SB0067 : GAP-43, Growth associated protein 43
[ CaMP Format ]
* Basic Information
| Organism | Rattus norvegicus (Norway rat) |
| Protein Names | neuromodulin [Rattus norvegicus]; neuromodulin; brain abundant, membrane attached signal protein 2; protein F1; growth-associated protein 43; axonal membrane protein GAP-43; growth accentuating protein 43; Neuromodulin; Axonal membrane protein GAP-43; Growth-associated protein 43; Protein F1 |
| Gene Names | Gap43; growth associated protein 43 |
| Gene Locus | 11q21; chromosome 11 |
| GO Function | Not available |
* Information From OMIM
Not Available.
* Structure Information
1. Primary Information
Length: 226 aa
Average Mass: 23.603 kDa
Monoisotopic Mass: 23.589 kDa
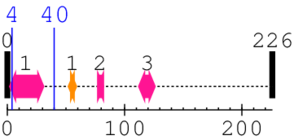
2. Domain Information
Annotated Domains: interpro / pfam / smart / prosite
Computationally Assigned Domains (Pfam+HMMER):
| domain name | begin | end | score | e-value |
|---|---|---|---|---|
| Gap junction protein N-terminal region 1. | 2 | 31 | 1.0 | 6.1 |
| --- cleavage 4 (inside Gap junction protein N-terminal region 2..31) --- | ||||
| --- cleavage 40 --- | ||||
| Neuromodulin 1. | 52 | 59 | 35.0 | 3.3 |
| Gap junction protein N-terminal region 2. | 77 | 82 | 9.0 | 0.3 |
| Gap junction protein N-terminal region 3. | 112 | 126 | 17.0 | 0.7 |
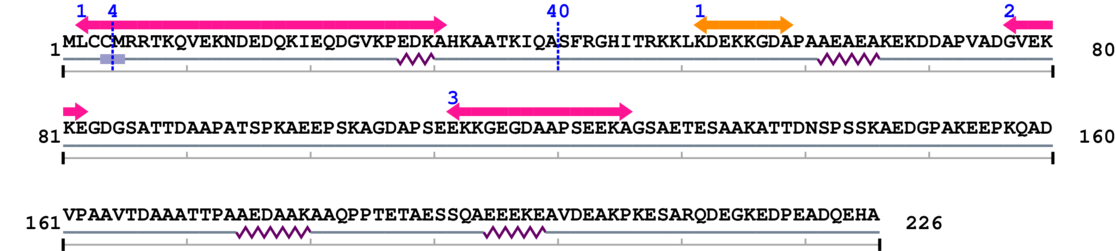
3. Sequence Information
Fasta Sequence: SB0067.fasta
Amino Acid Sequence and Secondary Structures (PsiPred):
4. 3D Information
Not Available.
* Cleavage Information
2 [sites] cleaved by Calpain 2
Source Reference: [PubMed ID: 11274743] Zakharov VV, Mosevitsky MI, Site-specific calcium-dependent proteolysis of neuronal protein GAP-43. Neurosci Res. 2001 Apr;39(4):447-53.
Cleavage sites (±10aa)
[Site 1] MLCC4-MRRTKQVEKN
Cys4  Met
Met
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| - | - | - | - | - | - | Met1 | Leu2 | Cys3 | Cys4 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Met5 | Arg6 | Arg7 | Thr8 | Lys9 | Gln10 | Val11 | Glu12 | Lys13 | Asn14 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| MLCCMRRTKQVEKNDEDQKIEQDGVKPEDKAHKA |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | Mus musculus | 76.30 | 1 | growth associated protein 43 |
| 2 | Macaca mulatta | 76.30 | 1 | PREDICTED: growth associated protein 43 isoform 2 |
| 3 | Rattus norvegicus | 76.30 | 1 | growth associated protein 43 |
| 4 | Bos taurus | 76.30 | 1 | growth associated protein 43 |
| 5 | Felis catus | 75.90 | 1 | growth associated protein 43 |
| 6 | synthetic construct | 73.20 | 2 | growth associated protein 43 |
| 7 | Homo sapiens | 73.20 | 1 | growth associated protein 43 |
| 8 | Lonchura striata | 67.80 | 1 | growth-associated protein 43 |
| 9 | N/A | 67.40 | 3 | NEUM_CHICK Neuromodulin (Axonal membrane protein |
| 10 | Xenopus laevis | 63.50 | 2 | growth associated protein 43 |
| 11 | Danio rerio | 56.60 | 1 | growth associated protein 43 |
| 12 | Tetraodon nigroviridis | 55.50 | 1 | unnamed protein product |
| 13 | Serinus canaria | 48.90 | 1 | NEUM_SERCA Neuromodulin (Axonal membrane protein |
| 14 | synthetic construct | 46.20 | 1 | LIBRA |
| 15 | Anguilla anguilla | 33.90 | 1 | growth-associated protein 43 |
Top-ranked sequences
| organism | matching |
|---|---|
| Mus musculus | Query 1 MLCC#MRRTKQVEKNDEDQKIEQDGVKPEDKAHKA 34 ||||#|||||||||||||||||||||||||||||| Query 1 MLCC#MRRTKQVEKNDEDQKIEQDGVKPEDKAHKA 34 |
| Macaca mulatta | Query 1 MLCC#MRRTKQVEKNDEDQKIEQDGVKPEDKAHKA 34 ||||#||||||||||||||||||||+||||||||| Sbjct 1 MLCC#MRRTKQVEKNDEDQKIEQDGIKPEDKAHKA 34 |
| Rattus norvegicus | Query 1 MLCC#MRRTKQVEKNDEDQKIEQDGVKPEDKAHKA 34 ||||#|||||||||||||||||||||||||||||| Query 1 MLCC#MRRTKQVEKNDEDQKIEQDGVKPEDKAHKA 34 |
| Bos taurus | Query 1 MLCC#MRRTKQVEKNDEDQKIEQDGVKPEDKAHKA 34 ||||#||||||||||||||||||||+||||||||| Sbjct 1 MLCC#MRRTKQVEKNDEDQKIEQDGIKPEDKAHKA 34 |
| Felis catus | Query 1 MLCC#MRRTKQVEKNDEDQKIEQDGVKPEDKAHKA 34 ||||#||||||||||||||||||||+||||||||| Sbjct 1 MLCC#MRRTKQVEKNDEDQKIEQDGIKPEDKAHKA 34 |
| synthetic construct | Query 1 MLCC#MRRTKQVEKNDEDQKIEQDGVKPEDKAHKA 34 ||||#|||||||||||+||||||||+||||||||| Sbjct 1 MLCC#MRRTKQVEKNDDDQKIEQDGIKPEDKAHKA 34 |
| Homo sapiens | Query 1 MLCC#MRRTKQVEKNDEDQKIEQDGVKPEDKAHKA 34 ||||#|||||||||||+||||||||+||||||||| Sbjct 1 MLCC#MRRTKQVEKNDDDQKIEQDGIKPEDKAHKA 34 |
| Lonchura striata | Query 1 MLCC#MRRTKQVEKN-DEDQKIEQDGVKPEDKAHKA 34 ||||#|||||||||| | ||||||||+||||||||| Sbjct 1 MLCC#MRRTKQVEKNEDGDQKIEQDGIKPEDKAHKA 35 |
| N/A | Query 1 MLCC#MRRTKQVEKN-DEDQKIEQDGVKPEDKAHKA 34 ||||#|||||||||| | ||||||||+||||||||| Sbjct 1 MLCC#MRRTKQVEKNEDGDQKIEQDGIKPEDKAHKA 35 |
| Xenopus laevis | Query 1 MLCC#MRRTKQVEKN-DEDQKIEQDGVKPEDKAHKA 34 ||||#|||||||||| | ||||+||| ||||||||| Sbjct 1 MLCC#MRRTKQVEKNEDGDQKIDQDGNKPEDKAHKA 35 |
| Danio rerio | Query 1 MLCC#MRRTKQVEKNDE-DQKIEQDGVKPEDKAHKA 34 ||||#+|||| ||||+| ||+|+|||+|||+ |||| Sbjct 1 MLCC#IRRTKPVEKNEEADQEIKQDGIKPEENAHKA 35 |
| Tetraodon nigroviridis | Query 1 MLCC#MRRTKQVEKNDE-DQKIEQDGV--KPEDKAHKA 34 ||||#+|||| ||||+| +||+|||| ||||||||| Sbjct 1 MLCC#IRRTKPVEKNEEAEQKVEQDGTTNKPEDKAHKA 37 |
| Serinus canaria | Query 8 TKQVEKN-DEDQKIEQDGVKPEDKAHKA 34 ||||||| | ||||||||+||||||||| Sbjct 1 TKQVEKNEDGDQKIEQDGIKPEDKAHKA 28 |
| synthetic construct | Query 1 MLCC#MRRTKQVEKNDEDQKI 20 ||||#|||||||||||||||| Query 1 MLCC#MRRTKQVEKNDEDQKI 20 |
| Anguilla anguilla | Query 7 RTKQVEKN-DEDQKIEQDGVKPEDKA 31 ||| |||| | | ||||| |||||| Sbjct 1 RTKPVEKNEDADLKIEQDANKPEDKA 26 |
[Site 2] AHKAATKIQA40-SFRGHITRKK
Ala40  Ser
Ser
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Ala31 | His32 | Lys33 | Ala34 | Ala35 | Thr36 | Lys37 | Ile38 | Gln39 | Ala40 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Ser41 | Phe42 | Arg43 | Gly44 | His45 | Ile46 | Thr47 | Arg48 | Lys49 | Lys50 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| VEKNDEDQKIEQDGVKPEDKAHKAATKIQASFRGHITRKKLKDEKKGDAPAAEAEAKEKD |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | Rattus norvegicus | 118.00 | 2 | growth associated protein 43 |
| 2 | Mus musculus | 115.00 | 1 | growth associated protein 43 |
| 3 | Felis catus | 107.00 | 1 | growth associated protein 43 |
| 4 | Homo sapiens | 99.80 | 2 | growth associated protein 43 |
| 5 | synthetic construct | 99.80 | 2 | growth associated protein 43 |
| 6 | Serinus canaria | 98.20 | 1 | NEUM_SERCA Neuromodulin (Axonal membrane protein |
| 7 | Lonchura striata | 98.20 | 1 | growth-associated protein 43 |
| 8 | N/A | 97.80 | 3 | NEUM_CHICK Neuromodulin (Axonal membrane protein |
| 9 | Bos taurus | 95.90 | 2 | growth associated protein 43 |
| 10 | Macaca mulatta | 90.90 | 2 | PREDICTED: growth associated protein 43 isoform 2 |
| 11 | Tetraodon nigroviridis | 75.90 | 3 | unnamed protein product |
| 12 | Danio rerio | 75.50 | 1 | growth associated protein 43 |
| 13 | Xenopus laevis | 67.40 | 2 | growth associated protein 43 |
| 14 | Felis catus | 65.50 | 1 | growth associated protein 43 |
| 15 | Danio rerio | 43.10 | 2 | PREDICTED: similar to neurogranin |
| 16 | Gillichthys mirabilis | 40.80 | 1 | AF419164_1 neurogranin |
| 17 | Canis familiaris | 37.00 | 2 | PREDICTED: similar to Sperm surface protein Sp17 |
| 18 | Serinus canaria | 35.40 | 1 | NEUG_SERCA Neurogranin (NG) (Canarigranin) gi |
| 19 | Apis mellifera | 35.00 | 1 | PREDICTED: similar to igloo CG18285-PA, isoform A |
| 20 | Xenopus tropicalis | 33.90 | 1 | sperm autoantigenic protein 17 |
| 21 | Bos taurus | 33.50 | 2 | sperm autoantigenic protein 17 |
| 22 | Mus musculus | 33.10 | 3 | unnamed protein product |
| 23 | Schistosoma japonicum | 33.10 | 1 | SJCHGC01496 protein |
| 24 | Homo sapiens | 32.30 | 5 | neurogranin |
| 25 | Drosophila pseudoobscura | 32.30 | 1 | GA14874-PA |
| 26 | Oryctolagus cuniculus | 32.30 | 1 | SP17 |
| 27 | Callithrix jacchus | 32.30 | 1 | AF134585_1 sperm protein Sp17 |
| 28 | Drosophila melanogaster | 32.00 | 2 | CG13544 CG13544-PA |
| 29 | Drosophila melanogaster | 32.00 | 2 | igloo CG18285-PA, isoform A |
| 30 | Cryptosporidium parvum Iowa II | 32.00 | 1 | myosin'myosin' |
| 31 | Ovis aries | 32.00 | 1 | SP17 protein |
| 32 | Cryptosporidium parvum | 32.00 | 1 | WD40 repeat myosin-like protein |
| 33 | Papio hamadryas | 31.60 | 1 | SP17_PAPHA Sperm surface protein Sp17 (Sperm auto |
| 34 | Macaca mulatta | 31.60 | 1 | AF334809_1 sperm protein 17 |
| 35 | Morone saxatilis | 31.20 | 1 | AF003249_1 myosin heavy chain FM3A |
| 36 | Triticum monococcum | 30.40 | 1 | putative calmodulin-binding protein |
| 37 | Arabidopsis thaliana | 30.40 | 1 | IQD20 (IQ-domain 20); calmodulin binding |
| 38 | Caenorhabditis briggsae AF16 | 30.00 | 1 | Hypothetical protein CBG08105 |
Top-ranked sequences
| organism | matching |
|---|---|
| Rattus norvegicus | Query 11 VEKNDEDQKIEQDGVKPEDKAHKAATKIQA#SFRGHITRKKLKDEKKGDAPAAEAEAKEKD 70 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 11 VEKNDEDQKIEQDGVKPEDKAHKAATKIQA#SFRGHITRKKLKDEKKGDAPAAEAEAKEKD 70 |
| Mus musculus | Query 11 VEKNDEDQKIEQDGVKPEDKAHKAATKIQA#SFRGHITRKKLKDEKKGDAPAAEAEAKEKD 70 ||||||||||||||||||||||||||||||#|||||||||||| ||||||||||||||||| Sbjct 11 VEKNDEDQKIEQDGVKPEDKAHKAATKIQA#SFRGHITRKKLKGEKKGDAPAAEAEAKEKD 70 |
| Felis catus | Query 11 VEKNDEDQKIEQDGVKPEDKAHKAATKIQA#SFRGHITRKKLKDEKKGDAPAAEAEAKEKD 70 ||||||||||||||+|||||||||||||||#|||||||||||| |||||| ||||| ||| Sbjct 11 VEKNDEDQKIEQDGIKPEDKAHKAATKIQA#SFRGHITRKKLKGEKKGDAQAAEAEGNEKD 70 |
| Homo sapiens | Query 11 VEKNDEDQKIEQDGVKPEDKAHKAATKIQA#SFRGHITRKKLKDEKKGDAPAAEAEAKEKD 70 |||||+||||||||+|||||||||||||||#|||||||||||| ||| | |||||| +|| Sbjct 11 VEKNDDDQKIEQDGIKPEDKAHKAATKIQA#SFRGHITRKKLKGEKKDDVQAAEAEANKKD 70 |
| synthetic construct | Query 11 VEKNDEDQKIEQDGVKPEDKAHKAATKIQA#SFRGHITRKKLKDEKKGDAPAAEAEAKEKD 70 |||||+||||||||+|||||||||||||||#|||||||||||| ||| | |||||| +|| Sbjct 11 VEKNDDDQKIEQDGIKPEDKAHKAATKIQA#SFRGHITRKKLKGEKKDDVQAAEAEANKKD 70 |
| Serinus canaria | Query 11 VEKN-DEDQKIEQDGVKPEDKAHKAATKIQA#SFRGHITRKKLKDEKKGDAPAAEAEAKEK 69 |||| | ||||||||+|||||||||||||||#|||||||||||| ||||||||+| +| +| Sbjct 4 VEKNEDGDQKIEQDGIKPEDKAHKAATKIQA#SFRGHITRKKLKGEKKGDAPASETDAADK 63 |
| Lonchura striata | Query 11 VEKN-DEDQKIEQDGVKPEDKAHKAATKIQA#SFRGHITRKKLKDEKKGDAPAAEAEAKEK 69 |||| | ||||||||+|||||||||||||||#|||||||||||| ||||||||+| +| +| Sbjct 11 VEKNEDGDQKIEQDGIKPEDKAHKAATKIQA#SFRGHITRKKLKGEKKGDAPASETDAADK 70 |
| N/A | Query 11 VEKN-DEDQKIEQDGVKPEDKAHKAATKIQA#SFRGHITRKKLKDEKKGDAPAAEAEAKEK 69 |||| | ||||||||+|||||||||||||||#|||||||||||| ||| ||||+|+|| +| Sbjct 11 VEKNEDGDQKIEQDGIKPEDKAHKAATKIQA#SFRGHITRKKLKGEKKADAPASESEAADK 70 |
| Bos taurus | Query 11 VEKNDEDQKIEQDGVKPEDKAHKAATKIQA#SFRGHITRKKLKDEKKGD 58 ||||||||||||||+|||||||||||||||#|||||||||||| ||||| Sbjct 11 VEKNDEDQKIEQDGIKPEDKAHKAATKIQA#SFRGHITRKKLKGEKKGD 58 |
| Macaca mulatta | Query 11 VEKNDEDQKIEQDGVKPEDKAHKAATKIQA#SFRGHITRKKLKDEKK 56 ||||||||||||||+|||||||||||||||#|||||||||||| ||| Sbjct 11 VEKNDEDQKIEQDGIKPEDKAHKAATKIQA#SFRGHITRKKLKGEKK 56 |
| Tetraodon nigroviridis | Query 11 VEKNDE-DQKIEQDGV--KPEDKAHKAATKIQA#SFRGHITRKKLKD---EKKGDAP 60 ||||+| +||+|||| |||||||||||||||#||||||||||+|| +| ||+| Sbjct 11 VEKNEEAEQKVEQDGTTNKPEDKAHKAATKIQA#SFRGHITRKKMKDGEKDKDGDSP 66 |
| Danio rerio | Query 11 VEKNDE-DQKIEQDGVKPEDKAHKAATKIQA#SFRGHITRKKLKD 53 ||||+| ||+|+|||+|||+ ||||||||||#||||||||||+|| Sbjct 11 VEKNEEADQEIKQDGIKPEENAHKAATKIQA#SFRGHITRKKMKD 54 |
| Xenopus laevis | Query 11 VEKN-DEDQKIEQDGVKPEDKAHKAATKIQA#SFRGHITR 48 |||| | ||||+||| |||||||||||||||#|||||| | Sbjct 11 VEKNEDGDQKIDQDGNKPEDKAHKAATKIQA#SFRGHIIR 49 |
| Felis catus | Query 33 KAATKIQA#SFRGHITRKKLKDEKKGDAPAAEAEAKEKD 70 ||||||||#|||||||||||| |||||| ||||| ||| Sbjct 1 KAATKIQA#SFRGHITRKKLKGEKKGDAQAAEAEGNEKD 38 |
| Danio rerio | Query 31 AHKAATKIQA#SFRGHITRKKLKDEK 55 |+||| ||||# ||||+||||+||+| Sbjct 26 ANKAAAKIQA#GFRGHMTRKKMKDDK 50 |
| Gillichthys mirabilis | Query 31 AHKAATKIQA#SFRGHITRKKLKDEKKGDA 59 |++|| ||||# ||||+||||+| | | + Sbjct 28 ANRAAAKIQA#GFRGHMTRKKMKPEDKSEG 56 |
| Canis familiaris | Query 34 AATKIQA#SFRGHITRKKLKDEKKGDA 59 || ||||#+||||+ |+++| | ||| Sbjct 117 AALKIQA#AFRGHLAREEVKKMKSGDA 142 |
| Serinus canaria | Query 37 KIQA#SFRGHITRKKLK----DEKKGDAPAAEA 64 ||||#|||||+||||+| | | || | + Sbjct 32 KIQA#SFRGHMTRKKIKGGEIDRKTKDAECANS 63 |
| Apis mellifera | Query 32 HKAATKIQA#SFRGHITRKKL 51 |||||||||#||||| |+++ Sbjct 97 HKAATKIQA#SFRGHKVRQEV 116 Query 34 AATKIQA#SFRGHITRK 49 |||||||#|||||++|| Sbjct 38 AATKIQA#SFRGHMSRK 53 |
| Xenopus tropicalis | Query 37 KIQA#SFRGHITRKKL---------KDEKKGDAPAAEAEA 66 ||||# ||||+||||+ || | | + | | Sbjct 456 KIQA#GFRGHMTRKKMKGGDKDLKSKDSKDGSSTGGENEG 494 |
| Bos taurus | Query 29 DKAHKAATKIQA#SFRGHITRKKLKDEKKGD 58 | || ||||#+||||+ |+++| | | Sbjct 108 DMEENAALKIQA#AFRGHLAREEVKKMKSSD 137 |
| Mus musculus | Query 34 AATKIQA#SFRGHITRKKLKDEKKGDA 59 ||| +||#+||||+ | || | |+ Sbjct 511 AATVLQA#AFRGHLARSKLVRSKVPDS 536 |
| Schistosoma japonicum | Query 34 AATKIQA#SFRGHITRKKLKDE 54 |||+|||#|+||++||| |+|+ Sbjct 99 AATRIQA#SYRGYLTRKALRDD 119 Query 29 DKAHKAATKIQA#SFRGHITRKK 50 || |||||||# +||+ |||| Sbjct 38 DKESLAATKIQA#GYRGYCTRKK 59 |
| Homo sapiens | Query 37 KIQA#SFRGHITRKKLKD 53 ||||#|||||+ |||+| Sbjct 32 KIQA#SFRGHMARKKIKS 48 |
| Drosophila pseudoobscura | Query 18 QKIEQDGVKPEDKAHKAATKIQA#SFRGHITRKKLKDE 54 ++++ | || +||||||||#||||| ||+ + | Sbjct 206 EELDIDLTDPE--LNKAATKIQA#SFRGHKTRRDTQPE 240 Query 28 EDKAHKAATKIQA#SFRGHITRKKLK 52 ||+| ||||||||# |||| |+ +| Sbjct 99 EDEA-KAATKIQA#VFRGHKVRETMK 122 |
| Oryctolagus cuniculus | Query 29 DKAHKAATKIQA#SFRGHITRKKLK 52 || || ||||#+||||+ |+ +| Sbjct 108 DKEEMAALKIQA#AFRGHLAREDVK 131 |
| Callithrix jacchus | Query 34 AATKIQA#SFRGHITRKKLK 52 || ||||#+||||| |+++| Sbjct 175 AAVKIQA#AFRGHIAREEVK 193 |
| Drosophila melanogaster | Query 28 EDKAHKAATKIQA#SFRGHITRKKLKDEKK 56 +| |+|||||||# ||| +|+ + | | Sbjct 113 QDHHHRAATKIQA#LFRGWKSRRTIHDHSK 141 |
| Drosophila melanogaster | Query 18 QKIEQDGVKPEDKAHKAATKIQA#SFRGHITRK 49 ++++ | || +||||||||#||||| ||| Sbjct 206 EELDIDLTDPE--LNKAATKIQA#SFRGHKTRK 235 Query 28 EDKAHKAATKIQA#SFRGHITRKKLK 52 ||+| ||||||||# |||| |+ +| Sbjct 97 EDEA-KAATKIQA#VFRGHKVRETMK 120 |
| Cryptosporidium parvum Iowa II | Query 33 KAATKIQA#SFRGHITRKKLK 52 ||| ||| # ||||+ |+||| Sbjct 980 KAALKIQT#IFRGHVARQKLK 999 |
| Ovis aries | Query 29 DKAHKAATKIQA#SFRGHITRKKLKDEKKGD 58 | || ||||#+||||+ |+++| | | Sbjct 108 DMEENAALKIQA#AFRGHLAREEVKKMKSID 137 |
| Cryptosporidium parvum | Query 33 KAATKIQA#SFRGHITRKKLK 52 ||| ||| # ||||+ |+||| Sbjct 880 KAALKIQT#IFRGHVARQKLK 899 |
| Papio hamadryas | Query 34 AATKIQA#SFRGHITRKKLK 52 || ||||#+||||+ |+++| Sbjct 117 AAVKIQA#AFRGHVAREEVK 135 |
| Macaca mulatta | Query 34 AATKIQA#SFRGHITRKKLK 52 || ||||#+||||+ |+++| Sbjct 111 AAVKIQA#AFRGHVAREEVK 129 |
| Morone saxatilis | Query 33 KAATKIQA#SFRGHITRKKLKDEKKGDAPAAEAEAK 67 |||| +|+#+|||| ||+|++| | || + +|| Sbjct 1377 KAATVLQS#NFRGHKERKRLQEE--GKIPAKKQKAK 1409 Query 33 KAATKIQA#SFRGHITRKKLKDEK 55 |||| +|+#+|||| ||||| |+ Sbjct 1527 KAATVLQS#NFRGHRDRKKLKAER 1549 Query 29 DKAHKAATKIQA#SFRGHITRKKLK 52 |+ +||||||||# +||| || | Sbjct 1172 DEKNKAATKIQA#RYRGHKERKSFK 1195 |
| Triticum monococcum | Query 26 KPEDKAHKAATKIQA#SFRGHITRKKLKD 53 +|+ + ||||+| # |+|| ||+ | | Sbjct 57 RPQAELDNAATKVQK#LFKGHRTRRNLAD 84 |
| Arabidopsis thaliana | Query 34 AATKIQA#SFRGHITRKKLKDEK 55 || ||||# ||||+ |+ | | Sbjct 39 AAVKIQA#FFRGHLARRAFKALK 60 |
| Caenorhabditis briggsae AF16 | Query 20 IEQDGVKPEDKAHKAATKIQA#SFRGHITRKKLKDEKKGDAPAA 62 || | |||+| |||||+# || +||| + || | || Sbjct 135 IEMD--TPEDRA---ATKIQS#EIRGFLTRKHVDKMKKEDTEAA 172 Query 26 KPEDKAHKAATKIQA#SFRGHITR 48 +| |||||||#+|+||+ | Sbjct 76 RPASPMDSAATKIQA#AFKGHLVR 98 |
* References
[PubMed ID: 23586486] Yoo S, Kim HH, Kim P, Donnelly CJ, Kalinski AL, Vuppalanchi D, Park M, Lee SJ, Merianda TT, Perrone-Bizzozero NI, Twiss JL, A HuD-ZBP1 ribonucleoprotein complex localizes GAP-43 mRNA into axons through its 3' untranslated region AU-rich regulatory element. J Neurochem. 2013 Sep;126(6):792-804. doi: 10.1111/jnc.12266. Epub 2013 Apr 30.
[PubMed ID: 23666783] ... Zhang W, Miao Y, Xing Z, Li H, Liu H, Li Z, Growth-associated protein-43 expression in cocultures of dorsal root ganglion neurons and skeletal muscle cells with different neurotrophins. Muscle Nerve. 2013 Jun;47(6):909-15. doi: 10.1002/mus.23689. Epub 2013 May 11.
[PubMed ID: 23426659] ... Donnelly CJ, Park M, Spillane M, Yoo S, Pacheco A, Gomes C, Vuppalanchi D, McDonald M, Kim HH, Merianda TT, Gallo G, Twiss JL, Axonally synthesized beta-actin and GAP-43 proteins support distinct modes of axonal growth. J Neurosci. 2013 Feb 20;33(8):3311-22. doi: 10.1523/JNEUROSCI.1722-12.2013.
[PubMed ID: 23938193] ... Chabot A, Meus MA, Hertig V, Duquette N, Calderone A, The neurogenic response of cardiac resident nestin(+) cells was associated with GAP43 upregulation and abrogated in a setting of type I diabetes. Cardiovasc Diabetol. 2013 Aug 12;12:114. doi: 10.1186/1475-2840-12-114.
[PubMed ID: 23316631] ... Zhang H, Fan S, [Expression and significance of growth-associated protein 43 in a rat model of intervertebral disc inflammation]. Zhongguo Xiu Fu Chong Jian Wai Ke Za Zhi. 2012 Dec;26(12):1435-41.
[PubMed ID: 1533624] ... Spencer SA, Schuh SM, Liu WS, Willard MB, GAP-43, a protein associated with axon growth, is phosphorylated at three sites in cultured neurons and rat brain. J Biol Chem. 1992 May 5;267(13):9059-64.
[PubMed ID: 1532026] ... Nedivi E, Basi GS, Akey IV, Skene JH, A neural-specific GAP-43 core promoter located between unusual DNA elements that interact to regulate its activity. J Neurosci. 1992 Mar;12(3):691-704.
[PubMed ID: 3428269] ... Rosenthal A, Chan SY, Henzel W, Haskell C, Kuang WJ, Chen E, Wilcox JN, Ullrich A, Goeddel DV, Routtenberg A, Primary structure and mRNA localization of protein F1, a growth-related protein kinase C substrate associated with synaptic plasticity. EMBO J. 1987 Dec 1;6(12):3641-6.
[PubMed ID: 3581170] ... Basi GS, Jacobson RD, Virag I, Schilling J, Skene JH, Primary structure and transcriptional regulation of GAP-43, a protein associated with nerve growth. Cell. 1987 Jun 19;49(6):785-91.
[PubMed ID: 2437653] ... Karns LR, Ng SC, Freeman JA, Fishman MC, Cloning of complementary DNA for GAP-43, a neuronal growth-related protein. Science. 1987 May 1;236(4801):597-600.