SB0072 : Glutamate receptor, ionotropic, NMDA 2A
[ CaMP Format ]
* Basic Information
| Organism | Rattus norvegicus (Norway rat) |
| Protein Names | glutamate receptor ionotropic, NMDA 2A precursor [Rattus norvegicus]; glutamate receptor ionotropic, NMDA 2A precursor; glutamate [NMDA] receptor subunit epsilon-1; glutamate receptor ionotropic, NMDA 2A; N-methyl-D-aspartate receptor subunit 2A; N-methyl D-aspartate receptor subtype 2A |
| Gene Names | Grin2a; glutamate receptor, ionotropic, N-methyl D-aspartate 2A |
| Gene Locus | 10q11; chromosome 10 |
| GO Function | Not available |
* Information From OMIM
Not Available.
* Structure Information
1. Primary Information
Length: 1464 aa
Average Mass: 165.481 kDa
Monoisotopic Mass: 165.377 kDa
2. Domain Information
Annotated Domains: Not Available.
Computationally Assigned Domains (Pfam+HMMER):
| domain name | begin | end | score | e-value |
|---|---|---|---|---|
| Receptor family ligand binding region 1. | 109 | 265 | 63.0 | 0.1 |
| Bacterial extracellular solute-binding proteins, family 3 1. | 457 | 797 | 22.0 | 0.0 |
| N-methyl D-aspartate receptor 2B3 C-terminus 1. | 839 | 1464 | 1.0 | 31.1 |
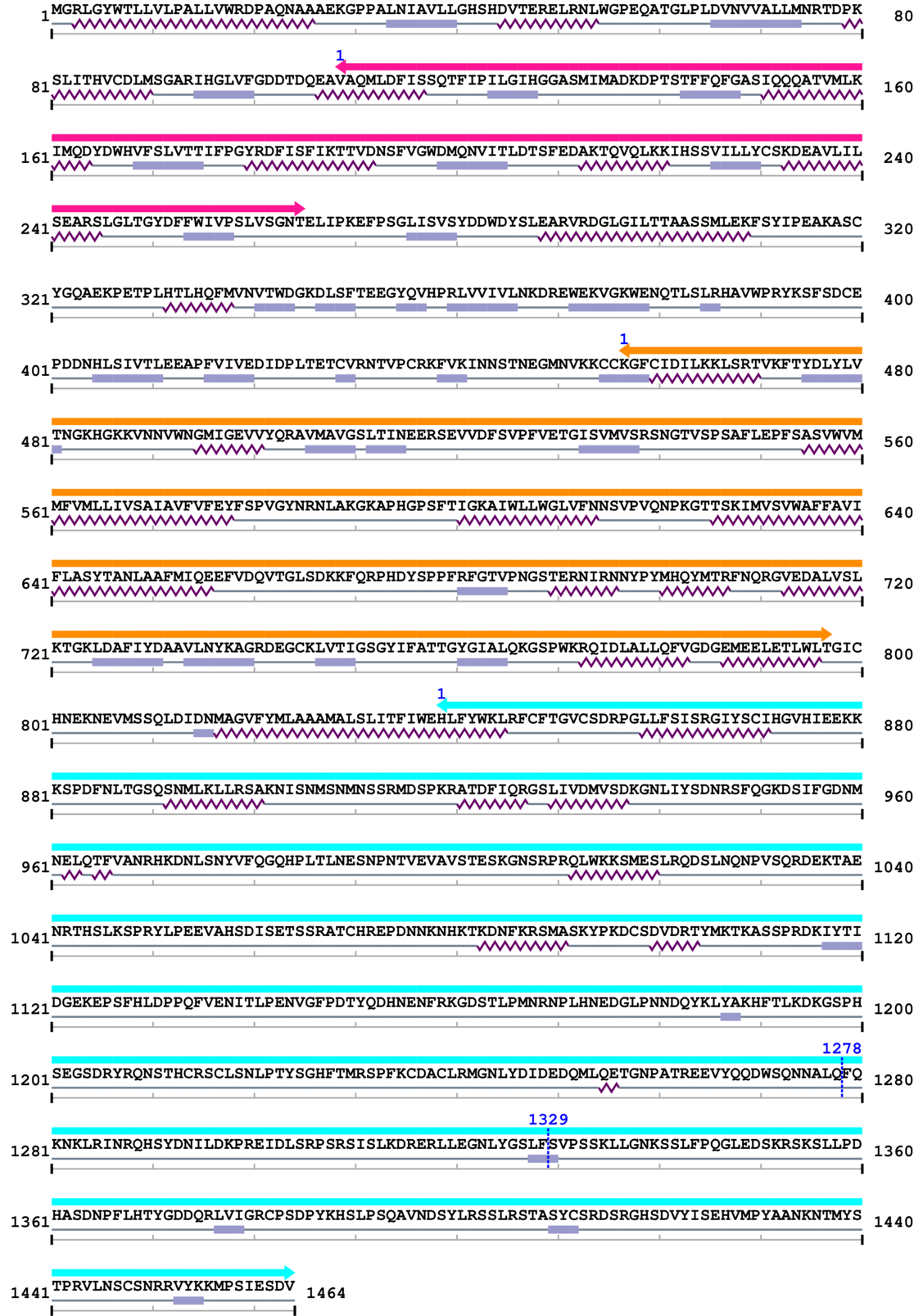
| --- cleavage 1278 (inside N-methyl D-aspartate receptor 2B3 C-terminus 839..1464) --- | ||||
| --- cleavage 1329 (inside N-methyl D-aspartate receptor 2B3 C-terminus 839..1464) --- | ||||
3. Sequence Information
Fasta Sequence: SB0072.fasta
Amino Acid Sequence and Secondary Structures (PsiPred):
4. 3D Information
Not Available.
* Cleavage Information
2 [sites] cleaved by Calpain 1
Source Reference: [PubMed ID: 11553682] Guttmann RP, Baker DL, Seifert KM, Cohen AS, Coulter DA, Lynch DR, Specific proteolysis of the NR2 subunit at multiple sites by calpain. J Neurochem. 2001 Sep;78(5):1083-93.
Cleavage sites (±10aa)
[Site 1] QDWSQNNALQ1278-FQKNKLRINR
Gln1278  Phe
Phe
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Gln1269 | Asp1270 | Trp1271 | Ser1272 | Gln1273 | Asn1274 | Asn1275 | Ala1276 | Leu1277 | Gln1278 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Phe1279 | Gln1280 | Lys1281 | Asn1282 | Lys1283 | Leu1284 | Arg1285 | Ile1286 | Asn1287 | Arg1288 |
 Sequence conservation (by blast)
Sequence conservation (by blast)
[Site 2] LEGNLYGSLF1329-SVPSSKLLGN
Phe1329  Ser
Ser
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Leu1320 | Glu1321 | Gly1322 | Asn1323 | Leu1324 | Tyr1325 | Gly1326 | Ser1327 | Leu1328 | Phe1329 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Ser1330 | Val1331 | Pro1332 | Ser1333 | Ser1334 | Lys1335 | Leu1336 | Leu1337 | Gly1338 | Asn1339 |
 Sequence conservation (by blast)
Sequence conservation (by blast)
* References
[PubMed ID: 24607230] Hansen KB, Ogden KK, Yuan H, Traynelis SF, Distinct functional and pharmacological properties of Triheteromeric GluN1/GluN2A/GluN2B NMDA receptors. Neuron. 2014 Mar 5;81(5):1084-96. doi: 10.1016/j.neuron.2014.01.035.