SB0037 : Vimentin
[ CaMP Format ]
* Basic Information
| Organism | Mus musculus (house mouse) |
| Protein Names | vimentin [Mus musculus]; vimentin; Vimentin |
| Gene Names | Vim; vimentin |
| Gene Locus | 2 10.04 cM; chromosome 2 |
| GO Function | Not available |
* Information From OMIM
Not Available.
* Structure Information
1. Primary Information
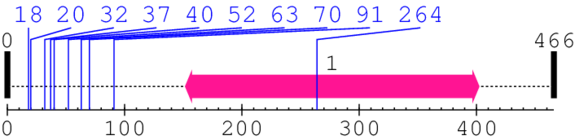
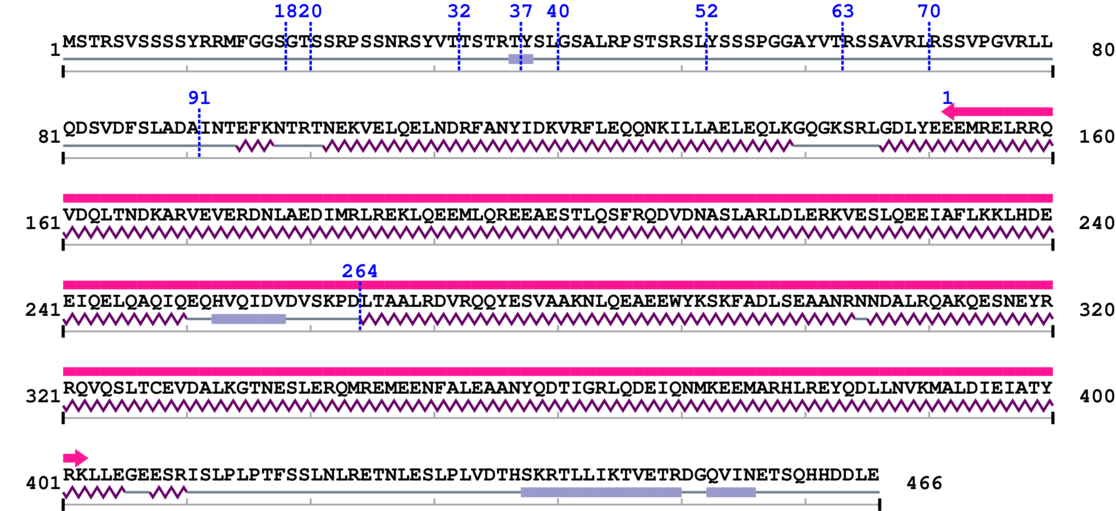
Length: 466 aa
Average Mass: 53.687 kDa
Monoisotopic Mass: 53.655 kDa
2. Domain Information
Annotated Domains: interpro / pfam / smart / prosite
Computationally Assigned Domains (Pfam+HMMER):
| domain name | begin | end | score | e-value |
|---|---|---|---|---|
| --- cleavage 18 --- | ||||
| --- cleavage 20 --- | ||||
| --- cleavage 32 --- | ||||
| --- cleavage 37 --- | ||||
| --- cleavage 40 --- | ||||
| --- cleavage 52 --- | ||||
| --- cleavage 63 --- | ||||
| --- cleavage 70 --- | ||||
| --- cleavage 91 --- | ||||
| AAA domain 1. | 152 | 402 | 743.0 | 34.7 |
| --- cleavage 264 (inside AAA domain 152..402) --- | ||||
3. Sequence Information
Fasta Sequence: SB0037.fasta
Amino Acid Sequence and Secondary Structures (PsiPred):
4. 3D Information
Not Available.
* Cleavage Information
10 [sites] cleaved by Calpain 2
Source Reference: [PubMed ID: 3028449] Fischer S, Vandekerckhove J, Ampe C, Traub P, Weber K, Protein-chemical identification of the major cleavage sites of the Ca2+ proteinase on murine vimentin, the mesenchymal intermediate filament protein. Biol Chem Hoppe Seyler. 1986 Nov;367(11):1147-52.
Cleavage sites (±10aa)
[Site 1] SSYRRMFGGS18-GTSSRPSSNR
Ser18  Gly
Gly
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Ser9 | Ser10 | Tyr11 | Arg12 | Arg13 | Met14 | Phe15 | Gly16 | Gly17 | Ser18 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Gly19 | Thr20 | Ser21 | Ser22 | Arg23 | Pro24 | Ser25 | Ser26 | Asn27 | Arg28 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| MSTRSVSSSSYRRMFGGSGTSSRPSSNRSYVTTSTRTYSLGSALRPST |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | Mus musculus | 87.80 | 10 | vimentin |
| 2 | N/A | 85.50 | 5 | vimentin protein |
| 3 | Rattus norvegicus | 85.50 | 1 | vimentin |
| 4 | Canis familiaris | 72.40 | 1 | PREDICTED: similar to Vimentin |
| 5 | Macaca fascicularis | 65.50 | 1 | VIME_MACFA Vimentin gi |
| 6 | synthetic construct | 63.50 | 1 | vimentin |
| 7 | Canis familiaris | 63.20 | 11 | PREDICTED: similar to Vimentin isoform 8 |
| 8 | Homo sapiens | 63.20 | 5 | vimentin |
| 9 | Pan troglodytes | 63.20 | 1 | vimentin |
| 10 | Ovis aries | 61.60 | 1 | VIME_SHEEP Vimentin gi |
| 11 | Gallus gallus | 53.10 | 2 | vimentin |
| 12 | Xenopus tropicalis | 43.50 | 1 | Vim protein |
| 13 | Xenopus laevis | 43.50 | 1 | VIM1_XENLA Vimentin-1/2 gi |
Top-ranked sequences
| organism | matching |
|---|---|
| Mus musculus | Query 1 MSTRSVSSSSYRRMFGGS#GTSSRPSSNRSYVTTSTRTYSLGSALRPST 48 ||||||||||||||||||#|||||||||||||||||||||||||||||| Query 1 MSTRSVSSSSYRRMFGGS#GTSSRPSSNRSYVTTSTRTYSLGSALRPST 48 |
| N/A | Query 1 MSTRSVSSSSYRRMFGGS#GTSSRPSSNRSYVTTSTRTYSLGSALRPST 48 ||||||||||||||||||#|||||||||||||||||||||||||||||| Query 1 MSTRSVSSSSYRRMFGGS#GTSSRPSSNRSYVTTSTRTYSLGSALRPST 48 |
| Rattus norvegicus | Query 1 MSTRSVSSSSYRRMFGGS#GTSSRPSSNRSYVTTSTRTYSLGSALRPST 48 ||||||||||||||||||#|||||||||||||||||||||||||||||| Query 1 MSTRSVSSSSYRRMFGGS#GTSSRPSSNRSYVTTSTRTYSLGSALRPST 48 |
| Canis familiaris | Query 1 MSTRSVSSSSYRRMFGGS#GTSSRPSSNRSYVTTSTRTYSLGSALRPST 48 ||||||| ||||||| #|| ||||| ||||||||||||||||+|||| Sbjct 1 MSTRSVSLSSYRRMFCVP#GTGSRPSSTRSYVTTSTRTYSLGSAMRPST 48 |
| Macaca fascicularis | Query 1 MSTRSVSSSSYRRMFGGS#GTSSRPSSNRSYVTTSTRTYSLGSALRPST 48 |+||||||||||||||| #||+|||||+||||||||||||||||||||| Sbjct 1 MTTRSVSSSSYRRMFGGP#GTASRPSSSRSYVTTSTRTYSLGSALRPST 48 |
| synthetic construct | Query 13 RMFGGS#GTSSRPSSNRSYVTTSTRTYSLGSALRPST 48 ||||| #||+|||||+||||||||||||||||||||| Sbjct 13 RMFGGP#GTASRPSSSRSYVTTSTRTYSLGSALRPST 48 |
| Canis familiaris | Query 13 RMFGGS#GTSSRPSSNRSYVTTSTRTYSLGSALRPST 48 ||||| #|| ||||| ||||||||||||||||||||| Sbjct 13 RMFGGP#GTGSRPSSTRSYVTTSTRTYSLGSALRPST 48 |
| Homo sapiens | Query 13 RMFGGS#GTSSRPSSNRSYVTTSTRTYSLGSALRPST 48 ||||| #||+|||||+||||||||||||||||||||| Sbjct 13 RMFGGP#GTASRPSSSRSYVTTSTRTYSLGSALRPST 48 |
| Pan troglodytes | Query 13 RMFGGS#GTSSRPSSNRSYVTTSTRTYSLGSALRPST 48 ||||| #||+|||||+||||||||||||||||||||| Sbjct 13 RMFGGP#GTASRPSSSRSYVTTSTRTYSLGSALRPST 48 |
| Ovis aries | Query 13 RMFGGS#GTSSRPSSNRSYVTTSTRTYSLGSALRPST 48 ||||| #||+||||| |||||||||||||||||||+| Sbjct 13 RMFGGP#GTASRPSSTRSYVTTSTRTYSLGSALRPTT 48 |
| Gallus gallus | Query 1 MSTRSVSSSSYRRMFGGS#GTSSRPSSNRSYVTTSTRTYSLGSALRPST 48 || | +||||||||| # ||||| |+|+||| ||||||||||+ Sbjct 1 MSFTSSKNSSYRRMFGG-#--GSRPSSGTRYITSSTR-YSLGSALRPSS 44 |
| Xenopus tropicalis | Query 2 STRSVSSSSYRRMFGGS#GTSSRPSSNRSYVTTSTRTYSLGSALRP 46 +| + + |||||+|||+# || || |||+||| ||||||+|| Sbjct 23 TTMATTKSSYRRIFGGN#PRSS--SSGNRYVTSSTR-YSLGSAMRP 64 |
| Xenopus laevis | Query 5 SVSSSSYRRMFGGS#GTSSRPSSNRSYVTTSTRTYSLGSALRPST 48 + + |||||+|||+# || || | |+||| |+||||+|||| Sbjct 2 ATTKSSYRRIFGGN#PRSS--SSGNRYATSSTR-YTLGSAMRPST 42 |
[Site 2] YRRMFGGSGT20-SSRPSSNRSY
Thr20  Ser
Ser
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Tyr11 | Arg12 | Arg13 | Met14 | Phe15 | Gly16 | Gly17 | Ser18 | Gly19 | Thr20 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Ser21 | Ser22 | Arg23 | Pro24 | Ser25 | Ser26 | Asn27 | Arg28 | Ser29 | Tyr30 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| MSTRSVSSSSYRRMFGGSGTSSRPSSNRSYVTTSTRTYSLGSALRPSTSR |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | Mus musculus | 90.50 | 10 | vimentin |
| 2 | N/A | 87.00 | 5 | vimentin protein |
| 3 | Rattus norvegicus | 87.00 | 1 | vimentin |
| 4 | Canis familiaris | 74.70 | 1 | PREDICTED: similar to Vimentin |
| 5 | Macaca fascicularis | 67.80 | 1 | VIME_MACFA Vimentin gi |
| 6 | Homo sapiens | 65.90 | 5 | unnamed protein product |
| 7 | synthetic construct | 65.90 | 1 | vimentin |
| 8 | Canis familiaris | 65.50 | 11 | PREDICTED: similar to Vimentin isoform 8 |
| 9 | Pan troglodytes | 65.50 | 1 | vimentin |
| 10 | Ovis aries | 64.70 | 1 | VIME_SHEEP Vimentin gi |
| 11 | Gallus gallus | 54.30 | 2 | vimentin |
| 12 | Xenopus laevis | 43.90 | 1 | VIM1_XENLA Vimentin-1/2 gi |
| 13 | Xenopus tropicalis | 42.70 | 1 | Vim protein |
Top-ranked sequences
| organism | matching |
|---|---|
| Mus musculus | Query 1 MSTRSVSSSSYRRMFGGSGT#SSRPSSNRSYVTTSTRTYSLGSALRPSTSR 50 ||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 1 MSTRSVSSSSYRRMFGGSGT#SSRPSSNRSYVTTSTRTYSLGSALRPSTSR 50 |
| N/A | Query 1 MSTRSVSSSSYRRMFGGSGT#SSRPSSNRSYVTTSTRTYSLGSALRPSTSR 50 ||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 1 MSTRSVSSSSYRRMFGGSGT#SSRPSSNRSYVTTSTRTYSLGSALRPSTSR 50 |
| Rattus norvegicus | Query 1 MSTRSVSSSSYRRMFGGSGT#SSRPSSNRSYVTTSTRTYSLGSALRPSTSR 50 ||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 1 MSTRSVSSSSYRRMFGGSGT#SSRPSSNRSYVTTSTRTYSLGSALRPSTSR 50 |
| Canis familiaris | Query 1 MSTRSVSSSSYRRMFGGSGT#SSRPSSNRSYVTTSTRTYSLGSALRPSTSR 50 ||||||| ||||||| ||# ||||| ||||||||||||||||+|||||| Sbjct 1 MSTRSVSLSSYRRMFCVPGT#GSRPSSTRSYVTTSTRTYSLGSAMRPSTSR 50 |
| Macaca fascicularis | Query 1 MSTRSVSSSSYRRMFGGSGT#SSRPSSNRSYVTTSTRTYSLGSALRPSTSR 50 |+||||||||||||||| ||#+|||||+||||||||||||||||||||||| Sbjct 1 MTTRSVSSSSYRRMFGGPGT#ASRPSSSRSYVTTSTRTYSLGSALRPSTSR 50 |
| Homo sapiens | Query 13 RMFGGSGT#SSRPSSNRSYVTTSTRTYSLGSALRPSTSR 50 ||||| ||#+|||||+||||||||||||||||||||||| Sbjct 13 RMFGGPGT#ASRPSSSRSYVTTSTRTYSLGSALRPSTSR 50 |
| synthetic construct | Query 13 RMFGGSGT#SSRPSSNRSYVTTSTRTYSLGSALRPSTSR 50 ||||| ||#+|||||+||||||||||||||||||||||| Sbjct 13 RMFGGPGT#ASRPSSSRSYVTTSTRTYSLGSALRPSTSR 50 |
| Canis familiaris | Query 13 RMFGGSGT#SSRPSSNRSYVTTSTRTYSLGSALRPSTSR 50 ||||| ||# ||||| ||||||||||||||||||||||| Sbjct 13 RMFGGPGT#GSRPSSTRSYVTTSTRTYSLGSALRPSTSR 50 |
| Pan troglodytes | Query 13 RMFGGSGT#SSRPSSNRSYVTTSTRTYSLGSALRPSTSR 50 ||||| ||#+|||||+||||||||||||||||||||||| Sbjct 13 RMFGGPGT#ASRPSSSRSYVTTSTRTYSLGSALRPSTSR 50 |
| Ovis aries | Query 13 RMFGGSGT#SSRPSSNRSYVTTSTRTYSLGSALRPSTSR 50 ||||| ||#+||||| |||||||||||||||||||+||| Sbjct 13 RMFGGPGT#ASRPSSTRSYVTTSTRTYSLGSALRPTTSR 50 |
| Gallus gallus | Query 1 MSTRSVSSSSYRRMFGGSGT#SSRPSSNRSYVTTSTRTYSLGSALRPSTSR 50 || | +||||||||| # ||||| |+|+||| ||||||||||++| Sbjct 1 MSFTSSKNSSYRRMFGGG--#-SRPSSGTRYITSSTR-YSLGSALRPSSAR 46 |
| Xenopus laevis | Query 5 SVSSSSYRRMFGGSGT#SSRPSSNRSYVTTSTRTYSLGSALRPSTS 49 + + |||||+|||+ #|| || | |+||| |+||||+||||| Sbjct 2 ATTKSSYRRIFGGNPR#SS--SSGNRYATSSTR-YTLGSAMRPSTS 43 |
| Xenopus tropicalis | Query 2 STRSVSSSSYRRMFGGSGT#SSRPSSNRSYVTTSTRTYSLGSALRP 46 +| + + |||||+|||+ #|| || |||+||| ||||||+|| Sbjct 23 TTMATTKSSYRRIFGGNPR#SS--SSGNRYVTSSTR-YSLGSAMRP 64 |
[Site 3] RPSSNRSYVT32-TSTRTYSLGS
Thr32  Thr
Thr
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Arg23 | Pro24 | Ser25 | Ser26 | Asn27 | Arg28 | Ser29 | Tyr30 | Val31 | Thr32 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Thr33 | Ser34 | Thr35 | Arg36 | Thr37 | Tyr38 | Ser39 | Leu40 | Gly41 | Ser42 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| TRSVSSSSYRRMFGGSGTSSRPSSNRSYVTTSTRTYSLGSALRPSTSRSLYSSSPGGAYV |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | Mus musculus | 117.00 | 11 | vimentin |
| 2 | N/A | 117.00 | 5 | vimentin protein |
| 3 | Rattus norvegicus | 117.00 | 1 | vimentin |
| 4 | Canis familiaris | 94.40 | 1 | PREDICTED: similar to Vimentin |
| 5 | Canis familiaris | 88.60 | 11 | PREDICTED: similar to vimentin isoform 1 |
| 6 | Homo sapiens | 88.20 | 5 | vimentin |
| 7 | synthetic construct | 88.20 | 1 | vimentin |
| 8 | Pan troglodytes | 88.20 | 1 | vimentin |
| 9 | Macaca fascicularis | 88.20 | 1 | VIME_MACFA Vimentin gi |
| 10 | Ovis aries | 81.60 | 1 | VIME_SHEEP Vimentin gi |
| 11 | Gallus gallus | 59.70 | 1 | vimentin |
| 12 | Xenopus tropicalis | 45.80 | 1 | Vim protein |
| 13 | Xenopus laevis | 45.10 | 1 | VIM1_XENLA Vimentin-1/2 gi |
Top-ranked sequences
| organism | matching |
|---|---|
| Mus musculus | Query 3 TRSVSSSSYRRMFGGSGTSSRPSSNRSYVT#TSTRTYSLGSALRPSTSRSLYSSSPGGAYV 62 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 3 TRSVSSSSYRRMFGGSGTSSRPSSNRSYVT#TSTRTYSLGSALRPSTSRSLYSSSPGGAYV 62 |
| N/A | Query 3 TRSVSSSSYRRMFGGSGTSSRPSSNRSYVT#TSTRTYSLGSALRPSTSRSLYSSSPGGAYV 62 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 3 TRSVSSSSYRRMFGGSGTSSRPSSNRSYVT#TSTRTYSLGSALRPSTSRSLYSSSPGGAYV 62 |
| Rattus norvegicus | Query 3 TRSVSSSSYRRMFGGSGTSSRPSSNRSYVT#TSTRTYSLGSALRPSTSRSLYSSSPGGAYV 62 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 3 TRSVSSSSYRRMFGGSGTSSRPSSNRSYVT#TSTRTYSLGSALRPSTSRSLYSSSPGGAYV 62 |
| Canis familiaris | Query 3 TRSVSSSSYRRMFGGSGTSSRPSSNRSYVT#TSTRTYSLGSALRPSTSRSLYSSSPGGAY 61 ||||| ||||||| || ||||| |||||#|||||||||||+|||||||||+||||| + Sbjct 3 TRSVSLSSYRRMFCVPGTGSRPSSTRSYVT#TSTRTYSLGSAMRPSTSRSLYASSPGGVF 61 |
| Canis familiaris | Query 13 RMFGGSGTSSRPSSNRSYVT#TSTRTYSLGSALRPSTSRSLYSSSPGGAY 61 ||||| || ||||| |||||#|||||||||||||||||||||+||||||| Sbjct 13 RMFGGPGTGSRPSSTRSYVT#TSTRTYSLGSALRPSTSRSLYASSPGGAY 61 |
| Homo sapiens | Query 13 RMFGGSGTSSRPSSNRSYVT#TSTRTYSLGSALRPSTSRSLYSSSPGGAY 61 ||||| ||+|||||+|||||#|||||||||||||||||||||+||||| | Sbjct 13 RMFGGPGTASRPSSSRSYVT#TSTRTYSLGSALRPSTSRSLYASSPGGVY 61 |
| synthetic construct | Query 13 RMFGGSGTSSRPSSNRSYVT#TSTRTYSLGSALRPSTSRSLYSSSPGGAY 61 ||||| ||+|||||+|||||#|||||||||||||||||||||+||||| | Sbjct 13 RMFGGPGTASRPSSSRSYVT#TSTRTYSLGSALRPSTSRSLYASSPGGVY 61 |
| Pan troglodytes | Query 13 RMFGGSGTSSRPSSNRSYVT#TSTRTYSLGSALRPSTSRSLYSSSPGGAY 61 ||||| ||+|||||+|||||#|||||||||||||||||||||+||||| | Sbjct 13 RMFGGPGTASRPSSSRSYVT#TSTRTYSLGSALRPSTSRSLYASSPGGVY 61 |
| Macaca fascicularis | Query 13 RMFGGSGTSSRPSSNRSYVT#TSTRTYSLGSALRPSTSRSLYSSSPGGAY 61 ||||| ||+|||||+|||||#|||||||||||||||||||||+||||| | Sbjct 13 RMFGGPGTASRPSSSRSYVT#TSTRTYSLGSALRPSTSRSLYASSPGGVY 61 |
| Ovis aries | Query 13 RMFGGSGTSSRPSSNRSYVT#TSTRTYSLGSALRPSTSRSLYSSSPGGAY 61 ||||| ||+||||| |||||#||||||||||||||+|||+||+||||| | Sbjct 13 RMFGGPGTASRPSSTRSYVT#TSTRTYSLGSALRPTTSRTLYTSSPGGVY 61 |
| Gallus gallus | Query 7 SSSSYRRMFGGSGTSSRPSSNRSYVT#TSTRTYSLGSALRPSTSRSLYSSSPGGAY 61 +||||||||| ||||| |+|#+||| ||||||||||++| + |+|||| | Sbjct 7 KNSSYRRMFGGG---SRPSSGTRYIT#SSTR-YSLGSALRPSSARYV-SASPGGVY 56 |
| Xenopus tropicalis | Query 3 TRSVSSSSYRRMFGGSGTSSRPSSNRSYVT#TSTRTYSLGSALRP----STSRSLYSSS 56 | + + |||||+|||+ || | || |||#+||| ||||||+|| |+|| +||+| Sbjct 24 TMATTKSSYRRIFGGNPRSS-SSGNR-YVT#SSTR-YSLGSAMRPGAGSSSSRVVYSTS 78 |
| Xenopus laevis | Query 7 SSSSYRRMFGGSGTSSRPSSNRSYVT#TSTRTYSLGSALRPST-SRSLYSSS 56 + |||||+|||+ || | || | |#+||| |+||||+|||| || +||+| Sbjct 4 TKSSYRRIFGGNPRSS-SSGNR-YAT#SSTR-YTLGSAMRPSTSSRMVYSTS 51 |
[Site 4] RSYVTTSTRT37-YSLGSALRPS
Thr37  Tyr
Tyr
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Arg28 | Ser29 | Tyr30 | Val31 | Thr32 | Thr33 | Ser34 | Thr35 | Arg36 | Thr37 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Tyr38 | Ser39 | Leu40 | Gly41 | Ser42 | Ala43 | Leu44 | Arg45 | Pro46 | Ser47 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| SSSYRRMFGGSGTSSRPSSNRSYVTTSTRTYSLGSALRPSTSRSLYSSSPGGAYVTRSSA |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | Mus musculus | 117.00 | 11 | unnamed protein product |
| 2 | N/A | 117.00 | 5 | vimentin protein |
| 3 | Rattus norvegicus | 117.00 | 1 | vimentin |
| 4 | Canis familiaris | 96.70 | 11 | PREDICTED: similar to vimentin isoform 1 |
| 5 | Homo sapiens | 96.30 | 5 | VIM |
| 6 | synthetic construct | 96.30 | 1 | vimentin |
| 7 | Macaca fascicularis | 96.30 | 1 | VIME_MACFA Vimentin gi |
| 8 | Pan troglodytes | 96.30 | 1 | vimentin |
| 9 | Canis familiaris | 94.70 | 1 | PREDICTED: similar to Vimentin |
| 10 | Ovis aries | 89.40 | 1 | VIME_SHEEP Vimentin gi |
| 11 | Gallus gallus | 60.80 | 2 | vimentin |
| 12 | Xenopus tropicalis | 45.10 | 1 | Vim protein |
| 13 | Xenopus laevis | 44.70 | 1 | VIM1_XENLA Vimentin-1/2 gi |
Top-ranked sequences
| organism | matching |
|---|---|
| Mus musculus | Query 8 SSSYRRMFGGSGTSSRPSSNRSYVTTSTRT#YSLGSALRPSTSRSLYSSSPGGAYVTRSSA 67 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 8 SSSYRRMFGGSGTSSRPSSNRSYVTTSTRT#YSLGSALRPSTSRSLYSSSPGGAYVTRSSA 67 |
| N/A | Query 8 SSSYRRMFGGSGTSSRPSSNRSYVTTSTRT#YSLGSALRPSTSRSLYSSSPGGAYVTRSSA 67 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 8 SSSYRRMFGGSGTSSRPSSNRSYVTTSTRT#YSLGSALRPSTSRSLYSSSPGGAYVTRSSA 67 |
| Rattus norvegicus | Query 8 SSSYRRMFGGSGTSSRPSSNRSYVTTSTRT#YSLGSALRPSTSRSLYSSSPGGAYVTRSSA 67 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 8 SSSYRRMFGGSGTSSRPSSNRSYVTTSTRT#YSLGSALRPSTSRSLYSSSPGGAYVTRSSA 67 |
| Canis familiaris | Query 14 MFGGSGTSSRPSSNRSYVTTSTRT#YSLGSALRPSTSRSLYSSSPGGAYVTRSSA 67 |||| || ||||| ||||||||||#||||||||||||||||+||||||| ||||| Sbjct 14 MFGGPGTGSRPSSTRSYVTTSTRT#YSLGSALRPSTSRSLYASSPGGAYATRSSA 67 |
| Homo sapiens | Query 14 MFGGSGTSSRPSSNRSYVTTSTRT#YSLGSALRPSTSRSLYSSSPGGAYVTRSSA 67 |||| ||+|||||+||||||||||#||||||||||||||||+||||| | ||||| Sbjct 14 MFGGPGTASRPSSSRSYVTTSTRT#YSLGSALRPSTSRSLYASSPGGVYATRSSA 67 |
| synthetic construct | Query 14 MFGGSGTSSRPSSNRSYVTTSTRT#YSLGSALRPSTSRSLYSSSPGGAYVTRSSA 67 |||| ||+|||||+||||||||||#||||||||||||||||+||||| | ||||| Sbjct 14 MFGGPGTASRPSSSRSYVTTSTRT#YSLGSALRPSTSRSLYASSPGGVYATRSSA 67 |
| Macaca fascicularis | Query 14 MFGGSGTSSRPSSNRSYVTTSTRT#YSLGSALRPSTSRSLYSSSPGGAYVTRSSA 67 |||| ||+|||||+||||||||||#||||||||||||||||+||||| | ||||| Sbjct 14 MFGGPGTASRPSSSRSYVTTSTRT#YSLGSALRPSTSRSLYASSPGGVYATRSSA 67 |
| Pan troglodytes | Query 14 MFGGSGTSSRPSSNRSYVTTSTRT#YSLGSALRPSTSRSLYSSSPGGAYVTRSSA 67 |||| ||+|||||+||||||||||#||||||||||||||||+||||| | ||||| Sbjct 14 MFGGPGTASRPSSSRSYVTTSTRT#YSLGSALRPSTSRSLYASSPGGVYATRSSA 67 |
| Canis familiaris | Query 9 SSYRRMFGGSGTSSRPSSNRSYVTTSTRT#YSLGSALRPSTSRSLYSSSPGGAYVTRSSA 67 ||||||| || ||||| ||||||||||#||||||+|||||||||+||||| + ||||| Sbjct 9 SSYRRMFCVPGTGSRPSSTRSYVTTSTRT#YSLGSAMRPSTSRSLYASSPGGVFTTRSSA 67 |
| Ovis aries | Query 14 MFGGSGTSSRPSSNRSYVTTSTRT#YSLGSALRPSTSRSLYSSSPGGAYVTRSSA 67 |||| ||+||||| ||||||||||#|||||||||+|||+||+||||| | ||||| Sbjct 14 MFGGPGTASRPSSTRSYVTTSTRT#YSLGSALRPTTSRTLYTSSPGGVYATRSSA 67 |
| Gallus gallus | Query 8 SSSYRRMFGGSGTSSRPSSNRSYVTTSTRT#YSLGSALRPSTSRSLYSSSPGGAYVTRSS 66 +||||||||| ||||| |+|+||| #||||||||||++| + |+|||| | |+++ Sbjct 8 NSSYRRMFGGG---SRPSSGTRYITSSTR-#YSLGSALRPSSARYV-SASPGGVYRTKAT 61 |
| Xenopus tropicalis | Query 9 SSYRRMFGGSGTSSRPSSNRSYVTTSTRT#YSLGSALRP----STSRSLYSSS 56 |||||+|||+ || | || |||+||| #||||||+|| |+|| +||+| Sbjct 30 SSYRRIFGGNPRSS-SSGNR-YVTSSTR-#YSLGSAMRPGAGSSSSRVVYSTS 78 |
| Xenopus laevis | Query 9 SSYRRMFGGSGTSSRPSSNRSYVTTSTRT#YSLGSALRPST-SRSLYSSS 56 |||||+|||+ || | || | |+||| #|+||||+|||| || +||+| Sbjct 6 SSYRRIFGGNPRSS-SSGNR-YATSSTR-#YTLGSAMRPSTSSRMVYSTS 51 |
[Site 5] VTTSTRTYSL40-GSALRPSTSR
Leu40  Gly
Gly
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Val31 | Thr32 | Thr33 | Ser34 | Thr35 | Arg36 | Thr37 | Tyr38 | Ser39 | Leu40 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Gly41 | Ser42 | Ala43 | Leu44 | Arg45 | Pro46 | Ser47 | Thr48 | Ser49 | Arg50 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| YRRMFGGSGTSSRPSSNRSYVTTSTRTYSLGSALRPSTSRSLYSSSPGGAYVTRSSAVRL |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | Mus musculus | 120.00 | 11 | unnamed protein product |
| 2 | N/A | 120.00 | 5 | vimentin protein |
| 3 | Rattus norvegicus | 119.00 | 1 | vimentin |
| 4 | Canis familiaris | 102.00 | 11 | PREDICTED: similar to vimentin isoform 1 |
| 5 | Homo sapiens | 102.00 | 5 | VIM |
| 6 | synthetic construct | 102.00 | 1 | vimentin |
| 7 | Pan troglodytes | 102.00 | 1 | vimentin |
| 8 | Macaca fascicularis | 102.00 | 1 | VIME_MACFA Vimentin gi |
| 9 | Canis familiaris | 97.10 | 1 | PREDICTED: similar to Vimentin |
| 10 | Ovis aries | 95.10 | 1 | VIME_SHEEP Vimentin gi |
| 11 | Gallus gallus | 64.30 | 2 | vimentin |
| 12 | Xenopus tropicalis | 43.10 | 1 | Vim protein |
| 13 | Xenopus laevis | 43.10 | 1 | VIM1_XENLA Vimentin-1/2 gi |
| 14 | Bos taurus | 37.40 | 1 | similar to vimentin |
Top-ranked sequences
| organism | matching |
|---|---|
| Mus musculus | Query 11 YRRMFGGSGTSSRPSSNRSYVTTSTRTYSL#GSALRPSTSRSLYSSSPGGAYVTRSSAVRL 70 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 11 YRRMFGGSGTSSRPSSNRSYVTTSTRTYSL#GSALRPSTSRSLYSSSPGGAYVTRSSAVRL 70 |
| N/A | Query 11 YRRMFGGSGTSSRPSSNRSYVTTSTRTYSL#GSALRPSTSRSLYSSSPGGAYVTRSSAVRL 70 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 11 YRRMFGGSGTSSRPSSNRSYVTTSTRTYSL#GSALRPSTSRSLYSSSPGGAYVTRSSAVRL 70 |
| Rattus norvegicus | Query 11 YRRMFGGSGTSSRPSSNRSYVTTSTRTYSL#GSALRPSTSRSLYSSSPGGAYVTRSSAVRL 70 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 11 YRRMFGGSGTSSRPSSNRSYVTTSTRTYSL#GSALRPSTSRSLYSSSPGGAYVTRSSAVRL 70 |
| Canis familiaris | Query 14 MFGGSGTSSRPSSNRSYVTTSTRTYSL#GSALRPSTSRSLYSSSPGGAYVTRSSAVRL 70 |||| || ||||| |||||||||||||#|||||||||||||+||||||| |||||||| Sbjct 14 MFGGPGTGSRPSSTRSYVTTSTRTYSL#GSALRPSTSRSLYASSPGGAYATRSSAVRL 70 |
| Homo sapiens | Query 14 MFGGSGTSSRPSSNRSYVTTSTRTYSL#GSALRPSTSRSLYSSSPGGAYVTRSSAVRL 70 |||| ||+|||||+|||||||||||||#|||||||||||||+||||| | |||||||| Sbjct 14 MFGGPGTASRPSSSRSYVTTSTRTYSL#GSALRPSTSRSLYASSPGGVYATRSSAVRL 70 |
| synthetic construct | Query 14 MFGGSGTSSRPSSNRSYVTTSTRTYSL#GSALRPSTSRSLYSSSPGGAYVTRSSAVRL 70 |||| ||+|||||+|||||||||||||#|||||||||||||+||||| | |||||||| Sbjct 14 MFGGPGTASRPSSSRSYVTTSTRTYSL#GSALRPSTSRSLYASSPGGVYATRSSAVRL 70 |
| Pan troglodytes | Query 14 MFGGSGTSSRPSSNRSYVTTSTRTYSL#GSALRPSTSRSLYSSSPGGAYVTRSSAVRL 70 |||| ||+|||||+|||||||||||||#|||||||||||||+||||| | |||||||| Sbjct 14 MFGGPGTASRPSSSRSYVTTSTRTYSL#GSALRPSTSRSLYASSPGGVYATRSSAVRL 70 |
| Macaca fascicularis | Query 14 MFGGSGTSSRPSSNRSYVTTSTRTYSL#GSALRPSTSRSLYSSSPGGAYVTRSSAVRL 70 |||| ||+|||||+|||||||||||||#|||||||||||||+||||| | |||||||| Sbjct 14 MFGGPGTASRPSSSRSYVTTSTRTYSL#GSALRPSTSRSLYASSPGGVYATRSSAVRL 70 |
| Canis familiaris | Query 11 YRRMFGGSGTSSRPSSNRSYVTTSTRTYSL#GSALRPSTSRSLYSSSPGGAYVTRSSAVRL 70 ||||| || ||||| |||||||||||||#|||+|||||||||+||||| + |||||+|| Sbjct 11 YRRMFCVPGTGSRPSSTRSYVTTSTRTYSL#GSAMRPSTSRSLYASSPGGVFTTRSSAMRL 70 |
| Ovis aries | Query 14 MFGGSGTSSRPSSNRSYVTTSTRTYSL#GSALRPSTSRSLYSSSPGGAYVTRSSAVRL 70 |||| ||+||||| |||||||||||||#||||||+|||+||+||||| | |||||||| Sbjct 14 MFGGPGTASRPSSTRSYVTTSTRTYSL#GSALRPTTSRTLYTSSPGGVYATRSSAVRL 70 |
| Gallus gallus | Query 11 YRRMFGGSGTSSRPSSNRSYVTTSTRTYSL#GSALRPSTSRSLYSSSPGGAYVTRSSAVRL 70 ||||||| ||||| |+|+||| |||#|||||||++| + |+|||| | |++++||| Sbjct 11 YRRMFGGG---SRPSSGTRYITSSTR-YSL#GSALRPSSARYV-SASPGGVYRTKATSVRL 65 |
| Xenopus tropicalis | Query 11 YRRMFGGSGTSSRPSSNRSYVTTSTRTYSL#GSALRP----STSRSLYSSS 56 |||+|||+ || | || |||+||| |||#|||+|| |+|| +||+| Sbjct 32 YRRIFGGNPRSS-SSGNR-YVTSSTR-YSL#GSAMRPGAGSSSSRVVYSTS 78 |
| Xenopus laevis | Query 11 YRRMFGGSGTSSRPSSNRSYVTTSTRTYSL#GSALRPST-SRSLYSSS 56 |||+|||+ || | || | |+||| |+|#|||+|||| || +||+| Sbjct 8 YRRIFGGNPRSS-SSGNR-YATSSTR-YTL#GSAMRPSTSSRMVYSTS 51 |
| Bos taurus | Query 50 RSLYSSSPGGAYVTRSSAVRL 70 |+||+||||| | |||||||| Sbjct 1 RTLYTSSPGGVYATRSSAVRL 21 |
[Site 6] ALRPSTSRSL52-YSSSPGGAYV
Leu52  Tyr
Tyr
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Ala43 | Leu44 | Arg45 | Pro46 | Ser47 | Thr48 | Ser49 | Arg50 | Ser51 | Leu52 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Tyr53 | Ser54 | Ser55 | Ser56 | Pro57 | Gly58 | Gly59 | Ala60 | Tyr61 | Val62 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| RPSSNRSYVTTSTRTYSLGSALRPSTSRSLYSSSPGGAYVTRSSAVRLRSSVPGVRLLQD |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | Mus musculus | 119.00 | 12 | unnamed protein product |
| 2 | N/A | 119.00 | 5 | vimentin protein |
| 3 | Rattus norvegicus | 117.00 | 1 | vimentin |
| 4 | Canis familiaris | 113.00 | 11 | PREDICTED: similar to vimentin isoform 1 |
| 5 | Homo sapiens | 111.00 | 5 | vimentin |
| 6 | Pan troglodytes | 111.00 | 1 | vimentin |
| 7 | synthetic construct | 111.00 | 1 | vimentin |
| 8 | Macaca fascicularis | 111.00 | 1 | VIME_MACFA Vimentin gi |
| 9 | Canis familiaris | 104.00 | 1 | PREDICTED: similar to Vimentin |
| 10 | Ovis aries | 103.00 | 1 | VIME_SHEEP Vimentin gi |
| 11 | Gallus gallus | 63.50 | 2 | vimentin |
| 12 | Bos taurus | 59.70 | 1 | similar to vimentin |
| 13 | Daboia russellii | 38.10 | 1 | AF447708_1 vimentin |
| 14 | Xenopus laevis | 33.50 | 1 | VIM1_XENLA Vimentin-1/2 gi |
Top-ranked sequences
| organism | matching |
|---|---|
| Mus musculus | Query 23 RPSSNRSYVTTSTRTYSLGSALRPSTSRSL#YSSSPGGAYVTRSSAVRLRSSVPGVRLLQD 82 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 23 RPSSNRSYVTTSTRTYSLGSALRPSTSRSL#YSSSPGGAYVTRSSAVRLRSSVPGVRLLQD 82 |
| N/A | Query 23 RPSSNRSYVTTSTRTYSLGSALRPSTSRSL#YSSSPGGAYVTRSSAVRLRSSVPGVRLLQD 82 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 23 RPSSNRSYVTTSTRTYSLGSALRPSTSRSL#YSSSPGGAYVTRSSAVRLRSSVPGVRLLQD 82 |
| Rattus norvegicus | Query 23 RPSSNRSYVTTSTRTYSLGSALRPSTSRSL#YSSSPGGAYVTRSSAVRLRSSVPGVRLLQD 82 ||||||||||||||||||||||||||||||#|||||||||||||||||||||+|||||||| Sbjct 23 RPSSNRSYVTTSTRTYSLGSALRPSTSRSL#YSSSPGGAYVTRSSAVRLRSSMPGVRLLQD 82 |
| Canis familiaris | Query 23 RPSSNRSYVTTSTRTYSLGSALRPSTSRSL#YSSSPGGAYVTRSSAVRLRSSVPGVRLLQD 82 |||| |||||||||||||||||||||||||#|+||||||| |||||||||||||||||||| Sbjct 23 RPSSTRSYVTTSTRTYSLGSALRPSTSRSL#YASSPGGAYATRSSAVRLRSSVPGVRLLQD 82 |
| Homo sapiens | Query 23 RPSSNRSYVTTSTRTYSLGSALRPSTSRSL#YSSSPGGAYVTRSSAVRLRSSVPGVRLLQD 82 ||||+|||||||||||||||||||||||||#|+||||| | |||||||||||||||||||| Sbjct 23 RPSSSRSYVTTSTRTYSLGSALRPSTSRSL#YASSPGGVYATRSSAVRLRSSVPGVRLLQD 82 |
| Pan troglodytes | Query 23 RPSSNRSYVTTSTRTYSLGSALRPSTSRSL#YSSSPGGAYVTRSSAVRLRSSVPGVRLLQD 82 ||||+|||||||||||||||||||||||||#|+||||| | |||||||||||||||||||| Sbjct 23 RPSSSRSYVTTSTRTYSLGSALRPSTSRSL#YASSPGGVYATRSSAVRLRSSVPGVRLLQD 82 |
| synthetic construct | Query 23 RPSSNRSYVTTSTRTYSLGSALRPSTSRSL#YSSSPGGAYVTRSSAVRLRSSVPGVRLLQD 82 ||||+|||||||||||||||||||||||||#|+||||| | |||||||||||||||||||| Sbjct 23 RPSSSRSYVTTSTRTYSLGSALRPSTSRSL#YASSPGGVYATRSSAVRLRSSVPGVRLLQD 82 |
| Macaca fascicularis | Query 23 RPSSNRSYVTTSTRTYSLGSALRPSTSRSL#YSSSPGGAYVTRSSAVRLRSSVPGVRLLQD 82 ||||+|||||||||||||||||||||||||#|+||||| | |||||||||||||||||||| Sbjct 23 RPSSSRSYVTTSTRTYSLGSALRPSTSRSL#YASSPGGVYATRSSAVRLRSSVPGVRLLQD 82 |
| Canis familiaris | Query 23 RPSSNRSYVTTSTRTYSLGSALRPSTSRSL#YSSSPGGAYVTRSSAVRLRSSVPGVRLLQD 82 |||| ||||||||||||||||+||||||||#|+||||| + |||||+||| ||||+||||| Sbjct 23 RPSSTRSYVTTSTRTYSLGSAMRPSTSRSL#YASSPGGVFTTRSSAMRLRGSVPGMRLLQD 82 |
| Ovis aries | Query 23 RPSSNRSYVTTSTRTYSLGSALRPSTSRSL#YSSSPGGAYVTRSSAVRLRSSVPGVRLLQD 82 |||| |||||||||||||||||||+|||+|#|+||||| | |||||||||| ||||||||| Sbjct 23 RPSSTRSYVTTSTRTYSLGSALRPTTSRTL#YTSSPGGVYATRSSAVRLRSGVPGVRLLQD 82 |
| Gallus gallus | Query 23 RPSSNRSYVTTSTRTYSLGSALRPSTSRSL#YSSSPGGAYVTRSSAVRLRSSVPGVRL 79 |||| |+|+||| ||||||||||++| +# |+|||| | |++++||||||+| +|+ Sbjct 20 RPSSGTRYITSSTR-YSLGSALRPSSARYV#-SASPGGVYRTKATSVRLRSSMPPMRM 74 |
| Bos taurus | Query 50 RSL#YSSSPGGAYVTRSSAVRLRSSVPGVRLLQD 82 |+|#|+||||| | |||||||||| ||||||||| Sbjct 1 RTL#YTSSPGGVYATRSSAVRLRSGVPGVRLLQD 33 |
| Daboia russellii | Query 38 YSLGSALRPSTSRSL#YSSSPGGAYVTRSSAVRLRSSVPGVR 78 |||||++|||+ | +# | ||| | | |+|+||| | +| Sbjct 36 YSLGSSIRPSSVRLV#--SCPGGIYA--SKAMRVRSSFPPMR 72 |
| Xenopus laevis | Query 25 SSNRSYVTTSTRTYSLGSALRPST-SRSL#YSSS 56 | || | |+||| |+||||+|||| || +#||+| Sbjct 21 SGNR-YATSSTR-YTLGSAMRPSTSSRMV#YSTS 51 |
[Site 7] SSSPGGAYVT63-RSSAVRLRSS
Thr63  Arg
Arg
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Ser54 | Ser55 | Ser56 | Pro57 | Gly58 | Gly59 | Ala60 | Tyr61 | Val62 | Thr63 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Arg64 | Ser65 | Ser66 | Ala67 | Val68 | Arg69 | Leu70 | Arg71 | Ser72 | Ser73 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| STRTYSLGSALRPSTSRSLYSSSPGGAYVTRSSAVRLRSSVPGVRLLQDSVDFSLADAIN |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | Mus musculus | 120.00 | 12 | unnamed protein product |
| 2 | N/A | 120.00 | 7 | vimentin protein |
| 3 | Rattus norvegicus | 119.00 | 1 | vimentin |
| 4 | Canis familiaris | 117.00 | 11 | PREDICTED: similar to vimentin isoform 1 |
| 5 | Homo sapiens | 110.00 | 5 | unnamed protein product |
| 6 | synthetic construct | 109.00 | 1 | vimentin |
| 7 | Pan troglodytes | 108.00 | 1 | vimentin |
| 8 | Ovis aries | 108.00 | 1 | VIME_SHEEP Vimentin gi |
| 9 | Macaca fascicularis | 108.00 | 1 | VIME_MACFA Vimentin gi |
| 10 | Canis familiaris | 103.00 | 1 | PREDICTED: similar to Vimentin |
| 11 | Bos taurus | 81.30 | 1 | similar to vimentin |
| 12 | Gallus gallus | 71.60 | 2 | vimentin |
| 13 | Daboia russellii | 54.70 | 1 | AF447708_1 vimentin |
| 14 | Acipenser baerii | 48.10 | 1 | vimentin |
| 15 | Xenopus laevis | 44.70 | 4 | VIM1_XENLA Vimentin-1/2 gi |
| 16 | Oncorhynchus mykiss | 43.50 | 1 | VIME_ONCMY Vimentin gi |
| 17 | Xenopus tropicalis | 42.00 | 1 | Vim protein |
| 18 | Danio rerio | 38.90 | 3 | vimentin |
| 19 | Cyprinus carpio | 34.70 | 1 | VIME_CYPCA Vimentin gi |
| 20 | Tetraodon nigroviridis | 32.70 | 1 | unnamed protein product |
| 21 | Synechococcus sp. CC9902 | 30.40 | 1 | hypothetical protein Syncc9902_1368 |
Top-ranked sequences
| organism | matching |
|---|---|
| Mus musculus | Query 34 STRTYSLGSALRPSTSRSLYSSSPGGAYVT#RSSAVRLRSSVPGVRLLQDSVDFSLADAIN 93 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 34 STRTYSLGSALRPSTSRSLYSSSPGGAYVT#RSSAVRLRSSVPGVRLLQDSVDFSLADAIN 93 |
| N/A | Query 34 STRTYSLGSALRPSTSRSLYSSSPGGAYVT#RSSAVRLRSSVPGVRLLQDSVDFSLADAIN 93 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 34 STRTYSLGSALRPSTSRSLYSSSPGGAYVT#RSSAVRLRSSVPGVRLLQDSVDFSLADAIN 93 |
| Rattus norvegicus | Query 34 STRTYSLGSALRPSTSRSLYSSSPGGAYVT#RSSAVRLRSSVPGVRLLQDSVDFSLADAIN 93 ||||||||||||||||||||||||||||||#||||||||||+||||||||||||||||||| Sbjct 34 STRTYSLGSALRPSTSRSLYSSSPGGAYVT#RSSAVRLRSSMPGVRLLQDSVDFSLADAIN 93 |
| Canis familiaris | Query 34 STRTYSLGSALRPSTSRSLYSSSPGGAYVT#RSSAVRLRSSVPGVRLLQDSVDFSLADAIN 93 ||||||||||||||||||||+||||||| |#|||||||||||||||||||||||||||||| Sbjct 34 STRTYSLGSALRPSTSRSLYASSPGGAYAT#RSSAVRLRSSVPGVRLLQDSVDFSLADAIN 93 |
| Homo sapiens | Query 34 STRTYSLGSALRPSTSRSLYSSSPGGAYVT#RSSAVRLRSSVPGVRLLQDSVDFSLADAIN 93 ||||||||||||||||||||+||||| | |#|||||||||||||||||||||||||||||| Sbjct 34 STRTYSLGSALRPSTSRSLYASSPGGVYAT#RSSAVRLRSSVPGVRLLQDSVDFSLADAIN 93 |
| synthetic construct | Query 34 STRTYSLGSALRPSTSRSLYSSSPGGAYVT#RSSAVRLRSSVPGVRLLQDSVDFSLADAIN 93 ||||||||||||||||||||+||||| | |#|||||||||||||||||||||||||||||| Sbjct 34 STRTYSLGSALRPSTSRSLYASSPGGVYAT#RSSAVRLRSSVPGVRLLQDSVDFSLADAIN 93 |
| Pan troglodytes | Query 34 STRTYSLGSALRPSTSRSLYSSSPGGAYVT#RSSAVRLRSSVPGVRLLQDSVDFSLADAIN 93 ||||||||||||||||||||+||||| | |#|||||||||||||||||||||||||||||| Sbjct 34 STRTYSLGSALRPSTSRSLYASSPGGVYAT#RSSAVRLRSSVPGVRLLQDSVDFSLADAIN 93 |
| Ovis aries | Query 34 STRTYSLGSALRPSTSRSLYSSSPGGAYVT#RSSAVRLRSSVPGVRLLQDSVDFSLADAIN 93 |||||||||||||+|||+||+||||| | |#||||||||| |||||||||||||||||||| Sbjct 34 STRTYSLGSALRPTTSRTLYTSSPGGVYAT#RSSAVRLRSGVPGVRLLQDSVDFSLADAIN 93 |
| Macaca fascicularis | Query 34 STRTYSLGSALRPSTSRSLYSSSPGGAYVT#RSSAVRLRSSVPGVRLLQDSVDFSLADAIN 93 ||||||||||||||||||||+||||| | |#|||||||||||||||||||||||||||||| Sbjct 34 STRTYSLGSALRPSTSRSLYASSPGGVYAT#RSSAVRLRSSVPGVRLLQDSVDFSLADAIN 93 |
| Canis familiaris | Query 34 STRTYSLGSALRPSTSRSLYSSSPGGAYVT#RSSAVRLRSSVPGVRLLQDSVDFSLADAIN 93 ||||||||||+|||||||||+||||| + |#||||+||| ||||+|||||||||||||||| Sbjct 34 STRTYSLGSAMRPSTSRSLYASSPGGVFTT#RSSAMRLRGSVPGMRLLQDSVDFSLADAIN 93 |
| Bos taurus | Query 50 RSLYSSSPGGAYVT#RSSAVRLRSSVPGVRLLQDSVDFSLADAIN 93 |+||+||||| | |#||||||||| |||||||||||||||||||| Sbjct 1 RTLYTSSPGGVYAT#RSSAVRLRSGVPGVRLLQDSVDFSLADAIN 44 |
| Gallus gallus | Query 34 STRTYSLGSALRPSTSRSLYSSSPGGAYVT#RSSAVRLRSSVPGVRLLQDSVDFSLADAIN 93 ||| ||||||||||++| + |+|||| | |#++++||||||+| +| + |+|||+|||||| Sbjct 11 STR-YSLGSALRPSSARYV-SASPGGVYAT#KATSVRLRSSMPPMR-MHDAVDFTLADAIN 67 |
| Daboia russellii | Query 38 YSLGSALRPSTSRSLYSSSPGGAYVT#RSSAVRLRSSVPGVRLLQDS-VDFSLADAIN 93 |||||++|||+ | + | ||| | # | |+|+||| | +| |+ |||||||||| Sbjct 36 YSLGSSIRPSSVRLV--SCPGGIYA-#-SKAMRVRSSFPPMRTGGDAMVDFSLADAIN 88 |
| Acipenser baerii | Query 34 STRTYSLGSALRPSTSRSLYSS-SPGGAYVT#RSSAVRLRSSVPGVRLLQDSVDFSLADAI 92 |||+|| | || |+| | | |#+||+ ||||| | ||| +++||+|+||| Sbjct 23 STRSYSSRQYSSPIRSRVSYTSHSAPTLYAT#KSSSTRLRSSAPQTRLLSETLDFALSDAI 82 Query 93 N 93 | Query 93 N 93 |
| Xenopus laevis | Query 34 STRTYSLGSALRPSTSRSLYSSSPGGAYVT#RSSAVRLRSSVPGVRLLQDSVDFSLADAIN 93 |+ |+||||+||||| + |+ | #+||+||||||+| | + |||||+||||+| Sbjct 28 SSTRYTLGSAMRPSTSSRMVYSTSSSPAVF#KSSSVRLRSSLPPAR-MADSVDFALADAVN 86 |
| Oncorhynchus mykiss | Query 34 STRTYSLGSALRPSTSRSLYS-SSPGGAYVT#RSSAVRLRSSVPGVRLLQDSVDFSLADAI 92 |+| || | +| +|| || |+| | +#++ |||||| | || |+|||+|+||| Sbjct 35 SSRQYS--SPVR--SSRMSYSVSAPPSIYAS#KN--VRLRSSAPMPRLSSDTVDFALSDAI 88 Query 93 N 93 | Query 93 N 93 |
| Xenopus tropicalis | Query 34 STRTYSLGSALRP----STSRSLYSSSPGGAYVT#RSSAVRLRSSVPGVRLLQDSVDFSLA 89 |+ ||||||+|| |+|| +||+| | | #+||+||||||+| | | |+|||+|| Sbjct 52 SSTRYSLGSAMRPGAGSSSSRVVYSTSASPA-VF#KSSSVRLRSSLPPAR-LADTVDFTLA 109 Query 90 DAIN 93 ||+| Sbjct 110 DAVN 113 |
| Danio rerio | Query 34 STRTYSLGSALRPSTSRSLYSSSPGGAYVT#RSSAVRLRSSVPGVRLLQDSVDFSLADAIN 93 |+| || + | + |||+| | # | +|+|| | || |++|| ||+||| Sbjct 27 SSRQYSSPVRVSSSRTSYNYSSAPPSVYA-#-SKGLRVRSGAPLPRLATDTLDFGLAEAIN 84 |
| Cyprinus carpio | Query 34 STRTYSLGSALRPSTSRSLYSSSPGGAYVT#RSSAVRLRSSVPGVRLLQDSVDFSLADAIN 93 |+| || | +| ++ | |+| + # | |+||| | || +++|| |||||| Sbjct 27 SSRQYS--SPVRTTSRVSYSSASSASPSIY#MSKGARVRSSGPLPRLATETLDFGLADAIN 84 |
| Tetraodon nigroviridis | Query 49 SRSLYSSSPGGAYVT#RSSAV------RLRSSVPGVRLLQDSVDFSLADAIN 93 ||+ ||| + ++# + || ||||| || +++||||+|||| Sbjct 22 SRTSYSSRQFSSPLS#SAPAVYALKTQRLRSSSAMPRLASENLDFSLSDAIN 72 |
| Synechococcus sp. CC9902 | Query 36 RTYSLGSALRPSTSRSLYSSSPGGAYVT#RSSAVRLRSSVPGVRLLQDSVDFSLAD 90 | + ++++ + | ++ +| |+|#| |++| + | + +++| +|+| Sbjct 92 RLFERSASIKDKSPRIIFDKTP--RYIT#RLRQVQIRFNQPAIAVIKDPRSLALSD 144 |
[Site 8] YVTRSSAVRL70-RSSVPGVRLL
Leu70  Arg
Arg
iTraq-117 Signal 8882.1 (

 ) for YVTRSSAVRLR
) for YVTRSSAVRLR
iTraq-117 Signal 351.4 (
 ) for TRSSAVRLRSSVPGVRLL
) for TRSSAVRLRSSVPGVRLL
iTraq-117 Signal 13888.7 (


 ) for SSVPGVRLL
) for SSVPGVRLL
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Tyr61 | Val62 | Thr63 | Arg64 | Ser65 | Ser66 | Ala67 | Val68 | Arg69 | Leu70 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Arg71 | Ser72 | Ser73 | Val74 | Pro75 | Gly76 | Val77 | Arg78 | Leu79 | Leu80 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| GSALRPSTSRSLYSSSPGGAYVTRSSAVRLRSSVPGVRLLQDSVDFSLADAINTEFKNTR |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | Mus musculus | 123.00 | 12 | vimentin |
| 2 | N/A | 122.00 | 9 | vimentin protein |
| 3 | Rattus norvegicus | 121.00 | 1 | vimentin |
| 4 | Canis familiaris | 120.00 | 11 | PREDICTED: similar to Vimentin isoform 3 |
| 5 | Homo sapiens | 118.00 | 5 | vimentin |
| 6 | synthetic construct | 118.00 | 1 | vimentin |
| 7 | Macaca fascicularis | 118.00 | 1 | VIME_MACFA Vimentin gi |
| 8 | Pan troglodytes | 117.00 | 1 | vimentin |
| 9 | Ovis aries | 111.00 | 2 | VIME_SHEEP Vimentin gi |
| 10 | Canis familiaris | 103.00 | 1 | PREDICTED: similar to Vimentin |
| 11 | Bos taurus | 95.90 | 1 | similar to vimentin |
| 12 | Gallus gallus | 75.50 | 2 | vimentin |
| 13 | Daboia russellii | 57.00 | 1 | AF447708_1 vimentin |
| 14 | Acipenser baerii | 56.20 | 2 | vimentin |
| 15 | Oncorhynchus mykiss | 48.90 | 1 | VIME_ONCMY Vimentin gi |
| 16 | Danio rerio | 45.80 | 5 | vimentin |
| 17 | Xenopus laevis | 45.10 | 4 | VIM1_XENLA Vimentin-1/2 gi |
| 18 | Xenopus tropicalis | 42.00 | 1 | Vim protein |
| 19 | Cyprinus carpio | 41.60 | 1 | VIME_CYPCA Vimentin gi |
| 20 | Tetraodon nigroviridis | 39.30 | 2 | unnamed protein product |
| 21 | Scyliorhinus stellaris | 35.40 | 1 | vimentin |
| 22 | Takifugu rubripes | 32.30 | 1 | TPA: desmin |
Top-ranked sequences
| organism | matching |
|---|---|
| Mus musculus | Query 41 GSALRPSTSRSLYSSSPGGAYVTRSSAVRL#RSSVPGVRLLQDSVDFSLADAINTEFKNTR 100 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 41 GSALRPSTSRSLYSSSPGGAYVTRSSAVRL#RSSVPGVRLLQDSVDFSLADAINTEFKNTR 100 |
| N/A | Query 41 GSALRPSTSRSLYSSSPGGAYVTRSSAVRL#RSSVPGVRLLQDSVDFSLADAINTEFKNTR 100 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 41 GSALRPSTSRSLYSSSPGGAYVTRSSAVRL#RSSVPGVRLLQDSVDFSLADAINTEFKNTR 100 |
| Rattus norvegicus | Query 41 GSALRPSTSRSLYSSSPGGAYVTRSSAVRL#RSSVPGVRLLQDSVDFSLADAINTEFKNTR 100 ||||||||||||||||||||||||||||||#|||+|||||||||||||||||||||||||| Sbjct 41 GSALRPSTSRSLYSSSPGGAYVTRSSAVRL#RSSMPGVRLLQDSVDFSLADAINTEFKNTR 100 |
| Canis familiaris | Query 41 GSALRPSTSRSLYSSSPGGAYVTRSSAVRL#RSSVPGVRLLQDSVDFSLADAINTEFKNTR 100 |||||||||||||+||||||| ||||||||#|||||||||||||||||||||||||||||| Sbjct 41 GSALRPSTSRSLYASSPGGAYATRSSAVRL#RSSVPGVRLLQDSVDFSLADAINTEFKNTR 100 |
| Homo sapiens | Query 41 GSALRPSTSRSLYSSSPGGAYVTRSSAVRL#RSSVPGVRLLQDSVDFSLADAINTEFKNTR 100 |||||||||||||+||||| | ||||||||#|||||||||||||||||||||||||||||| Sbjct 41 GSALRPSTSRSLYASSPGGVYATRSSAVRL#RSSVPGVRLLQDSVDFSLADAINTEFKNTR 100 |
| synthetic construct | Query 41 GSALRPSTSRSLYSSSPGGAYVTRSSAVRL#RSSVPGVRLLQDSVDFSLADAINTEFKNTR 100 |||||||||||||+||||| | ||||||||#|||||||||||||||||||||||||||||| Sbjct 41 GSALRPSTSRSLYASSPGGVYATRSSAVRL#RSSVPGVRLLQDSVDFSLADAINTEFKNTR 100 |
| Macaca fascicularis | Query 41 GSALRPSTSRSLYSSSPGGAYVTRSSAVRL#RSSVPGVRLLQDSVDFSLADAINTEFKNTR 100 |||||||||||||+||||| | ||||||||#|||||||||||||||||||||||||||||| Sbjct 41 GSALRPSTSRSLYASSPGGVYATRSSAVRL#RSSVPGVRLLQDSVDFSLADAINTEFKNTR 100 |
| Pan troglodytes | Query 41 GSALRPSTSRSLYSSSPGGAYVTRSSAVRL#RSSVPGVRLLQDSVDFSLADAINTEFKNTR 100 |||||||||||||+||||| | ||||||||#|||||||||||||||||||||||||||||| Sbjct 41 GSALRPSTSRSLYASSPGGVYATRSSAVRL#RSSVPGVRLLQDSVDFSLADAINTEFKNTR 100 |
| Ovis aries | Query 41 GSALRPSTSRSLYSSSPGGAYVTRSSAVRL#RSSVPGVRLLQDSVDFSLADAINTEFKNTR 100 ||||||+|||+||+||||| | ||||||||#|| ||||||||||||||||||||||||||| Sbjct 41 GSALRPTTSRTLYTSSPGGVYATRSSAVRL#RSGVPGVRLLQDSVDFSLADAINTEFKNTR 100 |
| Canis familiaris | Query 41 GSALRPSTSRSLYSSSPGGAYVTRSSAVRL#RSSVPGVRLLQDSVDFSLADAINTEFKNTR 100 |||+|||||||||+||||| + |||||+||#| ||||+||||||||||||||||||||||| Sbjct 41 GSAMRPSTSRSLYASSPGGVFTTRSSAMRL#RGSVPGMRLLQDSVDFSLADAINTEFKNTR 100 |
| Bos taurus | Query 50 RSLYSSSPGGAYVTRSSAVRL#RSSVPGVRLLQDSVDFSLADAINTEFKNTR 100 |+||+||||| | ||||||||#|| ||||||||||||||||||||||||||| Sbjct 1 RTLYTSSPGGVYATRSSAVRL#RSGVPGVRLLQDSVDFSLADAINTEFKNTR 51 |
| Gallus gallus | Query 41 GSALRPSTSRSLYSSSPGGAYVTRSSAVRL#RSSVPGVRLLQDSVDFSLADAINTEFKNTR 100 |||||||++| + |+|||| | |++++|||#|||+| +| + |+|||+|||||||||| | Sbjct 17 GSALRPSSARYV-SASPGGVYATKATSVRL#RSSMPPMR-MHDAVDFTLADAINTEFKANR 74 |
| Daboia russellii | Query 41 GSALRPSTSRSLYSSSPGGAYVTRSSAVRL#RSSVPGVRLLQDS-VDFSLADAINTEFKNT 99 ||++|||+ | + | ||| | | |+|+#||| | +| |+ |||||||||||||| Sbjct 39 GSSIRPSSVRLV--SCPGGIYA--SKAMRV#RSSFPPMRTGGDAMVDFSLADAINTEFKAN 94 Query 100 R 100 | Query 100 R 100 |
| Acipenser baerii | Query 46 PSTSRSLYSS-SPGGAYVTRSSAVRL#RSSVPGVRLLQDSVDFSLADAINTEFKNTR 100 | || |+| | | |+||+ ||#||| | ||| +++||+|+|||||||| | Sbjct 35 PIRSRVSYTSHSAPTLYATKSSSTRL#RSSAPQTRLLSETLDFALSDAINTEFKANR 90 |
| Oncorhynchus mykiss | Query 48 TSRSLYS-SSPGGAYVTRSSAVRL#RSSVPGVRLLQDSVDFSLADAINTEFKNTR 100 +|| || |+| | +++ |||#||| | || |+|||+|+||||+||| | Sbjct 45 SSRMSYSVSAPPSIYASKN--VRL#RSSAPMPRLSSDTVDFALSDAINSEFKANR 96 |
| Danio rerio | Query 53 YSSSPGGAYVTRSSAVRL#RSSVPGVRLLQDSVDFSLADAINTEFKNTR 100 |||+| | | +|+#|| | || |++|| ||+||||||| | Sbjct 46 YSSAPPSVYA--SKGLRV#RSGAPLPRLATDTLDFGLAEAINTEFKANR 91 |
| Xenopus laevis | Query 41 GSALRPSTSRSLYSSSPGGAYVTRSSAVRL#RSSVPGVRLLQDSVDFSLADAINTEFKNTR 100 |||+||||| + |+ | +||+|||#|||+| | + |||||+||||+| ||| | Sbjct 35 GSAMRPSTSSRMVYSTSSSPAVFKSSSVRL#RSSLPPAR-MADSVDFALADAVNLEFKANR 93 |
| Xenopus tropicalis | Query 41 GSALRP----STSRSLYSSSPGGAYVTRSSAVRL#RSSVPGVRLLQDSVDFSLADAINTEF 96 |||+|| |+|| +||+| | | +||+|||#|||+| || |+|||+||||+| |+ Sbjct 59 GSAMRPGAGSSSSRVVYSTSASPA-VFKSSSVRL#RSSLPPARLA-DTVDFTLADAVNLEY 116 Query 97 KNTR 100 | | Sbjct 117 KANR 120 |
| Cyprinus carpio | Query 65 SSAVRL#RSSVPGVRLLQDSVDFSLADAINTEFKNTR 100 | |+#||| | || +++|| |||||||||| | Sbjct 56 SKGARV#RSSGPLPRLATETLDFGLADAINTEFKTNR 91 |
| Tetraodon nigroviridis | Query 49 SRSLYSSSPGGAYVTRSSAV------RL#RSSVPGVRLLQDSVDFSLADAINTEFKNTR 100 ||+ ||| + ++ + || ||#||| || +++||||+||||||| | Sbjct 22 SRTSYSSRQFSSPLSSAPAVYALKTQRL#RSSSAMPRLASENLDFSLSDAINTEFITNR 79 |
| Scyliorhinus stellaris | Query 47 STSRSLYSSSPGGAYVTRSSAVRL#RSSVPGVRLLQDSVDFSLADAINTEFKNTR 100 | ||+ || + + | | #||| ||+ + +|++||| ||+|+||| | Sbjct 37 SVSRTSYSQP---SMIKRRSVRVN#RSSAPGIAM-GNSLNFSLVDAMNSEFKVNR 86 |
| Takifugu rubripes | Query 46 PSTSRSLYSSSPGGAYVTRSSAVRL#RSSVPGVRLLQDSVDFSLADAINTEFKNTR 100 || | || || + |+|+| # | + +||+||||+| +| ||| Sbjct 33 PSYSSYRVSSGSMGAGLGSSTALRT#YSG--------EKLDFNLADAMNQDFLNTR 79 |
[Site 9] DSVDFSLADA91-INTEFKNTRT
Ala91  Ile
Ile
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Asp82 | Ser83 | Val84 | Asp85 | Phe86 | Ser87 | Leu88 | Ala89 | Asp90 | Ala91 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Ile92 | Asn93 | Thr94 | Glu95 | Phe96 | Lys97 | Asn98 | Thr99 | Arg100 | Thr101 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| VTRSSAVRLRSSVPGVRLLQDSVDFSLADAINTEFKNTRTNEKVELQELNDRFANYIDKV |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | Mus musculus | 123.00 | 24 | unnamed protein product |
| 2 | N/A | 122.00 | 18 | vimentin protein |
| 3 | Rattus norvegicus | 122.00 | 6 | vimentin |
| 4 | Homo sapiens | 121.00 | 25 | vimentin |
| 5 | Canis familiaris | 121.00 | 16 | PREDICTED: similar to vimentin isoform 1 |
| 6 | synthetic construct | 121.00 | 2 | vimentin |
| 7 | Pan troglodytes | 121.00 | 1 | vimentin |
| 8 | Macaca fascicularis | 121.00 | 1 | VIME_MACFA Vimentin gi |
| 9 | Ovis aries | 120.00 | 2 | VIME_SHEEP Vimentin gi |
| 10 | Bos taurus | 117.00 | 6 | similar to vimentin |
| 11 | Canis familiaris | 112.00 | 2 | PREDICTED: similar to Vimentin |
| 12 | Gallus gallus | 94.40 | 3 | vimentin |
| 13 | Acipenser baerii | 90.10 | 2 | vimentin |
| 14 | Daboia russellii | 85.90 | 1 | AF447708_1 vimentin |
| 15 | Oncorhynchus mykiss | 83.60 | 2 | VIME_ONCMY Vimentin gi |
| 16 | Cyprinus carpio | 80.50 | 1 | VIME_CYPCA Vimentin gi |
| 17 | Danio rerio | 80.10 | 6 | vimentin |
| 18 | Sus scrofa | 72.80 | 3 | desmin |
| 19 | Cricetulus griseus | 72.40 | 1 | vimentin |
| 20 | Xenopus tropicalis | 70.90 | 3 | Vim protein |
| 21 | Tetraodon nigroviridis | 70.50 | 6 | unnamed protein product |
| 22 | Canis lupus familiaris | 69.70 | 1 | desmin |
| 23 | Xenopus laevis | 69.30 | 12 | desmin |
| 24 | Lampetra fluviatilis | 69.30 | 1 | intermediate filament protein type III |
| 25 | Mesocricetus auratus | 69.30 | 1 | DESM_MESAU Desmin gi |
| 26 | Scyliorhinus stellaris | 64.30 | 2 | vimentin |
| 27 | Takifugu rubripes | 63.90 | 1 | TPA: desmin |
| 28 | Bos taurus | 60.10 | 2 | glial fibrillary acidic protein |
| 29 | Pongo pygmaeus | 60.10 | 1 | GFAP_PONPY Glial fibrillary acidic protein (GFAP) |
| 30 | Homo sapiens | 59.70 | 1 | glial fibrillary acidic protein |
| 31 | Rattus norvegicus | 58.50 | 1 | glial fibrillary acidic protein |
| 32 | Mus musculus | 58.20 | 2 | glial fibrillary acidic protein |
| 33 | Torpedo californica | 57.00 | 1 | IF3T_TORCA Type III intermediate filament gi |
| 34 | Danio rerio | 53.10 | 1 | PREDICTED: similar to gefiltin |
| 35 | Xenopus laevis | 50.10 | 2 | neurofilament protein |
| 36 | Petromyzon marinus | 48.90 | 2 | neurofilament subunit NF-95 |
Top-ranked sequences
| organism | matching |
|---|---|
| Mus musculus | Query 62 VTRSSAVRLRSSVPGVRLLQDSVDFSLADA#INTEFKNTRTNEKVELQELNDRFANYIDKV 121 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 62 VTRSSAVRLRSSVPGVRLLQDSVDFSLADA#INTEFKNTRTNEKVELQELNDRFANYIDKV 121 |
| N/A | Query 62 VTRSSAVRLRSSVPGVRLLQDSVDFSLADA#INTEFKNTRTNEKVELQELNDRFANYIDKV 121 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 62 VTRSSAVRLRSSVPGVRLLQDSVDFSLADA#INTEFKNTRTNEKVELQELNDRFANYIDKV 121 |
| Rattus norvegicus | Query 62 VTRSSAVRLRSSVPGVRLLQDSVDFSLADA#INTEFKNTRTNEKVELQELNDRFANYIDKV 121 ||||||||||||+|||||||||||||||||#|||||||||||||||||||||||||||||| Sbjct 62 VTRSSAVRLRSSMPGVRLLQDSVDFSLADA#INTEFKNTRTNEKVELQELNDRFANYIDKV 121 |
| Homo sapiens | Query 62 VTRSSAVRLRSSVPGVRLLQDSVDFSLADA#INTEFKNTRTNEKVELQELNDRFANYIDKV 121 |||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Sbjct 62 ATRSSAVRLRSSVPGVRLLQDSVDFSLADA#INTEFKNTRTNEKVELQELNDRFANYIDKV 121 |
| Canis familiaris | Query 62 VTRSSAVRLRSSVPGVRLLQDSVDFSLADA#INTEFKNTRTNEKVELQELNDRFANYIDKV 121 |||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Sbjct 62 ATRSSAVRLRSSVPGVRLLQDSVDFSLADA#INTEFKNTRTNEKVELQELNDRFANYIDKV 121 |
| synthetic construct | Query 62 VTRSSAVRLRSSVPGVRLLQDSVDFSLADA#INTEFKNTRTNEKVELQELNDRFANYIDKV 121 |||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Sbjct 62 ATRSSAVRLRSSVPGVRLLQDSVDFSLADA#INTEFKNTRTNEKVELQELNDRFANYIDKV 121 |
| Pan troglodytes | Query 62 VTRSSAVRLRSSVPGVRLLQDSVDFSLADA#INTEFKNTRTNEKVELQELNDRFANYIDKV 121 |||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Sbjct 62 ATRSSAVRLRSSVPGVRLLQDSVDFSLADA#INTEFKNTRTNEKVELQELNDRFANYIDKV 121 |
| Macaca fascicularis | Query 62 VTRSSAVRLRSSVPGVRLLQDSVDFSLADA#INTEFKNTRTNEKVELQELNDRFANYIDKV 121 |||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Sbjct 62 ATRSSAVRLRSSVPGVRLLQDSVDFSLADA#INTEFKNTRTNEKVELQELNDRFANYIDKV 121 |
| Ovis aries | Query 62 VTRSSAVRLRSSVPGVRLLQDSVDFSLADA#INTEFKNTRTNEKVELQELNDRFANYIDKV 121 |||||||||| ||||||||||||||||||#|||||||||||||||||||||||||||||| Sbjct 62 ATRSSAVRLRSGVPGVRLLQDSVDFSLADA#INTEFKNTRTNEKVELQELNDRFANYIDKV 121 |
| Bos taurus | Query 63 TRSSAVRLRSSVPGVRLLQDSVDFSLADA#INTEFKNTRTNEKVELQELNDRFANYIDKV 121 |||||||||| ||||||||||||||||||#|||||||||||||||||||||||||||||| Sbjct 14 TRSSAVRLRSGVPGVRLLQDSVDFSLADA#INTEFKNTRTNEKVELQELNDRFANYIDKV 72 |
| Canis familiaris | Query 62 VTRSSAVRLRSSVPGVRLLQDSVDFSLADA#INTEFKNTRTNEKVELQELNDRFANYIDKV 121 |||||+||| ||||+||||||||||||||#||||||||||||||||||||| | |||||| Sbjct 62 TTRSSAMRLRGSVPGMRLLQDSVDFSLADA#INTEFKNTRTNEKVELQELNDLFTNYIDKV 121 |
| Gallus gallus | Query 63 TRSSAVRLRSSVPGVRLLQDSVDFSLADA#INTEFKNTRTNEKVELQELNDRFANYIDKV 121 |++++||||||+| +|+ |+|||+||||#|||||| |||||||||||||||||||||| Sbjct 58 TKATSVRLRSSMPPMRM-HDAVDFTLADA#INTEFKANRTNEKVELQELNDRFANYIDKV 115 |
| Acipenser baerii | Query 62 VTRSSAVRLRSSVPGVRLLQDSVDFSLADA#INTEFKNTRTNEKVELQELNDRFANYIDKV 121 |+||+ ||||| | ||| +++||+|+||#|||||| ||||| |+| ||||||+||||| Sbjct 52 ATKSSSTRLRSSAPQTRLLSETLDFALSDA#INTEFKANRTNEKAEMQHLNDRFASYIDKV 111 |
| Daboia russellii | Query 65 SSAVRLRSSVPGVRLLQDS-VDFSLADA#INTEFKNTRTNEKVELQELNDRFANYIDKV 121 | |+|+||| | +| |+ ||||||||#|||||| ||||| |||||||||||||||| Sbjct 59 SKAMRVRSSFPPMRTGGDAMVDFSLADA#INTEFKANRTNEKAELQELNDRFANYIDKV 116 |
| Oncorhynchus mykiss | Query 65 SSAVRLRSSVPGVRLLQDSVDFSLADA#INTEFKNTRTNEKVELQELNDRFANYIDKV 121 | |||||| | || |+|||+|+||#||+||| ||||| |+| ||||||+||||| Sbjct 61 SKNVRLRSSAPMPRLSSDTVDFALSDA#INSEFKANRTNEKAEMQHLNDRFASYIDKV 117 |
| Cyprinus carpio | Query 65 SSAVRLRSSVPGVRLLQDSVDFSLADA#INTEFKNTRTNEKVELQELNDRFANYIDKV 121 | |+||| | || +++|| ||||#|||||| ||||| |+| ||||||+||||| Sbjct 56 SKGARVRSSGPLPRLATETLDFGLADA#INTEFKTNRTNEKAEMQHLNDRFASYIDKV 112 |
| Danio rerio | Query 62 VTRSSAVRLRSSVPGVRLLQDSVDFSLADA#INTEFKNTRTNEKVELQELNDRFANYIDKV 121 | | +|+|| | || |++|| ||+|#|||||| ||||| |+| ||||||+||||| Sbjct 53 VYASKGLRVRSGAPLPRLATDTLDFGLAEA#INTEFKANRTNEKAEMQHLNDRFASYIDKV 112 |
| Sus scrofa | Query 62 VTRSSAVRLRSSVPGVRLLQDSVDFSLADA#INTEFKNTRTNEKVELQELNDRFANYIDKV 121 +| | |+ || | | +|||||||#+| || ||||||||||||||||||||+|| Sbjct 62 ASRLGATRVPSSSYGAGEL---LDFSLADA#VNQEFLTTRTNEKVELQELNDRFANYIEKV 118 |
| Cricetulus griseus | Query 87 SLADA#INTEFKNTRTNEKVELQELNDRFANYIDKV 121 |||||# ||||||||||||||||||||||||||||| Sbjct 1 SLADA#CNTEFKNTRTNEKVELQELNDRFANYIDKV 35 |
| Xenopus tropicalis | Query 75 PGVRLLQDSVDFSLADA#INTEFKNTRTNEKVELQELNDRFANYIDKV 121 | || |+|||+||||#+| |+| ||||| |+|||||||||+|||| Sbjct 96 PPARLA-DTVDFTLADA#VNLEYKANRTNEKAEMQELNDRFANFIDKV 141 |
| Tetraodon nigroviridis | Query 69 RLRSSVPGVRLLQDSVDFSLADA#INTEFKNTRTNEKVELQELNDRFANYIDKV 121 ||||| || +++||||+||#||||| ||||| ++| ||||||+||+|| Sbjct 48 RLRSSSAMPRLASENLDFSLSDA#INTEFITNRTNEKAQMQSLNDRFASYIEKV 100 |
| Canis lupus familiaris | Query 84 VDFSLADA#INTEFKNTRTNEKVELQELNDRFANYIDKV 121 +|||||||#+| || ||||||||||||||||||||+|| Sbjct 88 LDFSLADA#VNQEFLTTRTNEKVELQELNDRFANYIEKV 125 |
| Xenopus laevis | Query 82 DSVDFSLADA#INTEFKNTRTNEKVELQELNDRFANYIDKV 121 | +|||||||#+| || ||||||||||+|||||||||+|| Sbjct 75 DVLDFSLADA#MNQEFLQTRTNEKVELQDLNDRFANYIEKV 114 |
| Lampetra fluviatilis | Query 84 VDFSLADA#INTEFKNTRTNEKVELQELNDRFANYIDKV 121 ||||||||#+| +++ |||||||||||||||||+||||| Sbjct 88 VDFSLADA#LNADYRTTRTNEKVELQELNDRFASYIDKV 125 |
| Mesocricetus auratus | Query 84 VDFSLADA#INTEFKNTRTNEKVELQELNDRFANYIDKV 121 +|||||||#+| || ||||||||||||||||||||+|| Sbjct 88 LDFSLADA#VNQEFLATRTNEKVELQELNDRFANYIEKV 125 |
| Scyliorhinus stellaris | Query 71 RSSVPGVRLLQDSVDFSLADA#INTEFKNTRTNEKVELQELNDRFANYIDKV 121 ||| ||+ + +|++||| ||#+|+||| |+||| |+ ||||| ||++||| Sbjct 58 RSSAPGI-AMGNSLNFSLVDA#MNSEFKVNRSNEKAEMIELNDRLANFLDKV 107 |
| Takifugu rubripes | Query 82 DSVDFSLADA#INTEFKNTRTNEKVELQELNDRFANYIDKV 121 + +||+||||#+| +| ||||||| ||| ||||||+||+|| Sbjct 61 EKLDFNLADA#MNQDFLNTRTNEKAELQHLNDRFASYIEKV 100 |
| Bos taurus | Query 75 PGVRL--------LQDSVDFSLADA#INTEFKNTRTNEKVELQELNDRFANYIDKV 121 || || | |||||| |#+|+ || || +|+ |+ |||||||+||+|| Sbjct 29 PGTRLSLARMPPPLPARVDFSLAGA#LNSGFKETRASERAEMMELNDRFASYIEKV 83 |
| Pongo pygmaeus | Query 75 PGVRL--------LQDSVDFSLADA#INTEFKNTRTNEKVELQELNDRFANYIDKV 121 || || | |||||| |#+| || || +|+ |+ |||||||+||+|| Sbjct 33 PGTRLSLARMPPPLPTRVDFSLAGA#LNAGFKETRASERAEMMELNDRFASYIEKV 87 |
| Homo sapiens | Query 75 PGVRL--------LQDSVDFSLADA#INTEFKNTRTNEKVELQELNDRFANYIDKV 121 || || | |||||| |#+| || || +|+ |+ |||||||+||+|| Sbjct 33 PGTRLSLARMPPPLPTRVDFSLAGA#LNAGFKETRASERAEMMELNDRFASYIEKV 87 |
| Rattus norvegicus | Query 62 VTRSSAVRLRSSVPGVRLLQDSVDFSLADA#INTEFKNTRTNEKVELQELNDRFANYIDKV 121 + | | |+ +| |||||| |#+| || || +|+ |+ |||||||+||+|| Sbjct 32 IPRLSLSRMTPPLPA------RVDFSLAGA#LNAGFKETRASERAEMMELNDRFASYIEKV 85 |
| Mus musculus | Query 80 LQDSVDFSLADA#INTEFKNTRTNEKVELQELNDRFANYIDKV 121 | |||||| |#+| || || +|+ |+ |||||||+||+|| Sbjct 43 LPARVDFSLAGA#LNAGFKETRASERAEMMELNDRFASYIEKV 84 |
| Torpedo californica | Query 65 SSAVRLRSSVPGVRLLQDSVDFSLADA#INTEFKNTRTNEKVELQELNDRFANYIDKV 121 | +||| | | ++||+| ||#+|+||| ||||| |+ ||||| ||++||| Sbjct 59 SRSVRLVRSSAPVVASSSNLDFTLVDA#MNSEFKVNRTNEKAEMIELNDRLANFLDKV 115 |
| Danio rerio | Query 82 DSVDFSLADA#INTEFKNTRTNEKVELQELNDRFANYIDKV 121 |||||| #+| ||| ||||| +|| |||||| +|+|| Sbjct 73 DSVDFSQTSV#LNNEFKIVRTNEKEQLQGLNDRFAMFIEKV 112 |
| Xenopus laevis | Query 82 DSVDFSLADA#INTEFKNTRTNEKVELQELNDRFANYIDKV 121 |++| | |#|+++ | || |||+||+|||||||+|++| Sbjct 63 DTMDLSQVAA#ISSDLKIVRTQEKVQLQDLNDRFANFIERV 102 |
| Petromyzon marinus | Query 73 SVPGVRLLQDSVDF-SLADA#INTEFKNTRTNEKVELQELNDRFANYIDKV 121 | | + ||+|| | + # + +||++| ||| ||+|||||| ||+|| Sbjct 69 STPALNSAADSLDFMSQSSV#FSGDFKHSRVNEKQLLQDLNDRFAGYIEKV 118 |
[Site 10] QIDVDVSKPD264-LTAALRDVRQ
Asp264  Leu
Leu
iTraq-117 Signal 22351.3 (


 ) for AALRDVRQ
) for AALRDVRQ
iTraq-117 Signal 4566.0 (

 ) for ALRDVRQ
) for ALRDVRQ
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Gln255 | Ile256 | Asp257 | Val258 | Asp259 | Val260 | Ser261 | Lys262 | Pro263 | Asp264 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Leu265 | Thr266 | Ala267 | Ala268 | Leu269 | Arg270 | Asp271 | Val272 | Arg273 | Gln274 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| KKLHDEEIQELQAQIQEQHVQIDVDVSKPDLTAALRDVRQQYESVAAKNLQEAEEWYKSK |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | Mus musculus | 123.00 | 22 | unnamed protein product |
| 2 | N/A | 123.00 | 20 | VIME_MESAU Vimentin gi |
| 3 | Rattus norvegicus | 123.00 | 5 | vimentin |
| 4 | Cricetulus griseus | 122.00 | 1 | vimentin |
| 5 | Homo sapiens | 120.00 | 27 | vimentin |
| 6 | Canis familiaris | 120.00 | 16 | PREDICTED: similar to vimentin isoform 1 |
| 7 | synthetic construct | 120.00 | 2 | vimentin |
| 8 | Macaca fascicularis | 120.00 | 1 | VIME_MACFA Vimentin gi |
| 9 | Sus scrofa domestica | 120.00 | 1 | vimentin |
| 10 | Pan troglodytes | 120.00 | 1 | vimentin |
| 11 | Gallus gallus | 118.00 | 4 | vimentin |
| 12 | Daboia russellii | 112.00 | 1 | AF447708_1 vimentin |
| 13 | Canis familiaris | 111.00 | 2 | PREDICTED: similar to Vimentin |
| 14 | Xenopus laevis | 110.00 | 11 | VIM1_XENLA Vimentin-1/2 gi |
| 15 | Xenopus tropicalis | 108.00 | 2 | Vim protein |
| 16 | Cyprinus carpio | 105.00 | 1 | VIME_CYPCA Vimentin gi |
| 17 | Oncorhynchus mykiss | 103.00 | 2 | VIME_ONCMY Vimentin gi |
| 18 | Scyliorhinus stellaris | 102.00 | 2 | desmin |
| 19 | Acipenser baerii | 102.00 | 2 | vimentin |
| 20 | Danio rerio | 101.00 | 8 | vimentin |
| 21 | Takifugu rubripes | 97.10 | 1 | TPA: desmin |
| 22 | Tetraodon nigroviridis | 90.90 | 6 | unnamed protein product |
| 23 | Lampetra fluviatilis | 90.90 | 1 | intermediate filament protein |
| 24 | Pongo pygmaeus | 87.80 | 4 | hypothetical protein |
| 25 | synthetic construct | 87.80 | 1 | neurofilament light polypeptide |
| 26 | Oreochromis mossambicus | 86.70 | 1 | glial fibrillary acidic protein |
| 27 | Coturnix coturnix | 85.90 | 1 | NFL_COTJA Neurofilament light polypeptide (NF-L) |
| 28 | Danio rerio | 85.10 | 1 | PREDICTED: similar to gefiltin |
| 29 | Bos taurus | 84.00 | 3 | neurofilament, light polypeptide 68kDa |
| 30 | Xenopus laevis | 83.20 | 1 | neurofilament protein |
| 31 | Homo sapiens | 83.20 | 1 | glial fibrillary acidic protein |
| 32 | Mus musculus | 80.90 | 1 | glial fibrillary acidic protein |
| 33 | Torpedo californica | 78.60 | 1 | IF3T_TORCA Type III intermediate filament gi |
| 34 | Sus scrofa | 78.20 | 1 | glial fibrillary acidic protein |
| 35 | Sus scrofa | 77.00 | 2 | muscle-specific intermediate filament desmin |
| 36 | Mesocricetus auratus | 77.00 | 1 | DESM_MESAU Desmin gi |
| 37 | Canis lupus familiaris | 76.60 | 1 | desmin |
| 38 | Ovis aries | 75.90 | 1 | glial fibrillary acidic protein |
| 39 | Capreolus capreolus | 73.60 | 1 | glial fibrillary acidic protein |
| 40 | Equus caballus | 73.20 | 1 | glial fibrillary acidic protein |
| 41 | Dama dama | 73.20 | 1 | glial fibrillary acidic protein |
Top-ranked sequences
| organism | matching |
|---|---|
| Mus musculus | Query 235 KKLHDEEIQELQAQIQEQHVQIDVDVSKPD#LTAALRDVRQQYESVAAKNLQEAEEWYKSK 294 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 235 KKLHDEEIQELQAQIQEQHVQIDVDVSKPD#LTAALRDVRQQYESVAAKNLQEAEEWYKSK 294 |
| N/A | Query 235 KKLHDEEIQELQAQIQEQHVQIDVDVSKPD#LTAALRDVRQQYESVAAKNLQEAEEWYKSK 294 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 235 KKLHDEEIQELQAQIQEQHVQIDVDVSKPD#LTAALRDVRQQYESVAAKNLQEAEEWYKSK 294 |
| Rattus norvegicus | Query 235 KKLHDEEIQELQAQIQEQHVQIDVDVSKPD#LTAALRDVRQQYESVAAKNLQEAEEWYKSK 294 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 235 KKLHDEEIQELQAQIQEQHVQIDVDVSKPD#LTAALRDVRQQYESVAAKNLQEAEEWYKSK 294 |
| Cricetulus griseus | Query 235 KKLHDEEIQELQAQIQEQHVQIDVDVSKPD#LTAALRDVRQQYESVAAKNLQEAEEWYKSK 294 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 235 KKLHDEEIQELQAQIQEQHVQIDVDVSKPD#LTAALRDVRQQYESVAAKNLQEAEEWYKSK 294 |
| Homo sapiens | Query 235 KKLHDEEIQELQAQIQEQHVQIDVDVSKPD#LTAALRDVRQQYESVAAKNLQEAEEWYKSK 294 ||||+|||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Sbjct 235 KKLHEEEIQELQAQIQEQHVQIDVDVSKPD#LTAALRDVRQQYESVAAKNLQEAEEWYKSK 294 |
| Canis familiaris | Query 235 KKLHDEEIQELQAQIQEQHVQIDVDVSKPD#LTAALRDVRQQYESVAAKNLQEAEEWYKSK 294 ||||||||||||||||+||||||+||||||#|||||||||||||||||||||||||||||| Sbjct 235 KKLHDEEIQELQAQIQDQHVQIDMDVSKPD#LTAALRDVRQQYESVAAKNLQEAEEWYKSK 294 |
| synthetic construct | Query 235 KKLHDEEIQELQAQIQEQHVQIDVDVSKPD#LTAALRDVRQQYESVAAKNLQEAEEWYKSK 294 ||||+|||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Sbjct 235 KKLHEEEIQELQAQIQEQHVQIDVDVSKPD#LTAALRDVRQQYESVAAKNLQEAEEWYKSK 294 |
| Macaca fascicularis | Query 235 KKLHDEEIQELQAQIQEQHVQIDVDVSKPD#LTAALRDVRQQYESVAAKNLQEAEEWYKSK 294 ||||+|||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Sbjct 235 KKLHEEEIQELQAQIQEQHVQIDVDVSKPD#LTAALRDVRQQYESVAAKNLQEAEEWYKSK 294 |
| Sus scrofa domestica | Query 235 KKLHDEEIQELQAQIQEQHVQIDVDVSKPD#LTAALRDVRQQYESVAAKNLQEAEEWYKSK 294 |||||||||||||||||||||||+||||||#|||||||||||||||||||||||||||||| Sbjct 105 KKLHDEEIQELQAQIQEQHVQIDMDVSKPD#LTAALRDVRQQYESVAAKNLQEAEEWYKSK 164 |
| Pan troglodytes | Query 235 KKLHDEEIQELQAQIQEQHVQIDVDVSKPD#LTAALRDVRQQYESVAAKNLQEAEEWYKSK 294 ||||+|||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Sbjct 235 KKLHEEEIQELQAQIQEQHVQIDVDVSKPD#LTAALRDVRQQYESVAAKNLQEAEEWYKSK 294 |
| Gallus gallus | Query 235 KKLHDEEIQELQAQIQEQHVQIDVDVSKPD#LTAALRDVRQQYESVAAKNLQEAEEWYKSK 294 ||||||||+|||||+||||+|||+||||||#|||||||||||||||||||||||||||||| Sbjct 229 KKLHDEEIRELQAQLQEQHIQIDMDVSKPD#LTAALRDVRQQYESVAAKNLQEAEEWYKSK 288 |
| Daboia russellii | Query 235 KKLHDEEIQELQAQIQEQHVQIDVDVSKPD#LTAALRDVRQQYESVAAKNLQEAEEWYKSK 294 ||||+|||+|||||||+||+|||+||+|||#|||||||||||||||| ||||||||||||| Sbjct 230 KKLHEEEIRELQAQIQDQHIQIDMDVAKPD#LTAALRDVRQQYESVATKNLQEAEEWYKSK 289 |
| Canis familiaris | Query 235 KKLHDEEIQELQAQIQEQHVQIDVDVSKPD#LTAALRDVRQQYESVAAKNLQEAEEWYKSK 294 ||||||||||||||||+ |||||+||||||#||||||||||||||+|||||| |+|||||| Sbjct 237 KKLHDEEIQELQAQIQDWHVQIDMDVSKPD#LTAALRDVRQQYESLAAKNLQVAKEWYKSK 296 |
| Xenopus laevis | Query 235 KKLHDEEIQELQAQIQEQHVQIDVDVSKPD#LTAALRDVRQQYESVAAKNLQEAEEWYKSK 294 ||||||||+||| |||| |+|+|+||||||#|||||||||||||+|||||| +|||||||| Sbjct 228 KKLHDEEIRELQLQIQESHIQVDMDVSKPD#LTAALRDVRQQYENVAAKNLSDAEEWYKSK 287 |
| Xenopus tropicalis | Query 235 KKLHDEEIQELQAQIQEQHVQIDVDVSKPD#LTAALRDVRQQYESVAAKNLQEAEEWYKSK 294 ||||||||+||| |||| |+|+|+||||||#|||||||||||||+|||||| +||+||||| Sbjct 255 KKLHDEEIRELQLQIQESHIQVDMDVSKPD#LTAALRDVRQQYENVAAKNLSDAEDWYKSK 314 |
| Cyprinus carpio | Query 235 KKLHDEEIQELQAQIQEQHVQIDVDVSKPD#LTAALRDVRQQYESVAAKNLQEAEEWYKSK 294 |||||||+ ||| ||||||||||++|+|||#|||||+|||||||++|++||||+||||||| Sbjct 226 KKLHDEELAELQIQIQEQHVQIDMEVAKPD#LTAALKDVRQQYETLASRNLQESEEWYKSK 285 |
| Oncorhynchus mykiss | Query 235 KKLHDEEIQELQAQIQEQHVQIDVDVSKPD#LTAALRDVRQQYESVAAKNLQEAEEWYKSK 294 +||||||+ |||||||+||||||+||+|||#||||||||| |||++|++|||++|+||||| Sbjct 231 RKLHDEEVAELQAQIQDQHVQIDMDVAKPD#LTAALRDVRVQYETLASRNLQDSEDWYKSK 290 |
| Scyliorhinus stellaris | Query 235 KKLHDEEIQELQAQIQEQHVQIDVDVSKPD#LTAALRDVRQQYESVAAKNLQEAEEWYKSK 294 ||+|+||++|||||+||||||+++|+||||#|||||||+| ||| ++|||+|||||||||| Sbjct 224 KKVHEEELRELQAQLQEQHVQVEMDLSKPD#LTAALRDIRAQYECISAKNVQEAEEWYKSK 283 |
| Acipenser baerii | Query 235 KKLHDEEIQELQAQIQEQHVQIDVDVSKPD#LTAALRDVRQQYESVAAKNLQEAEEWYKSK 294 +||||||+ ||| ||||||+|||+||+|||#||||||||| |||++|+||+ |+|+||||| Sbjct 225 RKLHDEELAELQVQIQEQHIQIDMDVAKPD#LTAALRDVRLQYENLASKNMSESEDWYKSK 284 |
| Danio rerio | Query 235 KKLHDEEIQELQAQIQEQHVQIDVDVSKPD#LTAALRDVRQQYESVAAKNLQEAEEWYKSK 294 |||||||+ |+| |+||||+|||++|+|||#|||||+|||||||++|++||||+|+||||| Sbjct 226 KKLHDEELAEMQIQMQEQHLQIDMEVTKPD#LTAALKDVRQQYETLASRNLQESEDWYKSK 285 |
| Takifugu rubripes | Query 235 KKLHDEEIQELQAQIQEQHVQIDVDVSKPD#LTAALRDVRQQYESVAAKNLQEAEEWYKSK 294 ||+|+|||+|||+|+|| ||| +|+||||#|||||||+| ||| +||||+ ||||||||| Sbjct 215 KKIHEEEIRELQSQMQETQVQIQMDMSKPD#LTAALRDIRVQYEGIAAKNISEAEEWYKSK 274 |
| Tetraodon nigroviridis | Query 235 KKLHDEEIQELQAQIQEQHVQIDVDVSKPD#LTAALRDVRQQYESVAAKNLQEAEEWYKSK 294 ||+|+|||+||| |+|| ||| +|+||||#|||||+|+| ||| +||||+ +|| ||||| Sbjct 242 KKIHEEEIRELQMQMQETQVQIQMDMSKPD#LTAALKDIRAQYEDIAAKNIADAENWYKSK 301 |
| Lampetra fluviatilis | Query 235 KKLHDEEIQELQAQIQEQHVQIDVDVSKPD#LTAALRDVRQQYESVAAKNLQEAEEWYKSK 294 || ||||++|||| +||| | ++|| |||#| ||||||| ||||+|+ |||||| |+||| Sbjct 132 KKAHDEEVRELQAMMQEQQVTVEVDTVKPD#LAAALRDVRAQYESIASSNLQEAETWFKSK 191 |
| Pongo pygmaeus | Query 235 KKLHDEEIQELQAQIQEQHVQIDVDVSKPD#LTAALRDVRQQYESVAAKNLQEAEEWYKSK 294 ||+|+||| ||||||| + +++||+|||#|+|||+|+| ||| +||||+| ||||+||+ Sbjct 224 KKVHEEEIAELQAQIQYAQISVEMDVTKPD#LSAALKDIRAQYEKLAAKNMQNAEEWFKSR 283 |
| synthetic construct | Query 235 KKLHDEEIQELQAQIQEQHVQIDVDVSKPD#LTAALRDVRQQYESVAAKNLQEAEEWYKSK 294 ||+|+||| ||||||| + +++||+|||#|+|||+|+| ||| +||||+| ||||+||+ Sbjct 224 KKVHEEEIAELQAQIQYAQISVEMDVTKPD#LSAALKDIRAQYEKLAAKNMQNAEEWFKSR 283 |
| Oreochromis mossambicus | Query 235 KKLHDEEIQELQAQIQEQHVQIDVDVSKPD#LTAALRDVRQQYESVAAKNLQEAEEWYKS 293 ||+|++|++||| || | | +|+||||||#|||||||+| |||++|+ |+|| ||||+| Sbjct 121 KKIHEDELRELQEQIMAQQVHVDLDVSKPD#LTAALRDIRVQYENMASSNMQETEEWYRS 179 |
| Coturnix coturnix | Query 235 KKLHDEEIQELQAQIQEQHVQIDVDVS-KPD#LTAALRDVRQQYESVAAKNLQEAEEWYKS 293 ||+|+||+ ||||||| |+ +++||| |||#|+|||||+| ||| +||+|+| ||||++| Sbjct 225 KKVHEEELAELQAQIQYAHLSVEMDVSAKPD#LSAALRDIRAQYEKLAARNMQNAEEWFRS 284 Query 294 K 294 + Sbjct 285 R 285 |
| Danio rerio | Query 235 KKLHDEEIQELQAQIQEQHVQIDVDVSKPD#LTAALRDVRQQYESVAAKNLQEAEEWYKSK 294 +|+||||+ || || + ++|++||||#||+||+|+| |||++|+|||| |||||||| Sbjct 228 RKVHDEEVTELTNMIQAAQISVEVELSKPD#LTSALKDIRGQYETLASKNLQSAEEWYKSK 287 |
| Bos taurus | Query 235 KKLHDEEIQELQAQIQEQHVQIDVDVS-KPD#LTAALRDVRQQYESVAAKNLQEAEEWYKS 293 ||+|+||| ||||||| + +++||| |||#|+|||+|+| ||| +||||+| ||||+|| Sbjct 224 KKVHEEEIAELQAQIQYAQISVEMDVSSKPD#LSAALKDIRAQYEKLAAKNMQNAEEWFKS 283 Query 294 K 294 + Sbjct 284 R 284 |
| Xenopus laevis | Query 235 KKLHDEEIQELQAQIQEQHVQIDVDVSKPD#LTAALRDVRQQYESVAAKNLQEAEEWYKSK 294 ||+|+||+ +||+|+| | ++|+|+|||#|++||||+| ||| +||||+| ||||+||+ Sbjct 218 KKVHEEELSQLQSQVQYAQVSLEVEVAKPD#LSSALRDIRGQYEKLAAKNMQSAEEWFKSR 277 |
| Homo sapiens | Query 235 KKLHDEEIQELQAQIQEQHVQIDVDVSKPD#LTAALRDVRQQYESVAAKNLQEAEEWYKSK 294 +|+|+||++||| |+ | | +++||+|||#|||||+++| |||++|+ |+ ||||||+|| Sbjct 201 RKIHEEEVRELQEQLARQQVHVELDVAKPD#LTAALKEIRTQYEAMASSNMHEAEEWYRSK 260 |
| Mus musculus | Query 235 KKLHDEEIQELQAQIQEQHVQIDVDVSKPD#LTAALRDVRQQYESVAAKNLQEAEEWYKSK 294 +|+++||+++|+ |+ +| | +++||+|||#||||||++| |||+|| |+|| ||||+|| Sbjct 186 RKIYEEEVRDLREQLAQQQVHVEMDVAKPD#LTAALREIRTQYEAVATSNMQETEEWYRSK 245 |
| Torpedo californica | Query 235 KKLHDEEIQELQAQIQEQHVQIDVDVSKPD#LTAALRDVRQQYESVAAKNLQEAEEWYKSK 294 ||||+||| ||||||++ ++++|| +||#|||||+||| |++ +|+||+ | || |||| Sbjct 229 KKLHEEEIIELQAQIRDSQFKVEMDVVRPD#LTAALQDVRSQFDKLASKNIAETEELYKSK 288 |
| Sus scrofa | Query 235 KKLHDEEIQELQAQIQEQHVQIDVDVSKPD#LTAALRDVRQQYESVAAKNLQEAEEWYKSK 294 +|+||||++||| |+ +| | +++||+|||#||||||++| |||+|| |+|||||||+|| Sbjct 45 RKIHDEEVRELQEQLAQQQVHVEMDVTKPD#LTAALREIRTQYEAVATSNMQEAEEWYRSK 104 |
| Sus scrofa | Query 235 KKLHDEEIQELQAQIQEQHVQIDVDVSKPD#LTAALRDVRQQYESVAAKNLQEAEEWYKSK 294 ||+|+|||+|||||+||| ||+++|+||||#|||||||+| |||++||||+ ||||||||| Sbjct 241 KKVHEEEIRELQAQLQEQQVQVEMDMSKPD#LTAALRDIRAQYETIAAKNISEAEEWYKSK 300 |
| Mesocricetus auratus | Query 235 KKLHDEEIQELQAQIQEQHVQIDVDVSKPD#LTAALRDVRQQYESVAAKNLQEAEEWYKSK 294 ||+|+|||+|||||+||| ||+++|+||||#|||||||+| |||++||||+ ||||||||| Sbjct 239 KKVHEEEIRELQAQLQEQQVQVEMDMSKPD#LTAALRDIRAQYETIAAKNISEAEEWYKSK 298 |
| Canis lupus familiaris | Query 235 KKLHDEEIQELQAQIQEQHVQIDVDVSKPD#LTAALRDVRQQYESVAAKNLQEAEEWYKSK 294 ||+|+|||+|||||+||| ||+++|+||||#|||||||+| |||++||||+ ||||||||| Sbjct 239 KKVHEEEIRELQAQLQEQQVQVEMDMSKPD#LTAALRDIRAQYETIAAKNISEAEEWYKSK 298 |
| Ovis aries | Query 235 KKLHDEEIQELQAQIQEQHVQIDVDVSKPD#LTAALRDVRQQYESVAAKNLQEAEEWYKSK 294 +|+|+||++||| |+ +| | +++||+|||#||||||++| |||+||+ |+|||||||+|| Sbjct 45 RKIHEEEVRELQEQLAQQQVHVEMDVAKPD#LTAALREIRTQYEAVASSNMQEAEEWYRSK 104 |
| Capreolus capreolus | Query 235 KKLHDEEIQELQAQIQEQHVQIDVDVSKPD#LTAALRDVRQQYESVAAKNLQEAEEWYKSK 294 +|+|+||++||| |+ +| | +++||+|||#||||||++| |||++|+ |+ ||||||+|| Sbjct 45 RKIHEEEVRELQEQLAQQQVHVEMDVAKPD#LTAALREIRTQYEAMASSNMHEAEEWYRSK 104 |
| Equus caballus | Query 235 KKLHDEEIQELQAQIQEQHVQIDVDVSKPD#LTAALRDVRQQYESVAAKNLQEAEEWYKSK 294 +|+|+||++||| |+ +| | +++||+|||#||||||++| |||++|+ |+ ||||||+|| Sbjct 45 RKIHEEEVRELQEQLAQQQVHVELDVAKPD#LTAALREIRTQYEAMASSNMHEAEEWYRSK 104 |
| Dama dama | Query 235 KKLHDEEIQELQAQIQEQHVQIDVDVSKPD#LTAALRDVRQQYESVAAKNLQEAEEWYKSK 294 +|+|+||++||| |+ +| | +++||+|||#||||||++| |||++|+ |+ ||||||+|| Sbjct 45 RKIHEEEVRELQEQLAQQQVHVEMDVAKPD#LTAALREIRTQYEAMASSNMHEAEEWYRSK 104 |
* References
[PubMed ID: 24142690] Matsuyama M, Tanaka H, Inoko A, Goto H, Yonemura S, Kobori K, Hayashi Y, Kondo E, Itohara S, Izawa I, Inagaki M, Defect of mitotic vimentin phosphorylation causes microophthalmia and cataract via aneuploidy and senescence in lens epithelial cells. J Biol Chem. 2013 Dec 13;288(50):35626-35. doi: 10.1074/jbc.M113.514737. Epub
[PubMed ID: 24173802] ... Janisch KM, Vock VM, Fleming MS, Shrestha A, Grimsley-Myers CM, Rasoul BA, Neale SA, Cupp TD, Kinchen JM, Liem KF Jr, Dwyer ND, The vertebrate-specific Kinesin-6, Kif20b, is required for normal cytokinesis of polarized cortical stem cells and cerebral cortex size. Development. 2013 Dec;140(23):4672-82. doi: 10.1242/dev.093286. Epub 2013 Oct 30.
[PubMed ID: 24048858] ... Noseda R, Belin S, Piguet F, Vaccari I, Scarlino S, Brambilla P, Martinelli Boneschi F, Feltri ML, Wrabetz L, Quattrini A, Feinstein E, Huganir RL, Bolino A, DDIT4/REDD1/RTP801 is a novel negative regulator of Schwann cell myelination. J Neurosci. 2013 Sep 18;33(38):15295-305. doi: 10.1523/JNEUROSCI.2408-13.2013.
[PubMed ID: 23991052] ... Thomsen R, Daugaard TF, Holm IE, Nielsen AL, Alternative mRNA splicing from the glial fibrillary acidic protein (GFAP) gene generates isoforms with distinct subcellular mRNA localization patterns in astrocytes. PLoS One. 2013 Aug 26;8(8):e72110. doi: 10.1371/journal.pone.0072110. eCollection
[PubMed ID: 23936241] ... Andersson D, Wilhelmsson U, Nilsson M, Kubista M, Stahlberg A, Pekna M, Pekny M, Plasticity response in the contralesional hemisphere after subtle neurotrauma: gene expression profiling after partial deafferentation of the hippocampus. PLoS One. 2013 Jul 25;8(7):e70699. doi: 10.1371/journal.pone.0070699. Print 2013.
[PubMed ID: 1385302] ... Salier JP, Simon D, Rouet P, Raguenez G, Muscatelli F, Gebhard W, Guenet JL, Mattei MG, Homologous chromosomal locations of the four genes for inter-alpha-inhibitor and pre-alpha-inhibitor family in human and mouse: assignment of the ancestral gene for the lipocalin superfamily. Genomics. 1992 Sep;14(1):83-8.
[PubMed ID: 1315305] ... Pilz A, Moseley H, Peters J, Abbott C, Comparative mapping of mouse chromosome 2 and human chromosome 9q: the genes for gelsolin and dopamine beta-hydroxylase map to mouse chromosome 2. Genomics. 1992 Apr;12(4):715-9.
[PubMed ID: 1372074] ... Porteus MH, Brice AE, Bulfone A, Usdin TB, Ciaranello RD, Rubenstein JL, Isolation and characterization of a library of cDNA clones that are preferentially expressed in the embryonic telencephalon. Brain Res Mol Brain Res. 1992 Jan;12(1-3):7-22.
[PubMed ID: 1611218] ... Bastian H, Gruss P, Duboule D, Izpisua-Belmonte JC, The murine even-skipped-like gene Evx-2 is closely linked to the Hox-4 complex, but is transcribed in the opposite direction. Mamm Genome. 1992;3(4):241-3.
[PubMed ID: 1655359] ... Threadgill DS, Womack JE, Mapping HSA10 homologous loci in cattle. Cytogenet Cell Genet. 1991;57(2-3):123-6.