SB0092 : Filamin C
[ CaMP Format ]
* Basic Information
| Organism | Homo sapiens (human) |
| Protein Names | filamin-C isoform a [Homo sapiens]; filamin-C isoform a; actin binding protein 280; ABP-L, gamma filamin; filamin-2; ABP-280-like protein; Filamin-C; FLN-C; FLNc; ABP-L; Actin-binding-like protein; Filamin-2; Gamma-filamin |
| Gene Names | FLNC; ABPL, FLN2; ABPL; FLN2; filamin C, gamma |
| Gene Locus | 7q32-q35; chromosome 7 |
| GO Function | Not available |
* Information From OMIM
Description: Filamins, such as FLNC, are a family of actin (see ACTA1; OMIM:102610)-binding proteins involved in reshaping of the cytoskeleton. See FLN1 (FLNA; OMIM:300017) for background information on the filamin gene family.
* Structure Information
1. Primary Information
Length: 2725 aa
Average Mass: 291.019 kDa
Monoisotopic Mass: 290.841 kDa
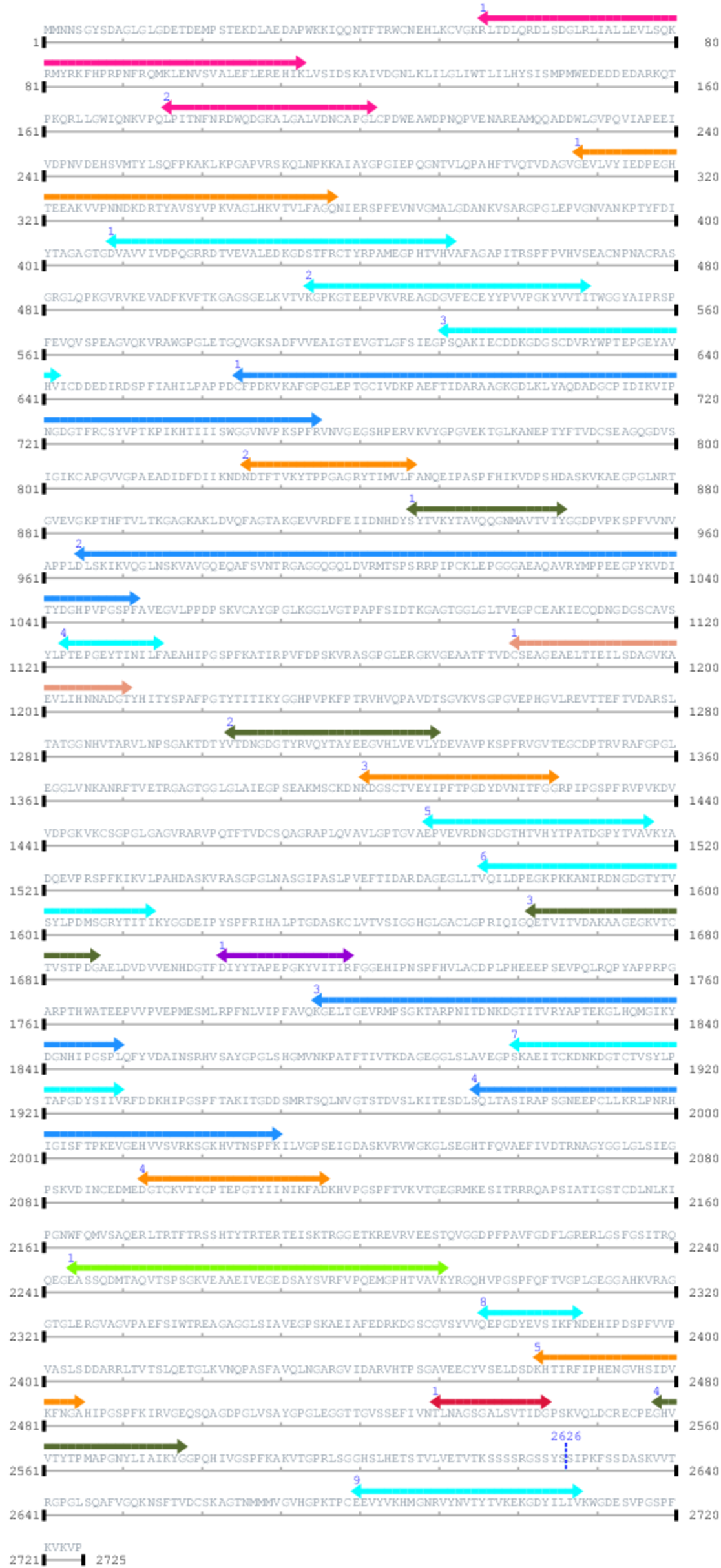
2. Domain Information
Annotated Domains: interpro / pfam / smart / prosite
Computationally Assigned Domains (Pfam+HMMER):
| domain name | begin | end | score | e-value |
|---|---|---|---|---|
| CAMSAP CH domain 1. | 56 | 113 | 12.0 | 0.0 |
| CAMSAP CH domain 2. | 176 | 202 | 11.0 | 0.0 |
| Bacterial Ig-like domain (group 3) 1. | 308 | 357 | 26.0 | 0.1 |
| YtkA-like 1. | 409 | 452 | 39.0 | 0.0 |
| YtkA-like 2. | 514 | 549 | 45.0 | 0.1 |
| YtkA-like 3. | 611 | 642 | 54.0 | 0.0 |
| Filamin/ABP280 repeat 1. | 665 | 755 | 2.0 | 0.0 |
| Bacterial Ig-like domain (group 3) 2. | 826 | 847 | 46.0 | 0.0 |
| Chagasin family peptidase inhibitor I42 1. | 927 | 946 | 53.0 | 0.0 |
| Filamin/ABP280 repeat 2. | 965 | 1052 | 3.0 | 0.0 |
| YtkA-like 4. | 1123 | 1135 | 63.0 | 0.0 |
| PapC N-terminal domain 1. | 1180 | 1211 | 54.0 | 0.0 |
| Chagasin family peptidase inhibitor I42 2. | 1304 | 1330 | 40.0 | 0.0 |
| Bacterial Ig-like domain (group 3) 3. | 1401 | 1425 | 45.0 | 0.0 |
| YtkA-like 5. | 1489 | 1517 | 57.0 | 0.0 |
| YtkA-like 6. | 1576 | 1614 | 42.0 | 0.0 |
| Chagasin family peptidase inhibitor I42 3. | 1662 | 1687 | 52.0 | 0.0 |
| Y_Y_Y domain 1. | 1703 | 1719 | 30.0 | 0.0 |
| Filamin/ABP280 repeat 3. | 1795 | 1850 | 41.0 | 0.0 |
| YtkA-like 7. | 1900 | 1930 | 55.0 | 0.0 |
| Filamin/ABP280 repeat 4. | 1975 | 2030 | 42.0 | 0.0 |
| Bacterial Ig-like domain (group 3) 4. | 2093 | 2116 | 46.0 | 0.0 |
| emp24/gp25L/p24 family/GOLD 1. | 2244 | 2291 | 34.0 | 0.0 |
| YtkA-like 8. | 2376 | 2388 | 64.0 | 0.0 |
| Bacterial Ig-like domain (group 3) 5. | 2463 | 2485 | 48.0 | 0.0 |
| Putative Ig domain 1. | 2530 | 2544 | 17.0 | 0.0 |
| Chagasin family peptidase inhibitor I42 4. | 2558 | 2578 | 53.0 | 0.0 |
| --- cleavage 2626 --- | ||||
| YtkA-like 9. | 2680 | 2708 | 57.0 | 0.2 |
3. Sequence Information
Fasta Sequence: SB0092.fasta
Amino Acid Sequence and Secondary Structures (PsiPred):
4. 3D Information
Known Structures in PDB: 1V05 (X-ray; 143 A; A=2633-2725), 2D7M (NMR; -; A=1536-1637), 2D7N (NMR; -; A=1782-1861), 2D7O (NMR; -; A=1856-1953), 2D7P (NMR; -; A=2405-2503), 2D7Q (NMR; -; A=2502-2599), 2K9U (NMR; -; A=2302-2415), 2NQC (X-ray; 205 A; A=2495-2598), 3V8O (X-ray; 280 A; A/B=569-761), 4MGX (X-ray; 316 A; A=572-756)
* Cleavage Information
1 [sites] cleaved by Calpain 1
Source Reference: [PubMed ID: 16297652] Raynaud F, Jond-Necand C, Marcilhac A, Furst D, Benyamin Y, Calpain 1-gamma filamin interaction in muscle cells: a possible in situ regulation by PKC-alpha. Int J Biochem Cell Biol. 2006 Mar;38(3):404-13. Epub 2005 Oct 27.
Cleavage sites (±10aa)
[Site 1] SSSSRGSSYS2626-SIPKFSSDAS
Ser2626  Ser
Ser
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Ser2617 | Ser2618 | Ser2619 | Ser2620 | Arg2621 | Gly2622 | Ser2623 | Ser2624 | Tyr2625 | Ser2626 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Ser2627 | Ile2628 | Pro2629 | Lys2630 | Phe2631 | Ser2632 | Ser2633 | Asp2634 | Ala2635 | Ser2636 |
 Sequence conservation (by blast)
Sequence conservation (by blast)
* References
[PubMed ID: 24469451] Sethi R, Seppala J, Tossavainen H, Ylilauri M, Ruskamo S, Pentikainen OT, Pentikainen U, Permi P, Ylanne J, A novel structural unit in the N-terminal region of filamins. J Biol Chem. 2014 Mar 21;289(12):8588-98. doi: 10.1074/jbc.M113.537456. Epub 2014