SB0075 : BAK, BCL2-antagonist/killer 1
[ CaMP Format ]
* Basic Information
| Organism | Homo sapiens (human) |
| Protein Names | bcl-2 homologous antagonist/killer [Homo sapiens]; bcl-2 homologous antagonist/killer; pro-apoptotic protein BAK; apoptosis regulator BAK; BCL2-like 7 protein; bcl2-L-7; bcl-2-like protein 7; Bcl-2 homologous antagonist/killer; Apoptosis regulator BAK; Bcl-2-like protein 7; Bcl2-L-7 |
| Gene Names | BAK1; BAK, BCL2L7, CDN1; BAK; BCL2L7; CDN1; BCL2-antagonist/killer 1 |
| Gene Locus | 6p21.3; chromosome 6 |
| GO Function | Not available |
* Information From OMIM
Function: Chittenden et al. (1995) and Kiefer et al. (1995) described the functional analysis of BAK, which promotes cell death and counteracts the protection from apoptosis provided by BCL2. Chittenden et al. (1995) found that enforced expression of BAK induced rapid and extensive apoptosis of serum-deprived fibroblasts. This suggested that BAK may be directly involved in activating the cell death machinery. Kiefer et al. (1995) pointed out that, like BAX (OMIM:600040), the BAK gene product primarily enhances apoptotic cell death following an appropriate stimulus. Unlike BAX, however, BAK can inhibit cell death in an Epstein-Barr virus-transformed cell line.
* Structure Information
1. Primary Information
Length: 211 aa
Average Mass: 23.408 kDa
Monoisotopic Mass: 23.394 kDa
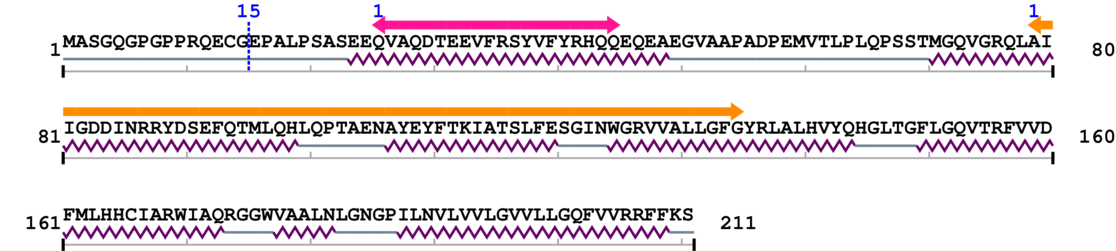
2. Domain Information
Annotated Domains: interpro / pfam / smart / prosite
Computationally Assigned Domains (Pfam+HMMER):
| domain name | begin | end | score | e-value |
|---|---|---|---|---|
| --- cleavage 15 --- | ||||
| Apoptosis regulator proteins, Bcl-2 family 1. | 26 | 45 | 7.0 | 0.1 |
| Apoptosis regulator M11, B cell 2 leukaemia/lymphoma like 1. | 79 | 135 | 35.0 | 0.0 |
3. Sequence Information
Fasta Sequence: SB0075.fasta
Amino Acid Sequence and Secondary Structures (PsiPred):
4. 3D Information
Known Structures in PDB: 1BXL (NMR; -; B=72-87), 2IMS (X-ray; 148 A; A=16-186), 2IMT (X-ray; 149 A; A=16-186), 2JBY (X-ray; 241 A; B=67-92), 2JCN (X-ray; 180 A; A=21-190), 2LP8 (NMR; -; B=72-87), 2M5B (NMR; -; A=18-186), 2XPX (X-ray; 205 A; B=67-92), 2YV6 (X-ray; 250 A; A=23-185), 3I1H (X-ray; 220 A; B=72-87), 3QBR (X-ray; 260 A; B/Y=63-96), 4D2L (X-ray; 290 A; B=67-91), 4U2U (X-ray; 290 A; A/B=23-186), 4U2V (X-ray; 230 A; A/B/C/D=68-148), 5AJK (X-ray; 255 A; B/D/F/H/J/L=67-92)
* Cleavage Information
1 [sites] cleaved by Calpain 1 and 2
Source Reference: [PubMed ID: 17157251] Moldoveanu T, Liu Q, Tocilj A, Watson M, Shore G, Gehring K, The X-ray structure of a BAK homodimer reveals an inhibitory zinc binding site. Mol Cell. 2006 Dec 8;24(5):677-88.
Cleavage sites (±10aa)
[Site 1] GPGPPRQECG15-EPALPSASEE
Gly15  Glu
Glu
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Gly6 | Pro7 | Gly8 | Pro9 | Pro10 | Arg11 | Gln12 | Glu13 | Cys14 | Gly15 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Glu16 | Pro17 | Ala18 | Leu19 | Pro20 | Ser21 | Ala22 | Ser23 | Glu24 | Glu25 |
 Sequence conservation (by blast)
Sequence conservation (by blast)
* References
[PubMed ID: 24265320] Dai H, Pang YP, Ramirez-Alvarado M, Kaufmann SH, Evaluation of the BH3-only protein Puma as a direct Bak activator. J Biol Chem. 2014 Jan 3;289(1):89-99. doi: 10.1074/jbc.M113.505701. Epub 2013 Nov