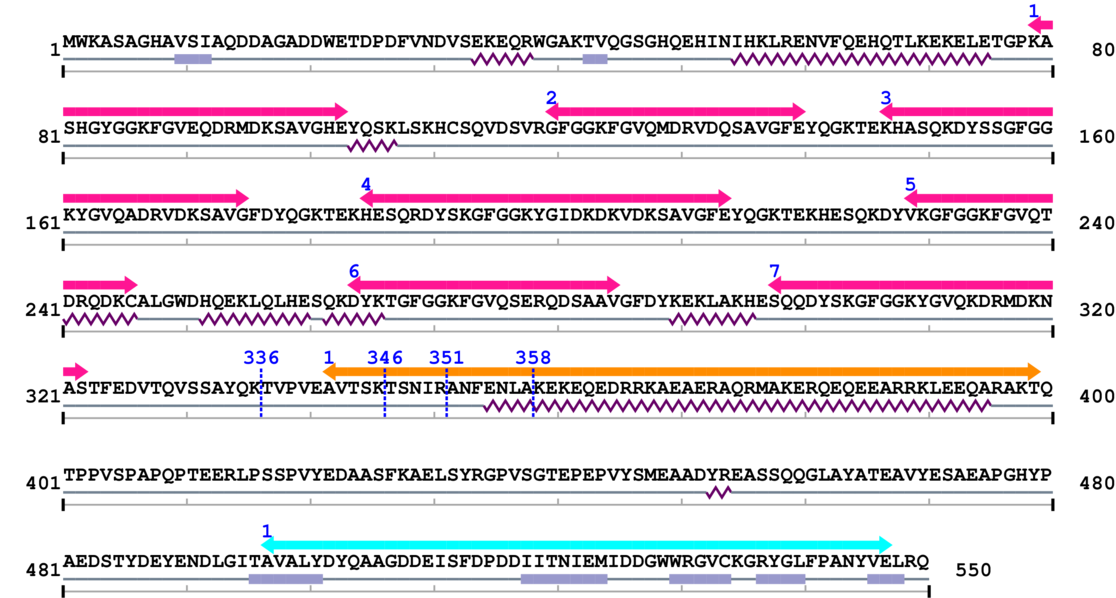
3. Sequence Information
Fasta Sequence: SB0022.fasta
Amino Acid Sequence and Secondary Structures (PsiPred):
* Cleavage Information
4 [sites] cleaved by Calpain 2
Source Reference: [PubMed ID: 16280362] Perrin BJ, Amann KJ, Huttenlocher A, Proteolysis of cortactin by calpain regulates membrane protrusion during cell migration. Mol Biol Cell. 2006 Jan;17(1):239-50. Epub 2005 Nov 9.
Cleavage sites (±10aa)
[Site 1] VTQVSSAYQK336-TVPVEAVTSK
Lys336  Thr
Thr
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Val327 | Thr328 | Gln329 | Val330 | Ser331 | Ser332 | Ala333 | Tyr334 | Gln335 | Lys336 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Thr337 | Val338 | Pro339 | Val340 | Glu341 | Ala342 | Val343 | Thr344 | Ser345 | Lys346 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| GGKYGVQKDRMDKNASTFEDVTQVSSAYQKTVPVEAVTSKTSNIRANFENLAKEKEQEDR |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | Tetraodon nigroviridis | 74.70 | 3 | unnamed protein product |
| 2 | Bos taurus | 32.00 | 1 | hematopoietic cell-specific Lyn substrate 1 |
| 3 | Rattus norvegicus | 31.60 | 3 | cortactin isoform C |
| 4 | Drosophila pseudoobscura | 31.60 | 1 | GA17577-PA |
| 5 | Homo sapiens | 31.20 | 5 | HCLS1_HUMAN Hematopoietic lineage cell-specific p |
| 6 | N/A | 31.20 | 2 | 2006363A protein SPY75 |
| 7 | synthetic construct | 31.20 | 2 | Homo sapiens hematopoietic cell-specific Lyn subs |
| 8 | Gallus gallus | 31.20 | 1 | cortactin |
| 9 | Schistosoma japonicum | 31.20 | 1 | SJCHGC00827 protein |
| 10 | Drosophila melanogaster | 31.20 | 1 | Cortactin CG3637-PA |
| 11 | Strongylocentrotus purpuratus | 31.20 | 1 | SRC8 protein |
| 12 | Xenopus laevis | 31.20 | 1 | cortactin |
| 13 | Drosophila melanogaster | 31.20 | 1 | Cortactin |
| 14 | Mus musculus | 30.80 | 6 | cortactin |
| 15 | Canis familiaris | 30.80 | 5 | PREDICTED: similar to cortactin isoform a isoform |
| 16 | Pongo pygmaeus | 30.80 | 1 | hypothetical protein |
| 17 | Suberites domuncula | 30.40 | 1 | cortactin |
| 18 | Danio rerio | 30.40 | 1 | cortactin |
| 19 | Plasmodium yoelii yoelii str. 17XNL | 30.40 | 1 | hypothetical protein PY01165 |
| 20 | mice, BALB/c 3T3 cells, Peptide, 546 aa | 30.40 | 1 | cortactin, p80/p85 |
Top-ranked sequences
| organism | matching |
|---|---|
| Tetraodon nigroviridis | Query 307 GGKYGVQKDRMDKNASTFEDVTQVSSAYQK#TVPVEAVTSKTSNIRANFENLAKEK 361 |||+||||||||| | |||+| + + +|||#| |||| | +|+| |||+||+| Sbjct 10 GGKFGVQKDRMDKAAGTFEEVEKPTPSYQK#TKPVEAAGSNAGSIKARFENMAKQK 64 |
| Bos taurus | Query 307 GGKYGVQKDRMDKNASTFEDVTQVSSAYQK#TVPVEAVTSKTSNIRANFENLA 358 ||+||+||||+||+| | ++ ++||+|#| |+|| +| ++| ||++| Sbjct 195 GGQYGIQKDRVDKSAVGFNEMEAPTTAYKK#TTPIEAASSGARGLKAKFESMA 246 Query 307 GGKYGVQKDRMDKNASTFEDVTQV 330 ||++||+||||||+| | | +| Sbjct 84 GGRFGVEKDRMDKSAVGHEYVAEV 107 Query 307 GGKYGVQKDRMDKNASTFE 325 ||||||+||| ||+| |+ Sbjct 121 GGKYGVEKDRADKSAVGFD 139 |
| Rattus norvegicus | Query 307 GGKYGVQKDRMDKNASTFEDVTQVSSAYQK#TVPVEAVTSKTSNIRANFENLAKEKEQEDR 366 |||+|||||||||||||||+| || |||||#|||+|||||||||||||||+||||+||||| Sbjct 233 GGKFGVQKDRMDKNASTFEEVVQVPSAYQK#TVPIEAVTSKTSNIRANFESLAKEREQEDR 292 Query 307 GGKYGVQKDRMDKNASTFE 325 ||||||| ||+||+| |+ Sbjct 159 GGKYGVQADRVDKSAVGFD 177 |
| Drosophila pseudoobscura | Query 307 GGKYGVQKDRMDKNASTFED----VTQVSSAYQK#TVPVEAVTSKTSNIRANFENLAK 359 |||+|||+|| ||+| ++ | |# || +| ||+|| |||||| Sbjct 195 GGKFGVQEDRKDKSAVGWDHKEAPQKHASQVDHK#VKPV-IEGAKPSNLRAKFENLAK 250 Query 307 GGKYGVQKDRMDKNA 321 |||+||+||||||+| Sbjct 84 GGKFGVEKDRMDKSA 98 |
| Homo sapiens | Query 307 GGKYGVQKDRMDKNASTFEDVTQVSSAYQK#TVPVEAVTSKTSNIRANFENLA 358 ||+||+||||+||+| | ++ ++||+|#| |+|| +| ++| ||++| Sbjct 195 GGQYGIQKDRVDKSAVGFNEMEAPTTAYKK#TTPIEAASSGARGLKAKFESMA 246 Query 307 GGKYGVQKDRMDKNASTFEDVTQV 330 ||++||++|||||+| | | +| Sbjct 84 GGRFGVERDRMDKSAVGHEYVAEV 107 Query 307 GGKYGVQKDRMDKNASTFE 325 ||||||++|| ||+| |+ Sbjct 121 GGKYGVERDRADKSAVGFD 139 |
| N/A | Query 307 GGKYGVQKDRMDKNASTFEDVTQVSSAYQK#TVPVEAVTSKTSNIRANFENLA 358 ||+||+||||+||+| | ++ ++||+|#| |+|| +| ++| ||++| Sbjct 195 GGQYGIQKDRVDKSAVGFNEMEAPTTAYKK#TTPIEAASSGARGLKAKFESMA 246 Query 307 GGKYGVQKDRMDKNASTFEDVTQV 330 ||++||++|||||+| | | +| Sbjct 84 GGRFGVERDRMDKSAVGHEYVAEV 107 Query 307 GGKYGVQKDRMDKNASTFE 325 ||||||++|| ||+| |+ Sbjct 121 GGKYGVERDRADKSAVGFD 139 |
| synthetic construct | Query 307 GGKYGVQKDRMDKNASTFEDVTQVSSAYQK#TVPVEAVTSKTSNIRANFENLA 358 ||+||+||||+||+| | ++ ++||+|#| |+|| +| ++| ||++| Sbjct 195 GGQYGIQKDRVDKSAVGFNEMEAPTTAYKK#TTPIEAASSGARGLKAKFESMA 246 Query 307 GGKYGVQKDRMDKNASTFEDVTQV 330 ||++||++|||||+| | | +| Sbjct 84 GGRFGVERDRMDKSAVGHEYVAEV 107 Query 307 GGKYGVQKDRMDKNASTFE 325 ||||||++|| ||+| |+ Sbjct 121 GGKYGVERDRADKSAVGFD 139 |
| Gallus gallus | Query 307 GGKYGVQKDRMDKNASTFEDVTQVSSAYQK#TVPVEAVTSKTSNIRANFENLAKEKEQEDR 366 |||||||||||||||+||||+ + +| |||#| ||| | +|||+|||| |||||||||||| Sbjct 316 GGKYGVQKDRMDKNAATFEDIEKPTSTYQK#TKPVERVANKTSSIRANLENLAKEKEQEDR 375 Query 307 GGKYGVQKDRMDKNASTFE 325 ||||||| ||+||+| |+ Sbjct 168 GGKYGVQADRVDKSAVGFD 186 Query 307 GGKYGVQKDRMDKNASTFE 325 |||||| ||++||+| || Sbjct 205 GGKYGVDKDKVDKSAVGFE 223 |
| Schistosoma japonicum | Query 307 GGKYGVQKDRMDKNASTFEDV 327 |||+||+||||||+| + + Sbjct 71 GGKFGVEKDRMDKSAVDWSHI 91 |
| Drosophila melanogaster | Query 307 GGKYGVQKDRMDKNASTFE----DVTQVSSAYQK#TVPVEAVTSKTSNIRANFENLAKEKE 362 |||+|||+|| ||+| ++ | |# || +| ||+|| |||||| | Sbjct 197 GGKFGVQEDRKDKSAVGWDHKEAPQKHASQVDHK#VKPV-IEGAKPSNLRAKFENLAKNSE 255 Query 363 QEDR 366 +| | Sbjct 256 EESR 259 Query 307 GGKYGVQKDRMDKNA 321 |||+||+||||||+| Sbjct 86 GGKFGVEKDRMDKSA 100 Query 307 GGKYGVQKDRMDKNASTFEDVTQV 330 |||+|||+|| ||+| ++ | +| Sbjct 123 GGKFGVQEDRKDKSAVGWDHVEKV 146 |
| Strongylocentrotus purpuratus | Query 307 GGKYGVQKDRMDKNASTFEDVTQVSSAYQK#TVPVEAVTSKTSNIRANFENLAKEKEQEDR 366 |||+|| |+ | | | |+ |||+|+|#| | | |+| || +|+ |+| | Sbjct 270 GGKFGVDKNAQDATAGGFGDMQGVSSSYKK#TRPQPPAKSGAGNMRNKFEQMAQAGEEESR 329 Query 307 GGKYGVQKDRMDKNA 321 |||+|||+|||||+| Sbjct 85 GGKFGVQQDRMDKSA 99 |
| Xenopus laevis | Query 307 GGKYGVQKDRMDKNASTFEDVTQVSSAYQK#TVPVEAVTSKTSNIRANFENLAKEKEQEDR 366 ||||||||||||| |++|||| +|||+|||#| ||| || |+||||||||||+|||||| Sbjct 305 GGKYGVQKDRMDKAAASFEDVEKVSSSYQK#TRPVEVEGSKASSIRANFENLAKDKEQEDR 364 Query 307 GGKYGVQKDRMDKNASTFEDVTQVS 331 |||+||+|||||++| | |++| Sbjct 83 GGKFGVEKDRMDRSAVGHEYQTKLS 107 Query 307 GGKYGVQKDRMDKNASTFE 325 ||||||| ||+||+| |+ Sbjct 157 GGKYGVQADRVDKSALGFD 175 |
| Drosophila melanogaster | Query 307 GGKYGVQKDRMDKNASTFE----DVTQVSSAYQK#TVPVEAVTSKTSNIRANFENLAKEKE 362 |||+|||+|| ||+| ++ | |# || +| ||+|| |||||| | Sbjct 197 GGKFGVQEDRKDKSAVGWDHKEAPQKHASQVDHK#VKPV-IEGAKPSNLRAKFENLAKNSE 255 Query 363 QEDR 366 +| | Sbjct 256 EESR 259 Query 307 GGKYGVQKDRMDKNA 321 |||+||+||||||+| Sbjct 86 GGKFGVEKDRMDKSA 100 Query 307 GGKYGVQKDRMDKNASTFEDVTQV 330 |||+|||+|| ||+| ++ | +| Sbjct 123 GGKFGVQEDRKDKSAVGWDHVEKV 146 |
| Mus musculus | Query 307 GGKYGVQKDRMDKNASTFEDVTQVSSAYQK#TVPVEAVTSKTSNIRANFENLAKEKEQEDR 366 |||||||||||||||||||+| || |||||#|||+||||||||||||||||||||+||||| Sbjct 307 GGKYGVQKDRMDKNASTFEEVVQVPSAYQK#TVPIEAVTSKTSNIRANFENLAKEREQEDR 366 Query 307 GGKYGVQKDRMDKNASTFE 325 ||||||| ||+||+| |+ Sbjct 159 GGKYGVQADRVDKSAVGFD 177 Query 307 GGKYGVQKDRMDKNASTFE 325 |||||+ ||++||+| || Sbjct 196 GGKYGIDKDKVDKSAVGFE 214 |
| Canis familiaris | Query 307 GGKYGVQKDRMDKNASTFEDVTQVSSAYQK#TVPVEAVTSKTSNIRANFENLAKEKEQEDR 366 ||||||||||||||||||||| ||+ ||||#||||||| |+|||||||||||||||||||| Sbjct 307 GGKYGVQKDRMDKNASTFEDVAQVAPAYQK#TVPVEAVNSRTSNIRANFENLAKEKEQEDR 366 Query 307 GGKYGVQKDRMDKNASTFE 325 ||||||| ||+||+| |+ Sbjct 159 GGKYGVQADRVDKSAVGFD 177 Query 307 GGKYGVQKDRMDKNASTFE 325 |||||+ ||++||+| || Sbjct 196 GGKYGIDKDKVDKSAVGFE 214 |
| Pongo pygmaeus | Query 307 GGKYGVQKDRMDKNASTFEDVTQVSSAYQK#TVPVEAVTSKTSNIRANFENLAKEKEQEDR 366 ||||||||||||||||||||||||||||||#||||||| |||||||||||||||||||||| Sbjct 270 GGKYGVQKDRMDKNASTFEDVTQVSSAYQK#TVPVEAVASKTSNIRANFENLAKEKEQEDR 329 Query 307 GGKYGVQKDRMDKNASTFE 325 ||||||| ||+||+| |+ Sbjct 159 GGKYGVQADRVDKSAVGFD 177 Query 307 GGKYGVQKDRMDKNASTFE 325 |||||+ ||++||+| || Sbjct 196 GGKYGIDKDKVDKSAVGFE 214 |
| Suberites domuncula | Query 307 GGKYGVQKDRMDKNASTFEDVTQVSSAYQK#TVPVEAVTSKTSNIRANFENLA 358 ||||||++ | +| ++|+ | | ++ # | +| +||+ |||+| Sbjct 271 GGKYGVEEGNQDSSAGGYDDMQAVKSDHRT#ERGVS--KGQTGSIRSKFENMA 320 Query 307 GGKYGVQKDRMDKNASTF---EDVTQVSS 332 ||||||| | ||||+ + | ++| || Sbjct 197 GGKYGVQTDSQDKNAAGWDYQEKLSQHSS 225 |
| Danio rerio | Query 307 GGKYGVQKDRMDKNASTFEDVTQVSSAYQK#TVPVEAVTSKTSNIRANFENLA 358 |||||||||||||+| |||+| + |+||||#| |||| +| |+|+| |||+| Sbjct 306 GGKYGVQKDRMDKSAGTFEEVQKPSAAYQK#TRPVEAASSSASSIKARFENIA 357 Query 307 GGKYGVQKDRMDKNASTFEDV 327 |||||+|+||+|++| || | Sbjct 121 GGKYGLQEDRVDQSAVGFEYV 141 Query 307 GGKYGVQKDRMDKNASTFEDVTQVS 331 |||+|+|+|||||+| | +++| Sbjct 84 GGKFGLQQDRMDKSAVGHEYQSKLS 108 Query 307 GGKYGVQKDRMDKNASTFE 325 ||+|||| ||+|++| || Sbjct 158 GGRYGVQADRVDQSAVGFE 176 |
| Plasmodium yoelii yoelii str. 17XNL | Query 311 GVQKDRMDKNASTFEDVTQVSSAYQK#TVP 339 +| +| || | + |+||++ || # +| Sbjct 28 AIQNERFIKNCSFYNDITQIN--YQN#NIP 54 |
| mice, BALB/c 3T3 cells, Peptide, 546 aa | Query 307 GGKYGVQKDRMDKNASTFEDVTQVSSAYQK#TVPVEAVTSKTSNIRANFENLAKEKEQEDR 366 |||||||||||||||||||+| || |||||#|||+||||||||||||||||||||+||||| Sbjct 307 GGKYGVQKDRMDKNASTFEEVVQVPSAYQK#TVPIEAVTSKTSNIRANFENLAKEREQEDR 366 Query 307 GGKYGVQKDRMDKNASTFE 325 ||||||| ||+||+| |+ Sbjct 159 GGKYGVQADRVDKSAVGFD 177 Query 307 GGKYGVQKDRMDKNASTFE 325 |||||+ ||++||+| || Sbjct 196 GGKYGIDKDKVDKSAVGFE 214 |
[Site 2] TVPVEAVTSK346-TSNIRANFEN
Lys346  Thr
Thr
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Thr337 | Val338 | Pro339 | Val340 | Glu341 | Ala342 | Val343 | Thr344 | Ser345 | Lys346 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Thr347 | Ser348 | Asn349 | Ile350 | Arg351 | Ala352 | Asn353 | Phe354 | Glu355 | Asn356 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| MDKNASTFEDVTQVSSAYQKTVPVEAVTSKTSNIRANFENLAKEKEQEDRRKAEAERAQR |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | Homo sapiens | 121.00 | 4 | SRC8_HUMAN Src substrate cortactin (Amplaxin) (On |
| 2 | synthetic construct | 121.00 | 2 | cortactin |
| 3 | Pongo pygmaeus | 119.00 | 1 | hypothetical protein |
| 4 | Mus musculus | 114.00 | 5 | cortactin |
| 5 | N/A | 114.00 | 2 | SRC8_MOUSE Src substrate cortactin gi |
| 6 | mice, BALB/c 3T3 cells, Peptide, 546 aa | 114.00 | 1 | cortactin, p80/p85 |
| 7 | Canis familiaris | 112.00 | 5 | PREDICTED: similar to cortactin isoform a isoform |
| 8 | Rattus norvegicus | 112.00 | 3 | cortactin isoform B |
| 9 | Gallus gallus | 96.30 | 1 | cortactin |
| 10 | Xenopus laevis | 83.60 | 1 | cortactin |
| 11 | Tetraodon nigroviridis | 55.10 | 3 | unnamed protein product |
| 12 | Strongylocentrotus purpuratus | 53.90 | 1 | SRC8 protein |
| 13 | Danio rerio | 50.10 | 1 | cortactin |
| 14 | Bos taurus | 40.00 | 1 | hematopoietic cell-specific Lyn substrate 1 |
| 15 | Drosophila melanogaster | 37.00 | 1 | Cortactin |
Top-ranked sequences
| organism | matching |
|---|---|
| Homo sapiens | Query 317 MDKNASTFEDVTQVSSAYQKTVPVEAVTSK#TSNIRANFENLAKEKEQEDRRKAEAERAQR 376 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 317 MDKNASTFEDVTQVSSAYQKTVPVEAVTSK#TSNIRANFENLAKEKEQEDRRKAEAERAQR 376 |
| synthetic construct | Query 317 MDKNASTFEDVTQVSSAYQKTVPVEAVTSK#TSNIRANFENLAKEKEQEDRRKAEAERAQR 376 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 317 MDKNASTFEDVTQVSSAYQKTVPVEAVTSK#TSNIRANFENLAKEKEQEDRRKAEAERAQR 376 |
| Pongo pygmaeus | Query 317 MDKNASTFEDVTQVSSAYQKTVPVEAVTSK#TSNIRANFENLAKEKEQEDRRKAEAERAQR 376 ||||||||||||||||||||||||||| ||#|||||||||||||||||||||||||||||| Sbjct 280 MDKNASTFEDVTQVSSAYQKTVPVEAVASK#TSNIRANFENLAKEKEQEDRRKAEAERAQR 339 |
| Mus musculus | Query 317 MDKNASTFEDVTQVSSAYQKTVPVEAVTSK#TSNIRANFENLAKEKEQEDRRKAEAERAQR 376 |||||||||+| || ||||||||+||||||#||||||||||||||+||||||||||||||| Sbjct 317 MDKNASTFEEVVQVPSAYQKTVPIEAVTSK#TSNIRANFENLAKEREQEDRRKAEAERAQR 376 |
| N/A | Query 317 MDKNASTFEDVTQVSSAYQKTVPVEAVTSK#TSNIRANFENLAKEKEQEDRRKAEAERAQR 376 |||||||||+| || ||||||||+||||||#||||||||||||||+||||||||||||||| Sbjct 317 MDKNASTFEEVVQVPSAYQKTVPIEAVTSK#TSNIRANFENLAKEREQEDRRKAEAERAQR 376 |
| mice, BALB/c 3T3 cells, Peptide, 546 aa | Query 317 MDKNASTFEDVTQVSSAYQKTVPVEAVTSK#TSNIRANFENLAKEKEQEDRRKAEAERAQR 376 |||||||||+| || ||||||||+||||||#||||||||||||||+||||||||||||||| Sbjct 317 MDKNASTFEEVVQVPSAYQKTVPIEAVTSK#TSNIRANFENLAKEREQEDRRKAEAERAQR 376 |
| Canis familiaris | Query 317 MDKNASTFEDVTQVSSAYQKTVPVEAVTSK#TSNIRANFENLAKEKEQEDRRKAEAERAQR 376 ||||||||||| ||+ ||||||||||| |+#||||||||||||||||||||||||||+||| Sbjct 317 MDKNASTFEDVAQVAPAYQKTVPVEAVNSR#TSNIRANFENLAKEKEQEDRRKAEAEKAQR 376 |
| Rattus norvegicus | Query 317 MDKNASTFEDVTQVSSAYQKTVPVEAVTSK#TSNIRANFENLAKEKEQEDRRKAEAERAQR 376 |||||||||+| || ||||||||+||||||#|||||||||+||||+||||||||||||||| Sbjct 280 MDKNASTFEEVVQVPSAYQKTVPIEAVTSK#TSNIRANFESLAKEREQEDRRKAEAERAQR 339 |
| Gallus gallus | Query 317 MDKNASTFEDVTQVSSAYQKTVPVEAVTSK#TSNIRANFENLAKEKEQEDRRKAEAERAQR 376 |||||+||||+ + +| |||| ||| | +|#||+|||| |||||||||||||||||||||| Sbjct 326 MDKNAATFEDIEKPTSTYQKTKPVERVANK#TSSIRANLENLAKEKEQEDRRKAEAERAQR 385 |
| Xenopus laevis | Query 317 MDKNASTFEDVTQVSSAYQKTVPVEAVTSK#TSNIRANFENLAKEKEQEDRRKAEA 371 ||| |++|||| +|||+|||| ||| ||# |+||||||||||+||||||+|||| Sbjct 315 MDKAAASFEDVEKVSSSYQKTRPVEVEGSK#ASSIRANFENLAKDKEQEDRKKAEA 369 |
| Tetraodon nigroviridis | Query 317 MDKNASTFEDVTQVSSAYQKTVPVEAVTSK#TSNIRANFENLAKEK 361 ||| | |||+| + + +|||| |||| | # +|+| |||+||+| Sbjct 325 MDKAAGTFEEVEKPTPSYQKTKPVEAAGSN#AGSIKARFENMAKQK 369 |
| Strongylocentrotus purpuratus | Query 318 DKNASTFEDVTQVSSAYQKTVPVEAVTSK#TSNIRANFENLAKEKEQEDRRKAEAERAQR 376 | | | |+ |||+|+|| | | # |+| || +|+ |+| ||||| || +| Sbjct 281 DATAGGFGDMQGVSSSYKKTRPQPPAKSG#AGNMRNKFEQMAQAGEEESRRKAEEERGRR 339 |
| Danio rerio | Query 317 MDKNASTFEDVTQVSSAYQKTVPVEAVTSK#TSNIRANFENLAK 359 |||+| |||+| + |+||||| |||| +| # |+|+| |||+|| Sbjct 316 MDKSAGTFEEVQKPSAAYQKTRPVEAASSS#ASSIKARFENIAK 358 |
| Bos taurus | Query 317 MDKNASTFEDVTQVSSAYQKTVPVEAVTSK#TSNIRANFENLA 358 +||+| | ++ ++||+|| |+|| +| # ++| ||++| Sbjct 205 VDKSAVGFNEMEAPTTAYKKTTPIEAASSG#ARGLKAKFESMA 246 |
| Drosophila melanogaster | Query 345 SK#TSNIRANFENLAKEKEQEDRRKAEAERAQR 376 +|# ||+|| |||||| |+| |++|| ++ | Sbjct 238 AK#PSNLRAKFENLAKNSEEESRKRAEEQKRLR 269 |
[Site 3] AVTSKTSNIR351-ANFENLAKEK
Arg351  Ala
Ala
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Ala342 | Val343 | Thr344 | Ser345 | Lys346 | Thr347 | Ser348 | Asn349 | Ile350 | Arg351 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Ala352 | Asn353 | Phe354 | Glu355 | Asn356 | Leu357 | Ala358 | Lys359 | Glu360 | Lys361 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| STFEDVTQVSSAYQKTVPVEAVTSKTSNIRANFENLAKEKEQEDRRKAEAERAQRMAKER |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | Homo sapiens | 120.00 | 4 | SRC8_HUMAN Src substrate cortactin (Amplaxin) (On |
| 2 | synthetic construct | 120.00 | 1 | cortactin |
| 3 | Pongo pygmaeus | 118.00 | 1 | hypothetical protein |
| 4 | Mus musculus | 113.00 | 5 | cortactin |
| 5 | N/A | 113.00 | 2 | SRC8_MOUSE Src substrate cortactin gi |
| 6 | mice, BALB/c 3T3 cells, Peptide, 546 aa | 113.00 | 1 | cortactin, p80/p85 |
| 7 | Canis familiaris | 111.00 | 5 | PREDICTED: similar to cortactin isoform a isoform |
| 8 | Rattus norvegicus | 109.00 | 3 | cortactin isoform B |
| 9 | Gallus gallus | 92.40 | 1 | cortactin |
| 10 | Xenopus laevis | 76.30 | 1 | cortactin |
| 11 | Strongylocentrotus purpuratus | 52.40 | 1 | SRC8 protein |
| 12 | Tetraodon nigroviridis | 48.90 | 3 | unnamed protein product |
| 13 | Danio rerio | 43.50 | 1 | cortactin |
| 14 | Drosophila melanogaster | 40.00 | 1 | Cortactin |
| 15 | Drosophila melanogaster | 40.00 | 1 | Cortactin CG3637-PA |
| 16 | Bos taurus | 34.70 | 1 | hematopoietic cell-specific Lyn substrate 1 |
Top-ranked sequences
| organism | matching |
|---|---|
| Homo sapiens | Query 322 STFEDVTQVSSAYQKTVPVEAVTSKTSNIR#ANFENLAKEKEQEDRRKAEAERAQRMAKER 381 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 322 STFEDVTQVSSAYQKTVPVEAVTSKTSNIR#ANFENLAKEKEQEDRRKAEAERAQRMAKER 381 |
| synthetic construct | Query 322 STFEDVTQVSSAYQKTVPVEAVTSKTSNIR#ANFENLAKEKEQEDRRKAEAERAQRMAKER 381 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 322 STFEDVTQVSSAYQKTVPVEAVTSKTSNIR#ANFENLAKEKEQEDRRKAEAERAQRMAKER 381 |
| Pongo pygmaeus | Query 322 STFEDVTQVSSAYQKTVPVEAVTSKTSNIR#ANFENLAKEKEQEDRRKAEAERAQRMAKER 381 |||||||||||||||||||||| |||||||#|||||||||||||||||||||||||||||| Sbjct 285 STFEDVTQVSSAYQKTVPVEAVASKTSNIR#ANFENLAKEKEQEDRRKAEAERAQRMAKER 344 |
| Mus musculus | Query 322 STFEDVTQVSSAYQKTVPVEAVTSKTSNIR#ANFENLAKEKEQEDRRKAEAERAQRMAKER 381 ||||+| || ||||||||+|||||||||||#|||||||||+|||||||||||||||||||| Sbjct 322 STFEEVVQVPSAYQKTVPIEAVTSKTSNIR#ANFENLAKEREQEDRRKAEAERAQRMAKER 381 |
| N/A | Query 322 STFEDVTQVSSAYQKTVPVEAVTSKTSNIR#ANFENLAKEKEQEDRRKAEAERAQRMAKER 381 ||||+| || ||||||||+|||||||||||#|||||||||+|||||||||||||||||||| Sbjct 322 STFEEVVQVPSAYQKTVPIEAVTSKTSNIR#ANFENLAKEREQEDRRKAEAERAQRMAKER 381 |
| mice, BALB/c 3T3 cells, Peptide, 546 aa | Query 322 STFEDVTQVSSAYQKTVPVEAVTSKTSNIR#ANFENLAKEKEQEDRRKAEAERAQRMAKER 381 ||||+| || ||||||||+|||||||||||#|||||||||+|||||||||||||||||||| Sbjct 322 STFEEVVQVPSAYQKTVPIEAVTSKTSNIR#ANFENLAKEREQEDRRKAEAERAQRMAKER 381 |
| Canis familiaris | Query 322 STFEDVTQVSSAYQKTVPVEAVTSKTSNIR#ANFENLAKEKEQEDRRKAEAERAQRMAKER 381 |||||| ||+ ||||||||||| |+|||||#|||||||||||||||||||||+|||||||| Sbjct 322 STFEDVAQVAPAYQKTVPVEAVNSRTSNIR#ANFENLAKEKEQEDRRKAEAEKAQRMAKER 381 |
| Rattus norvegicus | Query 322 STFEDVTQVSSAYQKTVPVEAVTSKTSNIR#ANFENLAKEKEQEDRRKAEAERAQRMAKER 381 ||||+| || ||||||||+|||||||||||#||||+||||+|||||||||||||||||+|| Sbjct 285 STFEEVVQVPSAYQKTVPIEAVTSKTSNIR#ANFESLAKEREQEDRRKAEAERAQRMAQER 344 |
| Gallus gallus | Query 322 STFEDVTQVSSAYQKTVPVEAVTSKTSNIR#ANFENLAKEKEQEDRRKAEAERAQRMAKER 381 +||||+ + +| |||| ||| | +|||+||#|| ||||||||||||||||||||||||+|+ Sbjct 331 ATFEDIEKPTSTYQKTKPVERVANKTSSIR#ANLENLAKEKEQEDRRKAEAERAQRMAREK 390 |
| Xenopus laevis | Query 322 STFEDVTQVSSAYQKTVPVEAVTSKTSNIR#ANFENLAKEKEQEDRRKAEA 371 ++|||| +|||+|||| ||| || |+||#||||||||+||||||+|||| Sbjct 320 ASFEDVEKVSSSYQKTRPVEVEGSKASSIR#ANFENLAKDKEQEDRKKAEA 369 |
| Strongylocentrotus purpuratus | Query 322 STFEDVTQVSSAYQKTVPVEAVTSKTSNIR#ANFENLAKEKEQEDRRKAEAERAQRMAK 379 | |+ |||+|+|| | | |+|# || +|+ |+| ||||| || +| |+ Sbjct 285 GGFGDMQGVSSSYKKTRPQPPAKSGAGNMR#NKFEQMAQAGEEESRRKAEEERGRRQAR 342 |
| Tetraodon nigroviridis | Query 322 STFEDVTQVSSAYQKTVPVEAVTSKTSNIR#ANFENLAKEKEQEDRRKAEAERAQRMAKER 381 |||+| + + +|||| |||| | +|+#| |||+||+|| ||||+|| || +| |||| Sbjct 330 GTFEEVEKPTPSYQKTKPVEAAGSNAGSIK#ARFENMAKQKEDEDRRRAEEERLRRQAKER 389 |
| Danio rerio | Query 322 STFEDVTQVSSAYQKTVPVEAVTSKTSNIR#ANFENLAKEKEQEDRRKAEAERAQRMAKER 381 |||+| + |+||||| |||| +| |+|+#| |||+||+||+|++++ | ||++| |||+ Sbjct 321 GTFEEVQKPSAAYQKTRPVEAASSSASSIK#ARFENIAKQKEEEEQQRLEEERSRRQAKEK 380 |
| Drosophila melanogaster | Query 345 SKTSNIR#ANFENLAKEKEQEDRRKAEAERAQRMAKER 381 +| ||+|#| |||||| |+| |++|| ++ | ||++ Sbjct 238 AKPSNLR#AKFENLAKNSEEESRKRAEEQKRLREAKDK 274 |
| Drosophila melanogaster | Query 345 SKTSNIR#ANFENLAKEKEQEDRRKAEAERAQRMAKER 381 +| ||+|#| |||||| |+| |++|| ++ | ||++ Sbjct 238 AKPSNLR#AKFENLAKNSEEESRKRAEEQKRLREAKDK 274 |
| Bos taurus | Query 324 FEDVTQVSSAYQKTVPVEAVTSKTSNIR#ANFENLA 358 | ++ ++||+|| |+|| +| ++#| ||++| Sbjct 212 FNEMEAPTTAYKKTTPIEAASSGARGLK#AKFESMA 246 |
[Site 4] NIRANFENLA358-KEKEQEDRRK
Ala358  Lys
Lys
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Asn349 | Ile350 | Arg351 | Ala352 | Asn353 | Phe354 | Glu355 | Asn356 | Leu357 | Ala358 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Lys359 | Glu360 | Lys361 | Glu362 | Gln363 | Glu364 | Asp365 | Arg366 | Arg367 | Lys368 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| QVSSAYQKTVPVEAVTSKTSNIRANFENLAKEKEQEDRRKAEAERAQRMAKERQEQEEAR |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | Homo sapiens | 118.00 | 4 | SRC8_HUMAN Src substrate cortactin (Amplaxin) (On |
| 2 | synthetic construct | 118.00 | 2 | cortactin |
| 3 | Mus musculus | 115.00 | 5 | cortactin |
| 4 | N/A | 115.00 | 2 | SRC8_MOUSE Src substrate cortactin gi |
| 5 | mice, BALB/c 3T3 cells, Peptide, 546 aa | 115.00 | 1 | cortactin, p80/p85 |
| 6 | Pongo pygmaeus | 115.00 | 1 | hypothetical protein |
| 7 | Canis familiaris | 110.00 | 5 | PREDICTED: similar to cortactin isoform a isoform |
| 8 | Rattus norvegicus | 105.00 | 3 | cortactin isoform B |
| 9 | Gallus gallus | 94.40 | 1 | cortactin |
| 10 | Xenopus laevis | 67.40 | 1 | cortactin |
| 11 | Tetraodon nigroviridis | 51.20 | 3 | unnamed protein product |
| 12 | Danio rerio | 47.80 | 1 | cortactin |
| 13 | Strongylocentrotus purpuratus | 45.80 | 1 | SRC8 protein |
| 14 | Drosophila melanogaster | 43.50 | 1 | Cortactin |
| 15 | Drosophila melanogaster | 43.50 | 1 | Cortactin CG3637-PA |
| 16 | Bos taurus | 36.60 | 1 | hematopoietic cell-specific Lyn substrate 1 |
| 17 | Drosophila pseudoobscura | 32.30 | 1 | GA17577-PA |
| 18 | Nicotiana tabacum | 31.20 | 1 | AF194022_1 sucrose-6-phosphate synthase A |
Top-ranked sequences
| organism | matching |
|---|---|
| Homo sapiens | Query 329 QVSSAYQKTVPVEAVTSKTSNIRANFENLA#KEKEQEDRRKAEAERAQRMAKERQEQEEAR 388 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 329 QVSSAYQKTVPVEAVTSKTSNIRANFENLA#KEKEQEDRRKAEAERAQRMAKERQEQEEAR 388 |
| synthetic construct | Query 329 QVSSAYQKTVPVEAVTSKTSNIRANFENLA#KEKEQEDRRKAEAERAQRMAKERQEQEEAR 388 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 329 QVSSAYQKTVPVEAVTSKTSNIRANFENLA#KEKEQEDRRKAEAERAQRMAKERQEQEEAR 388 |
| Mus musculus | Query 329 QVSSAYQKTVPVEAVTSKTSNIRANFENLA#KEKEQEDRRKAEAERAQRMAKERQEQEEAR 388 || ||||||||+||||||||||||||||||#||+||||||||||||||||||||||||||| Sbjct 329 QVPSAYQKTVPIEAVTSKTSNIRANFENLA#KEREQEDRRKAEAERAQRMAKERQEQEEAR 388 |
| N/A | Query 329 QVSSAYQKTVPVEAVTSKTSNIRANFENLA#KEKEQEDRRKAEAERAQRMAKERQEQEEAR 388 || ||||||||+||||||||||||||||||#||+||||||||||||||||||||||||||| Sbjct 329 QVPSAYQKTVPIEAVTSKTSNIRANFENLA#KEREQEDRRKAEAERAQRMAKERQEQEEAR 388 |
| mice, BALB/c 3T3 cells, Peptide, 546 aa | Query 329 QVSSAYQKTVPVEAVTSKTSNIRANFENLA#KEKEQEDRRKAEAERAQRMAKERQEQEEAR 388 || ||||||||+||||||||||||||||||#||+||||||||||||||||||||||||||| Sbjct 329 QVPSAYQKTVPIEAVTSKTSNIRANFENLA#KEREQEDRRKAEAERAQRMAKERQEQEEAR 388 |
| Pongo pygmaeus | Query 329 QVSSAYQKTVPVEAVTSKTSNIRANFENLA#KEKEQEDRRKAEAERAQRMAKERQEQEEAR 388 ||||||||||||||| ||||||||||||||#|||||||||||||||||||||||||||||| Sbjct 292 QVSSAYQKTVPVEAVASKTSNIRANFENLA#KEKEQEDRRKAEAERAQRMAKERQEQEEAR 351 |
| Canis familiaris | Query 329 QVSSAYQKTVPVEAVTSKTSNIRANFENLA#KEKEQEDRRKAEAERAQRMAKERQEQEEAR 388 ||+ ||||||||||| |+||||||||||||#||||||||||||||+||||||||||||||| Sbjct 329 QVAPAYQKTVPVEAVNSRTSNIRANFENLA#KEKEQEDRRKAEAEKAQRMAKERQEQEEAR 388 |
| Rattus norvegicus | Query 329 QVSSAYQKTVPVEAVTSKTSNIRANFENLA#KEKEQEDRRKAEAERAQRMAKERQEQE 385 || ||||||||+|||||||||||||||+||#||+|||||||||||||||||+|||||| Sbjct 292 QVPSAYQKTVPIEAVTSKTSNIRANFESLA#KEREQEDRRKAEAERAQRMAQERQEQE 348 |
| Gallus gallus | Query 331 SSAYQKTVPVEAVTSKTSNIRANFENLA#KEKEQEDRRKAEAERAQRMAKERQEQEEAR 388 +| |||| ||| | +|||+|||| ||||#||||||||||||||||||||+|+||||||| Sbjct 340 TSTYQKTKPVERVANKTSSIRANLENLA#KEKEQEDRRKAEAERAQRMAREKQEQEEAR 397 |
| Xenopus laevis | Query 329 QVSSAYQKTVPVEAVTSKTSNIRANFENLA#KEKEQEDRRKAEA 371 +|||+|||| ||| || |+|||||||||#|+||||||+|||| Sbjct 327 KVSSSYQKTRPVEVEGSKASSIRANFENLA#KDKEQEDRKKAEA 369 |
| Tetraodon nigroviridis | Query 331 SSAYQKTVPVEAVTSKTSNIRANFENLA#KEKEQEDRRKAEAERAQRMAKERQEQEEAR 388 + +|||| |||| | +|+| |||+|#|+|| ||||+|| || +| ||||||||||+ Sbjct 339 TPSYQKTKPVEAAGSNAGSIKARFENMA#KQKEDEDRRRAEEERLRRQAKERQEQEEAQ 396 |
| Danio rerio | Query 331 SSAYQKTVPVEAVTSKTSNIRANFENLA#KEKEQEDRRKAEAERAQRMAKERQEQEEAR 388 |+||||| |||| +| |+|+| |||+|#|+||+|++++ | ||++| |||+||||||| Sbjct 330 SAAYQKTRPVEAASSSASSIKARFENIA#KQKEEEEQQRLEEERSRRQAKEKQEQEEAR 387 |
| Strongylocentrotus purpuratus | Query 330 VSSAYQKTVPVEAVTSKTSNIRANFENLA#KEKEQEDRRKAEAERAQRMAK 379 |||+|+|| | | |+| || +|#+ |+| ||||| || +| |+ Sbjct 293 VSSSYKKTRPQPPAKSGAGNMRNKFEQMA#QAGEEESRRKAEEERGRRQAR 342 |
| Drosophila melanogaster | Query 345 SKTSNIRANFENLA#KEKEQEDRRKAEAERAQRMAKERQEQEEA 387 +| ||+|| |||||#| |+| |++|| ++ | ||+++++||| Sbjct 238 AKPSNLRAKFENLA#KNSEEESRKRAEEQKRLREAKDKRDREEA 280 |
| Drosophila melanogaster | Query 345 SKTSNIRANFENLA#KEKEQEDRRKAEAERAQRMAKERQEQEEA 387 +| ||+|| |||||#| |+| |++|| ++ | ||+++++||| Sbjct 238 AKPSNLRAKFENLA#KNSEEESRKRAEEQKRLREAKDKRDREEA 280 |
| Bos taurus | Query 331 SSAYQKTVPVEAVTSKTSNIRANFENLA#KEKEQEDRRKAEAERAQRMAKERQE 383 ++||+|| |+|| +| ++| ||++|#+|| + + | |+||+||+++|| Sbjct 219 TTAYKKTTPIEAASSGARGLKAKFESMA#EEKRK----REEEEKAQQMARQQQE 267 |
| Drosophila pseudoobscura | Query 345 SKTSNIRANFENLA#KEKEQEDRRKAEAERAQRMAKERQEQEEA 387 +| ||+|| |||||#| |+| |+++| ++ | ||+++++||| Sbjct 236 AKPSNLRAKFENLA#KNSEEESRKRSEEQKRLREAKDKRDREEA 278 |
| Nicotiana tabacum | Query 335 QKTVPVEAVTSKTSNIRANFENLA#KEKEQEDRRKAEAERAQRMAKERQEQEEAR 388 | | + ++ |+ |||#++|+| | |+|| ||| |||+|+ | Sbjct 63 QATRSPQERNTRLENMCWRIWNLA#RQKKQ-----LEGEQAQWMAKRRQEREKGR 111 |
* References
[PubMed ID: 24469447] Han SP, Gambin Y, Gomez GA, Verma S, Giles N, Michael M, Wu SK, Guo Z, Johnston W, Sierecki E, Parton RG, Alexandrov K, Yap AS, Cortactin scaffolds Arp2/3 and WAVE2 at the epithelial zonula adherens. J Biol Chem. 2014 Mar 14;289(11):7764-75. doi: 10.1074/jbc.M113.544478. Epub 2014
[PubMed ID: 24265417] ... Majumder S, Sowden MP, Gerber SA, Thomas T, Christie CK, Mohan A, Yin G, Lord EM, Berk BC, Pang J, G-protein-coupled receptor-2-interacting protein-1 is required for endothelial cell directional migration and tumor angiogenesis via cortactin-dependent lamellipodia formation. Arterioscler Thromb Vasc Biol. 2014 Feb;34(2):419-26. doi:
[PubMed ID: 23991004] ... Lin YC, Tsai PH, Lin CY, Cheng CH, Lin TH, Lee KP, Huang KY, Chen SH, Hwang JJ, Kandaswami CC, Lee MT, Impact of flavonoids on matrix metalloproteinase secretion and invadopodia formation in highly invasive A431-III cancer cells. PLoS One. 2013 Aug 21;8(8):e71903. doi: 10.1371/journal.pone.0071903. eCollection
[PubMed ID: 23702379] ... Noh SJ, Baek HA, Park HS, Jang KY, Moon WS, Kang MJ, Lee DG, Kim MH, Lee JH, Chung MJ, Expression of SIRT1 and cortactin is associated with progression of non-small cell lung cancer. Pathol Res Pract. 2013 Jun;209(6):365-70. doi: 10.1016/j.prp.2013.03.011. Epub
[PubMed ID: 24409670] ... Gang Z, Ya-Lin K, Dong-Qing W, Yu L, Li R, Hong-Yi Z, Combining cortactin and CTTN detection with clinicopathologic features increases effectiveness of survival predictions for patients with resectable hepatocellular carcinoma. Clin Lab. 2013;59(11-12):1343-52.
[PubMed ID: 18083107] ... Rikova K, Guo A, Zeng Q, Possemato A, Yu J, Haack H, Nardone J, Lee K, Reeves C, Li Y, Hu Y, Tan Z, Stokes M, Sullivan L, Mitchell J, Wetzel R, Macneill J, Ren JM, Yuan J, Bakalarski CE, Villen J, Kornhauser JM, Smith B, Li D, Zhou X, Gygi SP, Gu TL, Polakiewicz RD, Rush J, Comb MJ, Global survey of phosphotyrosine signaling identifies oncogenic kinases in lung cancer. Cell. 2007 Dec 14;131(6):1190-203.
[PubMed ID: 17322306] ... Sugiyama N, Masuda T, Shinoda K, Nakamura A, Tomita M, Ishihama Y, Phosphopeptide enrichment by aliphatic hydroxy acid-modified metal oxide chromatography for nano-LC-MS/MS in proteomics applications. Mol Cell Proteomics. 2007 Jun;6(6):1103-9. Epub 2007 Feb 23.
[PubMed ID: 17081983] ... Olsen JV, Blagoev B, Gnad F, Macek B, Kumar C, Mortensen P, Mann M, Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell. 2006 Nov 3;127(3):635-48.
[PubMed ID: 16825425] ... Martin KH, Jeffery ED, Grigera PR, Shabanowitz J, Hunt DF, Parsons JT, Cortactin phosphorylation sites mapped by mass spectrometry. J Cell Sci. 2006 Jul 15;119(Pt 14):2851-3.
[PubMed ID: 16212419] ... Amanchy R, Kalume DE, Iwahori A, Zhong J, Pandey A, Phosphoproteome analysis of HeLa cells using stable isotope labeling with amino acids in cell culture (SILAC). J Proteome Res. 2005 Sep-Oct;4(5):1661-71.
[PubMed ID: 15951569] ... Zhang Y, Wolf-Yadlin A, Ross PL, Pappin DJ, Rush J, Lauffenburger DA, White FM, Time-resolved mass spectrometry of tyrosine phosphorylation sites in the epidermal growth factor receptor signaling network reveals dynamic modules. Mol Cell Proteomics. 2005 Sep;4(9):1240-50. Epub 2005 Jun 11.
[PubMed ID: 15302935] ... Beausoleil SA, Jedrychowski M, Schwartz D, Elias JE, Villen J, Li J, Cohn MA, Cantley LC, Gygi SP, Large-scale characterization of HeLa cell nuclear phosphoproteins. Proc Natl Acad Sci U S A. 2004 Aug 17;101(33):12130-5. Epub 2004 Aug 9.
[PubMed ID: 15144186] ... Brill LM, Salomon AR, Ficarro SB, Mukherji M, Stettler-Gill M, Peters EC, Robust phosphoproteomic profiling of tyrosine phosphorylation sites from human T cells using immobilized metal affinity chromatography and tandem mass spectrometry. Anal Chem. 2004 May 15;76(10):2763-72.
[PubMed ID: 10921917] ... Kapus A, Di Ciano C, Sun J, Zhan X, Kim L, Wong TW, Rotstein OD, Cell volume-dependent phosphorylation of proteins of the cortical cytoskeleton and cell-cell contact sites. The role of Fyn and FER kinases. J Biol Chem. 2000 Oct 13;275(41):32289-98.
[PubMed ID: 9748248] ... Huang C, Liu J, Haudenschild CC, Zhan X, The role of tyrosine phosphorylation of cortactin in the locomotion of endothelial cells. J Biol Chem. 1998 Oct 2;273(40):25770-6.
[PubMed ID: 7566098] ... Adams MD, Kerlavage AR, Fleischmann RD, Fuldner RA, Bult CJ, Lee NH, Kirkness EF, Weinstock KG, Gocayne JD, White O, et al., Initial assessment of human gene diversity and expression patterns based upon 83 million nucleotides of cDNA sequence. Nature. 1995 Sep 28;377(6547 Suppl):3-174.
[PubMed ID: 8474448] ... Schuuring E, Verhoeven E, Litvinov S, Michalides RJ, The product of the EMS1 gene, amplified and overexpressed in human carcinomas, is homologous to a v-src substrate and is located in cell-substratum contact sites. Mol Cell Biol. 1993 May;13(5):2891-98.
[PubMed ID: 7685625] ... Brookes S, Lammie GA, Schuuring E, de Boer C, Michalides R, Dickson C, Peters G, Amplified region of chromosome band 11q13 in breast and squamous cell carcinomas encompasses three CpG islands telomeric of FGF3, including the expressed gene EMS1. Genes Chromosomes Cancer. 1993 Apr;6(4):222-31.
[PubMed ID: 7680654] ... Wu H, Parsons JT, Cortactin, an 80/85-kilodalton pp60src substrate, is a filamentous actin-binding protein enriched in the cell cortex. J Cell Biol. 1993 Mar;120(6):1417-26.
[PubMed ID: 1532244] ... Schuuring E, Verhoeven E, Mooi WJ, Michalides RJ, Identification and cloning of two overexpressed genes, U21B31/PRAD1 and EMS1, within the amplified chromosome 11q13 region in human carcinomas. Oncogene. 1992 Feb;7(2):355-61.