SB0047 : Protamine-Z
[ CaMP Format ]
* Basic Information
| Organism | Clupea harengus (Atlantic herring) |
| Protein Names | RecName: Full=Protamine-Z; AltName: Full=Clupeine-Z; Protamine-Z; Clupeine-Z |
| Gene Names | Not available |
| Gene Locus | Not available |
| GO Function | Not available |
* Information From OMIM
Not Available.
* Structure Information
1. Primary Information
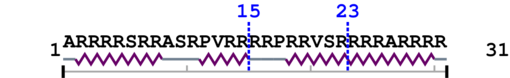
Length: 31 aa
Average Mass: 4.165 kDa
Monoisotopic Mass: 4.163 kDa
2. Domain Information
Annotated Domains: interpro / pfam / smart / prosite
Computationally Assigned Domains (Pfam+HMMER):
| domain name | begin | end | score | e-value |
|---|
3. Sequence Information
Fasta Sequence: SB0047.fasta
Amino Acid Sequence and Secondary Structures (PsiPred):
4. 3D Information
Known Structures in PDB: 7INS (X-ray; 200 A; G=-)
* Cleavage Information
2 [sites]
Source Reference: [PubMed ID: 2993267] Hayashi M, Inomata M, Nakamura M, Imahori K, Kawashima S, Hydrolysis of protamine by calcium-activated neutral protease (CANP). J Biochem. 1985 May;97(5):1363-70.
Cleavage sites (±10aa)
[Site 1] SRRASRPVRR15-RRPRRVSRRR
Arg15  Arg
Arg
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Ser6 | Arg7 | Arg8 | Ala9 | Ser10 | Arg11 | Pro12 | Val13 | Arg14 | Arg15 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Arg16 | Arg17 | Pro18 | Arg19 | Arg20 | Val21 | Ser22 | Arg23 | Arg24 | Arg25 |
 Sequence conservation (by blast)
Sequence conservation (by blast)
[Site 2] RRRRPRRVSR23-RRRARRRR
Arg23  Arg
Arg
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Arg14 | Arg15 | Arg16 | Arg17 | Pro18 | Arg19 | Arg20 | Val21 | Ser22 | Arg23 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Arg24 | Arg25 | Arg26 | Ala27 | Arg28 | Arg29 | Arg30 | Arg31 | - | - |
 Sequence conservation (by blast)
Sequence conservation (by blast)
* References
[PubMed ID: 1772633] Balschmidt P, Hansen FB, Dodson EJ, Dodson GG, Korber F, Structure of porcine insulin cocrystallized with clupeine Z. Acta Crystallogr B. 1991 Dec 1;47 ( Pt 6):975-86.