SB0066 : Troponin T2, cardiac, TnTc
[ CaMP Format ]
* Basic Information
| Organism | Mus musculus (house mouse) |
| Protein Names | troponin T, cardiac muscle isoform f [Mus musculus]; troponin T, cardiac muscle isoform f; troponin T, cardiac muscle; tnTc; cardiac muscle troponin T; cardiac TnT; Troponin T, cardiac muscle; TnTc; Cardiac muscle troponin T; cTnT |
| Gene Names | Tnnt2; troponin T2, cardiac |
| Gene Locus | 1 59.32 cM; chromosome 1 |
| GO Function | Not available |
* Information From OMIM
Not Available.
* Structure Information
1. Primary Information
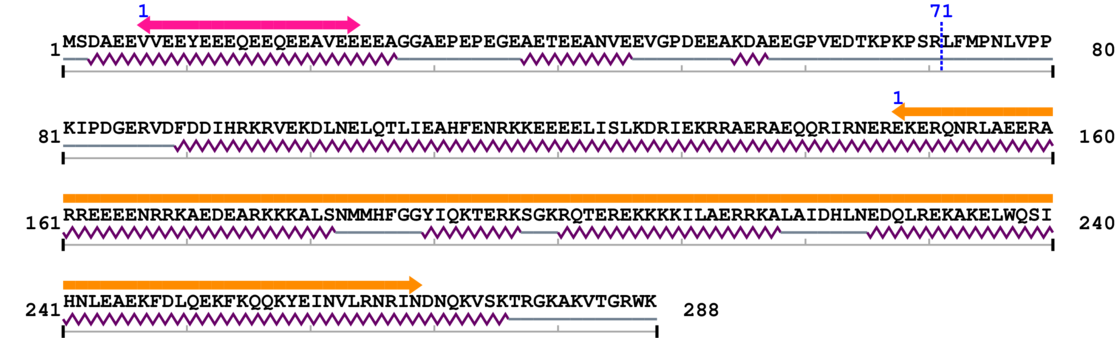
Length: 288 aa
Average Mass: 34.219 kDa
Monoisotopic Mass: 34.199 kDa
2. Domain Information
Annotated Domains: interpro / pfam / smart / prosite
Computationally Assigned Domains (Pfam+HMMER):
| domain name | begin | end | score | e-value |
|---|---|---|---|---|
| Troponin 1. | 7 | 24 | 16.0 | 3.8 |
| --- cleavage 71 --- | ||||
| Protein of unknown function (DUF1800) 1. | 148 | 269 | 58.0 | 7.8 |
3. Sequence Information
Fasta Sequence: SB0066.fasta
Amino Acid Sequence and Secondary Structures (PsiPred):
4. 3D Information
Not Available.
* Cleavage Information
1 [sites] cleaved by Calpain 1
Source Reference: [PubMed ID: 16981728] Zhang Z, Biesiadecki BJ, Jin JP, Selective deletion of the NH2-terminal variable region of cardiac troponin T in ischemia reperfusion by myofibril-associated mu-calpain cleavage. Biochemistry. 2006 Sep 26;45(38):11681-94.
Cleavage sites (±10aa)
[Site 1] VEDTKPKPSR71-LFMPNLVPPK
Arg71  Leu
Leu
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Val62 | Glu63 | Asp64 | Thr65 | Lys66 | Pro67 | Lys68 | Pro69 | Ser70 | Arg71 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Leu72 | Phe73 | Met74 | Pro75 | Asn76 | Leu77 | Val78 | Pro79 | Pro80 | Lys81 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| EANVEEVGPDEEAKDAEEGPVEDTKPKPSRLFMPNLVPPKIPDGERVDFDDIHRKRVEKD |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | Mus musculus | 124.00 | 19 | troponin T2, cardiac |
| 2 | Mus musculus | 124.00 | 6 | cardiac troponin T isoform A2a |
| 3 | Rattus norvegicus | 108.00 | 14 | troponin T gi |
| 4 | Felis catus | 90.90 | 1 | cardiac troponin T |
| 5 | Felis catus | 90.50 | 1 | troponin T type 2, cardiac |
| 6 | Canis familiaris | 88.20 | 6 | troponin T isoform 2 |
| 7 | Canis lupus familiaris | 88.20 | 1 | troponin T type 2, cardiac |
| 8 | Canis familiaris | 88.20 | 1 | cardiac troponin T isoform 1 |
| 9 | Cavia porcellus | 79.00 | 6 | cardiac troponin 2 adult #1 isoform |
| 10 | Homo sapiens | 77.80 | 16 | troponin T; TnT |
| 11 | N/A | 77.40 | 6 | TNNT2_HUMAN Troponin T, cardiac muscle (TnTc) (Ca |
| 12 | Homo sapiens | 77.40 | 3 | truncated cardiac troponin T |
| 13 | Oryctolagus cuniculus | 77.00 | 1 | troponin T cardiac isoform |
| 14 | Bos taurus | 72.40 | 3 | troponin T type 2, cardiac |
| 15 | Pan troglodytes | 72.40 | 1 | PREDICTED: similar to troponin T type 2, cardiac |
| 16 | Gallus gallus | 66.20 | 1 | troponin T type 2, cardiac |
| 17 | Xenopus laevis | 65.90 | 3 | Unknown (protein for MGC:132283) |
| 18 | Meleagris gallopavo | 65.90 | 1 | cardiac troponin T isoform |
| 19 | Meleagris gallopavo | 65.90 | 1 | AF274301_1 low Mr mutant cardiac troponin T |
| 20 | Tetraodon nigroviridis | 65.10 | 4 | unnamed protein product |
| 21 | Danio rerio | 64.70 | 3 | troponin T1, skeletal, slow |
| 22 | Danio rerio | 62.00 | 5 | troponin T2, cardiac |
| 23 | Bos taurus | 60.50 | 8 | troponin T slow skeletal muscle type |
| 24 | Sparus aurata | 60.50 | 3 | slow troponin T 2 |
| 25 | Sus scrofa | 60.50 | 2 | troponin T slow type isoform sTnT2 |
| 26 | synthetic construct | 60.50 | 2 | troponin T1, skeletal, slow |
| 27 | Sus scrofa | 60.50 | 1 | troponin T1, skeletal, slow |
| 28 | Rattus norvegicus | 59.70 | 33 | troponin T1, skeletal, slow |
| 29 | Salmo trutta | 57.40 | 1 | slow myotomal muscle troponin-T isoform 1S |
| 30 | Bufo marinus | 56.20 | 2 | low molecular weight slow troponin T |
| 31 | Gallus gallus | 50.80 | 1 | slow muscle troponin T |
| 32 | human, familial hypertrophic cardiomyopathy, family AW, Peptide Partial Mutant, 25 aa | 49.70 | 1 | troponin T {Ile79Asn} |
| 33 | Halocynthia roretzi | 47.80 | 1 | troponin T |
| 34 | Xenopus laevis | 43.90 | 4 | troponin T3, skeletal, fast |
| 35 | Gadus morhua | 43.50 | 1 | AF500272_1 fast skeletal muscle troponin T |
| 36 | Hyla chrysoscelis | 43.10 | 1 | troponin T |
| 37 | Xenopus tropicalis | 42.40 | 1 | troponin T type 3 (skeletal, fast) |
| 38 | rabbits, skeletal muscle, Peptide, 249 aa | 42.40 | 1 | troponin T beta isoform, TnT beta isoform, TnT-5 |
| 39 | rabbits, skeletal muscle, Peptide Partial, 234 aa | 42.40 | 1 | troponin T alpha isoform, TnT alpha isoform, TnT- |
| 40 | Xenopus tropicalis | 42.40 | 1 | troponin T3, skeletal, fast |
Top-ranked sequences
| organism | matching |
|---|---|
| Mus musculus | Query 42 EANVEEVGPDEEAKDAEEGPVEDTKPKPSR#LFMPNLVPPKIPDGERVDFDDIHRKRVEKD 101 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 42 EANVEEVGPDEEAKDAEEGPVEDTKPKPSR#LFMPNLVPPKIPDGERVDFDDIHRKRVEKD 101 |
| Mus musculus | Query 42 EANVEEVGPDEEAKDAEEGPVEDTKPKPSR#LFMPNLVPPKIPDGERVDFDDIHRKRVEKD 101 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 42 EANVEEVGPDEEAKDAEEGPVEDTKPKPSR#LFMPNLVPPKIPDGERVDFDDIHRKRVEKD 101 |
| Rattus norvegicus | Query 48 VGPDEEAKDAEEGPVEDTKPKPSR#LFMPNLVPPKIPDGERVDFDDIHRKRVEKD 101 |||||||+|||+|||||+||||||#||||||||||||||||||||||||||+||| Sbjct 47 VGPDEEARDAEDGPVEDSKPKPSR#LFMPNLVPPKIPDGERVDFDDIHRKRMEKD 100 |
| Felis catus | Query 43 ANVEEVGPDEEAKDAEEGPVEDTKPKPSR#LFMPNLVPPKIPDGERVDFDDIHRKRVEKD 101 ||| | +||||+ |+||||++|||| |# |||||||||||||||||||||||||+||| Sbjct 50 TNVEGDGQEEEAKETEDGPVEESKPKP-R#PFMPNLVPPKIPDGERVDFDDIHRKRMEKD 107 |
| Felis catus | Query 44 NVEEVGPDEEAKDAEEGPVEDTKPKPSR#LFMPNLVPPKIPDGERVDFDDIHRKRVEKD 101 ||| | +||||+ |+||||++|||| |# |||||||||||||||||||||||||+||| Sbjct 41 NVEGDGQEEEAKETEDGPVEESKPKP-R#PFMPNLVPPKIPDGERVDFDDIHRKRMEKD 97 |
| Canis familiaris | Query 44 NVEEVGPDEEAKDAEEGPVEDTKPKPSR#LFMPNLVPPKIPDGERVDFDDIHRKRVEKD 101 | | | + |||+||+||||++|||| |# |||||||||||||||||||||||||+||| Sbjct 46 NAEGDGQEGEAKEAEDGPVEESKPKP-R#PFMPNLVPPKIPDGERVDFDDIHRKRMEKD 102 |
| Canis lupus familiaris | Query 44 NVEEVGPDEEAKDAEEGPVEDTKPKPSR#LFMPNLVPPKIPDGERVDFDDIHRKRVEKD 101 | | | + |||+||+||||++|||| |# |||||||||||||||||||||||||+||| Sbjct 37 NAEGDGQEGEAKEAEDGPVEESKPKP-R#PFMPNLVPPKIPDGERVDFDDIHRKRMEKD 93 |
| Canis familiaris | Query 44 NVEEVGPDEEAKDAEEGPVEDTKPKPSR#LFMPNLVPPKIPDGERVDFDDIHRKRVEKD 101 | | | + |||+||+||||++|||| |# |||||||||||||||||||||||||+||| Sbjct 42 NAEGDGQEGEAKEAEDGPVEESKPKP-R#PFMPNLVPPKIPDGERVDFDDIHRKRMEKD 98 |
| Cavia porcellus | Query 61 PVEDTKPKPSR#LFMPNLVPPKIPDGERVDFDDIHRKRVEKD 101 |||++|||| |#||||||||||||||||||||||||||+||| Sbjct 60 PVEESKPKP-R#LFMPNLVPPKIPDGERVDFDDIHRKRMEKD 99 |
| Homo sapiens | Query 60 GPVEDTKPKPSR#LFMPNLVPPKIPDGERVDFDDIHRKRVEKD 101 ||+|++|||| |# |||||||||||||||||||||||||+||| Sbjct 58 GPMEESKPKP-R#SFMPNLVPPKIPDGERVDFDDIHRKRMEKD 98 |
| N/A | Query 60 GPVEDTKPKPSR#LFMPNLVPPKIPDGERVDFDDIHRKRVEKD 101 ||+|++|||| |# |||||||||||||||||||||||||+||| Sbjct 68 GPMEESKPKP-R#SFMPNLVPPKIPDGERVDFDDIHRKRMEKD 108 |
| Homo sapiens | Query 60 GPVEDTKPKPSR#LFMPNLVPPKIPDGERVDFDDIHRKRVEKD 101 ||+|++|||| |# |||||||||||||||||||||||||+||| Sbjct 58 GPMEESKPKP-R#SFMPNLVPPKIPDGERVDFDDIHRKRMEKD 98 |
| Oryctolagus cuniculus | Query 60 GPVEDTKPKPSR#LFMPNLVPPKIPDGERVDFDDIHRKRVEKD 101 ||||++|||| |# |||||||||||||||||||||||||+||| Sbjct 78 GPVEESKPKP-R#PFMPNLVPPKIPDGERVDFDDIHRKRMEKD 118 |
| Bos taurus | Query 61 PVEDTKPKPSR#LFMPNLVPPKIPDGERVDFDDIHRKRVEKD 101 |||+ |||| |# |||||||||||||||||||||||||+||| Sbjct 56 PVEEFKPKP-R#PFMPNLVPPKIPDGERVDFDDIHRKRMEKD 95 |
| Pan troglodytes | Query 62 VEDTKPKPSR#LFMPNLVPPKIPDGERVDFDDIHRKRVEKD 101 +|++|||| |# |||||||||||||||||||||||||+||| Sbjct 1 MEESKPKP-R#SFMPNLVPPKIPDGERVDFDDIHRKRMEKD 39 |
| Gallus gallus | Query 56 DAEEGPVE-DTKPKPSR#LFMPNLVPPKIPDGERVDFDDIHRKRVEKD 101 | |+ | | ++|||| +# |||||||||||||||+|||||||||+||| Sbjct 67 DREQEPGEGESKPKP-K#PFMPNLVPPKIPDGERLDFDDIHRKRMEKD 112 |
| Xenopus laevis | Query 65 TKPKPSR#LFMPNLVPPKIPDGERVDFDDIHRKRVEKD 101 +|||| +#||||||+| ||||||+||||||||||+||| Sbjct 44 SKPKP-K#LFMPNLIPLKIPDGEKVDFDDIHRKRMEKD 79 |
| Meleagris gallopavo | Query 64 DTKPKPSR#LFMPNLVPPKIPDGERVDFDDIHRKRVEKD 101 ++|||| +# |||||||||||||||+|||||||||+||| Sbjct 67 ESKPKP-K#PFMPNLVPPKIPDGERLDFDDIHRKRMEKD 103 |
| Meleagris gallopavo | Query 58 EEGPVEDTKPKPSR#LFMPNLVPPKIPDGERVDFDDIHRKRVEKD 101 ||| ++|||| +# |||||||||||||||+|||||||||+||| Sbjct 52 EEG---ESKPKP-K#PFMPNLVPPKIPDGERLDFDDIHRKRMEKD 91 |
| Tetraodon nigroviridis | Query 56 DAEE--GPVEDTKPKPSR#LFMPNLVPPKIPDGERVDFDDIHRKRVEKD 101 |+|| | |+++|+| # ++||+ |||+||||+|||||+||||+||| Sbjct 3 DSEEIKGEHEESRPRPKA#TYVPNMAPPKLPDGEKVDFDDLHRKRLEKD 50 |
| Danio rerio | Query 65 TKPKPSR#LFMPNLVPPKIPDGERVDFDDIHRKRVEKD 101 ||||| +#+|+||++|||+||||+|||||+|||||||| Sbjct 71 TKPKP-K#MFVPNIIPPKLPDGEKVDFDDLHRKRVEKD 106 |
| Danio rerio | Query 65 TKPKPSR#LFM-PNLVPPKIPDGERVDFDDIHRKRVEKD 101 ||| +# || |||||||||||||||||||||||+||| Sbjct 63 AKPKFLK#PFMLPNLVPPKIPDGERVDFDDIHRKRMEKD 100 |
| Bos taurus | Query 66 KPKPSR#LFMPNLVPPKIPDGERVDFDDIHRKRVEKD 101 +|||||# +| |+|||||+|||||||||||||+||| Sbjct 29 RPKPSR#PVVPPLIPPKIPEGERVDFDDIHRKRMEKD 64 |
| Sparus aurata | Query 43 ANVEEVGPDEEAKDAEEGPVEDTKPKPSR#LFMPNLVPPKIPDGERVDFDDIHRKRVEKD 101 ++ ||| +|| +| |+ ++|||| # || |+ |||||||+|||||||||| ||| Sbjct 2 SDTEEV-LEEEVQDGED----ESKPKPK-#-FMTNISAPKIPDGEKVDFDDIHRKRQEKD 53 |
| Sus scrofa | Query 66 KPKPSR#LFMPNLVPPKIPDGERVDFDDIHRKRVEKD 101 +|||||# +| |+|||||+|||||||||||||+||| Sbjct 28 RPKPSR#PVVPPLIPPKIPEGERVDFDDIHRKRMEKD 63 |
| synthetic construct | Query 66 KPKPSR#LFMPNLVPPKIPDGERVDFDDIHRKRVEKD 101 +|||||# +| |+|||||+|||||||||||||+||| Sbjct 39 RPKPSR#PVVPPLIPPKIPEGERVDFDDIHRKRMEKD 74 |
| Sus scrofa | Query 66 KPKPSR#LFMPNLVPPKIPDGERVDFDDIHRKRVEKD 101 +|||||# +| |+|||||+|||||||||||||+||| Sbjct 39 RPKPSR#PVVPPLIPPKIPEGERVDFDDIHRKRMEKD 74 |
| Rattus norvegicus | Query 67 PKPSR#LFMPNLVPPKIPDGERVDFDDIHRKRVEKD 101 |||||# +| |+|||||+|||||||||||||+||| Sbjct 39 PKPSR#PVVPPLIPPKIPEGERVDFDDIHRKRMEKD 73 |
| Salmo trutta | Query 45 VEEVGPDEEAKDAEEGPVEDTKPKPSR#LFMPNLVPPKIPDGE-RVDFDDIHRKRVEKD 101 ||| | ||| | +|+| || +# ||||+ |||+|+|+ +|||||+|||| ||| Sbjct 41 VEET-PAEEA----SGETQDSKAKP-K#SFMPNVAPPKLPEGDGKVDFDDLHRKRQEKD 92 |
| Bufo marinus | Query 73 FMPNLVPPKIPDGERVDFDDIHRKRVEKD 101 || | |||||+|||||||||||||+||| Sbjct 31 MMPRLAPPKIPEGERVDFDDIHRKRMEKD 59 |
| Gallus gallus | Query 76 NLVPPKIPDGERVDFDDIHRKRVEKD 101 | |||||+|||||||||||||+||| Sbjct 53 QLAPPKIPEGERVDFDDIHRKRMEKD 78 |
| human, familial hypertrophic cardiomyopathy, family AW, Peptide Partial Mutant, 25 aa | Query 74 MPNLVPPKIPDGERVDFDDIHRKRV 98 |||||||| |||||||||||||||+ Sbjct 1 MPNLVPPKNPDGERVDFDDIHRKRM 25 |
| Halocynthia roretzi | Query 71 R#LFMPNLVPPKIPDGERVDFDDIHRKRVEK 100 +# |||+ |||||||| +| +||||||++| Sbjct 9 K#FTMPNITPPKIPDGEVIDLEDIHRKRMDK 38 |
| Xenopus laevis | Query 75 PNLVPPKIPDGERVDFDDIHRKRVEKD 101 | | |||||||+|||||| +|| || Sbjct 60 PKLTAPKIPDGEKVDFDDIQKKRQNKD 86 |
| Gadus morhua | Query 61 PVEDTKPKPSR#LFMPNLVPPKIPDGERVDFDDIHRKRVEKD 101 ||| ||| # | |+ |||||||+|||||| +|| || Sbjct 9 QVEDEKPK---#-FKPS--APKIPDGEKVDFDDIQKKRQNKD 43 |
| Hyla chrysoscelis | Query 75 PNLVPPKIPDGERVDFDDIHRKRVEKD 101 | + |||||||+|||||| +|| || Sbjct 34 PKITAPKIPDGEKVDFDDIQKKRQNKD 60 |
| Xenopus tropicalis | Query 75 PNLVPPKIPDGERVDFDDIHRKRVEKD 101 | | ||||+||+|||||| +|| || Sbjct 28 PKLTAPKIPEGEKVDFDDIQKKRQNKD 54 |
| rabbits, skeletal muscle, Peptide, 249 aa | Query 75 PNLVPPKIPDGERVDFDDIHRKRVEKD 101 | | ||||+||+|||||| +|| || Sbjct 32 PKLTAPKIPEGEKVDFDDIQKKRQNKD 58 |
| rabbits, skeletal muscle, Peptide Partial, 234 aa | Query 75 PNLVPPKIPDGERVDFDDIHRKRVEKD 101 | | ||||+||+|||||| +|| || Sbjct 17 PKLTAPKIPEGEKVDFDDIQKKRQNKD 43 |
| Xenopus tropicalis | Query 75 PNLVPPKIPDGERVDFDDIHRKRVEKD 101 | | ||||+||+|||||| +|| || Sbjct 54 PKLTAPKIPEGEKVDFDDIQKKRQNKD 80 |
* References
[PubMed ID: 23897817] Kobayashi M, Debold EP, Turner MA, Kobayashi T, Cardiac muscle activation blunted by a mutation to the regulatory component, troponin T. J Biol Chem. 2013 Sep 6;288(36):26335-49. doi: 10.1074/jbc.M113.494096. Epub 2013
[PubMed ID: 23748972] ... Mamidi R, Michael JJ, Muthuchamy M, Chandra M, Interplay between the overlapping ends of tropomyosin and the N terminus of cardiac troponin T affects tropomyosin states on actin. FASEB J. 2013 Sep;27(9):3848-59. doi: 10.1096/fj.13-232363. Epub 2013 Jun 7.
[PubMed ID: 23987512] ... Park JH, Kang HJ, Kang SI, Lee JE, Hur J, Ge K, Mueller E, Li H, Lee BC, Lee SB, A multifunctional protein, EWS, is essential for early brown fat lineage determination. Dev Cell. 2013 Aug 26;26(4):393-404. doi: 10.1016/j.devcel.2013.07.002.
[PubMed ID: 23595884] ... Rog-Zielinska EA, Thomson A, Kenyon CJ, Brownstein DG, Moran CM, Szumska D, Michailidou Z, Richardson J, Owen E, Watt A, Morrison H, Forrester LM, Bhattacharya S, Holmes MC, Chapman KE, Glucocorticoid receptor is required for foetal heart maturation. Hum Mol Genet. 2013 Aug 15;22(16):3269-82. doi: 10.1093/hmg/ddt182. Epub 2013 Apr
[PubMed ID: 23743334] ... Liang X, Wang G, Lin L, Lowe J, Zhang Q, Bu L, Chen Y, Chen J, Sun Y, Evans SM, HCN4 dynamically marks the first heart field and conduction system precursors. Circ Res. 2013 Aug 2;113(4):399-407. doi: 10.1161/CIRCRESAHA.113.301588. Epub
[PubMed ID: 10021346] ... Wang DZ, Reiter RS, Lin JL, Wang Q, Williams HS, Krob SL, Schultheiss TM, Evans S, Lin JJ, Requirement of a novel gene, Xin, in cardiac morphogenesis. Development. 1999 Mar;126(6):1281-94.
[PubMed ID: 9637714] ... Tardiff JC, Factor SM, Tompkins BD, Hewett TE, Palmer BM, Moore RL, Schwartz S, Robbins J, Leinwand LA, A truncated cardiac troponin T molecule in transgenic mice suggests multiple cellular mechanisms for familial hypertrophic cardiomyopathy. J Clin Invest. 1998 Jun 15;101(12):2800-11.
[PubMed ID: 8654947] ... Jin JP, Wang J, Zhang J, Expression of cDNAs encoding mouse cardiac troponin T isoforms: characterization of a large sample of independent clones. Gene. 1996 Feb 12;168(2):217-21.
[PubMed ID: 7575526] ... Jin JP, Zhang J, Wang J, An embryonic alternative transcription initiation site and the 5'-upstream structure of mouse cardiac troponin T gene. Biochem Biophys Res Commun. 1995 Sep 25;214(3):1168-74.
[PubMed ID: 7958896] ... Chen Z, Friedrich GA, Soriano P, Transcriptional enhancer factor 1 disruption by a retroviral gene trap leads to heart defects and embryonic lethality in mice. Genes Dev. 1994 Oct 1;8(19):2293-301.