SB0082 : Crystallin, [beta]B2
[ CaMP Format ]
* Basic Information
| Organism | Bos taurus (cattle) |
| Protein Names | beta-crystallin B2 [Bos taurus]; beta-crystallin B2; beta-B2 crystallin; beta-crystallin Bp; Beta-crystallin B2; Beta-B2 crystallin; Beta-crystallin Bp |
| Gene Names | CRYBB2; crystallin, beta B2 |
| Gene Locus | 17; chromosome 17 |
| GO Function | Not available |
| Entrez Protein | Entrez Nucleotide | Entrez Gene | UniProt | OMIM | HGNC | HPRD | KEGG |
|---|---|---|---|---|---|---|---|
| NP_777232 | NM_174807 | 287011 | P02522 | N/A | N/A | N/A | bta:287011 |
* Information From OMIM
Not Available.
* Structure Information
1. Primary Information
Length: 205 aa
Average Mass: 23.298 kDa
Monoisotopic Mass: 23.283 kDa
2. Domain Information
Annotated Domains: interpro / pfam / smart / prosite
Computationally Assigned Domains (Pfam+HMMER):
| domain name | begin | end | score | e-value |
|---|---|---|---|---|
| --- cleavage 8 --- | ||||
| Peptidase inhibitor family I36 1. | 21 | 96 | 12.0 | 0.0 |
| Peptidase inhibitor family I36 2. | 108 | 191 | 9.0 | 0.0 |
3. Sequence Information
Fasta Sequence: SB0082.fasta
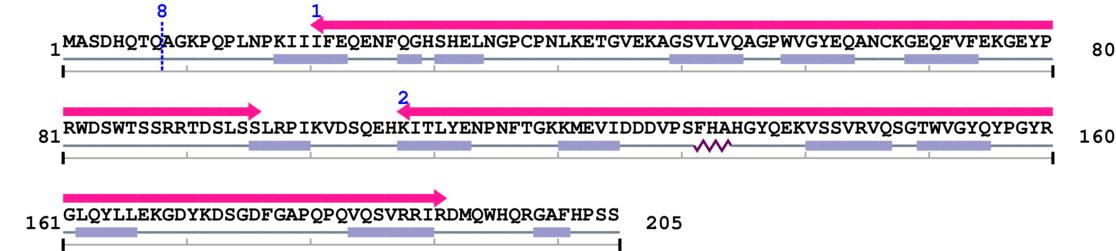
Amino Acid Sequence and Secondary Structures (PsiPred):
* Cleavage Information
1 [sites] cleaved by Calpain 2
Source Reference: [PubMed ID: 9485487] Shih M, Lampi KJ, Shearer TR, David LL, Cleavage of beta crystallins during maturation of bovine lens. Mol Vis. 1998 Feb 27;4:4.
Cleavage sites (±10aa)
[Site 1] MASDHQTQ8-AGKPQPLNPK
Gln8  Ala
Ala
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| - | - | Met1 | Ala2 | Ser3 | Asp4 | His5 | Gln6 | Thr7 | Gln8 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Ala9 | Gly10 | Lys11 | Pro12 | Gln13 | Pro14 | Leu15 | Asn16 | Pro17 | Lys18 |
 Sequence conservation (by blast)
Sequence conservation (by blast)
* References
[PubMed ID: 19206113] Diemer H, Atmanene C, Sanglier S, Morrissey B, Van Dorsselaer A, Downard KM, Detection and structural features of the betaB2-B3-crystallin heterodimer by radical probe mass spectrometry (RP-MS). J Mass Spectrom. 2009 May;44(5):803-12. doi: 10.1002/jms.1560.