SB0084 : BID, BH3 interacting domain death agonist
[ CaMP Format ]
* Basic Information
| Organism | Homo sapiens (human) |
| Protein Names | BH3-interacting domain death agonist isoform 2 [Homo sapiens]; BH3-interacting domain death agonist isoform 2; BH3-interacting domain death agonist; apoptic death agonist; Human BID coding sequence; desmocollin type 4; BID isoform ES(1b); BID isoform L(2); BID isoform Si6; p22 BID; BID; BH3-interacting domain death agonist p15; p15 BID; BH3-interacting domain death agonist p13; p13 BID; BH3-interacting domain death agonist p11; p11 BID |
| Gene Names | BID; BH3 interacting domain death agonist |
| Gene Locus | 22q11.1; chromosome 22 |
| GO Function | Not available |
* Information From OMIM
Function: Luo et al. (1998) reported the purification of a cytosolic protein that induces cytochrome c release from mitochondria in response to caspase-8 (CASP8; OMIM:601763), the apical caspase activated by cell surface death receptors such as FAS (OMIM:134637) and TNF (OMIM:191160). Peptide mass fingerprinting identified this protein as BID. CASP8 cleaves BID, and the COOH-terminal part translocates to mitochondria where it triggers cytochrome c release. Immunodepletion of BID from cell extracts eliminated the cytochrome c releasing activity. The cytochrome c releasing activity of BID was antagonized by BCL2. A mutation at the BH3 domain diminished its cytochrome c releasing activity. BID, therefore, relays an apoptotic signal from the cell surface to mitochondria.
* Structure Information
1. Primary Information
Length: 195 aa
Average Mass: 21.994 kDa
Monoisotopic Mass: 21.981 kDa
2. Domain Information
Annotated Domains: interpro / pfam / smart / prosite
Computationally Assigned Domains (Pfam+HMMER):
| domain name | begin | end | score | e-value |
|---|---|---|---|---|
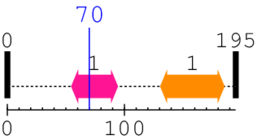
| Apoptosis regulator proteins, Bcl-2 family 1. | 55 | 94 | 6.0 | 0.0 |
| --- cleavage 70 (inside Apoptosis regulator proteins, Bcl-2 family 55..94) --- | ||||
| CRM1 C terminal 1. | 131 | 185 | 73.0 | 0.0 |
3. Sequence Information
Fasta Sequence: SB0084.fasta
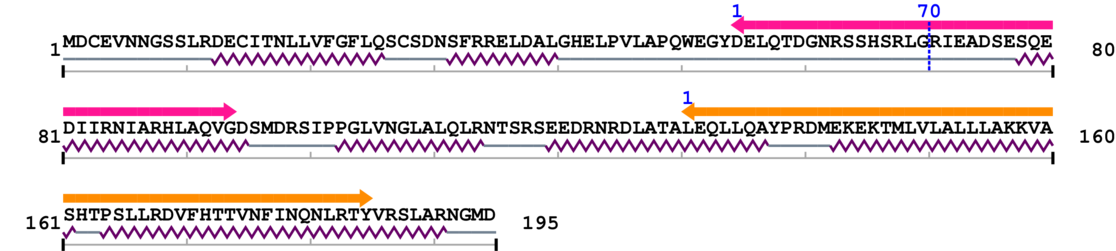
Amino Acid Sequence and Secondary Structures (PsiPred):
* Cleavage Information
1 [sites] cleaved by Calpain 2
Source Reference: [PubMed ID: 11940658] Mandic A, Viktorsson K, Strandberg L, Heiden T, Hansson J, Linder S, Shoshan MC, Calpain-mediated Bid cleavage and calpain-independent Bak modulation: two separate pathways in cisplatin-induced apoptosis. Mol Cell Biol. 2002 May;22(9):3003-13.
Cleavage sites (±10aa)
[Site 1] GNRSSHSRLG70-RIEADSESQE
Gly70  Arg
Arg
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Gly61 | Asn62 | Arg63 | Ser64 | Ser65 | His66 | Ser67 | Arg68 | Leu69 | Gly70 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Arg71 | Ile72 | Glu73 | Ala74 | Asp75 | Ser76 | Glu77 | Ser78 | Gln79 | Glu80 |
 Sequence conservation (by blast)
Sequence conservation (by blast)
* References
[PubMed ID: 24158446] Wang Y, Tjandra N, Structural insights of tBid, the caspase-8-activated Bid, and its BH3 domain. J Biol Chem. 2013 Dec 13;288(50):35840-51. doi: 10.1074/jbc.M113.503680. Epub