SB0086 : c-jun
[ CaMP Format ]
* Basic Information
| Organism | Rattus norvegicus (Norway rat) |
| Protein Names | transcription factor AP-1 [Rattus norvegicus]; transcription factor AP-1; AP1; activator protein 1; proto-oncogene c-jun; V-jun avian sarcoma virus 17 oncogene homolog; v-jun sarcoma virus 17 oncogene homolog; Avian sarcoma virus 17 (v-jun) oncogene homolog; Jun oncogene; Transcription factor AP-1; Activator protein 1; Proto-oncogene c-Jun |
| Gene Names | Jun; Rjg-9; jun proto-oncogene |
| Gene Locus | 5q34.1; chromosome 5 |
| GO Function | Not available |
* Information From OMIM
Not Available.
* Structure Information
1. Primary Information
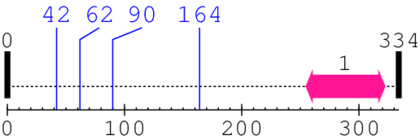
Length: 334 aa
Average Mass: 36.000 kDa
Monoisotopic Mass: 35.978 kDa
2. Domain Information
Annotated Domains: interpro / pfam / smart / prosite
Computationally Assigned Domains (Pfam+HMMER):
| domain name | begin | end | score | e-value |
|---|---|---|---|---|
| --- cleavage 42 --- | ||||
| --- cleavage 62 --- | ||||
| --- cleavage 90 --- | ||||
| --- cleavage 164 --- | ||||
| LMBR1-like membrane protein 1. | 255 | 322 | 180.0 | 1.4 |
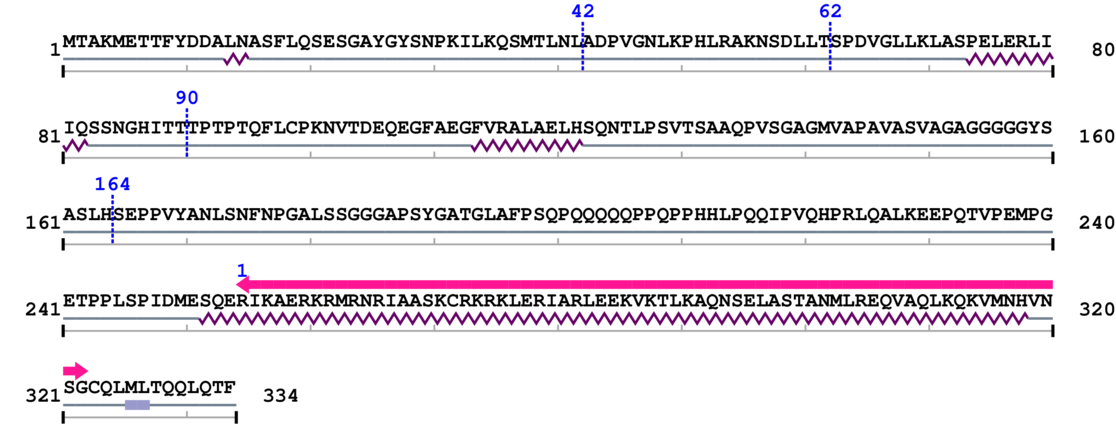
3. Sequence Information
Fasta Sequence: SB0086.fasta
Amino Acid Sequence and Secondary Structures (PsiPred):
4. 3D Information
Not Available.
* Cleavage Information
4 [sites] cleaved by Calpain 2
Source Reference: [PubMed ID: 10600648] Pariat M, Salvat C, Bebien M, Brockly F, Altieri E, Carillo S, Jariel-Encontre I, Piechaczyk M, The sensitivity of c-Jun and c-Fos proteins to calpains depends on conformational determinants of the monomers and not on formation of dimers. Biochem J. 2000 Jan 1;345 Pt 1:129-38.
Cleavage sites (±10aa)
[Site 1] ILKQSMTLNL42-ADPVGNLKPH
Leu42  Ala
Ala
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Ile33 | Leu34 | Lys35 | Gln36 | Ser37 | Met38 | Thr39 | Leu40 | Asn41 | Leu42 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Ala43 | Asp44 | Pro45 | Val46 | Gly47 | Asn48 | Leu49 | Lys50 | Pro51 | His52 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| ALNASFLQSESGAYGYSNPKILKQSMTLNLADPVGNLKPHLRAKNSDLLTSPDVGLLKLA |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | Rattus norvegicus | 122.00 | 1 | v-jun sarcoma virus 17 oncogene homolog |
| 2 | Mus musculus | 120.00 | 4 | unnamed protein product |
| 3 | Sus scrofa | 119.00 | 1 | C-JUN protein |
| 4 | Bos taurus | 119.00 | 1 | c-jun |
| 5 | Macaca fuscata | 117.00 | 1 | transcription factor c-Jun |
| 6 | N/A | 115.00 | 5 | 1404381A c-jun oncogene |
| 7 | Homo sapiens | 115.00 | 3 | JUN |
| 8 | Ovis aries | 115.00 | 2 | c-Jun protein |
| 9 | synthetic construct | 115.00 | 2 | v-jun sarcoma virus 17 oncogene-like |
| 10 | Gallus gallus | 92.40 | 1 | jun oncogene |
| 11 | Serinus canaria | 92.40 | 1 | JUN_SERCA Transcription factor AP-1 (Proto-oncogen |
| 12 | Coturnix coturnix | 90.90 | 1 | JUN_COTJA Transcription factor AP-1 (Proto-oncogen |
| 13 | Takifugu rubripes | 89.00 | 5 | c-Jun protein |
| 14 | Carassius auratus | 86.30 | 1 | c-Jun protein |
| 15 | Danio rerio | 85.50 | 3 | c-Jun |
| 16 | Tetraodon nigroviridis | 84.00 | 4 | unnamed protein product |
| 17 | Ctenopharyngodon idella | 84.00 | 2 | c-Jun protein |
| 18 | Xenopus laevis | 78.20 | 3 | c-Jun protein |
| 19 | Oncorhynchus mykiss | 77.00 | 1 | transcription factor AP-1 |
| 20 | Xiphophorus maculatus | 59.70 | 2 | c-JUN |
| 21 | Oryctolagus cuniculus | 40.00 | 1 | c-jun transcription factor |
Top-ranked sequences
| organism | matching |
|---|---|
| Rattus norvegicus | Query 13 ALNASFLQSESGAYGYSNPKILKQSMTLNL#ADPVGNLKPHLRAKNSDLLTSPDVGLLKLA 72 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 13 ALNASFLQSESGAYGYSNPKILKQSMTLNL#ADPVGNLKPHLRAKNSDLLTSPDVGLLKLA 72 |
| Mus musculus | Query 13 ALNASFLQSESGAYGYSNPKILKQSMTLNL#ADPVGNLKPHLRAKNSDLLTSPDVGLLKLA 72 ||||||||||||||||||||||||||||||#|||||+|||||||||||||||||||||||| Sbjct 13 ALNASFLQSESGAYGYSNPKILKQSMTLNL#ADPVGSLKPHLRAKNSDLLTSPDVGLLKLA 72 |
| Sus scrofa | Query 13 ALNASFLQSESGAYGYSNPKILKQSMTLNL#ADPVGNLKPHLRAKNSDLLTSPDVGLLKLA 72 ||||||||||||||||||||||||||||||#|||||||||||||++||||||||||||||| Sbjct 13 ALNASFLQSESGAYGYSNPKILKQSMTLNL#ADPVGNLKPHLRAQDSDLLTSPDVGLLKLA 72 |
| Bos taurus | Query 13 ALNASFLQSESGAYGYSNPKILKQSMTLNL#ADPVGNLKPHLRAKNSDLLTSPDVGLLKLA 72 ||||||||||||||||||||||||||||||#|||||+|||||||||||||||||||||||| Sbjct 8 ALNASFLQSESGAYGYSNPKILKQSMTLNL#ADPVGSLKPHLRAKNSDLLTSPDVGLLKLA 67 |
| Macaca fuscata | Query 13 ALNASFLQSESGAYGYSNPKILKQSMTLNL#ADPVGNLKPHLRAKNSDLLTSPDVGLLKLA 72 ||||||| ||||||||||||||||||||||#|||||+|||||||||||||||||||||||| Sbjct 13 ALNASFLPSESGAYGYSNPKILKQSMTLNL#ADPVGSLKPHLRAKNSDLLTSPDVGLLKLA 72 |
| N/A | Query 13 ALNASFLQSESGAYGYSNPKILKQSMTLNL#ADPVGNLKPHLRAKNSDLLTSPDVGLLKLA 72 ||||||| |||| |||||||||||||||||#|||||+|||||||||||||||||||||||| Sbjct 13 ALNASFLPSESGPYGYSNPKILKQSMTLNL#ADPVGSLKPHLRAKNSDLLTSPDVGLLKLA 72 |
| Homo sapiens | Query 13 ALNASFLQSESGAYGYSNPKILKQSMTLNL#ADPVGNLKPHLRAKNSDLLTSPDVGLLKLA 72 ||||||| |||| |||||||||||||||||#|||||+|||||||||||||||||||||||| Sbjct 13 ALNASFLPSESGPYGYSNPKILKQSMTLNL#ADPVGSLKPHLRAKNSDLLTSPDVGLLKLA 72 |
| Ovis aries | Query 15 NASFLQSESGAYGYSNPKILKQSMTLNL#ADPVGNLKPHLRAKNSDLLTSPDVGLLKLA 72 ||||||||||||||||||||||||||||#|||||+|||||||||||||||||||||||| Sbjct 1 NASFLQSESGAYGYSNPKILKQSMTLNL#ADPVGSLKPHLRAKNSDLLTSPDVGLLKLA 58 |
| synthetic construct | Query 13 ALNASFLQSESGAYGYSNPKILKQSMTLNL#ADPVGNLKPHLRAKNSDLLTSPDVGLLKLA 72 ||||||| |||| |||||||||||||||||#|||||+|||||||||||||||||||||||| Sbjct 13 ALNASFLPSESGPYGYSNPKILKQSMTLNL#ADPVGSLKPHLRAKNSDLLTSPDVGLLKLA 72 |
| Gallus gallus | Query 13 ALNASFLQSESGAYGYSNPKILKQSMTLNL#ADPVGNLKPHLRAKNSDLLTSPDVGLLKLA 72 |||||| ||| |||+| |+|||||||||#+| +|||||| ||+|+|||||||||||| Sbjct 9 ALNASFAPPESGGYGYNNAKVLKQSMTLNL#SDAASSLKPHLRNKNADILTSPDVGLLKLA 68 |
| Serinus canaria | Query 13 ALNASFLQSESGAYGYSNPKILKQSMTLNL#ADPVGNLKPHLRAKNSDLLTSPDVGLLKLA 72 ||+| | ||| |||+| |+|||+|||||#+|| ||||||| ||+|+|||||||||||| Sbjct 13 ALSAGFAPPESGGYGYNNAKVLKQNMTLNL#SDPSSNLKPHLRNKNADILTSPDVGLLKLA 72 |
| Coturnix coturnix | Query 13 ALNASFLQSESGAYGYSNPKILKQSMTLNL#ADPVGNLKPHLRAKNSDLLTSPDVGLLKLA 72 |||||| || ||||+| |+|||||||||#+|| +|||||| ||+|+|||||||||||| Sbjct 13 ALNASFAPPES-AYGYNNAKVLKQSMTLNL#SDPASSLKPHLRNKNADILTSPDVGLLKLA 71 |
| Takifugu rubripes | Query 13 ALNASFLQSESGAYGYSNPKILKQSMTLNL#ADPVGNLKPHLRAKNSDLLTSPDVGLLKLA 72 +||| | | ++ ||||||| || +|||||#+|| | |||||||| |+||||||||||||| Sbjct 13 SLNA-FSQHDTAGYGYSNPKALKHNMTLNL#SDPTGTLKPHLRAKASELLTSPDVGLLKLA 71 |
| Carassius auratus | Query 13 ALNASFLQSESGAYGYSNPKILKQSMTLNL#ADPVGNLKPHLRAKNSDLLTSPDVGLLKLA 72 +||++| | +| |+|| | | ||+||||||# || |||||||||| +|+|||||||||||| Sbjct 9 SLNSAFSQHDSAAFGY-NHKALKRSMTLNL#TDPSGNLKPHLRAKANDILTSPDVGLLKLA 67 |
| Danio rerio | Query 13 ALNASFLQSESGAYGYSNPKILKQSMTLNL#ADPVGNLKPHLRAKNSDLLTSPDVGLLKLA 72 +||++| | +| +|| | | || +|||||#+|| |||||||||| ||+|||||||||||| Sbjct 13 SLNSAFSQHDSATFGY-NHKALKHNMTLNL#SDPAGNLKPHLRAKASDILTSPDVGLLKLA 71 |
| Tetraodon nigroviridis | Query 13 ALNASFLQSESGAYGYSNPKILKQSMTLNL#ADPVGNLKPHLRAKNSDLLTSPDVGLLKLA 72 +||| | | ++ +|| +|| || +|||||# || |+|||||||| |+||||||||||||| Sbjct 13 SLNA-FSQHDTAGFGYGHPKALKHNMTLNL#CDPAGSLKPHLRAKASELLTSPDVGLLKLA 71 |
| Ctenopharyngodon idella | Query 13 ALNASFLQSESGAYGYSNPKILKQSMTLNL#ADPVGNLKPHLRAKNSDLLTSPDVGLLKLA 72 ++|++| | +| +|| | | || +|||||# || |||||||||| ||+|||||||||||| Sbjct 13 SVNSAFSQHDSAPFGY-NHKALKHNMTLNL#TDPAGNLKPHLRAKASDILTSPDVGLLKLA 71 |
| Xenopus laevis | Query 13 ALNASFLQSESGAYGYSNPKILKQSMTLNL#ADPVGNLKPHLRAKNSDLLTSPDVGLLKLA 72 ||+|+| | ++ |||+ |+|||||||||#+|| +||||| | ++||||||||||||| Sbjct 13 ALSAAFAQHDATPYGYN--KVLKQSMTLNL#SDPSSAIKPHLRNKAAELLTSPDVGLLKLA 70 |
| Oncorhynchus mykiss | Query 17 SFLQSESGAYGYSNPKILKQSMTLNL#ADPVGNLKPHLRAKNSDLLTSPDVGLLKLA 72 +| | ++ ||| ||| || ||||||#||| |||||+|| ||+|||||| ||||| Sbjct 16 AFSQHQNAGYGY-NPKALKHSMTLNL#ADPTITLKPHLQAKASDILTSPDVQLLKLA 70 |
| Xiphophorus maculatus | Query 13 ALNASFLQSESGA-YGYSNPKILKQSMTLNL#ADPVGNLKPHLRAKNSDLLTSPDVGLLKL 71 |+||| | + | ||+ ||| |||+|||||# || | || | + |+||||||||||| Sbjct 16 AVNASNSQHDGAAVYGF-NPKTLKQTMTLNL#NDP-KNFKPQLGGR-LDILTSPDVGLLKL 72 Query 72 A 72 | Query 72 A 72 |
| Oryctolagus cuniculus | Query 54 RAKNSDLLTSPDVGLLKLA 72 ||||||||||||||||||| Query 54 RAKNSDLLTSPDVGLLKLA 72 |
[Site 2] LRAKNSDLLT62-SPDVGLLKLA
Thr62  Ser
Ser
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Leu53 | Arg54 | Ala55 | Lys56 | Asn57 | Ser58 | Asp59 | Leu60 | Leu61 | Thr62 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Ser63 | Pro64 | Asp65 | Val66 | Gly67 | Leu68 | Leu69 | Lys70 | Leu71 | Ala72 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| ILKQSMTLNLADPVGNLKPHLRAKNSDLLTSPDVGLLKLASPELERLIIQSSNGHITTTP |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | Rattus norvegicus | 121.00 | 4 | v-jun sarcoma virus 17 oncogene homolog |
| 2 | Mus musculus | 119.00 | 11 | unnamed protein product |
| 3 | Homo sapiens | 119.00 | 8 | JUN |
| 4 | N/A | 119.00 | 7 | 1404381A c-jun oncogene |
| 5 | synthetic construct | 119.00 | 5 | v-jun sarcoma virus 17 oncogene-like |
| 6 | Macaca fuscata | 119.00 | 1 | transcription factor c-Jun |
| 7 | Ovis aries | 118.00 | 2 | c-Jun protein |
| 8 | Sus scrofa | 118.00 | 1 | C-JUN protein |
| 9 | Bos taurus | 118.00 | 1 | c-jun |
| 10 | Danio rerio | 105.00 | 3 | c-Jun |
| 11 | Ctenopharyngodon idella | 104.00 | 2 | c-Jun protein |
| 12 | Carassius auratus | 104.00 | 1 | c-Jun protein |
| 13 | Coturnix coturnix | 104.00 | 1 | JUN_COTJA Transcription factor AP-1 (Proto-oncogen |
| 14 | Serinus canaria | 104.00 | 1 | JUN_SERCA Transcription factor AP-1 (Proto-oncogen |
| 15 | Takifugu rubripes | 101.00 | 6 | c-Jun protein |
| 16 | Tetraodon nigroviridis | 101.00 | 5 | unnamed protein product |
| 17 | Gallus gallus | 100.00 | 2 | jun oncogene |
| 18 | Xenopus laevis | 99.40 | 3 | c-Jun protein |
| 19 | Oncorhynchus mykiss | 95.50 | 1 | transcription factor AP-1 |
| 20 | Oryctolagus cuniculus | 79.70 | 1 | c-jun transcription factor |
| 21 | Xiphophorus maculatus | 73.90 | 2 | c-JUN |
| 22 | Cyprinus carpio | 56.60 | 1 | JUNB_CYPCA Transcription factor jun-B gi |
| 23 | Canis familiaris | 54.30 | 4 | PREDICTED: similar to Transcription factor jun-D |
| 24 | Danio rerio | 49.30 | 1 | jun B proto-oncogene, like |
| 25 | Strongylocentrotus purpuratus | 48.10 | 1 | PREDICTED: similar to transcription factor AP-1 |
Top-ranked sequences
| organism | matching |
|---|---|
| Rattus norvegicus | Query 33 ILKQSMTLNLADPVGNLKPHLRAKNSDLLT#SPDVGLLKLASPELERLIIQSSNGHITTTP 92 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 33 ILKQSMTLNLADPVGNLKPHLRAKNSDLLT#SPDVGLLKLASPELERLIIQSSNGHITTTP 92 |
| Mus musculus | Query 33 ILKQSMTLNLADPVGNLKPHLRAKNSDLLT#SPDVGLLKLASPELERLIIQSSNGHITTTP 92 |||||||||||||||+||||||||||||||#|||||||||||||||||||||||||||||| Sbjct 33 ILKQSMTLNLADPVGSLKPHLRAKNSDLLT#SPDVGLLKLASPELERLIIQSSNGHITTTP 92 |
| Homo sapiens | Query 33 ILKQSMTLNLADPVGNLKPHLRAKNSDLLT#SPDVGLLKLASPELERLIIQSSNGHITTTP 92 |||||||||||||||+||||||||||||||#|||||||||||||||||||||||||||||| Sbjct 33 ILKQSMTLNLADPVGSLKPHLRAKNSDLLT#SPDVGLLKLASPELERLIIQSSNGHITTTP 92 |
| N/A | Query 33 ILKQSMTLNLADPVGNLKPHLRAKNSDLLT#SPDVGLLKLASPELERLIIQSSNGHITTTP 92 |||||||||||||||+||||||||||||||#|||||||||||||||||||||||||||||| Sbjct 33 ILKQSMTLNLADPVGSLKPHLRAKNSDLLT#SPDVGLLKLASPELERLIIQSSNGHITTTP 92 |
| synthetic construct | Query 33 ILKQSMTLNLADPVGNLKPHLRAKNSDLLT#SPDVGLLKLASPELERLIIQSSNGHITTTP 92 |||||||||||||||+||||||||||||||#|||||||||||||||||||||||||||||| Sbjct 33 ILKQSMTLNLADPVGSLKPHLRAKNSDLLT#SPDVGLLKLASPELERLIIQSSNGHITTTP 92 |
| Macaca fuscata | Query 33 ILKQSMTLNLADPVGNLKPHLRAKNSDLLT#SPDVGLLKLASPELERLIIQSSNGHITTTP 92 |||||||||||||||+||||||||||||||#|||||||||||||||||||||||||||||| Sbjct 33 ILKQSMTLNLADPVGSLKPHLRAKNSDLLT#SPDVGLLKLASPELERLIIQSSNGHITTTP 92 |
| Ovis aries | Query 33 ILKQSMTLNLADPVGNLKPHLRAKNSDLLT#SPDVGLLKLASPELERLIIQSSNGHITTTP 92 |||||||||||||||+||||||||||||||#|||||||||||||||||||||||||||||| Sbjct 19 ILKQSMTLNLADPVGSLKPHLRAKNSDLLT#SPDVGLLKLASPELERLIIQSSNGHITTTP 78 |
| Sus scrofa | Query 33 ILKQSMTLNLADPVGNLKPHLRAKNSDLLT#SPDVGLLKLASPELERLIIQSSNGHITTTP 92 |||||||||||||||||||||||++|||||#|||||||||||||||||||||||||||||| Sbjct 33 ILKQSMTLNLADPVGNLKPHLRAQDSDLLT#SPDVGLLKLASPELERLIIQSSNGHITTTP 92 |
| Bos taurus | Query 33 ILKQSMTLNLADPVGNLKPHLRAKNSDLLT#SPDVGLLKLASPELERLIIQSSNGHITTTP 92 |||||||||||||||+||||||||||||||#|||||||||||||||||||||||||||||| Sbjct 28 ILKQSMTLNLADPVGSLKPHLRAKNSDLLT#SPDVGLLKLASPELERLIIQSSNGHITTTP 87 |
| Danio rerio | Query 34 LKQSMTLNLADPVGNLKPHLRAKNSDLLT#SPDVGLLKLASPELERLIIQSSNGHITTTP 92 || +|||||+|| |||||||||| ||+||#|||||||||||||||||||||||| ||||| Sbjct 33 LKHNMTLNLSDPAGNLKPHLRAKASDILT#SPDVGLLKLASPELERLIIQSSNGMITTTP 91 |
| Ctenopharyngodon idella | Query 34 LKQSMTLNLADPVGNLKPHLRAKNSDLLT#SPDVGLLKLASPELERLIIQSSNGHITTTP 92 || +||||| || |||||||||| ||+||#|||||||||||||||||||||||| ||||| Sbjct 33 LKHNMTLNLTDPAGNLKPHLRAKASDILT#SPDVGLLKLASPELERLIIQSSNGLITTTP 91 |
| Carassius auratus | Query 34 LKQSMTLNLADPVGNLKPHLRAKNSDLLT#SPDVGLLKLASPELERLIIQSSNGHITTTP 92 ||+|||||| || |||||||||| +|+||#|||||||||||||||||||||||| ||||| Sbjct 29 LKRSMTLNLTDPSGNLKPHLRAKANDILT#SPDVGLLKLASPELERLIIQSSNGMITTTP 87 |
| Coturnix coturnix | Query 33 ILKQSMTLNLADPVGNLKPHLRAKNSDLLT#SPDVGLLKLASPELERLIIQSSNGHITTTP 92 +|||||||||+|| +|||||| ||+|+||#|||||||||||||||||||||||| ||||| Sbjct 32 VLKQSMTLNLSDPASSLKPHLRNKNADILT#SPDVGLLKLASPELERLIIQSSNGLITTTP 91 |
| Serinus canaria | Query 33 ILKQSMTLNLADPVGNLKPHLRAKNSDLLT#SPDVGLLKLASPELERLIIQSSNGHITTTP 92 +|||+|||||+|| ||||||| ||+|+||#|||||||||||||||||||||||| ||||| Sbjct 33 VLKQNMTLNLSDPSSNLKPHLRNKNADILT#SPDVGLLKLASPELERLIIQSSNGLITTTP 92 |
| Takifugu rubripes | Query 34 LKQSMTLNLADPVGNLKPHLRAKNSDLLT#SPDVGLLKLASPELERLIIQSSNGHITTTP 92 || +|||||+|| | |||||||| |+|||#|||||||||||||||||||||||| ||||| Sbjct 33 LKHNMTLNLSDPTGTLKPHLRAKASELLT#SPDVGLLKLASPELERLIIQSSNGLITTTP 91 |
| Tetraodon nigroviridis | Query 34 LKQSMTLNLADPVGNLKPHLRAKNSDLLT#SPDVGLLKLASPELERLIIQSSNGHITTTP 92 || +||||| || |+|||||||| |+|||#|||||||||||||||||||||||| ||||| Sbjct 33 LKHNMTLNLCDPAGSLKPHLRAKASELLT#SPDVGLLKLASPELERLIIQSSNGLITTTP 91 |
| Gallus gallus | Query 33 ILKQSMTLNLADPVGNLKPHLRAKNSDLLT#SPDVGLLKLASPELERLIIQSSNGHITTTP 92 +|||||||||+| +|||||| ||+|+||#|||||||||||||||||||||||| ||||| Sbjct 29 VLKQSMTLNLSDAASSLKPHLRNKNADILT#SPDVGLLKLASPELERLIIQSSNGLITTTP 88 |
| Xenopus laevis | Query 33 ILKQSMTLNLADPVGNLKPHLRAKNSDLLT#SPDVGLLKLASPELERLIIQSSNGHITTTP 92 +|||||||||+|| +||||| | ++|||#|||||||||||||||||||||||| ||||| Sbjct 31 VLKQSMTLNLSDPSSAIKPHLRNKAAELLT#SPDVGLLKLASPELERLIIQSSNGMITTTP 90 |
| Oncorhynchus mykiss | Query 34 LKQSMTLNLADPVGNLKPHLRAKNSDLLT#SPDVGLLKLASPELERLIIQSSNGHITTTP 92 || ||||||||| |||||+|| ||+||#|||| ||||||||||||||||||| ||||| Sbjct 32 LKHSMTLNLADPTITLKPHLQAKASDILT#SPDVQLLKLASPELERLIIQSSNGLITTTP 90 |
| Oryctolagus cuniculus | Query 54 RAKNSDLLT#SPDVGLLKLASPELERLIIQSSNGHITTTP 92 |||||||||#|||||||||||||||||||||||||||||| Query 54 RAKNSDLLT#SPDVGLLKLASPELERLIIQSSNGHITTTP 92 |
| Xiphophorus maculatus | Query 34 LKQSMTLNLADPVGNLKPHLRAKNSDLLT#SPDVGLLKLASPELERLIIQSSNGHITTTP 92 |||+||||| || | || | + |+||#||||||||||||||||||||| +| | || Sbjct 37 LKQTMTLNLNDPK-NFKPQLGGR-LDILT#SPDVGLLKLASPELERLIIQSCSGLTTPTP 93 |
| Cyprinus carpio | Query 35 KQSMTLNLADPVGNLKPHLRAKNSDLL--T#SPDVGLLKLASPELERLIIQSSNGHITT 90 |||| ||| +| ||| ||| +# | | ||||||||||||||+||| +|| Sbjct 33 KQSMNLNLTEPYRNLK-------SDLYHTS#GADSGSLKLASPELERLIIQTSNGVLTT 83 |
| Canis familiaris | Query 60 LLT#SPDVGLLKLASPELERLIIQSSNGHITTTP 92 ||+#|||+|||||||||||||||| ||| +|||| Sbjct 44 LLS#SPDLGLLKLASPELERLIIQ-SNGLVTTTP 75 |
| Danio rerio | Query 35 KQSMTLNLADPVGNLKPHLRAKNSDLL--T#SPDVGLLKLASPELERLIIQSSNGHITT 90 | | ||+ +| |+ |+ ||| +#| ||| ||||||||||||||+ || +|| Sbjct 33 KPGMNLNVTEP-----PY-RSLKSDLYQAS#SADVGSLKLASPELERLIIQTGNGVLTT 84 |
| Strongylocentrotus purpuratus | Query 35 KQSMTLNLADPVGNLKPHLRAKNSDLLT#SPDVGLLKLASPELERLIIQSSNGHITTTP 92 |+ |||+| || | | | ||+#+||| +|||||||||++|| | |+| ||| Sbjct 17 KKCMTLDL--DVGMGKRKLTAA---LLS#TPDVQMLKLASPELEKMII-SQQGNICTTP 68 |
[Site 3] IQSSNGHITT90-TPTPTQFLCP
Thr90  Thr
Thr
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Ile81 | Gln82 | Ser83 | Ser84 | Asn85 | Gly86 | His87 | Ile88 | Thr89 | Thr90 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Thr91 | Pro92 | Thr93 | Pro94 | Thr95 | Gln96 | Phe97 | Leu98 | Cys99 | Pro100 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| LTSPDVGLLKLASPELERLIIQSSNGHITTTPTPTQFLCPKNVTDEQEGFAEGFVRALAE |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | Mus musculus | 125.00 | 11 | unnamed protein product |
| 2 | Homo sapiens | 125.00 | 9 | jun oncogene |
| 3 | N/A | 125.00 | 7 | 1411298A c-jun gene |
| 4 | synthetic construct | 125.00 | 5 | v-jun sarcoma virus 17 oncogene-like |
| 5 | Rattus norvegicus | 125.00 | 4 | v-jun sarcoma virus 17 oncogene homolog |
| 6 | Oryctolagus cuniculus | 125.00 | 1 | c-jun transcription factor |
| 7 | Sus scrofa | 125.00 | 1 | C-JUN protein |
| 8 | Macaca fuscata | 125.00 | 1 | transcription factor c-Jun |
| 9 | Danio rerio | 122.00 | 3 | c-Jun |
| 10 | Xenopus laevis | 121.00 | 3 | jun oncogene |
| 11 | Ctenopharyngodon idella | 121.00 | 2 | c-Jun protein |
| 12 | Gallus gallus | 121.00 | 2 | jun oncogene |
| 13 | Serinus canaria | 121.00 | 1 | JUN_SERCA Transcription factor AP-1 (Proto-oncogen |
| 14 | Coturnix coturnix | 121.00 | 1 | JUN_COTJA Transcription factor AP-1 (Proto-oncogen |
| 15 | Carassius auratus | 120.00 | 1 | c-Jun protein |
| 16 | Tetraodon nigroviridis | 117.00 | 6 | unnamed protein product |
| 17 | Oncorhynchus mykiss | 117.00 | 1 | transcription factor AP-1 |
| 18 | Takifugu rubripes | 115.00 | 6 | c-Jun protein |
| 19 | Xiphophorus maculatus | 105.00 | 2 | c-JUN |
| 20 | Bos taurus | 105.00 | 1 | c-jun |
| 21 | Ovis aries | 91.70 | 2 | c-Jun protein |
| 22 | Canis familiaris | 84.70 | 4 | PREDICTED: similar to Transcription factor jun-D |
| 23 | Danio rerio | 84.00 | 1 | jun B proto-oncogene, like |
| 24 | Cyprinus carpio | 81.60 | 1 | JUNB_CYPCA Transcription factor jun-B gi |
| 25 | Strongylocentrotus purpuratus | 81.30 | 1 | PREDICTED: similar to transcription factor AP-1 |
| 26 | Xenopus tropicalis | 47.80 | 1 | jun B proto-oncogene |
| 27 | Ciona intestinalis | 47.40 | 1 | transcription factor protein |
| 28 | Drosophila pseudoobscura | 37.00 | 1 | GA15338-PA |
| 29 | Schistosoma japonicum | 36.20 | 2 | SJCHGC03684 protein |
| 30 | Anabaena variabilis ATCC 29413 | 32.00 | 1 | Response regulator receiver domain protein (CheY) |
| 31 | Nostoc sp. PCC 7120 | 31.60 | 1 | two-component response regulator |
| 32 | Sinorhizobium meliloti 1021 | 31.60 | 1 | putative enzyme protein |
| 33 | Dictyostelium discoideum AX4 | 31.20 | 1 | hypothetical protein DDBDRAFT_0216866 |
| 34 | Dictyostelium discoideum | 30.80 | 1 | similar to Homo sapiens (Human). Myotubularin-rela |
Top-ranked sequences
| organism | matching |
|---|---|
| Mus musculus | Query 61 LTSPDVGLLKLASPELERLIIQSSNGHITT#TPTPTQFLCPKNVTDEQEGFAEGFVRALAE 120 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 61 LTSPDVGLLKLASPELERLIIQSSNGHITT#TPTPTQFLCPKNVTDEQEGFAEGFVRALAE 120 |
| Homo sapiens | Query 61 LTSPDVGLLKLASPELERLIIQSSNGHITT#TPTPTQFLCPKNVTDEQEGFAEGFVRALAE 120 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 61 LTSPDVGLLKLASPELERLIIQSSNGHITT#TPTPTQFLCPKNVTDEQEGFAEGFVRALAE 120 |
| N/A | Query 61 LTSPDVGLLKLASPELERLIIQSSNGHITT#TPTPTQFLCPKNVTDEQEGFAEGFVRALAE 120 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 61 LTSPDVGLLKLASPELERLIIQSSNGHITT#TPTPTQFLCPKNVTDEQEGFAEGFVRALAE 120 |
| synthetic construct | Query 61 LTSPDVGLLKLASPELERLIIQSSNGHITT#TPTPTQFLCPKNVTDEQEGFAEGFVRALAE 120 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 61 LTSPDVGLLKLASPELERLIIQSSNGHITT#TPTPTQFLCPKNVTDEQEGFAEGFVRALAE 120 |
| Rattus norvegicus | Query 61 LTSPDVGLLKLASPELERLIIQSSNGHITT#TPTPTQFLCPKNVTDEQEGFAEGFVRALAE 120 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 61 LTSPDVGLLKLASPELERLIIQSSNGHITT#TPTPTQFLCPKNVTDEQEGFAEGFVRALAE 120 |
| Oryctolagus cuniculus | Query 61 LTSPDVGLLKLASPELERLIIQSSNGHITT#TPTPTQFLCPKNVTDEQEGFAEGFVRALAE 120 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 61 LTSPDVGLLKLASPELERLIIQSSNGHITT#TPTPTQFLCPKNVTDEQEGFAEGFVRALAE 120 |
| Sus scrofa | Query 61 LTSPDVGLLKLASPELERLIIQSSNGHITT#TPTPTQFLCPKNVTDEQEGFAEGFVRALAE 120 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 61 LTSPDVGLLKLASPELERLIIQSSNGHITT#TPTPTQFLCPKNVTDEQEGFAEGFVRALAE 120 |
| Macaca fuscata | Query 61 LTSPDVGLLKLASPELERLIIQSSNGHITT#TPTPTQFLCPKNVTDEQEGFAEGFVRALAE 120 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 61 LTSPDVGLLKLASPELERLIIQSSNGHITT#TPTPTQFLCPKNVTDEQEGFAEGFVRALAE 120 |
| Danio rerio | Query 61 LTSPDVGLLKLASPELERLIIQSSNGHITT#TPTPTQFLCPKNVTDEQEGFAEGFVRALAE 120 |||||||||||||||||||||||||| |||#|||||||||||||||||||||||||||||| Sbjct 60 LTSPDVGLLKLASPELERLIIQSSNGMITT#TPTPTQFLCPKNVTDEQEGFAEGFVRALAE 119 |
| Xenopus laevis | Query 61 LTSPDVGLLKLASPELERLIIQSSNGHITT#TPTPTQFLCPKNVTDEQEGFAEGFVRALAE 120 |||||||||||||||||||||||||| |||#|||||||||||||||||||||+|||||||| Sbjct 59 LTSPDVGLLKLASPELERLIIQSSNGMITT#TPTPTQFLCPKNVTDEQEGFADGFVRALAE 118 |
| Ctenopharyngodon idella | Query 61 LTSPDVGLLKLASPELERLIIQSSNGHITT#TPTPTQFLCPKNVTDEQEGFAEGFVRALAE 120 |||||||||||||||||||||||||| |||#|||||||||||||||||||||||||||||| Sbjct 60 LTSPDVGLLKLASPELERLIIQSSNGLITT#TPTPTQFLCPKNVTDEQEGFAEGFVRALAE 119 |
| Gallus gallus | Query 61 LTSPDVGLLKLASPELERLIIQSSNGHITT#TPTPTQFLCPKNVTDEQEGFAEGFVRALAE 120 |||||||||||||||||||||||||| |||#|||||||||||||||||||||||||||||| Sbjct 57 LTSPDVGLLKLASPELERLIIQSSNGLITT#TPTPTQFLCPKNVTDEQEGFAEGFVRALAE 116 |
| Serinus canaria | Query 61 LTSPDVGLLKLASPELERLIIQSSNGHITT#TPTPTQFLCPKNVTDEQEGFAEGFVRALAE 120 |||||||||||||||||||||||||| |||#|||||||||||||||||||||||||||||| Sbjct 61 LTSPDVGLLKLASPELERLIIQSSNGLITT#TPTPTQFLCPKNVTDEQEGFAEGFVRALAE 120 |
| Coturnix coturnix | Query 61 LTSPDVGLLKLASPELERLIIQSSNGHITT#TPTPTQFLCPKNVTDEQEGFAEGFVRALAE 120 |||||||||||||||||||||||||| |||#|||||||||||||||||||||||||||||| Sbjct 60 LTSPDVGLLKLASPELERLIIQSSNGLITT#TPTPTQFLCPKNVTDEQEGFAEGFVRALAE 119 |
| Carassius auratus | Query 61 LTSPDVGLLKLASPELERLIIQSSNGHITT#TPTPTQFLCPKNVTDEQEGFAEGFVRALAE 120 |||||||||||||||||||||||||| |||#|||||||||||||||||||||||| ||||| Sbjct 56 LTSPDVGLLKLASPELERLIIQSSNGMITT#TPTPTQFLCPKNVTDEQEGFAEGFARALAE 115 |
| Tetraodon nigroviridis | Query 61 LTSPDVGLLKLASPELERLIIQSSNGHITT#TPTPTQFLCPKNVTDEQEGFAEGFVRALAE 120 |||||||||||||||||||||||||| |||#|||||||+||||||+||||||||||||||| Sbjct 60 LTSPDVGLLKLASPELERLIIQSSNGLITT#TPTPTQFICPKNVTEEQEGFAEGFVRALAE 119 |
| Oncorhynchus mykiss | Query 61 LTSPDVGLLKLASPELERLIIQSSNGHITT#TPTPTQFLCPKNVTDEQEGFAEGFVRALAE 120 |||||| ||||||||||||||||||| |||#||||||||||+||||||||||||||||||| Sbjct 59 LTSPDVQLLKLASPELERLIIQSSNGLITT#TPTPTQFLCPRNVTDEQEGFAEGFVRALAE 118 |
| Takifugu rubripes | Query 61 LTSPDVGLLKLASPELERLIIQSSNGHITT#TPTPTQFLCPKNVTDEQEGFAEGFVRALAE 120 |||||||||||||||||||||||||| |||#|||||||+ |||||+||||||||||||||| Sbjct 60 LTSPDVGLLKLASPELERLIIQSSNGLITT#TPTPTQFISPKNVTEEQEGFAEGFVRALAE 119 |
| Xiphophorus maculatus | Query 61 LTSPDVGLLKLASPELERLIIQSSNGHITT#TPTPTQFLCPKNVTDEQEGFAEGFVRALAE 120 ||||||||||||||||||||||| +| |#|||||||+|||||+|||||||||||||||| Sbjct 62 LTSPDVGLLKLASPELERLIIQSCSG--LT#TPTPTQFICPKNVSDEQEGFAEGFVRALAE 119 |
| Bos taurus | Query 61 LTSPDVGLLKLASPELERLIIQSSNGHITT#TPTPTQFLCPKNVTDEQEGF 110 ||||||||||||||||||||||||||||||#|||||||||||||||||||| Query 61 LTSPDVGLLKLASPELERLIIQSSNGHITT#TPTPTQFLCPKNVTDEQEGF 110 |
| Ovis aries | Query 61 LTSPDVGLLKLASPELERLIIQSSNGHITT#TPTPTQFLCPKNVT 104 ||||||||||||||||||||||||||||||#|||||||||||||| Query 61 LTSPDVGLLKLASPELERLIIQSSNGHITT#TPTPTQFLCPKNVT 104 |
| Canis familiaris | Query 61 LTSPDVGLLKLASPELERLIIQSSNGHITT#TPTPTQFLCPKNVTDEQEGFAEGFVRAL 118 |+|||+||||||||||||||||| || +||#||| |||| || |++ ||||||+|| Sbjct 45 LSSPDLGLLKLASPELERLIIQS-NGLVTT#TPTSTQFLYPKVAASEEQEFAEGFVKAL 101 |
| Danio rerio | Query 62 TSPDVGLLKLASPELERLIIQSSNGHITT#TPTPTQFLCPKNVTDEQEGFAEGFVRALAE 120 +| ||| ||||||||||||||+ || + |#|||| |+| + +||||||||||||+|| | Sbjct 56 SSADVGSLKLASPELERLIIQTGNG-VLT#TPTPGQYLYGRGITDEQEGFAEGFVKALDE 113 |
| Cyprinus carpio | Query 65 DVGLLKLASPELERLIIQSSNGHITT#TPTPTQFLCPKNVTDEQEGFAEGFVRALAE 120 | | ||||||||||||||+||| + |#|||| |+| + +||||||||||||+|| + Sbjct 58 DSGSLKLASPELERLIIQTSNG-VLT#TPTPGQYLYSRGITDEQEGFAEGFVKALDD 112 |
| Strongylocentrotus purpuratus | Query 61 LTSPDVGLLKLASPELERLIIQSSNGHITT#TPTPTQFLCPKNVTDEQEGFAEGFVRAL 118 |++||| +|||||||||++|| | |+| |#|||| ||+ |||||+|| ||+||| || Sbjct 38 LSTPDVQMLKLASPELEKMII-SQQGNICT#TPTPGQFISPKNVTEEQAAFAQGFVEAL 94 |
| Xenopus tropicalis | Query 69 LKLASPELERLIIQSSNGHITT#TPTPTQFLCP---KNVTDEQEGFAEGFVRAL 118 ||+|+|||||+|| | | + +# |+ | +|+||||| +|||+|| Sbjct 66 LKMAAPELERMIIHSGPG-VIS#GPSGGQLYYSSRGNGITEEQEGFVDGFVKAL 117 |
| Ciona intestinalis | Query 61 LTSPD-VGLLKLASPELERLIIQSSNGHITT#TPTPTQFLCPKNVTDEQEGFAEGFVRAL 118 |++|| | +||+ ||+ +|+++ || #| +|| +| | ||| +|+|||| | Sbjct 134 LSTPDLVNMLKVGSPDFDRMLVHSSQA---N#TQNNSQFFQ-RNATQEQEIYAQGFVREL 188 |
| Drosophila pseudoobscura | Query 61 LTSPDVGLLKLASPELERLIIQSSNGHITT#TPTPTQFLCPK--NVTDEQEGFAEGFVRAL 118 + |||+ + +|+||++++ + |+ #|| | + | || ||| | +|| || Sbjct 82 INSPDLQAKTVNTPDLEKILLSN---HMMQ#TPQPGKVFPTKVGPVTSEQEAFGKGFEEAL 138 |
| Schistosoma japonicum | Query 75 ELERLIIQSSNGHITT#TP-TPTQFLCPKNVTDEQEGFAEGFVRALAE 120 |+|+++ |+ #+| || || ||+||+||| || | + | | Sbjct 84 EVEKIL-----QHLKA#SPQTPGSFLNPKHVTEEQERFANVFTQKLNE 125 |
| Anabaena variabilis ATCC 29413 | Query 64 PDVGLLKLASPELERLI-IQSSNGHITT#TPTPTQFLCPKNVTDEQEGFAEGFVRAL 118 ||| || + |+++ + |+ + |# | | | | || |||| |+|+ Sbjct 49 PDVILLDVMMPDIDGIATFQALQANPVT#QSIPVILLTAKTQTAEQRGFAELGVQAI 104 |
| Nostoc sp. PCC 7120 | Query 64 PDVGLLKLASPELERLI-IQSSNGHITT#TPTPTQFLCPKNVTDEQEGFAEGFVRAL 118 ||| || + |+++ + |+ + |# | | | | | || ||| ++|+ Sbjct 49 PDVILLDVMMPDMDGIATFQALQANPVT#QPIPVILLTAKTQTAEQRRFAELGIQAI 104 |
| Sinorhizobium meliloti 1021 | Query 64 PDVGLLKLASPELERLIIQSSNGHITT#TPTPTQFLCPKNVTDEQEGFAEGFVRALAE 120 | | ||+|+ ||+ + + |#+ | | | + +|| || +| +|++|+ Sbjct 69 PGVRWLKIAA-------IQAHDAQVVT#SGITTVFDCLRLGSDEDSGFPKGEMRSMAD 118 |
| Dictyostelium discoideum AX4 | Query 75 ELERLIIQSSNGHITT-------#TPT-PTQFLCPKNVTDEQEGFAEGF 114 | ||| ||| # || |+||+ | ||+||| || Sbjct 1262 EFERLQFPRDKWRITTLNSDYSL#CPTYPSQFIVPVNVSDEQLKKVSGF 1309 |
| Dictyostelium discoideum | Query 75 ELERLIIQSSNGHITT-------#TPT-PTQFLCPKNVTDEQEGFAEGF 114 | ||| ||| # || |+||+ | ||+||| || Sbjct 1217 EFERLQFPRDKWRITTLNSDYSL#CPTYPSQFIVPVNVSDEQLKKVSGF 1264 |
[Site 4] GGGGYSASLH164-SEPPVYANLS
His164  Ser
Ser
iTraq-117 Signal 3055.6 (

 ) for GGGGYSASLH
) for GGGGYSASLH
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Gly155 | Gly156 | Gly157 | Gly158 | Tyr159 | Ser160 | Ala161 | Ser162 | Leu163 | His164 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Ser165 | Glu166 | Pro167 | Pro168 | Val169 | Tyr170 | Ala171 | Asn172 | Leu173 | Ser174 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| QPVSGAGMVAPAVASVAGAGGGGGYSASLHSEPPVYANLSNFNPGALSSGGGAPSYGATG |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | Rattus norvegicus | 116.00 | 1 | v-jun sarcoma virus 17 oncogene homolog |
| 2 | Mus musculus | 113.00 | 4 | Jun oncogene |
| 3 | Homo sapiens | 81.30 | 4 | JUN protein |
| 4 | N/A | 80.90 | 5 | 1411298A c-jun gene |
| 5 | synthetic construct | 80.90 | 2 | v-jun sarcoma virus 17 oncogene homolog (avian) |
| 6 | Sus scrofa | 80.90 | 1 | C-JUN protein |
| 7 | Oryctolagus cuniculus | 70.90 | 1 | c-jun transcription factor |
| 8 | Coturnix coturnix | 63.50 | 1 | JUN_COTJA Transcription factor AP-1 (Proto-oncogen |
| 9 | Gallus gallus | 63.50 | 1 | jun oncogene |
| 10 | Serinus canaria | 57.80 | 1 | JUN_SERCA Transcription factor AP-1 (Proto-oncogen |
| 11 | Xenopus laevis | 51.60 | 3 | c-Jun protein |
| 12 | Ctenopharyngodon idella | 44.70 | 1 | c-Jun protein |
| 13 | Carassius auratus | 42.00 | 1 | c-Jun protein |
| 14 | Danio rerio | 40.40 | 2 | c-Jun protein |
Top-ranked sequences
| organism | matching |
|---|---|
| Rattus norvegicus | Query 135 QPVSGAGMVAPAVASVAGAGGGGGYSASLH#SEPPVYANLSNFNPGALSSGGGAPSYGATG 194 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 135 QPVSGAGMVAPAVASVAGAGGGGGYSASLH#SEPPVYANLSNFNPGALSSGGGAPSYGATG 194 |
| Mus musculus | Query 135 QPVSGAGMVAPAVASVAGAGGGGGYSASLH#SEPPVYANLSNFNPGALSSGGGAPSYGATG 194 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||| | Sbjct 135 QPVSGAGMVAPAVASVAGAGGGGGYSASLH#SEPPVYANLSNFNPGALSSGGGAPSYGAAG 194 |
| Homo sapiens | Query 135 QPVSGAGMVAPAVASVAGAGGGGGYSASLH#SEPPVYANLSNFNPGALSSGGGAPSYGATG 194 |||+|||||||||||||| | ||+|||||#|||||||||||||||||||||||||||| | Sbjct 35 QPVNGAGMVAPAVASVAGGSGSGGFSASLH#SEPPVYANLSNFNPGALSSGGGAPSYGAAG 94 |
| N/A | Query 135 QPVSGAGMVAPAVASVAGAGGGGGYSASLH#SEPPVYANLSNFNPGALSSGGGAPSYGATG 194 |||+|||||||||||||| | ||+|||||#|||||||||||||||||||||||||||| | Sbjct 135 QPVNGAGMVAPAVASVAGGSGSGGFSASLH#SEPPVYANLSNFNPGALSSGGGAPSYGAAG 194 |
| synthetic construct | Query 135 QPVSGAGMVAPAVASVAGAGGGGGYSASLH#SEPPVYANLSNFNPGALSSGGGAPSYGATG 194 |||+|||||||||||||| | ||+|||||#|||||||||||||||||||||||||||| | Sbjct 135 QPVNGAGMVAPAVASVAGGSGSGGFSASLH#SEPPVYANLSNFNPGALSSGGGAPSYGAAG 194 |
| Sus scrofa | Query 135 QPVSGAGMVAPAVASVAGAGGGGGYSASLH#SEPPVYANLSNFNPGALSSGGGAPSYGATG 194 |||||||+|||||||||| | ||+|||||#|||||||||||||||||||||||||||| | Sbjct 135 QPVSGAGLVAPAVASVAGGSGSGGFSASLH#SEPPVYANLSNFNPGALSSGGGAPSYGAAG 194 |
| Oryctolagus cuniculus | Query 135 QPVSGAGMVAPAVASVAGAGGGGGYSAS-LH#SEPPVYANLSNFNPGALSSGGGAPSYGAT 193 |||||||| ||||||||| ||| | ++ ||#|||||||||||||| ||||||||||||| Sbjct 82 QPVSGAGMAAPAVASVAGGGGGSGGFSASLH#SEPPVYANLSNFNPSALSSGGGAPSYGAA 141 Query 194 G 194 | Query 194 G 194 |
| Coturnix coturnix | Query 135 QPVSGAGMVAPAVASVAGAGGGGGYSASLH#SEPPVYANLSNFNPGALSSGGGAPSYGATG 194 ||||| || | |+ ||| ++ |||#|||||||||||||| ||+| ||+| | | Sbjct 134 QPVSG-GM-----APVSSMAGGGSFNTSLH#SEPPVYANLSNFNPNALNS---APNYNANG 184 |
| Gallus gallus | Query 135 QPVSGAGMVAPAVASVAGAGGGGGYSASLH#SEPPVYANLSNFNPGALSSGGGAPSYGATG 194 ||||| || | |+ ||| ++ |||#|||||||||||||| ||+| ||+| | | Sbjct 131 QPVSG-GM-----APVSSMAGGGSFNTSLH#SEPPVYANLSNFNPNALNS---APNYNANG 181 |
| Serinus canaria | Query 137 VSGAGMVAPAVASVAGAGGGGGYSASLH#SEPPVYANLSNFNPGALSSGGGAPSYGA 192 | | | +| |+ | ++ |||#|||||||||||||| |||| ||+| | Sbjct 131 TSAAQPVTSGMAPVSSMAGSTSFNTSLH#SEPPVYANLSNFNPNALSS---APNYNA 183 |
| Xenopus laevis | Query 147 VASVAGAGGGGGYSASLH#SEPPVYANLSNFNPGALSSGG 185 + ||+ | |+| +||#+||||||||||||| +++ Sbjct 139 LTSVSTIAGNTGFSNNLH#NEPPVYANLSNFNPSTITTSA 177 |
| Ctenopharyngodon idella | Query 146 AVASVAGAGGGGGYSASLH#SEPPVYANLSNFNPGALSSGGGAPSYGA 192 ++| |+ || ||+|+ #|+|||||+|+ ||| |+|+ | || + Sbjct 139 SMAPVSSIAGGAVYSSSMR#SDPPVYADLNTFNP-AISTANPAMSYTS 184 |
| Carassius auratus | Query 146 AVASVAGAGGGGGYSASLH#SEPPVYANLSNFNPGALSSGGGAPSYGA 192 ++| |+ || ||+|+ #|+|||||+|+ ||| +|+ | +| + Sbjct 135 SMAPVSSMAGGAVYSSSMR#SDPPVYADLNTFNP-VISTANPAMNYTS 180 |
| Danio rerio | Query 146 AVASVAGAGGGGGYSASLH#SEPPVYANLSNFNPGALSSGGGAP 188 ++| |+ || ||+++ #++|||||+|+ ||| |+|| | Sbjct 139 SMAPVSSIAGGAVYSSAMR#ADPPVYADLNTFNP-AISSSSANP 180 |
* References
[PubMed ID: 24492282] Dorn C, Engelmann JC, Saugspier M, Koch A, Hartmann A, Muller M, Spang R, Bosserhoff A, Hellerbrand C, Increased expression of c-Jun in nonalcoholic fatty liver disease. Lab Invest. 2014 Apr;94(4):394-408. doi: 10.1038/labinvest.2014.3. Epub 2014 Feb
[PubMed ID: 24421392] ... Ferreira DM, Afonso MB, Rodrigues PM, Simao AL, Pereira DM, Borralho PM, Rodrigues CM, Castro RE, c-Jun N-terminal kinase 1/c-Jun activation of the p53/microRNA 34a/sirtuin 1 pathway contributes to apoptosis induced by deoxycholic acid in rat liver. Mol Cell Biol. 2014 Mar;34(6):1100-20. doi: 10.1128/MCB.00420-13. Epub 2014 Jan
[PubMed ID: 23870463] ... Shi LQ, Ruan CL, Expression and significance of MMP-7, c-Jun and c-Fos in rats skin photoaging. Asian Pac J Trop Med. 2013 Oct;6(10):768-70. doi: 10.1016/S1995-7645(13)60135-2.
[PubMed ID: 23506737] ... Cheng X, Fu R, Gao M, Liu S, Li YQ, Song FH, Bruce IC, Zhou LH, Wu W, Intrathecal application of short interfering RNA knocks down c-jun expression and augments spinal motoneuron death after root avulsion in adult rats. Neuroscience. 2013 Jun 25;241:268-79. doi: 10.1016/j.neuroscience.2013.03.006.
[PubMed ID: 23818969] ... Holderness Parker N, Donninger H, Birrer MJ, Leaner VD, p21-activated kinase 3 (PAK3) is an AP-1 regulated gene contributing to actin organisation and migration of transformed fibroblasts. PLoS One. 2013 Jun 20;8(6):e66892. doi: 10.1371/journal.pone.0066892. Print 2013.
[PubMed ID: 7560090] ... Pravenec M, Gauguier D, Schott JJ, Buard J, Kren V, Bila V, Szpirer C, Szpirer J, Wang JM, Huang H, et al., Mapping of quantitative trait loci for blood pressure and cardiac mass in the rat by genome scanning of recombinant inbred strains. J Clin Invest. 1995 Oct;96(4):1973-8.
[PubMed ID: 7721748] ... Hyder SM, Nawaz Z, Chiappetta C, Yokoyama K, Stancel GM, The protooncogene c-jun contains an unusual estrogen-inducible enhancer within the coding sequence. J Biol Chem. 1995 Apr 14;270(15):8506-13.
[PubMed ID: 1310930] ... Kitabayashi I, Kawakami Z, Chiu R, Ozawa K, Matsuoka T, Toyoshima S, Umesono K, Evans RM, Gachelin G, Yokoyama K, Transcriptional regulation of the c-jun gene by retinoic acid and E1A during differentiation of F9 cells. EMBO J. 1992 Jan;11(1):167-75.
[PubMed ID: 2113275] ... Kitabayashi I, Saka F, Gachelin G, Yokoyama K, Nucleotide sequence of rat c-jun protooncogene. Nucleic Acids Res. 1990 Jun 11;18(11):3400.
[PubMed ID: 2507134] ... Sakai M, Okuda A, Hatayama I, Sato K, Nishi S, Muramatsu M, Structure and expression of the rat c-jun messenger RNA: tissue distribution and increase during chemical hepatocarcinogenesis. Cancer Res. 1989 Oct 15;49(20):5633-7.