SB0083 : p19INK4d, Cyclin-dependent kinase inhibitor 2D
[ CaMP Format ]
* Basic Information
| Organism | Homo sapiens (human) |
| Protein Names | cyclin-dependent kinase 4 inhibitor D [Homo sapiens]; cyclin-dependent kinase 4 inhibitor D; CDK inhibitor p19INK4d; cyclin-dependent kinase 4 inhibitor D p19; inhibitor of cyclin-dependent kinase 4d; cell cycle inhibitor, Nur77 associating protein; Cyclin-dependent kinase 4 inhibitor D; p19-INK4d |
| Gene Names | CDKN2D; cyclin-dependent kinase inhibitor 2D (p19, inhibits CDK4) |
| Gene Locus | 19p13; chromosome 19 |
| GO Function | Not available |
* Information From OMIM
Description: Cyclins are important in regulating the cell cycle through their formation of enzymatic complexes with various cyclin-dependent kinases. The D type cyclins complex with CDK4 (OMIM:123829) and CDK6 (OMIM:603368) to govern progression through the G1 phase of the cell cycle and later are involved with inactivating phosphorylation of the RB1 protein (OMIM:614041), which results in release of RB1-associated transcription factors that are needed for entry into S phase (Okuda et al., 1995). The activity of the cyclin D-dependent kinases is, in part, controlled by inhibitors such as the INK4 family, which includes INK4a (CDKN2A; OMIM:600160), 4b (CDKN2B; OMIM:600431), 4c (CDKN2C; OMIM:603369), and 4d (CDKN2D). INK4a has been shown to act by competing with CDK4 and CDK6 and functions as a tumor suppressor in a variety of cancers.
Function: The mouse INK4d protein interacts with Cdk6. In fibroblasts and macrophages it is rapidly induced at the G1-to-S transition. Overexpression of INK4d caused NIH 3T3 cells to arrest in G1 phase and inhibited cyclin D1-CDK4 kinase activity (Hirai et al., 1995).
* Structure Information
1. Primary Information
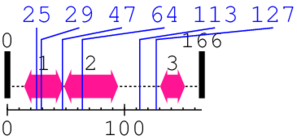
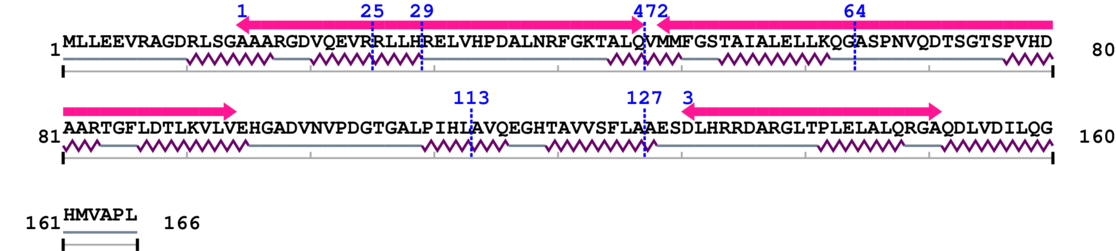
Length: 166 aa
Average Mass: 17.700 kDa
Monoisotopic Mass: 17.689 kDa
2. Domain Information
Annotated Domains: interpro / pfam / smart / prosite
Computationally Assigned Domains (Pfam+HMMER):
| domain name | begin | end | score | e-value |
|---|---|---|---|---|
| Ankyrin repeats (many copies) 1. | 15 | 47 | 23.0 | 0.0 |
| --- cleavage 25 (inside Ankyrin repeats (many copies) 15..47) --- | ||||
| --- cleavage 29 (inside Ankyrin repeats (many copies) 15..47) --- | ||||
| --- cleavage 47 (inside Ankyrin repeats (many copies) 15..47) --- | ||||
| --- cleavage 47 --- | ||||
| Ankyrin repeats (many copies) 2. | 49 | 94 | 9.0 | 0.0 |
| --- cleavage 64 (inside Ankyrin repeats (many copies) 49..94) --- | ||||
| --- cleavage 113 --- | ||||
| --- cleavage 127 --- | ||||
| Ankyrin repeats (many copies) 3. | 131 | 151 | 26.0 | 0.0 |
3. Sequence Information
Fasta Sequence: SB0083.fasta
Amino Acid Sequence and Secondary Structures (PsiPred):
* Cleavage Information
6 [sites] cleaved by Calpain 1
Source Reference: [PubMed ID: 16542156] Joy J, Nalabothula N, Ghosh M, Popp O, Jochum M, Machleidt W, Gil-Parrado S, Holak TA, Identification of calpain cleavage sites in the G1 cyclin-dependent kinase inhibitor p19(INK4d). Biol Chem. 2006 Mar;387(3):329-35.
Cleavage sites (±10aa)
[Site 1] AARGDVQEVR25-RLLHRELVHP
Arg25  Arg
Arg
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Ala16 | Ala17 | Arg18 | Gly19 | Asp20 | Val21 | Gln22 | Glu23 | Val24 | Arg25 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Arg26 | Leu27 | Leu28 | His29 | Arg30 | Glu31 | Leu32 | Val33 | His34 | Pro35 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| MLLEEVRAGDRLSGAAARGDVQEVRRLLHRELVHPDALNRFGKTALQVMMFGSTA |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | Homo sapiens | 111.00 | 17 | cyclin-dependent kinase inhibitor 2D |
| 2 | N/A | 111.00 | 14 | p19 protein |
| 3 | synthetic construct | 111.00 | 4 | cyclin-dependent kinase inhibitor 2D (p19, inhibi |
| 4 | Canis familiaris | 108.00 | 2 | PREDICTED: similar to Cyclin-dependent kinase 4 i |
| 5 | Rattus norvegicus | 106.00 | 5 | similar to cyclin-dependent kinase inhibitor 2D |
| 6 | Mus musculus | 105.00 | 14 | cyclin-dependent kinase inhibitor 2D |
| 7 | Bos taurus | 102.00 | 5 | cyclin-dependent kinase inhibitor 2D (p19, inhibi |
| 8 | Monodelphis domestica | 47.00 | 1 | cyclin-dependent kinase inhibitor 2A isoform 1 |
| 9 | Monodelphis domestica | 47.00 | 1 | tumor suppressor CDKN2A |
| 10 | Mesocricetus auratus | 46.60 | 3 | p15INK4b cyclin-dependent kinase inhibitor |
| 11 | Tetraodon nigroviridis | 46.20 | 4 | unnamed protein product |
| 12 | human, brain tumors, Peptide, 156 aa | 42.70 | 1 | cyclin D-dependent kinases 4 and 6-binding protei |
| 13 | Danio rerio | 41.60 | 1 | PREDICTED: similar to Chain A, Structure Of P18in |
| 14 | Sus scrofa | 41.20 | 4 | cyclin dependant kinase inhibitor |
| 15 | Takifugu rubripes | 40.80 | 2 | p19 |
| 16 | Gallus gallus | 37.00 | 1 | cyclin-dependent kinase inhibitor 2B |
| 17 | Xenopus tropicalis | 35.80 | 1 | cyclin-dependent kinase inhibitor 2B (p15, inhibi |
| 18 | Xiphophorus maculatus | 35.40 | 1 | 13CDKN2X protein |
| 19 | Xiphophorus helleri | 35.40 | 1 | p13CDKN2X |
| 20 | Apis mellifera | 34.30 | 1 | PREDICTED: similar to MYPT-75D CG6896-PA |
| 21 | Populus tremula x Populus tremuloides | 33.90 | 1 | potassium channel 2 |
| 22 | Canis lupus familiaris | 33.10 | 1 | uveal autoantigen with coiled-coil domains and an |
| 23 | Artemisia desertorum | 32.00 | 1 | ring zinc finger protein |
| 24 | Mus musculus | 31.20 | 1 | cyclin-dependent kinase inhibitor |
| 25 | Leishmania major strain Friedlin | 30.40 | 1 | hypothetical protein, conserved |
Top-ranked sequences
| organism | matching |
|---|---|
| Homo sapiens | Query 1 MLLEEVRAGDRLSGAAARGDVQEVR#RLLHRELVHPDALNRFGKTALQVMMFGSTA 55 |||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 1 MLLEEVRAGDRLSGAAARGDVQEVR#RLLHRELVHPDALNRFGKTALQVMMFGSTA 55 |
| N/A | Query 1 MLLEEVRAGDRLSGAAARGDVQEVR#RLLHRELVHPDALNRFGKTALQVMMFGSTA 55 |||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 1 MLLEEVRAGDRLSGAAARGDVQEVR#RLLHRELVHPDALNRFGKTALQVMMFGSTA 55 |
| synthetic construct | Query 1 MLLEEVRAGDRLSGAAARGDVQEVR#RLLHRELVHPDALNRFGKTALQVMMFGSTA 55 |||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 1 MLLEEVRAGDRLSGAAARGDVQEVR#RLLHRELVHPDALNRFGKTALQVMMFGSTA 55 |
| Canis familiaris | Query 1 MLLEEVRAGDRLSGAAARGDVQEVR#RLLHRELVHPDALNRFGKTALQVMMFGS 53 |||||||||||||||||||||||||#|||||||||||||||||||||||||||| Query 1 MLLEEVRAGDRLSGAAARGDVQEVR#RLLHRELVHPDALNRFGKTALQVMMFGS 53 |
| Rattus norvegicus | Query 1 MLLEEVRAGDRLSGAAARGDVQEVR#RLLHRELVHPDALNRFGKTALQVMMFGSTA 55 |||||| |||||||||||||||||#|||||||||||||||||||||||||||| | Sbjct 1 MLLEEVSVGDRLSGAAARGDVQEVR#RLLHRELVHPDALNRFGKTALQVMMFGSPA 55 |
| Mus musculus | Query 1 MLLEEVRAGDRLSGAAARGDVQEVR#RLLHRELVHPDALNRFGKTALQVMMFGSTA 55 |||||| |||||||||||||||||#|||||||||||||||||||||||||||| | Sbjct 1 MLLEEVCVGDRLSGAAARGDVQEVR#RLLHRELVHPDALNRFGKTALQVMMFGSPA 55 |
| Bos taurus | Query 1 MLLEEVRAGDRLSGAAARGDVQEVR#RLLHRELVHPDALNRFGKTALQVMMFGS 53 |||||| ||||||||||||||||||#|||| |||||| |||||||||||||||| Sbjct 1 MLLEEVHAGDRLSGAAARGDVQEVR#RLLHSELVHPDVLNRFGKTALQVMMFGS 53 |
| Monodelphis domestica | Query 4 EEVRAGDRLSGAAARGDVQEVR#RLLHREL-VHPDALNRFGKTALQVMMFGS 53 || +|++|+ ||||| + | # || || +|+|+||||++|+|||| |+ Sbjct 42 EESFSGEKLTEAAARGRTEVVT#ELL--ELGTNPNAVNRFGRSAIQVMMMGN 90 |
| Monodelphis domestica | Query 4 EEVRAGDRLSGAAARGDVQEVR#RLLHREL-VHPDALNRFGKTALQVMMFGS 53 || +|++|+ ||||| + | # || || +|+|+||||++|+|||| |+ Sbjct 8 EESFSGEKLTEAAARGRTEVVT#ELL--ELGTNPNAVNRFGRSAIQVMMMGN 56 |
| Mesocricetus auratus | Query 12 LSGAAARGDVQEVR#RLLHRELVHPDALNRFGKTALQVMMFGST 54 |+ ||||| |+ ||#+|| | |+|+||||+ +|||| ||| Sbjct 10 LATAAARGQVETVR#QLLEAG-VDPNAVNRFGRRPIQVMMMGST 51 |
| Tetraodon nigroviridis | Query 10 DRLSGAAARGDVQEVR#RLLHRELVHPDALNRFGKTALQVMMFGSTA 55 | |+ |||+|| +||# || + +||||+||||||| |||+ Sbjct 5 DELTAAAAKGDTAQVR#LLLGAG-AQVNGVNRFGRTALQVMMMGSTS 49 |
| human, brain tumors, Peptide, 156 aa | Query 8 AGDRLSGAAARGDVQEVR#RLLHRELVHPDALNRFGKTALQVMMFGST 54 + | |+ ||||| |+|||# || + |+| | +|+ +|||| || Sbjct 12 SADWLATAAARGRVEEVR#ALLE-AVALPNAPNSYGRRPIQVMMMGSA 57 |
| Danio rerio | Query 3 LEEVRAGDRLSGAAARGDVQEVR#RLLHRELVHPDALNRFGKTALQVMMFGSTA 55 + | | |||| ||| ||+ || #+ | |+ + |++|+|||||| | + Sbjct 1 MAEDTALDRLSTAAAIGDLMEVE#QTLQSN-VNVNEKNKYGRTALQVMKLGCPS 52 |
| Sus scrofa | Query 8 AGDRLSGAAARGDVQEVR#RLLHRELVHPDALNRFGKTALQVMMFGST 54 + | |+ ||||| |||# || + +| ||+|+| +|||| ||| Sbjct 4 SADWLASAAARGREGEVR#ALLEAGAL-ANAPNRYGRTPIQVMMMGST 49 |
| Takifugu rubripes | Query 23 EVR#RLLHRELVHPDALNRFGKTALQVMMFGST 54 ||+#|+| | || | |||||||||| |+ Sbjct 23 EVQ#RILEECRVPPDTRNEFGKTALQVMMLGNC 54 |
| Gallus gallus | Query 18 RGDVQEVR#RLLHRELVHPDALNRFGKTALQVMMFGST 54 |||+ |+# || |+|+| ||+| +|||| || Sbjct 19 RGDLLRVK#ELLD-GAADPNAVNSFGRTPIQVMMLGSP 54 |
| Xenopus tropicalis | Query 10 DRLSGAAARGDVQEVR#RLLHRELVHPDALNRFGKTALQVMMFGST 54 + | | ||||| |#++| + +| | |+| +|||| || Sbjct 6 NTLCSACARGDVDLAR#QMLQSG-IPVNATNSHGRTPIQVMMMGSP 49 |
| Xiphophorus maculatus | Query 10 DRLSGAAARGDVQEVR#RLLHRELVHPDALNRFGKTALQVMMFGST 54 | |+ |||+| || # || + + +| ||+ |+|||| ||+ Sbjct 5 DELTTAAAKGHTAEVE#ALLLQG-APVNGVNSFGRRAIQVMMMGSS 48 |
| Xiphophorus helleri | Query 10 DRLSGAAARGDVQEVR#RLLHRELVHPDALNRFGKTALQVMMFGST 54 | |+ |||+| || # || + + +| ||+ |+|||| ||+ Sbjct 5 DELTTAAAKGHTAEVE#ALLLQG-APVNGVNSFGRRAIQVMMMGSS 48 |
| Apis mellifera | Query 15 AAARGDVQEVR#RLLHRELVHPDALNRFGKTALQ 47 |||| |+ |||#||| + |+||+ | | ||| Sbjct 68 AAARNDIDEVR#RLLKKG-VNPDSTNEDGLTALH 99 |
| Populus tremula x Populus tremuloides | Query 8 AGDRLSGAAARGDVQEVR#RLLHRELVHPDALNRFGKTALQVMM 50 ||| | || + |+ ++# || + | + |+ +| ||||||| | Sbjct 623 AGDLLCTAAKQNDLMVMK#ELLKQGL-NVDSKDRHGKTALQVAM 664 |
| Canis lupus familiaris | Query 10 DRLSGAAARGDVQEVR#RLLHRELVHPDALNRFGKTALQVM 49 ||| || ||||++| # +| ++ ++| |+ |++| |+ Sbjct 38 DRLMKAAERGDVEKVS#SILAKKGINPGKLDVEGRSAFHVV 77 |
| Artemisia desertorum | Query 15 AAARGDVQEVR#RLLHRELVHPDALNRFGKTALQV-MMFGSTA 55 ||| | ++ | # || + |+|||||| +| | + | | | Sbjct 54 AAANGQIEIVN#LLLDKSSVNPDALNRRKQTPLMLAAMHGKIA 95 |
| Mus musculus | Query 8 AGDRLSGAAARGDVQEVR#RLLHRELVHPDALNRFGKTALQ 47 | |||+ ||+| | +||# || | |+| | ||+| +| Sbjct 4 AADRLA-RAAQGRVHDVR#ALLEAG-VSPNAPNSFGRTPIQ 41 |
| Leishmania major strain Friedlin | Query 10 DRLSGAAARGDVQEVR#RLLHRELVHPDALNRFGKTALQVMM-FGS 53 +++ || +| |||#||+ | | |||| ||| + || Sbjct 18 EKIHVAARKGQTDEVR#RLIETG-VSPTIQNRFGCTALHLACKFGC 61 |
[Site 2] DVQEVRRLLH29-RELVHPDALN
His29  Arg
Arg
iTraq-117 Signal 623.5 (
 ) for DVQEVRRLLH
) for DVQEVRRLLH
iTraq-117 Signal 5802.3 (

 ) for RELVHPDALN
) for RELVHPDALN
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Asp20 | Val21 | Gln22 | Glu23 | Val24 | Arg25 | Arg26 | Leu27 | Leu28 | His29 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Arg30 | Glu31 | Leu32 | Val33 | His34 | Pro35 | Asp36 | Ala37 | Leu38 | Asn39 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| MLLEEVRAGDRLSGAAARGDVQEVRRLLHRELVHPDALNRFGKTALQVMMFGSTAIALE |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | Homo sapiens | 118.00 | 17 | cyclin-dependent kinase inhibitor 2D |
| 2 | N/A | 118.00 | 14 | p19 protein |
| 3 | synthetic construct | 118.00 | 4 | cyclin-dependent kinase inhibitor 2D (p19, inhibi |
| 4 | Canis familiaris | 114.00 | 2 | PREDICTED: similar to Cyclin-dependent kinase 4 i |
| 5 | Rattus norvegicus | 112.00 | 5 | similar to cyclin-dependent kinase inhibitor 2D |
| 6 | Mus musculus | 111.00 | 13 | cyclin-dependent kinase inhibitor 2D |
| 7 | Bos taurus | 109.00 | 5 | cyclin-dependent kinase inhibitor 2D (p19, inhibi |
| 8 | Tetraodon nigroviridis | 48.90 | 4 | unnamed protein product |
| 9 | Mesocricetus auratus | 48.90 | 3 | p15INK4b cyclin-dependent kinase inhibitor |
| 10 | Monodelphis domestica | 48.10 | 1 | cyclin-dependent kinase inhibitor 2A isoform 1 |
| 11 | Monodelphis domestica | 47.80 | 1 | tumor suppressor CDKN2A |
| 12 | Danio rerio | 44.70 | 1 | PREDICTED: similar to Chain A, Structure Of P18in |
| 13 | Takifugu rubripes | 43.90 | 2 | p19 |
| 14 | human, brain tumors, Peptide, 156 aa | 43.90 | 1 | cyclin D-dependent kinases 4 and 6-binding protei |
| 15 | Sus scrofa | 42.70 | 4 | cyclin dependant kinase inhibitor |
| 16 | Gallus gallus | 38.50 | 1 | cyclin-dependent kinase inhibitor 2B |
| 17 | Xenopus tropicalis | 37.40 | 1 | cyclin-dependent kinase inhibitor 2B (p15, inhibi |
| 18 | Xiphophorus helleri | 37.00 | 1 | p13CDKN2X |
| 19 | Xiphophorus maculatus | 37.00 | 1 | 13CDKN2X protein |
| 20 | Apis mellifera | 34.30 | 1 | PREDICTED: similar to MYPT-75D CG6896-PA |
| 21 | Populus tremula x Populus tremuloides | 33.90 | 1 | potassium channel 2 |
| 22 | Canis lupus familiaris | 33.10 | 1 | uveal autoantigen with coiled-coil domains and an |
| 23 | Artemisia desertorum | 32.00 | 1 | ring zinc finger protein |
| 24 | Gallus gallus | 31.60 | 1 | ankyrin repeat domain 52 |
| 25 | Mus musculus | 31.20 | 1 | cyclin-dependent kinase inhibitor |
| 26 | Leishmania major strain Friedlin | 31.20 | 1 | hypothetical protein, conserved |
| 27 | Equus caballus | 29.30 | 1 | cyclin-dependent kinase 2A inhibitor |
Top-ranked sequences
| organism | matching |
|---|---|
| Homo sapiens | Query 1 MLLEEVRAGDRLSGAAARGDVQEVRRLLH#RELVHPDALNRFGKTALQVMMFGSTAIALE 59 |||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 1 MLLEEVRAGDRLSGAAARGDVQEVRRLLH#RELVHPDALNRFGKTALQVMMFGSTAIALE 59 |
| N/A | Query 1 MLLEEVRAGDRLSGAAARGDVQEVRRLLH#RELVHPDALNRFGKTALQVMMFGSTAIALE 59 |||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 1 MLLEEVRAGDRLSGAAARGDVQEVRRLLH#RELVHPDALNRFGKTALQVMMFGSTAIALE 59 |
| synthetic construct | Query 1 MLLEEVRAGDRLSGAAARGDVQEVRRLLH#RELVHPDALNRFGKTALQVMMFGSTAIALE 59 |||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 1 MLLEEVRAGDRLSGAAARGDVQEVRRLLH#RELVHPDALNRFGKTALQVMMFGSTAIALE 59 |
| Canis familiaris | Query 1 MLLEEVRAGDRLSGAAARGDVQEVRRLLH#RELVHPDALNRFGKTALQVMMFGSTAIALE 59 |||||||||||||||||||||||||||||#|||||||||||||||||||||||| |||| Sbjct 1 MLLEEVRAGDRLSGAAARGDVQEVRRLLH#RELVHPDALNRFGKTALQVMMFGSPTIALE 59 |
| Rattus norvegicus | Query 1 MLLEEVRAGDRLSGAAARGDVQEVRRLLH#RELVHPDALNRFGKTALQVMMFGSTAIALE 59 |||||| |||||||||||||||||||||#|||||||||||||||||||||||| |+||| Sbjct 1 MLLEEVSVGDRLSGAAARGDVQEVRRLLH#RELVHPDALNRFGKTALQVMMFGSPAVALE 59 |
| Mus musculus | Query 1 MLLEEVRAGDRLSGAAARGDVQEVRRLLH#RELVHPDALNRFGKTALQVMMFGSTAIALE 59 |||||| |||||||||||||||||||||#|||||||||||||||||||||||| |+||| Sbjct 1 MLLEEVCVGDRLSGAAARGDVQEVRRLLH#RELVHPDALNRFGKTALQVMMFGSPAVALE 59 |
| Bos taurus | Query 1 MLLEEVRAGDRLSGAAARGDVQEVRRLLH#RELVHPDALNRFGKTALQVMMFGSTAIALE 59 |||||| ||||||||||||||||||||||# |||||| |||||||||||||||| |||| Sbjct 1 MLLEEVHAGDRLSGAAARGDVQEVRRLLH#SELVHPDVLNRFGKTALQVMMFGSPTIALE 59 |
| Tetraodon nigroviridis | Query 10 DRLSGAAARGDVQEVRRLLH#RELVHPDALNRFGKTALQVMMFGSTAIA 57 | |+ |||+|| +|| || # + +||||+||||||| |||++| Sbjct 5 DELTAAAAKGDTAQVRLLLG#AG-AQVNGVNRFGRTALQVMMMGSTSVA 51 |
| Mesocricetus auratus | Query 12 LSGAAARGDVQEVRRLLH#RELVHPDALNRFGKTALQVMMFGSTAIA 57 |+ ||||| |+ ||+|| # | |+|+||||+ +|||| ||| +| Sbjct 10 LATAAARGQVETVRQLLE#AG-VDPNAVNRFGRRPIQVMMMGSTQVA 54 |
| Monodelphis domestica | Query 4 EEVRAGDRLSGAAARGDVQEVRRLLH#REL-VHPDALNRFGKTALQVMMFGSTAIA 57 || +|++|+ ||||| + | || # || +|+|+||||++|+|||| |+ +| Sbjct 42 EESFSGEKLTEAAARGRTEVVTELL-#-ELGTNPNAVNRFGRSAIQVMMMGNVRLA 94 |
| Monodelphis domestica | Query 4 EEVRAGDRLSGAAARGDVQEVRRLLH#REL-VHPDALNRFGKTALQVMMFGSTAIA 57 || +|++|+ ||||| + | || # || +|+|+||||++|+|||| |+ +| Sbjct 8 EESFSGEKLTEAAARGRTEVVTELL-#-ELGTNPNAVNRFGRSAIQVMMMGNVRLA 60 |
| Danio rerio | Query 3 LEEVRAGDRLSGAAARGDVQEVRRLLH#RELVHPDALNRFGKTALQVMMFGSTAIA 57 + | | |||| ||| ||+ || + | # |+ + |++|+|||||| | +|| Sbjct 1 MAEDTALDRLSTAAAIGDLMEVEQTLQ#SN-VNVNEKNKYGRTALQVMKLGCPSIA 54 |
| Takifugu rubripes | Query 23 EVRRLLH#RELVHPDALNRFGKTALQVMMFGSTAIA 57 ||+|+| # | || | |||||||||| |+ || Sbjct 23 EVQRILE#ECRVPPDTRNEFGKTALQVMMLGNCKIA 57 |
| human, brain tumors, Peptide, 156 aa | Query 8 AGDRLSGAAARGDVQEVRRLLH#RELVHPDALNRFGKTALQVMMFGSTAIA 57 + | |+ ||||| |+||| || # + |+| | +|+ +|||| || +| Sbjct 12 SADWLATAAARGRVEEVRALLE#-AVALPNAPNSYGRRPIQVMMMGSARVA 60 |
| Sus scrofa | Query 8 AGDRLSGAAARGDVQEVRRLLH#RELVHPDALNRFGKTALQVMMFGSTAIA 57 + | |+ ||||| ||| || # + +| ||+|+| +|||| ||| +| Sbjct 4 SADWLASAAARGREGEVRALLE#AGAL-ANAPNRYGRTPIQVMMMGSTRVA 52 |
| Gallus gallus | Query 18 RGDVQEVRRLLH#RELVHPDALNRFGKTALQVMMFGSTAIA 57 |||+ |+ || # |+|+| ||+| +|||| || +| Sbjct 19 RGDLLRVKELLD#-GAADPNAVNSFGRTPIQVMMLGSPRVA 57 |
| Xenopus tropicalis | Query 10 DRLSGAAARGDVQEVRRLLH#RELVHPDALNRFGKTALQVMMFGSTAIA 57 + | | ||||| |++| # + +| | |+| +|||| || +| Sbjct 6 NTLCSACARGDVDLARQMLQ#SGIPV-NATNSHGRTPIQVMMMGSPKMA 52 |
| Xiphophorus helleri | Query 10 DRLSGAAARGDVQEVRRLLH#RELVHPDALNRFGKTALQVMMFGSTAIA 57 | |+ |||+| || || #+ + +| ||+ |+|||| ||+ +| Sbjct 5 DELTTAAAKGHTAEVEALLL#QG-APVNGVNSFGRRAIQVMMMGSSEVA 51 |
| Xiphophorus maculatus | Query 10 DRLSGAAARGDVQEVRRLLH#RELVHPDALNRFGKTALQVMMFGSTAIA 57 | |+ |||+| || || #+ + +| ||+ |+|||| ||+ +| Sbjct 5 DELTTAAAKGHTAEVEALLL#QG-APVNGVNSFGRRAIQVMMMGSSEVA 51 |
| Apis mellifera | Query 15 AAARGDVQEVRRLLH#RELVHPDALNRFGKTALQ 47 |||| |+ |||||| #++ |+||+ | | ||| Sbjct 68 AAARNDIDEVRRLL-#KKGVNPDSTNEDGLTALH 99 |
| Populus tremula x Populus tremuloides | Query 8 AGDRLSGAAARGDVQEVRRLLH#RELVHPDALNRFGKTALQVMM 50 ||| | || + |+ ++ || #+ | + |+ +| ||||||| | Sbjct 623 AGDLLCTAAKQNDLMVMKELLK#QGL-NVDSKDRHGKTALQVAM 664 |
| Canis lupus familiaris | Query 10 DRLSGAAARGDVQEVRRLLH#RELVHPDALNRFGKTALQVM 49 ||| || ||||++| +| #++ ++| |+ |++| |+ Sbjct 38 DRLMKAAERGDVEKVSSILA#KKGINPGKLDVEGRSAFHVV 77 |
| Artemisia desertorum | Query 15 AAARGDVQEVRRLLH#RELVHPDALNRFGKTALQV-MMFGSTA 55 ||| | ++ | || #+ |+|||||| +| | + | | | Sbjct 54 AAANGQIEIVNLLLD#KSSVNPDALNRRKQTPLMLAAMHGKIA 95 |
| Gallus gallus | Query 12 LSGAAARGDVQEVRRLLH#RELVHPDALNRFGKTALQVMMF-GSTAIALE 59 | ||| | ++ || || #| | | | || ||| + + | |+| | Sbjct 210 LHTAAASGQIEVVRHLL-#RLGVEIDEPNSFGNTALHIACYMGQDAVANE 257 |
| Mus musculus | Query 8 AGDRLSGAAARGDVQEVRRLLH#RELVHPDALNRFGKTALQ 47 | |||+ ||+| | +|| || # | |+| | ||+| +| Sbjct 4 AADRLA-RAAQGRVHDVRALLE#-AGVSPNAPNSFGRTPIQ 41 |
| Leishmania major strain Friedlin | Query 10 DRLSGAAARGDVQEVRRLLH#RELVHPDALNRFGKTALQV-MMFGSTAIA 57 +++ || +| |||||+ # | | |||| ||| + || | Sbjct 18 EKIHVAARKGQTDEVRRLIE#TG-VSPTIQNRFGCTALHLACKFGCVDTA 65 |
| Equus caballus | Query 35 PDALNRFGKTALQVMMFGSTAIA 57 |+ +| ||+ +|||| || +| Sbjct 5 PNGVNGFGRRPIQVMMMGSVHVA 27 |
[Site 3] LNRFGKTALQ47-VMMFGSTAIA
Gln47  Val
Val
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Leu38 | Asn39 | Arg40 | Phe41 | Gly42 | Lys43 | Thr44 | Ala45 | Leu46 | Gln47 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Val48 | Met49 | Met50 | Phe51 | Gly52 | Ser53 | Thr54 | Ala55 | Ile56 | Ala57 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| RGDVQEVRRLLHRELVHPDALNRFGKTALQVMMFGSTAIALELLKQGASPNVQDTSGTSP |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | N/A | 124.00 | 17 | p19 protein |
| 2 | Homo sapiens | 124.00 | 10 | cyclin-dependent kinase inhibitor 2D |
| 3 | synthetic construct | 124.00 | 4 | cyclin-dependent kinase inhibitor 2D (p19, inhibi |
| 4 | Mus musculus | 119.00 | 18 | cyclin-dependent kinase inhibitor 2D |
| 5 | Rattus norvegicus | 119.00 | 8 | similar to cyclin-dependent kinase inhibitor 2D |
| 6 | Canis familiaris | 116.00 | 3 | PREDICTED: similar to Cyclin-dependent kinase 4 i |
| 7 | Bos taurus | 113.00 | 3 | cyclin-dependent kinase inhibitor 2D (p19, inhibi |
| 8 | Tetraodon nigroviridis | 67.00 | 3 | unnamed protein product |
| 9 | Takifugu rubripes | 65.10 | 2 | p19 |
| 10 | Mesocricetus auratus | 54.30 | 3 | p15INK4b cyclin-dependent kinase inhibitor |
| 11 | Gallus gallus | 49.70 | 1 | cyclin-dependent kinase inhibitor 2B |
| 12 | Monodelphis domestica | 48.50 | 1 | cyclin-dependent kinase inhibitor 2A isoform 1 |
| 13 | human, brain tumors, Peptide, 156 aa | 48.50 | 1 | cyclin D-dependent kinases 4 and 6-binding protei |
| 14 | Monodelphis domestica | 48.50 | 1 | tumor suppressor CDKN2A |
| 15 | Sus scrofa | 47.80 | 2 | cyclin dependant kinase inhibitor |
| 16 | Danio rerio | 47.80 | 1 | PREDICTED: similar to Chain A, Structure Of P18in |
| 17 | Xiphophorus maculatus | 45.10 | 1 | 13CDKN2X protein |
| 18 | Xiphophorus helleri | 45.10 | 1 | p13CDKN2X |
| 19 | Xenopus tropicalis | 42.40 | 1 | cyclin-dependent kinase inhibitor 2B (p15, inhibi |
| 20 | Solibacter usitatus Ellin6076 | 39.30 | 1 | ankyrin-related protein |
| 21 | Apis mellifera | 38.90 | 1 | PREDICTED: similar to MYPT-75D CG6896-PA |
| 22 | Equus caballus | 38.90 | 1 | cyclin-dependent kinase 2A inhibitor |
| 23 | Schistosoma japonicum | 38.90 | 1 | SJCHGC04635 protein |
| 24 | Drosophila melanogaster | 38.50 | 1 | CG3104 CG3104-PA, isoform A |
| 25 | Gallus gallus | 37.40 | 1 | ankyrin repeat domain 52 |
| 26 | Cryptosporidium hominis TU502 | 37.40 | 1 | proteasome 26S subunit |
| 27 | Aspergillus oryzae RIB40 | 36.60 | 1 | hypothetical protein |
| 28 | Egeria densa | 36.60 | 1 | inward potassium channel alpha subunit |
| 29 | Rickettsia felis URRWXCal2 | 36.20 | 1 | Ankyrin repeat |
| 30 | Bordetella parapertussis 12822 | 36.20 | 1 | hypothetical protein BPP0952 |
| 31 | Bordetella pertussis Tohama I | 36.20 | 1 | hypothetical protein BP2881 |
| 32 | Xenopus laevis | 35.80 | 3 | AF465262_1 inversin |
| 33 | Rattus norvegicus | 35.80 | 1 | 190 kDa ankyrin isoform; AnkG190 |
| 34 | Methanosarcina barkeri str. Fusaro | 35.40 | 1 | hypothetical protein Mbar_A2380 |
| 35 | Dictyostelium discoideum AX4 | 35.00 | 1 | ankyrin repeat-containing protein |
| 36 | Caenorhabditis briggsae AF16 | 34.70 | 1 | Hypothetical protein CBG03811 |
| 37 | Orf virus | 34.30 | 1 | Ankyrin/F-box protein |
| 38 | Wolbachia endosymbiont of Drosophila melanogaster | 34.30 | 1 | ankyrin repeat domain protein |
| 39 | Caenorhabditis elegans | 33.90 | 2 | GEI-4(Four) Interacting protein family member (gf |
| 40 | Neurospora crassa OR74A | 33.90 | 1 | hypothetical protein NCU04881 |
Top-ranked sequences
| organism | matching |
|---|---|
| N/A | Query 18 RGDVQEVRRLLHRELVHPDALNRFGKTALQ#VMMFGSTAIALELLKQGASPNVQDTSGTSP 77 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 18 RGDVQEVRRLLHRELVHPDALNRFGKTALQ#VMMFGSTAIALELLKQGASPNVQDTSGTSP 77 |
| Homo sapiens | Query 18 RGDVQEVRRLLHRELVHPDALNRFGKTALQ#VMMFGSTAIALELLKQGASPNVQDTSGTSP 77 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 18 RGDVQEVRRLLHRELVHPDALNRFGKTALQ#VMMFGSTAIALELLKQGASPNVQDTSGTSP 77 |
| synthetic construct | Query 18 RGDVQEVRRLLHRELVHPDALNRFGKTALQ#VMMFGSTAIALELLKQGASPNVQDTSGTSP 77 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 18 RGDVQEVRRLLHRELVHPDALNRFGKTALQ#VMMFGSTAIALELLKQGASPNVQDTSGTSP 77 |
| Mus musculus | Query 18 RGDVQEVRRLLHRELVHPDALNRFGKTALQ#VMMFGSTAIALELLKQGASPNVQDTSGTSP 77 ||||||||||||||||||||||||||||||#|||||| |+||||||||||||||| ||||| Sbjct 18 RGDVQEVRRLLHRELVHPDALNRFGKTALQ#VMMFGSPAVALELLKQGASPNVQDASGTSP 77 |
| Rattus norvegicus | Query 18 RGDVQEVRRLLHRELVHPDALNRFGKTALQ#VMMFGSTAIALELLKQGASPNVQDTSGTSP 77 ||||||||||||||||||||||||||||||#|||||| |+||||||||||||||| ||||| Sbjct 18 RGDVQEVRRLLHRELVHPDALNRFGKTALQ#VMMFGSPAVALELLKQGASPNVQDASGTSP 77 |
| Canis familiaris | Query 18 RGDVQEVRRLLHRELVHPDALNRFGKTALQ#VMMFGSTAIALELLKQGASPNVQDTSGTSP 77 ||||||||||||||||||||||||||||||#|||||| |||||||||||||||| +||+| Sbjct 18 RGDVQEVRRLLHRELVHPDALNRFGKTALQ#VMMFGSPTIALELLKQGASPNVQDATGTTP 77 |
| Bos taurus | Query 18 RGDVQEVRRLLHRELVHPDALNRFGKTALQ#VMMFGSTAIALELLKQGASPNVQDTSGTSP 77 |||||||||||| |||||| ||||||||||#|||||| |||||||||||||||| |||+| Sbjct 18 RGDVQEVRRLLHSELVHPDVLNRFGKTALQ#VMMFGSPTIALELLKQGASPNVQDASGTTP 77 |
| Tetraodon nigroviridis | Query 23 EVRRLLHRELVHPDALNRFGKTALQ#VMMFGSTAIALELLKQGASPNVQDTSGTSP 77 ||||+| | || | |||||||#||| |+ || ||++|| ||||| | +| Sbjct 23 EVRRILEECRVPPDTPNEFGKTALQ#VMMLGNCKIASLLLEKGADPNVQDKHGIAP 77 |
| Takifugu rubripes | Query 23 EVRRLLHRELVHPDALNRFGKTALQ#VMMFGSTAIALELLKQGASPNVQDTSGTSP 77 ||+|+| | || | |||||||#||| |+ || ||++|| ||||| | +| Sbjct 23 EVQRILEECRVPPDTRNEFGKTALQ#VMMLGNCKIASLLLEKGADPNVQDKHGIAP 77 |
| Mesocricetus auratus | Query 18 RGDVQEVRRLLHRELVHPDALNRFGKTALQ#VMMFGSTAIALELLKQGASPNVQD 71 || |+ ||+|| | |+|+||||+ +|#||| ||| +| || || || | Sbjct 16 RGQVETVRQLLEAG-VDPNAVNRFGRRPIQ#VMMMGSTQVAELLLLHGAEPNCAD 68 |
| Gallus gallus | Query 18 RGDVQEVRRLLHRELVHPDALNRFGKTALQ#VMMFGSTAIALELLKQGASPNVQDT-SGTS 76 |||+ |+ || |+|+| ||+| +|#||| || +| ||++|| || | +| Sbjct 19 RGDLLRVKELLD-GAADPNAVNSFGRTPIQ#VMMLGSPRVAELLLQRGADPNRPDPRTGCR 77 Query 77 P 77 | Query 77 P 77 |
| Monodelphis domestica | Query 18 RGDVQEVRRLLHREL-VHPDALNRFGKTALQ#VMMFGSTAIALELLKQGASPNVQD 71 || + | || || +|+|+||||++|+|#||| |+ +| ||+ || || | Sbjct 56 RGRTEVVTELL--ELGTNPNAVNRFGRSAIQ#VMMMGNVRLAAILLQYGAEPNTPD 108 |
| human, brain tumors, Peptide, 156 aa | Query 18 RGDVQEVRRLLHRELVHPDALNRFGKTALQ#VMMFGSTAIALELLKQGASPNVQDTS 73 || |+||| || + |+| | +|+ +|#||| || +| || || || | + Sbjct 22 RGRVEEVRALLE-AVALPNAPNSYGRRPIQ#VMMMGSARVAELLLLHGAEPNCADPA 76 |
| Monodelphis domestica | Query 18 RGDVQEVRRLLHREL-VHPDALNRFGKTALQ#VMMFGSTAIALELLKQGASPNVQD 71 || + | || || +|+|+||||++|+|#||| |+ +| ||+ || || | Sbjct 22 RGRTEVVTELL--ELGTNPNAVNRFGRSAIQ#VMMMGNVRLAAILLQYGAEPNTPD 74 |
| Sus scrofa | Query 18 RGDVQEVRRLLHRELVHPDALNRFGKTALQ#VMMFGSTAIALELLKQGASPNVQDTS 73 || ||| || + +| ||+|+| +|#||| ||| +| || || || +| + Sbjct 14 RGREGEVRALLEAGAL-ANAPNRYGRTPIQ#VMMMGSTRVAELLLLHGADPNCEDPA 68 |
| Danio rerio | Query 19 GDVQEVRRLLHRELVHPDALNRFGKTALQ#VMMFGSTAIALELLKQGASPNVQD 71 ||+ || + | |+ + |++|+||||#|| | +|| ||+ || |||+| Sbjct 17 GDLMEVEQTLQSN-VNVNEKNKYGRTALQ#VMKLGCPSIAETLLQAGADPNVRD 68 |
| Xiphophorus maculatus | Query 36 DALNRFGKTALQ#VMMFGSTAIALELLKQGASPNVQDTS-GTSP 77 + +| ||+ |+|#||| ||+ +| || +|| ||| | | | +| Sbjct 30 NGVNSFGRRAIQ#VMMMGSSEVARLLLTRGADPNVTDKSTGATP 72 |
| Xiphophorus helleri | Query 36 DALNRFGKTALQ#VMMFGSTAIALELLKQGASPNVQDTS-GTSP 77 + +| ||+ |+|#||| ||+ +| || +|| ||| | | | +| Sbjct 30 NGVNSFGRRAIQ#VMMMGSSEVARLLLTRGADPNVTDKSTGATP 72 |
| Xenopus tropicalis | Query 18 RGDVQEVRRLLHRELVHPDALNRFGKTALQ#VMMFGSTAIALELLKQGASPNVQD-TSGTS 76 |||| |++| + +| | |+| +|#||| || +| || || | + | +| Sbjct 14 RGDVDLARQMLQSG-IPVNATNSHGRTPIQ#VMMMGSPKMAQLLLDHGADPKLPDPCTGAC 72 Query 77 P 77 | Query 77 P 77 |
| Solibacter usitatus Ellin6076 | Query 18 RGDVQEVRRLLHRELVHPDALNRFGKTALQ#VMMFGSTAIALELLKQ-GASPNVQDTSGTS 76 | | |+ ||| + +|+| ||+| | | #+ | +||| | || || | + Sbjct 62 RQDDLEMSRLLLKAGANPNAANRYGMTVLA#LACTNGNAAIVELLLQAGADPNAALPGGET 121 Query 77 P 77 | Query 77 P 77 |
| Apis mellifera | Query 18 RGDVQEVRRLLHRELVHPDALNRFGKTALQ#VMMFGSTAIALELLKQ-GASPNVQDTSGTS 76 | |+ |||||| ++ |+||+ | | ||| # ++|| + ||+ | +|+ + Sbjct 71 RNDIDEVRRLL-KKGVNPDSTNEDGLTALH#QCCIDDNEEMMKLLIEFGANVNAEDSEKWT 129 Query 77 P 77 | Query 77 P 77 |
| Equus caballus | Query 35 PDALNRFGKTALQ#VMMFGSTAIALELLKQGASPNVQD 71 |+ +| ||+ +|#||| || +| || || || | Sbjct 5 PNGVNGFGRRPIQ#VMMMGSVHVAELLLLHGADPNRAD 41 |
| Schistosoma japonicum | Query 18 RGDVQEVRRLLHRELVHPDALNRFGKTAL-Q#VMMFGSTAIALELLKQGASPNVQDTSGTS 76 |||+ ||| || | || | | |+| |# + + + ||+ ||+|| +|| + Sbjct 94 RGDLDEVRELLSSG-VCPDVANEDGLTSLHQ#CCIDNNLEMCRLLLRYGANPNSRDTELWT 152 Query 77 P 77 | Query 77 P 77 |
| Drosophila melanogaster | Query 18 RGDVQEVRRLLHRELVHPDALNRFGKTALQ#VMMFGS-TAIALELLKQGASPNVQDTSGTS 76 ||| + |+| || | + | | | #+ | | +||| ||| || + +||+ Sbjct 19 RGDEVALLRVLDSGKVHVDCKDEDGTTPLI#LAAAGGHTYCVMELLDQGADPNSRRLTGTT 78 Query 77 P 77 | Query 77 P 77 |
| Gallus gallus | Query 19 GDVQEVRRLLHRELVHPDALNRFGKTALQ#VMMF-GSTAIALELLKQGASPNVQDTSGTSP 77 | ++ || || | | | | || ||| #+ + | |+| ||+ ||+ | + | +| Sbjct 217 GQIEVVRHLL-RLGVEIDEPNSFGNTALH#IACYMGQDAVANELVNYGANVNQPNEKGFTP 275 |
| Cryptosporidium hominis TU502 | Query 18 RGDVQEVRRLLHRELVHPDALNRFGKTALQ#VMM-FGSTAIALELLKQGASPNVQDTSGTS 76 + +|+ ++++| | ++ + | || #+ | + ||| ||| ||+|||+| | | Sbjct 159 KNEVEFLKKILERYPFLSNSRSSEGSYALH#LACDKGYSEIALLLLKNGANPNVKDNFGDS 218 Query 77 P 77 | Query 77 P 77 |
| Aspergillus oryzae RIB40 | Query 23 EVRRLLHRELVHPDALNRFGKTALQ#V-MMFGSTAIALELLKQGASPNVQDTSGTSP 77 | ||| |+|+| | ||| #| +| |+ + |||++|| | + + +| | Sbjct 167 SVMRLLLSYGADPNAVNSEGATALH#VGVMNGNYTMVAELLQRGADPTLTNAAGWLP 222 |
| Egeria densa | Query 18 RGDVQEVRRLLHRELVHPDALNRFGKTALQ#VMM-FGSTAIALELLKQGASPNVQDTSGTS 76 ||| | +|| |+ + |+||| #+ || || || +||+|| +|+ |++ Sbjct 433 RGDYMLVDKLLECG-SDPNEADSKGRTALH#IAARNGSERCALLLLDRGANPNSKDSDGST 491 Query 77 P 77 | Query 77 P 77 |
| Rickettsia felis URRWXCal2 | Query 19 GDVQEVRRLLHRELVHPDALNRFGKTALQ#VMMF-GSTAIALELLKQGASPNVQDTSGTS 76 ||+ ||+ || + + | |+||| # +| | | ||| ||||+ | || + Sbjct 17 GDLSEVQALLKSGVNIDEHKNERGETALY#NTLFTGYMDRAAFLLKHKASPNIPDNSGQT 75 |
| Bordetella parapertussis 12822 | Query 18 RGDVQEVRRLLHRELVHPDALNRFGKTA-LQ#VMMFGST-----AIALELLKQGASPNVQD 71 || |+ | || | | || +|| | | |+# ++ | || |++ || |+ | Sbjct 145 RGHVEVVDMLL-RAGVDPDHVNRLGWTGLLE#AIILGDGGTRHQAIVARLIEGGADVNLAD 203 Query 72 TSGTSP 77 | || Sbjct 204 GDGVSP 209 |
| Bordetella pertussis Tohama I | Query 18 RGDVQEVRRLLHRELVHPDALNRFGKTA-LQ#VMMFGST-----AIALELLKQGASPNVQD 71 || |+ | || | | || +|| | | |+# ++ | || |++ || |+ | Sbjct 139 RGHVEVVDMLL-RAGVDPDHVNRLGWTGLLE#AIILGDGGTRHQAIVARLIEGGADVNLAD 197 Query 72 TSGTSP 77 | || Sbjct 198 GDGVSP 203 |
| Xenopus laevis | Query 24 VRRLLHRELVHPDALNRFGKTALQ#VMMFGSTAIALE-LLKQGASPNVQDTSG 74 | ++| ++||| + |+| ||# +| +| |++ | ||+|| +| Sbjct 434 VCQILIENNINPDAQDYEGRTPLQ#CAAYGGYIGCMEVLMENKADPNIQDKNG 485 |
| Rattus norvegicus | Query 23 EVRRLLHRELVHPDALNRFGKTALQ#VMM-FGSTAIALELLKQGASPNVQDTSGTSP 77 || || ++ ||| + | | | #| + + +|| || |||||+ +| +| Sbjct 573 EVASLLLQKSASPDAAGKSGLTPLH#VAAHYDNQKVALLLLDQGASPHAAAKNGYTP 628 |
| Methanosarcina barkeri str. Fusaro | Query 19 GDVQEVRRLLHRELVHPDALNRFGKTALQ#VMMFGSTAIALELL-KQGASPNVQDTSGTS 76 | ++ | + | + | ++ + +||||| # + +||| | || ||+|| +| + Sbjct 23 GQIENVEKFLKSDKVDINSQDAYGKTALN#YAVNKGYRDVVELLIKGGADPNIQDENGDT 81 |
| Dictyostelium discoideum AX4 | Query 19 GDVQEVRRLLHRELVHPDALNRFGKTALQ#VMMF-GSTAIALELLKQGASPNVQDTSGTSP 77 ||++++ ||+ || + +| | # | || | || + |+ |++|++| +| Sbjct 46 GDIEKLSNLLNNSATSPDTPDSEKRTPLH#HAAFCGSAACVNFLLDKKANANIKDSAGNTP 105 |
| Caenorhabditis briggsae AF16 | Query 18 RGDVQEVRRLLHRELVHPDALNRFGKTAL-Q#VMMFGSTAIALELLKQGASPNVQDTSGTS 76 | | || |||+ | +|+ | | | | |# + + | | || +||+ | ||| + Sbjct 67 RADYAEVERLLN-ENANPNLHNDDGLTPLHQ#CAIDDNQQIMLLLLDRGANVNAQDTEQWT 125 Query 77 P 77 | Query 77 P 77 |
| Orf virus | Query 34 HPDALNRFGKTALQ#VMM---FGSTAIALELLKQGASPNVQDTSGTSP 77 | +| + |+| | #+ + | | +|| |+ ||+|| | | +| Sbjct 143 HVNAKDDLGRTPLH#IYLSGFFVSAPVALALIALGANPNATDAYGRTP 189 |
| Wolbachia endosymbiont of Drosophila melanogaster | Query 18 RGDVQEVRRLLHRELVHPDALNR--FGKTALQ-#VMMFGSTAIALELLKQGASPNVQDTSG 74 +| + +| |+ | |+ +||+| | + || # | + ||++||+||++| | Sbjct 77 KGCLDIIRFLIEEEKVNINALDRNAFKRIALHH#AAGEGHLEVIKFLLEKGANPNIRDIDG 136 Query 75 TSP 77 +| Sbjct 137 KNP 139 |
| Caenorhabditis elegans | Query 18 RGDVQEVRRLLHRELVHPDALNRFGKTAL-Q#VMMFGSTAIALELLKQGASPNVQDTSGTS 76 | | || |||+ + +|+ | | | | |# + + | | ||++||+ | ||| + Sbjct 67 RADYAEVERLLN-DNANPNLHNDDGLTPLHQ#CAIDDNQQIMLLLLERGANVNAQDTEQWT 125 Query 77 P 77 | Query 77 P 77 |
| Neurospora crassa OR74A | Query 36 DALNRFGKTAL-Q#VMMFGSTAIALELLKQGASPNVQDTSGTS 76 | |++|+||| |# + |+| |+++|| ||+|| | | Sbjct 939 DMANKYGETALMQ#AVSTRRWALAKLLIEKGADPNLQDGQGES 980 |
[Site 4] AIALELLKQG64-ASPNVQDTSG
Gly64  Ala
Ala
iTraq-117 Signal 302.2 (
 ) for AIALELLKQG
) for AIALELLKQG
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Ala55 | Ile56 | Ala57 | Leu58 | Glu59 | Leu60 | Leu61 | Lys62 | Gln63 | Gly64 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Ala65 | Ser66 | Pro67 | Asn68 | Val69 | Gln70 | Asp71 | Thr72 | Ser73 | Gly74 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| PDALNRFGKTALQVMMFGSTAIALELLKQGASPNVQDTSGTSPVHDAARTGFLDTLKVLV |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | N/A | 123.00 | 20 | A Chain A, Structure Of Cdk Inhibitor P19ink4d |
| 2 | Homo sapiens | 123.00 | 20 | cyclin-dependent kinase inhibitor 2D |
| 3 | synthetic construct | 123.00 | 4 | cyclin-dependent kinase inhibitor 2D (p19, inhibi |
| 4 | Mus musculus | 118.00 | 21 | cyclin-dependent kinase inhibitor 2D |
| 5 | Rattus norvegicus | 118.00 | 5 | similar to cyclin-dependent kinase inhibitor 2D |
| 6 | Canis familiaris | 112.00 | 8 | PREDICTED: similar to Cyclin-dependent kinase 4 i |
| 7 | Bos taurus | 112.00 | 3 | cyclin-dependent kinase inhibitor 2D (p19, inhibi |
| 8 | Takifugu rubripes | 90.90 | 2 | p19 |
| 9 | Tetraodon nigroviridis | 90.10 | 6 | unnamed protein product |
| 10 | Xiphophorus maculatus | 73.60 | 1 | 13CDKN2X protein |
| 11 | Xiphophorus helleri | 73.20 | 1 | p13CDKN2X |
| 12 | Monodelphis domestica | 73.20 | 1 | tumor suppressor CDKN2A |
| 13 | Mesocricetus auratus | 72.80 | 3 | p15INK4b cyclin-dependent kinase inhibitor |
| 14 | Monodelphis domestica | 72.80 | 1 | cyclin-dependent kinase inhibitor 2A isoform 1 |
| 15 | Sus scrofa | 70.90 | 4 | cyclin dependant kinase inhibitor |
| 16 | Gallus gallus | 70.10 | 1 | cyclin-dependent kinase inhibitor 2B |
| 17 | human, brain tumors, Peptide, 156 aa | 66.60 | 1 | cyclin D-dependent kinases 4 and 6-binding protei |
| 18 | Danio rerio | 65.10 | 1 | PREDICTED: similar to Chain A, Structure Of P18in |
| 19 | Xenopus tropicalis | 63.90 | 3 | cyclin-dependent kinase inhibitor 2B (p15, inhibi |
| 20 | Equus caballus | 56.20 | 1 | cyclin-dependent kinase 2A inhibitor |
| 21 | Felis catus | 53.50 | 2 | p16/CDKN2A/MTS1 |
| 22 | Mus spretus | 51.20 | 1 | cyclin-dependent kinase inhibitor protein |
| 23 | Rattus sp. | 45.80 | 1 | AAP31998.1 |
| 24 | Arabidopsis thaliana | 44.70 | 1 | EMB506 (EMBRYO DEFECTIVE 506); protein binding |
| 25 | Ustilago maydis 521 | 44.30 | 1 | hypothetical protein UM02100.1 |
| 26 | Theileria annulata strain Ankara | 43.90 | 1 | Ankyrin repeat protein, putative |
| 27 | Dictyostelium discoideum | 43.50 | 1 | homeobox-containing protein Wariai |
| 28 | Drosophila melanogaster | 43.10 | 1 | CG3104 CG3104-PA, isoform A |
| 29 | Samanea saman | 43.10 | 1 | pulvinus inward-rectifying channel for potassium |
| 30 | Dictyostelium discoideum AX4 | 43.10 | 1 | putative homeobox transcription factor |
| 31 | Oryza sativa Japonica Group | 43.10 | 1 | ankyrin repeat protein chloroplast precursor-like |
| 32 | Leifsonia xyli subsp. xyli str. CTCB07 | 42.00 | 1 | hypothetical protein Lxx18600 |
| 33 | Canis lupus familiaris | 35.40 | 1 | inversin |
| 34 | Wolbachia endosymbiont of Drosophila melanogaster | 35.00 | 1 | ankyrin repeat domain protein |
| 35 | Wolbachia pipientis | 34.70 | 1 | ankyrin domain protein |
| 36 | Xenopus laevis | 33.50 | 4 | inv2 |
| 37 | Apis mellifera | 33.50 | 1 | PREDICTED: similar to no mechanoreceptor potentia |
| 38 | Caenorhabditis elegans | 33.10 | 7 | AO13 ankyrin |
| 39 | synthetic construct | 33.10 | 3 | ankyrin repeat protein E2_17 |
| 40 | Treponema denticola ATCC 35405 | 33.10 | 1 | ankyrin repeat protein |
| 41 | Caenorhabditis briggsae AF16 | 32.30 | 1 | Hypothetical protein CBG09456 |
| 42 | Hemicentrotus pulcherrimus | 31.60 | 1 | inv-like protein |
Top-ranked sequences
| organism | matching |
|---|---|
| N/A | Query 35 PDALNRFGKTALQVMMFGSTAIALELLKQG#ASPNVQDTSGTSPVHDAARTGFLDTLKVLV 94 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 35 PDALNRFGKTALQVMMFGSTAIALELLKQG#ASPNVQDTSGTSPVHDAARTGFLDTLKVLV 94 |
| Homo sapiens | Query 35 PDALNRFGKTALQVMMFGSTAIALELLKQG#ASPNVQDTSGTSPVHDAARTGFLDTLKVLV 94 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 35 PDALNRFGKTALQVMMFGSTAIALELLKQG#ASPNVQDTSGTSPVHDAARTGFLDTLKVLV 94 |
| synthetic construct | Query 35 PDALNRFGKTALQVMMFGSTAIALELLKQG#ASPNVQDTSGTSPVHDAARTGFLDTLKVLV 94 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 35 PDALNRFGKTALQVMMFGSTAIALELLKQG#ASPNVQDTSGTSPVHDAARTGFLDTLKVLV 94 |
| Mus musculus | Query 35 PDALNRFGKTALQVMMFGSTAIALELLKQG#ASPNVQDTSGTSPVHDAARTGFLDTLKVLV 94 ||||||||||||||||||| |+||||||||#||||||| |||||||||||||||||||||| Sbjct 35 PDALNRFGKTALQVMMFGSPAVALELLKQG#ASPNVQDASGTSPVHDAARTGFLDTLKVLV 94 |
| Rattus norvegicus | Query 35 PDALNRFGKTALQVMMFGSTAIALELLKQG#ASPNVQDTSGTSPVHDAARTGFLDTLKVLV 94 ||||||||||||||||||| |+||||||||#||||||| |||||||||||||||||||||| Sbjct 35 PDALNRFGKTALQVMMFGSPAVALELLKQG#ASPNVQDASGTSPVHDAARTGFLDTLKVLV 94 |
| Canis familiaris | Query 35 PDALNRFGKTALQVMMFGSTAIALELLKQG#ASPNVQDTSGTSPVHDAARTGFLDTLKVLV 94 ||||||||||||||||||| |||||||||#||||||| +||+| |||||||||||||||| Sbjct 35 PDALNRFGKTALQVMMFGSPTIALELLKQG#ASPNVQDATGTTPAHDAARTGFLDTLKVLV 94 |
| Bos taurus | Query 35 PDALNRFGKTALQVMMFGSTAIALELLKQG#ASPNVQDTSGTSPVHDAARTGFLDTLKVLV 94 || |||||||||||||||| |||||||||#||||||| |||+| |||||||||||||||| Sbjct 35 PDVLNRFGKTALQVMMFGSPTIALELLKQG#ASPNVQDASGTTPAHDAARTGFLDTLKVLV 94 |
| Takifugu rubripes | Query 35 PDALNRFGKTALQVMMFGSTAIALELLKQG#ASPNVQDTSGTSPVHDAARTGFLDTLKVLV 94 || | |||||||||| |+ || ||++|#| ||||| | +||||||||||||||+||| Sbjct 35 PDTRNEFGKTALQVMMLGNCKIASLLLEKG#ADPNVQDKHGIAPVHDAARTGFLDTLQVLV 94 |
| Tetraodon nigroviridis | Query 35 PDALNRFGKTALQVMMFGSTAIALELLKQG#ASPNVQDTSGTSPVHDAARTGFLDTLKVLV 94 || | |||||||||| |+ || ||++|#| ||||| | +||||||||||||||+||| Sbjct 35 PDTPNEFGKTALQVMMLGNCKIASLLLEKG#ADPNVQDKHGIAPVHDAARTGFLDTLQVLV 94 |
| Xiphophorus maculatus | Query 36 DALNRFGKTALQVMMFGSTAIALELLKQG#ASPNVQDTS-GTSPVHDAARTGFLDTLKVLV 94 + +| ||+ |+|||| ||+ +| || +|#| ||| | | | +|+|||||||||||+++|| Sbjct 30 NGVNSFGRRAIQVMMMGSSEVARLLLTRG#ADPNVTDKSTGATPLHDAARTGFLDTVQLLV 89 |
| Xiphophorus helleri | Query 36 DALNRFGKTALQVMMFGSTAIALELLKQG#ASPNVQDTS-GTSPVHDAARTGFLDTLKVLV 94 + +| ||+ |+|||| ||+ +| || +|#| ||| | | | +|+|||||||||||+++|| Sbjct 30 NGVNSFGRRAIQVMMMGSSEVARLLLTRG#ADPNVTDKSTGATPLHDAARTGFLDTVQLLV 89 |
| Monodelphis domestica | Query 35 PDALNRFGKTALQVMMFGSTAIALELLKQG#ASPNVQD-TSGTSPVHDAARTGFLDTLKVL 93 |+|+||||++|+|||| |+ +| ||+ |#| || | |+ | ||||||| |||||| +| Sbjct 38 PNAVNRFGRSAIQVMMMGNVRLAAILLQYG#AEPNTPDPTTLTLPVHDAAREGFLDTLMLL 97 |
| Mesocricetus auratus | Query 35 PDALNRFGKTALQVMMFGSTAIALELLKQG#ASPNVQDTSG-TSPVHDAARTGFLDTLKVL 93 |+|+||||+ +|||| ||| +| || |#| || | + | ||||||| |||||| || Sbjct 32 PNAVNRFGRRPIQVMMMGSTQVAELLLLHG#AEPNCADPNTLTRPVHDAAREGFLDTLVVL 91 |
| Monodelphis domestica | Query 35 PDALNRFGKTALQVMMFGSTAIALELLKQG#ASPNVQD-TSGTSPVHDAARTGFLDTLKVL 93 |+|+||||++|+|||| |+ +| ||+ |#| || | |+ | ||||||| |||||| +| Sbjct 72 PNAVNRFGRSAIQVMMMGNVRLAAILLQYG#AEPNTPDPTTLTLPVHDAAREGFLDTLMLL 131 |
| Sus scrofa | Query 36 DALNRFGKTALQVMMFGSTAIALELLKQG#ASPNVQDTSG-TSPVHDAARTGFLDTLKVL 93 +| ||+|+| +|||| ||| +| || |#| || +| + | ||||||| |||||| || Sbjct 31 NAPNRYGRTPIQVMMMGSTRVAELLLLHG#ADPNCEDPATLTRPVHDAAREGFLDTLVVL 89 |
| Gallus gallus | Query 35 PDALNRFGKTALQVMMFGSTAIALELLKQG#ASPNVQDT-SGTSPVHDAARTGFLDTLKVL 93 |+|+| ||+| +|||| || +| ||++|#| || | +| | ||||| |||||| | Sbjct 35 PNAVNSFGRTPIQVMMLGSPRVAELLLQRG#ADPNRPDPRTGCRPAHDAARAGFLDTLAAL 94 |
| human, brain tumors, Peptide, 156 aa | Query 35 PDALNRFGKTALQVMMFGSTAIALELLKQG#ASPNVQDTSG-TSPVHDAARTGFLDTLKVL 93 |+| | +|+ +|||| || +| || |#| || | + | ||||||| |||||| || Sbjct 38 PNAPNSYGRRPIQVMMMGSARVAELLLLHG#AEPNCADPATLTRPVHDAAREGFLDTLVVL 97 |
| Danio rerio | Query 39 NRFGKTALQVMMFGSTAIALELLKQG#ASPNVQD-TSGTSPVHDAARTGFLDTLKVL 93 |++|+|||||| | +|| ||+ |#| |||+| | + +||||| |+|||| || Sbjct 36 NKYGRTALQVMKLGCPSIAETLLQAG#ADPNVRDPILGLTVIHDAARDGYLDTLHVL 91 |
| Xenopus tropicalis | Query 36 DALNRFGKTALQVMMFGSTAIALELLKQG#ASPNVQD-TSGTSPVHDAARTGFLDTLKVLV 94 +| | |+| +|||| || +| || |#| | + | +| ||||||| |||||| ||+ Sbjct 31 NATNSHGRTPIQVMMMGSPKMAQLLLDHG#ADPKLPDPCTGACPVHDAAREGFLDTLLVLL 90 |
| Equus caballus | Query 35 PDALNRFGKTALQVMMFGSTAIALELLKQG#ASPNVQDTSG-TSPVHDAARTGFL 87 |+ +| ||+ +|||| || +| || |#| || | | ||||||| ||| Sbjct 5 PNGVNGFGRRPIQVMMMGSVHVAELLLLHG#ADPNRADPDTLTRPVHDAAREGFL 58 |
| Felis catus | Query 48 VMMFGSTAIALELLKQG#ASPNVQDTSG-TSPVHDAARTGFLDTLKVL 93 ||| || +| || |#| || | + | ||||||| |||||| || Sbjct 1 VMMMGSARVAELLLLHG#ADPNCADPATLTRPVHDAAREGFLDTLVVL 47 |
| Mus spretus | Query 48 VMMFGSTAIALELLKQG#ASPNVQD-TSGTSPVHDAARTGFLDTLKVL 93 ||| |+ +| || |#| | +| |+ + ||||||| |||||| || Sbjct 1 VMMMGNVHVAALLLNYG#ADSNCEDPTTFSRPVHDAAREGFLDTLVVL 47 |
| Rattus sp. | Query 49 MMFGSTAIALELLKQG#ASPNVQDTSG-TSPVHDAARTGFLD 88 || || +| || |#| || | + | ||||||| |||| Sbjct 1 MMMGSAQVAELLLLHG#AEPNCADPATLTRPVHDAAREGFLD 41 |
| Arabidopsis thaliana | Query 36 DALNRFGKTALQVMMFGST-AIALELLKQG#ASPNVQDTSGTSPVHDAARTGFLDTLKVL 93 | +++ +||| + | |+ ||++|#|+|++|| | +|+| | + | | |+|+| Sbjct 179 DDVDKDNQTALHKAIIGKKEAVISHLLRKG#ANPHLQDRDGAAPIHYAVQVGALQTVKLL 237 |
| Ustilago maydis 521 | Query 39 NRFGKTAL-QVMMFGSTAIALELLKQG#ASPNVQDTSGTSPVHDAARTGFLDTLKVLV 94 +| |+||| + | | ++| || |#| | || | + +|+| |+|| +++|| Sbjct 398 DREGETALHKAAMAGKLSVASLLLSHG#ADANAQDADGWTALHNACSRGYLDLVRLLV 454 |
| Theileria annulata strain Ankara | Query 52 GSTAIALELLKQG#ASPNVQDTSGTSPVHDAARTGFLDTLKVLV 94 |+| + +||+ |#| ||+ |+ | +|+| |+ +|+| |+|+ Sbjct 253 GNTGLVKKLLENG#ADPNICDSQGWTPLHCASESGYLSVCKLLI 295 |
| Dictyostelium discoideum | Query 36 DALNRFGKTALQVM-MFGSTAIALELLKQG#ASPNVQDTSGTSPVHDAARTGFLDTLKVLV 94 +|++ | | | + | | ||+ |#| ||+||+ | +|+| | | ++|+| |+ Sbjct 532 NAIDIDGHTPLHTSSLMGQYLITRLLLENG#ADPNIQDSEGYTPIHYAVRESRIETVKFLI 591 |
| Drosophila melanogaster | Query 52 GSTAIALELLKQG#ASPNVQDTSGTSPVHDAARTGFLDTLKVLV 94 | | +||| ||#| || + +||+|+ ||+ | || +|+|+ Sbjct 54 GHTYCVMELLDQG#ADPNSRRLTGTTPLFFAAQGGHLDVVKILI 96 |
| Samanea saman | Query 46 LQVMMFGSTAIALELLKQG#ASPNVQDTSGTSPVHDAARTGFLDTLKVLV 94 | | |+ | |||| |# |+| |+ | +|+| || | | +|||+ Sbjct 548 LTVASTGNAAFLEELLKAG#LDPDVGDSKGKTPLHIAASNGHEDCVKVLL 596 |
| Dictyostelium discoideum AX4 | Query 36 DALNRFGKTALQVM-MFGSTAIALELLKQG#ASPNVQDTSGTSPVHDAARTGFLDTLKVLV 94 +|++ | | | + | | ||+ |#| ||+||+ | +|+| | | ++|+| |+ Sbjct 535 NAIDIDGHTPLHTSSLMGHDLITRLLLENG#ADPNIQDSEGYTPIHYAVRESRIETVKFLI 594 |
| Oryza sativa Japonica Group | Query 38 LNRFGKTALQVMMFGS-TAIALELLKQG#ASPNVQDTSGTSPVHDAARTGFLDTLKVLV 94 |++ | | | + | |+ ||++|#|+|+|+| | +|+| | + | | |+|+|+ Sbjct 190 LDKDGFTPLHKAVIGKKEAVISHLLRRG#ANPHVRDRDGATPLHYAVQVGALQTVKLLI 247 |
| Leifsonia xyli subsp. xyli str. CTCB07 | Query 36 DALNRFGKTALQVMMF------GSTAIALELLKQG#ASPNVQDTSGTSPVHDAARTGF 86 | +|| | |||| + |+ + +|| |#| |+++| | +|+ +| | || Sbjct 179 DHVNRLGGTALQEAIVMGDGGRGAQDVVRQLLTAG#ADPDIRDADGRTPLCNAERNGF 235 |
| Canis lupus familiaris | Query 35 PDALNRFGKTALQVMMFGSTAIALE-LLKQG#ASPNVQDTSGTSPVHDAARTGFLDTLKVL 93 |+ + |+| || +| + |++ #| ||+|| | + +| + |+|| +|+| Sbjct 449 PNVQDYAGRTPLQCAAYGGYINCMAVLMENN#ADPNIQDKEGRTALHWSCNNGYLDAIKLL 508 Query 94 V 94 + Sbjct 509 L 509 Query 36 DALNRFGKTALQVMMFGSTAIALELLKQG#A-SPNVQDTSGTSPVHDAARTGFLDTLKVLV 94 | +++ | + | | | ++| + # +||||| +| +|+ || |+++ + ||+ Sbjct 417 DLVDQDGHSLLHWAALGGNADVCQILIEN#KINPNVQDYAGRTPLQCAAYGGYINCMAVLM 476 |
| Wolbachia endosymbiont of Drosophila melanogaster | Query 36 DALNRFGKTALQ-VMMFGSTAIALELLKQG#ASPNVQDTSGTSPVHDAARTGFLDTLKVLV 94 | ||+|+| | +| | + || + # + ||| | +|+|||| | ++ +| |+ Sbjct 140 DLKNRYGETPLHYAAKYGHTQVLENLLGRS#TNVNVQSEVGRTPLHDAANNGHIEVVKHLI 199 Query 52 GSTAIALELLKQG#ASPNVQDTSGTSPVHDAARTGFLDTLKVLV 94 | + |+|+|#| ||| | +|+|+||+ | ++||+ Sbjct 190 GHIEVVKHLIKKG#ADVNVQSKVGRTPLHNAAKHGHTQVVEVLL 232 Query 39 NRFGKTALQ-VMMFGSTAIALELLKQG#ASPNVQDTSGTSPVHDAARTGFLDTLKVLV 94 ++ |+| | | | + |||+|#| |+|| | +|+| | + + |+|+ Sbjct 209 SKVGRTPLHNAAKHGHTQVVEVLLKKG#ADVNIQDRGGRTPLHYAVQRRYPKLAKLLL 265 |
| Wolbachia pipientis | Query 36 DALNRFGKTALQ-VMMFGSTAIALELLKQG#ASPNVQDTSGTSPVHDAARTGFLDTLKVLV 94 | ||+|+| | +| | + || + # + ||| | +|+|||| | ++ +| |+ Sbjct 139 DLKNRYGETPLHYAAKYGHTQVLENLLGRS#TNVNVQSEVGRTPLHDAANNGHIEVVKHLI 198 Query 52 GSTAIALELLKQG#ASPNVQDTSGTSPVHDAARTGFLDTLKVLV 94 | + |+|+|#| ||| | +|+|+|| |+++ +| |+ Sbjct 189 GHIEVVKHLIKKG#ADVNVQSKVGRTPLHNAANNGYIEVVKHLI 231 Query 39 NRFGKTALQ-VMMFGSTAIALELLKQG#ASPNVQDTSGTSPVHDAARTGFLDTLKVLV 94 ++ |+| | | | + |||+|#| |+|| | +|+| | + |+ |+|+ Sbjct 274 SKVGRTPLHNAAKHGHTQVVEVLLKKG#ADVNIQDRGGRTPLHYAVQRGYPKLAKLLL 330 Query 36 DALNRFGKTALQ-VMMFGSTAIALELLKQG#ASPNVQDTSGTSPVHDAARTGFLDTLKVLV 94 + ++++|+| | | + |+++ #| ||| | +|+|+||+ | ++||+ Sbjct 238 NVVDQYGRTPLHDAAKHGRIEVVKHLIEKE#ADVNVQSKVGRTPLHNAAKHGHTQVVEVLL 297 Query 39 NRFGKTALQ-VMMFGSTAIALELLKQG#ASPNVQDTSGTSPVHDAARTGFLDTLKVLV 94 ++ |+| | | + |+|+ #| || | | +|+||||+ | ++ +| |+ Sbjct 208 SKVGRTPLHNAANNGYIEVVKHLIKKE#ADVNVVDQYGRTPLHDAAKHGRIEVVKHLI 264 |
| Xenopus laevis | Query 35 PDALNRFGKTALQVMMFGSTAIALE-LLKQG#ASPNVQDTSGTSPVHDAARTGFLDTLKVL 93 ||| + |+| || +| +| |++ #| ||+|| +| + +| + |+|| +|+| Sbjct 445 PDAQDYEGRTPLQCAAYGGYIGCMEVLMENK#ADPNIQDKNGRTALHWSCNNGYLDAVKLL 504 Query 94 V 94 + Sbjct 505 L 505 Query 50 MFGSTAIALELLKQG#ASPNV-QDTSGTSPVHDAARTGFLDTLKVLV 94 + | | || ||++ # |||+ |+ | +|+| ||+ ||++||+ Sbjct 260 LLGHTPIAHLLLERN#NSPNIPSDSQGATPLHYAAQGNCPDTVRVLL 305 Query 36 DALNRFGKTALQVMMFGST-AIALELLKQG#ASPNVQDTSGTSPVHDAARTG 85 | ++ |+| | + | + | ||| |#| | | || + +| ||+|| Sbjct 38 DQEDQLGRTPLMYSVLGDRRSCAEALLKHG#AKVNRPDRSGRTALHLAAQTG 88 |
| Apis mellifera | Query 39 NRFGKTALQVM-MFGSTAIALELLKQG#ASPNVQDTSGTSPVHDAARTGFLDTLKVLV 94 +|+||| | + | + || ||#| | | +| +|+| ||| |+|| +|+|| Sbjct 1025 DRYGKTGLHIAATHGHYQMVEVLLGQG#AEINATDKNGWTPLHCAARAGYLDVVKLLV 1081 Query 56 IALELLKQG#ASPNVQDTSGTSPVHDAARTGFLDT 89 || ||| |#| ||+ | +||| || | | | Sbjct 468 CALMLLKSG#AGPNLTTDDGQTPVHVAASHGNLAT 501 |
| Caenorhabditis elegans | Query 36 DALNRFGKTALQVMMF-GSTAIALELLKQG#ASPNVQDTSGTSPVHDAARTGFLDTLKVLV 94 +| | | | | | |+ | + ||+||#|+|+|+ | +|+| ||| | ++||+ Sbjct 419 EATTESGLTPLHVAAFMGAINIVIYLLQQG#ANPDVETVRGETPLHLAARANQTDVVRVLI 478 Query 36 DALNRFGKTALQVM-MFGSTAIALELLKQG#ASPNVQDTSGTSPVHDAARTGFLDTLKVLV 94 || | | ||| + + | + | |++ |#|+ ||| +| +|++ ||+ + +| |+ Sbjct 93 DAATRKGNTALHIASLAGQSLIVTILVENG#ANVNVQSVNGFTPLYMAAQENHEEVVKYLL 152 Query 60 LLKQG#ASPNVQDTSGTSPVHDAARTGFLDTLKVL 93 ||+ |#|||| | +| +|+ | | |++ ++ | Sbjct 774 LLENG#ASPNEQTATGQTPLSIAQRLGYVSVVETL 807 |
| synthetic construct | Query 36 DALNRFGKTALQVMMF-GSTAIALELLKQG#ASPNVQDTSGTSPVHDAARTGFLDTLKVLV 94 +|++ +| | | + + | | ||| |#| | +| | +|+| || | |+ ++||+ Sbjct 41 NAMDYYGSTPLHLAAYNGHLEIVEVLLKNG#ADVNAKDFQGETPLHLAANNGHLEIVEVLL 100 Query 41 FGKTALQVMMFGSTAIALELLKQG#ASPNVQDTSGTSPVHDAARTGFLDTLKVLV 94 || |+ | |+ |#| | | |++|+| || | |+ ++||+ Sbjct 14 LGKKLLEAARAGQDDEVRILMANG#ADVNAMDYYGSTPLHLAAYNGHLEIVEVLL 67 |
| Treponema denticola ATCC 35405 | Query 35 PDALNRFGKTALQVMMFGSTAIAL--ELLKQG#ASPNVQDTSGTSPVHDAARTGFLDTL 90 |++ | | ||| ++| ++ | ||+ |#|+||++| | +|+| ||| | | + Sbjct 307 PNSRNSSGNTALHLVMPEASRSKLFNELITAG#ANPNLKDNYGETPLHVAARIGMNDDI 364 Query 44 TALQVMMF-GSTAIALELLKQG#ASPNVQDTSGTSPVH----DAARTGFLDTL 90 ||+| ++ |+| |+ ||+ |#| || +++|| + +| +|+|+ + | Sbjct 283 TAMQEAVYNGNTEAAILLLQAG#ADPNSRNSSGNTALHLVMPEASRSKLFNEL 334 |
| Caenorhabditis briggsae AF16 | Query 36 DALNRFGKTALQVMMF-GSTAIALELLKQG#ASPNVQDTSGTSPVHDAARTGFLDTLKVLV 94 +| | | | | | |+ | + ||+||#|+|+|+ | +|+| ||| | ++||+ Sbjct 420 EATTESGLTPLHVAAFMGAINIVIYLLQQG#ANPDVETVRGETPLHLAARANQTDVVRVLI 479 Query 36 DALNRFGKTALQVM-MFGSTAIALELLKQG#ASPNVQDTSGTSPVHDAARTGFLDTLKVLV 94 || | | ||| + + | + | |++ |#|+ ||| +| +|++ ||+ | ++ |+ Sbjct 94 DAATRKGNTALHIASLAGQSLIVTILVENG#ANVNVQSVNGFTPLYMAAQENHEDVVRYLL 153 Query 35 PDALNRFGKTALQVM-MFGSTAIALELLKQG#ASPNVQDTSGTSPVHDAARTGFLDTLKVL 93 || ++ | | | + +| + || +|#|+ | | ||+| | + | ++ || Sbjct 221 PDVTSKSGFTPLHIAAHYGHENVGTLLLDKG#ANVNYQARHNISPLHVATKWGRINMANVL 280 Query 94 V 94 + Sbjct 281 L 281 Query 47 QVMMFGSTAIALELLKQG#ASPNVQDTSGTSPVHDAARTGFLDTLKVL 93 | | ||+ |#|||| | +| +|+ | | |++ ++ | Sbjct 762 QAAQQGHNNCVRYLLENG#ASPNEQTATGQTPLAIAQRLGYVSVVETL 808 |
| Hemicentrotus pulcherrimus | Query 35 PDALNRFGKTALQVMMFGSTAIALELL-KQG#ASPNVQDTSGTSPVHDAARTGFLDTLKVL 93 |+ + |+| || +| + || + |#| ||+|| | + +| | ||+|| ++| Sbjct 499 PNVQDNAGRTPLQCAAYGGFIRCMTLLLEHG#ADPNLQDNEGMTALHWACSTGYLDATRLL 558 Query 94 V 94 + Sbjct 559 L 559 Query 52 GSTAIALELLKQG#ASPNVQDTSGTSPVHDAARTGFLDTLKVLV 94 | | + |+ # |||||| +| +|+ || ||+ + +|+ Sbjct 484 GHTCVCYHLMTHD#ISPNVQDNAGRTPLQCAAYGGFIRCMTLLL 526 Query 36 DALNRFGKTALQVMMFGSTAIALE----LLKQG#ASPNVQDTSGTSPVHDAARTGFLDTLK 91 | ++||+| | || | |+ |+|+|#|+ + +| | + +| | | | Sbjct 92 DQADQFGRTPL---MFAVLADRLDCAEVLIKKG#ANVDAKDNGGRTALHWATYKGVFRLCK 148 Query 92 VLV 94 +|| Sbjct 149 LLV 151 |
[Site 5] DGTGALPIHL113-AVQEGHTAVV
Leu113  Ala
Ala
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Asp104 | Gly105 | Thr106 | Gly107 | Ala108 | Leu109 | Pro110 | Ile111 | His112 | Leu113 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Ala114 | Val115 | Gln116 | Glu117 | Gly118 | His119 | Thr120 | Ala121 | Val122 | Val123 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| TGFLDTLKVLVEHGADVNVPDGTGALPIHLAVQEGHTAVVSFLAAESDLHRRDARGLTPL |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | N/A | 121.00 | 16 | A Chain A, Structure Of Cdk Inhibitor P19ink4d |
| 2 | Homo sapiens | 121.00 | 9 | cyclin-dependent kinase inhibitor 2D |
| 3 | synthetic construct | 121.00 | 2 | cyclin-dependent kinase inhibitor 2D (p19, inhibi |
| 4 | Canis familiaris | 113.00 | 7 | PREDICTED: similar to Cyclin-dependent kinase 4 i |
| 5 | Bos taurus | 109.00 | 4 | cyclin-dependent kinase inhibitor 2D (p19, inhibi |
| 6 | Mus musculus | 100.00 | 14 | cyclin-dependent kinase inhibitor 2D |
| 7 | Rattus norvegicus | 100.00 | 3 | similar to cyclin-dependent kinase inhibitor 2D |
| 8 | Takifugu rubripes | 77.40 | 1 | p19 |
| 9 | Tetraodon nigroviridis | 60.10 | 3 | unnamed protein product |
| 10 | Danio rerio | 56.60 | 1 | PREDICTED: similar to Chain A, Structure Of P18in |
| 11 | Caenorhabditis elegans | 47.40 | 1 | TRPA cation channel homolog family member (trpa-1 |
| 12 | Plasmodium berghei strain ANKA | 47.40 | 1 | hypothetical protein PB000903.01.0 |
| 13 | Plasmodium falciparum 3D7 | 47.40 | 1 | hypothetical protein PF11_0197 |
| 14 | synthetic construct | 43.90 | 6 | ankyrin repeat protein E4_8 |
| 15 | Strongylocentrotus purpuratus | 33.50 | 3 | PREDICTED: similar to ankyrin 2,3/unc44 |
| 16 | Wolbachia endosymbiont of Drosophila ananassae | 32.70 | 1 | ankyrin repeat domain protein |
| 17 | Arabidopsis thaliana | 31.20 | 3 | SPIK (SHAKER POLLEN INWARD K+ CHANNEL); cyclic nu |
| 18 | Xiphophorus maculatus | 31.20 | 1 | 13CDKN2X protein |
| 19 | Xiphophorus helleri | 30.80 | 1 | p13CDKN2X |
Top-ranked sequences
| organism | matching |
|---|---|
| N/A | Query 84 TGFLDTLKVLVEHGADVNVPDGTGALPIHL#AVQEGHTAVVSFLAAESDLHRRDARGLTPL 143 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 84 TGFLDTLKVLVEHGADVNVPDGTGALPIHL#AVQEGHTAVVSFLAAESDLHRRDARGLTPL 143 |
| Homo sapiens | Query 84 TGFLDTLKVLVEHGADVNVPDGTGALPIHL#AVQEGHTAVVSFLAAESDLHRRDARGLTPL 143 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 84 TGFLDTLKVLVEHGADVNVPDGTGALPIHL#AVQEGHTAVVSFLAAESDLHRRDARGLTPL 143 |
| synthetic construct | Query 84 TGFLDTLKVLVEHGADVNVPDGTGALPIHL#AVQEGHTAVVSFLAAESDLHRRDARGLTPL 143 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 84 TGFLDTLKVLVEHGADVNVPDGTGALPIHL#AVQEGHTAVVSFLAAESDLHRRDARGLTPL 143 |
| Canis familiaris | Query 84 TGFLDTLKVLVEHGADVNVPDGTGALPIHL#AVQEGHTAVVSFLAAESDLHRRDARGLTPL 143 |||||||||||||||||| |||||||||||#||+||||||||||| ||||| ||||||||| Sbjct 84 TGFLDTLKVLVEHGADVNAPDGTGALPIHL#AVREGHTAVVSFLATESDLHHRDARGLTPL 143 |
| Bos taurus | Query 84 TGFLDTLKVLVEHGADVNVPDGTGALPIHL#AVQEGHTAVVSFLAAESDLHRRDARGLTPL 143 |||||||||||||||||| |||||||||||#||+||||+|||||| ||||| ||| ||||| Sbjct 84 TGFLDTLKVLVEHGADVNAPDGTGALPIHL#AVREGHTSVVSFLATESDLHHRDATGLTPL 143 |
| Mus musculus | Query 84 TGFLDTLKVLVEHGADVNVPDGTGALPIHL#AVQEGHTAVVSFLAAESDLHRRDARGLTPL 143 |||||||||||||||||| | ||+|||||#|++|||++|||||| ||||| ||| ||||| Sbjct 84 TGFLDTLKVLVEHGADVNALDSTGSLPIHL#AIREGHSSVVSFLAPESDLHHRDASGLTPL 143 |
| Rattus norvegicus | Query 84 TGFLDTLKVLVEHGADVNVPDGTGALPIHL#AVQEGHTAVVSFLAAESDLHRRDARGLTPL 143 |||||||||||||||||| | ||+|||||#|++|||++|||||| ||||| +|| ||||| Sbjct 84 TGFLDTLKVLVEHGADVNTLDSTGSLPIHL#AIREGHSSVVSFLAPESDLHHKDASGLTPL 143 |
| Takifugu rubripes | Query 84 TGFLDTLKVLVEHGADVNVPDGTGALPIHL#AVQEGHTAVVSFLAAESDLHRRDARGLT 141 |||||||+||||+|| ||+|| +||||||+#|++||| || ||| ||| + | | Sbjct 84 TGFLDTLQVLVEYGASVNLPDQSGALPIHI#AIREGHRDVVEFLAPRSDLKHANKSGQT 141 |
| Tetraodon nigroviridis | Query 84 TGFLDTLKVLVEHGADVNVPDGTGALPI-HL#AVQEGHTAVVSFLA 127 |||||||+||||+|| ||+|| +||||| | # ++|| || ||| Sbjct 84 TGFLDTLQVLVEYGASVNLPDQSGALPIPHC#QSEKGHRDVVEFLA 128 |
| Danio rerio | Query 85 GFLDTLKVLVEHGADVNVPDGTGALPIHL#AVQEGHTAVVSFLAAE--SDLHRRDARGLTP 142 |+|||| || ++|||||+ | | ||+||#| +||| || || + +|+| || Sbjct 83 GYLDTLHVLAQNGADVNLLDNDGNLPLHL#AAREGHLDVVQFLVTHCVTQPFLANAKGYTP 142 |
| Caenorhabditis elegans | Query 85 GFLDTLKVLVEHGADVNVPDGTGALPIHL#AVQEGHTAVVSFLAAES--DLHRRDARGLTP 142 |||+ || ||| | + | + |+|+|+#| | |||+| | ++ | +|||| Sbjct 305 GFLEVLKALVEAGGNKNELNEVKAVPLHV#AAQMNQLEVVSYLIEEEKDNIDVVDEQGLTP 364 Query 143 L 143 | Query 143 L 143 |
| Plasmodium berghei strain ANKA | Query 85 GFLDTLKVLVEHGADVNVPDGTGALPIHL#AVQEGHTAVVSFLA-AESDLHRRDARGLT 141 |||| +| |++ ||++| | | +|+#| | ++ || | ++|+++| ||| Sbjct 251 GFLDIVKFLIKAGANINEEDSLGDSVLHI#AAYSGKMEIIKFLINAGVNIHKKNAEGLT 308 |
| Plasmodium falciparum 3D7 | Query 85 GFLDTLKVLVEHGADVNVPDGTGALPIHL#AVQEGHTAVVSFL-AAESDLHRRDARGLT 141 |++| +| || ||++|| | | |+|+#| || || + +|+++ ++ ||| Sbjct 306 GYIDIVKYLVNQGANINVEDSYGDTPLHM#AAYSEKLNVVDFLKSVGADINKTNSEGLT 363 |
| synthetic construct | Query 85 GFLDTLKVLVEHGADVNVPDGTGALPIHL#AVQEGHTAVVSFLAAE-SDLHRRDARGLTPL 143 | |+ ++||+++||||| | | |+||#| || +| | +|++ | |+||| Sbjct 91 GHLEIVEVLLKNGADVNAIDTIGYTPLHL#AANNGHLEIVEVLLKNGADVNAHDTNGVTPL 150 Query 85 GFLDTLKVLVEHGADVNVPDGTGALPIHL#AVQEGHTAVVSFLAAE-SDLHRRDARGLTPL 143 | |+ ++||+++||||| | || |+||#| || +| | +|++ | | ||| Sbjct 58 GHLEIVEVLLKNGADVNADDVTGFTPLHL#AAVWGHLEIVEVLLKNGADVNAIDTIGYTPL 117 Query 84 TGFLDTLKVLVEHGADVNVPDGTGALPIHL#AVQEGHTAVVS-FLAAESDLHRRDARGLTP 142 | |+ ++||+++||||| | | |+||#| ||| +| | +|++ +| | | Sbjct 123 NGHLEIVEVLLKNGADVNAHDTNGVTPLHL#AAHEGHLEIVEVLLKYGADVNAQDKFGKTA 182 Query 143 L 143 Sbjct 183 F 183 Query 88 DTLKVLVEHGADVNVPDGTGALPIHL#AVQEGHTAVVSFLAAE-SDLHRRDARGLTPL 143 | +++|+ +||||| | | |+||#| || +| | +|++ | | ||| Sbjct 28 DEVRILMANGADVNARDRDGNTPLHL#AADMGHLEIVEVLLKNGADVNADDVTGFTPL 84 |
| Strongylocentrotus purpuratus | Query 84 TGFLDTLKVLVEHGADVNVPDGTGALPIHL#AVQEGHTAVVSFLAAE-SDLHRRDARGLTP 142 +| +|++| |||| | | |+|+#| +||| +| || |+ |++|+ |+|| Sbjct 67 SGHASIVKLVVSKGADVEGIDDDGLTPLHV#ACREGHLEIVQFLFAKGGDINRQTFDGMTP 126 Query 143 L 143 | Query 143 L 143 Query 87 LDTLKVLVEHGADVNVPDGTGALPIHL#AVQEGHTAVVSFLAAE-SDLHRRDARGLTPL 143 || +| |+ |||+| | | |+|+#| | ++ +| ++ +|| + + | | | Sbjct 202 LDVIKSLLSAGADLNWLDKDGTTPLHV#ACYNYRTQILKYLLSKGADLQKAELDGTTSL 259 Query 85 GFLDTLKVLVEHGADVNVPDGTGALPIHL#AVQEGHTAVVSFLAAES-DLHRRDARGLTPL 143 | |+ ++ | | |+| | |+ +#| + || |+ +| ++ ++ | | +| || Sbjct 101 GHLEIVQFLFAKGGDINRQTFDGMTPLAM#ATKRGHVEVLKYLISKGVEIERSDKKGCPPL 160 Query 97 GADVNVPDGTGALPIHL#AVQEGHTAVVSFLAAESDLHRR 135 | || || | |+|+#| | |+++ | + |+| Sbjct 272 GELVNRPDARGETPLHV#ATSNGFTSIIDILVSRGGSHQR 310 |
| Wolbachia endosymbiont of Drosophila ananassae | Query 85 GFLDTLKVLVEHGADVNVPDGTGALPIHL#AVQEGHTAVVSFLAAESDLHRRDARGLTPL 143 | | +| |+|| || || | |+| #|+ || | + ++ +| | ||| Sbjct 35 GDLVGVKSLLEHEADFNVVDNENRNPLHY#AIMHGHKKVAKLFVNQLTINSKDKNGFTPL 93 Query 93 LVEHGADVNVPDGTGALPIHL#AVQEGH-TAVVSFLAAESDLHRRDARGLTPL 143 |||+| |+| | +| +| # +|| | | | |+| + | || Sbjct 10 LVENGFDINSKDASGITLLHK#FTKEGDLVGVKSLLEHEADFNVVDNENRNPL 61 |
| Arabidopsis thaliana | Query 87 LDTLKVLVEHGADVNVPDGTGALPIHL#AVQEGHTAVVSFLAAE-SDLHRRDARGLTP 142 || || ++++| || +||| | +| #|| ||| +| || + +|| |+ | || Sbjct 654 LDALKDIIKYGGDVTLPDGNGTTALHR#AVSEGHLEIVKFLLDQGADLDWPDSYGWTP 710 Query 92 VLVEHGADVNVPDGTGALPIHL#AVQEGHTAVVSFLA 127 +|+||||| |+ | | +|+ #|+ | + || Sbjct 595 LLLEHGADPNIRDSEGNVPLWE#AIIGRHREIAKLLA 630 Query 85 GFLDTLKVLVEHGADVNVPDGTGALPIHL#AVQEGHTAVVSFL 126 | |+ +| |++ |||++ || | | |#| +|+ + + Sbjct 685 GHLEIVKFLLDQGADLDWPDSYGWTPRGL#ADHQGNEEIKTLF 726 |
| Xiphophorus maculatus | Query 84 TGFLDTLKVLVEHGADVNVPDGTGALPIHL#AVQEGHTAVVSFL 126 ||||||+++||+ ||| | ||| |#| | ||| ||+ | Sbjct 79 TGFLDTVQLLVKAGADPQARDKDNCLPIDL#ARQNGHTDVVAVL 121 Query 85 GFLDTLKVLVEHGADVNVPD-GTGALPIHL#AVQEGHTAVVSFL-AAESDLHRRDARGLTP 142 | + ++|+ ||| || | ||| |+| #| + | | | | +| || | Sbjct 46 GSSEVARLLLTRGADPNVTDKSTGATPLHD#AARTGFLDTVQLLVKAGADPQARDKDNCLP 105 Query 143 L 143 + Sbjct 106 I 106 |
| Xiphophorus helleri | Query 84 TGFLDTLKVLVEHGADVNVPDGTGALPIHL#AVQEGHTAVVSFL 126 ||||||+++||| ||| | ||| |#| | ||| ||+ | Sbjct 79 TGFLDTVQLLVEAGADPQARDKDNCLPIDL#ARQNGHTDVVAVL 121 Query 85 GFLDTLKVLVEHGADVNVPD-GTGALPIHL#AVQEGHTAVVSFLA-AESDLHRRDARGLTP 142 | + ++|+ ||| || | ||| |+| #| + | | | | +| || | Sbjct 46 GSSEVARLLLTRGADPNVTDKSTGATPLHD#AARTGFLDTVQLLVEAGADPQARDKDNCLP 105 Query 143 L 143 + Sbjct 106 I 106 |
[Site 6] GHTAVVSFLA127-AESDLHRRDA
Ala127  Ala
Ala
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Gly118 | His119 | Thr120 | Ala121 | Val122 | Val123 | Ser124 | Phe125 | Leu126 | Ala127 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Ala128 | Glu129 | Ser130 | Asp131 | Leu132 | His133 | Arg134 | Arg135 | Asp136 | Ala137 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| ADVNVPDGTGALPIHLAVQEGHTAVVSFLAAESDLHRRDARGLTPLELALQRGAQDLVDI |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | N/A | 119.00 | 14 | p19 protein |
| 2 | Homo sapiens | 119.00 | 10 | cyclin-dependent kinase inhibitor 2D |
| 3 | synthetic construct | 119.00 | 2 | cyclin-dependent kinase inhibitor 2D (p19, inhibi |
| 4 | Canis familiaris | 102.00 | 9 | PREDICTED: similar to Cyclin-dependent kinase 4 i |
| 5 | Bos taurus | 100.00 | 2 | cyclin-dependent kinase inhibitor 2D (p19, inhibi |
| 6 | Mus musculus | 94.70 | 15 | unnamed protein product |
| 7 | Rattus norvegicus | 94.00 | 4 | similar to cyclin-dependent kinase inhibitor 2D |
| 8 | Takifugu rubripes | 56.60 | 1 | p19 |
| 9 | Drosophila pseudoobscura | 48.90 | 1 | GA19105-PA |
| 10 | Drosophila melanogaster | 48.90 | 1 | ANKTM1 |
| 11 | Arabidopsis thaliana | 46.60 | 3 | AKT6_ARATH Potassium channel AKT6 (Shaker pollen |
| 12 | Tetraodon nigroviridis | 45.80 | 3 | unnamed protein product |
| 13 | synthetic construct | 44.30 | 7 | ankyrin repeat protein mbp3_16 |
| 14 | Wolbachia endosymbiont of Drosophila melanogaster | 43.90 | 2 | ankyrin repeat domain protein |
| 15 | Wolbachia endosymbiont of Drosophila ananassae | 43.90 | 1 | ankyrin repeat domain protein |
| 16 | Egeria densa | 43.90 | 1 | inward potassium channel alpha subunit |
| 17 | Gallus gallus | 43.90 | 1 | mindbomb homolog 2 |
| 18 | Danio rerio | 43.10 | 1 | PREDICTED: similar to Chain A, Structure Of P18in |
| 19 | Paramecium bursaria Chlorella virus 1 | 43.10 | 1 | hypothetical protein PBCV1_A8L |
| 20 | Xenopus tropicalis | 40.80 | 1 | HECT domain and ankyrin repeat containing, E3 ubi |
| 21 | Wolbachia pipientis | 33.90 | 2 | ankyrin domain protein |
| 22 | Wolbachia endosymbiont of Drosophila simulans | 33.90 | 1 | ankyrin repeat domain protein |
| 23 | Aspergillus nidulans FGSC A4 | 33.10 | 1 | hypothetical protein AN8767.2 |
| 24 | Strongylocentrotus purpuratus | 32.30 | 3 | PREDICTED: similar to ankyrin 2,3/unc44, partial |
| 25 | Xenopus laevis | 32.30 | 1 | putative transient receptor potential channel |
| 26 | Geobacter metallireducens GS-15 | 32.30 | 1 | Ankyrin |
| 27 | Caenorhabditis elegans | 31.60 | 1 | TRP (transient receptor potential) channel family |
| 28 | Caenorhabditis briggsae AF16 | 30.80 | 1 | Hypothetical protein CBG13617 |
Top-ranked sequences
| organism | matching |
|---|---|
| N/A | Query 98 ADVNVPDGTGALPIHLAVQEGHTAVVSFLA#AESDLHRRDARGLTPLELALQRGAQDLVDI 157 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 98 ADVNVPDGTGALPIHLAVQEGHTAVVSFLA#AESDLHRRDARGLTPLELALQRGAQDLVDI 157 |
| Homo sapiens | Query 98 ADVNVPDGTGALPIHLAVQEGHTAVVSFLA#AESDLHRRDARGLTPLELALQRGAQDLVDI 157 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 98 ADVNVPDGTGALPIHLAVQEGHTAVVSFLA#AESDLHRRDARGLTPLELALQRGAQDLVDI 157 |
| synthetic construct | Query 98 ADVNVPDGTGALPIHLAVQEGHTAVVSFLA#AESDLHRRDARGLTPLELALQRGAQDLVDI 157 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 98 ADVNVPDGTGALPIHLAVQEGHTAVVSFLA#AESDLHRRDARGLTPLELALQRGAQDLVDI 157 |
| Canis familiaris | Query 98 ADVNVPDGTGALPIHLAVQEGHTAVVSFLA#AESDLHRRDARGLTPLELALQRGAQDLVDI 157 |||| |||||||||||||+|||||||||||# ||||| |||||||||||| |||||+|| Sbjct 98 ADVNAPDGTGALPIHLAVREGHTAVVSFLA#TESDLHHRDARGLTPLELAQGIGAQDLMDI 157 |
| Bos taurus | Query 98 ADVNVPDGTGALPIHLAVQEGHTAVVSFLA#AESDLHRRDARGLTPLELALQRGAQDLVDI 157 |||| |||||||||||||+||||+||||||# ||||| ||| |||||||| ||||+|+|| Sbjct 98 ADVNAPDGTGALPIHLAVREGHTSVVSFLA#TESDLHHRDATGLTPLELARGRGAQELMDI 157 |
| Mus musculus | Query 98 ADVNVPDGTGALPIHLAVQEGHTAVVSFLA#AESDLHRRDARGLTPLELALQRGAQDLVDI 157 |||| | ||+||||||++|||++||||||# ||||| ||| |||||||| |||||+|+|| Sbjct 77 ADVNALDSTGSLPIHLAIREGHSSVVSFLA#PESDLHHRDASGLTPLELARQRGAQNLMDI 136 |
| Rattus norvegicus | Query 98 ADVNVPDGTGALPIHLAVQEGHTAVVSFLA#AESDLHRRDARGLTPLELALQRGAQDLVDI 157 |||| | ||+||||||++|||++||||||# ||||| +|| |||||||| |||||+|+|| Sbjct 98 ADVNTLDSTGSLPIHLAIREGHSSVVSFLA#PESDLHHKDASGLTPLELARQRGAQNLMDI 157 |
| Takifugu rubripes | Query 98 ADVNVPDGTGALPIHLAVQEGHTAVVSFLA#AESDLHRRDARGLTPLELALQRGAQDLVDI 157 | ||+|| +||||||+|++||| || |||# ||| + | | ++| |++|+ Sbjct 98 ASVNLPDQSGALPIHIAIREGHRDVVEFLA#PRSDLKHANKSGQTAADVARASRVPDMMDL 157 |
| Drosophila pseudoobscura | Query 100 VNVPDGTGALPIHLAVQEGHTAVVSFLA#AESDLHRRDARGLTPLELALQRGAQDLVDI 157 +| || | |+|++ |+||| || | # | || | ||+|| | + +++ Sbjct 519 INESDGAGMTPLHISSQQGHTRVVQLLL#NRGALLHRDHTGRNPLQLAAMSGYTETIEL 576 |
| Drosophila melanogaster | Query 100 VNVPDGTGALPIHLAVQEGHTAVVSFLA#AESDLHRRDARGLTPLELALQRGAQDLVDI 157 +| || | |+|++ |+||| || | # | || | ||+|| | + +++ Sbjct 506 INESDGAGMTPLHISSQQGHTRVVQLLL#NRGALLHRDHTGRNPLQLAAMSGYTETIEL 563 |
| Arabidopsis thaliana | Query 98 ADVNVPDGTGALPIHLAVQEGHTAVVSFLA#AE-SDLHRRDARGLTPLELALQRGAQDL 154 || +||| | +| || ||| +| || # + +|| |+ | || || +| +++ Sbjct 665 GDVTLPDGNGTTALHRAVSEGHLEIVKFLL#DQGADLDWPDSYGWTPRGLADHQGNEEI 722 |
| Tetraodon nigroviridis | Query 98 ADVNVPDGTGALPIHLAVQEGHTAVVSFLA#AESDL-HRRDARGLTPLELALQRGAQDLV 155 || |+ | | ||+||| +||| || | # ++ |+ +| | |+|| +|| | | Sbjct 95 ADANMADRRGNLPLHLAAEEGHPEVVRLLM#EHTENPQSRNKQGATALQLAGRRGRVDTV 153 |
| synthetic construct | Query 98 ADVNVPDGTGALPIHLAVQEGHTAVVS-FLA#AESDLHRRDARGLTPLELALQRGAQDLVD 156 |||| | || |+||| + |+ +| | # +|++ +| | | ++++ | +|| + Sbjct 71 ADVNASDATGDTPLHLAAKWGYLGIVEVLLK#YGADVNAQDKFGKTAFDISIDNGNEDLAE 130 Query 157 I 157 | Query 157 I 157 Query 98 ADVNVPDGTGALPIHLAVQEGHTAVVS-FLA#AESDLHRRDARGLTPLELALQRGAQDLVD 156 |||| | | |+||| || +| | # +|++ || | ||| || + | +|+ Sbjct 38 ADVNAMDNFGVTPLHLAAYWGHFEIVEVLLK#YGADVNASDATGDTPLHLAAKWGYLGIVE 97 Query 157 I 157 + Sbjct 98 V 98 |
| Wolbachia endosymbiont of Drosophila melanogaster | Query 98 ADVNVPDGTGALPIHLAVQEGHTAVVSFLA#AESDLHRRDARGLTPLELALQRGAQDLVD 156 || || | |+| |+ || | # + ++ +| | ||| || + +|+| Sbjct 48 ADFNVVDNENRNPLHYAIMHGHKKVAKLFV#NQLTINSKDKNGFTPLHLAALQDDTELID 106 |
| Wolbachia endosymbiont of Drosophila ananassae | Query 98 ADVNVPDGTGALPIHLAVQEGHTAVVSFLA#AESDLHRRDARGLTPLELALQRGAQDLVD 156 || || | |+| |+ || | # + ++ +| | ||| || + +|+| Sbjct 48 ADFNVVDNENRNPLHYAIMHGHKKVAKLFV#NQLTINSKDKNGFTPLHLAALQDDTELID 106 |
| Egeria densa | Query 98 ADVNVPDGTGALPIHLAVQEGHTAVVSFLA#AE-SDLHRRDARGLTPLELALQRGAQDL 154 ||+|+| +|+ +| || +|+ +|||| # +| | | | || | | |+| |+ Sbjct 544 ADINLPKRSGSTALHSAVCDGNVEIVSFLL#QHGADPDRPDINGWTPREYADQQGHHDI 601 |
| Gallus gallus | Query 99 DVNVPDGTGALPIHLAVQEGHTAVVSFLA#AE-SDLHRRDARGLTPLELALQR 149 |||| + |+|||+ +|| +| | #+| ||++ | | | + +||+| Sbjct 658 DVNVKNNRNQTPLHLAIIQGHVGLVQLLV#SEGSDVNAEDEDGDTAMHIALER 709 |
| Danio rerio | Query 98 ADVNVPDGTGALPIHLAVQEGHTAVVSFLA#AE--SDLHRRDARGLTPLELAL 147 ||||+ | | ||+||| +||| || || # + +|+| || +|| Sbjct 96 ADVNLLDNDGNLPLHLAAREGHLDVVQFLV#THCVTQPFLANAKGYTPRDLAF 147 |
| Paramecium bursaria Chlorella virus 1 | Query 98 ADVNVPDGTGALPIHLAVQEGHTAVVSFLA-#AESDLHRRDARGLTPLELALQRGAQDLV 155 |++|| | | |+|+|+ | || | #| ++|+ | || ||+ +| | | Sbjct 57 ANLNVRDDEGRTPLHIAIIEWHTICFKLLVN#AGANLNVGKKYGFAPLHLAITKGCTDYV 115 |
| Xenopus tropicalis | Query 99 DVNVPDGTGALPIHLAVQEGH-TAVVSFLA#AESDLHRRDARGLTPLELALQRGAQDLVDI 157 +|+| | | +|+| | || | |+ | #+ +|++| + | ||| | | +| | Sbjct 156 NVDVEDAMGQTALHVACQNGHKTTVLCLLD#SGADINRPNVSGATPLYFACSHGQRDTAQI 215 Query 98 ADVNVPDGTGALPIHLAVQEGHTAVVSFLA#AESDLHRRDARGLTPLELALQRGAQDLVDI 157 ||+| |+ +|| |++ | | | # + | |+|||+| +| | + || Sbjct 188 ADINRPNVSGATPLYFACSHGQRDTAQILL#LRGAKYLPDKNGVTPLDLCVQGGYGETCDI 247 |
| Wolbachia pipientis | Query 98 ADVNVPDGTGALPIHLAVQEGHTAVVSFLA#AESDLHRRDARGLTPLELALQRGAQDLVD 156 || || | |+| |+ || | # + ++ +| | ||| || + +|+| Sbjct 30 ADFNVVDNENRNPLHYAIMHGHKKVAKLFV#NQLTINSKDKNGFTPLHLAALQDDTELID 88 Query 110 PIHLAVQEGHTAVVS-FLA#AESDLHRRDARGLTPLELALQRGAQDLVDI 157 |+ |||++||| +| | # ++++ + +| || ||+|+ |+ + Sbjct 289 PLLLAVKKGHTQIVKMLLE#VRANMNICEQQGFAPLHLAVQKDYFDIAQL 337 |
| Wolbachia endosymbiont of Drosophila simulans | Query 98 ADVNVPDGTGALPIHLAVQEGHTAVVSFLA#AESDLHRRDARGLTPLELALQRGAQDLVD 156 || || | |+| |+ || | # + ++ +| | ||| || + +|+| Sbjct 48 ADFNVVDNENRNPLHYAIMHGHKKVAKLFV#NQLTINSKDKNGFTPLHLAALQDDTELID 106 Query 110 PIHLAVQEGHTAVVS-FLA#AESDLHRRDARGLTPLELALQRGAQDLVDI 157 |+ |||++||| +| | # ++++ + +| || ||+|+ |+ + Sbjct 307 PLLLAVKKGHTQIVKMLLE#VRANMNICEQQGFAPLHLAVQKDYFDIAQL 355 |
| Aspergillus nidulans FGSC A4 | Query 98 ADVNVPDGTGALPIHLAVQEGHTAVVSFLA#AE-SDLHRRDARGLTPLELALQRGAQDLVD 156 ||||+|| | |+ ||+ || ||| | # |+ + | | ||| |+ +| |++| Sbjct 106 ADVNIPDSKGQTPLSWAVENGHQAVVQLLL#GHGSNPNTPDPGGQTPLSCAVSKGNQEIVK 165 Query 157 I 157 + Sbjct 166 L 166 Query 98 ADVNVPDGTGALPIHLAVQEGHTAVVSFLA#AESDLHRRD--ARGLTPLELALQRGAQDLV 155 ++ | || | |+ || +|+ +| | #+ ||| ||||| |+ | +++| Sbjct 139 SNPNTPDPGGQTPLSCAVSKGNQEIVKLLL#SSSDLECNTPHPNGLTPLCWAVNEGQEEIV 198 Query 156 DI 157 + Sbjct 199 QL 200 Query 99 DVNVPDGTGALPIHLAVQEGHTAVVSFLA#AESDL--HRRDARGLTPLELALQRGAQDLV 155 + | | | |+ || || +| | # ||+ ++ | | || |+++ + +| Sbjct 174 ECNTPHPNGLTPLCWAVNEGQEEIVQLLL#DRSDVDPNKPDTDGYAPLSRAVEKNSLAMV 232 |
| Strongylocentrotus purpuratus | Query 98 ADVNVPDGTGALPIHLAVQEGHTAVVSFLA#AE-SDLHRRDARGLTPLELALQRG 150 |||| | | +|+| | +|+ |+ +| # + ||+++ || | || |+| | Sbjct 556 ADVNEKDEEGEIPLHGAANDGNVEVIKYLI#QQGSDVNKMDAEGWTPFNAAVQEG 609 Query 98 ADVNVPDGTGALPIHLAVQEGHTAVVSFLA#AE-SDLHRRDARGLTPLELALQRG 150 |||| | +|+| | ||| |+ +| # + || ++ || | || |+| | Sbjct 944 ADVNEEHDKGMIPLHGAACEGHLEVMEYLI#QQGSDTNKCDAEGWTPFNAAVQYG 997 Query 98 ADVNVPDGTGALPIHLAVQEGHTAVVSFLA#AE-SDLHRRDARGLTPLELALQRG 150 |||| | +|+| | ||| |+ +| # + || ++ || | || |+| | Sbjct 653 ADVNEEDDRRKIPLHGAACEGHLEVMEYLI#QQGSDTNKCDAEGWTPFNAAVQYG 706 Query 98 ADVNVPDGTGALPIHLAVQEGHTAVVSFLA#AE-SDLHRRDARGLTPLELALQRG 150 |||| | | +|+| | +|+ |+++| # + || ++ || | || |+| | Sbjct 459 ADVNEEDEEGEIPLHGAAIDGNVEVMAYLI#QQGSDTNKCDADGWTPFNAAIQYG 512 Query 98 ADVNVPDGTGALPIHLAVQEGHTAVVSFLA#AE-SDLHRRDARGLTPLELALQRGAQDLVD 156 |||| | +|+| | || |+ +| # + +|++++| | ||| |+ | ++| Sbjct 1332 ADVNEEHDKGMIPLHGAAHRGHLEVMEYLI#QQGADVNKKDNTGWTPLHAAVSNGHLEVVK 1391 Query 157 I 157 + Sbjct 1392 V 1392 Query 98 ADVNVPDGTGALPIHLAVQEGHTAVVSFLA#AE-SDLHRRDARGLTPLELALQRG 150 |||| | | +|+| | || |+ +| # + |++++ ++ | || |+| | Sbjct 264 ADVNEEDEKGEIPLHGAASGGHLEVMKYLI#QQGSNVNKANSEGWTPFNAAVQYG 317 Query 98 ADVNVPDGTGALPIHLAVQEGHTAVVSFLA#AESDLHRRDARGLTPLELALQRGAQDLVDI 157 +||| | | | + ||||| | +| # + |+ |+||| | | | |+| Sbjct 589 SDVNKMDAEGWTPFNAAVQEGQLEAVKYLM#TKGAKQNRN-DGMTPLYAAAQSGRLDIVKF 647 Query 98 ADVNVPDGTGALPIHLAVQEGHTAVVSFLA#AE-SDLHRRDARGLTPLELALQRGAQDLVD 156 | ||| | | |+ | |||| || |||# + +| | || ||+ | | + |+ Sbjct 1448 AHVNVQDDEGWTPLEAAAQEGHEDVVDFLA#LDGADTDVNDIDGLAPLQAAANAGHPNAVE 1507 Query 98 ADVNVPDGTGALPIHLAVQEGHTAVVSFLA#AESDLHRRDARGLTPLELALQRGAQDLVDI 157 |||| | | + ||| || | +| # + | |+||| | | | |+| Sbjct 880 ADVNKADAKGGTSFNAAVQGGHLEAVEYLM#TKGAKQNR-YDGMTPLYAAAQSGCLDIVKF 938 Query 98 ADVNVPDGTGALPIHLAVQEGHTAVVSFLA#AESDLHRRDARGLTPLELALQRGAQDLVDI 157 |||| | | + ||| || | +| # + | |+||| | | | |+| Sbjct 1074 ADVNKADAKGGTSFNAAVQGGHLEAVEYLM#TKGAKQNR-YDGMTPLYAAAQSGCLDIVKF 1132 Query 98 ADVNVPDGTGALPIHLAVQEGHTAVVSFLA#AESDLHRRDARGLTPLELALQRGAQDLVDI 157 |||| | | + ||| || | +| # + | |+||| | | | |+| Sbjct 1268 ADVNKADAKGGTSFNAAVQGGHLEAVEYLM#TKGAKQNR-YDGMTPLYAAAQSGCLDIVKF 1326 Query 98 ADVNVPDGTGALPIHLAVQEGHTAVVSFLA#AESDLHRRDARGLTPLELALQRGAQDLVDI 157 +| | | | | + ||| || | +| # + | | |+||| | | | |+| Sbjct 686 SDTNKCDAEGWTPFNAAVQYGHLESVKYLI#TKGAKRNRYA-GMTPLYAAAQSGHLDIVKF 744 Query 98 ADVNVPDGTGALPIHLAVQEGHTAVVSFLA#AESDLHRRDARGLTPLELALQRGAQDLVDI 157 +| | | | | + ||| || | +| # + | | |+||| | | | |+| Sbjct 977 SDTNKCDAEGWTPFNAAVQYGHLESVKYLI#TKGAKRNRYA-GMTPLYAAAQSGHLDIVKF 1035 Query 98 ADVNVPDGTGALPIHLAVQEGHTAVVSFLA#AESDLHRRDARGLTPLELALQRGAQDLVDI 157 |||| | | + ||| || | +| # + | |+||| | | | |+| Sbjct 1171 ADVNKADAKGGTSFNAAVQGGHLEAVEYLM#TKGAKQNR-FDGMTPLYAAAQSGCLDIVKF 1229 Query 98 ADVNVPDGTGALPIHLAVQEGHTAVVSFLA#AE-SDLHRRDARGLTPLELALQRGAQDLVD 156 |||| | +|+| | || |+ +| # + +|+++ ||+| | |+| | + |+ Sbjct 1235 ADVNEEHDKGMIPLHGAAHRGHLEVMEYLI#QQGADVNKADAKGGTSFNAAVQGGHLEAVE 1294 Query 157 I 157 Sbjct 1295 Y 1295 Query 98 ADVNVPDGTGALPIHLAVQEGHTAVVSFLA#AESDLHRRDARGLTPLELALQRGAQDLVDI 157 +||| | | + ||| || | +| # + | |+||| | | | |+| Sbjct 783 SDVNKADAKGGTSFNAAVQGGHLEAVEYLM#TKGAKQNR-YDGMTPLYAAAQSGCLDIVKF 841 Query 98 ADVNVPDGTGALPIHLAVQEGHTAVVSFLA#AESDLHRRDARGLTPLELALQRGAQDLVDI 157 | || | | | + ||| || | +| # + | | | ||| | | | |+| Sbjct 200 AYVNKADSKGCTPFNAAVQYGHLEAVEYLM#TKGAKQNRYA-GRTPLYAAAQLGHLDIVKF 258 Query 98 ADVNVPDGTGALPIHLAVQEGHTAVVSFLA#AES------------------DLHRRDARG 139 |||| | || |+| || || || | #|+ |++ |+ | Sbjct 1365 ADVNKKDNTGWTPLHAAVSNGHLEVVKVLL#AKGAQGTMFEGLTLVLVSNGFDVNERNECG 1424 Query 140 LTPLELALQRGAQDLVDI 157 +|| | |++ + Sbjct 1425 KSPLHAGCYNGNIDILKL 1442 Query 98 ADVNVPDGTGALPIHLAVQEGHTAVVSFLA#AESDLHRRDARGLTPLELALQRGAQDLVDI 157 ++|| + | | + ||| || | +| # + | |+||| | | | ++| Sbjct 297 SNVNKANSEGWTPFNAAVQYGHLEAVKYLM#TKGATQNR-YNGMTPLYAAAQSGHLNIVQF 355 Query 98 ADVNVPDGTGALPIHLAVQEGHTAVVSFLA#AESDLHRRDARGLTPLELALQRGAQDLV 155 +| | | | | + |+| || | +| # + | | | ||| | | | |+| Sbjct 492 SDTNKCDADGWTPFNAAIQYGHLESVKYLI#TKGAKQNRYA-GRTPLYAAAQLGHLDIV 548 Query 98 ADVNVPDGTGALPIHLAVQEGHTAVVSFLA#AE-SDLHRRDARGLTPLELALQRGAQDLVD 156 |||| | | +|+ | | |+ +| # + ||+++ ||+| | |+| | + |+ Sbjct 750 ADVNEEDEEGMIPLRGAAAGGQLEVMEYLI#QQGSDVNKADAKGGTSFNAAVQGGHLEAVE 809 Query 157 I 157 Sbjct 810 Y 810 Query 98 ADVNVPDGTGALPIHLAVQEGHTAVVSFLA#AE-SDLHRRDARGLTPLELALQRGAQDLVD 156 |||| | | +|+ | | |+ +| # + +|+++ ||+| | |+| | + |+ Sbjct 1041 ADVNEEDEEGMIPLRGAAAGGQLEVMEYLI#QQGADVNKADAKGGTSFNAAVQGGHLEAVE 1100 Query 157 I 157 Sbjct 1101 Y 1101 Query 107 GALPIHLAVQEGHTAVVSFLA#AE-SDLHRRDARGLTPLELALQRGAQDLVDI 157 | |++ | | || +| | #+| +|++ | |+ || | | ++++ Sbjct 726 GMTPLYAAAQSGHLDIVKFFI#SEGADVNEEDEEGMIPLRGAAAGGQLEVMEY 777 Query 107 GALPIHLAVQEGHTAVVSFLA#AE-SDLHRRDARGLTPLELALQRGAQDLVDI 157 | |++ | | || +| | #+| +|++ | |+ || | | ++++ Sbjct 1017 GMTPLYAAAQSGHLDIVKFFI#SEGADVNEEDEEGMIPLRGAAAGGQLEVMEY 1068 Query 98 ADVNVPDGTGALPIHLAVQEGHTAVVSFLA#AE-SDLHRRDARGLTPLELALQRGAQDLVD 156 |||| +|+| | | |+ +| # + +|+++ ||+| | |+| | + |+ Sbjct 1138 ADVNEEHARRMIPLHGAAHRGQLEVMEYLI#QQGADVNKADAKGGTSFNAAVQGGHLEAVE 1197 Query 157 I 157 Sbjct 1198 Y 1198 |
| Xenopus laevis | Query 107 GALPIHLAVQEGHTAVVSFLA#AE--SDLHRRDARGLTPLELALQRGAQDLV 155 |+ ||||| | |||||| | #++ | || +| || | | || | +++ Sbjct 857 GSTPIHLAAQNGHTAVVGLLL#SKSTSQLHMKDKRGRTCLHLAAANGHIEMM 907 Query 98 ADVNVPDGTGALPIHLAVQEGHTAVVSFL---A#AESDLHRRDARGLTPLELALQRGAQDL 154 |++|| | | |+| | + | | || #| | +| | | ++ | + ||+ Sbjct 915 AEINVTDKNGWCPLHFAARSGFLDTVRFLVECG#ANPTLECKD--GKTAIQYAAAKNHQDV 972 Query 155 VDI 157 | Sbjct 973 VSF 975 Query 98 ADVNVPDGTGALPIHLAVQEGHTAVVS-FLA#AESDLHRRDARGLTPLELALQRGAQDLVD 156 | |+| | | +||| + || + | # ++ ++ + ||||| | | | || Sbjct 528 ARVDVFDEHGKAALHLAAENGHDKIADILLK#HKAFVNAKTKLGLTPLHLCAQNGFNHLVK 587 Query 157 I 157 + Sbjct 588 L 588 Query 98 ADVNVPDGTGALPIHLAVQEGHTAVVSFL-A#AESDLHRRDARGLTPLELALQRGAQDLVD 156 |+|+ + | |+|+ | ++ || # +++ + | ||| +| +|| ++|+ Sbjct 136 ANVDCQNDEGQTPLHIVAWAGDEMMLKFLHQ#CKTNANITDKMERTPLHVAAERGNTNVVE 195 Query 157 I 157 | Query 157 I 157 |
| Geobacter metallireducens GS-15 | Query 98 ADVNVPDGTGALPIHLAVQEGHTAVVS-FLA#AESDLHRRDARGLTPLELALQRGAQDLVD 156 ||+|+ | |+| || |+ | |||# +| |+ ||||| +|| +|| +|+ Sbjct 218 ADINIAGRYGDTPLHYAVGLGYEEFVRLFLA#RGADPTIRNDRGLTPRQLAEERGFASIVE 277 Query 157 I 157 + Sbjct 278 L 278 Query 98 ADVNVPDGTGALPIHLAVQEGHTAVVSFLA-#AESDLHRRDARGLTPLELALQRGAQDLV 155 | || +| | |+ || ||| | | #| +|+ ++ ||| | +|| +| Sbjct 119 ARVNAVNGEGNTPLFAAVHEGHDDVARILLD#AGADVTIANSYWYTPLREACRRGNTGMV 177 Query 98 ADVNVPDGTGALPIHLAVQ-EGHTAVVSFLA#AESDLHRRDARGLTPLELALQRGAQDLV 155 ||||+ | | |+| || | | | #+ +|++ | ||| |+ | ++ | Sbjct 185 ADVNITDNKGFTPLHAAVSAEALEAATLILD#SGADINIAGRYGDTPLHYAVGLGYEEFV 243 |
| Caenorhabditis elegans | Query 109 LPIHLAVQEGHTAVVSFLA#AES--DLHRRDARGLTPLELALQRGAQDLVDI 157 +|+||| |+|| ||| | #+ | | +| || ||| || | | ++| + Sbjct 1194 IPLHLAAQQGHIAVVGMLL#SRSTQQQHAKDWRGRTPLHLAAQNGHYEMVSL 1244 Query 104 DGTGALPIHLAVQEGHTAVVSFLA#AE-SDLHRRDARGLTPLELALQRGAQDLVDI 157 | | |+||| | || +|| | #|+ |+++ | | | | | + | +| + Sbjct 1223 DWRGRTPLHLAAQNGHYEMVSLLI#AQGSNINVMDQNGWTGLHFATRAGHLSVVKL 1277 Query 104 DGTGALPIHLAVQEGHTAVVS-FLA#AESDLHRR--DARGLTPLELALQRG 150 || |+|||||| + |+ +| |+# || | | | | | || + | Sbjct 407 DGDGSLPIHLAFKFGNVNIVELLLS#GPSDEQTRKADGNGDTLLHLAARSG 456 Query 98 ADVNVPDGTGALPIHLAVQEGHTAVVSFL 126 | || || |+||| | || ||+ | Sbjct 897 AFVNSKSKTGEAPLHLAAQHGHVKVVNVL 925 |
| Caenorhabditis briggsae AF16 | Query 109 LPIHLAVQEGHTAVVSFLA#AES--DLHRRDARGLTPLELALQRGAQDLVDI 157 +|+||| |+|| ||| | #+ | | +| || ||| || | | ++| + Sbjct 1211 IPLHLAAQQGHIAVVGMLL#SRSTQQQHAKDWRGRTPLHLAAQNGHYEMVSL 1261 Query 104 DGTGALPIHLAVQEGHTAVVSFLA#AE-SDLHRRDARGLTPLELALQRGAQDLVDI 157 | | |+||| | || +|| | #|+ |+++ | | | | | + | +| + Sbjct 1240 DWRGRTPLHLAAQNGHYEMVSLLI#AQGSNINVMDQNGWTGLHFATRAGHLSVVKL 1294 Query 104 DGTGALPIHLAVQEGHTAVVS-FLA#AESDLHRR--DARGLTPLELALQRGAQDLV 155 || |+|||||| + |+ +| |+# || | | | | | || + |+ + | Sbjct 423 DGEGSLPIHLAFKFGNVNIVELLLS#GPSDEQTRKADGNGDTLLHLAARSGSIEAV 477 Query 98 ADVNVPDGTGALPIHLAVQEGHTAVVSFL 126 | || || |+||| | || ||+ | Sbjct 914 AFVNSKSKTGEAPLHLAAQHGHVKVVNVL 942 Query 98 ADVNVPDGTGALPIHLAVQEGHTAVVSFLA#A-ESDLHRRDARGLTPLELALQRGAQDLVD 156 | ++| | | +||| || ++| | # ++ ++ + | || || | | +|+ Sbjct 881 ARIDVFDEMGRTALHLAAFNGHLSIVHLLL#QHKAFVNSKSKTGEAPLHLAAQHGHVKVVN 940 Query 157 I 157 + Sbjct 941 V 941 |
* References
[PubMed ID: 23536791] Lin S, Wang MJ, Tseng KY, Polypyrimidine tract-binding protein induces p19(Ink4d) expression and inhibits the proliferation of H1299 cells. PLoS One. 2013;8(3):e58227. doi: 10.1371/journal.pone.0058227. Epub 2013 Mar 11.
[PubMed ID: 22747700] ... Miranda-Sayago JM, Fernandez-Arcas N, Reyes-Engel A, Benito C, Narbona I, Alonso A, Changes in CDKN2D , TP53, and miR125a expression: potential role in the evaluation of human amniotic fluid-derived mesenchymal stromal cell fitness. Genes Cells. 2012 Aug;17(8):673-87. doi: 10.1111/j.1365-2443.2012.01623.x. Epub
[PubMed ID: 22476863] ... Carcagno AL, Giono LE, Marazita MC, Castillo DS, Pregi N, Canepa ET, E2F1 induces p19INK4d, a protein involved in the DNA damage response, following UV irradiation. Mol Cell Biochem. 2012 Jul;366(1-2):123-9. doi: 10.1007/s11010-012-1289-8. Epub
[PubMed ID: 22558186] ... Marazita MC, Ogara MF, Sonzogni SV, Marti M, Dusetti NJ, Pignataro OP, Canepa ET, CDK2 and PKA mediated-sequential phosphorylation is critical for p19INK4d function in the DNA damage response. PLoS One. 2012;7(4):e35638. doi: 10.1371/journal.pone.0035638. Epub 2012 Apr 25.
[PubMed ID: 21901251] ... Morishita A, Gong J, Deguchi A, Tani J, Miyoshi H, Yoshida H, Himoto T, Yoneyama H, Mori H, Kato K, Kurokohchi K, Deguchi K, Izuishi K, Suzuki Y, Kushida Y, Haba R, Iwama H, Watanabe S, D'Armiento J, Masaki T, Frequent loss of p19INK4D expression in hepatocellular carcinoma: relationship to tumor differentiation and patient survival. Oncol Rep. 2011 Dec;26(6):1363-8. doi: 10.3892/or.2011.1452. Epub 2011 Sep 7.
[PubMed ID: 10734227] ... Thullberg M, Bartkova J, Khan S, Hansen K, Ronnstrand L, Lukas J, Strauss M, Bartek J, Distinct versus redundant properties among members of the INK4 family of cyclin-dependent kinase inhibitors. FEBS Lett. 2000 Mar 24;470(2):161-6.
[PubMed ID: 9001419] ... Schwaller J, Pabst T, Koeffler HP, Niklaus G, Loetscher P, Fey MF, Tobler A, Expression and regulation of G1 cell-cycle inhibitors (p16INK4A, p15INK4B, p18INK4C, p19INK4D) in human acute myeloid leukemia and normal myeloid cells. Leukemia. 1997 Jan;11(1):54-63.
[PubMed ID: 8741839] ... Guan KL, Jenkins CW, Li Y, O'Keefe CL, Noh S, Wu X, Zariwala M, Matera AG, Xiong Y, Isolation and characterization of p19INK4d, a p16-related inhibitor specific to CDK6 and CDK4. Mol Biol Cell. 1996 Jan;7(1):57-70.
[PubMed ID: 8575754] ... Okuda T, Hirai H, Valentine VA, Shurtleff SA, Kidd VJ, Lahti JM, Sherr CJ, Downing JR, Molecular cloning, expression pattern, and chromosomal localization of human CDKN2D/INK4d, an inhibitor of cyclin D-dependent kinases. Genomics. 1995 Oct 10;29(3):623-30.
[PubMed ID: 7739548] ... Chan FK, Zhang J, Cheng L, Shapiro DN, Winoto A, Identification of human and mouse p19, a novel CDK4 and CDK6 inhibitor with homology to p16ink4. Mol Cell Biol. 1995 May;15(5):2682-8.
[PubMed ID: 7739547] ... Hirai H, Roussel MF, Kato JY, Ashmun RA, Sherr CJ, Novel INK4 proteins, p19 and p18, are specific inhibitors of the cyclin D-dependent kinases CDK4 and CDK6. Mol Cell Biol. 1995 May;15(5):2672-81.