SB0079 : Integrin [beta]1 isoform 1D
[ CaMP Format ]
* Basic Information
| Organism | Homo sapiens (human) |
| Protein Names | integrin beta-1 isoform 1D precursor [Homo sapiens]; integrin beta-1 isoform 1D precursor; integrin VLA-4 beta subunit; very late activation protein, beta polypeptide; glycoprotein IIa; Integrin beta-1; Fibronectin receptor subunit beta; Glycoprotein IIa; GPIIA; VLA-4 subunit beta |
| Gene Names | ITGB1; FNRB, MDF2, MSK12; FNRB; MDF2; MSK12; integrin, beta 1 (fibronectin receptor, beta polypeptide, antigen CD29 includes MDF2, MSK12) |
| Gene Locus | 10p11.2; chromosome 10 |
| GO Function | Not available |
* Information From OMIM
Function: Arregui et al. (2000) demonstrated that cell-permeable (Trojan) peptides containing the third helix of the antennapedia homeodomain fused to a peptide mimicking the juxtamembrane (JMP) region of the cytoplasmic domain of N-cadherin (CDH2; OMIM:114020) result in the inhibition of both CDH2 and ITGB1 function. Microscopic analysis showed that expression of JMP, which binds to the cytoplasmic domain of CDH2, results in a reduction of neurite outgrowth on cadherin substrates. Treatment of cells with JMP resulted in the release of FER (OMIM:176942) from the cadherin complex and its accumulation in the integrin complex. The accumulation of FER in the integrin complex and the inhibitory effects of JMP could be reversed with a peptide that mimics the first coiled-coil domain of FER. The results suggested that FER mediates crosstalk between CDH2 and ITGB1.
* Structure Information
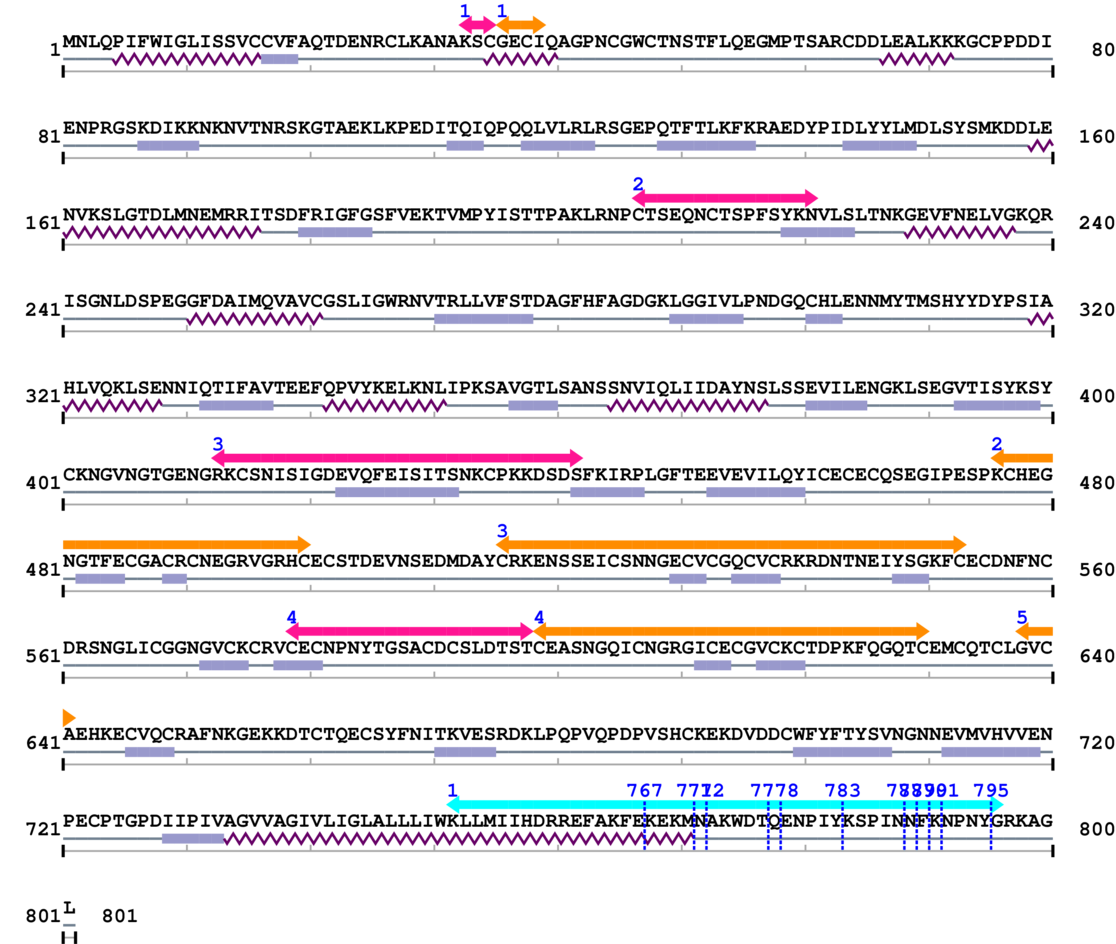
1. Primary Information
Length: 801 aa
Average Mass: 88.883 kDa
Monoisotopic Mass: 88.825 kDa
2. Domain Information
Annotated Domains: interpro / pfam / smart / prosite
Computationally Assigned Domains (Pfam+HMMER):
| domain name | begin | end | score | e-value |
|---|---|---|---|---|
| Integrin beta tail domain 1. | 33 | 35 | 5.0 | 2.2 |
| EGF-like domain 1. | 36 | 39 | 12.0 | 1.7 |
| Integrin beta tail domain 2. | 207 | 221 | 48.0 | 0.0 |
| Integrin beta tail domain 3. | 413 | 442 | 25.0 | 1.4 |
| EGF-like domain 2. | 476 | 500 | 7.0 | 18.7 |
| EGF-like domain 3. | 516 | 553 | 1.0 | 13.5 |
| Integrin beta tail domain 4. | 579 | 598 | 13.0 | 14.8 |
| EGF-like domain 4. | 599 | 630 | 1.0 | 16.5 |
| EGF-like domain 5. | 638 | 641 | 12.0 | 5.8 |
| Integrin beta cytoplasmic domain 1. | 752 | 796 | 1.0 | 2.3 |
| --- cleavage 767 (inside Integrin beta cytoplasmic domain 752..796) --- | ||||
| --- cleavage 771 (inside Integrin beta cytoplasmic domain 752..796) --- | ||||
| --- cleavage 772 (inside Integrin beta cytoplasmic domain 752..796) --- | ||||
| --- cleavage 777 (inside Integrin beta cytoplasmic domain 752..796) --- | ||||
| --- cleavage 778 (inside Integrin beta cytoplasmic domain 752..796) --- | ||||
| --- cleavage 783 (inside Integrin beta cytoplasmic domain 752..796) --- | ||||
| --- cleavage 788 (inside Integrin beta cytoplasmic domain 752..796) --- | ||||
| --- cleavage 789 (inside Integrin beta cytoplasmic domain 752..796) --- | ||||
| --- cleavage 790 (inside Integrin beta cytoplasmic domain 752..796) --- | ||||
| --- cleavage 791 (inside Integrin beta cytoplasmic domain 752..796) --- | ||||
| --- cleavage 795 (inside Integrin beta cytoplasmic domain 752..796) --- | ||||
3. Sequence Information
Fasta Sequence: SB0079.fasta
Amino Acid Sequence and Secondary Structures (PsiPred):
4. 3D Information
Known Structures in PDB: 1K11 (Model; -; B=786-797), 1LHA (Model; -; A=86-543), 3G9W (X-ray; 216 A; C/D=752-785), 3T9K (X-ray; 230 A; A/B=758-769), 3VI3 (X-ray; 290 A; B/D=21-465), 3VI4 (X-ray; 290 A; B/D=21-465), 4DX9 (X-ray; 300 A; 6/7/8/9/B/D/F/H/J/L/N/P/R/T/V/X/Z/b/d/f/h/j/l/n/p/r/t/v/x/z=784-798), 4WJK (X-ray; 185 A; B=21-465), 4WK0 (X-ray; 178 A; B=21-465), 4WK2 (X-ray; 250 A; B=21-465), 4WK4 (X-ray; 250 A; B=21-465)
* Cleavage Information
11 [sites] cleaved by Calpain 2
Source Reference: [PubMed ID: 10571053] Pfaff M, Du X, Ginsberg MH, Calpain cleavage of integrin beta cytoplasmic domains. FEBS Lett. 1999 Oct 22;460(1):17-22.
Cleavage sites (±10aa)
[Site 1] HDRREFAKFE767-KEKMNAKWDT
Glu767  Lys
Lys
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| His758 | Asp759 | Arg760 | Arg761 | Glu762 | Phe763 | Ala764 | Lys765 | Phe766 | Glu767 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Lys768 | Glu769 | Lys770 | Met771 | Asn772 | Ala773 | Lys774 | Trp775 | Asp776 | Thr777 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| AGIVLIGLALLLIWKLLMIIHDRREFAKFEKEKMNAKWDTQENPIYKSPINNFKNPNYGR |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | Canis familiaris | 135.00 | 10 | PREDICTED: similar to integrin beta 1 isoform 1D |
| 2 | Bos taurus | 135.00 | 6 | ITGB1 protein |
| 3 | N/A | 125.00 | 26 | S66396 integrin beta-1 chain isoform D - human (f |
| 4 | Mus musculus | 125.00 | 12 | beta 1 integrin |
| 5 | Gallus gallus | 113.00 | 3 | integrin beta1D |
| 6 | Ictalurus punctatus | 107.00 | 1 | beta-1 integrin |
| 7 | Pseudoplusia includens | 93.20 | 1 | integrin beta 1 |
| 8 | Anopheles gambiae | 87.80 | 2 | integrin beta subunit |
| 9 | Anopheles gambiae str. PEST | 87.80 | 1 | integrin beta subunit (AGAP000815-PA) |
| 10 | Crassostrea gigas | 86.70 | 1 | integrin beta cgh |
| 11 | Tetraodon nigroviridis | 85.50 | 6 | unnamed protein product |
| 12 | Sus scrofa | 85.50 | 3 | integrin beta-1 subunit |
| 13 | Xenopus laevis | 85.50 | 2 | integrin beta-1 subunit |
| 14 | Pongo pygmaeus | 85.50 | 1 | ITB1_PONPY Integrin beta-1 precursor (Fibronectin |
| 15 | Homo sapiens | 85.10 | 11 | integrin beta 1 isoform 1A precursor |
| 16 | Danio rerio | 85.10 | 5 | integrin, beta 1a |
| 17 | Oryctolagus cuniculus | 84.70 | 1 | glycoprotein IIIa |
| 18 | Rattus norvegicus | 84.00 | 4 | integrin beta 3 |
| 19 | synthetic construct | 84.00 | 3 | AF239959_1 hybrid integrin beta 3 subunit precurs |
| 20 | Mus musculus | 84.00 | 1 | integrin beta 3 precursor |
| 21 | rats, Peptide Partial, 723 aa | 84.00 | 1 | beta 3 integrin, GPIIIA |
| 22 | Sus scrofa | 84.00 | 1 | integrin beta chain, beta 3 |
| 23 | Canis lupus familiaris | 84.00 | 1 | integrin beta chain, beta 3 |
| 24 | Oryctolagus cuniculus | 83.20 | 1 | integrin beta 1 |
| 25 | mice, Peptide Partial, 680 aa | 82.40 | 1 | beta 3 integrin, GPIIIA |
| 26 | Felis catus | 82.40 | 1 | integrin beta 1 |
| 27 | Drosophila melanogaster | 78.20 | 1 | myospheroid CG1560-PA |
| 28 | Xenopus laevis | 74.70 | 1 | hypothetical protein LOC379708 |
| 29 | Ovis aries | 74.30 | 2 | ITB6_SHEEP Integrin beta-6 precursor gi |
| 30 | Biomphalaria glabrata | 73.60 | 1 | beta integrin subunit |
| 31 | Gallus gallus | 69.70 | 2 | integrin, beta 3 precursor |
| 32 | Podocoryne carnea | 69.70 | 1 | AF308652_1 integrin beta chain |
| 33 | Pan troglodytes | 65.90 | 2 | integrin, beta 2 |
| 34 | Homo sapiens | 65.90 | 1 | integrin, beta 2 precursor |
| 35 | Halocynthia roretzi | 64.30 | 2 | integrin beta Hr1 precursor |
| 36 | Strongylocentrotus purpuratus | 63.20 | 3 | integrin beta-C subunit |
| 37 | Mus sp. | 62.40 | 1 | beta 7 integrin |
| 38 | Caenorhabditis briggsae | 60.80 | 1 | Hypothetical protein CBG03601 |
| 39 | Macaca mulatta | 59.30 | 1 | integrin beta 1 |
| 40 | Caenorhabditis elegans | 58.90 | 1 | Paralysed Arrest at Two-fold family member (pat-3 |
| 41 | Lytechinus variegatus | 55.80 | 1 | beta-C integrin subunit |
| 42 | Anopheles gambiae str. PEST | 55.50 | 1 | AGAP010233-PA |
| 43 | Schistosoma japonicum | 54.30 | 1 | SJCHGC06221 protein |
| 44 | Acropora millepora | 51.20 | 1 | integrin subunit betaCn1 |
| 45 | Pacifastacus leniusculus | 50.10 | 1 | integrin |
| 46 | Ovis canadensis | 48.50 | 1 | ITB2_OVICA Integrin beta-2 precursor (Cell surfac |
| 47 | Sigmodon hispidus | 46.20 | 1 | AF445415_1 integrin beta-2 precursor |
Top-ranked sequences
| organism | matching |
|---|---|
| Canis familiaris | Query 738 AGIVLIGLALLLIWKLLMIIHDRREFAKFE#KEKMNAKWDTQENPIYKSPINNFKNPNYGR 797 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 738 AGIVLIGLALLLIWKLLMIIHDRREFAKFE#KEKMNAKWDTQENPIYKSPINNFKNPNYGR 797 |
| Bos taurus | Query 738 AGIVLIGLALLLIWKLLMIIHDRREFAKFE#KEKMNAKWDTQENPIYKSPINNFKNPNYGR 797 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 738 AGIVLIGLALLLIWKLLMIIHDRREFAKFE#KEKMNAKWDTQENPIYKSPINNFKNPNYGR 797 |
| N/A | Query 738 AGIVLIGLALLLIWKLLMIIHDRREFAKFE#KEKMNAKWDTQENPIYKSPINNFKNPNYGR 797 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 738 AGIVLIGLALLLIWKLLMIIHDRREFAKFE#KEKMNAKWDTQENPIYKSPINNFKNPNYGR 797 |
| Mus musculus | Query 738 AGIVLIGLALLLIWKLLMIIHDRREFAKFE#KEKMNAKWDTQENPIYKSPINNFKNPNYGR 797 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 738 AGIVLIGLALLLIWKLLMIIHDRREFAKFE#KEKMNAKWDTQENPIYKSPINNFKNPNYGR 797 |
| Gallus gallus | Query 744 GLALLLIWKLLMIIHDRREFAKFE#KEKMNAKWDTQENPIYKSPINNFKNPNYGR 797 ||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 744 GLALLLIWKLLMIIHDRREFAKFE#KEKMNAKWDTQENPIYKSPINNFKNPNYGR 797 |
| Ictalurus punctatus | Query 738 AGIVLIGLALLLIWKLLMIIHDRREFAKFE#KEKMNAKWDTQENPIYKSPINNFKNPNY 795 ||||||||||| ||||||||||||||||||#||||||||| ||||||| + || | Sbjct 747 AGIVLIGLALLFIWKLLMIIHDRREFAKFE#KEKMNAKWDAGENPIYKSAVTTVVNPKY 804 |
| Pseudoplusia includens | Query 738 AGIVLIGLALLLIWKLLMIIHDRREFAKFE#KEKMNAKWDTQENPIYKSPINNFKNPNYG 796 | |||+|||||++||+ |||||||+||#||+| ||||| |||||| + |||| | Sbjct 777 AAIVLVGLALLMLWKMATTSHDRREFARFE#KERMMAKWDTGENPIYKQATSTFKNPTYA 835 |
| Anopheles gambiae | Query 744 GLALLLIWKLLMIIHDRREFAKFE#KEKMNAKWDTQENPIYKSPINNFKNPNYG 796 |+|+||+||+| ||||||||+||#||+| ||||| |||||| |||| | Sbjct 783 GMAVLLLWKVLTSIHDRREFARFE#KERMMAKWDTGENPIYKQATTTFKNPTYA 835 |
| Anopheles gambiae str. PEST | Query 744 GLALLLIWKLLMIIHDRREFAKFE#KEKMNAKWDTQENPIYKSPINNFKNPNYG 796 |+|+||+||+| ||||||||+||#||+| ||||| |||||| |||| | Sbjct 783 GMAVLLLWKVLTSIHDRREFARFE#KERMMAKWDTGENPIYKQATTTFKNPTYA 835 |
| Crassostrea gigas | Query 738 AGIVLIGLALLLIWKLLMIIHDRREFAKFE#KEKMNAKWDTQENPIYKSPINNFKNPNY 795 |||| |+ ||||||| | |+|| |+||#|| |||+||| |||||| + | || | Sbjct 740 AGIVFFGIILLLIWKLFTTISDKRELARFE#KEAMNARWDTGENPIYKQATSTFVNPTY 797 |
| Tetraodon nigroviridis | Query 738 AGIVLIGLALLLIWKLLMIIHDRREFAKFE#KEKMNAKWDTQENPIYKSPINNFKNPNY 795 ||||||||||||||||||||||||||||||#|||||||||| ||||||| + || | Sbjct 731 AGIVLIGLALLLIWKLLMIIHDRREFAKFE#KEKMNAKWDTGENPIYKSAVTTVVNPKY 788 |
| Sus scrofa | Query 738 AGIVLIGLALLLIWKLLMIIHDRREFAKFE#KEKMNAKWDTQENPIYKSPINNFKNPNY 795 ||||||||||||||||||||||||||||||#|||||||||| ||||||| + || | Sbjct 738 AGIVLIGLALLLIWKLLMIIHDRREFAKFE#KEKMNAKWDTGENPIYKSAVTTVVNPKY 795 |
| Xenopus laevis | Query 738 AGIVLIGLALLLIWKLLMIIHDRREFAKFE#KEKMNAKWDTQENPIYKSPINNFKNPNY 795 ||||||||||||||||||||||||||||||#|||||||||| ||||||| + || | Sbjct 738 AGIVLIGLALLLIWKLLMIIHDRREFAKFE#KEKMNAKWDTGENPIYKSAVTTVVNPKY 795 |
| Pongo pygmaeus | Query 738 AGIVLIGLALLLIWKLLMIIHDRREFAKFE#KEKMNAKWDTQENPIYKSPINNFKNPNY 795 ||||||||||||||||||||||||||||||#|||||||||| ||||||| + || | Sbjct 738 AGIVLIGLALLLIWKLLMIIHDRREFAKFE#KEKMNAKWDTGENPIYKSAVTTVVNPKY 795 |
| Homo sapiens | Query 738 AGIVLIGLALLLIWKLLMIIHDRREFAKFE#KEKMNAKWDTQENPIYKSPINNFKNPNY 795 ||||||||||||||||||||||||||||||#|||||||||| ||||||| + || | Sbjct 738 AGIVLIGLALLLIWKLLMIIHDRREFAKFE#KEKMNAKWDTGENPIYKSAVTTVVNPKY 795 |
| Danio rerio | Query 738 AGIVLIGLALLLIWKLLMIIHDRREFAKFE#KEKMNAKWDTQENPIYKSPINNFKNPNY 795 ||||||||||||||||||||||||||||||#|||||||||| ||||||| + || | Sbjct 738 AGIVLIGLALLLIWKLLMIIHDRREFAKFE#KEKMNAKWDTGENPIYKSAVTTVINPKY 795 |
| Oryctolagus cuniculus | Query 738 AGIVLIGLALLLIWKLLMIIHDRREFAKFE#KEKMNAKWDTQENPIYKSPINNFKNPNY 795 |+||| |||||||||+ ||||+||||||#+|+ ||||| ||+|| + | | | Sbjct 728 GAILLIGFALLLIWKLLITIHDRKEFAKFE#EERARAKWDTANNPLYKEATSTFTNITY 785 |
| Rattus norvegicus | Query 738 AGIVLIGLALLLIWKLLMIIHDRREFAKFE#KEKMNAKWDTQENPIYKSPINNFKNPNY 795 |+||||| |||||||+ ||||+||||||#+|+ ||||| ||+|| + | | | Sbjct 727 GAILLIGLATLLIWKLLITIHDRKEFAKFE#EERARAKWDTANNPLYKEATSTFTNITY 784 |
| synthetic construct | Query 738 AGIVLIGLALLLIWKLLMIIHDRREFAKFE#KEKMNAKWDTQENPIYKSPINNFKNPNY 795 |+||||| |||||||+ ||||+||||||#+|+ ||||| ||+|| + | | | Sbjct 728 GAILLIGLATLLIWKLLITIHDRKEFAKFE#EERARAKWDTANNPLYKEATSTFTNITY 785 |
| Mus musculus | Query 738 AGIVLIGLALLLIWKLLMIIHDRREFAKFE#KEKMNAKWDTQENPIYKSPINNFKNPNY 795 |+||||| |||||||+ ||||+||||||#+|+ ||||| ||+|| + | | | Sbjct 727 GAILLIGLATLLIWKLLITIHDRKEFAKFE#EERARAKWDTANNPLYKEATSTFTNITY 784 |
| rats, Peptide Partial, 723 aa | Query 738 AGIVLIGLALLLIWKLLMIIHDRREFAKFE#KEKMNAKWDTQENPIYKSPINNFKNPNY 795 |+||||| |||||||+ ||||+||||||#+|+ ||||| ||+|| + | | | Sbjct 663 GAILLIGLATLLIWKLLITIHDRKEFAKFE#EERARAKWDTANNPLYKEATSTFTNITY 720 |
| Sus scrofa | Query 738 AGIVLIGLALLLIWKLLMIIHDRREFAKFE#KEKMNAKWDTQENPIYKSPINNFKNPNY 795 |+||||| |||||||+ ||||+||||||#+|+ ||||| ||+|| + | | | Sbjct 724 GAILLIGLATLLIWKLLITIHDRKEFAKFE#EERARAKWDTANNPLYKEATSTFTNITY 781 |
| Canis lupus familiaris | Query 738 AGIVLIGLALLLIWKLLMIIHDRREFAKFE#KEKMNAKWDTQENPIYKSPINNFKNPNY 795 |+||||| |||||||+ ||||+||||||#+|+ ||||| ||+|| + | | | Sbjct 724 GAILLIGLATLLIWKLLITIHDRKEFAKFE#EERARAKWDTANNPLYKEATSTFTNITY 781 |
| Oryctolagus cuniculus | Query 738 AGIVLIGLALLLIWKLLMIIHDRREFAKFE#KEKMNAKWDTQENPIYKSPINNFKNPNY 795 ||||||||||||||||||||||||||||||#|||||||||| ||||||| + || | Sbjct 3 AGIVLIGLALLLIWKLLMIIHDRREFAKFE#KEKMNAKWDTGENPIYKSAVTTVVNPKY 60 |
| mice, Peptide Partial, 680 aa | Query 738 AGIVLIGLALLLIWKLLMIIHDRREFAKFE#KEKMNAKWDTQENPIYKSPINNFKN 792 |+||||| |||||||+ ||||+||||||#+|+ ||||| ||+|| + | | Sbjct 624 GAILLIGLATLLIWKLLITIHDRKEFAKFE#EERARAKWDTANNPLYKEATSTFTN 678 |
| Felis catus | Query 738 AGIVLIGLALLLIWKLLMIIHDRREFAKFE#KEKMNAKWDTQENPIYKSPINNFKNPNY 795 |||||||||||||||||||||| |||||||#|||||||||| ||||||| + || | Sbjct 738 AGIVLIGLALLLIWKLLMIIHDTREFAKFE#KEKMNAKWDTGENPIYKSAVTTVVNPKY 795 |
| Drosophila melanogaster | Query 738 AGIVLIGLALLLIWKLLMIIHDRREFAKFE#KEKMNAKWDTQENPIYKSPINNFKNPNYG 796 | |||+|||+||+|||| ||||||||+||#||+||||||| |||||| + |||| | Sbjct 786 AAIVLVGLAILLLWKLLTTIHDRREFARFE#KERMNAKWDTGENPIYKQATSTFKNPMYA 844 |
| Xenopus laevis | Query 738 AGIVLIGLALLLIWKLLMIIHDRREFAKFE#KEKMNAKWDTQENPIYKSPINNFKNPNY 795 ||++ |||| |+|||++ | || |+ +||#||+||+||+ ||+| + +|||+ Sbjct 712 AGVIFIGLACLIIWKIITEIKDRNEYQRFE#KERMNSKWNPGHNPLYHNATTTVQNPNF 769 |
| Ovis aries | Query 739 GIVLIGLALLLIWKLLMIIHDRREFAKFE#KEKMNAKWDTQENPIYKSPINNFKNPNY 795 |+|||+||| |||||+ |||+| ||||# |+ ||| | ||+|+ + ||| | Sbjct 717 AILLIGVALLCIWKLLVSFHDRKEVAKFE#AERSKAKWQTGTNPLYRGSTSTFKNVTY 773 |
| Biomphalaria glabrata | Query 757 IHDRREFAKFE#KEKMNAKWDTQENPIYKSPINNFKNPNYG 796 ||| |||||||#||+ |||||| |||||| + |||| || Sbjct 747 IHDTREFAKFE#KERQNAKWDTGENPIYKQATSTFKNPTYG 786 |
| Gallus gallus | Query 751 WKLLMIIHDRREFAKFE#KEKMNAKWDTQENPIYKSPINNFKNPNY 795 ||||+ ||||||||+||#+|| ||||| ||+|| + | | | Sbjct 733 WKLLITIHDRREFARFE#EEKARAKWDTGNNPLYKEATSTFTNITY 777 |
| Podocoryne carnea | Query 738 AGIVLIGLALLLIWKLLMIIHDRREFAKFE#KEKMNAKWDTQENPIYKSPINNFKNPNYG 796 |||| |||||||||||| | |||||||||#|++ | |||+ |||||| + |+|| || Sbjct 721 AGIVGIGLALLLIWKLLATIQDRREFAKFE#KDRQNPKWDSGENPIYKKATSTFQNPMYG 779 |
| Pan troglodytes | Query 738 AGIVLIGLALLLIWKLLMIIHDRREFAKFE#KEKMNAKWDTQENPIYKSPINNFKNPNYG 796 |||||||+ ||+||| |+ + | ||+ +||#|||+ ++|+ +||++|| || + Sbjct 710 AGIVLIGILLLVIWKALIHLSDLREYRRFE#KEKLKSQWN-NDNPLFKSATTTVMNPKFA 767 |
| Homo sapiens | Query 738 AGIVLIGLALLLIWKLLMIIHDRREFAKFE#KEKMNAKWDTQENPIYKSPINNFKNPNYG 796 |||||||+ ||+||| |+ + | ||+ +||#|||+ ++|+ +||++|| || + Sbjct 710 AGIVLIGILLLVIWKALIHLSDLREYRRFE#KEKLKSQWN-NDNPLFKSATTTVMNPKFA 767 |
| Halocynthia roretzi | Query 738 AGIVLIGLALLLIWKLLMIIHDRREFAKFE#KEKMNAKWDTQENPIYKSPINNFKNPNYG 796 |||||||| | +||+ + |++|+ ||#+| ++| ||++ +| |+||+|| Sbjct 791 AGIVLIGLIALAVWKVYQTMRDKKEWENFE#REMKQSRWTKDINPVFVNPSAKFENPSYG 849 |
| Strongylocentrotus purpuratus | Query 739 GIVLIGLALLLIWKLLMIIHDRREFAKFE#KEKMNAKWDTQENPIYKSPINNFKNPNY 795 |+ |+||||||||+|| ||||||| ||#||+ || |+ |||||| + |||| | Sbjct 747 GVFLVGLALLLIWRLLTYIHDRREFQNFE#KERANATWEGGENPIYKPSTSVFKNPTY 803 |
| Mus sp. | Query 738 AGIVLIGLALLLIWKLLMIIHDRREFAKFE#KEKMNAKWDTQENPIYKSPINNFKNPNY 795 ||| +|| |+| ++| + |+||||+ +||#||+ | ||+||| | || + Sbjct 732 GGIVAVGLGLVLAYRLSVEIYDRREYRRFE#KEQQQLNWKQDNNPLYKSAITTTVNPRF 789 |
| Caenorhabditis briggsae | Query 758 HDRREFAKFE#KEKMNAKWDTQENPIYKSPINNFKNPNYG 796 ||| |+||| # |++ ||||| |||||| |||| | Sbjct 768 HDRAEYAKFN#NERLMAKWDTNENPIYKQATTTFKNPVYA 806 |
| Macaca mulatta | Query 738 AGIVLIGLALLLIWKLLMIIHDRREFAKFE#KEKMNAKW 775 ||||||||||||||||||||||||||||||#|||||||| Query 738 AGIVLIGLALLLIWKLLMIIHDRREFAKFE#KEKMNAKW 775 |
| Caenorhabditis elegans | Query 758 HDRREFAKFE#KEKMNAKWDTQENPIYKSPINNFKNPNYG 796 ||| |+| | # |++ ||||| |||||| |||| | Sbjct 767 HDRSEYATFN#NERLMAKWDTNENPIYKQATTTFKNPVYA 805 |
| Lytechinus variegatus | Query 747 LLLIWKLLMIIHDRREFAKFE#KEKMNAKWDTQENPIYKSPINNFKNPNY 795 |||||+|| ||||||| ||#||+ || |+ |||||| + |||| | Sbjct 755 LLLIWRLLTYIHDRREFQNFE#KERANATWEGGENPIYKPSTSVFKNPTY 803 |
| Anopheles gambiae str. PEST | Query 738 AGIVLIGLALLLIWKLLMIIHDRREFAKFE#KEKMNAKWDTQENPIYKSPINNFKNP 793 | |++ || +| ++ ++ ||+ |||||#||+ |+|+|||||+||| | Sbjct 740 ASIIIGGLLMLFCYRCKIMYDDRKMFAKFE#KEREQETKYQMESPLYKSPISNFKVP 795 |
| Schistosoma japonicum | Query 751 WKLLMIIHDRREFAKFE#KEKMNAKWDTQENPIYKSPINNFKNPNY 795 +||++ | |||| | |+#++ | +|+ ||||++|| | || + Sbjct 239 YKLVITIDDRRELANFK#QQGENMRWEMAENPIFESPTTNVLNPTF 283 |
| Acropora millepora | Query 752 KLLMIIHDRREFAKFE#KEKMNAKWDTQENPIYKSPINNFKNPNYG 796 | | + || |+ |||#+|+|++|| ++||+|++ |+|| | Sbjct 744 KGLFTMVDRIEYQKFE#RERMHSKWTREKNPLYQAAKTTFENPTYA 788 |
| Pacifastacus leniusculus | Query 738 AGIVLIGLALLLIWKLLMIIHDRREFAKFE#KEKMNAKWDTQENPIYKSPINNFKNPNYGR 797 | || ||| ||||||| +|||||+||||#||+ |||+||| |+|| + + Sbjct 740 AAIVAIGLLTLLIWKLLTTLHDRREYAKFE#KERKLPSGKRAENPLYKSAKTTFQNPAFAQ 799 |
| Ovis canadensis | Query 750 IWKLLMIIHDRREFAKFE#KEKMNAKWDTQENPIYKSPINNFKNPNYG 796 ||| | + | ||+ +||#|||+ ++|+ +||++|| || + Sbjct 723 IWKALTHLSDLREYHRFE#KEKLKSQWN-NDNPLFKSATTTVMNPKFA 768 |
| Sigmodon hispidus | Query 748 LLIWKLLMIIHDRREFAKFE#KEKMNAKWDTQENPIYKSPINNFKNPNYG 796 |+||| | + | ||+ ||#|||+ ++|+ +||++|| || + Sbjct 720 LVIWKALTHLTDLREYRHFE#KEKLKSQWN-NDNPLFKSATTTVMNPKFA 767 |
[Site 2] EFAKFEKEKM771-NAKWDTQENP
Met771  Asn
Asn
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Glu762 | Phe763 | Ala764 | Lys765 | Phe766 | Glu767 | Lys768 | Glu769 | Lys770 | Met771 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Asn772 | Ala773 | Lys774 | Trp775 | Asp776 | Thr777 | Gln778 | Glu779 | Asn780 | Pro781 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| LIGLALLLIWKLLMIIHDRREFAKFEKEKMNAKWDTQENPIYKSPINNFKNPNYGRKAGL |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | Canis familiaris | 135.00 | 10 | PREDICTED: similar to integrin beta 1 isoform 1D |
| 2 | Bos taurus | 135.00 | 6 | ITGB1 protein |
| 3 | N/A | 125.00 | 26 | S66396 integrin beta-1 chain isoform D - human (f |
| 4 | Mus musculus | 124.00 | 12 | beta 1 integrin |
| 5 | Gallus gallus | 120.00 | 3 | integrin beta1D |
| 6 | Ictalurus punctatus | 99.80 | 1 | beta-1 integrin |
| 7 | Pseudoplusia includens | 89.40 | 1 | integrin beta 1 |
| 8 | Anopheles gambiae | 88.60 | 2 | integrin beta subunit |
| 9 | Anopheles gambiae str. PEST | 88.60 | 1 | integrin beta subunit (AGAP000815-PA) |
| 10 | Oryctolagus cuniculus | 82.40 | 1 | glycoprotein IIIa |
| 11 | Rattus norvegicus | 82.00 | 4 | integrin beta 3 |
| 12 | synthetic construct | 82.00 | 3 | AF239959_1 hybrid integrin beta 3 subunit precurs |
| 13 | Sus scrofa | 82.00 | 1 | integrin beta chain, beta 3 |
| 14 | Mus musculus | 82.00 | 1 | integrin beta 3 precursor |
| 15 | Canis lupus familiaris | 82.00 | 1 | integrin beta chain, beta 3 |
| 16 | rats, Peptide Partial, 723 aa | 82.00 | 1 | beta 3 integrin, GPIIIA |
| 17 | Xenopus laevis | 81.60 | 2 | integrin beta-3 subunit |
| 18 | Sus scrofa | 80.50 | 3 | AF170527_1 glycoprotein IIIa; GPIIIa |
| 19 | mice, Peptide Partial, 680 aa | 80.10 | 1 | beta 3 integrin, GPIIIA |
| 20 | Crassostrea gigas | 79.30 | 1 | integrin beta cgh |
| 21 | Tetraodon nigroviridis | 78.60 | 6 | unnamed protein product |
| 22 | Danio rerio | 78.60 | 5 | integrin beta 3b |
| 23 | Homo sapiens | 78.20 | 11 | ITB1_HUMAN Integrin beta-1 precursor (Fibronectin |
| 24 | Pongo pygmaeus | 78.20 | 1 | ITB1_PONPY Integrin beta-1 precursor (Fibronectin |
| 25 | Oryctolagus cuniculus | 76.30 | 1 | integrin beta 1 |
| 26 | Felis catus | 75.50 | 1 | integrin beta 1 |
| 27 | Drosophila melanogaster | 75.50 | 1 | myospheroid CG1560-PA |
| 28 | Ovis aries | 73.60 | 2 | ITB6_SHEEP Integrin beta-6 precursor gi |
| 29 | Biomphalaria glabrata | 73.60 | 1 | beta integrin subunit |
| 30 | Gallus gallus | 69.70 | 2 | integrin, beta 3 precursor |
| 31 | Xenopus laevis | 68.60 | 1 | hypothetical protein LOC379708 |
| 32 | Podocoryne carnea | 67.80 | 1 | AF308652_1 integrin beta chain |
| 33 | Caenorhabditis briggsae | 63.50 | 1 | Hypothetical protein CBG03601 |
| 34 | Strongylocentrotus purpuratus | 61.60 | 3 | integrin beta L subunit |
| 35 | Caenorhabditis elegans | 61.60 | 1 | Paralysed Arrest at Two-fold family member (pat-3 |
| 36 | Halocynthia roretzi | 60.10 | 2 | integrin beta Hr2 precursor |
| 37 | Pan troglodytes | 58.90 | 2 | integrin, beta 2 |
| 38 | Homo sapiens | 58.90 | 1 | integrin, beta 2 precursor |
| 39 | Lytechinus variegatus | 58.20 | 1 | beta-C integrin subunit |
| 40 | Mus sp. | 57.40 | 1 | beta 7 integrin |
| 41 | Schistosoma japonicum | 54.30 | 1 | SJCHGC06221 protein |
| 42 | Anopheles gambiae str. PEST | 52.00 | 1 | AGAP010233-PA |
| 43 | Macaca mulatta | 51.60 | 1 | integrin beta 1 |
| 44 | Acropora millepora | 51.20 | 1 | integrin subunit betaCn1 |
| 45 | Pacifastacus leniusculus | 48.90 | 1 | integrin |
| 46 | Ovis canadensis | 48.50 | 1 | ITB2_OVICA Integrin beta-2 precursor (Cell surfac |
| 47 | Sigmodon hispidus | 46.20 | 1 | AF445415_1 integrin beta-2 precursor |
Top-ranked sequences
| organism | matching |
|---|---|
| Canis familiaris | Query 742 LIGLALLLIWKLLMIIHDRREFAKFEKEKM#NAKWDTQENPIYKSPINNFKNPNYGRKAGL 801 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 742 LIGLALLLIWKLLMIIHDRREFAKFEKEKM#NAKWDTQENPIYKSPINNFKNPNYGRKAGL 801 |
| Bos taurus | Query 742 LIGLALLLIWKLLMIIHDRREFAKFEKEKM#NAKWDTQENPIYKSPINNFKNPNYGRKAGL 801 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 742 LIGLALLLIWKLLMIIHDRREFAKFEKEKM#NAKWDTQENPIYKSPINNFKNPNYGRKAGL 801 |
| N/A | Query 742 LIGLALLLIWKLLMIIHDRREFAKFEKEKM#NAKWDTQENPIYKSPINNFKNPNYGRKAGL 801 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 742 LIGLALLLIWKLLMIIHDRREFAKFEKEKM#NAKWDTQENPIYKSPINNFKNPNYGRKAGL 801 |
| Mus musculus | Query 742 LIGLALLLIWKLLMIIHDRREFAKFEKEKM#NAKWDTQENPIYKSPINNFKNPNYGRKAGL 801 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 742 LIGLALLLIWKLLMIIHDRREFAKFEKEKM#NAKWDTQENPIYKSPINNFKNPNYGRKAGL 801 |
| Gallus gallus | Query 744 GLALLLIWKLLMIIHDRREFAKFEKEKM#NAKWDTQENPIYKSPINNFKNPNYGRKAGL 801 ||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 744 GLALLLIWKLLMIIHDRREFAKFEKEKM#NAKWDTQENPIYKSPINNFKNPNYGRKAGL 801 |
| Ictalurus punctatus | Query 742 LIGLALLLIWKLLMIIHDRREFAKFEKEKM#NAKWDTQENPIYKSPINNFKNPNYGRK 798 ||||||| ||||||||||||||||||||||#||||| ||||||| + || | | Sbjct 751 LIGLALLFIWKLLMIIHDRREFAKFEKEKM#NAKWDAGENPIYKSAVTTVVNPKYEGK 807 |
| Pseudoplusia includens | Query 742 LIGLALLLIWKLLMIIHDRREFAKFEKEKM#NAKWDTQENPIYKSPINNFKNPNYGRK 798 |+|||||++||+ |||||||+||||+|# ||||| |||||| + |||| | | Sbjct 781 LVGLALLMLWKMATTSHDRREFARFEKERM#MAKWDTGENPIYKQATSTFKNPTYAGK 837 |
| Anopheles gambiae | Query 744 GLALLLIWKLLMIIHDRREFAKFEKEKM#NAKWDTQENPIYKSPINNFKNPNYGRK 798 |+|+||+||+| ||||||||+||||+|# ||||| |||||| |||| | | Sbjct 783 GMAVLLLWKVLTSIHDRREFARFEKERM#MAKWDTGENPIYKQATTTFKNPTYAGK 837 |
| Anopheles gambiae str. PEST | Query 744 GLALLLIWKLLMIIHDRREFAKFEKEKM#NAKWDTQENPIYKSPINNFKNPNYGRK 798 |+|+||+||+| ||||||||+||||+|# ||||| |||||| |||| | | Sbjct 783 GMAVLLLWKVLTSIHDRREFARFEKERM#MAKWDTGENPIYKQATTTFKNPTYAGK 837 |
| Oryctolagus cuniculus | Query 742 LIGLALLLIWKLLMIIHDRREFAKFEKEKM#NAKWDTQENPIYKSPINNFKNPNY 795 ||| |||||||||+ ||||+||||||+|+ # ||||| ||+|| + | | | Sbjct 732 LIGFALLLIWKLLITIHDRKEFAKFEEERA#RAKWDTANNPLYKEATSTFTNITY 785 |
| Rattus norvegicus | Query 742 LIGLALLLIWKLLMIIHDRREFAKFEKEKM#NAKWDTQENPIYKSPINNFKNPNY 795 ||||| |||||||+ ||||+||||||+|+ # ||||| ||+|| + | | | Sbjct 731 LIGLATLLIWKLLITIHDRKEFAKFEEERA#RAKWDTANNPLYKEATSTFTNITY 784 |
| synthetic construct | Query 742 LIGLALLLIWKLLMIIHDRREFAKFEKEKM#NAKWDTQENPIYKSPINNFKNPNY 795 ||||| |||||||+ ||||+||||||+|+ # ||||| ||+|| + | | | Sbjct 732 LIGLATLLIWKLLITIHDRKEFAKFEEERA#RAKWDTANNPLYKEATSTFTNITY 785 |
| Sus scrofa | Query 742 LIGLALLLIWKLLMIIHDRREFAKFEKEKM#NAKWDTQENPIYKSPINNFKNPNY 795 ||||| |||||||+ ||||+||||||+|+ # ||||| ||+|| + | | | Sbjct 728 LIGLATLLIWKLLITIHDRKEFAKFEEERA#RAKWDTANNPLYKEATSTFTNITY 781 |
| Mus musculus | Query 742 LIGLALLLIWKLLMIIHDRREFAKFEKEKM#NAKWDTQENPIYKSPINNFKNPNY 795 ||||| |||||||+ ||||+||||||+|+ # ||||| ||+|| + | | | Sbjct 731 LIGLATLLIWKLLITIHDRKEFAKFEEERA#RAKWDTANNPLYKEATSTFTNITY 784 |
| Canis lupus familiaris | Query 742 LIGLALLLIWKLLMIIHDRREFAKFEKEKM#NAKWDTQENPIYKSPINNFKNPNY 795 ||||| |||||||+ ||||+||||||+|+ # ||||| ||+|| + | | | Sbjct 728 LIGLATLLIWKLLITIHDRKEFAKFEEERA#RAKWDTANNPLYKEATSTFTNITY 781 |
| rats, Peptide Partial, 723 aa | Query 742 LIGLALLLIWKLLMIIHDRREFAKFEKEKM#NAKWDTQENPIYKSPINNFKNPNY 795 ||||| |||||||+ ||||+||||||+|+ # ||||| ||+|| + | | | Sbjct 667 LIGLATLLIWKLLITIHDRKEFAKFEEERA#RAKWDTANNPLYKEATSTFTNITY 720 |
| Xenopus laevis | Query 742 LIGLALLLIWKLLMIIHDRREFAKFEKEKM#NAKWDTQENPIYKSPINNFKNPNY 795 |||| |||||||+ |||||||||||+|+ # ||||| ||+|| + | | | Sbjct 730 LIGLVALLIWKLLITIHDRREFAKFEEERA#KAKWDTAHNPLYKGATSTFTNITY 783 |
| Sus scrofa | Query 742 LIGLALLLIWKLLMIIHDRREFAKFEKEKM#NAKWDTQENPIYKSPINNFKNPNY 795 ||| | |||||||+ ||||+||||||+|+ # ||||| ||+|| + | | | Sbjct 728 LIGFATLLIWKLLITIHDRKEFAKFEEERA#RAKWDTANNPLYKEATSTFTNITY 781 |
| mice, Peptide Partial, 680 aa | Query 742 LIGLALLLIWKLLMIIHDRREFAKFEKEKM#NAKWDTQENPIYKSPINNFKN 792 ||||| |||||||+ ||||+||||||+|+ # ||||| ||+|| + | | Sbjct 628 LIGLATLLIWKLLITIHDRKEFAKFEEERA#RAKWDTANNPLYKEATSTFTN 678 |
| Crassostrea gigas | Query 742 LIGLALLLIWKLLMIIHDRREFAKFEKEKM#NAKWDTQENPIYKSPINNFKNPNY 795 |+ ||||||| | |+|| |+|||| |#||+||| |||||| + | || | Sbjct 744 FFGIILLLIWKLFTTISDKRELARFEKEAM#NARWDTGENPIYKQATSTFVNPTY 797 |
| Tetraodon nigroviridis | Query 742 LIGLALLLIWKLLMIIHDRREFAKFEKEKM#NAKWDTQENPIYKSPINNFKNPNY 795 +||| |||||||+ |||||||||||+|+ # ||||| ||+|| + | | + Sbjct 747 FLGLAALLIWKLLITIHDRREFAKFEEERA#RAKWDTGHNPLYKGATSTFTNITF 800 |
| Danio rerio | Query 742 LIGLALLLIWKLLMIIHDRREFAKFEKEKM#NAKWDTQENPIYKSPINNFKNPNY 795 +||| |||||||+ |||||||||||+|+ # |||+| ||+|| + | | | Sbjct 733 FLGLAALLIWKLLITIHDRREFAKFEEERA#RAKWETGHNPLYKGATSTFTNITY 786 |
| Homo sapiens | Query 755 MIIHDRREFAKFEKEKM#NAKWDTQENPIYKSPINNFKNPNYGRK 798 |||||||||||||||||#|||||| ||||||| + || | | Sbjct 755 MIIHDRREFAKFEKEKM#NAKWDTGENPIYKSAVTTVVNPKYEGK 798 |
| Pongo pygmaeus | Query 755 MIIHDRREFAKFEKEKM#NAKWDTQENPIYKSPINNFKNPNYGRK 798 |||||||||||||||||#|||||| ||||||| + || | | Sbjct 755 MIIHDRREFAKFEKEKM#NAKWDTGENPIYKSAVTTVVNPKYEGK 798 |
| Oryctolagus cuniculus | Query 742 LIGLALLLIWKLLMIIHDRREFAKFEKEKM#NAKWDTQENPIYKSPINNFKNPNY 795 ||||||||||||||||||||||||||||||#|||||| ||||||| + || | Sbjct 7 LIGLALLLIWKLLMIIHDRREFAKFEKEKM#NAKWDTGENPIYKSAVTTVVNPKY 60 |
| Felis catus | Query 755 MIIHDRREFAKFEKEKM#NAKWDTQENPIYKSPINNFKNPNYGRK 798 ||||| |||||||||||#|||||| ||||||| + || | | Sbjct 755 MIIHDTREFAKFEKEKM#NAKWDTGENPIYKSAVTTVVNPKYEGK 798 |
| Drosophila melanogaster | Query 757 IHDRREFAKFEKEKM#NAKWDTQENPIYKSPINNFKNPNYGRK 798 ||||||||+||||+|#|||||| |||||| + |||| | | Sbjct 805 IHDRREFARFEKERM#NAKWDTGENPIYKQATSTFKNPMYAGK 846 |
| Ovis aries | Query 742 LIGLALLLIWKLLMIIHDRREFAKFEKEKM#NAKWDTQENPIYKSPINNFKNPNYGRK 798 |||+||| |||||+ |||+| |||| |+ # ||| | ||+|+ + ||| | | Sbjct 720 LIGVALLCIWKLLVSFHDRKEVAKFEAERS#KAKWQTGTNPLYRGSTSTFKNVTYKHK 776 |
| Biomphalaria glabrata | Query 757 IHDRREFAKFEKEKM#NAKWDTQENPIYKSPINNFKNPNYGRK 798 ||| |||||||||+ #|||||| |||||| + |||| || + Sbjct 747 IHDTREFAKFEKERQ#NAKWDTGENPIYKQATSTFKNPTYGSQ 788 |
| Gallus gallus | Query 751 WKLLMIIHDRREFAKFEKEKM#NAKWDTQENPIYKSPINNFKNPNY 795 ||||+ ||||||||+||+|| # ||||| ||+|| + | | | Sbjct 733 WKLLITIHDRREFARFEEEKA#RAKWDTGNNPLYKEATSTFTNITY 777 |
| Xenopus laevis | Query 742 LIGLALLLIWKLLMIIHDRREFAKFEKEKM#NAKWDTQENPIYKSPINNFKNPNY 795 |||| |+|||++ | || |+ +||||+|#|+||+ ||+| + +|||+ Sbjct 716 FIGLACLIIWKIITEIKDRNEYQRFEKERM#NSKWNPGHNPLYHNATTTVQNPNF 769 |
| Podocoryne carnea | Query 757 IHDRREFAKFEKEKM#NAKWDTQENPIYKSPINNFKNPNYGRK 798 | ||||||||||++ #| |||+ |||||| + |+|| || | Sbjct 740 IQDRREFAKFEKDRQ#NPKWDSGENPIYKKATSTFQNPMYGNK 781 |
| Caenorhabditis briggsae | Query 758 HDRREFAKFEKEKM#NAKWDTQENPIYKSPINNFKNPNYGRKA 799 ||| |+||| |++# ||||| |||||| |||| | || Sbjct 768 HDRAEYAKFNNERL#MAKWDTNENPIYKQATTTFKNPVYAGKA 809 |
| Strongylocentrotus purpuratus | Query 751 WKLLMIIHDRREFAKFEKEKM#NAKWDTQENPIYKSPINNFKNPNYGR 797 |+| + |+||+|++| + # |+|| +|||||| |||| ||+ Sbjct 756 WRLYTYVQDKREYAQWENDCK#KAQWDQSDNPIYKSSTTTFKNPTYGK 802 |
| Caenorhabditis elegans | Query 758 HDRREFAKFEKEKM#NAKWDTQENPIYKSPINNFKNPNYGRKA 799 ||| |+| | |++# ||||| |||||| |||| | || Sbjct 767 HDRSEYATFNNERL#MAKWDTNENPIYKQATTTFKNPVYAGKA 808 |
| Halocynthia roretzi | Query 742 LIGLALLLIWKLLMIIHDRREFAKFEKEKM#NAKWDTQENPIYKSPINNFKNPNYG 796 |||+ |+|||++ + |+||+ ||+||+ # || |||+| + + | || +| Sbjct 778 LIGIIALIIWKVIQTLRDKREYEKFKKEED#LRKWTKGENPVYVNASSKFDNPMFG 832 |
| Pan troglodytes | Query 742 LIGLALLLIWKLLMIIHDRREFAKFEKEKM#NAKWDTQENPIYKSPINNFKNPNYG 796 |||+ ||+||| |+ + | ||+ +|||||+# ++|+ +||++|| || + Sbjct 714 LIGILLLVIWKALIHLSDLREYRRFEKEKL#KSQWN-NDNPLFKSATTTVMNPKFA 767 |
| Homo sapiens | Query 742 LIGLALLLIWKLLMIIHDRREFAKFEKEKM#NAKWDTQENPIYKSPINNFKNPNYG 796 |||+ ||+||| |+ + | ||+ +|||||+# ++|+ +||++|| || + Sbjct 714 LIGILLLVIWKALIHLSDLREYRRFEKEKL#KSQWN-NDNPLFKSATTTVMNPKFA 767 |
| Lytechinus variegatus | Query 750 IWKLLMIIHDRREFAKFEKEKM#NAKWDTQENPIYKSPINNFKNPNYGRK 798 ||+|| ||||||| ||||+ #|| |+ |||||| + |||| | | Sbjct 758 IWRLLTYIHDRREFQNFEKERA#NATWEGGENPIYKPSTSVFKNPTYNIK 806 |
| Mus sp. | Query 743 IGLALLLIWKLLMIIHDRREFAKFEKEKM#NAKWDTQENPIYKSPINNFKNPNY 795 +|| |+| ++| + |+||||+ +||||+ # | ||+||| | || + Sbjct 737 VGLGLVLAYRLSVEIYDRREYRRFEKEQQ#QLNWKQDNNPLYKSAITTTVNPRF 789 |
| Schistosoma japonicum | Query 751 WKLLMIIHDRREFAKFEKEKM#NAKWDTQENPIYKSPINNFKNPNY 795 +||++ | |||| | |+++ #| +|+ ||||++|| | || + Sbjct 239 YKLVITIDDRRELANFKQQGE#NMRWEMAENPIFESPTTNVLNPTF 283 |
| Anopheles gambiae str. PEST | Query 744 GLALLLIWKLLMIIHDRREFAKFEKEKM#NAKWDTQENPIYKSPINNFKNP 793 || +| ++ ++ ||+ |||||||+ # |+|+|||||+||| | Sbjct 746 GLLMLFCYRCKIMYDDRKMFAKFEKERE#QETKYQMESPLYKSPISNFKVP 795 |
| Macaca mulatta | Query 742 LIGLALLLIWKLLMIIHDRREFAKFEKEKM#NAKW 775 ||||||||||||||||||||||||||||||#|||| Query 742 LIGLALLLIWKLLMIIHDRREFAKFEKEKM#NAKW 775 |
| Acropora millepora | Query 752 KLLMIIHDRREFAKFEKEKM#NAKWDTQENPIYKSPINNFKNPNY--GRK 798 | | + || |+ |||+|+|#++|| ++||+|++ |+|| | ||+ Sbjct 744 KGLFTMVDRIEYQKFERERM#HSKWTREKNPLYQAAKTTFENPTYAGGRQ 792 |
| Pacifastacus leniusculus | Query 758 HDRREFAKFEKEKM#NAKWDTQENPIYKSPINNFKNPNYGR 797 |||||+||||||+ # |||+||| |+|| + + Sbjct 760 HDRREYAKFEKERK#LPSGKRAENPLYKSAKTTFQNPAFAQ 799 |
| Ovis canadensis | Query 750 IWKLLMIIHDRREFAKFEKEKM#NAKWDTQENPIYKSPINNFKNPNYG 796 ||| | + | ||+ +|||||+# ++|+ +||++|| || + Sbjct 723 IWKALTHLSDLREYHRFEKEKL#KSQWN-NDNPLFKSATTTVMNPKFA 768 |
| Sigmodon hispidus | Query 748 LLIWKLLMIIHDRREFAKFEKEKM#NAKWDTQENPIYKSPINNFKNPNYG 796 |+||| | + | ||+ |||||+# ++|+ +||++|| || + Sbjct 720 LVIWKALTHLTDLREYRHFEKEKL#KSQWN-NDNPLFKSATTTVMNPKFA 767 |
[Site 3] FAKFEKEKMN772-AKWDTQENPI
Asn772  Ala
Ala
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Phe763 | Ala764 | Lys765 | Phe766 | Glu767 | Lys768 | Glu769 | Lys770 | Met771 | Asn772 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Ala773 | Lys774 | Trp775 | Asp776 | Thr777 | Gln778 | Glu779 | Asn780 | Pro781 | Ile782 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| IGLALLLIWKLLMIIHDRREFAKFEKEKMNAKWDTQENPIYKSPINNFKNPNYGRKAGL |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | Canis familiaris | 133.00 | 10 | PREDICTED: similar to integrin beta 1 isoform 1D |
| 2 | Bos taurus | 133.00 | 7 | ITGB1 protein |
| 3 | N/A | 123.00 | 25 | S66396 integrin beta-1 chain isoform D - human (f |
| 4 | Mus musculus | 123.00 | 12 | beta 1 integrin |
| 5 | Gallus gallus | 120.00 | 3 | integrin beta1D |
| 6 | Ictalurus punctatus | 98.20 | 1 | beta-1 integrin |
| 7 | Anopheles gambiae | 88.60 | 2 | integrin beta subunit |
| 8 | Anopheles gambiae str. PEST | 88.60 | 1 | integrin beta subunit (AGAP000815-PA) |
| 9 | Pseudoplusia includens | 87.80 | 1 | integrin beta 1 |
| 10 | Oryctolagus cuniculus | 80.90 | 1 | glycoprotein IIIa |
| 11 | Rattus norvegicus | 80.10 | 4 | integrin beta 3 |
| 12 | synthetic construct | 80.10 | 3 | AF239959_1 hybrid integrin beta 3 subunit precurs |
| 13 | Xenopus laevis | 80.10 | 2 | integrin beta-3 subunit |
| 14 | Sus scrofa | 80.10 | 1 | integrin beta chain, beta 3 |
| 15 | Canis lupus familiaris | 80.10 | 1 | integrin beta chain, beta 3 |
| 16 | Mus musculus | 80.10 | 1 | integrin beta 3 precursor |
| 17 | rats, Peptide Partial, 723 aa | 80.10 | 1 | beta 3 integrin, GPIIIA |
| 18 | Crassostrea gigas | 79.30 | 1 | integrin beta cgh |
| 19 | Sus scrofa | 79.00 | 3 | AF170527_1 glycoprotein IIIa; GPIIIa |
| 20 | Tetraodon nigroviridis | 78.60 | 5 | unnamed protein product |
| 21 | mice, Peptide Partial, 680 aa | 78.60 | 1 | beta 3 integrin, GPIIIA |
| 22 | Homo sapiens | 78.20 | 11 | integrin beta 1 isoform 1A precursor |
| 23 | Danio rerio | 78.20 | 5 | integrin, beta 1a |
| 24 | Pongo pygmaeus | 78.20 | 1 | ITB1_PONPY Integrin beta-1 precursor (Fibronectin |
| 25 | Felis catus | 75.50 | 1 | integrin beta 1 |
| 26 | Drosophila melanogaster | 75.50 | 1 | myospheroid CG1560-PA |
| 27 | Oryctolagus cuniculus | 74.30 | 1 | integrin beta 1 |
| 28 | Biomphalaria glabrata | 73.60 | 1 | beta integrin subunit |
| 29 | Ovis aries | 72.00 | 2 | ITB6_SHEEP Integrin beta-6 precursor gi |
| 30 | Gallus gallus | 69.70 | 2 | integrin, beta 3 precursor |
| 31 | Xenopus laevis | 68.60 | 1 | hypothetical protein LOC379708 |
| 32 | Podocoryne carnea | 67.80 | 1 | AF308652_1 integrin beta chain |
| 33 | Caenorhabditis briggsae | 63.50 | 1 | Hypothetical protein CBG03601 |
| 34 | Strongylocentrotus purpuratus | 61.60 | 3 | integrin beta L subunit |
| 35 | Caenorhabditis elegans | 61.60 | 1 | Paralysed Arrest at Two-fold family member (pat-3 |
| 36 | Halocynthia roretzi | 58.50 | 2 | integrin beta Hr2 precursor |
| 37 | Pan troglodytes | 58.20 | 2 | PREDICTED: integrin, beta 7 isoform 5 |
| 38 | Lytechinus variegatus | 58.20 | 1 | beta-C integrin subunit |
| 39 | Mus sp. | 57.40 | 1 | beta 7 integrin |
| 40 | Homo sapiens | 57.00 | 1 | integrin, beta 2 precursor |
| 41 | Schistosoma japonicum | 54.30 | 1 | SJCHGC06221 protein |
| 42 | Anopheles gambiae str. PEST | 52.00 | 1 | AGAP010233-PA |
| 43 | Acropora millepora | 51.20 | 1 | integrin subunit betaCn1 |
| 44 | Macaca mulatta | 49.70 | 1 | integrin beta 1 |
| 45 | Pacifastacus leniusculus | 48.90 | 1 | integrin |
| 46 | Ovis canadensis | 48.50 | 1 | ITB2_OVICA Integrin beta-2 precursor (Cell surfac |
| 47 | Sigmodon hispidus | 47.40 | 1 | AF445415_1 integrin beta-2 precursor |
Top-ranked sequences
| organism | matching |
|---|---|
| Canis familiaris | Query 743 IGLALLLIWKLLMIIHDRREFAKFEKEKMN#AKWDTQENPIYKSPINNFKNPNYGRKAGL 801 ||||||||||||||||||||||||||||||#||||||||||||||||||||||||||||| Query 743 IGLALLLIWKLLMIIHDRREFAKFEKEKMN#AKWDTQENPIYKSPINNFKNPNYGRKAGL 801 |
| Bos taurus | Query 743 IGLALLLIWKLLMIIHDRREFAKFEKEKMN#AKWDTQENPIYKSPINNFKNPNYGRKAGL 801 ||||||||||||||||||||||||||||||#||||||||||||||||||||||||||||| Query 743 IGLALLLIWKLLMIIHDRREFAKFEKEKMN#AKWDTQENPIYKSPINNFKNPNYGRKAGL 801 |
| N/A | Query 743 IGLALLLIWKLLMIIHDRREFAKFEKEKMN#AKWDTQENPIYKSPINNFKNPNYGRKAGL 801 ||||||||||||||||||||||||||||||#||||||||||||||||||||||||||||| Query 743 IGLALLLIWKLLMIIHDRREFAKFEKEKMN#AKWDTQENPIYKSPINNFKNPNYGRKAGL 801 |
| Mus musculus | Query 743 IGLALLLIWKLLMIIHDRREFAKFEKEKMN#AKWDTQENPIYKSPINNFKNPNYGRKAGL 801 ||||||||||||||||||||||||||||||#||||||||||||||||||||||||||||| Query 743 IGLALLLIWKLLMIIHDRREFAKFEKEKMN#AKWDTQENPIYKSPINNFKNPNYGRKAGL 801 |
| Gallus gallus | Query 744 GLALLLIWKLLMIIHDRREFAKFEKEKMN#AKWDTQENPIYKSPINNFKNPNYGRKAGL 801 |||||||||||||||||||||||||||||#||||||||||||||||||||||||||||| Query 744 GLALLLIWKLLMIIHDRREFAKFEKEKMN#AKWDTQENPIYKSPINNFKNPNYGRKAGL 801 |
| Ictalurus punctatus | Query 743 IGLALLLIWKLLMIIHDRREFAKFEKEKMN#AKWDTQENPIYKSPINNFKNPNYGRK 798 |||||| |||||||||||||||||||||||#|||| ||||||| + || | | Sbjct 752 IGLALLFIWKLLMIIHDRREFAKFEKEKMN#AKWDAGENPIYKSAVTTVVNPKYEGK 807 |
| Anopheles gambiae | Query 744 GLALLLIWKLLMIIHDRREFAKFEKEKMN#AKWDTQENPIYKSPINNFKNPNYGRK 798 |+|+||+||+| ||||||||+||||+| #||||| |||||| |||| | | Sbjct 783 GMAVLLLWKVLTSIHDRREFARFEKERMM#AKWDTGENPIYKQATTTFKNPTYAGK 837 |
| Anopheles gambiae str. PEST | Query 744 GLALLLIWKLLMIIHDRREFAKFEKEKMN#AKWDTQENPIYKSPINNFKNPNYGRK 798 |+|+||+||+| ||||||||+||||+| #||||| |||||| |||| | | Sbjct 783 GMAVLLLWKVLTSIHDRREFARFEKERMM#AKWDTGENPIYKQATTTFKNPTYAGK 837 |
| Pseudoplusia includens | Query 743 IGLALLLIWKLLMIIHDRREFAKFEKEKMN#AKWDTQENPIYKSPINNFKNPNYGRK 798 +|||||++||+ |||||||+||||+| #||||| |||||| + |||| | | Sbjct 782 VGLALLMLWKMATTSHDRREFARFEKERMM#AKWDTGENPIYKQATSTFKNPTYAGK 837 |
| Oryctolagus cuniculus | Query 743 IGLALLLIWKLLMIIHDRREFAKFEKEKMN#AKWDTQENPIYKSPINNFKNPNY 795 || |||||||||+ ||||+||||||+|+ #||||| ||+|| + | | | Sbjct 733 IGFALLLIWKLLITIHDRKEFAKFEEERAR#AKWDTANNPLYKEATSTFTNITY 785 |
| Rattus norvegicus | Query 743 IGLALLLIWKLLMIIHDRREFAKFEKEKMN#AKWDTQENPIYKSPINNFKNPNY 795 |||| |||||||+ ||||+||||||+|+ #||||| ||+|| + | | | Sbjct 732 IGLATLLIWKLLITIHDRKEFAKFEEERAR#AKWDTANNPLYKEATSTFTNITY 784 |
| synthetic construct | Query 743 IGLALLLIWKLLMIIHDRREFAKFEKEKMN#AKWDTQENPIYKSPINNFKNPNY 795 |||| |||||||+ ||||+||||||+|+ #||||| ||+|| + | | | Sbjct 733 IGLATLLIWKLLITIHDRKEFAKFEEERAR#AKWDTANNPLYKEATSTFTNITY 785 |
| Xenopus laevis | Query 743 IGLALLLIWKLLMIIHDRREFAKFEKEKMN#AKWDTQENPIYKSPINNFKNPNY 795 ||| |||||||+ |||||||||||+|+ #||||| ||+|| + | | | Sbjct 731 IGLVALLIWKLLITIHDRREFAKFEEERAK#AKWDTAHNPLYKGATSTFTNITY 783 |
| Sus scrofa | Query 743 IGLALLLIWKLLMIIHDRREFAKFEKEKMN#AKWDTQENPIYKSPINNFKNPNY 795 |||| |||||||+ ||||+||||||+|+ #||||| ||+|| + | | | Sbjct 729 IGLATLLIWKLLITIHDRKEFAKFEEERAR#AKWDTANNPLYKEATSTFTNITY 781 |
| Canis lupus familiaris | Query 743 IGLALLLIWKLLMIIHDRREFAKFEKEKMN#AKWDTQENPIYKSPINNFKNPNY 795 |||| |||||||+ ||||+||||||+|+ #||||| ||+|| + | | | Sbjct 729 IGLATLLIWKLLITIHDRKEFAKFEEERAR#AKWDTANNPLYKEATSTFTNITY 781 |
| Mus musculus | Query 743 IGLALLLIWKLLMIIHDRREFAKFEKEKMN#AKWDTQENPIYKSPINNFKNPNY 795 |||| |||||||+ ||||+||||||+|+ #||||| ||+|| + | | | Sbjct 732 IGLATLLIWKLLITIHDRKEFAKFEEERAR#AKWDTANNPLYKEATSTFTNITY 784 |
| rats, Peptide Partial, 723 aa | Query 743 IGLALLLIWKLLMIIHDRREFAKFEKEKMN#AKWDTQENPIYKSPINNFKNPNY 795 |||| |||||||+ ||||+||||||+|+ #||||| ||+|| + | | | Sbjct 668 IGLATLLIWKLLITIHDRKEFAKFEEERAR#AKWDTANNPLYKEATSTFTNITY 720 |
| Crassostrea gigas | Query 744 GLALLLIWKLLMIIHDRREFAKFEKEKMN#AKWDTQENPIYKSPINNFKNPNY 795 |+ ||||||| | |+|| |+|||| ||#|+||| |||||| + | || | Sbjct 746 GIILLLIWKLFTTISDKRELARFEKEAMN#ARWDTGENPIYKQATSTFVNPTY 797 |
| Sus scrofa | Query 743 IGLALLLIWKLLMIIHDRREFAKFEKEKMN#AKWDTQENPIYKSPINNFKNPNY 795 || | |||||||+ ||||+||||||+|+ #||||| ||+|| + | | | Sbjct 729 IGFATLLIWKLLITIHDRKEFAKFEEERAR#AKWDTANNPLYKEATSTFTNITY 781 |
| Tetraodon nigroviridis | Query 743 IGLALLLIWKLLMIIHDRREFAKFEKEKMN#AKWDTQENPIYKSPINNFKNPNY 795 +||| |||||||+ |||||||||||+|+ #||||| ||+|| + | | + Sbjct 748 LGLAALLIWKLLITIHDRREFAKFEEERAR#AKWDTGHNPLYKGATSTFTNITF 800 |
| mice, Peptide Partial, 680 aa | Query 743 IGLALLLIWKLLMIIHDRREFAKFEKEKMN#AKWDTQENPIYKSPINNFKN 792 |||| |||||||+ ||||+||||||+|+ #||||| ||+|| + | | Sbjct 629 IGLATLLIWKLLITIHDRKEFAKFEEERAR#AKWDTANNPLYKEATSTFTN 678 |
| Homo sapiens | Query 755 MIIHDRREFAKFEKEKMN#AKWDTQENPIYKSPINNFKNPNYGRK 798 ||||||||||||||||||#||||| ||||||| + || | | Sbjct 755 MIIHDRREFAKFEKEKMN#AKWDTGENPIYKSAVTTVVNPKYEGK 798 |
| Danio rerio | Query 755 MIIHDRREFAKFEKEKMN#AKWDTQENPIYKSPINNFKNPNYGRK 798 ||||||||||||||||||#||||| ||||||| + || | | Sbjct 755 MIIHDRREFAKFEKEKMN#AKWDTGENPIYKSAVTTVINPKYEGK 798 |
| Pongo pygmaeus | Query 755 MIIHDRREFAKFEKEKMN#AKWDTQENPIYKSPINNFKNPNYGRK 798 ||||||||||||||||||#||||| ||||||| + || | | Sbjct 755 MIIHDRREFAKFEKEKMN#AKWDTGENPIYKSAVTTVVNPKYEGK 798 |
| Felis catus | Query 755 MIIHDRREFAKFEKEKMN#AKWDTQENPIYKSPINNFKNPNYGRK 798 ||||| ||||||||||||#||||| ||||||| + || | | Sbjct 755 MIIHDTREFAKFEKEKMN#AKWDTGENPIYKSAVTTVVNPKYEGK 798 |
| Drosophila melanogaster | Query 757 IHDRREFAKFEKEKMN#AKWDTQENPIYKSPINNFKNPNYGRK 798 ||||||||+||||+||#||||| |||||| + |||| | | Sbjct 805 IHDRREFARFEKERMN#AKWDTGENPIYKQATSTFKNPMYAGK 846 |
| Oryctolagus cuniculus | Query 743 IGLALLLIWKLLMIIHDRREFAKFEKEKMN#AKWDTQENPIYKSPINNFKNPNY 795 ||||||||||||||||||||||||||||||#||||| ||||||| + || | Sbjct 8 IGLALLLIWKLLMIIHDRREFAKFEKEKMN#AKWDTGENPIYKSAVTTVVNPKY 60 |
| Biomphalaria glabrata | Query 757 IHDRREFAKFEKEKMN#AKWDTQENPIYKSPINNFKNPNYGRK 798 ||| |||||||||+ |#||||| |||||| + |||| || + Sbjct 747 IHDTREFAKFEKERQN#AKWDTGENPIYKQATSTFKNPTYGSQ 788 |
| Ovis aries | Query 743 IGLALLLIWKLLMIIHDRREFAKFEKEKMN#AKWDTQENPIYKSPINNFKNPNYGRK 798 ||+||| |||||+ |||+| |||| |+ #||| | ||+|+ + ||| | | Sbjct 721 IGVALLCIWKLLVSFHDRKEVAKFEAERSK#AKWQTGTNPLYRGSTSTFKNVTYKHK 776 |
| Gallus gallus | Query 751 WKLLMIIHDRREFAKFEKEKMN#AKWDTQENPIYKSPINNFKNPNY 795 ||||+ ||||||||+||+|| #||||| ||+|| + | | | Sbjct 733 WKLLITIHDRREFARFEEEKAR#AKWDTGNNPLYKEATSTFTNITY 777 |
| Xenopus laevis | Query 743 IGLALLLIWKLLMIIHDRREFAKFEKEKMN#AKWDTQENPIYKSPINNFKNPNY 795 |||| |+|||++ | || |+ +||||+||#+||+ ||+| + +|||+ Sbjct 717 IGLACLIIWKIITEIKDRNEYQRFEKERMN#SKWNPGHNPLYHNATTTVQNPNF 769 |
| Podocoryne carnea | Query 757 IHDRREFAKFEKEKMN#AKWDTQENPIYKSPINNFKNPNYGRK 798 | ||||||||||++ |# |||+ |||||| + |+|| || | Sbjct 740 IQDRREFAKFEKDRQN#PKWDSGENPIYKKATSTFQNPMYGNK 781 |
| Caenorhabditis briggsae | Query 758 HDRREFAKFEKEKMN#AKWDTQENPIYKSPINNFKNPNYGRKA 799 ||| |+||| |++ #||||| |||||| |||| | || Sbjct 768 HDRAEYAKFNNERLM#AKWDTNENPIYKQATTTFKNPVYAGKA 809 |
| Strongylocentrotus purpuratus | Query 751 WKLLMIIHDRREFAKFEKEKMN#AKWDTQENPIYKSPINNFKNPNYGR 797 |+| + |+||+|++| + #|+|| +|||||| |||| ||+ Sbjct 756 WRLYTYVQDKREYAQWENDCKK#AQWDQSDNPIYKSSTTTFKNPTYGK 802 |
| Caenorhabditis elegans | Query 758 HDRREFAKFEKEKMN#AKWDTQENPIYKSPINNFKNPNYGRKA 799 ||| |+| | |++ #||||| |||||| |||| | || Sbjct 767 HDRSEYATFNNERLM#AKWDTNENPIYKQATTTFKNPVYAGKA 808 |
| Halocynthia roretzi | Query 743 IGLALLLIWKLLMIIHDRREFAKFEKEKMN#AKWDTQENPIYKSPINNFKNPNYG 796 ||+ |+|||++ + |+||+ ||+||+ # || |||+| + + | || +| Sbjct 779 IGIIALIIWKVIQTLRDKREYEKFKKEEDL#RKWTKGENPVYVNASSKFDNPMFG 832 |
| Pan troglodytes | Query 743 IGLALLLIWKLLMIIHDRREFAKFEKEKMN#AKWDTQENPIYKSPINNFKNPNY 795 +|| |+| ++| + |+||||+++||||+ # | ||+||| | || + Sbjct 738 VGLGLVLAYRLSVEIYDRREYSRFEKEQQQ#LNWKQDSNPLYKSAITTTINPRF 790 |
| Lytechinus variegatus | Query 750 IWKLLMIIHDRREFAKFEKEKMN#AKWDTQENPIYKSPINNFKNPNYGRK 798 ||+|| ||||||| ||||+ |#| |+ |||||| + |||| | | Sbjct 758 IWRLLTYIHDRREFQNFEKERAN#ATWEGGENPIYKPSTSVFKNPTYNIK 806 |
| Mus sp. | Query 743 IGLALLLIWKLLMIIHDRREFAKFEKEKMN#AKWDTQENPIYKSPINNFKNPNY 795 +|| |+| ++| + |+||||+ +||||+ # | ||+||| | || + Sbjct 737 VGLGLVLAYRLSVEIYDRREYRRFEKEQQQ#LNWKQDNNPLYKSAITTTVNPRF 789 |
| Homo sapiens | Query 743 IGLALLLIWKLLMIIHDRREFAKFEKEKMN#AKWDTQENPIYKSPINNFKNPNYG 796 ||+ ||+||| |+ + | ||+ +|||||+ #++|+ +||++|| || + Sbjct 715 IGILLLVIWKALIHLSDLREYRRFEKEKLK#SQWN-NDNPLFKSATTTVMNPKFA 767 |
| Schistosoma japonicum | Query 751 WKLLMIIHDRREFAKFEKEKMN#AKWDTQENPIYKSPINNFKNPNY 795 +||++ | |||| | |+++ |# +|+ ||||++|| | || + Sbjct 239 YKLVITIDDRRELANFKQQGEN#MRWEMAENPIFESPTTNVLNPTF 283 |
| Anopheles gambiae str. PEST | Query 744 GLALLLIWKLLMIIHDRREFAKFEKEKMN#AKWDTQENPIYKSPINNFKNP 793 || +| ++ ++ ||+ |||||||+ # |+|+|||||+||| | Sbjct 746 GLLMLFCYRCKIMYDDRKMFAKFEKEREQ#ETKYQMESPLYKSPISNFKVP 795 |
| Acropora millepora | Query 752 KLLMIIHDRREFAKFEKEKMN#AKWDTQENPIYKSPINNFKNPNY--GRK 798 | | + || |+ |||+|+|+#+|| ++||+|++ |+|| | ||+ Sbjct 744 KGLFTMVDRIEYQKFERERMH#SKWTREKNPLYQAAKTTFENPTYAGGRQ 792 |
| Macaca mulatta | Query 743 IGLALLLIWKLLMIIHDRREFAKFEKEKMN#AKW 775 ||||||||||||||||||||||||||||||#||| Query 743 IGLALLLIWKLLMIIHDRREFAKFEKEKMN#AKW 775 |
| Pacifastacus leniusculus | Query 758 HDRREFAKFEKEKMN#AKWDTQENPIYKSPINNFKNPNYGR 797 |||||+||||||+ # |||+||| |+|| + + Sbjct 760 HDRREYAKFEKERKL#PSGKRAENPLYKSAKTTFQNPAFAQ 799 |
| Ovis canadensis | Query 750 IWKLLMIIHDRREFAKFEKEKMN#AKWDTQENPIYKSPINNFKNPNYG 796 ||| | + | ||+ +|||||+ #++|+ +||++|| || + Sbjct 723 IWKALTHLSDLREYHRFEKEKLK#SQWN-NDNPLFKSATTTVMNPKFA 768 |
| Sigmodon hispidus | Query 750 IWKLLMIIHDRREFAKFEKEKMN#AKWDTQENPIYKSPINNFKNPNYG 796 ||| | + | ||+ |||||+ #++|+ +||++|| || + Sbjct 722 IWKALTHLTDLREYRHFEKEKLK#SQWN-NDNPLFKSATTTVMNPKFA 767 |
[Site 4] KEKMNAKWDT777-QENPIYKSPI
Thr777  Gln
Gln
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Lys768 | Glu769 | Lys770 | Met771 | Asn772 | Ala773 | Lys774 | Trp775 | Asp776 | Thr777 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Gln778 | Glu779 | Asn780 | Pro781 | Ile782 | Tyr783 | Lys784 | Ser785 | Pro786 | Ile787 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| LLIWKLLMIIHDRREFAKFEKEKMNAKWDTQENPIYKSPINNFKNPNYGRKAGL |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | Canis familiaris | 124.00 | 10 | PREDICTED: similar to integrin beta 1 isoform 1D |
| 2 | Bos taurus | 124.00 | 7 | ITGB1 protein |
| 3 | N/A | 114.00 | 27 | S66396 integrin beta-1 chain isoform D - human (f |
| 4 | Mus musculus | 114.00 | 12 | beta 1 integrin |
| 5 | Gallus gallus | 113.00 | 3 | integrin beta1D |
| 6 | Ictalurus punctatus | 89.40 | 1 | beta-1 integrin |
| 7 | Anopheles gambiae | 83.60 | 2 | integrin beta subunit |
| 8 | Anopheles gambiae str. PEST | 83.60 | 1 | integrin beta subunit (AGAP000815-PA) |
| 9 | Pseudoplusia includens | 79.30 | 1 | integrin beta 1 |
| 10 | Homo sapiens | 78.20 | 11 | integrin beta 1 isoform 1A precursor |
| 11 | Danio rerio | 78.20 | 5 | integrin, beta 1a |
| 12 | Tetraodon nigroviridis | 78.20 | 5 | unnamed protein product |
| 13 | Sus scrofa | 78.20 | 3 | integrin beta-1 subunit |
| 14 | Xenopus laevis | 78.20 | 2 | integrin beta-1 subunit |
| 15 | Pongo pygmaeus | 78.20 | 1 | ITB1_PONPY Integrin beta-1 precursor (Fibronectin |
| 16 | Felis catus | 75.50 | 1 | integrin beta 1 |
| 17 | Crassostrea gigas | 75.50 | 1 | integrin beta cgh |
| 18 | Drosophila melanogaster | 75.50 | 1 | myospheroid CG1560-PA |
| 19 | Oryctolagus cuniculus | 74.30 | 1 | integrin beta 1 |
| 20 | Rattus norvegicus | 73.60 | 4 | integrin beta 3 |
| 21 | synthetic construct | 73.60 | 3 | AF239959_1 hybrid integrin beta 3 subunit precurs |
| 22 | Canis lupus familiaris | 73.60 | 1 | integrin beta chain, beta 3 |
| 23 | rats, Peptide Partial, 723 aa | 73.60 | 1 | beta 3 integrin, GPIIIA |
| 24 | Oryctolagus cuniculus | 73.60 | 1 | glycoprotein IIIa |
| 25 | Mus musculus | 73.60 | 1 | integrin beta 3 precursor |
| 26 | Biomphalaria glabrata | 73.60 | 1 | beta integrin subunit |
| 27 | Sus scrofa | 73.60 | 1 | integrin beta chain, beta 3 |
| 28 | mice, Peptide Partial, 680 aa | 72.00 | 1 | beta 3 integrin, GPIIIA |
| 29 | Gallus gallus | 69.70 | 2 | integrin, beta 3 precursor |
| 30 | Podocoryne carnea | 67.80 | 1 | AF308652_1 integrin beta chain |
| 31 | Ovis aries | 64.30 | 2 | ITB6_SHEEP Integrin beta-6 precursor gi |
| 32 | Caenorhabditis briggsae | 63.50 | 1 | Hypothetical protein CBG03601 |
| 33 | Strongylocentrotus purpuratus | 62.00 | 3 | integrin beta-C subunit |
| 34 | Lytechinus variegatus | 62.00 | 1 | beta-C integrin subunit |
| 35 | Xenopus laevis | 61.60 | 1 | hypothetical protein LOC379708 |
| 36 | Caenorhabditis elegans | 61.60 | 1 | Paralysed Arrest at Two-fold family member (pat-3 |
| 37 | Halocynthia roretzi | 55.10 | 2 | integrin beta Hr2 precursor |
| 38 | Schistosoma japonicum | 54.30 | 1 | SJCHGC06221 protein |
| 39 | Pan troglodytes | 51.60 | 2 | PREDICTED: integrin, beta 7 isoform 5 |
| 40 | Macaca mulatta | 51.60 | 1 | integrin beta 1 |
| 41 | Homo sapiens | 51.60 | 1 | integrin, beta 2 precursor |
| 42 | Acropora millepora | 51.20 | 1 | integrin subunit betaCn1 |
| 43 | Mus sp. | 50.40 | 1 | beta 7 integrin |
| 44 | Pacifastacus leniusculus | 48.90 | 1 | integrin |
| 45 | Ovis canadensis | 48.50 | 1 | ITB2_OVICA Integrin beta-2 precursor (Cell surfac |
| 46 | Anopheles gambiae str. PEST | 47.80 | 1 | AGAP010233-PA |
| 47 | Sigmodon hispidus | 47.40 | 1 | AF445415_1 integrin beta-2 precursor |
Top-ranked sequences
| organism | matching |
|---|---|
| Canis familiaris | Query 748 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#QENPIYKSPINNFKNPNYGRKAGL 801 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||| Query 748 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#QENPIYKSPINNFKNPNYGRKAGL 801 |
| Bos taurus | Query 748 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#QENPIYKSPINNFKNPNYGRKAGL 801 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||| Query 748 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#QENPIYKSPINNFKNPNYGRKAGL 801 |
| N/A | Query 748 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#QENPIYKSPINNFKNPNYGRKAGL 801 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||| Query 748 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#QENPIYKSPINNFKNPNYGRKAGL 801 |
| Mus musculus | Query 748 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#QENPIYKSPINNFKNPNYGRKAGL 801 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||| Query 748 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#QENPIYKSPINNFKNPNYGRKAGL 801 |
| Gallus gallus | Query 748 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#QENPIYKSPINNFKNPNYGRKAGL 801 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||| Query 748 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#QENPIYKSPINNFKNPNYGRKAGL 801 |
| Ictalurus punctatus | Query 748 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#QENPIYKSPINNFKNPNYGRK 798 | ||||||||||||||||||||||||||| # ||||||| + || | | Sbjct 757 LFIWKLLMIIHDRREFAKFEKEKMNAKWDA#GENPIYKSAVTTVVNPKYEGK 807 |
| Anopheles gambiae | Query 748 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#QENPIYKSPINNFKNPNYGRK 798 ||+||+| ||||||||+||||+| |||||# |||||| |||| | | Sbjct 787 LLLWKVLTSIHDRREFARFEKERMMAKWDT#GENPIYKQATTTFKNPTYAGK 837 |
| Anopheles gambiae str. PEST | Query 748 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#QENPIYKSPINNFKNPNYGRK 798 ||+||+| ||||||||+||||+| |||||# |||||| |||| | | Sbjct 787 LLLWKVLTSIHDRREFARFEKERMMAKWDT#GENPIYKQATTTFKNPTYAGK 837 |
| Pseudoplusia includens | Query 748 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#QENPIYKSPINNFKNPNYGRK 798 |++||+ |||||||+||||+| |||||# |||||| + |||| | | Sbjct 787 LMLWKMATTSHDRREFARFEKERMMAKWDT#GENPIYKQATSTFKNPTYAGK 837 |
| Homo sapiens | Query 755 MIIHDRREFAKFEKEKMNAKWDT#QENPIYKSPINNFKNPNYGRK 798 |||||||||||||||||||||||# ||||||| + || | | Sbjct 755 MIIHDRREFAKFEKEKMNAKWDT#GENPIYKSAVTTVVNPKYEGK 798 |
| Danio rerio | Query 755 MIIHDRREFAKFEKEKMNAKWDT#QENPIYKSPINNFKNPNYGRK 798 |||||||||||||||||||||||# ||||||| + || | | Sbjct 755 MIIHDRREFAKFEKEKMNAKWDT#GENPIYKSAVTTVINPKYEGK 798 |
| Tetraodon nigroviridis | Query 755 MIIHDRREFAKFEKEKMNAKWDT#QENPIYKSPINNFKNPNYGRK 798 |||||||||||||||||||||||# ||||||| + || | | Sbjct 748 MIIHDRREFAKFEKEKMNAKWDT#GENPIYKSAVTTVVNPKYEGK 791 |
| Sus scrofa | Query 755 MIIHDRREFAKFEKEKMNAKWDT#QENPIYKSPINNFKNPNYGRK 798 |||||||||||||||||||||||# ||||||| + || | | Sbjct 755 MIIHDRREFAKFEKEKMNAKWDT#GENPIYKSAVTTVVNPKYEGK 798 |
| Xenopus laevis | Query 755 MIIHDRREFAKFEKEKMNAKWDT#QENPIYKSPINNFKNPNYGRK 798 |||||||||||||||||||||||# ||||||| + || | | Sbjct 755 MIIHDRREFAKFEKEKMNAKWDT#GENPIYKSAVTTVVNPKYEGK 798 |
| Pongo pygmaeus | Query 755 MIIHDRREFAKFEKEKMNAKWDT#QENPIYKSPINNFKNPNYGRK 798 |||||||||||||||||||||||# ||||||| + || | | Sbjct 755 MIIHDRREFAKFEKEKMNAKWDT#GENPIYKSAVTTVVNPKYEGK 798 |
| Felis catus | Query 755 MIIHDRREFAKFEKEKMNAKWDT#QENPIYKSPINNFKNPNYGRK 798 ||||| |||||||||||||||||# ||||||| + || | | Sbjct 755 MIIHDTREFAKFEKEKMNAKWDT#GENPIYKSAVTTVVNPKYEGK 798 |
| Crassostrea gigas | Query 748 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#QENPIYKSPINNFKNPNY 795 |||||| | |+|| |+|||| |||+|||# |||||| + | || | Sbjct 750 LLIWKLFTTISDKRELARFEKEAMNARWDT#GENPIYKQATSTFVNPTY 797 |
| Drosophila melanogaster | Query 757 IHDRREFAKFEKEKMNAKWDT#QENPIYKSPINNFKNPNYGRK 798 ||||||||+||||+|||||||# |||||| + |||| | | Sbjct 805 IHDRREFARFEKERMNAKWDT#GENPIYKQATSTFKNPMYAGK 846 |
| Oryctolagus cuniculus | Query 755 MIIHDRREFAKFEKEKMNAKWDT#QENPIYKSPINNFKNPNY 795 |||||||||||||||||||||||# ||||||| + || | Sbjct 20 MIIHDRREFAKFEKEKMNAKWDT#GENPIYKSAVTTVVNPKY 60 |
| Rattus norvegicus | Query 748 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#QENPIYKSPINNFKNPNY 795 |||||||+ ||||+||||||+|+ |||||# ||+|| + | | | Sbjct 737 LLIWKLLITIHDRKEFAKFEEERARAKWDT#ANNPLYKEATSTFTNITY 784 |
| synthetic construct | Query 748 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#QENPIYKSPINNFKNPNY 795 |||||||+ ||||+||||||+|+ |||||# ||+|| + | | | Sbjct 738 LLIWKLLITIHDRKEFAKFEEERARAKWDT#ANNPLYKEATSTFTNITY 785 |
| Canis lupus familiaris | Query 748 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#QENPIYKSPINNFKNPNY 795 |||||||+ ||||+||||||+|+ |||||# ||+|| + | | | Sbjct 734 LLIWKLLITIHDRKEFAKFEEERARAKWDT#ANNPLYKEATSTFTNITY 781 |
| rats, Peptide Partial, 723 aa | Query 748 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#QENPIYKSPINNFKNPNY 795 |||||||+ ||||+||||||+|+ |||||# ||+|| + | | | Sbjct 673 LLIWKLLITIHDRKEFAKFEEERARAKWDT#ANNPLYKEATSTFTNITY 720 |
| Oryctolagus cuniculus | Query 748 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#QENPIYKSPINNFKNPNY 795 |||||||+ ||||+||||||+|+ |||||# ||+|| + | | | Sbjct 738 LLIWKLLITIHDRKEFAKFEEERARAKWDT#ANNPLYKEATSTFTNITY 785 |
| Mus musculus | Query 748 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#QENPIYKSPINNFKNPNY 795 |||||||+ ||||+||||||+|+ |||||# ||+|| + | | | Sbjct 737 LLIWKLLITIHDRKEFAKFEEERARAKWDT#ANNPLYKEATSTFTNITY 784 |
| Biomphalaria glabrata | Query 757 IHDRREFAKFEKEKMNAKWDT#QENPIYKSPINNFKNPNYGRK 798 ||| |||||||||+ ||||||# |||||| + |||| || + Sbjct 747 IHDTREFAKFEKERQNAKWDT#GENPIYKQATSTFKNPTYGSQ 788 |
| Sus scrofa | Query 748 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#QENPIYKSPINNFKNPNY 795 |||||||+ ||||+||||||+|+ |||||# ||+|| + | | | Sbjct 734 LLIWKLLITIHDRKEFAKFEEERARAKWDT#ANNPLYKEATSTFTNITY 781 |
| mice, Peptide Partial, 680 aa | Query 748 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#QENPIYKSPINNFKN 792 |||||||+ ||||+||||||+|+ |||||# ||+|| + | | Sbjct 634 LLIWKLLITIHDRKEFAKFEEERARAKWDT#ANNPLYKEATSTFTN 678 |
| Gallus gallus | Query 751 WKLLMIIHDRREFAKFEKEKMNAKWDT#QENPIYKSPINNFKNPNY 795 ||||+ ||||||||+||+|| |||||# ||+|| + | | | Sbjct 733 WKLLITIHDRREFARFEEEKARAKWDT#GNNPLYKEATSTFTNITY 777 |
| Podocoryne carnea | Query 757 IHDRREFAKFEKEKMNAKWDT#QENPIYKSPINNFKNPNYGRK 798 | ||||||||||++ | |||+# |||||| + |+|| || | Sbjct 740 IQDRREFAKFEKDRQNPKWDS#GENPIYKKATSTFQNPMYGNK 781 |
| Ovis aries | Query 748 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#QENPIYKSPINNFKNPNYGRK 798 | |||||+ |||+| |||| |+ ||| |# ||+|+ + ||| | | Sbjct 726 LCIWKLLVSFHDRKEVAKFEAERSKAKWQT#GTNPLYRGSTSTFKNVTYKHK 776 |
| Caenorhabditis briggsae | Query 758 HDRREFAKFEKEKMNAKWDT#QENPIYKSPINNFKNPNYGRKA 799 ||| |+||| |++ |||||# |||||| |||| | || Sbjct 768 HDRAEYAKFNNERLMAKWDT#NENPIYKQATTTFKNPVYAGKA 809 |
| Strongylocentrotus purpuratus | Query 757 IHDRREFAKFEKEKMNAKWDT#QENPIYKSPINNFKNPNYGRK 798 ||||||| ||||+ || |+ # |||||| + |||| | | Sbjct 765 IHDRREFQNFEKERANATWEG#GENPIYKPSTSVFKNPTYNIK 806 |
| Lytechinus variegatus | Query 757 IHDRREFAKFEKEKMNAKWDT#QENPIYKSPINNFKNPNYGRK 798 ||||||| ||||+ || |+ # |||||| + |||| | | Sbjct 765 IHDRREFQNFEKERANATWEG#GENPIYKPSTSVFKNPTYNIK 806 |
| Xenopus laevis | Query 748 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#QENPIYKSPINNFKNPNY 795 |+|||++ | || |+ +||||+||+||+ # ||+| + +|||+ Sbjct 722 LIIWKIITEIKDRNEYQRFEKERMNSKWNP#GHNPLYHNATTTVQNPNF 769 |
| Caenorhabditis elegans | Query 758 HDRREFAKFEKEKMNAKWDT#QENPIYKSPINNFKNPNYGRKA 799 ||| |+| | |++ |||||# |||||| |||| | || Sbjct 767 HDRSEYATFNNERLMAKWDT#NENPIYKQATTTFKNPVYAGKA 808 |
| Halocynthia roretzi | Query 748 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#QENPIYKSPINNFKNPNYG 796 |+|||++ + |+||+ ||+||+ || # |||+| + + | || +| Sbjct 784 LIIWKVIQTLRDKREYEKFKKEEDLRKWTK#GENPVYVNASSKFDNPMFG 832 |
| Schistosoma japonicum | Query 751 WKLLMIIHDRREFAKFEKEKMNAKWDT#QENPIYKSPINNFKNPNY 795 +||++ | |||| | |+++ | +|+ # ||||++|| | || + Sbjct 239 YKLVITIDDRRELANFKQQGENMRWEM#AENPIFESPTTNVLNPTF 283 |
| Pan troglodytes | Query 748 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#QENPIYKSPINNFKNPNY 795 +| ++| + |+||||+++||||+ | # ||+||| | || + Sbjct 743 VLAYRLSVEIYDRREYSRFEKEQQQLNWKQ#DSNPLYKSAITTTINPRF 790 |
| Macaca mulatta | Query 755 MIIHDRREFAKFEKEKMNAKW 775 ||||||||||||||||||||| Query 755 MIIHDRREFAKFEKEKMNAKW 775 |
| Homo sapiens | Query 748 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#QENPIYKSPINNFKNPNYG 796 |+||| |+ + | ||+ +|||||+ ++|+ # +||++|| || + Sbjct 720 LVIWKALIHLSDLREYRRFEKEKLKSQWN-#NDNPLFKSATTTVMNPKFA 767 |
| Acropora millepora | Query 752 KLLMIIHDRREFAKFEKEKMNAKWDT#QENPIYKSPINNFKNPNY--GRK 798 | | + || |+ |||+|+|++|| #++||+|++ |+|| | ||+ Sbjct 744 KGLFTMVDRIEYQKFERERMHSKWTR#EKNPLYQAAKTTFENPTYAGGRQ 792 |
| Mus sp. | Query 748 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#QENPIYKSPINNFKNPNY 795 +| ++| + |+||||+ +||||+ | # ||+||| | || + Sbjct 742 VLAYRLSVEIYDRREYRRFEKEQQQLNWKQ#DNNPLYKSAITTTVNPRF 789 |
| Pacifastacus leniusculus | Query 758 HDRREFAKFEKEKMNAKWDT#QENPIYKSPINNFKNPNYGR 797 |||||+||||||+ # |||+||| |+|| + + Sbjct 760 HDRREYAKFEKERKLPSGKR#AENPLYKSAKTTFQNPAFAQ 799 |
| Ovis canadensis | Query 750 IWKLLMIIHDRREFAKFEKEKMNAKWDT#QENPIYKSPINNFKNPNYG 796 ||| | + | ||+ +|||||+ ++|+ # +||++|| || + Sbjct 723 IWKALTHLSDLREYHRFEKEKLKSQWN-#NDNPLFKSATTTVMNPKFA 768 |
| Anopheles gambiae str. PEST | Query 748 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#QENPIYKSPINNFKNP 793 | ++ ++ ||+ |||||||+ # |+|+|||||+||| | Sbjct 750 LFCYRCKIMYDDRKMFAKFEKEREQETKYQ#MESPLYKSPISNFKVP 795 |
| Sigmodon hispidus | Query 750 IWKLLMIIHDRREFAKFEKEKMNAKWDT#QENPIYKSPINNFKNPNYG 796 ||| | + | ||+ |||||+ ++|+ # +||++|| || + Sbjct 722 IWKALTHLTDLREYRHFEKEKLKSQWN-#NDNPLFKSATTTVMNPKFA 767 |
[Site 5] EKMNAKWDTQ778-ENPIYKSPIN
Gln778  Glu
Glu
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Glu769 | Lys770 | Met771 | Asn772 | Ala773 | Lys774 | Trp775 | Asp776 | Thr777 | Gln778 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Glu779 | Asn780 | Pro781 | Ile782 | Tyr783 | Lys784 | Ser785 | Pro786 | Ile787 | Asn788 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| LIWKLLMIIHDRREFAKFEKEKMNAKWDTQENPIYKSPINNFKNPNYGRKAGL |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | Canis familiaris | 123.00 | 10 | PREDICTED: similar to integrin beta 1 isoform 1D |
| 2 | Bos taurus | 123.00 | 7 | ITGB1 protein |
| 3 | N/A | 113.00 | 28 | S66396 integrin beta-1 chain isoform D - human (f |
| 4 | Mus musculus | 112.00 | 12 | beta 1 integrin |
| 5 | Gallus gallus | 111.00 | 3 | integrin beta1D |
| 6 | Ictalurus punctatus | 87.80 | 1 | beta-1 integrin |
| 7 | Anopheles gambiae | 82.00 | 2 | integrin beta subunit |
| 8 | Anopheles gambiae str. PEST | 82.00 | 1 | integrin beta subunit (AGAP000815-PA) |
| 9 | Homo sapiens | 78.20 | 11 | ITB1_HUMAN Integrin beta-1 precursor (Fibronectin |
| 10 | Danio rerio | 78.20 | 5 | integrin, beta 1a |
| 11 | Tetraodon nigroviridis | 78.20 | 5 | unnamed protein product |
| 12 | Sus scrofa | 78.20 | 3 | integrin beta-1 subunit |
| 13 | Xenopus laevis | 78.20 | 2 | integrin beta-1 subunit |
| 14 | Pongo pygmaeus | 78.20 | 1 | ITB1_PONPY Integrin beta-1 precursor (Fibronectin |
| 15 | Pseudoplusia includens | 77.80 | 1 | integrin beta 1 |
| 16 | Felis catus | 75.50 | 1 | integrin beta 1 |
| 17 | Drosophila melanogaster | 75.50 | 1 | myospheroid CG1560-PA |
| 18 | Oryctolagus cuniculus | 73.90 | 1 | integrin beta 1 |
| 19 | Crassostrea gigas | 73.90 | 1 | integrin beta cgh |
| 20 | Biomphalaria glabrata | 73.60 | 1 | beta integrin subunit |
| 21 | Rattus norvegicus | 72.00 | 4 | integrin beta 3 |
| 22 | synthetic construct | 72.00 | 3 | AF239959_1 hybrid integrin beta 3 subunit precurs |
| 23 | Sus scrofa | 72.00 | 1 | integrin beta chain, beta 3 |
| 24 | Oryctolagus cuniculus | 72.00 | 1 | glycoprotein IIIa |
| 25 | Mus musculus | 72.00 | 1 | integrin beta 3 precursor |
| 26 | Canis lupus familiaris | 72.00 | 1 | integrin beta chain, beta 3 |
| 27 | rats, Peptide Partial, 723 aa | 72.00 | 1 | beta 3 integrin, GPIIIA |
| 28 | mice, Peptide Partial, 680 aa | 70.50 | 1 | beta 3 integrin, GPIIIA |
| 29 | Gallus gallus | 69.70 | 2 | integrin, beta 3 precursor |
| 30 | Podocoryne carnea | 67.80 | 1 | AF308652_1 integrin beta chain |
| 31 | Caenorhabditis briggsae | 63.50 | 1 | Hypothetical protein CBG03601 |
| 32 | Ovis aries | 63.20 | 2 | ITB6_SHEEP Integrin beta-6 precursor gi |
| 33 | Strongylocentrotus purpuratus | 62.00 | 3 | integrin beta-C subunit |
| 34 | Lytechinus variegatus | 62.00 | 1 | beta-C integrin subunit |
| 35 | Caenorhabditis elegans | 61.60 | 1 | Paralysed Arrest at Two-fold family member (pat-3 |
| 36 | Xenopus laevis | 60.10 | 1 | hypothetical protein LOC379708 |
| 37 | Schistosoma japonicum | 54.30 | 1 | SJCHGC06221 protein |
| 38 | Halocynthia roretzi | 53.10 | 2 | integrin beta Hr2 precursor |
| 39 | Macaca mulatta | 51.60 | 1 | integrin beta 1 |
| 40 | Pan troglodytes | 51.20 | 2 | PREDICTED: integrin, beta 7 isoform 5 |
| 41 | Acropora millepora | 51.20 | 1 | integrin subunit betaCn1 |
| 42 | Mus sp. | 50.10 | 1 | beta 7 integrin |
| 43 | Homo sapiens | 50.10 | 1 | integrin, beta 2 precursor |
| 44 | Pacifastacus leniusculus | 48.90 | 1 | integrin |
| 45 | Ovis canadensis | 48.50 | 1 | ITB2_OVICA Integrin beta-2 precursor (Cell surfac |
| 46 | Sigmodon hispidus | 47.40 | 1 | AF445415_1 integrin beta-2 precursor |
| 47 | Anopheles gambiae str. PEST | 46.60 | 1 | AGAP010233-PA |
Top-ranked sequences
| organism | matching |
|---|---|
| Canis familiaris | Query 749 LIWKLLMIIHDRREFAKFEKEKMNAKWDTQ#ENPIYKSPINNFKNPNYGRKAGL 801 ||||||||||||||||||||||||||||||#||||||||||||||||||||||| Query 749 LIWKLLMIIHDRREFAKFEKEKMNAKWDTQ#ENPIYKSPINNFKNPNYGRKAGL 801 |
| Bos taurus | Query 749 LIWKLLMIIHDRREFAKFEKEKMNAKWDTQ#ENPIYKSPINNFKNPNYGRKAGL 801 ||||||||||||||||||||||||||||||#||||||||||||||||||||||| Query 749 LIWKLLMIIHDRREFAKFEKEKMNAKWDTQ#ENPIYKSPINNFKNPNYGRKAGL 801 |
| N/A | Query 749 LIWKLLMIIHDRREFAKFEKEKMNAKWDTQ#ENPIYKSPINNFKNPNYGRKAGL 801 ||||||||||||||||||||||||||||||#||||||||||||||||||||||| Query 749 LIWKLLMIIHDRREFAKFEKEKMNAKWDTQ#ENPIYKSPINNFKNPNYGRKAGL 801 |
| Mus musculus | Query 749 LIWKLLMIIHDRREFAKFEKEKMNAKWDTQ#ENPIYKSPINNFKNPNYGRKAGL 801 ||||||||||||||||||||||||||||||#||||||||||||||||||||||| Query 749 LIWKLLMIIHDRREFAKFEKEKMNAKWDTQ#ENPIYKSPINNFKNPNYGRKAGL 801 |
| Gallus gallus | Query 749 LIWKLLMIIHDRREFAKFEKEKMNAKWDTQ#ENPIYKSPINNFKNPNYGRKAGL 801 ||||||||||||||||||||||||||||||#||||||||||||||||||||||| Query 749 LIWKLLMIIHDRREFAKFEKEKMNAKWDTQ#ENPIYKSPINNFKNPNYGRKAGL 801 |
| Ictalurus punctatus | Query 749 LIWKLLMIIHDRREFAKFEKEKMNAKWDTQ#ENPIYKSPINNFKNPNYGRK 798 ||||||||||||||||||||||||||| #||||||| + || | | Sbjct 758 FIWKLLMIIHDRREFAKFEKEKMNAKWDAG#ENPIYKSAVTTVVNPKYEGK 807 |
| Anopheles gambiae | Query 749 LIWKLLMIIHDRREFAKFEKEKMNAKWDTQ#ENPIYKSPINNFKNPNYGRK 798 |+||+| ||||||||+||||+| ||||| #|||||| |||| | | Sbjct 788 LLWKVLTSIHDRREFARFEKERMMAKWDTG#ENPIYKQATTTFKNPTYAGK 837 |
| Anopheles gambiae str. PEST | Query 749 LIWKLLMIIHDRREFAKFEKEKMNAKWDTQ#ENPIYKSPINNFKNPNYGRK 798 |+||+| ||||||||+||||+| ||||| #|||||| |||| | | Sbjct 788 LLWKVLTSIHDRREFARFEKERMMAKWDTG#ENPIYKQATTTFKNPTYAGK 837 |
| Homo sapiens | Query 755 MIIHDRREFAKFEKEKMNAKWDTQ#ENPIYKSPINNFKNPNYGRK 798 ||||||||||||||||||||||| #||||||| + || | | Sbjct 755 MIIHDRREFAKFEKEKMNAKWDTG#ENPIYKSAVTTVVNPKYEGK 798 |
| Danio rerio | Query 755 MIIHDRREFAKFEKEKMNAKWDTQ#ENPIYKSPINNFKNPNYGRK 798 ||||||||||||||||||||||| #||||||| + || | | Sbjct 755 MIIHDRREFAKFEKEKMNAKWDTG#ENPIYKSAVTTVINPKYEGK 798 |
| Tetraodon nigroviridis | Query 755 MIIHDRREFAKFEKEKMNAKWDTQ#ENPIYKSPINNFKNPNYGRK 798 ||||||||||||||||||||||| #||||||| + || | | Sbjct 748 MIIHDRREFAKFEKEKMNAKWDTG#ENPIYKSAVTTVVNPKYEGK 791 |
| Sus scrofa | Query 755 MIIHDRREFAKFEKEKMNAKWDTQ#ENPIYKSPINNFKNPNYGRK 798 ||||||||||||||||||||||| #||||||| + || | | Sbjct 755 MIIHDRREFAKFEKEKMNAKWDTG#ENPIYKSAVTTVVNPKYEGK 798 |
| Xenopus laevis | Query 755 MIIHDRREFAKFEKEKMNAKWDTQ#ENPIYKSPINNFKNPNYGRK 798 ||||||||||||||||||||||| #||||||| + || | | Sbjct 755 MIIHDRREFAKFEKEKMNAKWDTG#ENPIYKSAVTTVVNPKYEGK 798 |
| Pongo pygmaeus | Query 755 MIIHDRREFAKFEKEKMNAKWDTQ#ENPIYKSPINNFKNPNYGRK 798 ||||||||||||||||||||||| #||||||| + || | | Sbjct 755 MIIHDRREFAKFEKEKMNAKWDTG#ENPIYKSAVTTVVNPKYEGK 798 |
| Pseudoplusia includens | Query 749 LIWKLLMIIHDRREFAKFEKEKMNAKWDTQ#ENPIYKSPINNFKNPNYGRK 798 ++||+ |||||||+||||+| ||||| #|||||| + |||| | | Sbjct 788 MLWKMATTSHDRREFARFEKERMMAKWDTG#ENPIYKQATSTFKNPTYAGK 837 |
| Felis catus | Query 755 MIIHDRREFAKFEKEKMNAKWDTQ#ENPIYKSPINNFKNPNYGRK 798 ||||| ||||||||||||||||| #||||||| + || | | Sbjct 755 MIIHDTREFAKFEKEKMNAKWDTG#ENPIYKSAVTTVVNPKYEGK 798 |
| Drosophila melanogaster | Query 757 IHDRREFAKFEKEKMNAKWDTQ#ENPIYKSPINNFKNPNYGRK 798 ||||||||+||||+||||||| #|||||| + |||| | | Sbjct 805 IHDRREFARFEKERMNAKWDTG#ENPIYKQATSTFKNPMYAGK 846 |
| Oryctolagus cuniculus | Query 755 MIIHDRREFAKFEKEKMNAKWDTQ#ENPIYKSPINNFKNPNY 795 ||||||||||||||||||||||| #||||||| + || | Sbjct 20 MIIHDRREFAKFEKEKMNAKWDTG#ENPIYKSAVTTVVNPKY 60 |
| Crassostrea gigas | Query 749 LIWKLLMIIHDRREFAKFEKEKMNAKWDTQ#ENPIYKSPINNFKNPNY 795 ||||| | |+|| |+|||| |||+||| #|||||| + | || | Sbjct 751 LIWKLFTTISDKRELARFEKEAMNARWDTG#ENPIYKQATSTFVNPTY 797 |
| Biomphalaria glabrata | Query 757 IHDRREFAKFEKEKMNAKWDTQ#ENPIYKSPINNFKNPNYGRK 798 ||| |||||||||+ |||||| #|||||| + |||| || + Sbjct 747 IHDTREFAKFEKERQNAKWDTG#ENPIYKQATSTFKNPTYGSQ 788 |
| Rattus norvegicus | Query 749 LIWKLLMIIHDRREFAKFEKEKMNAKWDTQ#ENPIYKSPINNFKNPNY 795 ||||||+ ||||+||||||+|+ ||||| # ||+|| + | | | Sbjct 738 LIWKLLITIHDRKEFAKFEEERARAKWDTA#NNPLYKEATSTFTNITY 784 |
| synthetic construct | Query 749 LIWKLLMIIHDRREFAKFEKEKMNAKWDTQ#ENPIYKSPINNFKNPNY 795 ||||||+ ||||+||||||+|+ ||||| # ||+|| + | | | Sbjct 739 LIWKLLITIHDRKEFAKFEEERARAKWDTA#NNPLYKEATSTFTNITY 785 |
| Sus scrofa | Query 749 LIWKLLMIIHDRREFAKFEKEKMNAKWDTQ#ENPIYKSPINNFKNPNY 795 ||||||+ ||||+||||||+|+ ||||| # ||+|| + | | | Sbjct 735 LIWKLLITIHDRKEFAKFEEERARAKWDTA#NNPLYKEATSTFTNITY 781 |
| Oryctolagus cuniculus | Query 749 LIWKLLMIIHDRREFAKFEKEKMNAKWDTQ#ENPIYKSPINNFKNPNY 795 ||||||+ ||||+||||||+|+ ||||| # ||+|| + | | | Sbjct 739 LIWKLLITIHDRKEFAKFEEERARAKWDTA#NNPLYKEATSTFTNITY 785 |
| Mus musculus | Query 749 LIWKLLMIIHDRREFAKFEKEKMNAKWDTQ#ENPIYKSPINNFKNPNY 795 ||||||+ ||||+||||||+|+ ||||| # ||+|| + | | | Sbjct 738 LIWKLLITIHDRKEFAKFEEERARAKWDTA#NNPLYKEATSTFTNITY 784 |
| Canis lupus familiaris | Query 749 LIWKLLMIIHDRREFAKFEKEKMNAKWDTQ#ENPIYKSPINNFKNPNY 795 ||||||+ ||||+||||||+|+ ||||| # ||+|| + | | | Sbjct 735 LIWKLLITIHDRKEFAKFEEERARAKWDTA#NNPLYKEATSTFTNITY 781 |
| rats, Peptide Partial, 723 aa | Query 749 LIWKLLMIIHDRREFAKFEKEKMNAKWDTQ#ENPIYKSPINNFKNPNY 795 ||||||+ ||||+||||||+|+ ||||| # ||+|| + | | | Sbjct 674 LIWKLLITIHDRKEFAKFEEERARAKWDTA#NNPLYKEATSTFTNITY 720 |
| mice, Peptide Partial, 680 aa | Query 749 LIWKLLMIIHDRREFAKFEKEKMNAKWDTQ#ENPIYKSPINNFKN 792 ||||||+ ||||+||||||+|+ ||||| # ||+|| + | | Sbjct 635 LIWKLLITIHDRKEFAKFEEERARAKWDTA#NNPLYKEATSTFTN 678 |
| Gallus gallus | Query 751 WKLLMIIHDRREFAKFEKEKMNAKWDTQ#ENPIYKSPINNFKNPNY 795 ||||+ ||||||||+||+|| ||||| # ||+|| + | | | Sbjct 733 WKLLITIHDRREFARFEEEKARAKWDTG#NNPLYKEATSTFTNITY 777 |
| Podocoryne carnea | Query 757 IHDRREFAKFEKEKMNAKWDTQ#ENPIYKSPINNFKNPNYGRK 798 | ||||||||||++ | |||+ #|||||| + |+|| || | Sbjct 740 IQDRREFAKFEKDRQNPKWDSG#ENPIYKKATSTFQNPMYGNK 781 |
| Caenorhabditis briggsae | Query 758 HDRREFAKFEKEKMNAKWDTQ#ENPIYKSPINNFKNPNYGRKA 799 ||| |+||| |++ ||||| #|||||| |||| | || Sbjct 768 HDRAEYAKFNNERLMAKWDTN#ENPIYKQATTTFKNPVYAGKA 809 |
| Ovis aries | Query 750 IWKLLMIIHDRREFAKFEKEKMNAKWDTQ#ENPIYKSPINNFKNPNYGRK 798 |||||+ |||+| |||| |+ ||| | # ||+|+ + ||| | | Sbjct 728 IWKLLVSFHDRKEVAKFEAERSKAKWQTG#TNPLYRGSTSTFKNVTYKHK 776 |
| Strongylocentrotus purpuratus | Query 757 IHDRREFAKFEKEKMNAKWDTQ#ENPIYKSPINNFKNPNYGRK 798 ||||||| ||||+ || |+ #|||||| + |||| | | Sbjct 765 IHDRREFQNFEKERANATWEGG#ENPIYKPSTSVFKNPTYNIK 806 |
| Lytechinus variegatus | Query 757 IHDRREFAKFEKEKMNAKWDTQ#ENPIYKSPINNFKNPNYGRK 798 ||||||| ||||+ || |+ #|||||| + |||| | | Sbjct 765 IHDRREFQNFEKERANATWEGG#ENPIYKPSTSVFKNPTYNIK 806 |
| Caenorhabditis elegans | Query 758 HDRREFAKFEKEKMNAKWDTQ#ENPIYKSPINNFKNPNYGRKA 799 ||| |+| | |++ ||||| #|||||| |||| | || Sbjct 767 HDRSEYATFNNERLMAKWDTN#ENPIYKQATTTFKNPVYAGKA 808 |
| Xenopus laevis | Query 749 LIWKLLMIIHDRREFAKFEKEKMNAKWDTQ#ENPIYKSPINNFKNPNY 795 +|||++ | || |+ +||||+||+||+ # ||+| + +|||+ Sbjct 723 IIWKIITEIKDRNEYQRFEKERMNSKWNPG#HNPLYHNATTTVQNPNF 769 |
| Schistosoma japonicum | Query 751 WKLLMIIHDRREFAKFEKEKMNAKWDTQ#ENPIYKSPINNFKNPNY 795 +||++ | |||| | |+++ | +|+ #||||++|| | || + Sbjct 239 YKLVITIDDRRELANFKQQGENMRWEMA#ENPIFESPTTNVLNPTF 283 |
| Halocynthia roretzi | Query 749 LIWKLLMIIHDRREFAKFEKEKMNAKWDTQ#ENPIYKSPINNFKNPNYG 796 +|||++ + |+||+ ||+||+ || #|||+| + + | || +| Sbjct 785 IIWKVIQTLRDKREYEKFKKEEDLRKWTKG#ENPVYVNASSKFDNPMFG 832 |
| Macaca mulatta | Query 755 MIIHDRREFAKFEKEKMNAKW 775 ||||||||||||||||||||| Query 755 MIIHDRREFAKFEKEKMNAKW 775 |
| Pan troglodytes | Query 749 LIWKLLMIIHDRREFAKFEKEKMNAKWDTQ#ENPIYKSPINNFKNPNY 795 | ++| + |+||||+++||||+ | # ||+||| | || + Sbjct 744 LAYRLSVEIYDRREYSRFEKEQQQLNWKQD#SNPLYKSAITTTINPRF 790 |
| Acropora millepora | Query 752 KLLMIIHDRREFAKFEKEKMNAKWDTQ#ENPIYKSPINNFKNPNY--GRK 798 | | + || |+ |||+|+|++|| +#+||+|++ |+|| | ||+ Sbjct 744 KGLFTMVDRIEYQKFERERMHSKWTRE#KNPLYQAAKTTFENPTYAGGRQ 792 |
| Mus sp. | Query 749 LIWKLLMIIHDRREFAKFEKEKMNAKWDTQ#ENPIYKSPINNFKNPNY 795 | ++| + |+||||+ +||||+ | # ||+||| | || + Sbjct 743 LAYRLSVEIYDRREYRRFEKEQQQLNWKQD#NNPLYKSAITTTVNPRF 789 |
| Homo sapiens | Query 749 LIWKLLMIIHDRREFAKFEKEKMNAKWDTQ#ENPIYKSPINNFKNPNYG 796 +||| |+ + | ||+ +|||||+ ++|+ #+||++|| || + Sbjct 721 VIWKALIHLSDLREYRRFEKEKLKSQWN-N#DNPLFKSATTTVMNPKFA 767 |
| Pacifastacus leniusculus | Query 758 HDRREFAKFEKEKMNAKWDTQ#ENPIYKSPINNFKNPNYGR 797 |||||+||||||+ #|||+||| |+|| + + Sbjct 760 HDRREYAKFEKERKLPSGKRA#ENPLYKSAKTTFQNPAFAQ 799 |
| Ovis canadensis | Query 750 IWKLLMIIHDRREFAKFEKEKMNAKWDTQ#ENPIYKSPINNFKNPNYG 796 ||| | + | ||+ +|||||+ ++|+ #+||++|| || + Sbjct 723 IWKALTHLSDLREYHRFEKEKLKSQWN-N#DNPLFKSATTTVMNPKFA 768 |
| Sigmodon hispidus | Query 750 IWKLLMIIHDRREFAKFEKEKMNAKWDTQ#ENPIYKSPINNFKNPNYG 796 ||| | + | ||+ |||||+ ++|+ #+||++|| || + Sbjct 722 IWKALTHLTDLREYRHFEKEKLKSQWN-N#DNPLFKSATTTVMNPKFA 767 |
| Anopheles gambiae str. PEST | Query 751 WKLLMIIHDRREFAKFEKEKMNAKWDTQ#ENPIYKSPINNFKNP 793 ++ ++ ||+ |||||||+ #|+|+|||||+||| | Sbjct 753 YRCKIMYDDRKMFAKFEKEREQETKYQM#ESPLYKSPISNFKVP 795 |
[Site 6] KWDTQENPIY783-KSPINNFKNP
Tyr783  Lys
Lys
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Lys774 | Trp775 | Asp776 | Thr777 | Gln778 | Glu779 | Asn780 | Pro781 | Ile782 | Tyr783 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Lys784 | Ser785 | Pro786 | Ile787 | Asn788 | Asn789 | Phe790 | Lys791 | Asn792 | Pro793 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| LMIIHDRREFAKFEKEKMNAKWDTQENPIYKSPINNFKNPNYGRKAGL |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | Canis familiaris | 110.00 | 10 | PREDICTED: similar to integrin beta 1 isoform 1D |
| 2 | Bos taurus | 110.00 | 7 | ITGB1 protein |
| 3 | N/A | 103.00 | 28 | S66396 integrin beta-1 chain isoform D - human (f |
| 4 | Mus musculus | 102.00 | 12 | beta 1 integrin |
| 5 | Gallus gallus | 102.00 | 3 | integrin beta1D |
| 6 | Tetraodon nigroviridis | 78.20 | 5 | unnamed protein product |
| 7 | Xenopus laevis | 78.20 | 2 | integrin beta-1 subunit |
| 8 | Ictalurus punctatus | 77.80 | 1 | beta-1 integrin |
| 9 | Sus scrofa | 77.40 | 3 | integrin beta-1 subunit |
| 10 | Pongo pygmaeus | 77.40 | 1 | ITB1_PONPY Integrin beta-1 precursor (Fibronectin |
| 11 | Homo sapiens | 77.00 | 11 | ITB1_HUMAN Integrin beta-1 precursor (Fibronectin |
| 12 | Danio rerio | 77.00 | 5 | integrin, beta 1a |
| 13 | Drosophila melanogaster | 75.50 | 1 | myospheroid CG1560-PA |
| 14 | Felis catus | 74.70 | 1 | integrin beta 1 |
| 15 | Oryctolagus cuniculus | 74.30 | 1 | integrin beta 1 |
| 16 | Anopheles gambiae | 73.20 | 2 | integrin beta subunit |
| 17 | Anopheles gambiae str. PEST | 73.20 | 1 | integrin beta subunit (AGAP000815-PA) |
| 18 | Biomphalaria glabrata | 72.40 | 1 | beta integrin subunit |
| 19 | Pseudoplusia includens | 71.20 | 1 | integrin beta 1 |
| 20 | Podocoryne carnea | 67.40 | 1 | AF308652_1 integrin beta chain |
| 21 | Gallus gallus | 66.20 | 2 | integrin beta 1A precursor |
| 22 | Caenorhabditis briggsae | 62.40 | 1 | Hypothetical protein CBG03601 |
| 23 | Crassostrea gigas | 62.40 | 1 | integrin beta cgh |
| 24 | Strongylocentrotus purpuratus | 62.00 | 3 | integrin beta-C subunit |
| 25 | Lytechinus variegatus | 62.00 | 1 | beta-C integrin subunit |
| 26 | Rattus norvegicus | 60.80 | 4 | integrin beta 3 |
| 27 | synthetic construct | 60.80 | 3 | AF239959_1 hybrid integrin beta 3 subunit precurs |
| 28 | Mus musculus | 60.80 | 1 | integrin beta 3 precursor |
| 29 | Canis lupus familiaris | 60.80 | 1 | integrin beta chain, beta 3 |
| 30 | rats, Peptide Partial, 723 aa | 60.80 | 1 | beta 3 integrin, GPIIIA |
| 31 | Caenorhabditis elegans | 60.80 | 1 | Paralysed Arrest at Two-fold family member (pat-3 |
| 32 | Oryctolagus cuniculus | 60.80 | 1 | glycoprotein IIIa |
| 33 | Sus scrofa | 60.80 | 1 | integrin beta chain, beta 3 |
| 34 | mice, Peptide Partial, 680 aa | 58.90 | 1 | beta 3 integrin, GPIIIA |
| 35 | Ovis aries | 53.50 | 2 | ITB6_SHEEP Integrin beta-6 precursor gi |
| 36 | Xenopus laevis | 51.60 | 1 | hypothetical protein LOC379708 |
| 37 | Macaca mulatta | 51.60 | 1 | integrin beta 1 |
| 38 | Acropora millepora | 50.80 | 1 | integrin subunit betaCn1 |
| 39 | Schistosoma japonicum | 50.10 | 1 | SJCHGC06221 protein |
| 40 | Pan troglodytes | 48.90 | 2 | PREDICTED: integrin, beta 7 isoform 5 |
| 41 | Pacifastacus leniusculus | 48.90 | 1 | integrin |
| 42 | Mus sp. | 47.80 | 1 | beta 7 integrin |
| 43 | Anopheles gambiae str. PEST | 46.20 | 1 | AGAP010233-PA |
| 44 | Ophlitaspongia tenuis | 45.10 | 1 | integrin subunit betaPo1 |
| 45 | Halocynthia roretzi | 44.30 | 2 | integrin beta Hr2 precursor |
| 46 | Homo sapiens | 42.00 | 1 | integrin, beta 2 precursor |
| 47 | Ovis canadensis | 41.20 | 1 | ITB2_OVICA Integrin beta-2 precursor (Cell surfac |
| 48 | Suberites domuncula | 40.40 | 1 | integrin beta subunit |
Top-ranked sequences
| organism | matching |
|---|---|
| Canis familiaris | Query 754 LMIIHDRREFAKFEKEKMNAKWDTQENPIY#KSPINNFKNPNYGRKAGL 801 ||||||||||||||||||||||||||||||#|||||||||||||||||| Query 754 LMIIHDRREFAKFEKEKMNAKWDTQENPIY#KSPINNFKNPNYGRKAGL 801 |
| Bos taurus | Query 754 LMIIHDRREFAKFEKEKMNAKWDTQENPIY#KSPINNFKNPNYGRKAGL 801 ||||||||||||||||||||||||||||||#|||||||||||||||||| Query 754 LMIIHDRREFAKFEKEKMNAKWDTQENPIY#KSPINNFKNPNYGRKAGL 801 |
| N/A | Query 754 LMIIHDRREFAKFEKEKMNAKWDTQENPIY#KSPINNFKNPNYGRKAGL 801 ||||||||||||||||||||||||||||||#|||||||||||||||||| Query 754 LMIIHDRREFAKFEKEKMNAKWDTQENPIY#KSPINNFKNPNYGRKAGL 801 |
| Mus musculus | Query 754 LMIIHDRREFAKFEKEKMNAKWDTQENPIY#KSPINNFKNPNYGRKAGL 801 ||||||||||||||||||||||||||||||#|||||||||||||||||| Query 754 LMIIHDRREFAKFEKEKMNAKWDTQENPIY#KSPINNFKNPNYGRKAGL 801 |
| Gallus gallus | Query 754 LMIIHDRREFAKFEKEKMNAKWDTQENPIY#KSPINNFKNPNYGRKAGL 801 ||||||||||||||||||||||||||||||#|||||||||||||||||| Query 754 LMIIHDRREFAKFEKEKMNAKWDTQENPIY#KSPINNFKNPNYGRKAGL 801 |
| Tetraodon nigroviridis | Query 755 MIIHDRREFAKFEKEKMNAKWDTQENPIY#KSPINNFKNPNYGRK 798 ||||||||||||||||||||||| |||||#|| + || | | Sbjct 828 MIIHDRREFAKFEKEKMNAKWDTGENPIY#KSAVTTVVNPKYEGK 871 |
| Xenopus laevis | Query 755 MIIHDRREFAKFEKEKMNAKWDTQENPIY#KSPINNFKNPNYGRK 798 ||||||||||||||||||||||| |||||#|| + || | | Sbjct 755 MIIHDRREFAKFEKEKMNAKWDTGENPIY#KSAVTTVVNPKYEGK 798 |
| Ictalurus punctatus | Query 754 LMIIHDRREFAKFEKEKMNAKWDTQENPIY#KSPINNFKNPNYGRK 798 ||||||||||||||||||||||| |||||#|| + || | | Sbjct 763 LMIIHDRREFAKFEKEKMNAKWDAGENPIY#KSAVTTVVNPKYEGK 807 |
| Sus scrofa | Query 755 MIIHDRREFAKFEKEKMNAKWDTQENPIY#KSPINNFKNPNYGRK 798 ||||||||||||||||||||||| |||||#|| + || | | Sbjct 755 MIIHDRREFAKFEKEKMNAKWDTGENPIY#KSAVTTVVNPKYEGK 798 |
| Pongo pygmaeus | Query 755 MIIHDRREFAKFEKEKMNAKWDTQENPIY#KSPINNFKNPNYGRK 798 ||||||||||||||||||||||| |||||#|| + || | | Sbjct 755 MIIHDRREFAKFEKEKMNAKWDTGENPIY#KSAVTTVVNPKYEGK 798 |
| Homo sapiens | Query 755 MIIHDRREFAKFEKEKMNAKWDTQENPIY#KSPINNFKNPNYGRK 798 ||||||||||||||||||||||| |||||#|| + || | | Sbjct 755 MIIHDRREFAKFEKEKMNAKWDTGENPIY#KSAVTTVVNPKYEGK 798 |
| Danio rerio | Query 755 MIIHDRREFAKFEKEKMNAKWDTQENPIY#KSPINNFKNPNYGRK 798 ||||||||||||||||||||||| |||||#|| + || | | Sbjct 755 MIIHDRREFAKFEKEKMNAKWDTGENPIY#KSAVTTVINPKYEGK 798 |
| Drosophila melanogaster | Query 757 IHDRREFAKFEKEKMNAKWDTQENPIY#KSPINNFKNPNYGRK 798 ||||||||+||||+||||||| |||||#| + |||| | | Sbjct 805 IHDRREFARFEKERMNAKWDTGENPIY#KQATSTFKNPMYAGK 846 |
| Felis catus | Query 755 MIIHDRREFAKFEKEKMNAKWDTQENPIY#KSPINNFKNPNYGRK 798 ||||| ||||||||||||||||| |||||#|| + || | | Sbjct 755 MIIHDTREFAKFEKEKMNAKWDTGENPIY#KSAVTTVVNPKYEGK 798 |
| Oryctolagus cuniculus | Query 755 MIIHDRREFAKFEKEKMNAKWDTQENPIY#KSPINNFKNPNY 795 ||||||||||||||||||||||| |||||#|| + || | Sbjct 20 MIIHDRREFAKFEKEKMNAKWDTGENPIY#KSAVTTVVNPKY 60 |
| Anopheles gambiae | Query 754 LMIIHDRREFAKFEKEKMNAKWDTQENPIY#KSPINNFKNPNYGRK 798 | ||||||||+||||+| ||||| |||||#| |||| | | Sbjct 793 LTSIHDRREFARFEKERMMAKWDTGENPIY#KQATTTFKNPTYAGK 837 |
| Anopheles gambiae str. PEST | Query 754 LMIIHDRREFAKFEKEKMNAKWDTQENPIY#KSPINNFKNPNYGRK 798 | ||||||||+||||+| ||||| |||||#| |||| | | Sbjct 793 LTSIHDRREFARFEKERMMAKWDTGENPIY#KQATTTFKNPTYAGK 837 |
| Biomphalaria glabrata | Query 757 IHDRREFAKFEKEKMNAKWDTQENPIY#KSPINNFKNPNYGRK 798 ||| |||||||||+ |||||| |||||#| + |||| || + Sbjct 747 IHDTREFAKFEKERQNAKWDTGENPIY#KQATSTFKNPTYGSQ 788 |
| Pseudoplusia includens | Query 758 HDRREFAKFEKEKMNAKWDTQENPIY#KSPINNFKNPNYGRK 798 |||||||+||||+| ||||| |||||#| + |||| | | Sbjct 797 HDRREFARFEKERMMAKWDTGENPIY#KQATSTFKNPTYAGK 837 |
| Podocoryne carnea | Query 757 IHDRREFAKFEKEKMNAKWDTQENPIY#KSPINNFKNPNYGRK 798 | ||||||||||++ | |||+ |||||#| + |+|| || | Sbjct 740 IQDRREFAKFEKDRQNPKWDSGENPIY#KKATSTFQNPMYGNK 781 |
| Gallus gallus | Query 755 MIIHDRREFAKFEKEKMNAKWDTQENPIY#KSPI 787 ||||||||||||||||||||||| |||||#|| + Sbjct 12 MIIHDRREFAKFEKEKMNAKWDTGENPIY#KSAV 44 |
| Caenorhabditis briggsae | Query 758 HDRREFAKFEKEKMNAKWDTQENPIY#KSPINNFKNPNYGRKA 799 ||| |+||| |++ ||||| |||||#| |||| | || Sbjct 768 HDRAEYAKFNNERLMAKWDTNENPIY#KQATTTFKNPVYAGKA 809 |
| Crassostrea gigas | Query 757 IHDRREFAKFEKEKMNAKWDTQENPIY#KSPINNFKNPNY 795 | |+|| |+|||| |||+||| |||||#| + | || | Sbjct 759 ISDKRELARFEKEAMNARWDTGENPIY#KQATSTFVNPTY 797 |
| Strongylocentrotus purpuratus | Query 757 IHDRREFAKFEKEKMNAKWDTQENPIY#KSPINNFKNPNYGRK 798 ||||||| ||||+ || |+ |||||#| + |||| | | Sbjct 765 IHDRREFQNFEKERANATWEGGENPIY#KPSTSVFKNPTYNIK 806 |
| Lytechinus variegatus | Query 757 IHDRREFAKFEKEKMNAKWDTQENPIY#KSPINNFKNPNYGRK 798 ||||||| ||||+ || |+ |||||#| + |||| | | Sbjct 765 IHDRREFQNFEKERANATWEGGENPIY#KPSTSVFKNPTYNIK 806 |
| Rattus norvegicus | Query 754 LMIIHDRREFAKFEKEKMNAKWDTQENPIY#KSPINNFKNPNY 795 |+ ||||+||||||+|+ ||||| ||+|#| + | | | Sbjct 743 LITIHDRKEFAKFEEERARAKWDTANNPLY#KEATSTFTNITY 784 |
| synthetic construct | Query 754 LMIIHDRREFAKFEKEKMNAKWDTQENPIY#KSPINNFKNPNY 795 |+ ||||+||||||+|+ ||||| ||+|#| + | | | Sbjct 744 LITIHDRKEFAKFEEERARAKWDTANNPLY#KEATSTFTNITY 785 |
| Mus musculus | Query 754 LMIIHDRREFAKFEKEKMNAKWDTQENPIY#KSPINNFKNPNY 795 |+ ||||+||||||+|+ ||||| ||+|#| + | | | Sbjct 743 LITIHDRKEFAKFEEERARAKWDTANNPLY#KEATSTFTNITY 784 |
| Canis lupus familiaris | Query 754 LMIIHDRREFAKFEKEKMNAKWDTQENPIY#KSPINNFKNPNY 795 |+ ||||+||||||+|+ ||||| ||+|#| + | | | Sbjct 740 LITIHDRKEFAKFEEERARAKWDTANNPLY#KEATSTFTNITY 781 |
| rats, Peptide Partial, 723 aa | Query 754 LMIIHDRREFAKFEKEKMNAKWDTQENPIY#KSPINNFKNPNY 795 |+ ||||+||||||+|+ ||||| ||+|#| + | | | Sbjct 679 LITIHDRKEFAKFEEERARAKWDTANNPLY#KEATSTFTNITY 720 |
| Caenorhabditis elegans | Query 758 HDRREFAKFEKEKMNAKWDTQENPIY#KSPINNFKNPNYGRKA 799 ||| |+| | |++ ||||| |||||#| |||| | || Sbjct 767 HDRSEYATFNNERLMAKWDTNENPIY#KQATTTFKNPVYAGKA 808 |
| Oryctolagus cuniculus | Query 754 LMIIHDRREFAKFEKEKMNAKWDTQENPIY#KSPINNFKNPNY 795 |+ ||||+||||||+|+ ||||| ||+|#| + | | | Sbjct 744 LITIHDRKEFAKFEEERARAKWDTANNPLY#KEATSTFTNITY 785 |
| Sus scrofa | Query 754 LMIIHDRREFAKFEKEKMNAKWDTQENPIY#KSPINNFKNPNY 795 |+ ||||+||||||+|+ ||||| ||+|#| + | | | Sbjct 740 LITIHDRKEFAKFEEERARAKWDTANNPLY#KEATSTFTNITY 781 |
| mice, Peptide Partial, 680 aa | Query 754 LMIIHDRREFAKFEKEKMNAKWDTQENPIY#KSPINNFKN 792 |+ ||||+||||||+|+ ||||| ||+|#| + | | Sbjct 640 LITIHDRKEFAKFEEERARAKWDTANNPLY#KEATSTFTN 678 |
| Ovis aries | Query 754 LMIIHDRREFAKFEKEKMNAKWDTQENPIY#KSPINNFKNPNYGRK 798 |+ |||+| |||| |+ ||| | ||+|#+ + ||| | | Sbjct 732 LVSFHDRKEVAKFEAERSKAKWQTGTNPLY#RGSTSTFKNVTYKHK 776 |
| Xenopus laevis | Query 757 IHDRREFAKFEKEKMNAKWDTQENPIY#KSPINNFKNPNY 795 | || |+ +||||+||+||+ ||+|# + +|||+ Sbjct 731 IKDRNEYQRFEKERMNSKWNPGHNPLY#HNATTTVQNPNF 769 |
| Macaca mulatta | Query 755 MIIHDRREFAKFEKEKMNAKW 775 ||||||||||||||||||||| Query 755 MIIHDRREFAKFEKEKMNAKW 775 |
| Acropora millepora | Query 754 LMIIHDRREFAKFEKEKMNAKWDTQENPIY#KSPINNFKNPNY--GRK 798 | + || |+ |||+|+|++|| ++||+|#++ |+|| | ||+ Sbjct 746 LFTMVDRIEYQKFERERMHSKWTREKNPLY#QAAKTTFENPTYAGGRQ 792 |
| Schistosoma japonicum | Query 754 LMIIHDRREFAKFEKEKMNAKWDTQENPIY#KSPINNFKNPNY 795 ++ | |||| | |+++ | +|+ ||||+#+|| | || + Sbjct 242 VITIDDRRELANFKQQGENMRWEMAENPIF#ESPTTNVLNPTF 283 |
| Pan troglodytes | Query 757 IHDRREFAKFEKEKMNAKWDTQENPIY#KSPINNFKNPNY 795 |+||||+++||||+ | ||+|#|| | || + Sbjct 752 IYDRREYSRFEKEQQQLNWKQDSNPLY#KSAITTTINPRF 790 |
| Pacifastacus leniusculus | Query 758 HDRREFAKFEKEKMNAKWDTQENPIY#KSPINNFKNPNYGR 797 |||||+||||||+ |||+|#|| |+|| + + Sbjct 760 HDRREYAKFEKERKLPSGKRAENPLY#KSAKTTFQNPAFAQ 799 |
| Mus sp. | Query 757 IHDRREFAKFEKEKMNAKWDTQENPIY#KSPINNFKNPNY 795 |+||||+ +||||+ | ||+|#|| | || + Sbjct 751 IYDRREYRRFEKEQQQLNWKQDNNPLY#KSAITTTVNPRF 789 |
| Anopheles gambiae str. PEST | Query 759 DRREFAKFEKEKMNAKWDTQENPIY#KSPINNFKNP 793 ||+ |||||||+ |+|+|#||||+||| | Sbjct 761 DRKMFAKFEKEREQETKYQMESPLY#KSPISNFKVP 795 |
| Ophlitaspongia tenuis | Query 759 DRREFAKFEKEKMNAKWDTQENPIY#KSPINNFKNPNYGR 797 | | |||+| |||+ |||+|#+| +++|| ||+ Sbjct 800 DVVEVRKFEREIKNAKYTKNENPLY#RSATKDYQNPLYGK 838 |
| Halocynthia roretzi | Query 754 LMIIHDRREFAKFEKEKMNAKWDTQENPIY#KSPINNFKNPNYG 796 + + |+||+ ||+||+ || |||+|# + + | || +| Sbjct 790 IQTLRDKREYEKFKKEEDLRKWTKGENPVY#VNASSKFDNPMFG 832 |
| Homo sapiens | Query 754 LMIIHDRREFAKFEKEKMNAKWDTQENPIY#KSPINNFKNPNYG 796 |+ + | ||+ +|||||+ ++|+ +||++#|| || + Sbjct 726 LIHLSDLREYRRFEKEKLKSQWN-NDNPLF#KSATTTVMNPKFA 767 |
| Ovis canadensis | Query 757 IHDRREFAKFEKEKMNAKWDTQENPIY#KSPINNFKNPNYG 796 + | ||+ +|||||+ ++|+ +||++#|| || + Sbjct 730 LSDLREYHRFEKEKLKSQWN-NDNPLF#KSATTTVMNPKFA 768 |
| Suberites domuncula | Query 757 IHDRREFAKFEKEKMNAKWDTQENPIY#KSPINNFKNPNYGR 797 | | | ||||| |+ +||+|#+| +++|| ||+ Sbjct 724 IIDVIEVKKFEKELAKTKYPKHQNPLY#RSAAKDYQNPIYGQ 764 |
[Site 7] ENPIYKSPIN788-NFKNPNYGRK
Asn788  Asn
Asn
iTraq-117 Signal 9143.1 (

 ) for NFKNPNYGRK
) for NFKNPNYGRK
iTraq-117 Signal 2019.9 (

 ) for FKNPNYGRK
) for FKNPNYGRK
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Glu779 | Asn780 | Pro781 | Ile782 | Tyr783 | Lys784 | Ser785 | Pro786 | Ile787 | Asn788 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Asn789 | Phe790 | Lys791 | Asn792 | Pro793 | Asn794 | Tyr795 | Gly796 | Arg797 | Lys798 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| DRREFAKFEKEKMNAKWDTQENPIYKSPINNFKNPNYGRKAGL |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | Canis familiaris | 97.40 | 10 | PREDICTED: similar to integrin beta 1 isoform 1D |
| 2 | Bos taurus | 97.10 | 7 | ITGB1 protein |
| 3 | N/A | 92.80 | 27 | S66396 integrin beta-1 chain isoform D - human (f |
| 4 | Mus musculus | 92.40 | 12 | beta 1 integrin |
| 5 | Gallus gallus | 91.30 | 3 | integrin beta1D |
| 6 | Drosophila melanogaster | 68.90 | 1 | myospheroid CG1560-PA |
| 7 | Tetraodon nigroviridis | 67.80 | 5 | unnamed protein product |
| 8 | Xenopus laevis | 67.80 | 2 | integrin beta-1 subunit |
| 9 | Pseudoplusia includens | 67.80 | 1 | integrin beta 1 |
| 10 | Homo sapiens | 66.60 | 10 | ITB1_HUMAN Integrin beta-1 precursor (Fibronectin |
| 11 | Sus scrofa | 66.60 | 3 | integrin beta-1 subunit |
| 12 | Anopheles gambiae | 66.60 | 2 | integrin beta subunit |
| 13 | Anopheles gambiae str. PEST | 66.60 | 1 | integrin beta subunit (AGAP000815-PA) |
| 14 | Pongo pygmaeus | 66.60 | 1 | ITB1_PONPY Integrin beta-1 precursor (Fibronectin |
| 15 | Danio rerio | 66.20 | 5 | integrin, beta 1a |
| 16 | Ictalurus punctatus | 65.90 | 1 | beta-1 integrin |
| 17 | Oryctolagus cuniculus | 65.50 | 1 | integrin beta 1 |
| 18 | Biomphalaria glabrata | 65.10 | 1 | beta integrin subunit |
| 19 | Felis catus | 64.30 | 1 | integrin beta 1 |
| 20 | Podocoryne carnea | 63.90 | 1 | AF308652_1 integrin beta chain |
| 21 | Crassostrea gigas | 59.30 | 1 | integrin beta cgh |
| 22 | Caenorhabditis briggsae | 57.80 | 1 | Hypothetical protein CBG03601 |
| 23 | Strongylocentrotus purpuratus | 57.40 | 3 | integrin beta-C subunit |
| 24 | Gallus gallus | 57.40 | 2 | integrin beta 1A precursor |
| 25 | Lytechinus variegatus | 56.60 | 1 | beta-C integrin subunit |
| 26 | Caenorhabditis elegans | 55.80 | 1 | Paralysed Arrest at Two-fold family member (pat-3 |
| 27 | Rattus norvegicus | 53.90 | 5 | integrin beta 3 |
| 28 | Mus musculus | 53.90 | 1 | integrin beta 3 precursor |
| 29 | rats, Peptide Partial, 723 aa | 53.90 | 1 | beta 3 integrin, GPIIIA |
| 30 | Oryctolagus cuniculus | 53.90 | 1 | glycoprotein IIIa |
| 31 | Canis lupus familiaris | 53.90 | 1 | integrin beta chain, beta 3 |
| 32 | Sus scrofa | 53.90 | 1 | integrin beta chain, beta 3 |
| 33 | synthetic construct | 53.50 | 3 | AF239959_1 hybrid integrin beta 3 subunit precurs |
| 34 | mice, Peptide Partial, 680 aa | 52.40 | 1 | beta 3 integrin, GPIIIA |
| 35 | Ovis aries | 49.70 | 2 | ITB6_SHEEP Integrin beta-6 precursor gi |
| 36 | Xenopus laevis | 48.90 | 1 | hypothetical protein LOC379708 |
| 37 | Acropora millepora | 48.50 | 1 | integrin subunit betaCn1 |
| 38 | Schistosoma japonicum | 47.40 | 1 | SJCHGC06221 protein |
| 39 | Pan troglodytes | 46.20 | 2 | PREDICTED: integrin, beta 7 isoform 5 |
| 40 | Mus sp. | 45.40 | 1 | beta 7 integrin |
| 41 | Ophlitaspongia tenuis | 45.10 | 1 | integrin subunit betaPo1 |
| 42 | Anopheles gambiae str. PEST | 45.10 | 1 | AGAP010233-PA |
| 43 | Pacifastacus leniusculus | 45.10 | 1 | integrin |
| 44 | Halocynthia roretzi | 42.70 | 2 | integrin beta Hr2 precursor |
| 45 | Macaca mulatta | 42.00 | 1 | integrin beta 1 |
| 46 | Homo sapiens | 40.80 | 1 | integrin, beta 2 precursor |
| 47 | Ovis canadensis | 40.40 | 1 | ITB2_OVICA Integrin beta-2 precursor (Cell surfac |
| 48 | Suberites domuncula | 40.40 | 1 | integrin beta subunit |
| 49 | Geodia cydonium | 39.30 | 1 | integrin beta subunit |
| 50 | Sigmodon hispidus | 39.30 | 1 | AF445415_1 integrin beta-2 precursor |
| 51 | Drosophila melanogaster | 38.90 | 1 | integrin beta subunit |
| 52 | nu | 38.90 | 1 | beta |
| 53 | Drosophila pseudoobscura | 38.10 | 1 | GA14581-PA |
Top-ranked sequences
| organism | matching |
|---|---|
| Canis familiaris | Query 759 DRREFAKFEKEKMNAKWDTQENPIYKSPIN#NFKNPNYGRKAGL 801 ||||||||||||||||||||||||||||||#||||||||||||| Query 759 DRREFAKFEKEKMNAKWDTQENPIYKSPIN#NFKNPNYGRKAGL 801 |
| Bos taurus | Query 759 DRREFAKFEKEKMNAKWDTQENPIYKSPIN#NFKNPNYGRKAGL 801 ||||||||||||||||||||||||||||||#||||||||||||| Query 759 DRREFAKFEKEKMNAKWDTQENPIYKSPIN#NFKNPNYGRKAGL 801 |
| N/A | Query 759 DRREFAKFEKEKMNAKWDTQENPIYKSPIN#NFKNPNYGRKAGL 801 ||||||||||||||||||||||||||||||#||||||||||||| Query 759 DRREFAKFEKEKMNAKWDTQENPIYKSPIN#NFKNPNYGRKAGL 801 |
| Mus musculus | Query 759 DRREFAKFEKEKMNAKWDTQENPIYKSPIN#NFKNPNYGRKAGL 801 ||||||||||||||||||||||||||||||#||||||||||||| Query 759 DRREFAKFEKEKMNAKWDTQENPIYKSPIN#NFKNPNYGRKAGL 801 |
| Gallus gallus | Query 759 DRREFAKFEKEKMNAKWDTQENPIYKSPIN#NFKNPNYGRKAGL 801 ||||||||||||||||||||||||||||||#||||||||||||| Query 759 DRREFAKFEKEKMNAKWDTQENPIYKSPIN#NFKNPNYGRKAGL 801 |
| Drosophila melanogaster | Query 759 DRREFAKFEKEKMNAKWDTQENPIYKSPIN#NFKNPNYGRK 798 ||||||+||||+||||||| |||||| +# |||| | | Sbjct 807 DRREFARFEKERMNAKWDTGENPIYKQATS#TFKNPMYAGK 846 |
| Tetraodon nigroviridis | Query 759 DRREFAKFEKEKMNAKWDTQENPIYKSPIN#NFKNPNYGRK 798 ||||||||||||||||||| ||||||| + # || | | Sbjct 832 DRREFAKFEKEKMNAKWDTGENPIYKSAVT#TVVNPKYEGK 871 |
| Xenopus laevis | Query 759 DRREFAKFEKEKMNAKWDTQENPIYKSPIN#NFKNPNYGRK 798 ||||||||||||||||||| ||||||| + # || | | Sbjct 759 DRREFAKFEKEKMNAKWDTGENPIYKSAVT#TVVNPKYEGK 798 |
| Pseudoplusia includens | Query 759 DRREFAKFEKEKMNAKWDTQENPIYKSPIN#NFKNPNYGRK 798 ||||||+||||+| ||||| |||||| +# |||| | | Sbjct 798 DRREFARFEKERMMAKWDTGENPIYKQATS#TFKNPTYAGK 837 |
| Homo sapiens | Query 759 DRREFAKFEKEKMNAKWDTQENPIYKSPIN#NFKNPNYGRK 798 ||||||||||||||||||| ||||||| + # || | | Sbjct 759 DRREFAKFEKEKMNAKWDTGENPIYKSAVT#TVVNPKYEGK 798 |
| Sus scrofa | Query 759 DRREFAKFEKEKMNAKWDTQENPIYKSPIN#NFKNPNYGRK 798 ||||||||||||||||||| ||||||| + # || | | Sbjct 759 DRREFAKFEKEKMNAKWDTGENPIYKSAVT#TVVNPKYEGK 798 |
| Anopheles gambiae | Query 759 DRREFAKFEKEKMNAKWDTQENPIYKSPIN#NFKNPNYGRK 798 ||||||+||||+| ||||| |||||| # |||| | | Sbjct 798 DRREFARFEKERMMAKWDTGENPIYKQATT#TFKNPTYAGK 837 |
| Anopheles gambiae str. PEST | Query 759 DRREFAKFEKEKMNAKWDTQENPIYKSPIN#NFKNPNYGRK 798 ||||||+||||+| ||||| |||||| # |||| | | Sbjct 798 DRREFARFEKERMMAKWDTGENPIYKQATT#TFKNPTYAGK 837 |
| Pongo pygmaeus | Query 759 DRREFAKFEKEKMNAKWDTQENPIYKSPIN#NFKNPNYGRK 798 ||||||||||||||||||| ||||||| + # || | | Sbjct 759 DRREFAKFEKEKMNAKWDTGENPIYKSAVT#TVVNPKYEGK 798 |
| Danio rerio | Query 759 DRREFAKFEKEKMNAKWDTQENPIYKSPIN#NFKNPNYGRK 798 ||||||||||||||||||| ||||||| + # || | | Sbjct 759 DRREFAKFEKEKMNAKWDTGENPIYKSAVT#TVINPKYEGK 798 |
| Ictalurus punctatus | Query 759 DRREFAKFEKEKMNAKWDTQENPIYKSPIN#NFKNPNYGRK 798 |||||||||||||||||| ||||||| + # || | | Sbjct 768 DRREFAKFEKEKMNAKWDAGENPIYKSAVT#TVVNPKYEGK 807 |
| Oryctolagus cuniculus | Query 759 DRREFAKFEKEKMNAKWDTQENPIYKSPIN#NFKNPNY 795 ||||||||||||||||||| ||||||| + # || | Sbjct 24 DRREFAKFEKEKMNAKWDTGENPIYKSAVT#TVVNPKY 60 |
| Biomphalaria glabrata | Query 759 DRREFAKFEKEKMNAKWDTQENPIYKSPIN#NFKNPNYGRK 798 | |||||||||+ |||||| |||||| +# |||| || + Sbjct 749 DTREFAKFEKERQNAKWDTGENPIYKQATS#TFKNPTYGSQ 788 |
| Felis catus | Query 759 DRREFAKFEKEKMNAKWDTQENPIYKSPIN#NFKNPNYGRK 798 | ||||||||||||||||| ||||||| + # || | | Sbjct 759 DTREFAKFEKEKMNAKWDTGENPIYKSAVT#TVVNPKYEGK 798 |
| Podocoryne carnea | Query 759 DRREFAKFEKEKMNAKWDTQENPIYKSPIN#NFKNPNYGRK 798 ||||||||||++ | |||+ |||||| +# |+|| || | Sbjct 742 DRREFAKFEKDRQNPKWDSGENPIYKKATS#TFQNPMYGNK 781 |
| Crassostrea gigas | Query 759 DRREFAKFEKEKMNAKWDTQENPIYKSPIN#NFKNPNY 795 |+|| |+|||| |||+||| |||||| +# | || | Sbjct 761 DKRELARFEKEAMNARWDTGENPIYKQATS#TFVNPTY 797 |
| Caenorhabditis briggsae | Query 759 DRREFAKFEKEKMNAKWDTQENPIYKSPIN#NFKNPNYGRKA 799 || |+||| |++ ||||| |||||| # |||| | || Sbjct 769 DRAEYAKFNNERLMAKWDTNENPIYKQATT#TFKNPVYAGKA 809 |
| Strongylocentrotus purpuratus | Query 759 DRREFAKFEKEKMNAKWDTQENPIYKSPIN#NFKNPNYGRK 798 ||||| ||||+ || |+ |||||| +# |||| | | Sbjct 767 DRREFQNFEKERANATWEGGENPIYKPSTS#VFKNPTYNIK 806 |
| Gallus gallus | Query 759 DRREFAKFEKEKMNAKWDTQENPIYKSPI 787 ||||||||||||||||||| ||||||| + Sbjct 16 DRREFAKFEKEKMNAKWDTGENPIYKSAV 44 |
| Lytechinus variegatus | Query 759 DRREFAKFEKEKMNAKWDTQENPIYKSPIN#NFKNPNYGRK 798 ||||| ||||+ || |+ |||||| +# |||| | | Sbjct 767 DRREFQNFEKERANATWEGGENPIYKPSTS#VFKNPTYNIK 806 |
| Caenorhabditis elegans | Query 759 DRREFAKFEKEKMNAKWDTQENPIYKSPIN#NFKNPNYGRKA 799 || |+| | |++ ||||| |||||| # |||| | || Sbjct 768 DRSEYATFNNERLMAKWDTNENPIYKQATT#TFKNPVYAGKA 808 |
| Rattus norvegicus | Query 759 DRREFAKFEKEKMNAKWDTQENPIYKSPIN#NFKNPNY 795 ||+||||||+|+ ||||| ||+|| +# | | | Sbjct 748 DRKEFAKFEEERARAKWDTANNPLYKEATS#TFTNITY 784 |
| Mus musculus | Query 759 DRREFAKFEKEKMNAKWDTQENPIYKSPIN#NFKNPNY 795 ||+||||||+|+ ||||| ||+|| +# | | | Sbjct 748 DRKEFAKFEEERARAKWDTANNPLYKEATS#TFTNITY 784 |
| rats, Peptide Partial, 723 aa | Query 759 DRREFAKFEKEKMNAKWDTQENPIYKSPIN#NFKNPNY 795 ||+||||||+|+ ||||| ||+|| +# | | | Sbjct 684 DRKEFAKFEEERARAKWDTANNPLYKEATS#TFTNITY 720 |
| Oryctolagus cuniculus | Query 759 DRREFAKFEKEKMNAKWDTQENPIYKSPIN#NFKNPNY 795 ||+||||||+|+ ||||| ||+|| +# | | | Sbjct 749 DRKEFAKFEEERARAKWDTANNPLYKEATS#TFTNITY 785 |
| Canis lupus familiaris | Query 759 DRREFAKFEKEKMNAKWDTQENPIYKSPIN#NFKNPNY 795 ||+||||||+|+ ||||| ||+|| +# | | | Sbjct 745 DRKEFAKFEEERARAKWDTANNPLYKEATS#TFTNITY 781 |
| Sus scrofa | Query 759 DRREFAKFEKEKMNAKWDTQENPIYKSPIN#NFKNPNY 795 ||+||||||+|+ ||||| ||+|| +# | | | Sbjct 745 DRKEFAKFEEERARAKWDTANNPLYKEATS#TFTNITY 781 |
| synthetic construct | Query 759 DRREFAKFEKEKMNAKWDTQENPIYKSPIN#NFKNPNY 795 ||+||||||+|+ ||||| ||+|| +# | | | Sbjct 749 DRKEFAKFEEERARAKWDTANNPLYKEATS#TFTNITY 785 |
| mice, Peptide Partial, 680 aa | Query 759 DRREFAKFEKEKMNAKWDTQENPIYKSPIN#NFKN 792 ||+||||||+|+ ||||| ||+|| +# | | Sbjct 645 DRKEFAKFEEERARAKWDTANNPLYKEATS#TFTN 678 |
| Ovis aries | Query 759 DRREFAKFEKEKMNAKWDTQENPIYKSPIN#NFKNPNYGRK 798 ||+| |||| |+ ||| | ||+|+ +# ||| | | Sbjct 737 DRKEVAKFEAERSKAKWQTGTNPLYRGSTS#TFKNVTYKHK 776 |
| Xenopus laevis | Query 759 DRREFAKFEKEKMNAKWDTQENPIYKSPIN#NFKNPNY 795 || |+ +||||+||+||+ ||+| + # +|||+ Sbjct 733 DRNEYQRFEKERMNSKWNPGHNPLYHNATT#TVQNPNF 769 |
| Acropora millepora | Query 759 DRREFAKFEKEKMNAKWDTQENPIYKSPIN#NFKNPNYG 796 || |+ |||+|+|++|| ++||+|++ # |+|| | Sbjct 751 DRIEYQKFERERMHSKWTREKNPLYQAAKT#TFENPTYA 788 |
| Schistosoma japonicum | Query 759 DRREFAKFEKEKMNAKWDTQENPIYKSPIN#NFKNPNY 795 |||| | |+++ | +|+ ||||++|| #| || + Sbjct 247 DRRELANFKQQGENMRWEMAENPIFESPTT#NVLNPTF 283 |
| Pan troglodytes | Query 759 DRREFAKFEKEKMNAKWDTQENPIYKSPIN#NFKNPNY 795 ||||+++||||+ | ||+||| | # || + Sbjct 754 DRREYSRFEKEQQQLNWKQDSNPLYKSAIT#TTINPRF 790 |
| Mus sp. | Query 759 DRREFAKFEKEKMNAKWDTQENPIYKSPIN#NFKNPNY 795 ||||+ +||||+ | ||+||| | # || + Sbjct 753 DRREYRRFEKEQQQLNWKQDNNPLYKSAIT#TTVNPRF 789 |
| Ophlitaspongia tenuis | Query 759 DRREFAKFEKEKMNAKWDTQENPIYKSPIN#NFKNPNYGR 797 | | |||+| |||+ |||+|+| #+++|| ||+ Sbjct 800 DVVEVRKFEREIKNAKYTKNENPLYRSATK#DYQNPLYGK 838 |
| Anopheles gambiae str. PEST | Query 759 DRREFAKFEKEKMNAKWDTQENPIYKSPIN#NFKNP 793 ||+ |||||||+ |+|+|||||+#||| | Sbjct 761 DRKMFAKFEKEREQETKYQMESPLYKSPIS#NFKVP 795 |
| Pacifastacus leniusculus | Query 759 DRREFAKFEKEKMNAKWDTQENPIYKSPIN#NFKNPNYGR 797 ||||+||||||+ |||+||| # |+|| + + Sbjct 761 DRREYAKFEKERKLPSGKRAENPLYKSAKT#TFQNPAFAQ 799 |
| Halocynthia roretzi | Query 759 DRREFAKFEKEKMNAKWDTQENPIYKSPIN#NFKNPNYG 796 |+||+ ||+||+ || |||+| + +# | || +| Sbjct 795 DKREYEKFKKEEDLRKWTKGENPVYVNASS#KFDNPMFG 832 |
| Macaca mulatta | Query 759 DRREFAKFEKEKMNAKW 775 ||||||||||||||||| Query 759 DRREFAKFEKEKMNAKW 775 |
| Homo sapiens | Query 759 DRREFAKFEKEKMNAKWDTQENPIYKSPIN#NFKNPNYG 796 | ||+ +|||||+ ++|+ +||++|| # || + Sbjct 731 DLREYRRFEKEKLKSQWN-NDNPLFKSATT#TVMNPKFA 767 |
| Ovis canadensis | Query 759 DRREFAKFEKEKMNAKWDTQENPIYKSPIN#NFKNPNYG 796 | ||+ +|||||+ ++|+ +||++|| # || + Sbjct 732 DLREYHRFEKEKLKSQWN-NDNPLFKSATT#TVMNPKFA 768 |
| Suberites domuncula | Query 759 DRREFAKFEKEKMNAKWDTQENPIYKSPIN#NFKNPNYGR 797 | | ||||| |+ +||+|+| #+++|| ||+ Sbjct 726 DVIEVKKFEKELAKTKYPKHQNPLYRSAAK#DYQNPIYGQ 764 |
| Geodia cydonium | Query 759 DRREFAKFEKEKMNAKWDTQENPIYKSPIN#NFKNPNYGR 797 | | ||| | |+ |||+| | # ||| ||+ Sbjct 840 DYTELKKFENELAKVKYTKNENPLYHSATT#EHKNPIYGQ 878 |
| Sigmodon hispidus | Query 759 DRREFAKFEKEKMNAKWDTQENPIYKSPIN#NFKNPNYG 796 | ||+ |||||+ ++|+ +||++|| # || + Sbjct 731 DLREYRHFEKEKLKSQWN-NDNPLFKSATT#TVMNPKFA 767 |
| Drosophila melanogaster | Query 759 DRREFAKFEKEKMNAKWDTQENPIYKSPIN#NFKNP 793 | ||+||||+++ |+ ||||||+ |+ # ++ | Sbjct 753 DAREYAKFEEDQKNSV--RQENPIYRDPVG#RYEVP 785 |
| nu | Query 759 DRREFAKFEKEKMNAKWDTQENPIYKSPIN#NFKNP 793 | ||+||||+++ |+ ||||||+ |+ # ++ | Sbjct 753 DAREYAKFEEDQKNSV--RQENPIYRDPVG#RYEVP 785 |
| Drosophila pseudoobscura | Query 759 DRREFAKFEKEKMNAKWDTQENPIYKSPIN#NFKNP 793 | ||+|+||+|+ || +||||+|+ |+ # + | Sbjct 726 DAREYARFEEEQ--AKRVSQENPLYRDPVG#RYVVP 758 |
[Site 8] NPIYKSPINN789-FKNPNYGRKA
Asn789  Phe
Phe
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Asn780 | Pro781 | Ile782 | Tyr783 | Lys784 | Ser785 | Pro786 | Ile787 | Asn788 | Asn789 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Phe790 | Lys791 | Asn792 | Pro793 | Asn794 | Tyr795 | Gly796 | Arg797 | Lys798 | Ala799 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| RREFAKFEKEKMNAKWDTQENPIYKSPINNFKNPNYGRKAGL |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | Canis familiaris | 95.10 | 10 | PREDICTED: similar to integrin beta 1 isoform 1D |
| 2 | Bos taurus | 95.10 | 7 | ITGB1 protein |
| 3 | N/A | 90.10 | 27 | S66396 integrin beta-1 chain isoform D - human (f |
| 4 | Mus musculus | 89.70 | 12 | beta 1 integrin |
| 5 | Gallus gallus | 89.00 | 3 | integrin beta1D |
| 6 | Drosophila melanogaster | 66.60 | 1 | myospheroid CG1560-PA |
| 7 | Pseudoplusia includens | 65.90 | 1 | integrin beta 1 |
| 8 | Tetraodon nigroviridis | 65.50 | 5 | unnamed protein product |
| 9 | Xenopus laevis | 65.50 | 2 | integrin beta-1 subunit |
| 10 | Sus scrofa | 64.70 | 3 | integrin beta-1 subunit |
| 11 | Homo sapiens | 64.30 | 10 | ITB1_HUMAN Integrin beta-1 precursor (Fibronectin |
| 12 | Anopheles gambiae | 64.30 | 2 | integrin beta subunit |
| 13 | Pongo pygmaeus | 64.30 | 1 | ITB1_PONPY Integrin beta-1 precursor (Fibronectin |
| 14 | Anopheles gambiae str. PEST | 64.30 | 1 | integrin beta subunit (AGAP000815-PA) |
| 15 | Danio rerio | 63.90 | 5 | integrin, beta 1a |
| 16 | Ictalurus punctatus | 63.50 | 1 | beta-1 integrin |
| 17 | Biomphalaria glabrata | 63.50 | 1 | beta integrin subunit |
| 18 | Oryctolagus cuniculus | 62.80 | 1 | integrin beta 1 |
| 19 | Felis catus | 62.40 | 1 | integrin beta 1 |
| 20 | Podocoryne carnea | 62.00 | 1 | AF308652_1 integrin beta chain |
| 21 | Crassostrea gigas | 57.40 | 1 | integrin beta cgh |
| 22 | Caenorhabditis briggsae | 55.80 | 1 | Hypothetical protein CBG03601 |
| 23 | Gallus gallus | 55.10 | 2 | integrin beta 1A precursor |
| 24 | Strongylocentrotus purpuratus | 54.70 | 3 | integrin beta-C subunit |
| 25 | Lytechinus variegatus | 54.70 | 1 | beta-C integrin subunit |
| 26 | Caenorhabditis elegans | 53.90 | 1 | Paralysed Arrest at Two-fold family member (pat-3 |
| 27 | Rattus norvegicus | 51.60 | 5 | integrin beta 3 |
| 28 | Mus musculus | 51.60 | 1 | integrin beta 3 precursor |
| 29 | rats, Peptide Partial, 723 aa | 51.60 | 1 | beta 3 integrin, GPIIIA |
| 30 | Canis lupus familiaris | 51.60 | 1 | integrin beta chain, beta 3 |
| 31 | Sus scrofa | 51.60 | 1 | integrin beta chain, beta 3 |
| 32 | Oryctolagus cuniculus | 51.60 | 1 | glycoprotein IIIa |
| 33 | synthetic construct | 51.20 | 3 | AF239959_1 hybrid integrin beta 3 subunit precurs |
| 34 | mice, Peptide Partial, 680 aa | 50.10 | 1 | beta 3 integrin, GPIIIA |
| 35 | Ovis aries | 47.00 | 2 | ITB6_SHEEP Integrin beta-6 precursor gi |
| 36 | Xenopus laevis | 46.60 | 1 | hypothetical protein LOC379708 |
| 37 | Acropora millepora | 46.20 | 1 | integrin subunit betaCn1 |
| 38 | Schistosoma japonicum | 45.40 | 1 | SJCHGC06221 protein |
| 39 | Ophlitaspongia tenuis | 44.70 | 1 | integrin subunit betaPo1 |
| 40 | Pan troglodytes | 43.90 | 2 | PREDICTED: integrin, beta 7 isoform 5 |
| 41 | Anopheles gambiae str. PEST | 43.10 | 1 | AGAP010233-PA |
| 42 | Mus sp. | 43.10 | 1 | beta 7 integrin |
| 43 | Pacifastacus leniusculus | 43.10 | 1 | integrin |
| 44 | Halocynthia roretzi | 40.40 | 2 | integrin beta Hr2 precursor |
| 45 | Suberites domuncula | 40.00 | 1 | integrin beta subunit |
| 46 | Macaca mulatta | 39.70 | 1 | integrin beta 1 |
| 47 | Homo sapiens | 39.70 | 1 | integrin, beta 2 precursor |
| 48 | Ovis canadensis | 38.90 | 1 | ITB2_OVICA Integrin beta-2 precursor (Cell surfac |
| 49 | Geodia cydonium | 38.10 | 1 | integrin beta subunit |
| 50 | Sigmodon hispidus | 37.70 | 1 | AF445415_1 integrin beta-2 precursor |
| 51 | nu | 37.40 | 1 | beta |
| 52 | Drosophila melanogaster | 37.40 | 1 | integrin beta subunit |
Top-ranked sequences
| organism | matching |
|---|---|
| Canis familiaris | Query 760 RREFAKFEKEKMNAKWDTQENPIYKSPINN#FKNPNYGRKAGL 801 ||||||||||||||||||||||||||||||#|||||||||||| Query 760 RREFAKFEKEKMNAKWDTQENPIYKSPINN#FKNPNYGRKAGL 801 |
| Bos taurus | Query 760 RREFAKFEKEKMNAKWDTQENPIYKSPINN#FKNPNYGRKAGL 801 ||||||||||||||||||||||||||||||#|||||||||||| Query 760 RREFAKFEKEKMNAKWDTQENPIYKSPINN#FKNPNYGRKAGL 801 |
| N/A | Query 760 RREFAKFEKEKMNAKWDTQENPIYKSPINN#FKNPNYGRKAGL 801 ||||||||||||||||||||||||||||||#|||||||||||| Query 760 RREFAKFEKEKMNAKWDTQENPIYKSPINN#FKNPNYGRKAGL 801 |
| Mus musculus | Query 760 RREFAKFEKEKMNAKWDTQENPIYKSPINN#FKNPNYGRKAGL 801 ||||||||||||||||||||||||||||||#|||||||||||| Query 760 RREFAKFEKEKMNAKWDTQENPIYKSPINN#FKNPNYGRKAGL 801 |
| Gallus gallus | Query 760 RREFAKFEKEKMNAKWDTQENPIYKSPINN#FKNPNYGRKAGL 801 ||||||||||||||||||||||||||||||#|||||||||||| Query 760 RREFAKFEKEKMNAKWDTQENPIYKSPINN#FKNPNYGRKAGL 801 |
| Drosophila melanogaster | Query 760 RREFAKFEKEKMNAKWDTQENPIYKSPINN#FKNPNYGRK 798 |||||+||||+||||||| |||||| + #|||| | | Sbjct 808 RREFARFEKERMNAKWDTGENPIYKQATST#FKNPMYAGK 846 |
| Pseudoplusia includens | Query 760 RREFAKFEKEKMNAKWDTQENPIYKSPINN#FKNPNYGRK 798 |||||+||||+| ||||| |||||| + #|||| | | Sbjct 799 RREFARFEKERMMAKWDTGENPIYKQATST#FKNPTYAGK 837 |
| Tetraodon nigroviridis | Query 760 RREFAKFEKEKMNAKWDTQENPIYKSPINN#FKNPNYGRK 798 |||||||||||||||||| ||||||| + # || | | Sbjct 833 RREFAKFEKEKMNAKWDTGENPIYKSAVTT#VVNPKYEGK 871 |
| Xenopus laevis | Query 760 RREFAKFEKEKMNAKWDTQENPIYKSPINN#FKNPNYGRK 798 |||||||||||||||||| ||||||| + # || | | Sbjct 760 RREFAKFEKEKMNAKWDTGENPIYKSAVTT#VVNPKYEGK 798 |
| Sus scrofa | Query 760 RREFAKFEKEKMNAKWDTQENPIYKSPINN#FKNPNYGRK 798 |||||||||||||||||| ||||||| + # || | | Sbjct 760 RREFAKFEKEKMNAKWDTGENPIYKSAVTT#VVNPKYEGK 798 |
| Homo sapiens | Query 760 RREFAKFEKEKMNAKWDTQENPIYKSPINN#FKNPNYGRK 798 |||||||||||||||||| ||||||| + # || | | Sbjct 760 RREFAKFEKEKMNAKWDTGENPIYKSAVTT#VVNPKYEGK 798 |
| Anopheles gambiae | Query 760 RREFAKFEKEKMNAKWDTQENPIYKSPINN#FKNPNYGRK 798 |||||+||||+| ||||| |||||| #|||| | | Sbjct 799 RREFARFEKERMMAKWDTGENPIYKQATTT#FKNPTYAGK 837 |
| Pongo pygmaeus | Query 760 RREFAKFEKEKMNAKWDTQENPIYKSPINN#FKNPNYGRK 798 |||||||||||||||||| ||||||| + # || | | Sbjct 760 RREFAKFEKEKMNAKWDTGENPIYKSAVTT#VVNPKYEGK 798 |
| Anopheles gambiae str. PEST | Query 760 RREFAKFEKEKMNAKWDTQENPIYKSPINN#FKNPNYGRK 798 |||||+||||+| ||||| |||||| #|||| | | Sbjct 799 RREFARFEKERMMAKWDTGENPIYKQATTT#FKNPTYAGK 837 |
| Danio rerio | Query 760 RREFAKFEKEKMNAKWDTQENPIYKSPINN#FKNPNYGRK 798 |||||||||||||||||| ||||||| + # || | | Sbjct 760 RREFAKFEKEKMNAKWDTGENPIYKSAVTT#VINPKYEGK 798 |
| Ictalurus punctatus | Query 760 RREFAKFEKEKMNAKWDTQENPIYKSPINN#FKNPNYGRK 798 ||||||||||||||||| ||||||| + # || | | Sbjct 769 RREFAKFEKEKMNAKWDAGENPIYKSAVTT#VVNPKYEGK 807 |
| Biomphalaria glabrata | Query 761 REFAKFEKEKMNAKWDTQENPIYKSPINN#FKNPNYGRK 798 |||||||||+ |||||| |||||| + #|||| || + Sbjct 751 REFAKFEKERQNAKWDTGENPIYKQATST#FKNPTYGSQ 788 |
| Oryctolagus cuniculus | Query 760 RREFAKFEKEKMNAKWDTQENPIYKSPINN#FKNPNY 795 |||||||||||||||||| ||||||| + # || | Sbjct 25 RREFAKFEKEKMNAKWDTGENPIYKSAVTT#VVNPKY 60 |
| Felis catus | Query 761 REFAKFEKEKMNAKWDTQENPIYKSPINN#FKNPNYGRK 798 ||||||||||||||||| ||||||| + # || | | Sbjct 761 REFAKFEKEKMNAKWDTGENPIYKSAVTT#VVNPKYEGK 798 |
| Podocoryne carnea | Query 760 RREFAKFEKEKMNAKWDTQENPIYKSPINN#FKNPNYGRK 798 |||||||||++ | |||+ |||||| + #|+|| || | Sbjct 743 RREFAKFEKDRQNPKWDSGENPIYKKATST#FQNPMYGNK 781 |
| Crassostrea gigas | Query 760 RREFAKFEKEKMNAKWDTQENPIYKSPINN#FKNPNY 795 +|| |+|||| |||+||| |||||| + #| || | Sbjct 762 KRELARFEKEAMNARWDTGENPIYKQATST#FVNPTY 797 |
| Caenorhabditis briggsae | Query 760 RREFAKFEKEKMNAKWDTQENPIYKSPINN#FKNPNYGRKA 799 | |+||| |++ ||||| |||||| #|||| | || Sbjct 770 RAEYAKFNNERLMAKWDTNENPIYKQATTT#FKNPVYAGKA 809 |
| Gallus gallus | Query 760 RREFAKFEKEKMNAKWDTQENPIYKSPI 787 |||||||||||||||||| ||||||| + Sbjct 17 RREFAKFEKEKMNAKWDTGENPIYKSAV 44 |
| Strongylocentrotus purpuratus | Query 760 RREFAKFEKEKMNAKWDTQENPIYKSPINN#FKNPNYGRK 798 |||| ||||+ || |+ |||||| + #|||| | | Sbjct 768 RREFQNFEKERANATWEGGENPIYKPSTSV#FKNPTYNIK 806 |
| Lytechinus variegatus | Query 760 RREFAKFEKEKMNAKWDTQENPIYKSPINN#FKNPNYGRK 798 |||| ||||+ || |+ |||||| + #|||| | | Sbjct 768 RREFQNFEKERANATWEGGENPIYKPSTSV#FKNPTYNIK 806 |
| Caenorhabditis elegans | Query 760 RREFAKFEKEKMNAKWDTQENPIYKSPINN#FKNPNYGRKA 799 | |+| | |++ ||||| |||||| #|||| | || Sbjct 769 RSEYATFNNERLMAKWDTNENPIYKQATTT#FKNPVYAGKA 808 |
| Rattus norvegicus | Query 760 RREFAKFEKEKMNAKWDTQENPIYKSPINN#FKNPNY 795 |+||||||+|+ ||||| ||+|| + #| | | Sbjct 749 RKEFAKFEEERARAKWDTANNPLYKEATST#FTNITY 784 |
| Mus musculus | Query 760 RREFAKFEKEKMNAKWDTQENPIYKSPINN#FKNPNY 795 |+||||||+|+ ||||| ||+|| + #| | | Sbjct 749 RKEFAKFEEERARAKWDTANNPLYKEATST#FTNITY 784 |
| rats, Peptide Partial, 723 aa | Query 760 RREFAKFEKEKMNAKWDTQENPIYKSPINN#FKNPNY 795 |+||||||+|+ ||||| ||+|| + #| | | Sbjct 685 RKEFAKFEEERARAKWDTANNPLYKEATST#FTNITY 720 |
| Canis lupus familiaris | Query 760 RREFAKFEKEKMNAKWDTQENPIYKSPINN#FKNPNY 795 |+||||||+|+ ||||| ||+|| + #| | | Sbjct 746 RKEFAKFEEERARAKWDTANNPLYKEATST#FTNITY 781 |
| Sus scrofa | Query 760 RREFAKFEKEKMNAKWDTQENPIYKSPINN#FKNPNY 795 |+||||||+|+ ||||| ||+|| + #| | | Sbjct 746 RKEFAKFEEERARAKWDTANNPLYKEATST#FTNITY 781 |
| Oryctolagus cuniculus | Query 760 RREFAKFEKEKMNAKWDTQENPIYKSPINN#FKNPNY 795 |+||||||+|+ ||||| ||+|| + #| | | Sbjct 750 RKEFAKFEEERARAKWDTANNPLYKEATST#FTNITY 785 |
| synthetic construct | Query 760 RREFAKFEKEKMNAKWDTQENPIYKSPINN#FKNPNY 795 |+||||||+|+ ||||| ||+|| + #| | | Sbjct 750 RKEFAKFEEERARAKWDTANNPLYKEATST#FTNITY 785 |
| mice, Peptide Partial, 680 aa | Query 760 RREFAKFEKEKMNAKWDTQENPIYKSPINN#FKN 792 |+||||||+|+ ||||| ||+|| + #| | Sbjct 646 RKEFAKFEEERARAKWDTANNPLYKEATST#FTN 678 |
| Ovis aries | Query 760 RREFAKFEKEKMNAKWDTQENPIYKSPINN#FKNPNYGRK 798 |+| |||| |+ ||| | ||+|+ + #||| | | Sbjct 738 RKEVAKFEAERSKAKWQTGTNPLYRGSTST#FKNVTYKHK 776 |
| Xenopus laevis | Query 760 RREFAKFEKEKMNAKWDTQENPIYKSPINN#FKNPNY 795 | |+ +||||+||+||+ ||+| + # +|||+ Sbjct 734 RNEYQRFEKERMNSKWNPGHNPLYHNATTT#VQNPNF 769 |
| Acropora millepora | Query 760 RREFAKFEKEKMNAKWDTQENPIYKSPINN#FKNPNYG 796 | |+ |||+|+|++|| ++||+|++ #|+|| | Sbjct 752 RIEYQKFERERMHSKWTREKNPLYQAAKTT#FENPTYA 788 |
| Schistosoma japonicum | Query 760 RREFAKFEKEKMNAKWDTQENPIYKSPINN#FKNPNY 795 ||| | |+++ | +|+ ||||++|| |# || + Sbjct 248 RRELANFKQQGENMRWEMAENPIFESPTTN#VLNPTF 283 |
| Ophlitaspongia tenuis | Query 762 EFAKFEKEKMNAKWDTQENPIYKSPINN#FKNPNYGR 797 | |||+| |||+ |||+|+| +#++|| ||+ Sbjct 803 EVRKFEREIKNAKYTKNENPLYRSATKD#YQNPLYGK 838 |
| Pan troglodytes | Query 760 RREFAKFEKEKMNAKWDTQENPIYKSPINN#FKNPNY 795 |||+++||||+ | ||+||| | # || + Sbjct 755 RREYSRFEKEQQQLNWKQDSNPLYKSAITT#TINPRF 790 |
| Anopheles gambiae str. PEST | Query 760 RREFAKFEKEKMNAKWDTQENPIYKSPINN#FKNP 793 |+ |||||||+ |+|+|||||+|#|| | Sbjct 762 RKMFAKFEKEREQETKYQMESPLYKSPISN#FKVP 795 |
| Mus sp. | Query 760 RREFAKFEKEKMNAKWDTQENPIYKSPINN#FKNPNY 795 |||+ +||||+ | ||+||| | # || + Sbjct 754 RREYRRFEKEQQQLNWKQDNNPLYKSAITT#TVNPRF 789 |
| Pacifastacus leniusculus | Query 760 RREFAKFEKEKMNAKWDTQENPIYKSPINN#FKNPNYGR 797 |||+||||||+ |||+||| #|+|| + + Sbjct 762 RREYAKFEKERKLPSGKRAENPLYKSAKTT#FQNPAFAQ 799 |
| Halocynthia roretzi | Query 760 RREFAKFEKEKMNAKWDTQENPIYKSPINN#FKNPNYG 796 +||+ ||+||+ || |||+| + + #| || +| Sbjct 796 KREYEKFKKEEDLRKWTKGENPVYVNASSK#FDNPMFG 832 |
| Suberites domuncula | Query 762 EFAKFEKEKMNAKWDTQENPIYKSPINN#FKNPNYGR 797 | ||||| |+ +||+|+| +#++|| ||+ Sbjct 729 EVKKFEKELAKTKYPKHQNPLYRSAAKD#YQNPIYGQ 764 |
| Macaca mulatta | Query 760 RREFAKFEKEKMNAKW 775 |||||||||||||||| Query 760 RREFAKFEKEKMNAKW 775 |
| Homo sapiens | Query 761 REFAKFEKEKMNAKWDTQENPIYKSPINN#FKNPNYG 796 ||+ +|||||+ ++|+ +||++|| # || + Sbjct 733 REYRRFEKEKLKSQWN-NDNPLFKSATTT#VMNPKFA 767 |
| Ovis canadensis | Query 761 REFAKFEKEKMNAKWDTQENPIYKSPINN#FKNPNYG 796 ||+ +|||||+ ++|+ +||++|| # || + Sbjct 734 REYHRFEKEKLKSQWN-NDNPLFKSATTT#VMNPKFA 768 |
| Geodia cydonium | Query 762 EFAKFEKEKMNAKWDTQENPIYKSPINN#FKNPNYGR 797 | ||| | |+ |||+| | # ||| ||+ Sbjct 843 ELKKFENELAKVKYTKNENPLYHSATTE#HKNPIYGQ 878 |
| Sigmodon hispidus | Query 761 REFAKFEKEKMNAKWDTQENPIYKSPINN#FKNPNYG 796 ||+ |||||+ ++|+ +||++|| # || + Sbjct 733 REYRHFEKEKLKSQWN-NDNPLFKSATTT#VMNPKFA 767 |
| nu | Query 761 REFAKFEKEKMNAKWDTQENPIYKSPINN#FKNP 793 ||+||||+++ |+ ||||||+ |+ #++ | Sbjct 755 REYAKFEEDQKNSV--RQENPIYRDPVGR#YEVP 785 |
| Drosophila melanogaster | Query 761 REFAKFEKEKMNAKWDTQENPIYKSPINN#FKNP 793 ||+||||+++ |+ ||||||+ |+ #++ | Sbjct 755 REYAKFEEDQKNSV--RQENPIYRDPVGR#YEVP 785 |
[Site 9] PIYKSPINNF790-KNPNYGRKAG
Phe790  Lys
Lys
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Pro781 | Ile782 | Tyr783 | Lys784 | Ser785 | Pro786 | Ile787 | Asn788 | Asn789 | Phe790 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Lys791 | Asn792 | Pro793 | Asn794 | Tyr795 | Gly796 | Arg797 | Lys798 | Ala799 | Gly800 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| REFAKFEKEKMNAKWDTQENPIYKSPINNFKNPNYGRKAGL |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | Canis familiaris | 92.40 | 10 | PREDICTED: similar to integrin beta 1 isoform 1D |
| 2 | Bos taurus | 92.40 | 7 | ITGB1 protein |
| 3 | N/A | 87.80 | 27 | S66396 integrin beta-1 chain isoform D - human (f |
| 4 | Mus musculus | 87.80 | 12 | beta 1 integrin |
| 5 | Gallus gallus | 86.70 | 3 | integrin beta1D |
| 6 | Drosophila melanogaster | 64.30 | 1 | myospheroid CG1560-PA |
| 7 | Pseudoplusia includens | 63.50 | 1 | integrin beta 1 |
| 8 | Tetraodon nigroviridis | 63.20 | 5 | unnamed protein product |
| 9 | Xenopus laevis | 63.20 | 2 | integrin beta-1 subunit |
| 10 | Biomphalaria glabrata | 63.20 | 1 | beta integrin subunit |
| 11 | Anopheles gambiae | 62.40 | 2 | integrin beta subunit |
| 12 | Anopheles gambiae str. PEST | 62.40 | 1 | integrin beta subunit (AGAP000815-PA) |
| 13 | Felis catus | 62.40 | 1 | integrin beta 1 |
| 14 | Homo sapiens | 62.00 | 10 | ITB1_HUMAN Integrin beta-1 precursor (Fibronectin |
| 15 | Danio rerio | 62.00 | 5 | integrin, beta 1a |
| 16 | Sus scrofa | 62.00 | 3 | integrin beta-1 subunit |
| 17 | Pongo pygmaeus | 62.00 | 1 | ITB1_PONPY Integrin beta-1 precursor (Fibronectin |
| 18 | Ictalurus punctatus | 61.20 | 1 | beta-1 integrin |
| 19 | Oryctolagus cuniculus | 60.80 | 1 | integrin beta 1 |
| 20 | Podocoryne carnea | 59.70 | 1 | AF308652_1 integrin beta chain |
| 21 | Crassostrea gigas | 56.20 | 1 | integrin beta cgh |
| 22 | Caenorhabditis briggsae | 53.90 | 1 | Hypothetical protein CBG03601 |
| 23 | Strongylocentrotus purpuratus | 52.80 | 3 | integrin beta-C subunit |
| 24 | Gallus gallus | 52.80 | 2 | integrin beta 1A precursor |
| 25 | Lytechinus variegatus | 52.40 | 1 | beta-C integrin subunit |
| 26 | Caenorhabditis elegans | 52.00 | 1 | Paralysed Arrest at Two-fold family member (pat-3 |
| 27 | Rattus norvegicus | 49.30 | 5 | integrin beta 3 |
| 28 | Mus musculus | 49.30 | 1 | integrin beta 3 precursor |
| 29 | rats, Peptide Partial, 723 aa | 49.30 | 1 | beta 3 integrin, GPIIIA |
| 30 | Canis lupus familiaris | 49.30 | 1 | integrin beta chain, beta 3 |
| 31 | Sus scrofa | 49.30 | 1 | integrin beta chain, beta 3 |
| 32 | Oryctolagus cuniculus | 49.30 | 1 | glycoprotein IIIa |
| 33 | synthetic construct | 48.90 | 3 | AF239959_1 hybrid integrin beta 3 subunit precurs |
| 34 | mice, Peptide Partial, 680 aa | 47.80 | 1 | beta 3 integrin, GPIIIA |
| 35 | Acropora millepora | 45.40 | 1 | integrin subunit betaCn1 |
| 36 | Ovis aries | 44.70 | 2 | ITB6_SHEEP Integrin beta-6 precursor gi |
| 37 | Ophlitaspongia tenuis | 44.70 | 1 | integrin subunit betaPo1 |
| 38 | Xenopus laevis | 44.30 | 1 | hypothetical protein LOC379708 |
| 39 | Schistosoma japonicum | 43.10 | 1 | SJCHGC06221 protein |
| 40 | Pan troglodytes | 41.60 | 2 | PREDICTED: integrin, beta 7 isoform 5 |
| 41 | Anopheles gambiae str. PEST | 40.80 | 1 | AGAP010233-PA |
| 42 | Mus sp. | 40.80 | 1 | beta 7 integrin |
| 43 | Pacifastacus leniusculus | 40.80 | 1 | integrin |
| 44 | Suberites domuncula | 40.00 | 1 | integrin beta subunit |
| 45 | Halocynthia roretzi | 39.70 | 2 | integrin beta Hr2 precursor |
| 46 | Homo sapiens | 39.70 | 1 | integrin, beta 2 precursor |
| 47 | Ovis canadensis | 38.90 | 1 | ITB2_OVICA Integrin beta-2 precursor (Cell surfac |
| 48 | Geodia cydonium | 38.10 | 1 | integrin beta subunit |
| 49 | Sigmodon hispidus | 37.70 | 1 | AF445415_1 integrin beta-2 precursor |
| 50 | Drosophila melanogaster | 37.40 | 1 | integrin beta subunit |
| 51 | Macaca mulatta | 37.40 | 1 | integrin beta 1 |
| 52 | nu | 37.40 | 1 | beta |
| 53 | Drosophila pseudoobscura | 37.00 | 1 | GA14581-PA |
Top-ranked sequences
| organism | matching |
|---|---|
| Canis familiaris | Query 761 REFAKFEKEKMNAKWDTQENPIYKSPINNF#KNPNYGRKAGL 801 ||||||||||||||||||||||||||||||#||||||||||| Query 761 REFAKFEKEKMNAKWDTQENPIYKSPINNF#KNPNYGRKAGL 801 |
| Bos taurus | Query 761 REFAKFEKEKMNAKWDTQENPIYKSPINNF#KNPNYGRKAGL 801 ||||||||||||||||||||||||||||||#||||||||||| Query 761 REFAKFEKEKMNAKWDTQENPIYKSPINNF#KNPNYGRKAGL 801 |
| N/A | Query 761 REFAKFEKEKMNAKWDTQENPIYKSPINNF#KNPNYGRKAGL 801 ||||||||||||||||||||||||||||||#||||||||||| Query 761 REFAKFEKEKMNAKWDTQENPIYKSPINNF#KNPNYGRKAGL 801 |
| Mus musculus | Query 761 REFAKFEKEKMNAKWDTQENPIYKSPINNF#KNPNYGRKAGL 801 ||||||||||||||||||||||||||||||#||||||||||| Query 761 REFAKFEKEKMNAKWDTQENPIYKSPINNF#KNPNYGRKAGL 801 |
| Gallus gallus | Query 761 REFAKFEKEKMNAKWDTQENPIYKSPINNF#KNPNYGRKAGL 801 ||||||||||||||||||||||||||||||#||||||||||| Query 761 REFAKFEKEKMNAKWDTQENPIYKSPINNF#KNPNYGRKAGL 801 |
| Drosophila melanogaster | Query 761 REFAKFEKEKMNAKWDTQENPIYKSPINNF#KNPNYGRK 798 ||||+||||+||||||| |||||| + |#||| | | Sbjct 809 REFARFEKERMNAKWDTGENPIYKQATSTF#KNPMYAGK 846 |
| Pseudoplusia includens | Query 761 REFAKFEKEKMNAKWDTQENPIYKSPINNF#KNPNYGRK 798 ||||+||||+| ||||| |||||| + |#||| | | Sbjct 800 REFARFEKERMMAKWDTGENPIYKQATSTF#KNPTYAGK 837 |
| Tetraodon nigroviridis | Query 761 REFAKFEKEKMNAKWDTQENPIYKSPINNF#KNPNYGRK 798 ||||||||||||||||| ||||||| + # || | | Sbjct 834 REFAKFEKEKMNAKWDTGENPIYKSAVTTV#VNPKYEGK 871 |
| Xenopus laevis | Query 761 REFAKFEKEKMNAKWDTQENPIYKSPINNF#KNPNYGRK 798 ||||||||||||||||| ||||||| + # || | | Sbjct 761 REFAKFEKEKMNAKWDTGENPIYKSAVTTV#VNPKYEGK 798 |
| Biomphalaria glabrata | Query 761 REFAKFEKEKMNAKWDTQENPIYKSPINNF#KNPNYGRK 798 |||||||||+ |||||| |||||| + |#||| || + Sbjct 751 REFAKFEKERQNAKWDTGENPIYKQATSTF#KNPTYGSQ 788 |
| Anopheles gambiae | Query 761 REFAKFEKEKMNAKWDTQENPIYKSPINNF#KNPNYGRK 798 ||||+||||+| ||||| |||||| |#||| | | Sbjct 800 REFARFEKERMMAKWDTGENPIYKQATTTF#KNPTYAGK 837 |
| Anopheles gambiae str. PEST | Query 761 REFAKFEKEKMNAKWDTQENPIYKSPINNF#KNPNYGRK 798 ||||+||||+| ||||| |||||| |#||| | | Sbjct 800 REFARFEKERMMAKWDTGENPIYKQATTTF#KNPTYAGK 837 |
| Felis catus | Query 761 REFAKFEKEKMNAKWDTQENPIYKSPINNF#KNPNYGRK 798 ||||||||||||||||| ||||||| + # || | | Sbjct 761 REFAKFEKEKMNAKWDTGENPIYKSAVTTV#VNPKYEGK 798 |
| Homo sapiens | Query 761 REFAKFEKEKMNAKWDTQENPIYKSPINNF#KNPNYGRK 798 ||||||||||||||||| ||||||| + # || | | Sbjct 761 REFAKFEKEKMNAKWDTGENPIYKSAVTTV#VNPKYEGK 798 |
| Danio rerio | Query 761 REFAKFEKEKMNAKWDTQENPIYKSPINNF#KNPNYGRK 798 ||||||||||||||||| ||||||| + # || | | Sbjct 761 REFAKFEKEKMNAKWDTGENPIYKSAVTTV#INPKYEGK 798 |
| Sus scrofa | Query 761 REFAKFEKEKMNAKWDTQENPIYKSPINNF#KNPNYGRK 798 ||||||||||||||||| ||||||| + # || | | Sbjct 761 REFAKFEKEKMNAKWDTGENPIYKSAVTTV#VNPKYEGK 798 |
| Pongo pygmaeus | Query 761 REFAKFEKEKMNAKWDTQENPIYKSPINNF#KNPNYGRK 798 ||||||||||||||||| ||||||| + # || | | Sbjct 761 REFAKFEKEKMNAKWDTGENPIYKSAVTTV#VNPKYEGK 798 |
| Ictalurus punctatus | Query 761 REFAKFEKEKMNAKWDTQENPIYKSPINNF#KNPNYGRK 798 |||||||||||||||| ||||||| + # || | | Sbjct 770 REFAKFEKEKMNAKWDAGENPIYKSAVTTV#VNPKYEGK 807 |
| Oryctolagus cuniculus | Query 761 REFAKFEKEKMNAKWDTQENPIYKSPINNF#KNPNY 795 ||||||||||||||||| ||||||| + # || | Sbjct 26 REFAKFEKEKMNAKWDTGENPIYKSAVTTV#VNPKY 60 |
| Podocoryne carnea | Query 761 REFAKFEKEKMNAKWDTQENPIYKSPINNF#KNPNYGRK 798 ||||||||++ | |||+ |||||| + |#+|| || | Sbjct 744 REFAKFEKDRQNPKWDSGENPIYKKATSTF#QNPMYGNK 781 |
| Crassostrea gigas | Query 761 REFAKFEKEKMNAKWDTQENPIYKSPINNF#KNPNY 795 || |+|||| |||+||| |||||| + |# || | Sbjct 763 RELARFEKEAMNARWDTGENPIYKQATSTF#VNPTY 797 |
| Caenorhabditis briggsae | Query 762 EFAKFEKEKMNAKWDTQENPIYKSPINNF#KNPNYGRKA 799 |+||| |++ ||||| |||||| |#||| | || Sbjct 772 EYAKFNNERLMAKWDTNENPIYKQATTTF#KNPVYAGKA 809 |
| Strongylocentrotus purpuratus | Query 761 REFAKFEKEKMNAKWDTQENPIYKSPINNF#KNPNYGRK 798 ||| ||||+ || |+ |||||| + |#||| | | Sbjct 769 REFQNFEKERANATWEGGENPIYKPSTSVF#KNPTYNIK 806 |
| Gallus gallus | Query 761 REFAKFEKEKMNAKWDTQENPIYKSPI 787 ||||||||||||||||| ||||||| + Sbjct 18 REFAKFEKEKMNAKWDTGENPIYKSAV 44 |
| Lytechinus variegatus | Query 761 REFAKFEKEKMNAKWDTQENPIYKSPINNF#KNPNYGRK 798 ||| ||||+ || |+ |||||| + |#||| | | Sbjct 769 REFQNFEKERANATWEGGENPIYKPSTSVF#KNPTYNIK 806 |
| Caenorhabditis elegans | Query 762 EFAKFEKEKMNAKWDTQENPIYKSPINNF#KNPNYGRKA 799 |+| | |++ ||||| |||||| |#||| | || Sbjct 771 EYATFNNERLMAKWDTNENPIYKQATTTF#KNPVYAGKA 808 |
| Rattus norvegicus | Query 761 REFAKFEKEKMNAKWDTQENPIYKSPINNF#KNPNY 795 +||||||+|+ ||||| ||+|| + |# | | Sbjct 750 KEFAKFEEERARAKWDTANNPLYKEATSTF#TNITY 784 |
| Mus musculus | Query 761 REFAKFEKEKMNAKWDTQENPIYKSPINNF#KNPNY 795 +||||||+|+ ||||| ||+|| + |# | | Sbjct 750 KEFAKFEEERARAKWDTANNPLYKEATSTF#TNITY 784 |
| rats, Peptide Partial, 723 aa | Query 761 REFAKFEKEKMNAKWDTQENPIYKSPINNF#KNPNY 795 +||||||+|+ ||||| ||+|| + |# | | Sbjct 686 KEFAKFEEERARAKWDTANNPLYKEATSTF#TNITY 720 |
| Canis lupus familiaris | Query 761 REFAKFEKEKMNAKWDTQENPIYKSPINNF#KNPNY 795 +||||||+|+ ||||| ||+|| + |# | | Sbjct 747 KEFAKFEEERARAKWDTANNPLYKEATSTF#TNITY 781 |
| Sus scrofa | Query 761 REFAKFEKEKMNAKWDTQENPIYKSPINNF#KNPNY 795 +||||||+|+ ||||| ||+|| + |# | | Sbjct 747 KEFAKFEEERARAKWDTANNPLYKEATSTF#TNITY 781 |
| Oryctolagus cuniculus | Query 761 REFAKFEKEKMNAKWDTQENPIYKSPINNF#KNPNY 795 +||||||+|+ ||||| ||+|| + |# | | Sbjct 751 KEFAKFEEERARAKWDTANNPLYKEATSTF#TNITY 785 |
| synthetic construct | Query 761 REFAKFEKEKMNAKWDTQENPIYKSPINNF#KNPNY 795 +||||||+|+ ||||| ||+|| + |# | | Sbjct 751 KEFAKFEEERARAKWDTANNPLYKEATSTF#TNITY 785 |
| mice, Peptide Partial, 680 aa | Query 761 REFAKFEKEKMNAKWDTQENPIYKSPINNF#KN 792 +||||||+|+ ||||| ||+|| + |# | Sbjct 647 KEFAKFEEERARAKWDTANNPLYKEATSTF#TN 678 |
| Acropora millepora | Query 762 EFAKFEKEKMNAKWDTQENPIYKSPINNF#KNPNYG 796 |+ |||+|+|++|| ++||+|++ |#+|| | Sbjct 754 EYQKFERERMHSKWTREKNPLYQAAKTTF#ENPTYA 788 |
| Ovis aries | Query 761 REFAKFEKEKMNAKWDTQENPIYKSPINNF#KNPNYGRK 798 +| |||| |+ ||| | ||+|+ + |#|| | | Sbjct 739 KEVAKFEAERSKAKWQTGTNPLYRGSTSTF#KNVTYKHK 776 |
| Ophlitaspongia tenuis | Query 762 EFAKFEKEKMNAKWDTQENPIYKSPINNF#KNPNYGR 797 | |||+| |||+ |||+|+| ++#+|| ||+ Sbjct 803 EVRKFEREIKNAKYTKNENPLYRSATKDY#QNPLYGK 838 |
| Xenopus laevis | Query 762 EFAKFEKEKMNAKWDTQENPIYKSPINNF#KNPNY 795 |+ +||||+||+||+ ||+| + #+|||+ Sbjct 736 EYQRFEKERMNSKWNPGHNPLYHNATTTV#QNPNF 769 |
| Schistosoma japonicum | Query 761 REFAKFEKEKMNAKWDTQENPIYKSPINNF#KNPNY 795 || | |+++ | +|+ ||||++|| | # || + Sbjct 249 RELANFKQQGENMRWEMAENPIFESPTTNV#LNPTF 283 |
| Pan troglodytes | Query 761 REFAKFEKEKMNAKWDTQENPIYKSPINNF#KNPNY 795 ||+++||||+ | ||+||| | # || + Sbjct 756 REYSRFEKEQQQLNWKQDSNPLYKSAITTT#INPRF 790 |
| Anopheles gambiae str. PEST | Query 761 REFAKFEKEKMNAKWDTQENPIYKSPINNF#KNP 793 + |||||||+ |+|+|||||+||#| | Sbjct 763 KMFAKFEKEREQETKYQMESPLYKSPISNF#KVP 795 |
| Mus sp. | Query 761 REFAKFEKEKMNAKWDTQENPIYKSPINNF#KNPNY 795 ||+ +||||+ | ||+||| | # || + Sbjct 755 REYRRFEKEQQQLNWKQDNNPLYKSAITTT#VNPRF 789 |
| Pacifastacus leniusculus | Query 761 REFAKFEKEKMNAKWDTQENPIYKSPINNF#KNPNYGR 797 ||+||||||+ |||+||| |#+|| + + Sbjct 763 REYAKFEKERKLPSGKRAENPLYKSAKTTF#QNPAFAQ 799 |
| Suberites domuncula | Query 762 EFAKFEKEKMNAKWDTQENPIYKSPINNF#KNPNYGR 797 | ||||| |+ +||+|+| ++#+|| ||+ Sbjct 729 EVKKFEKELAKTKYPKHQNPLYRSAAKDY#QNPIYGQ 764 |
| Halocynthia roretzi | Query 761 REFAKFEKEKMNAKWDTQENPIYKSPINNF#KNPNYG 796 ||+ ||+||+ || |||+| + + |# || +| Sbjct 797 REYEKFKKEEDLRKWTKGENPVYVNASSKF#DNPMFG 832 |
| Homo sapiens | Query 761 REFAKFEKEKMNAKWDTQENPIYKSPINNF#KNPNYG 796 ||+ +|||||+ ++|+ +||++|| # || + Sbjct 733 REYRRFEKEKLKSQWN-NDNPLFKSATTTV#MNPKFA 767 |
| Ovis canadensis | Query 761 REFAKFEKEKMNAKWDTQENPIYKSPINNF#KNPNYG 796 ||+ +|||||+ ++|+ +||++|| # || + Sbjct 734 REYHRFEKEKLKSQWN-NDNPLFKSATTTV#MNPKFA 768 |
| Geodia cydonium | Query 762 EFAKFEKEKMNAKWDTQENPIYKSPINNF#KNPNYGR 797 | ||| | |+ |||+| | #||| ||+ Sbjct 843 ELKKFENELAKVKYTKNENPLYHSATTEH#KNPIYGQ 878 |
| Sigmodon hispidus | Query 761 REFAKFEKEKMNAKWDTQENPIYKSPINNF#KNPNYG 796 ||+ |||||+ ++|+ +||++|| # || + Sbjct 733 REYRHFEKEKLKSQWN-NDNPLFKSATTTV#MNPKFA 767 |
| Drosophila melanogaster | Query 761 REFAKFEKEKMNAKWDTQENPIYKSPINNF#KNP 793 ||+||||+++ |+ ||||||+ |+ +#+ | Sbjct 755 REYAKFEEDQKNSV--RQENPIYRDPVGRY#EVP 785 |
| Macaca mulatta | Query 761 REFAKFEKEKMNAKW 775 ||||||||||||||| Query 761 REFAKFEKEKMNAKW 775 |
| nu | Query 761 REFAKFEKEKMNAKWDTQENPIYKSPINNF#KNP 793 ||+||||+++ |+ ||||||+ |+ +#+ | Sbjct 755 REYAKFEEDQKNSV--RQENPIYRDPVGRY#EVP 785 |
| Drosophila pseudoobscura | Query 761 REFAKFEKEKMNAKWDTQENPIYKSPINNF#KNP 793 ||+|+||+|+ || +||||+|+ |+ +# | Sbjct 728 REYARFEEEQ--AKRVSQENPLYRDPVGRY#VVP 758 |
[Site 10] IYKSPINNFK791-NPNYGRKAGL
Lys791  Asn
Asn
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Ile782 | Tyr783 | Lys784 | Ser785 | Pro786 | Ile787 | Asn788 | Asn789 | Phe790 | Lys791 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Asn792 | Pro793 | Asn794 | Tyr795 | Gly796 | Arg797 | Lys798 | Ala799 | Gly800 | Leu801 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| EFAKFEKEKMNAKWDTQENPIYKSPINNFKNPNYGRKAGL |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | Canis familiaris | 90.10 | 10 | PREDICTED: similar to integrin beta 1 isoform 1D |
| 2 | Bos taurus | 89.70 | 7 | ITGB1 protein |
| 3 | N/A | 85.90 | 27 | S66396 integrin beta-1 chain isoform D - human (f |
| 4 | Mus musculus | 85.50 | 12 | beta 1 integrin |
| 5 | Gallus gallus | 84.70 | 3 | integrin beta1D |
| 6 | Drosophila melanogaster | 62.00 | 1 | myospheroid CG1560-PA |
| 7 | Tetraodon nigroviridis | 61.20 | 5 | unnamed protein product |
| 8 | Biomphalaria glabrata | 60.80 | 1 | beta integrin subunit |
| 9 | Pseudoplusia includens | 60.80 | 1 | integrin beta 1 |
| 10 | Xenopus laevis | 60.50 | 2 | integrin beta-1 subunit |
| 11 | Anopheles gambiae | 60.10 | 2 | integrin beta subunit |
| 12 | Anopheles gambiae str. PEST | 60.10 | 1 | integrin beta subunit (AGAP000815-PA) |
| 13 | Danio rerio | 59.70 | 5 | integrin, beta 1a |
| 14 | Sus scrofa | 59.70 | 3 | integrin beta-1 subunit |
| 15 | Felis catus | 59.70 | 1 | integrin beta 1 |
| 16 | Homo sapiens | 59.30 | 10 | ITB1_HUMAN Integrin beta-1 precursor (Fibronectin |
| 17 | Pongo pygmaeus | 59.30 | 1 | ITB1_PONPY Integrin beta-1 precursor (Fibronectin |
| 18 | Ictalurus punctatus | 58.90 | 1 | beta-1 integrin |
| 19 | Oryctolagus cuniculus | 58.20 | 1 | integrin beta 1 |
| 20 | Podocoryne carnea | 57.40 | 1 | AF308652_1 integrin beta chain |
| 21 | Caenorhabditis briggsae | 53.90 | 1 | Hypothetical protein CBG03601 |
| 22 | Crassostrea gigas | 53.90 | 1 | integrin beta cgh |
| 23 | Caenorhabditis elegans | 52.00 | 1 | Paralysed Arrest at Two-fold family member (pat-3 |
| 24 | Gallus gallus | 50.80 | 2 | integrin beta 1A precursor |
| 25 | Strongylocentrotus purpuratus | 50.10 | 3 | integrin beta-C subunit |
| 26 | Lytechinus variegatus | 50.10 | 1 | beta-C integrin subunit |
| 27 | Rattus norvegicus | 48.50 | 5 | integrin beta 3 |
| 28 | Mus musculus | 48.50 | 1 | integrin beta 3 precursor |
| 29 | Oryctolagus cuniculus | 48.50 | 1 | glycoprotein IIIa |
| 30 | Sus scrofa | 48.50 | 1 | integrin beta chain, beta 3 |
| 31 | Canis lupus familiaris | 48.50 | 1 | integrin beta chain, beta 3 |
| 32 | rats, Peptide Partial, 723 aa | 48.50 | 1 | beta 3 integrin, GPIIIA |
| 33 | synthetic construct | 48.10 | 3 | AF239959_1 hybrid integrin beta 3 subunit precurs |
| 34 | mice, Peptide Partial, 680 aa | 47.00 | 1 | beta 3 integrin, GPIIIA |
| 35 | Acropora millepora | 45.10 | 1 | integrin subunit betaCn1 |
| 36 | Ophlitaspongia tenuis | 44.70 | 1 | integrin subunit betaPo1 |
| 37 | Xenopus laevis | 44.30 | 1 | hypothetical protein LOC379708 |
| 38 | Ovis aries | 43.90 | 2 | ITB6_SHEEP Integrin beta-6 precursor gi |
| 39 | Anopheles gambiae str. PEST | 40.80 | 1 | AGAP010233-PA |
| 40 | Schistosoma japonicum | 40.80 | 1 | SJCHGC06221 protein |
| 41 | Suberites domuncula | 40.00 | 1 | integrin beta subunit |
| 42 | Pan troglodytes | 39.30 | 2 | PREDICTED: integrin, beta 7 isoform 5 |
| 43 | Mus sp. | 38.50 | 1 | beta 7 integrin |
| 44 | Pacifastacus leniusculus | 38.50 | 1 | integrin |
| 45 | Halocynthia roretzi | 38.10 | 2 | integrin beta Hr1 precursor |
| 46 | Geodia cydonium | 38.10 | 1 | integrin beta subunit |
| 47 | Homo sapiens | 37.40 | 1 | integrin, beta 2 precursor |
| 48 | Ovis canadensis | 37.00 | 1 | ITB2_OVICA Integrin beta-2 precursor (Cell surfac |
| 49 | Sigmodon hispidus | 35.40 | 1 | AF445415_1 integrin beta-2 precursor |
| 50 | nu | 35.00 | 1 | beta |
| 51 | Drosophila melanogaster | 35.00 | 1 | integrin beta subunit |
| 52 | Macaca mulatta | 34.70 | 1 | integrin beta 1 |
| 53 | Drosophila pseudoobscura | 34.70 | 1 | GA14581-PA |
Top-ranked sequences
| organism | matching |
|---|---|
| Canis familiaris | Query 762 EFAKFEKEKMNAKWDTQENPIYKSPINNFK#NPNYGRKAGL 801 ||||||||||||||||||||||||||||||#|||||||||| Query 762 EFAKFEKEKMNAKWDTQENPIYKSPINNFK#NPNYGRKAGL 801 |
| Bos taurus | Query 762 EFAKFEKEKMNAKWDTQENPIYKSPINNFK#NPNYGRKAGL 801 ||||||||||||||||||||||||||||||#|||||||||| Query 762 EFAKFEKEKMNAKWDTQENPIYKSPINNFK#NPNYGRKAGL 801 |
| N/A | Query 762 EFAKFEKEKMNAKWDTQENPIYKSPINNFK#NPNYGRKAGL 801 ||||||||||||||||||||||||||||||#|||||||||| Query 762 EFAKFEKEKMNAKWDTQENPIYKSPINNFK#NPNYGRKAGL 801 |
| Mus musculus | Query 762 EFAKFEKEKMNAKWDTQENPIYKSPINNFK#NPNYGRKAGL 801 ||||||||||||||||||||||||||||||#|||||||||| Query 762 EFAKFEKEKMNAKWDTQENPIYKSPINNFK#NPNYGRKAGL 801 |
| Gallus gallus | Query 762 EFAKFEKEKMNAKWDTQENPIYKSPINNFK#NPNYGRKAGL 801 ||||||||||||||||||||||||||||||#|||||||||| Query 762 EFAKFEKEKMNAKWDTQENPIYKSPINNFK#NPNYGRKAGL 801 |
| Drosophila melanogaster | Query 762 EFAKFEKEKMNAKWDTQENPIYKSPINNFK#NPNYGRK 798 |||+||||+||||||| |||||| + ||#|| | | Sbjct 810 EFARFEKERMNAKWDTGENPIYKQATSTFK#NPMYAGK 846 |
| Tetraodon nigroviridis | Query 762 EFAKFEKEKMNAKWDTQENPIYKSPINNFK#NPNYGRK 798 |||||||||||||||| ||||||| + #|| | | Sbjct 835 EFAKFEKEKMNAKWDTGENPIYKSAVTTVV#NPKYEGK 871 |
| Biomphalaria glabrata | Query 762 EFAKFEKEKMNAKWDTQENPIYKSPINNFK#NPNYGRK 798 ||||||||+ |||||| |||||| + ||#|| || + Sbjct 752 EFAKFEKERQNAKWDTGENPIYKQATSTFK#NPTYGSQ 788 |
| Pseudoplusia includens | Query 762 EFAKFEKEKMNAKWDTQENPIYKSPINNFK#NPNYGRK 798 |||+||||+| ||||| |||||| + ||#|| | | Sbjct 801 EFARFEKERMMAKWDTGENPIYKQATSTFK#NPTYAGK 837 |
| Xenopus laevis | Query 762 EFAKFEKEKMNAKWDTQENPIYKSPINNFK#NPNYGRK 798 |||||||||||||||| ||||||| + #|| | | Sbjct 762 EFAKFEKEKMNAKWDTGENPIYKSAVTTVV#NPKYEGK 798 |
| Anopheles gambiae | Query 762 EFAKFEKEKMNAKWDTQENPIYKSPINNFK#NPNYGRK 798 |||+||||+| ||||| |||||| ||#|| | | Sbjct 801 EFARFEKERMMAKWDTGENPIYKQATTTFK#NPTYAGK 837 |
| Anopheles gambiae str. PEST | Query 762 EFAKFEKEKMNAKWDTQENPIYKSPINNFK#NPNYGRK 798 |||+||||+| ||||| |||||| ||#|| | | Sbjct 801 EFARFEKERMMAKWDTGENPIYKQATTTFK#NPTYAGK 837 |
| Danio rerio | Query 762 EFAKFEKEKMNAKWDTQENPIYKSPINNFK#NPNYGRK 798 |||||||||||||||| ||||||| + #|| | | Sbjct 762 EFAKFEKEKMNAKWDTGENPIYKSAVTTVI#NPKYEGK 798 |
| Sus scrofa | Query 762 EFAKFEKEKMNAKWDTQENPIYKSPINNFK#NPNYGRK 798 |||||||||||||||| ||||||| + #|| | | Sbjct 762 EFAKFEKEKMNAKWDTGENPIYKSAVTTVV#NPKYEGK 798 |
| Felis catus | Query 762 EFAKFEKEKMNAKWDTQENPIYKSPINNFK#NPNYGRK 798 |||||||||||||||| ||||||| + #|| | | Sbjct 762 EFAKFEKEKMNAKWDTGENPIYKSAVTTVV#NPKYEGK 798 |
| Homo sapiens | Query 762 EFAKFEKEKMNAKWDTQENPIYKSPINNFK#NPNYGRK 798 |||||||||||||||| ||||||| + #|| | | Sbjct 762 EFAKFEKEKMNAKWDTGENPIYKSAVTTVV#NPKYEGK 798 |
| Pongo pygmaeus | Query 762 EFAKFEKEKMNAKWDTQENPIYKSPINNFK#NPNYGRK 798 |||||||||||||||| ||||||| + #|| | | Sbjct 762 EFAKFEKEKMNAKWDTGENPIYKSAVTTVV#NPKYEGK 798 |
| Ictalurus punctatus | Query 762 EFAKFEKEKMNAKWDTQENPIYKSPINNFK#NPNYGRK 798 ||||||||||||||| ||||||| + #|| | | Sbjct 771 EFAKFEKEKMNAKWDAGENPIYKSAVTTVV#NPKYEGK 807 |
| Oryctolagus cuniculus | Query 762 EFAKFEKEKMNAKWDTQENPIYKSPINNFK#NPNY 795 |||||||||||||||| ||||||| + #|| | Sbjct 27 EFAKFEKEKMNAKWDTGENPIYKSAVTTVV#NPKY 60 |
| Podocoryne carnea | Query 762 EFAKFEKEKMNAKWDTQENPIYKSPINNFK#NPNYGRK 798 |||||||++ | |||+ |||||| + |+#|| || | Sbjct 745 EFAKFEKDRQNPKWDSGENPIYKKATSTFQ#NPMYGNK 781 |
| Caenorhabditis briggsae | Query 762 EFAKFEKEKMNAKWDTQENPIYKSPINNFK#NPNYGRKA 799 |+||| |++ ||||| |||||| ||#|| | || Sbjct 772 EYAKFNNERLMAKWDTNENPIYKQATTTFK#NPVYAGKA 809 |
| Crassostrea gigas | Query 762 EFAKFEKEKMNAKWDTQENPIYKSPINNFK#NPNY 795 | |+|||| |||+||| |||||| + | #|| | Sbjct 764 ELARFEKEAMNARWDTGENPIYKQATSTFV#NPTY 797 |
| Caenorhabditis elegans | Query 762 EFAKFEKEKMNAKWDTQENPIYKSPINNFK#NPNYGRKA 799 |+| | |++ ||||| |||||| ||#|| | || Sbjct 771 EYATFNNERLMAKWDTNENPIYKQATTTFK#NPVYAGKA 808 |
| Gallus gallus | Query 762 EFAKFEKEKMNAKWDTQENPIYKSPI 787 |||||||||||||||| ||||||| + Sbjct 19 EFAKFEKEKMNAKWDTGENPIYKSAV 44 |
| Strongylocentrotus purpuratus | Query 762 EFAKFEKEKMNAKWDTQENPIYKSPINNFK#NPNYGRK 798 || ||||+ || |+ |||||| + ||#|| | | Sbjct 770 EFQNFEKERANATWEGGENPIYKPSTSVFK#NPTYNIK 806 |
| Lytechinus variegatus | Query 762 EFAKFEKEKMNAKWDTQENPIYKSPINNFK#NPNYGRK 798 || ||||+ || |+ |||||| + ||#|| | | Sbjct 770 EFQNFEKERANATWEGGENPIYKPSTSVFK#NPTYNIK 806 |
| Rattus norvegicus | Query 762 EFAKFEKEKMNAKWDTQENPIYKSPINNFK#NPNY 795 ||||||+|+ ||||| ||+|| + | #| | Sbjct 751 EFAKFEEERARAKWDTANNPLYKEATSTFT#NITY 784 |
| Mus musculus | Query 762 EFAKFEKEKMNAKWDTQENPIYKSPINNFK#NPNY 795 ||||||+|+ ||||| ||+|| + | #| | Sbjct 751 EFAKFEEERARAKWDTANNPLYKEATSTFT#NITY 784 |
| Oryctolagus cuniculus | Query 762 EFAKFEKEKMNAKWDTQENPIYKSPINNFK#NPNY 795 ||||||+|+ ||||| ||+|| + | #| | Sbjct 752 EFAKFEEERARAKWDTANNPLYKEATSTFT#NITY 785 |
| Sus scrofa | Query 762 EFAKFEKEKMNAKWDTQENPIYKSPINNFK#NPNY 795 ||||||+|+ ||||| ||+|| + | #| | Sbjct 748 EFAKFEEERARAKWDTANNPLYKEATSTFT#NITY 781 |
| Canis lupus familiaris | Query 762 EFAKFEKEKMNAKWDTQENPIYKSPINNFK#NPNY 795 ||||||+|+ ||||| ||+|| + | #| | Sbjct 748 EFAKFEEERARAKWDTANNPLYKEATSTFT#NITY 781 |
| rats, Peptide Partial, 723 aa | Query 762 EFAKFEKEKMNAKWDTQENPIYKSPINNFK#NPNY 795 ||||||+|+ ||||| ||+|| + | #| | Sbjct 687 EFAKFEEERARAKWDTANNPLYKEATSTFT#NITY 720 |
| synthetic construct | Query 762 EFAKFEKEKMNAKWDTQENPIYKSPINNFK#NPNY 795 ||||||+|+ ||||| ||+|| + | #| | Sbjct 752 EFAKFEEERARAKWDTANNPLYKEATSTFT#NITY 785 |
| mice, Peptide Partial, 680 aa | Query 762 EFAKFEKEKMNAKWDTQENPIYKSPINNFK#N 792 ||||||+|+ ||||| ||+|| + | #| Sbjct 648 EFAKFEEERARAKWDTANNPLYKEATSTFT#N 678 |
| Acropora millepora | Query 762 EFAKFEKEKMNAKWDTQENPIYKSPINNFK#NPNYG 796 |+ |||+|+|++|| ++||+|++ |+#|| | Sbjct 754 EYQKFERERMHSKWTREKNPLYQAAKTTFE#NPTYA 788 |
| Ophlitaspongia tenuis | Query 762 EFAKFEKEKMNAKWDTQENPIYKSPINNFK#NPNYGR 797 | |||+| |||+ |||+|+| +++#|| ||+ Sbjct 803 EVRKFEREIKNAKYTKNENPLYRSATKDYQ#NPLYGK 838 |
| Xenopus laevis | Query 762 EFAKFEKEKMNAKWDTQENPIYKSPINNFK#NPNY 795 |+ +||||+||+||+ ||+| + +#|||+ Sbjct 736 EYQRFEKERMNSKWNPGHNPLYHNATTTVQ#NPNF 769 |
| Ovis aries | Query 762 EFAKFEKEKMNAKWDTQENPIYKSPINNFK#NPNYGRK 798 | |||| |+ ||| | ||+|+ + ||#| | | Sbjct 740 EVAKFEAERSKAKWQTGTNPLYRGSTSTFK#NVTYKHK 776 |
| Anopheles gambiae str. PEST | Query 763 FAKFEKEKMNAKWDTQENPIYKSPINNFK#NP 793 |||||||+ |+|+|||||+|||# | Sbjct 765 FAKFEKEREQETKYQMESPLYKSPISNFK#VP 795 |
| Schistosoma japonicum | Query 762 EFAKFEKEKMNAKWDTQENPIYKSPINNFK#NPNY 795 | | |+++ | +|+ ||||++|| | #|| + Sbjct 250 ELANFKQQGENMRWEMAENPIFESPTTNVL#NPTF 283 |
| Suberites domuncula | Query 762 EFAKFEKEKMNAKWDTQENPIYKSPINNFK#NPNYGR 797 | ||||| |+ +||+|+| +++#|| ||+ Sbjct 729 EVKKFEKELAKTKYPKHQNPLYRSAAKDYQ#NPIYGQ 764 |
| Pan troglodytes | Query 762 EFAKFEKEKMNAKWDTQENPIYKSPINNFK#NPNY 795 |+++||||+ | ||+||| | #|| + Sbjct 757 EYSRFEKEQQQLNWKQDSNPLYKSAITTTI#NPRF 790 |
| Mus sp. | Query 762 EFAKFEKEKMNAKWDTQENPIYKSPINNFK#NPNY 795 |+ +||||+ | ||+||| | #|| + Sbjct 756 EYRRFEKEQQQLNWKQDNNPLYKSAITTTV#NPRF 789 |
| Pacifastacus leniusculus | Query 762 EFAKFEKEKMNAKWDTQENPIYKSPINNFK#NPNYGR 797 |+||||||+ |||+||| |+#|| + + Sbjct 764 EYAKFEKERKLPSGKRAENPLYKSAKTTFQ#NPAFAQ 799 |
| Halocynthia roretzi | Query 762 EFAKFEKEKMNAKWDTQENPIYKSPINNFK#NPNYG 796 |+ ||+| ++| ||++ +| |+#||+|| Sbjct 815 EWENFEREMKQSRWTKDINPVFVNPSAKFE#NPSYG 849 |
| Geodia cydonium | Query 762 EFAKFEKEKMNAKWDTQENPIYKSPINNFK#NPNYGR 797 | ||| | |+ |||+| | |#|| ||+ Sbjct 843 ELKKFENELAKVKYTKNENPLYHSATTEHK#NPIYGQ 878 |
| Homo sapiens | Query 762 EFAKFEKEKMNAKWDTQENPIYKSPINNFK#NPNYG 796 |+ +|||||+ ++|+ +||++|| #|| + Sbjct 734 EYRRFEKEKLKSQWN-NDNPLFKSATTTVM#NPKFA 767 |
| Ovis canadensis | Query 762 EFAKFEKEKMNAKWDTQENPIYKSPINNFK#NPNYG 796 |+ +|||||+ ++|+ +||++|| #|| + Sbjct 735 EYHRFEKEKLKSQWN-NDNPLFKSATTTVM#NPKFA 768 |
| Sigmodon hispidus | Query 762 EFAKFEKEKMNAKWDTQENPIYKSPINNFK#NPNYG 796 |+ |||||+ ++|+ +||++|| #|| + Sbjct 734 EYRHFEKEKLKSQWN-NDNPLFKSATTTVM#NPKFA 767 |
| nu | Query 762 EFAKFEKEKMNAKWDTQENPIYKSPINNFK#NP 793 |+||||+++ |+ ||||||+ |+ ++# | Sbjct 756 EYAKFEEDQKNSV--RQENPIYRDPVGRYE#VP 785 |
| Drosophila melanogaster | Query 762 EFAKFEKEKMNAKWDTQENPIYKSPINNFK#NP 793 |+||||+++ |+ ||||||+ |+ ++# | Sbjct 756 EYAKFEEDQKNSV--RQENPIYRDPVGRYE#VP 785 |
| Macaca mulatta | Query 762 EFAKFEKEKMNAKW 775 |||||||||||||| Query 762 EFAKFEKEKMNAKW 775 |
| Drosophila pseudoobscura | Query 762 EFAKFEKEKMNAKWDTQENPIYKSPINNFK#NP 793 |+|+||+|+ || +||||+|+ |+ + # | Sbjct 729 EYARFEEEQ--AKRVSQENPLYRDPVGRYV#VP 758 |
[Site 11] PINNFKNPNY795-GRKAGL
Tyr795  Gly
Gly
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Pro786 | Ile787 | Asn788 | Asn789 | Phe790 | Lys791 | Asn792 | Pro793 | Asn794 | Tyr795 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Gly796 | Arg797 | Lys798 | Ala799 | Gly800 | Leu801 | - | - | - | - |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| FEKEKMNAKWDTQENPIYKSPINNFKNPNYGRKAGL |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | Canis familiaris | 81.60 | 9 | PREDICTED: similar to integrin beta 1 isoform 1D |
| 2 | Bos taurus | 81.30 | 5 | ITGB1 protein |
| 3 | N/A | 77.80 | 22 | S66396 integrin beta-1 chain isoform D - human (f |
| 4 | Mus musculus | 77.80 | 9 | beta 1 integrin |
| 5 | Gallus gallus | 77.40 | 3 | integrin beta1D |
| 6 | Drosophila melanogaster | 54.70 | 1 | myospheroid CG1560-PA |
| 7 | Pseudoplusia includens | 53.50 | 1 | integrin beta 1 |
| 8 | Biomphalaria glabrata | 53.50 | 1 | beta integrin subunit |
| 9 | Tetraodon nigroviridis | 52.80 | 5 | unnamed protein product |
| 10 | Xenopus laevis | 52.80 | 2 | integrin beta-1 subunit |
| 11 | Anopheles gambiae | 52.80 | 2 | integrin beta subunit |
| 12 | Anopheles gambiae str. PEST | 52.80 | 1 | integrin beta subunit (AGAP000815-PA) |
| 13 | Homo sapiens | 51.60 | 9 | ITB1_HUMAN Integrin beta-1 precursor (Fibronectin |
| 14 | Danio rerio | 51.60 | 5 | integrin, beta 1a |
| 15 | Sus scrofa | 51.60 | 3 | integrin beta-1 subunit |
| 16 | Felis catus | 51.60 | 1 | integrin beta 1 |
| 17 | Pongo pygmaeus | 51.60 | 1 | ITB1_PONPY Integrin beta-1 precursor (Fibronectin |
| 18 | Oryctolagus cuniculus | 50.80 | 1 | integrin beta 1 |
| 19 | Ictalurus punctatus | 50.40 | 1 | beta-1 integrin |
| 20 | Podocoryne carnea | 49.30 | 1 | AF308652_1 integrin beta chain |
| 21 | Crassostrea gigas | 48.90 | 1 | integrin beta cgh |
| 22 | Caenorhabditis briggsae | 47.40 | 1 | Hypothetical protein CBG03601 |
| 23 | Caenorhabditis elegans | 47.40 | 1 | Paralysed Arrest at Two-fold family member (pat-3 |
| 24 | Strongylocentrotus purpuratus | 45.80 | 3 | integrin beta-C subunit |
| 25 | Lytechinus variegatus | 45.40 | 1 | beta-C integrin subunit |
| 26 | Gallus gallus | 43.50 | 2 | integrin beta 1A precursor |
| 27 | Ophlitaspongia tenuis | 41.60 | 1 | integrin subunit betaPo1 |
| 28 | Xenopus laevis | 40.40 | 1 | hypothetical protein LOC379708 |
| 29 | Canis lupus familiaris | 40.40 | 1 | integrin beta chain, beta 3 |
| 30 | rats, Peptide Partial, 723 aa | 40.40 | 1 | beta 3 integrin, GPIIIA |
| 31 | Oryctolagus cuniculus | 40.40 | 1 | glycoprotein IIIa |
| 32 | Acropora millepora | 40.40 | 1 | integrin subunit betaCn1 |
| 33 | Rattus norvegicus | 40.00 | 5 | integrin beta 3 |
| 34 | Sus scrofa | 40.00 | 1 | integrin beta chain, beta 3 |
| 35 | Mus musculus | 40.00 | 1 | integrin beta 3 precursor |
| 36 | synthetic construct | 39.70 | 2 | AF239959_1 hybrid integrin beta 3 subunit precurs |
| 37 | Ovis aries | 38.90 | 2 | ITB6_SHEEP Integrin beta-6 precursor gi |
| 38 | mice, Peptide Partial, 680 aa | 38.50 | 1 | beta 3 integrin, GPIIIA |
| 39 | Schistosoma japonicum | 37.00 | 1 | SJCHGC06221 protein |
| 40 | Suberites domuncula | 36.60 | 1 | integrin beta subunit |
| 41 | Halocynthia roretzi | 35.80 | 2 | integrin beta Hr1 precursor |
| 42 | Pan troglodytes | 34.70 | 2 | PREDICTED: integrin, beta 7 isoform 5 |
| 43 | Anopheles gambiae str. PEST | 34.70 | 1 | AGAP010233-PA |
| 44 | Mus sp. | 34.70 | 1 | beta 7 integrin |
| 45 | Geodia cydonium | 34.30 | 1 | integrin beta subunit |
| 46 | Homo sapiens | 33.50 | 1 | integrin, beta 2 precursor |
| 47 | Ovis canadensis | 33.10 | 1 | ITB2_OVICA Integrin beta-2 precursor (Cell surfac |
| 48 | Sigmodon hispidus | 33.10 | 1 | AF445415_1 integrin beta-2 precursor |
Top-ranked sequences
| organism | matching |
|---|---|
| Canis familiaris | Query 766 FEKEKMNAKWDTQENPIYKSPINNFKNPNY#GRKAGL 801 ||||||||||||||||||||||||||||||#|||||| Query 766 FEKEKMNAKWDTQENPIYKSPINNFKNPNY#GRKAGL 801 |
| Bos taurus | Query 766 FEKEKMNAKWDTQENPIYKSPINNFKNPNY#GRKAGL 801 ||||||||||||||||||||||||||||||#|||||| Query 766 FEKEKMNAKWDTQENPIYKSPINNFKNPNY#GRKAGL 801 |
| N/A | Query 766 FEKEKMNAKWDTQENPIYKSPINNFKNPNY#GRKAGL 801 ||||||||||||||||||||||||||||||#|||||| Query 766 FEKEKMNAKWDTQENPIYKSPINNFKNPNY#GRKAGL 801 |
| Mus musculus | Query 766 FEKEKMNAKWDTQENPIYKSPINNFKNPNY#GRKAGL 801 ||||||||||||||||||||||||||||||#|||||| Query 766 FEKEKMNAKWDTQENPIYKSPINNFKNPNY#GRKAGL 801 |
| Gallus gallus | Query 766 FEKEKMNAKWDTQENPIYKSPINNFKNPNY#GRKAGL 801 ||||||||||||||||||||||||||||||#|||||| Query 766 FEKEKMNAKWDTQENPIYKSPINNFKNPNY#GRKAGL 801 |
| Drosophila melanogaster | Query 766 FEKEKMNAKWDTQENPIYKSPINNFKNPNY#GRK 798 ||||+||||||| |||||| + |||| |# | Sbjct 814 FEKERMNAKWDTGENPIYKQATSTFKNPMY#AGK 846 |
| Pseudoplusia includens | Query 766 FEKEKMNAKWDTQENPIYKSPINNFKNPNY#GRK 798 ||||+| ||||| |||||| + |||| |# | Sbjct 805 FEKERMMAKWDTGENPIYKQATSTFKNPTY#AGK 837 |
| Biomphalaria glabrata | Query 766 FEKEKMNAKWDTQENPIYKSPINNFKNPNY#GRK 798 ||||+ |||||| |||||| + |||| |#| + Sbjct 756 FEKERQNAKWDTGENPIYKQATSTFKNPTY#GSQ 788 |
| Tetraodon nigroviridis | Query 766 FEKEKMNAKWDTQENPIYKSPINNFKNPNY#GRK 798 |||||||||||| ||||||| + || |# | Sbjct 839 FEKEKMNAKWDTGENPIYKSAVTTVVNPKY#EGK 871 |
| Xenopus laevis | Query 766 FEKEKMNAKWDTQENPIYKSPINNFKNPNY#GRK 798 |||||||||||| ||||||| + || |# | Sbjct 766 FEKEKMNAKWDTGENPIYKSAVTTVVNPKY#EGK 798 |
| Anopheles gambiae | Query 766 FEKEKMNAKWDTQENPIYKSPINNFKNPNY#GRK 798 ||||+| ||||| |||||| |||| |# | Sbjct 805 FEKERMMAKWDTGENPIYKQATTTFKNPTY#AGK 837 |
| Anopheles gambiae str. PEST | Query 766 FEKEKMNAKWDTQENPIYKSPINNFKNPNY#GRK 798 ||||+| ||||| |||||| |||| |# | Sbjct 805 FEKERMMAKWDTGENPIYKQATTTFKNPTY#AGK 837 |
| Homo sapiens | Query 766 FEKEKMNAKWDTQENPIYKSPINNFKNPNY#GRK 798 |||||||||||| ||||||| + || |# | Sbjct 766 FEKEKMNAKWDTGENPIYKSAVTTVVNPKY#EGK 798 |
| Danio rerio | Query 766 FEKEKMNAKWDTQENPIYKSPINNFKNPNY#GRK 798 |||||||||||| ||||||| + || |# | Sbjct 766 FEKEKMNAKWDTGENPIYKSAVTTVINPKY#EGK 798 |
| Sus scrofa | Query 766 FEKEKMNAKWDTQENPIYKSPINNFKNPNY#GRK 798 |||||||||||| ||||||| + || |# | Sbjct 766 FEKEKMNAKWDTGENPIYKSAVTTVVNPKY#EGK 798 |
| Felis catus | Query 766 FEKEKMNAKWDTQENPIYKSPINNFKNPNY#GRK 798 |||||||||||| ||||||| + || |# | Sbjct 766 FEKEKMNAKWDTGENPIYKSAVTTVVNPKY#EGK 798 |
| Pongo pygmaeus | Query 766 FEKEKMNAKWDTQENPIYKSPINNFKNPNY#GRK 798 |||||||||||| ||||||| + || |# | Sbjct 766 FEKEKMNAKWDTGENPIYKSAVTTVVNPKY#EGK 798 |
| Oryctolagus cuniculus | Query 766 FEKEKMNAKWDTQENPIYKSPINNFKNPNY# 795 |||||||||||| ||||||| + || |# Sbjct 31 FEKEKMNAKWDTGENPIYKSAVTTVVNPKY# 60 |
| Ictalurus punctatus | Query 766 FEKEKMNAKWDTQENPIYKSPINNFKNPNY#GRK 798 ||||||||||| ||||||| + || |# | Sbjct 775 FEKEKMNAKWDAGENPIYKSAVTTVVNPKY#EGK 807 |
| Podocoryne carnea | Query 766 FEKEKMNAKWDTQENPIYKSPINNFKNPNY#GRK 798 |||++ | |||+ |||||| + |+|| |#| | Sbjct 749 FEKDRQNPKWDSGENPIYKKATSTFQNPMY#GNK 781 |
| Crassostrea gigas | Query 766 FEKEKMNAKWDTQENPIYKSPINNFKNPNY# 795 |||| |||+||| |||||| + | || |# Sbjct 768 FEKEAMNARWDTGENPIYKQATSTFVNPTY# 797 |
| Caenorhabditis briggsae | Query 766 FEKEKMNAKWDTQENPIYKSPINNFKNPNY#GRKA 799 | |++ ||||| |||||| |||| |# || Sbjct 776 FNNERLMAKWDTNENPIYKQATTTFKNPVY#AGKA 809 |
| Caenorhabditis elegans | Query 766 FEKEKMNAKWDTQENPIYKSPINNFKNPNY#GRKA 799 | |++ ||||| |||||| |||| |# || Sbjct 775 FNNERLMAKWDTNENPIYKQATTTFKNPVY#AGKA 808 |
| Strongylocentrotus purpuratus | Query 766 FEKEKMNAKWDTQENPIYKSPINNFKNPNY#GRK 798 ||||+ || |+ |||||| + |||| |# | Sbjct 774 FEKERANATWEGGENPIYKPSTSVFKNPTY#NIK 806 |
| Lytechinus variegatus | Query 766 FEKEKMNAKWDTQENPIYKSPINNFKNPNY#GRK 798 ||||+ || |+ |||||| + |||| |# | Sbjct 774 FEKERANATWEGGENPIYKPSTSVFKNPTY#NIK 806 |
| Gallus gallus | Query 766 FEKEKMNAKWDTQENPIYKSPI 787 |||||||||||| ||||||| + Sbjct 23 FEKEKMNAKWDTGENPIYKSAV 44 |
| Ophlitaspongia tenuis | Query 766 FEKEKMNAKWDTQENPIYKSPINNFKNPNY#GR 797 ||+| |||+ |||+|+| +++|| |#|+ Sbjct 807 FEREIKNAKYTKNENPLYRSATKDYQNPLY#GK 838 |
| Xenopus laevis | Query 766 FEKEKMNAKWDTQENPIYKSPINNFKNPNY# 795 ||||+||+||+ ||+| + +|||+# Sbjct 740 FEKERMNSKWNPGHNPLYHNATTTVQNPNF# 769 |
| Canis lupus familiaris | Query 766 FEKEKMNAKWDTQENPIYKSPINNFKNPNY# 795 ||+|+ ||||| ||+|| + | | |# Sbjct 752 FEEERARAKWDTANNPLYKEATSTFTNITY# 781 |
| rats, Peptide Partial, 723 aa | Query 766 FEKEKMNAKWDTQENPIYKSPINNFKNPNY# 795 ||+|+ ||||| ||+|| + | | |# Sbjct 691 FEEERARAKWDTANNPLYKEATSTFTNITY# 720 |
| Oryctolagus cuniculus | Query 766 FEKEKMNAKWDTQENPIYKSPINNFKNPNY# 795 ||+|+ ||||| ||+|| + | | |# Sbjct 756 FEEERARAKWDTANNPLYKEATSTFTNITY# 785 |
| Acropora millepora | Query 766 FEKEKMNAKWDTQENPIYKSPINNFKNPNY#G 796 ||+|+|++|| ++||+|++ |+|| |# Sbjct 758 FERERMHSKWTREKNPLYQAAKTTFENPTY#A 788 |
| Rattus norvegicus | Query 766 FEKEKMNAKWDTQENPIYKSPINNFKNPNY# 795 ||+|+ ||||| ||+|| + | | |# Sbjct 755 FEEERARAKWDTANNPLYKEATSTFTNITY# 784 |
| Sus scrofa | Query 766 FEKEKMNAKWDTQENPIYKSPINNFKNPNY# 795 ||+|+ ||||| ||+|| + | | |# Sbjct 752 FEEERARAKWDTANNPLYKEATSTFTNITY# 781 |
| Mus musculus | Query 766 FEKEKMNAKWDTQENPIYKSPINNFKNPNY# 795 ||+|+ ||||| ||+|| + | | |# Sbjct 755 FEEERARAKWDTANNPLYKEATSTFTNITY# 784 |
| synthetic construct | Query 766 FEKEKMNAKWDTQENPIYKSPINNFKNPNY# 795 ||+|+ ||||| ||+|| + | | |# Sbjct 756 FEEERARAKWDTANNPLYKEATSTFTNITY# 785 |
| Ovis aries | Query 766 FEKEKMNAKWDTQENPIYKSPINNFKNPNY#GRK 798 || |+ ||| | ||+|+ + ||| |# | Sbjct 744 FEAERSKAKWQTGTNPLYRGSTSTFKNVTY#KHK 776 |
| mice, Peptide Partial, 680 aa | Query 766 FEKEKMNAKWDTQENPIYKSPINNFKN 792 ||+|+ ||||| ||+|| + | | Sbjct 652 FEEERARAKWDTANNPLYKEATSTFTN 678 |
| Schistosoma japonicum | Query 766 FEKEKMNAKWDTQENPIYKSPINNFKNPNY# 795 |+++ | +|+ ||||++|| | || +# Sbjct 254 FKQQGENMRWEMAENPIFESPTTNVLNPTF# 283 |
| Suberites domuncula | Query 766 FEKEKMNAKWDTQENPIYKSPINNFKNPNY#GR 797 |||| |+ +||+|+| +++|| |#|+ Sbjct 733 FEKELAKTKYPKHQNPLYRSAAKDYQNPIY#GQ 764 |
| Halocynthia roretzi | Query 766 FEKEKMNAKWDTQENPIYKSPINNFKNPNY#G 796 ||+| ++| ||++ +| |+||+|#| Sbjct 819 FEREMKQSRWTKDINPVFVNPSAKFENPSY#G 849 |
| Pan troglodytes | Query 766 FEKEKMNAKWDTQENPIYKSPINNFKNPNY# 795 ||||+ | ||+||| | || +# Sbjct 761 FEKEQQQLNWKQDSNPLYKSAITTTINPRF# 790 |
| Anopheles gambiae str. PEST | Query 766 FEKEKMNAKWDTQENPIYKSPINNFKNP 793 ||||+ |+|+|||||+||| | Sbjct 768 FEKEREQETKYQMESPLYKSPISNFKVP 795 |
| Mus sp. | Query 766 FEKEKMNAKWDTQENPIYKSPINNFKNPNY# 795 ||||+ | ||+||| | || +# Sbjct 760 FEKEQQQLNWKQDNNPLYKSAITTTVNPRF# 789 |
| Geodia cydonium | Query 766 FEKEKMNAKWDTQENPIYKSPINNFKNPNY#GR 797 || | |+ |||+| | ||| |#|+ Sbjct 847 FENELAKVKYTKNENPLYHSATTEHKNPIY#GQ 878 |
| Homo sapiens | Query 766 FEKEKMNAKWDTQENPIYKSPINNFKNPNY#G 796 |||||+ ++|+ +||++|| || +# Sbjct 738 FEKEKLKSQWN-NDNPLFKSATTTVMNPKF#A 767 |
| Ovis canadensis | Query 766 FEKEKMNAKWDTQENPIYKSPINNFKNPNY#G 796 |||||+ ++|+ +||++|| || +# Sbjct 739 FEKEKLKSQWN-NDNPLFKSATTTVMNPKF#A 768 |
| Sigmodon hispidus | Query 766 FEKEKMNAKWDTQENPIYKSPINNFKNPNY#G 796 |||||+ ++|+ +||++|| || +# Sbjct 738 FEKEKLKSQWN-NDNPLFKSATTTVMNPKF#A 767 |
* References
[PubMed ID: 24510996] Lee C, Lee C, Lee S, Siu A, Ramos DM, The cytoplasmic extension of the integrin beta6 subunit regulates epithelial-to-mesenchymal transition. Anticancer Res. 2014 Feb;34(2):659-64.
[PubMed ID: 24455846] ... Wojcik-Krowiranda K, Forma E, Zaczek A, Brys M, Anna MK, Bienkiewicz A, [Expression of E-cadherin and beta1-integrin mRNA in endometrial cancer]. Ginekol Pol. 2013 Nov;84(11):910-4.
[PubMed ID: 16157583] ... Seales EC, Shaikh FM, Woodard-Grice AV, Aggarwal P, McBrayer AC, Hennessy KM, Bellis SL, A protein kinase C/Ras/ERK signaling pathway activates myeloid fibronectin receptors by altering beta1 integrin sialylation. J Biol Chem. 2005 Nov 11;280(45):37610-5. Epub 2005 Sep 12.
[PubMed ID: 11779688] ... Armulik A, Splice variants of human beta 1 integrins: origin, biosynthesis and functions. Front Biosci. 2002 Jan 1;7:d219-27.
[PubMed ID: 9494094] ... Svineng G, Fassler R, Johansson S, Identification of beta1C-2, a novel variant of the integrin beta1 subunit generated by utilization of an alternative splice acceptor site in exon C. Biochem J. 1998 Mar 15;330 ( Pt 3):1255-63.
[PubMed ID: 8567725] ... Belkin AM, Zhidkova NI, Balzac F, Altruda F, Tomatis D, Maier A, Tarone G, Koteliansky VE, Burridge K, Beta 1D integrin displaces the beta 1A isoform in striated muscles: localization at junctional structures and signaling potential in nonmuscle cells. J Cell Biol. 1996 Jan;132(1-2):211-26.
[PubMed ID: 7545396] ... Zhidkova NI, Belkin AM, Mayne R, Novel isoform of beta 1 integrin expressed in skeletal and cardiac muscle. Biochem Biophys Res Commun. 1995 Sep 5;214(1):279-85.
[PubMed ID: 7544298] ... van der Flier A, Kuikman I, Baudoin C, van der Neut R, Sonnenberg A, A novel beta 1 integrin isoform produced by alternative splicing: unique expression in cardiac and skeletal muscle. FEBS Lett. 1995 Aug 7;369(2-3):340-4.
[PubMed ID: 8444890] ... Cervella P, Silengo L, Pastore C, Altruda F, Human beta 1-integrin gene expression is regulated by two promoter regions. J Biol Chem. 1993 Mar 5;268(7):5148-55.
[PubMed ID: 1551917] ... Languino LR, Ruoslahti E, An alternative form of the integrin beta 1 subunit with a variant cytoplasmic domain. J Biol Chem. 1992 Apr 5;267(10):7116-20.
[PubMed ID: 1690718] ... Bodary SC, McLean JW, The integrin beta 1 subunit associates with the vitronectin receptor alpha v subunit to form a novel vitronectin receptor in a human embryonic kidney cell line. J Biol Chem. 1990 Apr 15;265(11):5938-41.