SB0048 : Annexin I
[ CaMP Format ]
* Basic Information
| Organism | Homo sapiens (human) |
| Protein Names | annexin A1 [Homo sapiens]; annexin A1; annexin I (lipocortin I); annexin-1; calpactin-2; calpactin II; chromobindin-9; phospholipase A2 inhibitory protein; Annexin A1; Annexin I; Annexin-1; Calpactin II; Calpactin-2; Chromobindin-9; Lipocortin I; Phospholipase A2 inhibitory protein; p35 |
| Gene Names | ANXA1; ANX1, LPC1; ANX1; LPC1; annexin A1 |
| Gene Locus | 9q21.13; chromosome 9 |
| GO Function | Not available |
* Information From OMIM
Function: Walther et al. (2000) showed that ANXA1 acts through the formyl peptide receptor (FPR; OMIM:136537) on human neutrophils. Peptides derived from the unique N-terminal domain of ANXA1 serve as FPR ligands and trigger different signaling pathways in a dose-dependent manner. Lower peptide concentrations possibly found in inflammatory situations elicit Ca(2+) transients without fully activating the mitogen-activated protein kinase pathway. This causes a specific inhibition of the transendothelial migration of neutrophils and a desensitization of neutrophils toward a chemoattractant challenge. These findings identified ANXA1 peptides as novel, endogenous FPR ligands and established a mechanistic basis of ANXA1-mediated antiinflammatory effects.
* Structure Information
1. Primary Information
Length: 346 aa
Average Mass: 38.714 kDa
Monoisotopic Mass: 38.690 kDa
2. Domain Information
Annotated Domains: interpro / pfam / smart / prosite
Computationally Assigned Domains (Pfam+HMMER):
| domain name | begin | end | score | e-value |
|---|---|---|---|---|
| --- cleavage 26 --- | ||||
| Myb DNA-binding like 1. | 50 | 103 | 18.0 | 0.0 |
| Myb DNA-binding like 2. | 125 | 151 | 21.0 | 0.0 |
| Myb DNA-binding like 3. | 213 | 263 | 25.0 | 0.0 |
| Myb DNA-binding like 4. | 284 | 305 | 21.0 | 0.0 |
3. Sequence Information
Fasta Sequence: SB0048.fasta
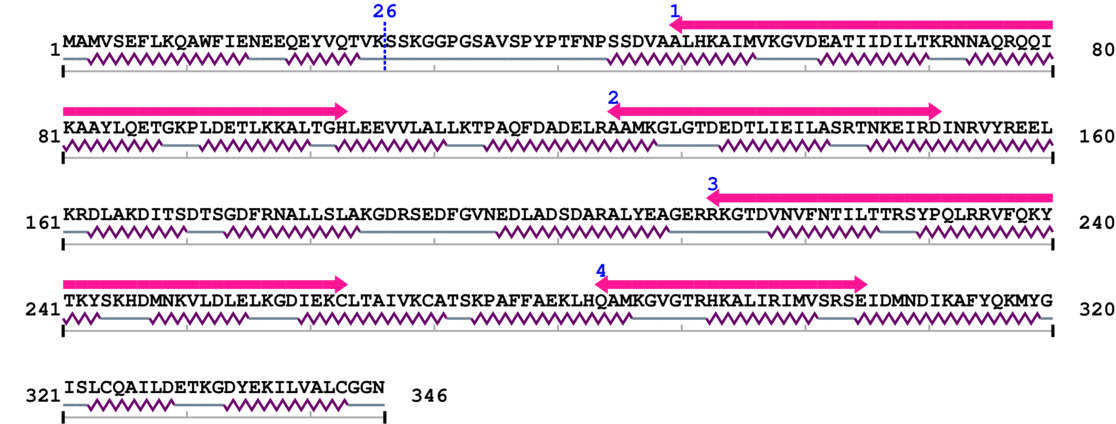
Amino Acid Sequence and Secondary Structures (PsiPred):
* Cleavage Information
1 [sites] cleaved by Calpain 1
Source Reference: [PubMed ID: 2540167] Ando Y, Imamura S, Hong YM, Owada MK, Kakunaga T, Kannagi R, Enhancement of calcium sensitivity of lipocortin I in phospholipid binding induced by limited proteolysis and phosphorylation at the amino terminus as analyzed by phospholipid affinity column chromatography. J Biol Chem. 1989 Apr 25;264(12):6948-55.
Cleavage sites (±10aa)
[Site 1] EEQEYVQTVK26-SSKGGPGSAV
Lys26  Ser
Ser
iTraq-117 Signal 721.2 (
 ) for EEQEYVQTVK
) for EEQEYVQTVK
iTraq-117 Signal 2656.4 (

 ) for SSKGGPGSAV
) for SSKGGPGSAV
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Glu17 | Glu18 | Gln19 | Glu20 | Tyr21 | Val22 | Gln23 | Thr24 | Val25 | Lys26 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Ser27 | Ser28 | Lys29 | Gly30 | Gly31 | Pro32 | Gly33 | Ser34 | Ala35 | Val36 |
 Sequence conservation (by blast)
Sequence conservation (by blast)
* References
[PubMed ID: 24480285] Yu CJ, Ko CJ, Hsieh CH, Chien CT, Huang LH, Lee CW, Jiang CC, Proteomic analysis of osteoarthritic chondrocyte reveals the hyaluronic acid-regulated proteins involved in chondroprotective effect under oxidative stress. J Proteomics. 2014 Mar 17;99:40-53. doi: 10.1016/j.jprot.2014.01.016. Epub 2014