SB0091 : Thioredoxin peroxidase 1
[ CaMP Format ]
* Basic Information
| Organism | Homo sapiens (human) |
| Protein Names | peroxiredoxin-2 [Homo sapiens]; peroxiredoxin-2; thiol-specific antioxidant 1; thioredoxin peroxidase 1; torin; thioredoxin-dependent peroxide reductase 1; natural killer cell-enhancing factor B; epididymis secretory sperm binding protein Li 2a; Peroxiredoxin-2; 1.11.1.15; Natural killer cell-enhancing factor B; NKEF-B; PRP; Thiol-specific antioxidant protein; TSA; Thioredoxin peroxidase 1; Thioredoxin-dependent peroxide reductase 1 |
| Gene Names | PRDX2; NKEFB, TDPX1; NKEFB; TDPX1; peroxiredoxin 2 |
| Gene Locus | 19p13.2; chromosome 19 |
| GO Function | Not available |
* Information From OMIM
Function: To clarify the physiologic relevance of peroxiredoxins, Lee et al. (2003) generated a mouse model deficient in PRDX2, which is abundantly expressed in all types of cells. The Prdx2 -/- mice were healthy in appearance and fertile. However, they had splenomegaly caused by the congestion of red pulp with hemosiderin accumulation. Heinz bodies were detected in their peripheral blood, and morphologically abnormal cells were increased in the dense red blood cell (RBC) fractions, which contained markedly higher levels of ROS. The null mice had significantly decreased hematocrit levels, but increased reticulocyte counts and erythropoietin levels, indicative of a compensatory action to maintain hematologic homeostasis. A labeling experiment in null mice showed that a variety of RBC proteins were highly oxidized. The results suggested that Prdx -/- mice have hemolytic anemia and that peroxiredoxin II plays a major role in protecting RBCs from oxidative stress in mice.
* Structure Information
1. Primary Information
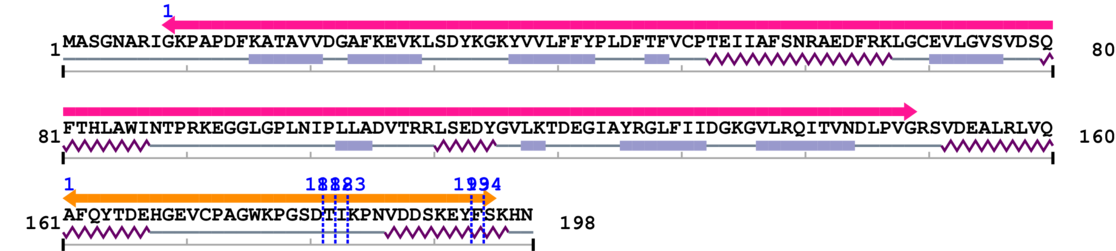
Length: 198 aa
Average Mass: 21.892 kDa
Monoisotopic Mass: 21.878 kDa
2. Domain Information
Annotated Domains: interpro / pfam / smart / prosite
Computationally Assigned Domains (Pfam+HMMER):
| domain name | begin | end | score | e-value |
|---|---|---|---|---|
| Redoxin 1. | 9 | 149 | 3.0 | 0.0 |
| C-terminal domain of 1-Cys peroxiredoxin 1. | 161 | 195 | 1.0 | 0.0 |
| --- cleavage 181 (inside C-terminal domain of 1-Cys peroxiredoxin 161..195) --- | ||||
| --- cleavage 182 (inside C-terminal domain of 1-Cys peroxiredoxin 161..195) --- | ||||
| --- cleavage 183 (inside C-terminal domain of 1-Cys peroxiredoxin 161..195) --- | ||||
| --- cleavage 193 (inside C-terminal domain of 1-Cys peroxiredoxin 161..195) --- | ||||
| --- cleavage 194 (inside C-terminal domain of 1-Cys peroxiredoxin 161..195) --- | ||||
3. Sequence Information
Fasta Sequence: SB0091.fasta
Amino Acid Sequence and Secondary Structures (PsiPred):
4. 3D Information
Known Structures in PDB: 1QMV (X-ray; 170 A; A/B/C/D/E/F/G/H/I/J=2-198)
* Cleavage Information
5 [sites] cleaved by Calpain 1
Source Reference: [PubMed ID: 9602152] Schroder E, Willis AC, Ponting CP, Porcine natural-killer-enhancing factor-B: oligomerisation and identification as a calpain substrate in vitro. Biochim Biophys Acta. 1998 Apr 2;1383(2):279-91.
Cleavage sites (±10aa)
[Site 1] CPAGWKPGSD181-TIKPNVDDSK
Asp181  Thr
Thr
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Cys172 | Pro173 | Ala174 | Gly175 | Trp176 | Lys177 | Pro178 | Gly179 | Ser180 | Asp181 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Thr182 | Ile183 | Lys184 | Pro185 | Asn186 | Val187 | Asp188 | Asp189 | Ser190 | Lys191 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| VDEALRLVQAFQYTDEHGEVCPAGWKPGSDTIKPNVDDSKEYFSKHN |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | N/A | 103.00 | 14 | enhancer protein |
| 2 | Mus musculus | 103.00 | 11 | unnamed protein product |
| 3 | Homo sapiens | 103.00 | 8 | TSA |
| 4 | synthetic construct | 103.00 | 6 | peroxiredoxin 2 |
| 5 | Rattus norvegicus | 103.00 | 5 | Peroxiredoxin 2 |
| 6 | synthetic construct | 103.00 | 1 | thioredoxin peroxidase 1-like |
| 7 | Cricetulus griseus | 103.00 | 1 | PRDX2_CRIGR Peroxiredoxin-2 gi |
| 8 | Homo sapiens | 103.00 | 1 | peroxiredoxin 2 isoform a |
| 9 | Bos taurus | 101.00 | 5 | peroxiredoxin 2 |
| 10 | Spermophilus tridecemlineatus | 90.10 | 1 | peroxiredoxin 2 |
| 11 | Canis familiaris | 88.20 | 8 | PREDICTED: similar to peroxiredoxin 1 |
| 12 | Xenopus tropicalis | 88.20 | 3 | peroxiredoxin 2 |
| 13 | Gallus gallus | 87.80 | 1 | PREDICTED: similar to natural killer cell enhanci |
| 14 | Tetraodon nigroviridis | 87.40 | 3 | unnamed protein product |
| 15 | Myotis lucifugus | 87.40 | 1 | PRDX1_MYOLU Peroxiredoxin-1 gi |
| 16 | Cricetulus griseus | 87.40 | 1 | PRDX1_CRIGR Peroxiredoxin-1 (Thioredoxin peroxida |
| 17 | Oncorhynchus mykiss | 86.30 | 1 | AF250193_1 natural killer cell enhancement factor |
| 18 | Danio rerio | 85.90 | 2 | hypothetical protein LOC541344 |
| 19 | Cyprinus carpio | 84.30 | 1 | natural killer cell enhancing factor |
| 20 | Ostertagia ostertagi | 84.30 | 1 | thioredoxin peroxidase |
| 21 | Cynops pyrrhogaster | 84.00 | 1 | TDX_CYNPY Peroxiredoxin (Thioredoxin peroxidase) |
| 22 | Paralichthys olivaceus | 84.00 | 1 | natural killer enhancing factor |
| 23 | Xenopus laevis | 82.80 | 3 | hypothetical protein LOC432262 |
| 24 | Caenorhabditis elegans | 82.80 | 2 | PeRoxireDoXin family member (prdx-2) |
| 25 | Caenorhabditis briggsae AF16 | 82.80 | 2 | Hypothetical protein CBG02380 |
| 26 | Onchocerca volvulus | 82.00 | 1 | peroxidoxin-2 |
| 27 | Onchocerca volvulus | 81.60 | 1 | thioredoxin peroxidase |
| 28 | Onchocerca ochengi | 81.60 | 1 | peroxidoxin-2 |
| 29 | Drosophila melanogaster | 80.50 | 3 | thioredoxin peroxidase 2 CG1274-PA, isoform A |
| 30 | Ascaris suum | 80.10 | 1 | PRDX_ASCSU Peroxiredoxin (AsPrx) (Thioredoxin per |
| 31 | Apis mellifera | 79.30 | 1 | PREDICTED: similar to thioredoxin peroxidase 1 CG |
| 32 | Dirofilaria immitis | 79.30 | 1 | thioredoxin peroxidase |
| 33 | Drosophila pseudoobscura | 78.20 | 3 | GA11781-PA |
| 34 | Biomphalaria glabrata | 78.20 | 1 | thioredoxin peroxidase BgTPx |
| 35 | Acanthocheilonema viteae | 77.40 | 1 | thiredoxin peroxidase |
| 36 | Dirofilaria immitis | 76.60 | 1 | peroxidoxin-1 |
| 37 | Ixodes scapularis | 73.90 | 1 | mitochondrial truncated thioredoxin-dependent per |
| 38 | Aedes aegypti | 73.20 | 1 | peroxiredoxins, prx-1, prx-2, prx-3 |
| 39 | Drosophila melanogaster | 72.80 | 1 | Peroxiredoxin 5037 CG5826-PA |
| 40 | Globodera rostochiensis | 72.40 | 1 | peroxiredoxin |
| 41 | Trypanosoma cruzi strain CL Brener | 72.00 | 1 | tryparedoxin peroxidase |
| 42 | Apis mellifera ligustica | 71.60 | 1 | thioredoxin peroxidase |
| 43 | Litomosoides sigmodontis | 71.20 | 1 | AF105258_1 peroxidoxin-2 |
| 44 | Trypanosoma cruzi strain CL Brener | 70.90 | 3 | tryparedoxin peroxidase, putative |
| 45 | Echinococcus multilocularis | 70.10 | 1 | thioredoxin peroxidase |
| 46 | Echinococcus granulosus | 70.10 | 1 | thioredoxin peroxidase |
| 47 | Candida albicans SC5314 | 69.70 | 1 | thioredoxin peroxidase |
| 48 | Schistosoma mansoni | 69.30 | 1 | AF301001_1 thioredoxin peroxidase 3 |
| 49 | Trypanosoma cruzi | 68.90 | 1 | AF320771_1 tryparedoxin peroxidase |
| 50 | Trypanosoma brucei TREU927 | 68.60 | 1 | tryparedoxin peroxidase |
| 51 | Yarrowia lipolytica | 67.80 | 1 | hypothetical protein |
| 52 | Candida glabrata | 67.40 | 1 | unnamed protein product |
| 53 | Nitrosococcus oceani ATCC 19707 | 65.90 | 1 | Alkyl hydroperoxide reductase/ Thiol specific ant |
| 54 | Magnetospirillum magneticum AMB-1 | 65.90 | 1 | Peroxiredoxin |
Top-ranked sequences
| organism | matching |
|---|---|
| N/A | Query 152 VDEALRLVQAFQYTDEHGEVCPAGWKPGSD#TIKPNVDDSKEYFSKHN 198 ||||||||||||||||||||||||||||||#||||||||||||||||| Query 152 VDEALRLVQAFQYTDEHGEVCPAGWKPGSD#TIKPNVDDSKEYFSKHN 198 |
| Mus musculus | Query 152 VDEALRLVQAFQYTDEHGEVCPAGWKPGSD#TIKPNVDDSKEYFSKHN 198 ||||||||||||||||||||||||||||||#||||||||||||||||| Query 152 VDEALRLVQAFQYTDEHGEVCPAGWKPGSD#TIKPNVDDSKEYFSKHN 198 |
| Homo sapiens | Query 152 VDEALRLVQAFQYTDEHGEVCPAGWKPGSD#TIKPNVDDSKEYFSKHN 198 ||||||||||||||||||||||||||||||#||||||||||||||||| Query 152 VDEALRLVQAFQYTDEHGEVCPAGWKPGSD#TIKPNVDDSKEYFSKHN 198 |
| synthetic construct | Query 152 VDEALRLVQAFQYTDEHGEVCPAGWKPGSD#TIKPNVDDSKEYFSKHN 198 ||||||||||||||||||||||||||||||#||||||||||||||||| Query 152 VDEALRLVQAFQYTDEHGEVCPAGWKPGSD#TIKPNVDDSKEYFSKHN 198 |
| Rattus norvegicus | Query 152 VDEALRLVQAFQYTDEHGEVCPAGWKPGSD#TIKPNVDDSKEYFSKHN 198 ||||||||||||||||||||||||||||||#||||||||||||||||| Query 152 VDEALRLVQAFQYTDEHGEVCPAGWKPGSD#TIKPNVDDSKEYFSKHN 198 |
| synthetic construct | Query 152 VDEALRLVQAFQYTDEHGEVCPAGWKPGSD#TIKPNVDDSKEYFSKHN 198 ||||||||||||||||||||||||||||||#||||||||||||||||| Query 152 VDEALRLVQAFQYTDEHGEVCPAGWKPGSD#TIKPNVDDSKEYFSKHN 198 |
| Cricetulus griseus | Query 152 VDEALRLVQAFQYTDEHGEVCPAGWKPGSD#TIKPNVDDSKEYFSKHN 198 ||||||||||||||||||||||||||||||#||||||||||||||||| Query 152 VDEALRLVQAFQYTDEHGEVCPAGWKPGSD#TIKPNVDDSKEYFSKHN 198 |
| Homo sapiens | Query 152 VDEALRLVQAFQYTDEHGEVCPAGWKPGSD#TIKPNVDDSKEYFSKHN 198 ||||||||||||||||||||||||||||||#||||||||||||||||| Query 152 VDEALRLVQAFQYTDEHGEVCPAGWKPGSD#TIKPNVDDSKEYFSKHN 198 |
| Bos taurus | Query 152 VDEALRLVQAFQYTDEHGEVCPAGWKPGSD#TIKPNVDDSKEYFSKHN 198 ||||||||||||||||||||||||| ||||#||||||||||||||||| Sbjct 153 VDEALRLVQAFQYTDEHGEVCPAGWTPGSD#TIKPNVDDSKEYFSKHN 199 |
| Spermophilus tridecemlineatus | Query 152 VDEALRLVQAFQYTDEHGEVCPAGWKPGSD#TIKPNVDDSKE 192 ||||||||||||||||||||||||||||||#||||||||||| Query 152 VDEALRLVQAFQYTDEHGEVCPAGWKPGSD#TIKPNVDDSKE 192 |
| Canis familiaris | Query 152 VDEALRLVQAFQYTDEHGEVCPAGWKPGSD#TIKPNVDDSKEYFSKHN 198 ||| ||||||||+||+||||||||||||||#||||+| ||||||| Sbjct 54 VDETLRLVQAFQFTDKHGEVCPAGWKPGSD#TIKPDVQKSKEYFSKQK 100 |
| Xenopus tropicalis | Query 152 VDEALRLVQAFQYTDEHGEVCPAGWKPGSD#TIKPNVDDSKEYFSK 196 |+| |||||||||||+||||||||||||| #|||||| ||||+||| Sbjct 160 VEETLRLVQAFQYTDQHGEVCPAGWKPGSS#TIKPNVKDSKEFFSK 204 |
| Gallus gallus | Query 152 VDEALRLVQAFQYTDEHGEVCPAGWKPGSD#TIKPNVDDSKEYFSKHN 198 ||| ||||||||+||+||||||||||||||#||||+| ||||||| Sbjct 153 VDETLRLVQAFQFTDKHGEVCPAGWKPGSD#TIKPDVQKSKEYFSKQK 199 |
| Tetraodon nigroviridis | Query 152 VDEALRLVQAFQYTDEHGEVCPAGWKPGSD#TIKPNVDDSKEYFSKH 197 |+| ||||||||+||+||||||||||||||#||||+| |||+|||| Sbjct 153 VEETLRLVQAFQFTDKHGEVCPAGWKPGSD#TIKPDVQKSKEFFSKH 198 |
| Myotis lucifugus | Query 152 VDEALRLVQAFQYTDEHGEVCPAGWKPGSD#TIKPNVDDSKEYFSKH 197 ||| ||||||||+||+||||||||||||||#||||+| ||||||| Sbjct 153 VDETLRLVQAFQFTDKHGEVCPAGWKPGSD#TIKPDVQKSKEYFSKQ 198 |
| Cricetulus griseus | Query 152 VDEALRLVQAFQYTDEHGEVCPAGWKPGSD#TIKPNVDDSKEYFSKH 197 ||| ||||||||+||+||||||||||||||#||||+| ||||||| Sbjct 153 VDEILRLVQAFQFTDKHGEVCPAGWKPGSD#TIKPDVQKSKEYFSKQ 198 |
| Oncorhynchus mykiss | Query 152 VDEALRLVQAFQYTDEHGEVCPAGWKPGSD#TIKPNVDDSKEYFSKH 197 ||| ||||||||+||+||||||||||||||#||||+| ||++||| Sbjct 153 VDETLRLVQAFQFTDKHGEVCPAGWKPGSD#TIKPDVQKSKDFFSKQ 198 |
| Danio rerio | Query 152 VDEALRLVQAFQYTDEHGEVCPAGWKPGSD#TIKPNVDDSKEYFSKHN 198 +|| ||||||||+||+|||||||||||| |#||||+|+ ||++||| | Sbjct 153 IDETLRLVQAFQFTDKHGEVCPAGWKPGKD#TIKPDVNQSKDFFSKQN 199 |
| Cyprinus carpio | Query 152 VDEALRLVQAFQYTDEHGEVCPAGWKPGSD#TIKPNVDDSKEYFSKHN 198 +|| ||||||||+||+|||||||||||| |#||||+| ||+|||| + Sbjct 153 IDETLRLVQAFQFTDKHGEVCPAGWKPGKD#TIKPDVQQSKDYFSKQH 199 |
| Ostertagia ostertagi | Query 152 VDEALRLVQAFQYTDEHGEVCPAGWKPGSD#TIKPNVDDSKEYFSKHN 198 ||| ||||||||| |+||||||||| || #|||| | |||||||| | Sbjct 147 VDETLRLVQAFQYVDKHGEVCPAGWTPGKA#TIKPGVKDSKEYFSKAN 193 |
| Cynops pyrrhogaster | Query 152 VDEALRLVQAFQYTDEHGEVCPAGWKPGSD#TIKPNVDDSKEYFSKHN 198 ||| ||||||||+||+ |||||||||||||#||||++ ||||||| Sbjct 153 VDETLRLVQAFQHTDKFGEVCPAGWKPGSD#TIKPDISKSKEYFSKQK 199 |
| Paralichthys olivaceus | Query 152 VDEALRLVQAFQYTDEHGEVCPAGWKPGSD#TIKPNVDDSKEYFSKH 197 |+| ||||||||+||+||||||||||||||#||||+| ||++||| Sbjct 153 VEETLRLVQAFQFTDKHGEVCPAGWKPGSD#TIKPDVQKSKDFFSKQ 198 |
| Xenopus laevis | Query 152 VDEALRLVQAFQYTDEHGEVCPAGWKPGSD#TIKPNVDDSKEYFSKHN 198 ||| ||||||||+||++||||||||+||||#||||+| |||||+| Sbjct 153 VDETLRLVQAFQFTDKYGEVCPAGWQPGSD#TIKPDVQKSKEYFNKQK 199 |
| Caenorhabditis elegans | Query 152 VDEALRLVQAFQYTDEHGEVCPAGWKPGSD#TIKPNVDDSKEYFSKH 197 ||| ||||||||+ ++||||||||| ||||#|||| | +|+||| || Sbjct 150 VDETLRLVQAFQFVEKHGEVCPAGWTPGSD#TIKPGVKESQEYFKKH 195 |
| Caenorhabditis briggsae AF16 | Query 152 VDEALRLVQAFQYTDEHGEVCPAGWKPGSD#TIKPNVDDSKEYFSKH 197 ||| ||||||||+ ++||||||||| ||||#||||+| |+||| || Sbjct 530 VDETLRLVQAFQFVEKHGEVCPAGWTPGSD#TIKPDVKKSQEYFGKH 575 |
| Onchocerca volvulus | Query 152 VDEALRLVQAFQYTDEHGEVCPAGWKPGSD#TIKPNVDDSKEYFSKH 197 ||| ||||||||+ | ||||||| |+|||+#|||| | +||||| || Sbjct 154 VDETLRLVQAFQFVDNHGEVCPANWQPGSE#TIKPEVKESKEYFGKH 199 |
| Onchocerca volvulus | Query 152 VDEALRLVQAFQYTDEHGEVCPAGWKPGSD#TIKPNVDDSKEYFSKH 197 ||| |||+||||+ | ||||||| |+|||+#|||| | +||||| || Sbjct 154 VDETLRLIQAFQFVDNHGEVCPANWQPGSE#TIKPEVKESKEYFGKH 199 |
| Onchocerca ochengi | Query 152 VDEALRLVQAFQYTDEHGEVCPAGWKPGSD#TIKPNVDDSKEYFSKH 197 ||| |||+||||+ | ||||||| |+|||+#|||| | +||||| || Sbjct 154 VDETLRLIQAFQFVDNHGEVCPANWQPGSE#TIKPEVKESKEYFGKH 199 |
| Drosophila melanogaster | Query 152 VDEALRLVQAFQYTDEHGEVCPAGWKPGSD#TIKPNVDDSKEYFSKHN 198 ||| +|||||||||| |||||||||+||+|#|| || ++ +||+|+| Sbjct 196 VDETIRLVQAFQYTDTHGEVCPAGWRPGAD#TIVPNPEEKTKYFAKNN 242 |
| Ascaris suum | Query 152 VDEALRLVQAFQYTDEHGEVCPAGWKPGSD#TIKPNVDDSKEYFSKH 197 | | ||||||||+ |+||||||||| ||+|#|||| | +|| || || Sbjct 150 VTETLRLVQAFQFVDKHGEVCPAGWTPGAD#TIKPGVKESKAYFEKH 195 |
| Apis mellifera | Query 152 VDEALRLVQAFQYTDEHGEVCPAGWKPGSD#TIKPNVDDSKEYF 194 ||| |||||||||||+|||||||||||| #|+||+| ||||| Sbjct 149 VDETLRLVQAFQYTDKHGEVCPAGWKPGKK#TMKPDVVGSKEYF 191 |
| Dirofilaria immitis | Query 152 VDEALRLVQAFQYTDEHGEVCPAGWKPGSD#TIKPNVDDSKEYFSKH 197 ||| |||+||||+ | ||||||| |+|||+# ||| | +|| || || Sbjct 154 VDETLRLIQAFQFVDNHGEVCPANWQPGSE#AIKPGVKESKAYFEKH 199 |
| Drosophila pseudoobscura | Query 152 VDEALRLVQAFQYTDEHGEVCPAGWKPGSD#TIKPNVDDSKEYFSKHN 198 ||| +|||||||||| |||||||||+||+|#|| |+ ++ +||+|+| Sbjct 197 VDETIRLVQAFQYTDTHGEVCPAGWRPGAD#TIVPDPEEKTKYFAKNN 243 |
| Biomphalaria glabrata | Query 152 VDEALRLVQAFQYTDEHGEVCPAGWKPGSD#TIKPNVDDSKEYFSKHN 198 ||| |||||||||||+||||||||||||| #|| |+ ||||| + + Sbjct 173 VDETLRLVQAFQYTDKHGEVCPAGWKPGSA#TIIPDPKKSKEYFKQQS 219 |
| Acanthocheilonema viteae | Query 152 VDEALRLVQAFQYTDEHGEVCPAGWKPGSD#TIKPNVDDSKEYF 194 ||| |||+||||+ |+||||||| | |||+#|||| | +|| || Sbjct 154 VDETLRLIQAFQFVDKHGEVCPANWHPGSE#TIKPGVKESKAYF 196 |
| Dirofilaria immitis | Query 152 VDEALRLVQAFQYTDEHGEVCPAGWKPGSD#TIKPNVDDSKEYFSKH 197 ||| |||+||||+ | ||||||| | |||+# ||| | +|| || || Sbjct 154 VDETLRLIQAFQFVDNHGEVCPANWHPGSE#AIKPGVKESKAYFEKH 199 |
| Ixodes scapularis | Query 152 VDEALRLVQAFQYTDEHGEVCPAGWKPGSD#TIKPNVDDSKEYFSKHN 198 ||| ||||+|||+ ++|||||||||+| | #||||+ +|+||||| | Sbjct 187 VDETLRLVKAFQFVEKHGEVCPAGWQPDSP#TIKPDPKNSQEYFSKVN 233 |
| Aedes aegypti | Query 152 VDEALRLVQAFQYTDEHGEVCPAGWKPGSD#TIKPNVDDSKEYFSKHN 198 ||| ||||||||+|||||||||| ||||| #|+ + |||||+ | Sbjct 150 VDETLRLVQAFQFTDEHGEVCPANWKPGSK#TMVADPQKSKEYFNAAN 196 |
| Drosophila melanogaster | Query 152 VDEALRLVQAFQYTDEHGEVCPAGWKPGSD--#TIKPNVDDSKEYFSKH 197 ||| |||++|||+ ++||||||| | | |+ #||||+|++||+||||| Sbjct 186 VDEVLRLIKAFQFVEQHGEVCPANWNPNSNPA#TIKPDVEESKKYFSKH 233 |
| Globodera rostochiensis | Query 152 VDEALRLVQAFQYTDEHGEVCPAGWKPGSD#TIKPNVDDSKEYFSK 196 ||| |||||||+||| ||||||| |+|| |#||||+ + |+ +| | Sbjct 153 VDETLRLVQAFKYTDTHGEVCPANWQPGED#TIKPDPEGSQTFFGK 197 |
| Trypanosoma cruzi strain CL Brener | Query 152 VDEALRLVQAFQYTDEHGEVCPAGWKPGSD#TIKPNVDDSKEYF 194 ||||||||+|||+ +|||||||| |||| #|+||+ + ||||| Sbjct 153 VDEALRLVKAFQFVEEHGEVCPANWKPGDK#TMKPDPEKSKEYF 195 |
| Apis mellifera ligustica | Query 152 VDEALRLVQAFQYTDEHGEVCPAGWKPGSD#TIKPNVDDSKEYFSKHN 198 ||| |||++|||+ ++||||||| |+| | #||||| |||+|| | Sbjct 196 VDETLRLIKAFQFVEKHGEVCPANWQPDSK#TIKPNPKDSKQYFESVN 242 |
| Litomosoides sigmodontis | Query 152 VDEALRLVQAFQYTDEHGEVCPAGWKPGSD#TIKPNVDDSK 191 ||| |||+||||+ |+|||+||| |+|||+#|||| | +|| Sbjct 154 VDETLRLIQAFQFVDKHGELCPANWQPGSE#TIKPGVKESK 193 |
| Trypanosoma cruzi strain CL Brener | Query 152 VDEALRLVQAFQYTDEHGEVCPAGWKPGSD#TIKPNVDDSKEYF 194 ||||||||+|||+ +|||||||| |||| #|+||+ + |||+| Sbjct 153 VDEALRLVKAFQFVEEHGEVCPANWKPGDK#TMKPDPEKSKEFF 195 |
| Echinococcus multilocularis | Query 152 VDEALRLVQAFQYTDEHGEVCPAGWKPGSD#TIKPNVDDSKEYFS 195 |||||||+ |||+||+||||||| | ||| #| ||+ | | + | Sbjct 149 VDEALRLLDAFQFTDKHGEVCPANWHPGSK#TFKPSAGDLKSFMS 192 |
| Echinococcus granulosus | Query 152 VDEALRLVQAFQYTDEHGEVCPAGWKPGSD#TIKPNVDDSKEYFS 195 |||||||+ |||+||+||||||| |+||| #| ||+ | | + | Sbjct 141 VDEALRLLDAFQFTDKHGEVCPANWQPGSK#TFKPSAGDLKSFMS 184 |
| Candida albicans SC5314 | Query 152 VDEALRLVQAFQYTDEHGEVCPAGWKPGSD#TIKPNVDDSKEYFSKHN 198 |+|+|||++|||+|+++|||||| | || +#||||+ + |||||+| | Sbjct 149 VEESLRLLEAFQFTEKYGEVCPANWHPGDE#TIKPSPEASKEYFNKVN 195 |
| Schistosoma mansoni | Query 152 VDEALRLVQAFQYTDEHGEVCPAGWKPGSD#TIKPNVDDSKEYFSKHN 198 ||| ||||+||||||++|||||| |+| #||||++ |||| | | Sbjct 173 VDEVLRLVRAFQYTDKYGEVCPADWQPKGP#TIKPDLKKYKEYFHKVN 219 |
| Trypanosoma cruzi | Query 152 VDEALRLVQAFQYTDEHGEVCPAGWKPGSD#TIKPNVDDSKEYF 194 ||||||||+|||+ ++||||||| |||| # +||+ + ||||| Sbjct 153 VDEALRLVKAFQFVEKHGEVCPANWKPGDK#AMKPDPEKSKEYF 195 |
| Trypanosoma brucei TREU927 | Query 152 VDEALRLVQAFQYTDEHGEVCPAGWKPGSD#TIKPNVDDSKEYFSKHN 198 ||| ||||+|||+ ++||||||| ||||| #|+| + + |++||| | Sbjct 153 VDETLRLVKAFQFVEKHGEVCPANWKPGSK#TMKADPNGSQDYFSSMN 199 |
| Yarrowia lipolytica | Query 152 VDEALRLVQAFQYTDEHGEVCPAGWKPGSD#TIKPNVDDSKEYFSKHN 198 |+| |||+ |||+|++||||||| |+ |||#||| + ++|||| | | Sbjct 149 VEETLRLIDAFQFTEKHGEVCPANWQKGSD#TIKADPVNAKEYFEKAN 195 |
| Candida glabrata | Query 152 VDEALRLVQAFQYTDEHGEVCPAGWKPGSD#TIKPNVDDSKEYFSKHN 198 |+|+||||+ ||+||++| | | | ||| #|||| |+|||||| + | Sbjct 149 VEESLRLVEGFQWTDKNGTVLPCNWTPGSA#TIKPTVEDSKEYFKEAN 195 |
| Nitrosococcus oceani ATCC 19707 | Query 152 VDEALRLVQAFQYTDEHGEVCPAGWKPGSD#TIKPNVDDSKEYFSKH 197 |+| ||+| | |+|+||||||||||+ | +# |+|+ + || ||| Sbjct 150 VEEMLRVVDALQFTEEHGEVCPAGWRKGEE#AIRPDAEGVAEYLSKH 195 |
| Magnetospirillum magneticum AMB-1 | Query 152 VDEALRLVQAFQYTDEHGEVCPAGWKPGSD#TIKPNVDDSKEYFSKH 197 |||||||+ | |+++|||||||||| | # +||| + || +|| Sbjct 158 VDEALRLIDALQFSEEHGEVCPAGWNKGKA#GMKPNPNGVAEYLAKH 203 |
[Site 2] PAGWKPGSDT182-IKPNVDDSKE
Thr182  Ile
Ile
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Pro173 | Ala174 | Gly175 | Trp176 | Lys177 | Pro178 | Gly179 | Ser180 | Asp181 | Thr182 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Ile183 | Lys184 | Pro185 | Asn186 | Val187 | Asp188 | Asp189 | Ser190 | Lys191 | Glu192 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| DEALRLVQAFQYTDEHGEVCPAGWKPGSDTIKPNVDDSKEYFSKHN |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | N/A | 102.00 | 14 | enhancer protein |
| 2 | Mus musculus | 102.00 | 11 | peroxiredoxin 2 |
| 3 | synthetic construct | 102.00 | 6 | peroxiredoxin 2 |
| 4 | Rattus norvegicus | 102.00 | 5 | Peroxiredoxin 2 |
| 5 | Cricetulus griseus | 102.00 | 1 | PRDX2_CRIGR Peroxiredoxin-2 gi |
| 6 | Homo sapiens | 102.00 | 1 | peroxiredoxin 2 isoform a |
| 7 | Homo sapiens | 101.00 | 8 | TSA |
| 8 | synthetic construct | 101.00 | 1 | thioredoxin peroxidase 1-like |
| 9 | Bos taurus | 99.80 | 5 | peroxiredoxin 2 |
| 10 | Spermophilus tridecemlineatus | 88.60 | 1 | peroxiredoxin 2 |
| 11 | Canis familiaris | 86.70 | 8 | PREDICTED: similar to peroxiredoxin 1 |
| 12 | Xenopus tropicalis | 86.70 | 3 | peroxiredoxin 2 |
| 13 | Gallus gallus | 86.30 | 1 | PREDICTED: similar to natural killer cell enhanci |
| 14 | Tetraodon nigroviridis | 85.90 | 3 | unnamed protein product |
| 15 | Myotis lucifugus | 85.90 | 1 | PRDX1_MYOLU Peroxiredoxin-1 gi |
| 16 | Cricetulus griseus | 85.50 | 1 | PRDX1_CRIGR Peroxiredoxin-1 (Thioredoxin peroxida |
| 17 | Oncorhynchus mykiss | 84.70 | 1 | AF250193_1 natural killer cell enhancement factor |
| 18 | Danio rerio | 84.30 | 2 | hypothetical protein LOC541344 |
| 19 | Cyprinus carpio | 82.80 | 1 | natural killer cell enhancing factor |
| 20 | Ostertagia ostertagi | 82.80 | 1 | thioredoxin peroxidase |
| 21 | Cynops pyrrhogaster | 82.00 | 1 | TDX_CYNPY Peroxiredoxin (Thioredoxin peroxidase) |
| 22 | Paralichthys olivaceus | 82.00 | 1 | natural killer enhancing factor |
| 23 | Xenopus laevis | 81.30 | 3 | hypothetical protein LOC432262 |
| 24 | Caenorhabditis briggsae AF16 | 81.30 | 2 | Hypothetical protein CBG02380 |
| 25 | Caenorhabditis elegans | 81.30 | 2 | PeRoxireDoXin family member (prdx-2) |
| 26 | Onchocerca volvulus | 80.50 | 1 | peroxidoxin-2 |
| 27 | Onchocerca volvulus | 80.10 | 1 | thioredoxin peroxidase |
| 28 | Onchocerca ochengi | 80.10 | 1 | peroxidoxin-2 |
| 29 | Ascaris suum | 79.00 | 1 | PRDX_ASCSU Peroxiredoxin (AsPrx) (Thioredoxin per |
| 30 | Drosophila melanogaster | 78.60 | 2 | thioredoxin peroxidase 2 CG1274-PA, isoform A |
| 31 | Apis mellifera | 77.80 | 1 | PREDICTED: similar to thioredoxin peroxidase 1 CG |
| 32 | Dirofilaria immitis | 77.80 | 1 | thioredoxin peroxidase |
| 33 | Drosophila pseudoobscura | 76.60 | 3 | GA11781-PA |
| 34 | Biomphalaria glabrata | 76.30 | 1 | thioredoxin peroxidase BgTPx |
| 35 | Acanthocheilonema viteae | 75.90 | 1 | thiredoxin peroxidase |
| 36 | Dirofilaria immitis | 74.70 | 1 | peroxidoxin-1 |
| 37 | Ixodes scapularis | 72.40 | 1 | mitochondrial truncated thioredoxin-dependent per |
| 38 | Aedes aegypti | 71.60 | 1 | peroxiredoxins, prx-1, prx-2, prx-3 |
| 39 | Drosophila melanogaster | 71.20 | 1 | Peroxiredoxin 5037 CG5826-PA |
| 40 | Globodera rostochiensis | 70.90 | 1 | peroxiredoxin |
| 41 | Trypanosoma cruzi strain CL Brener | 70.50 | 1 | tryparedoxin peroxidase |
| 42 | Apis mellifera ligustica | 70.10 | 1 | thioredoxin peroxidase |
| 43 | Litomosoides sigmodontis | 69.70 | 1 | AF105258_1 peroxidoxin-2 |
| 44 | Trypanosoma cruzi strain CL Brener | 69.30 | 3 | tryparedoxin peroxidase, putative |
| 45 | Echinococcus multilocularis | 68.60 | 1 | thioredoxin peroxidase |
| 46 | Echinococcus granulosus | 68.20 | 1 | thioredoxin peroxidase |
| 47 | Candida albicans SC5314 | 68.20 | 1 | thioredoxin peroxidase |
| 48 | Schistosoma mansoni | 67.80 | 1 | AF301001_1 thioredoxin peroxidase 3 |
| 49 | Trypanosoma cruzi | 67.00 | 1 | AF320771_1 tryparedoxin peroxidase |
| 50 | Trypanosoma brucei TREU927 | 67.00 | 1 | tryparedoxin peroxidase |
| 51 | Yarrowia lipolytica | 66.20 | 1 | hypothetical protein |
| 52 | Candida glabrata | 65.90 | 1 | unnamed protein product |
| 53 | Entamoeba moshkovskii | 65.10 | 3 | peroxiredoxin |
Top-ranked sequences
| organism | matching |
|---|---|
| N/A | Query 153 DEALRLVQAFQYTDEHGEVCPAGWKPGSDT#IKPNVDDSKEYFSKHN 198 ||||||||||||||||||||||||||||||#|||||||||||||||| Query 153 DEALRLVQAFQYTDEHGEVCPAGWKPGSDT#IKPNVDDSKEYFSKHN 198 |
| Mus musculus | Query 153 DEALRLVQAFQYTDEHGEVCPAGWKPGSDT#IKPNVDDSKEYFSKHN 198 ||||||||||||||||||||||||||||||#|||||||||||||||| Query 153 DEALRLVQAFQYTDEHGEVCPAGWKPGSDT#IKPNVDDSKEYFSKHN 198 |
| synthetic construct | Query 153 DEALRLVQAFQYTDEHGEVCPAGWKPGSDT#IKPNVDDSKEYFSKHN 198 ||||||||||||||||||||||||||||||#|||||||||||||||| Query 153 DEALRLVQAFQYTDEHGEVCPAGWKPGSDT#IKPNVDDSKEYFSKHN 198 |
| Rattus norvegicus | Query 153 DEALRLVQAFQYTDEHGEVCPAGWKPGSDT#IKPNVDDSKEYFSKHN 198 ||||||||||||||||||||||||||||||#|||||||||||||||| Query 153 DEALRLVQAFQYTDEHGEVCPAGWKPGSDT#IKPNVDDSKEYFSKHN 198 |
| Cricetulus griseus | Query 153 DEALRLVQAFQYTDEHGEVCPAGWKPGSDT#IKPNVDDSKEYFSKHN 198 ||||||||||||||||||||||||||||||#|||||||||||||||| Query 153 DEALRLVQAFQYTDEHGEVCPAGWKPGSDT#IKPNVDDSKEYFSKHN 198 |
| Homo sapiens | Query 153 DEALRLVQAFQYTDEHGEVCPAGWKPGSDT#IKPNVDDSKEYFSKHN 198 ||||||||||||||||||||||||||||||#|||||||||||||||| Query 153 DEALRLVQAFQYTDEHGEVCPAGWKPGSDT#IKPNVDDSKEYFSKHN 198 |
| Homo sapiens | Query 153 DEALRLVQAFQYTDEHGEVCPAGWKPGSDT#IKPNVDDSKEYFSKHN 198 ||||||||||||||||||||||||||||||#|||||||||||||||| Query 153 DEALRLVQAFQYTDEHGEVCPAGWKPGSDT#IKPNVDDSKEYFSKHN 198 |
| synthetic construct | Query 153 DEALRLVQAFQYTDEHGEVCPAGWKPGSDT#IKPNVDDSKEYFSKHN 198 ||||||||||||||||||||||||||||||#|||||||||||||||| Query 153 DEALRLVQAFQYTDEHGEVCPAGWKPGSDT#IKPNVDDSKEYFSKHN 198 |
| Bos taurus | Query 153 DEALRLVQAFQYTDEHGEVCPAGWKPGSDT#IKPNVDDSKEYFSKHN 198 |||||||||||||||||||||||| |||||#|||||||||||||||| Sbjct 154 DEALRLVQAFQYTDEHGEVCPAGWTPGSDT#IKPNVDDSKEYFSKHN 199 |
| Spermophilus tridecemlineatus | Query 153 DEALRLVQAFQYTDEHGEVCPAGWKPGSDT#IKPNVDDSKE 192 ||||||||||||||||||||||||||||||#|||||||||| Query 153 DEALRLVQAFQYTDEHGEVCPAGWKPGSDT#IKPNVDDSKE 192 |
| Canis familiaris | Query 153 DEALRLVQAFQYTDEHGEVCPAGWKPGSDT#IKPNVDDSKEYFSKHN 198 || ||||||||+||+|||||||||||||||#|||+| ||||||| Sbjct 55 DETLRLVQAFQFTDKHGEVCPAGWKPGSDT#IKPDVQKSKEYFSKQK 100 |
| Xenopus tropicalis | Query 153 DEALRLVQAFQYTDEHGEVCPAGWKPGSDT#IKPNVDDSKEYFSK 196 +| |||||||||||+||||||||||||| |#||||| ||||+||| Sbjct 161 EETLRLVQAFQYTDQHGEVCPAGWKPGSST#IKPNVKDSKEFFSK 204 |
| Gallus gallus | Query 153 DEALRLVQAFQYTDEHGEVCPAGWKPGSDT#IKPNVDDSKEYFSKHN 198 || ||||||||+||+|||||||||||||||#|||+| ||||||| Sbjct 154 DETLRLVQAFQFTDKHGEVCPAGWKPGSDT#IKPDVQKSKEYFSKQK 199 |
| Tetraodon nigroviridis | Query 153 DEALRLVQAFQYTDEHGEVCPAGWKPGSDT#IKPNVDDSKEYFSKH 197 +| ||||||||+||+|||||||||||||||#|||+| |||+|||| Sbjct 154 EETLRLVQAFQFTDKHGEVCPAGWKPGSDT#IKPDVQKSKEFFSKH 198 |
| Myotis lucifugus | Query 153 DEALRLVQAFQYTDEHGEVCPAGWKPGSDT#IKPNVDDSKEYFSKH 197 || ||||||||+||+|||||||||||||||#|||+| ||||||| Sbjct 154 DETLRLVQAFQFTDKHGEVCPAGWKPGSDT#IKPDVQKSKEYFSKQ 198 |
| Cricetulus griseus | Query 153 DEALRLVQAFQYTDEHGEVCPAGWKPGSDT#IKPNVDDSKEYFSKH 197 || ||||||||+||+|||||||||||||||#|||+| ||||||| Sbjct 154 DEILRLVQAFQFTDKHGEVCPAGWKPGSDT#IKPDVQKSKEYFSKQ 198 |
| Oncorhynchus mykiss | Query 153 DEALRLVQAFQYTDEHGEVCPAGWKPGSDT#IKPNVDDSKEYFSKH 197 || ||||||||+||+|||||||||||||||#|||+| ||++||| Sbjct 154 DETLRLVQAFQFTDKHGEVCPAGWKPGSDT#IKPDVQKSKDFFSKQ 198 |
| Danio rerio | Query 153 DEALRLVQAFQYTDEHGEVCPAGWKPGSDT#IKPNVDDSKEYFSKHN 198 || ||||||||+||+|||||||||||| ||#|||+|+ ||++||| | Sbjct 154 DETLRLVQAFQFTDKHGEVCPAGWKPGKDT#IKPDVNQSKDFFSKQN 199 |
| Cyprinus carpio | Query 153 DEALRLVQAFQYTDEHGEVCPAGWKPGSDT#IKPNVDDSKEYFSKHN 198 || ||||||||+||+|||||||||||| ||#|||+| ||+|||| + Sbjct 154 DETLRLVQAFQFTDKHGEVCPAGWKPGKDT#IKPDVQQSKDYFSKQH 199 |
| Ostertagia ostertagi | Query 153 DEALRLVQAFQYTDEHGEVCPAGWKPGSDT#IKPNVDDSKEYFSKHN 198 || ||||||||| |+||||||||| || |#||| | |||||||| | Sbjct 148 DETLRLVQAFQYVDKHGEVCPAGWTPGKAT#IKPGVKDSKEYFSKAN 193 |
| Cynops pyrrhogaster | Query 153 DEALRLVQAFQYTDEHGEVCPAGWKPGSDT#IKPNVDDSKEYFSKHN 198 || ||||||||+||+ ||||||||||||||#|||++ ||||||| Sbjct 154 DETLRLVQAFQHTDKFGEVCPAGWKPGSDT#IKPDISKSKEYFSKQK 199 |
| Paralichthys olivaceus | Query 153 DEALRLVQAFQYTDEHGEVCPAGWKPGSDT#IKPNVDDSKEYFSKH 197 +| ||||||||+||+|||||||||||||||#|||+| ||++||| Sbjct 154 EETLRLVQAFQFTDKHGEVCPAGWKPGSDT#IKPDVQKSKDFFSKQ 198 |
| Xenopus laevis | Query 153 DEALRLVQAFQYTDEHGEVCPAGWKPGSDT#IKPNVDDSKEYFSKHN 198 || ||||||||+||++||||||||+|||||#|||+| |||||+| Sbjct 154 DETLRLVQAFQFTDKYGEVCPAGWQPGSDT#IKPDVQKSKEYFNKQK 199 |
| Caenorhabditis briggsae AF16 | Query 153 DEALRLVQAFQYTDEHGEVCPAGWKPGSDT#IKPNVDDSKEYFSKH 197 || ||||||||+ ++||||||||| |||||#|||+| |+||| || Sbjct 531 DETLRLVQAFQFVEKHGEVCPAGWTPGSDT#IKPDVKKSQEYFGKH 575 |
| Caenorhabditis elegans | Query 153 DEALRLVQAFQYTDEHGEVCPAGWKPGSDT#IKPNVDDSKEYFSKH 197 || ||||||||+ ++||||||||| |||||#||| | +|+||| || Sbjct 151 DETLRLVQAFQFVEKHGEVCPAGWTPGSDT#IKPGVKESQEYFKKH 195 |
| Onchocerca volvulus | Query 153 DEALRLVQAFQYTDEHGEVCPAGWKPGSDT#IKPNVDDSKEYFSKH 197 || ||||||||+ | ||||||| |+|||+|#||| | +||||| || Sbjct 155 DETLRLVQAFQFVDNHGEVCPANWQPGSET#IKPEVKESKEYFGKH 199 |
| Onchocerca volvulus | Query 153 DEALRLVQAFQYTDEHGEVCPAGWKPGSDT#IKPNVDDSKEYFSKH 197 || |||+||||+ | ||||||| |+|||+|#||| | +||||| || Sbjct 155 DETLRLIQAFQFVDNHGEVCPANWQPGSET#IKPEVKESKEYFGKH 199 |
| Onchocerca ochengi | Query 153 DEALRLVQAFQYTDEHGEVCPAGWKPGSDT#IKPNVDDSKEYFSKH 197 || |||+||||+ | ||||||| |+|||+|#||| | +||||| || Sbjct 155 DETLRLIQAFQFVDNHGEVCPANWQPGSET#IKPEVKESKEYFGKH 199 |
| Ascaris suum | Query 154 EALRLVQAFQYTDEHGEVCPAGWKPGSDT#IKPNVDDSKEYFSKH 197 | ||||||||+ |+||||||||| ||+||#||| | +|| || || Sbjct 152 ETLRLVQAFQFVDKHGEVCPAGWTPGADT#IKPGVKESKAYFEKH 195 |
| Drosophila melanogaster | Query 153 DEALRLVQAFQYTDEHGEVCPAGWKPGSDT#IKPNVDDSKEYFSKHN 198 || +|||||||||| |||||||||+||+||#| || ++ +||+|+| Sbjct 197 DETIRLVQAFQYTDTHGEVCPAGWRPGADT#IVPNPEEKTKYFAKNN 242 |
| Apis mellifera | Query 153 DEALRLVQAFQYTDEHGEVCPAGWKPGSDT#IKPNVDDSKEYF 194 || |||||||||||+|||||||||||| |#+||+| ||||| Sbjct 150 DETLRLVQAFQYTDKHGEVCPAGWKPGKKT#MKPDVVGSKEYF 191 |
| Dirofilaria immitis | Query 153 DEALRLVQAFQYTDEHGEVCPAGWKPGSDT#IKPNVDDSKEYFSKH 197 || |||+||||+ | ||||||| |+|||+ #||| | +|| || || Sbjct 155 DETLRLIQAFQFVDNHGEVCPANWQPGSEA#IKPGVKESKAYFEKH 199 |
| Drosophila pseudoobscura | Query 153 DEALRLVQAFQYTDEHGEVCPAGWKPGSDT#IKPNVDDSKEYFSKHN 198 || +|||||||||| |||||||||+||+||#| |+ ++ +||+|+| Sbjct 198 DETIRLVQAFQYTDTHGEVCPAGWRPGADT#IVPDPEEKTKYFAKNN 243 |
| Biomphalaria glabrata | Query 153 DEALRLVQAFQYTDEHGEVCPAGWKPGSDT#IKPNVDDSKEYFSKHN 198 || |||||||||||+||||||||||||| |#| |+ ||||| + + Sbjct 174 DETLRLVQAFQYTDKHGEVCPAGWKPGSAT#IIPDPKKSKEYFKQQS 219 |
| Acanthocheilonema viteae | Query 153 DEALRLVQAFQYTDEHGEVCPAGWKPGSDT#IKPNVDDSKEYF 194 || |||+||||+ |+||||||| | |||+|#||| | +|| || Sbjct 155 DETLRLIQAFQFVDKHGEVCPANWHPGSET#IKPGVKESKAYF 196 |
| Dirofilaria immitis | Query 153 DEALRLVQAFQYTDEHGEVCPAGWKPGSDT#IKPNVDDSKEYFSKH 197 || |||+||||+ | ||||||| | |||+ #||| | +|| || || Sbjct 155 DETLRLIQAFQFVDNHGEVCPANWHPGSEA#IKPGVKESKAYFEKH 199 |
| Ixodes scapularis | Query 153 DEALRLVQAFQYTDEHGEVCPAGWKPGSDT#IKPNVDDSKEYFSKHN 198 || ||||+|||+ ++|||||||||+| | |#|||+ +|+||||| | Sbjct 188 DETLRLVKAFQFVEKHGEVCPAGWQPDSPT#IKPDPKNSQEYFSKVN 233 |
| Aedes aegypti | Query 153 DEALRLVQAFQYTDEHGEVCPAGWKPGSDT#IKPNVDDSKEYFSKHN 198 || ||||||||+|||||||||| ||||| |#+ + |||||+ | Sbjct 151 DETLRLVQAFQFTDEHGEVCPANWKPGSKT#MVADPQKSKEYFNAAN 196 |
| Drosophila melanogaster | Query 153 DEALRLVQAFQYTDEHGEVCPAGWKPGSD--T#IKPNVDDSKEYFSKH 197 || |||++|||+ ++||||||| | | |+ |#|||+|++||+||||| Sbjct 187 DEVLRLIKAFQFVEQHGEVCPANWNPNSNPAT#IKPDVEESKKYFSKH 233 |
| Globodera rostochiensis | Query 153 DEALRLVQAFQYTDEHGEVCPAGWKPGSDT#IKPNVDDSKEYFSK 196 || |||||||+||| ||||||| |+|| ||#|||+ + |+ +| | Sbjct 154 DETLRLVQAFKYTDTHGEVCPANWQPGEDT#IKPDPEGSQTFFGK 197 |
| Trypanosoma cruzi strain CL Brener | Query 153 DEALRLVQAFQYTDEHGEVCPAGWKPGSDT#IKPNVDDSKEYF 194 |||||||+|||+ +|||||||| |||| |#+||+ + ||||| Sbjct 154 DEALRLVKAFQFVEEHGEVCPANWKPGDKT#MKPDPEKSKEYF 195 |
| Apis mellifera ligustica | Query 153 DEALRLVQAFQYTDEHGEVCPAGWKPGSDT#IKPNVDDSKEYFSKHN 198 || |||++|||+ ++||||||| |+| | |#|||| |||+|| | Sbjct 197 DETLRLIKAFQFVEKHGEVCPANWQPDSKT#IKPNPKDSKQYFESVN 242 |
| Litomosoides sigmodontis | Query 153 DEALRLVQAFQYTDEHGEVCPAGWKPGSDT#IKPNVDDSK 191 || |||+||||+ |+|||+||| |+|||+|#||| | +|| Sbjct 155 DETLRLIQAFQFVDKHGELCPANWQPGSET#IKPGVKESK 193 |
| Trypanosoma cruzi strain CL Brener | Query 153 DEALRLVQAFQYTDEHGEVCPAGWKPGSDT#IKPNVDDSKEYF 194 |||||||+|||+ +|||||||| |||| |#+||+ + |||+| Sbjct 154 DEALRLVKAFQFVEEHGEVCPANWKPGDKT#MKPDPEKSKEFF 195 |
| Echinococcus multilocularis | Query 153 DEALRLVQAFQYTDEHGEVCPAGWKPGSDT#IKPNVDDSKEYFS 195 ||||||+ |||+||+||||||| | ||| |# ||+ | | + | Sbjct 150 DEALRLLDAFQFTDKHGEVCPANWHPGSKT#FKPSAGDLKSFMS 192 |
| Echinococcus granulosus | Query 153 DEALRLVQAFQYTDEHGEVCPAGWKPGSDT#IKPNVDDSKEYFS 195 ||||||+ |||+||+||||||| |+||| |# ||+ | | + | Sbjct 142 DEALRLLDAFQFTDKHGEVCPANWQPGSKT#FKPSAGDLKSFMS 184 |
| Candida albicans SC5314 | Query 153 DEALRLVQAFQYTDEHGEVCPAGWKPGSDT#IKPNVDDSKEYFSKHN 198 +|+|||++|||+|+++|||||| | || +|#|||+ + |||||+| | Sbjct 150 EESLRLLEAFQFTEKYGEVCPANWHPGDET#IKPSPEASKEYFNKVN 195 |
| Schistosoma mansoni | Query 153 DEALRLVQAFQYTDEHGEVCPAGWKPGSDT#IKPNVDDSKEYFSKHN 198 || ||||+||||||++|||||| |+| |#|||++ |||| | | Sbjct 174 DEVLRLVRAFQYTDKYGEVCPADWQPKGPT#IKPDLKKYKEYFHKVN 219 |
| Trypanosoma cruzi | Query 153 DEALRLVQAFQYTDEHGEVCPAGWKPGSDT#IKPNVDDSKEYF 194 |||||||+|||+ ++||||||| |||| #+||+ + ||||| Sbjct 154 DEALRLVKAFQFVEKHGEVCPANWKPGDKA#MKPDPEKSKEYF 195 |
| Trypanosoma brucei TREU927 | Query 153 DEALRLVQAFQYTDEHGEVCPAGWKPGSDT#IKPNVDDSKEYFSKHN 198 || ||||+|||+ ++||||||| ||||| |#+| + + |++||| | Sbjct 154 DETLRLVKAFQFVEKHGEVCPANWKPGSKT#MKADPNGSQDYFSSMN 199 |
| Yarrowia lipolytica | Query 153 DEALRLVQAFQYTDEHGEVCPAGWKPGSDT#IKPNVDDSKEYFSKHN 198 +| |||+ |||+|++||||||| |+ ||||#|| + ++|||| | | Sbjct 150 EETLRLIDAFQFTEKHGEVCPANWQKGSDT#IKADPVNAKEYFEKAN 195 |
| Candida glabrata | Query 153 DEALRLVQAFQYTDEHGEVCPAGWKPGSDT#IKPNVDDSKEYFSKHN 198 +|+||||+ ||+||++| | | | ||| |#||| |+|||||| + | Sbjct 150 EESLRLVEGFQWTDKNGTVLPCNWTPGSAT#IKPTVEDSKEYFKEAN 195 |
| Entamoeba moshkovskii | Query 153 DEALRLVQAFQYTDEHGEVCPAGWKPGSDT#IKPNVDDSKEYFSKH 197 || +|+|+| |+||+|| ||| ||||+||#|+|+ | |+| | | Sbjct 173 DETIRIVKAIQFTDQHGAVCPLNWKPGNDT#IEPSHDGIKKYLSSH 217 |
[Site 3] AGWKPGSDTI183-KPNVDDSKEY
Ile183  Lys
Lys
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Ala174 | Gly175 | Trp176 | Lys177 | Pro178 | Gly179 | Ser180 | Asp181 | Thr182 | Ile183 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Lys184 | Pro185 | Asn186 | Val187 | Asp188 | Asp189 | Ser190 | Lys191 | Glu192 | Tyr193 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| EALRLVQAFQYTDEHGEVCPAGWKPGSDTIKPNVDDSKEYFSKHN |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | N/A | 100.00 | 14 | enhancer protein |
| 2 | Mus musculus | 100.00 | 11 | unnamed protein product |
| 3 | synthetic construct | 100.00 | 6 | peroxiredoxin 2 |
| 4 | Cricetulus griseus | 100.00 | 1 | PRDX2_CRIGR Peroxiredoxin-2 gi |
| 5 | Homo sapiens | 100.00 | 1 | peroxiredoxin 2 isoform a |
| 6 | Homo sapiens | 99.80 | 8 | TSA |
| 7 | Rattus norvegicus | 99.80 | 5 | peroxiredoxin 2 |
| 8 | synthetic construct | 99.40 | 1 | thioredoxin peroxidase 1-like |
| 9 | Bos taurus | 97.80 | 5 | peroxiredoxin 2 |
| 10 | Spermophilus tridecemlineatus | 86.70 | 1 | peroxiredoxin 2 |
| 11 | Xenopus tropicalis | 85.90 | 3 | peroxiredoxin 2 |
| 12 | Tetraodon nigroviridis | 85.50 | 3 | unnamed protein product |
| 13 | Canis familiaris | 84.70 | 8 | PREDICTED: similar to peroxiredoxin 1 |
| 14 | Gallus gallus | 84.00 | 1 | PREDICTED: similar to natural killer cell enhanci |
| 15 | Myotis lucifugus | 83.60 | 1 | PRDX1_MYOLU Peroxiredoxin-1 gi |
| 16 | Cricetulus griseus | 83.60 | 1 | PRDX1_CRIGR Peroxiredoxin-1 (Thioredoxin peroxida |
| 17 | Oncorhynchus mykiss | 82.80 | 1 | AF250193_1 natural killer cell enhancement factor |
| 18 | Danio rerio | 82.40 | 2 | hypothetical protein LOC541344 |
| 19 | Paralichthys olivaceus | 82.40 | 1 | natural killer enhancing factor |
| 20 | Cyprinus carpio | 80.90 | 1 | natural killer cell enhancing factor |
| 21 | Ostertagia ostertagi | 80.50 | 1 | thioredoxin peroxidase |
| 22 | Cynops pyrrhogaster | 80.10 | 1 | TDX_CYNPY Peroxiredoxin (Thioredoxin peroxidase) |
| 23 | Caenorhabditis briggsae AF16 | 79.30 | 2 | Hypothetical protein CBG02380 |
| 24 | Xenopus laevis | 79.00 | 3 | hypothetical protein LOC432262 |
| 25 | Ascaris suum | 79.00 | 1 | PRDX_ASCSU Peroxiredoxin (AsPrx) (Thioredoxin per |
| 26 | Caenorhabditis elegans | 79.00 | 1 | PeRoxireDoXin family member (prdx-2) |
| 27 | Onchocerca volvulus | 78.20 | 1 | peroxidoxin-2 |
| 28 | Onchocerca ochengi | 77.80 | 1 | peroxidoxin-2 |
| 29 | Onchocerca volvulus | 77.80 | 1 | thioredoxin peroxidase |
| 30 | Drosophila melanogaster | 76.60 | 3 | thioredoxin peroxidase 2 CG1274-PA, isoform A |
| 31 | Dirofilaria immitis | 75.90 | 1 | thioredoxin peroxidase |
| 32 | Apis mellifera | 75.50 | 1 | PREDICTED: similar to thioredoxin peroxidase 1 CG |
| 33 | Dirofilaria immitis | 75.10 | 1 | peroxidoxin-1 |
| 34 | Drosophila pseudoobscura | 74.70 | 3 | GA11781-PA |
| 35 | Biomphalaria glabrata | 74.30 | 1 | thioredoxin peroxidase BgTPx |
| 36 | Acanthocheilonema viteae | 73.60 | 1 | thiredoxin peroxidase |
| 37 | Ixodes scapularis | 70.10 | 1 | mitochondrial truncated thioredoxin-dependent per |
| 38 | Aedes aegypti | 69.30 | 1 | peroxiredoxins, prx-1, prx-2, prx-3 |
| 39 | Drosophila melanogaster | 68.90 | 1 | Peroxiredoxin 5037 CG5826-PA |
| 40 | Globodera rostochiensis | 68.60 | 1 | peroxiredoxin |
| 41 | Trypanosoma cruzi strain CL Brener | 68.20 | 1 | tryparedoxin peroxidase |
| 42 | Apis mellifera ligustica | 67.80 | 1 | thioredoxin peroxidase |
| 43 | Litomosoides sigmodontis | 67.80 | 1 | AF105258_1 peroxidoxin-2 |
| 44 | Candida albicans SC5314 | 67.40 | 1 | thioredoxin peroxidase |
| 45 | Trypanosoma cruzi strain CL Brener | 67.00 | 3 | tryparedoxin peroxidase, putative |
| 46 | Echinococcus multilocularis | 66.60 | 1 | thioredoxin peroxidase |
| 47 | Echinococcus granulosus | 66.60 | 1 | thioredoxin peroxidase |
| 48 | Yarrowia lipolytica | 65.90 | 1 | hypothetical protein |
| 49 | Candida glabrata | 65.50 | 2 | unnamed protein product |
| 50 | Schistosoma mansoni | 65.50 | 1 | AF301001_1 thioredoxin peroxidase 3 |
| 51 | Trypanosoma cruzi | 65.10 | 1 | AF320771_1 tryparedoxin peroxidase |
| 52 | Trypanosoma brucei TREU927 | 64.70 | 1 | tryparedoxin peroxidase |
| 53 | Nitrosococcus oceani ATCC 19707 | 64.30 | 1 | Alkyl hydroperoxide reductase/ Thiol specific ant |
| 54 | Fasciola hepatica | 63.20 | 2 | thiol-specific antioxidant protein |
| 55 | Entamoeba histolytica HM-1:IMSS | 63.20 | 1 | Truncated peroxiredoxin 1 |
| 56 | Entamoeba moshkovskii | 63.20 | 1 | peroxiredoxin |
Top-ranked sequences
| organism | matching |
|---|---|
| N/A | Query 154 EALRLVQAFQYTDEHGEVCPAGWKPGSDTI#KPNVDDSKEYFSKHN 198 ||||||||||||||||||||||||||||||#||||||||||||||| Query 154 EALRLVQAFQYTDEHGEVCPAGWKPGSDTI#KPNVDDSKEYFSKHN 198 |
| Mus musculus | Query 154 EALRLVQAFQYTDEHGEVCPAGWKPGSDTI#KPNVDDSKEYFSKHN 198 ||||||||||||||||||||||||||||||#||||||||||||||| Query 154 EALRLVQAFQYTDEHGEVCPAGWKPGSDTI#KPNVDDSKEYFSKHN 198 |
| synthetic construct | Query 154 EALRLVQAFQYTDEHGEVCPAGWKPGSDTI#KPNVDDSKEYFSKHN 198 ||||||||||||||||||||||||||||||#||||||||||||||| Query 154 EALRLVQAFQYTDEHGEVCPAGWKPGSDTI#KPNVDDSKEYFSKHN 198 |
| Cricetulus griseus | Query 154 EALRLVQAFQYTDEHGEVCPAGWKPGSDTI#KPNVDDSKEYFSKHN 198 ||||||||||||||||||||||||||||||#||||||||||||||| Query 154 EALRLVQAFQYTDEHGEVCPAGWKPGSDTI#KPNVDDSKEYFSKHN 198 |
| Homo sapiens | Query 154 EALRLVQAFQYTDEHGEVCPAGWKPGSDTI#KPNVDDSKEYFSKHN 198 ||||||||||||||||||||||||||||||#||||||||||||||| Query 154 EALRLVQAFQYTDEHGEVCPAGWKPGSDTI#KPNVDDSKEYFSKHN 198 |
| Homo sapiens | Query 154 EALRLVQAFQYTDEHGEVCPAGWKPGSDTI#KPNVDDSKEYFSKHN 198 ||||||||||||||||||||||||||||||#||||||||||||||| Query 154 EALRLVQAFQYTDEHGEVCPAGWKPGSDTI#KPNVDDSKEYFSKHN 198 |
| Rattus norvegicus | Query 154 EALRLVQAFQYTDEHGEVCPAGWKPGSDTI#KPNVDDSKEYFSKHN 198 ||||||||||||||||||||||||||||||#||||||||||||||| Query 154 EALRLVQAFQYTDEHGEVCPAGWKPGSDTI#KPNVDDSKEYFSKHN 198 |
| synthetic construct | Query 154 EALRLVQAFQYTDEHGEVCPAGWKPGSDTI#KPNVDDSKEYFSKHN 198 ||||||||||||||||||||||||||||||#||||||||||||||| Query 154 EALRLVQAFQYTDEHGEVCPAGWKPGSDTI#KPNVDDSKEYFSKHN 198 |
| Bos taurus | Query 154 EALRLVQAFQYTDEHGEVCPAGWKPGSDTI#KPNVDDSKEYFSKHN 198 ||||||||||||||||||||||| ||||||#||||||||||||||| Sbjct 155 EALRLVQAFQYTDEHGEVCPAGWTPGSDTI#KPNVDDSKEYFSKHN 199 |
| Spermophilus tridecemlineatus | Query 154 EALRLVQAFQYTDEHGEVCPAGWKPGSDTI#KPNVDDSKE 192 ||||||||||||||||||||||||||||||#||||||||| Query 154 EALRLVQAFQYTDEHGEVCPAGWKPGSDTI#KPNVDDSKE 192 |
| Xenopus tropicalis | Query 154 EALRLVQAFQYTDEHGEVCPAGWKPGSDTI#KPNVDDSKEYFSK 196 | |||||||||||+||||||||||||| ||#|||| ||||+||| Sbjct 162 ETLRLVQAFQYTDQHGEVCPAGWKPGSSTI#KPNVKDSKEFFSK 204 |
| Tetraodon nigroviridis | Query 154 EALRLVQAFQYTDEHGEVCPAGWKPGSDTI#KPNVDDSKEYFSKH 197 | ||||||||+||+||||||||||||||||#||+| |||+|||| Sbjct 155 ETLRLVQAFQFTDKHGEVCPAGWKPGSDTI#KPDVQKSKEFFSKH 198 |
| Canis familiaris | Query 154 EALRLVQAFQYTDEHGEVCPAGWKPGSDTI#KPNVDDSKEYFSKH 197 | ||||||||+||+||||||||||||||||#||+| ||||||| Sbjct 56 ETLRLVQAFQFTDKHGEVCPAGWKPGSDTI#KPDVQKSKEYFSKQ 99 |
| Gallus gallus | Query 154 EALRLVQAFQYTDEHGEVCPAGWKPGSDTI#KPNVDDSKEYFSKHN 198 | ||||||||+||+||||||||||||||||#||+| ||||||| Sbjct 155 ETLRLVQAFQFTDKHGEVCPAGWKPGSDTI#KPDVQKSKEYFSKQK 199 |
| Myotis lucifugus | Query 154 EALRLVQAFQYTDEHGEVCPAGWKPGSDTI#KPNVDDSKEYFSKH 197 | ||||||||+||+||||||||||||||||#||+| ||||||| Sbjct 155 ETLRLVQAFQFTDKHGEVCPAGWKPGSDTI#KPDVQKSKEYFSKQ 198 |
| Cricetulus griseus | Query 154 EALRLVQAFQYTDEHGEVCPAGWKPGSDTI#KPNVDDSKEYFSKH 197 | ||||||||+||+||||||||||||||||#||+| ||||||| Sbjct 155 EILRLVQAFQFTDKHGEVCPAGWKPGSDTI#KPDVQKSKEYFSKQ 198 |
| Oncorhynchus mykiss | Query 154 EALRLVQAFQYTDEHGEVCPAGWKPGSDTI#KPNVDDSKEYFSKH 197 | ||||||||+||+||||||||||||||||#||+| ||++||| Sbjct 155 ETLRLVQAFQFTDKHGEVCPAGWKPGSDTI#KPDVQKSKDFFSKQ 198 |
| Danio rerio | Query 154 EALRLVQAFQYTDEHGEVCPAGWKPGSDTI#KPNVDDSKEYFSKHN 198 | ||||||||+||+|||||||||||| |||#||+|+ ||++||| | Sbjct 155 ETLRLVQAFQFTDKHGEVCPAGWKPGKDTI#KPDVNQSKDFFSKQN 199 |
| Paralichthys olivaceus | Query 154 EALRLVQAFQYTDEHGEVCPAGWKPGSDTI#KPNVDDSKEYFSKH 197 | ||||||||+||+||||||||||||||||#||+| ||++||| Sbjct 155 ETLRLVQAFQFTDKHGEVCPAGWKPGSDTI#KPDVQKSKDFFSKQ 198 |
| Cyprinus carpio | Query 154 EALRLVQAFQYTDEHGEVCPAGWKPGSDTI#KPNVDDSKEYFSKHN 198 | ||||||||+||+|||||||||||| |||#||+| ||+|||| + Sbjct 155 ETLRLVQAFQFTDKHGEVCPAGWKPGKDTI#KPDVQQSKDYFSKQH 199 |
| Ostertagia ostertagi | Query 154 EALRLVQAFQYTDEHGEVCPAGWKPGSDTI#KPNVDDSKEYFSKHN 198 | ||||||||| |+||||||||| || ||#|| | |||||||| | Sbjct 149 ETLRLVQAFQYVDKHGEVCPAGWTPGKATI#KPGVKDSKEYFSKAN 193 |
| Cynops pyrrhogaster | Query 154 EALRLVQAFQYTDEHGEVCPAGWKPGSDTI#KPNVDDSKEYFSKHN 198 | ||||||||+||+ |||||||||||||||#||++ ||||||| Sbjct 155 ETLRLVQAFQHTDKFGEVCPAGWKPGSDTI#KPDISKSKEYFSKQK 199 |
| Caenorhabditis briggsae AF16 | Query 154 EALRLVQAFQYTDEHGEVCPAGWKPGSDTI#KPNVDDSKEYFSKH 197 | ||||||||+ ++||||||||| ||||||#||+| |+||| || Sbjct 532 ETLRLVQAFQFVEKHGEVCPAGWTPGSDTI#KPDVKKSQEYFGKH 575 |
| Xenopus laevis | Query 154 EALRLVQAFQYTDEHGEVCPAGWKPGSDTI#KPNVDDSKEYFSKHN 198 | ||||||||+||++||||||||+||||||#||+| |||||+| Sbjct 155 ETLRLVQAFQFTDKYGEVCPAGWQPGSDTI#KPDVQKSKEYFNKQK 199 |
| Ascaris suum | Query 154 EALRLVQAFQYTDEHGEVCPAGWKPGSDTI#KPNVDDSKEYFSKH 197 | ||||||||+ |+||||||||| ||+|||#|| | +|| || || Sbjct 152 ETLRLVQAFQFVDKHGEVCPAGWTPGADTI#KPGVKESKAYFEKH 195 |
| Caenorhabditis elegans | Query 154 EALRLVQAFQYTDEHGEVCPAGWKPGSDTI#KPNVDDSKEYFSKH 197 | ||||||||+ ++||||||||| ||||||#|| | +|+||| || Sbjct 152 ETLRLVQAFQFVEKHGEVCPAGWTPGSDTI#KPGVKESQEYFKKH 195 |
| Onchocerca volvulus | Query 154 EALRLVQAFQYTDEHGEVCPAGWKPGSDTI#KPNVDDSKEYFSKH 197 | ||||||||+ | ||||||| |+|||+||#|| | +||||| || Sbjct 156 ETLRLVQAFQFVDNHGEVCPANWQPGSETI#KPEVKESKEYFGKH 199 |
| Onchocerca ochengi | Query 154 EALRLVQAFQYTDEHGEVCPAGWKPGSDTI#KPNVDDSKEYFSKH 197 | |||+||||+ | ||||||| |+|||+||#|| | +||||| || Sbjct 156 ETLRLIQAFQFVDNHGEVCPANWQPGSETI#KPEVKESKEYFGKH 199 |
| Onchocerca volvulus | Query 154 EALRLVQAFQYTDEHGEVCPAGWKPGSDTI#KPNVDDSKEYFSKH 197 | |||+||||+ | ||||||| |+|||+||#|| | +||||| || Sbjct 156 ETLRLIQAFQFVDNHGEVCPANWQPGSETI#KPEVKESKEYFGKH 199 |
| Drosophila melanogaster | Query 154 EALRLVQAFQYTDEHGEVCPAGWKPGSDTI#KPNVDDSKEYFSKHN 198 | +|||||||||| |||||||||+||+|||# || ++ +||+|+| Sbjct 198 ETIRLVQAFQYTDTHGEVCPAGWRPGADTI#VPNPEEKTKYFAKNN 242 |
| Dirofilaria immitis | Query 154 EALRLVQAFQYTDEHGEVCPAGWKPGSDTI#KPNVDDSKEYFSKH 197 | |||+||||+ | ||||||| |+|||+ |#|| | +|| || || Sbjct 156 ETLRLIQAFQFVDNHGEVCPANWQPGSEAI#KPGVKESKAYFEKH 199 |
| Apis mellifera | Query 154 EALRLVQAFQYTDEHGEVCPAGWKPGSDTI#KPNVDDSKEYF 194 | |||||||||||+|||||||||||| |+#||+| ||||| Sbjct 151 ETLRLVQAFQYTDKHGEVCPAGWKPGKKTM#KPDVVGSKEYF 191 |
| Dirofilaria immitis | Query 154 EALRLVQAFQYTDEHGEVCPAGWKPGSDTI#KPNVDDSKEYFSKH 197 | |||+||||+ | ||||||| | |||+ |#|| | +|| || || Sbjct 156 ETLRLIQAFQFVDNHGEVCPANWHPGSEAI#KPGVKESKAYFEKH 199 |
| Drosophila pseudoobscura | Query 154 EALRLVQAFQYTDEHGEVCPAGWKPGSDTI#KPNVDDSKEYFSKHN 198 | +|||||||||| |||||||||+||+|||# |+ ++ +||+|+| Sbjct 199 ETIRLVQAFQYTDTHGEVCPAGWRPGADTI#VPDPEEKTKYFAKNN 243 |
| Biomphalaria glabrata | Query 154 EALRLVQAFQYTDEHGEVCPAGWKPGSDTI#KPNVDDSKEYFSKHN 198 | |||||||||||+||||||||||||| ||# |+ ||||| + + Sbjct 175 ETLRLVQAFQYTDKHGEVCPAGWKPGSATI#IPDPKKSKEYFKQQS 219 |
| Acanthocheilonema viteae | Query 154 EALRLVQAFQYTDEHGEVCPAGWKPGSDTI#KPNVDDSKEYF 194 | |||+||||+ |+||||||| | |||+||#|| | +|| || Sbjct 156 ETLRLIQAFQFVDKHGEVCPANWHPGSETI#KPGVKESKAYF 196 |
| Ixodes scapularis | Query 154 EALRLVQAFQYTDEHGEVCPAGWKPGSDTI#KPNVDDSKEYFSKHN 198 | ||||+|||+ ++|||||||||+| | ||#||+ +|+||||| | Sbjct 189 ETLRLVKAFQFVEKHGEVCPAGWQPDSPTI#KPDPKNSQEYFSKVN 233 |
| Aedes aegypti | Query 154 EALRLVQAFQYTDEHGEVCPAGWKPGSDTI#KPNVDDSKEYFSKHN 198 | ||||||||+|||||||||| ||||| |+# + |||||+ | Sbjct 152 ETLRLVQAFQFTDEHGEVCPANWKPGSKTM#VADPQKSKEYFNAAN 196 |
| Drosophila melanogaster | Query 154 EALRLVQAFQYTDEHGEVCPAGWKPGSD--TI#KPNVDDSKEYFSKH 197 | |||++|||+ ++||||||| | | |+ ||#||+|++||+||||| Sbjct 188 EVLRLIKAFQFVEQHGEVCPANWNPNSNPATI#KPDVEESKKYFSKH 233 |
| Globodera rostochiensis | Query 154 EALRLVQAFQYTDEHGEVCPAGWKPGSDTI#KPNVDDSKEYFSK 196 | |||||||+||| ||||||| |+|| |||#||+ + |+ +| | Sbjct 155 ETLRLVQAFKYTDTHGEVCPANWQPGEDTI#KPDPEGSQTFFGK 197 |
| Trypanosoma cruzi strain CL Brener | Query 154 EALRLVQAFQYTDEHGEVCPAGWKPGSDTI#KPNVDDSKEYF 194 ||||||+|||+ +|||||||| |||| |+#||+ + ||||| Sbjct 155 EALRLVKAFQFVEEHGEVCPANWKPGDKTM#KPDPEKSKEYF 195 |
| Apis mellifera ligustica | Query 154 EALRLVQAFQYTDEHGEVCPAGWKPGSDTI#KPNVDDSKEYFSKHN 198 | |||++|||+ ++||||||| |+| | ||#||| |||+|| | Sbjct 198 ETLRLIKAFQFVEKHGEVCPANWQPDSKTI#KPNPKDSKQYFESVN 242 |
| Litomosoides sigmodontis | Query 154 EALRLVQAFQYTDEHGEVCPAGWKPGSDTI#KPNVDDSK 191 | |||+||||+ |+|||+||| |+|||+||#|| | +|| Sbjct 156 ETLRLIQAFQFVDKHGELCPANWQPGSETI#KPGVKESK 193 |
| Candida albicans SC5314 | Query 154 EALRLVQAFQYTDEHGEVCPAGWKPGSDTI#KPNVDDSKEYFSKHN 198 |+|||++|||+|+++|||||| | || +||#||+ + |||||+| | Sbjct 151 ESLRLLEAFQFTEKYGEVCPANWHPGDETI#KPSPEASKEYFNKVN 195 |
| Trypanosoma cruzi strain CL Brener | Query 154 EALRLVQAFQYTDEHGEVCPAGWKPGSDTI#KPNVDDSKEYF 194 ||||||+|||+ +|||||||| |||| |+#||+ + |||+| Sbjct 155 EALRLVKAFQFVEEHGEVCPANWKPGDKTM#KPDPEKSKEFF 195 |
| Echinococcus multilocularis | Query 154 EALRLVQAFQYTDEHGEVCPAGWKPGSDTI#KPNVDDSKEYFS 195 |||||+ |||+||+||||||| | ||| | #||+ | | + | Sbjct 151 EALRLLDAFQFTDKHGEVCPANWHPGSKTF#KPSAGDLKSFMS 192 |
| Echinococcus granulosus | Query 154 EALRLVQAFQYTDEHGEVCPAGWKPGSDTI#KPNVDDSKEYFS 195 |||||+ |||+||+||||||| |+||| | #||+ | | + | Sbjct 143 EALRLLDAFQFTDKHGEVCPANWQPGSKTF#KPSAGDLKSFMS 184 |
| Yarrowia lipolytica | Query 154 EALRLVQAFQYTDEHGEVCPAGWKPGSDTI#KPNVDDSKEYFSKHN 198 | |||+ |||+|++||||||| |+ |||||#| + ++|||| | | Sbjct 151 ETLRLIDAFQFTEKHGEVCPANWQKGSDTI#KADPVNAKEYFEKAN 195 |
| Candida glabrata | Query 154 EALRLVQAFQYTDEHGEVCPAGWKPGSDTI#KPNVDDSKEYFSKHN 198 |+||||+ ||+||++| | | | ||| ||#|| |+|||||| + | Sbjct 151 ESLRLVEGFQWTDKNGTVLPCNWTPGSATI#KPTVEDSKEYFKEAN 195 |
| Schistosoma mansoni | Query 154 EALRLVQAFQYTDEHGEVCPAGWKPGSDTI#KPNVDDSKEYFSKHN 198 | ||||+||||||++|||||| |+| ||#||++ |||| | | Sbjct 175 EVLRLVRAFQYTDKYGEVCPADWQPKGPTI#KPDLKKYKEYFHKVN 219 |
| Trypanosoma cruzi | Query 154 EALRLVQAFQYTDEHGEVCPAGWKPGSDTI#KPNVDDSKEYF 194 ||||||+|||+ ++||||||| |||| +#||+ + ||||| Sbjct 155 EALRLVKAFQFVEKHGEVCPANWKPGDKAM#KPDPEKSKEYF 195 |
| Trypanosoma brucei TREU927 | Query 154 EALRLVQAFQYTDEHGEVCPAGWKPGSDTI#KPNVDDSKEYFSKHN 198 | ||||+|||+ ++||||||| ||||| |+#| + + |++||| | Sbjct 155 ETLRLVKAFQFVEKHGEVCPANWKPGSKTM#KADPNGSQDYFSSMN 199 |
| Nitrosococcus oceani ATCC 19707 | Query 154 EALRLVQAFQYTDEHGEVCPAGWKPGSDTI#KPNVDDSKEYFSKH 197 | ||+| | |+|+||||||||||+ | + |#+|+ + || ||| Sbjct 152 EMLRVVDALQFTEEHGEVCPAGWRKGEEAI#RPDAEGVAEYLSKH 195 |
| Fasciola hepatica | Query 154 EALRLVQAFQYTDEHGEVCPAGWKPGSDTI#KPNVDDSKEYFSKHN 198 |||||+ || + +|||||||| ||| | ||# | | || ||| | Sbjct 150 EALRLLDAFIFHEEHGEVCPANWKPKSKTI#VPTPDGSKAYFSSAN 194 |
| Entamoeba histolytica HM-1:IMSS | Query 154 EALRLVQAFQYTDEHGEVCPAGWKPGSDTI#KPNVDDSKEYFSKH 197 | +|+|+| |++|||| ||| |||| |||#+| | |+| + | Sbjct 118 ETIRIVKAIQFSDEHGAVCPLNWKPGKDTI#EPTPDGIKKYLTAH 161 |
| Entamoeba moshkovskii | Query 154 EALRLVQAFQYTDEHGEVCPAGWKPGSDTI#KPNVDDSKEYFSKH 197 | +|+|+| |+||+|| ||| ||||+|||#+|+ | |+| | | Sbjct 174 ETIRIVKAIQFTDQHGAVCPLNWKPGNDTI#EPSHDGIKKYLSSH 217 |
[Site 4] KPNVDDSKEY193-FSKHN
Tyr193  Phe
Phe
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Lys184 | Pro185 | Asn186 | Val187 | Asp188 | Asp189 | Ser190 | Lys191 | Glu192 | Tyr193 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Phe194 | Ser195 | Lys196 | His197 | Asn198 | - | - | - | - | - |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| YTDEHGEVCPAGWKPGSDTIKPNVDDSKEYFSKHN |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | N/A | 79.70 | 13 | A Chain A, Thioredoxin Peroxidase B From Red Bloo |
| 2 | Mus musculus | 79.70 | 11 | unnamed protein product |
| 3 | synthetic construct | 79.70 | 6 | peroxiredoxin 2 |
| 4 | Homo sapiens | 79.30 | 8 | TSA |
| 5 | Rattus norvegicus | 79.30 | 5 | peroxiredoxin 2 |
| 6 | Homo sapiens | 79.30 | 1 | peroxiredoxin 2 isoform a |
| 7 | Cricetulus griseus | 79.30 | 1 | PRDX2_CRIGR Peroxiredoxin-2 gi |
| 8 | synthetic construct | 79.00 | 1 | thioredoxin peroxidase 1-like |
| 9 | Bos taurus | 77.40 | 4 | peroxiredoxin 2 |
| 10 | Tetraodon nigroviridis | 68.20 | 3 | unnamed protein product |
| 11 | Xenopus tropicalis | 67.80 | 3 | peroxiredoxin 2 |
| 12 | Canis familiaris | 66.20 | 8 | PREDICTED: similar to peroxiredoxin 1 |
| 13 | Spermophilus tridecemlineatus | 66.20 | 1 | peroxiredoxin 2 |
| 14 | Gallus gallus | 65.90 | 1 | PREDICTED: similar to natural killer cell enhanci |
| 15 | Cricetulus griseus | 65.50 | 1 | PRDX1_CRIGR Peroxiredoxin-1 (Thioredoxin peroxida |
| 16 | Myotis lucifugus | 65.50 | 1 | PRDX1_MYOLU Peroxiredoxin-1 gi |
| 17 | Oncorhynchus mykiss | 65.10 | 1 | AF250193_1 natural killer cell enhancement factor |
| 18 | Paralichthys olivaceus | 64.70 | 1 | natural killer enhancing factor |
| 19 | Danio rerio | 64.30 | 2 | hypothetical protein LOC541344 |
| 20 | Caenorhabditis briggsae AF16 | 63.20 | 2 | Hypothetical protein CBG02380 |
| 21 | Cyprinus carpio | 62.80 | 1 | natural killer cell enhancing factor |
| 22 | Cynops pyrrhogaster | 62.80 | 1 | TDX_CYNPY Peroxiredoxin (Thioredoxin peroxidase) |
| 23 | Ostertagia ostertagi | 62.00 | 1 | thioredoxin peroxidase |
| 24 | Xenopus laevis | 61.20 | 2 | hypothetical protein LOC432262 |
| 25 | Caenorhabditis elegans | 60.80 | 1 | PeRoxireDoXin family member (prdx-2) |
| 26 | Ascaris suum | 60.50 | 1 | PRDX_ASCSU Peroxiredoxin (AsPrx) (Thioredoxin per |
| 27 | Onchocerca volvulus | 60.10 | 1 | peroxidoxin-2 |
| 28 | Onchocerca ochengi | 60.10 | 1 | peroxidoxin-2 |
| 29 | Onchocerca volvulus | 59.70 | 1 | thioredoxin peroxidase |
| 30 | Drosophila melanogaster | 58.50 | 1 | thioredoxin peroxidase 2 CG1274-PA, isoform A |
| 31 | Dirofilaria immitis | 58.50 | 1 | thioredoxin peroxidase |
| 32 | Apis mellifera | 58.20 | 1 | PREDICTED: similar to thioredoxin peroxidase 1 CG |
| 33 | Drosophila pseudoobscura | 56.60 | 3 | GA19159-PA |
| 34 | Acanthocheilonema viteae | 56.20 | 1 | thiredoxin peroxidase |
| 35 | Ixodes scapularis | 55.80 | 1 | mitochondrial truncated thioredoxin-dependent per |
| 36 | Biomphalaria glabrata | 55.80 | 1 | thioredoxin peroxidase BgTPx |
| 37 | Drosophila melanogaster | 55.50 | 1 | Peroxiredoxin 5037 CG5826-PA |
| 38 | Dirofilaria immitis | 54.70 | 1 | peroxidoxin-1 |
| 39 | Apis mellifera ligustica | 54.30 | 1 | thioredoxin peroxidase |
| 40 | Nitrosococcus oceani ATCC 19707 | 53.10 | 1 | Alkyl hydroperoxide reductase/ Thiol specific ant |
| 41 | Aedes aegypti | 52.80 | 1 | peroxiredoxins, prx-1, prx-2, prx-3 |
| 42 | Globodera rostochiensis | 51.60 | 1 | peroxiredoxin |
| 43 | Schistosoma mansoni | 51.60 | 1 | AF301001_1 thioredoxin peroxidase 3 |
| 44 | Candida albicans SC5314 | 51.20 | 1 | thioredoxin peroxidase |
| 45 | Echinococcus granulosus | 50.40 | 1 | thioredoxin peroxidase |
| 46 | Yarrowia lipolytica | 50.40 | 1 | hypothetical protein |
| 47 | Echinococcus multilocularis | 50.40 | 1 | thioredoxin peroxidase |
| 48 | Entamoeba moshkovskii | 50.10 | 3 | peroxiredoxin |
| 49 | Litomosoides sigmodontis | 50.10 | 1 | AF105258_1 peroxidoxin-2 |
| 50 | Trypanosoma cruzi strain CL Brener | 50.10 | 1 | tryparedoxin peroxidase |
| 51 | Candida glabrata | 49.30 | 2 | unnamed protein product |
| 52 | Fasciola hepatica | 49.30 | 2 | thiol-specific antioxidant protein |
| 53 | Tetrahymena thermophila SB210 | 49.30 | 1 | AhpC/TSA family protein |
| 54 | Trypanosoma cruzi strain CL Brener | 48.90 | 3 | tryparedoxin peroxidase, putative |
| 55 | Entamoeba histolytica HM-1:IMSS | 48.50 | 1 | Truncated peroxiredoxin 1 |
| 56 | Entamoeba histolytica | 48.50 | 1 | 30,000-Mr antigen |
Top-ranked sequences
| organism | matching |
|---|---|
| N/A | Query 164 YTDEHGEVCPAGWKPGSDTIKPNVDDSKEY#FSKHN 198 ||||||||||||||||||||||||||||||#||||| Query 164 YTDEHGEVCPAGWKPGSDTIKPNVDDSKEY#FSKHN 198 |
| Mus musculus | Query 164 YTDEHGEVCPAGWKPGSDTIKPNVDDSKEY#FSKHN 198 ||||||||||||||||||||||||||||||#||||| Query 164 YTDEHGEVCPAGWKPGSDTIKPNVDDSKEY#FSKHN 198 |
| synthetic construct | Query 164 YTDEHGEVCPAGWKPGSDTIKPNVDDSKEY#FSKHN 198 ||||||||||||||||||||||||||||||#||||| Query 164 YTDEHGEVCPAGWKPGSDTIKPNVDDSKEY#FSKHN 198 |
| Homo sapiens | Query 164 YTDEHGEVCPAGWKPGSDTIKPNVDDSKEY#FSKHN 198 ||||||||||||||||||||||||||||||#||||| Query 164 YTDEHGEVCPAGWKPGSDTIKPNVDDSKEY#FSKHN 198 |
| Rattus norvegicus | Query 164 YTDEHGEVCPAGWKPGSDTIKPNVDDSKEY#FSKHN 198 ||||||||||||||||||||||||||||||#||||| Query 164 YTDEHGEVCPAGWKPGSDTIKPNVDDSKEY#FSKHN 198 |
| Homo sapiens | Query 164 YTDEHGEVCPAGWKPGSDTIKPNVDDSKEY#FSKHN 198 ||||||||||||||||||||||||||||||#||||| Query 164 YTDEHGEVCPAGWKPGSDTIKPNVDDSKEY#FSKHN 198 |
| Cricetulus griseus | Query 164 YTDEHGEVCPAGWKPGSDTIKPNVDDSKEY#FSKHN 198 ||||||||||||||||||||||||||||||#||||| Query 164 YTDEHGEVCPAGWKPGSDTIKPNVDDSKEY#FSKHN 198 |
| synthetic construct | Query 164 YTDEHGEVCPAGWKPGSDTIKPNVDDSKEY#FSKHN 198 ||||||||||||||||||||||||||||||#||||| Query 164 YTDEHGEVCPAGWKPGSDTIKPNVDDSKEY#FSKHN 198 |
| Bos taurus | Query 164 YTDEHGEVCPAGWKPGSDTIKPNVDDSKEY#FSKHN 198 ||||||||||||| ||||||||||||||||#||||| Sbjct 165 YTDEHGEVCPAGWTPGSDTIKPNVDDSKEY#FSKHN 199 |
| Tetraodon nigroviridis | Query 164 YTDEHGEVCPAGWKPGSDTIKPNVDDSKEY#FSKH 197 +||+||||||||||||||||||+| |||+#|||| Sbjct 165 FTDKHGEVCPAGWKPGSDTIKPDVQKSKEF#FSKH 198 |
| Xenopus tropicalis | Query 164 YTDEHGEVCPAGWKPGSDTIKPNVDDSKEY#FSK 196 |||+||||||||||||| |||||| ||||+#||| Sbjct 172 YTDQHGEVCPAGWKPGSSTIKPNVKDSKEF#FSK 204 |
| Canis familiaris | Query 164 YTDEHGEVCPAGWKPGSDTIKPNVDDSKEY#FSKHN 198 +||+||||||||||||||||||+| ||||#||| Sbjct 66 FTDKHGEVCPAGWKPGSDTIKPDVQKSKEY#FSKQK 100 |
| Spermophilus tridecemlineatus | Query 164 YTDEHGEVCPAGWKPGSDTIKPNVDDSKE 192 ||||||||||||||||||||||||||||| Query 164 YTDEHGEVCPAGWKPGSDTIKPNVDDSKE 192 |
| Gallus gallus | Query 164 YTDEHGEVCPAGWKPGSDTIKPNVDDSKEY#FSKHN 198 +||+||||||||||||||||||+| ||||#||| Sbjct 165 FTDKHGEVCPAGWKPGSDTIKPDVQKSKEY#FSKQK 199 |
| Cricetulus griseus | Query 164 YTDEHGEVCPAGWKPGSDTIKPNVDDSKEY#FSKH 197 +||+||||||||||||||||||+| ||||#||| Sbjct 165 FTDKHGEVCPAGWKPGSDTIKPDVQKSKEY#FSKQ 198 |
| Myotis lucifugus | Query 164 YTDEHGEVCPAGWKPGSDTIKPNVDDSKEY#FSKH 197 +||+||||||||||||||||||+| ||||#||| Sbjct 165 FTDKHGEVCPAGWKPGSDTIKPDVQKSKEY#FSKQ 198 |
| Oncorhynchus mykiss | Query 164 YTDEHGEVCPAGWKPGSDTIKPNVDDSKEY#FSKH 197 +||+||||||||||||||||||+| ||++#||| Sbjct 165 FTDKHGEVCPAGWKPGSDTIKPDVQKSKDF#FSKQ 198 |
| Paralichthys olivaceus | Query 164 YTDEHGEVCPAGWKPGSDTIKPNVDDSKEY#FSKH 197 +||+||||||||||||||||||+| ||++#||| Sbjct 165 FTDKHGEVCPAGWKPGSDTIKPDVQKSKDF#FSKQ 198 |
| Danio rerio | Query 164 YTDEHGEVCPAGWKPGSDTIKPNVDDSKEY#FSKHN 198 +||+|||||||||||| |||||+|+ ||++#||| | Sbjct 165 FTDKHGEVCPAGWKPGKDTIKPDVNQSKDF#FSKQN 199 |
| Caenorhabditis briggsae AF16 | Query 164 YTDEHGEVCPAGWKPGSDTIKPNVDDSKEY#FSKH 197 + ++||||||||| ||||||||+| |+||#| || Sbjct 542 FVEKHGEVCPAGWTPGSDTIKPDVKKSQEY#FGKH 575 |
| Cyprinus carpio | Query 164 YTDEHGEVCPAGWKPGSDTIKPNVDDSKEY#FSKHN 198 +||+|||||||||||| |||||+| ||+|#||| + Sbjct 165 FTDKHGEVCPAGWKPGKDTIKPDVQQSKDY#FSKQH 199 |
| Cynops pyrrhogaster | Query 164 YTDEHGEVCPAGWKPGSDTIKPNVDDSKEY#FSKH 197 +||+ |||||||||||||||||++ ||||#||| Sbjct 165 HTDKFGEVCPAGWKPGSDTIKPDISKSKEY#FSKQ 198 |
| Ostertagia ostertagi | Query 164 YTDEHGEVCPAGWKPGSDTIKPNVDDSKEY#FSKHN 198 | |+||||||||| || |||| | |||||#||| | Sbjct 159 YVDKHGEVCPAGWTPGKATIKPGVKDSKEY#FSKAN 193 |
| Xenopus laevis | Query 164 YTDEHGEVCPAGWKPGSDTIKPNVDDSKEY#FSKHN 198 +||++||||||||+||||||||+| ||||#|+| Sbjct 165 FTDKYGEVCPAGWQPGSDTIKPDVQKSKEY#FNKQK 199 |
| Caenorhabditis elegans | Query 164 YTDEHGEVCPAGWKPGSDTIKPNVDDSKEY#FSKH 197 + ++||||||||| |||||||| | +|+||#| || Sbjct 162 FVEKHGEVCPAGWTPGSDTIKPGVKESQEY#FKKH 195 |
| Ascaris suum | Query 164 YTDEHGEVCPAGWKPGSDTIKPNVDDSKEY#FSKH 197 + |+||||||||| ||+||||| | +|| |#| || Sbjct 162 FVDKHGEVCPAGWTPGADTIKPGVKESKAY#FEKH 195 |
| Onchocerca volvulus | Query 164 YTDEHGEVCPAGWKPGSDTIKPNVDDSKEY#FSKH 197 + | ||||||| |+|||+|||| | +||||#| || Sbjct 166 FVDNHGEVCPANWQPGSETIKPEVKESKEY#FGKH 199 |
| Onchocerca ochengi | Query 164 YTDEHGEVCPAGWKPGSDTIKPNVDDSKEY#FSKH 197 + | ||||||| |+|||+|||| | +||||#| || Sbjct 166 FVDNHGEVCPANWQPGSETIKPEVKESKEY#FGKH 199 |
| Onchocerca volvulus | Query 164 YTDEHGEVCPAGWKPGSDTIKPNVDDSKEY#FSKH 197 + | ||||||| |+|||+|||| | +||||#| || Sbjct 166 FVDNHGEVCPANWQPGSETIKPEVKESKEY#FGKH 199 |
| Drosophila melanogaster | Query 164 YTDEHGEVCPAGWKPGSDTIKPNVDDSKEY#FSKHN 198 ||| |||||||||+||+||| || ++ +|#|+|+| Sbjct 208 YTDTHGEVCPAGWRPGADTIVPNPEEKTKY#FAKNN 242 |
| Dirofilaria immitis | Query 164 YTDEHGEVCPAGWKPGSDTIKPNVDDSKEY#FSKH 197 + | ||||||| |+|||+ ||| | +|| |#| || Sbjct 166 FVDNHGEVCPANWQPGSEAIKPGVKESKAY#FEKH 199 |
| Apis mellifera | Query 164 YTDEHGEVCPAGWKPGSDTIKPNVDDSKEY#F 194 |||+|||||||||||| |+||+| ||||#| Sbjct 161 YTDKHGEVCPAGWKPGKKTMKPDVVGSKEY#F 191 |
| Drosophila pseudoobscura | Query 164 YTDEHGEVCPAGWKPGSD--TIKPNVDDSKEY#FSKH 197 + ++||||||| | | ++ ||||+|+|||+|#|||| Sbjct 197 FVEQHGEVCPANWNPKTNPATIKPDVEDSKQY#FSKH 232 |
| Acanthocheilonema viteae | Query 164 YTDEHGEVCPAGWKPGSDTIKPNVDDSKEY#F 194 + |+||||||| | |||+|||| | +|| |#| Sbjct 166 FVDKHGEVCPANWHPGSETIKPGVKESKAY#F 196 |
| Ixodes scapularis | Query 164 YTDEHGEVCPAGWKPGSDTIKPNVDDSKEY#FSKHN 198 + ++|||||||||+| | ||||+ +|+||#||| | Sbjct 199 FVEKHGEVCPAGWQPDSPTIKPDPKNSQEY#FSKVN 233 |
| Biomphalaria glabrata | Query 164 YTDEHGEVCPAGWKPGSDTIKPNVDDSKEY#FSKHN 198 |||+||||||||||||| || |+ ||||#| + + Sbjct 185 YTDKHGEVCPAGWKPGSATIIPDPKKSKEY#FKQQS 219 |
| Drosophila melanogaster | Query 164 YTDEHGEVCPAGWKPGSD--TIKPNVDDSKEY#FSKH 197 + ++||||||| | | |+ ||||+|++||+|#|||| Sbjct 198 FVEQHGEVCPANWNPNSNPATIKPDVEESKKY#FSKH 233 |
| Dirofilaria immitis | Query 164 YTDEHGEVCPAGWKPGSDTIKPNVDDSKEY#FSKH 197 + | ||||||| | |||+ ||| | +|| |#| || Sbjct 166 FVDNHGEVCPANWHPGSEAIKPGVKESKAY#FEKH 199 |
| Apis mellifera ligustica | Query 164 YTDEHGEVCPAGWKPGSDTIKPNVDDSKEY#FSKHN 198 + ++||||||| |+| | ||||| |||+|#| | Sbjct 208 FVEKHGEVCPANWQPDSKTIKPNPKDSKQY#FESVN 242 |
| Nitrosococcus oceani ATCC 19707 | Query 164 YTDEHGEVCPAGWKPGSDTIKPNVDDSKEY#FSKH 197 +|+||||||||||+ | + |+|+ + ||# ||| Sbjct 162 FTEEHGEVCPAGWRKGEEAIRPDAEGVAEY#LSKH 195 |
| Aedes aegypti | Query 164 YTDEHGEVCPAGWKPGSDTIKPNVDDSKEY#FSKHN 198 +|||||||||| ||||| |+ + ||||#|+ | Sbjct 162 FTDEHGEVCPANWKPGSKTMVADPQKSKEY#FNAAN 196 |
| Globodera rostochiensis | Query 164 YTDEHGEVCPAGWKPGSDTIKPNVDDSKEY#FSK 196 ||| ||||||| |+|| |||||+ + |+ +#| | Sbjct 165 YTDTHGEVCPANWQPGEDTIKPDPEGSQTF#FGK 197 |
| Schistosoma mansoni | Query 164 YTDEHGEVCPAGWKPGSDTIKPNVDDSKEY#FSKHN 198 |||++|||||| |+| ||||++ |||#| | | Sbjct 185 YTDKYGEVCPADWQPKGPTIKPDLKKYKEY#FHKVN 219 |
| Candida albicans SC5314 | Query 164 YTDEHGEVCPAGWKPGSDTIKPNVDDSKEY#FSKHN 198 +|+++|||||| | || +||||+ + ||||#|+| | Sbjct 161 FTEKYGEVCPANWHPGDETIKPSPEASKEY#FNKVN 195 |
| Echinococcus granulosus | Query 164 YTDEHGEVCPAGWKPGSDTIKPNVDDSKEY#FS 195 +||+||||||| |+||| | ||+ | | +# | Sbjct 153 FTDKHGEVCPANWQPGSKTFKPSAGDLKSF#MS 184 |
| Yarrowia lipolytica | Query 164 YTDEHGEVCPAGWKPGSDTIKPNVDDSKEY#FSKHN 198 +|++||||||| |+ |||||| + ++|||#| | | Sbjct 161 FTEKHGEVCPANWQKGSDTIKADPVNAKEY#FEKAN 195 |
| Echinococcus multilocularis | Query 164 YTDEHGEVCPAGWKPGSDTIKPNVDDSKEY#FS 195 +||+||||||| | ||| | ||+ | | +# | Sbjct 161 FTDKHGEVCPANWHPGSKTFKPSAGDLKSF#MS 192 |
| Entamoeba moshkovskii | Query 164 YTDEHGEVCPAGWKPGSDTIKPNVDDSKEY#FSKH 197 +||+|| ||| ||||+|||+|+ | |+|# | | Sbjct 184 FTDQHGAVCPLNWKPGNDTIEPSHDGIKKY#LSSH 217 |
| Litomosoides sigmodontis | Query 164 YTDEHGEVCPAGWKPGSDTIKPNVDDSK 191 + |+|||+||| |+|||+|||| | +|| Sbjct 166 FVDKHGELCPANWQPGSETIKPGVKESK 193 |
| Trypanosoma cruzi strain CL Brener | Query 164 YTDEHGEVCPAGWKPGSDTIKPNVDDSKEY#F 194 + +|||||||| |||| |+||+ + ||||#| Sbjct 165 FVEEHGEVCPANWKPGDKTMKPDPEKSKEY#F 195 |
| Candida glabrata | Query 164 YTDEHGEVCPAGWKPGSDTIKPNVDDSKEY#FSKHN 198 +||++| | | | ||| |||| |+|||||#| + | Sbjct 161 WTDKNGTVLPCNWTPGSATIKPTVEDSKEY#FKEAN 195 |
| Fasciola hepatica | Query 164 YTDEHGEVCPAGWKPGSDTIKPNVDDSKEY#FSKHN 198 + +|||||||| ||| | || | | || |#|| | Sbjct 160 FHEEHGEVCPANWKPKSKTIVPTPDGSKAY#FSSAN 194 |
| Tetrahymena thermophila SB210 | Query 164 YTDEHGEVCPAGWKPGSDTIKPNVDDSKEY#FS 195 +|| ||||||| |+|| || |+ | +|#|| Sbjct 196 HTDTHGEVCPANWQPGQKTIIPDQDQKIKY#FS 227 |
| Trypanosoma cruzi strain CL Brener | Query 164 YTDEHGEVCPAGWKPGSDTIKPNVDDSKEY#F 194 + +|||||||| |||| |+||+ + |||+#| Sbjct 165 FVEEHGEVCPANWKPGDKTMKPDPEKSKEF#F 195 |
| Entamoeba histolytica HM-1:IMSS | Query 164 YTDEHGEVCPAGWKPGSDTIKPNVDDSKEY#FSKH 197 ++|||| ||| |||| |||+| | |+|# + | Sbjct 128 FSDEHGAVCPLNWKPGKDTIEPTPDGIKKY#LTAH 161 |
| Entamoeba histolytica | Query 164 YTDEHGEVCPAGWKPGSDTIKPNVDDSKEY#FSKH 197 ++|||| ||| |||| |||+| | |+|# + | Sbjct 185 FSDEHGAVCPLNWKPGKDTIEPTPDGIKKY#LTAH 218 |
[Site 5] PNVDDSKEYF194-SKHN
Phe194  Ser
Ser
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Pro185 | Asn186 | Val187 | Asp188 | Asp189 | Ser190 | Lys191 | Glu192 | Tyr193 | Phe194 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Ser195 | Lys196 | His197 | Asn198 | - | - | - | - | - | - |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| TDEHGEVCPAGWKPGSDTIKPNVDDSKEYFSKHN |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | N/A | 77.00 | 13 | enhancer protein |
| 2 | Mus musculus | 77.00 | 11 | unnamed protein product |
| 3 | synthetic construct | 77.00 | 6 | peroxiredoxin 2 |
| 4 | Rattus norvegicus | 77.00 | 5 | Peroxiredoxin 2 |
| 5 | Cricetulus griseus | 77.00 | 1 | PRDX2_CRIGR Peroxiredoxin-2 gi |
| 6 | Homo sapiens | 77.00 | 1 | peroxiredoxin 2 isoform a |
| 7 | Homo sapiens | 76.60 | 8 | TSA |
| 8 | synthetic construct | 76.30 | 1 | thioredoxin peroxidase 1-like |
| 9 | Bos taurus | 74.70 | 4 | peroxiredoxin 2 |
| 10 | Tetraodon nigroviridis | 65.50 | 3 | unnamed protein product |
| 11 | Xenopus tropicalis | 65.10 | 3 | peroxiredoxin 2 |
| 12 | Canis familiaris | 64.70 | 8 | PREDICTED: similar to peroxiredoxin 1 |
| 13 | Gallus gallus | 64.30 | 1 | PREDICTED: similar to natural killer cell enhanci |
| 14 | Myotis lucifugus | 63.90 | 1 | PRDX1_MYOLU Peroxiredoxin-1 gi |
| 15 | Cricetulus griseus | 63.90 | 1 | PRDX1_CRIGR Peroxiredoxin-1 (Thioredoxin peroxida |
| 16 | Oncorhynchus mykiss | 63.50 | 1 | AF250193_1 natural killer cell enhancement factor |
| 17 | Spermophilus tridecemlineatus | 63.50 | 1 | peroxiredoxin 2 |
| 18 | Paralichthys olivaceus | 63.20 | 1 | natural killer enhancing factor |
| 19 | Danio rerio | 62.40 | 2 | hypothetical protein LOC541344 |
| 20 | Caenorhabditis briggsae AF16 | 62.00 | 2 | Hypothetical protein CBG02380 |
| 21 | Cyprinus carpio | 61.20 | 1 | natural killer cell enhancing factor |
| 22 | Cynops pyrrhogaster | 61.20 | 1 | TDX_CYNPY Peroxiredoxin (Thioredoxin peroxidase) |
| 23 | Xenopus laevis | 60.10 | 2 | hypothetical protein LOC432262 |
| 24 | Ostertagia ostertagi | 59.70 | 1 | thioredoxin peroxidase |
| 25 | Caenorhabditis elegans | 59.30 | 1 | PeRoxireDoXin family member (prdx-2) |
| 26 | Ascaris suum | 58.90 | 1 | PRDX_ASCSU Peroxiredoxin (AsPrx) (Thioredoxin per |
| 27 | Onchocerca ochengi | 58.50 | 1 | peroxidoxin-2 |
| 28 | Onchocerca volvulus | 58.50 | 1 | thioredoxin peroxidase |
| 29 | Onchocerca volvulus | 58.50 | 1 | peroxidoxin-2 |
| 30 | Drosophila melanogaster | 56.20 | 1 | thioredoxin peroxidase 2 CG1274-PA, isoform A |
| 31 | Drosophila pseudoobscura | 55.50 | 2 | GA19159-PA |
| 32 | Apis mellifera | 55.50 | 1 | PREDICTED: similar to thioredoxin peroxidase 1 CG |
| 33 | Acanthocheilonema viteae | 55.10 | 1 | thiredoxin peroxidase |
| 34 | Ixodes scapularis | 54.70 | 1 | mitochondrial truncated thioredoxin-dependent per |
| 35 | Dirofilaria immitis | 54.70 | 1 | thioredoxin peroxidase |
| 36 | Drosophila melanogaster | 53.90 | 1 | Peroxiredoxin 5037 CG5826-PA |
| 37 | Biomphalaria glabrata | 53.10 | 1 | thioredoxin peroxidase BgTPx |
| 38 | Dirofilaria immitis | 53.10 | 1 | peroxidoxin-1 |
| 39 | Apis mellifera ligustica | 52.80 | 1 | thioredoxin peroxidase |
| 40 | Nitrosococcus oceani ATCC 19707 | 52.00 | 1 | Alkyl hydroperoxide reductase/ Thiol specific ant |
| 41 | Aedes aegypti | 50.10 | 1 | peroxiredoxins, prx-1, prx-2, prx-3 |
| 42 | Candida albicans SC5314 | 49.70 | 1 | thioredoxin peroxidase |
| 43 | Schistosoma mansoni | 49.30 | 1 | AF301001_1 thioredoxin peroxidase 3 |
| 44 | Fasciola hepatica | 48.90 | 2 | thiol-specific antioxidant protein |
| 45 | Litomosoides sigmodontis | 48.90 | 1 | AF105258_1 peroxidoxin-2 |
| 46 | Yarrowia lipolytica | 48.90 | 1 | hypothetical protein |
| 47 | Echinococcus granulosus | 48.90 | 1 | thioredoxin peroxidase |
| 48 | Tetrahymena thermophila SB210 | 48.90 | 1 | AhpC/TSA family protein |
| 49 | Globodera rostochiensis | 48.90 | 1 | peroxiredoxin |
| 50 | Echinococcus multilocularis | 48.90 | 1 | thioredoxin peroxidase |
| 51 | Entamoeba moshkovskii | 48.50 | 3 | peroxiredoxin |
| 52 | Candida glabrata | 48.50 | 2 | unnamed protein product |
| 53 | Trypanosoma cruzi strain CL Brener | 48.50 | 1 | tryparedoxin peroxidase |
| 54 | Chlamydomonas reinhardtii | 47.80 | 1 | 2-cys peroxiredoxin, chloroplastic |
| 55 | Trypanosoma cruzi strain CL Brener | 47.40 | 3 | tryparedoxin peroxidase, putative |
| 56 | Entamoeba histolytica HM-1:IMSS | 47.00 | 1 | Truncated peroxiredoxin 1 |
| 57 | Entamoeba histolytica | 47.00 | 1 | 30,000-Mr antigen |
| 58 | Oryza sativa (japonica cultivar-group) | 46.60 | 1 | OJ991214_12.15 |
Top-ranked sequences
| organism | matching |
|---|---|
| N/A | Query 165 TDEHGEVCPAGWKPGSDTIKPNVDDSKEYF#SKHN 198 ||||||||||||||||||||||||||||||#|||| Query 165 TDEHGEVCPAGWKPGSDTIKPNVDDSKEYF#SKHN 198 |
| Mus musculus | Query 165 TDEHGEVCPAGWKPGSDTIKPNVDDSKEYF#SKHN 198 ||||||||||||||||||||||||||||||#|||| Query 165 TDEHGEVCPAGWKPGSDTIKPNVDDSKEYF#SKHN 198 |
| synthetic construct | Query 165 TDEHGEVCPAGWKPGSDTIKPNVDDSKEYF#SKHN 198 ||||||||||||||||||||||||||||||#|||| Query 165 TDEHGEVCPAGWKPGSDTIKPNVDDSKEYF#SKHN 198 |
| Rattus norvegicus | Query 165 TDEHGEVCPAGWKPGSDTIKPNVDDSKEYF#SKHN 198 ||||||||||||||||||||||||||||||#|||| Query 165 TDEHGEVCPAGWKPGSDTIKPNVDDSKEYF#SKHN 198 |
| Cricetulus griseus | Query 165 TDEHGEVCPAGWKPGSDTIKPNVDDSKEYF#SKHN 198 ||||||||||||||||||||||||||||||#|||| Query 165 TDEHGEVCPAGWKPGSDTIKPNVDDSKEYF#SKHN 198 |
| Homo sapiens | Query 165 TDEHGEVCPAGWKPGSDTIKPNVDDSKEYF#SKHN 198 ||||||||||||||||||||||||||||||#|||| Query 165 TDEHGEVCPAGWKPGSDTIKPNVDDSKEYF#SKHN 198 |
| Homo sapiens | Query 165 TDEHGEVCPAGWKPGSDTIKPNVDDSKEYF#SKHN 198 ||||||||||||||||||||||||||||||#|||| Query 165 TDEHGEVCPAGWKPGSDTIKPNVDDSKEYF#SKHN 198 |
| synthetic construct | Query 165 TDEHGEVCPAGWKPGSDTIKPNVDDSKEYF#SKHN 198 ||||||||||||||||||||||||||||||#|||| Query 165 TDEHGEVCPAGWKPGSDTIKPNVDDSKEYF#SKHN 198 |
| Bos taurus | Query 165 TDEHGEVCPAGWKPGSDTIKPNVDDSKEYF#SKHN 198 |||||||||||| |||||||||||||||||#|||| Sbjct 166 TDEHGEVCPAGWTPGSDTIKPNVDDSKEYF#SKHN 199 |
| Tetraodon nigroviridis | Query 165 TDEHGEVCPAGWKPGSDTIKPNVDDSKEYF#SKH 197 ||+||||||||||||||||||+| |||+|#||| Sbjct 166 TDKHGEVCPAGWKPGSDTIKPDVQKSKEFF#SKH 198 |
| Xenopus tropicalis | Query 165 TDEHGEVCPAGWKPGSDTIKPNVDDSKEYF#SK 196 ||+||||||||||||| |||||| ||||+|#|| Sbjct 173 TDQHGEVCPAGWKPGSSTIKPNVKDSKEFF#SK 204 |
| Canis familiaris | Query 165 TDEHGEVCPAGWKPGSDTIKPNVDDSKEYF#SKH 197 ||+||||||||||||||||||+| |||||#|| Sbjct 67 TDKHGEVCPAGWKPGSDTIKPDVQKSKEYF#SKQ 99 |
| Gallus gallus | Query 165 TDEHGEVCPAGWKPGSDTIKPNVDDSKEYF#SKHN 198 ||+||||||||||||||||||+| |||||#|| Sbjct 166 TDKHGEVCPAGWKPGSDTIKPDVQKSKEYF#SKQK 199 |
| Myotis lucifugus | Query 165 TDEHGEVCPAGWKPGSDTIKPNVDDSKEYF#SKH 197 ||+||||||||||||||||||+| |||||#|| Sbjct 166 TDKHGEVCPAGWKPGSDTIKPDVQKSKEYF#SKQ 198 |
| Cricetulus griseus | Query 165 TDEHGEVCPAGWKPGSDTIKPNVDDSKEYF#SKH 197 ||+||||||||||||||||||+| |||||#|| Sbjct 166 TDKHGEVCPAGWKPGSDTIKPDVQKSKEYF#SKQ 198 |
| Oncorhynchus mykiss | Query 165 TDEHGEVCPAGWKPGSDTIKPNVDDSKEYF#SKH 197 ||+||||||||||||||||||+| ||++|#|| Sbjct 166 TDKHGEVCPAGWKPGSDTIKPDVQKSKDFF#SKQ 198 |
| Spermophilus tridecemlineatus | Query 165 TDEHGEVCPAGWKPGSDTIKPNVDDSKE 192 |||||||||||||||||||||||||||| Query 165 TDEHGEVCPAGWKPGSDTIKPNVDDSKE 192 |
| Paralichthys olivaceus | Query 165 TDEHGEVCPAGWKPGSDTIKPNVDDSKEYF#SKH 197 ||+||||||||||||||||||+| ||++|#|| Sbjct 166 TDKHGEVCPAGWKPGSDTIKPDVQKSKDFF#SKQ 198 |
| Danio rerio | Query 165 TDEHGEVCPAGWKPGSDTIKPNVDDSKEYF#SKHN 198 ||+|||||||||||| |||||+|+ ||++|#|| | Sbjct 166 TDKHGEVCPAGWKPGKDTIKPDVNQSKDFF#SKQN 199 |
| Caenorhabditis briggsae AF16 | Query 166 DEHGEVCPAGWKPGSDTIKPNVDDSKEYF#SKH 197 ++||||||||| ||||||||+| |+|||# || Sbjct 544 EKHGEVCPAGWTPGSDTIKPDVKKSQEYF#GKH 575 |
| Cyprinus carpio | Query 165 TDEHGEVCPAGWKPGSDTIKPNVDDSKEYF#SKHN 198 ||+|||||||||||| |||||+| ||+||#|| + Sbjct 166 TDKHGEVCPAGWKPGKDTIKPDVQQSKDYF#SKQH 199 |
| Cynops pyrrhogaster | Query 165 TDEHGEVCPAGWKPGSDTIKPNVDDSKEYF#SKHN 198 ||+ |||||||||||||||||++ |||||#|| Sbjct 166 TDKFGEVCPAGWKPGSDTIKPDISKSKEYF#SKQK 199 |
| Xenopus laevis | Query 165 TDEHGEVCPAGWKPGSDTIKPNVDDSKEYF#SKH 197 ||++||||||||+||||||||+| |||||#+| Sbjct 166 TDKYGEVCPAGWQPGSDTIKPDVQKSKEYF#NKQ 198 |
| Ostertagia ostertagi | Query 165 TDEHGEVCPAGWKPGSDTIKPNVDDSKEYF#SKHN 198 |+||||||||| || |||| | ||||||#|| | Sbjct 160 VDKHGEVCPAGWTPGKATIKPGVKDSKEYF#SKAN 193 |
| Caenorhabditis elegans | Query 165 TDEHGEVCPAGWKPGSDTIKPNVDDSKEYF#SKH 197 ++||||||||| |||||||| | +|+|||# || Sbjct 163 VEKHGEVCPAGWTPGSDTIKPGVKESQEYF#KKH 195 |
| Ascaris suum | Query 165 TDEHGEVCPAGWKPGSDTIKPNVDDSKEYF#SKH 197 |+||||||||| ||+||||| | +|| ||# || Sbjct 163 VDKHGEVCPAGWTPGADTIKPGVKESKAYF#EKH 195 |
| Onchocerca ochengi | Query 165 TDEHGEVCPAGWKPGSDTIKPNVDDSKEYF#SKH 197 | ||||||| |+|||+|||| | +|||||# || Sbjct 167 VDNHGEVCPANWQPGSETIKPEVKESKEYF#GKH 199 |
| Onchocerca volvulus | Query 165 TDEHGEVCPAGWKPGSDTIKPNVDDSKEYF#SKH 197 | ||||||| |+|||+|||| | +|||||# || Sbjct 167 VDNHGEVCPANWQPGSETIKPEVKESKEYF#GKH 199 |
| Onchocerca volvulus | Query 165 TDEHGEVCPAGWKPGSDTIKPNVDDSKEYF#SKH 197 | ||||||| |+|||+|||| | +|||||# || Sbjct 167 VDNHGEVCPANWQPGSETIKPEVKESKEYF#GKH 199 |
| Drosophila melanogaster | Query 165 TDEHGEVCPAGWKPGSDTIKPNVDDSKEYF#SKHN 198 || |||||||||+||+||| || ++ +||#+|+| Sbjct 209 TDTHGEVCPAGWRPGADTIVPNPEEKTKYF#AKNN 242 |
| Drosophila pseudoobscura | Query 166 DEHGEVCPAGWKPGSD--TIKPNVDDSKEYF#SKH 197 ++||||||| | | ++ ||||+|+|||+||#||| Sbjct 199 EQHGEVCPANWNPKTNPATIKPDVEDSKQYF#SKH 232 |
| Apis mellifera | Query 165 TDEHGEVCPAGWKPGSDTIKPNVDDSKEYF# 194 ||+|||||||||||| |+||+| |||||# Sbjct 162 TDKHGEVCPAGWKPGKKTMKPDVVGSKEYF# 191 |
| Acanthocheilonema viteae | Query 166 DEHGEVCPAGWKPGSDTIKPNVDDSKEYF# 194 |+||||||| | |||+|||| | +|| ||# Sbjct 168 DKHGEVCPANWHPGSETIKPGVKESKAYF# 196 |
| Ixodes scapularis | Query 166 DEHGEVCPAGWKPGSDTIKPNVDDSKEYF#SKHN 198 ++|||||||||+| | ||||+ +|+|||#|| | Sbjct 201 EKHGEVCPAGWQPDSPTIKPDPKNSQEYF#SKVN 233 |
| Dirofilaria immitis | Query 165 TDEHGEVCPAGWKPGSDTIKPNVDDSKEYF#SKH 197 | ||||||| |+|||+ ||| | +|| ||# || Sbjct 167 VDNHGEVCPANWQPGSEAIKPGVKESKAYF#EKH 199 |
| Drosophila melanogaster | Query 166 DEHGEVCPAGWKPGSD--TIKPNVDDSKEYF#SKH 197 ++||||||| | | |+ ||||+|++||+||#||| Sbjct 200 EQHGEVCPANWNPNSNPATIKPDVEESKKYF#SKH 233 |
| Biomphalaria glabrata | Query 165 TDEHGEVCPAGWKPGSDTIKPNVDDSKEYF#SKHN 198 ||+||||||||||||| || |+ |||||# + + Sbjct 186 TDKHGEVCPAGWKPGSATIIPDPKKSKEYF#KQQS 219 |
| Dirofilaria immitis | Query 165 TDEHGEVCPAGWKPGSDTIKPNVDDSKEYF#SKH 197 | ||||||| | |||+ ||| | +|| ||# || Sbjct 167 VDNHGEVCPANWHPGSEAIKPGVKESKAYF#EKH 199 |
| Apis mellifera ligustica | Query 166 DEHGEVCPAGWKPGSDTIKPNVDDSKEYF#SKHN 198 ++||||||| |+| | ||||| |||+||# | Sbjct 210 EKHGEVCPANWQPDSKTIKPNPKDSKQYF#ESVN 242 |
| Nitrosococcus oceani ATCC 19707 | Query 165 TDEHGEVCPAGWKPGSDTIKPNVDDSKEYF#SKH 197 |+||||||||||+ | + |+|+ + || #||| Sbjct 163 TEEHGEVCPAGWRKGEEAIRPDAEGVAEYL#SKH 195 |
| Aedes aegypti | Query 165 TDEHGEVCPAGWKPGSDTIKPNVDDSKEYF#SKHN 198 |||||||||| ||||| |+ + |||||#+ | Sbjct 163 TDEHGEVCPANWKPGSKTMVADPQKSKEYF#NAAN 196 |
| Candida albicans SC5314 | Query 165 TDEHGEVCPAGWKPGSDTIKPNVDDSKEYF#SKHN 198 |+++|||||| | || +||||+ + |||||#+| | Sbjct 162 TEKYGEVCPANWHPGDETIKPSPEASKEYF#NKVN 195 |
| Schistosoma mansoni | Query 165 TDEHGEVCPAGWKPGSDTIKPNVDDSKEYF#SKHN 198 ||++|||||| |+| ||||++ ||||# | | Sbjct 186 TDKYGEVCPADWQPKGPTIKPDLKKYKEYF#HKVN 219 |
| Fasciola hepatica | Query 166 DEHGEVCPAGWKPGSDTIKPNVDDSKEYF#SKHN 198 +|||||||| ||| | || | | || ||#| | Sbjct 162 EEHGEVCPANWKPKSKTIVPTPDGSKAYF#SSAN 194 |
| Litomosoides sigmodontis | Query 166 DEHGEVCPAGWKPGSDTIKPNVDDSK 191 |+|||+||| |+|||+|||| | +|| Sbjct 168 DKHGELCPANWQPGSETIKPGVKESK 193 |
| Yarrowia lipolytica | Query 165 TDEHGEVCPAGWKPGSDTIKPNVDDSKEYF#SKHN 198 |++||||||| |+ |||||| + ++||||# | | Sbjct 162 TEKHGEVCPANWQKGSDTIKADPVNAKEYF#EKAN 195 |
| Echinococcus granulosus | Query 165 TDEHGEVCPAGWKPGSDTIKPNVDDSKEYF#S 195 ||+||||||| |+||| | ||+ | | + #| Sbjct 154 TDKHGEVCPANWQPGSKTFKPSAGDLKSFM#S 184 |
| Tetrahymena thermophila SB210 | Query 165 TDEHGEVCPAGWKPGSDTIKPNVDDSKEYF#S 195 || ||||||| |+|| || |+ | +||#| Sbjct 197 TDTHGEVCPANWQPGQKTIIPDQDQKIKYF#S 227 |
| Globodera rostochiensis | Query 165 TDEHGEVCPAGWKPGSDTIKPNVDDSKEYF#SK 196 || ||||||| |+|| |||||+ + |+ +|# | Sbjct 166 TDTHGEVCPANWQPGEDTIKPDPEGSQTFF#GK 197 |
| Echinococcus multilocularis | Query 165 TDEHGEVCPAGWKPGSDTIKPNVDDSKEYF#S 195 ||+||||||| | ||| | ||+ | | + #| Sbjct 162 TDKHGEVCPANWHPGSKTFKPSAGDLKSFM#S 192 |
| Entamoeba moshkovskii | Query 165 TDEHGEVCPAGWKPGSDTIKPNVDDSKEYF#SKH 197 ||+|| ||| ||||+|||+|+ | |+| #| | Sbjct 185 TDQHGAVCPLNWKPGNDTIEPSHDGIKKYL#SSH 217 |
| Candida glabrata | Query 165 TDEHGEVCPAGWKPGSDTIKPNVDDSKEYF#SKHN 198 ||++| | | | ||| |||| |+||||||# + | Sbjct 162 TDKNGTVLPCNWTPGSATIKPTVEDSKEYF#KEAN 195 |
| Trypanosoma cruzi strain CL Brener | Query 165 TDEHGEVCPAGWKPGSDTIKPNVDDSKEYF# 194 +|||||||| |||| |+||+ + |||||# Sbjct 166 VEEHGEVCPANWKPGDKTMKPDPEKSKEYF# 195 |
| Chlamydomonas reinhardtii | Query 170 EVCPAGWKPGSDTIKPNVDDSKEYF#S 195 |||||||||| |+||+ |||||#| Sbjct 208 EVCPAGWKPGDKTMKPDPKGSKEYF#S 233 |
| Trypanosoma cruzi strain CL Brener | Query 165 TDEHGEVCPAGWKPGSDTIKPNVDDSKEYF# 194 +|||||||| |||| |+||+ + |||+|# Sbjct 166 VEEHGEVCPANWKPGDKTMKPDPEKSKEFF# 195 |
| Entamoeba histolytica HM-1:IMSS | Query 165 TDEHGEVCPAGWKPGSDTIKPNVDDSKEYF#SKH 197 +|||| ||| |||| |||+| | |+| #+ | Sbjct 129 SDEHGAVCPLNWKPGKDTIEPTPDGIKKYL#TAH 161 |
| Entamoeba histolytica | Query 165 TDEHGEVCPAGWKPGSDTIKPNVDDSKEYF#SKH 197 +|||| ||| |||| |||+| | |+| #+ | Sbjct 186 SDEHGAVCPLNWKPGKDTIEPTPDGIKKYL#TAH 218 |
| Oryza sativa (japonica cultivar-group) | Query 170 EVCPAGWKPGSDTIKPNVDDSKEYF#S 195 |||||||||| ++||+ ||||||#+ Sbjct 140 EVCPAGWKPGEKSMKPDPKDSKEYF#A 165 |
* References
[PubMed ID: 24152995] Klebe S, Callahan T, Power JH, Peroxiredoxin I and II in human eyes: cellular distribution and association with pterygium and DNA damage. J Histochem Cytochem. 2014 Jan;62(1):85-96. doi: 10.1369/0022155413508409. Epub
[PubMed ID: 24222118] ... Suenaga S, Kuramitsu Y, Wang Y, Baron B, Kitagawa T, Akada J, Tokuda K, Kaino S, Maehara S, Maehara Y, Sakaida I, Nakamura K, Human pancreatic cancer cells with acquired gemcitabine resistance exhibit significant up-regulation of peroxiredoxin-2 compared to sensitive parental cells. Anticancer Res. 2013 Nov;33(11):4821-6.
[PubMed ID: 24003226] ... Haynes AC, Qian J, Reisz JA, Furdui CM, Lowther WT, Molecular basis for the resistance of human mitochondrial 2-Cys peroxiredoxin 3 to hyperoxidation. J Biol Chem. 2013 Oct 11;288(41):29714-23. doi: 10.1074/jbc.M113.473470. Epub
[PubMed ID: 22960070] ... Han YH, Kim SU, Kwon TH, Lee DS, Ha HL, Park DS, Woo EJ, Lee SH, Kim JM, Chae HB, Lee SY, Kim BY, Yoon do Y, Rhee SG, Fibach E, Yu DY, Peroxiredoxin II is essential for preventing hemolytic anemia from oxidative stress through maintaining hemoglobin stability. Biochem Biophys Res Commun. 2012 Sep 28;426(3):427-32. doi:
[PubMed ID: 18479207] ... Low FM, Hampton MB, Winterbourn CC, Peroxiredoxin 2 and peroxide metabolism in the erythrocyte. Antioxid Redox Signal. 2008 Sep;10(9):1621-30. doi: 10.1089/ars.2008.2081.
[PubMed ID: 7607688] ... Pahl P, Berger R, Hart I, Chae HZ, Rhee SG, Patterson D, Localization of TDPX1, a human homologue of the yeast thioredoxin-dependent peroxide reductase gene (TPX), to chromosome 13q12. Genomics. 1995 Apr 10;26(3):602-6.
[PubMed ID: 8041738] ... Chae HZ, Robison K, Poole LB, Church G, Storz G, Rhee SG, Cloning and sequencing of thiol-specific antioxidant from mammalian brain: alkyl hydroperoxide reductase and thiol-specific antioxidant define a large family of antioxidant enzymes. Proc Natl Acad Sci U S A. 1994 Jul 19;91(15):7017-21.
[PubMed ID: 8144038] ... Lim YS, Cha MK, Kim HK, Kim IH, The thiol-specific antioxidant protein from human brain: gene cloning and analysis of conserved cysteine regions. Gene. 1994 Mar 25;140(2):279-84.
[PubMed ID: 8123012] ... Lim YS, Cha MK, Yun CH, Kim HK, Kim K, Kim IH, Purification and characterization of thiol-specific antioxidant protein from human red blood cell: a new type of antioxidant protein. Biochem Biophys Res Commun. 1994 Feb 28;199(1):199-206.
[PubMed ID: 8026862] ... Shau H, Butterfield LH, Chiu R, Kim A, Cloning and sequence analysis of candidate human natural killer-enhancing factor genes. Immunogenetics. 1994;40(2):129-34.