SB0043 : Integrin [beta]3
[ CaMP Format ]
* Basic Information
| Organism | Homo sapiens (human) |
| Protein Names | integrin beta-3 precursor [Homo sapiens]; integrin beta-3 precursor; platelet membrane glycoprotein IIIa; Integrin beta-3; Platelet membrane glycoprotein IIIa; GPIIIa |
| Gene Names | ITGB3; GP3A; integrin, beta 3 (platelet glycoprotein IIIa, antigen CD61) |
| Gene Locus | 17q21.32; chromosome 17 |
| GO Function | Not available |
* Information From OMIM
Description: The ITGB3 gene encodes glycoprotein IIIa (GP IIIa), the beta subunit of the platelet membrane adhesive protein receptor complex GP IIb/IIIa. The alpha subunit, GP IIb, is encoded by the ITGA2B gene (OMIM:607759). The GP IIb/IIIa complex belongs to the integrin class of cell adhesion molecule receptors that share a common heterodimeric structure with alpha and beta subunits (summary by Bray et al., 1987 and Bajt et al., 1992).
Function: The GP IIb/IIIa complex mediates platelet aggregation by acting as a receptor for fibrinogen. The complex also acts as a receptor for von Willebrand factor and fibronectin (Prandini et al., 1988).
* Structure Information
1. Primary Information
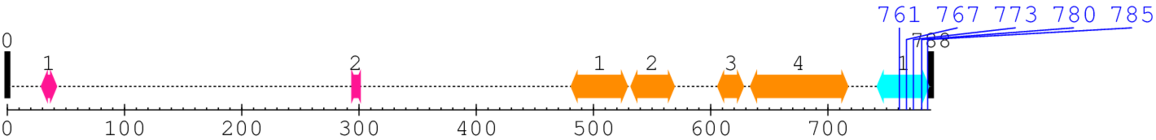
Length: 788 aa
Average Mass: 87.057 kDa
Monoisotopic Mass: 87.000 kDa
2. Domain Information
Annotated Domains: interpro / pfam / smart / prosite
Computationally Assigned Domains (Pfam+HMMER):
| domain name | begin | end | score | e-value |
|---|---|---|---|---|
| EGF-like domain 1. | 29 | 42 | 6.0 | 1.1 |
| EGF-like domain 2. | 296 | 299 | 11.0 | 0.1 |
| Integrin beta tail domain 1. | 481 | 529 | 8.0 | 12.3 |
| Integrin beta tail domain 2. | 532 | 569 | 7.0 | 18.2 |
| Integrin beta tail domain 3. | 606 | 628 | 4.0 | 10.4 |
| Integrin beta tail domain 4. | 634 | 717 | 1.0 | 11.2 |
| Integrin beta cytoplasmic domain 1. | 742 | 786 | 1.0 | 1.4 |
| --- cleavage 761 (inside Integrin beta cytoplasmic domain 742..786) --- | ||||
| --- cleavage 767 (inside Integrin beta cytoplasmic domain 742..786) --- | ||||
| --- cleavage 773 (inside Integrin beta cytoplasmic domain 742..786) --- | ||||
| --- cleavage 780 (inside Integrin beta cytoplasmic domain 742..786) --- | ||||
| --- cleavage 785 (inside Integrin beta cytoplasmic domain 742..786) --- | ||||
3. Sequence Information
Fasta Sequence: SB0043.fasta
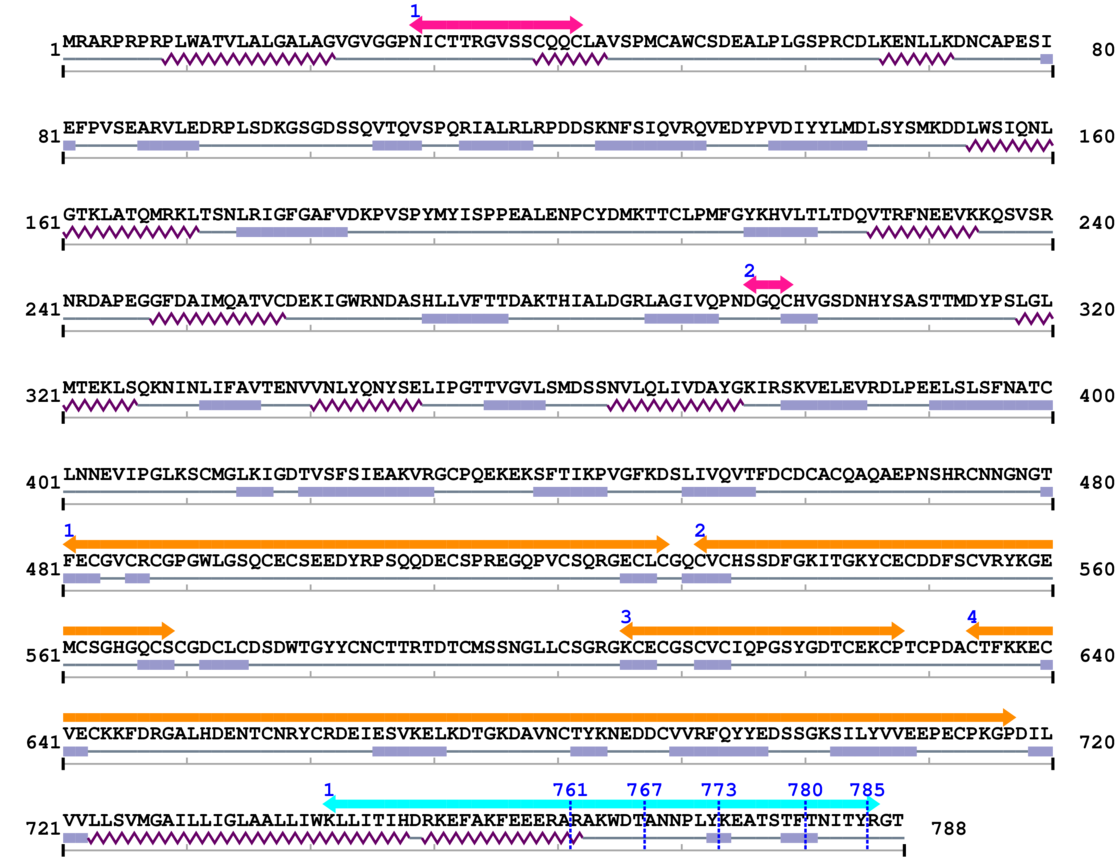
Amino Acid Sequence and Secondary Structures (PsiPred):
4. 3D Information
Known Structures in PDB: 1JV2 (X-ray; 310 A; B=27-718), 1KUP (NMR; -; B=742-766), 1KUZ (NMR; -; B=742-766), 1L5G (X-ray; 320 A; B=27-718), 1M1X (X-ray; 330 A; B=27-718), 1M8O (NMR; -; B=742-788), 1MIZ (X-ray; 190 A; A=765-769), 1MK7 (X-ray; 220 A; A/C=765-775), 1MK9 (X-ray; 280 A; A/C/E/G=765-776), 1RN0 (Model; -; B=135-378), 1S4X (NMR; -; A=742-788), 1TYE (X-ray; 290 A; B/D/F=27-466), 1U8C (X-ray; 310 A; B=27-718), 2INI (Model; -; B=81-460, B=558-716), 2K9J (NMR; -; B=711-753), 2KNC (NMR; -; B=715-788), 2KV9 (NMR; -; B=739-788), 2L1C (NMR; -; B=762-788), 2L91 (NMR; -; A=711-753), 2LJD (NMR; -; A=742-788), 2LJE (NMR; -; A=742-788), 2LJF (NMR; -; A=742-788), 2MTP (NMR; -; C=742-788), 2Q6W (X-ray; 225 A; C/F=50-61), 2RMZ (NMR; -; A=711-753), 2RN0 (NMR; -; A=711-753), 2VC2 (X-ray; 310 A; B=27-487), 2VDK (X-ray; 280 A; B=27-487), 2VDL (X-ray; 275 A; B=27-487), 2VDM (X-ray; 290 A; B=27-487), 2VDN (X-ray; 290 A; B=27-487), 2VDO (X-ray; 251 A; B=27-487), 2VDP (X-ray; 280 A; B=27-487), 2VDQ (X-ray; 259 A; B=27-487), 2VDR (X-ray; 240 A; B=27-487), 3FCS (X-ray; 255 A; B/D=27-716), 3FCU (X-ray; 290 A; B/D/F=27-487), 3IJE (X-ray; 290 A; B=27-721), 3NID (X-ray; 230 A; B/D=27-497), 3NIF (X-ray; 240 A; B/D=27-497), 3NIG (X-ray; 225 A; B/D=27-497), 3T3M (X-ray; 260 A; B/D=27-498), 3T3P (X-ray; 220 A; B/D=27-498), 3ZDX (X-ray; 245 A; B/D=27-498), 3ZDY (X-ray; 245 A; B/D=27-498), 3ZDZ (X-ray; 275 A; B/D=27-498), 3ZE0 (X-ray; 295 A; B/D=27-498), 3ZE1 (X-ray; 300 A; B/D=27-498), 3ZE2 (X-ray; 235 A; B/D=27-498), 4CAK (EM; 2050 A; B=27-716), 4G1E (X-ray; 300 A; B=27-717), 4G1M (X-ray; 290 A; B=27-718), 4MMX (X-ray; 332 A; B=27-718), 4MMY (X-ray; 318 A; B=27-718), 4MMZ (X-ray; 310 A; B=27-718), 4O02 (X-ray; 360 A; B=27-718)
* Cleavage Information
5 [sites] cleaved by Calpain 1
Source Reference: [PubMed ID: 7592818] Du X, Saido TC, Tsubuki S, Indig FE, Williams MJ, Ginsberg MH, Calpain cleavage of the cytoplasmic domain of the integrin beta 3 subunit. J Biol Chem. 1995 Nov 3;270(44):26146-51.
Cleavage sites (±10aa)
[Site 1] EFAKFEEERA761-RAKWDTANNP
Ala761  Arg
Arg
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Glu752 | Phe753 | Ala754 | Lys755 | Phe756 | Glu757 | Glu758 | Glu759 | Arg760 | Ala761 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Arg762 | Ala763 | Lys764 | Trp765 | Asp766 | Thr767 | Ala768 | Asn769 | Asn770 | Pro771 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| LIGLAALLIWKLLITIHDRKEFAKFEEERARAKWDTANNPLYKEATSTFTNITYRGT |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | N/A | 124.00 | 26 | glycoprotein IIIa |
| 2 | Mus musculus | 123.00 | 12 | ITB3_MOUSE Integrin beta-3 precursor (Platelet me |
| 3 | Canis familiaris | 123.00 | 10 | platelet glycoprotein IIIa |
| 4 | Bos taurus | 123.00 | 6 | PREDICTED: similar to platelet glycoprotein IIIa |
| 5 | Rattus norvegicus | 123.00 | 4 | integrin beta 3 |
| 6 | synthetic construct | 123.00 | 3 | AF239959_1 hybrid integrin beta 3 subunit precurs |
| 7 | Sus scrofa | 123.00 | 1 | integrin beta chain, beta 3 |
| 8 | rats, Peptide Partial, 723 aa | 123.00 | 1 | beta 3 integrin, GPIIIA |
| 9 | Canis lupus familiaris | 123.00 | 1 | integrin beta chain, beta 3 |
| 10 | Mus musculus | 123.00 | 1 | integrin beta 3 precursor |
| 11 | Sus scrofa | 121.00 | 3 | AF170527_1 glycoprotein IIIa; GPIIIa |
| 12 | Oryctolagus cuniculus | 120.00 | 1 | glycoprotein IIIa |
| 13 | Xenopus laevis | 113.00 | 2 | integrin beta-3 subunit |
| 14 | mice, Peptide Partial, 680 aa | 113.00 | 1 | beta 3 integrin, GPIIIA |
| 15 | Tetraodon nigroviridis | 111.00 | 8 | unnamed protein product |
| 16 | Danio rerio | 110.00 | 5 | integrin beta 3b |
| 17 | Homo sapiens | 109.00 | 11 | glycoprotein IIIa |
| 18 | Gallus gallus | 102.00 | 2 | integrin, beta 3 precursor |
| 19 | Ovis aries | 87.40 | 2 | ITB6_SHEEP Integrin beta-6 precursor gi |
| 20 | Anopheles gambiae | 87.40 | 1 | integrin beta subunit |
| 21 | Pseudoplusia includens | 87.40 | 1 | integrin beta 1 |
| 22 | Anopheles gambiae str. PEST | 87.40 | 1 | integrin beta subunit (AGAP000815-PA) |
| 23 | Ictalurus punctatus | 82.80 | 2 | beta-1 integrin |
| 24 | Crassostrea gigas | 77.80 | 1 | integrin beta cgh |
| 25 | Drosophila melanogaster | 71.60 | 1 | myospheroid CG1560-PA |
| 26 | Biomphalaria glabrata | 68.20 | 1 | beta integrin subunit |
| 27 | Xenopus laevis | 67.80 | 1 | hypothetical protein LOC379708 |
| 28 | Gallus gallus | 63.50 | 3 | integrin, beta 1 (fibronectin receptor, beta poly |
| 29 | Oryctolagus cuniculus | 63.50 | 1 | integrin beta 1 |
| 30 | Pongo pygmaeus | 63.50 | 1 | ITB1_PONPY Integrin beta-1 precursor (Fibronectin |
| 31 | Podocoryne carnea | 62.80 | 1 | AF308652_1 integrin beta chain |
| 32 | Caenorhabditis briggsae | 61.60 | 1 | Hypothetical protein CBG03601 |
| 33 | Felis catus | 60.80 | 1 | integrin beta 1 |
| 34 | Caenorhabditis elegans | 59.70 | 1 | Paralysed Arrest at Two-fold family member (pat-3 |
| 35 | Halocynthia roretzi | 58.90 | 2 | integrin beta Hr2 precursor |
| 36 | Mus sp. | 58.50 | 1 | beta 7 integrin |
| 37 | Strongylocentrotus purpuratus | 57.00 | 3 | integrin beta-C subunit |
| 38 | Pan troglodytes | 55.80 | 2 | integrin, beta 2 |
| 39 | Homo sapiens | 55.80 | 1 | integrin, beta 2 precursor |
| 40 | Lytechinus variegatus | 53.90 | 1 | beta-C integrin subunit |
| 41 | Acropora millepora | 53.50 | 1 | integrin subunit betaCn1 |
| 42 | Pacifastacus leniusculus | 47.00 | 1 | integrin |
| 43 | Schistosoma japonicum | 46.60 | 1 | SJCHGC06221 protein |
| 44 | Ovis canadensis | 45.40 | 1 | ITB2_OVICA Integrin beta-2 precursor (Cell surfac |
| 45 | Sigmodon hispidus | 43.90 | 1 | AF445415_1 integrin beta-2 precursor |
Top-ranked sequences
| organism | matching |
|---|---|
| N/A | Query 732 LIGLAALLIWKLLITIHDRKEFAKFEEERA#RAKWDTANNPLYKEATSTFTNITYRGT 788 ||||||||||||||||||||||||||||||#||||||||||||||||||||||||||| Query 732 LIGLAALLIWKLLITIHDRKEFAKFEEERA#RAKWDTANNPLYKEATSTFTNITYRGT 788 |
| Mus musculus | Query 732 LIGLAALLIWKLLITIHDRKEFAKFEEERA#RAKWDTANNPLYKEATSTFTNITYRGT 788 ||||| ||||||||||||||||||||||||#||||||||||||||||||||||||||| Sbjct 731 LIGLATLLIWKLLITIHDRKEFAKFEEERA#RAKWDTANNPLYKEATSTFTNITYRGT 787 |
| Canis familiaris | Query 732 LIGLAALLIWKLLITIHDRKEFAKFEEERA#RAKWDTANNPLYKEATSTFTNITYRGT 788 ||||| ||||||||||||||||||||||||#||||||||||||||||||||||||||| Sbjct 728 LIGLATLLIWKLLITIHDRKEFAKFEEERA#RAKWDTANNPLYKEATSTFTNITYRGT 784 |
| Bos taurus | Query 732 LIGLAALLIWKLLITIHDRKEFAKFEEERA#RAKWDTANNPLYKEATSTFTNITYRGT 788 ||||| ||||||||||||||||||||||||#||||||||||||||||||||||||||| Sbjct 728 LIGLATLLIWKLLITIHDRKEFAKFEEERA#RAKWDTANNPLYKEATSTFTNITYRGT 784 |
| Rattus norvegicus | Query 732 LIGLAALLIWKLLITIHDRKEFAKFEEERA#RAKWDTANNPLYKEATSTFTNITYRGT 788 ||||| ||||||||||||||||||||||||#||||||||||||||||||||||||||| Sbjct 731 LIGLATLLIWKLLITIHDRKEFAKFEEERA#RAKWDTANNPLYKEATSTFTNITYRGT 787 |
| synthetic construct | Query 732 LIGLAALLIWKLLITIHDRKEFAKFEEERA#RAKWDTANNPLYKEATSTFTNITYRGT 788 ||||| ||||||||||||||||||||||||#||||||||||||||||||||||||||| Sbjct 732 LIGLATLLIWKLLITIHDRKEFAKFEEERA#RAKWDTANNPLYKEATSTFTNITYRGT 788 |
| Sus scrofa | Query 732 LIGLAALLIWKLLITIHDRKEFAKFEEERA#RAKWDTANNPLYKEATSTFTNITYRGT 788 ||||| ||||||||||||||||||||||||#||||||||||||||||||||||||||| Sbjct 728 LIGLATLLIWKLLITIHDRKEFAKFEEERA#RAKWDTANNPLYKEATSTFTNITYRGT 784 |
| rats, Peptide Partial, 723 aa | Query 732 LIGLAALLIWKLLITIHDRKEFAKFEEERA#RAKWDTANNPLYKEATSTFTNITYRGT 788 ||||| ||||||||||||||||||||||||#||||||||||||||||||||||||||| Sbjct 667 LIGLATLLIWKLLITIHDRKEFAKFEEERA#RAKWDTANNPLYKEATSTFTNITYRGT 723 |
| Canis lupus familiaris | Query 732 LIGLAALLIWKLLITIHDRKEFAKFEEERA#RAKWDTANNPLYKEATSTFTNITYRGT 788 ||||| ||||||||||||||||||||||||#||||||||||||||||||||||||||| Sbjct 728 LIGLATLLIWKLLITIHDRKEFAKFEEERA#RAKWDTANNPLYKEATSTFTNITYRGT 784 |
| Mus musculus | Query 732 LIGLAALLIWKLLITIHDRKEFAKFEEERA#RAKWDTANNPLYKEATSTFTNITYRGT 788 ||||| ||||||||||||||||||||||||#||||||||||||||||||||||||||| Sbjct 731 LIGLATLLIWKLLITIHDRKEFAKFEEERA#RAKWDTANNPLYKEATSTFTNITYRGT 787 |
| Sus scrofa | Query 732 LIGLAALLIWKLLITIHDRKEFAKFEEERA#RAKWDTANNPLYKEATSTFTNITYRGT 788 ||| | ||||||||||||||||||||||||#||||||||||||||||||||||||||| Sbjct 728 LIGFATLLIWKLLITIHDRKEFAKFEEERA#RAKWDTANNPLYKEATSTFTNITYRGT 784 |
| Oryctolagus cuniculus | Query 732 LIGLAALLIWKLLITIHDRKEFAKFEEERA#RAKWDTANNPLYKEATSTFTNITYRGT 788 ||| | ||||||||||||||||||||||||#||||||||||||||||||||||||||| Sbjct 732 LIGFALLLIWKLLITIHDRKEFAKFEEERA#RAKWDTANNPLYKEATSTFTNITYRGT 788 |
| Xenopus laevis | Query 732 LIGLAALLIWKLLITIHDRKEFAKFEEERA#RAKWDTANNPLYKEATSTFTNITYRG 787 |||| ||||||||||||||+||||||||||#+||||||+||||| |||||||||||| Sbjct 730 LIGLVALLIWKLLITIHDRREFAKFEEERA#KAKWDTAHNPLYKGATSTFTNITYRG 785 |
| mice, Peptide Partial, 680 aa | Query 732 LIGLAALLIWKLLITIHDRKEFAKFEEERA#RAKWDTANNPLYKEATSTFTNIT 784 ||||| ||||||||||||||||||||||||#||||||||||||||||||||||| Sbjct 628 LIGLATLLIWKLLITIHDRKEFAKFEEERA#RAKWDTANNPLYKEATSTFTNIT 680 |
| Tetraodon nigroviridis | Query 732 LIGLAALLIWKLLITIHDRKEFAKFEEERA#RAKWDTANNPLYKEATSTFTNITYRG 787 +|||||||||||||||||+||||||||||#|||||| +||||| |||||||||+|| Sbjct 747 FLGLAALLIWKLLITIHDRREFAKFEEERA#RAKWDTGHNPLYKGATSTFTNITFRG 802 |
| Danio rerio | Query 732 LIGLAALLIWKLLITIHDRKEFAKFEEERA#RAKWDTANNPLYKEATSTFTNITYRG 787 +|||||||||||||||||+||||||||||#||||+| +||||| |||||||||||| Sbjct 733 FLGLAALLIWKLLITIHDRREFAKFEEERA#RAKWETGHNPLYKGATSTFTNITYRG 788 |
| Homo sapiens | Query 732 LIGLAALLIWKLLITIHDRKEFAKFEEERA#RAKWDTANNPLYKEATSTFTNITYRGT 788 ||||| ||||||||||||||||||||#||||||||||||||||||||||||||| Sbjct 706 LIGLAPCSSGKLLITIHDRKEFAKFEEERA#RAKWDTANNPLYKEATSTFTNITYRGT 762 |
| Gallus gallus | Query 741 WKLLITIHDRKEFAKFEEERA#RAKWDTANNPLYKEATSTFTNITYRG 787 ||||||||||+|||+||||+|#|||||| ||||||||||||||||||| Sbjct 733 WKLLITIHDRREFARFEEEKA#RAKWDTGNNPLYKEATSTFTNITYRG 779 |
| Ovis aries | Query 732 LIGLAALLIWKLLITIHDRKEFAKFEEERA#RAKWDTANNPLYKEATSTFTNITYR 786 |||+| | |||||++ ||||| |||| ||+#+||| | ||||+ +|||| |+||+ Sbjct 720 LIGVALLCIWKLLVSFHDRKEVAKFEAERS#KAKWQTGTNPLYRGSTSTFKNVTYK 774 |
| Anopheles gambiae | Query 734 GLAALLIWKLLITIHDRKEFAKFEEERA#RAKWDTANNPLYKEATSTFTNITYRG 787 |+| ||+||+| +||||+|||+||+|| # ||||| ||+||+||+|| | || | Sbjct 783 GMAVLLLWKVLTSIHDRREFARFEKERM#MAKWDTGENPIYKQATTTFKNPTYAG 836 |
| Pseudoplusia includens | Query 732 LIGLAALLIWKLLITIHDRKEFAKFEEERA#RAKWDTANNPLYKEATSTFTNITYRG 787 |+||| |++||+ | |||+|||+||+|| # ||||| ||+||+||||| | || | Sbjct 781 LVGLALLMLWKMATTSHDRREFARFEKERM#MAKWDTGENPIYKQATSTFKNPTYAG 836 |
| Anopheles gambiae str. PEST | Query 734 GLAALLIWKLLITIHDRKEFAKFEEERA#RAKWDTANNPLYKEATSTFTNITYRG 787 |+| ||+||+| +||||+|||+||+|| # ||||| ||+||+||+|| | || | Sbjct 783 GMAVLLLWKVLTSIHDRREFARFEKERM#MAKWDTGENPIYKQATTTFKNPTYAG 836 |
| Ictalurus punctatus | Query 732 LIGLAALLIWKLLITIHDRKEFAKFEEERA#RAKWDTANNPLYKEATSTFTNITYRG 787 ||||| | |||||+ ||||+||||||+|+ # |||| ||+|| | +| | | | Sbjct 751 LIGLALLFIWKLLMIIHDRREFAKFEKEKM#NAKWDAGENPIYKSAVTTVVNPKYEG 806 |
| Crassostrea gigas | Query 732 LIGLAALLIWKLLITIHDRKEFAKFEEERA#RAKWDTANNPLYKEATSTFTNITYR 786 |+ |||||| || |++| |+||+| # |+||| ||+||+||||| | ||| Sbjct 744 FFGIILLLIWKLFTTISDKRELARFEKEAM#NARWDTGENPIYKQATSTFVNPTYR 798 |
| Drosophila melanogaster | Query 746 TIHDRKEFAKFEEERA#RAKWDTANNPLYKEATSTFTNITYRG 787 |||||+|||+||+|| # ||||| ||+||+||||| | | | Sbjct 804 TIHDRREFARFEKERM#NAKWDTGENPIYKQATSTFKNPMYAG 845 |
| Biomphalaria glabrata | Query 747 IHDRKEFAKFEEERA#RAKWDTANNPLYKEATSTFTNITY 785 ||| +||||||+|| # ||||| ||+||+||||| | || Sbjct 747 IHDTREFAKFEKERQ#NAKWDTGENPIYKQATSTFKNPTY 785 |
| Xenopus laevis | Query 732 LIGLAALLIWKLLITIHDRKEFAKFEEERA#RAKWDTANNPLYKEATSTFTNITYRG 787 |||| |+|||++ | || |+ +||+|| # +||+ +|||| ||+| | + | Sbjct 716 FIGLACLIIWKIITEIKDRNEYQRFEKERM#NSKWNPGHNPLYHNATTTVQNPNFSG 771 |
| Gallus gallus | Query 732 LIGLAALLIWKLLITIHDRKEFAKFEEERA#RAKWDTANNPLYKEATSTFTNITYRG 787 ||||| |||||||+ ||||+||||||+|+ # ||||| ||+|| | +| | | | Sbjct 747 LIGLALLLIWKLLMIIHDRREFAKFEKEKM#NAKWDTGENPIYKSAVTTVVNPKYEG 802 |
| Oryctolagus cuniculus | Query 732 LIGLAALLIWKLLITIHDRKEFAKFEEERA#RAKWDTANNPLYKEATSTFTNITYRG 787 ||||| |||||||+ ||||+||||||+|+ # ||||| ||+|| | +| | | | Sbjct 7 LIGLALLLIWKLLMIIHDRREFAKFEKEKM#NAKWDTGENPIYKSAVTTVVNPKYEG 62 |
| Pongo pygmaeus | Query 732 LIGLAALLIWKLLITIHDRKEFAKFEEERA#RAKWDTANNPLYKEATSTFTNITYRG 787 ||||| |||||||+ ||||+||||||+|+ # ||||| ||+|| | +| | | | Sbjct 742 LIGLALLLIWKLLMIIHDRREFAKFEKEKM#NAKWDTGENPIYKSAVTTVVNPKYEG 797 |
| Podocoryne carnea | Query 746 TIHDRKEFAKFEEERA#RAKWDTANNPLYKEATSTFTNITY 785 || ||+||||||++| # |||+ ||+||+||||| | | Sbjct 739 TIQDRREFAKFEKDRQ#NPKWDSGENPIYKKATSTFQNPMY 778 |
| Caenorhabditis briggsae | Query 748 HDRKEFAKFEEERA#RAKWDTANNPLYKEATSTFTNITYRG 787 ||| |+||| || # ||||| ||+||+||+|| | | | Sbjct 768 HDRAEYAKFNNERL#MAKWDTNENPIYKQATTTFKNPVYAG 807 |
| Felis catus | Query 732 LIGLAALLIWKLLITIHDRKEFAKFEEERA#RAKWDTANNPLYKEATSTFTNITYRG 787 ||||| |||||||+ ||| +||||||+|+ # ||||| ||+|| | +| | | | Sbjct 742 LIGLALLLIWKLLMIIHDTREFAKFEKEKM#NAKWDTGENPIYKSAVTTVVNPKYEG 797 |
| Caenorhabditis elegans | Query 748 HDRKEFAKFEEERA#RAKWDTANNPLYKEATSTFTNITYRG 787 ||| |+| | || # ||||| ||+||+||+|| | | | Sbjct 767 HDRSEYATFNNERL#MAKWDTNENPIYKQATTTFKNPVYAG 806 |
| Halocynthia roretzi | Query 732 LIGLAALLIWKLLITIHDRKEFAKFEEERA#RAKWDTANNPLYKEATSTFTNITYRG 787 |||+ ||+|||++ |+ |++|+ ||++| # || ||+| |+| | | + | Sbjct 778 LIGIIALIIWKVIQTLRDKREYEKFKKEED#LRKWTKGENPVYVNASSKFDNPMFGG 833 |
| Mus sp. | Query 733 IGLAALLIWKLLITIHDRKEFAKFEEERA#RAKWDTANNPLYKEATSTFTNITYRGT 788 +|| +| ++| + |+||+|+ +||+|+ #+ | |||||| | +| | ++|| Sbjct 737 VGLGLVLAYRLSVEIYDRREYRRFEKEQQ#QLNWKQDNNPLYKSAITTTVNPRFQGT 792 |
| Strongylocentrotus purpuratus | Query 732 LIGLAALLIWKLLITIHDRKEFAKFEEERA#RAKWDTANNPLYKEATSTFTNITY 785 |+||| ||||+|| ||||+|| ||+|||# | |+ ||+|| +|| | | || Sbjct 750 LVGLALLLIWRLLTYIHDRREFQNFEKERA#NATWEGGENPIYKPSTSVFKNPTY 803 |
| Pan troglodytes | Query 732 LIGLAALLIWKLLITIHDRKEFAKFEEERA#RAKWDTANNPLYKEATSTFTN 782 |||+ |+||| || + | +|+ +||+|+ #+++|+ +|||+| ||+| | Sbjct 714 LIGILLLVIWKALIHLSDLREYRRFEKEKL#KSQWNN-DNPLFKSATTTVMN 763 |
| Homo sapiens | Query 732 LIGLAALLIWKLLITIHDRKEFAKFEEERA#RAKWDTANNPLYKEATSTFTN 782 |||+ |+||| || + | +|+ +||+|+ #+++|+ +|||+| ||+| | Sbjct 714 LIGILLLVIWKALIHLSDLREYRRFEKEKL#KSQWNN-DNPLFKSATTTVMN 763 |
| Lytechinus variegatus | Query 740 IWKLLITIHDRKEFAKFEEERA#RAKWDTANNPLYKEATSTFTNITY 785 ||+|| ||||+|| ||+|||# | |+ ||+|| +|| | | || Sbjct 758 IWRLLTYIHDRREFQNFEKERA#NATWEGGENPIYKPSTSVFKNPTY 803 |
| Acropora millepora | Query 742 KLLITIHDRKEFAKFEEERA#RAKWDTANNPLYKEATSTFTNITYRG 787 | | |+ || |+ ||| || # +|| ||||+ | +|| | || | Sbjct 744 KGLFTMVDRIEYQKFERERM#HSKWTREKNPLYQAAKTTFENPTYAG 789 |
| Pacifastacus leniusculus | Query 746 TIHDRKEFAKFEEERA#RAKWDTANNPLYKEATSTFTNITY 785 |+|||+|+||||+|| # | ||||| | +|| | + Sbjct 758 TLHDRREYAKFEKERK#LPSGKRAENPLYKSAKTTFQNPAF 797 |
| Schistosoma japonicum | Query 741 WKLLITIHDRKEFAKFEEERA#RAKWDTANNPLYKEATSTFTNITY 785 +||+||| ||+| | |+++ # +|+ | ||+++ |+ | |+ Sbjct 239 YKLVITIDDRRELANFKQQGE#NMRWEMAENPIFESPTTNVLNPTF 283 |
| Ovis canadensis | Query 738 LLIWKLLITIHDRKEFAKFEEERA#RAKWDTANNPLYKEATSTFTN 782 | ||| | + | +|+ +||+|+ #+++|+ +|||+| ||+| | Sbjct 721 LAIWKALTHLSDLREYHRFEKEKL#KSQWNN-DNPLFKSATTTVMN 764 |
| Sigmodon hispidus | Query 738 LLIWKLLITIHDRKEFAKFEEERA#RAKWDTANNPLYKEATSTFTN 782 |+||| | + | +|+ ||+|+ #+++|+ +|||+| ||+| | Sbjct 720 LVIWKALTHLTDLREYRHFEKEKL#KSQWNN-DNPLFKSATTTVMN 763 |
[Site 2] EERARAKWDT767-ANNPLYKEAT
Thr767  Ala
Ala
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Glu758 | Glu759 | Arg760 | Ala761 | Arg762 | Ala763 | Lys764 | Trp765 | Asp766 | Thr767 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Ala768 | Asn769 | Asn770 | Pro771 | Leu772 | Tyr773 | Lys774 | Glu775 | Ala776 | Thr777 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| LLIWKLLITIHDRKEFAKFEEERARAKWDTANNPLYKEATSTFTNITYRGT |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | N/A | 114.00 | 28 | glycoprotein IIIa precursor |
| 2 | Mus musculus | 114.00 | 12 | ITB3_MOUSE Integrin beta-3 precursor (Platelet me |
| 3 | Canis familiaris | 114.00 | 10 | platelet glycoprotein IIIa |
| 4 | Bos taurus | 114.00 | 7 | PREDICTED: similar to platelet glycoprotein IIIa |
| 5 | Rattus norvegicus | 114.00 | 5 | integrin beta 3 |
| 6 | Sus scrofa | 114.00 | 3 | AF170527_1 glycoprotein IIIa; GPIIIa |
| 7 | synthetic construct | 114.00 | 3 | AF239959_1 hybrid integrin beta 3 subunit precurs |
| 8 | Sus scrofa | 114.00 | 1 | integrin beta chain, beta 3 |
| 9 | Oryctolagus cuniculus | 114.00 | 1 | glycoprotein IIIa |
| 10 | rats, Peptide Partial, 723 aa | 114.00 | 1 | beta 3 integrin, GPIIIA |
| 11 | Mus musculus | 114.00 | 1 | integrin beta 3 precursor |
| 12 | Canis lupus familiaris | 114.00 | 1 | integrin beta chain, beta 3 |
| 13 | Homo sapiens | 104.00 | 11 | glycoprotein IIIa |
| 14 | Xenopus laevis | 104.00 | 2 | integrin beta-3 subunit |
| 15 | mice, Peptide Partial, 680 aa | 104.00 | 1 | beta 3 integrin, GPIIIA |
| 16 | Tetraodon nigroviridis | 103.00 | 5 | unnamed protein product |
| 17 | Danio rerio | 102.00 | 5 | integrin beta 3b |
| 18 | Gallus gallus | 102.00 | 2 | integrin, beta 3 precursor |
| 19 | Anopheles gambiae | 82.80 | 1 | integrin beta subunit |
| 20 | Anopheles gambiae str. PEST | 82.80 | 1 | integrin beta subunit (AGAP000815-PA) |
| 21 | Ovis aries | 80.10 | 2 | ITB6_SHEEP Integrin beta-6 precursor gi |
| 22 | Pseudoplusia includens | 80.10 | 1 | integrin beta 1 |
| 23 | Crassostrea gigas | 75.90 | 1 | integrin beta cgh |
| 24 | Ictalurus punctatus | 74.30 | 1 | beta-1 integrin |
| 25 | Drosophila melanogaster | 71.60 | 1 | myospheroid CG1560-PA |
| 26 | Biomphalaria glabrata | 68.20 | 1 | beta integrin subunit |
| 27 | Gallus gallus | 63.50 | 3 | integrin, beta 1 (fibronectin receptor, beta poly |
| 28 | Pongo pygmaeus | 63.50 | 1 | ITB1_PONPY Integrin beta-1 precursor (Fibronectin |
| 29 | Podocoryne carnea | 62.80 | 1 | AF308652_1 integrin beta chain |
| 30 | Caenorhabditis briggsae | 61.60 | 1 | Hypothetical protein CBG03601 |
| 31 | Oryctolagus cuniculus | 60.80 | 1 | integrin beta 1 |
| 32 | Xenopus laevis | 60.50 | 1 | hypothetical protein LOC379708 |
| 33 | Felis catus | 60.50 | 1 | integrin beta 1 |
| 34 | Caenorhabditis elegans | 59.70 | 1 | Paralysed Arrest at Two-fold family member (pat-3 |
| 35 | Strongylocentrotus purpuratus | 58.20 | 3 | integrin beta-C subunit |
| 36 | Lytechinus variegatus | 58.20 | 1 | beta-C integrin subunit |
| 37 | Mus sp. | 54.30 | 1 | beta 7 integrin |
| 38 | Acropora millepora | 53.50 | 1 | integrin subunit betaCn1 |
| 39 | Halocynthia roretzi | 51.60 | 2 | integrin beta Hr2 precursor |
| 40 | Pan troglodytes | 50.80 | 2 | integrin, beta 2 |
| 41 | Homo sapiens | 50.80 | 1 | integrin, beta 2 precursor |
| 42 | Pacifastacus leniusculus | 47.00 | 1 | integrin |
| 43 | Schistosoma japonicum | 46.60 | 1 | SJCHGC06221 protein |
| 44 | Ovis canadensis | 46.60 | 1 | ITB2_OVICA Integrin beta-2 precursor (Cell surfac |
| 45 | Sigmodon hispidus | 45.40 | 1 | AF445415_1 integrin beta-2 precursor |
Top-ranked sequences
| organism | matching |
|---|---|
| N/A | Query 738 LLIWKLLITIHDRKEFAKFEEERARAKWDT#ANNPLYKEATSTFTNITYRGT 788 ||||||||||||||||||||||||||||||#||||||||||||||||||||| Query 738 LLIWKLLITIHDRKEFAKFEEERARAKWDT#ANNPLYKEATSTFTNITYRGT 788 |
| Mus musculus | Query 738 LLIWKLLITIHDRKEFAKFEEERARAKWDT#ANNPLYKEATSTFTNITYRGT 788 ||||||||||||||||||||||||||||||#||||||||||||||||||||| Query 738 LLIWKLLITIHDRKEFAKFEEERARAKWDT#ANNPLYKEATSTFTNITYRGT 788 |
| Canis familiaris | Query 738 LLIWKLLITIHDRKEFAKFEEERARAKWDT#ANNPLYKEATSTFTNITYRGT 788 ||||||||||||||||||||||||||||||#||||||||||||||||||||| Query 738 LLIWKLLITIHDRKEFAKFEEERARAKWDT#ANNPLYKEATSTFTNITYRGT 788 |
| Bos taurus | Query 738 LLIWKLLITIHDRKEFAKFEEERARAKWDT#ANNPLYKEATSTFTNITYRGT 788 ||||||||||||||||||||||||||||||#||||||||||||||||||||| Query 738 LLIWKLLITIHDRKEFAKFEEERARAKWDT#ANNPLYKEATSTFTNITYRGT 788 |
| Rattus norvegicus | Query 738 LLIWKLLITIHDRKEFAKFEEERARAKWDT#ANNPLYKEATSTFTNITYRGT 788 ||||||||||||||||||||||||||||||#||||||||||||||||||||| Query 738 LLIWKLLITIHDRKEFAKFEEERARAKWDT#ANNPLYKEATSTFTNITYRGT 788 |
| Sus scrofa | Query 738 LLIWKLLITIHDRKEFAKFEEERARAKWDT#ANNPLYKEATSTFTNITYRGT 788 ||||||||||||||||||||||||||||||#||||||||||||||||||||| Query 738 LLIWKLLITIHDRKEFAKFEEERARAKWDT#ANNPLYKEATSTFTNITYRGT 788 |
| synthetic construct | Query 738 LLIWKLLITIHDRKEFAKFEEERARAKWDT#ANNPLYKEATSTFTNITYRGT 788 ||||||||||||||||||||||||||||||#||||||||||||||||||||| Query 738 LLIWKLLITIHDRKEFAKFEEERARAKWDT#ANNPLYKEATSTFTNITYRGT 788 |
| Sus scrofa | Query 738 LLIWKLLITIHDRKEFAKFEEERARAKWDT#ANNPLYKEATSTFTNITYRGT 788 ||||||||||||||||||||||||||||||#||||||||||||||||||||| Query 738 LLIWKLLITIHDRKEFAKFEEERARAKWDT#ANNPLYKEATSTFTNITYRGT 788 |
| Oryctolagus cuniculus | Query 738 LLIWKLLITIHDRKEFAKFEEERARAKWDT#ANNPLYKEATSTFTNITYRGT 788 ||||||||||||||||||||||||||||||#||||||||||||||||||||| Query 738 LLIWKLLITIHDRKEFAKFEEERARAKWDT#ANNPLYKEATSTFTNITYRGT 788 |
| rats, Peptide Partial, 723 aa | Query 738 LLIWKLLITIHDRKEFAKFEEERARAKWDT#ANNPLYKEATSTFTNITYRGT 788 ||||||||||||||||||||||||||||||#||||||||||||||||||||| Query 738 LLIWKLLITIHDRKEFAKFEEERARAKWDT#ANNPLYKEATSTFTNITYRGT 788 |
| Mus musculus | Query 738 LLIWKLLITIHDRKEFAKFEEERARAKWDT#ANNPLYKEATSTFTNITYRGT 788 ||||||||||||||||||||||||||||||#||||||||||||||||||||| Query 738 LLIWKLLITIHDRKEFAKFEEERARAKWDT#ANNPLYKEATSTFTNITYRGT 788 |
| Canis lupus familiaris | Query 738 LLIWKLLITIHDRKEFAKFEEERARAKWDT#ANNPLYKEATSTFTNITYRGT 788 ||||||||||||||||||||||||||||||#||||||||||||||||||||| Query 738 LLIWKLLITIHDRKEFAKFEEERARAKWDT#ANNPLYKEATSTFTNITYRGT 788 |
| Homo sapiens | Query 742 KLLITIHDRKEFAKFEEERARAKWDT#ANNPLYKEATSTFTNITYRGT 788 ||||||||||||||||||||||||||#||||||||||||||||||||| Query 742 KLLITIHDRKEFAKFEEERARAKWDT#ANNPLYKEATSTFTNITYRGT 788 |
| Xenopus laevis | Query 738 LLIWKLLITIHDRKEFAKFEEERARAKWDT#ANNPLYKEATSTFTNITYRG 787 |||||||||||||+||||||||||+|||||#|+||||| |||||||||||| Sbjct 736 LLIWKLLITIHDRREFAKFEEERAKAKWDT#AHNPLYKGATSTFTNITYRG 785 |
| mice, Peptide Partial, 680 aa | Query 738 LLIWKLLITIHDRKEFAKFEEERARAKWDT#ANNPLYKEATSTFTNIT 784 ||||||||||||||||||||||||||||||#||||||||||||||||| Query 738 LLIWKLLITIHDRKEFAKFEEERARAKWDT#ANNPLYKEATSTFTNIT 784 |
| Tetraodon nigroviridis | Query 738 LLIWKLLITIHDRKEFAKFEEERARAKWDT#ANNPLYKEATSTFTNITYRG 787 |||||||||||||+||||||||||||||||# +||||| |||||||||+|| Sbjct 753 LLIWKLLITIHDRREFAKFEEERARAKWDT#GHNPLYKGATSTFTNITFRG 802 |
| Danio rerio | Query 738 LLIWKLLITIHDRKEFAKFEEERARAKWDT#ANNPLYKEATSTFTNITYRG 787 |||||||||||||+||||||||||||||+|# +||||| |||||||||||| Sbjct 739 LLIWKLLITIHDRREFAKFEEERARAKWET#GHNPLYKGATSTFTNITYRG 788 |
| Gallus gallus | Query 741 WKLLITIHDRKEFAKFEEERARAKWDT#ANNPLYKEATSTFTNITYRG 787 ||||||||||+|||+||||+|||||||# ||||||||||||||||||| Sbjct 733 WKLLITIHDRREFARFEEEKARAKWDT#GNNPLYKEATSTFTNITYRG 779 |
| Anopheles gambiae | Query 738 LLIWKLLITIHDRKEFAKFEEERARAKWDT#ANNPLYKEATSTFTNITYRG 787 ||+||+| +||||+|||+||+|| |||||# ||+||+||+|| | || | Sbjct 787 LLLWKVLTSIHDRREFARFEKERMMAKWDT#GENPIYKQATTTFKNPTYAG 836 |
| Anopheles gambiae str. PEST | Query 738 LLIWKLLITIHDRKEFAKFEEERARAKWDT#ANNPLYKEATSTFTNITYRG 787 ||+||+| +||||+|||+||+|| |||||# ||+||+||+|| | || | Sbjct 787 LLLWKVLTSIHDRREFARFEKERMMAKWDT#GENPIYKQATTTFKNPTYAG 836 |
| Ovis aries | Query 738 LLIWKLLITIHDRKEFAKFEEERARAKWDT#ANNPLYKEATSTFTNITYR 786 | |||||++ ||||| |||| ||++||| |# ||||+ +|||| |+||+ Sbjct 726 LCIWKLLVSFHDRKEVAKFEAERSKAKWQT#GTNPLYRGSTSTFKNVTYK 774 |
| Pseudoplusia includens | Query 738 LLIWKLLITIHDRKEFAKFEEERARAKWDT#ANNPLYKEATSTFTNITYRG 787 |++||+ | |||+|||+||+|| |||||# ||+||+||||| | || | Sbjct 787 LMLWKMATTSHDRREFARFEKERMMAKWDT#GENPIYKQATSTFKNPTYAG 836 |
| Crassostrea gigas | Query 738 LLIWKLLITIHDRKEFAKFEEERARAKWDT#ANNPLYKEATSTFTNITYR 786 |||||| || |++| |+||+| |+|||# ||+||+||||| | ||| Sbjct 750 LLIWKLFTTISDKRELARFEKEAMNARWDT#GENPIYKQATSTFVNPTYR 798 |
| Ictalurus punctatus | Query 738 LLIWKLLITIHDRKEFAKFEEERARAKWDT#ANNPLYKEATSTFTNITYRG 787 | |||||+ ||||+||||||+|+ |||| # ||+|| | +| | | | Sbjct 757 LFIWKLLMIIHDRREFAKFEKEKMNAKWDA#GENPIYKSAVTTVVNPKYEG 806 |
| Drosophila melanogaster | Query 746 TIHDRKEFAKFEEERARAKWDT#ANNPLYKEATSTFTNITYRG 787 |||||+|||+||+|| |||||# ||+||+||||| | | | Sbjct 804 TIHDRREFARFEKERMNAKWDT#GENPIYKQATSTFKNPMYAG 845 |
| Biomphalaria glabrata | Query 747 IHDRKEFAKFEEERARAKWDT#ANNPLYKEATSTFTNITY 785 ||| +||||||+|| |||||# ||+||+||||| | || Sbjct 747 IHDTREFAKFEKERQNAKWDT#GENPIYKQATSTFKNPTY 785 |
| Gallus gallus | Query 745 ITIHDRKEFAKFEEERARAKWDT#ANNPLYKEATSTFTNITYRG 787 + ||||+||||||+|+ |||||# ||+|| | +| | | | Sbjct 760 MIIHDRREFAKFEKEKMNAKWDT#GENPIYKSAVTTVVNPKYEG 802 |
| Pongo pygmaeus | Query 745 ITIHDRKEFAKFEEERARAKWDT#ANNPLYKEATSTFTNITYRG 787 + ||||+||||||+|+ |||||# ||+|| | +| | | | Sbjct 755 MIIHDRREFAKFEKEKMNAKWDT#GENPIYKSAVTTVVNPKYEG 797 |
| Podocoryne carnea | Query 746 TIHDRKEFAKFEEERARAKWDT#ANNPLYKEATSTFTNITY 785 || ||+||||||++| |||+# ||+||+||||| | | Sbjct 739 TIQDRREFAKFEKDRQNPKWDS#GENPIYKKATSTFQNPMY 778 |
| Caenorhabditis briggsae | Query 748 HDRKEFAKFEEERARAKWDT#ANNPLYKEATSTFTNITYRG 787 ||| |+||| || |||||# ||+||+||+|| | | | Sbjct 768 HDRAEYAKFNNERLMAKWDT#NENPIYKQATTTFKNPVYAG 807 |
| Oryctolagus cuniculus | Query 745 ITIHDRKEFAKFEEERARAKWDT#ANNPLYKEATSTFTNITYRG 787 + ||||+||||||+|+ |||||# ||+|| | +| | | | Sbjct 20 MIIHDRREFAKFEKEKMNAKWDT#GENPIYKSAVTTVVNPKYEG 62 |
| Xenopus laevis | Query 738 LLIWKLLITIHDRKEFAKFEEERARAKWDT#ANNPLYKEATSTFTNITYRG 787 |+|||++ | || |+ +||+|| +||+ # +|||| ||+| | + | Sbjct 722 LIIWKIITEIKDRNEYQRFEKERMNSKWNP#GHNPLYHNATTTVQNPNFSG 771 |
| Felis catus | Query 745 ITIHDRKEFAKFEEERARAKWDT#ANNPLYKEATSTFTNITYRG 787 + ||| +||||||+|+ |||||# ||+|| | +| | | | Sbjct 755 MIIHDTREFAKFEKEKMNAKWDT#GENPIYKSAVTTVVNPKYEG 797 |
| Caenorhabditis elegans | Query 748 HDRKEFAKFEEERARAKWDT#ANNPLYKEATSTFTNITYRG 787 ||| |+| | || |||||# ||+||+||+|| | | | Sbjct 767 HDRSEYATFNNERLMAKWDT#NENPIYKQATTTFKNPVYAG 806 |
| Strongylocentrotus purpuratus | Query 747 IHDRKEFAKFEEERARAKWDT#ANNPLYKEATSTFTNITY 785 ||||+|| ||+||| | |+ # ||+|| +|| | | || Sbjct 765 IHDRREFQNFEKERANATWEG#GENPIYKPSTSVFKNPTY 803 |
| Lytechinus variegatus | Query 747 IHDRKEFAKFEEERARAKWDT#ANNPLYKEATSTFTNITY 785 ||||+|| ||+||| | |+ # ||+|| +|| | | || Sbjct 765 IHDRREFQNFEKERANATWEG#GENPIYKPSTSVFKNPTY 803 |
| Mus sp. | Query 738 LLIWKLLITIHDRKEFAKFEEERARAKWDT#ANNPLYKEATSTFTNITYRGT 788 +| ++| + |+||+|+ +||+|+ + | # |||||| | +| | ++|| Sbjct 742 VLAYRLSVEIYDRREYRRFEKEQQQLNWKQ#DNNPLYKSAITTTVNPRFQGT 792 |
| Acropora millepora | Query 742 KLLITIHDRKEFAKFEEERARAKWDT#ANNPLYKEATSTFTNITYRG 787 | | |+ || |+ ||| || +|| # ||||+ | +|| | || | Sbjct 744 KGLFTMVDRIEYQKFERERMHSKWTR#EKNPLYQAAKTTFENPTYAG 789 |
| Halocynthia roretzi | Query 738 LLIWKLLITIHDRKEFAKFEEERARAKWDT#ANNPLYKEATSTFTNITYRG 787 |+|||++ |+ |++|+ ||++| || # ||+| |+| | | + | Sbjct 784 LIIWKVIQTLRDKREYEKFKKEEDLRKWTK#GENPVYVNASSKFDNPMFGG 833 |
| Pan troglodytes | Query 738 LLIWKLLITIHDRKEFAKFEEERARAKWDT#ANNPLYKEATSTFTN 782 |+||| || + | +|+ +||+|+ +++|+ # +|||+| ||+| | Sbjct 720 LVIWKALIHLSDLREYRRFEKEKLKSQWNN#-DNPLFKSATTTVMN 763 |
| Homo sapiens | Query 738 LLIWKLLITIHDRKEFAKFEEERARAKWDT#ANNPLYKEATSTFTN 782 |+||| || + | +|+ +||+|+ +++|+ # +|||+| ||+| | Sbjct 720 LVIWKALIHLSDLREYRRFEKEKLKSQWNN#-DNPLFKSATTTVMN 763 |
| Pacifastacus leniusculus | Query 746 TIHDRKEFAKFEEERARAKWDT#ANNPLYKEATSTFTNITY 785 |+|||+|+||||+|| #| ||||| | +|| | + Sbjct 758 TLHDRREYAKFEKERKLPSGKR#AENPLYKSAKTTFQNPAF 797 |
| Schistosoma japonicum | Query 741 WKLLITIHDRKEFAKFEEERARAKWDT#ANNPLYKEATSTFTNITY 785 +||+||| ||+| | |+++ +|+ #| ||+++ |+ | |+ Sbjct 239 YKLVITIDDRRELANFKQQGENMRWEM#AENPIFESPTTNVLNPTF 283 |
| Ovis canadensis | Query 740 IWKLLITIHDRKEFAKFEEERARAKWDT#ANNPLYKEATSTFTN 782 ||| | + | +|+ +||+|+ +++|+ # +|||+| ||+| | Sbjct 723 IWKALTHLSDLREYHRFEKEKLKSQWNN#-DNPLFKSATTTVMN 764 |
| Sigmodon hispidus | Query 740 IWKLLITIHDRKEFAKFEEERARAKWDT#ANNPLYKEATSTFTN 782 ||| | + | +|+ ||+|+ +++|+ # +|||+| ||+| | Sbjct 722 IWKALTHLTDLREYRHFEKEKLKSQWNN#-DNPLFKSATTTVMN 763 |
[Site 3] KWDTANNPLY773-KEATSTFTNI
Tyr773  Lys
Lys
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Lys764 | Trp765 | Asp766 | Thr767 | Ala768 | Asn769 | Asn770 | Pro771 | Leu772 | Tyr773 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Lys774 | Glu775 | Ala776 | Thr777 | Ser778 | Thr779 | Phe780 | Thr781 | Asn782 | Ile783 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| LITIHDRKEFAKFEEERARAKWDTANNPLYKEATSTFTNITYRGT |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | N/A | 101.00 | 28 | glycoprotein IIIa precursor |
| 2 | Mus musculus | 101.00 | 12 | ITB3_MOUSE Integrin beta-3 precursor (Platelet me |
| 3 | Homo sapiens | 101.00 | 11 | glycoprotein IIIa |
| 4 | Canis familiaris | 101.00 | 10 | platelet glycoprotein IIIa |
| 5 | Bos taurus | 101.00 | 7 | PREDICTED: similar to platelet glycoprotein IIIa |
| 6 | Rattus norvegicus | 101.00 | 4 | integrin beta 3 |
| 7 | synthetic construct | 101.00 | 3 | AF239959_1 hybrid integrin beta 3 subunit precurs |
| 8 | Sus scrofa | 101.00 | 3 | AF170527_1 glycoprotein IIIa; GPIIIa |
| 9 | Mus musculus | 101.00 | 1 | integrin beta 3 precursor |
| 10 | Canis lupus familiaris | 101.00 | 1 | integrin beta chain, beta 3 |
| 11 | Sus scrofa | 101.00 | 1 | integrin beta chain, beta 3 |
| 12 | Oryctolagus cuniculus | 101.00 | 1 | glycoprotein IIIa |
| 13 | rats, Peptide Partial, 723 aa | 101.00 | 1 | beta 3 integrin, GPIIIA |
| 14 | Gallus gallus | 94.40 | 2 | integrin, beta 3 precursor |
| 15 | Xenopus laevis | 92.00 | 2 | integrin beta-3 subunit |
| 16 | mice, Peptide Partial, 680 aa | 92.00 | 1 | beta 3 integrin, GPIIIA |
| 17 | Tetraodon nigroviridis | 90.10 | 5 | unnamed protein product |
| 18 | Danio rerio | 90.10 | 5 | integrin beta 3b |
| 19 | Anopheles gambiae str. PEST | 72.00 | 1 | integrin beta subunit (AGAP000815-PA) |
| 20 | Anopheles gambiae | 72.00 | 1 | integrin beta subunit |
| 21 | Drosophila melanogaster | 71.60 | 1 | myospheroid CG1560-PA |
| 22 | Pseudoplusia includens | 70.50 | 1 | integrin beta 1 |
| 23 | Ovis aries | 69.70 | 2 | ITB6_SHEEP Integrin beta-6 precursor gi |
| 24 | Biomphalaria glabrata | 67.80 | 1 | beta integrin subunit |
| 25 | Gallus gallus | 63.50 | 3 | integrin, beta 1 (fibronectin receptor, beta poly |
| 26 | Pongo pygmaeus | 63.50 | 1 | ITB1_PONPY Integrin beta-1 precursor (Fibronectin |
| 27 | Ictalurus punctatus | 63.20 | 2 | beta-1 integrin |
| 28 | Crassostrea gigas | 62.80 | 1 | integrin beta cgh |
| 29 | Podocoryne carnea | 62.40 | 1 | AF308652_1 integrin beta chain |
| 30 | Caenorhabditis briggsae | 61.20 | 1 | Hypothetical protein CBG03601 |
| 31 | Oryctolagus cuniculus | 60.80 | 1 | integrin beta 1 |
| 32 | Felis catus | 60.50 | 1 | integrin beta 1 |
| 33 | Caenorhabditis elegans | 59.30 | 1 | Paralysed Arrest at Two-fold family member (pat-3 |
| 34 | Strongylocentrotus purpuratus | 58.20 | 3 | integrin beta-C subunit |
| 35 | Lytechinus variegatus | 58.20 | 1 | beta-C integrin subunit |
| 36 | Acropora millepora | 53.10 | 1 | integrin subunit betaCn1 |
| 37 | Mus sp. | 50.40 | 1 | beta 7 integrin |
| 38 | Xenopus laevis | 49.70 | 1 | hypothetical protein LOC379708 |
| 39 | Pacifastacus leniusculus | 47.80 | 1 | integrin |
| 40 | Pan troglodytes | 44.70 | 2 | PREDICTED: integrin, beta 7 isoform 5 |
| 41 | Schistosoma japonicum | 42.00 | 1 | SJCHGC06221 protein |
| 42 | Homo sapiens | 41.60 | 1 | integrin, beta 2 precursor |
| 43 | Halocynthia roretzi | 41.20 | 1 | integrin beta Hr2 precursor |
| 44 | Ovis canadensis | 39.30 | 1 | ITB2_OVICA Integrin beta-2 precursor (Cell surfac |
| 45 | Macaca mulatta | 38.50 | 1 | integrin beta 1 |
Top-ranked sequences
| organism | matching |
|---|---|
| N/A | Query 744 LITIHDRKEFAKFEEERARAKWDTANNPLY#KEATSTFTNITYRGT 788 ||||||||||||||||||||||||||||||#||||||||||||||| Query 744 LITIHDRKEFAKFEEERARAKWDTANNPLY#KEATSTFTNITYRGT 788 |
| Mus musculus | Query 744 LITIHDRKEFAKFEEERARAKWDTANNPLY#KEATSTFTNITYRGT 788 ||||||||||||||||||||||||||||||#||||||||||||||| Query 744 LITIHDRKEFAKFEEERARAKWDTANNPLY#KEATSTFTNITYRGT 788 |
| Homo sapiens | Query 744 LITIHDRKEFAKFEEERARAKWDTANNPLY#KEATSTFTNITYRGT 788 ||||||||||||||||||||||||||||||#||||||||||||||| Query 744 LITIHDRKEFAKFEEERARAKWDTANNPLY#KEATSTFTNITYRGT 788 |
| Canis familiaris | Query 744 LITIHDRKEFAKFEEERARAKWDTANNPLY#KEATSTFTNITYRGT 788 ||||||||||||||||||||||||||||||#||||||||||||||| Query 744 LITIHDRKEFAKFEEERARAKWDTANNPLY#KEATSTFTNITYRGT 788 |
| Bos taurus | Query 744 LITIHDRKEFAKFEEERARAKWDTANNPLY#KEATSTFTNITYRGT 788 ||||||||||||||||||||||||||||||#||||||||||||||| Query 744 LITIHDRKEFAKFEEERARAKWDTANNPLY#KEATSTFTNITYRGT 788 |
| Rattus norvegicus | Query 744 LITIHDRKEFAKFEEERARAKWDTANNPLY#KEATSTFTNITYRGT 788 ||||||||||||||||||||||||||||||#||||||||||||||| Query 744 LITIHDRKEFAKFEEERARAKWDTANNPLY#KEATSTFTNITYRGT 788 |
| synthetic construct | Query 744 LITIHDRKEFAKFEEERARAKWDTANNPLY#KEATSTFTNITYRGT 788 ||||||||||||||||||||||||||||||#||||||||||||||| Query 744 LITIHDRKEFAKFEEERARAKWDTANNPLY#KEATSTFTNITYRGT 788 |
| Sus scrofa | Query 744 LITIHDRKEFAKFEEERARAKWDTANNPLY#KEATSTFTNITYRGT 788 ||||||||||||||||||||||||||||||#||||||||||||||| Query 744 LITIHDRKEFAKFEEERARAKWDTANNPLY#KEATSTFTNITYRGT 788 |
| Mus musculus | Query 744 LITIHDRKEFAKFEEERARAKWDTANNPLY#KEATSTFTNITYRGT 788 ||||||||||||||||||||||||||||||#||||||||||||||| Query 744 LITIHDRKEFAKFEEERARAKWDTANNPLY#KEATSTFTNITYRGT 788 |
| Canis lupus familiaris | Query 744 LITIHDRKEFAKFEEERARAKWDTANNPLY#KEATSTFTNITYRGT 788 ||||||||||||||||||||||||||||||#||||||||||||||| Query 744 LITIHDRKEFAKFEEERARAKWDTANNPLY#KEATSTFTNITYRGT 788 |
| Sus scrofa | Query 744 LITIHDRKEFAKFEEERARAKWDTANNPLY#KEATSTFTNITYRGT 788 ||||||||||||||||||||||||||||||#||||||||||||||| Query 744 LITIHDRKEFAKFEEERARAKWDTANNPLY#KEATSTFTNITYRGT 788 |
| Oryctolagus cuniculus | Query 744 LITIHDRKEFAKFEEERARAKWDTANNPLY#KEATSTFTNITYRGT 788 ||||||||||||||||||||||||||||||#||||||||||||||| Query 744 LITIHDRKEFAKFEEERARAKWDTANNPLY#KEATSTFTNITYRGT 788 |
| rats, Peptide Partial, 723 aa | Query 744 LITIHDRKEFAKFEEERARAKWDTANNPLY#KEATSTFTNITYRGT 788 ||||||||||||||||||||||||||||||#||||||||||||||| Query 744 LITIHDRKEFAKFEEERARAKWDTANNPLY#KEATSTFTNITYRGT 788 |
| Gallus gallus | Query 744 LITIHDRKEFAKFEEERARAKWDTANNPLY#KEATSTFTNITYRG 787 |||||||+|||+||||+||||||| |||||#|||||||||||||| Sbjct 736 LITIHDRREFARFEEEKARAKWDTGNNPLY#KEATSTFTNITYRG 779 |
| Xenopus laevis | Query 744 LITIHDRKEFAKFEEERARAKWDTANNPLY#KEATSTFTNITYRG 787 |||||||+||||||||||+||||||+||||#| |||||||||||| Sbjct 742 LITIHDRREFAKFEEERAKAKWDTAHNPLY#KGATSTFTNITYRG 785 |
| mice, Peptide Partial, 680 aa | Query 744 LITIHDRKEFAKFEEERARAKWDTANNPLY#KEATSTFTNIT 784 ||||||||||||||||||||||||||||||#||||||||||| Query 744 LITIHDRKEFAKFEEERARAKWDTANNPLY#KEATSTFTNIT 784 |
| Tetraodon nigroviridis | Query 744 LITIHDRKEFAKFEEERARAKWDTANNPLY#KEATSTFTNITYRG 787 |||||||+|||||||||||||||| +||||#| |||||||||+|| Sbjct 759 LITIHDRREFAKFEEERARAKWDTGHNPLY#KGATSTFTNITFRG 802 |
| Danio rerio | Query 744 LITIHDRKEFAKFEEERARAKWDTANNPLY#KEATSTFTNITYRG 787 |||||||+||||||||||||||+| +||||#| |||||||||||| Sbjct 745 LITIHDRREFAKFEEERARAKWETGHNPLY#KGATSTFTNITYRG 788 |
| Anopheles gambiae str. PEST | Query 744 LITIHDRKEFAKFEEERARAKWDTANNPLY#KEATSTFTNITYRG 787 | +||||+|||+||+|| ||||| ||+|#|+||+|| | || | Sbjct 793 LTSIHDRREFARFEKERMMAKWDTGENPIY#KQATTTFKNPTYAG 836 |
| Anopheles gambiae | Query 744 LITIHDRKEFAKFEEERARAKWDTANNPLY#KEATSTFTNITYRG 787 | +||||+|||+||+|| ||||| ||+|#|+||+|| | || | Sbjct 793 LTSIHDRREFARFEKERMMAKWDTGENPIY#KQATTTFKNPTYAG 836 |
| Drosophila melanogaster | Query 746 TIHDRKEFAKFEEERARAKWDTANNPLY#KEATSTFTNITYRG 787 |||||+|||+||+|| ||||| ||+|#|+||||| | | | Sbjct 804 TIHDRREFARFEKERMNAKWDTGENPIY#KQATSTFKNPMYAG 845 |
| Pseudoplusia includens | Query 746 TIHDRKEFAKFEEERARAKWDTANNPLY#KEATSTFTNITYRG 787 | |||+|||+||+|| ||||| ||+|#|+||||| | || | Sbjct 795 TSHDRREFARFEKERMMAKWDTGENPIY#KQATSTFKNPTYAG 836 |
| Ovis aries | Query 744 LITIHDRKEFAKFEEERARAKWDTANNPLY#KEATSTFTNITYR 786 |++ ||||| |||| ||++||| | ||||#+ +|||| |+||+ Sbjct 732 LVSFHDRKEVAKFEAERSKAKWQTGTNPLY#RGSTSTFKNVTYK 774 |
| Biomphalaria glabrata | Query 747 IHDRKEFAKFEEERARAKWDTANNPLY#KEATSTFTNITY 785 ||| +||||||+|| ||||| ||+|#|+||||| | || Sbjct 747 IHDTREFAKFEKERQNAKWDTGENPIY#KQATSTFKNPTY 785 |
| Gallus gallus | Query 745 ITIHDRKEFAKFEEERARAKWDTANNPLY#KEATSTFTNITYRG 787 + ||||+||||||+|+ ||||| ||+|#| | +| | | | Sbjct 760 MIIHDRREFAKFEKEKMNAKWDTGENPIY#KSAVTTVVNPKYEG 802 |
| Pongo pygmaeus | Query 745 ITIHDRKEFAKFEEERARAKWDTANNPLY#KEATSTFTNITYRG 787 + ||||+||||||+|+ ||||| ||+|#| | +| | | | Sbjct 755 MIIHDRREFAKFEKEKMNAKWDTGENPIY#KSAVTTVVNPKYEG 797 |
| Ictalurus punctatus | Query 744 LITIHDRKEFAKFEEERARAKWDTANNPLY#KEATSTFTNITYRG 787 |+ ||||+||||||+|+ |||| ||+|#| | +| | | | Sbjct 763 LMIIHDRREFAKFEKEKMNAKWDAGENPIY#KSAVTTVVNPKYEG 806 |
| Crassostrea gigas | Query 746 TIHDRKEFAKFEEERARAKWDTANNPLY#KEATSTFTNITYR 786 || |++| |+||+| |+||| ||+|#|+||||| | ||| Sbjct 758 TISDKRELARFEKEAMNARWDTGENPIY#KQATSTFVNPTYR 798 |
| Podocoryne carnea | Query 746 TIHDRKEFAKFEEERARAKWDTANNPLY#KEATSTFTNITY 785 || ||+||||||++| |||+ ||+|#|+||||| | | Sbjct 739 TIQDRREFAKFEKDRQNPKWDSGENPIY#KKATSTFQNPMY 778 |
| Caenorhabditis briggsae | Query 748 HDRKEFAKFEEERARAKWDTANNPLY#KEATSTFTNITYRG 787 ||| |+||| || ||||| ||+|#|+||+|| | | | Sbjct 768 HDRAEYAKFNNERLMAKWDTNENPIY#KQATTTFKNPVYAG 807 |
| Oryctolagus cuniculus | Query 745 ITIHDRKEFAKFEEERARAKWDTANNPLY#KEATSTFTNITYRG 787 + ||||+||||||+|+ ||||| ||+|#| | +| | | | Sbjct 20 MIIHDRREFAKFEKEKMNAKWDTGENPIY#KSAVTTVVNPKYEG 62 |
| Felis catus | Query 745 ITIHDRKEFAKFEEERARAKWDTANNPLY#KEATSTFTNITYRG 787 + ||| +||||||+|+ ||||| ||+|#| | +| | | | Sbjct 755 MIIHDTREFAKFEKEKMNAKWDTGENPIY#KSAVTTVVNPKYEG 797 |
| Caenorhabditis elegans | Query 748 HDRKEFAKFEEERARAKWDTANNPLY#KEATSTFTNITYRG 787 ||| |+| | || ||||| ||+|#|+||+|| | | | Sbjct 767 HDRSEYATFNNERLMAKWDTNENPIY#KQATTTFKNPVYAG 806 |
| Strongylocentrotus purpuratus | Query 747 IHDRKEFAKFEEERARAKWDTANNPLY#KEATSTFTNITY 785 ||||+|| ||+||| | |+ ||+|#| +|| | | || Sbjct 765 IHDRREFQNFEKERANATWEGGENPIY#KPSTSVFKNPTY 803 |
| Lytechinus variegatus | Query 747 IHDRKEFAKFEEERARAKWDTANNPLY#KEATSTFTNITY 785 ||||+|| ||+||| | |+ ||+|#| +|| | | || Sbjct 765 IHDRREFQNFEKERANATWEGGENPIY#KPSTSVFKNPTY 803 |
| Acropora millepora | Query 744 LITIHDRKEFAKFEEERARAKWDTANNPLY#KEATSTFTNITYRG 787 | |+ || |+ ||| || +|| ||||#+ | +|| | || | Sbjct 746 LFTMVDRIEYQKFERERMHSKWTREKNPLY#QAAKTTFENPTYAG 789 |
| Mus sp. | Query 745 ITIHDRKEFAKFEEERARAKWDTANNPLY#KEATSTFTNITYRGT 788 + |+||+|+ +||+|+ + | |||||#| | +| | ++|| Sbjct 749 VEIYDRREYRRFEKEQQQLNWKQDNNPLY#KSAITTTVNPRFQGT 792 |
| Xenopus laevis | Query 747 IHDRKEFAKFEEERARAKWDTANNPLY#KEATSTFTNITYRG 787 | || |+ +||+|| +||+ +||||# ||+| | + | Sbjct 731 IKDRNEYQRFEKERMNSKWNPGHNPLY#HNATTTVQNPNFSG 771 |
| Pacifastacus leniusculus | Query 748 HDRKEFAKFEEERARAKWDTANNPLY#KEATSTFTNITY 785 |||+|+||||+|| | ||||#| | +|| | + Sbjct 760 HDRREYAKFEKERKLPSGKRAENPLY#KSAKTTFQNPAF 797 |
| Pan troglodytes | Query 745 ITIHDRKEFAKFEEERARAKWDTANNPLY#KEATSTFTNITYR 786 + |+||+|+++||+|+ + | +||||#| | +| | ++ Sbjct 750 VEIYDRREYSRFEKEQQQLNWKQDSNPLY#KSAITTTINPRFQ 791 |
| Schistosoma japonicum | Query 744 LITIHDRKEFAKFEEERARAKWDTANNPLY#KEATSTFTNITY 785 +||| ||+| | |+++ +|+ | ||++#+ |+ | |+ Sbjct 242 VITIDDRRELANFKQQGENMRWEMAENPIF#ESPTTNVLNPTF 283 |
| Homo sapiens | Query 744 LITIHDRKEFAKFEEERARAKWDTANNPLY#KEATSTFTN 782 || + | +|+ +||+|+ +++|+ +|||+#| ||+| | Sbjct 726 LIHLSDLREYRRFEKEKLKSQWNN-DNPLF#KSATTTVMN 763 |
| Halocynthia roretzi | Query 746 TIHDRKEFAKFEEERARAKWDTANNPLY#KEATSTFTNITYRG 787 |+ |++|+ ||++| || ||+|# |+| | | + | Sbjct 792 TLRDKREYEKFKKEEDLRKWTKGENPVY#VNASSKFDNPMFGG 833 |
| Ovis canadensis | Query 744 LITIHDRKEFAKFEEERARAKWDTANNPLY#KEATSTFTN 782 | + | +|+ +||+|+ +++|+ +|||+#| ||+| | Sbjct 727 LTHLSDLREYHRFEKEKLKSQWNN-DNPLF#KSATTTVMN 764 |
| Macaca mulatta | Query 745 ITIHDRKEFAKFEEERARAKW 765 + ||||+||||||+|+ ||| Sbjct 144 MIIHDRREFAKFEKEKMNAKW 164 |
[Site 4] PLYKEATSTF780-TNITYRGT
Phe780  Thr
Thr
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Pro771 | Leu772 | Tyr773 | Lys774 | Glu775 | Ala776 | Thr777 | Ser778 | Thr779 | Phe780 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Thr781 | Asn782 | Ile783 | Thr784 | Tyr785 | Arg786 | Gly787 | Thr788 | - | - |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| KEFAKFEEERARAKWDTANNPLYKEATSTFTNITYRGT |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | N/A | 86.70 | 27 | glycoprotein IIIa precursor |
| 2 | Mus musculus | 86.70 | 12 | ITB3_MOUSE Integrin beta-3 precursor (Platelet me |
| 3 | Homo sapiens | 86.70 | 11 | glycoprotein IIIa |
| 4 | Canis familiaris | 86.70 | 10 | platelet glycoprotein IIIa |
| 5 | Bos taurus | 86.70 | 7 | PREDICTED: similar to platelet glycoprotein IIIa |
| 6 | Rattus norvegicus | 86.70 | 5 | integrin beta 3 |
| 7 | Sus scrofa | 86.70 | 3 | AF170527_1 glycoprotein IIIa; GPIIIa |
| 8 | rats, Peptide Partial, 723 aa | 86.70 | 1 | beta 3 integrin, GPIIIA |
| 9 | Mus musculus | 86.70 | 1 | integrin beta 3 precursor |
| 10 | Sus scrofa | 86.70 | 1 | integrin beta chain, beta 3 |
| 11 | Oryctolagus cuniculus | 86.70 | 1 | glycoprotein IIIa |
| 12 | Canis lupus familiaris | 86.70 | 1 | integrin beta chain, beta 3 |
| 13 | synthetic construct | 86.30 | 3 | AF239959_1 hybrid integrin beta 3 subunit precurs |
| 14 | Gallus gallus | 79.70 | 2 | integrin, beta 3 precursor |
| 15 | mice, Peptide Partial, 680 aa | 77.40 | 1 | beta 3 integrin, GPIIIA |
| 16 | Xenopus laevis | 77.00 | 2 | integrin beta-3 subunit |
| 17 | Tetraodon nigroviridis | 76.60 | 5 | unnamed protein product |
| 18 | Danio rerio | 75.10 | 5 | integrin beta 3b |
| 19 | Pseudoplusia includens | 61.60 | 1 | integrin beta 1 |
| 20 | Anopheles gambiae | 59.70 | 1 | integrin beta subunit |
| 21 | Anopheles gambiae str. PEST | 59.30 | 1 | integrin beta subunit (AGAP000815-PA) |
| 22 | Biomphalaria glabrata | 58.90 | 1 | beta integrin subunit |
| 23 | Drosophila melanogaster | 58.90 | 1 | myospheroid CG1560-PA |
| 24 | Ovis aries | 58.50 | 2 | ITB6_SHEEP Integrin beta-6 precursor gi |
| 25 | Crassostrea gigas | 55.10 | 1 | integrin beta cgh |
| 26 | Felis catus | 53.10 | 1 | integrin beta 1 |
| 27 | Gallus gallus | 52.80 | 3 | integrin, beta 1 (fibronectin receptor, beta poly |
| 28 | Podocoryne carnea | 52.80 | 1 | AF308652_1 integrin beta chain |
| 29 | Pongo pygmaeus | 52.80 | 1 | ITB1_PONPY Integrin beta-1 precursor (Fibronectin |
| 30 | Caenorhabditis briggsae | 52.40 | 1 | Hypothetical protein CBG03601 |
| 31 | Ictalurus punctatus | 51.60 | 2 | beta-1 integrin |
| 32 | Oryctolagus cuniculus | 50.40 | 1 | integrin beta 1 |
| 33 | Caenorhabditis elegans | 50.40 | 1 | Paralysed Arrest at Two-fold family member (pat-3 |
| 34 | Strongylocentrotus purpuratus | 48.90 | 3 | integrin beta G subunit |
| 35 | Lytechinus variegatus | 48.50 | 1 | beta-C integrin subunit |
| 36 | Acropora millepora | 45.40 | 1 | integrin subunit betaCn1 |
| 37 | Xenopus laevis | 43.50 | 1 | hypothetical protein LOC379708 |
| 38 | Mus sp. | 42.70 | 1 | beta 7 integrin |
| 39 | Pacifastacus leniusculus | 40.00 | 1 | integrin |
| 40 | Pan troglodytes | 37.00 | 2 | PREDICTED: integrin, beta 7 isoform 5 |
| 41 | Ovis canadensis | 37.00 | 1 | ITB2_OVICA Integrin beta-2 precursor (Cell surfac |
| 42 | Homo sapiens | 37.00 | 1 | integrin, beta 2 precursor |
| 43 | Sigmodon hispidus | 35.80 | 1 | AF445415_1 integrin beta-2 precursor |
| 44 | Halocynthia roretzi | 35.00 | 1 | integrin beta Hr2 precursor |
| 45 | Suberites domuncula | 34.30 | 1 | integrin beta subunit |
| 46 | Cyprinus carpio | 33.90 | 2 | integrin beta2-chain |
| 47 | Geodia cydonium | 33.90 | 1 | integrin beta subunit |
Top-ranked sequences
| organism | matching |
|---|---|
| N/A | Query 751 KEFAKFEEERARAKWDTANNPLYKEATSTF#TNITYRGT 788 ||||||||||||||||||||||||||||||#|||||||| Query 751 KEFAKFEEERARAKWDTANNPLYKEATSTF#TNITYRGT 788 |
| Mus musculus | Query 751 KEFAKFEEERARAKWDTANNPLYKEATSTF#TNITYRGT 788 ||||||||||||||||||||||||||||||#|||||||| Query 751 KEFAKFEEERARAKWDTANNPLYKEATSTF#TNITYRGT 788 |
| Homo sapiens | Query 751 KEFAKFEEERARAKWDTANNPLYKEATSTF#TNITYRGT 788 ||||||||||||||||||||||||||||||#|||||||| Query 751 KEFAKFEEERARAKWDTANNPLYKEATSTF#TNITYRGT 788 |
| Canis familiaris | Query 751 KEFAKFEEERARAKWDTANNPLYKEATSTF#TNITYRGT 788 ||||||||||||||||||||||||||||||#|||||||| Query 751 KEFAKFEEERARAKWDTANNPLYKEATSTF#TNITYRGT 788 |
| Bos taurus | Query 751 KEFAKFEEERARAKWDTANNPLYKEATSTF#TNITYRGT 788 ||||||||||||||||||||||||||||||#|||||||| Query 751 KEFAKFEEERARAKWDTANNPLYKEATSTF#TNITYRGT 788 |
| Rattus norvegicus | Query 751 KEFAKFEEERARAKWDTANNPLYKEATSTF#TNITYRGT 788 ||||||||||||||||||||||||||||||#|||||||| Query 751 KEFAKFEEERARAKWDTANNPLYKEATSTF#TNITYRGT 788 |
| Sus scrofa | Query 751 KEFAKFEEERARAKWDTANNPLYKEATSTF#TNITYRGT 788 ||||||||||||||||||||||||||||||#|||||||| Query 751 KEFAKFEEERARAKWDTANNPLYKEATSTF#TNITYRGT 788 |
| rats, Peptide Partial, 723 aa | Query 751 KEFAKFEEERARAKWDTANNPLYKEATSTF#TNITYRGT 788 ||||||||||||||||||||||||||||||#|||||||| Query 751 KEFAKFEEERARAKWDTANNPLYKEATSTF#TNITYRGT 788 |
| Mus musculus | Query 751 KEFAKFEEERARAKWDTANNPLYKEATSTF#TNITYRGT 788 ||||||||||||||||||||||||||||||#|||||||| Query 751 KEFAKFEEERARAKWDTANNPLYKEATSTF#TNITYRGT 788 |
| Sus scrofa | Query 751 KEFAKFEEERARAKWDTANNPLYKEATSTF#TNITYRGT 788 ||||||||||||||||||||||||||||||#|||||||| Query 751 KEFAKFEEERARAKWDTANNPLYKEATSTF#TNITYRGT 788 |
| Oryctolagus cuniculus | Query 751 KEFAKFEEERARAKWDTANNPLYKEATSTF#TNITYRGT 788 ||||||||||||||||||||||||||||||#|||||||| Query 751 KEFAKFEEERARAKWDTANNPLYKEATSTF#TNITYRGT 788 |
| Canis lupus familiaris | Query 751 KEFAKFEEERARAKWDTANNPLYKEATSTF#TNITYRGT 788 ||||||||||||||||||||||||||||||#|||||||| Query 751 KEFAKFEEERARAKWDTANNPLYKEATSTF#TNITYRGT 788 |
| synthetic construct | Query 751 KEFAKFEEERARAKWDTANNPLYKEATSTF#TNITYRGT 788 ||||||||||||||||||||||||||||||#|||||||| Query 751 KEFAKFEEERARAKWDTANNPLYKEATSTF#TNITYRGT 788 |
| Gallus gallus | Query 751 KEFAKFEEERARAKWDTANNPLYKEATSTF#TNITYRG 787 +|||+||||+||||||| ||||||||||||#||||||| Sbjct 743 REFARFEEEKARAKWDTGNNPLYKEATSTF#TNITYRG 779 |
| mice, Peptide Partial, 680 aa | Query 751 KEFAKFEEERARAKWDTANNPLYKEATSTF#TNIT 784 ||||||||||||||||||||||||||||||#|||| Query 751 KEFAKFEEERARAKWDTANNPLYKEATSTF#TNIT 784 |
| Xenopus laevis | Query 751 KEFAKFEEERARAKWDTANNPLYKEATSTF#TNITYRG 787 +||||||||||+||||||+||||| |||||#||||||| Sbjct 749 REFAKFEEERAKAKWDTAHNPLYKGATSTF#TNITYRG 785 |
| Tetraodon nigroviridis | Query 751 KEFAKFEEERARAKWDTANNPLYKEATSTF#TNITYRG 787 ||||||||||||||||| |||||| |||||#||| |+| Sbjct 715 KEFAKFEEERARAKWDTGNNPLYKGATSTF#TNIAYKG 751 |
| Danio rerio | Query 751 KEFAKFEEERARAKWDTANNPLYKEATSTF#TNITYRG 787 +||||||||||||||+| +||||| |||||#||||||| Sbjct 752 REFAKFEEERARAKWETGHNPLYKGATSTF#TNITYRG 788 |
| Pseudoplusia includens | Query 751 KEFAKFEEERARAKWDTANNPLYKEATSTF#TNITYRG 787 +|||+||+|| ||||| ||+||+|||||# | || | Sbjct 800 REFARFEKERMMAKWDTGENPIYKQATSTF#KNPTYAG 836 |
| Anopheles gambiae | Query 751 KEFAKFEEERARAKWDTANNPLYKEATSTF#TNITYRG 787 +|||+||+|| ||||| ||+||+||+||# | || | Sbjct 800 REFARFEKERMMAKWDTGENPIYKQATTTF#KNPTYAG 836 |
| Anopheles gambiae str. PEST | Query 751 KEFAKFEEERARAKWDTANNPLYKEATSTF#TNITYRG 787 +|||+||+|| ||||| ||+||+||+||# | || | Sbjct 800 REFARFEKERMMAKWDTGENPIYKQATTTF#KNPTYAG 836 |
| Biomphalaria glabrata | Query 751 KEFAKFEEERARAKWDTANNPLYKEATSTF#TNITY 785 +||||||+|| ||||| ||+||+|||||# | || Sbjct 751 REFAKFEKERQNAKWDTGENPIYKQATSTF#KNPTY 785 |
| Drosophila melanogaster | Query 751 KEFAKFEEERARAKWDTANNPLYKEATSTF#TNITYRG 787 +|||+||+|| ||||| ||+||+|||||# | | | Sbjct 809 REFARFEKERMNAKWDTGENPIYKQATSTF#KNPMYAG 845 |
| Ovis aries | Query 751 KEFAKFEEERARAKWDTANNPLYKEATSTF#TNITYR 786 || |||| ||++||| | ||||+ +||||# |+||+ Sbjct 739 KEVAKFEAERSKAKWQTGTNPLYRGSTSTF#KNVTYK 774 |
| Crassostrea gigas | Query 751 KEFAKFEEERARAKWDTANNPLYKEATSTF#TNITYR 786 +| |+||+| |+||| ||+||+|||||# | ||| Sbjct 763 RELARFEKEAMNARWDTGENPIYKQATSTF#VNPTYR 798 |
| Felis catus | Query 751 KEFAKFEEERARAKWDTANNPLYKEATSTF#TNITYRG 787 +||||||+|+ ||||| ||+|| | +| # | | | Sbjct 761 REFAKFEKEKMNAKWDTGENPIYKSAVTTV#VNPKYEG 797 |
| Gallus gallus | Query 751 KEFAKFEEERARAKWDTANNPLYKEATSTF#TNITYRG 787 +||||||+|+ ||||| ||+|| | +| # | | | Sbjct 766 REFAKFEKEKMNAKWDTGENPIYKSAVTTV#VNPKYEG 802 |
| Podocoryne carnea | Query 751 KEFAKFEEERARAKWDTANNPLYKEATSTF#TNITY 785 +||||||++| |||+ ||+||+|||||# | | Sbjct 744 REFAKFEKDRQNPKWDSGENPIYKKATSTF#QNPMY 778 |
| Pongo pygmaeus | Query 751 KEFAKFEEERARAKWDTANNPLYKEATSTF#TNITYRG 787 +||||||+|+ ||||| ||+|| | +| # | | | Sbjct 761 REFAKFEKEKMNAKWDTGENPIYKSAVTTV#VNPKYEG 797 |
| Caenorhabditis briggsae | Query 752 EFAKFEEERARAKWDTANNPLYKEATSTF#TNITYRG 787 |+||| || ||||| ||+||+||+||# | | | Sbjct 772 EYAKFNNERLMAKWDTNENPIYKQATTTF#KNPVYAG 807 |
| Ictalurus punctatus | Query 751 KEFAKFEEERARAKWDTANNPLYKEATSTF#TNITYRG 787 +||||||+|+ |||| ||+|| | +| # | | | Sbjct 770 REFAKFEKEKMNAKWDAGENPIYKSAVTTV#VNPKYEG 806 |
| Oryctolagus cuniculus | Query 751 KEFAKFEEERARAKWDTANNPLYKEATSTF#TNITYRG 787 +||||||+|+ ||||| ||+|| | +| # | | | Sbjct 26 REFAKFEKEKMNAKWDTGENPIYKSAVTTV#VNPKYEG 62 |
| Caenorhabditis elegans | Query 752 EFAKFEEERARAKWDTANNPLYKEATSTF#TNITYRG 787 |+| | || ||||| ||+||+||+||# | | | Sbjct 771 EYATFNNERLMAKWDTNENPIYKQATTTF#KNPVYAG 806 |
| Strongylocentrotus purpuratus | Query 751 KEFAKFEEERARAKWDTANNPLYKEATSTF#TNITYR 786 +||| ||+||| | ||+|| +||||# | ||+ Sbjct 747 REFASFEKERAGTHWGQNENPIYKPSTSTF#KNPTYQ 782 |
| Lytechinus variegatus | Query 751 KEFAKFEEERARAKWDTANNPLYKEATSTF#TNITY 785 +|| ||+||| | |+ ||+|| +|| |# | || Sbjct 769 REFQNFEKERANATWEGGENPIYKPSTSVF#KNPTY 803 |
| Acropora millepora | Query 752 EFAKFEEERARAKWDTANNPLYKEATSTF#TNITYRG 787 |+ ||| || +|| ||||+ | +||# | || | Sbjct 754 EYQKFERERMHSKWTREKNPLYQAAKTTF#ENPTYAG 789 |
| Xenopus laevis | Query 752 EFAKFEEERARAKWDTANNPLYKEATSTF#TNITYRG 787 |+ +||+|| +||+ +|||| ||+| # | + | Sbjct 736 EYQRFEKERMNSKWNPGHNPLYHNATTTV#QNPNFSG 771 |
| Mus sp. | Query 751 KEFAKFEEERARAKWDTANNPLYKEATSTF#TNITYRGT 788 +|+ +||+|+ + | |||||| | +| # | ++|| Sbjct 755 REYRRFEKEQQQLNWKQDNNPLYKSAITTT#VNPRFQGT 792 |
| Pacifastacus leniusculus | Query 751 KEFAKFEEERARAKWDTANNPLYKEATSTF#TNITY 785 +|+||||+|| | ||||| | +||# | + Sbjct 763 REYAKFEKERKLPSGKRAENPLYKSAKTTF#QNPAF 797 |
| Pan troglodytes | Query 751 KEFAKFEEERARAKWDTANNPLYKEATSTF#TNITYR 786 +|+++||+|+ + | +||||| | +| # | ++ Sbjct 756 REYSRFEKEQQQLNWKQDSNPLYKSAITTT#INPRFQ 791 |
| Ovis canadensis | Query 751 KEFAKFEEERARAKWDTANNPLYKEATSTF#TN 782 +|+ +||+|+ +++|+ +|||+| ||+| # | Sbjct 734 REYHRFEKEKLKSQWNN-DNPLFKSATTTV#MN 764 |
| Homo sapiens | Query 751 KEFAKFEEERARAKWDTANNPLYKEATSTF#TN 782 +|+ +||+|+ +++|+ +|||+| ||+| # | Sbjct 733 REYRRFEKEKLKSQWNN-DNPLFKSATTTV#MN 763 |
| Sigmodon hispidus | Query 751 KEFAKFEEERARAKWDTANNPLYKEATSTF#TN 782 +|+ ||+|+ +++|+ +|||+| ||+| # | Sbjct 733 REYRHFEKEKLKSQWNN-DNPLFKSATTTV#MN 763 |
| Halocynthia roretzi | Query 751 KEFAKFEEERARAKWDTANNPLYKEATSTF#TNITYRG 787 +|+ ||++| || ||+| |+| |# | + | Sbjct 797 REYEKFKKEEDLRKWTKGENPVYVNASSKF#DNPMFGG 833 |
| Suberites domuncula | Query 752 EFAKFEEERARAKWDTANNPLYKEATSTF#TNITY 785 | |||+| |+ |+ ||||+ | +# | | Sbjct 729 EVKKFEKELAKTKYPKHQNPLYRSAAKDY#QNPIY 762 |
| Cyprinus carpio | Query 751 KEFAKFEEERARAKWDTANNPLYKEATSTF#TNITYRG 787 +|+ +||++| | + |||++ ||+| # | |+ | Sbjct 731 REWKRFEKDRKHEK-TSGTNPLFQNATTTI#QNPTFSG 766 |
| Geodia cydonium | Query 752 EFAKFEEERARAKWDTANNPLYKEATSTF#TNITY 785 | ||| | |+ |+ |||| ||+ # | | Sbjct 843 ELKKFENELAKVKYTKNENPLYHSATTEH#KNPIY 876 |
[Site 5] ATSTFTNITY785-RGT
Tyr785  Arg
Arg
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Ala776 | Thr777 | Ser778 | Thr779 | Phe780 | Thr781 | Asn782 | Ile783 | Thr784 | Tyr785 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Arg786 | Gly787 | Thr788 | - | - | - | - | - | - | - |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| FEEERARAKWDTANNPLYKEATSTFTNITYRGT |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | N/A | 76.60 | 27 | plate glycoprotein IIIa (GPIIIa) |
| 2 | Mus musculus | 76.60 | 12 | ITB3_MOUSE Integrin beta-3 precursor (Platelet me |
| 3 | Canis familiaris | 76.60 | 10 | platelet glycoprotein IIIa |
| 4 | Rattus norvegicus | 76.60 | 5 | integrin beta 3 |
| 5 | Sus scrofa | 76.60 | 3 | AF170527_1 glycoprotein IIIa; GPIIIa |
| 6 | Oryctolagus cuniculus | 76.60 | 1 | glycoprotein IIIa |
| 7 | rats, Peptide Partial, 723 aa | 76.60 | 1 | beta 3 integrin, GPIIIA |
| 8 | Sus scrofa | 76.60 | 1 | integrin beta chain, beta 3 |
| 9 | Canis lupus familiaris | 76.60 | 1 | integrin beta chain, beta 3 |
| 10 | Homo sapiens | 76.30 | 11 | glycoprotein IIIa |
| 11 | Bos taurus | 76.30 | 7 | PREDICTED: similar to platelet glycoprotein IIIa |
| 12 | Mus musculus | 76.30 | 1 | integrin beta 3 precursor |
| 13 | synthetic construct | 75.90 | 3 | AF239959_1 hybrid integrin beta 3 subunit precurs |
| 14 | Gallus gallus | 71.60 | 2 | integrin, beta 3 precursor |
| 15 | Xenopus laevis | 67.80 | 2 | integrin beta-3 subunit |
| 16 | mice, Peptide Partial, 680 aa | 67.00 | 1 | beta 3 integrin, GPIIIA |
| 17 | Tetraodon nigroviridis | 66.60 | 5 | unnamed protein product |
| 18 | Danio rerio | 66.20 | 5 | integrin beta 3b |
| 19 | Pseudoplusia includens | 53.90 | 1 | integrin beta 1 |
| 20 | Ovis aries | 51.60 | 2 | ITB6_SHEEP Integrin beta-6 precursor gi |
| 21 | Anopheles gambiae str. PEST | 51.20 | 1 | integrin beta subunit (AGAP000815-PA) |
| 22 | Anopheles gambiae | 51.20 | 1 | integrin beta subunit |
| 23 | Drosophila melanogaster | 51.20 | 1 | myospheroid CG1560-PA |
| 24 | Biomphalaria glabrata | 50.80 | 1 | beta integrin subunit |
| 25 | Crassostrea gigas | 49.30 | 1 | integrin beta cgh |
| 26 | Caenorhabditis briggsae | 45.80 | 1 | Hypothetical protein CBG03601 |
| 27 | Caenorhabditis elegans | 45.80 | 1 | Paralysed Arrest at Two-fold family member (pat-3 |
| 28 | Felis catus | 44.30 | 1 | integrin beta 1 |
| 29 | Gallus gallus | 43.90 | 3 | integrin, beta 1 (fibronectin receptor, beta poly |
| 30 | Pongo pygmaeus | 43.90 | 1 | ITB1_PONPY Integrin beta-1 precursor (Fibronectin |
| 31 | Podocoryne carnea | 43.90 | 1 | AF308652_1 integrin beta chain |
| 32 | Strongylocentrotus purpuratus | 43.50 | 3 | integrin beta-C subunit |
| 33 | Lytechinus variegatus | 43.10 | 1 | beta-C integrin subunit |
| 34 | Ictalurus punctatus | 42.70 | 2 | beta-1 integrin |
| 35 | Oryctolagus cuniculus | 42.40 | 1 | integrin beta 1 |
| 36 | Acropora millepora | 41.20 | 1 | integrin subunit betaCn1 |
| 37 | Xenopus laevis | 39.70 | 1 | hypothetical protein LOC379708 |
| 38 | Mus sp. | 38.10 | 1 | beta 7 integrin |
| 39 | Pan troglodytes | 32.30 | 2 | integrin, beta 2 |
| 40 | Ovis canadensis | 32.30 | 1 | ITB2_OVICA Integrin beta-2 precursor (Cell surfac |
| 41 | Pacifastacus leniusculus | 32.30 | 1 | integrin |
| 42 | Homo sapiens | 32.30 | 1 | integrin, beta 2 precursor |
| 43 | Sigmodon hispidus | 32.30 | 1 | AF445415_1 integrin beta-2 precursor |
| 44 | Suberites domuncula | 30.80 | 1 | integrin beta subunit |
| 45 | Ophlitaspongia tenuis | 30.40 | 1 | integrin subunit betaPo1 |
| 46 | Geodia cydonium | 30.00 | 1 | integrin beta subunit |
Top-ranked sequences
| organism | matching |
|---|---|
| N/A | Query 756 FEEERARAKWDTANNPLYKEATSTFTNITY#RGT 788 ||||||||||||||||||||||||||||||#||| Query 756 FEEERARAKWDTANNPLYKEATSTFTNITY#RGT 788 |
| Mus musculus | Query 756 FEEERARAKWDTANNPLYKEATSTFTNITY#RGT 788 ||||||||||||||||||||||||||||||#||| Query 756 FEEERARAKWDTANNPLYKEATSTFTNITY#RGT 788 |
| Canis familiaris | Query 756 FEEERARAKWDTANNPLYKEATSTFTNITY#RGT 788 ||||||||||||||||||||||||||||||#||| Query 756 FEEERARAKWDTANNPLYKEATSTFTNITY#RGT 788 |
| Rattus norvegicus | Query 756 FEEERARAKWDTANNPLYKEATSTFTNITY#RGT 788 ||||||||||||||||||||||||||||||#||| Query 756 FEEERARAKWDTANNPLYKEATSTFTNITY#RGT 788 |
| Sus scrofa | Query 756 FEEERARAKWDTANNPLYKEATSTFTNITY#RGT 788 ||||||||||||||||||||||||||||||#||| Query 756 FEEERARAKWDTANNPLYKEATSTFTNITY#RGT 788 |
| Oryctolagus cuniculus | Query 756 FEEERARAKWDTANNPLYKEATSTFTNITY#RGT 788 ||||||||||||||||||||||||||||||#||| Query 756 FEEERARAKWDTANNPLYKEATSTFTNITY#RGT 788 |
| rats, Peptide Partial, 723 aa | Query 756 FEEERARAKWDTANNPLYKEATSTFTNITY#RGT 788 ||||||||||||||||||||||||||||||#||| Query 756 FEEERARAKWDTANNPLYKEATSTFTNITY#RGT 788 |
| Sus scrofa | Query 756 FEEERARAKWDTANNPLYKEATSTFTNITY#RGT 788 ||||||||||||||||||||||||||||||#||| Query 756 FEEERARAKWDTANNPLYKEATSTFTNITY#RGT 788 |
| Canis lupus familiaris | Query 756 FEEERARAKWDTANNPLYKEATSTFTNITY#RGT 788 ||||||||||||||||||||||||||||||#||| Query 756 FEEERARAKWDTANNPLYKEATSTFTNITY#RGT 788 |
| Homo sapiens | Query 756 FEEERARAKWDTANNPLYKEATSTFTNITY#RGT 788 ||||||||||||||||||||||||||||||#||| Query 756 FEEERARAKWDTANNPLYKEATSTFTNITY#RGT 788 |
| Bos taurus | Query 756 FEEERARAKWDTANNPLYKEATSTFTNITY#RGT 788 ||||||||||||||||||||||||||||||#||| Query 756 FEEERARAKWDTANNPLYKEATSTFTNITY#RGT 788 |
| Mus musculus | Query 756 FEEERARAKWDTANNPLYKEATSTFTNITY#RGT 788 ||||||||||||||||||||||||||||||#||| Query 756 FEEERARAKWDTANNPLYKEATSTFTNITY#RGT 788 |
| synthetic construct | Query 756 FEEERARAKWDTANNPLYKEATSTFTNITY#RGT 788 ||||||||||||||||||||||||||||||#||| Query 756 FEEERARAKWDTANNPLYKEATSTFTNITY#RGT 788 |
| Gallus gallus | Query 756 FEEERARAKWDTANNPLYKEATSTFTNITY#RG 787 ||||+||||||| |||||||||||||||||#|| Sbjct 748 FEEEKARAKWDTGNNPLYKEATSTFTNITY#RG 779 |
| Xenopus laevis | Query 756 FEEERARAKWDTANNPLYKEATSTFTNITY#RG 787 ||||||+||||||+||||| ||||||||||#|| Sbjct 754 FEEERAKAKWDTAHNPLYKGATSTFTNITY#RG 785 |
| mice, Peptide Partial, 680 aa | Query 756 FEEERARAKWDTANNPLYKEATSTFTNIT 784 ||||||||||||||||||||||||||||| Query 756 FEEERARAKWDTANNPLYKEATSTFTNIT 784 |
| Tetraodon nigroviridis | Query 756 FEEERARAKWDTANNPLYKEATSTFTNITY#RG 787 |||||||||||| +||||| |||||||||+#|| Sbjct 771 FEEERARAKWDTGHNPLYKGATSTFTNITF#RG 802 |
| Danio rerio | Query 756 FEEERARAKWDTANNPLYKEATSTFTNITY#RG 787 ||||||||||+| +||||| ||||||||||#|| Sbjct 757 FEEERARAKWETGHNPLYKGATSTFTNITY#RG 788 |
| Pseudoplusia includens | Query 756 FEEERARAKWDTANNPLYKEATSTFTNITY#RG 787 ||+|| ||||| ||+||+||||| | ||# | Sbjct 805 FEKERMMAKWDTGENPIYKQATSTFKNPTY#AG 836 |
| Ovis aries | Query 756 FEEERARAKWDTANNPLYKEATSTFTNITY#R 786 || ||++||| | ||||+ +|||| |+||#+ Sbjct 744 FEAERSKAKWQTGTNPLYRGSTSTFKNVTY#K 774 |
| Anopheles gambiae str. PEST | Query 756 FEEERARAKWDTANNPLYKEATSTFTNITY#RG 787 ||+|| ||||| ||+||+||+|| | ||# | Sbjct 805 FEKERMMAKWDTGENPIYKQATTTFKNPTY#AG 836 |
| Anopheles gambiae | Query 756 FEEERARAKWDTANNPLYKEATSTFTNITY#RG 787 ||+|| ||||| ||+||+||+|| | ||# | Sbjct 805 FEKERMMAKWDTGENPIYKQATTTFKNPTY#AG 836 |
| Drosophila melanogaster | Query 756 FEEERARAKWDTANNPLYKEATSTFTNITY#RG 787 ||+|| ||||| ||+||+||||| | |# | Sbjct 814 FEKERMNAKWDTGENPIYKQATSTFKNPMY#AG 845 |
| Biomphalaria glabrata | Query 756 FEEERARAKWDTANNPLYKEATSTFTNITY# 785 ||+|| ||||| ||+||+||||| | ||# Sbjct 756 FEKERQNAKWDTGENPIYKQATSTFKNPTY# 785 |
| Crassostrea gigas | Query 756 FEEERARAKWDTANNPLYKEATSTFTNITY#R 786 ||+| |+||| ||+||+||||| | ||#| Sbjct 768 FEKEAMNARWDTGENPIYKQATSTFVNPTY#R 798 |
| Caenorhabditis briggsae | Query 756 FEEERARAKWDTANNPLYKEATSTFTNITY#RG 787 | || ||||| ||+||+||+|| | |# | Sbjct 776 FNNERLMAKWDTNENPIYKQATTTFKNPVY#AG 807 |
| Caenorhabditis elegans | Query 756 FEEERARAKWDTANNPLYKEATSTFTNITY#RG 787 | || ||||| ||+||+||+|| | |# | Sbjct 775 FNNERLMAKWDTNENPIYKQATTTFKNPVY#AG 806 |
| Felis catus | Query 756 FEEERARAKWDTANNPLYKEATSTFTNITY#RG 787 ||+|+ ||||| ||+|| | +| | |# | Sbjct 766 FEKEKMNAKWDTGENPIYKSAVTTVVNPKY#EG 797 |
| Gallus gallus | Query 756 FEEERARAKWDTANNPLYKEATSTFTNITY#RG 787 ||+|+ ||||| ||+|| | +| | |# | Sbjct 771 FEKEKMNAKWDTGENPIYKSAVTTVVNPKY#EG 802 |
| Pongo pygmaeus | Query 756 FEEERARAKWDTANNPLYKEATSTFTNITY#RG 787 ||+|+ ||||| ||+|| | +| | |# | Sbjct 766 FEKEKMNAKWDTGENPIYKSAVTTVVNPKY#EG 797 |
| Podocoryne carnea | Query 756 FEEERARAKWDTANNPLYKEATSTFTNITY# 785 ||++| |||+ ||+||+||||| | |# Sbjct 749 FEKDRQNPKWDSGENPIYKKATSTFQNPMY# 778 |
| Strongylocentrotus purpuratus | Query 756 FEEERARAKWDTANNPLYKEATSTFTNITY# 785 ||+||| | |+ ||+|| +|| | | ||# Sbjct 774 FEKERANATWEGGENPIYKPSTSVFKNPTY# 803 |
| Lytechinus variegatus | Query 756 FEEERARAKWDTANNPLYKEATSTFTNITY# 785 ||+||| | |+ ||+|| +|| | | ||# Sbjct 774 FEKERANATWEGGENPIYKPSTSVFKNPTY# 803 |
| Ictalurus punctatus | Query 756 FEEERARAKWDTANNPLYKEATSTFTNITY#RG 787 ||+|+ |||| ||+|| | +| | |# | Sbjct 775 FEKEKMNAKWDAGENPIYKSAVTTVVNPKY#EG 806 |
| Oryctolagus cuniculus | Query 756 FEEERARAKWDTANNPLYKEATSTFTNITY#RG 787 ||+|+ ||||| ||+|| | +| | |# | Sbjct 31 FEKEKMNAKWDTGENPIYKSAVTTVVNPKY#EG 62 |
| Acropora millepora | Query 756 FEEERARAKWDTANNPLYKEATSTFTNITY#RG 787 || || +|| ||||+ | +|| | ||# | Sbjct 758 FERERMHSKWTREKNPLYQAAKTTFENPTY#AG 789 |
| Xenopus laevis | Query 756 FEEERARAKWDTANNPLYKEATSTFTNITY#RG 787 ||+|| +||+ +|||| ||+| | +# | Sbjct 740 FEKERMNSKWNPGHNPLYHNATTTVQNPNF#SG 771 |
| Mus sp. | Query 756 FEEERARAKWDTANNPLYKEATSTFTNITY#RGT 788 ||+|+ + | |||||| | +| | +#+|| Sbjct 760 FEKEQQQLNWKQDNNPLYKSAITTTVNPRF#QGT 792 |
| Pan troglodytes | Query 756 FEEERARAKWDTANNPLYKEATSTFTN 782 ||+|+ +++|+ +|||+| ||+| | Sbjct 738 FEKEKLKSQWNN-DNPLFKSATTTVMN 763 |
| Ovis canadensis | Query 756 FEEERARAKWDTANNPLYKEATSTFTN 782 ||+|+ +++|+ +|||+| ||+| | Sbjct 739 FEKEKLKSQWNN-DNPLFKSATTTVMN 764 |
| Pacifastacus leniusculus | Query 756 FEEERARAKWDTANNPLYKEATSTFTNITY# 785 ||+|| | ||||| | +|| | +# Sbjct 768 FEKERKLPSGKRAENPLYKSAKTTFQNPAF# 797 |
| Homo sapiens | Query 756 FEEERARAKWDTANNPLYKEATSTFTN 782 ||+|+ +++|+ +|||+| ||+| | Sbjct 738 FEKEKLKSQWNN-DNPLFKSATTTVMN 763 |
| Sigmodon hispidus | Query 756 FEEERARAKWDTANNPLYKEATSTFTN 782 ||+|+ +++|+ +|||+| ||+| | Sbjct 738 FEKEKLKSQWNN-DNPLFKSATTTVMN 763 |
| Suberites domuncula | Query 756 FEEERARAKWDTANNPLYKEATSTFTNITY# 785 ||+| |+ |+ ||||+ | + | |# Sbjct 733 FEKELAKTKYPKHQNPLYRSAAKDYQNPIY# 762 |
| Ophlitaspongia tenuis | Query 756 FEEERARAKWDTANNPLYKEATSTFTNITY# 785 || | ||+ ||||+ || + | |# Sbjct 807 FEREIKNAKYTKNENPLYRSATKDYQNPLY# 836 |
| Geodia cydonium | Query 756 FEEERARAKWDTANNPLYKEATSTFTNITY# 785 || | |+ |+ |||| ||+ | |# Sbjct 847 FENELAKVKYTKNENPLYHSATTEHKNPIY# 876 |
* References
[PubMed ID: 24176823] Zhao F, Li L, Guan L, Yang H, Wu C, Liu Y, Roles for GP IIb/IIIa and alphavbeta3 integrins in MDA-MB-231 cell invasion and shear flow-induced cancer cell mechanotransduction. Cancer Lett. 2014 Mar 1;344(1):62-73. doi: 10.1016/j.canlet.2013.10.019. Epub
[PubMed ID: 24136164] ... Choi WS, Rice WJ, Stokes DL, Coller BS, Three-dimensional reconstruction of intact human integrin alphaIIbbeta3: new implications for activation-dependent ligand binding. Blood. 2013 Dec 19;122(26):4165-71. doi: 10.1182/blood-2013-04-499194. Epub 2013
[PubMed ID: 24109241] ... Gianni T, Leoni V, Campadelli-Fiume G, Type I interferon and NF-kappaB activation elicited by herpes simplex virus gH/gL via alphavbeta3 integrin in epithelial and neuronal cell lines. J Virol. 2013 Dec;87(24):13911-6. doi: 10.1128/JVI.01894-13. Epub 2013 Oct 9.
[PubMed ID: 24437181] ... Rebrova TIu, Muslimova EF, Afanas'ev SA, Sergienko TN, Repin AN, [Resistance to clopidogrel and polymorphism of P2RY12 and GPIIIA genes in patients with chronic coronary heart disease]. Klin Med (Mosk). 2013;91(8):29-31.
[PubMed ID: 24437179] ... Zotova TIu, Miandina GI, Frolov VA, Komarova AG, Zotov AK, [The influence of ITGB3 gene polymorphism on the frequency of arterial hypertension in patients with acute coronary syndrome]. Klin Med (Mosk). 2013;91(8):22-4.
[PubMed ID: 11723131] ... Datta A, Huber F, Boettiger D, Phosphorylation of beta3 integrin controls ligand binding strength. J Biol Chem. 2002 Feb 8;277(6):3943-9. Epub 2001 Nov 26.
[PubMed ID: 10896934] ... Kirk RI, Sanderson MR, Lerea KM, Threonine phosphorylation of the beta 3 integrin cytoplasmic tail, at a site recognized by PDK1 and Akt/PKB in vitro, regulates Shc binding. J Biol Chem. 2000 Oct 6;275(40):30901-6.
[PubMed ID: 7592818] ... Du X, Saido TC, Tsubuki S, Indig FE, Williams MJ, Ginsberg MH, Calpain cleavage of the cytoplasmic domain of the integrin beta 3 subunit. J Biol Chem. 1995 Nov 3;270(44):26146-51.
[PubMed ID: 1429573] ... Bajt ML, Loftus JC, Gawaz MP, Ginsberg MH, Characterization of a gain of function mutation of integrin alpha IIb beta 3 (platelet glycoprotein IIb-IIIa). J Biol Chem. 1992 Nov 5;267(31):22211-6.
[PubMed ID: 1438206] ... Chen YP, Djaffar I, Pidard D, Steiner B, Cieutat AM, Caen JP, Rosa JP, Ser-752-->Pro mutation in the cytoplasmic domain of integrin beta 3 subunit and defective activation of platelet integrin alpha IIb beta 3 (glycoprotein IIb-IIIa) in a variant of Glanzmann thrombasthenia. Proc Natl Acad Sci U S A. 1992 Nov 1;89(21):10169-73.
[PubMed ID: 1430225] ... Wang R, Furihata K, McFarland JG, Friedman K, Aster RH, Newman PJ, An amino acid polymorphism within the RGD binding domain of platelet membrane glycoprotein IIIa is responsible for the formation of the Pena/Penb alloantigen system. J Clin Invest. 1992 Nov;90(5):2038-43.
[PubMed ID: 1382574] ... Jiang WM, Jenkins D, Yuan Q, Leung E, Choo KH, Watson JD, Krissansen GW, The gene organization of the human beta 7 subunit, the common beta subunit of the leukocyte integrins HML-1 and LPAM-1. Int Immunol. 1992 Sep;4(9):1031-40.
[PubMed ID: 1371279] ... Bajt ML, Ginsberg MH, Frelinger AL 3rd, Berndt MC, Loftus JC, A spontaneous mutation of integrin alpha IIb beta 3 (platelet glycoprotein IIb-IIIa) helps define a ligand binding site. J Biol Chem. 1992 Feb 25;267(6):3789-94.