SB0055 : Protein kinase C[alpha]
[ CaMP Format ]
* Basic Information
| Organism | Rattus norvegicus (Norway rat) |
| Protein Names | PREDICTED: similar to Protein kinase C alpha type (PKC-alpha) (PKC-A) [Rattus norvegicus]; similar to Protein kinase C alpha type (PKC-alpha) (PKC-A); Protein kinase C alpha type; PKC-A; PKC-alpha; 2.7.11.13 |
| Gene Names | Prkca; Pkca; Derived by automated computational analysis using gene prediction method: GNOMON. Supporting evidence includes similarity to: 4 mRNAs, 51 ESTs, 19 Proteins |
| Gene Locus | Not available |
| GO Function | Not available |
| Entrez Protein | Entrez Nucleotide | Entrez Gene | UniProt | OMIM | HGNC | HPRD | KEGG |
|---|---|---|---|---|---|---|---|
| XP_001081588 | XM_001081588 | 24680 | P05696 | N/A | N/A | N/A | rno:24680 |
* Information From OMIM
Not Available.
* Structure Information
1. Primary Information
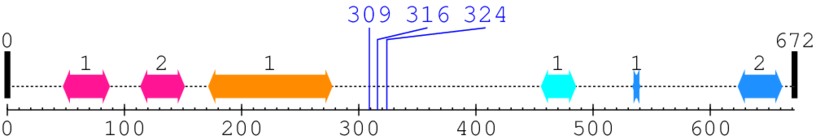
Length: 672 aa
Average Mass: 76.791 kDa
Monoisotopic Mass: 76.742 kDa
2. Domain Information
Annotated Domains: interpro / pfam / smart / prosite
Computationally Assigned Domains (Pfam+HMMER):
| domain name | begin | end | score | e-value |
|---|---|---|---|---|
| Putative zinc-RING and/or ribbon 1. | 48 | 87 | 156.0 | 9.0 |
| Putative zinc-RING and/or ribbon 2. | 114 | 151 | 157.0 | 2.7 |
| C2 domain 1. | 172 | 277 | 2.0 | 0.1 |
| --- cleavage 309 --- | ||||
| --- cleavage 316 --- | ||||
| --- cleavage 324 --- | ||||
| Phosphotransferase enzyme family 1. | 456 | 485 | 165.0 | 0.0 |
| Protein kinase C terminal domain 1. | 534 | 540 | 18.0 | 0.0 |
| Protein kinase C terminal domain 2. | 624 | 661 | 8.0 | 0.4 |
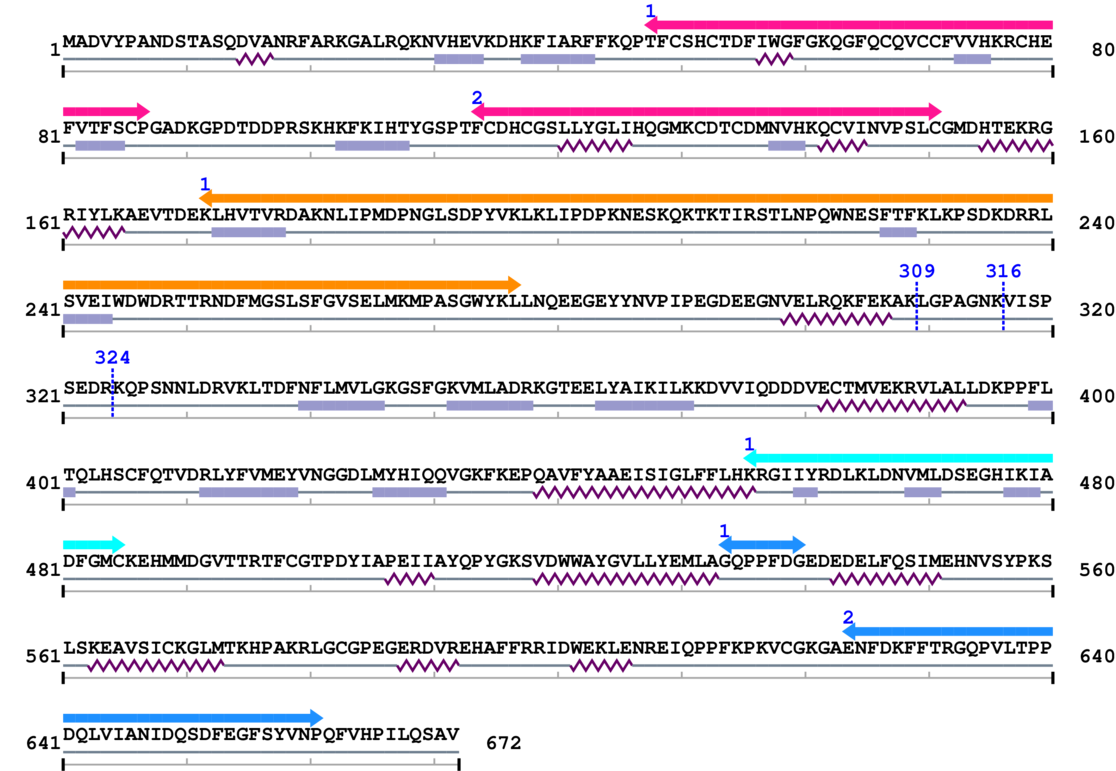
3. Sequence Information
Fasta Sequence: SB0055.fasta
Amino Acid Sequence and Secondary Structures (PsiPred):
* Cleavage Information
3 [sites] cleaved by Calpain 1 and 2
Source Reference: [PubMed ID: 2537303] Kishimoto A, Mikawa K, Hashimoto K, Yasuda I, Tanaka S, Tominaga M, Kuroda T, Nishizuka Y, Limited proteolysis of protein kinase C subspecies by calcium-dependent neutral protease (calpain). J Biol Chem. 1989 Mar 5;264(7):4088-92.
Cleavage sites (±10aa)
[Site 1] ELRQKFEKAK309-LGPAGNKVIS
Lys309  Leu
Leu
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Glu300 | Leu301 | Arg302 | Gln303 | Lys304 | Phe305 | Glu306 | Lys307 | Ala308 | Lys309 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Leu310 | Gly311 | Pro312 | Ala313 | Gly314 | Asn315 | Lys316 | Val317 | Ile318 | Ser319 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| QEEGEYYNVPIPEGDEEGNVELRQKFEKAKLGPAGNKVISPSEDRKQPSNNLDRVKLTDF |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | Rattus rattus | 124.00 | 1 | KPCA_RAT Protein kinase C alpha type (PKC-alpha) |
| 2 | N/A | 123.00 | 14 | protein kinase C alpha-polypeptide |
| 3 | Homo sapiens | 123.00 | 8 | protein kinase C, alpha |
| 4 | Mus musculus | 123.00 | 6 | protein kinase C, alpha |
| 5 | Oryctolagus cuniculus | 123.00 | 4 | protein kinase C, alpha |
| 6 | Canis familiaris | 123.00 | 3 | PREDICTED: similar to Protein kinase C, alpha typ |
| 7 | Bos taurus | 123.00 | 2 | protein kinase C, alpha |
| 8 | synthetic construct | 123.00 | 1 | protein kinase C alpha |
| 9 | Homo sapiens | 123.00 | 1 | aging-associated gene 6 protein |
| 10 | Takifugu rubripes | 79.30 | 1 | protein kinase C, alpha type |
| 11 | Scyliorhinus canicula | 62.40 | 1 | protein kinase C |
| 12 | Petromyzon marinus | 58.90 | 1 | protein kinase C |
| 13 | Rattus norvegicus | 57.40 | 3 | protein kinase C, beta |
| 14 | Danio rerio | 54.70 | 2 | protein kinase C, beta 1, like |
| 15 | Tetraodon nigroviridis | 53.50 | 4 | unnamed protein product |
| 16 | Danio rerio | 53.10 | 1 | protein kinase C |
| 17 | Myxine glutinosa | 50.40 | 1 | protein kinase C |
| 18 | Branchiostoma lanceolatum | 38.10 | 1 | protein kinase C |
| 19 | Bombyx mori | 36.20 | 1 | conventional protein kinase C |
| 20 | Drosophila melanogaster | 34.70 | 4 | Protein C kinase 53E CG6622-PA, isoform A |
| 21 | Aplysia californica | 31.60 | 1 | KPC1_APLCA Calcium-dependent protein kinase C (AP |
| 22 | Drosophila pseudoobscura | 30.80 | 1 | GA19732-PA |
Top-ranked sequences
| organism | matching |
|---|---|
| Rattus rattus | Query 280 QEEGEYYNVPIPEGDEEGNVELRQKFEKAK#LGPAGNKVISPSEDRKQPSNNLDRVKLTDF 339 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 280 QEEGEYYNVPIPEGDEEGNVELRQKFEKAK#LGPAGNKVISPSEDRKQPSNNLDRVKLTDF 339 |
| N/A | Query 280 QEEGEYYNVPIPEGDEEGNVELRQKFEKAK#LGPAGNKVISPSEDRKQPSNNLDRVKLTDF 339 |||||||||||||||||||+||||||||||#|||||||||||||||||||||||||||||| Sbjct 266 QEEGEYYNVPIPEGDEEGNMELRQKFEKAK#LGPAGNKVISPSEDRKQPSNNLDRVKLTDF 325 |
| Homo sapiens | Query 280 QEEGEYYNVPIPEGDEEGNVELRQKFEKAK#LGPAGNKVISPSEDRKQPSNNLDRVKLTDF 339 |||||||||||||||||||+||||||||||#|||||||||||||||||||||||||||||| Sbjct 280 QEEGEYYNVPIPEGDEEGNMELRQKFEKAK#LGPAGNKVISPSEDRKQPSNNLDRVKLTDF 339 |
| Mus musculus | Query 280 QEEGEYYNVPIPEGDEEGNVELRQKFEKAK#LGPAGNKVISPSEDRKQPSNNLDRVKLTDF 339 |||||||||||||||||||+||||||||||#|||||||||||||||||||||||||||||| Sbjct 280 QEEGEYYNVPIPEGDEEGNMELRQKFEKAK#LGPAGNKVISPSEDRKQPSNNLDRVKLTDF 339 |
| Oryctolagus cuniculus | Query 280 QEEGEYYNVPIPEGDEEGNVELRQKFEKAK#LGPAGNKVISPSEDRKQPSNNLDRVKLTDF 339 ||||||||||||||||+|||||||||||||#|||||||||||||||||||||||||||||| Sbjct 280 QEEGEYYNVPIPEGDEDGNVELRQKFEKAK#LGPAGNKVISPSEDRKQPSNNLDRVKLTDF 339 |
| Canis familiaris | Query 280 QEEGEYYNVPIPEGDEEGNVELRQKFEKAK#LGPAGNKVISPSEDRKQPSNNLDRVKLTDF 339 ||||||||||||||||||||||||||||||#|||||||||||||||+|||||||||||||| Sbjct 198 QEEGEYYNVPIPEGDEEGNVELRQKFEKAK#LGPAGNKVISPSEDRRQPSNNLDRVKLTDF 257 |
| Bos taurus | Query 280 QEEGEYYNVPIPEGDEEGNVELRQKFEKAK#LGPAGNKVISPSEDRKQPSNNLDRVKLTDF 339 ||||||||||||||||||||||||||||||#|||||||||||||||+|||||||||||||| Sbjct 280 QEEGEYYNVPIPEGDEEGNVELRQKFEKAK#LGPAGNKVISPSEDRRQPSNNLDRVKLTDF 339 |
| synthetic construct | Query 280 QEEGEYYNVPIPEGDEEGNVELRQKFEKAK#LGPAGNKVISPSEDRKQPSNNLDRVKLTDF 339 |||||||||||||||||||+||||||||||#|||||||||||||||||||||||||||||| Sbjct 280 QEEGEYYNVPIPEGDEEGNMELRQKFEKAK#LGPAGNKVISPSEDRKQPSNNLDRVKLTDF 339 |
| Homo sapiens | Query 280 QEEGEYYNVPIPEGDEEGNVELRQKFEKAK#LGPAGNKVISPSEDRKQPSNNLDRVKLTDF 339 |||||||||||||||||||+||||||||||#|||||||||||||||||||||||||||||| Sbjct 280 QEEGEYYNVPIPEGDEEGNMELRQKFEKAK#LGPAGNKVISPSEDRKQPSNNLDRVKLTDF 339 |
| Takifugu rubripes | Query 280 QEEGEYYNVPIPEGDEEGNVELRQKFEKAK#LGPAGNKVISPSEDRK--QPSNNLDRVKLT 337 ||||||||||||| |+ |+||||||||||#|| | |||+||+ |+ || |+|||+| Sbjct 278 QEEGEYYNVPIPEVDDV-NLELRQKFEKAK#LG-QGKKVITPSDHRRFSLPSGNMDRVRLN 335 Query 338 DF 339 || Query 338 DF 339 |
| Scyliorhinus canicula | Query 280 QEEGEYYNVPIP-EGDEEGNVELRQKFEKAK#LGPAGNKVI---SPSEDRKQPSNNLDRVK 335 ||||||||||+| | | ||| |||||||+|+#+|| |+| | | + | ||+| Sbjct 200 QEEGEYYNVPVPPEEDSEGNEELRQKFERAR#IGP-GSKAAEEKRSSSWSKIDNGNRDRMK 258 Query 336 LTDF 339 | || Sbjct 259 LADF 262 |
| Petromyzon marinus | Query 280 QEEGEYYNVPIPEGDEEGNVELRQKFEKAK#LGPAGNKVISP--SEDRKQPS-------NN 330 ||||||||||| |+|+ |+|+|||||+#|||+ + | |++|+ | | Sbjct 210 QEEGEYYNVPIAPDIEDGDTEMRRKFEKAR#LGPS----VKPRASDERRANSLSAILNNAN 265 Query 331 LDRVKLTDF 339 +|||| || Sbjct 266 VDRVKADDF 274 |
| Rattus norvegicus | Query 280 QEEGEYYNVPIPEGDEEGNVELRQKFEKAK#LGPAGNKV----ISPSEDRKQPSNNLDRVK 335 ||||||+|||+| ||| |||||||+||#+| | | + + + + | ||+| Sbjct 280 QEEGEYFNVPVPPEGSEGNEELRQKFERAK#IG-QGTKAPEEKTANTISKFDNNGNRDRMK 338 Query 336 LTDF 339 |||| Query 336 LTDF 339 |
| Danio rerio | Query 281 EEGEYYNVPIPEGDEEGNVELRQKFEKAK#LGPAGNKVISPSED---RKQPSNNLDRVKLT 337 ||| ++|||+ +|||| || ++||+||#+|| | |++ + + | ||+|| Sbjct 277 EEGVFFNVPVYVDEEEGNKELSKRFERAK#IGPGTNNTDGASKNDLSKYDANGNQDRMKLA 336 Query 338 DF 339 || Query 338 DF 339 |
| Tetraodon nigroviridis | Query 280 QEEGEYYNVPIPEGDEEGNVELRQKFEKAK#LGPAGNK--VISPSEDRKQPSNNLDRVKLT 337 ||||||+|||+| |||| |||||||+||#+|| | + + | + | ||+|| Sbjct 216 QEEGEYFNVPVPPEGEEGNEELRQKFERAK#IGPGKNTDGKSANAASRFDSNGNQDRMKLA 275 Query 338 DF 339 || Query 338 DF 339 |
| Danio rerio | Query 280 QEEGEYYNVPIPEGDEEGNVELRQKFEKAK#LGPAGNKVISPSEDRKQPS-NNLDRVKLTD 338 ||||||+|||+| |||| |||||||+||#+||+ | + | | | ||+||+| Sbjct 211 QEEGEYFNVPVPPEGEEGNEELRQKFERAK#IGPSKTDGSSSNAISKFDSYGNRDRMKLSD 270 Query 339 F 339 | Query 339 F 339 |
| Myxine glutinosa | Query 280 QEEGEYYNVPI-PEGDEEGNVELRQKFEKAK#LGPAGNKVISPSEDRKQPSNN-----LDR 333 ||||||||||+ |||+| +||||+ ++++#+ + ||++| |||++ ||| Sbjct 201 QEEGEYYNVPVAPEGEE--GLELRQRLQRSQ#IARSDKNKQSPAKD--QPSDSASLSGLDR 256 Query 334 VKLTDF 339 | | || Sbjct 257 VNLEDF 262 |
| Branchiostoma lanceolatum | Query 280 QEEGEYYNVPIPEGDEEGNV-ELRQKFEKAK#LGPAGNKVISPSEDRKQPSN--------N 330 ||||||||||| ||| || +|++ +| |#| | ||| | + Sbjct 211 QEEGEYYNVPI--ADEEENVAKLKEHLQKQK#L----------DEQRKQKSKRSEECNIGS 258 Query 331 LDRVKLTDF 339 || +| || Sbjct 259 LDHMKAADF 267 |
| Bombyx mori | Query 280 QEEGEYYNVPIPEGDEEGNVELRQKFEKAK#LGPAGNKVISPSEDRKQPSN--NLDRVKLT 337 |||||+||||+| ||| +| | + +# | + | ||+ | | | ++ | Sbjct 283 QEEGEFYNVPVP---EEG-ADLVQLKNQMR#ATTVGARRPPPPPDREVPHNVAAADVIRAT 338 Query 338 DF 339 || Query 338 DF 339 |
| Drosophila melanogaster | Query 280 QEEGEYYNVPIPEGDEEGNVELRQKFEKAK#LGPAGNK-VISPSEDRKQPSNNLDRVKLTD 338 |+|||||||| + ||+ ++|+|| # |+ | ++ |+ |+ | ++ || Sbjct 289 QDEGEYYNVPCAD-DEQDLLKLKQK-----#--PSQKKPMVMRSDTNTHTSSKKDMIRATD 340 Query 339 F 339 | Query 339 F 339 |
| Aplysia californica | Query 280 QEEGEYYNVPIPEGDEEGNVELRQKFEKAK#LGPAGNKVISPSEDRKQPSNNLDRVKLTDF 339 |||||+| ||+ + | |++ | ++ #+ | |+ | + | |+ +|| Sbjct 264 QEEGEFYGVPVTDDITESIQEIKSKMHRSS#ISSEKR---YPEPDKVQNMSKQDIVRASDF 320 |
| Drosophila pseudoobscura | Query 280 QEEGEYYNVPIPEGDEEGNVELRQKFEKAK#LGPAGNKVISPSEDRKQPS--NNLDRVKLT 337 |+|||||||| + ||+ ++|+|| # |+ | + | | + | ++ | Sbjct 175 QDEGEYYNVPCAD-DEQDLLKLKQK-----#--PSQKKPLVMRSDTNTHSSISKKDMIRAT 226 Query 338 DF 339 || Query 338 DF 339 |
[Site 2] KAKLGPAGNK316-VISPSEDRKQ
Lys316  Val
Val
iTraq-117 Signal 1206.9 (

 ) for KVISPSEDRKQ
) for KVISPSEDRKQ
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Lys307 | Ala308 | Lys309 | Leu310 | Gly311 | Pro312 | Ala313 | Gly314 | Asn315 | Lys316 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Val317 | Ile318 | Ser319 | Pro320 | Ser321 | Glu322 | Asp323 | Arg324 | Lys325 | Gln326 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| NVPIPEGDEEGNVELRQKFEKAKLGPAGNKVISPSEDRKQPSNNLDRVKLTDFNFLMVLG |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | Rattus rattus | 124.00 | 1 | KPCA_RAT Protein kinase C alpha type (PKC-alpha) |
| 2 | N/A | 123.00 | 11 | protein kinase C alpha-polypeptide |
| 3 | Mus musculus | 123.00 | 4 | protein kinase C, alpha |
| 4 | Bos taurus | 123.00 | 2 | protein kinase C, alpha |
| 5 | Homo sapiens | 122.00 | 6 | protein kinase C, alpha |
| 6 | Canis familiaris | 122.00 | 5 | PREDICTED: similar to Protein kinase C, alpha typ |
| 7 | Oryctolagus cuniculus | 122.00 | 4 | protein kinase C, alpha |
| 8 | synthetic construct | 122.00 | 1 | protein kinase C alpha |
| 9 | Homo sapiens | 122.00 | 1 | aging-associated gene 6 protein |
| 10 | Takifugu rubripes | 75.10 | 1 | protein kinase C, alpha type |
| 11 | Scyliorhinus canicula | 64.30 | 1 | protein kinase C |
| 12 | Danio rerio | 61.20 | 2 | protein kinase C, beta 1, like |
| 13 | Rattus norvegicus | 58.90 | 3 | unnamed protein product |
| 14 | Petromyzon marinus | 57.00 | 1 | protein kinase C |
| 15 | Tetraodon nigroviridis | 53.90 | 3 | unnamed protein product |
| 16 | Danio rerio | 53.10 | 1 | protein kinase C |
| 17 | Myxine glutinosa | 45.10 | 1 | protein kinase C |
| 18 | Branchiostoma lanceolatum | 38.90 | 1 | protein kinase C |
| 19 | Bombyx mori | 35.40 | 1 | conventional protein kinase C |
| 20 | Drosophila melanogaster | 31.20 | 4 | Protein C kinase 53E CG6622-PA, isoform A |
| 21 | Pongo pygmaeus | 31.20 | 1 | LASP1_PONPY LIM and SH3 domain protein 1 (LASP-1) |
Top-ranked sequences
| organism | matching |
|---|---|
| Rattus rattus | Query 287 NVPIPEGDEEGNVELRQKFEKAKLGPAGNK#VISPSEDRKQPSNNLDRVKLTDFNFLMVLG 346 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 287 NVPIPEGDEEGNVELRQKFEKAKLGPAGNK#VISPSEDRKQPSNNLDRVKLTDFNFLMVLG 346 |
| N/A | Query 287 NVPIPEGDEEGNVELRQKFEKAKLGPAGNK#VISPSEDRKQPSNNLDRVKLTDFNFLMVLG 346 ||||||||||||+|||||||||||||||||#|||||||||||||||||||||||||||||| Sbjct 273 NVPIPEGDEEGNMELRQKFEKAKLGPAGNK#VISPSEDRKQPSNNLDRVKLTDFNFLMVLG 332 |
| Mus musculus | Query 287 NVPIPEGDEEGNVELRQKFEKAKLGPAGNK#VISPSEDRKQPSNNLDRVKLTDFNFLMVLG 346 ||||||||||||+|||||||||||||||||#|||||||||||||||||||||||||||||| Sbjct 287 NVPIPEGDEEGNMELRQKFEKAKLGPAGNK#VISPSEDRKQPSNNLDRVKLTDFNFLMVLG 346 |
| Bos taurus | Query 287 NVPIPEGDEEGNVELRQKFEKAKLGPAGNK#VISPSEDRKQPSNNLDRVKLTDFNFLMVLG 346 ||||||||||||||||||||||||||||||#||||||||+||||||||||||||||||||| Sbjct 287 NVPIPEGDEEGNVELRQKFEKAKLGPAGNK#VISPSEDRRQPSNNLDRVKLTDFNFLMVLG 346 |
| Homo sapiens | Query 287 NVPIPEGDEEGNVELRQKFEKAKLGPAGNK#VISPSEDRKQPSNNLDRVKLTDFNFLMVLG 346 ||||||||||||+|||||||||||||||||#|||||||||||||||||||||||||||||| Sbjct 287 NVPIPEGDEEGNMELRQKFEKAKLGPAGNK#VISPSEDRKQPSNNLDRVKLTDFNFLMVLG 346 |
| Canis familiaris | Query 287 NVPIPEGDEEGNVELRQKFEKAKLGPAGNK#VISPSEDRKQPSNNLDRVKLTDFNFLMVLG 346 ||||||||||||||||||||||||||||||#||||||||+||||||||||||||||||||| Sbjct 205 NVPIPEGDEEGNVELRQKFEKAKLGPAGNK#VISPSEDRRQPSNNLDRVKLTDFNFLMVLG 264 |
| Oryctolagus cuniculus | Query 287 NVPIPEGDEEGNVELRQKFEKAKLGPAGNK#VISPSEDRKQPSNNLDRVKLTDFNFLMVLG 346 |||||||||+||||||||||||||||||||#|||||||||||||||||||||||||||||| Sbjct 287 NVPIPEGDEDGNVELRQKFEKAKLGPAGNK#VISPSEDRKQPSNNLDRVKLTDFNFLMVLG 346 |
| synthetic construct | Query 287 NVPIPEGDEEGNVELRQKFEKAKLGPAGNK#VISPSEDRKQPSNNLDRVKLTDFNFLMVLG 346 ||||||||||||+|||||||||||||||||#|||||||||||||||||||||||||||||| Sbjct 287 NVPIPEGDEEGNMELRQKFEKAKLGPAGNK#VISPSEDRKQPSNNLDRVKLTDFNFLMVLG 346 |
| Homo sapiens | Query 287 NVPIPEGDEEGNVELRQKFEKAKLGPAGNK#VISPSEDRKQPSNNLDRVKLTDFNFLMVLG 346 ||||||||||||+|||||||||||||||||#|||||||||||||||||||||||||||||| Sbjct 287 NVPIPEGDEEGNMELRQKFEKAKLGPAGNK#VISPSEDRKQPSNNLDRVKLTDFNFLMVLG 346 |
| Takifugu rubripes | Query 287 NVPIPEGDEEGNVELRQKFEKAKLGPAGNK#VISPSEDRK--QPSNNLDRVKLTDFNFLMV 344 |||||| |+ |+|||||||||||| | |#||+||+ |+ || |+|||+| ||||| + Sbjct 285 NVPIPEVDDV-NLELRQKFEKAKLG-QGKK#VITPSDHRRFSLPSGNMDRVRLNDFNFLAL 342 Query 345 LG 346 || Query 345 LG 346 |
| Scyliorhinus canicula | Query 287 NVPIP-EGDEEGNVELRQKFEKAKLGPAGNK#VISPSEDRKQPS------NNLDRVKLTDF 339 |||+| | | ||| |||||||+|++|| |+|# +|+++ | | ||+|| || Sbjct 207 NVPVPPEEDSEGNEELRQKFERARIGP-GSK#A---AEEKRSSSWSKIDNGNRDRMKLADF 262 Query 340 NFLMVLG 346 ||||||| Query 340 NFLMVLG 346 |
| Danio rerio | Query 287 NVPIPEGDEEGNVELRQKFEKAKLGPAGNK#VISPSED---RKQPSNNLDRVKLTDFNFLM 343 |||+ +|||| || ++||+||+|| | # |++ + + | ||+|| ||||+| Sbjct 283 NVPVYVDEEEGNKELSKRFERAKIGPGTNN#TDGASKNDLSKYDANGNQDRMKLADFNFIM 342 Query 344 VLG 346 ||| Query 344 VLG 346 |
| Rattus norvegicus | Query 287 NVPIPEGDEEGNVELRQKFEKAKLGPAGNK#VISPSEDRKQP------SNNLDRVKLTDFN 340 |||+| ||| |||||||+||+| | |# +| | + | ||+|||||| Sbjct 287 NVPVPPEGSEGNEELRQKFERAKIG-QGTK#--APEEKTANTISKFDNNGNRDRMKLTDFN 343 Query 341 FLMVLG 346 |||||| Query 341 FLMVLG 346 |
| Petromyzon marinus | Query 287 NVPIPEGDEEGNVELRQKFEKAKLGPAGNK#VISP--SEDRKQPS-------NNLDRVKLT 337 |||| |+|+ |+|+|||||+|||+ # + | |++|+ | |+|||| Sbjct 217 NVPIAPDIEDGDTEMRRKFEKARLGPS---#-VKPRASDERRANSLSAILNNANVDRVKAD 272 Query 338 DFNFLMVLG 346 ||||||||| Query 338 DFNFLMVLG 346 |
| Tetraodon nigroviridis | Query 287 NVPIPEGDEEGNVELRQKFEKAKLGPAGNK--#VISPSEDRKQPSNNLDRVKLTDFNFLMV 344 |||+| |||| |||||||+||+|| | # + + | + | ||+|| ||||||| Sbjct 223 NVPVPPEGEEGNEELRQKFERAKIGPGKNTDG#KSANAASRFDSNGNQDRMKLADFNFLMV 282 Query 345 LG 346 || Query 345 LG 346 |
| Danio rerio | Query 287 NVPIPEGDEEGNVELRQKFEKAKLGPAGNK#VISPSEDRKQPS-NNLDRVKLTDFNFLMVL 345 |||+| |||| |||||||+||+||+ # | + | | | ||+||+|||||||| Sbjct 218 NVPVPPEGEEGNEELRQKFERAKIGPSKTD#GSSSNAISKFDSYGNRDRMKLSDFNFLMVL 277 Query 346 G 346 | Query 346 G 346 |
| Myxine glutinosa | Query 287 NVPI-PEGDEEGNVELRQKFEKAKLGPAGNK#VISPSEDRKQPSNN-----LDRVKLTDFN 340 |||+ |||+| +||||+ +++++ + # ||++| |||++ |||| | || Sbjct 208 NVPVAPEGEE--GLELRQRLQRSQIARSDKN#KQSPAKD--QPSDSASLSGLDRVNLEDFV 263 Query 341 FLMVLG 346 || ||| Sbjct 264 FLTVLG 269 |
| Branchiostoma lanceolatum | Query 287 NVPIPEGDEEGNV-ELRQKFEKAKLGPAGNK#VISPSEDRKQPSN--------NLDRVKLT 337 |||| ||| || +|++ +| || # | ||| | +|| +| Sbjct 218 NVPI--ADEEENVAKLKEHLQKQKL------#----DEQRKQKSKRSEECNIGSLDHMKAA 265 Query 338 DFNFLMVLG 346 ||||||||| Query 338 DFNFLMVLG 346 |
| Bombyx mori | Query 287 NVPIPEGDEEGNVELRQKFEKAKLGPAGNK#VISPSEDRKQPSN--NLDRVKLTDFNFLMV 344 |||+|| || +| | + + | +# | ||+ | | | ++ |||||+|| Sbjct 290 NVPVPE---EG-ADLVQLKNQMRATTVGAR#RPPPPPDREVPHNVAAADVIRATDFNFIMV 345 Query 345 LG 346 || Query 345 LG 346 |
| Drosophila melanogaster | Query 287 NVPIPEGDEEGNVELRQKFEKAKLGPAGNK-#VISPSEDRKQPSNNLDRVKLTDFNFLMVL 345 ||| + ||+ ++|+|| |+ | #++ |+ |+ | ++ |||||+ || Sbjct 296 NVPCAD-DEQDLLKLKQK-------PSQKKP#MVMRSDTNTHTSSKKDMIRATDFNFIKVL 347 Query 346 G 346 | Query 346 G 346 |
| Pongo pygmaeus | Query 294 DEEGNVELRQKFEKAKLGPAGNK#VISPSEDRKQPSNNLDR 333 |+ |++ ++|||+++||+| +# + | | |++ | Sbjct 115 DQISNIKYHEEFEKSRMGPSGGE#GMEPERRDSQDSSSYRR 154 |
[Site 3] NKVISPSEDR324-KQPSNNLDRV
Arg324  Lys
Lys
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Asn315 | Lys316 | Val317 | Ile318 | Ser319 | Pro320 | Ser321 | Glu322 | Asp323 | Arg324 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Lys325 | Gln326 | Pro327 | Ser328 | Asn329 | Asn330 | Leu331 | Asp332 | Arg333 | Val334 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| EEGNVELRQKFEKAKLGPAGNKVISPSEDRKQPSNNLDRVKLTDFNFLMVLGKGSFGKVM |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | Rattus rattus | 123.00 | 1 | KPCA_RAT Protein kinase C alpha type (PKC-alpha) |
| 2 | N/A | 122.00 | 15 | protein kinase C alpha-polypeptide |
| 3 | Canis familiaris | 122.00 | 10 | PREDICTED: similar to Protein kinase C, alpha typ |
| 4 | Mus musculus | 122.00 | 9 | protein kinase C, alpha |
| 5 | Oryctolagus cuniculus | 122.00 | 5 | protein kinase C, alpha |
| 6 | Bos taurus | 122.00 | 2 | protein kinase C, alpha |
| 7 | Homo sapiens | 121.00 | 12 | protein kinase C, alpha |
| 8 | Homo sapiens | 121.00 | 2 | aging-associated gene 6 protein |
| 9 | synthetic construct | 121.00 | 1 | protein kinase C alpha |
| 10 | Takifugu rubripes | 82.00 | 1 | protein kinase C, alpha type |
| 11 | Scyliorhinus canicula | 68.90 | 1 | protein kinase C |
| 12 | Danio rerio | 68.60 | 2 | protein kinase C, beta 1, like |
| 13 | Petromyzon marinus | 67.80 | 1 | protein kinase C |
| 14 | Tetraodon nigroviridis | 66.60 | 5 | unnamed protein product |
| 15 | Rattus norvegicus | 66.60 | 3 | unnamed protein product |
| 16 | Danio rerio | 63.50 | 1 | protein kinase C |
| 17 | Myxine glutinosa | 55.80 | 1 | protein kinase C |
| 18 | Branchiostoma lanceolatum | 48.10 | 1 | protein kinase C |
| 19 | Bombyx mori | 46.20 | 1 | conventional protein kinase C |
| 20 | Apis mellifera | 44.70 | 1 | PREDICTED: similar to protein kinase C, delta |
| 21 | Drosophila melanogaster | 42.40 | 6 | protein kinase C |
| 22 | Aplysia californica | 42.00 | 2 | KPC1_APLCA Calcium-dependent protein kinase C (AP |
| 23 | Magnaporthe grisea | 41.60 | 1 | AF136600_1 protein kinase C |
| 24 | Caenorhabditis briggsae AF16 | 41.20 | 2 | Hypothetical protein CBG16150 |
| 25 | Caenorhabditis elegans | 40.80 | 5 | Protein Kinase C family member (pkc-2) |
| 26 | Caenorhabditis elegans | 40.80 | 2 | protein kinase C2 A isoform |
| 27 | Botryotinia fuckeliana | 40.00 | 1 | protein kinase C |
| 28 | Schizosaccharomyces pombe | 39.30 | 2 | protein kinase |
| 29 | Schizosaccharomyces pombe 972h- | 39.30 | 2 | protein kinase C (PKC)-like Pck1 |
| 30 | Drosophila pseudoobscura | 38.90 | 2 | GA19732-PA |
| 31 | Asparagus officinalis | 38.90 | 1 | S6 ribosomal protein kinase |
| 32 | Rattus norvegicus | 38.50 | 1 | protein kinase C, epsilon |
| 33 | Leptosphaeria maculans | 38.50 | 1 | AF487263_9 protein kinase C |
| 34 | Yarrowia lipolytica | 38.50 | 1 | hypothetical protein |
| 35 | Suberites domuncula | 38.10 | 2 | Serine/Threonine protein kinase |
| 36 | Sycon raphanus | 37.70 | 2 | Serine/Threonine protein kinase |
| 37 | Calliphora vicina | 37.70 | 1 | eye-specific protein kinase C |
| 38 | Cryptococcus neoformans var. neoformans JEC21 | 37.70 | 1 | protein kinase C |
| 39 | Cryptococcus neoformans var. neoformans B-3501A | 37.70 | 1 | hypothetical protein CNBC3910 |
| 40 | Cryptococcus neoformans var. grubii | 37.70 | 1 | protein kinase C 1 |
| 41 | Cryptococcus neoformans var. neoformans | 37.70 | 1 | protein kinase C 1 |
| 42 | synthetic construct | 37.40 | 1 | serum/glucocorticoid regulated kinase-like |
| 43 | Squalus acanthias | 37.40 | 1 | s-sgk1 |
| 44 | Drosophila melanogaster | 37.40 | 1 | Protein C kinase 98E CG1954-PA |
| 45 | Sporothrix schenckii | 37.00 | 1 | AF124792_1 protein kinase C; serine/threonine pro |
| 46 | Bos taurus | 36.20 | 1 | PREDICTED: similar to Serum/glucocorticoid regula |
| 47 | Cochliobolus heterostrophus | 36.20 | 1 | KPC1_COCHE Protein kinase C-like gi |
| 48 | Ancylostoma caninum | 33.90 | 1 | AF301263_1 protein kinase C |
Top-ranked sequences
| organism | matching |
|---|---|
| Rattus rattus | Query 295 EEGNVELRQKFEKAKLGPAGNKVISPSEDR#KQPSNNLDRVKLTDFNFLMVLGKGSFGKVM 354 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 295 EEGNVELRQKFEKAKLGPAGNKVISPSEDR#KQPSNNLDRVKLTDFNFLMVLGKGSFGKVM 354 |
| N/A | Query 295 EEGNVELRQKFEKAKLGPAGNKVISPSEDR#KQPSNNLDRVKLTDFNFLMVLGKGSFGKVM 354 ||||+|||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Sbjct 281 EEGNMELRQKFEKAKLGPAGNKVISPSEDR#KQPSNNLDRVKLTDFNFLMVLGKGSFGKVM 340 |
| Canis familiaris | Query 295 EEGNVELRQKFEKAKLGPAGNKVISPSEDR#KQPSNNLDRVKLTDFNFLMVLGKGSFGKVM 354 ||||||||||||||||||||||||||||||#+||||||||||||||||||||||||||||| Sbjct 213 EEGNVELRQKFEKAKLGPAGNKVISPSEDR#RQPSNNLDRVKLTDFNFLMVLGKGSFGKVM 272 |
| Mus musculus | Query 295 EEGNVELRQKFEKAKLGPAGNKVISPSEDR#KQPSNNLDRVKLTDFNFLMVLGKGSFGKVM 354 ||||+|||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Sbjct 295 EEGNMELRQKFEKAKLGPAGNKVISPSEDR#KQPSNNLDRVKLTDFNFLMVLGKGSFGKVM 354 |
| Oryctolagus cuniculus | Query 295 EEGNVELRQKFEKAKLGPAGNKVISPSEDR#KQPSNNLDRVKLTDFNFLMVLGKGSFGKVM 354 |+||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Sbjct 295 EDGNVELRQKFEKAKLGPAGNKVISPSEDR#KQPSNNLDRVKLTDFNFLMVLGKGSFGKVM 354 |
| Bos taurus | Query 295 EEGNVELRQKFEKAKLGPAGNKVISPSEDR#KQPSNNLDRVKLTDFNFLMVLGKGSFGKVM 354 ||||||||||||||||||||||||||||||#+||||||||||||||||||||||||||||| Sbjct 295 EEGNVELRQKFEKAKLGPAGNKVISPSEDR#RQPSNNLDRVKLTDFNFLMVLGKGSFGKVM 354 |
| Homo sapiens | Query 295 EEGNVELRQKFEKAKLGPAGNKVISPSEDR#KQPSNNLDRVKLTDFNFLMVLGKGSFGKVM 354 ||||+|||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Sbjct 295 EEGNMELRQKFEKAKLGPAGNKVISPSEDR#KQPSNNLDRVKLTDFNFLMVLGKGSFGKVM 354 |
| Homo sapiens | Query 295 EEGNVELRQKFEKAKLGPAGNKVISPSEDR#KQPSNNLDRVKLTDFNFLMVLGKGSFGKVM 354 ||||+|||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Sbjct 295 EEGNMELRQKFEKAKLGPAGNKVISPSEDR#KQPSNNLDRVKLTDFNFLMVLGKGSFGKVM 354 |
| synthetic construct | Query 295 EEGNVELRQKFEKAKLGPAGNKVISPSEDR#KQPSNNLDRVKLTDFNFLMVLGKGSFGKVM 354 ||||+|||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Sbjct 295 EEGNMELRQKFEKAKLGPAGNKVISPSEDR#KQPSNNLDRVKLTDFNFLMVLGKGSFGKVM 354 |
| Takifugu rubripes | Query 298 NVELRQKFEKAKLGPAGNKVISPSEDR#K--QPSNNLDRVKLTDFNFLMVLGKGSFGKVM 354 |+|||||||||||| | |||+||+ |#+ || |+|||+| ||||| +|||||||||| Sbjct 295 NLELRQKFEKAKLG-QGKKVITPSDHR#RFSLPSGNMDRVRLNDFNFLALLGKGSFGKVM 352 |
| Scyliorhinus canicula | Query 296 EGNVELRQKFEKAKLGPAGNKVIS---PSEDR#KQPSNNLDRVKLTDFNFLMVLGKGSFGK 352 ||| |||||||+|++|| |+| | #| + | ||+|| ||||||||||||||| Sbjct 217 EGNEELRQKFERARIGP-GSKAAEEKRSSSWS#KIDNGNRDRMKLADFNFLMVLGKGSFGK 275 Query 353 VM 354 || Query 353 VM 354 |
| Danio rerio | Query 295 EEGNVELRQKFEKAKLGPAGNKVISPSED---R#KQPSNNLDRVKLTDFNFLMVLGKGSFG 351 |||| || ++||+||+|| | |++ +# + | ||+|| ||||+||||||||| Sbjct 291 EEGNKELSKRFERAKIGPGTNNTDGASKNDLSK#YDANGNQDRMKLADFNFIMVLGKGSFG 350 Query 352 KVM 354 ||| Query 352 KVM 354 |
| Petromyzon marinus | Query 295 EEGNVELRQKFEKAKLGPAGNKVISP--SEDR#KQPS-------NNLDRVKLTDFNFLMVL 345 |+|+ |+|+|||||+|||+ + | |++|#+ | |+|||| |||||||| Sbjct 225 EDGDTEMRRKFEKARLGPS----VKPRASDER#RANSLSAILNNANVDRVKADDFNFLMVL 280 Query 346 GKGSFGKVM 354 ||||||||| Query 346 GKGSFGKVM 354 |
| Tetraodon nigroviridis | Query 301 LRQKFEKAKLGPAGNK--VISPSEDR#KQPSNNLDRVKLTDFNFLMVLGKGSFGKVM 354 ||||||+||+|| | + + |# + | ||+|| ||||||||||||||||| Sbjct 237 LRQKFERAKIGPGKNTDGKSANAASR#FDSNGNQDRMKLADFNFLMVLGKGSFGKVM 292 |
| Rattus norvegicus | Query 296 EGNVELRQKFEKAKLGPAGNKVISPSEDR#KQP------SNNLDRVKLTDFNFLMVLGKGS 349 ||| |||||||+||+| | | +| | # + | ||+||||||||||||||| Sbjct 296 EGNEELRQKFERAKIG-QGTK--APEEKT#ANTISKFDNNGNRDRMKLTDFNFLMVLGKGS 352 Query 350 FGKVM 354 ||||| Query 350 FGKVM 354 |
| Danio rerio | Query 301 LRQKFEKAKLGPAGNKVISPSEDR#KQPS-NNLDRVKLTDFNFLMVLGKGSFGKVM 354 ||||||+||+||+ | + #| | | ||+||+||||||||||||||||| Sbjct 232 LRQKFERAKIGPSKTDGSSSNAIS#KFDSYGNRDRMKLSDFNFLMVLGKGSFGKVM 286 |
| Myxine glutinosa | Query 295 EEGNVELRQKFEKAKLGPAGNKVISPSEDR#KQPSNN-----LDRVKLTDFNFLMVLGKGS 349 ||| +||||+ +++++ + ||++| # |||++ |||| | || || |||||| Sbjct 216 EEG-LELRQRLQRSQIARSDKNKQSPAKD-#-QPSDSASLSGLDRVNLEDFVFLTVLGKGS 272 Query 350 FGKVM 354 ||||| Query 350 FGKVM 354 |
| Branchiostoma lanceolatum | Query 295 EEGNVELRQKFEKAKLGPAGNKVISPSEDR#KQPSN--------NLDRVKLTDFNFLMVLG 346 || +|++ +| || | |#|| | +|| +| ||||||||| Sbjct 225 EENVAKLKEHLQKQKL----------DEQR#KQKSKRSEECNIGSLDHMKAADFNFLMVLG 274 Query 347 KGSFGKVM 354 |||||||| Query 347 KGSFGKVM 354 |
| Bombyx mori | Query 295 EEGNVELRQKFEKAKLGPAGNKVISPSEDR#KQPSN--NLDRVKLTDFNFLMVLGKGSFGK 352 ||| +| | + + | + | ||#+ | | | ++ |||||+|||||||||| Sbjct 295 EEG-ADLVQLKNQMRATTVGARRPPPPPDR#EVPHNVAAADVIRATDFNFIMVLGKGSFGK 353 Query 353 VM 354 || Query 353 VM 354 |
| Apis mellifera | Query 320 PSEDR#KQPSNNLDRVK---LTDFNFLMVLGKGSFGKVM 354 || # |+ || | | +|||||| ||||||||||+ Sbjct 280 PSGRI#TPPATNLPRFKKYNVTDFNFLKVLGKGSFGKVL 317 |
| Drosophila melanogaster | Query 303 QKFEKAKLGPAGNK-VISPSEDR#KQPSNNLDRVKLTDFNFLMVLGKGSFGKVM 354 | | | |+ | ++ |+ # |+ | ++ |||||+ ||||||||||+ Sbjct 304 QDLLKLKQKPSQKKPMVMRSDTN#THTSSKKDMIRATDFNFIKVLGKGSFGKVL 356 |
| Aplysia californica | Query 320 PSEDR#KQPSNNLDRVKLTDFNFLMVLGKGSFGKVM 354 | |+# | + | |+ +||||| ||||||||||+ Sbjct 301 PEPDK#VQNMSKQDIVRASDFNFLTVLGKGSFGKVV 335 |
| Magnaporthe grisea | Query 302 RQKFEKAKLGPAGNKVISPSEDR#KQPS----NNLDRVKLTDFNFLMVLGKGSFGKVM 354 +| |+ + | || | |# || |+ | |||| |||||+||||| Sbjct 816 QQHQEQQIISPTAGTVIPTSARR#PLPSATDPGTGQRIGLDHFNFLAVLGKGNFGKVM 872 |
| Caenorhabditis briggsae AF16 | Query 328 SNNLDRVKLTDFNFLMVLGKGSFGKVM 354 + | | +| +||||| ||||||||||+ Sbjct 404 NTNRDVIKASDFNFLTVLGKGSFGKVL 430 |
| Caenorhabditis elegans | Query 328 SNNLDRVKLTDFNFLMVLGKGSFGKVM 354 |+| + +| +||||| ||||||||||+ Sbjct 337 SSNHNVIKASDFNFLTVLGKGSFGKVL 363 |
| Caenorhabditis elegans | Query 328 SNNLDRVKLTDFNFLMVLGKGSFGKVM 354 |+| + +| +||||| ||||||||||+ Sbjct 337 SSNHNVIKASDFNFLTVLGKGSFGKVL 363 |
| Botryotinia fuckeliana | Query 312 PAGNKVISPSEDR#KQ-PSNNL----DRVKLTDFNFLMVLGKGSFGKVM 354 | +|+ + |#|| |+ | |+ | |||| |||||+||||| Sbjct 813 PGSQQVVPQGQQR#KQVPAANDAGTGRRIGLDHFNFLAVLGKGNFGKVM 860 |
| Schizosaccharomyces pombe | Query 320 PSEDR#KQPSNNLDRVKLTDFNFLMVLGKGSFGKVM 354 | |#++ | || | || || |||||+||||| Sbjct 648 PESPR#REKKN---RVTLDDFTFLAVLGKGNFGKVM 679 |
| Schizosaccharomyces pombe 972h- | Query 320 PSEDR#KQPSNNLDRVKLTDFNFLMVLGKGSFGKVM 354 | |#++ | || | || || |||||+||||| Sbjct 648 PESPR#REKKN---RVTLDDFTFLAVLGKGNFGKVM 679 |
| Drosophila pseudoobscura | Query 332 DRVKLTDFNFLMVLGKGSFGKVM 354 | ++ |||||+ ||||||||||+ Sbjct 221 DMIRATDFNFIKVLGKGSFGKVL 243 |
| Asparagus officinalis | Query 321 SEDR#KQPSN-NLDRVKLTDFNFLMVLGKGSFGKVM 354 +|+ #+ ||+ |++|| | ||+|| |+|+|+|||| Sbjct 106 AEEH#EVPSSENVERVGLEDFDFLKVVGQGAFGKVF 140 |
| Rattus norvegicus | Query 316 KVISPSEDR#KQPSNNLDRVKLTDFNFLMVLGKGSFGKVM 354 ++ || |+ #+ |+ | +|||+ ||||||||||| Sbjct 385 QLASPGENG#EVRQGQAKRLGLDEFNFIKVLGKGSFGKVM 423 |
| Leptosphaeria maculans | Query 308 AKLGPAGNKVISPSEDR#KQPSNNLD----RVKLTDFNFLMVLGKGSFGKVM 354 |++ |+ + |++# || | |+ | |||| |||||+||||| Sbjct 825 AQIPPSKQLPPANPENK#VTPSANTQGSGKRIGLDHFNFLAVLGKGNFGKVM 875 |
| Yarrowia lipolytica | Query 333 RVKLTDFNFLMVLGKGSFGKVM 354 |+ | ||||| |||||+||||| Sbjct 919 RIGLDDFNFLAVLGKGNFGKVM 940 |
| Suberites domuncula | Query 328 SNNLDRVKLTDFNFLMVLGKGSFGKV 353 + ++ ++|| ||| |+|||||||||| Sbjct 332 AKDMPKMKLEDFNLLVVLGKGSFGKV 357 |
| Sycon raphanus | Query 335 KLTDFNFLMVLGKGSFGKVM 354 ||+||+|| ||||||||||| Sbjct 362 KLSDFSFLKVLGKGSFGKVM 381 |
| Calliphora vicina | Query 320 PSEDR#KQPSNNL---DRVKLTDFNFLMVLGKGSFGKVM 354 | | #| +|+ | ++ ||||+ ||||||+||++ Sbjct 333 PRIDN#KDMPHNMSKRDMIRAADFNFIKVLGKGSYGKIL 370 |
| Cryptococcus neoformans var. neoformans JEC21 | Query 333 RVKLTDFNFLMVLGKGSFGKVM 354 +| | ||||| |||||+||||| Sbjct 751 KVGLDDFNFLAVLGKGNFGKVM 772 |
| Cryptococcus neoformans var. neoformans B-3501A | Query 333 RVKLTDFNFLMVLGKGSFGKVM 354 +| | ||||| |||||+||||| Sbjct 754 KVGLDDFNFLAVLGKGNFGKVM 775 |
| Cryptococcus neoformans var. grubii | Query 333 RVKLTDFNFLMVLGKGSFGKVM 354 +| | ||||| |||||+||||| Sbjct 751 KVGLDDFNFLAVLGKGNFGKVM 772 |
| Cryptococcus neoformans var. neoformans | Query 333 RVKLTDFNFLMVLGKGSFGKVM 354 +| | ||||| |||||+||||| Sbjct 751 KVGLDDFNFLAVLGKGNFGKVM 772 |
| synthetic construct | Query 295 EEGNVELRQKFEKAKLGPAGNKVISPSEDR#KQPSNNLDRVKLTDFNFLMVLGKGSFGKVM 354 | + +| + |||+|| | # |||+|| |+||||||||+ Sbjct 134 ERSSQKLHSTSQNINLGPSGNPHAKP----#------------TDFDFLKVIGKGSFGKVL 177 |
| Squalus acanthias | Query 327 PSNNLDRVKLTDFNFLMVLGKGSFGKVM 354 ||+| + | +||||| |+||||||||+ Sbjct 89 PSSN-PQAKPSDFNFLKVIGKGSFGKVL 115 |
| Drosophila melanogaster | Query 333 RVKLTDFNFLMVLGKGSFGKVM 354 + | ||||+ ||||||||||| Sbjct 402 KCSLLDFNFIKVLGKGSFGKVM 423 |
| Sporothrix schenckii | Query 333 RVKLTDFNFLMVLGKGSFGKVM 354 |+ | +|||| |||||+||||| Sbjct 863 RIGLDNFNFLAVLGKGNFGKVM 884 |
| Bos taurus | Query 310 LGPAGNKVISPSEDR#KQPSNNLDRVKLTDFNFLMVLGKGSFGKVM 354 |||+|| | # |||+|| |+||||||||+ Sbjct 143 LGPSGNPHAKP----#------------TDFDFLKVIGKGSFGKVL 171 |
| Cochliobolus heterostrophus | Query 308 AKLGPAGNKVISPSEDR#KQPSNNLDRVKLTDFNFLMVLGKGSFGKVM 354 + +| | ++|+ + #+ |+ | |||| |||||+||||| Sbjct 821 SAVGEVPGKKVTPAANT#QGTGK---RIGLDHFNFLAVLGKGNFGKVM 864 |
| Ancylostoma caninum | Query 334 VKLTDFNFLMVLGKGSFGKV 353 || +|| |+ |||||||||| Sbjct 26 VKESDFTFITVLGKGSFGKV 45 |
* References
None.