SB0077 : Histone H2A type 1
[ CaMP Format ]
* Basic Information
| Organism | Bos taurus (cattle) |
| Protein Names | PREDICTED: histone H2A type 1 [Bos taurus]; histone H2A type 1; Histone H2A type 1; H2A.1b |
| Gene Names | HIST1H2AM; Derived by automated computational analysis using gene prediction method: Gnomon. Supporting evidence includes similarity to: 1 EST, 80 Proteins, and 100% coverage of the annotated genomic feature by RNAseq alignments |
| Gene Locus | Not available |
| GO Function | Not available |
| Entrez Protein | Entrez Nucleotide | Entrez Gene | UniProt | OMIM | HGNC | HPRD | KEGG |
|---|---|---|---|---|---|---|---|
| XP_876240 | XM_871147 | 618824 | P0C0S9 | N/A | N/A | N/A | bta:618824 |
* Information From OMIM
Not Available.
* Structure Information
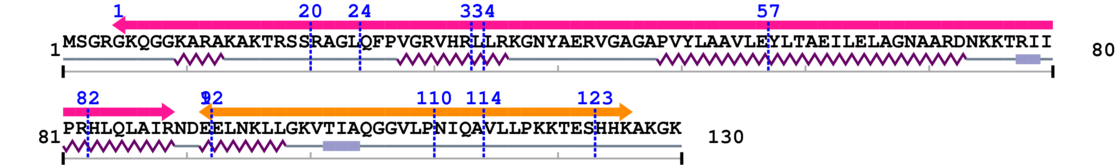
1. Primary Information
Length: 130 aa
Average Mass: 14.091 kDa
Monoisotopic Mass: 14.083 kDa
2. Domain Information
Annotated Domains: interpro / pfam / smart / prosite
Computationally Assigned Domains (Pfam+HMMER):
| domain name | begin | end | score | e-value |
|---|---|---|---|---|
| Core histone H2A/H2B/H3/H4 1. | 5 | 89 | 45.0 | 0.0 |
| --- cleavage 20 (inside Core histone H2A/H2B/H3/H4 5..89) --- | ||||
| --- cleavage 24 (inside Core histone H2A/H2B/H3/H4 5..89) --- | ||||
| --- cleavage 33 (inside Core histone H2A/H2B/H3/H4 5..89) --- | ||||
| --- cleavage 34 (inside Core histone H2A/H2B/H3/H4 5..89) --- | ||||
| --- cleavage 57 (inside Core histone H2A/H2B/H3/H4 5..89) --- | ||||
| --- cleavage 82 (inside Core histone H2A/H2B/H3/H4 5..89) --- | ||||
| --- cleavage 92 --- | ||||
| C-terminus of histone H2A 1. | 92 | 126 | 1.0 | 2.4 |
| --- cleavage 92 (inside C-terminus of histone H2A 92..126) --- | ||||
| --- cleavage 110 (inside C-terminus of histone H2A 92..126) --- | ||||
| --- cleavage 114 (inside C-terminus of histone H2A 92..126) --- | ||||
| --- cleavage 123 (inside C-terminus of histone H2A 92..126) --- | ||||
3. Sequence Information
Fasta Sequence: SB0077.fasta
Amino Acid Sequence and Secondary Structures (PsiPred):
4. 3D Information
Not Available.
* Cleavage Information
10 [sites]
Source Reference: [PubMed ID: 3038857] Sakai K, Akanuma H, Imahori K, Kawashima S, A unique specificity of a calcium activated neutral protease indicated in histone hydrolysis. J Biochem. 1987 Apr;101(4):911-8.
Cleavage sites (±10aa)
[Site 1] ARAKAKTRSS20-RAGLQFPVGR
Ser20  Arg
Arg
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Ala11 | Arg12 | Ala13 | Lys14 | Ala15 | Lys16 | Thr17 | Arg18 | Ser19 | Ser20 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Arg21 | Ala22 | Gly23 | Leu24 | Gln25 | Phe26 | Pro27 | Val28 | Gly29 | Arg30 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| MSGRGKQGGKARAKAKTRSSRAGLQFPVGRVHRLLRKGNYAERVGAGAPV |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | Danio rerio | 103.00 | 2 | hypothetical protein LOC393453 |
| 2 | Canis familiaris | 100.00 | 14 | PREDICTED: similar to Histone H2A.l (H2A/l) |
| 3 | Homo sapiens | 99.80 | 15 | histone cluster 1, H2ai |
| 4 | Mus musculus | 98.60 | 6 | histone cluster 1, H2ak |
| 5 | Xenopus laevis | 98.60 | 5 | hypothetical protein LOC734746 |
| 6 | Xenopus tropicalis | 98.60 | 4 | MGC69325 protein |
| 7 | Rattus norvegicus | 98.60 | 2 | H2A1F_RAT Histone H2A type 1-F gi |
| 8 | Gallus gallus | 98.20 | 6 | H2A3_CHICK Histone H2A-III gi |
| 9 | Bos taurus | 98.20 | 4 | PREDICTED: similar to homeostatic thymus hormone |
| 10 | synthetic construct | 98.20 | 2 | histone 2 H2aa |
| 11 | Cairina moschata | 98.20 | 1 | H2A_CAIMO Histone H2A gi |
| 12 | Gallus gallus | 98.20 | 1 | histone cluster 2, H2ac |
| 13 | Tetraodon nigroviridis | 97.40 | 9 | unnamed protein product |
| 14 | Bos taurus | 97.40 | 3 | PREDICTED: similar to Chain K, 2.9 Angstrom X-Ray |
| 15 | Oncorhynchus mykiss | 97.40 | 1 | H2A_ONCMY Histone H2A gi |
| 16 | N/A | 97.10 | 14 | 1109175A homeostatic thymus hormone alpha |
| 17 | Danio rerio | 97.10 | 6 | PREDICTED: similar to Mid1ip1 protein |
| 18 | Mus musculus | 97.10 | 2 | H2A histone family, member J |
| 19 | Homo sapiens | 97.10 | 2 | H2A histone family, member J |
| 20 | Bufo bufo gagarizans | 97.10 | 1 | AF255739_1 replication-dependent histone H2A |
| 21 | Bufo bufo gagarizans | 96.70 | 2 | replication-dependent histone H2A |
| 22 | Mus musculus domesticus | 95.90 | 1 | histone H2A |
| 23 | Contains: Hipposin | 93.20 | 1 | H2A_HIPHI Histone H2A |
| 24 | Rattus norvegicus | 91.30 | 1 | histone cluster 1, H2aa |
| 25 | Strongylocentrotus purpuratus | 88.60 | 2 | late histone L3 H2a |
| 26 | Plasmodium berghei strain ANKA | 87.80 | 1 | histone h2a |
| 27 | Acropora formosa | 80.50 | 1 | H2A_ACRFO Histone H2A gi |
| 28 | Contains: Buforin-1 (Buforin I); Buforin-2 (Buforin II) | 73.90 | 1 | H2A_BUFBG Histone H2A |
| 29 | Ashbya gossypii ATCC 10895 | 72.40 | 2 | AEL003Cp |
| 30 | Ashbya gossypii | 72.40 | 1 | AF384989_1 histone 2A |
| 31 | Pan troglodytes | 72.40 | 1 | PREDICTED: similar to none |
| 32 | Platynereis dumerilii | 71.60 | 1 | H2A_PLADU Histone H2A gi |
| 33 | Apis mellifera | 71.60 | 1 | PREDICTED: similar to CG31618-PA |
| 34 | Ustilago maydis 521 | 71.20 | 1 | histone H2A |
| 35 | Schizosaccharomyces pombe 972h- | 70.90 | 1 | histone H2A beta |
| 36 | Schizosaccharomyces pombe 972h- | 70.90 | 1 | histone H2A alpha (PMID 3018512) |
Top-ranked sequences
| organism | matching |
|---|---|
| Danio rerio | Query 1 MSGRGKQGGKARAKAKTRSS#RAGLQFPVGRVHRLLRKGNYAERVGAGAPV 50 |||||| |||||||||||||#|||||||||||||||||||||||||||||| Sbjct 1 MSGRGKTGGKARAKAKTRSS#RAGLQFPVGRVHRLLRKGNYAERVGAGAPV 50 |
| Canis familiaris | Query 1 MSGRGKQGGKARAKAKTRSS#RAGLQFPVGRVHRLLRKGNYAERVGAGAPV 50 ||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 1 MSGRGKQGGKARAKAKTRSS#RAGLQFPVGRVHRLLRKGNYAERVGAGAPV 50 |
| Homo sapiens | Query 1 MSGRGKQGGKARAKAKTRSS#RAGLQFPVGRVHRLLRKGNYAERVGAGAPV 50 ||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 1 MSGRGKQGGKARAKAKTRSS#RAGLQFPVGRVHRLLRKGNYAERVGAGAPV 50 |
| Mus musculus | Query 1 MSGRGKQGGKARAKAKTRSS#RAGLQFPVGRVHRLLRKGNYAERVGAGAPV 50 ||||||||||||||||||||#||||||||||||||||||||+||||||||| Sbjct 1 MSGRGKQGGKARAKAKTRSS#RAGLQFPVGRVHRLLRKGNYSERVGAGAPV 50 |
| Xenopus laevis | Query 1 MSGRGKQGGKARAKAKTRSS#RAGLQFPVGRVHRLLRKGNYAERVGAGAPV 50 |||||||||| |||||||||#|||||||||||||||||||||||||||||| Sbjct 1 MSGRGKQGGKTRAKAKTRSS#RAGLQFPVGRVHRLLRKGNYAERVGAGAPV 50 |
| Xenopus tropicalis | Query 1 MSGRGKQGGKARAKAKTRSS#RAGLQFPVGRVHRLLRKGNYAERVGAGAPV 50 |||||||||| |||||||||#|||||||||||||||||||||||||||||| Sbjct 1 MSGRGKQGGKVRAKAKTRSS#RAGLQFPVGRVHRLLRKGNYAERVGAGAPV 50 |
| Rattus norvegicus | Query 1 MSGRGKQGGKARAKAKTRSS#RAGLQFPVGRVHRLLRKGNYAERVGAGAPV 50 ||||||||||||||||||||#||||||||||||||||||||+||||||||| Sbjct 1 MSGRGKQGGKARAKAKTRSS#RAGLQFPVGRVHRLLRKGNYSERVGAGAPV 50 |
| Gallus gallus | Query 1 MSGRGKQGGKARAKAKTRSS#RAGLQFPVGRVHRLLRKGNYAERVGAGAPV 50 ||||||||||||||||+|||#|||||||||||||||||||||||||||||| Sbjct 1 MSGRGKQGGKARAKAKSRSS#RAGLQFPVGRVHRLLRKGNYAERVGAGAPV 50 |
| Bos taurus | Query 1 MSGRGKQGGKARAKAKTRSS#RAGLQFPVGRVHRLLRKGNYAERVGAGAPV 50 |||||||||||||||+||||#|||||||||||||||||||||||||||||| Sbjct 1 MSGRGKQGGKARAKARTRSS#RAGLQFPVGRVHRLLRKGNYAERVGAGAPV 50 |
| synthetic construct | Query 1 MSGRGKQGGKARAKAKTRSS#RAGLQFPVGRVHRLLRKGNYAERVGAGAPV 50 ||||||||||||||||+|||#|||||||||||||||||||||||||||||| Sbjct 1 MSGRGKQGGKARAKAKSRSS#RAGLQFPVGRVHRLLRKGNYAERVGAGAPV 50 |
| Cairina moschata | Query 1 MSGRGKQGGKARAKAKTRSS#RAGLQFPVGRVHRLLRKGNYAERVGAGAPV 50 ||||||||||||||||+|||#|||||||||||||||||||||||||||||| Sbjct 1 MSGRGKQGGKARAKAKSRSS#RAGLQFPVGRVHRLLRKGNYAERVGAGAPV 50 |
| Gallus gallus | Query 1 MSGRGKQGGKARAKAKTRSS#RAGLQFPVGRVHRLLRKGNYAERVGAGAPV 50 ||||||||||||||||+|||#|||||||||||||||||||||||||||||| Sbjct 1 MSGRGKQGGKARAKAKSRSS#RAGLQFPVGRVHRLLRKGNYAERVGAGAPV 50 |
| Tetraodon nigroviridis | Query 1 MSGRGKQGGKARAKAKTRSS#RAGLQFPVGRVHRLLRKGNYAERVGAGAPV 50 |||||| |||||||||||||#|||||||||||||||||||||||||||||| Sbjct 1 MSGRGKTGGKARAKAKTRSS#RAGLQFPVGRVHRLLRKGNYAERVGAGAPV 50 |
| Bos taurus | Query 1 MSGRGKQGGKARAKAKTRSS#RAGLQFPVGRVHRLLRKGNYAERVGAGAPV 50 |||||||||| |||||||||#||||||||||||||||||||+||||||||| Sbjct 1 MSGRGKQGGKTRAKAKTRSS#RAGLQFPVGRVHRLLRKGNYSERVGAGAPV 50 |
| Oncorhynchus mykiss | Query 1 MSGRGKQGGKARAKAKTRSS#RAGLQFPVGRVHRLLRKGNYAERVGAGAPV 50 |||||| |||||||||||||#|||||||||||||||||||||||||||||| Sbjct 1 MSGRGKTGGKARAKAKTRSS#RAGLQFPVGRVHRLLRKGNYAERVGAGAPV 50 |
| N/A | Query 2 SGRGKQGGKARAKAKTRSS#RAGLQFPVGRVHRLLRKGNYAERVGAGAPV 50 |||||||||||||||||||#|||||||||||||||||||||||||||||| Query 2 SGRGKQGGKARAKAKTRSS#RAGLQFPVGRVHRLLRKGNYAERVGAGAPV 50 |
| Danio rerio | Query 1 MSGRGKQGGKARAKAKTRSS#RAGLQFPVGRVHRLLRKGNYAERVGAGAPV 50 |||||| |||||||||||||#|||||||||||||||||||||||||||||| Sbjct 1 MSGRGKTGGKARAKAKTRSS#RAGLQFPVGRVHRLLRKGNYAERVGAGAPV 50 |
| Mus musculus | Query 1 MSGRGKQGGKARAKAKTRSS#RAGLQFPVGRVHRLLRKGNYAERVGAGAPV 50 |||||||||| |||||+|||#|||||||||||||||||||||||||||||| Sbjct 1 MSGRGKQGGKVRAKAKSRSS#RAGLQFPVGRVHRLLRKGNYAERVGAGAPV 50 |
| Homo sapiens | Query 1 MSGRGKQGGKARAKAKTRSS#RAGLQFPVGRVHRLLRKGNYAERVGAGAPV 50 |||||||||| |||||+|||#|||||||||||||||||||||||||||||| Sbjct 1 MSGRGKQGGKVRAKAKSRSS#RAGLQFPVGRVHRLLRKGNYAERVGAGAPV 50 |
| Bufo bufo gagarizans | Query 1 MSGRGKQGGKARAKAKTRSS#RAGLQFPVGRVHRLLRKGNYAERVGAGAPV 50 |||||||||| |||||||||#|||||||||||||||||||||+|||||||| Sbjct 1 MSGRGKQGGKVRAKAKTRSS#RAGLQFPVGRVHRLLRKGNYAQRVGAGAPV 50 |
| Bufo bufo gagarizans | Query 1 MSGRGKQGGKARAKAKTRSS#RAGLQFPVGRVHRLLRKGNYAERVGAGAPV 50 |||||||||| |||||||||#|||||||||||||||||||||+|||||||| Sbjct 1 MSGRGKQGGKVRAKAKTRSS#RAGLQFPVGRVHRLLRKGNYAQRVGAGAPV 50 |
| Mus musculus domesticus | Query 1 MSGRGKQGGKARAKAKTRSS#RAGLQFPVGRVHRLLRKGNYAERVGAGAPV 50 ||| ||||||||||||+|||#|||||||||||||||||||||||||||||| Sbjct 8 MSGLGKQGGKARAKAKSRSS#RAGLQFPVGRVHRLLRKGNYAERVGAGAPV 57 |
| Contains: Hipposin | Query 1 MSGRGKQGGKARAKAKTRSS#RAGLQFPVGRVHRLLRKGNYAERVGAGAPV 50 |||||| |||||||||||||#||||||||||||||||||||| |||||||| Sbjct 1 MSGRGKTGGKARAKAKTRSS#RAGLQFPVGRVHRLLRKGNYAHRVGAGAPV 50 |
| Rattus norvegicus | Query 1 MSGRGKQGGKARAKAKTRSS#RAGLQFPVGRVHRLLRKGNYAERVGAGAPV 50 |||| |||||||||||+|| #||||||||||||||||+||||||+||| || Sbjct 1 MSGRAKQGGKARAKAKSRSF#RAGLQFPVGRVHRLLRQGNYAERIGAGTPV 50 |
| Strongylocentrotus purpuratus | Query 1 MSGRGKQGGKARAKAKTRSS#RAGLQFPVGRVHRLLRKGNYAERVGAGAPV 50 |||||| ||||||||+||+#||||||||||||| |||||||+|||||||| Sbjct 1 MSGRGKGAGKARAKAKSRSA#RAGLQFPVGRVHRFLRKGNYAQRVGAGAPV 50 |
| Plasmodium berghei strain ANKA | Query 6 KQGGKARAKAKTRSS#RAGLQFPVGRVHRLLRKGNYAERVGAGAPV 50 |||||||||||+|||#|||||||||||||||||||||||||+|||| Sbjct 1 KQGGKARAKAKSRSS#RAGLQFPVGRVHRLLRKGNYAERVGSGAPV 45 |
| Acropora formosa | Query 1 MSGRGKQGGKAR-AKAKTRSS#RAGLQFPVGRVHRLLRKGNYAERVGAGAPV 50 |||||| |||+ |+|+|||#||||||||||+||||||||||||||||||| Sbjct 1 MSGRGK--GKAQGTKSKSRSS#RAGLQFPVGRIHRLLRKGNYAERVGAGAPV 49 |
| Contains: Buforin-1 (Buforin I); Buforin-2 (Buforin II) | Query 2 SGRGKQGGKARAKAKTRSS#RAGLQFPVGRVHRLLRKGNY 40 +|||||||| |||||||||#|||||||||||||||||||| Sbjct 1 AGRGKQGGKVRAKAKTRSS#RAGLQFPVGRVHRLLRKGNY 39 |
| Ashbya gossypii ATCC 10895 | Query 1 MSGRGKQGGKARAKAKTRSS#RAGLQFPVGRVHRLLRKGNYAERVGAGAPV 50 |||+| + | | +++||+#+||| ||||||||||||||||+|+|+|||| Sbjct 1 MSGKGGKAGSAAKASQSRSA#KAGLTFPVGRVHRLLRKGNYAQRIGSGAPV 50 |
| Ashbya gossypii | Query 1 MSGRGKQGGKARAKAKTRSS#RAGLQFPVGRVHRLLRKGNYAERVGAGAPV 50 |||+| + | | +++||+#+||| ||||||||||||||||+|+|+|||| Sbjct 1 MSGKGGKAGSAAKASQSRSA#KAGLTFPVGRVHRLLRKGNYAQRIGSGAPV 50 |
| Pan troglodytes | Query 7 QGGKARAKAKTRSS#RAGLQFPVGRVHRLLRKGNYAERVGAGAPV 50 || |||||||| ||#|||||| |||+||||||||||+ | ||||| Sbjct 2 QGSKARAKAKTHSS#RAGLQFSVGRMHRLLRKGNYAKPVRAGAPV 45 |
| Platynereis dumerilii | Query 17 TRSS#RAGLQFPVGRVHRLLRKGNYAERVGAGAPV 50 ||||#||||||||||+||||||||||||||||||| Sbjct 16 TRSS#RAGLQFPVGRIHRLLRKGNYAERVGAGAPV 49 |
| Apis mellifera | Query 17 TRSS#RAGLQFPVGRVHRLLRKGNYAERVGAGAPV 50 ||||#||||||||||+||||||||||||||||||| Sbjct 16 TRSS#RAGLQFPVGRIHRLLRKGNYAERVGAGAPV 49 |
| Ustilago maydis 521 | Query 10 KARAKAKTRSS#RAGLQFPVGRVHRLLRKGNYAERVGAGAPV 50 | +||++||+#+|||||||||+||||||||||+|||||||| Sbjct 13 DASSKAQSRSA#KAGLQFPVGRIHRLLRKGNYAQRVGAGAPV 53 |
| Schizosaccharomyces pombe 972h- | Query 1 MSGRGKQGGKARA--KAKTRSS#RAGLQFPVGRVHRLLRKGNYAERVGAGAPV 50 ||| || |||| |++||+#+||| ||||||||||||||||+|||||||| Sbjct 1 MSG-GKSGGKAAVAKSAQSRSA#KAGLAFPVGRVHRLLRKGNYAQRVGAGAPV 51 |
| Schizosaccharomyces pombe 972h- | Query 1 MSGRGKQGGKARA--KAKTRSS#RAGLQFPVGRVHRLLRKGNYAERVGAGAPV 50 ||| || |||| |++||+#+||| ||||||||||||||||+|||||||| Sbjct 1 MSG-GKSGGKAAVAKSAQSRSA#KAGLAFPVGRVHRLLRKGNYAQRVGAGAPV 51 |
[Site 2] AKTRSSRAGL24-QFPVGRVHRL
Leu24  Gln
Gln
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Ala15 | Lys16 | Thr17 | Arg18 | Ser19 | Ser20 | Arg21 | Ala22 | Gly23 | Leu24 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Gln25 | Phe26 | Pro27 | Val28 | Gly29 | Arg30 | Val31 | His32 | Arg33 | Leu34 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| MSGRGKQGGKARAKAKTRSSRAGLQFPVGRVHRLLRKGNYAERVGAGAPVYLAA |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | Danio rerio | 112.00 | 2 | hypothetical protein LOC393453 |
| 2 | Homo sapiens | 107.00 | 15 | histone cluster 1, H2ai |
| 3 | Canis familiaris | 107.00 | 14 | PREDICTED: similar to Histone H2A.l (H2A/l) |
| 4 | Mus musculus | 106.00 | 6 | histone cluster 1, H2ak |
| 5 | Xenopus laevis | 106.00 | 5 | hypothetical protein LOC734746 |
| 6 | Xenopus tropicalis | 106.00 | 4 | MGC69325 protein |
| 7 | Rattus norvegicus | 106.00 | 2 | H2A1F_RAT Histone H2A type 1-F gi |
| 8 | Tetraodon nigroviridis | 105.00 | 9 | unnamed protein product |
| 9 | Gallus gallus | 105.00 | 6 | H2A3_CHICK Histone H2A-III gi |
| 10 | Bos taurus | 105.00 | 4 | PREDICTED: similar to homeostatic thymus hormone |
| 11 | Bos taurus | 105.00 | 3 | PREDICTED: similar to Chain K, 2.9 Angstrom X-Ray |
| 12 | synthetic construct | 105.00 | 2 | histone 1 H2ac |
| 13 | Homo sapiens | 105.00 | 2 | H2A histone family, member J |
| 14 | Oncorhynchus mykiss | 105.00 | 1 | H2A_ONCMY Histone H2A gi |
| 15 | Cairina moschata | 105.00 | 1 | H2A_CAIMO Histone H2A gi |
| 16 | Gallus gallus | 105.00 | 1 | histone cluster 2, H2ac |
| 17 | N/A | 104.00 | 15 | 1109175A homeostatic thymus hormone alpha |
| 18 | Danio rerio | 104.00 | 6 | PREDICTED: similar to Mid1ip1 protein |
| 19 | Mus musculus | 104.00 | 2 | H2A histone family, member J |
| 20 | Bufo bufo gagarizans | 104.00 | 2 | replication-dependent histone H2A |
| 21 | Bufo bufo gagarizans | 104.00 | 1 | AF255739_1 replication-dependent histone H2A |
| 22 | Mus musculus domesticus | 102.00 | 1 | histone H2A |
| 23 | Rattus norvegicus | 99.00 | 1 | histone cluster 1, H2aa |
| 24 | Contains: Hipposin | 97.80 | 1 | H2A_HIPHI Histone H2A |
| 25 | Strongylocentrotus purpuratus | 95.90 | 2 | late histone L3 H2a |
| 26 | Plasmodium berghei strain ANKA | 94.40 | 1 | histone h2a |
| 27 | Acropora formosa | 87.40 | 1 | H2A_ACRFO Histone H2A gi |
| 28 | Platynereis dumerilii | 79.30 | 1 | H2A_PLADU Histone H2A gi |
| 29 | Apis mellifera | 79.00 | 1 | PREDICTED: similar to CG31618-PA |
| 30 | Ashbya gossypii ATCC 10895 | 78.60 | 1 | AEL003Cp |
| 31 | Ustilago maydis 521 | 78.60 | 1 | histone H2A |
| 32 | Schizosaccharomyces pombe 972h- | 78.20 | 1 | histone H2A beta |
| 33 | Arabidopsis thaliana | 78.20 | 1 | Unknown protein |
| 34 | Schizosaccharomyces pombe 972h- | 78.20 | 1 | histone H2A alpha (PMID 3018512) |
| 35 | Ashbya gossypii | 78.20 | 1 | AF384989_1 histone 2A |
| 36 | Asellus aquaticus | 77.80 | 1 | putative H2A histone |
| 37 | Pan troglodytes | 77.80 | 1 | PREDICTED: similar to none |
Top-ranked sequences
| organism | matching |
|---|---|
| Danio rerio | Query 1 MSGRGKQGGKARAKAKTRSSRAGL#QFPVGRVHRLLRKGNYAERVGAGAPVYLAA 54 |||||| |||||||||||||||||#|||||||||||||||||||||||||||||| Sbjct 1 MSGRGKTGGKARAKAKTRSSRAGL#QFPVGRVHRLLRKGNYAERVGAGAPVYLAA 54 |
| Homo sapiens | Query 1 MSGRGKQGGKARAKAKTRSSRAGL#QFPVGRVHRLLRKGNYAERVGAGAPVYLAA 54 ||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 1 MSGRGKQGGKARAKAKTRSSRAGL#QFPVGRVHRLLRKGNYAERVGAGAPVYLAA 54 |
| Canis familiaris | Query 1 MSGRGKQGGKARAKAKTRSSRAGL#QFPVGRVHRLLRKGNYAERVGAGAPVYLAA 54 ||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 1 MSGRGKQGGKARAKAKTRSSRAGL#QFPVGRVHRLLRKGNYAERVGAGAPVYLAA 54 |
| Mus musculus | Query 1 MSGRGKQGGKARAKAKTRSSRAGL#QFPVGRVHRLLRKGNYAERVGAGAPVYLAA 54 ||||||||||||||||||||||||#||||||||||||||||+||||||||||||| Sbjct 1 MSGRGKQGGKARAKAKTRSSRAGL#QFPVGRVHRLLRKGNYSERVGAGAPVYLAA 54 |
| Xenopus laevis | Query 1 MSGRGKQGGKARAKAKTRSSRAGL#QFPVGRVHRLLRKGNYAERVGAGAPVYLAA 54 |||||||||| |||||||||||||#|||||||||||||||||||||||||||||| Sbjct 1 MSGRGKQGGKTRAKAKTRSSRAGL#QFPVGRVHRLLRKGNYAERVGAGAPVYLAA 54 |
| Xenopus tropicalis | Query 1 MSGRGKQGGKARAKAKTRSSRAGL#QFPVGRVHRLLRKGNYAERVGAGAPVYLAA 54 |||||||||| |||||||||||||#|||||||||||||||||||||||||||||| Sbjct 1 MSGRGKQGGKVRAKAKTRSSRAGL#QFPVGRVHRLLRKGNYAERVGAGAPVYLAA 54 |
| Rattus norvegicus | Query 1 MSGRGKQGGKARAKAKTRSSRAGL#QFPVGRVHRLLRKGNYAERVGAGAPVYLAA 54 ||||||||||||||||||||||||#||||||||||||||||+||||||||||||| Sbjct 1 MSGRGKQGGKARAKAKTRSSRAGL#QFPVGRVHRLLRKGNYSERVGAGAPVYLAA 54 |
| Tetraodon nigroviridis | Query 1 MSGRGKQGGKARAKAKTRSSRAGL#QFPVGRVHRLLRKGNYAERVGAGAPVYLAA 54 |||||| |||||||||||||||||#|||||||||||||||||||||||||||||| Sbjct 1 MSGRGKTGGKARAKAKTRSSRAGL#QFPVGRVHRLLRKGNYAERVGAGAPVYLAA 54 |
| Gallus gallus | Query 1 MSGRGKQGGKARAKAKTRSSRAGL#QFPVGRVHRLLRKGNYAERVGAGAPVYLAA 54 ||||||||||||||||+|||||||#|||||||||||||||||||||||||||||| Sbjct 1 MSGRGKQGGKARAKAKSRSSRAGL#QFPVGRVHRLLRKGNYAERVGAGAPVYLAA 54 |
| Bos taurus | Query 1 MSGRGKQGGKARAKAKTRSSRAGL#QFPVGRVHRLLRKGNYAERVGAGAPVYLAA 54 |||||||||||||||+||||||||#|||||||||||||||||||||||||||||| Sbjct 1 MSGRGKQGGKARAKARTRSSRAGL#QFPVGRVHRLLRKGNYAERVGAGAPVYLAA 54 |
| Bos taurus | Query 1 MSGRGKQGGKARAKAKTRSSRAGL#QFPVGRVHRLLRKGNYAERVGAGAPVYLAA 54 |||||||||| |||||||||||||#||||||||||||||||+||||||||||||| Sbjct 1 MSGRGKQGGKTRAKAKTRSSRAGL#QFPVGRVHRLLRKGNYSERVGAGAPVYLAA 54 |
| synthetic construct | Query 1 MSGRGKQGGKARAKAKTRSSRAGL#QFPVGRVHRLLRKGNYAERVGAGAPVYLAA 54 ||||||||||||||||+|||||||#|||||||||||||||||||||||||||||| Sbjct 1 MSGRGKQGGKARAKAKSRSSRAGL#QFPVGRVHRLLRKGNYAERVGAGAPVYLAA 54 |
| Homo sapiens | Query 1 MSGRGKQGGKARAKAKTRSSRAGL#QFPVGRVHRLLRKGNYAERVGAGAPVYLAA 54 |||||||||| |||||+|||||||#|||||||||||||||||||||||||||||| Sbjct 1 MSGRGKQGGKVRAKAKSRSSRAGL#QFPVGRVHRLLRKGNYAERVGAGAPVYLAA 54 |
| Oncorhynchus mykiss | Query 1 MSGRGKQGGKARAKAKTRSSRAGL#QFPVGRVHRLLRKGNYAERVGAGAPVYLAA 54 |||||| |||||||||||||||||#|||||||||||||||||||||||||||||| Sbjct 1 MSGRGKTGGKARAKAKTRSSRAGL#QFPVGRVHRLLRKGNYAERVGAGAPVYLAA 54 |
| Cairina moschata | Query 1 MSGRGKQGGKARAKAKTRSSRAGL#QFPVGRVHRLLRKGNYAERVGAGAPVYLAA 54 ||||||||||||||||+|||||||#|||||||||||||||||||||||||||||| Sbjct 1 MSGRGKQGGKARAKAKSRSSRAGL#QFPVGRVHRLLRKGNYAERVGAGAPVYLAA 54 |
| Gallus gallus | Query 1 MSGRGKQGGKARAKAKTRSSRAGL#QFPVGRVHRLLRKGNYAERVGAGAPVYLAA 54 ||||||||||||||||+|||||||#|||||||||||||||||||||||||||||| Sbjct 1 MSGRGKQGGKARAKAKSRSSRAGL#QFPVGRVHRLLRKGNYAERVGAGAPVYLAA 54 |
| N/A | Query 2 SGRGKQGGKARAKAKTRSSRAGL#QFPVGRVHRLLRKGNYAERVGAGAPVYLAA 54 |||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 2 SGRGKQGGKARAKAKTRSSRAGL#QFPVGRVHRLLRKGNYAERVGAGAPVYLAA 54 |
| Danio rerio | Query 1 MSGRGKQGGKARAKAKTRSSRAGL#QFPVGRVHRLLRKGNYAERVGAGAPVYLAA 54 |||||| |||||||||||||||||#|||||||||||||||||||||||||||||| Sbjct 1 MSGRGKTGGKARAKAKTRSSRAGL#QFPVGRVHRLLRKGNYAERVGAGAPVYLAA 54 |
| Mus musculus | Query 1 MSGRGKQGGKARAKAKTRSSRAGL#QFPVGRVHRLLRKGNYAERVGAGAPVYLAA 54 |||||||||| |||||+|||||||#|||||||||||||||||||||||||||||| Sbjct 1 MSGRGKQGGKVRAKAKSRSSRAGL#QFPVGRVHRLLRKGNYAERVGAGAPVYLAA 54 |
| Bufo bufo gagarizans | Query 1 MSGRGKQGGKARAKAKTRSSRAGL#QFPVGRVHRLLRKGNYAERVGAGAPVYLAA 54 |||||||||| |||||||||||||#|||||||||||||||||+|||||||||||| Sbjct 1 MSGRGKQGGKVRAKAKTRSSRAGL#QFPVGRVHRLLRKGNYAQRVGAGAPVYLAA 54 |
| Bufo bufo gagarizans | Query 1 MSGRGKQGGKARAKAKTRSSRAGL#QFPVGRVHRLLRKGNYAERVGAGAPVYLAA 54 |||||||||| |||||||||||||#|||||||||||||||||+|||||||||||| Sbjct 1 MSGRGKQGGKVRAKAKTRSSRAGL#QFPVGRVHRLLRKGNYAQRVGAGAPVYLAA 54 |
| Mus musculus domesticus | Query 1 MSGRGKQGGKARAKAKTRSSRAGL#QFPVGRVHRLLRKGNYAERVGAGAPVYLAA 54 ||| ||||||||||||+|||||||#|||||||||||||||||||||||||||+|| Sbjct 8 MSGLGKQGGKARAKAKSRSSRAGL#QFPVGRVHRLLRKGNYAERVGAGAPVYMAA 61 |
| Rattus norvegicus | Query 1 MSGRGKQGGKARAKAKTRSSRAGL#QFPVGRVHRLLRKGNYAERVGAGAPVYLAA 54 |||| |||||||||||+|| ||||#||||||||||||+||||||+||| |||||| Sbjct 1 MSGRAKQGGKARAKAKSRSFRAGL#QFPVGRVHRLLRQGNYAERIGAGTPVYLAA 54 |
| Contains: Hipposin | Query 1 MSGRGKQGGKARAKAKTRSSRAGL#QFPVGRVHRLLRKGNYAERVGAGAPVYL 52 |||||| |||||||||||||||||#||||||||||||||||| |||||||||| Sbjct 1 MSGRGKTGGKARAKAKTRSSRAGL#QFPVGRVHRLLRKGNYAHRVGAGAPVYL 52 |
| Strongylocentrotus purpuratus | Query 1 MSGRGKQGGKARAKAKTRSSRAGL#QFPVGRVHRLLRKGNYAERVGAGAPVYLAA 54 |||||| ||||||||+||+||||#||||||||| |||||||+|||||||||||| Sbjct 1 MSGRGKGAGKARAKAKSRSARAGL#QFPVGRVHRFLRKGNYAQRVGAGAPVYLAA 54 |
| Plasmodium berghei strain ANKA | Query 6 KQGGKARAKAKTRSSRAGL#QFPVGRVHRLLRKGNYAERVGAGAPVYLAA 54 |||||||||||+|||||||#|||||||||||||||||||||+|||||+|| Sbjct 1 KQGGKARAKAKSRSSRAGL#QFPVGRVHRLLRKGNYAERVGSGAPVYMAA 49 |
| Acropora formosa | Query 1 MSGRGKQGGKAR-AKAKTRSSRAGL#QFPVGRVHRLLRKGNYAERVGAGAPVYLAA 54 |||||| |||+ |+|+|||||||#||||||+||||||||||||||||||||+|| Sbjct 1 MSGRGK--GKAQGTKSKSRSSRAGL#QFPVGRIHRLLRKGNYAERVGAGAPVYMAA 53 |
| Platynereis dumerilii | Query 17 TRSSRAGL#QFPVGRVHRLLRKGNYAERVGAGAPVYLAA 54 ||||||||#||||||+||||||||||||||||||||||| Sbjct 16 TRSSRAGL#QFPVGRIHRLLRKGNYAERVGAGAPVYLAA 53 |
| Apis mellifera | Query 17 TRSSRAGL#QFPVGRVHRLLRKGNYAERVGAGAPVYLAA 54 ||||||||#||||||+||||||||||||||||||||||| Sbjct 16 TRSSRAGL#QFPVGRIHRLLRKGNYAERVGAGAPVYLAA 53 |
| Ashbya gossypii ATCC 10895 | Query 1 MSGRGKQGGKARAKAKTRSSRAGL#QFPVGRVHRLLRKGNYAERVGAGAPVYLAA 54 |||+| + | | +++||++|||# ||||||||||||||||+|+|+|||||| | Sbjct 1 MSGKGGKAGSAAKASQSRSAKAGL#TFPVGRVHRLLRKGNYAQRIGSGAPVYLTA 54 |
| Ustilago maydis 521 | Query 10 KARAKAKTRSSRAGL#QFPVGRVHRLLRKGNYAERVGAGAPVYLAA 54 | +||++||++|||#||||||+||||||||||+|||||||||||| Sbjct 13 DASSKAQSRSAKAGL#QFPVGRIHRLLRKGNYAQRVGAGAPVYLAA 57 |
| Schizosaccharomyces pombe 972h- | Query 1 MSGRGKQGGKARA--KAKTRSSRAGL#QFPVGRVHRLLRKGNYAERVGAGAPVYLAA 54 ||| || |||| |++||++|||# ||||||||||||||||+|||||||||||| Sbjct 1 MSG-GKSGGKAAVAKSAQSRSAKAGL#AFPVGRVHRLLRKGNYAQRVGAGAPVYLAA 55 |
| Arabidopsis thaliana | Query 17 TRSSRAGL#QFPVGRVHRLLRKGNYAERVGAGAPVYLAA 54 +|||||||#||||||+||||||||||||||||||||||| Sbjct 16 SRSSRAGL#QFPVGRIHRLLRKGNYAERVGAGAPVYLAA 53 |
| Schizosaccharomyces pombe 972h- | Query 1 MSGRGKQGGKARA--KAKTRSSRAGL#QFPVGRVHRLLRKGNYAERVGAGAPVYLAA 54 ||| || |||| |++||++|||# ||||||||||||||||+|||||||||||| Sbjct 1 MSG-GKSGGKAAVAKSAQSRSAKAGL#AFPVGRVHRLLRKGNYAQRVGAGAPVYLAA 55 |
| Ashbya gossypii | Query 1 MSGRGKQGGKARAKAKTRSSRAGL#QFPVGRVHRLLRKGNYAERVGAGAPVYLAA 54 |||+| + | | +++||++|||# ||||||||||||||||+|+|+|||||| | Sbjct 1 MSGKGGKAGSAAKASQSRSAKAGL#TFPVGRVHRLLRKGNYAQRIGSGAPVYLTA 54 |
| Asellus aquaticus | Query 17 TRSSRAGL#QFPVGRVHRLLRKGNYAERVGAGAPVYLAA 54 +|||||||#||||||+||||||||||||||||||||||| Sbjct 16 SRSSRAGL#QFPVGRIHRLLRKGNYAERVGAGAPVYLAA 53 |
| Pan troglodytes | Query 7 QGGKARAKAKTRSSRAGL#QFPVGRVHRLLRKGNYAERVGAGAPVYLAA 54 || |||||||| ||||||#|| |||+||||||||||+ | ||||||| | Sbjct 2 QGSKARAKAKTHSSRAGL#QFSVGRMHRLLRKGNYAKPVRAGAPVYLEA 49 |
[Site 3] LQFPVGRVHR33-LLRKGNYAER
Arg33  Leu
Leu
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Leu24 | Gln25 | Phe26 | Pro27 | Val28 | Gly29 | Arg30 | Val31 | His32 | Arg33 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Leu34 | Leu35 | Arg36 | Lys37 | Gly38 | Asn39 | Tyr40 | Ala41 | Glu42 | Arg43 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| RGKQGGKARAKAKTRSSRAGLQFPVGRVHRLLRKGNYAERVGAGAPVYLAAVLEYLTAEI |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | Danio rerio | 120.00 | 2 | hypothetical protein LOC393453 |
| 2 | Homo sapiens | 119.00 | 15 | histone cluster 1, H2ai |
| 3 | N/A | 119.00 | 14 | 1109175A homeostatic thymus hormone alpha |
| 4 | Canis familiaris | 119.00 | 14 | PREDICTED: similar to Histone H2A.l (H2A/l) |
| 5 | Mus musculus | 118.00 | 6 | histone cluster 1, H2ak |
| 6 | Xenopus laevis | 118.00 | 5 | hypothetical protein LOC494591 |
| 7 | Bos taurus | 118.00 | 3 | PREDICTED: similar to Chain K, 2.9 Angstrom X-Ray |
| 8 | Xenopus tropicalis | 118.00 | 3 | MGC69325 protein |
| 9 | Rattus norvegicus | 118.00 | 2 | H2A1F_RAT Histone H2A type 1-F gi |
| 10 | Cairina moschata | 118.00 | 1 | H2A_CAIMO Histone H2A gi |
| 11 | Gallus gallus | 118.00 | 1 | histone cluster 2, H2ac |
| 12 | Gallus gallus | 117.00 | 6 | H2A3_CHICK Histone H2A-III gi |
| 13 | Bos taurus | 117.00 | 4 | PREDICTED: similar to homeostatic thymus hormone |
| 14 | synthetic construct | 117.00 | 2 | histone 1 H2ac |
| 15 | Oncorhynchus mykiss | 117.00 | 1 | H2A_ONCMY Histone H2A gi |
| 16 | Tetraodon nigroviridis | 116.00 | 7 | unnamed protein product |
| 17 | Danio rerio | 116.00 | 6 | PREDICTED: similar to Mid1ip1 protein |
| 18 | Homo sapiens | 116.00 | 2 | H2A histone family, member J |
| 19 | Bufo bufo gagarizans | 116.00 | 2 | replication-dependent histone H2A |
| 20 | Mus musculus | 116.00 | 2 | H2A histone family, member J |
| 21 | Bufo bufo gagarizans | 116.00 | 1 | AF255739_1 replication-dependent histone H2A |
| 22 | Mus musculus domesticus | 115.00 | 1 | histone H2A |
| 23 | Plasmodium berghei strain ANKA | 112.00 | 1 | histone h2a |
| 24 | Rattus norvegicus | 110.00 | 1 | histone cluster 1, H2aa |
| 25 | Strongylocentrotus purpuratus | 106.00 | 2 | late histone L3 H2a |
| 26 | Acropora formosa | 98.20 | 1 | H2A_ACRFO Histone H2A gi |
| 27 | Ustilago maydis 521 | 94.40 | 1 | histone H2A |
| 28 | Pan troglodytes | 94.00 | 1 | PREDICTED: similar to none |
| 29 | Strongylocentrotus purpuratus | 93.60 | 1 | PREDICTED: similar to histone L3 H2a |
| 30 | Oikopleura dioica | 93.20 | 1 | histone h2A.3 |
| 31 | Schizosaccharomyces pombe 972h- | 92.80 | 1 | histone H2A beta |
| 32 | Contains: Hipposin | 92.80 | 1 | H2A_HIPHI Histone H2A |
| 33 | Schizosaccharomyces pombe 972h- | 92.80 | 1 | histone H2A alpha (PMID 3018512) |
| 34 | Platynereis dumerilii | 92.80 | 1 | H2A_PLADU Histone H2A gi |
| 35 | Schistosoma japonicum | 92.40 | 1 | SJCHGC05779 protein |
| 36 | Apis mellifera | 92.40 | 1 | PREDICTED: similar to CG31618-PA |
| 37 | Arabidopsis thaliana | 92.00 | 1 | Unknown protein |
| 38 | Asellus aquaticus | 91.30 | 1 | putative H2A histone |
| 39 | Tribolium castaneum | 91.30 | 1 | PREDICTED: similar to CG31618-PA |
Top-ranked sequences
| organism | matching |
|---|---|
| Danio rerio | Query 4 RGKQGGKARAKAKTRSSRAGLQFPVGRVHR#LLRKGNYAERVGAGAPVYLAAVLEYLTAEI 63 ||| ||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Sbjct 4 RGKTGGKARAKAKTRSSRAGLQFPVGRVHR#LLRKGNYAERVGAGAPVYLAAVLEYLTAEI 63 |
| Homo sapiens | Query 4 RGKQGGKARAKAKTRSSRAGLQFPVGRVHR#LLRKGNYAERVGAGAPVYLAAVLEYLTAEI 63 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 4 RGKQGGKARAKAKTRSSRAGLQFPVGRVHR#LLRKGNYAERVGAGAPVYLAAVLEYLTAEI 63 |
| N/A | Query 4 RGKQGGKARAKAKTRSSRAGLQFPVGRVHR#LLRKGNYAERVGAGAPVYLAAVLEYLTAEI 63 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 4 RGKQGGKARAKAKTRSSRAGLQFPVGRVHR#LLRKGNYAERVGAGAPVYLAAVLEYLTAEI 63 |
| Canis familiaris | Query 4 RGKQGGKARAKAKTRSSRAGLQFPVGRVHR#LLRKGNYAERVGAGAPVYLAAVLEYLTAEI 63 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 4 RGKQGGKARAKAKTRSSRAGLQFPVGRVHR#LLRKGNYAERVGAGAPVYLAAVLEYLTAEI 63 |
| Mus musculus | Query 4 RGKQGGKARAKAKTRSSRAGLQFPVGRVHR#LLRKGNYAERVGAGAPVYLAAVLEYLTAEI 63 ||||||||||||||||||||||||||||||#|||||||+|||||||||||||||||||||| Sbjct 4 RGKQGGKARAKAKTRSSRAGLQFPVGRVHR#LLRKGNYSERVGAGAPVYLAAVLEYLTAEI 63 |
| Xenopus laevis | Query 4 RGKQGGKARAKAKTRSSRAGLQFPVGRVHR#LLRKGNYAERVGAGAPVYLAAVLEYLTAEI 63 ||||||| ||||||||||||||||||||||#|||||||||||||||||||||||||||||| Sbjct 4 RGKQGGKTRAKAKTRSSRAGLQFPVGRVHR#LLRKGNYAERVGAGAPVYLAAVLEYLTAEI 63 |
| Bos taurus | Query 4 RGKQGGKARAKAKTRSSRAGLQFPVGRVHR#LLRKGNYAERVGAGAPVYLAAVLEYLTAEI 63 ||||||||||||||||||||||||||||||#|||||||+|||||||||||||||||||||| Sbjct 4 RGKQGGKARAKAKTRSSRAGLQFPVGRVHR#LLRKGNYSERVGAGAPVYLAAVLEYLTAEI 63 |
| Xenopus tropicalis | Query 4 RGKQGGKARAKAKTRSSRAGLQFPVGRVHR#LLRKGNYAERVGAGAPVYLAAVLEYLTAEI 63 ||||||| ||||||||||||||||||||||#|||||||||||||||||||||||||||||| Sbjct 4 RGKQGGKVRAKAKTRSSRAGLQFPVGRVHR#LLRKGNYAERVGAGAPVYLAAVLEYLTAEI 63 |
| Rattus norvegicus | Query 4 RGKQGGKARAKAKTRSSRAGLQFPVGRVHR#LLRKGNYAERVGAGAPVYLAAVLEYLTAEI 63 ||||||||||||||||||||||||||||||#|||||||+|||||||||||||||||||||| Sbjct 4 RGKQGGKARAKAKTRSSRAGLQFPVGRVHR#LLRKGNYSERVGAGAPVYLAAVLEYLTAEI 63 |
| Cairina moschata | Query 4 RGKQGGKARAKAKTRSSRAGLQFPVGRVHR#LLRKGNYAERVGAGAPVYLAAVLEYLTAEI 63 |||||||||||||+||||||||||||||||#|||||||||||||||||||||||||||||| Sbjct 4 RGKQGGKARAKAKSRSSRAGLQFPVGRVHR#LLRKGNYAERVGAGAPVYLAAVLEYLTAEI 63 |
| Gallus gallus | Query 4 RGKQGGKARAKAKTRSSRAGLQFPVGRVHR#LLRKGNYAERVGAGAPVYLAAVLEYLTAEI 63 |||||||||||||+||||||||||||||||#|||||||||||||||||||||||||||||| Sbjct 4 RGKQGGKARAKAKSRSSRAGLQFPVGRVHR#LLRKGNYAERVGAGAPVYLAAVLEYLTAEI 63 |
| Gallus gallus | Query 4 RGKQGGKARAKAKTRSSRAGLQFPVGRVHR#LLRKGNYAERVGAGAPVYLAAVLEYLTAEI 63 |||||||||||||+||||||||||||||||#|||||||||||||||||||||||||||||| Sbjct 4 RGKQGGKARAKAKSRSSRAGLQFPVGRVHR#LLRKGNYAERVGAGAPVYLAAVLEYLTAEI 63 |
| Bos taurus | Query 4 RGKQGGKARAKAKTRSSRAGLQFPVGRVHR#LLRKGNYAERVGAGAPVYLAAVLEYLTAEI 63 ||||||||||||+|||||||||||||||||#|||||||||||||||||||||||||||||| Sbjct 4 RGKQGGKARAKARTRSSRAGLQFPVGRVHR#LLRKGNYAERVGAGAPVYLAAVLEYLTAEI 63 |
| synthetic construct | Query 4 RGKQGGKARAKAKTRSSRAGLQFPVGRVHR#LLRKGNYAERVGAGAPVYLAAVLEYLTAEI 63 |||||||||||||+||||||||||||||||#|||||||||||||||||||||||||||||| Sbjct 4 RGKQGGKARAKAKSRSSRAGLQFPVGRVHR#LLRKGNYAERVGAGAPVYLAAVLEYLTAEI 63 |
| Oncorhynchus mykiss | Query 4 RGKQGGKARAKAKTRSSRAGLQFPVGRVHR#LLRKGNYAERVGAGAPVYLAAVLEYLTAEI 63 ||| ||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Sbjct 4 RGKTGGKARAKAKTRSSRAGLQFPVGRVHR#LLRKGNYAERVGAGAPVYLAAVLEYLTAEI 63 |
| Tetraodon nigroviridis | Query 4 RGKQGGKARAKAKTRSSRAGLQFPVGRVHR#LLRKGNYAERVGAGAPVYLAAVLEYLTAEI 63 ||| ||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Sbjct 4 RGKTGGKARAKAKTRSSRAGLQFPVGRVHR#LLRKGNYAERVGAGAPVYLAAVLEYLTAEI 63 |
| Danio rerio | Query 4 RGKQGGKARAKAKTRSSRAGLQFPVGRVHR#LLRKGNYAERVGAGAPVYLAAVLEYLTAEI 63 ||| ||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Sbjct 4 RGKTGGKARAKAKTRSSRAGLQFPVGRVHR#LLRKGNYAERVGAGAPVYLAAVLEYLTAEI 63 |
| Homo sapiens | Query 4 RGKQGGKARAKAKTRSSRAGLQFPVGRVHR#LLRKGNYAERVGAGAPVYLAAVLEYLTAEI 63 ||||||| |||||+||||||||||||||||#|||||||||||||||||||||||||||||| Sbjct 4 RGKQGGKVRAKAKSRSSRAGLQFPVGRVHR#LLRKGNYAERVGAGAPVYLAAVLEYLTAEI 63 |
| Bufo bufo gagarizans | Query 4 RGKQGGKARAKAKTRSSRAGLQFPVGRVHR#LLRKGNYAERVGAGAPVYLAAVLEYLTAEI 63 ||||||| ||||||||||||||||||||||#||||||||+||||||||||||||||||||| Sbjct 4 RGKQGGKVRAKAKTRSSRAGLQFPVGRVHR#LLRKGNYAQRVGAGAPVYLAAVLEYLTAEI 63 |
| Mus musculus | Query 4 RGKQGGKARAKAKTRSSRAGLQFPVGRVHR#LLRKGNYAERVGAGAPVYLAAVLEYLTAEI 63 ||||||| |||||+||||||||||||||||#|||||||||||||||||||||||||||||| Sbjct 4 RGKQGGKVRAKAKSRSSRAGLQFPVGRVHR#LLRKGNYAERVGAGAPVYLAAVLEYLTAEI 63 |
| Bufo bufo gagarizans | Query 4 RGKQGGKARAKAKTRSSRAGLQFPVGRVHR#LLRKGNYAERVGAGAPVYLAAVLEYLTAEI 63 ||||||| ||||||||||||||||||||||#||||||||+||||||||||||||||||||| Sbjct 4 RGKQGGKVRAKAKTRSSRAGLQFPVGRVHR#LLRKGNYAQRVGAGAPVYLAAVLEYLTAEI 63 |
| Mus musculus domesticus | Query 5 GKQGGKARAKAKTRSSRAGLQFPVGRVHR#LLRKGNYAERVGAGAPVYLAAVLEYLTAEI 63 ||||||||||||+||||||||||||||||#||||||||||||||||||+||||||||||| Sbjct 12 GKQGGKARAKAKSRSSRAGLQFPVGRVHR#LLRKGNYAERVGAGAPVYMAAVLEYLTAEI 70 |
| Plasmodium berghei strain ANKA | Query 6 KQGGKARAKAKTRSSRAGLQFPVGRVHR#LLRKGNYAERVGAGAPVYLAAVLEYLTAEI 63 |||||||||||+||||||||||||||||#||||||||||||+|||||+||||||||||| Sbjct 1 KQGGKARAKAKSRSSRAGLQFPVGRVHR#LLRKGNYAERVGSGAPVYMAAVLEYLTAEI 58 |
| Rattus norvegicus | Query 4 RGKQGGKARAKAKTRSSRAGLQFPVGRVHR#LLRKGNYAERVGAGAPVYLAAVLEYLTAEI 63 | |||||||||||+|| |||||||||||||#|||+||||||+||| ||||||||||||||| Sbjct 4 RAKQGGKARAKAKSRSFRAGLQFPVGRVHR#LLRQGNYAERIGAGTPVYLAAVLEYLTAEI 63 |
| Strongylocentrotus purpuratus | Query 4 RGKQGGKARAKAKTRSSRAGLQFPVGRVHR#LLRKGNYAERVGAGAPVYLAAVLEYLTAEI 63 ||| ||||||||+||+|||||||||||||# |||||||+||||||||||||||||| ||| Sbjct 4 RGKGAGKARAKAKSRSARAGLQFPVGRVHR#FLRKGNYAQRVGAGAPVYLAAVLEYLAAEI 63 |
| Acropora formosa | Query 4 RGKQGGKAR-AKAKTRSSRAGLQFPVGRVHR#LLRKGNYAERVGAGAPVYLAAVLEYLTAE 62 ||| |||+ |+|+|||||||||||||+||#||||||||||||||||||+|||||||+|| Sbjct 4 RGK--GKAQGTKSKSRSSRAGLQFPVGRIHR#LLRKGNYAERVGAGAPVYMAAVLEYLSAE 61 Query 63 I 63 | Query 63 I 63 |
| Ustilago maydis 521 | Query 10 KARAKAKTRSSRAGLQFPVGRVHR#LLRKGNYAERVGAGAPVYLAAVLEYLTAEI 63 | +||++||++|||||||||+||#||||||||+||||||||||||||||| ||| Sbjct 13 DASSKAQSRSAKAGLQFPVGRIHR#LLRKGNYAQRVGAGAPVYLAAVLEYLAAEI 66 |
| Pan troglodytes | Query 7 QGGKARAKAKTRSSRAGLQFPVGRVHR#LLRKGNYAERVGAGAPVYLAAVLEYLTAEI 63 || |||||||| |||||||| |||+||#||||||||+ | ||||||| ||||||||+| Sbjct 2 QGSKARAKAKTHSSRAGLQFSVGRMHR#LLRKGNYAKPVRAGAPVYLEAVLEYLTAKI 58 |
| Strongylocentrotus purpuratus | Query 13 AKAKTRSSRAGLQFPVGRVHR#LLRKGNYAERVGAGAPVYLAAVLEYLTAEI 63 +|||+||+|||||||||||||# ||||||| ||||||||||||||||| ||| Sbjct 13 SKAKSRSARAGLQFPVGRVHR#FLRKGNYAARVGAGAPVYLAAVLEYLAAEI 63 |
| Oikopleura dioica | Query 12 RAKAKTRSSRAGLQFPVGRVHR#LLRKGNYAERVGAGAPVYLAAVLEYLTAEI 63 +| |+||||||||||||||||#+|| ||+|+||||||||||||||||||||+ Sbjct 13 KAGKKSRSSRAGLQFPVGRVHR#MLRNGNWADRVGAGAPVYLAAVLEYLTAEV 64 |
| Schizosaccharomyces pombe 972h- | Query 5 GKQGGKARA--KAKTRSSRAGLQFPVGRVHR#LLRKGNYAERVGAGAPVYLAAVLEYLTAE 62 || |||| |++||++||| ||||||||#||||||||+||||||||||||||||| || Sbjct 4 GKSGGKAAVAKSAQSRSAKAGLAFPVGRVHR#LLRKGNYAQRVGAGAPVYLAAVLEYLAAE 63 Query 63 I 63 | Query 63 I 63 |
| Contains: Hipposin | Query 4 RGKQGGKARAKAKTRSSRAGLQFPVGRVHR#LLRKGNYAERVGAGAPVYL 52 ||| ||||||||||||||||||||||||||#|||||||| |||||||||| Sbjct 4 RGKTGGKARAKAKTRSSRAGLQFPVGRVHR#LLRKGNYAHRVGAGAPVYL 52 |
| Schizosaccharomyces pombe 972h- | Query 5 GKQGGKARA--KAKTRSSRAGLQFPVGRVHR#LLRKGNYAERVGAGAPVYLAAVLEYLTAE 62 || |||| |++||++||| ||||||||#||||||||+||||||||||||||||| || Sbjct 4 GKSGGKAAVAKSAQSRSAKAGLAFPVGRVHR#LLRKGNYAQRVGAGAPVYLAAVLEYLAAE 63 Query 63 I 63 | Query 63 I 63 |
| Platynereis dumerilii | Query 17 TRSSRAGLQFPVGRVHR#LLRKGNYAERVGAGAPVYLAAVLEYLTAEI 63 ||||||||||||||+||#||||||||||||||||||||||+||| ||+ Sbjct 16 TRSSRAGLQFPVGRIHR#LLRKGNYAERVGAGAPVYLAAVMEYLAAEV 62 |
| Schistosoma japonicum | Query 18 RSSRAGLQFPVGRVHR#LLRKGNYAERVGAGAPVYLAAVLEYLTAEI 63 ||||||||||| ||||#|||||||||||||||||||||||||||||| Sbjct 1 RSSRAGLQFPVARVHR#LLRKGNYAERVGAGAPVYLAAVLEYLTAEI 46 |
| Apis mellifera | Query 17 TRSSRAGLQFPVGRVHR#LLRKGNYAERVGAGAPVYLAAVLEYLTAEI 63 ||||||||||||||+||#||||||||||||||||||||||+||| ||+ Sbjct 16 TRSSRAGLQFPVGRIHR#LLRKGNYAERVGAGAPVYLAAVMEYLAAEV 62 |
| Arabidopsis thaliana | Query 17 TRSSRAGLQFPVGRVHR#LLRKGNYAERVGAGAPVYLAAVLEYLTAEI 63 +|||||||||||||+||#||||||||||||||||||||||+||| ||+ Sbjct 16 SRSSRAGLQFPVGRIHR#LLRKGNYAERVGAGAPVYLAAVMEYLAAEV 62 |
| Asellus aquaticus | Query 17 TRSSRAGLQFPVGRVHR#LLRKGNYAERVGAGAPVYLAAVLEYLTAEI 63 +|||||||||||||+||#||||||||||||||||||||||+||| ||+ Sbjct 16 SRSSRAGLQFPVGRIHR#LLRKGNYAERVGAGAPVYLAAVMEYLAAEV 62 |
| Tribolium castaneum | Query 17 TRSSRAGLQFPVGRVHR#LLRKGNYAERVGAGAPVYLAAVLEYLTAEI 63 +|||||||||||||+||#||||||||||||||||||||||+||| ||+ Sbjct 16 SRSSRAGLQFPVGRIHR#LLRKGNYAERVGAGAPVYLAAVMEYLAAEV 62 |
[Site 4] QFPVGRVHRL34-LRKGNYAERV
Leu34  Leu
Leu
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Gln25 | Phe26 | Pro27 | Val28 | Gly29 | Arg30 | Val31 | His32 | Arg33 | Leu34 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Leu35 | Arg36 | Lys37 | Gly38 | Asn39 | Tyr40 | Ala41 | Glu42 | Arg43 | Val44 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| GKQGGKARAKAKTRSSRAGLQFPVGRVHRLLRKGNYAERVGAGAPVYLAAVLEYLTAEIL |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | Danio rerio | 120.00 | 2 | hypothetical protein LOC393453 |
| 2 | Homo sapiens | 119.00 | 15 | histone cluster 1, H2ai |
| 3 | N/A | 119.00 | 14 | 1109175A homeostatic thymus hormone alpha |
| 4 | Canis familiaris | 119.00 | 14 | PREDICTED: similar to Histone H2A.l (H2A/l) |
| 5 | Mus musculus | 118.00 | 6 | histone cluster 1, H2ak |
| 6 | Xenopus laevis | 118.00 | 5 | hypothetical protein LOC734746 |
| 7 | Rattus norvegicus | 118.00 | 2 | H2A1F_RAT Histone H2A type 1-F gi |
| 8 | Cairina moschata | 118.00 | 1 | H2A_CAIMO Histone H2A gi |
| 9 | Gallus gallus | 118.00 | 1 | histone cluster 2, H2ac |
| 10 | Gallus gallus | 117.00 | 6 | H2A3_CHICK Histone H2A-III gi |
| 11 | Bos taurus | 117.00 | 4 | PREDICTED: similar to homeostatic thymus hormone |
| 12 | Xenopus tropicalis | 117.00 | 3 | MGC69325 protein |
| 13 | Bos taurus | 117.00 | 3 | PREDICTED: similar to Chain K, 2.9 Angstrom X-Ray |
| 14 | synthetic construct | 117.00 | 2 | histone 1 H2ac |
| 15 | Mus musculus domesticus | 117.00 | 1 | histone H2A |
| 16 | Tetraodon nigroviridis | 116.00 | 7 | unnamed protein product |
| 17 | Danio rerio | 116.00 | 6 | PREDICTED: similar to Mid1ip1 protein |
| 18 | Mus musculus | 116.00 | 2 | H2A histone family, member J |
| 19 | Homo sapiens | 116.00 | 2 | H2A histone family, member J |
| 20 | Bufo bufo gagarizans | 116.00 | 2 | replication-dependent histone H2A |
| 21 | Oncorhynchus mykiss | 116.00 | 1 | H2A_ONCMY Histone H2A gi |
| 22 | Bufo bufo gagarizans | 116.00 | 1 | AF255739_1 replication-dependent histone H2A |
| 23 | Plasmodium berghei strain ANKA | 114.00 | 1 | histone h2a |
| 24 | Rattus norvegicus | 110.00 | 1 | histone cluster 1, H2aa |
| 25 | Strongylocentrotus purpuratus | 106.00 | 2 | late histone L3 H2a |
| 26 | Acropora formosa | 99.80 | 1 | H2A_ACRFO Histone H2A gi |
| 27 | Ustilago maydis 521 | 95.90 | 1 | histone H2A |
| 28 | Pan troglodytes | 95.50 | 1 | PREDICTED: similar to none |
| 29 | Strongylocentrotus purpuratus | 95.10 | 1 | PREDICTED: similar to histone L3 H2a |
| 30 | Schizosaccharomyces pombe 972h- | 94.40 | 1 | histone H2A alpha (PMID 3018512) |
| 31 | Schizosaccharomyces pombe 972h- | 94.40 | 1 | histone H2A beta |
| 32 | Platynereis dumerilii | 94.40 | 1 | H2A_PLADU Histone H2A gi |
| 33 | Apis mellifera | 94.00 | 1 | PREDICTED: similar to CG31618-PA |
| 34 | Schistosoma japonicum | 94.00 | 1 | SJCHGC05779 protein |
| 35 | Arabidopsis thaliana | 93.60 | 1 | Unknown protein |
| 36 | Tribolium castaneum | 93.20 | 1 | PREDICTED: similar to CG31618-PA |
| 37 | Asellus aquaticus | 93.20 | 1 | putative H2A histone |
| 38 | Oikopleura dioica | 93.20 | 1 | histone h2A.3 |
| 39 | Litopenaeus vannamei | 91.30 | 1 | histone H2A |
| 40 | Contains: Hipposin | 90.90 | 1 | H2A_HIPHI Histone H2A |
Top-ranked sequences
| organism | matching |
|---|---|
| Danio rerio | Query 5 GKQGGKARAKAKTRSSRAGLQFPVGRVHRL#LRKGNYAERVGAGAPVYLAAVLEYLTAEIL 64 || |||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Sbjct 5 GKTGGKARAKAKTRSSRAGLQFPVGRVHRL#LRKGNYAERVGAGAPVYLAAVLEYLTAEIL 64 |
| Homo sapiens | Query 5 GKQGGKARAKAKTRSSRAGLQFPVGRVHRL#LRKGNYAERVGAGAPVYLAAVLEYLTAEIL 64 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 5 GKQGGKARAKAKTRSSRAGLQFPVGRVHRL#LRKGNYAERVGAGAPVYLAAVLEYLTAEIL 64 |
| N/A | Query 5 GKQGGKARAKAKTRSSRAGLQFPVGRVHRL#LRKGNYAERVGAGAPVYLAAVLEYLTAEIL 64 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 5 GKQGGKARAKAKTRSSRAGLQFPVGRVHRL#LRKGNYAERVGAGAPVYLAAVLEYLTAEIL 64 |
| Canis familiaris | Query 5 GKQGGKARAKAKTRSSRAGLQFPVGRVHRL#LRKGNYAERVGAGAPVYLAAVLEYLTAEIL 64 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 5 GKQGGKARAKAKTRSSRAGLQFPVGRVHRL#LRKGNYAERVGAGAPVYLAAVLEYLTAEIL 64 |
| Mus musculus | Query 5 GKQGGKARAKAKTRSSRAGLQFPVGRVHRL#LRKGNYAERVGAGAPVYLAAVLEYLTAEIL 64 ||||||||||||||||||||||||||||||#||||||+||||||||||||||||||||||| Sbjct 5 GKQGGKARAKAKTRSSRAGLQFPVGRVHRL#LRKGNYSERVGAGAPVYLAAVLEYLTAEIL 64 |
| Xenopus laevis | Query 5 GKQGGKARAKAKTRSSRAGLQFPVGRVHRL#LRKGNYAERVGAGAPVYLAAVLEYLTAEIL 64 |||||| |||||||||||||||||||||||#|||||||||||||||||||||||||||||| Sbjct 5 GKQGGKTRAKAKTRSSRAGLQFPVGRVHRL#LRKGNYAERVGAGAPVYLAAVLEYLTAEIL 64 |
| Rattus norvegicus | Query 5 GKQGGKARAKAKTRSSRAGLQFPVGRVHRL#LRKGNYAERVGAGAPVYLAAVLEYLTAEIL 64 ||||||||||||||||||||||||||||||#||||||+||||||||||||||||||||||| Sbjct 5 GKQGGKARAKAKTRSSRAGLQFPVGRVHRL#LRKGNYSERVGAGAPVYLAAVLEYLTAEIL 64 |
| Cairina moschata | Query 5 GKQGGKARAKAKTRSSRAGLQFPVGRVHRL#LRKGNYAERVGAGAPVYLAAVLEYLTAEIL 64 ||||||||||||+|||||||||||||||||#|||||||||||||||||||||||||||||| Sbjct 5 GKQGGKARAKAKSRSSRAGLQFPVGRVHRL#LRKGNYAERVGAGAPVYLAAVLEYLTAEIL 64 |
| Gallus gallus | Query 5 GKQGGKARAKAKTRSSRAGLQFPVGRVHRL#LRKGNYAERVGAGAPVYLAAVLEYLTAEIL 64 ||||||||||||+|||||||||||||||||#|||||||||||||||||||||||||||||| Sbjct 5 GKQGGKARAKAKSRSSRAGLQFPVGRVHRL#LRKGNYAERVGAGAPVYLAAVLEYLTAEIL 64 |
| Gallus gallus | Query 5 GKQGGKARAKAKTRSSRAGLQFPVGRVHRL#LRKGNYAERVGAGAPVYLAAVLEYLTAEIL 64 ||||||||||||+|||||||||||||||||#|||||||||||||||||||||||||||||| Sbjct 5 GKQGGKARAKAKSRSSRAGLQFPVGRVHRL#LRKGNYAERVGAGAPVYLAAVLEYLTAEIL 64 |
| Bos taurus | Query 5 GKQGGKARAKAKTRSSRAGLQFPVGRVHRL#LRKGNYAERVGAGAPVYLAAVLEYLTAEIL 64 |||||||||||+||||||||||||||||||#|||||||||||||||||||||||||||||| Sbjct 5 GKQGGKARAKARTRSSRAGLQFPVGRVHRL#LRKGNYAERVGAGAPVYLAAVLEYLTAEIL 64 |
| Xenopus tropicalis | Query 5 GKQGGKARAKAKTRSSRAGLQFPVGRVHRL#LRKGNYAERVGAGAPVYLAAVLEYLTAEIL 64 |||||| |||||||||||||||||||||||#|||||||||||||||||||||||||||||| Sbjct 5 GKQGGKVRAKAKTRSSRAGLQFPVGRVHRL#LRKGNYAERVGAGAPVYLAAVLEYLTAEIL 64 |
| Bos taurus | Query 5 GKQGGKARAKAKTRSSRAGLQFPVGRVHRL#LRKGNYAERVGAGAPVYLAAVLEYLTAEIL 64 ||||||||||||||||||||||||||||||#||||||+||||||||||||||||||||||| Sbjct 5 GKQGGKARAKAKTRSSRAGLQFPVGRVHRL#LRKGNYSERVGAGAPVYLAAVLEYLTAEIL 64 |
| synthetic construct | Query 5 GKQGGKARAKAKTRSSRAGLQFPVGRVHRL#LRKGNYAERVGAGAPVYLAAVLEYLTAEIL 64 ||||||||||||+|||||||||||||||||#|||||||||||||||||||||||||||||| Sbjct 5 GKQGGKARAKAKSRSSRAGLQFPVGRVHRL#LRKGNYAERVGAGAPVYLAAVLEYLTAEIL 64 |
| Mus musculus domesticus | Query 5 GKQGGKARAKAKTRSSRAGLQFPVGRVHRL#LRKGNYAERVGAGAPVYLAAVLEYLTAEIL 64 ||||||||||||+|||||||||||||||||#|||||||||||||||||+|||||||||||| Sbjct 12 GKQGGKARAKAKSRSSRAGLQFPVGRVHRL#LRKGNYAERVGAGAPVYMAAVLEYLTAEIL 71 |
| Tetraodon nigroviridis | Query 5 GKQGGKARAKAKTRSSRAGLQFPVGRVHRL#LRKGNYAERVGAGAPVYLAAVLEYLTAEIL 64 || |||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Sbjct 5 GKTGGKARAKAKTRSSRAGLQFPVGRVHRL#LRKGNYAERVGAGAPVYLAAVLEYLTAEIL 64 |
| Danio rerio | Query 5 GKQGGKARAKAKTRSSRAGLQFPVGRVHRL#LRKGNYAERVGAGAPVYLAAVLEYLTAEIL 64 || |||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Sbjct 5 GKTGGKARAKAKTRSSRAGLQFPVGRVHRL#LRKGNYAERVGAGAPVYLAAVLEYLTAEIL 64 |
| Mus musculus | Query 5 GKQGGKARAKAKTRSSRAGLQFPVGRVHRL#LRKGNYAERVGAGAPVYLAAVLEYLTAEIL 64 |||||| |||||+|||||||||||||||||#|||||||||||||||||||||||||||||| Sbjct 5 GKQGGKVRAKAKSRSSRAGLQFPVGRVHRL#LRKGNYAERVGAGAPVYLAAVLEYLTAEIL 64 |
| Homo sapiens | Query 5 GKQGGKARAKAKTRSSRAGLQFPVGRVHRL#LRKGNYAERVGAGAPVYLAAVLEYLTAEIL 64 |||||| |||||+|||||||||||||||||#|||||||||||||||||||||||||||||| Sbjct 5 GKQGGKVRAKAKSRSSRAGLQFPVGRVHRL#LRKGNYAERVGAGAPVYLAAVLEYLTAEIL 64 |
| Bufo bufo gagarizans | Query 5 GKQGGKARAKAKTRSSRAGLQFPVGRVHRL#LRKGNYAERVGAGAPVYLAAVLEYLTAEIL 64 |||||| |||||||||||||||||||||||#|||||||+|||||||||||||||||||||| Sbjct 5 GKQGGKVRAKAKTRSSRAGLQFPVGRVHRL#LRKGNYAQRVGAGAPVYLAAVLEYLTAEIL 64 |
| Oncorhynchus mykiss | Query 5 GKQGGKARAKAKTRSSRAGLQFPVGRVHRL#LRKGNYAERVGAGAPVYLAAVLEYLTAEIL 64 || |||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Sbjct 5 GKTGGKARAKAKTRSSRAGLQFPVGRVHRL#LRKGNYAERVGAGAPVYLAAVLEYLTAEIL 64 |
| Bufo bufo gagarizans | Query 5 GKQGGKARAKAKTRSSRAGLQFPVGRVHRL#LRKGNYAERVGAGAPVYLAAVLEYLTAEIL 64 |||||| |||||||||||||||||||||||#|||||||+|||||||||||||||||||||| Sbjct 5 GKQGGKVRAKAKTRSSRAGLQFPVGRVHRL#LRKGNYAQRVGAGAPVYLAAVLEYLTAEIL 64 |
| Plasmodium berghei strain ANKA | Query 6 KQGGKARAKAKTRSSRAGLQFPVGRVHRL#LRKGNYAERVGAGAPVYLAAVLEYLTAEIL 64 |||||||||||+|||||||||||||||||#|||||||||||+|||||+|||||||||||| Sbjct 1 KQGGKARAKAKSRSSRAGLQFPVGRVHRL#LRKGNYAERVGSGAPVYMAAVLEYLTAEIL 59 |
| Rattus norvegicus | Query 6 KQGGKARAKAKTRSSRAGLQFPVGRVHRL#LRKGNYAERVGAGAPVYLAAVLEYLTAEIL 64 |||||||||||+|| ||||||||||||||#||+||||||+||| |||||||||||||||| Sbjct 6 KQGGKARAKAKSRSFRAGLQFPVGRVHRL#LRQGNYAERIGAGTPVYLAAVLEYLTAEIL 64 |
| Strongylocentrotus purpuratus | Query 5 GKQGGKARAKAKTRSSRAGLQFPVGRVHRL#LRKGNYAERVGAGAPVYLAAVLEYLTAEIL 64 || ||||||||+||+||||||||||||| #|||||||+||||||||||||||||| |||| Sbjct 5 GKGAGKARAKAKSRSARAGLQFPVGRVHRF#LRKGNYAQRVGAGAPVYLAAVLEYLAAEIL 64 |
| Acropora formosa | Query 5 GKQGGKAR-AKAKTRSSRAGLQFPVGRVHRL#LRKGNYAERVGAGAPVYLAAVLEYLTAEI 63 |+ |||+ |+|+|||||||||||||+|||#|||||||||||||||||+|||||||+||| Sbjct 3 GRGKGKAQGTKSKSRSSRAGLQFPVGRIHRL#LRKGNYAERVGAGAPVYMAAVLEYLSAEI 62 Query 64 L 64 | Query 64 L 64 |
| Ustilago maydis 521 | Query 10 KARAKAKTRSSRAGLQFPVGRVHRL#LRKGNYAERVGAGAPVYLAAVLEYLTAEIL 64 | +||++||++|||||||||+|||#|||||||+||||||||||||||||| |||| Sbjct 13 DASSKAQSRSAKAGLQFPVGRIHRL#LRKGNYAQRVGAGAPVYLAAVLEYLAAEIL 67 |
| Pan troglodytes | Query 7 QGGKARAKAKTRSSRAGLQFPVGRVHRL#LRKGNYAERVGAGAPVYLAAVLEYLTAEIL 64 || |||||||| |||||||| |||+|||#|||||||+ | ||||||| ||||||||+|| Sbjct 2 QGSKARAKAKTHSSRAGLQFSVGRMHRL#LRKGNYAKPVRAGAPVYLEAVLEYLTAKIL 59 |
| Strongylocentrotus purpuratus | Query 13 AKAKTRSSRAGLQFPVGRVHRL#LRKGNYAERVGAGAPVYLAAVLEYLTAEIL 64 +|||+||+||||||||||||| #||||||| ||||||||||||||||| |||| Sbjct 13 SKAKSRSARAGLQFPVGRVHRF#LRKGNYAARVGAGAPVYLAAVLEYLAAEIL 64 |
| Schizosaccharomyces pombe 972h- | Query 5 GKQGGKARA--KAKTRSSRAGLQFPVGRVHRL#LRKGNYAERVGAGAPVYLAAVLEYLTAE 62 || |||| |++||++||| |||||||||#|||||||+||||||||||||||||| || Sbjct 4 GKSGGKAAVAKSAQSRSAKAGLAFPVGRVHRL#LRKGNYAQRVGAGAPVYLAAVLEYLAAE 63 Query 63 IL 64 || Query 63 IL 64 |
| Schizosaccharomyces pombe 972h- | Query 5 GKQGGKARA--KAKTRSSRAGLQFPVGRVHRL#LRKGNYAERVGAGAPVYLAAVLEYLTAE 62 || |||| |++||++||| |||||||||#|||||||+||||||||||||||||| || Sbjct 4 GKSGGKAAVAKSAQSRSAKAGLAFPVGRVHRL#LRKGNYAQRVGAGAPVYLAAVLEYLAAE 63 Query 63 IL 64 || Query 63 IL 64 |
| Platynereis dumerilii | Query 17 TRSSRAGLQFPVGRVHRL#LRKGNYAERVGAGAPVYLAAVLEYLTAEIL 64 ||||||||||||||+|||#|||||||||||||||||||||+||| ||+| Sbjct 16 TRSSRAGLQFPVGRIHRL#LRKGNYAERVGAGAPVYLAAVMEYLAAEVL 63 |
| Apis mellifera | Query 17 TRSSRAGLQFPVGRVHRL#LRKGNYAERVGAGAPVYLAAVLEYLTAEIL 64 ||||||||||||||+|||#|||||||||||||||||||||+||| ||+| Sbjct 16 TRSSRAGLQFPVGRIHRL#LRKGNYAERVGAGAPVYLAAVMEYLAAEVL 63 |
| Schistosoma japonicum | Query 18 RSSRAGLQFPVGRVHRL#LRKGNYAERVGAGAPVYLAAVLEYLTAEIL 64 ||||||||||| |||||#|||||||||||||||||||||||||||||| Sbjct 1 RSSRAGLQFPVARVHRL#LRKGNYAERVGAGAPVYLAAVLEYLTAEIL 47 |
| Arabidopsis thaliana | Query 17 TRSSRAGLQFPVGRVHRL#LRKGNYAERVGAGAPVYLAAVLEYLTAEIL 64 +|||||||||||||+|||#|||||||||||||||||||||+||| ||+| Sbjct 16 SRSSRAGLQFPVGRIHRL#LRKGNYAERVGAGAPVYLAAVMEYLAAEVL 63 |
| Tribolium castaneum | Query 17 TRSSRAGLQFPVGRVHRL#LRKGNYAERVGAGAPVYLAAVLEYLTAEIL 64 +|||||||||||||+|||#|||||||||||||||||||||+||| ||+| Sbjct 16 SRSSRAGLQFPVGRIHRL#LRKGNYAERVGAGAPVYLAAVMEYLAAEVL 63 |
| Asellus aquaticus | Query 17 TRSSRAGLQFPVGRVHRL#LRKGNYAERVGAGAPVYLAAVLEYLTAEIL 64 +|||||||||||||+|||#|||||||||||||||||||||+||| ||+| Sbjct 16 SRSSRAGLQFPVGRIHRL#LRKGNYAERVGAGAPVYLAAVMEYLAAEVL 63 |
| Oikopleura dioica | Query 12 RAKAKTRSSRAGLQFPVGRVHRL#LRKGNYAERVGAGAPVYLAAVLEYLTAEIL 64 +| |+||||||||||||||||+#|| ||+|+||||||||||||||||||||+ Sbjct 13 KAGKKSRSSRAGLQFPVGRVHRM#LRNGNWADRVGAGAPVYLAAVLEYLTAEVC 65 |
| Litopenaeus vannamei | Query 18 RSSRAGLQFPVGRVHRL#LRKGNYAERVGAGAPVYLAAVLEYLTAEIL 64 |||||||||||||+|||#|||||||||||||||||||||+||| ||+| Sbjct 17 RSSRAGLQFPVGRIHRL#LRKGNYAERVGAGAPVYLAAVMEYLAAEVL 63 |
| Contains: Hipposin | Query 5 GKQGGKARAKAKTRSSRAGLQFPVGRVHRL#LRKGNYAERVGAGAPVYL 52 || |||||||||||||||||||||||||||#||||||| |||||||||| Sbjct 5 GKTGGKARAKAKTRSSRAGLQFPVGRVHRL#LRKGNYAHRVGAGAPVYL 52 |
[Site 5] APVYLAAVLE57-YLTAEILELA
Glu57  Tyr
Tyr
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Ala48 | Pro49 | Val50 | Tyr51 | Leu52 | Ala53 | Ala54 | Val55 | Leu56 | Glu57 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Tyr58 | Leu59 | Thr60 | Ala61 | Glu62 | Ile63 | Leu64 | Glu65 | Leu66 | Ala67 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| VGRVHRLLRKGNYAERVGAGAPVYLAAVLEYLTAEILELAGNAARDNKKTRIIPRHLQLA |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | N/A | 121.00 | 15 | C Chain C, Structure Of The 4_601_167 Tetranucleos |
| 2 | Xenopus laevis | 121.00 | 4 | hypothetical protein LOC734746 |
| 3 | Xenopus tropicalis | 121.00 | 3 | MGC69325 protein |
| 4 | Danio rerio | 121.00 | 1 | H2A histone family, member X |
| 5 | Canis familiaris | 120.00 | 14 | PREDICTED: similar to Histone H2A.l (H2A/l) |
| 6 | Homo sapiens | 120.00 | 12 | histone cluster 2, H2ab |
| 7 | Danio rerio | 120.00 | 6 | PREDICTED: similar to Mid1ip1 protein |
| 8 | Gallus gallus | 120.00 | 5 | histone H2A |
| 9 | Tetraodon nigroviridis | 120.00 | 5 | unnamed protein product |
| 10 | Bos taurus | 120.00 | 3 | PREDICTED: similar to homeostatic thymus hormone |
| 11 | synthetic construct | 120.00 | 2 | histone 1 H2ac |
| 12 | Homo sapiens | 120.00 | 2 | H2A histone family, member J |
| 13 | Mus musculus | 120.00 | 2 | H2A histone family, member J |
| 14 | Gallus gallus | 120.00 | 1 | histone cluster 2, H2ac |
| 15 | Cairina moschata | 120.00 | 1 | H2A_CAIMO Histone H2A gi |
| 16 | Mus musculus domesticus | 120.00 | 1 | histone H2A |
| 17 | Oncorhynchus mykiss | 120.00 | 1 | H2A_ONCMY Histone H2A gi |
| 18 | Mus musculus | 119.00 | 6 | histone cluster 1, H2af |
| 19 | Bos taurus | 119.00 | 2 | PREDICTED: similar to Chain K, 2.9 Angstrom X-Ray |
| 20 | Bufo bufo gagarizans | 119.00 | 1 | replication-dependent histone H2A |
| 21 | Plasmodium berghei strain ANKA | 119.00 | 1 | histone h2a |
| 22 | Bufo bufo gagarizans | 119.00 | 1 | AF255739_1 replication-dependent histone H2A |
| 23 | Oikopleura dioica | 117.00 | 2 | histone h2A.1 |
| 24 | Schistosoma japonicum | 117.00 | 2 | similar to GenBank Accession Number X01064 histon |
| 25 | Magnaporthe grisea 70-15 | 117.00 | 1 | hypothetical protein MGG_03577 |
| 26 | Neosartorya fischeri NRRL 181 | 117.00 | 1 | histone h2a |
| 27 | Arabidopsis thaliana | 117.00 | 1 | Unknown protein |
| 28 | Aspergillus nidulans FGSC A4 | 117.00 | 1 | H2A_EMENI Histone H2A |
| 29 | Neurospora crassa OR74A | 117.00 | 1 | histone H2A |
| 30 | Aspergillus niger | 117.00 | 1 | histone H2A httA-Aspergillus niger gi |
| 31 | Gibberella zeae PH-1 | 117.00 | 1 | H2A_NEUCR Histone H2A |
| 32 | Podospora anserina | 117.00 | 1 | H2A_PODAN Histone H2A gi |
| 33 | Rattus norvegicus | 117.00 | 1 | histone cluster 1, H2aa |
| 34 | Botryotinia fuckeliana | 117.00 | 1 | histone H2A |
| 35 | Schizosaccharomyces pombe 972h- | 117.00 | 1 | histone H2A alpha (PMID 3018512) |
| 36 | Schizosaccharomyces pombe 972h- | 117.00 | 1 | histone H2A beta |
| 37 | Acropora formosa | 117.00 | 1 | H2A_ACRFO Histone H2A gi |
| 38 | Strongylocentrotus purpuratus | 116.00 | 2 | late histone L3 H2a |
| 39 | Drosophila melanogaster | 116.00 | 2 | His2A:CG31618 CG31618-PA |
| 40 | Rattus norvegicus | 116.00 | 1 | H2A1F_RAT Histone H2A type 1-F gi |
| 41 | Tribolium castaneum | 116.00 | 1 | PREDICTED: similar to CG31618-PA |
| 42 | Asellus aquaticus | 116.00 | 1 | putative H2A histone |
| 43 | Urechis caupo | 116.00 | 1 | H2A_URECA Histone H2A gi |
| 44 | Apis mellifera | 116.00 | 1 | PREDICTED: similar to CG31618-PA |
| 45 | Platynereis dumerilii | 116.00 | 1 | H2A_PLADU Histone H2A gi |
| 46 | Anopheles gambiae str. PEST | 116.00 | 1 | AGAP003913-PA |
| 47 | Ustilago maydis 521 | 115.00 | 1 | histone H2A |
| 48 | Ashbya gossypii ATCC 10895 | 115.00 | 1 | AEL003Cp |
| 49 | Chironomus thummi | 115.00 | 1 | H2A_CHITH Histone H2A gi |
| 50 | Mytilus edulis | 115.00 | 1 | H2A_MYTCA Histone H2A gi |
| 51 | Litopenaeus vannamei | 114.00 | 1 | histone H2A |
| 52 | Ashbya gossypii | 114.00 | 1 | AF384989_1 histone 2A |
| 53 | Strongylocentrotus purpuratus | 114.00 | 1 | PREDICTED: similar to histone L3 H2a |
Top-ranked sequences
| organism | matching |
|---|---|
| N/A | Query 28 VGRVHRLLRKGNYAERVGAGAPVYLAAVLE#YLTAEILELAGNAARDNKKTRIIPRHLQLA 87 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 28 VGRVHRLLRKGNYAERVGAGAPVYLAAVLE#YLTAEILELAGNAARDNKKTRIIPRHLQLA 87 |
| Xenopus laevis | Query 28 VGRVHRLLRKGNYAERVGAGAPVYLAAVLE#YLTAEILELAGNAARDNKKTRIIPRHLQLA 87 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 28 VGRVHRLLRKGNYAERVGAGAPVYLAAVLE#YLTAEILELAGNAARDNKKTRIIPRHLQLA 87 |
| Xenopus tropicalis | Query 28 VGRVHRLLRKGNYAERVGAGAPVYLAAVLE#YLTAEILELAGNAARDNKKTRIIPRHLQLA 87 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 28 VGRVHRLLRKGNYAERVGAGAPVYLAAVLE#YLTAEILELAGNAARDNKKTRIIPRHLQLA 87 |
| Danio rerio | Query 28 VGRVHRLLRKGNYAERVGAGAPVYLAAVLE#YLTAEILELAGNAARDNKKTRIIPRHLQLA 87 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 28 VGRVHRLLRKGNYAERVGAGAPVYLAAVLE#YLTAEILELAGNAARDNKKTRIIPRHLQLA 87 |
| Canis familiaris | Query 28 VGRVHRLLRKGNYAERVGAGAPVYLAAVLE#YLTAEILELAGNAARDNKKTRIIPRHLQLA 87 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 28 VGRVHRLLRKGNYAERVGAGAPVYLAAVLE#YLTAEILELAGNAARDNKKTRIIPRHLQLA 87 |
| Homo sapiens | Query 28 VGRVHRLLRKGNYAERVGAGAPVYLAAVLE#YLTAEILELAGNAARDNKKTRIIPRHLQLA 87 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 28 VGRVHRLLRKGNYAERVGAGAPVYLAAVLE#YLTAEILELAGNAARDNKKTRIIPRHLQLA 87 |
| Danio rerio | Query 28 VGRVHRLLRKGNYAERVGAGAPVYLAAVLE#YLTAEILELAGNAARDNKKTRIIPRHLQLA 87 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 28 VGRVHRLLRKGNYAERVGAGAPVYLAAVLE#YLTAEILELAGNAARDNKKTRIIPRHLQLA 87 |
| Gallus gallus | Query 28 VGRVHRLLRKGNYAERVGAGAPVYLAAVLE#YLTAEILELAGNAARDNKKTRIIPRHLQLA 87 ||||||||||||||||||||||||+|||||#|||||||||||||||||||||||||||||| Sbjct 28 VGRVHRLLRKGNYAERVGAGAPVYMAAVLE#YLTAEILELAGNAARDNKKTRIIPRHLQLA 87 |
| Tetraodon nigroviridis | Query 28 VGRVHRLLRKGNYAERVGAGAPVYLAAVLE#YLTAEILELAGNAARDNKKTRIIPRHLQLA 87 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 28 VGRVHRLLRKGNYAERVGAGAPVYLAAVLE#YLTAEILELAGNAARDNKKTRIIPRHLQLA 87 |
| Bos taurus | Query 28 VGRVHRLLRKGNYAERVGAGAPVYLAAVLE#YLTAEILELAGNAARDNKKTRIIPRHLQLA 87 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 28 VGRVHRLLRKGNYAERVGAGAPVYLAAVLE#YLTAEILELAGNAARDNKKTRIIPRHLQLA 87 |
| synthetic construct | Query 28 VGRVHRLLRKGNYAERVGAGAPVYLAAVLE#YLTAEILELAGNAARDNKKTRIIPRHLQLA 87 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 28 VGRVHRLLRKGNYAERVGAGAPVYLAAVLE#YLTAEILELAGNAARDNKKTRIIPRHLQLA 87 |
| Homo sapiens | Query 28 VGRVHRLLRKGNYAERVGAGAPVYLAAVLE#YLTAEILELAGNAARDNKKTRIIPRHLQLA 87 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 28 VGRVHRLLRKGNYAERVGAGAPVYLAAVLE#YLTAEILELAGNAARDNKKTRIIPRHLQLA 87 |
| Mus musculus | Query 28 VGRVHRLLRKGNYAERVGAGAPVYLAAVLE#YLTAEILELAGNAARDNKKTRIIPRHLQLA 87 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 28 VGRVHRLLRKGNYAERVGAGAPVYLAAVLE#YLTAEILELAGNAARDNKKTRIIPRHLQLA 87 |
| Gallus gallus | Query 28 VGRVHRLLRKGNYAERVGAGAPVYLAAVLE#YLTAEILELAGNAARDNKKTRIIPRHLQLA 87 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 28 VGRVHRLLRKGNYAERVGAGAPVYLAAVLE#YLTAEILELAGNAARDNKKTRIIPRHLQLA 87 |
| Cairina moschata | Query 28 VGRVHRLLRKGNYAERVGAGAPVYLAAVLE#YLTAEILELAGNAARDNKKTRIIPRHLQLA 87 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 28 VGRVHRLLRKGNYAERVGAGAPVYLAAVLE#YLTAEILELAGNAARDNKKTRIIPRHLQLA 87 |
| Mus musculus domesticus | Query 28 VGRVHRLLRKGNYAERVGAGAPVYLAAVLE#YLTAEILELAGNAARDNKKTRIIPRHLQLA 87 ||||||||||||||||||||||||+|||||#|||||||||||||||||||||||||||||| Sbjct 35 VGRVHRLLRKGNYAERVGAGAPVYMAAVLE#YLTAEILELAGNAARDNKKTRIIPRHLQLA 94 |
| Oncorhynchus mykiss | Query 28 VGRVHRLLRKGNYAERVGAGAPVYLAAVLE#YLTAEILELAGNAARDNKKTRIIPRHLQLA 87 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 28 VGRVHRLLRKGNYAERVGAGAPVYLAAVLE#YLTAEILELAGNAARDNKKTRIIPRHLQLA 87 |
| Mus musculus | Query 28 VGRVHRLLRKGNYAERVGAGAPVYLAAVLE#YLTAEILELAGNAARDNKKTRIIPRHLQLA 87 |||||||||||||+||||||||||||||||#|||||||||||||||||||||||||||||| Sbjct 28 VGRVHRLLRKGNYSERVGAGAPVYLAAVLE#YLTAEILELAGNAARDNKKTRIIPRHLQLA 87 |
| Bos taurus | Query 28 VGRVHRLLRKGNYAERVGAGAPVYLAAVLE#YLTAEILELAGNAARDNKKTRIIPRHLQLA 87 |||||||||||||+||||||||||||||||#|||||||||||||||||||||||||||||| Sbjct 28 VGRVHRLLRKGNYSERVGAGAPVYLAAVLE#YLTAEILELAGNAARDNKKTRIIPRHLQLA 87 |
| Bufo bufo gagarizans | Query 28 VGRVHRLLRKGNYAERVGAGAPVYLAAVLE#YLTAEILELAGNAARDNKKTRIIPRHLQLA 87 ||||||||||||||+|||||||||||||||#|||||||||||||||||||||||||||||| Sbjct 28 VGRVHRLLRKGNYAQRVGAGAPVYLAAVLE#YLTAEILELAGNAARDNKKTRIIPRHLQLA 87 |
| Plasmodium berghei strain ANKA | Query 28 VGRVHRLLRKGNYAERVGAGAPVYLAAVLE#YLTAEILELAGNAARDNKKTRIIPRHLQLA 87 ||||||||||||||||||+|||||+|||||#|||||||||||||||||||||||||||||| Sbjct 23 VGRVHRLLRKGNYAERVGSGAPVYMAAVLE#YLTAEILELAGNAARDNKKTRIIPRHLQLA 82 |
| Bufo bufo gagarizans | Query 28 VGRVHRLLRKGNYAERVGAGAPVYLAAVLE#YLTAEILELAGNAARDNKKTRIIPRHLQLA 87 ||||||||||||||+|||||||||||||||#|||||||||||||||||||||||||||||| Sbjct 28 VGRVHRLLRKGNYAQRVGAGAPVYLAAVLE#YLTAEILELAGNAARDNKKTRIIPRHLQLA 87 |
| Oikopleura dioica | Query 28 VGRVHRLLRKGNYAERVGAGAPVYLAAVLE#YLTAEILELAGNAARDNKKTRIIPRHLQLA 87 ||||||+|||||||||||||||||||||||#||+||+|||||||||||||||||||||||| Sbjct 27 VGRVHRMLRKGNYAERVGAGAPVYLAAVLE#YLSAEVLELAGNAARDNKKTRIIPRHLQLA 86 |
| Schistosoma japonicum | Query 28 VGRVHRLLRKGNYAERVGAGAPVYLAAVLE#YLTAEILELAGNAARDNKKTRIIPRHLQLA 87 | ||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Sbjct 28 VARVHRLLRKGNYAERVGAGAPVYLAAVLE#YLTAEILELAGNAARDNKKTRIIPRHLQLA 87 |
| Magnaporthe grisea 70-15 | Query 28 VGRVHRLLRKGNYAERVGAGAPVYLAAVLE#YLTAEILELAGNAARDNKKTRIIPRHLQLA 87 ||||||||||||||+|||||||||||||||#|| ||||||||||||||||||||||||||| Sbjct 30 VGRVHRLLRKGNYAQRVGAGAPVYLAAVLE#YLAAEILELAGNAARDNKKTRIIPRHLQLA 89 |
| Neosartorya fischeri NRRL 181 | Query 28 VGRVHRLLRKGNYAERVGAGAPVYLAAVLE#YLTAEILELAGNAARDNKKTRIIPRHLQLA 87 ||||||||||||||+|||||||||||||||#|| ||||||||||||||||||||||||||| Sbjct 29 VGRVHRLLRKGNYAQRVGAGAPVYLAAVLE#YLAAEILELAGNAARDNKKTRIIPRHLQLA 88 |
| Arabidopsis thaliana | Query 28 VGRVHRLLRKGNYAERVGAGAPVYLAAVLE#YLTAEILELAGNAARDNKKTRIIPRHLQLA 87 |||+||||||||||||||||||||||||+|#|| ||+|||||||||||||||||||||||| Sbjct 27 VGRIHRLLRKGNYAERVGAGAPVYLAAVME#YLAAEVLELAGNAARDNKKTRIIPRHLQLA 86 |
| Aspergillus nidulans FGSC A4 | Query 28 VGRVHRLLRKGNYAERVGAGAPVYLAAVLE#YLTAEILELAGNAARDNKKTRIIPRHLQLA 87 ||||||||||||||+|||||||||||||||#|| ||||||||||||||||||||||||||| Sbjct 29 VGRVHRLLRKGNYAQRVGAGAPVYLAAVLE#YLAAEILELAGNAARDNKKTRIIPRHLQLA 88 |
| Neurospora crassa OR74A | Query 28 VGRVHRLLRKGNYAERVGAGAPVYLAAVLE#YLTAEILELAGNAARDNKKTRIIPRHLQLA 87 ||||||||||||||+|||||||||||||||#|| ||||||||||||||||||||||||||| Sbjct 30 VGRVHRLLRKGNYAQRVGAGAPVYLAAVLE#YLAAEILELAGNAARDNKKTRIIPRHLQLA 89 |
| Aspergillus niger | Query 28 VGRVHRLLRKGNYAERVGAGAPVYLAAVLE#YLTAEILELAGNAARDNKKTRIIPRHLQLA 87 ||||||||||||||+|||||||||||||||#|| ||||||||||||||||||||||||||| Sbjct 29 VGRVHRLLRKGNYAQRVGAGAPVYLAAVLE#YLAAEILELAGNAARDNKKTRIIPRHLQLA 88 |
| Gibberella zeae PH-1 | Query 28 VGRVHRLLRKGNYAERVGAGAPVYLAAVLE#YLTAEILELAGNAARDNKKTRIIPRHLQLA 87 ||||||||||||||+|||||||||||||||#|| ||||||||||||||||||||||||||| Sbjct 30 VGRVHRLLRKGNYAQRVGAGAPVYLAAVLE#YLAAEILELAGNAARDNKKTRIIPRHLQLA 89 |
| Podospora anserina | Query 28 VGRVHRLLRKGNYAERVGAGAPVYLAAVLE#YLTAEILELAGNAARDNKKTRIIPRHLQLA 87 ||||||||||||||+|||||||||||||||#|| ||||||||||||||||||||||||||| Sbjct 30 VGRVHRLLRKGNYAQRVGAGAPVYLAAVLE#YLAAEILELAGNAARDNKKTRIIPRHLQLA 89 |
| Rattus norvegicus | Query 28 VGRVHRLLRKGNYAERVGAGAPVYLAAVLE#YLTAEILELAGNAARDNKKTRIIPRHLQLA 87 |||||||||+||||||+||| |||||||||#|||||||||||||||||||||||||||||| Sbjct 28 VGRVHRLLRQGNYAERIGAGTPVYLAAVLE#YLTAEILELAGNAARDNKKTRIIPRHLQLA 87 |
| Botryotinia fuckeliana | Query 28 VGRVHRLLRKGNYAERVGAGAPVYLAAVLE#YLTAEILELAGNAARDNKKTRIIPRHLQLA 87 ||||||||||||||+|||||||||||||||#|| ||||||||||||||||||||||||||| Sbjct 31 VGRVHRLLRKGNYAQRVGAGAPVYLAAVLE#YLAAEILELAGNAARDNKKTRIIPRHLQLA 90 |
| Schizosaccharomyces pombe 972h- | Query 28 VGRVHRLLRKGNYAERVGAGAPVYLAAVLE#YLTAEILELAGNAARDNKKTRIIPRHLQLA 87 ||||||||||||||+|||||||||||||||#|| ||||||||||||||||||||||||||| Sbjct 29 VGRVHRLLRKGNYAQRVGAGAPVYLAAVLE#YLAAEILELAGNAARDNKKTRIIPRHLQLA 88 |
| Schizosaccharomyces pombe 972h- | Query 28 VGRVHRLLRKGNYAERVGAGAPVYLAAVLE#YLTAEILELAGNAARDNKKTRIIPRHLQLA 87 ||||||||||||||+|||||||||||||||#|| ||||||||||||||||||||||||||| Sbjct 29 VGRVHRLLRKGNYAQRVGAGAPVYLAAVLE#YLAAEILELAGNAARDNKKTRIIPRHLQLA 88 |
| Acropora formosa | Query 28 VGRVHRLLRKGNYAERVGAGAPVYLAAVLE#YLTAEILELAGNAARDNKKTRIIPRHLQLA 87 |||+||||||||||||||||||||+|||||#||+||||||||||||||||+|||||||||| Sbjct 27 VGRIHRLLRKGNYAERVGAGAPVYMAAVLE#YLSAEILELAGNAARDNKKSRIIPRHLQLA 86 |
| Strongylocentrotus purpuratus | Query 28 VGRVHRLLRKGNYAERVGAGAPVYLAAVLE#YLTAEILELAGNAARDNKKTRIIPRHLQLA 87 |||||| |||||||+|||||||||||||||#|| ||||||||||||||||||||||||||| Sbjct 28 VGRVHRFLRKGNYAQRVGAGAPVYLAAVLE#YLAAEILELAGNAARDNKKTRIIPRHLQLA 87 |
| Drosophila melanogaster | Query 28 VGRVHRLLRKGNYAERVGAGAPVYLAAVLE#YLTAEILELAGNAARDNKKTRIIPRHLQLA 87 |||+||||||||||||||||||||||||+|#|| ||+|||||||||||||||||||||||| Sbjct 27 VGRIHRLLRKGNYAERVGAGAPVYLAAVME#YLAAEVLELAGNAARDNKKTRIIPRHLQLA 86 |
| Rattus norvegicus | Query 28 VGRVHRLLRKGNYAERVGAGAPVYLAAVLE#YLTAEILELAGNAARDNKKTRIIPRHLQLA 87 |||||||||||||+||||||||||||||||#|||||||||| ||||||||||||||||||| Sbjct 28 VGRVHRLLRKGNYSERVGAGAPVYLAAVLE#YLTAEILELAANAARDNKKTRIIPRHLQLA 87 |
| Tribolium castaneum | Query 28 VGRVHRLLRKGNYAERVGAGAPVYLAAVLE#YLTAEILELAGNAARDNKKTRIIPRHLQLA 87 |||+||||||||||||||||||||||||+|#|| ||+|||||||||||||||||||||||| Sbjct 27 VGRIHRLLRKGNYAERVGAGAPVYLAAVME#YLAAEVLELAGNAARDNKKTRIIPRHLQLA 86 |
| Asellus aquaticus | Query 28 VGRVHRLLRKGNYAERVGAGAPVYLAAVLE#YLTAEILELAGNAARDNKKTRIIPRHLQLA 87 |||+||||||||||||||||||||||||+|#|| ||+|||||||||||||||||||||||| Sbjct 27 VGRIHRLLRKGNYAERVGAGAPVYLAAVME#YLAAEVLELAGNAARDNKKTRIIPRHLQLA 86 |
| Urechis caupo | Query 28 VGRVHRLLRKGNYAERVGAGAPVYLAAVLE#YLTAEILELAGNAARDNKKTRIIPRHLQLA 87 |||+||||||||||||+|||||||||||+|#|| ||+|||||||||||||||||||||||| Sbjct 27 VGRIHRLLRKGNYAERIGAGAPVYLAAVME#YLAAEVLELAGNAARDNKKTRIIPRHLQLA 86 |
| Apis mellifera | Query 28 VGRVHRLLRKGNYAERVGAGAPVYLAAVLE#YLTAEILELAGNAARDNKKTRIIPRHLQLA 87 |||+||||||||||||||||||||||||+|#|| ||+|||||||||||||||||||||||| Sbjct 27 VGRIHRLLRKGNYAERVGAGAPVYLAAVME#YLAAEVLELAGNAARDNKKTRIIPRHLQLA 86 |
| Platynereis dumerilii | Query 28 VGRVHRLLRKGNYAERVGAGAPVYLAAVLE#YLTAEILELAGNAARDNKKTRIIPRHLQLA 87 |||+||||||||||||||||||||||||+|#|| ||+|||||||||||||||||||||||| Sbjct 27 VGRIHRLLRKGNYAERVGAGAPVYLAAVME#YLAAEVLELAGNAARDNKKTRIIPRHLQLA 86 |
| Anopheles gambiae str. PEST | Query 28 VGRVHRLLRKGNYAERVGAGAPVYLAAVLE#YLTAEILELAGNAARDNKKTRIIPRHLQLA 87 |||+||||||||||||||||||||||||+|#|| ||+|||||||||||||||||||||||| Sbjct 27 VGRIHRLLRKGNYAERVGAGAPVYLAAVME#YLAAEVLELAGNAARDNKKTRIIPRHLQLA 86 |
| Ustilago maydis 521 | Query 28 VGRVHRLLRKGNYAERVGAGAPVYLAAVLE#YLTAEILELAGNAARDNKKTRIIPRHLQLA 87 |||+||||||||||+|||||||||||||||#|| ||||||||||||||||+|||||||||| Sbjct 31 VGRIHRLLRKGNYAQRVGAGAPVYLAAVLE#YLAAEILELAGNAARDNKKSRIIPRHLQLA 90 |
| Ashbya gossypii ATCC 10895 | Query 28 VGRVHRLLRKGNYAERVGAGAPVYLAAVLE#YLTAEILELAGNAARDNKKTRIIPRHLQLA 87 ||||||||||||||+|+|+|||||| ||||#|| ||||||||||||||||||||||||||| Sbjct 28 VGRVHRLLRKGNYAQRIGSGAPVYLTAVLE#YLAAEILELAGNAARDNKKTRIIPRHLQLA 87 |
| Chironomus thummi | Query 28 VGRVHRLLRKGNYAERVGAGAPVYLAAVLE#YLTAEILELAGNAARDNKKTRIIPRHLQLA 87 |||+||||||||| ||||||||||||||+|#|| ||+|||||||||||||||||||||||| Sbjct 27 VGRIHRLLRKGNYGERVGAGAPVYLAAVME#YLAAEVLELAGNAARDNKKTRIIPRHLQLA 86 |
| Mytilus edulis | Query 28 VGRVHRLLRKGNYAERVGAGAPVYLAAVLE#YLTAEILELAGNAARDNKKTRIIPRHLQLA 87 |||+||||||||||||||||||||||||||#|| ||+|||||||||||||+|||||||||| Sbjct 27 VGRIHRLLRKGNYAERVGAGAPVYLAAVLE#YLAAEVLELAGNAARDNKKSRIIPRHLQLA 86 |
| Litopenaeus vannamei | Query 28 VGRVHRLLRKGNYAERVGAGAPVYLAAVLE#YLTAEILELAGNAARDNKKTRIIPRHLQLA 87 |||+||||||||||||||||||||||||+|#|| ||+||||||||||||||||+||||||| Sbjct 27 VGRIHRLLRKGNYAERVGAGAPVYLAAVME#YLAAEVLELAGNAARDNKKTRIVPRHLQLA 86 |
| Ashbya gossypii | Query 28 VGRVHRLLRKGNYAERVGAGAPVYLAAVLE#YLTAEILELAGNAARDNKKTRIIPRHLQLA 87 ||||||||||||||+|+|+|||||| ||||#|| ||||||||||||||||||||||||||| Sbjct 28 VGRVHRLLRKGNYAQRIGSGAPVYLTAVLE#YLAAEILELAGNAARDNKKTRIIPRHLQLA 87 |
| Strongylocentrotus purpuratus | Query 28 VGRVHRLLRKGNYAERVGAGAPVYLAAVLE#YLTAEILELAGNAARDNKKTRIIPRHLQLA 87 |||||| ||||||| |||||||||||||||#|| ||||||||||||||||||||||||||| Sbjct 28 VGRVHRFLRKGNYAARVGAGAPVYLAAVLE#YLAAEILELAGNAARDNKKTRIIPRHLQLA 87 |
[Site 6] DNKKTRIIPR82-HLQLAIRNDE
Arg82  His
His
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Asp73 | Asn74 | Lys75 | Lys76 | Thr77 | Arg78 | Ile79 | Ile80 | Pro81 | Arg82 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| His83 | Leu84 | Gln85 | Leu86 | Ala87 | Ile88 | Arg89 | Asn90 | Asp91 | Glu92 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| AAVLEYLTAEILELAGNAARDNKKTRIIPRHLQLAIRNDEELNKLLGKVTIAQGGVLPNI |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | N/A | 120.00 | 22 | A Chain A, Histone Octamer (Chicken), Chromosomal |
| 2 | Canis familiaris | 120.00 | 14 | PREDICTED: similar to histone 2, H2ac |
| 3 | Homo sapiens | 120.00 | 13 | histone cluster 2, H2aa3 |
| 4 | Gallus gallus | 120.00 | 3 | histone H2A |
| 5 | synthetic construct | 120.00 | 2 | histone 2 H2aa |
| 6 | Homo sapiens | 120.00 | 2 | H2A histone family, member J |
| 7 | Plasmodium berghei strain ANKA | 120.00 | 1 | histone h2a |
| 8 | Gallus gallus | 120.00 | 1 | histone cluster 2, H2ac |
| 9 | Mus musculus domesticus | 120.00 | 1 | histone H2A |
| 10 | Cairina moschata | 120.00 | 1 | H2A_CAIMO Histone H2A gi |
| 11 | Mus musculus | 119.00 | 6 | histone cluster 1, H2ak |
| 12 | Xenopus laevis | 119.00 | 4 | hypothetical protein LOC494591 |
| 13 | Bos taurus | 119.00 | 3 | PREDICTED: similar to homeostatic thymus hormone |
| 14 | Bos taurus | 119.00 | 3 | PREDICTED: similar to Chain K, 2.9 Angstrom X-Ray |
| 15 | Mus musculus | 119.00 | 2 | H2A histone family, member J |
| 16 | Rattus norvegicus | 119.00 | 2 | Histone H2a |
| 17 | Rattus norvegicus | 119.00 | 1 | histone cluster 1, H2aa |
| 18 | Tetraodon nigroviridis | 118.00 | 5 | unnamed protein product |
| 19 | Xenopus tropicalis | 118.00 | 3 | H2A histone family, member X |
| 20 | Danio rerio | 118.00 | 1 | H2A histone family, member X |
| 21 | Aspergillus fumigatus Af293 | 118.00 | 1 | histone H2A |
| 22 | Danio rerio | 117.00 | 6 | histone 2, H2a |
| 23 | Bufo bufo gagarizans | 117.00 | 1 | replication-dependent histone H2A |
| 24 | Oncorhynchus mykiss | 117.00 | 1 | H2A_ONCMY Histone H2A gi |
| 25 | Bufo bufo gagarizans | 117.00 | 1 | AF255739_1 replication-dependent histone H2A |
| 26 | Strongylocentrotus purpuratus | 116.00 | 2 | histone H2A |
| 27 | Schistosoma japonicum | 116.00 | 2 | similar to GenBank Accession Number X01064 histon |
| 28 | Gibberella zeae PH-1 | 116.00 | 1 | H2A_NEUCR Histone H2A |
| 29 | Oikopleura dioica | 115.00 | 2 | histone h2A.1 |
| 30 | Aspergillus nidulans FGSC A4 | 115.00 | 1 | H2A_EMENI Histone H2A |
| 31 | Magnaporthe grisea 70-15 | 115.00 | 1 | hypothetical protein MGG_03577 |
| 32 | Podospora anserina | 115.00 | 1 | H2A_PODAN Histone H2A gi |
| 33 | Botryotinia fuckeliana | 115.00 | 1 | histone H2A |
| 34 | Schizosaccharomyces pombe 972h- | 115.00 | 1 | histone H2A alpha (PMID 3018512) |
| 35 | Aspergillus niger | 115.00 | 1 | histone H2A httA-Aspergillus niger gi |
| 36 | Neurospora crassa OR74A | 115.00 | 1 | histone H2A |
| 37 | Neosartorya fischeri NRRL 181 | 115.00 | 1 | histone h2a |
| 38 | Strongylocentrotus purpuratus | 114.00 | 4 | PREDICTED: similar to histone H2A-2 |
| 39 | Schizosaccharomyces pombe 972h- | 114.00 | 1 | histone H2A beta |
| 40 | Ashbya gossypii ATCC 10895 | 113.00 | 2 | AEL003Cp |
| 41 | Arabidopsis thaliana | 113.00 | 1 | Unknown protein |
| 42 | Candida albicans SC5314 | 112.00 | 2 | histone H2A |
| 43 | Griffithsia japonica | 112.00 | 2 | histone H2A protein |
| 44 | Acropora formosa | 112.00 | 1 | H2A_ACRFO Histone H2A gi |
| 45 | Chironomus thummi | 111.00 | 2 | H2A_CHITH Histone H2A gi |
| 46 | Drosophila melanogaster | 111.00 | 1 | His2A:CG31618 CG31618-PA |
| 47 | Urechis caupo | 111.00 | 1 | H2A_URECA Histone H2A gi |
| 48 | Candida glabrata | 111.00 | 1 | unnamed protein product |
| 49 | Asellus aquaticus | 111.00 | 1 | putative H2A histone |
| 50 | Tribolium castaneum | 111.00 | 1 | PREDICTED: similar to CG31618-PA |
| 51 | Platynereis dumerilii | 111.00 | 1 | H2A_PLADU Histone H2A gi |
Top-ranked sequences
| organism | matching |
|---|---|
| N/A | Query 53 AAVLEYLTAEILELAGNAARDNKKTRIIPR#HLQLAIRNDEELNKLLGKVTIAQGGVLPNI 112 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 53 AAVLEYLTAEILELAGNAARDNKKTRIIPR#HLQLAIRNDEELNKLLGKVTIAQGGVLPNI 112 |
| Canis familiaris | Query 53 AAVLEYLTAEILELAGNAARDNKKTRIIPR#HLQLAIRNDEELNKLLGKVTIAQGGVLPNI 112 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 53 AAVLEYLTAEILELAGNAARDNKKTRIIPR#HLQLAIRNDEELNKLLGKVTIAQGGVLPNI 112 |
| Homo sapiens | Query 53 AAVLEYLTAEILELAGNAARDNKKTRIIPR#HLQLAIRNDEELNKLLGKVTIAQGGVLPNI 112 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 53 AAVLEYLTAEILELAGNAARDNKKTRIIPR#HLQLAIRNDEELNKLLGKVTIAQGGVLPNI 112 |
| Gallus gallus | Query 53 AAVLEYLTAEILELAGNAARDNKKTRIIPR#HLQLAIRNDEELNKLLGKVTIAQGGVLPNI 112 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 53 AAVLEYLTAEILELAGNAARDNKKTRIIPR#HLQLAIRNDEELNKLLGKVTIAQGGVLPNI 112 |
| synthetic construct | Query 53 AAVLEYLTAEILELAGNAARDNKKTRIIPR#HLQLAIRNDEELNKLLGKVTIAQGGVLPNI 112 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 53 AAVLEYLTAEILELAGNAARDNKKTRIIPR#HLQLAIRNDEELNKLLGKVTIAQGGVLPNI 112 |
| Homo sapiens | Query 53 AAVLEYLTAEILELAGNAARDNKKTRIIPR#HLQLAIRNDEELNKLLGKVTIAQGGVLPNI 112 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 53 AAVLEYLTAEILELAGNAARDNKKTRIIPR#HLQLAIRNDEELNKLLGKVTIAQGGVLPNI 112 |
| Plasmodium berghei strain ANKA | Query 53 AAVLEYLTAEILELAGNAARDNKKTRIIPR#HLQLAIRNDEELNKLLGKVTIAQGGVLPNI 112 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 53 AAVLEYLTAEILELAGNAARDNKKTRIIPR#HLQLAIRNDEELNKLLGKVTIAQGGVLPNI 112 |
| Gallus gallus | Query 53 AAVLEYLTAEILELAGNAARDNKKTRIIPR#HLQLAIRNDEELNKLLGKVTIAQGGVLPNI 112 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 53 AAVLEYLTAEILELAGNAARDNKKTRIIPR#HLQLAIRNDEELNKLLGKVTIAQGGVLPNI 112 |
| Mus musculus domesticus | Query 53 AAVLEYLTAEILELAGNAARDNKKTRIIPR#HLQLAIRNDEELNKLLGKVTIAQGGVLPNI 112 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 53 AAVLEYLTAEILELAGNAARDNKKTRIIPR#HLQLAIRNDEELNKLLGKVTIAQGGVLPNI 112 |
| Cairina moschata | Query 53 AAVLEYLTAEILELAGNAARDNKKTRIIPR#HLQLAIRNDEELNKLLGKVTIAQGGVLPNI 112 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 53 AAVLEYLTAEILELAGNAARDNKKTRIIPR#HLQLAIRNDEELNKLLGKVTIAQGGVLPNI 112 |
| Mus musculus | Query 53 AAVLEYLTAEILELAGNAARDNKKTRIIPR#HLQLAIRNDEELNKLLGKVTIAQGGVLPNI 112 ||||||||||||||||||||||||||||||#|||||||||||||||||+|||||||||||| Sbjct 53 AAVLEYLTAEILELAGNAARDNKKTRIIPR#HLQLAIRNDEELNKLLGRVTIAQGGVLPNI 112 |
| Xenopus laevis | Query 53 AAVLEYLTAEILELAGNAARDNKKTRIIPR#HLQLAIRNDEELNKLLGKVTIAQGGVLPNI 112 ||||||||||||||||||||||||||||||#|||||+|||||||||||+|||||||||||| Sbjct 53 AAVLEYLTAEILELAGNAARDNKKTRIIPR#HLQLAVRNDEELNKLLGRVTIAQGGVLPNI 112 |
| Bos taurus | Query 53 AAVLEYLTAEILELAGNAARDNKKTRIIPR#HLQLAIRNDEELNKLLGKVTIAQGGVLPNI 112 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 53 AAVLEYLTAEILELAGNAARDNKKTRIIPR#HLQLAIRNDEELNKLLGKVTIAQGGVLPNI 112 |
| Bos taurus | Query 53 AAVLEYLTAEILELAGNAARDNKKTRIIPR#HLQLAIRNDEELNKLLGKVTIAQGGVLPNI 112 ||||||||||||||||||||||||||||||#|||||||||||||||||+|||||||||||| Sbjct 53 AAVLEYLTAEILELAGNAARDNKKTRIIPR#HLQLAIRNDEELNKLLGRVTIAQGGVLPNI 112 |
| Mus musculus | Query 53 AAVLEYLTAEILELAGNAARDNKKTRIIPR#HLQLAIRNDEELNKLLGKVTIAQGGVLPNI 112 ||||||||||||||||||||||||||||||#|||||||||||||||||+|||||||||||| Sbjct 53 AAVLEYLTAEILELAGNAARDNKKTRIIPR#HLQLAIRNDEELNKLLGRVTIAQGGVLPNI 112 |
| Rattus norvegicus | Query 53 AAVLEYLTAEILELAGNAARDNKKTRIIPR#HLQLAIRNDEELNKLLGKVTIAQGGVLPNI 112 ||||||||||||||||||||||||||||||#|||||||||||||||||+|||||||||||| Sbjct 53 AAVLEYLTAEILELAGNAARDNKKTRIIPR#HLQLAIRNDEELNKLLGRVTIAQGGVLPNI 112 |
| Rattus norvegicus | Query 53 AAVLEYLTAEILELAGNAARDNKKTRIIPR#HLQLAIRNDEELNKLLGKVTIAQGGVLPNI 112 ||||||||||||||||||||||||||||||#|||||||||||||||||+|||||||||||| Sbjct 53 AAVLEYLTAEILELAGNAARDNKKTRIIPR#HLQLAIRNDEELNKLLGRVTIAQGGVLPNI 112 |
| Tetraodon nigroviridis | Query 53 AAVLEYLTAEILELAGNAARDNKKTRIIPR#HLQLAIRNDEELNKLLGKVTIAQGGVLPNI 112 ||||||||||||||||||||||||||||||#|||||+|||||||||||+|||||||||||| Sbjct 53 AAVLEYLTAEILELAGNAARDNKKTRIIPR#HLQLAVRNDEELNKLLGRVTIAQGGVLPNI 112 |
| Xenopus tropicalis | Query 53 AAVLEYLTAEILELAGNAARDNKKTRIIPR#HLQLAIRNDEELNKLLGKVTIAQGGVLPNI 112 ||||||||||||||||||||||||||||||#|||||+||||||||||| |||||||||||| Sbjct 53 AAVLEYLTAEILELAGNAARDNKKTRIIPR#HLQLAVRNDEELNKLLGGVTIAQGGVLPNI 112 |
| Danio rerio | Query 53 AAVLEYLTAEILELAGNAARDNKKTRIIPR#HLQLAIRNDEELNKLLGKVTIAQGGVLPNI 112 ||||||||||||||||||||||||||||||#|||||+||||||||||| |||||||||||| Sbjct 53 AAVLEYLTAEILELAGNAARDNKKTRIIPR#HLQLAVRNDEELNKLLGGVTIAQGGVLPNI 112 |
| Aspergillus fumigatus Af293 | Query 53 AAVLEYLTAEILELAGNAARDNKKTRIIPR#HLQLAIRNDEELNKLLGKVTIAQGGVLPNI 112 ||||||| ||||||||||||||||||||||#||||||||||||||||| |||||||||||| Sbjct 54 AAVLEYLAAEILELAGNAARDNKKTRIIPR#HLQLAIRNDEELNKLLGHVTIAQGGVLPNI 113 |
| Danio rerio | Query 53 AAVLEYLTAEILELAGNAARDNKKTRIIPR#HLQLAIRNDEELNKLLGKVTIAQGGVLPNI 112 ||||||||||||||||||||||||||||||#|||||+||||||||||| |||||||||||| Sbjct 53 AAVLEYLTAEILELAGNAARDNKKTRIIPR#HLQLAVRNDEELNKLLGGVTIAQGGVLPNI 112 |
| Bufo bufo gagarizans | Query 53 AAVLEYLTAEILELAGNAARDNKKTRIIPR#HLQLAIRNDEELNKLLGKVTIAQGGVLPNI 112 ||||||||||||||||||||||||||||||#|||||+||||||||||| |||||||||||| Sbjct 53 AAVLEYLTAEILELAGNAARDNKKTRIIPR#HLQLAVRNDEELNKLLGGVTIAQGGVLPNI 112 |
| Oncorhynchus mykiss | Query 53 AAVLEYLTAEILELAGNAARDNKKTRIIPR#HLQLAIRNDEELNKLLGKVTIAQGGVLPNI 112 ||||||||||||||||||||||||||||||#|||||+||||||||||| |||||||||||| Sbjct 53 AAVLEYLTAEILELAGNAARDNKKTRIIPR#HLQLAVRNDEELNKLLGGVTIAQGGVLPNI 112 |
| Bufo bufo gagarizans | Query 53 AAVLEYLTAEILELAGNAARDNKKTRIIPR#HLQLAIRNDEELNKLLGKVTIAQGGVLPNI 112 ||||||||||||||||||||||||||||||#|||||+||||||||||| |||||||||||| Sbjct 53 AAVLEYLTAEILELAGNAARDNKKTRIIPR#HLQLAVRNDEELNKLLGGVTIAQGGVLPNI 112 |
| Strongylocentrotus purpuratus | Query 53 AAVLEYLTAEILELAGNAARDNKKTRIIPR#HLQLAIRNDEELNKLLGKVTIAQGGVLPNI 112 ||||||||||||||||||||||||+|||||#|||||+||||||||||| |||||||||||| Sbjct 52 AAVLEYLTAEILELAGNAARDNKKSRIIPR#HLQLAVRNDEELNKLLGGVTIAQGGVLPNI 111 |
| Schistosoma japonicum | Query 53 AAVLEYLTAEILELAGNAARDNKKTRIIPR#HLQLAIRNDEELNKLLGKVTIAQGGVLPNI 112 ||||||||||||||||||||||||||||||#|||||+||||||||||| |||||||||||| Sbjct 53 AAVLEYLTAEILELAGNAARDNKKTRIIPR#HLQLAVRNDEELNKLLGGVTIAQGGVLPNI 112 |
| Gibberella zeae PH-1 | Query 53 AAVLEYLTAEILELAGNAARDNKKTRIIPR#HLQLAIRNDEELNKLLGKVTIAQGGVLPNI 112 ||||||| ||||||||||||||||||||||#||||||||||||||||| |||||||||||| Sbjct 55 AAVLEYLAAEILELAGNAARDNKKTRIIPR#HLQLAIRNDEELNKLLGHVTIAQGGVLPNI 114 |
| Oikopleura dioica | Query 53 AAVLEYLTAEILELAGNAARDNKKTRIIPR#HLQLAIRNDEELNKLLGKVTIAQGGVLPNI 112 |||||||+||+|||||||||||||||||||#||||||||||||||||| |||||||||||| Sbjct 52 AAVLEYLSAEVLELAGNAARDNKKTRIIPR#HLQLAIRNDEELNKLLGGVTIAQGGVLPNI 111 |
| Aspergillus nidulans FGSC A4 | Query 53 AAVLEYLTAEILELAGNAARDNKKTRIIPR#HLQLAIRNDEELNKLLGKVTIAQGGVLPNI 112 ||||||| ||||||||||||||||||||||#||||||||||||||||| |||||||||||| Sbjct 54 AAVLEYLAAEILELAGNAARDNKKTRIIPR#HLQLAIRNDEELNKLLGHVTIAQGGVLPNI 113 |
| Magnaporthe grisea 70-15 | Query 53 AAVLEYLTAEILELAGNAARDNKKTRIIPR#HLQLAIRNDEELNKLLGKVTIAQGGVLPNI 112 ||||||| ||||||||||||||||||||||#||||||||||||||||| |||||||||||| Sbjct 55 AAVLEYLAAEILELAGNAARDNKKTRIIPR#HLQLAIRNDEELNKLLGHVTIAQGGVLPNI 114 |
| Podospora anserina | Query 53 AAVLEYLTAEILELAGNAARDNKKTRIIPR#HLQLAIRNDEELNKLLGKVTIAQGGVLPNI 112 ||||||| ||||||||||||||||||||||#||||||||||||||||| |||||||||||| Sbjct 55 AAVLEYLAAEILELAGNAARDNKKTRIIPR#HLQLAIRNDEELNKLLGHVTIAQGGVLPNI 114 |
| Botryotinia fuckeliana | Query 53 AAVLEYLTAEILELAGNAARDNKKTRIIPR#HLQLAIRNDEELNKLLGKVTIAQGGVLPNI 112 ||||||| ||||||||||||||||||||||#||||||||||||||||| |||||||||||| Sbjct 56 AAVLEYLAAEILELAGNAARDNKKTRIIPR#HLQLAIRNDEELNKLLGHVTIAQGGVLPNI 115 |
| Schizosaccharomyces pombe 972h- | Query 53 AAVLEYLTAEILELAGNAARDNKKTRIIPR#HLQLAIRNDEELNKLLGKVTIAQGGVLPNI 112 ||||||| ||||||||||||||||||||||#||||||||||||||||| ||||||||+||| Sbjct 54 AAVLEYLAAEILELAGNAARDNKKTRIIPR#HLQLAIRNDEELNKLLGHVTIAQGGVVPNI 113 |
| Aspergillus niger | Query 53 AAVLEYLTAEILELAGNAARDNKKTRIIPR#HLQLAIRNDEELNKLLGKVTIAQGGVLPNI 112 ||||||| ||||||||||||||||||||||#||||||||||||||||| |||||||||||| Sbjct 54 AAVLEYLAAEILELAGNAARDNKKTRIIPR#HLQLAIRNDEELNKLLGHVTIAQGGVLPNI 113 |
| Neurospora crassa OR74A | Query 53 AAVLEYLTAEILELAGNAARDNKKTRIIPR#HLQLAIRNDEELNKLLGKVTIAQGGVLPNI 112 ||||||| ||||||||||||||||||||||#||||||||||||||||| |||||||||||| Sbjct 55 AAVLEYLAAEILELAGNAARDNKKTRIIPR#HLQLAIRNDEELNKLLGHVTIAQGGVLPNI 114 |
| Neosartorya fischeri NRRL 181 | Query 53 AAVLEYLTAEILELAGNAARDNKKTRIIPR#HLQLAIRNDEELNKLLGKVTIAQGGVLPNI 112 ||||||| ||||||||||||||||||||||#||||||||||||||||| |||||||||||| Sbjct 54 AAVLEYLAAEILELAGNAARDNKKTRIIPR#HLQLAIRNDEELNKLLGHVTIAQGGVLPNI 113 |
| Strongylocentrotus purpuratus | Query 53 AAVLEYLTAEILELAGNAARDNKKTRIIPR#HLQLAIRNDEELNKLLGKVTIAQGGVLPNI 112 +||||||||||||||||||||||| |||||#|||||+||||||||||| |||||||||||| Sbjct 52 SAVLEYLTAEILELAGNAARDNKKARIIPR#HLQLAVRNDEELNKLLGGVTIAQGGVLPNI 111 |
| Schizosaccharomyces pombe 972h- | Query 53 AAVLEYLTAEILELAGNAARDNKKTRIIPR#HLQLAIRNDEELNKLLGKVTIAQGGVLPNI 112 ||||||| ||||||||||||||||||||||#||||||||||||||||| ||||||||+||| Sbjct 54 AAVLEYLAAEILELAGNAARDNKKTRIIPR#HLQLAIRNDEELNKLLGHVTIAQGGVVPNI 113 |
| Ashbya gossypii ATCC 10895 | Query 54 AVLEYLTAEILELAGNAARDNKKTRIIPR#HLQLAIRNDEELNKLLGKVTIAQGGVLPNI 112 |||||| ||||||||||||||||||||||#|||||||||+||||||| |||||||||||| Sbjct 54 AVLEYLAAEILELAGNAARDNKKTRIIPR#HLQLAIRNDDELNKLLGNVTIAQGGVLPNI 112 |
| Arabidopsis thaliana | Query 53 AAVLEYLTAEILELAGNAARDNKKTRIIPR#HLQLAIRNDEELNKLLGKVTIAQGGVLPNI 112 |||+||| ||+|||||||||||||||||||#|||||||||||||||| |||||||||||| Sbjct 52 AAVMEYLAAEVLELAGNAARDNKKTRIIPR#HLQLAIRNDEELNKLLSGVTIAQGGVLPNI 111 |
| Candida albicans SC5314 | Query 54 AVLEYLTAEILELAGNAARDNKKTRIIPR#HLQLAIRNDEELNKLLGKVTIAQGGVLPNI 112 +||||| ||||||||||||||||+|||||#||||||||||||||||| |||||||||||| Sbjct 54 SVLEYLAAEILELAGNAARDNKKSRIIPR#HLQLAIRNDEELNKLLGDVTIAQGGVLPNI 112 |
| Griffithsia japonica | Query 53 AAVLEYLTAEILELAGNAARDNKKTRIIPR#HLQLAIRNDEELNKLLGKVTIAQGGVLPNI 112 |||+||||||+|||||||||||||+|||||#|+|||+||||||||||| |||| ||||||| Sbjct 47 AAVMEYLTAEVLELAGNAARDNKKSRIIPR#HIQLAVRNDEELNKLLGGVTIASGGVLPNI 106 |
| Acropora formosa | Query 53 AAVLEYLTAEILELAGNAARDNKKTRIIPR#HLQLAIRNDEELNKLLGKVTIAQGGVLPNI 112 |||||||+||||||||||||||||+|||||#|||||+|||||||||| |||||||||||| Sbjct 52 AAVLEYLSAEILELAGNAARDNKKSRIIPR#HLQLAVRNDEELNKLLAGVTIAQGGVLPNI 111 |
| Chironomus thummi | Query 53 AAVLEYLTAEILELAGNAARDNKKTRIIPR#HLQLAIRNDEELNKLLGKVTIAQGGVLPNI 112 |||+||| ||+|||||||||||||||||||#|||||||||||||||| |||||||||||| Sbjct 52 AAVMEYLAAEVLELAGNAARDNKKTRIIPR#HLQLAIRNDEELNKLLSGVTIAQGGVLPNI 111 |
| Drosophila melanogaster | Query 53 AAVLEYLTAEILELAGNAARDNKKTRIIPR#HLQLAIRNDEELNKLLGKVTIAQGGVLPNI 112 |||+||| ||+|||||||||||||||||||#|||||||||||||||| |||||||||||| Sbjct 52 AAVMEYLAAEVLELAGNAARDNKKTRIIPR#HLQLAIRNDEELNKLLSGVTIAQGGVLPNI 111 |
| Urechis caupo | Query 53 AAVLEYLTAEILELAGNAARDNKKTRIIPR#HLQLAIRNDEELNKLLGKVTIAQGGVLPNI 112 |||+||| ||+|||||||||||||||||||#|||||||||||||||| |||||||||||| Sbjct 52 AAVMEYLAAEVLELAGNAARDNKKTRIIPR#HLQLAIRNDEELNKLLSGVTIAQGGVLPNI 111 |
| Candida glabrata | Query 54 AVLEYLTAEILELAGNAARDNKKTRIIPR#HLQLAIRNDEELNKLLGKVTIAQGGVLPNI 112 |||||| ||||||||||||||||+|||||#|||||||||+||||||| |||||||||||| Sbjct 55 AVLEYLAAEILELAGNAARDNKKSRIIPR#HLQLAIRNDDELNKLLGNVTIAQGGVLPNI 113 |
| Asellus aquaticus | Query 53 AAVLEYLTAEILELAGNAARDNKKTRIIPR#HLQLAIRNDEELNKLLGKVTIAQGGVLPNI 112 |||+||| ||+|||||||||||||||||||#|||||||||||||||| |||||||||||| Sbjct 52 AAVMEYLAAEVLELAGNAARDNKKTRIIPR#HLQLAIRNDEELNKLLSGVTIAQGGVLPNI 111 |
| Tribolium castaneum | Query 53 AAVLEYLTAEILELAGNAARDNKKTRIIPR#HLQLAIRNDEELNKLLGKVTIAQGGVLPNI 112 |||+||| ||+|||||||||||||||||||#|||||||||||||||| |||||||||||| Sbjct 52 AAVMEYLAAEVLELAGNAARDNKKTRIIPR#HLQLAIRNDEELNKLLSGVTIAQGGVLPNI 111 |
| Platynereis dumerilii | Query 53 AAVLEYLTAEILELAGNAARDNKKTRIIPR#HLQLAIRNDEELNKLLGKVTIAQGGVLPNI 112 |||+||| ||+|||||||||||||||||||#|||||||||||||||| |||||||||||| Sbjct 52 AAVMEYLAAEVLELAGNAARDNKKTRIIPR#HLQLAIRNDEELNKLLSGVTIAQGGVLPNI 111 |
[Site 7] HLQLAIRNDE92-ELNKLLGKVT
Glu92  Glu
Glu
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| His83 | Leu84 | Gln85 | Leu86 | Ala87 | Ile88 | Arg89 | Asn90 | Asp91 | Glu92 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Glu93 | Leu94 | Asn95 | Lys96 | Leu97 | Leu98 | Gly99 | Lys100 | Val101 | Thr102 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| ILELAGNAARDNKKTRIIPRHLQLAIRNDEELNKLLGKVTIAQGGVLPNIQAVLLPKKTE |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | Homo sapiens | 120.00 | 13 | histone cluster 1, H2ai |
| 2 | Canis familiaris | 120.00 | 13 | PREDICTED: similar to histone 2, H2ac |
| 3 | Bos taurus | 120.00 | 3 | PREDICTED: similar to Hist2h2aa1 protein |
| 4 | Gallus gallus | 120.00 | 3 | histone H2A |
| 5 | synthetic construct | 120.00 | 2 | histone 2 H2aa |
| 6 | Homo sapiens | 120.00 | 2 | H2A histone family, member J |
| 7 | Mus musculus domesticus | 120.00 | 1 | histone H2A |
| 8 | Plasmodium berghei strain ANKA | 120.00 | 1 | histone h2a |
| 9 | N/A | 119.00 | 17 | histone H2A.1 |
| 10 | Mus musculus | 119.00 | 6 | histone cluster 1, H2ak |
| 11 | Bos taurus | 119.00 | 3 | PREDICTED: similar to Chain K, 2.9 Angstrom X-Ray |
| 12 | Rattus norvegicus | 119.00 | 2 | Histone H2a |
| 13 | Mus musculus | 119.00 | 2 | H2A histone family, member J |
| 14 | Rattus norvegicus | 119.00 | 1 | histone cluster 1, H2aa |
| 15 | Cairina moschata | 119.00 | 1 | H2A_CAIMO Histone H2A gi |
| 16 | Tetraodon nigroviridis | 118.00 | 5 | unnamed protein product |
| 17 | Xenopus laevis | 118.00 | 4 | hypothetical protein LOC494591 |
| 18 | Gallus gallus | 118.00 | 1 | histone cluster 2, H2ac |
| 19 | Danio rerio | 117.00 | 6 | histone 2, H2a |
| 20 | Bufo bufo gagarizans | 117.00 | 1 | AF255739_1 replication-dependent histone H2A |
| 21 | Oncorhynchus mykiss | 117.00 | 1 | H2A_ONCMY Histone H2A gi |
| 22 | Xenopus tropicalis | 116.00 | 3 | MGC69325 protein |
| 23 | Schistosoma japonicum | 116.00 | 2 | SJCHGC05779 protein |
| 24 | Danio rerio | 116.00 | 1 | H2A histone family, member X |
| 25 | Chironomus thummi | 115.00 | 2 | H2AO_CHITH Histone H2A, orphon gi |
| 26 | Oikopleura dioica | 114.00 | 2 | histone h2A.1 |
| 27 | Drosophila melanogaster | 114.00 | 2 | His2A:CG33853 CG33853-PA |
| 28 | Strongylocentrotus purpuratus | 114.00 | 2 | histone H2A |
| 29 | Tribolium castaneum | 114.00 | 1 | PREDICTED: similar to CG31618-PA |
| 30 | Anopheles gambiae str. PEST | 114.00 | 1 | AGAP003913-PA |
| 31 | Asellus aquaticus | 114.00 | 1 | putative H2A histone |
| 32 | Arabidopsis thaliana | 114.00 | 1 | Unknown protein |
| 33 | Acropora formosa | 114.00 | 1 | H2A_ACRFO Histone H2A gi |
| 34 | Strongylocentrotus purpuratus | 113.00 | 4 | PREDICTED: similar to histone L3 H2a |
| 35 | Platynereis dumerilii | 112.00 | 1 | H2A_PLADU Histone H2A gi |
| 36 | Chaetopterus variopedatus | 112.00 | 1 | histone H2A |
| 37 | Bufo bufo gagarizans | 111.00 | 2 | replication-dependent histone H2A |
| 38 | Bombyx mori | 111.00 | 1 | histone H2A-like protein |
| 39 | Mytilus trossulus | 111.00 | 1 | H2A_MYTTR Histone H2A gi |
| 40 | Mytilus edulis | 111.00 | 1 | H2A_MYTCA Histone H2A gi |
| 41 | Urechis caupo | 111.00 | 1 | H2A_URECA Histone H2A gi |
| 42 | Caenorhabditis briggsae AF16 | 110.00 | 2 | Hypothetical protein CBG19303 |
| 43 | Caenorhabditis elegans | 110.00 | 2 | HIStone family member (his-35) |
| 44 | Drosophila melanogaster | 110.00 | 1 | H2A histone |
| 45 | Botryotinia fuckeliana | 109.00 | 1 | histone H2A |
| 46 | Ashbya gossypii ATCC 10895 | 109.00 | 1 | AEL003Cp |
| 47 | Gibberella zeae PH-1 | 108.00 | 1 | H2A_NEUCR Histone H2A |
| 48 | Neurospora crassa OR74A | 108.00 | 1 | histone H2A |
| 49 | Magnaporthe grisea 70-15 | 108.00 | 1 | hypothetical protein MGG_03577 |
Top-ranked sequences
| organism | matching |
|---|---|
| Homo sapiens | Query 63 ILELAGNAARDNKKTRIIPRHLQLAIRNDE#ELNKLLGKVTIAQGGVLPNIQAVLLPKKTE 122 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 63 ILELAGNAARDNKKTRIIPRHLQLAIRNDE#ELNKLLGKVTIAQGGVLPNIQAVLLPKKTE 122 |
| Canis familiaris | Query 63 ILELAGNAARDNKKTRIIPRHLQLAIRNDE#ELNKLLGKVTIAQGGVLPNIQAVLLPKKTE 122 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 63 ILELAGNAARDNKKTRIIPRHLQLAIRNDE#ELNKLLGKVTIAQGGVLPNIQAVLLPKKTE 122 |
| Bos taurus | Query 63 ILELAGNAARDNKKTRIIPRHLQLAIRNDE#ELNKLLGKVTIAQGGVLPNIQAVLLPKKTE 122 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 63 ILELAGNAARDNKKTRIIPRHLQLAIRNDE#ELNKLLGKVTIAQGGVLPNIQAVLLPKKTE 122 |
| Gallus gallus | Query 63 ILELAGNAARDNKKTRIIPRHLQLAIRNDE#ELNKLLGKVTIAQGGVLPNIQAVLLPKKTE 122 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 63 ILELAGNAARDNKKTRIIPRHLQLAIRNDE#ELNKLLGKVTIAQGGVLPNIQAVLLPKKTE 122 |
| synthetic construct | Query 63 ILELAGNAARDNKKTRIIPRHLQLAIRNDE#ELNKLLGKVTIAQGGVLPNIQAVLLPKKTE 122 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 63 ILELAGNAARDNKKTRIIPRHLQLAIRNDE#ELNKLLGKVTIAQGGVLPNIQAVLLPKKTE 122 |
| Homo sapiens | Query 63 ILELAGNAARDNKKTRIIPRHLQLAIRNDE#ELNKLLGKVTIAQGGVLPNIQAVLLPKKTE 122 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 63 ILELAGNAARDNKKTRIIPRHLQLAIRNDE#ELNKLLGKVTIAQGGVLPNIQAVLLPKKTE 122 |
| Mus musculus domesticus | Query 63 ILELAGNAARDNKKTRIIPRHLQLAIRNDE#ELNKLLGKVTIAQGGVLPNIQAVLLPKKTE 122 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 63 ILELAGNAARDNKKTRIIPRHLQLAIRNDE#ELNKLLGKVTIAQGGVLPNIQAVLLPKKTE 122 |
| Plasmodium berghei strain ANKA | Query 63 ILELAGNAARDNKKTRIIPRHLQLAIRNDE#ELNKLLGKVTIAQGGVLPNIQAVLLPKKTE 122 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 63 ILELAGNAARDNKKTRIIPRHLQLAIRNDE#ELNKLLGKVTIAQGGVLPNIQAVLLPKKTE 122 |
| N/A | Query 63 ILELAGNAARDNKKTRIIPRHLQLAIRNDE#ELNKLLGKVTIAQGGVLPNIQAVLLPKKTE 122 ||||||||||||||||||||||||||||||#|||||||+|||||||||||||||||||||| Sbjct 63 ILELAGNAARDNKKTRIIPRHLQLAIRNDE#ELNKLLGRVTIAQGGVLPNIQAVLLPKKTE 122 |
| Mus musculus | Query 63 ILELAGNAARDNKKTRIIPRHLQLAIRNDE#ELNKLLGKVTIAQGGVLPNIQAVLLPKKTE 122 ||||||||||||||||||||||||||||||#|||||||+|||||||||||||||||||||| Sbjct 63 ILELAGNAARDNKKTRIIPRHLQLAIRNDE#ELNKLLGRVTIAQGGVLPNIQAVLLPKKTE 122 |
| Bos taurus | Query 63 ILELAGNAARDNKKTRIIPRHLQLAIRNDE#ELNKLLGKVTIAQGGVLPNIQAVLLPKKTE 122 ||||||||||||||||||||||||||||||#|||||||+|||||||||||||||||||||| Sbjct 63 ILELAGNAARDNKKTRIIPRHLQLAIRNDE#ELNKLLGRVTIAQGGVLPNIQAVLLPKKTE 122 |
| Rattus norvegicus | Query 63 ILELAGNAARDNKKTRIIPRHLQLAIRNDE#ELNKLLGKVTIAQGGVLPNIQAVLLPKKTE 122 ||||||||||||||||||||||||||||||#|||||||+|||||||||||||||||||||| Sbjct 63 ILELAGNAARDNKKTRIIPRHLQLAIRNDE#ELNKLLGRVTIAQGGVLPNIQAVLLPKKTE 122 |
| Mus musculus | Query 63 ILELAGNAARDNKKTRIIPRHLQLAIRNDE#ELNKLLGKVTIAQGGVLPNIQAVLLPKKTE 122 ||||||||||||||||||||||||||||||#|||||||+|||||||||||||||||||||| Sbjct 63 ILELAGNAARDNKKTRIIPRHLQLAIRNDE#ELNKLLGRVTIAQGGVLPNIQAVLLPKKTE 122 |
| Rattus norvegicus | Query 63 ILELAGNAARDNKKTRIIPRHLQLAIRNDE#ELNKLLGKVTIAQGGVLPNIQAVLLPKKTE 122 ||||||||||||||||||||||||||||||#|||||||+|||||||||||||||||||||| Sbjct 63 ILELAGNAARDNKKTRIIPRHLQLAIRNDE#ELNKLLGRVTIAQGGVLPNIQAVLLPKKTE 122 |
| Cairina moschata | Query 63 ILELAGNAARDNKKTRIIPRHLQLAIRNDE#ELNKLLGKVTIAQGGVLPNIQAVLLPKKTE 122 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||+ Sbjct 63 ILELAGNAARDNKKTRIIPRHLQLAIRNDE#ELNKLLGKVTIAQGGVLPNIQAVLLPKKTD 122 |
| Tetraodon nigroviridis | Query 63 ILELAGNAARDNKKTRIIPRHLQLAIRNDE#ELNKLLGKVTIAQGGVLPNIQAVLLPKKTE 122 |||||||||||||||||||||||||+||||#|||||||+|||||||||||||||||||||| Sbjct 63 ILELAGNAARDNKKTRIIPRHLQLAVRNDE#ELNKLLGRVTIAQGGVLPNIQAVLLPKKTE 122 |
| Xenopus laevis | Query 63 ILELAGNAARDNKKTRIIPRHLQLAIRNDE#ELNKLLGKVTIAQGGVLPNIQAVLLPKKTE 122 |||||||||||||||||||||||||+||||#|||||||+|||||||||||||+|||||||| Sbjct 63 ILELAGNAARDNKKTRIIPRHLQLAVRNDE#ELNKLLGRVTIAQGGVLPNIQSVLLPKKTE 122 |
| Gallus gallus | Query 63 ILELAGNAARDNKKTRIIPRHLQLAIRNDE#ELNKLLGKVTIAQGGVLPNIQAVLLPKKTE 122 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||+ Sbjct 63 ILELAGNAARDNKKTRIIPRHLQLAIRNDE#ELNKLLGKVTIAQGGVLPNIQAVLLPKKTD 122 |
| Danio rerio | Query 63 ILELAGNAARDNKKTRIIPRHLQLAIRNDE#ELNKLLGKVTIAQGGVLPNIQAVLLPKKTE 122 |||||||||||||||||||||||||+||||#||||||| |||||||||||||||||||||| Sbjct 63 ILELAGNAARDNKKTRIIPRHLQLAVRNDE#ELNKLLGGVTIAQGGVLPNIQAVLLPKKTE 122 |
| Bufo bufo gagarizans | Query 63 ILELAGNAARDNKKTRIIPRHLQLAIRNDE#ELNKLLGKVTIAQGGVLPNIQAVLLPKKTE 122 |||||||||||||||||||||||||+||||#||||||| |||||||||||||||||||||| Sbjct 63 ILELAGNAARDNKKTRIIPRHLQLAVRNDE#ELNKLLGGVTIAQGGVLPNIQAVLLPKKTE 122 |
| Oncorhynchus mykiss | Query 63 ILELAGNAARDNKKTRIIPRHLQLAIRNDE#ELNKLLGKVTIAQGGVLPNIQAVLLPKKTE 122 |||||||||||||||||||||||||+||||#||||||| |||||||||||||||||||||| Sbjct 63 ILELAGNAARDNKKTRIIPRHLQLAVRNDE#ELNKLLGGVTIAQGGVLPNIQAVLLPKKTE 122 |
| Xenopus tropicalis | Query 63 ILELAGNAARDNKKTRIIPRHLQLAIRNDE#ELNKLLGKVTIAQGGVLPNIQAVLLPKKTE 122 |||||||||||||||||||||||||+||||#||||||| |||||||||||||+|||||||| Sbjct 63 ILELAGNAARDNKKTRIIPRHLQLAVRNDE#ELNKLLGGVTIAQGGVLPNIQSVLLPKKTE 122 |
| Schistosoma japonicum | Query 63 ILELAGNAARDNKKTRIIPRHLQLAIRNDE#ELNKLLGKVTIAQGGVLPNIQAVLLPKKTE 122 |||||||||||||||||||||||||+||||#||||||| |||||||||||||||||||||| Sbjct 46 ILELAGNAARDNKKTRIIPRHLQLAVRNDE#ELNKLLGGVTIAQGGVLPNIQAVLLPKKTE 105 |
| Danio rerio | Query 63 ILELAGNAARDNKKTRIIPRHLQLAIRNDE#ELNKLLGKVTIAQGGVLPNIQAVLLPKKT 121 |||||||||||||||||||||||||+||||#||||||| ||||||||||||||||||||| Sbjct 63 ILELAGNAARDNKKTRIIPRHLQLAVRNDE#ELNKLLGGVTIAQGGVLPNIQAVLLPKKT 121 |
| Chironomus thummi | Query 63 ILELAGNAARDNKKTRIIPRHLQLAIRNDE#ELNKLLGKVTIAQGGVLPNIQAVLLPKKTE 122 ||||||||||||||||||||||||||||||#|||||| |||||||||||||||||||||| Sbjct 62 ILELAGNAARDNKKTRIIPRHLQLAIRNDE#ELNKLLSGVTIAQGGVLPNIQAVLLPKKTE 121 |
| Oikopleura dioica | Query 63 ILELAGNAARDNKKTRIIPRHLQLAIRNDE#ELNKLLGKVTIAQGGVLPNIQAVLLPKKT 121 +|||||||||||||||||||||||||||||#||||||| ||||||||||||||+|||||| Sbjct 62 VLELAGNAARDNKKTRIIPRHLQLAIRNDE#ELNKLLGGVTIAQGGVLPNIQAILLPKKT 120 |
| Drosophila melanogaster | Query 63 ILELAGNAARDNKKTRIIPRHLQLAIRNDE#ELNKLLGKVTIAQGGVLPNIQAVLLPKKTE 122 +|||||||||||||||||||||||||||||#|||||| |||||||||||||||||||||| Sbjct 62 VLELAGNAARDNKKTRIIPRHLQLAIRNDE#ELNKLLSGVTIAQGGVLPNIQAVLLPKKTE 121 |
| Strongylocentrotus purpuratus | Query 63 ILELAGNAARDNKKTRIIPRHLQLAIRNDE#ELNKLLGKVTIAQGGVLPNIQAVLLPKKT 121 ||||||||||||||+||||||||||+||||#||||||| ||||||||||||||||||||| Sbjct 62 ILELAGNAARDNKKSRIIPRHLQLAVRNDE#ELNKLLGGVTIAQGGVLPNIQAVLLPKKT 120 |
| Tribolium castaneum | Query 63 ILELAGNAARDNKKTRIIPRHLQLAIRNDE#ELNKLLGKVTIAQGGVLPNIQAVLLPKKTE 122 +|||||||||||||||||||||||||||||#|||||| |||||||||||||||||||||| Sbjct 62 VLELAGNAARDNKKTRIIPRHLQLAIRNDE#ELNKLLSGVTIAQGGVLPNIQAVLLPKKTE 121 |
| Anopheles gambiae str. PEST | Query 63 ILELAGNAARDNKKTRIIPRHLQLAIRNDE#ELNKLLGKVTIAQGGVLPNIQAVLLPKKTE 122 +|||||||||||||||||||||||||||||#|||||| |||||||||||||||||||||| Sbjct 62 VLELAGNAARDNKKTRIIPRHLQLAIRNDE#ELNKLLSGVTIAQGGVLPNIQAVLLPKKTE 121 |
| Asellus aquaticus | Query 63 ILELAGNAARDNKKTRIIPRHLQLAIRNDE#ELNKLLGKVTIAQGGVLPNIQAVLLPKKTE 122 +|||||||||||||||||||||||||||||#|||||| |||||||||||||||||||||| Sbjct 62 VLELAGNAARDNKKTRIIPRHLQLAIRNDE#ELNKLLSGVTIAQGGVLPNIQAVLLPKKTE 121 |
| Arabidopsis thaliana | Query 63 ILELAGNAARDNKKTRIIPRHLQLAIRNDE#ELNKLLGKVTIAQGGVLPNIQAVLLPKKTE 122 +|||||||||||||||||||||||||||||#|||||| ||||||||||||||||||||| Sbjct 62 VLELAGNAARDNKKTRIIPRHLQLAIRNDE#ELNKLLSGVTIAQGGVLPNIQAVLLPKKTS 121 |
| Acropora formosa | Query 63 ILELAGNAARDNKKTRIIPRHLQLAIRNDE#ELNKLLGKVTIAQGGVLPNIQAVLLPKKTE 122 ||||||||||||||+||||||||||+||||#|||||| |||||||||||||||||||||| Sbjct 62 ILELAGNAARDNKKSRIIPRHLQLAVRNDE#ELNKLLAGVTIAQGGVLPNIQAVLLPKKTE 121 |
| Strongylocentrotus purpuratus | Query 63 ILELAGNAARDNKKTRIIPRHLQLAIRNDE#ELNKLLGKVTIAQGGVLPNIQAVLLPKKTE 122 |||||||||||||||||||||||||+||||#|||||| ||||||||||||||||||||| Sbjct 63 ILELAGNAARDNKKTRIIPRHLQLAVRNDE#ELNKLLSGVTIAQGGVLPNIQAVLLPKKTS 122 |
| Platynereis dumerilii | Query 63 ILELAGNAARDNKKTRIIPRHLQLAIRNDE#ELNKLLGKVTIAQGGVLPNIQAVLLPKKT 121 +|||||||||||||||||||||||||||||#|||||| ||||||||||||||||||||| Sbjct 62 VLELAGNAARDNKKTRIIPRHLQLAIRNDE#ELNKLLSGVTIAQGGVLPNIQAVLLPKKT 120 |
| Chaetopterus variopedatus | Query 63 ILELAGNAARDNKKTRIIPRHLQLAIRNDE#ELNKLLGKVTIAQGGVLPNIQAVLLPKKTE 122 +|||||||||||||+|||||||||||||||#|||||| |||||||||||||||||||||+ Sbjct 62 VLELAGNAARDNKKSRIIPRHLQLAIRNDE#ELNKLLSGVTIAQGGVLPNIQAVLLPKKTQ 121 |
| Bufo bufo gagarizans | Query 63 ILELAGNAARDNKKTRIIPRHLQLAIRNDE#ELNKLLGKVTIAQGGVLPNIQAVLLPK 119 |||||||||||||||||||||||||+||||#||||||| ||||||||||||||||||| Sbjct 63 ILELAGNAARDNKKTRIIPRHLQLAVRNDE#ELNKLLGGVTIAQGGVLPNIQAVLLPK 119 |
| Bombyx mori | Query 63 ILELAGNAARDNKKTRIIPRHLQLAIRNDE#ELNKLLGKVTIAQGGVLPNIQAVLLPKKTE 122 +|||||||||||||||||||||||||||||#||||| |||||||||||||||||||||| Sbjct 62 VLELAGNAARDNKKTRIIPRHLQLAIRNDE#ELNKLPSGVTIAQGGVLPNIQAVLLPKKTE 121 |
| Mytilus trossulus | Query 63 ILELAGNAARDNKKTRIIPRHLQLAIRNDE#ELNKLLGKVTIAQGGVLPNIQAVLLPKKTE 122 +|||||||||||||+|||||||||||||||#|||||| |||||||||||||||||||||+ Sbjct 62 VLELAGNAARDNKKSRIIPRHLQLAIRNDE#ELNKLLSGVTIAQGGVLPNIQAVLLPKKTQ 121 |
| Mytilus edulis | Query 63 ILELAGNAARDNKKTRIIPRHLQLAIRNDE#ELNKLLGKVTIAQGGVLPNIQAVLLPKKTE 122 +|||||||||||||+|||||||||||||||#|||||| |||||||||||||||||||||+ Sbjct 62 VLELAGNAARDNKKSRIIPRHLQLAIRNDE#ELNKLLSGVTIAQGGVLPNIQAVLLPKKTQ 121 |
| Urechis caupo | Query 63 ILELAGNAARDNKKTRIIPRHLQLAIRNDE#ELNKLLGKVTIAQGGVLPNIQAVLLPKKTE 122 +|||||||||||||||||||||||||||||#|||||| ||||||||||||||||||||+ Sbjct 62 VLELAGNAARDNKKTRIIPRHLQLAIRNDE#ELNKLLSGVTIAQGGVLPNIQAVLLPKKSS 121 |
| Caenorhabditis briggsae AF16 | Query 63 ILELAGNAARDNKKTRIIPRHLQLAIRNDE#ELNKLLGKVTIAQGGVLPNIQAVLLPKKT 121 +|||||||||||||||| |||||||+||||#|||||| ||||||||||||||||||||| Sbjct 64 VLELAGNAARDNKKTRIAPRHLQLAVRNDE#ELNKLLAGVTIAQGGVLPNIQAVLLPKKT 122 |
| Caenorhabditis elegans | Query 63 ILELAGNAARDNKKTRIIPRHLQLAIRNDE#ELNKLLGKVTIAQGGVLPNIQAVLLPKKT 121 +|||||||||||||||| |||||||+||||#|||||| ||||||||||||||||||||| Sbjct 64 VLELAGNAARDNKKTRIAPRHLQLAVRNDE#ELNKLLAGVTIAQGGVLPNIQAVLLPKKT 122 |
| Drosophila melanogaster | Query 65 ELAGNAARDNKKTRIIPRHLQLAIRNDE#ELNKLLGKVTIAQGGVLPNIQAVLLPKKTE 122 ||||||||||||||||||||||||||||#|||||| |||||||||||||||||||||| Sbjct 1 ELAGNAARDNKKTRIIPRHLQLAIRNDE#ELNKLLSGVTIAQGGVLPNIQAVLLPKKTE 58 |
| Botryotinia fuckeliana | Query 63 ILELAGNAARDNKKTRIIPRHLQLAIRNDE#ELNKLLGKVTIAQGGVLPNIQAVLLPKKT 121 ||||||||||||||||||||||||||||||#||||||| |||||||||||| |||||| Sbjct 66 ILELAGNAARDNKKTRIIPRHLQLAIRNDE#ELNKLLGHVTIAQGGVLPNIHQSLLPKKT 124 |
| Ashbya gossypii ATCC 10895 | Query 63 ILELAGNAARDNKKTRIIPRHLQLAIRNDE#ELNKLLGKVTIAQGGVLPNIQAVLLPKKT 121 |||||||||||||||||||||||||||||+#||||||| |||||||||||| | |||||+ Sbjct 63 ILELAGNAARDNKKTRIIPRHLQLAIRNDD#ELNKLLGNVTIAQGGVLPNIHANLLPKKS 121 |
| Gibberella zeae PH-1 | Query 63 ILELAGNAARDNKKTRIIPRHLQLAIRNDE#ELNKLLGKVTIAQGGVLPNIQAVLLPKKT 121 ||||||||||||||||||||||||||||||#||||||| |||||||||||| |||||| Sbjct 65 ILELAGNAARDNKKTRIIPRHLQLAIRNDE#ELNKLLGHVTIAQGGVLPNIHQNLLPKKT 123 |
| Neurospora crassa OR74A | Query 63 ILELAGNAARDNKKTRIIPRHLQLAIRNDE#ELNKLLGKVTIAQGGVLPNIQAVLLPKKT 121 ||||||||||||||||||||||||||||||#||||||| |||||||||||| |||||| Sbjct 65 ILELAGNAARDNKKTRIIPRHLQLAIRNDE#ELNKLLGHVTIAQGGVLPNIHQNLLPKKT 123 |
| Magnaporthe grisea 70-15 | Query 63 ILELAGNAARDNKKTRIIPRHLQLAIRNDE#ELNKLLGKVTIAQGGVLPNIQAVLLPKKT 121 ||||||||||||||||||||||||||||||#||||||| |||||||||||| |||||| Sbjct 65 ILELAGNAARDNKKTRIIPRHLQLAIRNDE#ELNKLLGHVTIAQGGVLPNIHQNLLPKKT 123 |
[Site 8] VTIAQGGVLP110-NIQAVLLPKK
Pro110  Asn
Asn
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Val101 | Thr102 | Ile103 | Ala104 | Gln105 | Gly106 | Gly107 | Val108 | Leu109 | Pro110 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Asn111 | Ile112 | Gln113 | Ala114 | Val115 | Leu116 | Leu117 | Pro118 | Lys119 | Lys120 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| PRHLQLAIRNDEELNKLLGKVTIAQGGVLPNIQAVLLPKKTESHHKAKGK |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | N/A | 102.00 | 15 | 1109175A homeostatic thymus hormone alpha |
| 2 | Homo sapiens | 102.00 | 13 | histone cluster 1, H2ai |
| 3 | Canis familiaris | 102.00 | 13 | PREDICTED: similar to Histone H2A.l (H2A/l) |
| 4 | Bos taurus | 102.00 | 3 | PREDICTED: similar to homeostatic thymus hormone |
| 5 | synthetic construct | 102.00 | 2 | histone 2 H2aa |
| 6 | Mus musculus domesticus | 102.00 | 1 | histone H2A |
| 7 | Plasmodium berghei strain ANKA | 102.00 | 1 | histone h2a |
| 8 | Mus musculus | 101.00 | 6 | unnamed protein product |
| 9 | Bos taurus | 101.00 | 3 | PREDICTED: similar to Chain K, 2.9 Angstrom X-Ray |
| 10 | Rattus norvegicus | 100.00 | 2 | Histone H2a |
| 11 | Rattus norvegicus | 96.30 | 1 | histone cluster 1, H2aa |
| 12 | Gallus gallus | 92.80 | 3 | histone H2A |
| 13 | Gallus gallus | 91.70 | 1 | histone cluster 2, H2ac |
| 14 | Cairina moschata | 91.30 | 1 | H2A_CAIMO Histone H2A gi |
| 15 | Xenopus laevis | 90.50 | 4 | hypothetical protein LOC494591 |
| 16 | Bufo bufo gagarizans | 89.00 | 2 | AF255740_2 histone H2A variant |
| 17 | Homo sapiens | 89.00 | 2 | H2A histone family, member J |
| 18 | Bufo bufo gagarizans | 89.00 | 1 | AF255739_1 replication-dependent histone H2A |
| 19 | Xenopus tropicalis | 88.20 | 3 | hypothetical protein LOC594922 |
| 20 | Mus musculus | 88.20 | 2 | H2A histone family, member J |
| 21 | Oncorhynchus mykiss | 87.00 | 1 | H2A_ONCMY Histone H2A gi |
| 22 | Danio rerio | 86.30 | 6 | PREDICTED: similar to Mid1ip1 protein |
| 23 | Schistosoma japonicum | 86.30 | 2 | SJCHGC05779 protein |
| 24 | Tetraodon nigroviridis | 85.50 | 6 | unnamed protein product |
| 25 | Chironomus thummi | 83.60 | 2 | H2AO_CHITH Histone H2A, orphon gi |
| 26 | Strongylocentrotus purpuratus | 82.40 | 2 | histone H2A |
| 27 | Danio rerio | 82.00 | 1 | H2A histone family, member X |
| 28 | Strongylocentrotus purpuratus | 81.60 | 4 | PREDICTED: similar to histone H2A-2 |
| 29 | Anopheles gambiae str. PEST | 81.60 | 2 | AGAP003913-PA |
| 30 | Oikopleura dioica | 81.60 | 2 | histone h2A.1 |
| 31 | Drosophila melanogaster | 81.60 | 2 | His2A:CG33853 CG33853-PA |
| 32 | Asellus aquaticus | 81.60 | 1 | putative H2A histone |
| 33 | Tribolium castaneum | 81.60 | 1 | PREDICTED: similar to CG31618-PA |
| 34 | Acropora formosa | 81.60 | 1 | H2A_ACRFO Histone H2A gi |
| 35 | Arabidopsis thaliana | 81.30 | 1 | Unknown protein |
| 36 | Drosophila melanogaster | 80.90 | 1 | H2A histone |
| 37 | Chaetopterus variopedatus | 80.50 | 1 | histone H2A |
| 38 | Mytilus edulis | 80.50 | 1 | H2A_MYTCA Histone H2A gi |
| 39 | Mytilus trossulus | 80.50 | 1 | H2A_MYTTR Histone H2A gi |
| 40 | Platynereis dumerilii | 80.10 | 1 | H2A_PLADU Histone H2A gi |
| 41 | Caenorhabditis briggsae AF16 | 79.70 | 3 | Hypothetical protein CBG19303 |
| 42 | Caenorhabditis elegans | 79.30 | 2 | HIStone family member (his-68) |
| 43 | Urechis caupo | 79.00 | 1 | H2A_URECA Histone H2A gi |
| 44 | Bombyx mori | 78.60 | 1 | histone H2A-like protein |
Top-ranked sequences
| organism | matching |
|---|---|
| N/A | Query 81 PRHLQLAIRNDEELNKLLGKVTIAQGGVLP#NIQAVLLPKKTESHHKAKGK 130 ||||||||||||||||||||||||||||||#|||||||||||||||||||| Query 81 PRHLQLAIRNDEELNKLLGKVTIAQGGVLP#NIQAVLLPKKTESHHKAKGK 130 |
| Homo sapiens | Query 81 PRHLQLAIRNDEELNKLLGKVTIAQGGVLP#NIQAVLLPKKTESHHKAKGK 130 ||||||||||||||||||||||||||||||#|||||||||||||||||||| Query 81 PRHLQLAIRNDEELNKLLGKVTIAQGGVLP#NIQAVLLPKKTESHHKAKGK 130 |
| Canis familiaris | Query 81 PRHLQLAIRNDEELNKLLGKVTIAQGGVLP#NIQAVLLPKKTESHHKAKGK 130 |||||||||||||||||||+||||||||||#|||||||||||||||||||| Sbjct 102 PRHLQLAIRNDEELNKLLGRVTIAQGGVLP#NIQAVLLPKKTESHHKAKGK 151 |
| Bos taurus | Query 81 PRHLQLAIRNDEELNKLLGKVTIAQGGVLP#NIQAVLLPKKTESHHKAKGK 130 ||||||||||||||||||||||||||||||#|||||||||||||||||||| Query 81 PRHLQLAIRNDEELNKLLGKVTIAQGGVLP#NIQAVLLPKKTESHHKAKGK 130 |
| synthetic construct | Query 81 PRHLQLAIRNDEELNKLLGKVTIAQGGVLP#NIQAVLLPKKTESHHKAKGK 130 ||||||||||||||||||||||||||||||#|||||||||||||||||||| Query 81 PRHLQLAIRNDEELNKLLGKVTIAQGGVLP#NIQAVLLPKKTESHHKAKGK 130 |
| Mus musculus domesticus | Query 81 PRHLQLAIRNDEELNKLLGKVTIAQGGVLP#NIQAVLLPKKTESHHKAKGK 130 ||||||||||||||||||||||||||||||#|||||||||||||||||||| Query 81 PRHLQLAIRNDEELNKLLGKVTIAQGGVLP#NIQAVLLPKKTESHHKAKGK 130 |
| Plasmodium berghei strain ANKA | Query 81 PRHLQLAIRNDEELNKLLGKVTIAQGGVLP#NIQAVLLPKKTESHHKAKGK 130 ||||||||||||||||||||||||||||||#|||||||||||||||||||| Query 81 PRHLQLAIRNDEELNKLLGKVTIAQGGVLP#NIQAVLLPKKTESHHKAKGK 130 |
| Mus musculus | Query 81 PRHLQLAIRNDEELNKLLGKVTIAQGGVLP#NIQAVLLPKKTESHHKAKGK 130 |||||||||||||||||||+||||||||||#|||||||||||||||||||| Sbjct 86 PRHLQLAIRNDEELNKLLGRVTIAQGGVLP#NIQAVLLPKKTESHHKAKGK 135 |
| Bos taurus | Query 81 PRHLQLAIRNDEELNKLLGKVTIAQGGVLP#NIQAVLLPKKTESHHKAKGK 130 |||||||||||||||||||+||||||||||#|||||||||||||||||||| Sbjct 81 PRHLQLAIRNDEELNKLLGRVTIAQGGVLP#NIQAVLLPKKTESHHKAKGK 130 |
| Rattus norvegicus | Query 81 PRHLQLAIRNDEELNKLLGKVTIAQGGVLP#NIQAVLLPKKTESHHKAKGK 130 |||||||||||||||||||+||||||||||#||||||||||||||||+||| Sbjct 81 PRHLQLAIRNDEELNKLLGRVTIAQGGVLP#NIQAVLLPKKTESHHKSKGK 130 |
| Rattus norvegicus | Query 81 PRHLQLAIRNDEELNKLLGKVTIAQGGVLP#NIQAVLLPKKTESHHKAKGK 130 |||||||||||||||||||+||||||||||#||||||||||||||||++ | Sbjct 81 PRHLQLAIRNDEELNKLLGRVTIAQGGVLP#NIQAVLLPKKTESHHKSQTK 130 |
| Gallus gallus | Query 81 PRHLQLAIRNDEELNKLLGKVTIAQGGVLP#NIQAVLLPKKTESHHKAKGK 130 ||||||||||||||||||||||||||||||#||||||||||||| |||| | Sbjct 81 PRHLQLAIRNDEELNKLLGKVTIAQGGVLP#NIQAVLLPKKTES-HKAKSK 129 |
| Gallus gallus | Query 81 PRHLQLAIRNDEELNKLLGKVTIAQGGVLP#NIQAVLLPKKTESHHKAKGK 130 ||||||||||||||||||||||||||||||#|||||||||||+| |||| | Sbjct 81 PRHLQLAIRNDEELNKLLGKVTIAQGGVLP#NIQAVLLPKKTDS-HKAKAK 129 |
| Cairina moschata | Query 81 PRHLQLAIRNDEELNKLLGKVTIAQGGVLP#NIQAVLLPKKTESHHKAKGK 130 ||||||||||||||||||||||||||||||#|||||||||||+| |||| | Sbjct 81 PRHLQLAIRNDEELNKLLGKVTIAQGGVLP#NIQAVLLPKKTDS-HKAKSK 129 |
| Xenopus laevis | Query 81 PRHLQLAIRNDEELNKLLGKVTIAQGGVLP#NIQAVLLPKKTESHHKAKGK 130 |||||||+|||||||||||+||||||||||#|||+||||||||| || | Sbjct 81 PRHLQLAVRNDEELNKLLGRVTIAQGGVLP#NIQSVLLPKKTESSKSAKSK 130 |
| Bufo bufo gagarizans | Query 81 PRHLQLAIRNDEELNKLLGKVTIAQGGVLP#NIQAVLLPKKTESHHKAKGK 130 |||||||+||||||||||| ||||||||||#||||||||||||| || | Sbjct 79 PRHLQLAVRNDEELNKLLGGVTIAQGGVLP#NIQAVLLPKKTESSKPAKSK 128 |
| Homo sapiens | Query 81 PRHLQLAIRNDEELNKLLGKVTIAQGGVLP#NIQAVLLPKKTESH 124 ||||||||||||||||||||||||||||||#||||||||||||| Sbjct 81 PRHLQLAIRNDEELNKLLGKVTIAQGGVLP#NIQAVLLPKKTESQ 124 |
| Bufo bufo gagarizans | Query 81 PRHLQLAIRNDEELNKLLGKVTIAQGGVLP#NIQAVLLPKKTESHHKAKGK 130 |||||||+||||||||||| ||||||||||#||||||||||||| || | Sbjct 81 PRHLQLAVRNDEELNKLLGGVTIAQGGVLP#NIQAVLLPKKTESSKPAKSK 130 |
| Xenopus tropicalis | Query 81 PRHLQLAIRNDEELNKLLGKVTIAQGGVLP#NIQAVLLPKKTESHHKAKGK 130 |||||||+||||||||||| ||||||||||#|||+||||||||| || | Sbjct 81 PRHLQLAVRNDEELNKLLGGVTIAQGGVLP#NIQSVLLPKKTESSKAAKSK 130 |
| Mus musculus | Query 81 PRHLQLAIRNDEELNKLLGKVTIAQGGVLP#NIQAVLLPKKTESH 124 |||||||||||||||||||+||||||||||#||||||||||||| Sbjct 81 PRHLQLAIRNDEELNKLLGRVTIAQGGVLP#NIQAVLLPKKTESQ 124 |
| Oncorhynchus mykiss | Query 81 PRHLQLAIRNDEELNKLLGKVTIAQGGVLP#NIQAVLLPKKTESHHKAK 128 |||||||+||||||||||| ||||||||||#|||||||||||| ||| Sbjct 81 PRHLQLAVRNDEELNKLLGGVTIAQGGVLP#NIQAVLLPKKTEKAVKAK 128 |
| Danio rerio | Query 81 PRHLQLAIRNDEELNKLLGKVTIAQGGVLP#NIQAVLLPKKTESHHKAK 128 |||||||+||||||||||| ||||||||||#|||||||||||| | | Sbjct 81 PRHLQLAVRNDEELNKLLGGVTIAQGGVLP#NIQAVLLPKKTEKAAKGK 128 |
| Schistosoma japonicum | Query 81 PRHLQLAIRNDEELNKLLGKVTIAQGGVLP#NIQAVLLPKKTESHHKAK 128 |||||||+||||||||||| ||||||||||#|||||||||||| ||| Sbjct 64 PRHLQLAVRNDEELNKLLGGVTIAQGGVLP#NIQAVLLPKKTEKAVKAK 111 |
| Tetraodon nigroviridis | Query 81 PRHLQLAIRNDEELNKLLGKVTIAQGGVLP#NIQAVLLPKKTE 122 |||||||+|||||||||||+||||||||||#|||||||||||| Sbjct 81 PRHLQLAVRNDEELNKLLGRVTIAQGGVLP#NIQAVLLPKKTE 122 |
| Chironomus thummi | Query 81 PRHLQLAIRNDEELNKLLGKVTIAQGGVLP#NIQAVLLPKKTES 123 |||||||||||||||||| ||||||||||#||||||||||||| Sbjct 80 PRHLQLAIRNDEELNKLLSGVTIAQGGVLP#NIQAVLLPKKTES 122 |
| Strongylocentrotus purpuratus | Query 81 PRHLQLAIRNDEELNKLLGKVTIAQGGVLP#NIQAVLLPKKT 121 |||||||+||||||||||| ||||||||||#||||||||||| Sbjct 80 PRHLQLAVRNDEELNKLLGGVTIAQGGVLP#NIQAVLLPKKT 120 |
| Danio rerio | Query 81 PRHLQLAIRNDEELNKLLGKVTIAQGGVLP#NIQAVLLPKKT 121 |||||||+||||||||||| ||||||||||#||||||||||| Sbjct 81 PRHLQLAVRNDEELNKLLGGVTIAQGGVLP#NIQAVLLPKKT 121 |
| Strongylocentrotus purpuratus | Query 81 PRHLQLAIRNDEELNKLLGKVTIAQGGVLP#NIQAVLLPKKT 121 |||||||+||||||||||| ||||||||||#||||||||||| Sbjct 80 PRHLQLAVRNDEELNKLLGGVTIAQGGVLP#NIQAVLLPKKT 120 |
| Anopheles gambiae str. PEST | Query 81 PRHLQLAIRNDEELNKLLGKVTIAQGGVLP#NIQAVLLPKKTESH 124 |||||||||||||||||| ||||||||||#|||||||||||| + Sbjct 80 PRHLQLAIRNDEELNKLLSGVTIAQGGVLP#NIQAVLLPKKTEKN 123 |
| Oikopleura dioica | Query 81 PRHLQLAIRNDEELNKLLGKVTIAQGGVLP#NIQAVLLPKKT 121 ||||||||||||||||||| ||||||||||#||||+|||||| Sbjct 80 PRHLQLAIRNDEELNKLLGGVTIAQGGVLP#NIQAILLPKKT 120 |
| Drosophila melanogaster | Query 81 PRHLQLAIRNDEELNKLLGKVTIAQGGVLP#NIQAVLLPKKTE 122 |||||||||||||||||| ||||||||||#|||||||||||| Sbjct 80 PRHLQLAIRNDEELNKLLSGVTIAQGGVLP#NIQAVLLPKKTE 121 |
| Asellus aquaticus | Query 81 PRHLQLAIRNDEELNKLLGKVTIAQGGVLP#NIQAVLLPKKTE 122 |||||||||||||||||| ||||||||||#|||||||||||| Sbjct 80 PRHLQLAIRNDEELNKLLSGVTIAQGGVLP#NIQAVLLPKKTE 121 |
| Tribolium castaneum | Query 81 PRHLQLAIRNDEELNKLLGKVTIAQGGVLP#NIQAVLLPKKTE 122 |||||||||||||||||| ||||||||||#|||||||||||| Sbjct 80 PRHLQLAIRNDEELNKLLSGVTIAQGGVLP#NIQAVLLPKKTE 121 |
| Acropora formosa | Query 81 PRHLQLAIRNDEELNKLLGKVTIAQGGVLP#NIQAVLLPKKTE 122 |||||||+|||||||||| ||||||||||#|||||||||||| Sbjct 80 PRHLQLAVRNDEELNKLLAGVTIAQGGVLP#NIQAVLLPKKTE 121 |
| Arabidopsis thaliana | Query 81 PRHLQLAIRNDEELNKLLGKVTIAQGGVLP#NIQAVLLPKKTES 123 |||||||||||||||||| ||||||||||#||||||||||| + Sbjct 80 PRHLQLAIRNDEELNKLLSGVTIAQGGVLP#NIQAVLLPKKTSA 122 |
| Drosophila melanogaster | Query 81 PRHLQLAIRNDEELNKLLGKVTIAQGGVLP#NIQAVLLPKKTE 122 |||||||||||||||||| ||||||||||#|||||||||||| Sbjct 17 PRHLQLAIRNDEELNKLLSGVTIAQGGVLP#NIQAVLLPKKTE 58 |
| Chaetopterus variopedatus | Query 81 PRHLQLAIRNDEELNKLLGKVTIAQGGVLP#NIQAVLLPKKTE 122 |||||||||||||||||| ||||||||||#|||||||||||+ Sbjct 80 PRHLQLAIRNDEELNKLLSGVTIAQGGVLP#NIQAVLLPKKTQ 121 |
| Mytilus edulis | Query 81 PRHLQLAIRNDEELNKLLGKVTIAQGGVLP#NIQAVLLPKKTESHHK 126 |||||||||||||||||| ||||||||||#|||||||||||+ | Sbjct 80 PRHLQLAIRNDEELNKLLSGVTIAQGGVLP#NIQAVLLPKKTQKAAK 125 |
| Mytilus trossulus | Query 81 PRHLQLAIRNDEELNKLLGKVTIAQGGVLP#NIQAVLLPKKTESHHK 126 |||||||||||||||||| ||||||||||#|||||||||||+ | Sbjct 80 PRHLQLAIRNDEELNKLLSGVTIAQGGVLP#NIQAVLLPKKTQKAAK 125 |
| Platynereis dumerilii | Query 81 PRHLQLAIRNDEELNKLLGKVTIAQGGVLP#NIQAVLLPKKT 121 |||||||||||||||||| ||||||||||#||||||||||| Sbjct 80 PRHLQLAIRNDEELNKLLSGVTIAQGGVLP#NIQAVLLPKKT 120 |
| Caenorhabditis briggsae AF16 | Query 81 PRHLQLAIRNDEELNKLLGKVTIAQGGVLP#NIQAVLLPKKTES 123 |||||||+|||||||||| ||||||||||#||||||||||| + Sbjct 82 PRHLQLAVRNDEELNKLLAGVTIAQGGVLP#NIQAVLLPKKTAA 124 |
| Caenorhabditis elegans | Query 81 PRHLQLAIRNDEELNKLLGKVTIAQGGVLP#NIQAVLLPKKT 121 |||||||+|||||||||| ||||||||||#||||||||||| Sbjct 82 PRHLQLAVRNDEELNKLLAGVTIAQGGVLP#NIQAVLLPKKT 122 |
| Urechis caupo | Query 81 PRHLQLAIRNDEELNKLLGKVTIAQGGVLP#NIQAVLLPKKTESHHK 126 |||||||||||||||||| ||||||||||#||||||||||+ | Sbjct 80 PRHLQLAIRNDEELNKLLSGVTIAQGGVLP#NIQAVLLPKKSSQKTK 125 |
| Bombyx mori | Query 81 PRHLQLAIRNDEELNKLLGKVTIAQGGVLP#NIQAVLLPKKTE 122 ||||||||||||||||| ||||||||||#|||||||||||| Sbjct 80 PRHLQLAIRNDEELNKLPSGVTIAQGGVLP#NIQAVLLPKKTE 121 |
[Site 9] QGGVLPNIQA114-VLLPKKTESH
Ala114  Val
Val
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Gln105 | Gly106 | Gly107 | Val108 | Leu109 | Pro110 | Asn111 | Ile112 | Gln113 | Ala114 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Val115 | Leu116 | Leu117 | Pro118 | Lys119 | Lys120 | Thr121 | Glu122 | Ser123 | His124 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| QLAIRNDEELNKLLGKVTIAQGGVLPNIQAVLLPKKTESHHKAKGK |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | Homo sapiens | 93.60 | 13 | histone cluster 2, H2aa3 |
| 2 | Bos taurus | 93.60 | 3 | PREDICTED: similar to Hist2h2aa1 protein |
| 3 | Mus musculus domesticus | 93.60 | 1 | histone H2A |
| 4 | N/A | 93.20 | 15 | 1109175A homeostatic thymus hormone alpha |
| 5 | synthetic construct | 93.20 | 2 | histone 2 H2aa |
| 6 | Plasmodium berghei strain ANKA | 93.20 | 1 | histone h2a |
| 7 | Canis familiaris | 92.80 | 13 | PREDICTED: similar to Histone H2A.l (H2A/l) |
| 8 | Mus musculus | 92.40 | 7 | unnamed protein product |
| 9 | Bos taurus | 92.40 | 3 | PREDICTED: similar to Chain K, 2.9 Angstrom X-Ray |
| 10 | Rattus norvegicus | 91.70 | 2 | Histone H2a |
| 11 | Rattus norvegicus | 87.40 | 1 | histone cluster 1, H2aa |
| 12 | Gallus gallus | 83.60 | 3 | histone H2A |
| 13 | Gallus gallus | 82.40 | 1 | histone cluster 2, H2ac |
| 14 | Cairina moschata | 82.00 | 1 | H2A_CAIMO Histone H2A gi |
| 15 | Xenopus laevis | 80.90 | 4 | hypothetical protein LOC494591 |
| 16 | Bufo bufo gagarizans | 79.30 | 2 | AF255740_2 histone H2A variant |
| 17 | Homo sapiens | 79.30 | 2 | H2A histone family, member J |
| 18 | Bufo bufo gagarizans | 79.30 | 1 | AF255739_1 replication-dependent histone H2A |
| 19 | Xenopus tropicalis | 79.00 | 3 | hypothetical protein LOC594922 |
| 20 | Mus musculus | 78.60 | 2 | H2A histone family, member J |
| 21 | Oncorhynchus mykiss | 77.00 | 1 | H2A_ONCMY Histone H2A gi |
| 22 | Schistosoma japonicum | 76.60 | 2 | SJCHGC05779 protein |
| 23 | Tetraodon nigroviridis | 76.30 | 6 | unnamed protein product |
| 24 | Danio rerio | 76.30 | 6 | PREDICTED: similar to Mid1ip1 protein |
| 25 | Chironomus thummi | 74.30 | 2 | H2A_CHITH Histone H2A gi |
| 26 | Strongylocentrotus purpuratus | 72.80 | 2 | histone H2A |
| 27 | Strongylocentrotus purpuratus | 72.40 | 4 | PREDICTED: similar to histone H2A-2 |
| 28 | Drosophila melanogaster | 72.40 | 2 | His2A:CG31618 CG31618-PA |
| 29 | Anopheles gambiae str. PEST | 72.40 | 2 | AGAP003913-PA |
| 30 | Danio rerio | 72.40 | 1 | H2A histone family, member X |
| 31 | Asellus aquaticus | 72.40 | 1 | putative H2A histone |
| 32 | Acropora formosa | 72.40 | 1 | H2A_ACRFO Histone H2A gi |
| 33 | Tribolium castaneum | 72.40 | 1 | PREDICTED: similar to CG31618-PA |
| 34 | Oikopleura dioica | 72.00 | 3 | histone h2A.1 |
| 35 | Arabidopsis thaliana | 71.60 | 1 | Unknown protein |
| 36 | Drosophila melanogaster | 71.60 | 1 | H2A histone |
| 37 | Chaetopterus variopedatus | 71.20 | 1 | histone H2A |
| 38 | Mytilus edulis | 70.90 | 1 | H2A_MYTCA Histone H2A gi |
| 39 | Mytilus trossulus | 70.90 | 1 | H2A_MYTTR Histone H2A gi |
| 40 | Caenorhabditis briggsae AF16 | 70.50 | 3 | Hypothetical protein CBG19303 |
| 41 | Platynereis dumerilii | 70.50 | 1 | H2A_PLADU Histone H2A gi |
| 42 | Caenorhabditis elegans | 70.10 | 2 | HIStone family member (his-35) |
| 43 | Bombyx mori | 69.70 | 1 | histone H2A-like protein |
| 44 | Urechis caupo | 69.70 | 1 | H2A_URECA Histone H2A gi |
| 45 | Drosophila virilis | 68.20 | 1 | histone H2A |
Top-ranked sequences
| organism | matching |
|---|---|
| Homo sapiens | Query 85 QLAIRNDEELNKLLGKVTIAQGGVLPNIQA#VLLPKKTESHHKAKGK 130 ||||||||||||||||||||||||||||||#|||||||||||||||| Query 85 QLAIRNDEELNKLLGKVTIAQGGVLPNIQA#VLLPKKTESHHKAKGK 130 |
| Bos taurus | Query 85 QLAIRNDEELNKLLGKVTIAQGGVLPNIQA#VLLPKKTESHHKAKGK 130 ||||||||||||||||||||||||||||||#|||||||||||||||| Query 85 QLAIRNDEELNKLLGKVTIAQGGVLPNIQA#VLLPKKTESHHKAKGK 130 |
| Mus musculus domesticus | Query 85 QLAIRNDEELNKLLGKVTIAQGGVLPNIQA#VLLPKKTESHHKAKGK 130 ||||||||||||||||||||||||||||||#|||||||||||||||| Query 85 QLAIRNDEELNKLLGKVTIAQGGVLPNIQA#VLLPKKTESHHKAKGK 130 |
| N/A | Query 85 QLAIRNDEELNKLLGKVTIAQGGVLPNIQA#VLLPKKTESHHKAKGK 130 ||||||||||||||||||||||||||||||#|||||||||||||||| Query 85 QLAIRNDEELNKLLGKVTIAQGGVLPNIQA#VLLPKKTESHHKAKGK 130 |
| synthetic construct | Query 85 QLAIRNDEELNKLLGKVTIAQGGVLPNIQA#VLLPKKTESHHKAKGK 130 ||||||||||||||||||||||||||||||#|||||||||||||||| Query 85 QLAIRNDEELNKLLGKVTIAQGGVLPNIQA#VLLPKKTESHHKAKGK 130 |
| Plasmodium berghei strain ANKA | Query 85 QLAIRNDEELNKLLGKVTIAQGGVLPNIQA#VLLPKKTESHHKAKGK 130 ||||||||||||||||||||||||||||||#|||||||||||||||| Query 85 QLAIRNDEELNKLLGKVTIAQGGVLPNIQA#VLLPKKTESHHKAKGK 130 |
| Canis familiaris | Query 85 QLAIRNDEELNKLLGKVTIAQGGVLPNIQA#VLLPKKTESHHKAKGK 130 |||||||||||||||+||||||||||||||#|||||||||||||||| Sbjct 106 QLAIRNDEELNKLLGRVTIAQGGVLPNIQA#VLLPKKTESHHKAKGK 151 |
| Mus musculus | Query 85 QLAIRNDEELNKLLGKVTIAQGGVLPNIQA#VLLPKKTESHHKAKGK 130 |||||||||||||||+||||||||||||||#|||||||||||||||| Sbjct 90 QLAIRNDEELNKLLGRVTIAQGGVLPNIQA#VLLPKKTESHHKAKGK 135 |
| Bos taurus | Query 85 QLAIRNDEELNKLLGKVTIAQGGVLPNIQA#VLLPKKTESHHKAKGK 130 |||||||||||||||+||||||||||||||#|||||||||||||||| Sbjct 85 QLAIRNDEELNKLLGRVTIAQGGVLPNIQA#VLLPKKTESHHKAKGK 130 |
| Rattus norvegicus | Query 85 QLAIRNDEELNKLLGKVTIAQGGVLPNIQA#VLLPKKTESHHKAKGK 130 |||||||||||||||+||||||||||||||#||||||||||||+||| Sbjct 85 QLAIRNDEELNKLLGRVTIAQGGVLPNIQA#VLLPKKTESHHKSKGK 130 |
| Rattus norvegicus | Query 85 QLAIRNDEELNKLLGKVTIAQGGVLPNIQA#VLLPKKTESHHKAKGK 130 |||||||||||||||+||||||||||||||#||||||||||||++ | Sbjct 85 QLAIRNDEELNKLLGRVTIAQGGVLPNIQA#VLLPKKTESHHKSQTK 130 |
| Gallus gallus | Query 85 QLAIRNDEELNKLLGKVTIAQGGVLPNIQA#VLLPKKTESHHKAKGK 130 ||||||||||||||||||||||||||||||#||||||||| |||| | Sbjct 85 QLAIRNDEELNKLLGKVTIAQGGVLPNIQA#VLLPKKTES-HKAKSK 129 |
| Gallus gallus | Query 85 QLAIRNDEELNKLLGKVTIAQGGVLPNIQA#VLLPKKTESHHKAKGK 130 ||||||||||||||||||||||||||||||#|||||||+| |||| | Sbjct 85 QLAIRNDEELNKLLGKVTIAQGGVLPNIQA#VLLPKKTDS-HKAKAK 129 |
| Cairina moschata | Query 85 QLAIRNDEELNKLLGKVTIAQGGVLPNIQA#VLLPKKTESHHKAKGK 130 ||||||||||||||||||||||||||||||#|||||||+| |||| | Sbjct 85 QLAIRNDEELNKLLGKVTIAQGGVLPNIQA#VLLPKKTDS-HKAKSK 129 |
| Xenopus laevis | Query 85 QLAIRNDEELNKLLGKVTIAQGGVLPNIQA#VLLPKKTESHHKAKGK 130 |||+|||||||||||+|||||||||||||+#||||||||| || | Sbjct 85 QLAVRNDEELNKLLGRVTIAQGGVLPNIQS#VLLPKKTESSKSAKSK 130 |
| Bufo bufo gagarizans | Query 85 QLAIRNDEELNKLLGKVTIAQGGVLPNIQA#VLLPKKTESHHKAKGK 130 |||+||||||||||| ||||||||||||||#||||||||| || | Sbjct 83 QLAVRNDEELNKLLGGVTIAQGGVLPNIQA#VLLPKKTESSKPAKSK 128 |
| Homo sapiens | Query 85 QLAIRNDEELNKLLGKVTIAQGGVLPNIQA#VLLPKKTESH 124 ||||||||||||||||||||||||||||||#||||||||| Sbjct 85 QLAIRNDEELNKLLGKVTIAQGGVLPNIQA#VLLPKKTESQ 124 |
| Bufo bufo gagarizans | Query 85 QLAIRNDEELNKLLGKVTIAQGGVLPNIQA#VLLPKKTESHHKAKGK 130 |||+||||||||||| ||||||||||||||#||||||||| || | Sbjct 85 QLAVRNDEELNKLLGGVTIAQGGVLPNIQA#VLLPKKTESSKPAKSK 130 |
| Xenopus tropicalis | Query 85 QLAIRNDEELNKLLGKVTIAQGGVLPNIQA#VLLPKKTESHHKAKGK 130 |||+||||||||||| |||||||||||||+#||||||||| || | Sbjct 85 QLAVRNDEELNKLLGGVTIAQGGVLPNIQS#VLLPKKTESSKAAKSK 130 |
| Mus musculus | Query 85 QLAIRNDEELNKLLGKVTIAQGGVLPNIQA#VLLPKKTESH 124 |||||||||||||||+||||||||||||||#||||||||| Sbjct 85 QLAIRNDEELNKLLGRVTIAQGGVLPNIQA#VLLPKKTESQ 124 |
| Oncorhynchus mykiss | Query 85 QLAIRNDEELNKLLGKVTIAQGGVLPNIQA#VLLPKKTESHHKAK 128 |||+||||||||||| ||||||||||||||#|||||||| ||| Sbjct 85 QLAVRNDEELNKLLGGVTIAQGGVLPNIQA#VLLPKKTEKAVKAK 128 |
| Schistosoma japonicum | Query 85 QLAIRNDEELNKLLGKVTIAQGGVLPNIQA#VLLPKKTESHHKAK 128 |||+||||||||||| ||||||||||||||#|||||||| ||| Sbjct 68 QLAVRNDEELNKLLGGVTIAQGGVLPNIQA#VLLPKKTEKAVKAK 111 |
| Tetraodon nigroviridis | Query 85 QLAIRNDEELNKLLGKVTIAQGGVLPNIQA#VLLPKKTE 122 |||+|||||||||||+||||||||||||||#|||||||| Sbjct 85 QLAVRNDEELNKLLGRVTIAQGGVLPNIQA#VLLPKKTE 122 |
| Danio rerio | Query 85 QLAIRNDEELNKLLGKVTIAQGGVLPNIQA#VLLPKKTESHHKAK 128 |||+||||||||||| ||||||||||||||#|||||||| | | Sbjct 85 QLAVRNDEELNKLLGGVTIAQGGVLPNIQA#VLLPKKTEKAAKGK 128 |
| Chironomus thummi | Query 85 QLAIRNDEELNKLLGKVTIAQGGVLPNIQA#VLLPKKTES 123 |||||||||||||| ||||||||||||||#||||||||| Sbjct 84 QLAIRNDEELNKLLSGVTIAQGGVLPNIQA#VLLPKKTES 122 |
| Strongylocentrotus purpuratus | Query 85 QLAIRNDEELNKLLGKVTIAQGGVLPNIQA#VLLPKKT 121 |||+||||||||||| ||||||||||||||#||||||| Sbjct 84 QLAVRNDEELNKLLGGVTIAQGGVLPNIQA#VLLPKKT 120 |
| Strongylocentrotus purpuratus | Query 85 QLAIRNDEELNKLLGKVTIAQGGVLPNIQA#VLLPKKT 121 |||+||||||||||| ||||||||||||||#||||||| Sbjct 84 QLAVRNDEELNKLLGGVTIAQGGVLPNIQA#VLLPKKT 120 |
| Drosophila melanogaster | Query 85 QLAIRNDEELNKLLGKVTIAQGGVLPNIQA#VLLPKKTE 122 |||||||||||||| ||||||||||||||#|||||||| Sbjct 84 QLAIRNDEELNKLLSGVTIAQGGVLPNIQA#VLLPKKTE 121 |
| Anopheles gambiae str. PEST | Query 85 QLAIRNDEELNKLLGKVTIAQGGVLPNIQA#VLLPKKTE 122 |||||||||||||| ||||||||||||||#|||||||| Sbjct 84 QLAIRNDEELNKLLSGVTIAQGGVLPNIQA#VLLPKKTE 121 |
| Danio rerio | Query 85 QLAIRNDEELNKLLGKVTIAQGGVLPNIQA#VLLPKKT 121 |||+||||||||||| ||||||||||||||#||||||| Sbjct 85 QLAVRNDEELNKLLGGVTIAQGGVLPNIQA#VLLPKKT 121 |
| Asellus aquaticus | Query 85 QLAIRNDEELNKLLGKVTIAQGGVLPNIQA#VLLPKKTE 122 |||||||||||||| ||||||||||||||#|||||||| Sbjct 84 QLAIRNDEELNKLLSGVTIAQGGVLPNIQA#VLLPKKTE 121 |
| Acropora formosa | Query 85 QLAIRNDEELNKLLGKVTIAQGGVLPNIQA#VLLPKKTE 122 |||+|||||||||| ||||||||||||||#|||||||| Sbjct 84 QLAVRNDEELNKLLAGVTIAQGGVLPNIQA#VLLPKKTE 121 |
| Tribolium castaneum | Query 85 QLAIRNDEELNKLLGKVTIAQGGVLPNIQA#VLLPKKTE 122 |||||||||||||| ||||||||||||||#|||||||| Sbjct 84 QLAIRNDEELNKLLSGVTIAQGGVLPNIQA#VLLPKKTE 121 |
| Oikopleura dioica | Query 85 QLAIRNDEELNKLLGKVTIAQGGVLPNIQA#VLLPKKT 121 ||||||||||||||| ||||||||||||||#+|||||| Sbjct 84 QLAIRNDEELNKLLGGVTIAQGGVLPNIQA#ILLPKKT 120 |
| Arabidopsis thaliana | Query 85 QLAIRNDEELNKLLGKVTIAQGGVLPNIQA#VLLPKKTES 123 |||||||||||||| ||||||||||||||#||||||| + Sbjct 84 QLAIRNDEELNKLLSGVTIAQGGVLPNIQA#VLLPKKTSA 122 |
| Drosophila melanogaster | Query 85 QLAIRNDEELNKLLGKVTIAQGGVLPNIQA#VLLPKKTE 122 |||||||||||||| ||||||||||||||#|||||||| Sbjct 21 QLAIRNDEELNKLLSGVTIAQGGVLPNIQA#VLLPKKTE 58 |
| Chaetopterus variopedatus | Query 85 QLAIRNDEELNKLLGKVTIAQGGVLPNIQA#VLLPKKTE 122 |||||||||||||| ||||||||||||||#|||||||+ Sbjct 84 QLAIRNDEELNKLLSGVTIAQGGVLPNIQA#VLLPKKTQ 121 |
| Mytilus edulis | Query 85 QLAIRNDEELNKLLGKVTIAQGGVLPNIQA#VLLPKKTESHHK 126 |||||||||||||| ||||||||||||||#|||||||+ | Sbjct 84 QLAIRNDEELNKLLSGVTIAQGGVLPNIQA#VLLPKKTQKAAK 125 |
| Mytilus trossulus | Query 85 QLAIRNDEELNKLLGKVTIAQGGVLPNIQA#VLLPKKTESHHK 126 |||||||||||||| ||||||||||||||#|||||||+ | Sbjct 84 QLAIRNDEELNKLLSGVTIAQGGVLPNIQA#VLLPKKTQKAAK 125 |
| Caenorhabditis briggsae AF16 | Query 85 QLAIRNDEELNKLLGKVTIAQGGVLPNIQA#VLLPKKTES 123 |||+|||||||||| ||||||||||||||#||||||| + Sbjct 86 QLAVRNDEELNKLLAGVTIAQGGVLPNIQA#VLLPKKTAA 124 |
| Platynereis dumerilii | Query 85 QLAIRNDEELNKLLGKVTIAQGGVLPNIQA#VLLPKKT 121 |||||||||||||| ||||||||||||||#||||||| Sbjct 84 QLAIRNDEELNKLLSGVTIAQGGVLPNIQA#VLLPKKT 120 |
| Caenorhabditis elegans | Query 85 QLAIRNDEELNKLLGKVTIAQGGVLPNIQA#VLLPKKT 121 |||+|||||||||| ||||||||||||||#||||||| Sbjct 86 QLAVRNDEELNKLLAGVTIAQGGVLPNIQA#VLLPKKT 122 |
| Bombyx mori | Query 85 QLAIRNDEELNKLLGKVTIAQGGVLPNIQA#VLLPKKTE 122 ||||||||||||| ||||||||||||||#|||||||| Sbjct 84 QLAIRNDEELNKLPSGVTIAQGGVLPNIQA#VLLPKKTE 121 |
| Urechis caupo | Query 85 QLAIRNDEELNKLLGKVTIAQGGVLPNIQA#VLLPKKTESHHK 126 |||||||||||||| ||||||||||||||#||||||+ | Sbjct 84 QLAIRNDEELNKLLSGVTIAQGGVLPNIQA#VLLPKKSSQKTK 125 |
| Drosophila virilis | Query 87 AIRNDEELNKLLGKVTIAQGGVLPNIQA#VLLPKKTE 122 |||||||||||| ||||||||||||||#|||||||| Sbjct 1 AIRNDEELNKLLSGVTIAQGGVLPNIQA#VLLPKKTE 36 |
[Site 10] AVLLPKKTES123-HHKAKGK
Ser123  His
His
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Ala114 | Val115 | Leu116 | Leu117 | Pro118 | Lys119 | Lys120 | Thr121 | Glu122 | Ser123 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| His124 | His125 | Lys126 | Ala127 | Lys128 | Gly129 | Lys130 | - | - | - |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| LNKLLGKVTIAQGGVLPNIQAVLLPKKTESHHKAKGK |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | Homo sapiens | 74.70 | 13 | histone cluster 2, H2aa3 |
| 2 | Bos taurus | 74.70 | 3 | PREDICTED: similar to Hist2h2aa1 protein |
| 3 | synthetic construct | 74.70 | 2 | histone 2 H2aa |
| 4 | Mus musculus domesticus | 74.70 | 1 | histone H2A |
| 5 | Plasmodium berghei strain ANKA | 74.70 | 1 | histone h2a |
| 6 | N/A | 74.30 | 15 | 1109175A homeostatic thymus hormone alpha |
| 7 | Canis familiaris | 74.30 | 13 | PREDICTED: similar to Histone H2A.l (H2A/l) |
| 8 | Mus musculus | 73.90 | 7 | unnamed protein product |
| 9 | Bos taurus | 73.90 | 3 | PREDICTED: similar to Chain K, 2.9 Angstrom X-Ray |
| 10 | Rattus norvegicus | 73.20 | 2 | Histone H2a |
| 11 | Rattus norvegicus | 68.90 | 1 | histone cluster 1, H2aa |
| 12 | Gallus gallus | 64.70 | 3 | histone H2A |
| 13 | Cairina moschata | 63.50 | 1 | H2A_CAIMO Histone H2A gi |
| 14 | Gallus gallus | 63.50 | 1 | histone cluster 2, H2ac |
| 15 | Xenopus laevis | 62.80 | 5 | hypothetical protein LOC494591 |
| 16 | Homo sapiens | 61.20 | 2 | H2A histone family, member J |
| 17 | Bufo bufo gagarizans | 61.20 | 2 | AF255740_2 histone H2A variant |
| 18 | Bufo bufo gagarizans | 61.20 | 1 | AF255739_1 replication-dependent histone H2A |
| 19 | Xenopus tropicalis | 60.50 | 3 | MGC69325 protein |
| 20 | Mus musculus | 60.50 | 2 | H2A histone family, member J |
| 21 | Schistosoma japonicum | 58.90 | 2 | SJCHGC05779 protein |
| 22 | Oncorhynchus mykiss | 58.90 | 1 | H2A_ONCMY Histone H2A gi |
| 23 | Tetraodon nigroviridis | 58.50 | 6 | unnamed protein product |
| 24 | Danio rerio | 58.20 | 6 | histone 2, H2a |
| 25 | Chironomus thummi | 55.80 | 2 | H2AO_CHITH Histone H2A, orphon gi |
| 26 | Strongylocentrotus purpuratus | 54.70 | 2 | histone H2A |
| 27 | Strongylocentrotus purpuratus | 54.30 | 4 | PREDICTED: similar to histone H2A-2 |
| 28 | Anopheles gambiae str. PEST | 54.30 | 2 | AGAP003913-PA |
| 29 | Drosophila melanogaster | 54.30 | 2 | His2A:CG33853 CG33853-PA |
| 30 | Pan troglodytes | 54.30 | 1 | PREDICTED: similar to Histone H2a |
| 31 | Danio rerio | 54.30 | 1 | H2A histone family, member X |
| 32 | Asellus aquaticus | 54.30 | 1 | putative H2A histone |
| 33 | Acropora formosa | 54.30 | 1 | H2A_ACRFO Histone H2A gi |
| 34 | Tribolium castaneum | 53.90 | 1 | PREDICTED: similar to CG31618-PA |
| 35 | Oikopleura dioica | 53.50 | 3 | histone h2A.1 |
| 36 | Drosophila virilis | 53.50 | 1 | histone H2A |
| 37 | Drosophila melanogaster | 53.50 | 1 | H2A histone |
| 38 | Drosophila sechellia | 52.80 | 1 | histone 2A |
| 39 | Chaetopterus variopedatus | 52.80 | 1 | histone H2A |
| 40 | Arabidopsis thaliana | 52.80 | 1 | Unknown protein |
| 41 | Mytilus edulis | 52.80 | 1 | H2A_MYTCA Histone H2A gi |
| 42 | Caenorhabditis briggsae AF16 | 52.40 | 3 | Hypothetical protein CBG19303 |
| 43 | Caenorhabditis elegans | 52.40 | 2 | HIStone family member (his-35) |
| 44 | Mytilus trossulus | 52.40 | 1 | H2A_MYTTR Histone H2A gi |
| 45 | Platynereis dumerilii | 52.00 | 1 | H2A_PLADU Histone H2A gi |
| 46 | Anopheles gambiae | 51.20 | 1 | histone H2a |
| 47 | Artemia sp. | 51.20 | 1 | unnamed protein product |
Top-ranked sequences
| organism | matching |
|---|---|
| Homo sapiens | Query 94 LNKLLGKVTIAQGGVLPNIQAVLLPKKTES#HHKAKGK 130 ||||||||||||||||||||||||||||||#||||||| Query 94 LNKLLGKVTIAQGGVLPNIQAVLLPKKTES#HHKAKGK 130 |
| Bos taurus | Query 94 LNKLLGKVTIAQGGVLPNIQAVLLPKKTES#HHKAKGK 130 ||||||||||||||||||||||||||||||#||||||| Query 94 LNKLLGKVTIAQGGVLPNIQAVLLPKKTES#HHKAKGK 130 |
| synthetic construct | Query 94 LNKLLGKVTIAQGGVLPNIQAVLLPKKTES#HHKAKGK 130 ||||||||||||||||||||||||||||||#||||||| Query 94 LNKLLGKVTIAQGGVLPNIQAVLLPKKTES#HHKAKGK 130 |
| Mus musculus domesticus | Query 94 LNKLLGKVTIAQGGVLPNIQAVLLPKKTES#HHKAKGK 130 ||||||||||||||||||||||||||||||#||||||| Query 94 LNKLLGKVTIAQGGVLPNIQAVLLPKKTES#HHKAKGK 130 |
| Plasmodium berghei strain ANKA | Query 94 LNKLLGKVTIAQGGVLPNIQAVLLPKKTES#HHKAKGK 130 ||||||||||||||||||||||||||||||#||||||| Query 94 LNKLLGKVTIAQGGVLPNIQAVLLPKKTES#HHKAKGK 130 |
| N/A | Query 94 LNKLLGKVTIAQGGVLPNIQAVLLPKKTES#HHKAKGK 130 ||||||||||||||||||||||||||||||#||||||| Query 94 LNKLLGKVTIAQGGVLPNIQAVLLPKKTES#HHKAKGK 130 |
| Canis familiaris | Query 94 LNKLLGKVTIAQGGVLPNIQAVLLPKKTES#HHKAKGK 130 ||||||+|||||||||||||||||||||||#||||||| Sbjct 115 LNKLLGRVTIAQGGVLPNIQAVLLPKKTES#HHKAKGK 151 |
| Mus musculus | Query 94 LNKLLGKVTIAQGGVLPNIQAVLLPKKTES#HHKAKGK 130 ||||||+|||||||||||||||||||||||#||||||| Sbjct 99 LNKLLGRVTIAQGGVLPNIQAVLLPKKTES#HHKAKGK 135 |
| Bos taurus | Query 94 LNKLLGKVTIAQGGVLPNIQAVLLPKKTES#HHKAKGK 130 ||||||+|||||||||||||||||||||||#||||||| Sbjct 94 LNKLLGRVTIAQGGVLPNIQAVLLPKKTES#HHKAKGK 130 |
| Rattus norvegicus | Query 94 LNKLLGKVTIAQGGVLPNIQAVLLPKKTES#HHKAKGK 130 ||||||+|||||||||||||||||||||||#|||+||| Sbjct 94 LNKLLGRVTIAQGGVLPNIQAVLLPKKTES#HHKSKGK 130 |
| Rattus norvegicus | Query 94 LNKLLGKVTIAQGGVLPNIQAVLLPKKTES#HHKAKGK 130 ||||||+|||||||||||||||||||||||#|||++ | Sbjct 94 LNKLLGRVTIAQGGVLPNIQAVLLPKKTES#HHKSQTK 130 |
| Gallus gallus | Query 94 LNKLLGKVTIAQGGVLPNIQAVLLPKKTES#HHKAKGK 130 ||||||||||||||||||||||||||||||# |||| | Sbjct 94 LNKLLGKVTIAQGGVLPNIQAVLLPKKTES#-HKAKSK 129 |
| Cairina moschata | Query 94 LNKLLGKVTIAQGGVLPNIQAVLLPKKTES#HHKAKGK 130 ||||||||||||||||||||||||||||+|# |||| | Sbjct 94 LNKLLGKVTIAQGGVLPNIQAVLLPKKTDS#-HKAKSK 129 |
| Gallus gallus | Query 94 LNKLLGKVTIAQGGVLPNIQAVLLPKKTES#HHKAKGK 130 ||||||||||||||||||||||||||||+|# |||| | Sbjct 94 LNKLLGKVTIAQGGVLPNIQAVLLPKKTDS#-HKAKAK 129 |
| Xenopus laevis | Query 94 LNKLLGKVTIAQGGVLPNIQAVLLPKKTES#HHKAKGK 130 ||||||+|||||||||||||+|||||||||# || | Sbjct 94 LNKLLGRVTIAQGGVLPNIQSVLLPKKTES#SKSAKSK 130 |
| Homo sapiens | Query 94 LNKLLGKVTIAQGGVLPNIQAVLLPKKTES#H 124 ||||||||||||||||||||||||||||||# Sbjct 94 LNKLLGKVTIAQGGVLPNIQAVLLPKKTES#Q 124 |
| Bufo bufo gagarizans | Query 94 LNKLLGKVTIAQGGVLPNIQAVLLPKKTES#HHKAKGK 130 |||||| |||||||||||||||||||||||# || | Sbjct 92 LNKLLGGVTIAQGGVLPNIQAVLLPKKTES#SKPAKSK 128 |
| Bufo bufo gagarizans | Query 94 LNKLLGKVTIAQGGVLPNIQAVLLPKKTES#HHKAKGK 130 |||||| |||||||||||||||||||||||# || | Sbjct 94 LNKLLGGVTIAQGGVLPNIQAVLLPKKTES#SKPAKSK 130 |
| Xenopus tropicalis | Query 94 LNKLLGKVTIAQGGVLPNIQAVLLPKKTES#HHKAKGK 130 |||||| |||||||||||||+|||||||||# || | Sbjct 94 LNKLLGGVTIAQGGVLPNIQSVLLPKKTES#SKAAKSK 130 |
| Mus musculus | Query 94 LNKLLGKVTIAQGGVLPNIQAVLLPKKTES#H 124 ||||||+|||||||||||||||||||||||# Sbjct 94 LNKLLGRVTIAQGGVLPNIQAVLLPKKTES#Q 124 |
| Schistosoma japonicum | Query 94 LNKLLGKVTIAQGGVLPNIQAVLLPKKTES#HHKAK 128 |||||| |||||||||||||||||||||| # ||| Sbjct 77 LNKLLGGVTIAQGGVLPNIQAVLLPKKTEK#AVKAK 111 |
| Oncorhynchus mykiss | Query 94 LNKLLGKVTIAQGGVLPNIQAVLLPKKTES#HHKAK 128 |||||| |||||||||||||||||||||| # ||| Sbjct 94 LNKLLGGVTIAQGGVLPNIQAVLLPKKTEK#AVKAK 128 |
| Tetraodon nigroviridis | Query 94 LNKLLGKVTIAQGGVLPNIQAVLLPKKTE 122 ||||||+|||||||||||||||||||||| Sbjct 94 LNKLLGRVTIAQGGVLPNIQAVLLPKKTE 122 |
| Danio rerio | Query 94 LNKLLGKVTIAQGGVLPNIQAVLLPKKTES#HHKAK 128 |||||| |||||||||||||||||||||| # | | Sbjct 94 LNKLLGGVTIAQGGVLPNIQAVLLPKKTEK#AAKGK 128 |
| Chironomus thummi | Query 94 LNKLLGKVTIAQGGVLPNIQAVLLPKKTES# 123 ||||| |||||||||||||||||||||||# Sbjct 93 LNKLLSGVTIAQGGVLPNIQAVLLPKKTES# 122 |
| Strongylocentrotus purpuratus | Query 94 LNKLLGKVTIAQGGVLPNIQAVLLPKKT 121 |||||| ||||||||||||||||||||| Sbjct 93 LNKLLGGVTIAQGGVLPNIQAVLLPKKT 120 |
| Strongylocentrotus purpuratus | Query 94 LNKLLGKVTIAQGGVLPNIQAVLLPKKT 121 |||||| ||||||||||||||||||||| Sbjct 93 LNKLLGGVTIAQGGVLPNIQAVLLPKKT 120 |
| Anopheles gambiae str. PEST | Query 94 LNKLLGKVTIAQGGVLPNIQAVLLPKKTES#H 124 ||||| |||||||||||||||||||||| #+ Sbjct 93 LNKLLSGVTIAQGGVLPNIQAVLLPKKTEK#N 123 |
| Drosophila melanogaster | Query 94 LNKLLGKVTIAQGGVLPNIQAVLLPKKTE 122 ||||| |||||||||||||||||||||| Sbjct 93 LNKLLSGVTIAQGGVLPNIQAVLLPKKTE 121 |
| Pan troglodytes | Query 94 LNKLLGKVTIAQGGVLPNIQAVLLPKKTES#HHKAKGK 130 +||||||| | | |||||||++|||||||#| | | | Sbjct 94 INKLLGKVIITQVTVLPNIQAMMLPKKTES#HLKVKEK 130 |
| Danio rerio | Query 94 LNKLLGKVTIAQGGVLPNIQAVLLPKKT 121 |||||| ||||||||||||||||||||| Sbjct 94 LNKLLGGVTIAQGGVLPNIQAVLLPKKT 121 |
| Asellus aquaticus | Query 94 LNKLLGKVTIAQGGVLPNIQAVLLPKKTE 122 ||||| |||||||||||||||||||||| Sbjct 93 LNKLLSGVTIAQGGVLPNIQAVLLPKKTE 121 |
| Acropora formosa | Query 94 LNKLLGKVTIAQGGVLPNIQAVLLPKKTE 122 ||||| |||||||||||||||||||||| Sbjct 93 LNKLLAGVTIAQGGVLPNIQAVLLPKKTE 121 |
| Tribolium castaneum | Query 94 LNKLLGKVTIAQGGVLPNIQAVLLPKKTE 122 ||||| |||||||||||||||||||||| Sbjct 93 LNKLLSGVTIAQGGVLPNIQAVLLPKKTE 121 |
| Oikopleura dioica | Query 94 LNKLLGKVTIAQGGVLPNIQAVLLPKKT 121 |||||| ||||||||||||||+|||||| Sbjct 93 LNKLLGGVTIAQGGVLPNIQAILLPKKT 120 |
| Drosophila virilis | Query 94 LNKLLGKVTIAQGGVLPNIQAVLLPKKTE 122 ||||| |||||||||||||||||||||| Sbjct 8 LNKLLSGVTIAQGGVLPNIQAVLLPKKTE 36 |
| Drosophila melanogaster | Query 94 LNKLLGKVTIAQGGVLPNIQAVLLPKKTE 122 ||||| |||||||||||||||||||||| Sbjct 30 LNKLLSGVTIAQGGVLPNIQAVLLPKKTE 58 |
| Drosophila sechellia | Query 94 LNKLLGKVTIAQGGVLPNIQAVLLPKKTE 122 ||||| |||||||||||||||||||||| Sbjct 2 LNKLLSGVTIAQGGVLPNIQAVLLPKKTE 30 |
| Chaetopterus variopedatus | Query 94 LNKLLGKVTIAQGGVLPNIQAVLLPKKTE 122 ||||| |||||||||||||||||||||+ Sbjct 93 LNKLLSGVTIAQGGVLPNIQAVLLPKKTQ 121 |
| Arabidopsis thaliana | Query 94 LNKLLGKVTIAQGGVLPNIQAVLLPKKTES# 123 ||||| ||||||||||||||||||||| +# Sbjct 93 LNKLLSGVTIAQGGVLPNIQAVLLPKKTSA# 122 |
| Mytilus edulis | Query 94 LNKLLGKVTIAQGGVLPNIQAVLLPKKTES#HHK 126 ||||| |||||||||||||||||||||+ # | Sbjct 93 LNKLLSGVTIAQGGVLPNIQAVLLPKKTQK#AAK 125 |
| Caenorhabditis briggsae AF16 | Query 94 LNKLLGKVTIAQGGVLPNIQAVLLPKKTES# 123 ||||| ||||||||||||||||||||| +# Sbjct 95 LNKLLAGVTIAQGGVLPNIQAVLLPKKTAA# 124 |
| Caenorhabditis elegans | Query 94 LNKLLGKVTIAQGGVLPNIQAVLLPKKT 121 ||||| ||||||||||||||||||||| Sbjct 95 LNKLLAGVTIAQGGVLPNIQAVLLPKKT 122 |
| Mytilus trossulus | Query 94 LNKLLGKVTIAQGGVLPNIQAVLLPKKTES#HHK 126 ||||| |||||||||||||||||||||+ # | Sbjct 93 LNKLLSGVTIAQGGVLPNIQAVLLPKKTQK#AAK 125 |
| Platynereis dumerilii | Query 94 LNKLLGKVTIAQGGVLPNIQAVLLPKKT 121 ||||| ||||||||||||||||||||| Sbjct 93 LNKLLSGVTIAQGGVLPNIQAVLLPKKT 120 |
| Anopheles gambiae | Query 95 NKLLGKVTIAQGGVLPNIQAVLLPKKT 121 |||| +|||||||||||||||||||+| Sbjct 79 NKLLRRVTIAQGGVLPNIQAVLLPKRT 105 |
| Artemia sp. | Query 96 KLLGKVTIAQGGVLPNIQAVLLPKKTES#HHKA 127 ||| |||||||||||||||||||||| # || Sbjct 1 KLLSGVTIAQGGVLPNIQAVLLPKKTEK#PAKA 32 |
* References
None.