SB0087 : Histone H3.2
[ CaMP Format ]
* Basic Information
| Organism | Bos taurus (cattle) |
| Protein Names | PREDICTED: histone H3.2 [Bos taurus]; histone H3.2; Histone H3.2 |
| Gene Names | LOC619141; Derived by automated computational analysis using gene prediction method: GNOMON. Supporting evidence includes similarity to: 1 mRNA, 13 Proteins |
| Gene Locus | Not available |
| GO Function | Not available |
| Entrez Protein | Entrez Nucleotide | Entrez Gene | UniProt | OMIM | HGNC | HPRD | KEGG |
|---|---|---|---|---|---|---|---|
| XP_876553 | XM_871460 | 619141 | P84227 | N/A | N/A | N/A | bta:619141 |
* Information From OMIM
Not Available.
* Structure Information
1. Primary Information
Length: 136 aa
Average Mass: 15.388 kDa
Monoisotopic Mass: 15.379 kDa
2. Domain Information
Annotated Domains: interpro / pfam / smart / prosite
Computationally Assigned Domains (Pfam+HMMER):
| domain name | begin | end | score | e-value |
|---|---|---|---|---|
| --- cleavage 10 --- | ||||
| Centromere kinetochore component CENP-T histone fold 1. | 20 | 28 | 15.0 | 0.0 |
| --- cleavage 23 (inside Centromere kinetochore component CENP-T histone fold 20..28) --- | ||||
| --- cleavage 28 (inside Centromere kinetochore component CENP-T histone fold 20..28) --- | ||||
| --- cleavage 28 --- | ||||
| --- cleavage 33 --- | ||||
| --- cleavage 34 --- | ||||
| --- cleavage 49 --- | ||||
| Kinetochore component CENP-S 1. | 70 | 130 | 13.0 | 0.0 |
| --- cleavage 119 (inside Kinetochore component CENP-S 70..130) --- | ||||
| --- cleavage 129 (inside Kinetochore component CENP-S 70..130) --- | ||||
3. Sequence Information
Fasta Sequence: SB0087.fasta
Amino Acid Sequence and Secondary Structures (PsiPred):
4. 3D Information
Not Available.
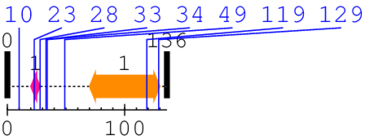
* Cleavage Information
8 [sites]
Source Reference: [PubMed ID: 3038857] Sakai K, Akanuma H, Imahori K, Kawashima S, A unique specificity of a calcium activated neutral protease indicated in histone hydrolysis. J Biochem. 1987 Apr;101(4):911-8.
Cleavage sites (±10aa)
[Site 1] MARTKQTARK10-STGGKAPRKQ
Lys10  Ser
Ser
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Met1 | Ala2 | Arg3 | Thr4 | Lys5 | Gln6 | Thr7 | Ala8 | Arg9 | Lys10 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Ser11 | Thr12 | Gly13 | Gly14 | Lys15 | Ala16 | Pro17 | Arg18 | Lys19 | Gln20 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| MARTKQTARKSTGGKAPRKQLATKAARKSAPATGGVKKPH |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | Oryza sativa (japonica cultivar-group) | 79.00 | 1 | histone H3 - maize |
| 2 | Canis familiaris | 78.60 | 8 | PREDICTED: similar to histone 1, H2ai (predicted) |
| 3 | Homo sapiens | 78.60 | 6 | Histone cluster 1, H3i |
| 4 | Mytilus chilensis | 78.60 | 1 | histone H3 |
| 5 | Acilius sylvanus | 78.60 | 1 | histone 3 |
| 6 | Psammechinus miliaris | 78.60 | 1 | cleavage stage histone H3 |
| 7 | N/A | 78.20 | 11 | H32_PEA Histone H3.2 gi |
| 8 | Mus musculus | 78.20 | 8 | histone cluster 2, H3c1 isoform 1 |
| 9 | Strongylocentrotus purpuratus | 78.20 | 3 | histone H3 |
| 10 | Danio rerio | 78.20 | 2 | hypothetical protein LOC794406 |
| 11 | Drosophila melanogaster | 78.20 | 2 | H3 histone |
| 12 | Oikopleura dioica | 78.20 | 2 | histone h3.1 |
| 13 | Cyathermia naticoides | 78.20 | 1 | histone H3 |
| 14 | Cricetulus longicaudatus | 78.20 | 1 | H32_CRILO Histone H3.2 gi |
| 15 | Attenella margarita | 78.20 | 1 | histone H3 |
| 16 | Strongylocentrotus purpuratus | 78.20 | 1 | late embryonic histone H3 |
| 17 | Styela plicata | 78.20 | 1 | H3 histone |
| 18 | Stygifloris sp. EP141 | 78.20 | 1 | histone H3 |
| 19 | Drosophila persimilis | 78.20 | 1 | histone 3 |
| 20 | Expression vector pET3-H3 | 78.20 | 1 | Xenopus laevis-like histone H3 |
| 21 | Asellus aquaticus | 78.20 | 1 | putative H3 histone |
| 22 | Thekalabis sp. DM16 | 78.20 | 1 | histone 3 |
| 23 | Mytilus californianus | 78.20 | 1 | histone H3 |
| 24 | Arabidopsis thaliana | 78.20 | 1 | histone H3 |
| 25 | Xenopus laevis | 78.20 | 1 | histone H3 |
| 26 | Mortierella alpina | 78.20 | 1 | H3_MORAP Histone H3 gi |
| 27 | Isonychia sp. EP142 | 78.20 | 1 | histone H3 |
| 28 | Lilium longiflorum | 78.20 | 1 | H32_LILLO Histone H3.2 gi |
| 29 | synthetic construct | 77.40 | 1 | histone 3 H3 |
| 30 | Drosophila virilis | 77.40 | 1 | histone H3 |
| 31 | Griffithsia japonica | 77.00 | 2 | putative histone H3 protein |
| 32 | Venerupis (Ruditapes) philippinarum | 77.00 | 2 | replacement histone H3.3 |
| 33 | Nematostella vectensis | 77.00 | 1 | predicted protein |
| 34 | Anopheles gambiae str. PEST | 77.00 | 1 | AGAP012197-PA |
| 35 | Gibbula zonata | 77.00 | 1 | histone H3 |
| 36 | Griffithsia japonica | 77.00 | 1 | H3_GRIJA Histone H3 gi |
| 37 | Kluyveromyces lactis | 77.00 | 1 | unnamed protein product |
| 38 | Drosophila simulans | 77.00 | 1 | Histone H3 |
| 39 | Caenorhabditis elegans | 77.00 | 1 | HIStone family member (his-42) |
| 40 | Callibaetis sp. EP020 | 77.00 | 1 | histone H3 |
| 41 | Scissurella cf. coronata CET-2005 | 77.00 | 1 | histone H3 |
| 42 | Rattus norvegicus | 77.00 | 1 | hypothetical protein LOC691496 |
| 43 | Pongo pygmaeus | 77.00 | 1 | H33_PONPY Histone H3.3 gi |
| 44 | Pan troglodytes | 77.00 | 1 | PREDICTED: hypothetical protein |
| 45 | Bos taurus | 76.60 | 2 | PREDICTED: similar to histone protein Hist2h3c1 |
| 46 | Siphlonella sp. EP083 | 76.60 | 1 | histone H3 |
| 47 | Neurospora crassa OR74A | 76.60 | 1 | histone H3 |
| 48 | Neurospora crassa | 76.60 | 1 | histone H3 |
| 49 | Drosophila erecta | 76.60 | 1 | histone H3 |
| 50 | Candida glabrata | 76.60 | 1 | AF520061_3 HHT2p |
| 51 | Mus pahari | 76.60 | 1 | histone H3.2 protein |
| 52 | Lepetodrilus ovalis | 76.60 | 1 | histone H3 |
| 53 | Malus x domestica | 76.30 | 1 | histone 3 |
| 54 | Tetrapygus niger | 76.30 | 1 | AF356516_2 early histone H3 |
Top-ranked sequences
| organism | matching |
|---|---|
| Oryza sativa (japonica cultivar-group) | Query 1 MARTKQTARK#STGGKAPRKQLATKAARKSAPATGGVKKPH 40 ||||||||||#|||||||||||||||||||||||||||||| Query 1 MARTKQTARK#STGGKAPRKQLATKAARKSAPATGGVKKPH 40 |
| Canis familiaris | Query 1 MARTKQTARK#STGGKAPRKQLATKAARKSAPATGGVKKPH 40 ||||||||||#|||||||||||||||||||||||||||||| Query 1 MARTKQTARK#STGGKAPRKQLATKAARKSAPATGGVKKPH 40 |
| Homo sapiens | Query 1 MARTKQTARK#STGGKAPRKQLATKAARKSAPATGGVKKPH 40 ||||||||||#|||||||||||||||||||||||||||||| Query 1 MARTKQTARK#STGGKAPRKQLATKAARKSAPATGGVKKPH 40 |
| Mytilus chilensis | Query 1 MARTKQTARK#STGGKAPRKQLATKAARKSAPATGGVKKPH 40 ||||||||||#|||||||||||||||||||||||||||||| Query 1 MARTKQTARK#STGGKAPRKQLATKAARKSAPATGGVKKPH 40 |
| Acilius sylvanus | Query 1 MARTKQTARK#STGGKAPRKQLATKAARKSAPATGGVKKPH 40 ||||||||||#|||||||||||||||||||||||||||||| Query 1 MARTKQTARK#STGGKAPRKQLATKAARKSAPATGGVKKPH 40 |
| Psammechinus miliaris | Query 1 MARTKQTARK#STGGKAPRKQLATKAARKSAPATGGVKKPH 40 ||||||||||#|||||||||||||||||||||||||||||| Query 1 MARTKQTARK#STGGKAPRKQLATKAARKSAPATGGVKKPH 40 |
| N/A | Query 1 MARTKQTARK#STGGKAPRKQLATKAARKSAPATGGVKKPH 40 ||||||||||#|||||||||||||||||||||||||||||| Query 1 MARTKQTARK#STGGKAPRKQLATKAARKSAPATGGVKKPH 40 |
| Mus musculus | Query 1 MARTKQTARK#STGGKAPRKQLATKAARKSAPATGGVKKPH 40 ||||||||||#|||||||||||||||||||||||||||||| Query 1 MARTKQTARK#STGGKAPRKQLATKAARKSAPATGGVKKPH 40 |
| Strongylocentrotus purpuratus | Query 1 MARTKQTARK#STGGKAPRKQLATKAARKSAPATGGVKKPH 40 ||||||||||#|||||||||||||||||||||||||||||| Query 1 MARTKQTARK#STGGKAPRKQLATKAARKSAPATGGVKKPH 40 |
| Danio rerio | Query 1 MARTKQTARK#STGGKAPRKQLATKAARKSAPATGGVKKPH 40 ||||||||||#|||||||||||||||||||||||||||||| Query 1 MARTKQTARK#STGGKAPRKQLATKAARKSAPATGGVKKPH 40 |
| Drosophila melanogaster | Query 1 MARTKQTARK#STGGKAPRKQLATKAARKSAPATGGVKKPH 40 ||||||||||#|||||||||||||||||||||||||||||| Query 1 MARTKQTARK#STGGKAPRKQLATKAARKSAPATGGVKKPH 40 |
| Oikopleura dioica | Query 1 MARTKQTARK#STGGKAPRKQLATKAARKSAPATGGVKKPH 40 ||||||||||#|||||||||||||||||||||||||||||| Query 1 MARTKQTARK#STGGKAPRKQLATKAARKSAPATGGVKKPH 40 |
| Cyathermia naticoides | Query 1 MARTKQTARK#STGGKAPRKQLATKAARKSAPATGGVKKPH 40 ||||||||||#|||||||||||||||||||||||||||||| Query 1 MARTKQTARK#STGGKAPRKQLATKAARKSAPATGGVKKPH 40 |
| Cricetulus longicaudatus | Query 1 MARTKQTARK#STGGKAPRKQLATKAARKSAPATGGVKKPH 40 ||||||||||#|||||||||||||||||||||||||||||| Query 1 MARTKQTARK#STGGKAPRKQLATKAARKSAPATGGVKKPH 40 |
| Attenella margarita | Query 1 MARTKQTARK#STGGKAPRKQLATKAARKSAPATGGVKKPH 40 ||||||||||#|||||||||||||||||||||||||||||| Query 1 MARTKQTARK#STGGKAPRKQLATKAARKSAPATGGVKKPH 40 |
| Strongylocentrotus purpuratus | Query 1 MARTKQTARK#STGGKAPRKQLATKAARKSAPATGGVKKPH 40 ||||||||||#|||||||||||||||||||||||||||||| Query 1 MARTKQTARK#STGGKAPRKQLATKAARKSAPATGGVKKPH 40 |
| Styela plicata | Query 1 MARTKQTARK#STGGKAPRKQLATKAARKSAPATGGVKKPH 40 ||||||||||#|||||||||||||||||||||||||||||| Query 1 MARTKQTARK#STGGKAPRKQLATKAARKSAPATGGVKKPH 40 |
| Stygifloris sp. EP141 | Query 1 MARTKQTARK#STGGKAPRKQLATKAARKSAPATGGVKKPH 40 ||||||||||#|||||||||||||||||||||||||||||| Query 1 MARTKQTARK#STGGKAPRKQLATKAARKSAPATGGVKKPH 40 |
| Drosophila persimilis | Query 1 MARTKQTARK#STGGKAPRKQLATKAARKSAPATGGVKKPH 40 ||||||||||#|||||||||||||||||||||||||||||| Query 1 MARTKQTARK#STGGKAPRKQLATKAARKSAPATGGVKKPH 40 |
| Expression vector pET3-H3 | Query 1 MARTKQTARK#STGGKAPRKQLATKAARKSAPATGGVKKPH 40 ||||||||||#|||||||||||||||||||||||||||||| Query 1 MARTKQTARK#STGGKAPRKQLATKAARKSAPATGGVKKPH 40 |
| Asellus aquaticus | Query 1 MARTKQTARK#STGGKAPRKQLATKAARKSAPATGGVKKPH 40 ||||||||||#|||||||||||||||||||||||||||||| Query 1 MARTKQTARK#STGGKAPRKQLATKAARKSAPATGGVKKPH 40 |
| Thekalabis sp. DM16 | Query 1 MARTKQTARK#STGGKAPRKQLATKAARKSAPATGGVKKPH 40 ||||||||||#|||||||||||||||||||||||||||||| Query 1 MARTKQTARK#STGGKAPRKQLATKAARKSAPATGGVKKPH 40 |
| Mytilus californianus | Query 1 MARTKQTARK#STGGKAPRKQLATKAARKSAPATGGVKKPH 40 ||||||||||#|||||||||||||||||||||||||||||| Query 1 MARTKQTARK#STGGKAPRKQLATKAARKSAPATGGVKKPH 40 |
| Arabidopsis thaliana | Query 1 MARTKQTARK#STGGKAPRKQLATKAARKSAPATGGVKKPH 40 ||||||||||#|||||||||||||||||||||||||||||| Query 1 MARTKQTARK#STGGKAPRKQLATKAARKSAPATGGVKKPH 40 |
| Xenopus laevis | Query 1 MARTKQTARK#STGGKAPRKQLATKAARKSAPATGGVKKPH 40 ||||||||||#|||||||||||||||||||||||||||||| Query 1 MARTKQTARK#STGGKAPRKQLATKAARKSAPATGGVKKPH 40 |
| Mortierella alpina | Query 1 MARTKQTARK#STGGKAPRKQLATKAARKSAPATGGVKKPH 40 ||||||||||#|||||||||||||||||||||||||||||| Query 1 MARTKQTARK#STGGKAPRKQLATKAARKSAPATGGVKKPH 40 |
| Isonychia sp. EP142 | Query 1 MARTKQTARK#STGGKAPRKQLATKAARKSAPATGGVKKPH 40 ||||||||||#|||||||||||||||||||||||||||||| Query 1 MARTKQTARK#STGGKAPRKQLATKAARKSAPATGGVKKPH 40 |
| Lilium longiflorum | Query 1 MARTKQTARK#STGGKAPRKQLATKAARKSAPATGGVKKPH 40 ||||||||||#|||||||||||||||||||||||||||||| Query 1 MARTKQTARK#STGGKAPRKQLATKAARKSAPATGGVKKPH 40 |
| synthetic construct | Query 1 MARTKQTARK#STGGKAPRKQLATKAARKSAPATGGVKKPH 40 ||||||||||#|||||||||||||| ||||||||||||||| Sbjct 1 MARTKQTARK#STGGKAPRKQLATKVARKSAPATGGVKKPH 40 |
| Drosophila virilis | Query 1 MARTKQTARK#STGGKAPRKQLATKAARKSAPATGGVKKPH 40 ||||||||||#|||||||||||||||||||||||||||||| Query 1 MARTKQTARK#STGGKAPRKQLATKAARKSAPATGGVKKPH 40 |
| Griffithsia japonica | Query 1 MARTKQTARK#STGGKAPRKQLATKAARKSAPATGGVKKPH 40 ||||||||||#||||||||||||||||||||||+||||||| Sbjct 1 MARTKQTARK#STGGKAPRKQLATKAARKSAPASGGVKKPH 40 |
| Venerupis (Ruditapes) philippinarum | Query 1 MARTKQTARK#STGGKAPRKQLATKAARKSAPATGGVKKPH 40 ||||||||||#|||||||||||||||||||||+|||||||| Sbjct 1 MARTKQTARK#STGGKAPRKQLATKAARKSAPSTGGVKKPH 40 |
| Nematostella vectensis | Query 1 MARTKQTARK#STGGKAPRKQLATKAARKSAPATGGVKKPH 40 ||||||||||#|||||||||||||||||||||+|||||||| Sbjct 1 MARTKQTARK#STGGKAPRKQLATKAARKSAPSTGGVKKPH 40 |
| Anopheles gambiae str. PEST | Query 1 MARTKQTARK#STGGKAPRKQLATKAARKSAPATGGVKKPH 40 ||||||||||#|||||||||||||||||||||+|||||||| Sbjct 1 MARTKQTARK#STGGKAPRKQLATKAARKSAPSTGGVKKPH 40 |
| Gibbula zonata | Query 1 MARTKQTARK#STGGKAPRKQLATKAARKSAPATGGVKKPH 40 | ||||||||#|||||||||||||||||||||||||||||| Sbjct 1 MVRTKQTARK#STGGKAPRKQLATKAARKSAPATGGVKKPH 40 |
| Griffithsia japonica | Query 1 MARTKQTARK#STGGKAPRKQLATKAARKSAPATGGVKKPH 40 ||||||||||#||||||||||||||||||||||+||||||| Sbjct 1 MARTKQTARK#STGGKAPRKQLATKAARKSAPASGGVKKPH 40 |
| Kluyveromyces lactis | Query 1 MARTKQTARK#STGGKAPRKQLATKAARKSAPATGGVKKPH 40 ||||||||||#||||||||||||+||||||||+|||||||| Sbjct 41 MARTKQTARK#STGGKAPRKQLASKAARKSAPSTGGVKKPH 80 |
| Drosophila simulans | Query 1 MARTKQTARK#STGGKAPRKQLATKAARKSAPATGGVKKPH 40 ||||||||||#||||||||||||||||||| |||||||||| Sbjct 1 MARTKQTARK#STGGKAPRKQLATKAARKSVPATGGVKKPH 40 |
| Caenorhabditis elegans | Query 1 MARTKQTARK#STGGKAPRKQLATKAARKSAPATGGVKKPH 40 ||||||||||#||||||||||||||||||||||+||||||| Sbjct 1 MARTKQTARK#STGGKAPRKQLATKAARKSAPASGGVKKPH 40 |
| Callibaetis sp. EP020 | Query 1 MARTKQTARK#STGGKAPRKQLATKAARKSAPATGGVKKPH 40 ||||||||||#|+|||||||||||||||||||||||||||| Sbjct 1 MARTKQTARK#SSGGKAPRKQLATKAARKSAPATGGVKKPH 40 |
| Scissurella cf. coronata CET-2005 | Query 1 MARTKQTARK#STGGKAPRKQLATKAARKSAPATGGVKKPH 40 | ||||||||#|||||||||||||||||||||||||||||| Sbjct 1 MVRTKQTARK#STGGKAPRKQLATKAARKSAPATGGVKKPH 40 |
| Rattus norvegicus | Query 1 MARTKQTARK#STGGKAPRKQLATKAARKSAPATGGVKKPH 40 ||||||||||#|||||||||||||| ||||||||||||||| Sbjct 1 MARTKQTARK#STGGKAPRKQLATKVARKSAPATGGVKKPH 40 |
| Pongo pygmaeus | Query 1 MARTKQTARK#STGGKAPRKQLATKAARKSAPATGGVKKPH 40 ||||||||||#|||||||||||||||||||||+|||||||| Sbjct 1 MARTKQTARK#STGGKAPRKQLATKAARKSAPSTGGVKKPH 40 |
| Pan troglodytes | Query 1 MARTKQTARK#STGGKAPRKQLATKAARKSAPATGGVKKPH 40 ||||||||||#|||||||||||||||||||||+|||||||| Sbjct 1 MARTKQTARK#STGGKAPRKQLATKAARKSAPSTGGVKKPH 40 |
| Bos taurus | Query 1 MARTKQTARK#STGGKAPRKQLATKAARKSAPATGGVKKPH 40 | ||||||||#|||||||||||||||||||||||||||||| Sbjct 1 MTRTKQTARK#STGGKAPRKQLATKAARKSAPATGGVKKPH 40 |
| Siphlonella sp. EP083 | Query 1 MARTKQTARK#STGGKAPRKQLATKAARKSAPATGGVKKPH 40 ||||||||| #|||||||||||||||||||||||||||||| Sbjct 1 MARTKQTARX#STGGKAPRKQLATKAARKSAPATGGVKKPH 40 |
| Neurospora crassa OR74A | Query 1 MARTKQTARK#STGGKAPRKQLATKAARKSAPATGGVKKPH 40 ||||||||||#||||||||||||+||||||||+|||||||| Sbjct 1 MARTKQTARK#STGGKAPRKQLASKAARKSAPSTGGVKKPH 40 |
| Neurospora crassa | Query 1 MARTKQTARK#STGGKAPRKQLATKAARKSAPATGGVKKPH 40 ||||||||||#||||||||||||+||||||||+|||||||| Sbjct 1 MARTKQTARK#STGGKAPRKQLASKAARKSAPSTGGVKKPH 40 |
| Drosophila erecta | Query 1 MARTKQTARK#STGGKAPRKQLATKAARKSAPATGGVKKPH 40 ||||||| ||#|||||||||||||||||||||||||||||| Sbjct 1 MARTKQTTRK#STGGKAPRKQLATKAARKSAPATGGVKKPH 40 |
| Candida glabrata | Query 1 MARTKQTARK#STGGKAPRKQLATKAARKSAPATGGVKKPH 40 ||||||||||#||||||||||||+||||||||+|||||||| Sbjct 1 MARTKQTARK#STGGKAPRKQLASKAARKSAPSTGGVKKPH 40 |
| Mus pahari | Query 1 MARTKQTARK#STGGKAPRKQLATKAARKSAPATGGVKKPH 40 ||||||||||#||||||||||||||||||||| |||||||| Sbjct 1 MARTKQTARK#STGGKAPRKQLATKAARKSAPTTGGVKKPH 40 |
| Lepetodrilus ovalis | Query 1 MARTKQTARK#STGGKAPRKQLATKAARKSAPATGGVKKPH 40 ||||||| ||#|||||||||||||||||||||||||||||| Sbjct 1 MARTKQTXRK#STGGKAPRKQLATKAARKSAPATGGVKKPH 40 |
| Malus x domestica | Query 1 MARTKQTARK#STGGKAPRKQLATKAARKSAPATGGVKKPH 40 ||||||||||#||||||||||||||||||||| |||||||| Sbjct 1 MARTKQTARK#STGGKAPRKQLATKAARKSAPTTGGVKKPH 40 |
| Tetrapygus niger | Query 1 MARTKQTARK#STGGKAPRKQLATKAARKSAPATGGVKKPH 40 ||||||||||#|||||||||||||||||||||||||||||| Query 1 MARTKQTARK#STGGKAPRKQLATKAARKSAPATGGVKKPH 40 |
[Site 2] GKAPRKQLAT23-KAARKSAPAT
Thr23  Lys
Lys
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Gly14 | Lys15 | Ala16 | Pro17 | Arg18 | Lys19 | Gln20 | Leu21 | Ala22 | Thr23 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Lys24 | Ala25 | Ala26 | Arg27 | Lys28 | Ser29 | Ala30 | Pro31 | Ala32 | Thr33 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| MARTKQTARKSTGGKAPRKQLATKAARKSAPATGGVKKPHRYRPGTVALREIR |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | Canis familiaris | 105.00 | 7 | PREDICTED: similar to histone 1, H2ai (predicted) |
| 2 | Homo sapiens | 105.00 | 6 | Histone cluster 1, H3i |
| 3 | Acilius sylvanus | 105.00 | 1 | histone 3 |
| 4 | Stygifloris sp. EP141 | 105.00 | 1 | histone H3 |
| 5 | N/A | 104.00 | 10 | histone H3 |
| 6 | Mus musculus | 104.00 | 8 | histone cluster 2, H3c1 isoform 1 |
| 7 | Strongylocentrotus purpuratus | 104.00 | 3 | PREDICTED: hypothetical protein |
| 8 | Oikopleura dioica | 104.00 | 2 | histone h3.1 |
| 9 | Drosophila melanogaster | 104.00 | 2 | H3 histone |
| 10 | Cyathermia naticoides | 104.00 | 1 | histone H3 |
| 11 | Expression vector pET3-H3 | 104.00 | 1 | Xenopus laevis-like histone H3 |
| 12 | Strongylocentrotus purpuratus | 104.00 | 1 | late embryonic histone H3 |
| 13 | Danio rerio | 104.00 | 1 | hypothetical protein LOC794406 |
| 14 | Xenopus laevis | 104.00 | 1 | histone H3 |
| 15 | Drosophila persimilis | 104.00 | 1 | histone 3 |
| 16 | Asellus aquaticus | 104.00 | 1 | putative H3 histone |
| 17 | Styela plicata | 104.00 | 1 | H3 histone |
| 18 | Oryza sativa (japonica cultivar-group) | 104.00 | 1 | histone H3 - maize |
| 19 | Attenella margarita | 104.00 | 1 | histone H3 |
| 20 | Mytilus chilensis | 104.00 | 1 | histone H3 |
| 21 | Thekalabis sp. DM16 | 104.00 | 1 | histone 3 |
| 22 | Isonychia sp. EP142 | 104.00 | 1 | histone H3 |
| 23 | Mytilus californianus | 104.00 | 1 | histone H3 |
| 24 | Mortierella alpina | 104.00 | 1 | H3_MORAP Histone H3 gi |
| 25 | Caenorhabditis elegans | 103.00 | 3 | HIStone family member (his-42) |
| 26 | Arabidopsis thaliana | 103.00 | 2 | histone H3 |
| 27 | Lilium longiflorum | 103.00 | 2 | H32_LILLO Histone H3.2 gi |
| 28 | Venerupis (Ruditapes) philippinarum | 103.00 | 2 | replacement histone H3.3 |
| 29 | synthetic construct | 103.00 | 1 | histone 3 H3 |
| 30 | Mus pahari | 103.00 | 1 | histone H3.2 protein |
| 31 | Drosophila simulans | 103.00 | 1 | Histone H3 |
| 32 | Nematostella vectensis | 103.00 | 1 | predicted protein |
| 33 | Callibaetis sp. EP020 | 103.00 | 1 | histone H3 |
| 34 | Pan troglodytes | 103.00 | 1 | PREDICTED: hypothetical protein |
| 35 | Pongo pygmaeus | 103.00 | 1 | H33_PONPY Histone H3.3 gi |
| 36 | Anopheles gambiae str. PEST | 103.00 | 1 | AGAP012197-PA |
| 37 | Psammechinus miliaris | 103.00 | 1 | cleavage stage histone H3 |
| 38 | Gibbula zonata | 103.00 | 1 | histone H3 |
| 39 | Scissurella cf. coronata CET-2005 | 103.00 | 1 | histone H3 |
| 40 | Siphlonella sp. EP083 | 103.00 | 1 | histone H3 |
| 41 | Drosophila virilis | 103.00 | 1 | histone H3 |
| 42 | Lepetodrilus ovalis | 103.00 | 1 | histone H3 |
| 43 | Caenorhabditis briggsae AF16 | 102.00 | 3 | Hypothetical protein CBG16863 |
| 44 | Griffithsia japonica | 102.00 | 2 | putative histone H3 protein |
| 45 | Bos taurus | 102.00 | 2 | PREDICTED: similar to histone protein Hist2h3c1 |
| 46 | Erythemis sp. OD024 | 102.00 | 1 | histone H3 |
| 47 | Gromphadorhina portentosa | 102.00 | 1 | histone 3 |
| 48 | Cricetulus longicaudatus | 102.00 | 1 | H32_CRILO Histone H3.2 gi |
| 49 | Caudatella hystrix | 102.00 | 1 | histone H3 |
| 50 | Grylloblatta campodeiformis | 102.00 | 1 | histone 3 |
| 51 | Malus x domestica | 102.00 | 1 | histone 3 |
| 52 | Griffithsia japonica | 102.00 | 1 | H3_GRIJA Histone H3 gi |
| 53 | Orxines macklottii | 102.00 | 1 | histone 3 |
| 54 | Tetrapygus niger | 102.00 | 1 | AF356516_2 early histone H3 |
| 55 | Oryza sativa | 102.00 | 1 | AF467728_1 disease-resistent-related protein |
| 56 | Picea abies | 102.00 | 1 | histone 3 |
| 57 | Progomphus obscurus | 102.00 | 1 | histone H3 |
| 58 | Acilius fraternus | 102.00 | 1 | histone 3 |
| 59 | Agathemera crassa | 102.00 | 1 | histone 3 |
| 60 | Drosophila erecta | 102.00 | 1 | histone H3 |
| 61 | Prosopistoma wouterae | 101.00 | 1 | histone H3 |
Top-ranked sequences
| organism | matching |
|---|---|
| Canis familiaris | Query 1 MARTKQTARKSTGGKAPRKQLAT#KAARKSAPATGGVKKPHRYRPGTVALREIR 53 |||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 1 MARTKQTARKSTGGKAPRKQLAT#KAARKSAPATGGVKKPHRYRPGTVALREIR 53 |
| Homo sapiens | Query 1 MARTKQTARKSTGGKAPRKQLAT#KAARKSAPATGGVKKPHRYRPGTVALREIR 53 |||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 1 MARTKQTARKSTGGKAPRKQLAT#KAARKSAPATGGVKKPHRYRPGTVALREIR 53 |
| Acilius sylvanus | Query 1 MARTKQTARKSTGGKAPRKQLAT#KAARKSAPATGGVKKPHRYRPGTVALREIR 53 |||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 1 MARTKQTARKSTGGKAPRKQLAT#KAARKSAPATGGVKKPHRYRPGTVALREIR 53 |
| Stygifloris sp. EP141 | Query 1 MARTKQTARKSTGGKAPRKQLAT#KAARKSAPATGGVKKPHRYRPGTVALREIR 53 |||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 1 MARTKQTARKSTGGKAPRKQLAT#KAARKSAPATGGVKKPHRYRPGTVALREIR 53 |
| N/A | Query 1 MARTKQTARKSTGGKAPRKQLAT#KAARKSAPATGGVKKPHRYRPGTVALREIR 53 |||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 1 MARTKQTARKSTGGKAPRKQLAT#KAARKSAPATGGVKKPHRYRPGTVALREIR 53 |
| Mus musculus | Query 1 MARTKQTARKSTGGKAPRKQLAT#KAARKSAPATGGVKKPHRYRPGTVALREIR 53 |||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 1 MARTKQTARKSTGGKAPRKQLAT#KAARKSAPATGGVKKPHRYRPGTVALREIR 53 |
| Strongylocentrotus purpuratus | Query 1 MARTKQTARKSTGGKAPRKQLAT#KAARKSAPATGGVKKPHRYRPGTVALREIR 53 |||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 1 MARTKQTARKSTGGKAPRKQLAT#KAARKSAPATGGVKKPHRYRPGTVALREIR 53 |
| Oikopleura dioica | Query 1 MARTKQTARKSTGGKAPRKQLAT#KAARKSAPATGGVKKPHRYRPGTVALREIR 53 |||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 1 MARTKQTARKSTGGKAPRKQLAT#KAARKSAPATGGVKKPHRYRPGTVALREIR 53 |
| Drosophila melanogaster | Query 1 MARTKQTARKSTGGKAPRKQLAT#KAARKSAPATGGVKKPHRYRPGTVALREIR 53 |||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 1 MARTKQTARKSTGGKAPRKQLAT#KAARKSAPATGGVKKPHRYRPGTVALREIR 53 |
| Cyathermia naticoides | Query 1 MARTKQTARKSTGGKAPRKQLAT#KAARKSAPATGGVKKPHRYRPGTVALREIR 53 |||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 1 MARTKQTARKSTGGKAPRKQLAT#KAARKSAPATGGVKKPHRYRPGTVALREIR 53 |
| Expression vector pET3-H3 | Query 1 MARTKQTARKSTGGKAPRKQLAT#KAARKSAPATGGVKKPHRYRPGTVALREIR 53 |||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 1 MARTKQTARKSTGGKAPRKQLAT#KAARKSAPATGGVKKPHRYRPGTVALREIR 53 |
| Strongylocentrotus purpuratus | Query 1 MARTKQTARKSTGGKAPRKQLAT#KAARKSAPATGGVKKPHRYRPGTVALREIR 53 |||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 1 MARTKQTARKSTGGKAPRKQLAT#KAARKSAPATGGVKKPHRYRPGTVALREIR 53 |
| Danio rerio | Query 1 MARTKQTARKSTGGKAPRKQLAT#KAARKSAPATGGVKKPHRYRPGTVALREIR 53 |||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 1 MARTKQTARKSTGGKAPRKQLAT#KAARKSAPATGGVKKPHRYRPGTVALREIR 53 |
| Xenopus laevis | Query 1 MARTKQTARKSTGGKAPRKQLAT#KAARKSAPATGGVKKPHRYRPGTVALREIR 53 |||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 1 MARTKQTARKSTGGKAPRKQLAT#KAARKSAPATGGVKKPHRYRPGTVALREIR 53 |
| Drosophila persimilis | Query 1 MARTKQTARKSTGGKAPRKQLAT#KAARKSAPATGGVKKPHRYRPGTVALREIR 53 |||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 1 MARTKQTARKSTGGKAPRKQLAT#KAARKSAPATGGVKKPHRYRPGTVALREIR 53 |
| Asellus aquaticus | Query 1 MARTKQTARKSTGGKAPRKQLAT#KAARKSAPATGGVKKPHRYRPGTVALREIR 53 |||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 1 MARTKQTARKSTGGKAPRKQLAT#KAARKSAPATGGVKKPHRYRPGTVALREIR 53 |
| Styela plicata | Query 1 MARTKQTARKSTGGKAPRKQLAT#KAARKSAPATGGVKKPHRYRPGTVALREIR 53 |||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 1 MARTKQTARKSTGGKAPRKQLAT#KAARKSAPATGGVKKPHRYRPGTVALREIR 53 |
| Oryza sativa (japonica cultivar-group) | Query 1 MARTKQTARKSTGGKAPRKQLAT#KAARKSAPATGGVKKPHRYRPGTVALREIR 53 |||||||||||||||||||||||#||||||||||||||||||+||||||||||| Sbjct 1 MARTKQTARKSTGGKAPRKQLAT#KAARKSAPATGGVKKPHRFRPGTVALREIR 53 |
| Attenella margarita | Query 1 MARTKQTARKSTGGKAPRKQLAT#KAARKSAPATGGVKKPHRYRPGTVALREIR 53 |||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 1 MARTKQTARKSTGGKAPRKQLAT#KAARKSAPATGGVKKPHRYRPGTVALREIR 53 |
| Mytilus chilensis | Query 1 MARTKQTARKSTGGKAPRKQLAT#KAARKSAPATGGVKKPHRYRPGTVALREIR 53 |||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 1 MARTKQTARKSTGGKAPRKQLAT#KAARKSAPATGGVKKPHRYRPGTVALREIR 53 |
| Thekalabis sp. DM16 | Query 1 MARTKQTARKSTGGKAPRKQLAT#KAARKSAPATGGVKKPHRYRPGTVALREIR 53 |||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 1 MARTKQTARKSTGGKAPRKQLAT#KAARKSAPATGGVKKPHRYRPGTVALREIR 53 |
| Isonychia sp. EP142 | Query 1 MARTKQTARKSTGGKAPRKQLAT#KAARKSAPATGGVKKPHRYRPGTVALREIR 53 |||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 1 MARTKQTARKSTGGKAPRKQLAT#KAARKSAPATGGVKKPHRYRPGTVALREIR 53 |
| Mytilus californianus | Query 1 MARTKQTARKSTGGKAPRKQLAT#KAARKSAPATGGVKKPHRYRPGTVALREIR 53 |||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 1 MARTKQTARKSTGGKAPRKQLAT#KAARKSAPATGGVKKPHRYRPGTVALREIR 53 |
| Mortierella alpina | Query 1 MARTKQTARKSTGGKAPRKQLAT#KAARKSAPATGGVKKPHRYRPGTVALREIR 53 |||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 1 MARTKQTARKSTGGKAPRKQLAT#KAARKSAPATGGVKKPHRYRPGTVALREIR 53 |
| Caenorhabditis elegans | Query 1 MARTKQTARKSTGGKAPRKQLAT#KAARKSAPATGGVKKPHRYRPGTVALREIR 53 |||||||||||||||||||||||#|||||||||+|||||||||||||||||||| Sbjct 1 MARTKQTARKSTGGKAPRKQLAT#KAARKSAPASGGVKKPHRYRPGTVALREIR 53 |
| Arabidopsis thaliana | Query 1 MARTKQTARKSTGGKAPRKQLAT#KAARKSAPATGGVKKPHRYRPGTVALREIR 53 |||||||||||||||||||||||#||||||||||||||||||+||||||||||| Sbjct 1 MARTKQTARKSTGGKAPRKQLAT#KAARKSAPATGGVKKPHRFRPGTVALREIR 53 |
| Lilium longiflorum | Query 1 MARTKQTARKSTGGKAPRKQLAT#KAARKSAPATGGVKKPHRYRPGTVALREIR 53 |||||||||||||||||||||||#||||||||||||||||||+||||||||||| Sbjct 1 MARTKQTARKSTGGKAPRKQLAT#KAARKSAPATGGVKKPHRFRPGTVALREIR 53 |
| Venerupis (Ruditapes) philippinarum | Query 1 MARTKQTARKSTGGKAPRKQLAT#KAARKSAPATGGVKKPHRYRPGTVALREIR 53 |||||||||||||||||||||||#||||||||+||||||||||||||||||||| Sbjct 1 MARTKQTARKSTGGKAPRKQLAT#KAARKSAPSTGGVKKPHRYRPGTVALREIR 53 |
| synthetic construct | Query 1 MARTKQTARKSTGGKAPRKQLAT#KAARKSAPATGGVKKPHRYRPGTVALREIR 53 |||||||||||||||||||||||#| |||||||||||||||||||||||||||| Sbjct 1 MARTKQTARKSTGGKAPRKQLAT#KVARKSAPATGGVKKPHRYRPGTVALREIR 53 |
| Mus pahari | Query 1 MARTKQTARKSTGGKAPRKQLAT#KAARKSAPATGGVKKPHRYRPGTVALREIR 53 |||||||||||||||||||||||#|||||||| ||||||||||||||||||||| Sbjct 1 MARTKQTARKSTGGKAPRKQLAT#KAARKSAPTTGGVKKPHRYRPGTVALREIR 53 |
| Drosophila simulans | Query 1 MARTKQTARKSTGGKAPRKQLAT#KAARKSAPATGGVKKPHRYRPGTVALREIR 53 |||||||||||||||||||||||#|||||| ||||||||||||||||||||||| Sbjct 1 MARTKQTARKSTGGKAPRKQLAT#KAARKSVPATGGVKKPHRYRPGTVALREIR 53 |
| Nematostella vectensis | Query 1 MARTKQTARKSTGGKAPRKQLAT#KAARKSAPATGGVKKPHRYRPGTVALREIR 53 |||||||||||||||||||||||#||||||||+||||||||||||||||||||| Sbjct 1 MARTKQTARKSTGGKAPRKQLAT#KAARKSAPSTGGVKKPHRYRPGTVALREIR 53 |
| Callibaetis sp. EP020 | Query 1 MARTKQTARKSTGGKAPRKQLAT#KAARKSAPATGGVKKPHRYRPGTVALREIR 53 |||||||||||+|||||||||||#|||||||||||||||||||||||||||||| Sbjct 1 MARTKQTARKSSGGKAPRKQLAT#KAARKSAPATGGVKKPHRYRPGTVALREIR 53 |
| Pan troglodytes | Query 1 MARTKQTARKSTGGKAPRKQLAT#KAARKSAPATGGVKKPHRYRPGTVALREIR 53 |||||||||||||||||||||||#||||||||+||||||||||||||||||||| Sbjct 1 MARTKQTARKSTGGKAPRKQLAT#KAARKSAPSTGGVKKPHRYRPGTVALREIR 53 |
| Pongo pygmaeus | Query 1 MARTKQTARKSTGGKAPRKQLAT#KAARKSAPATGGVKKPHRYRPGTVALREIR 53 |||||||||||||||||||||||#||||||||+||||||||||||||||||||| Sbjct 1 MARTKQTARKSTGGKAPRKQLAT#KAARKSAPSTGGVKKPHRYRPGTVALREIR 53 |
| Anopheles gambiae str. PEST | Query 1 MARTKQTARKSTGGKAPRKQLAT#KAARKSAPATGGVKKPHRYRPGTVALREIR 53 |||||||||||||||||||||||#||||||||+||||||||||||||||||||| Sbjct 1 MARTKQTARKSTGGKAPRKQLAT#KAARKSAPSTGGVKKPHRYRPGTVALREIR 53 |
| Psammechinus miliaris | Query 1 MARTKQTARKSTGGKAPRKQLAT#KAARKSAPATGGVKKPHRYRPGTVALREIR 53 |||||||||||||||||||||||#||||||||||||||||||+||||||||||| Sbjct 1 MARTKQTARKSTGGKAPRKQLAT#KAARKSAPATGGVKKPHRFRPGTVALREIR 53 |
| Gibbula zonata | Query 1 MARTKQTARKSTGGKAPRKQLAT#KAARKSAPATGGVKKPHRYRPGTVALREIR 53 | |||||||||||||||||||||#|||||||||||||||||||||||||||||| Sbjct 1 MVRTKQTARKSTGGKAPRKQLAT#KAARKSAPATGGVKKPHRYRPGTVALREIR 53 |
| Scissurella cf. coronata CET-2005 | Query 1 MARTKQTARKSTGGKAPRKQLAT#KAARKSAPATGGVKKPHRYRPGTVALREIR 53 | |||||||||||||||||||||#|||||||||||||||||||||||||||||| Sbjct 1 MVRTKQTARKSTGGKAPRKQLAT#KAARKSAPATGGVKKPHRYRPGTVALREIR 53 |
| Siphlonella sp. EP083 | Query 1 MARTKQTARKSTGGKAPRKQLAT#KAARKSAPATGGVKKPHRYRPGTVALREIR 53 ||||||||| |||||||||||||#|||||||||||||||||||||||||||||| Sbjct 1 MARTKQTARXSTGGKAPRKQLAT#KAARKSAPATGGVKKPHRYRPGTVALREIR 53 |
| Drosophila virilis | Query 1 MARTKQTARKSTGGKAPRKQLAT#KAARKSAPATGGVKKPHRYRPGTVALREIR 53 |||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 1 MARTKQTARKSTGGKAPRKQLAT#KAARKSAPATGGVKKPHRYRPGTVALREIR 53 |
| Lepetodrilus ovalis | Query 1 MARTKQTARKSTGGKAPRKQLAT#KAARKSAPATGGVKKPHRYRPGTVALREIR 53 ||||||| |||||||||||||||#|||||||||||||||||||||||||||||| Sbjct 1 MARTKQTXRKSTGGKAPRKQLAT#KAARKSAPATGGVKKPHRYRPGTVALREIR 53 |
| Caenorhabditis briggsae AF16 | Query 1 MARTKQTARKSTGGKAPRKQLAT#KAARKSAPATGGVKKPHRYRPGTVALREIR 53 |||||||||||||||||||||||#|||||||| ||||||||||||||||||||| Sbjct 1 MARTKQTARKSTGGKAPRKQLAT#KAARKSAPTTGGVKKPHRYRPGTVALREIR 53 |
| Griffithsia japonica | Query 1 MARTKQTARKSTGGKAPRKQLAT#KAARKSAPATGGVKKPHRYRPGTVALREIR 53 |||||||||||||||||||||||#|||||||||+||||||||+||||||||||| Sbjct 1 MARTKQTARKSTGGKAPRKQLAT#KAARKSAPASGGVKKPHRFRPGTVALREIR 53 |
| Bos taurus | Query 1 MARTKQTARKSTGGKAPRKQLAT#KAARKSAPATGGVKKPHRYRPGTVALREIR 53 | |||||||||||||||||||||#|||||||||||||||||||||||||||||| Sbjct 1 MTRTKQTARKSTGGKAPRKQLAT#KAARKSAPATGGVKKPHRYRPGTVALREIR 53 |
| Erythemis sp. OD024 | Query 2 ARTKQTARKSTGGKAPRKQLAT#KAARKSAPATGGVKKPHRYRPGTVALREIR 53 ||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 2 ARTKQTARKSTGGKAPRKQLAT#KAARKSAPATGGVKKPHRYRPGTVALREIR 53 |
| Gromphadorhina portentosa | Query 2 ARTKQTARKSTGGKAPRKQLAT#KAARKSAPATGGVKKPHRYRPGTVALREIR 53 ||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 2 ARTKQTARKSTGGKAPRKQLAT#KAARKSAPATGGVKKPHRYRPGTVALREIR 53 |
| Cricetulus longicaudatus | Query 1 MARTKQTARKSTGGKAPRKQLAT#KAARKSAPATGGVKKPHRYRPGTVALREIR 53 |||||||||||||||||||||||#|||||||||||||||||||||||| ||||| Sbjct 1 MARTKQTARKSTGGKAPRKQLAT#KAARKSAPATGGVKKPHRYRPGTVTLREIR 53 |
| Caudatella hystrix | Query 1 MARTKQTARKSTGGKAPRKQLAT#KAARKSAPATGGVKKPHRYRPGTVALREIR 53 |||||||||||||||||||| ||#|||||||||||||||||||||||||||||| Sbjct 1 MARTKQTARKSTGGKAPRKQXAT#KAARKSAPATGGVKKPHRYRPGTVALREIR 53 |
| Grylloblatta campodeiformis | Query 2 ARTKQTARKSTGGKAPRKQLAT#KAARKSAPATGGVKKPHRYRPGTVALREIR 53 ||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 2 ARTKQTARKSTGGKAPRKQLAT#KAARKSAPATGGVKKPHRYRPGTVALREIR 53 |
| Malus x domestica | Query 1 MARTKQTARKSTGGKAPRKQLAT#KAARKSAPATGGVKKPHRYRPGTVALREIR 53 |||||||||||||||||||||||#|||||||| ||||||||||||||||||||| Sbjct 1 MARTKQTARKSTGGKAPRKQLAT#KAARKSAPTTGGVKKPHRYRPGTVALREIR 53 |
| Griffithsia japonica | Query 1 MARTKQTARKSTGGKAPRKQLAT#KAARKSAPATGGVKKPHRYRPGTVALREIR 53 |||||||||||||||||||||||#|||||||||+||||||||+||||||||||| Sbjct 1 MARTKQTARKSTGGKAPRKQLAT#KAARKSAPASGGVKKPHRFRPGTVALREIR 53 |
| Orxines macklottii | Query 2 ARTKQTARKSTGGKAPRKQLAT#KAARKSAPATGGVKKPHRYRPGTVALREIR 53 ||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 2 ARTKQTARKSTGGKAPRKQLAT#KAARKSAPATGGVKKPHRYRPGTVALREIR 53 |
| Tetrapygus niger | Query 1 MARTKQTARKSTGGKAPRKQLAT#KAARKSAPATGGVKKPHRYRPGTVALREIR 53 |||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 1 MARTKQTARKSTGGKAPRKQLAT#KAARKSAPATGGVKKPHRYRPGTVALREIR 53 |
| Oryza sativa | Query 1 MARTKQTARKSTGGKAPRKQLAT#KAARKSAPATGGVKKPHRYRPGTVALREIR 53 |||||||||||||||||||||||#|||||||| ||||||||||||||||||||| Sbjct 1 MARTKQTARKSTGGKAPRKQLAT#KAARKSAPTTGGVKKPHRYRPGTVALREIR 53 |
| Picea abies | Query 1 MARTKQTARKSTGGKAPRKQLAT#KAARKSAPATGGVKKPHRYRPGTVALREIR 53 |||||||||||||||||||||||#|||||||| ||||||||||||||||||||| Sbjct 1 MARTKQTARKSTGGKAPRKQLAT#KAARKSAPTTGGVKKPHRYRPGTVALREIR 53 |
| Progomphus obscurus | Query 2 ARTKQTARKSTGGKAPRKQLAT#KAARKSAPATGGVKKPHRYRPGTVALREIR 53 ||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 2 ARTKQTARKSTGGKAPRKQLAT#KAARKSAPATGGVKKPHRYRPGTVALREIR 53 |
| Acilius fraternus | Query 1 MARTKQTARKSTGGKAPRKQLAT#KAARKSAPATGGVKKPHRYRPGTVALREIR 53 |||||||| ||||||||||||||#|||||||||||||||||||||||||||||| Sbjct 1 MARTKQTAXKSTGGKAPRKQLAT#KAARKSAPATGGVKKPHRYRPGTVALREIR 53 |
| Agathemera crassa | Query 2 ARTKQTARKSTGGKAPRKQLAT#KAARKSAPATGGVKKPHRYRPGTVALREIR 53 ||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 2 ARTKQTARKSTGGKAPRKQLAT#KAARKSAPATGGVKKPHRYRPGTVALREIR 53 |
| Drosophila erecta | Query 1 MARTKQTARKSTGGKAPRKQLAT#KAARKSAPATGGVKKPHRYRPGTVALREIR 53 ||||||| |||||||||||||||#|||||||||||||||||||||||||||||| Sbjct 1 MARTKQTTRKSTGGKAPRKQLAT#KAARKSAPATGGVKKPHRYRPGTVALREIR 53 |
| Prosopistoma wouterae | Query 2 ARTKQTARKSTGGKAPRKQLAT#KAARKSAPATGGVKKPHRYRPGTVALREIR 53 ||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 2 ARTKQTARKSTGGKAPRKQLAT#KAARKSAPATGGVKKPHRYRPGTVALREIR 53 |
[Site 3] KQLATKAARK28-SAPATGGVKK
Lys28  Ser
Ser
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Lys19 | Gln20 | Leu21 | Ala22 | Thr23 | Lys24 | Ala25 | Ala26 | Arg27 | Lys28 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Ser29 | Ala30 | Pro31 | Ala32 | Thr33 | Gly34 | Gly35 | Val36 | Lys37 | Lys38 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| MARTKQTARKSTGGKAPRKQLATKAARKSAPATGGVKKPHRYRPGTVALREIRRYQKS |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | Canis familiaris | 115.00 | 8 | PREDICTED: similar to histone 1, H2ai (predicted) |
| 2 | Homo sapiens | 115.00 | 6 | Histone cluster 1, H3i |
| 3 | Stygifloris sp. EP141 | 115.00 | 1 | histone H3 |
| 4 | Acilius sylvanus | 115.00 | 1 | histone 3 |
| 5 | Expression vector pET3-H3 | 115.00 | 1 | Xenopus laevis-like histone H3 |
| 6 | N/A | 114.00 | 10 | histone H3 |
| 7 | Mus musculus | 114.00 | 8 | histone cluster 2, H3c1 isoform 1 |
| 8 | Strongylocentrotus purpuratus | 114.00 | 3 | histone H3 |
| 9 | Drosophila melanogaster | 114.00 | 2 | H3 histone |
| 10 | Oikopleura dioica | 114.00 | 2 | histone h3.1 |
| 11 | synthetic construct | 114.00 | 1 | histone 3 H3 |
| 12 | Asellus aquaticus | 114.00 | 1 | putative H3 histone |
| 13 | Xenopus laevis | 114.00 | 1 | histone H3 |
| 14 | Psammechinus miliaris | 114.00 | 1 | cleavage stage histone H3 |
| 15 | Attenella margarita | 114.00 | 1 | histone H3 |
| 16 | Danio rerio | 114.00 | 1 | hypothetical protein LOC794406 |
| 17 | Drosophila persimilis | 114.00 | 1 | histone 3 |
| 18 | Cyathermia naticoides | 114.00 | 1 | histone H3 |
| 19 | Thekalabis sp. DM16 | 114.00 | 1 | histone 3 |
| 20 | Strongylocentrotus purpuratus | 114.00 | 1 | late embryonic histone H3 |
| 21 | Isonychia sp. EP142 | 114.00 | 1 | histone H3 |
| 22 | Styela plicata | 114.00 | 1 | H3 histone |
| 23 | Mytilus chilensis | 114.00 | 1 | histone H3 |
| 24 | Mytilus californianus | 114.00 | 1 | histone H3 |
| 25 | Mortierella alpina | 114.00 | 1 | H3_MORAP Histone H3 gi |
| 26 | Caenorhabditis elegans | 113.00 | 3 | HIStone family member (his-42) |
| 27 | Bos taurus | 113.00 | 2 | PREDICTED: similar to histone protein Hist2h3c1 |
| 28 | Venerupis (Ruditapes) philippinarum | 113.00 | 2 | replacement histone H3.3 |
| 29 | Anopheles gambiae str. PEST | 113.00 | 2 | AGAP012197-PA |
| 30 | Gibbula zonata | 113.00 | 1 | histone H3 |
| 31 | Lepetodrilus ovalis | 113.00 | 1 | histone H3 |
| 32 | Callibaetis sp. EP020 | 113.00 | 1 | histone H3 |
| 33 | Mus pahari | 113.00 | 1 | histone H3.2 protein |
| 34 | Nematostella vectensis | 113.00 | 1 | predicted protein |
| 35 | Siphlonella sp. EP083 | 113.00 | 1 | histone H3 |
| 36 | Cricetulus longicaudatus | 113.00 | 1 | H32_CRILO Histone H3.2 gi |
| 37 | Drosophila virilis | 113.00 | 1 | histone H3 |
| 38 | Drosophila simulans | 113.00 | 1 | Histone H3 |
| 39 | Oryza sativa (japonica cultivar-group) | 113.00 | 1 | histone H3 - maize |
| 40 | Pan troglodytes | 113.00 | 1 | PREDICTED: hypothetical protein |
| 41 | Scissurella cf. coronata CET-2005 | 113.00 | 1 | histone H3 |
| 42 | Drosophila erecta | 113.00 | 1 | histone H3 |
| 43 | Pongo pygmaeus | 113.00 | 1 | H33_PONPY Histone H3.3 gi |
| 44 | Caenorhabditis briggsae AF16 | 112.00 | 4 | Hypothetical protein CBG01505 |
| 45 | Progomphus obscurus | 112.00 | 1 | histone H3 |
| 46 | Craterostigma plantagineum | 112.00 | 1 | histone H3.3 |
| 47 | Caudatella hystrix | 112.00 | 1 | histone H3 |
| 48 | Gromphadorhina portentosa | 112.00 | 1 | histone 3 |
| 49 | Grylloblatta campodeiformis | 112.00 | 1 | histone 3 |
| 50 | Drosophila pseudoobscura | 112.00 | 1 | histone 3 |
| 51 | Lilium longiflorum | 112.00 | 1 | H32_LILLO Histone H3.2 gi |
| 52 | Orxines macklottii | 112.00 | 1 | histone 3 |
| 53 | Erythemis sp. OD024 | 112.00 | 1 | histone H3 |
| 54 | Scutus unguis | 112.00 | 1 | histone H3 |
| 55 | Arabidopsis thaliana | 112.00 | 1 | histone H3 |
| 56 | Tetrapygus niger | 112.00 | 1 | AF356516_2 early histone H3 |
| 57 | Anatoma euglypta | 112.00 | 1 | histone H3 |
| 58 | Agathemera crassa | 112.00 | 1 | histone 3 |
| 59 | Acropora formosa | 112.00 | 1 | H3_ACRFO Histone H3 gi |
| 60 | Ustilago maydis 521 | 112.00 | 1 | histone H3 |
| 61 | Acilius fraternus | 112.00 | 1 | histone 3 |
Top-ranked sequences
| organism | matching |
|---|---|
| Canis familiaris | Query 1 MARTKQTARKSTGGKAPRKQLATKAARK#SAPATGGVKKPHRYRPGTVALREIRRYQKS 58 ||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 1 MARTKQTARKSTGGKAPRKQLATKAARK#SAPATGGVKKPHRYRPGTVALREIRRYQKS 58 |
| Homo sapiens | Query 1 MARTKQTARKSTGGKAPRKQLATKAARK#SAPATGGVKKPHRYRPGTVALREIRRYQKS 58 ||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 1 MARTKQTARKSTGGKAPRKQLATKAARK#SAPATGGVKKPHRYRPGTVALREIRRYQKS 58 |
| Stygifloris sp. EP141 | Query 1 MARTKQTARKSTGGKAPRKQLATKAARK#SAPATGGVKKPHRYRPGTVALREIRRYQKS 58 ||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 1 MARTKQTARKSTGGKAPRKQLATKAARK#SAPATGGVKKPHRYRPGTVALREIRRYQKS 58 |
| Acilius sylvanus | Query 1 MARTKQTARKSTGGKAPRKQLATKAARK#SAPATGGVKKPHRYRPGTVALREIRRYQKS 58 ||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 1 MARTKQTARKSTGGKAPRKQLATKAARK#SAPATGGVKKPHRYRPGTVALREIRRYQKS 58 |
| Expression vector pET3-H3 | Query 1 MARTKQTARKSTGGKAPRKQLATKAARK#SAPATGGVKKPHRYRPGTVALREIRRYQKS 58 ||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 1 MARTKQTARKSTGGKAPRKQLATKAARK#SAPATGGVKKPHRYRPGTVALREIRRYQKS 58 |
| N/A | Query 1 MARTKQTARKSTGGKAPRKQLATKAARK#SAPATGGVKKPHRYRPGTVALREIRRYQKS 58 ||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 1 MARTKQTARKSTGGKAPRKQLATKAARK#SAPATGGVKKPHRYRPGTVALREIRRYQKS 58 |
| Mus musculus | Query 1 MARTKQTARKSTGGKAPRKQLATKAARK#SAPATGGVKKPHRYRPGTVALREIRRYQKS 58 ||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 1 MARTKQTARKSTGGKAPRKQLATKAARK#SAPATGGVKKPHRYRPGTVALREIRRYQKS 58 |
| Strongylocentrotus purpuratus | Query 1 MARTKQTARKSTGGKAPRKQLATKAARK#SAPATGGVKKPHRYRPGTVALREIRRYQKS 58 ||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 1 MARTKQTARKSTGGKAPRKQLATKAARK#SAPATGGVKKPHRYRPGTVALREIRRYQKS 58 |
| Drosophila melanogaster | Query 1 MARTKQTARKSTGGKAPRKQLATKAARK#SAPATGGVKKPHRYRPGTVALREIRRYQKS 58 ||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 1 MARTKQTARKSTGGKAPRKQLATKAARK#SAPATGGVKKPHRYRPGTVALREIRRYQKS 58 |
| Oikopleura dioica | Query 1 MARTKQTARKSTGGKAPRKQLATKAARK#SAPATGGVKKPHRYRPGTVALREIRRYQKS 58 ||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 1 MARTKQTARKSTGGKAPRKQLATKAARK#SAPATGGVKKPHRYRPGTVALREIRRYQKS 58 |
| synthetic construct | Query 1 MARTKQTARKSTGGKAPRKQLATKAARK#SAPATGGVKKPHRYRPGTVALREIRRYQKS 58 |||||||||||||||||||||||| |||#|||||||||||||||||||||||||||||| Sbjct 1 MARTKQTARKSTGGKAPRKQLATKVARK#SAPATGGVKKPHRYRPGTVALREIRRYQKS 58 |
| Asellus aquaticus | Query 1 MARTKQTARKSTGGKAPRKQLATKAARK#SAPATGGVKKPHRYRPGTVALREIRRYQKS 58 ||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 1 MARTKQTARKSTGGKAPRKQLATKAARK#SAPATGGVKKPHRYRPGTVALREIRRYQKS 58 |
| Xenopus laevis | Query 1 MARTKQTARKSTGGKAPRKQLATKAARK#SAPATGGVKKPHRYRPGTVALREIRRYQKS 58 ||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 1 MARTKQTARKSTGGKAPRKQLATKAARK#SAPATGGVKKPHRYRPGTVALREIRRYQKS 58 |
| Psammechinus miliaris | Query 1 MARTKQTARKSTGGKAPRKQLATKAARK#SAPATGGVKKPHRYRPGTVALREIRRYQKS 58 ||||||||||||||||||||||||||||#|||||||||||||+|||||||||||||||| Sbjct 1 MARTKQTARKSTGGKAPRKQLATKAARK#SAPATGGVKKPHRFRPGTVALREIRRYQKS 58 |
| Attenella margarita | Query 1 MARTKQTARKSTGGKAPRKQLATKAARK#SAPATGGVKKPHRYRPGTVALREIRRYQKS 58 ||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 1 MARTKQTARKSTGGKAPRKQLATKAARK#SAPATGGVKKPHRYRPGTVALREIRRYQKS 58 |
| Danio rerio | Query 1 MARTKQTARKSTGGKAPRKQLATKAARK#SAPATGGVKKPHRYRPGTVALREIRRYQKS 58 ||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 1 MARTKQTARKSTGGKAPRKQLATKAARK#SAPATGGVKKPHRYRPGTVALREIRRYQKS 58 |
| Drosophila persimilis | Query 1 MARTKQTARKSTGGKAPRKQLATKAARK#SAPATGGVKKPHRYRPGTVALREIRRYQKS 58 ||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 1 MARTKQTARKSTGGKAPRKQLATKAARK#SAPATGGVKKPHRYRPGTVALREIRRYQKS 58 |
| Cyathermia naticoides | Query 1 MARTKQTARKSTGGKAPRKQLATKAARK#SAPATGGVKKPHRYRPGTVALREIRRYQKS 58 ||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 1 MARTKQTARKSTGGKAPRKQLATKAARK#SAPATGGVKKPHRYRPGTVALREIRRYQKS 58 |
| Thekalabis sp. DM16 | Query 1 MARTKQTARKSTGGKAPRKQLATKAARK#SAPATGGVKKPHRYRPGTVALREIRRYQKS 58 ||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 1 MARTKQTARKSTGGKAPRKQLATKAARK#SAPATGGVKKPHRYRPGTVALREIRRYQKS 58 |
| Strongylocentrotus purpuratus | Query 1 MARTKQTARKSTGGKAPRKQLATKAARK#SAPATGGVKKPHRYRPGTVALREIRRYQKS 58 ||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 1 MARTKQTARKSTGGKAPRKQLATKAARK#SAPATGGVKKPHRYRPGTVALREIRRYQKS 58 |
| Isonychia sp. EP142 | Query 1 MARTKQTARKSTGGKAPRKQLATKAARK#SAPATGGVKKPHRYRPGTVALREIRRYQKS 58 ||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 1 MARTKQTARKSTGGKAPRKQLATKAARK#SAPATGGVKKPHRYRPGTVALREIRRYQKS 58 |
| Styela plicata | Query 1 MARTKQTARKSTGGKAPRKQLATKAARK#SAPATGGVKKPHRYRPGTVALREIRRYQKS 58 ||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 1 MARTKQTARKSTGGKAPRKQLATKAARK#SAPATGGVKKPHRYRPGTVALREIRRYQKS 58 |
| Mytilus chilensis | Query 1 MARTKQTARKSTGGKAPRKQLATKAARK#SAPATGGVKKPHRYRPGTVALREIRRYQKS 58 ||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 1 MARTKQTARKSTGGKAPRKQLATKAARK#SAPATGGVKKPHRYRPGTVALREIRRYQKS 58 |
| Mytilus californianus | Query 1 MARTKQTARKSTGGKAPRKQLATKAARK#SAPATGGVKKPHRYRPGTVALREIRRYQKS 58 ||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 1 MARTKQTARKSTGGKAPRKQLATKAARK#SAPATGGVKKPHRYRPGTVALREIRRYQKS 58 |
| Mortierella alpina | Query 1 MARTKQTARKSTGGKAPRKQLATKAARK#SAPATGGVKKPHRYRPGTVALREIRRYQKS 58 ||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 1 MARTKQTARKSTGGKAPRKQLATKAARK#SAPATGGVKKPHRYRPGTVALREIRRYQKS 58 |
| Caenorhabditis elegans | Query 1 MARTKQTARKSTGGKAPRKQLATKAARK#SAPATGGVKKPHRYRPGTVALREIRRYQKS 58 ||||||||||||||||||||||||||||#||||+||||||||||||||||||||||||| Sbjct 1 MARTKQTARKSTGGKAPRKQLATKAARK#SAPASGGVKKPHRYRPGTVALREIRRYQKS 58 |
| Bos taurus | Query 1 MARTKQTARKSTGGKAPRKQLATKAARK#SAPATGGVKKPHRYRPGTVALREIRRYQKS 58 | ||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Sbjct 1 MTRTKQTARKSTGGKAPRKQLATKAARK#SAPATGGVKKPHRYRPGTVALREIRRYQKS 58 |
| Venerupis (Ruditapes) philippinarum | Query 1 MARTKQTARKSTGGKAPRKQLATKAARK#SAPATGGVKKPHRYRPGTVALREIRRYQKS 58 ||||||||||||||||||||||||||||#|||+|||||||||||||||||||||||||| Sbjct 1 MARTKQTARKSTGGKAPRKQLATKAARK#SAPSTGGVKKPHRYRPGTVALREIRRYQKS 58 |
| Anopheles gambiae str. PEST | Query 1 MARTKQTARKSTGGKAPRKQLATKAARK#SAPATGGVKKPHRYRPGTVALREIRRYQKS 58 ||||||||||||||||||||||||||||#|||+|||||||||||||||||||||||||| Sbjct 1 MARTKQTARKSTGGKAPRKQLATKAARK#SAPSTGGVKKPHRYRPGTVALREIRRYQKS 58 |
| Gibbula zonata | Query 1 MARTKQTARKSTGGKAPRKQLATKAARK#SAPATGGVKKPHRYRPGTVALREIRRYQKS 58 | ||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Sbjct 1 MVRTKQTARKSTGGKAPRKQLATKAARK#SAPATGGVKKPHRYRPGTVALREIRRYQKS 58 |
| Lepetodrilus ovalis | Query 1 MARTKQTARKSTGGKAPRKQLATKAARK#SAPATGGVKKPHRYRPGTVALREIRRYQKS 58 ||||||| ||||||||||||||||||||#|||||||||||||||||||||||||||||| Sbjct 1 MARTKQTXRKSTGGKAPRKQLATKAARK#SAPATGGVKKPHRYRPGTVALREIRRYQKS 58 |
| Callibaetis sp. EP020 | Query 1 MARTKQTARKSTGGKAPRKQLATKAARK#SAPATGGVKKPHRYRPGTVALREIRRYQKS 58 |||||||||||+||||||||||||||||#|||||||||||||||||||||||||||||| Sbjct 1 MARTKQTARKSSGGKAPRKQLATKAARK#SAPATGGVKKPHRYRPGTVALREIRRYQKS 58 |
| Mus pahari | Query 1 MARTKQTARKSTGGKAPRKQLATKAARK#SAPATGGVKKPHRYRPGTVALREIRRYQKS 58 ||||||||||||||||||||||||||||#||| |||||||||||||||||||||||||| Sbjct 1 MARTKQTARKSTGGKAPRKQLATKAARK#SAPTTGGVKKPHRYRPGTVALREIRRYQKS 58 |
| Nematostella vectensis | Query 1 MARTKQTARKSTGGKAPRKQLATKAARK#SAPATGGVKKPHRYRPGTVALREIRRYQKS 58 ||||||||||||||||||||||||||||#|||+|||||||||||||||||||||||||| Sbjct 1 MARTKQTARKSTGGKAPRKQLATKAARK#SAPSTGGVKKPHRYRPGTVALREIRRYQKS 58 |
| Siphlonella sp. EP083 | Query 1 MARTKQTARKSTGGKAPRKQLATKAARK#SAPATGGVKKPHRYRPGTVALREIRRYQKS 58 ||||||||| ||||||||||||||||||#|||||||||||||||||||||||||||||| Sbjct 1 MARTKQTARXSTGGKAPRKQLATKAARK#SAPATGGVKKPHRYRPGTVALREIRRYQKS 58 |
| Cricetulus longicaudatus | Query 1 MARTKQTARKSTGGKAPRKQLATKAARK#SAPATGGVKKPHRYRPGTVALREIRRYQKS 58 ||||||||||||||||||||||||||||#||||||||||||||||||| |||||||||| Sbjct 1 MARTKQTARKSTGGKAPRKQLATKAARK#SAPATGGVKKPHRYRPGTVTLREIRRYQKS 58 |
| Drosophila virilis | Query 1 MARTKQTARKSTGGKAPRKQLATKAARK#SAPATGGVKKPHRYRPGTVALREIRRYQKS 58 ||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 1 MARTKQTARKSTGGKAPRKQLATKAARK#SAPATGGVKKPHRYRPGTVALREIRRYQKS 58 |
| Drosophila simulans | Query 1 MARTKQTARKSTGGKAPRKQLATKAARK#SAPATGGVKKPHRYRPGTVALREIRRYQKS 58 ||||||||||||||||||||||||||||#| |||||||||||||||||||||||||||| Sbjct 1 MARTKQTARKSTGGKAPRKQLATKAARK#SVPATGGVKKPHRYRPGTVALREIRRYQKS 58 |
| Oryza sativa (japonica cultivar-group) | Query 1 MARTKQTARKSTGGKAPRKQLATKAARK#SAPATGGVKKPHRYRPGTVALREIRRYQKS 58 ||||||||||||||||||||||||||||#|||||||||||||+|||||||||||+|||| Sbjct 1 MARTKQTARKSTGGKAPRKQLATKAARK#SAPATGGVKKPHRFRPGTVALREIRKYQKS 58 |
| Pan troglodytes | Query 1 MARTKQTARKSTGGKAPRKQLATKAARK#SAPATGGVKKPHRYRPGTVALREIRRYQKS 58 ||||||||||||||||||||||||||||#|||+|||||||||||||||||||||||||| Sbjct 1 MARTKQTARKSTGGKAPRKQLATKAARK#SAPSTGGVKKPHRYRPGTVALREIRRYQKS 58 |
| Scissurella cf. coronata CET-2005 | Query 1 MARTKQTARKSTGGKAPRKQLATKAARK#SAPATGGVKKPHRYRPGTVALREIRRYQKS 58 | ||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Sbjct 1 MVRTKQTARKSTGGKAPRKQLATKAARK#SAPATGGVKKPHRYRPGTVALREIRRYQKS 58 |
| Drosophila erecta | Query 1 MARTKQTARKSTGGKAPRKQLATKAARK#SAPATGGVKKPHRYRPGTVALREIRRYQKS 58 ||||||| ||||||||||||||||||||#|||||||||||||||||||||||||||||| Sbjct 1 MARTKQTTRKSTGGKAPRKQLATKAARK#SAPATGGVKKPHRYRPGTVALREIRRYQKS 58 |
| Pongo pygmaeus | Query 1 MARTKQTARKSTGGKAPRKQLATKAARK#SAPATGGVKKPHRYRPGTVALREIRRYQKS 58 ||||||||||||||||||||||||||||#|||+|||||||||||||||||||||||||| Sbjct 1 MARTKQTARKSTGGKAPRKQLATKAARK#SAPSTGGVKKPHRYRPGTVALREIRRYQKS 58 |
| Caenorhabditis briggsae AF16 | Query 1 MARTKQTARKSTGGKAPRKQLATKAARK#SAPATGGVKKPHRYRPGTVALREIRRYQKS 58 ||||||||||||||||||||||||||||#|||| ||||||||||||||||||||||||| Sbjct 1 MARTKQTARKSTGGKAPRKQLATKAARK#SAPAQGGVKKPHRYRPGTVALREIRRYQKS 58 |
| Progomphus obscurus | Query 2 ARTKQTARKSTGGKAPRKQLATKAARK#SAPATGGVKKPHRYRPGTVALREIRRYQKS 58 |||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 2 ARTKQTARKSTGGKAPRKQLATKAARK#SAPATGGVKKPHRYRPGTVALREIRRYQKS 58 |
| Craterostigma plantagineum | Query 1 MARTKQTARKSTGGKAPRKQLATKAARK#SAPATGGVKKPHRYRPGTVALREIRRYQKS 58 ||||||||||||||||||||||||||||#|||++||||||||||||||||||||||||| Sbjct 1 MARTKQTARKSTGGKAPRKQLATKAARK#SAPSSGGVKKPHRYRPGTVALREIRRYQKS 58 |
| Caudatella hystrix | Query 1 MARTKQTARKSTGGKAPRKQLATKAARK#SAPATGGVKKPHRYRPGTVALREIRRYQKS 58 |||||||||||||||||||| |||||||#|||||||||||||||||||||||||||||| Sbjct 1 MARTKQTARKSTGGKAPRKQXATKAARK#SAPATGGVKKPHRYRPGTVALREIRRYQKS 58 |
| Gromphadorhina portentosa | Query 2 ARTKQTARKSTGGKAPRKQLATKAARK#SAPATGGVKKPHRYRPGTVALREIRRYQKS 58 |||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 2 ARTKQTARKSTGGKAPRKQLATKAARK#SAPATGGVKKPHRYRPGTVALREIRRYQKS 58 |
| Grylloblatta campodeiformis | Query 2 ARTKQTARKSTGGKAPRKQLATKAARK#SAPATGGVKKPHRYRPGTVALREIRRYQKS 58 |||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 2 ARTKQTARKSTGGKAPRKQLATKAARK#SAPATGGVKKPHRYRPGTVALREIRRYQKS 58 |
| Drosophila pseudoobscura | Query 1 MARTKQTARKSTGGKAPRKQLATKAARK#SAPATGGVKKPHRYRPGTVALREIRRYQKS 58 ||||||||||||||||||||||||||||# ||||||||||||||||||||||||||||| Sbjct 1 MARTKQTARKSTGGKAPRKQLATKAARK#GAPATGGVKKPHRYRPGTVALREIRRYQKS 58 |
| Lilium longiflorum | Query 1 MARTKQTARKSTGGKAPRKQLATKAARK#SAPATGGVKKPHRYRPGTVALREIRRYQKS 58 ||||||||||||||||||||||||||||#|||||||||||||+|||||||||||+|||| Sbjct 1 MARTKQTARKSTGGKAPRKQLATKAARK#SAPATGGVKKPHRFRPGTVALREIRKYQKS 58 |
| Orxines macklottii | Query 2 ARTKQTARKSTGGKAPRKQLATKAARK#SAPATGGVKKPHRYRPGTVALREIRRYQKS 58 |||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 2 ARTKQTARKSTGGKAPRKQLATKAARK#SAPATGGVKKPHRYRPGTVALREIRRYQKS 58 |
| Erythemis sp. OD024 | Query 2 ARTKQTARKSTGGKAPRKQLATKAARK#SAPATGGVKKPHRYRPGTVALREIRRYQKS 58 |||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 2 ARTKQTARKSTGGKAPRKQLATKAARK#SAPATGGVKKPHRYRPGTVALREIRRYQKS 58 |
| Scutus unguis | Query 1 MARTKQTARKSTGGKAPRKQLATKAARK#SAPATGGVKKPHRYRPGTVALREIRRYQKS 58 ||||||||||||| ||||||||||||||#|||||||||||||||||||||||||||||| Sbjct 1 MARTKQTARKSTGVKAPRKQLATKAARK#SAPATGGVKKPHRYRPGTVALREIRRYQKS 58 |
| Arabidopsis thaliana | Query 1 MARTKQTARKSTGGKAPRKQLATKAARK#SAPATGGVKKPHRYRPGTVALREIRRYQKS 58 ||||||||||||||||||||||||||||#|||||||||||||+|||||||||||+|||| Sbjct 1 MARTKQTARKSTGGKAPRKQLATKAARK#SAPATGGVKKPHRFRPGTVALREIRKYQKS 58 |
| Tetrapygus niger | Query 1 MARTKQTARKSTGGKAPRKQLATKAARK#SAPATGGVKKPHRYRPGTVALREIRRYQKS 58 ||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 1 MARTKQTARKSTGGKAPRKQLATKAARK#SAPATGGVKKPHRYRPGTVALREIRRYQKS 58 |
| Anatoma euglypta | Query 1 MARTKQTARKSTGGKAPRKQLATKAARK#SAPATGGVKKPHRYRPGTVALREIRRYQKS 58 || |||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Sbjct 1 MACTKQTARKSTGGKAPRKQLATKAARK#SAPATGGVKKPHRYRPGTVALREIRRYQKS 58 |
| Agathemera crassa | Query 2 ARTKQTARKSTGGKAPRKQLATKAARK#SAPATGGVKKPHRYRPGTVALREIRRYQKS 58 |||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 2 ARTKQTARKSTGGKAPRKQLATKAARK#SAPATGGVKKPHRYRPGTVALREIRRYQKS 58 |
| Acropora formosa | Query 1 MARTKQTARKSTGGKAPRKQLATKAARK#SAPATGGVKKPHRYRPGTVALREIRRYQKS 58 |||||||||||||||||||||||||| |#|||||||||||||||||||||||||||||| Sbjct 1 MARTKQTARKSTGGKAPRKQLATKAAAK#SAPATGGVKKPHRYRPGTVALREIRRYQKS 58 |
| Ustilago maydis 521 | Query 1 MARTKQTARKSTGGKAPRKQLATKAARK#SAPATGGVKKPHRYRPGTVALREIRRYQKS 58 ||||||||||||||||||||||||||||#|||+ ||||||||||||||||||||||||| Sbjct 1 MARTKQTARKSTGGKAPRKQLATKAARK#SAPSAGGVKKPHRYRPGTVALREIRRYQKS 58 |
| Acilius fraternus | Query 1 MARTKQTARKSTGGKAPRKQLATKAARK#SAPATGGVKKPHRYRPGTVALREIRRYQKS 58 |||||||| |||||||||||||||||||#|||||||||||||||||||||||||||||| Sbjct 1 MARTKQTAXKSTGGKAPRKQLATKAARK#SAPATGGVKKPHRYRPGTVALREIRRYQKS 58 |
[Site 4] KAARKSAPAT33-GGVKKPHRYR
Thr33  Gly
Gly
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Lys24 | Ala25 | Ala26 | Arg27 | Lys28 | Ser29 | Ala30 | Pro31 | Ala32 | Thr33 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Gly34 | Gly35 | Val36 | Lys37 | Lys38 | Pro39 | His40 | Arg41 | Tyr42 | Arg43 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| TKQTARKSTGGKAPRKQLATKAARKSAPATGGVKKPHRYRPGTVALREIRRYQKSTELLI |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | N/A | 119.00 | 6 | histone H3 |
| 2 | Canis familiaris | 119.00 | 5 | PREDICTED: similar to histone 1, H2ai (predicted) |
| 3 | Homo sapiens | 119.00 | 4 | histone H3 |
| 4 | Mus musculus | 119.00 | 4 | histone cluster 1, H3f |
| 5 | Strongylocentrotus purpuratus | 119.00 | 2 | histone H3 |
| 6 | Acilius sylvanus | 119.00 | 1 | histone 3 |
| 7 | Tropidoderus childrenii | 119.00 | 1 | histone 3 |
| 8 | Cyathermia naticoides | 119.00 | 1 | histone H3 |
| 9 | Stygifloris sp. EP141 | 119.00 | 1 | histone H3 |
| 10 | Clypeosectus sp. CET-2005 | 119.00 | 1 | histone H3 |
| 11 | Thekalabis sp. DM16 | 119.00 | 1 | histone 3 |
| 12 | Strongylocentrotus purpuratus | 119.00 | 1 | late embryonic histone H3 |
| 13 | Grylloblatta campodeiformis | 119.00 | 1 | histone 3 |
| 14 | Gromphadorhina portentosa | 119.00 | 1 | histone 3 |
| 15 | Erythemis sp. OD024 | 119.00 | 1 | histone H3 |
| 16 | Expression vector pET3-H3 | 119.00 | 1 | Xenopus laevis-like histone H3 |
| 17 | Progomphus obscurus | 119.00 | 1 | histone H3 |
| 18 | Attenella margarita | 119.00 | 1 | histone H3 |
| 19 | Orxines macklottii | 119.00 | 1 | histone 3 |
| 20 | Agathemera crassa | 119.00 | 1 | histone 3 |
| 21 | Danio rerio | 119.00 | 1 | hypothetical protein LOC794406 |
| 22 | Neohirasea sp. WS29 | 119.00 | 1 | histone 3 |
| 23 | Anatoma euglypta | 119.00 | 1 | histone H3 |
| 24 | Mytilus chilensis | 119.00 | 1 | histone H3 |
| 25 | Mytilus californianus | 119.00 | 1 | histone H3 |
| 26 | Fissurella virescens | 119.00 | 1 | histone H3 |
| 27 | Gibbula zonata | 119.00 | 1 | histone H3 |
| 28 | Extatosoma tiaratum | 119.00 | 1 | histone 3 |
| 29 | Bos taurus | 118.00 | 2 | PREDICTED: similar to histone protein Hist2h3c1 |
| 30 | Oikopleura dioica | 118.00 | 2 | histone h3.2 |
| 31 | Scissurella cf. coronata CET-2005 | 118.00 | 2 | histone H3 |
| 32 | Anopheles gambiae str. PEST | 118.00 | 2 | AGAP012709-PA |
| 33 | Sungaya inexpectata | 118.00 | 1 | histone 3 |
| 34 | Griseargiolestes albescens | 118.00 | 1 | histone H3 |
| 35 | Mortierella alpina | 118.00 | 1 | H3_MORAP Histone H3 gi |
| 36 | Xenopus laevis | 118.00 | 1 | histone H3 |
| 37 | Oreophoetes peruana | 118.00 | 1 | histone 3 |
| 38 | Asellus aquaticus | 118.00 | 1 | putative H3 histone |
| 39 | Aretaon asperrimus | 118.00 | 1 | histone 3 |
| 40 | Elassoneuria sp. EP167 | 118.00 | 1 | histone H3 |
| 41 | Styela plicata | 118.00 | 1 | H3 histone |
| 42 | Sukaschitrochus atkinsoni | 118.00 | 1 | histone H3 |
| 43 | Drosophila persimilis | 118.00 | 1 | histone 3 |
| 44 | Drosophila melanogaster | 118.00 | 1 | H3 histone |
| 45 | Isonychia sp. EP142 | 118.00 | 1 | histone H3 |
| 46 | Drosophila simulans | 117.00 | 1 | Histone H3 |
| 47 | Drosophila virilis | 117.00 | 1 | histone H3 |
| 48 | Lepetodrilus ovalis | 117.00 | 1 | histone H3 |
| 49 | Callibaetis sp. EP020 | 117.00 | 1 | histone H3 |
| 50 | Caenorhabditis elegans | 117.00 | 1 | HIStone family member (his-42) |
| 51 | Machilis sp. BYU_ACAR003 | 117.00 | 1 | histone H3 |
| 52 | Austrophlebiodes sp. EP027 | 117.00 | 1 | histone H3 |
| 53 | Venerupis (Ruditapes) philippinarum | 117.00 | 1 | replacement histone H3.3 |
| 54 | synthetic construct | 117.00 | 1 | histone 3 H3 |
| 55 | Psammechinus miliaris | 117.00 | 1 | cleavage stage histone H3 |
| 56 | Siphlonella sp. EP083 | 117.00 | 1 | histone H3 |
| 57 | Phyllophaga sp. BYU_ACCO040 | 117.00 | 1 | histone H3 |
| 58 | Tetrapygus niger | 116.00 | 1 | AF356516_2 early histone H3 |
| 59 | Prosopistoma wouterae | 116.00 | 1 | histone H3 |
Top-ranked sequences
| organism | matching |
|---|---|
| N/A | Query 4 TKQTARKSTGGKAPRKQLATKAARKSAPAT#GGVKKPHRYRPGTVALREIRRYQKSTELLI 63 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 4 TKQTARKSTGGKAPRKQLATKAARKSAPAT#GGVKKPHRYRPGTVALREIRRYQKSTELLI 63 |
| Canis familiaris | Query 4 TKQTARKSTGGKAPRKQLATKAARKSAPAT#GGVKKPHRYRPGTVALREIRRYQKSTELLI 63 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 4 TKQTARKSTGGKAPRKQLATKAARKSAPAT#GGVKKPHRYRPGTVALREIRRYQKSTELLI 63 |
| Homo sapiens | Query 4 TKQTARKSTGGKAPRKQLATKAARKSAPAT#GGVKKPHRYRPGTVALREIRRYQKSTELLI 63 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 4 TKQTARKSTGGKAPRKQLATKAARKSAPAT#GGVKKPHRYRPGTVALREIRRYQKSTELLI 63 |
| Mus musculus | Query 4 TKQTARKSTGGKAPRKQLATKAARKSAPAT#GGVKKPHRYRPGTVALREIRRYQKSTELLI 63 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 4 TKQTARKSTGGKAPRKQLATKAARKSAPAT#GGVKKPHRYRPGTVALREIRRYQKSTELLI 63 |
| Strongylocentrotus purpuratus | Query 4 TKQTARKSTGGKAPRKQLATKAARKSAPAT#GGVKKPHRYRPGTVALREIRRYQKSTELLI 63 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 4 TKQTARKSTGGKAPRKQLATKAARKSAPAT#GGVKKPHRYRPGTVALREIRRYQKSTELLI 63 |
| Acilius sylvanus | Query 4 TKQTARKSTGGKAPRKQLATKAARKSAPAT#GGVKKPHRYRPGTVALREIRRYQKSTELLI 63 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 4 TKQTARKSTGGKAPRKQLATKAARKSAPAT#GGVKKPHRYRPGTVALREIRRYQKSTELLI 63 |
| Tropidoderus childrenii | Query 4 TKQTARKSTGGKAPRKQLATKAARKSAPAT#GGVKKPHRYRPGTVALREIRRYQKSTELLI 63 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 4 TKQTARKSTGGKAPRKQLATKAARKSAPAT#GGVKKPHRYRPGTVALREIRRYQKSTELLI 63 |
| Cyathermia naticoides | Query 4 TKQTARKSTGGKAPRKQLATKAARKSAPAT#GGVKKPHRYRPGTVALREIRRYQKSTELLI 63 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 4 TKQTARKSTGGKAPRKQLATKAARKSAPAT#GGVKKPHRYRPGTVALREIRRYQKSTELLI 63 |
| Stygifloris sp. EP141 | Query 4 TKQTARKSTGGKAPRKQLATKAARKSAPAT#GGVKKPHRYRPGTVALREIRRYQKSTELLI 63 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 4 TKQTARKSTGGKAPRKQLATKAARKSAPAT#GGVKKPHRYRPGTVALREIRRYQKSTELLI 63 |
| Clypeosectus sp. CET-2005 | Query 4 TKQTARKSTGGKAPRKQLATKAARKSAPAT#GGVKKPHRYRPGTVALREIRRYQKSTELLI 63 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 4 TKQTARKSTGGKAPRKQLATKAARKSAPAT#GGVKKPHRYRPGTVALREIRRYQKSTELLI 63 |
| Thekalabis sp. DM16 | Query 4 TKQTARKSTGGKAPRKQLATKAARKSAPAT#GGVKKPHRYRPGTVALREIRRYQKSTELLI 63 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 4 TKQTARKSTGGKAPRKQLATKAARKSAPAT#GGVKKPHRYRPGTVALREIRRYQKSTELLI 63 |
| Strongylocentrotus purpuratus | Query 4 TKQTARKSTGGKAPRKQLATKAARKSAPAT#GGVKKPHRYRPGTVALREIRRYQKSTELLI 63 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 4 TKQTARKSTGGKAPRKQLATKAARKSAPAT#GGVKKPHRYRPGTVALREIRRYQKSTELLI 63 |
| Grylloblatta campodeiformis | Query 4 TKQTARKSTGGKAPRKQLATKAARKSAPAT#GGVKKPHRYRPGTVALREIRRYQKSTELLI 63 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 4 TKQTARKSTGGKAPRKQLATKAARKSAPAT#GGVKKPHRYRPGTVALREIRRYQKSTELLI 63 |
| Gromphadorhina portentosa | Query 4 TKQTARKSTGGKAPRKQLATKAARKSAPAT#GGVKKPHRYRPGTVALREIRRYQKSTELLI 63 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 4 TKQTARKSTGGKAPRKQLATKAARKSAPAT#GGVKKPHRYRPGTVALREIRRYQKSTELLI 63 |
| Erythemis sp. OD024 | Query 4 TKQTARKSTGGKAPRKQLATKAARKSAPAT#GGVKKPHRYRPGTVALREIRRYQKSTELLI 63 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 4 TKQTARKSTGGKAPRKQLATKAARKSAPAT#GGVKKPHRYRPGTVALREIRRYQKSTELLI 63 |
| Expression vector pET3-H3 | Query 4 TKQTARKSTGGKAPRKQLATKAARKSAPAT#GGVKKPHRYRPGTVALREIRRYQKSTELLI 63 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 4 TKQTARKSTGGKAPRKQLATKAARKSAPAT#GGVKKPHRYRPGTVALREIRRYQKSTELLI 63 |
| Progomphus obscurus | Query 4 TKQTARKSTGGKAPRKQLATKAARKSAPAT#GGVKKPHRYRPGTVALREIRRYQKSTELLI 63 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 4 TKQTARKSTGGKAPRKQLATKAARKSAPAT#GGVKKPHRYRPGTVALREIRRYQKSTELLI 63 |
| Attenella margarita | Query 4 TKQTARKSTGGKAPRKQLATKAARKSAPAT#GGVKKPHRYRPGTVALREIRRYQKSTELLI 63 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 4 TKQTARKSTGGKAPRKQLATKAARKSAPAT#GGVKKPHRYRPGTVALREIRRYQKSTELLI 63 |
| Orxines macklottii | Query 4 TKQTARKSTGGKAPRKQLATKAARKSAPAT#GGVKKPHRYRPGTVALREIRRYQKSTELLI 63 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 4 TKQTARKSTGGKAPRKQLATKAARKSAPAT#GGVKKPHRYRPGTVALREIRRYQKSTELLI 63 |
| Agathemera crassa | Query 4 TKQTARKSTGGKAPRKQLATKAARKSAPAT#GGVKKPHRYRPGTVALREIRRYQKSTELLI 63 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 4 TKQTARKSTGGKAPRKQLATKAARKSAPAT#GGVKKPHRYRPGTVALREIRRYQKSTELLI 63 |
| Danio rerio | Query 4 TKQTARKSTGGKAPRKQLATKAARKSAPAT#GGVKKPHRYRPGTVALREIRRYQKSTELLI 63 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 4 TKQTARKSTGGKAPRKQLATKAARKSAPAT#GGVKKPHRYRPGTVALREIRRYQKSTELLI 63 |
| Neohirasea sp. WS29 | Query 4 TKQTARKSTGGKAPRKQLATKAARKSAPAT#GGVKKPHRYRPGTVALREIRRYQKSTELLI 63 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 4 TKQTARKSTGGKAPRKQLATKAARKSAPAT#GGVKKPHRYRPGTVALREIRRYQKSTELLI 63 |
| Anatoma euglypta | Query 4 TKQTARKSTGGKAPRKQLATKAARKSAPAT#GGVKKPHRYRPGTVALREIRRYQKSTELLI 63 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 4 TKQTARKSTGGKAPRKQLATKAARKSAPAT#GGVKKPHRYRPGTVALREIRRYQKSTELLI 63 |
| Mytilus chilensis | Query 4 TKQTARKSTGGKAPRKQLATKAARKSAPAT#GGVKKPHRYRPGTVALREIRRYQKSTELLI 63 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 4 TKQTARKSTGGKAPRKQLATKAARKSAPAT#GGVKKPHRYRPGTVALREIRRYQKSTELLI 63 |
| Mytilus californianus | Query 4 TKQTARKSTGGKAPRKQLATKAARKSAPAT#GGVKKPHRYRPGTVALREIRRYQKSTELLI 63 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 4 TKQTARKSTGGKAPRKQLATKAARKSAPAT#GGVKKPHRYRPGTVALREIRRYQKSTELLI 63 |
| Fissurella virescens | Query 4 TKQTARKSTGGKAPRKQLATKAARKSAPAT#GGVKKPHRYRPGTVALREIRRYQKSTELLI 63 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 4 TKQTARKSTGGKAPRKQLATKAARKSAPAT#GGVKKPHRYRPGTVALREIRRYQKSTELLI 63 |
| Gibbula zonata | Query 4 TKQTARKSTGGKAPRKQLATKAARKSAPAT#GGVKKPHRYRPGTVALREIRRYQKSTELLI 63 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 4 TKQTARKSTGGKAPRKQLATKAARKSAPAT#GGVKKPHRYRPGTVALREIRRYQKSTELLI 63 |
| Extatosoma tiaratum | Query 4 TKQTARKSTGGKAPRKQLATKAARKSAPAT#GGVKKPHRYRPGTVALREIRRYQKSTELLI 63 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 4 TKQTARKSTGGKAPRKQLATKAARKSAPAT#GGVKKPHRYRPGTVALREIRRYQKSTELLI 63 |
| Bos taurus | Query 4 TKQTARKSTGGKAPRKQLATKAARKSAPAT#GGVKKPHRYRPGTVALREIRRYQKSTELLI 63 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 4 TKQTARKSTGGKAPRKQLATKAARKSAPAT#GGVKKPHRYRPGTVALREIRRYQKSTELLI 63 |
| Oikopleura dioica | Query 4 TKQTARKSTGGKAPRKQLATKAARKSAPAT#GGVKKPHRYRPGTVALREIRRYQKSTELLI 63 ||||||||||||||||||||||||||||||#||+||||||||||||||||||||||||||| Sbjct 4 TKQTARKSTGGKAPRKQLATKAARKSAPAT#GGIKKPHRYRPGTVALREIRRYQKSTELLI 63 |
| Scissurella cf. coronata CET-2005 | Query 4 TKQTARKSTGGKAPRKQLATKAARKSAPAT#GGVKKPHRYRPGTVALREIRRYQKSTELLI 63 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 4 TKQTARKSTGGKAPRKQLATKAARKSAPAT#GGVKKPHRYRPGTVALREIRRYQKSTELLI 63 |
| Anopheles gambiae str. PEST | Query 4 TKQTARKSTGGKAPRKQLATKAARKSAPAT#GGVKKPHRYRPGTVALREIRRYQKSTELLI 63 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 4 TKQTARKSTGGKAPRKQLATKAARKSAPAT#GGVKKPHRYRPGTVALREIRRYQKSTELLI 63 |
| Sungaya inexpectata | Query 4 TKQTARKSTGGKAPRKQLATKAARKSAPAT#GGVKKPHRYRPGTVALREIRRYQKSTELLI 63 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 4 TKQTARKSTGGKAPRKQLATKAARKSAPAT#GGVKKPHRYRPGTVALREIRRYQKSTELLI 63 |
| Griseargiolestes albescens | Query 4 TKQTARKSTGGKAPRKQLATKAARKSAPAT#GGVKKPHRYRPGTVALREIRRYQKSTELLI 63 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 4 TKQTARKSTGGKAPRKQLATKAARKSAPAT#GGVKKPHRYRPGTVALREIRRYQKSTELLI 63 |
| Mortierella alpina | Query 4 TKQTARKSTGGKAPRKQLATKAARKSAPAT#GGVKKPHRYRPGTVALREIRRYQKSTELLI 63 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 4 TKQTARKSTGGKAPRKQLATKAARKSAPAT#GGVKKPHRYRPGTVALREIRRYQKSTELLI 63 |
| Xenopus laevis | Query 4 TKQTARKSTGGKAPRKQLATKAARKSAPAT#GGVKKPHRYRPGTVALREIRRYQKSTELLI 63 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 4 TKQTARKSTGGKAPRKQLATKAARKSAPAT#GGVKKPHRYRPGTVALREIRRYQKSTELLI 63 |
| Oreophoetes peruana | Query 4 TKQTARKSTGGKAPRKQLATKAARKSAPAT#GGVKKPHRYRPGTVALREIRRYQKSTELLI 63 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 4 TKQTARKSTGGKAPRKQLATKAARKSAPAT#GGVKKPHRYRPGTVALREIRRYQKSTELLI 63 |
| Asellus aquaticus | Query 4 TKQTARKSTGGKAPRKQLATKAARKSAPAT#GGVKKPHRYRPGTVALREIRRYQKSTELLI 63 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 4 TKQTARKSTGGKAPRKQLATKAARKSAPAT#GGVKKPHRYRPGTVALREIRRYQKSTELLI 63 |
| Aretaon asperrimus | Query 4 TKQTARKSTGGKAPRKQLATKAARKSAPAT#GGVKKPHRYRPGTVALREIRRYQKSTELLI 63 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 4 TKQTARKSTGGKAPRKQLATKAARKSAPAT#GGVKKPHRYRPGTVALREIRRYQKSTELLI 63 |
| Elassoneuria sp. EP167 | Query 4 TKQTARKSTGGKAPRKQLATKAARKSAPAT#GGVKKPHRYRPGTVALREIRRYQKSTELLI 63 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 4 TKQTARKSTGGKAPRKQLATKAARKSAPAT#GGVKKPHRYRPGTVALREIRRYQKSTELLI 63 |
| Styela plicata | Query 4 TKQTARKSTGGKAPRKQLATKAARKSAPAT#GGVKKPHRYRPGTVALREIRRYQKSTELLI 63 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 4 TKQTARKSTGGKAPRKQLATKAARKSAPAT#GGVKKPHRYRPGTVALREIRRYQKSTELLI 63 |
| Sukaschitrochus atkinsoni | Query 4 TKQTARKSTGGKAPRKQLATKAARKSAPAT#GGVKKPHRYRPGTVALREIRRYQKSTELLI 63 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 4 TKQTARKSTGGKAPRKQLATKAARKSAPAT#GGVKKPHRYRPGTVALREIRRYQKSTELLI 63 |
| Drosophila persimilis | Query 4 TKQTARKSTGGKAPRKQLATKAARKSAPAT#GGVKKPHRYRPGTVALREIRRYQKSTELLI 63 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 4 TKQTARKSTGGKAPRKQLATKAARKSAPAT#GGVKKPHRYRPGTVALREIRRYQKSTELLI 63 |
| Drosophila melanogaster | Query 4 TKQTARKSTGGKAPRKQLATKAARKSAPAT#GGVKKPHRYRPGTVALREIRRYQKSTELLI 63 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 4 TKQTARKSTGGKAPRKQLATKAARKSAPAT#GGVKKPHRYRPGTVALREIRRYQKSTELLI 63 |
| Isonychia sp. EP142 | Query 4 TKQTARKSTGGKAPRKQLATKAARKSAPAT#GGVKKPHRYRPGTVALREIRRYQKSTELLI 63 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 4 TKQTARKSTGGKAPRKQLATKAARKSAPAT#GGVKKPHRYRPGTVALREIRRYQKSTELLI 63 |
| Drosophila simulans | Query 4 TKQTARKSTGGKAPRKQLATKAARKSAPAT#GGVKKPHRYRPGTVALREIRRYQKSTELLI 63 |||||||||||||||||||||||||| |||#|||||||||||||||||||||||||||||| Sbjct 4 TKQTARKSTGGKAPRKQLATKAARKSVPAT#GGVKKPHRYRPGTVALREIRRYQKSTELLI 63 |
| Drosophila virilis | Query 4 TKQTARKSTGGKAPRKQLATKAARKSAPAT#GGVKKPHRYRPGTVALREIRRYQKSTELLI 63 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 4 TKQTARKSTGGKAPRKQLATKAARKSAPAT#GGVKKPHRYRPGTVALREIRRYQKSTELLI 63 |
| Lepetodrilus ovalis | Query 4 TKQTARKSTGGKAPRKQLATKAARKSAPAT#GGVKKPHRYRPGTVALREIRRYQKSTELLI 63 |||| |||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Sbjct 4 TKQTXRKSTGGKAPRKQLATKAARKSAPAT#GGVKKPHRYRPGTVALREIRRYQKSTELLI 63 |
| Callibaetis sp. EP020 | Query 4 TKQTARKSTGGKAPRKQLATKAARKSAPAT#GGVKKPHRYRPGTVALREIRRYQKSTELLI 63 ||||||||+|||||||||||||||||||||#|||||||||||||||||||||||||||||| Sbjct 4 TKQTARKSSGGKAPRKQLATKAARKSAPAT#GGVKKPHRYRPGTVALREIRRYQKSTELLI 63 |
| Caenorhabditis elegans | Query 4 TKQTARKSTGGKAPRKQLATKAARKSAPAT#GGVKKPHRYRPGTVALREIRRYQKSTELLI 63 |||||||||||||||||||||||||||||+#|||||||||||||||||||||||||||||| Sbjct 4 TKQTARKSTGGKAPRKQLATKAARKSAPAS#GGVKKPHRYRPGTVALREIRRYQKSTELLI 63 |
| Machilis sp. BYU_ACAR003 | Query 5 KQTARKSTGGKAPRKQLATKAARKSAPAT#GGVKKPHRYRPGTVALREIRRYQKSTELLI 63 |||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 5 KQTARKSTGGKAPRKQLATKAARKSAPAT#GGVKKPHRYRPGTVALREIRRYQKSTELLI 63 |
| Austrophlebiodes sp. EP027 | Query 5 KQTARKSTGGKAPRKQLATKAARKSAPAT#GGVKKPHRYRPGTVALREIRRYQKSTELLI 63 |||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 5 KQTARKSTGGKAPRKQLATKAARKSAPAT#GGVKKPHRYRPGTVALREIRRYQKSTELLI 63 |
| Venerupis (Ruditapes) philippinarum | Query 4 TKQTARKSTGGKAPRKQLATKAARKSAPAT#GGVKKPHRYRPGTVALREIRRYQKSTELLI 63 ||||||||||||||||||||||||||||+|#|||||||||||||||||||||||||||||| Sbjct 4 TKQTARKSTGGKAPRKQLATKAARKSAPST#GGVKKPHRYRPGTVALREIRRYQKSTELLI 63 |
| synthetic construct | Query 4 TKQTARKSTGGKAPRKQLATKAARKSAPAT#GGVKKPHRYRPGTVALREIRRYQKSTELLI 63 ||||||||||||||||||||| ||||||||#|||||||||||||||||||||||||||||| Sbjct 4 TKQTARKSTGGKAPRKQLATKVARKSAPAT#GGVKKPHRYRPGTVALREIRRYQKSTELLI 63 |
| Psammechinus miliaris | Query 4 TKQTARKSTGGKAPRKQLATKAARKSAPAT#GGVKKPHRYRPGTVALREIRRYQKSTELLI 63 ||||||||||||||||||||||||||||||#||||||||+||||||||||||||||||||| Sbjct 4 TKQTARKSTGGKAPRKQLATKAARKSAPAT#GGVKKPHRFRPGTVALREIRRYQKSTELLI 63 |
| Siphlonella sp. EP083 | Query 4 TKQTARKSTGGKAPRKQLATKAARKSAPAT#GGVKKPHRYRPGTVALREIRRYQKSTELLI 63 |||||| |||||||||||||||||||||||#|||||||||||||||||||||||||||||| Sbjct 4 TKQTARXSTGGKAPRKQLATKAARKSAPAT#GGVKKPHRYRPGTVALREIRRYQKSTELLI 63 |
| Phyllophaga sp. BYU_ACCO040 | Query 5 KQTARKSTGGKAPRKQLATKAARKSAPAT#GGVKKPHRYRPGTVALREIRRYQKSTELLI 63 |||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 5 KQTARKSTGGKAPRKQLATKAARKSAPAT#GGVKKPHRYRPGTVALREIRRYQKSTELLI 63 |
| Tetrapygus niger | Query 4 TKQTARKSTGGKAPRKQLATKAARKSAPAT#GGVKKPHRYRPGTVALREIRRYQKSTELLI 63 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 4 TKQTARKSTGGKAPRKQLATKAARKSAPAT#GGVKKPHRYRPGTVALREIRRYQKSTELLI 63 |
| Prosopistoma wouterae | Query 4 TKQTARKSTGGKAPRKQLATKAARKSAPAT#GGVKKPHRYRPGTVALREIRRYQKSTELLI 63 ||||||||||||||||||||||||||||||#||||||||||||||||||||||| |||||| Sbjct 3 TKQTARKSTGGKAPRKQLATKAARKSAPAT#GGVKKPHRYRPGTVALREIRRYQXSTELLI 62 |
[Site 5] AARKSAPATG34-GVKKPHRYRP
Gly34  Gly
Gly
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Ala25 | Ala26 | Arg27 | Lys28 | Ser29 | Ala30 | Pro31 | Ala32 | Thr33 | Gly34 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Gly35 | Val36 | Lys37 | Lys38 | Pro39 | His40 | Arg41 | Tyr42 | Arg43 | Pro44 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| KQTARKSTGGKAPRKQLATKAARKSAPATGGVKKPHRYRPGTVALREIRRYQKSTELLIR |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | Canis familiaris | 119.00 | 4 | PREDICTED: similar to histone 1, H2ai (predicted) |
| 2 | Homo sapiens | 119.00 | 4 | Histone cluster 1, H3i |
| 3 | Expression vector pET3-H3 | 119.00 | 1 | Xenopus laevis-like histone H3 |
| 4 | Thekalabis sp. DM16 | 119.00 | 1 | histone 3 |
| 5 | Agathemera crassa | 119.00 | 1 | histone 3 |
| 6 | Clypeosectus sp. CET-2005 | 119.00 | 1 | histone H3 |
| 7 | Cyathermia naticoides | 119.00 | 1 | histone H3 |
| 8 | Stygifloris sp. EP141 | 119.00 | 1 | histone H3 |
| 9 | Attenella margarita | 119.00 | 1 | histone H3 |
| 10 | Tropidoderus childrenii | 119.00 | 1 | histone 3 |
| 11 | Strongylocentrotus purpuratus | 119.00 | 1 | late embryonic histone H3 |
| 12 | Gibbula zonata | 119.00 | 1 | histone H3 |
| 13 | Anatoma euglypta | 119.00 | 1 | histone H3 |
| 14 | Gromphadorhina portentosa | 119.00 | 1 | histone 3 |
| 15 | Progomphus obscurus | 119.00 | 1 | histone H3 |
| 16 | Grylloblatta campodeiformis | 119.00 | 1 | histone 3 |
| 17 | Orxines macklottii | 119.00 | 1 | histone 3 |
| 18 | Acilius sylvanus | 119.00 | 1 | histone 3 |
| 19 | Mytilus californianus | 119.00 | 1 | histone H3 |
| 20 | Mus musculus | 118.00 | 4 | histone cluster 1, H3f |
| 21 | N/A | 118.00 | 3 | histone H3 |
| 22 | Strongylocentrotus purpuratus | 118.00 | 2 | histone H3 |
| 23 | Bos taurus | 118.00 | 2 | PREDICTED: similar to histone protein Hist2h3c1 |
| 24 | Oikopleura dioica | 118.00 | 2 | histone h3.1 |
| 25 | Scissurella cf. coronata CET-2005 | 118.00 | 2 | histone H3 |
| 26 | Mortierella alpina | 118.00 | 1 | H3_MORAP Histone H3 gi |
| 27 | Machilis sp. BYU_ACAR003 | 118.00 | 1 | histone H3 |
| 28 | Asellus aquaticus | 118.00 | 1 | putative H3 histone |
| 29 | Aretaon asperrimus | 118.00 | 1 | histone 3 |
| 30 | Oreophoetes peruana | 118.00 | 1 | histone 3 |
| 31 | Phyllophaga sp. BYU_ACCO040 | 118.00 | 1 | histone H3 |
| 32 | Anopheles gambiae str. PEST | 118.00 | 1 | AGAP012709-PA |
| 33 | Griseargiolestes albescens | 118.00 | 1 | histone H3 |
| 34 | Neohirasea sp. WS29 | 118.00 | 1 | histone 3 |
| 35 | Fissurella virescens | 118.00 | 1 | histone H3 |
| 36 | Extatosoma tiaratum | 118.00 | 1 | histone 3 |
| 37 | Mytilus chilensis | 118.00 | 1 | histone H3 |
| 38 | Erythemis sp. OD024 | 118.00 | 1 | histone H3 |
| 39 | Elassoneuria sp. EP167 | 118.00 | 1 | histone H3 |
| 40 | Styela plicata | 118.00 | 1 | H3 histone |
| 41 | Drosophila persimilis | 118.00 | 1 | histone 3 |
| 42 | Drosophila melanogaster | 118.00 | 1 | H3 histone |
| 43 | Danio rerio | 118.00 | 1 | hypothetical protein LOC794406 |
| 44 | Sukaschitrochus atkinsoni | 118.00 | 1 | histone H3 |
| 45 | Sungaya inexpectata | 118.00 | 1 | histone 3 |
| 46 | Xenopus laevis | 118.00 | 1 | histone H3 |
| 47 | Austrophlebiodes sp. EP027 | 118.00 | 1 | histone H3 |
| 48 | Isonychia sp. EP142 | 118.00 | 1 | histone H3 |
| 49 | synthetic construct | 117.00 | 1 | histone 3 H3 |
| 50 | Callibaetis sp. EP020 | 117.00 | 1 | histone H3 |
| 51 | Drosophila virilis | 117.00 | 1 | histone H3 |
| 52 | Labidiosticta vallisi | 117.00 | 1 | histone H3 |
| 53 | Psammechinus miliaris | 117.00 | 1 | cleavage stage histone H3 |
| 54 | Murphyella sp. EP082 | 117.00 | 1 | histone H3 |
| 55 | Prosopistoma wouterae | 116.00 | 1 | histone H3 |
| 56 | Tetrapygus niger | 116.00 | 1 | AF356516_2 early histone H3 |
| 57 | Battigrassiella wheeleri | 116.00 | 1 | histone H3 |
Top-ranked sequences
| organism | matching |
|---|---|
| Canis familiaris | Query 5 KQTARKSTGGKAPRKQLATKAARKSAPATG#GVKKPHRYRPGTVALREIRRYQKSTELLIR 64 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 5 KQTARKSTGGKAPRKQLATKAARKSAPATG#GVKKPHRYRPGTVALREIRRYQKSTELLIR 64 |
| Homo sapiens | Query 5 KQTARKSTGGKAPRKQLATKAARKSAPATG#GVKKPHRYRPGTVALREIRRYQKSTELLIR 64 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 5 KQTARKSTGGKAPRKQLATKAARKSAPATG#GVKKPHRYRPGTVALREIRRYQKSTELLIR 64 |
| Expression vector pET3-H3 | Query 5 KQTARKSTGGKAPRKQLATKAARKSAPATG#GVKKPHRYRPGTVALREIRRYQKSTELLIR 64 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 5 KQTARKSTGGKAPRKQLATKAARKSAPATG#GVKKPHRYRPGTVALREIRRYQKSTELLIR 64 |
| Thekalabis sp. DM16 | Query 5 KQTARKSTGGKAPRKQLATKAARKSAPATG#GVKKPHRYRPGTVALREIRRYQKSTELLIR 64 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 5 KQTARKSTGGKAPRKQLATKAARKSAPATG#GVKKPHRYRPGTVALREIRRYQKSTELLIR 64 |
| Agathemera crassa | Query 5 KQTARKSTGGKAPRKQLATKAARKSAPATG#GVKKPHRYRPGTVALREIRRYQKSTELLIR 64 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 5 KQTARKSTGGKAPRKQLATKAARKSAPATG#GVKKPHRYRPGTVALREIRRYQKSTELLIR 64 |
| Clypeosectus sp. CET-2005 | Query 5 KQTARKSTGGKAPRKQLATKAARKSAPATG#GVKKPHRYRPGTVALREIRRYQKSTELLIR 64 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 5 KQTARKSTGGKAPRKQLATKAARKSAPATG#GVKKPHRYRPGTVALREIRRYQKSTELLIR 64 |
| Cyathermia naticoides | Query 5 KQTARKSTGGKAPRKQLATKAARKSAPATG#GVKKPHRYRPGTVALREIRRYQKSTELLIR 64 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 5 KQTARKSTGGKAPRKQLATKAARKSAPATG#GVKKPHRYRPGTVALREIRRYQKSTELLIR 64 |
| Stygifloris sp. EP141 | Query 5 KQTARKSTGGKAPRKQLATKAARKSAPATG#GVKKPHRYRPGTVALREIRRYQKSTELLIR 64 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 5 KQTARKSTGGKAPRKQLATKAARKSAPATG#GVKKPHRYRPGTVALREIRRYQKSTELLIR 64 |
| Attenella margarita | Query 5 KQTARKSTGGKAPRKQLATKAARKSAPATG#GVKKPHRYRPGTVALREIRRYQKSTELLIR 64 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 5 KQTARKSTGGKAPRKQLATKAARKSAPATG#GVKKPHRYRPGTVALREIRRYQKSTELLIR 64 |
| Tropidoderus childrenii | Query 5 KQTARKSTGGKAPRKQLATKAARKSAPATG#GVKKPHRYRPGTVALREIRRYQKSTELLIR 64 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 5 KQTARKSTGGKAPRKQLATKAARKSAPATG#GVKKPHRYRPGTVALREIRRYQKSTELLIR 64 |
| Strongylocentrotus purpuratus | Query 5 KQTARKSTGGKAPRKQLATKAARKSAPATG#GVKKPHRYRPGTVALREIRRYQKSTELLIR 64 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 5 KQTARKSTGGKAPRKQLATKAARKSAPATG#GVKKPHRYRPGTVALREIRRYQKSTELLIR 64 |
| Gibbula zonata | Query 5 KQTARKSTGGKAPRKQLATKAARKSAPATG#GVKKPHRYRPGTVALREIRRYQKSTELLIR 64 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 5 KQTARKSTGGKAPRKQLATKAARKSAPATG#GVKKPHRYRPGTVALREIRRYQKSTELLIR 64 |
| Anatoma euglypta | Query 5 KQTARKSTGGKAPRKQLATKAARKSAPATG#GVKKPHRYRPGTVALREIRRYQKSTELLIR 64 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 5 KQTARKSTGGKAPRKQLATKAARKSAPATG#GVKKPHRYRPGTVALREIRRYQKSTELLIR 64 |
| Gromphadorhina portentosa | Query 5 KQTARKSTGGKAPRKQLATKAARKSAPATG#GVKKPHRYRPGTVALREIRRYQKSTELLIR 64 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 5 KQTARKSTGGKAPRKQLATKAARKSAPATG#GVKKPHRYRPGTVALREIRRYQKSTELLIR 64 |
| Progomphus obscurus | Query 5 KQTARKSTGGKAPRKQLATKAARKSAPATG#GVKKPHRYRPGTVALREIRRYQKSTELLIR 64 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 5 KQTARKSTGGKAPRKQLATKAARKSAPATG#GVKKPHRYRPGTVALREIRRYQKSTELLIR 64 |
| Grylloblatta campodeiformis | Query 5 KQTARKSTGGKAPRKQLATKAARKSAPATG#GVKKPHRYRPGTVALREIRRYQKSTELLIR 64 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 5 KQTARKSTGGKAPRKQLATKAARKSAPATG#GVKKPHRYRPGTVALREIRRYQKSTELLIR 64 |
| Orxines macklottii | Query 5 KQTARKSTGGKAPRKQLATKAARKSAPATG#GVKKPHRYRPGTVALREIRRYQKSTELLIR 64 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 5 KQTARKSTGGKAPRKQLATKAARKSAPATG#GVKKPHRYRPGTVALREIRRYQKSTELLIR 64 |
| Acilius sylvanus | Query 5 KQTARKSTGGKAPRKQLATKAARKSAPATG#GVKKPHRYRPGTVALREIRRYQKSTELLIR 64 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 5 KQTARKSTGGKAPRKQLATKAARKSAPATG#GVKKPHRYRPGTVALREIRRYQKSTELLIR 64 |
| Mytilus californianus | Query 5 KQTARKSTGGKAPRKQLATKAARKSAPATG#GVKKPHRYRPGTVALREIRRYQKSTELLIR 64 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 5 KQTARKSTGGKAPRKQLATKAARKSAPATG#GVKKPHRYRPGTVALREIRRYQKSTELLIR 64 |
| Mus musculus | Query 5 KQTARKSTGGKAPRKQLATKAARKSAPATG#GVKKPHRYRPGTVALREIRRYQKSTELLIR 64 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 5 KQTARKSTGGKAPRKQLATKAARKSAPATG#GVKKPHRYRPGTVALREIRRYQKSTELLIR 64 |
| N/A | Query 5 KQTARKSTGGKAPRKQLATKAARKSAPATG#GVKKPHRYRPGTVALREIRRYQKSTELLIR 64 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 5 KQTARKSTGGKAPRKQLATKAARKSAPATG#GVKKPHRYRPGTVALREIRRYQKSTELLIR 64 |
| Strongylocentrotus purpuratus | Query 5 KQTARKSTGGKAPRKQLATKAARKSAPATG#GVKKPHRYRPGTVALREIRRYQKSTELLIR 64 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 5 KQTARKSTGGKAPRKQLATKAARKSAPATG#GVKKPHRYRPGTVALREIRRYQKSTELLIR 64 |
| Bos taurus | Query 5 KQTARKSTGGKAPRKQLATKAARKSAPATG#GVKKPHRYRPGTVALREIRRYQKSTELLIR 64 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 5 KQTARKSTGGKAPRKQLATKAARKSAPATG#GVKKPHRYRPGTVALREIRRYQKSTELLIR 64 |
| Oikopleura dioica | Query 5 KQTARKSTGGKAPRKQLATKAARKSAPATG#GVKKPHRYRPGTVALREIRRYQKSTELLIR 64 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 5 KQTARKSTGGKAPRKQLATKAARKSAPATG#GVKKPHRYRPGTVALREIRRYQKSTELLIR 64 |
| Scissurella cf. coronata CET-2005 | Query 5 KQTARKSTGGKAPRKQLATKAARKSAPATG#GVKKPHRYRPGTVALREIRRYQKSTELLIR 64 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 5 KQTARKSTGGKAPRKQLATKAARKSAPATG#GVKKPHRYRPGTVALREIRRYQKSTELLIR 64 |
| Mortierella alpina | Query 5 KQTARKSTGGKAPRKQLATKAARKSAPATG#GVKKPHRYRPGTVALREIRRYQKSTELLIR 64 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 5 KQTARKSTGGKAPRKQLATKAARKSAPATG#GVKKPHRYRPGTVALREIRRYQKSTELLIR 64 |
| Machilis sp. BYU_ACAR003 | Query 5 KQTARKSTGGKAPRKQLATKAARKSAPATG#GVKKPHRYRPGTVALREIRRYQKSTELLIR 64 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 5 KQTARKSTGGKAPRKQLATKAARKSAPATG#GVKKPHRYRPGTVALREIRRYQKSTELLIR 64 |
| Asellus aquaticus | Query 5 KQTARKSTGGKAPRKQLATKAARKSAPATG#GVKKPHRYRPGTVALREIRRYQKSTELLIR 64 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 5 KQTARKSTGGKAPRKQLATKAARKSAPATG#GVKKPHRYRPGTVALREIRRYQKSTELLIR 64 |
| Aretaon asperrimus | Query 5 KQTARKSTGGKAPRKQLATKAARKSAPATG#GVKKPHRYRPGTVALREIRRYQKSTELLIR 64 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 5 KQTARKSTGGKAPRKQLATKAARKSAPATG#GVKKPHRYRPGTVALREIRRYQKSTELLIR 64 |
| Oreophoetes peruana | Query 5 KQTARKSTGGKAPRKQLATKAARKSAPATG#GVKKPHRYRPGTVALREIRRYQKSTELLIR 64 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 5 KQTARKSTGGKAPRKQLATKAARKSAPATG#GVKKPHRYRPGTVALREIRRYQKSTELLIR 64 |
| Phyllophaga sp. BYU_ACCO040 | Query 5 KQTARKSTGGKAPRKQLATKAARKSAPATG#GVKKPHRYRPGTVALREIRRYQKSTELLIR 64 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 5 KQTARKSTGGKAPRKQLATKAARKSAPATG#GVKKPHRYRPGTVALREIRRYQKSTELLIR 64 |
| Anopheles gambiae str. PEST | Query 5 KQTARKSTGGKAPRKQLATKAARKSAPATG#GVKKPHRYRPGTVALREIRRYQKSTELLIR 64 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 5 KQTARKSTGGKAPRKQLATKAARKSAPATG#GVKKPHRYRPGTVALREIRRYQKSTELLIR 64 |
| Griseargiolestes albescens | Query 5 KQTARKSTGGKAPRKQLATKAARKSAPATG#GVKKPHRYRPGTVALREIRRYQKSTELLIR 64 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 5 KQTARKSTGGKAPRKQLATKAARKSAPATG#GVKKPHRYRPGTVALREIRRYQKSTELLIR 64 |
| Neohirasea sp. WS29 | Query 5 KQTARKSTGGKAPRKQLATKAARKSAPATG#GVKKPHRYRPGTVALREIRRYQKSTELLIR 64 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 5 KQTARKSTGGKAPRKQLATKAARKSAPATG#GVKKPHRYRPGTVALREIRRYQKSTELLIR 64 |
| Fissurella virescens | Query 5 KQTARKSTGGKAPRKQLATKAARKSAPATG#GVKKPHRYRPGTVALREIRRYQKSTELLIR 64 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 5 KQTARKSTGGKAPRKQLATKAARKSAPATG#GVKKPHRYRPGTVALREIRRYQKSTELLIR 64 |
| Extatosoma tiaratum | Query 5 KQTARKSTGGKAPRKQLATKAARKSAPATG#GVKKPHRYRPGTVALREIRRYQKSTELLIR 64 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 5 KQTARKSTGGKAPRKQLATKAARKSAPATG#GVKKPHRYRPGTVALREIRRYQKSTELLIR 64 |
| Mytilus chilensis | Query 5 KQTARKSTGGKAPRKQLATKAARKSAPATG#GVKKPHRYRPGTVALREIRRYQKSTELLIR 64 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 5 KQTARKSTGGKAPRKQLATKAARKSAPATG#GVKKPHRYRPGTVALREIRRYQKSTELLIR 64 |
| Erythemis sp. OD024 | Query 5 KQTARKSTGGKAPRKQLATKAARKSAPATG#GVKKPHRYRPGTVALREIRRYQKSTELLIR 64 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 5 KQTARKSTGGKAPRKQLATKAARKSAPATG#GVKKPHRYRPGTVALREIRRYQKSTELLIR 64 |
| Elassoneuria sp. EP167 | Query 5 KQTARKSTGGKAPRKQLATKAARKSAPATG#GVKKPHRYRPGTVALREIRRYQKSTELLIR 64 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 5 KQTARKSTGGKAPRKQLATKAARKSAPATG#GVKKPHRYRPGTVALREIRRYQKSTELLIR 64 |
| Styela plicata | Query 5 KQTARKSTGGKAPRKQLATKAARKSAPATG#GVKKPHRYRPGTVALREIRRYQKSTELLIR 64 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 5 KQTARKSTGGKAPRKQLATKAARKSAPATG#GVKKPHRYRPGTVALREIRRYQKSTELLIR 64 |
| Drosophila persimilis | Query 5 KQTARKSTGGKAPRKQLATKAARKSAPATG#GVKKPHRYRPGTVALREIRRYQKSTELLIR 64 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 5 KQTARKSTGGKAPRKQLATKAARKSAPATG#GVKKPHRYRPGTVALREIRRYQKSTELLIR 64 |
| Drosophila melanogaster | Query 5 KQTARKSTGGKAPRKQLATKAARKSAPATG#GVKKPHRYRPGTVALREIRRYQKSTELLIR 64 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 5 KQTARKSTGGKAPRKQLATKAARKSAPATG#GVKKPHRYRPGTVALREIRRYQKSTELLIR 64 |
| Danio rerio | Query 5 KQTARKSTGGKAPRKQLATKAARKSAPATG#GVKKPHRYRPGTVALREIRRYQKSTELLIR 64 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 5 KQTARKSTGGKAPRKQLATKAARKSAPATG#GVKKPHRYRPGTVALREIRRYQKSTELLIR 64 |
| Sukaschitrochus atkinsoni | Query 5 KQTARKSTGGKAPRKQLATKAARKSAPATG#GVKKPHRYRPGTVALREIRRYQKSTELLIR 64 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 5 KQTARKSTGGKAPRKQLATKAARKSAPATG#GVKKPHRYRPGTVALREIRRYQKSTELLIR 64 |
| Sungaya inexpectata | Query 5 KQTARKSTGGKAPRKQLATKAARKSAPATG#GVKKPHRYRPGTVALREIRRYQKSTELLIR 64 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 5 KQTARKSTGGKAPRKQLATKAARKSAPATG#GVKKPHRYRPGTVALREIRRYQKSTELLIR 64 |
| Xenopus laevis | Query 5 KQTARKSTGGKAPRKQLATKAARKSAPATG#GVKKPHRYRPGTVALREIRRYQKSTELLIR 64 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 5 KQTARKSTGGKAPRKQLATKAARKSAPATG#GVKKPHRYRPGTVALREIRRYQKSTELLIR 64 |
| Austrophlebiodes sp. EP027 | Query 5 KQTARKSTGGKAPRKQLATKAARKSAPATG#GVKKPHRYRPGTVALREIRRYQKSTELLIR 64 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 5 KQTARKSTGGKAPRKQLATKAARKSAPATG#GVKKPHRYRPGTVALREIRRYQKSTELLIR 64 |
| Isonychia sp. EP142 | Query 5 KQTARKSTGGKAPRKQLATKAARKSAPATG#GVKKPHRYRPGTVALREIRRYQKSTELLIR 64 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 5 KQTARKSTGGKAPRKQLATKAARKSAPATG#GVKKPHRYRPGTVALREIRRYQKSTELLIR 64 |
| synthetic construct | Query 5 KQTARKSTGGKAPRKQLATKAARKSAPATG#GVKKPHRYRPGTVALREIRRYQKSTELLIR 64 |||||||||||||||||||| |||||||||#|||||||||||||||||||||||||||||| Sbjct 5 KQTARKSTGGKAPRKQLATKVARKSAPATG#GVKKPHRYRPGTVALREIRRYQKSTELLIR 64 |
| Callibaetis sp. EP020 | Query 5 KQTARKSTGGKAPRKQLATKAARKSAPATG#GVKKPHRYRPGTVALREIRRYQKSTELLIR 64 |||||||+||||||||||||||||||||||#|||||||||||||||||||||||||||||| Sbjct 5 KQTARKSSGGKAPRKQLATKAARKSAPATG#GVKKPHRYRPGTVALREIRRYQKSTELLIR 64 |
| Drosophila virilis | Query 5 KQTARKSTGGKAPRKQLATKAARKSAPATG#GVKKPHRYRPGTVALREIRRYQKSTELLIR 64 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 5 KQTARKSTGGKAPRKQLATKAARKSAPATG#GVKKPHRYRPGTVALREIRRYQKSTELLIR 64 |
| Labidiosticta vallisi | Query 6 QTARKSTGGKAPRKQLATKAARKSAPATG#GVKKPHRYRPGTVALREIRRYQKSTELLIR 64 |||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 6 QTARKSTGGKAPRKQLATKAARKSAPATG#GVKKPHRYRPGTVALREIRRYQKSTELLIR 64 |
| Psammechinus miliaris | Query 5 KQTARKSTGGKAPRKQLATKAARKSAPATG#GVKKPHRYRPGTVALREIRRYQKSTELLIR 64 ||||||||||||||||||||||||||||||#|||||||+|||||||||||||||||||||| Sbjct 5 KQTARKSTGGKAPRKQLATKAARKSAPATG#GVKKPHRFRPGTVALREIRRYQKSTELLIR 64 |
| Murphyella sp. EP082 | Query 6 QTARKSTGGKAPRKQLATKAARKSAPATG#GVKKPHRYRPGTVALREIRRYQKSTELLIR 64 |||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 6 QTARKSTGGKAPRKQLATKAARKSAPATG#GVKKPHRYRPGTVALREIRRYQKSTELLIR 64 |
| Prosopistoma wouterae | Query 5 KQTARKSTGGKAPRKQLATKAARKSAPATG#GVKKPHRYRPGTVALREIRRYQKSTELLIR 64 ||||||||||||||||||||||||||||||#|||||||||||||||||||||| ||||||| Sbjct 4 KQTARKSTGGKAPRKQLATKAARKSAPATG#GVKKPHRYRPGTVALREIRRYQXSTELLIR 63 |
| Tetrapygus niger | Query 5 KQTARKSTGGKAPRKQLATKAARKSAPATG#GVKKPHRYRPGTVALREIRRYQKSTELLIR 64 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 5 KQTARKSTGGKAPRKQLATKAARKSAPATG#GVKKPHRYRPGTVALREIRRYQKSTELLIR 64 |
| Battigrassiella wheeleri | Query 5 KQTARKSTGGKAPRKQLATKAARKSAPATG#GVKKPHRYRPGTVALREIRRYQKSTELLIR 64 || |||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Sbjct 1 KQAARKSTGGKAPRKQLATKAARKSAPATG#GVKKPHRYRPGTVALREIRRYQKSTELLIR 60 |
[Site 6] HRYRPGTVAL49-REIRRYQKST
Leu49  Arg
Arg
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| His40 | Arg41 | Tyr42 | Arg43 | Pro44 | Gly45 | Thr46 | Val47 | Ala48 | Leu49 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Arg50 | Glu51 | Ile52 | Arg53 | Arg54 | Tyr55 | Gln56 | Lys57 | Ser58 | Thr59 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| QLATKAARKSAPATGGVKKPHRYRPGTVALREIRRYQKSTELLIRKLPFQRLVREIAQDF |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | Zeus faber | 122.00 | 1 | histone H3 |
| 2 | Uleiota sp. BYU_ACCO072 | 122.00 | 1 | histone H3 |
| 3 | Trachyscorpia cristulata | 122.00 | 1 | histone H3 |
| 4 | Tidarren sisyphoides | 122.00 | 1 | histone H3A |
| 5 | Tettigarcta crinita | 122.00 | 1 | histone 3 |
| 6 | Synanceia verrucosa | 122.00 | 1 | histone H3 |
| 7 | Sukaschitrochus atkinsoni | 122.00 | 1 | histone H3 |
| 8 | Stauroteuthis syrtensis | 122.00 | 1 | histone H3 |
| 9 | Scutus unguis | 122.00 | 1 | histone H3 |
| 10 | Scomberoides lysan | 122.00 | 1 | histone H3 |
| 11 | Scoloplos simplex | 122.00 | 1 | AF185261_1 histone H3 |
| 12 | Acilius sylvanus | 122.00 | 1 | histone 3 |
| 13 | Pterois volitans | 122.00 | 1 | histone H3 |
| 14 | Protopelagonemertes sp. 544 | 122.00 | 1 | histone H3 |
| 15 | Polyplocia sp. Eph132 | 122.00 | 1 | histone 3 |
| 16 | Polycentropsis abbreviata | 122.00 | 1 | histone H3a |
| 17 | Pintalia alta | 122.00 | 1 | histone 3 |
| 18 | Phyllophaga sp. BYU_ACCO040 | 122.00 | 1 | histone H3 |
| 19 | Phyllochaetopterus sp. AMW24402 | 122.00 | 1 | AF185246_1 histone H3 |
| 20 | Phascolion strombi | 122.00 | 1 | histone H3 |
| 21 | Petrobiinae gen. sp. | 122.00 | 1 | AF110865_1 histone H3 |
| 22 | Pelagonemertes sp. 545 | 122.00 | 1 | histone H3 |
| 23 | Pectinariophyes reticulata | 122.00 | 1 | histone 3 |
| 24 | Panulirus regius | 122.00 | 1 | histone H3 |
| 25 | Palaemonetes paludosus | 122.00 | 1 | histone H3 |
| 26 | Owenia fusiformis | 122.00 | 1 | AF185234_1 histone H3 |
| 27 | Orxines macklottii | 122.00 | 1 | histone 3 |
| 28 | Oroperipatus corrodai | 122.00 | 1 | AF110848_1 histone H3 |
| 29 | Oreophoetes peruana | 122.00 | 1 | histone 3 |
| 30 | Oncotophasma martini | 122.00 | 1 | histone 3 |
| 31 | Aetapcus maculatus | 122.00 | 1 | histone H3 |
| 32 | Notomastus torquatus | 122.00 | 1 | AF185258_1 histone H3 |
| 33 | Normanichthys crockeri | 122.00 | 1 | histone H3 |
| 34 | Nipponentomon sp. | 122.00 | 1 | AF110861_1 histone H3 |
| 35 | Agathemera crassa | 122.00 | 1 | histone 3 |
| 36 | Neohirasea sp. WS29 | 122.00 | 1 | histone 3 |
| 37 | Marukawichthys ambulator | 122.00 | 1 | histone H3 |
| 38 | Amphicteis dalmatica | 122.00 | 1 | AF342700_1 histone H3 |
| 39 | Liorhina sp. JRC-2004 | 122.00 | 1 | histone 3 |
| 40 | Lepyronia quadrangularis | 122.00 | 1 | histone 3 |
| 41 | Leiognathus fasciatus | 122.00 | 1 | histone H3 |
| 42 | Lancetes varius | 122.00 | 1 | histone H3 |
| 43 | Labrisomus multiporosus | 122.00 | 1 | histone H3 |
| 44 | Anatoma euglypta | 122.00 | 1 | histone H3 |
| 45 | Aphredoderus sayanus | 122.00 | 1 | histone H3 |
| 46 | Hygrobia maculata | 122.00 | 1 | histone H3 |
| 47 | Hygrobia hermanni | 122.00 | 1 | histone H3 |
| 48 | Hygrobia australasiae | 122.00 | 1 | histone H3 |
| 49 | Hubrechtella dubia | 122.00 | 1 | histone H3 |
| 50 | Homalodisca elongata | 122.00 | 1 | histone H3 |
| 51 | Haplognathia gubbarnorum | 122.00 | 1 | histone H3 |
| 52 | Gratidia hispidula | 122.00 | 1 | histone 3 |
| 53 | Gobiocichla ethelwynnae | 122.00 | 1 | histone H3a |
| 54 | Glycera sp. AMW196835 | 122.00 | 1 | AF185236_1 histone H3 |
| 55 | Architeuthis dux | 122.00 | 1 | histone H3 |
| 56 | Euchlanis alata | 122.00 | 1 | histone H3 |
| 57 | Erythemis sp. OD024 | 122.00 | 1 | histone H3 |
| 58 | Elates ransonnetii | 122.00 | 1 | histone H3 |
| 59 | Elassoneuria sp. EP167 | 122.00 | 1 | histone H3 |
| 60 | Austrophlebiodes sp. EP027 | 122.00 | 1 | histone H3 |
| 61 | Cyphophthalmus minutus | 122.00 | 1 | histone H3 |
| 62 | Battigrassiella wheeleri | 122.00 | 1 | histone H3 |
| 63 | Ctenomorphodes briareus | 122.00 | 1 | histone 3 |
| 64 | Craterostigmus tasmanianus | 122.00 | 1 | AF110850_1 histone H3 |
| 65 | Cottus poecilopus | 122.00 | 1 | histone H3 |
| 66 | Cosmoscarta sp. JRC-2004 | 122.00 | 1 | histone 3 |
| 67 | Coloburiscoides sp. EP094 | 122.00 | 1 | histone H3 |
| 68 | Clypeosectus sp. CET-2005 | 122.00 | 1 | histone H3 |
| 69 | Clastoptera obtusa | 122.00 | 1 | histone 3 |
| 70 | Chilara taylori | 122.00 | 1 | histone H3 |
| 71 | Chaquihua sp. EP080 | 122.00 | 1 | histone 3 |
| 72 | Cuerna gladiola | 122.00 | 1 | histone H3 |
| 73 | Drosophila virilis | 121.00 | 1 | histone H3 |
| 74 | Aspidosiphon fischeri | 121.00 | 1 | histone H3 |
| 75 | Evexus n. sp. JRC-2004-r | 121.00 | 1 | histone 3 |
| 76 | Aplysia cf. juliana | 121.00 | 1 | histone H3 |
| 77 | Idanthyrsus pennatus | 121.00 | 1 | AF185231_1 histone H3 |
| 78 | Ischnochiton australis | 121.00 | 1 | histone H3 |
| 79 | Lysmata debelius | 121.00 | 1 | histone H3 |
| 80 | Nerita atramentosa | 121.00 | 1 | histone H3 |
| 81 | Onchidium sp. | 121.00 | 1 | histone H3 |
| 82 | Aegla abtao | 121.00 | 1 | histone H3 |
| 83 | Robertus neglectus | 121.00 | 1 | histone H3A |
Top-ranked sequences
| organism | matching |
|---|---|
| Zeus faber | Query 20 QLATKAARKSAPATGGVKKPHRYRPGTVAL#REIRRYQKSTELLIRKLPFQRLVREIAQDF 79 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 20 QLATKAARKSAPATGGVKKPHRYRPGTVAL#REIRRYQKSTELLIRKLPFQRLVREIAQDF 79 |
| Uleiota sp. BYU_ACCO072 | Query 20 QLATKAARKSAPATGGVKKPHRYRPGTVAL#REIRRYQKSTELLIRKLPFQRLVREIAQDF 79 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 20 QLATKAARKSAPATGGVKKPHRYRPGTVAL#REIRRYQKSTELLIRKLPFQRLVREIAQDF 79 |
| Trachyscorpia cristulata | Query 20 QLATKAARKSAPATGGVKKPHRYRPGTVAL#REIRRYQKSTELLIRKLPFQRLVREIAQDF 79 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 20 QLATKAARKSAPATGGVKKPHRYRPGTVAL#REIRRYQKSTELLIRKLPFQRLVREIAQDF 79 |
| Tidarren sisyphoides | Query 20 QLATKAARKSAPATGGVKKPHRYRPGTVAL#REIRRYQKSTELLIRKLPFQRLVREIAQDF 79 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 20 QLATKAARKSAPATGGVKKPHRYRPGTVAL#REIRRYQKSTELLIRKLPFQRLVREIAQDF 79 |
| Tettigarcta crinita | Query 20 QLATKAARKSAPATGGVKKPHRYRPGTVAL#REIRRYQKSTELLIRKLPFQRLVREIAQDF 79 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 20 QLATKAARKSAPATGGVKKPHRYRPGTVAL#REIRRYQKSTELLIRKLPFQRLVREIAQDF 79 |
| Synanceia verrucosa | Query 20 QLATKAARKSAPATGGVKKPHRYRPGTVAL#REIRRYQKSTELLIRKLPFQRLVREIAQDF 79 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 20 QLATKAARKSAPATGGVKKPHRYRPGTVAL#REIRRYQKSTELLIRKLPFQRLVREIAQDF 79 |
| Sukaschitrochus atkinsoni | Query 20 QLATKAARKSAPATGGVKKPHRYRPGTVAL#REIRRYQKSTELLIRKLPFQRLVREIAQDF 79 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 20 QLATKAARKSAPATGGVKKPHRYRPGTVAL#REIRRYQKSTELLIRKLPFQRLVREIAQDF 79 |
| Stauroteuthis syrtensis | Query 20 QLATKAARKSAPATGGVKKPHRYRPGTVAL#REIRRYQKSTELLIRKLPFQRLVREIAQDF 79 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 20 QLATKAARKSAPATGGVKKPHRYRPGTVAL#REIRRYQKSTELLIRKLPFQRLVREIAQDF 79 |
| Scutus unguis | Query 20 QLATKAARKSAPATGGVKKPHRYRPGTVAL#REIRRYQKSTELLIRKLPFQRLVREIAQDF 79 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 20 QLATKAARKSAPATGGVKKPHRYRPGTVAL#REIRRYQKSTELLIRKLPFQRLVREIAQDF 79 |
| Scomberoides lysan | Query 20 QLATKAARKSAPATGGVKKPHRYRPGTVAL#REIRRYQKSTELLIRKLPFQRLVREIAQDF 79 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 20 QLATKAARKSAPATGGVKKPHRYRPGTVAL#REIRRYQKSTELLIRKLPFQRLVREIAQDF 79 |
| Scoloplos simplex | Query 20 QLATKAARKSAPATGGVKKPHRYRPGTVAL#REIRRYQKSTELLIRKLPFQRLVREIAQDF 79 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 20 QLATKAARKSAPATGGVKKPHRYRPGTVAL#REIRRYQKSTELLIRKLPFQRLVREIAQDF 79 |
| Acilius sylvanus | Query 20 QLATKAARKSAPATGGVKKPHRYRPGTVAL#REIRRYQKSTELLIRKLPFQRLVREIAQDF 79 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 20 QLATKAARKSAPATGGVKKPHRYRPGTVAL#REIRRYQKSTELLIRKLPFQRLVREIAQDF 79 |
| Pterois volitans | Query 20 QLATKAARKSAPATGGVKKPHRYRPGTVAL#REIRRYQKSTELLIRKLPFQRLVREIAQDF 79 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 20 QLATKAARKSAPATGGVKKPHRYRPGTVAL#REIRRYQKSTELLIRKLPFQRLVREIAQDF 79 |
| Protopelagonemertes sp. 544 | Query 20 QLATKAARKSAPATGGVKKPHRYRPGTVAL#REIRRYQKSTELLIRKLPFQRLVREIAQDF 79 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 20 QLATKAARKSAPATGGVKKPHRYRPGTVAL#REIRRYQKSTELLIRKLPFQRLVREIAQDF 79 |
| Polyplocia sp. Eph132 | Query 20 QLATKAARKSAPATGGVKKPHRYRPGTVAL#REIRRYQKSTELLIRKLPFQRLVREIAQDF 79 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 20 QLATKAARKSAPATGGVKKPHRYRPGTVAL#REIRRYQKSTELLIRKLPFQRLVREIAQDF 79 |
| Polycentropsis abbreviata | Query 20 QLATKAARKSAPATGGVKKPHRYRPGTVAL#REIRRYQKSTELLIRKLPFQRLVREIAQDF 79 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 20 QLATKAARKSAPATGGVKKPHRYRPGTVAL#REIRRYQKSTELLIRKLPFQRLVREIAQDF 79 |
| Pintalia alta | Query 20 QLATKAARKSAPATGGVKKPHRYRPGTVAL#REIRRYQKSTELLIRKLPFQRLVREIAQDF 79 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 20 QLATKAARKSAPATGGVKKPHRYRPGTVAL#REIRRYQKSTELLIRKLPFQRLVREIAQDF 79 |
| Phyllophaga sp. BYU_ACCO040 | Query 20 QLATKAARKSAPATGGVKKPHRYRPGTVAL#REIRRYQKSTELLIRKLPFQRLVREIAQDF 79 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 20 QLATKAARKSAPATGGVKKPHRYRPGTVAL#REIRRYQKSTELLIRKLPFQRLVREIAQDF 79 |
| Phyllochaetopterus sp. AMW24402 | Query 20 QLATKAARKSAPATGGVKKPHRYRPGTVAL#REIRRYQKSTELLIRKLPFQRLVREIAQDF 79 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 20 QLATKAARKSAPATGGVKKPHRYRPGTVAL#REIRRYQKSTELLIRKLPFQRLVREIAQDF 79 |
| Phascolion strombi | Query 20 QLATKAARKSAPATGGVKKPHRYRPGTVAL#REIRRYQKSTELLIRKLPFQRLVREIAQDF 79 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 20 QLATKAARKSAPATGGVKKPHRYRPGTVAL#REIRRYQKSTELLIRKLPFQRLVREIAQDF 79 |
| Petrobiinae gen. sp. | Query 20 QLATKAARKSAPATGGVKKPHRYRPGTVAL#REIRRYQKSTELLIRKLPFQRLVREIAQDF 79 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 20 QLATKAARKSAPATGGVKKPHRYRPGTVAL#REIRRYQKSTELLIRKLPFQRLVREIAQDF 79 |
| Pelagonemertes sp. 545 | Query 20 QLATKAARKSAPATGGVKKPHRYRPGTVAL#REIRRYQKSTELLIRKLPFQRLVREIAQDF 79 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 20 QLATKAARKSAPATGGVKKPHRYRPGTVAL#REIRRYQKSTELLIRKLPFQRLVREIAQDF 79 |
| Pectinariophyes reticulata | Query 20 QLATKAARKSAPATGGVKKPHRYRPGTVAL#REIRRYQKSTELLIRKLPFQRLVREIAQDF 79 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 20 QLATKAARKSAPATGGVKKPHRYRPGTVAL#REIRRYQKSTELLIRKLPFQRLVREIAQDF 79 |
| Panulirus regius | Query 20 QLATKAARKSAPATGGVKKPHRYRPGTVAL#REIRRYQKSTELLIRKLPFQRLVREIAQDF 79 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 20 QLATKAARKSAPATGGVKKPHRYRPGTVAL#REIRRYQKSTELLIRKLPFQRLVREIAQDF 79 |
| Palaemonetes paludosus | Query 20 QLATKAARKSAPATGGVKKPHRYRPGTVAL#REIRRYQKSTELLIRKLPFQRLVREIAQDF 79 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 20 QLATKAARKSAPATGGVKKPHRYRPGTVAL#REIRRYQKSTELLIRKLPFQRLVREIAQDF 79 |
| Owenia fusiformis | Query 20 QLATKAARKSAPATGGVKKPHRYRPGTVAL#REIRRYQKSTELLIRKLPFQRLVREIAQDF 79 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 20 QLATKAARKSAPATGGVKKPHRYRPGTVAL#REIRRYQKSTELLIRKLPFQRLVREIAQDF 79 |
| Orxines macklottii | Query 20 QLATKAARKSAPATGGVKKPHRYRPGTVAL#REIRRYQKSTELLIRKLPFQRLVREIAQDF 79 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 20 QLATKAARKSAPATGGVKKPHRYRPGTVAL#REIRRYQKSTELLIRKLPFQRLVREIAQDF 79 |
| Oroperipatus corrodai | Query 20 QLATKAARKSAPATGGVKKPHRYRPGTVAL#REIRRYQKSTELLIRKLPFQRLVREIAQDF 79 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 20 QLATKAARKSAPATGGVKKPHRYRPGTVAL#REIRRYQKSTELLIRKLPFQRLVREIAQDF 79 |
| Oreophoetes peruana | Query 20 QLATKAARKSAPATGGVKKPHRYRPGTVAL#REIRRYQKSTELLIRKLPFQRLVREIAQDF 79 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 20 QLATKAARKSAPATGGVKKPHRYRPGTVAL#REIRRYQKSTELLIRKLPFQRLVREIAQDF 79 |
| Oncotophasma martini | Query 20 QLATKAARKSAPATGGVKKPHRYRPGTVAL#REIRRYQKSTELLIRKLPFQRLVREIAQDF 79 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 20 QLATKAARKSAPATGGVKKPHRYRPGTVAL#REIRRYQKSTELLIRKLPFQRLVREIAQDF 79 |
| Aetapcus maculatus | Query 20 QLATKAARKSAPATGGVKKPHRYRPGTVAL#REIRRYQKSTELLIRKLPFQRLVREIAQDF 79 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 20 QLATKAARKSAPATGGVKKPHRYRPGTVAL#REIRRYQKSTELLIRKLPFQRLVREIAQDF 79 |
| Notomastus torquatus | Query 20 QLATKAARKSAPATGGVKKPHRYRPGTVAL#REIRRYQKSTELLIRKLPFQRLVREIAQDF 79 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 20 QLATKAARKSAPATGGVKKPHRYRPGTVAL#REIRRYQKSTELLIRKLPFQRLVREIAQDF 79 |
| Normanichthys crockeri | Query 20 QLATKAARKSAPATGGVKKPHRYRPGTVAL#REIRRYQKSTELLIRKLPFQRLVREIAQDF 79 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 20 QLATKAARKSAPATGGVKKPHRYRPGTVAL#REIRRYQKSTELLIRKLPFQRLVREIAQDF 79 |
| Nipponentomon sp. | Query 20 QLATKAARKSAPATGGVKKPHRYRPGTVAL#REIRRYQKSTELLIRKLPFQRLVREIAQDF 79 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 20 QLATKAARKSAPATGGVKKPHRYRPGTVAL#REIRRYQKSTELLIRKLPFQRLVREIAQDF 79 |
| Agathemera crassa | Query 20 QLATKAARKSAPATGGVKKPHRYRPGTVAL#REIRRYQKSTELLIRKLPFQRLVREIAQDF 79 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 20 QLATKAARKSAPATGGVKKPHRYRPGTVAL#REIRRYQKSTELLIRKLPFQRLVREIAQDF 79 |
| Neohirasea sp. WS29 | Query 20 QLATKAARKSAPATGGVKKPHRYRPGTVAL#REIRRYQKSTELLIRKLPFQRLVREIAQDF 79 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 20 QLATKAARKSAPATGGVKKPHRYRPGTVAL#REIRRYQKSTELLIRKLPFQRLVREIAQDF 79 |
| Marukawichthys ambulator | Query 20 QLATKAARKSAPATGGVKKPHRYRPGTVAL#REIRRYQKSTELLIRKLPFQRLVREIAQDF 79 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 20 QLATKAARKSAPATGGVKKPHRYRPGTVAL#REIRRYQKSTELLIRKLPFQRLVREIAQDF 79 |
| Amphicteis dalmatica | Query 20 QLATKAARKSAPATGGVKKPHRYRPGTVAL#REIRRYQKSTELLIRKLPFQRLVREIAQDF 79 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 20 QLATKAARKSAPATGGVKKPHRYRPGTVAL#REIRRYQKSTELLIRKLPFQRLVREIAQDF 79 |
| Liorhina sp. JRC-2004 | Query 20 QLATKAARKSAPATGGVKKPHRYRPGTVAL#REIRRYQKSTELLIRKLPFQRLVREIAQDF 79 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 20 QLATKAARKSAPATGGVKKPHRYRPGTVAL#REIRRYQKSTELLIRKLPFQRLVREIAQDF 79 |
| Lepyronia quadrangularis | Query 20 QLATKAARKSAPATGGVKKPHRYRPGTVAL#REIRRYQKSTELLIRKLPFQRLVREIAQDF 79 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 20 QLATKAARKSAPATGGVKKPHRYRPGTVAL#REIRRYQKSTELLIRKLPFQRLVREIAQDF 79 |
| Leiognathus fasciatus | Query 20 QLATKAARKSAPATGGVKKPHRYRPGTVAL#REIRRYQKSTELLIRKLPFQRLVREIAQDF 79 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 20 QLATKAARKSAPATGGVKKPHRYRPGTVAL#REIRRYQKSTELLIRKLPFQRLVREIAQDF 79 |
| Lancetes varius | Query 20 QLATKAARKSAPATGGVKKPHRYRPGTVAL#REIRRYQKSTELLIRKLPFQRLVREIAQDF 79 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 20 QLATKAARKSAPATGGVKKPHRYRPGTVAL#REIRRYQKSTELLIRKLPFQRLVREIAQDF 79 |
| Labrisomus multiporosus | Query 20 QLATKAARKSAPATGGVKKPHRYRPGTVAL#REIRRYQKSTELLIRKLPFQRLVREIAQDF 79 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 20 QLATKAARKSAPATGGVKKPHRYRPGTVAL#REIRRYQKSTELLIRKLPFQRLVREIAQDF 79 |
| Anatoma euglypta | Query 20 QLATKAARKSAPATGGVKKPHRYRPGTVAL#REIRRYQKSTELLIRKLPFQRLVREIAQDF 79 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 20 QLATKAARKSAPATGGVKKPHRYRPGTVAL#REIRRYQKSTELLIRKLPFQRLVREIAQDF 79 |
| Aphredoderus sayanus | Query 20 QLATKAARKSAPATGGVKKPHRYRPGTVAL#REIRRYQKSTELLIRKLPFQRLVREIAQDF 79 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 20 QLATKAARKSAPATGGVKKPHRYRPGTVAL#REIRRYQKSTELLIRKLPFQRLVREIAQDF 79 |
| Hygrobia maculata | Query 20 QLATKAARKSAPATGGVKKPHRYRPGTVAL#REIRRYQKSTELLIRKLPFQRLVREIAQDF 79 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 20 QLATKAARKSAPATGGVKKPHRYRPGTVAL#REIRRYQKSTELLIRKLPFQRLVREIAQDF 79 |
| Hygrobia hermanni | Query 20 QLATKAARKSAPATGGVKKPHRYRPGTVAL#REIRRYQKSTELLIRKLPFQRLVREIAQDF 79 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 20 QLATKAARKSAPATGGVKKPHRYRPGTVAL#REIRRYQKSTELLIRKLPFQRLVREIAQDF 79 |
| Hygrobia australasiae | Query 20 QLATKAARKSAPATGGVKKPHRYRPGTVAL#REIRRYQKSTELLIRKLPFQRLVREIAQDF 79 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 20 QLATKAARKSAPATGGVKKPHRYRPGTVAL#REIRRYQKSTELLIRKLPFQRLVREIAQDF 79 |
| Hubrechtella dubia | Query 20 QLATKAARKSAPATGGVKKPHRYRPGTVAL#REIRRYQKSTELLIRKLPFQRLVREIAQDF 79 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 20 QLATKAARKSAPATGGVKKPHRYRPGTVAL#REIRRYQKSTELLIRKLPFQRLVREIAQDF 79 |
| Homalodisca elongata | Query 20 QLATKAARKSAPATGGVKKPHRYRPGTVAL#REIRRYQKSTELLIRKLPFQRLVREIAQDF 79 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 20 QLATKAARKSAPATGGVKKPHRYRPGTVAL#REIRRYQKSTELLIRKLPFQRLVREIAQDF 79 |
| Haplognathia gubbarnorum | Query 20 QLATKAARKSAPATGGVKKPHRYRPGTVAL#REIRRYQKSTELLIRKLPFQRLVREIAQDF 79 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 20 QLATKAARKSAPATGGVKKPHRYRPGTVAL#REIRRYQKSTELLIRKLPFQRLVREIAQDF 79 |
| Gratidia hispidula | Query 20 QLATKAARKSAPATGGVKKPHRYRPGTVAL#REIRRYQKSTELLIRKLPFQRLVREIAQDF 79 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 20 QLATKAARKSAPATGGVKKPHRYRPGTVAL#REIRRYQKSTELLIRKLPFQRLVREIAQDF 79 |
| Gobiocichla ethelwynnae | Query 20 QLATKAARKSAPATGGVKKPHRYRPGTVAL#REIRRYQKSTELLIRKLPFQRLVREIAQDF 79 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 20 QLATKAARKSAPATGGVKKPHRYRPGTVAL#REIRRYQKSTELLIRKLPFQRLVREIAQDF 79 |
| Glycera sp. AMW196835 | Query 20 QLATKAARKSAPATGGVKKPHRYRPGTVAL#REIRRYQKSTELLIRKLPFQRLVREIAQDF 79 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 20 QLATKAARKSAPATGGVKKPHRYRPGTVAL#REIRRYQKSTELLIRKLPFQRLVREIAQDF 79 |
| Architeuthis dux | Query 20 QLATKAARKSAPATGGVKKPHRYRPGTVAL#REIRRYQKSTELLIRKLPFQRLVREIAQDF 79 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 20 QLATKAARKSAPATGGVKKPHRYRPGTVAL#REIRRYQKSTELLIRKLPFQRLVREIAQDF 79 |
| Euchlanis alata | Query 20 QLATKAARKSAPATGGVKKPHRYRPGTVAL#REIRRYQKSTELLIRKLPFQRLVREIAQDF 79 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 20 QLATKAARKSAPATGGVKKPHRYRPGTVAL#REIRRYQKSTELLIRKLPFQRLVREIAQDF 79 |
| Erythemis sp. OD024 | Query 20 QLATKAARKSAPATGGVKKPHRYRPGTVAL#REIRRYQKSTELLIRKLPFQRLVREIAQDF 79 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 20 QLATKAARKSAPATGGVKKPHRYRPGTVAL#REIRRYQKSTELLIRKLPFQRLVREIAQDF 79 |
| Elates ransonnetii | Query 20 QLATKAARKSAPATGGVKKPHRYRPGTVAL#REIRRYQKSTELLIRKLPFQRLVREIAQDF 79 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 20 QLATKAARKSAPATGGVKKPHRYRPGTVAL#REIRRYQKSTELLIRKLPFQRLVREIAQDF 79 |
| Elassoneuria sp. EP167 | Query 20 QLATKAARKSAPATGGVKKPHRYRPGTVAL#REIRRYQKSTELLIRKLPFQRLVREIAQDF 79 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 20 QLATKAARKSAPATGGVKKPHRYRPGTVAL#REIRRYQKSTELLIRKLPFQRLVREIAQDF 79 |
| Austrophlebiodes sp. EP027 | Query 20 QLATKAARKSAPATGGVKKPHRYRPGTVAL#REIRRYQKSTELLIRKLPFQRLVREIAQDF 79 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 20 QLATKAARKSAPATGGVKKPHRYRPGTVAL#REIRRYQKSTELLIRKLPFQRLVREIAQDF 79 |
| Cyphophthalmus minutus | Query 20 QLATKAARKSAPATGGVKKPHRYRPGTVAL#REIRRYQKSTELLIRKLPFQRLVREIAQDF 79 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 20 QLATKAARKSAPATGGVKKPHRYRPGTVAL#REIRRYQKSTELLIRKLPFQRLVREIAQDF 79 |
| Battigrassiella wheeleri | Query 20 QLATKAARKSAPATGGVKKPHRYRPGTVAL#REIRRYQKSTELLIRKLPFQRLVREIAQDF 79 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 20 QLATKAARKSAPATGGVKKPHRYRPGTVAL#REIRRYQKSTELLIRKLPFQRLVREIAQDF 79 |
| Ctenomorphodes briareus | Query 20 QLATKAARKSAPATGGVKKPHRYRPGTVAL#REIRRYQKSTELLIRKLPFQRLVREIAQDF 79 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 20 QLATKAARKSAPATGGVKKPHRYRPGTVAL#REIRRYQKSTELLIRKLPFQRLVREIAQDF 79 |
| Craterostigmus tasmanianus | Query 20 QLATKAARKSAPATGGVKKPHRYRPGTVAL#REIRRYQKSTELLIRKLPFQRLVREIAQDF 79 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 20 QLATKAARKSAPATGGVKKPHRYRPGTVAL#REIRRYQKSTELLIRKLPFQRLVREIAQDF 79 |
| Cottus poecilopus | Query 20 QLATKAARKSAPATGGVKKPHRYRPGTVAL#REIRRYQKSTELLIRKLPFQRLVREIAQDF 79 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 20 QLATKAARKSAPATGGVKKPHRYRPGTVAL#REIRRYQKSTELLIRKLPFQRLVREIAQDF 79 |
| Cosmoscarta sp. JRC-2004 | Query 20 QLATKAARKSAPATGGVKKPHRYRPGTVAL#REIRRYQKSTELLIRKLPFQRLVREIAQDF 79 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 20 QLATKAARKSAPATGGVKKPHRYRPGTVAL#REIRRYQKSTELLIRKLPFQRLVREIAQDF 79 |
| Coloburiscoides sp. EP094 | Query 20 QLATKAARKSAPATGGVKKPHRYRPGTVAL#REIRRYQKSTELLIRKLPFQRLVREIAQDF 79 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 20 QLATKAARKSAPATGGVKKPHRYRPGTVAL#REIRRYQKSTELLIRKLPFQRLVREIAQDF 79 |
| Clypeosectus sp. CET-2005 | Query 20 QLATKAARKSAPATGGVKKPHRYRPGTVAL#REIRRYQKSTELLIRKLPFQRLVREIAQDF 79 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 20 QLATKAARKSAPATGGVKKPHRYRPGTVAL#REIRRYQKSTELLIRKLPFQRLVREIAQDF 79 |
| Clastoptera obtusa | Query 20 QLATKAARKSAPATGGVKKPHRYRPGTVAL#REIRRYQKSTELLIRKLPFQRLVREIAQDF 79 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 20 QLATKAARKSAPATGGVKKPHRYRPGTVAL#REIRRYQKSTELLIRKLPFQRLVREIAQDF 79 |
| Chilara taylori | Query 20 QLATKAARKSAPATGGVKKPHRYRPGTVAL#REIRRYQKSTELLIRKLPFQRLVREIAQDF 79 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 20 QLATKAARKSAPATGGVKKPHRYRPGTVAL#REIRRYQKSTELLIRKLPFQRLVREIAQDF 79 |
| Chaquihua sp. EP080 | Query 20 QLATKAARKSAPATGGVKKPHRYRPGTVAL#REIRRYQKSTELLIRKLPFQRLVREIAQDF 79 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 20 QLATKAARKSAPATGGVKKPHRYRPGTVAL#REIRRYQKSTELLIRKLPFQRLVREIAQDF 79 |
| Cuerna gladiola | Query 20 QLATKAARKSAPATGGVKKPHRYRPGTVAL#REIRRYQKSTELLIRKLPFQRLVREIAQDF 79 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 20 QLATKAARKSAPATGGVKKPHRYRPGTVAL#REIRRYQKSTELLIRKLPFQRLVREIAQDF 79 |
| Drosophila virilis | Query 20 QLATKAARKSAPATGGVKKPHRYRPGTVAL#REIRRYQKSTELLIRKLPFQRLVREIAQDF 79 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 20 QLATKAARKSAPATGGVKKPHRYRPGTVAL#REIRRYQKSTELLIRKLPFQRLVREIAQDF 79 |
| Aspidosiphon fischeri | Query 20 QLATKAARKSAPATGGVKKPHRYRPGTVAL#REIRRYQKSTELLIRKLPFQRLVREIAQDF 79 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 20 QLATKAARKSAPATGGVKKPHRYRPGTVAL#REIRRYQKSTELLIRKLPFQRLVREIAQDF 79 |
| Evexus n. sp. JRC-2004-r | Query 20 QLATKAARKSAPATGGVKKPHRYRPGTVAL#REIRRYQKSTELLIRKLPFQRLVREIAQDF 79 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 20 QLATKAARKSAPATGGVKKPHRYRPGTVAL#REIRRYQKSTELLIRKLPFQRLVREIAQDF 79 |
| Aplysia cf. juliana | Query 20 QLATKAARKSAPATGGVKKPHRYRPGTVAL#REIRRYQKSTELLIRKLPFQRLVREIAQDF 79 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 20 QLATKAARKSAPATGGVKKPHRYRPGTVAL#REIRRYQKSTELLIRKLPFQRLVREIAQDF 79 |
| Idanthyrsus pennatus | Query 20 QLATKAARKSAPATGGVKKPHRYRPGTVAL#REIRRYQKSTELLIRKLPFQRLVREIAQDF 79 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 20 QLATKAARKSAPATGGVKKPHRYRPGTVAL#REIRRYQKSTELLIRKLPFQRLVREIAQDF 79 |
| Ischnochiton australis | Query 20 QLATKAARKSAPATGGVKKPHRYRPGTVAL#REIRRYQKSTELLIRKLPFQRLVREIAQDF 79 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 20 QLATKAARKSAPATGGVKKPHRYRPGTVAL#REIRRYQKSTELLIRKLPFQRLVREIAQDF 79 |
| Lysmata debelius | Query 20 QLATKAARKSAPATGGVKKPHRYRPGTVAL#REIRRYQKSTELLIRKLPFQRLVREIAQDF 79 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 20 QLATKAARKSAPATGGVKKPHRYRPGTVAL#REIRRYQKSTELLIRKLPFQRLVREIAQDF 79 |
| Nerita atramentosa | Query 20 QLATKAARKSAPATGGVKKPHRYRPGTVAL#REIRRYQKSTELLIRKLPFQRLVREIAQDF 79 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 20 QLATKAARKSAPATGGVKKPHRYRPGTVAL#REIRRYQKSTELLIRKLPFQRLVREIAQDF 79 |
| Onchidium sp. | Query 20 QLATKAARKSAPATGGVKKPHRYRPGTVAL#REIRRYQKSTELLIRKLPFQRLVREIAQDF 79 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 20 QLATKAARKSAPATGGVKKPHRYRPGTVAL#REIRRYQKSTELLIRKLPFQRLVREIAQDF 79 |
| Aegla abtao | Query 20 QLATKAARKSAPATGGVKKPHRYRPGTVAL#REIRRYQKSTELLIRKLPFQRLVREIAQDF 79 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 20 QLATKAARKSAPATGGVKKPHRYRPGTVAL#REIRRYQKSTELLIRKLPFQRLVREIAQDF 79 |
| Robertus neglectus | Query 20 QLATKAARKSAPATGGVKKPHRYRPGTVAL#REIRRYQKSTELLIRKLPFQRLVREIAQDF 79 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 20 QLATKAARKSAPATGGVKKPHRYRPGTVAL#REIRRYQKSTELLIRKLPFQRLVREIAQDF 79 |
[Site 7] LCAIHAKRVT119-IMPKDIQLAR
Thr119  Ile
Ile
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Leu110 | Cys111 | Ala112 | Ile113 | His114 | Ala115 | Lys116 | Arg117 | Val118 | Thr119 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Ile120 | Met121 | Pro122 | Lys123 | Asp124 | Ile125 | Gln126 | Leu127 | Ala128 | Arg129 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| VMALQEASEAYLVGLFEDTNLCAIHAKRVTIMPKDIQLARRIRGERA |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | N/A | 100.00 | 26 | histone H3 |
| 2 | Mus musculus | 99.80 | 10 | histone cluster 1, H3f |
| 3 | Bos taurus | 99.80 | 4 | PREDICTED: similar to histone protein Hist2h3c1 |
| 4 | Strongylocentrotus purpuratus | 99.80 | 3 | PREDICTED: hypothetical protein |
| 5 | Anopheles gambiae str. PEST | 99.80 | 2 | AGAP012709-PA |
| 6 | Drosophila erecta | 99.80 | 1 | histone H3 |
| 7 | Cricetulus longicaudatus | 99.80 | 1 | H32_CRILO Histone H3.2 gi |
| 8 | Drosophila pseudoobscura | 99.40 | 1 | histone 3 |
| 9 | Canis familiaris | 99.00 | 5 | PREDICTED: similar to H3 histone, family 2 isofor |
| 10 | Danio rerio | 99.00 | 2 | hypothetical protein LOC794406 |
| 11 | Drosophila melanogaster | 99.00 | 1 | H3 histone |
| 12 | Xenopus laevis | 98.60 | 3 | unnamed protein product |
| 13 | Tetraodon nigroviridis | 98.60 | 1 | unnamed protein product |
| 14 | Mus musculus domesticus | 98.60 | 1 | histone H3 (partial) |
| 15 | Caenorhabditis elegans | 98.20 | 4 | HIStone family member (his-42) |
| 16 | Acropora formosa | 98.20 | 1 | H3_ACRFO Histone H3 gi |
| 17 | Drosophila simulans | 98.20 | 1 | Histone H3 |
| 18 | Mus pahari | 97.80 | 1 | histone H3.2 protein |
| 19 | Mytilus chilensis | 97.80 | 1 | histone H3 |
| 20 | Plasmodium yoelii yoelii str. 17XNL | 97.40 | 2 | histone 3 |
| 21 | Oikopleura dioica | 97.40 | 1 | histone h3.1 |
| 22 | Homo sapiens | 97.10 | 4 | histone cluster 1, H3a |
| 23 | Caenorhabditis briggsae AF16 | 97.10 | 4 | Hypothetical protein CBG01505 |
| 24 | Expression vector pET3-H3 | 97.10 | 1 | Xenopus laevis-like histone H3 |
| 25 | Drosophila persimilis | 97.10 | 1 | histone 3 |
| 26 | Lilium longiflorum | 96.70 | 5 | H33A_LILLO Histone H3.3a (Somatic-like histone H3 |
| 27 | Arabidopsis thaliana | 96.70 | 3 | histone H3.2 |
| 28 | Chlamydomonas reinhardtii | 96.70 | 1 | histone H3 |
| 29 | Trichinella spiralis | 96.70 | 1 | histone H3 |
| 30 | Medicago sativa | 96.30 | 5 | histone H3.2 |
| 31 | Xenopus tropicalis | 96.30 | 1 | H3 histone |
| 32 | Strongylocentrotus purpuratus | 96.30 | 1 | late embryonic histone H3 |
| 33 | Styela plicata | 96.30 | 1 | H3 histone |
| 34 | Arabidopsis thaliana | 95.90 | 3 | histone H3.2, putative |
| 35 | Venerupis (Ruditapes) philippinarum | 95.90 | 2 | replacement histone H3.3 |
| 36 | Tortula ruralis | 95.90 | 1 | AF093108_1 histone H3 |
| 37 | Gallus gallus | 95.90 | 1 | H3 histone, family 3A |
| 38 | Drosophila virilis | 95.90 | 1 | histone H3 |
| 39 | Pongo pygmaeus | 95.90 | 1 | H33_PONPY Histone H3.3 gi |
| 40 | Rattus norvegicus | 95.90 | 1 | hypothetical protein LOC691496 |
| 41 | Eimeria tenella | 95.50 | 1 | AF361949_1 histone 3 |
| 42 | Toxoplasma gondii | 95.50 | 1 | histone H3 |
| 43 | Cichorium intybus | 95.50 | 1 | H32_CICIN Histone H3.2 gi |
| 44 | Beta vulgaris subsp. vulgaris | 95.50 | 1 | histone H3 |
| 45 | Craterostigma plantagineum | 95.10 | 1 | histone H3.3 |
| 46 | Mytilus californianus | 95.10 | 1 | histone H3 |
| 47 | Toxoplasma gondii | 95.10 | 1 | histone H3.3 variant; TgH3.3 |
| 48 | Oryza sativa | 95.10 | 1 | AF467728_1 disease-resistent-related protein |
| 49 | Entodinium caudatum | 94.70 | 2 | histone H3 |
| 50 | Fucus serratus | 94.70 | 1 | histone H3 |
| 51 | Griffithsia japonica | 94.70 | 1 | putative histone H3 protein |
| 52 | Platichthys flesus | 94.70 | 1 | histone H3.3 |
| 53 | Griffithsia japonica | 94.70 | 1 | H3_GRIJA Histone H3 gi |
| 54 | Picea abies | 94.40 | 1 | histone 3 |
| 55 | Nematostella vectensis | 94.40 | 1 | predicted protein |
| 56 | Theileria annulata strain Ankara | 94.00 | 1 | histone H3, putative |
| 57 | synthetic construct | 94.00 | 1 | histone 3 H3 |
| 58 | Theileria parva strain Muguga | 94.00 | 1 | histone H3 |
| 59 | Cryptosporidium parvum Iowa II | 94.00 | 1 | histone H3 |
| 60 | Cryptosporidium hominis TU502 | 93.60 | 1 | H3 histone, family 2; histone 2, H3ca1 |
| 61 | Rheum australe | 93.60 | 1 | histone 3 |
| 62 | Aquilegia formosa | 92.80 | 1 | histone H3 |
Top-ranked sequences
| organism | matching |
|---|---|
| N/A | Query 90 VMALQEASEAYLVGLFEDTNLCAIHAKRVT#IMPKDIQLARRIRGERA 136 ||||||||||||||||||||||||||||||#||||||||||||||||| Query 90 VMALQEASEAYLVGLFEDTNLCAIHAKRVT#IMPKDIQLARRIRGERA 136 |
| Mus musculus | Query 90 VMALQEASEAYLVGLFEDTNLCAIHAKRVT#IMPKDIQLARRIRGERA 136 ||||||||||||||||||||||||||||||#||||||||||||||||| Query 90 VMALQEASEAYLVGLFEDTNLCAIHAKRVT#IMPKDIQLARRIRGERA 136 |
| Bos taurus | Query 90 VMALQEASEAYLVGLFEDTNLCAIHAKRVT#IMPKDIQLARRIRGERA 136 ||||||||||||||||||||||||||||||#||||||||||||||||| Query 90 VMALQEASEAYLVGLFEDTNLCAIHAKRVT#IMPKDIQLARRIRGERA 136 |
| Strongylocentrotus purpuratus | Query 90 VMALQEASEAYLVGLFEDTNLCAIHAKRVT#IMPKDIQLARRIRGERA 136 ||||||||||||||||||||||||||||||#||||||||||||||||| Query 90 VMALQEASEAYLVGLFEDTNLCAIHAKRVT#IMPKDIQLARRIRGERA 136 |
| Anopheles gambiae str. PEST | Query 90 VMALQEASEAYLVGLFEDTNLCAIHAKRVT#IMPKDIQLARRIRGERA 136 ||||||||||||||||||||||||||||||#||||||||||||||||| Query 90 VMALQEASEAYLVGLFEDTNLCAIHAKRVT#IMPKDIQLARRIRGERA 136 |
| Drosophila erecta | Query 90 VMALQEASEAYLVGLFEDTNLCAIHAKRVT#IMPKDIQLARRIRGERA 136 ||||||||||||||||||||||||||||||#||||||||||||||||| Query 90 VMALQEASEAYLVGLFEDTNLCAIHAKRVT#IMPKDIQLARRIRGERA 136 |
| Cricetulus longicaudatus | Query 90 VMALQEASEAYLVGLFEDTNLCAIHAKRVT#IMPKDIQLARRIRGERA 136 ||||||||||||||||||||||||||||||#||||||||||||||||| Query 90 VMALQEASEAYLVGLFEDTNLCAIHAKRVT#IMPKDIQLARRIRGERA 136 |
| Drosophila pseudoobscura | Query 90 VMALQEASEAYLVGLFEDTNLCAIHAKRVT#IMPKDIQLARRIRGERA 136 ||||||||||||||||||||||||||||||#||||||||||||||||| Query 90 VMALQEASEAYLVGLFEDTNLCAIHAKRVT#IMPKDIQLARRIRGERA 136 |
| Canis familiaris | Query 90 VMALQEASEAYLVGLFEDTNLCAIHAKRVT#IMPKDIQLARRIRGERA 136 ||||||||||||||||||||||||||||||#||||||||||||||||| Query 90 VMALQEASEAYLVGLFEDTNLCAIHAKRVT#IMPKDIQLARRIRGERA 136 |
| Danio rerio | Query 90 VMALQEASEAYLVGLFEDTNLCAIHAKRVT#IMPKDIQLARRIRGERA 136 ||||||+|||||||||||||||||||||||#||||||||||||||||| Sbjct 90 VMALQESSEAYLVGLFEDTNLCAIHAKRVT#IMPKDIQLARRIRGERA 136 |
| Drosophila melanogaster | Query 90 VMALQEASEAYLVGLFEDTNLCAIHAKRVT#IMPKDIQLARRIRGERA 136 ||||||||||||||||||||||||||||+|#||||||||||||||||| Sbjct 90 VMALQEASEAYLVGLFEDTNLCAIHAKRIT#IMPKDIQLARRIRGERA 136 |
| Xenopus laevis | Query 90 VMALQEASEAYLVGLFEDTNLCAIHAKRVT#IMPKDIQLARRIRGERA 136 ||||||||||||||||||||||||||||||#||||||||||||||||| Query 90 VMALQEASEAYLVGLFEDTNLCAIHAKRVT#IMPKDIQLARRIRGERA 136 |
| Tetraodon nigroviridis | Query 90 VMALQEASEAYLVGLFEDTNLCAIHAKRVT#IMPKDIQLARRIRGERA 136 ||||||||||||||||||||||||||||||#||||||||||||||||| Query 90 VMALQEASEAYLVGLFEDTNLCAIHAKRVT#IMPKDIQLARRIRGERA 136 |
| Mus musculus domesticus | Query 90 VMALQEASEAYLVGLFEDTNLCAIHAKRVT#IMPKDIQLARRIRGERA 136 ||||||||||||||||||||||||||||||#||||||||||||||||| Query 90 VMALQEASEAYLVGLFEDTNLCAIHAKRVT#IMPKDIQLARRIRGERA 136 |
| Caenorhabditis elegans | Query 90 VMALQEASEAYLVGLFEDTNLCAIHAKRVT#IMPKDIQLARRIRGERA 136 |||||||+||||||||||||||||||||||#||||||||||||||||| Sbjct 90 VMALQEAAEAYLVGLFEDTNLCAIHAKRVT#IMPKDIQLARRIRGERA 136 |
| Acropora formosa | Query 90 VMALQEASEAYLVGLFEDTNLCAIHAKRVT#IMPKDIQLARRIRGERA 136 |+||||||||||||||||||||||||||||#||||||||||||||||| Sbjct 90 VLALQEASEAYLVGLFEDTNLCAIHAKRVT#IMPKDIQLARRIRGERA 136 |
| Drosophila simulans | Query 90 VMALQEASEAYLVGLFEDTNLCAIHAKRVT#IMPKDIQLARRIRGERA 136 ||||||||| ||||||||||||||||||||#||||||||||||||||| Sbjct 90 VMALQEASETYLVGLFEDTNLCAIHAKRVT#IMPKDIQLARRIRGERA 136 |
| Mus pahari | Query 90 VMALQEASEAYLVGLFEDTNLCAIHAKRVT#IMPKDIQLARRIRGERA 136 ||||||||||||||||||||||||||||||#|||||||| |||||||| Sbjct 90 VMALQEASEAYLVGLFEDTNLCAIHAKRVT#IMPKDIQLVRRIRGERA 136 |
| Mytilus chilensis | Query 90 VMALQEASEAYLVGLFEDTNLCAIHAKRVT#IMPKDIQLARRIRGERA 136 ||||||||||||||||||||||||||||||#|||||||| |||||||| Sbjct 90 VMALQEASEAYLVGLFEDTNLCAIHAKRVT#IMPKDIQLVRRIRGERA 136 |
| Plasmodium yoelii yoelii str. 17XNL | Query 90 VMALQEASEAYLVGLFEDTNLCAIHAKRVT#IMPKDIQLARRIRGERA 136 |||||||+||||||||||||||||||||||#||||||||||||||||+ Sbjct 90 VMALQEAAEAYLVGLFEDTNLCAIHAKRVT#IMPKDIQLARRIRGERS 136 |
| Oikopleura dioica | Query 90 VMALQEASEAYLVGLFEDTNLCAIHAKRVT#IMPKDIQLARRIRGERA 136 |||||||||||||||||||+||||||||||#||||||||||||||||| Sbjct 90 VMALQEASEAYLVGLFEDTHLCAIHAKRVT#IMPKDIQLARRIRGERA 136 |
| Homo sapiens | Query 90 VMALQEASEAYLVGLFEDTNLCAIHAKRVT#IMPKDIQLARRIRGERA 136 ||||||| ||||||||||||||||||||||#||||||||||||||||| Sbjct 90 VMALQEACEAYLVGLFEDTNLCAIHAKRVT#IMPKDIQLARRIRGERA 136 |
| Caenorhabditis briggsae AF16 | Query 90 VMALQEASEAYLVGLFEDTNLCAIHAKRVT#IMPKDIQLARRIRGERA 136 ||||||++||||||||||||||||||||||#||||||||||||||||| Sbjct 90 VMALQESAEAYLVGLFEDTNLCAIHAKRVT#IMPKDIQLARRIRGERA 136 |
| Expression vector pET3-H3 | Query 90 VMALQEASEAYLVGLFEDTNLCAIHAKRVT#IMPKDIQLARRIRGERA 136 ||||||||||||| ||||||||||||||||#||||||||||||||||| Sbjct 90 VMALQEASEAYLVALFEDTNLCAIHAKRVT#IMPKDIQLARRIRGERA 136 |
| Drosophila persimilis | Query 90 VMALQEASEAYLVGLFEDTNLCAIHAKRVT#IMPKDIQLARRIRGERA 136 ||||||||||||||||||||||||||||||#|||| |||||||||||| Sbjct 90 VMALQEASEAYLVGLFEDTNLCAIHAKRVT#IMPKGIQLARRIRGERA 136 |
| Lilium longiflorum | Query 90 VMALQEASEAYLVGLFEDTNLCAIHAKRVT#IMPKDIQLARRIRGERA 136 |+|||||+||||||||||||||||||||||#||||||||||||||||| Sbjct 90 VLALQEAAEAYLVGLFEDTNLCAIHAKRVT#IMPKDIQLARRIRGERA 136 |
| Arabidopsis thaliana | Query 90 VMALQEASEAYLVGLFEDTNLCAIHAKRVT#IMPKDIQLARRIRGERA 136 |+|||||+||||||||||||||||||||||#||||||||||||||||| Sbjct 90 VLALQEAAEAYLVGLFEDTNLCAIHAKRVT#IMPKDIQLARRIRGERA 136 |
| Chlamydomonas reinhardtii | Query 90 VMALQEASEAYLVGLFEDTNLCAIHAKRVT#IMPKDIQLARRIRGERA 136 |+|||||+||||||||||||||||||||||#||||||||||||||||| Sbjct 89 VLALQEAAEAYLVGLFEDTNLCAIHAKRVT#IMPKDIQLARRIRGERA 135 |
| Trichinella spiralis | Query 90 VMALQEASEAYLVGLFEDTNLCAIHAKRVT#IMPKDIQLARRIRGERA 136 |+|||||+||||||||||||||||||||||#||||||||||||||||| Sbjct 90 VLALQEAAEAYLVGLFEDTNLCAIHAKRVT#IMPKDIQLARRIRGERA 136 |
| Medicago sativa | Query 90 VMALQEASEAYLVGLFEDTNLCAIHAKRVT#IMPKDIQLARRIRGERA 136 |+|||||+||||||||||||||||||||||#||||||||||||||||| Sbjct 81 VLALQEAAEAYLVGLFEDTNLCAIHAKRVT#IMPKDIQLARRIRGERA 127 |
| Xenopus tropicalis | Query 90 VMALQEASEAYLVGLFEDTNLCAIHAKRVT#IMPKDIQLARRIRGERA 136 + ||||||||||||||||||||||||||||#||||||||||||||||| Sbjct 90 IGALQEASEAYLVGLFEDTNLCAIHAKRVT#IMPKDIQLARRIRGERA 136 |
| Strongylocentrotus purpuratus | Query 90 VMALQEASEAYLVGLFEDTNLCAIHAKRVT#IMPKDIQLARRIRGERA 136 ||||||||||||| ||||||||||||||||#||||||||||||||||| Sbjct 90 VMALQEASEAYLVRLFEDTNLCAIHAKRVT#IMPKDIQLARRIRGERA 136 |
| Styela plicata | Query 90 VMALQEASEAYLVGLFEDTNLCAIHAKRVT#IMPKDIQLARRIRGERA 136 ||||||||| ||||||||||||||||||||#|||||||| |||||||| Sbjct 90 VMALQEASERYLVGLFEDTNLCAIHAKRVT#IMPKDIQLGRRIRGERA 136 |
| Arabidopsis thaliana | Query 90 VMALQEASEAYLVGLFEDTNLCAIHAKRVT#IMPKDIQLARRIRGERA 136 |+|||||+||||||||||||||||||||||#|||||+||||||||||| Sbjct 90 VLALQEAAEAYLVGLFEDTNLCAIHAKRVT#IMPKDVQLARRIRGERA 136 |
| Venerupis (Ruditapes) philippinarum | Query 90 VMALQEASEAYLVGLFEDTNLCAIHAKRVT#IMPKDIQLARRIRGERA 136 + ||||||||||||||||||||||||||||#||||||||||||||||| Sbjct 90 IGALQEASEAYLVGLFEDTNLCAIHAKRVT#IMPKDIQLARRIRGERA 136 |
| Tortula ruralis | Query 90 VMALQEASEAYLVGLFEDTNLCAIHAKRVT#IMPKDIQLARRIRGERA 136 |+|||||+||||||||||||||||||||||#||||||||||||||||| Sbjct 71 VLALQEAAEAYLVGLFEDTNLCAIHAKRVT#IMPKDIQLARRIRGERA 117 |
| Gallus gallus | Query 90 VMALQEASEAYLVGLFEDTNLCAIHAKRVT#IMPKDIQLARRIRGERA 136 + ||||||||||||||||||||||||||||#||||||||||||||||| Sbjct 90 IGALQEASEAYLVGLFEDTNLCAIHAKRVT#IMPKDIQLARRIRGERA 136 |
| Drosophila virilis | Query 90 VMALQEASEAYLVGLFEDTNLCAIHAKRVT#IMPKDIQLARRIRGERA 136 ||||||||| ||||||||||||||||||||#||||||||||||||||| Sbjct 3 VMALQEASERYLVGLFEDTNLCAIHAKRVT#IMPKDIQLARRIRGERA 49 |
| Pongo pygmaeus | Query 90 VMALQEASEAYLVGLFEDTNLCAIHAKRVT#IMPKDIQLARRIRGERA 136 + ||||||||||||||||||||||||||||#||||||||||||||||| Sbjct 90 IGALQEASEAYLVGLFEDTNLCAIHAKRVT#IMPKDIQLARRIRGERA 136 |
| Rattus norvegicus | Query 90 VMALQEASEAYLVGLFEDTNLCAIHAKRVT#IMPKDIQLARRIRGERA 136 ||||||| |+||||||||||||||||||||#||||||||||||||||| Sbjct 90 VMALQEACESYLVGLFEDTNLCAIHAKRVT#IMPKDIQLARRIRGERA 136 |
| Eimeria tenella | Query 90 VMALQEASEAYLVGLFEDTNLCAIHAKRVT#IMPKDIQLARRIRGERA 136 |+|||||+||||||||||||||||||||||#||||||||||||||||+ Sbjct 90 VLALQEAAEAYLVGLFEDTNLCAIHAKRVT#IMPKDIQLARRIRGERS 136 |
| Toxoplasma gondii | Query 90 VMALQEASEAYLVGLFEDTNLCAIHAKRVT#IMPKDIQLARRIRGERA 136 |+|||||+||||||||||||||||||||||#||||||||||||||||+ Sbjct 90 VLALQEAAEAYLVGLFEDTNLCAIHAKRVT#IMPKDIQLARRIRGERS 136 |
| Cichorium intybus | Query 90 VMALQEASEAYLVGLFEDTNLCAIHAKRVT#IMPKDIQLARRIRGERA 136 | ||||||||||||||||||||||||||||#|||||+||||||||||| Sbjct 90 VAALQEASEAYLVGLFEDTNLCAIHAKRVT#IMPKDMQLARRIRGERA 136 |
| Beta vulgaris subsp. vulgaris | Query 90 VMALQEASEAYLVGLFEDTNLCAIHAKRVT#IMPKDIQLARRIRGERA 136 |+|||||+||||||||||||||||||||||#||||||||||||||||| Sbjct 52 VLALQEAAEAYLVGLFEDTNLCAIHAKRVT#IMPKDIQLARRIRGERA 98 |
| Craterostigma plantagineum | Query 90 VMALQEASEAYLVGLFEDTNLCAIHAKRVT#IMPKDIQLARRIRGERA 136 | |||||+||||||||||||||||||||||#||||||||||||||||| Sbjct 90 VGALQEAAEAYLVGLFEDTNLCAIHAKRVT#IMPKDIQLARRIRGERA 136 |
| Mytilus californianus | Query 90 VMALQEASEAYLVGLFEDTNLCAIHAKRVT#IMPKDIQLARRIRGERA 136 ||||||||||||||||||||| ||||||||#||||||||||||||||| Sbjct 90 VMALQEASEAYLVGLFEDTNLRAIHAKRVT#IMPKDIQLARRIRGERA 136 |
| Toxoplasma gondii | Query 90 VMALQEASEAYLVGLFEDTNLCAIHAKRVT#IMPKDIQLARRIRGERA 136 ++|||||+||||||||||||||||||||||#||||||||||||||||+ Sbjct 90 ILALQEAAEAYLVGLFEDTNLCAIHAKRVT#IMPKDIQLARRIRGERS 136 |
| Oryza sativa | Query 90 VMALQEASEAYLVGLFEDTNLCAIHAKRVT#IMPKDIQLARRIRGERA 136 |+|||||+||||||||||||||||||||||#||||||+|||||||||| Sbjct 90 VLALQEAAEAYLVGLFEDTNLCAIHAKRVT#IMPKDIKLARRIRGERA 136 |
| Entodinium caudatum | Query 90 VMALQEASEAYLVGLFEDTNLCAIHAKRVT#IMPKDIQLARRIRGER 135 |+|||||||+||||||||||||||||||||#|||||||||||||||+ Sbjct 89 VLALQEASESYLVGLFEDTNLCAIHAKRVT#IMPKDIQLARRIRGEK 134 |
| Fucus serratus | Query 90 VMALQEASEAYLVGLFEDTNLCAIHAKRVT#IMPKDIQLARRIRGERA 136 |+|||||+||||||||||||||||||||||#||||||||||||||||+ Sbjct 62 VLALQEAAEAYLVGLFEDTNLCAIHAKRVT#IMPKDIQLARRIRGERS 108 |
| Griffithsia japonica | Query 90 VMALQEASEAYLVGLFEDTNLCAIHAKRVT#IMPKDIQLARRIRGER 135 |+|||||+|||+||||||||||||||||||#|||||||||||||||| Sbjct 90 VLALQEAAEAYMVGLFEDTNLCAIHAKRVT#IMPKDIQLARRIRGER 135 |
| Platichthys flesus | Query 90 VMALQEASEAYLVGLFEDTNLCAIHAKRVT#IMPKDIQLARRIRGERA 136 + ||||||||||||||||||||||||||||#||||||||||||||||| Sbjct 50 IGALQEASEAYLVGLFEDTNLCAIHAKRVT#IMPKDIQLARRIRGERA 96 |
| Griffithsia japonica | Query 90 VMALQEASEAYLVGLFEDTNLCAIHAKRVT#IMPKDIQLARRIRGER 135 |+|||||+|||+||||||||||||||||||#|||||||||||||||| Sbjct 90 VLALQEAAEAYMVGLFEDTNLCAIHAKRVT#IMPKDIQLARRIRGER 135 |
| Picea abies | Query 90 VMALQEASEAYLVGLFEDTNLCAIHAKRVT#IMPKDIQLARRIRGERA 136 |+|||||+||||||||||||||||||||||#||||| ||||||||||| Sbjct 90 VLALQEAAEAYLVGLFEDTNLCAIHAKRVT#IMPKDXQLARRIRGERA 136 |
| Nematostella vectensis | Query 90 VMALQEASEAYLVGLFEDTNLCAIHAKRVT#IMPKDIQLARRIRGERA 136 + |||||+||||||||||||||||||||||#||||||||||||||||| Sbjct 90 IGALQEAAEAYLVGLFEDTNLCAIHAKRVT#IMPKDIQLARRIRGERA 136 |
| Theileria annulata strain Ankara | Query 90 VMALQEASEAYLVGLFEDTNLCAIHAKRVT#IMPKDIQLARRIRGERA 136 |+|||||+||||||||||||||||||||||#|||||+ |||||||||| Sbjct 90 VLALQEAAEAYLVGLFEDTNLCAIHAKRVT#IMPKDVHLARRIRGERA 136 |
| synthetic construct | Query 90 VMALQEASEAYLVGLFEDTNLCAIHAKRVT#IMPKDIQLARRIRGERA 136 ||||||| |+|||||||||||| |||||||#||||||||||||||||| Sbjct 90 VMALQEACESYLVGLFEDTNLCVIHAKRVT#IMPKDIQLARRIRGERA 136 |
| Theileria parva strain Muguga | Query 90 VMALQEASEAYLVGLFEDTNLCAIHAKRVT#IMPKDIQLARRIRGERA 136 |+|||||+||||||||||||||||||||||#|||||+ |||||||||| Sbjct 90 VLALQEAAEAYLVGLFEDTNLCAIHAKRVT#IMPKDVHLARRIRGERA 136 |
| Cryptosporidium parvum Iowa II | Query 90 VMALQEASEAYLVGLFEDTNLCAIHAKRVT#IMPKDIQLARRIRGER 135 |||||||+|||||||||||||||||| |||#|||||||||||||||| Sbjct 90 VMALQEAAEAYLVGLFEDTNLCAIHAHRVT#IMPKDIQLARRIRGER 135 |
| Cryptosporidium hominis TU502 | Query 90 VMALQEASEAYLVGLFEDTNLCAIHAKRVT#IMPKDIQLARRIRGER 135 |||||||+|||||||||||||||||| |||#|||||+|||||||||| Sbjct 90 VMALQEAAEAYLVGLFEDTNLCAIHAHRVT#IMPKDVQLARRIRGER 135 |
| Rheum australe | Query 90 VMALQEASEAYLVGLFEDTNLCAIHAKRVT#IMPKDIQLARRIRGERA 136 | |||||+|+||||||||||||||||||||#|||||+||||||||||| Sbjct 90 VAALQEAAESYLVGLFEDTNLCAIHAKRVT#IMPKDMQLARRIRGERA 136 |
| Aquilegia formosa | Query 90 VMALQEASEAYLVGLFEDTNLCAIHAKRVT#IMPKDIQLARRIRGERA 136 | ||||| ||||||||||||||||||||||#||||||||||||||||| Sbjct 26 VSALQEALEAYLVGLFEDTNLCAIHAKRVT#IMPKDIQLARRIRGERA 72 |
[Site 8] IMPKDIQLAR129-RIRGERA
Arg129  Arg
Arg
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Ile120 | Met121 | Pro122 | Lys123 | Asp124 | Ile125 | Gln126 | Leu127 | Ala128 | Arg129 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Arg130 | Ile131 | Arg132 | Gly133 | Glu134 | Arg135 | Ala136 | - | - | - |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| YLVGLFEDTNLCAIHAKRVTIMPKDIQLARRIRGERA |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | N/A | 81.30 | 23 | histone H3 |
| 2 | Xenopus laevis | 81.30 | 3 | histone H3 |
| 3 | Mus musculus | 80.90 | 10 | PREDICTED: hypothetical protein |
| 4 | Lilium longiflorum | 80.90 | 5 | H32_LILLO Histone H3.2 gi |
| 5 | Homo sapiens | 80.90 | 5 | H3 histone, family 3A |
| 6 | Caenorhabditis briggsae AF16 | 80.90 | 4 | Hypothetical protein CBG16863 |
| 7 | Caenorhabditis elegans | 80.90 | 4 | HIStone family member (his-42) |
| 8 | Bos taurus | 80.90 | 4 | PREDICTED: similar to histone protein Hist2h3c1 |
| 9 | Arabidopsis thaliana | 80.90 | 4 | histone H3 |
| 10 | Strongylocentrotus purpuratus | 80.90 | 3 | PREDICTED: similar to H3.3 histone |
| 11 | Danio rerio | 80.90 | 2 | hypothetical protein LOC794406 |
| 12 | Venerupis (Ruditapes) philippinarum | 80.90 | 2 | replacement histone H3.3 |
| 13 | Anopheles gambiae str. PEST | 80.90 | 2 | AGAP012197-PA |
| 14 | Craterostigma plantagineum | 80.90 | 1 | histone H3.3 |
| 15 | Drosophila erecta | 80.90 | 1 | histone H3 |
| 16 | Drosophila simulans | 80.90 | 1 | Histone H3 |
| 17 | Cricetulus longicaudatus | 80.90 | 1 | H32_CRILO Histone H3.2 gi |
| 18 | Trichinella spiralis | 80.90 | 1 | histone H3 |
| 19 | Xenopus tropicalis | 80.90 | 1 | H3 histone |
| 20 | Acropora formosa | 80.90 | 1 | H3_ACRFO Histone H3 gi |
| 21 | Pongo pygmaeus | 80.90 | 1 | H33_PONPY Histone H3.3 gi |
| 22 | Medicago sativa | 80.50 | 5 | histone H3.2 |
| 23 | Canis familiaris | 80.50 | 4 | PREDICTED: similar to histone 1, H2ai (predicted) |
| 24 | Chlamydomonas reinhardtii | 80.50 | 1 | histone H3 |
| 25 | Gallus gallus | 80.50 | 1 | H3 histone, family 3A |
| 26 | Nematostella vectensis | 80.50 | 1 | predicted protein |
| 27 | Drosophila pseudoobscura | 80.50 | 1 | histone 3 |
| 28 | Mastigamoeba balamuthi | 80.50 | 1 | H3_MASBA Histone H3 gi |
| 29 | Rattus norvegicus | 80.50 | 1 | hypothetical protein LOC691496 |
| 30 | Arabidopsis thaliana | 80.10 | 3 | histone H3.2, putative |
| 31 | Plasmodium yoelii yoelii str. 17XNL | 80.10 | 2 | histone 3 |
| 32 | Tortula ruralis | 80.10 | 1 | AF093108_1 histone H3 |
| 33 | Drosophila melanogaster | 80.10 | 1 | H3 histone |
| 34 | Mus musculus domesticus | 80.10 | 1 | histone H3 (partial) |
| 35 | Rheum australe | 80.10 | 1 | histone 3 |
| 36 | Tetraodon nigroviridis | 80.10 | 1 | unnamed protein product |
| 37 | Aquilegia formosa | 80.10 | 1 | histone H3 |
| 38 | Drosophila virilis | 79.70 | 1 | histone H3 |
| 39 | Cichorium intybus | 79.70 | 1 | H32_CICIN Histone H3.2 gi |
| 40 | Platichthys flesus | 79.70 | 1 | histone H3.3 |
| 41 | Eimeria tenella | 79.70 | 1 | AF361949_1 histone 3 |
| 42 | Toxoplasma gondii | 79.70 | 1 | histone H3 |
| 43 | Toxoplasma gondii | 79.70 | 1 | histone H3.3 variant; TgH3.3 |
| 44 | Beta vulgaris subsp. vulgaris | 79.30 | 1 | histone H3 |
| 45 | Styela plicata | 79.30 | 1 | H3 histone |
| 46 | Fucus serratus | 79.00 | 1 | histone H3 |
| 47 | Entamoeba histolytica HM-1:IMSS | 79.00 | 1 | histone H3, putative |
| 48 | Mus pahari | 79.00 | 1 | histone H3.2 protein |
| 49 | Mytilus chilensis | 79.00 | 1 | histone H3 |
| 50 | Oryza sativa | 79.00 | 1 | AF467728_1 disease-resistent-related protein |
| 51 | Griffithsia japonica | 79.00 | 1 | putative histone H3 protein |
| 52 | Griffithsia japonica | 79.00 | 1 | H3_GRIJA Histone H3 gi |
| 53 | Entodinium caudatum | 78.60 | 2 | histone H3 |
| 54 | Oikopleura dioica | 78.60 | 1 | histone h3.1 |
| 55 | synthetic construct | 78.60 | 1 | histone 3 H3 |
| 56 | Picea abies | 78.60 | 1 | histone 3 |
| 57 | Expression vector pET3-H3 | 78.60 | 1 | Xenopus laevis-like histone H3 |
| 58 | Drosophila yakuba | 78.60 | 1 | His3 |
| 59 | Narcissus pseudonarcissus | 78.20 | 1 | histone H3 |
| 60 | Drosophila persimilis | 78.20 | 1 | histone 3 |
| 61 | Theileria parva strain Muguga | 77.80 | 1 | histone H3 |
| 62 | Dictyostelium discoideum AX4 | 77.80 | 1 | histone H3 |
| 63 | Theileria annulata strain Ankara | 77.80 | 1 | histone H3, putative |
| 64 | Strongylocentrotus purpuratus | 77.40 | 1 | late embryonic histone H3 |
Top-ranked sequences
| organism | matching |
|---|---|
| N/A | Query 100 YLVGLFEDTNLCAIHAKRVTIMPKDIQLAR#RIRGERA 136 ||||||||||||||||||||||||||||||#||||||| Query 100 YLVGLFEDTNLCAIHAKRVTIMPKDIQLAR#RIRGERA 136 |
| Xenopus laevis | Query 100 YLVGLFEDTNLCAIHAKRVTIMPKDIQLAR#RIRGERA 136 ||||||||||||||||||||||||||||||#||||||| Query 100 YLVGLFEDTNLCAIHAKRVTIMPKDIQLAR#RIRGERA 136 |
| Mus musculus | Query 100 YLVGLFEDTNLCAIHAKRVTIMPKDIQLAR#RIRGERA 136 ||||||||||||||||||||||||||||||#||||||| Query 100 YLVGLFEDTNLCAIHAKRVTIMPKDIQLAR#RIRGERA 136 |
| Lilium longiflorum | Query 100 YLVGLFEDTNLCAIHAKRVTIMPKDIQLAR#RIRGERA 136 ||||||||||||||||||||||||||||||#||||||| Query 100 YLVGLFEDTNLCAIHAKRVTIMPKDIQLAR#RIRGERA 136 |
| Homo sapiens | Query 100 YLVGLFEDTNLCAIHAKRVTIMPKDIQLAR#RIRGERA 136 ||||||||||||||||||||||||||||||#||||||| Query 100 YLVGLFEDTNLCAIHAKRVTIMPKDIQLAR#RIRGERA 136 |
| Caenorhabditis briggsae AF16 | Query 100 YLVGLFEDTNLCAIHAKRVTIMPKDIQLAR#RIRGERA 136 ||||||||||||||||||||||||||||||#||||||| Query 100 YLVGLFEDTNLCAIHAKRVTIMPKDIQLAR#RIRGERA 136 |
| Caenorhabditis elegans | Query 100 YLVGLFEDTNLCAIHAKRVTIMPKDIQLAR#RIRGERA 136 ||||||||||||||||||||||||||||||#||||||| Query 100 YLVGLFEDTNLCAIHAKRVTIMPKDIQLAR#RIRGERA 136 |
| Bos taurus | Query 100 YLVGLFEDTNLCAIHAKRVTIMPKDIQLAR#RIRGERA 136 ||||||||||||||||||||||||||||||#||||||| Query 100 YLVGLFEDTNLCAIHAKRVTIMPKDIQLAR#RIRGERA 136 |
| Arabidopsis thaliana | Query 100 YLVGLFEDTNLCAIHAKRVTIMPKDIQLAR#RIRGERA 136 ||||||||||||||||||||||||||||||#||||||| Query 100 YLVGLFEDTNLCAIHAKRVTIMPKDIQLAR#RIRGERA 136 |
| Strongylocentrotus purpuratus | Query 100 YLVGLFEDTNLCAIHAKRVTIMPKDIQLAR#RIRGERA 136 ||||||||||||||||||||||||||||||#||||||| Query 100 YLVGLFEDTNLCAIHAKRVTIMPKDIQLAR#RIRGERA 136 |
| Danio rerio | Query 100 YLVGLFEDTNLCAIHAKRVTIMPKDIQLAR#RIRGERA 136 ||||||||||||||||||||||||||||||#||||||| Query 100 YLVGLFEDTNLCAIHAKRVTIMPKDIQLAR#RIRGERA 136 |
| Venerupis (Ruditapes) philippinarum | Query 100 YLVGLFEDTNLCAIHAKRVTIMPKDIQLAR#RIRGERA 136 ||||||||||||||||||||||||||||||#||||||| Query 100 YLVGLFEDTNLCAIHAKRVTIMPKDIQLAR#RIRGERA 136 |
| Anopheles gambiae str. PEST | Query 100 YLVGLFEDTNLCAIHAKRVTIMPKDIQLAR#RIRGERA 136 ||||||||||||||||||||||||||||||#||||||| Query 100 YLVGLFEDTNLCAIHAKRVTIMPKDIQLAR#RIRGERA 136 |
| Craterostigma plantagineum | Query 100 YLVGLFEDTNLCAIHAKRVTIMPKDIQLAR#RIRGERA 136 ||||||||||||||||||||||||||||||#||||||| Query 100 YLVGLFEDTNLCAIHAKRVTIMPKDIQLAR#RIRGERA 136 |
| Drosophila erecta | Query 100 YLVGLFEDTNLCAIHAKRVTIMPKDIQLAR#RIRGERA 136 ||||||||||||||||||||||||||||||#||||||| Query 100 YLVGLFEDTNLCAIHAKRVTIMPKDIQLAR#RIRGERA 136 |
| Drosophila simulans | Query 100 YLVGLFEDTNLCAIHAKRVTIMPKDIQLAR#RIRGERA 136 ||||||||||||||||||||||||||||||#||||||| Query 100 YLVGLFEDTNLCAIHAKRVTIMPKDIQLAR#RIRGERA 136 |
| Cricetulus longicaudatus | Query 100 YLVGLFEDTNLCAIHAKRVTIMPKDIQLAR#RIRGERA 136 ||||||||||||||||||||||||||||||#||||||| Query 100 YLVGLFEDTNLCAIHAKRVTIMPKDIQLAR#RIRGERA 136 |
| Trichinella spiralis | Query 100 YLVGLFEDTNLCAIHAKRVTIMPKDIQLAR#RIRGERA 136 ||||||||||||||||||||||||||||||#||||||| Query 100 YLVGLFEDTNLCAIHAKRVTIMPKDIQLAR#RIRGERA 136 |
| Xenopus tropicalis | Query 100 YLVGLFEDTNLCAIHAKRVTIMPKDIQLAR#RIRGERA 136 ||||||||||||||||||||||||||||||#||||||| Query 100 YLVGLFEDTNLCAIHAKRVTIMPKDIQLAR#RIRGERA 136 |
| Acropora formosa | Query 100 YLVGLFEDTNLCAIHAKRVTIMPKDIQLAR#RIRGERA 136 ||||||||||||||||||||||||||||||#||||||| Query 100 YLVGLFEDTNLCAIHAKRVTIMPKDIQLAR#RIRGERA 136 |
| Pongo pygmaeus | Query 100 YLVGLFEDTNLCAIHAKRVTIMPKDIQLAR#RIRGERA 136 ||||||||||||||||||||||||||||||#||||||| Query 100 YLVGLFEDTNLCAIHAKRVTIMPKDIQLAR#RIRGERA 136 |
| Medicago sativa | Query 100 YLVGLFEDTNLCAIHAKRVTIMPKDIQLAR#RIRGERA 136 ||||||||||||||||||||||||||||||#||||||| Query 100 YLVGLFEDTNLCAIHAKRVTIMPKDIQLAR#RIRGERA 136 |
| Canis familiaris | Query 100 YLVGLFEDTNLCAIHAKRVTIMPKDIQLAR#RIRGERA 136 ||||||||||||||||||||||||||||||#||||||| Query 100 YLVGLFEDTNLCAIHAKRVTIMPKDIQLAR#RIRGERA 136 |
| Chlamydomonas reinhardtii | Query 100 YLVGLFEDTNLCAIHAKRVTIMPKDIQLAR#RIRGERA 136 ||||||||||||||||||||||||||||||#||||||| Query 100 YLVGLFEDTNLCAIHAKRVTIMPKDIQLAR#RIRGERA 136 |
| Gallus gallus | Query 100 YLVGLFEDTNLCAIHAKRVTIMPKDIQLAR#RIRGERA 136 ||||||||||||||||||||||||||||||#||||||| Query 100 YLVGLFEDTNLCAIHAKRVTIMPKDIQLAR#RIRGERA 136 |
| Nematostella vectensis | Query 100 YLVGLFEDTNLCAIHAKRVTIMPKDIQLAR#RIRGERA 136 ||||||||||||||||||||||||||||||#||||||| Query 100 YLVGLFEDTNLCAIHAKRVTIMPKDIQLAR#RIRGERA 136 |
| Drosophila pseudoobscura | Query 100 YLVGLFEDTNLCAIHAKRVTIMPKDIQLAR#RIRGERA 136 ||||||||||||||||||||||||||||||#||||||| Query 100 YLVGLFEDTNLCAIHAKRVTIMPKDIQLAR#RIRGERA 136 |
| Mastigamoeba balamuthi | Query 100 YLVGLFEDTNLCAIHAKRVTIMPKDIQLAR#RIRGERA 136 ||||||||||||||||||||||||||||||#||||||| Query 100 YLVGLFEDTNLCAIHAKRVTIMPKDIQLAR#RIRGERA 136 |
| Rattus norvegicus | Query 100 YLVGLFEDTNLCAIHAKRVTIMPKDIQLAR#RIRGERA 136 ||||||||||||||||||||||||||||||#||||||| Query 100 YLVGLFEDTNLCAIHAKRVTIMPKDIQLAR#RIRGERA 136 |
| Arabidopsis thaliana | Query 100 YLVGLFEDTNLCAIHAKRVTIMPKDIQLAR#RIRGERA 136 |||||||||||||||||||||||||+||||#||||||| Sbjct 100 YLVGLFEDTNLCAIHAKRVTIMPKDVQLAR#RIRGERA 136 |
| Plasmodium yoelii yoelii str. 17XNL | Query 100 YLVGLFEDTNLCAIHAKRVTIMPKDIQLAR#RIRGERA 136 ||||||||||||||||||||||||||||||#||||||+ Sbjct 100 YLVGLFEDTNLCAIHAKRVTIMPKDIQLAR#RIRGERS 136 |
| Tortula ruralis | Query 100 YLVGLFEDTNLCAIHAKRVTIMPKDIQLAR#RIRGERA 136 ||||||||||||||||||||||||||||||#||||||| Query 100 YLVGLFEDTNLCAIHAKRVTIMPKDIQLAR#RIRGERA 136 |
| Drosophila melanogaster | Query 100 YLVGLFEDTNLCAIHAKRVTIMPKDIQLAR#RIRGERA 136 ||||||||||||||||||+|||||||||||#||||||| Sbjct 100 YLVGLFEDTNLCAIHAKRITIMPKDIQLAR#RIRGERA 136 |
| Mus musculus domesticus | Query 100 YLVGLFEDTNLCAIHAKRVTIMPKDIQLAR#RIRGERA 136 ||||||||||||||||||||||||||||||#||||||| Query 100 YLVGLFEDTNLCAIHAKRVTIMPKDIQLAR#RIRGERA 136 |
| Rheum australe | Query 100 YLVGLFEDTNLCAIHAKRVTIMPKDIQLAR#RIRGERA 136 |||||||||||||||||||||||||+||||#||||||| Sbjct 100 YLVGLFEDTNLCAIHAKRVTIMPKDMQLAR#RIRGERA 136 |
| Tetraodon nigroviridis | Query 100 YLVGLFEDTNLCAIHAKRVTIMPKDIQLAR#RIRGERA 136 ||||||||||||||||||||||||||||||#||||||| Query 100 YLVGLFEDTNLCAIHAKRVTIMPKDIQLAR#RIRGERA 136 |
| Aquilegia formosa | Query 100 YLVGLFEDTNLCAIHAKRVTIMPKDIQLAR#RIRGERA 136 ||||||||||||||||||||||||||||||#||||||| Query 100 YLVGLFEDTNLCAIHAKRVTIMPKDIQLAR#RIRGERA 136 |
| Drosophila virilis | Query 100 YLVGLFEDTNLCAIHAKRVTIMPKDIQLAR#RIRGERA 136 ||||||||||||||||||||||||||||||#||||||| Query 100 YLVGLFEDTNLCAIHAKRVTIMPKDIQLAR#RIRGERA 136 |
| Cichorium intybus | Query 100 YLVGLFEDTNLCAIHAKRVTIMPKDIQLAR#RIRGERA 136 |||||||||||||||||||||||||+||||#||||||| Sbjct 100 YLVGLFEDTNLCAIHAKRVTIMPKDMQLAR#RIRGERA 136 |
| Platichthys flesus | Query 100 YLVGLFEDTNLCAIHAKRVTIMPKDIQLAR#RIRGERA 136 ||||||||||||||||||||||||||||||#||||||| Query 100 YLVGLFEDTNLCAIHAKRVTIMPKDIQLAR#RIRGERA 136 |
| Eimeria tenella | Query 100 YLVGLFEDTNLCAIHAKRVTIMPKDIQLAR#RIRGERA 136 ||||||||||||||||||||||||||||||#||||||+ Sbjct 100 YLVGLFEDTNLCAIHAKRVTIMPKDIQLAR#RIRGERS 136 |
| Toxoplasma gondii | Query 100 YLVGLFEDTNLCAIHAKRVTIMPKDIQLAR#RIRGERA 136 ||||||||||||||||||||||||||||||#||||||+ Sbjct 100 YLVGLFEDTNLCAIHAKRVTIMPKDIQLAR#RIRGERS 136 |
| Toxoplasma gondii | Query 100 YLVGLFEDTNLCAIHAKRVTIMPKDIQLAR#RIRGERA 136 ||||||||||||||||||||||||||||||#||||||+ Sbjct 100 YLVGLFEDTNLCAIHAKRVTIMPKDIQLAR#RIRGERS 136 |
| Beta vulgaris subsp. vulgaris | Query 100 YLVGLFEDTNLCAIHAKRVTIMPKDIQLAR#RIRGERA 136 ||||||||||||||||||||||||||||||#||||||| Query 100 YLVGLFEDTNLCAIHAKRVTIMPKDIQLAR#RIRGERA 136 |
| Styela plicata | Query 100 YLVGLFEDTNLCAIHAKRVTIMPKDIQLAR#RIRGERA 136 |||||||||||||||||||||||||||| |#||||||| Sbjct 100 YLVGLFEDTNLCAIHAKRVTIMPKDIQLGR#RIRGERA 136 |
| Fucus serratus | Query 100 YLVGLFEDTNLCAIHAKRVTIMPKDIQLAR#RIRGERA 136 ||||||||||||||||||||||||||||||#||||||+ Sbjct 72 YLVGLFEDTNLCAIHAKRVTIMPKDIQLAR#RIRGERS 108 |
| Entamoeba histolytica HM-1:IMSS | Query 100 YLVGLFEDTNLCAIHAKRVTIMPKDIQLAR#RIRGER 135 ||||||||||||||||||+||||||+||||#|||||| Sbjct 99 YLVGLFEDTNLCAIHAKRITIMPKDMQLAR#RIRGER 134 |
| Mus pahari | Query 100 YLVGLFEDTNLCAIHAKRVTIMPKDIQLAR#RIRGERA 136 |||||||||||||||||||||||||||| |#||||||| Sbjct 100 YLVGLFEDTNLCAIHAKRVTIMPKDIQLVR#RIRGERA 136 |
| Mytilus chilensis | Query 100 YLVGLFEDTNLCAIHAKRVTIMPKDIQLAR#RIRGERA 136 |||||||||||||||||||||||||||| |#||||||| Sbjct 100 YLVGLFEDTNLCAIHAKRVTIMPKDIQLVR#RIRGERA 136 |
| Oryza sativa | Query 100 YLVGLFEDTNLCAIHAKRVTIMPKDIQLAR#RIRGERA 136 ||||||||||||||||||||||||||+|||#||||||| Sbjct 100 YLVGLFEDTNLCAIHAKRVTIMPKDIKLAR#RIRGERA 136 |
| Griffithsia japonica | Query 100 YLVGLFEDTNLCAIHAKRVTIMPKDIQLAR#RIRGER 135 |+||||||||||||||||||||||||||||#|||||| Sbjct 100 YMVGLFEDTNLCAIHAKRVTIMPKDIQLAR#RIRGER 135 |
| Griffithsia japonica | Query 100 YLVGLFEDTNLCAIHAKRVTIMPKDIQLAR#RIRGER 135 |+||||||||||||||||||||||||||||#|||||| Sbjct 100 YMVGLFEDTNLCAIHAKRVTIMPKDIQLAR#RIRGER 135 |
| Entodinium caudatum | Query 100 YLVGLFEDTNLCAIHAKRVTIMPKDIQLAR#RIRGER 135 ||||||||||||||||||||||||||||||#|||||+ Sbjct 99 YLVGLFEDTNLCAIHAKRVTIMPKDIQLAR#RIRGEK 134 |
| Oikopleura dioica | Query 100 YLVGLFEDTNLCAIHAKRVTIMPKDIQLAR#RIRGERA 136 |||||||||+||||||||||||||||||||#||||||| Sbjct 100 YLVGLFEDTHLCAIHAKRVTIMPKDIQLAR#RIRGERA 136 |
| synthetic construct | Query 100 YLVGLFEDTNLCAIHAKRVTIMPKDIQLAR#RIRGERA 136 |||||||||||| |||||||||||||||||#||||||| Sbjct 100 YLVGLFEDTNLCVIHAKRVTIMPKDIQLAR#RIRGERA 136 |
| Picea abies | Query 100 YLVGLFEDTNLCAIHAKRVTIMPKDIQLAR#RIRGERA 136 ||||||||||||||||||||||||| ||||#||||||| Sbjct 100 YLVGLFEDTNLCAIHAKRVTIMPKDXQLAR#RIRGERA 136 |
| Expression vector pET3-H3 | Query 100 YLVGLFEDTNLCAIHAKRVTIMPKDIQLAR#RIRGERA 136 ||| ||||||||||||||||||||||||||#||||||| Sbjct 100 YLVALFEDTNLCAIHAKRVTIMPKDIQLAR#RIRGERA 136 |
| Drosophila yakuba | Query 100 YLVGLFEDTNLCAIHAKRVTIMPKDIQLAR#RIRGERA 136 ||||||||||||||||||||||||||||||#||||||| Query 100 YLVGLFEDTNLCAIHAKRVTIMPKDIQLAR#RIRGERA 136 |
| Narcissus pseudonarcissus | Query 100 YLVGLFEDTNLCAIHAKRVTIMPKDIQLAR#RIRGERA 136 |||||||||| |||||||||||||||||||#||||||| Sbjct 92 YLVGLFEDTNXCAIHAKRVTIMPKDIQLAR#RIRGERA 128 |
| Drosophila persimilis | Query 100 YLVGLFEDTNLCAIHAKRVTIMPKDIQLAR#RIRGERA 136 |||||||||||||||||||||||| |||||#||||||| Sbjct 100 YLVGLFEDTNLCAIHAKRVTIMPKGIQLAR#RIRGERA 136 |
| Theileria parva strain Muguga | Query 100 YLVGLFEDTNLCAIHAKRVTIMPKDIQLAR#RIRGERA 136 |||||||||||||||||||||||||+ |||#||||||| Sbjct 100 YLVGLFEDTNLCAIHAKRVTIMPKDVHLAR#RIRGERA 136 |
| Dictyostelium discoideum AX4 | Query 100 YLVGLFEDTNLCAIHAKRVTIMPKDIQLAR#RIRGERA 136 |||||||||||||||||||||||||| |||#||||||+ Sbjct 103 YLVGLFEDTNLCAIHAKRVTIMPKDIHLAR#RIRGERS 139 |
| Theileria annulata strain Ankara | Query 100 YLVGLFEDTNLCAIHAKRVTIMPKDIQLAR#RIRGERA 136 |||||||||||||||||||||||||+ |||#||||||| Sbjct 100 YLVGLFEDTNLCAIHAKRVTIMPKDVHLAR#RIRGERA 136 |
| Strongylocentrotus purpuratus | Query 100 YLVGLFEDTNLCAIHAKRVTIMPKDIQLAR#RIRGERA 136 ||| ||||||||||||||||||||||||||#||||||| Sbjct 100 YLVRLFEDTNLCAIHAKRVTIMPKDIQLAR#RIRGERA 136 |
* References
None.