SB0056 : Plasma membrane calcium ATPase 2
[ CaMP Format ]
* Basic Information
| Organism | Homo sapiens (human) |
| Protein Names | plasma membrane calcium-transporting ATPase 2 isoform 1 [Homo sapiens]; plasma membrane calcium-transporting ATPase 2 isoform 1; plasma membrane Ca2+ pump 2; plasma membrane calcium pump; plasma membrane calcium-transporting ATPase 2; plasma membrane calcium ATPase; plasma membrane Ca(2+)-ATPase; Plasma membrane calcium-transporting ATPase 2; PMCA2; 3.6.3.8; Plasma membrane calcium ATPase isoform 2; Plasma membrane calcium pump isoform 2 |
| Gene Names | ATP2B2; PMCA2; ATPase, Ca++ transporting, plasma membrane 2 |
| Gene Locus | 3p25.3; chromosome 3 |
| GO Function | Not available |
| Entrez Protein | Entrez Nucleotide | Entrez Gene | UniProt | OMIM | HGNC | HPRD | KEGG |
|---|---|---|---|---|---|---|---|
| NP_001001331 | NM_001001331 | 491 | Q01814 | 108733 | HGNC:815 | 00160 | hsa:491 |
* Information From OMIM
Description: The Ca(2+)-ATPases are a family of plasma membrane pumps encoded by at least 4 genes: ATP2B1 (OMIM:108731) on chromosome 12q21; ATP2B2; ATP2B3 (OMIM:300014) on Xq28; and ATP2B4 (OMIM:108732) on 1q25.
* Structure Information
1. Primary Information
Length: 1243 aa
Average Mass: 136.875 kDa
Monoisotopic Mass: 136.789 kDa
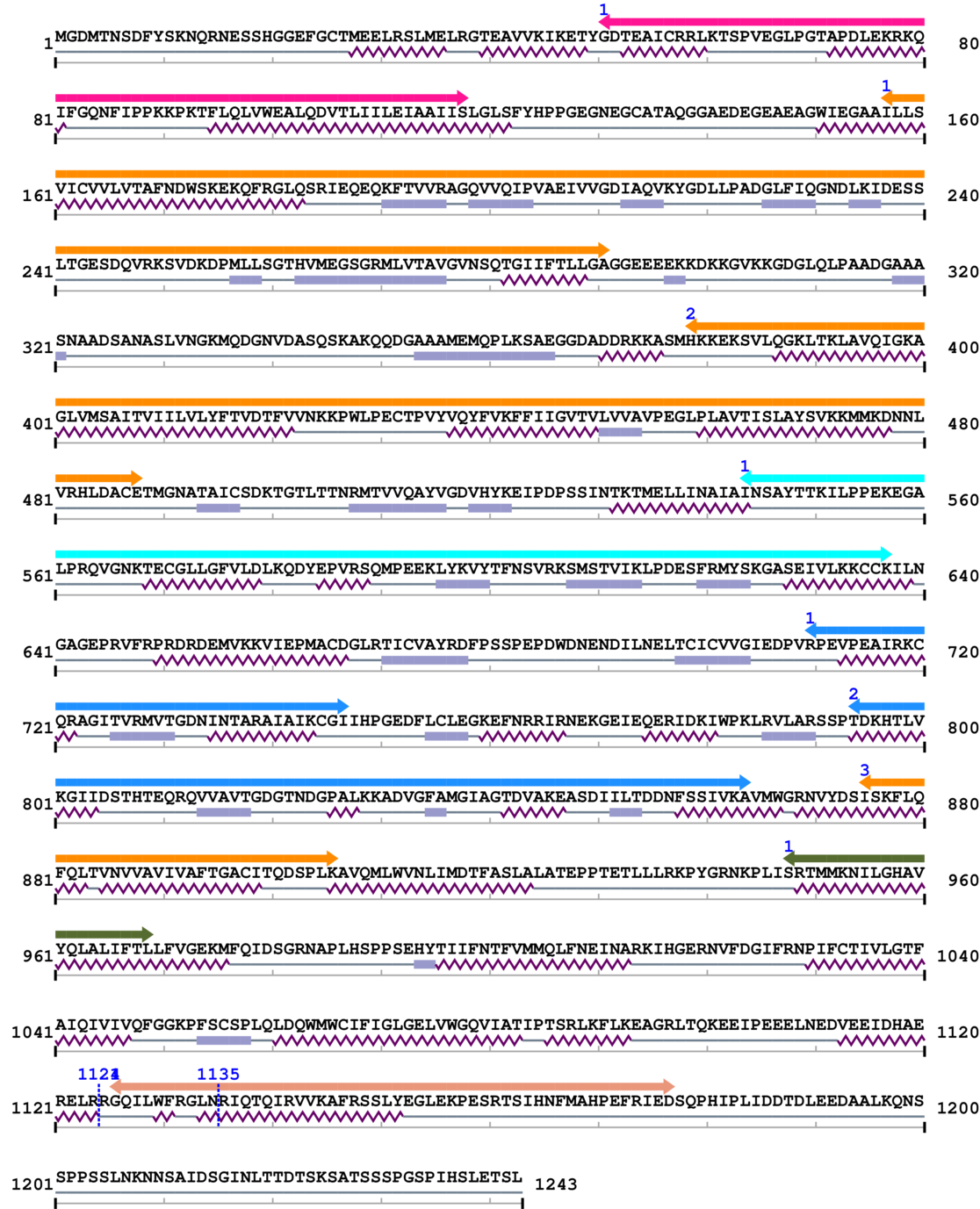
2. Domain Information
Annotated Domains: interpro / pfam / smart / prosite
Computationally Assigned Domains (Pfam+HMMER):
| domain name | begin | end | score | e-value |
|---|---|---|---|---|
| Cation transporter/ATPase, N-terminus 1. | 51 | 118 | 4.0 | 0.0 |
| E1-E2 ATPase 1. | 157 | 291 | 3.0 | 1.0 |
| E1-E2 ATPase 2. | 379 | 488 | 133.0 | 0.7 |
| Cation transport ATPase (P-type) 1. | 544 | 637 | 3.0 | 0.0 |
| haloacid dehalogenase-like hydrolase 1. | 710 | 747 | 17.0 | 0.1 |
| haloacid dehalogenase-like hydrolase 2. | 794 | 864 | 186.0 | 0.5 |
| E1-E2 ATPase 3. | 875 | 906 | 144.0 | 0.4 |
| Integral membrane protein DUF106 1. | 948 | 969 | 88.0 | 0.1 |
| --- cleavage 1124 --- | ||||
| Plasma membrane calcium transporter ATPase C terminal 1. | 1126 | 1177 | 1.0 | 0.1 |
| --- cleavage 1135 (inside Plasma membrane calcium transporter ATPase C terminal 1126..1177) --- | ||||
3. Sequence Information
Fasta Sequence: SB0056.fasta
Amino Acid Sequence and Secondary Structures (PsiPred):
4. 3D Information
Not Available.
* Cleavage Information
2 [sites] cleaved by Calpain 1
Source Reference: [PubMed ID: 2542272] James P, Vorherr T, Krebs J, Morelli A, Castello G, McCormick DJ, Penniston JT, De Flora A, Carafoli E, Modulation of erythrocyte Ca2+-ATPase by selective calpain cleavage of the calmodulin-binding domain. J Biol Chem. 1989 May 15;264(14):8289-96.
Cleavage sites (±10aa)
[Site 1] EIDHAERELR1124-RGQILWFRGL
Arg1124  Arg
Arg
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Glu1115 | Ile1116 | Asp1117 | His1118 | Ala1119 | Glu1120 | Arg1121 | Glu1122 | Leu1123 | Arg1124 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Arg1125 | Gly1126 | Gln1127 | Ile1128 | Leu1129 | Trp1130 | Phe1131 | Arg1132 | Gly1133 | Leu1134 |
 Sequence conservation (by blast)
Sequence conservation (by blast)
[Site 2] GQILWFRGLN1135-RIQTQIRVVK
Asn1135  Arg
Arg
iTraq-117 Signal 8790.4 (

 ) for RIQTQIRVVK
) for RIQTQIRVVK
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Gly1126 | Gln1127 | Ile1128 | Leu1129 | Trp1130 | Phe1131 | Arg1132 | Gly1133 | Leu1134 | Asn1135 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Arg1136 | Ile1137 | Gln1138 | Thr1139 | Gln1140 | Ile1141 | Arg1142 | Val1143 | Val1144 | Lys1145 |
 Sequence conservation (by blast)
Sequence conservation (by blast)
* References
[PubMed ID: 23620727] Yang W, Liu J, Zheng F, Jia M, Zhao L, Lu T, Ruan Y, Zhang J, Yue W, Zhang D, Wang L, The evidence for association of ATP2B2 polymorphisms with autism in Chinese Han population. PLoS One. 2013 Apr 19;8(4):e61021. doi: 10.1371/journal.pone.0061021. Print 2013.